Abstract
Cholesterol metabolism is paramount to cells. Aberrations to cholesterol metabolism affects cholesterol homeostasis, which may impact the risk of several diseases. Recent evidence has suggested that vascular smooth muscle cell (VSMC) cholesterol metabolism may play a role in atherosclerosis. However, there is scant in vitro mechanistic data involving primary VSMC that directly tests how VSMC cholesterol metabolism may impact atherosclerosis. One reason for this lack of data is due to the impracticality of gene manipulation studies in primary VSMC, as cultured primary VSMC become senescent and lose their morphology rapidly. However, there are no immortalized VSMC lines known to be suitable for studying VSMC cholesterol metabolism. The purpose of this study was to determine whether MOVAS cells, a commercially available VSMC line, are suitable to use for studying VSMC cholesterol metabolism. Using immunoblotting and immunofluorescence, we showed that MOVAS cells express ABCA1, ABCG1, and SREBP-2. We also determined that MOVAS cells efflux cholesterol to apoAI and HDL, which indicates functionality of ABCA1/ABCG1. In serum-starved MOVAS cells, SREBP-2 target gene expression was increased, confirming SREBP-2 functionality. We detected miR-33a expression in MOVAS cells and determined this microRNA can silence ABCA1 and ABCG1 via identifying conserved miR-33a binding sites within ABCA1/ABCG1 3’UTRs in MOVAS cells. We showed that cholesterol-loading MOVAS cells results in this cell line to transdifferentiate into a macrophage-like cell, which also occurs when VSMC accumulate cholesterol. Our characterization of MOVAS cells sufficiently demonstrates that they are suitable to use for studying VSMC cholesterol metabolism in the context of atherosclerosis.
Keywords: Lipoprotein, Phenotypic switching, Primary cells, Reverse cholesterol transport
Introduction
Atherosclerosis causes more deaths than any other disease globally (Rodriguez-Saldana et al. 2014; Khan et al. 2020). Atherosclerosis is responsible for causing narrowing of the arteries, mainly due to cholesterol accumulation within the arterial wall, and this may lead to death from either myocardial infarction or ischemic strokes (Insull 2009). Since cholesterol accumulation within arteries drives atherosclerosis (Lusis 2000), removing cholesterol from arterial cells may prevent or retard atherosclerosis (Wacker et al. 2017). Two genes involved in removing intracellular cholesterol are ABCA1 and ABCG1, which efflux cholesterol to apoAI and HDL, respectively (Yvan-Charvet et al. 2010). ABCA1 and ABCG1 assist in regulating cholesterol homeostasis (Seeree et al. 2019), along with the transcription factor SREBP-2 and its intronic microRNA, miR-33a (Najafi-Shoushtari et al. 2010). SREBP-2 functions to activate genes involved in cholesterol biosynthesis (Horton et al. 2002), while miR-33a is co-expressed with SREBP-2 and aids in retaining intracellular cholesterol levels via silencing ABCA1/ABCG1 expression (Najafi-Shoushtari et al. 2010; Rayner et al. 2010). All four of the above-mentioned genes are critical to cholesterol metabolism and altering the expression of these genes in arterial cells may either promote or protect against atherosclerosis.
Both the pathogenesis and pathophysiology of atherosclerosis is complex and not entirely understood (Milioti et al. 2008; Milutinovic et al. 2020). For instance, lipid-laden macrophages have been well-recognized to be the primary cell type found within atherosclerotic lesions (Insull 2009). However, this has been recently challenged (Bennett et al. 2016), as a number of studies have identified vascular smooth muscle cells (VSMC) to be the major cell type in atherosclerotic plaques (Allahverdian et al. 2014; Feil et al. 2014; Shankman et al. 2015; Wang et al. 2019). This incertitude may arise from the plasticity VSMC demonstrate (Rosenfeld 2015), as they are able to convert into a macrophage-like cell (MLC) phenotype upon cholesterol-loading (Rong et al. 2003; Vengrenyuk et al. 2015). This has been shown to result in VSMC losing expression of classical VSMC markers, while triggering expression of classical macrophage markers in these cells (Vengrenyuk et al. 2015; Shankman et al. 2015; Feil et al. 2014). Interestingly, MLC of VSMC origin have been shown to have decreased ABCA1 expression, which may exacerbate atherosclerosis (Allahverdian et al. 2014; Choi et al. 2009).
Genetic manipulation of ABCA1, ABCG1, SREBP-2, and/or miR-33a within cultured VSMC may prove to be extremely informative in understanding the relevant importance of these genes in relation to cholesterol homeostasis and metabolism. Furthermore, identifying what potential roles these four genes demonstrate within VSMC (in the context of atherosclerosis) may better determine whether ablation of these genes may be atheroprotective or atherogenic. A robust approach to assess function of the four above-mentioned genes within VSMC would be to perform CRISPR-Cas9-mediated gene editing (Ran et al. 2013). Primary VSMC though are notorious for becoming senescent and losing their morphology at a low passage number (Sedding D.G. 2005), making primary VSMC an impractical choice for CRISPR-Cas9. However, a possible viable alternative would be to perform gene editing in an immortalized VSMC line. The potential drawback to this option though is that immortalized cell lines may lack crucial functions and characteristics typically found in their primary cell counterparts (Kaur & Dufour 2012). Moreover, to the authors’ knowledge, no immortalized VSMC line has been assessed to precisely determine whether the cell line may be useful in studying cholesterol homeostasis/metabolism in respect to atherosclerosis.
In this study, we extensively characterized MOVAS cells, which are a commercially available immortalized VSMC line derived from the aortic smooth muscle cells (AoSMC) of C57BL/6 mice (Afroze & Husain 2000). When compared to primary mouse AoSMC, our results show that MOVAS cells display similar characteristics to primary VSMC, as MOVAS cells express miR-33a-5p and miR-33a-3p mature strands; express ABCA1, ABCG1, and SREBP-2 protein; efflux cholesterol to apoAI and HDL; contain conserved miR-33a binding sites within the 3’UTR of both ABCA1 and ABCG1 genes; express SREBP-2 target genes when serum-starved; and accumulate lipid and transdifferentiate into MLC when loaded with cholesterol. Therefore, we conclude that MOVAS cells are indeed a suitable cell line to use when examining VSMC cholesterol homeostasis and metabolism in the context of atherosclerosis.
Materials and Methods
Cell Culture
MOVAS cells were obtained from American Type Culture Collection (Manassas, VA) and primary mouse aortic smooth muscle cells (AoSMC) were purchased from Cell Biologics Inc. (Chicago, IL). Cultured cells were maintained in standard growth medium consisting of high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Corning, NY) supplemented with FB Essence (10%; VWR Life Science Seradigm, Radnor, PA), and penicillin-streptomycin (P/S; 1%; Corning). Medium for MOVAS cells was also supplemented with G418 (500 μg/mL; VWR Life Science Seradigm; Radnor, PA). AoSMC were used between passages 2 and 6. Cells were incubated at 37°C with 5% CO2 in 10 cm tissue culture (TC) dishes and standard growth medium was replenished every 2–3 days. For experiments, cells were first seeded into TC plates and allowed to grow to 70–80% confluency before beginning respective treatments.
RT-qPCR
MOVAS cells and AoSMC, cultured in 6-well TC plates, were first washed with phosphate-buffered saline (PBS; Corning) and either replenished with standard growth medium to maintain basal conditions, or cultured with standard growth medium minus serum, to induce serum-starvation. For both conditions, cells were also supplemented with fatty acid-free bovine serum albumin (BSA-FAFA; 2 mg/mL; Sigma-Aldrich, St. Louis, MO). After 72 h, cells were washed with PBS and treated with TRI Reagent (Zymo Research, Irvine, CA). We isolated total RNA using Direct-zol RNA purification kits (Zymo Research). We quantified total RNA using a SpectraMax® QuickDrop™ Micro-Volume Spectrophotometer (Molecular Devices, LLC., San Jose, CA) and converted 100 ng of total RNA into cDNA using a Quantabio qScript® cDNA SuperMix kit (Beverly, MA). We then amplified cDNA using a Quantabio PerfeCTa SYBR Green FastMix kit and analyzed our qPCR data using the ΔΔCT method (Schmittgen & Livak 2008). The reference gene we used for normalization was GADPH and the primer pairs we used for RT-qPCR are listed in Table 1.
Table 1.
Primer Pairs
| Target | Sequence (5´–3´) | |
|---|---|---|
| GAPDH | forward: | AGGTCGGTGTGAACGGATTTG |
| reverse: | GGGGTCGTTGATGGCAACA | |
| HMGCR | forward: | AGAGCGAGTGCATTAGCAAAG |
| reverse: | GATTGCCATTCCACGAGCTAT | |
| LDLR | forward: | AGGCAGACTGCAAGGACAAG |
| reverse: | CCGTGAATGCAGGAGCCATC | |
| SQLE | forward: | GCTGGGCCTTGGAGATACAG |
| reverse: | CAGTGGGTACGGAATTTGAACT | |
| MiR-33a-5p | forward: | CGCGTGCATTGTAGTTGCATTGC |
| reverse: | GCATAGACCTGAATGGCGGTA | |
| MiR-33a-3p | forward: | CAATGTTTCCACAGTGCATCA |
| reverse: | GCATAGACCTGAATGGCGGTA | |
| ABCA1 3′ UTR | forward: | AAGAGCGAGGTCTTCCTTTG |
| reverse: | TGGCTTAATGGACGAGGATG | |
| ABCG1 3′ UTR | forward: | CAGGGACTAACGCAACGCAATG |
| reverse: | CCCTCAAGTTGGAGGGATACAC |
cDNA Analysis
To assess miR-33a-5p and miR-33a-3p expression, we first seeded MOVAS and AoSMC into 10 cm culture dishes, washed cells with PBS, lysed cells with TRI reagent, and isolated total cellular RNA using a Direct-zol RNA kit (Zymo Research). We quantified total RNA using QuickDrop and then used 100 ng of total RNA to convert mature microRNA strands into cDNA using a Quantabio qScript™ microRNA cDNA Synthesis kit. Using the cDNA as template, we performed end-point PCR by using forward primers and a universal reverse primer (Table 1) to amplify either miR-33a-5p or miR-33a-3p. We digested the PCR products with either BsrDI or TspRI (New England Biolabs, Ipswich, MA) and then assessed the amplicons and digested fragments via TBE-agarose gel electrophoresis using a GelDoc system (Analytik Jena US, Upland, California).
For 3’UTR sequencing of ABCA1 and ABCG1, AoSMC and MOVAS cells maintained in 10 cm TC dishes were washed with PBS, treated with TRI Reagent LS (Molecular Research Center, Inc.; Cincannati, OH), and then total cellular RNA was isolated as previously described (Rio et al. 2010). We quantified total RNA with QuickDrop and used 1 μg of cellular RNA to both convert total RNA into cDNA and use newly synthesized cDNA for end-point RT-PCR via using a Quantabio qScript XLT 1-Step RT-PCR Kit. The primer pairs (Table 1) used for end-point RT-PCR reactions targeted the 3’UTR of ABCA1 and ABCG1 which have been shown to contain miR-33a binding sites (Rayner et al. 2010). PCR products generated from these reactions were then sequenced by Eton Bioscience, Inc. (San Diego, CA).
Cholesterol Efflux Assays
MOVAS cells and AoSMC were seeded and maintained in 48-well TC plates. To measure apoAI/HDL-mediated cholesterol efflux in AoSMC and MOVAS cells, we removed standard growth medium, washed cells with PBS, and then treated cells with DMEM containing BSA-FAFA (2 mg/mL), P/S (1%), and [3H] cholesterol (1 μCi/mL; PerkinElmer, Waltham, MA) for 72 h. During these conditions, a sub-set of cells were also incubated with cholesterol–methyl-β-cyclodextrin (MβCD:Chol) (10 μg/mL; Sigma-Aldrich), to attempt to trigger VSMC transdifferentiation into MLC (Vengrenyuk et al. 2015). After cholesterol-loading, we removed the medium, washed cells with PBS, and treated cells with 100 μg/mL of either apoAI or HDL (Academy Bio-Medical Company, Houston, TX), or vehicle-treated cells (i.e. no cholesterol acceptors), for 72 h. Cholesterol acceptors were diluted in DMEM containing 2 mg/mL BSA-FAFA and 1% P/S. After treatments, medium was filtered to remove non-adherent cells, [3H] was counted in medium and cells using a liquid scintillation counter (LS 6500; Beckman Coulter, Brea, CA), and apoAI/HDL-mediated cholesterol efflux was calculated as previously described (Stamatikos et al. 2019; Stamatikos et al. 2020).
Immunoblotting
We cultured MOVAS cells and AoSMC in 6-well TC plates using standard growth medium, washed cells with PBS, harvested protein from cells using a mammalian protease inhibitor cocktail diluted in RIPA lysis buffer (VWR Life Science), and quantified cell lysate protein by using a BCA assay (BioVision, Milpitas, California.). Using equal amounts of protein per sample, we used SDS-PAGE for protein separation, and then transferred proteins onto PVDF membranes (Merck Millipore Ltd., Burlington, Massachusetts, United States). We blocked membranes with blocking buffer (Stamatikos et al. 2019; Stamatikos et al. 2020) and then probed for ABCA1 (1:1,000 dilution, sc-58219; Santa Cruz Biotechnology, Dallas, TX) and ABCG1 (1:5,000 dilution, NB400–132; Novus Biologicals, Littleton, CO). We also probed for HSP90 (1:5,000 dilution, 610419; BD Biosciences, San Jose, CA), which served as a loading control.
To detect SREBP-2 expression, AoSMC and MOVAS were seeded into 6-well TC plates cultured in standard growth medium supplemented with BSA-FAFA (2 mg/mL) to reflect basal conditions, or serum-starved by culturing cells in standard growth medium, minus serum, and supplemented with BSA-FAFA (2 mg/mL). After 72 h, protein was extracted and quantified, then SDS-PAGE, transfer, and blocking was performed as described above. After blocking, membranes were probed for SREBP-2 (1:500 dilution, sc-271616; Santa Cruz Biotechnology) and the loading control, GAPDH (1:1,000 dilution, sc-365062; Santa Cruz Biotechnology). Horseradish peroxidase (HRP)-conjugated secondary antibodies used were HRP-conjugated goat anti-rabbit IgG (1:10,000 dilution, HAF008; Novus Biologicals) and HRP-conjugated goat anti-mouse IgG (1:10,000 dilution, AP181P; Sigma-Aldrich). ECL substrate (Immobilon ECL Ultra Western HRP Substrate; MilliporeSigma, Billerica, MA) was used to detect HRP and imaging was performed with a ChemiDoc system (Analytik Jena US).
Fluorescence Imaging
For all fluorescence imaging analyses, MOVAS cells and AoSMC were seeded into sterile 4-well chamber TC slides (Corning) and grown in standard growth medium before beginning respective treatments. Cells maintained in basal conditions were stained for ABCA1, ABCG1, and SREBP-2. To assess VSMC transdifferentiation and cholesterol/lipid accumulation, cells were first washed with PBS and then provided DMEM supplemented with BSA-FAFA (2 mg/mL) and P/S (1%), and containing either MβCD:Chol (10 μg/mL) or vehicle only. After 72 h, cells were stained with Oil Red O (ORO) or stained for ACTA2 and CD68. For experiments involving restoring VSMC phenotype, cells were first loaded with MβCD:Chol for 72 h as described above, washed with PBS, and then treated with serum-free medium containing 100 μg/mL of either apoAI or HDL, or vehicle only. After 72 h, cells were stained for ACTA2 and CD68.
To prepare cells for staining, we first washed cells in PBS, fixed cells in 4% paraformaldehyde for 10 minutes, and then washed cells in PBS. For ORO stains, we prepared a staining solution by mixing 3 parts ORO stock solution (Sigma-Aldrich) with 2 parts H2O, and filtered this solution. Cells were first washed with H2O and then with 60% isopropanol. Cells were then incubated with the ORO staining solution for 15 minutes and washed with H2O to remove excess ORO. For immmunostaining, cells were blocked with 20 mM glycine for 10 minutes and then permeabilized in Triton-X-100 (0.2% v/v in PBS) for 20 minutes. Cells were then washed in PBS and incubated in blocking solution (3% BSA and 10% goat serum diluted in PBS) for 1 h. Cells were incubated in primary antibodies diluted 1:50 in PBS containing 1% (w/v) BSA overnight at 4°C. For this incubation step, we used the following primary antibodies: ABCA1 (sc-58219; Santa Cruz Biotechnology), ABCG1 (ST1606; Calbiochem, San Diego, CA), ACTA2 (sc-32251; Santa Cruz Biotechnology), CD68 (sc-20060; Santa Cruz Biotechnology), and SREBP-2 (sc-271616; Santa Cruz Biotechnology). Following incubation, we washed cells with PBS containing 1% (w/v) BSA, incubated cells with Alexa Fluor 546 goat anti-mouse IgG2a and/or Alexa Fluor 488 goat anti-mouse IgG1 secondary antibodies (10 μg/mL; Invitrogen, Carlsbad, CA) for 1 h, and then washed cells with PBS. Following all stains, cells were counterstained with DAPI (5 μg/mL; Invitrogen), and subsequently washed with PBS. For ORO, ACTA2, and CD68 stains, cells were mounted in PBS:Glycerol (50/50 v/v). For ABCA1, ABCG1, and SREBP-2 staining, cells were mounted in ProLong Gold (Thermo Fisher Scientific, Waltham, MA). All imaging was conducted using a Leica SP8X MP Confocal System equipped with HyD detectors, a 405 nm laser, a tunable white light laser, and time gating capabilities (Leica Microsystems, Buffalo Grove, IL). Leica LAS-X software (Leica Microsystems Version 3.5.5.19976) was utilized for image capture and export.
Statistical Analyses
We used SigmaPlot (Systat Software Inc, San Jose, CA) to analyze statistics. Normality and equal variance assumptions were assessed using a Shapiro-Wilk test and a Brown-Forsythe test, respectively. When both assumptions were met, we performed a Student’s t-test. When normality was violated, we performed a Mann-Whitney rank-sum test. When equal variances were not assumed, we performed a Welch’s t-test. The level of statistical significance was set at P<0.05.
Results
MOVAS Cells Express Functional ABCA1 and ABCG1
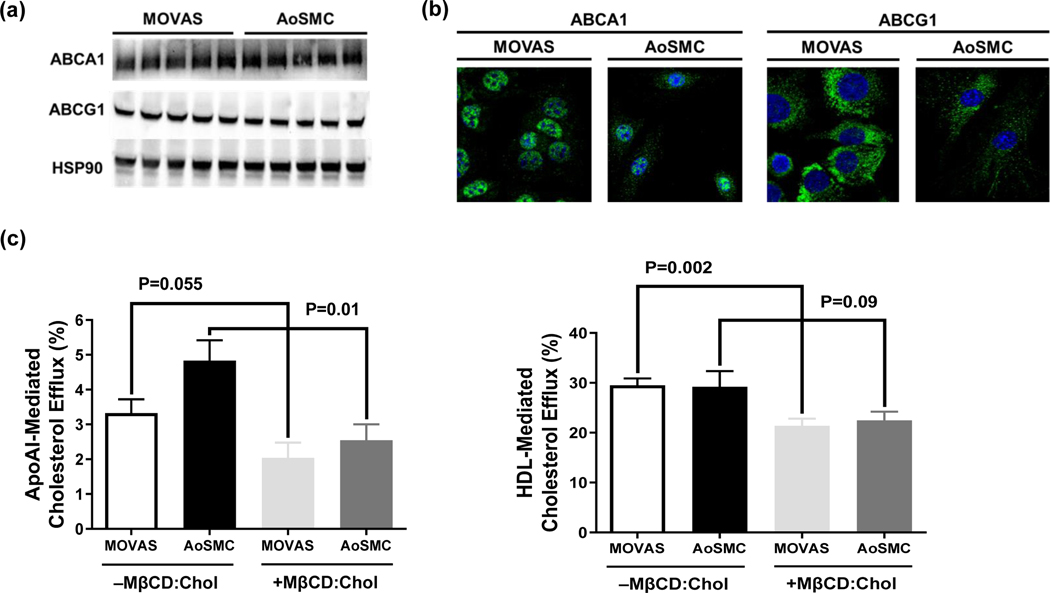
ABCA1 and ABCG1 are essential to the two main active pathways involving cholesterol efflux (Phillips 2014), making both proteins crucial to cholesterol metabolism. Therefore, we assessed ABCA1 and ABCG1 protein expression in MOVAS cells and detected both proteins via immunoblotting (Fig. 1a) and immunofluorescence (Fig. 1b). To determine if these proteins are functional, we performed cholesterol efflux assays to assess ABCA1 and ABCG1-dependent cholesterol efflux by using apoAI and HDL as cholesterol acceptors. Our results show that MOVAS cells efflux cholesterol to both apoAI and HDL (Fig. 1c), which implies that ABCA1/ABCG1 protein in MOVAS cells is functional. Furthermore, when MOVAS cells were co-incubated with [3H] cholesterol and MβCD:Chol before introducing apoAI and HDL, cholesterol efflux was shown to be reduced (Fig. 1c). This finding is similar to other observations which show impaired cholesterol efflux in cholesterol-filled VSMC that have transdifferentiated into MLC (Choi et al. 2009) and suggests that MOVAS cells are capable of converting into MLC upon cholesterol-loading.
Fig. 1.
MOVAS Cells Demonstrate ABCA1/ABCG1-dependent Cholesterol Efflux. ABCA1 and ABCG1 protein detected in basal MOVAS cells and primary mouse aortic smooth muscle cells (AoSMC) via immunoblotting (a) and immunofluorescence staining (b). ApoAI- and HDL-mediated cholesterol efflux (c) measured in serum-starved, [3H] cholesterol-loaded MOVAS cells and AoSMC, cholesterol–methyl-β-cyclodextrin (MβCD:Chol). (a) Five biological replicates per cell type. (b) ABCA1 and ABCG1 protein (green), cell nuclei counterstained with DAPI (blue). (c) Two independent experiments with three biological replicates per treatment for each experiment. Data are mean ± SEM.
MOVAS Cells Transdifferentiate into MLC
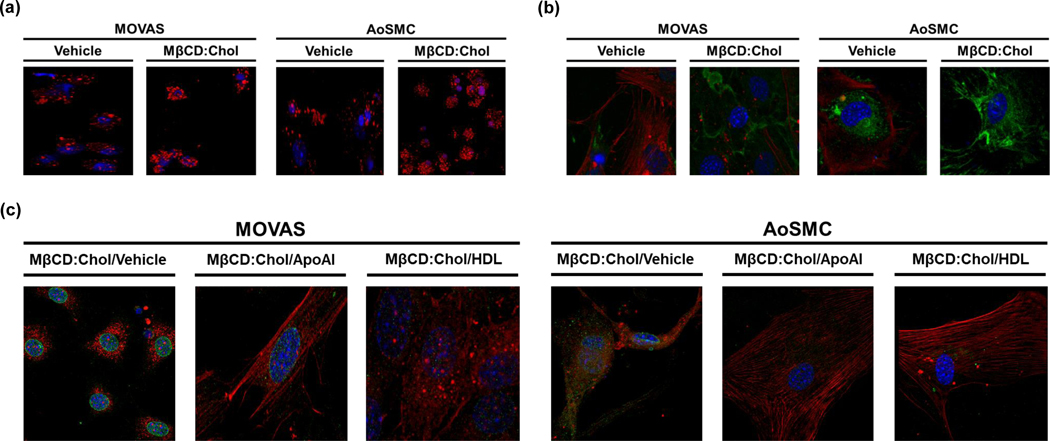
It is well-established that cholesterol-loading induces VSMC transdifferentiation into MLC (Vengrenyuk et al. 2015; Rong et al. 2003) and it is postulated that this phenotypic change is critical for atherosclerosis progression (Basatemur et al. 2019). Therefore, we used epifluorescence and immunofluorescence to determine whether MOVAS cells are capable of transdifferentiating into MLC when loaded with cholesterol. By using an established MβCD:Chol loading protocol for VSMC, we stained cells for ORO to detect lipid droplets and stained for the classical SMC marker ACTA2 and the classical macrophage marker CD68. Our results show that MOVAS cells loaded with MβCD:Chol contain enriched lipid droplets, suppress ACTA2 expression, and demonstrate robust CD68 expression when compared to MOVAS cells not loaded with MβCD:Chol (Fig. 2a, b). However, when MβCD:Chol-loaded MOVAS cells are treated with either apoAI or HDL, VSMC phenotype in these cells appears to be restored based on reestablishing VSMC morphology and positively altering ACTA2 and CD68 expression patterns, when compared to MβCD:Chol-loaded MOVAS cells treated with vehicle only (Fig. 2c). These findings involving VSMC transdifferentiation into MLC by MβCD:Chol-loading VSMC and subsequently restoring VSMC morphology upon apoAI/HDL-treatment are in parallel with other observations using primary AoSMC (Vengrenyuk et al. 2015). Moreover, these results further indicate that MOVAS cells are able to remove cholesterol by apoAI/HDL-mediated mechanisms, and that this efflux likely occurs via an ABCA1/ABCG1-dependent process.
Fig. 2.
Cholesterol Accumulation in MOVAS Cells Triggers Macrophage-like Cell Transdifferentiation. Vehicle-treated and cholesterol–methyl-β-cyclodextrin (MβCD:Chol)-treated MOVAS cells and primary mouse aortic smooth muscle cells (AoSMC) were stained with Oil Red O (a) to detect lipid or co-stained to detect ACTA2 and CD68 (b). (c) MβCD:Chol-treated MOVAS cells and AoSMC were incubated with vehicle, apoAI, or HDL, and then co-stained to detect ACTA2 and CD68. (a-c) Cell nuclei counterstained with DAPI (blue); (b, c) ACTA2 protein (red), CD68 protein (green).
MOVAS Cells Express Functional SREBP-2 and Mature miR-33a-5p/3p
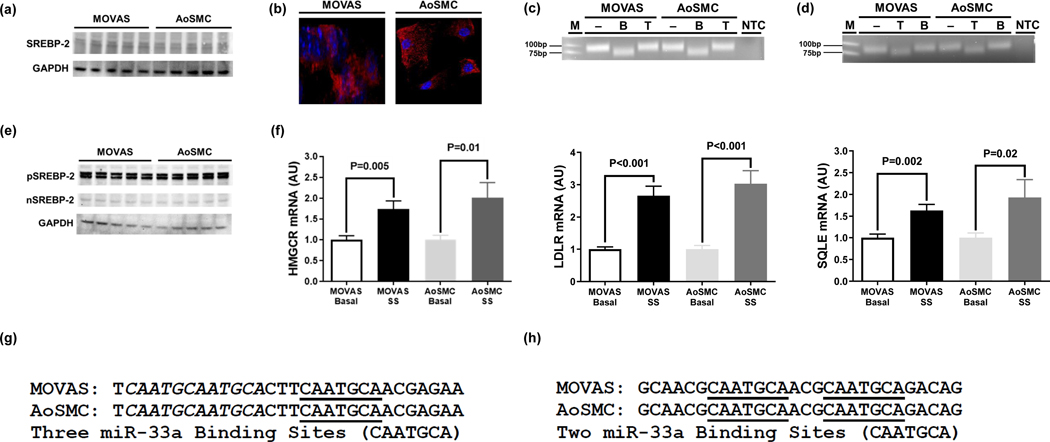
Cholesterol homeostasis is largely controlled by the transcription factor SREBP-2 and its intronic microRNA miR-33a (Najafi-Shoushtari et al. 2010), making these two genes imperative to cholesterol metabolism. We first assessed whether SREBP-2 protein and the mature (i.e. functional) strands of miR-33a are expressed in MOVAS cells. We detected SREBP-2 protein expression in MOVAS cells using both immunoblotting and immunofluorescence, as well as determined MOVAS cells express miR-33a-5p and miR-33a-3p mature strands via end-point RT-PCR and restriction digest (Fig. 3a-d). Since SREBP-2 is a transcription factor that is activated by low intracellular cholesterol levels (Horton et al. 2002), we sterol-depleted MOVAS cells using serum-starvation to induce robust activation of SREBP-2 (Brovkovych et al. 2019), and then measured gene expression of the three well-established SREBP-2 targets HMG-CoA reductase, low density lipoprotein receptor, and squalene epoxidase (Horton et al. 2002). Our results show a significant increase in both the precursor (inactive) and nuclear (active) forms of SREBP-2 protein expression in serum-starved MOVAS cells (Fig. 3e), in addition to a significant increase in the expression of all three SREBP-2 target genes in these cells, when compared to MOVAS cells cultured in basal conditions (Fig. 3f), which implies that SREBP-2 is functional in MOVAS cells. And to determine whether miR-33a is capable of silencing expression of ABCA1/ABCG1, we performed end-point RT-PCR to amplify a segment of the 3’UTR within respective ABCA1 and ABCG1 genes that have previously shown to contain conserved miR-33a binding sites (Rayner et al. 2010), and sequenced these PCR products. Sequencing results revealed miR-33a binding sites in MOVAS cells that are identical to conserved miR-33a binding sites previously reported in the literature (Fig. 3g, h) (Rayner et al. 2010), which confirms miR-33a is capable of downregulating ABCA1/ABCG1 expression in MOVAS cells.
Fig. 3.
Cholesterol Homeostasis in MOVAS Cells is Regulated by SREBP-2/miR-33a. SREBP-2 protein detected in basal MOVAS cells and primary mouse aortic smooth muscle cells (AoSMC) using immunoblotting (a) and immunofluorescent staining (b). End-point RT-PCR and amplicon restriction digestion analysis by agarose gel electrophoresis for the detection of miR-33a-5p (c) and miR-33a-3p (d) in basal MOVAS cells and AoSMC. (e) Inactive (pSREBP-2) and active (nSREBP-2) forms of SREBP-2 detected in serum-starved MOVAS cells and AoSMC via immunoblotting. (f) Expression of the SREBP-2 target genes HMG-CoA reductase (HMGCR), low density lipoprotein receptor (LDLR), and squalene epoxidase (SQLE) in basal and serum-starved (SS) MOVAS cells and AoSMC measured with RT-qPCR. End-Point RT-PCR and sequencing of ABCA1 (g) and ABCG1 (h) 3’UTR in basal MOVAS cells and AoSMC. (a, e) Five biological replicates per cell type. (b) SREBP-2 protein (red), cell nuclei counterstained with DAPI (blue). (c, d) MiR-33a-5p cDNA contains one BsrDI restriction site, but no TspRI restriction sites and miR-33a-3p contains one TspRI restriction site, but no BsrDI restriction sites. M, DNA marker/ladder; NTC, non-template control PCR reactions; minus (–) identifies undigested amplicons; B, BsrDI-digested amplicons; T, TspRI-digested amplicons. (f) AU, arbitrary units; three independent experiments with three biological replicates per condition in each experiment. Data are mean ± SEM. (g, h) MiR-33a binding sites (CAATGCA) that are isolated are underlined and are italicized if they are overlapping.
Discussion
In our study, we assessed whether MOVAS cells may be used to study VSMC cholesterol metabolism (in the context of atherosclerosis) in vitro. By using cultured MOVAS cells and cultured primary mouse AoSMC as a positive technical control, MOVAS cells demonstrate similar characteristics as VSMC. In our main findings using MOVAS cells, we report that MOVAS cells express functional SREBP-2/miR-33a, ABCA1, and ABCG1, MOVAS cells transdifferentiate into MLC when loaded with MβCD:Chol, and MβCD:Chol-loaded MOVAS cells restore VSMC phenotype when treated with the cholesterol acceptors apoAI and HDL.
In the field of atherosclerosis research, there has recently been a shift towards focusing on the possible roles VSMC may have in exacerbating atherosclerosis (Allahverdian et al. 2018). Traditionally, it has been recognized that VSMC play a protective role against atherosclerosis by preventing plaque rupture (Schwartz et al. 2000), and while this assumption is still acknowledged, VSMC are now considered to also play a pro-atherogenic role, particularly in the later, more advanced stages of atherosclerosis progression (Chistiakov et al. 2015). VSMC are capable of switching to various cell phenotypes (Sorokin et al. 2020), one being MLC. For this reason, studying atherogenic mechanisms VSMC may display in vitro has heavily relied upon using cultured primary VSMC. However, primary VSMC are known to be finicky and are confined to a very low passage number, so thereby cannot be used for gene editing purposes. To evaluate SREBP-2/miR-33a, ABCA1, and ABCG1 function in cultured VSMC, utilizing an immortalized VSMC line will likely be needed.
There have been several reports which implies VSMC/MLC ABCA1 expression is pertinent to atherosclerotic disease (Allahverdian et al. 2014; Allahverdian et al. 2012; Choi et al. 2009; Wang et al. 2019). Indeed, research has shown that ABCA1 expression is reduced in VSMC that have trans differentiation into MLC, and it has been postulated that this effect may at least partially drive atherosclerosis via impairing apoAI-mediated cholesterol efflux (Allahverdian et al. 2014; Wang et al. 2019). And while VSMC/MLC ABCG1 expression has been examined less when compared to ABCA1, it has been speculated that reductions in ABCG1 expression in MLC will also exacerbate atherosclerosis via impairing HDL-mediated cholesterol efflux (Allahverdian et al. 2012). Our findings do show that when MOVAS cells and cultured AoSMC are MβCD:Chol-loaded, apoAI/HDL-mediated cholesterol efflux is impaired in these cells, which indicates that MLC do have a reduced capacity to efflux cholesterol to apoAI/HDL via ABCA1/ABCG1-dependent processes. Since cholesterol accumulation within VSMC triggers transdifferentiation of these cells into a MLC phenotype, it is possible that overexpressing ABCA1/ABCG1 may prevent or at least delay MLC transdifferentiation, while ablating VSMC ABCA1/ABCG1 may hasten this process. However, to the authors’ knowledge, this has yet to be investigated. Therefore, utilizing MOVAS cells to directly study VSMC ABCA1/ABCG1 function may be useful to the atherosclerosis field.
The miRNA miR-33a has been extensively characterized in macrophages and hepatocytes, while very little is known about its function in VSMC. Moreover, miR-33a’s impact on atherosclerosis is controversial, as some research indicates expression of this miRNA is atherogenic, while other reports show no atheroprotective effect from miR-33a inhibition, and there are even some findings which demonstrate atherogenic effects from ablating miR-33a expression. The initial promise of miR-33a inhibition as a potential therapy for atherosclerosis arose from miR-33a’s ability to silence ABCA1/ABCG1 expression (Horie et al. 2010; Mao et al. 2014). However, while short-term miR-33a inhibition has shown to effectively treat atherosclerosis (Rayner et al. 2011), long-term miR-33a has been shown to be ineffective against treating atherosclerosis (Marquart et al. 2013). Furthermore, long-term miR-33a ablation results in deleterious consequences (Naar 2018), which include hepatic steatosis, dyslipidemia, glucose intolerance, insulin resistance, hyperphagia, and obesity (Goedeke et al. 2014; Horie et al. 2013; Price et al. 2017; Price et al. 2018). Most research involving miR-33a’s impact on atherosclerosis excludes directly assessing miR-33a-3p, as there is only one published study to the authors’ knowledge which rigorously analyzes the role of miR-33a-3p in regulating cholesterol/lipid metabolism (Goedeke et al. 2013). Moreover, there has only been one published report which directly assesses VSMC miR-33a-5p expression modulating cholesterol efflux (Stamatikos et al. 2020). Based on our characterization data using MOVAS cells, studies may be performed using these cells to alter expression of miR-33a-3p and/or miR-33a-5p to determine whether manipulating expression of miR-33a-5p/3p influences VSMC transdifferentiation into MLC via impacting cholesterol efflux.
In conclusion, MOVAS cells are a suitable immortalized cell line to study VSMC cholesterol metabolism in the context of atherosclerosis. This versatile cell line is economical, robust, and commercially available. By utilizing MOVAS cells, studies may be performed to analyze VSMC ABCA1, ABCG1, and miR-33a function through either gene editing (i.e. CRISPR-Cas9) or stable overexpression. The implementation of these studies may provide insight on the importance of these genes to VSMC cholesterol metabolism in respect to atherosclerosis.
Acknowledgments
This work was supported in part by an American Heart Association Institutional Research Enhancement Award 20AIREA35120024 and the Clemson University’s R-Initiative Program through a Clemson University Core Incentivized Access Award. Imaging analyses was conducted in the Clemson Light Imaging Facility, which is supported in part by NIH Eukaryotic Pathogens Innovation Center Centers of Biomedical Research Excellence grant P20GM109094, NIH SC BioCRAFT Centers of Biomedical Research Excellence grant 5P30GM131959, and the Clemson University Division of Research. The Leica SP8X confocal microscope system housed in the Clemson Light Imaging Facility is partially supported by NSF MRI grants 1126407 and 1920095. We thank Dr. Kimberly Paul for access to her laboratory to conduct cholesterol efflux assays.
List of Abbreviations
- ABCA1
ATP-Binding Cassette Transporter A1
- ABCG1
ATP Binding Cassette Transporter G1
- ACTA2
Smooth Muscle Alpha (α)-2 Actin
- ApoAI
Apolipoprotein A-I
- AoSMC
Aortic Smooth Muscle Cells
- BSA
Bovine Serum Albumin
- BSA-FAFA
Fatty Acid-Free Bovine Serum Albumin
- CD68
Cluster of Differentiation 68
- DAPI
4′,6-Diamidino-2-Phenylindole
- DMEM
Dulbecco’s Modified Eagle’s Medium
- ECL
Enhanced Chemiluminescence
- GAPDH
Glyceraldehyde 3-Phosphate Dehydrogenase
- HDL
High-Density Lipoprotein
- HMG-CoA reductase
3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase
- HRP
Horseradish Peroxidase
- HSP90
Heat Shock Protein 90
- MβCD:Chol
Cholesterol–Methyl-β-Cyclodextrin
- MLC
Macrophage-Like Cells
- ORO
Oil Red O
- P/S
Penicillin-Streptomycin
- PBS
Phosphate-Buffered Saline
- SREBP-2
Sterol Regulatory Element–Binding Protein-2
- TC
Tissue Culture
- VSMC
Vascular Smooth Muscle Cells
Footnotes
Conflict of Interest
None.
References
- Afroze T. and Husain M. (2000) c-Myb-binding sites mediate G(1)/S-associated repression of the plasma membrane Ca(2+)-ATPase-1 promoter. J Biol Chem 275, 9062–9069. [DOI] [PubMed] [Google Scholar]
- Allahverdian S, Chaabane C, Boukais K, Francis GA and Bochaton-Piallat ML (2018) Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res 114, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdian S, Chehroudi AC, McManus BM, Abraham T. and Francis GA (2014) Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 129, 1551–1559. [DOI] [PubMed] [Google Scholar]
- Allahverdian S, Pannu PS and Francis GA (2012) Contribution of monocyte-derived macrophages and smooth muscle cells to arterial foam cell formation. Cardiovasc Res 95, 165–172. [DOI] [PubMed] [Google Scholar]
- Basatemur GL, Jorgensen HF, Clarke MCH, Bennett MR and Mallat Z. (2019) Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol 16, 727–744. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Sinha S. and Owens GK (2016) Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res 118, 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brovkovych V, Aldrich A, Li N, Atilla-Gokcumen GE and Frasor J. (2019) Removal of Serum Lipids and Lipid-Derived Metabolites to Investigate Breast Cancer Cell Biology. Proteomics 19, e1800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Orekhov AN and Bobryshev YV (2015) Vascular smooth muscle cell in atherosclerosis. Acta Physiol (Oxf) 214, 33–50. [DOI] [PubMed] [Google Scholar]
- Choi HY, Rahmani M, Wong BW, Allahverdian S, McManus BM, Pickering JG, Chan T. and Francis GA (2009) ATP-binding cassette transporter A1 expression and apolipoprotein A-I binding are impaired in intima-type arterial smooth muscle cells. Circulation 119, 3223–3231. [DOI] [PubMed] [Google Scholar]
- Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M. and Feil R. (2014) Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res 115, 662–667. [DOI] [PubMed] [Google Scholar]
- Goedeke L, Salerno A, Ramirez CM et al. (2014) Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med 6, 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Vales-Lara FM, Fenstermaker M. et al. (2013) A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol 33, 2339–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Nishino T, Baba O. et al. (2013) MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun 4, 2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Ono K, Horiguchi M. et al. (2010) MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A 107, 17321–17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL and Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insull W Jr. (2009) The pathology of atherosclerosis: plaque development and plaque responses to medical treatment. Am J Med 122, S3–S14. [DOI] [PubMed] [Google Scholar]
- Kaur G. and Dufour JM (2012) Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Hashim MJ, Mustafa H. et al. (2020) Global Epidemiology of Ischemic Heart Disease: Results from the Global Burden of Disease Study. Cureus 12, e9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ (2000) Atherosclerosis. Nature 407, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Lei H, Liu Q. et al. (2014) Effects of miR-33a-5P on ABCA1/G1-mediated cholesterol efflux under inflammatory stress in THP-1 macrophages. PLoS One 9, e109722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart TJ, Wu J, Lusis AJ and Baldan A. (2013) Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol 33, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioti N, Bermudez-Fajardo A, Penichet ML and Oviedo-Orta E. (2008) Antigen-induced immunomodulation in the pathogenesis of atherosclerosis. Clin Dev Immunol 2008, 723539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic A, Suput D. and Zorc-Pleskovic R. (2020) Pathogenesis of atherosclerosis in the tunica intima, media, and adventitia of coronary arteries: An updated review. Bosn J Basic Med Sci 20, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM (2018) miR-33: A Metabolic Conundrum. Trends Endocrinol Metab 29, 667–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE and Naar AM (2010) MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MC (2014) Molecular mechanisms of cellular cholesterol efflux. J Biol Chem 289, 24020–24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Rotllan N, Canfran-Duque A. et al. (2017) Genetic Dissection of the Impact of miR-33a and miR-33b during the Progression of Atherosclerosis. Cell Rep 21, 1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Singh AK, Rotllan N. et al. (2018) Genetic Ablation of miR-33 Increases Food Intake, Enhances Adipose Tissue Expansion, and Promotes Obesity and Insulin Resistance. Cell Rep 22, 2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Sheedy FJ, Esau CC et al. (2011) Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 121, 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ and Fernandez-Hernando C. (2010) MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC, Ares M Jr., Hannon GJ and Nilsen TW (2010) Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010, pdb prot5439. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Saldana J, Rodriguez-Flores M, Cantu-Brito C. and Aguirre-Garcia J. (2014) A pathological study of the epidemiology of atherosclerosis in Mexico city. Cardiol Res Pract 2014, 264205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong JX, Shapiro M, Trogan E. and Fisher EA (2003) Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci U S A 100, 13531–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld ME (2015) Converting smooth muscle cells to macrophage-like cells with KLF4 in atherosclerotic plaques. Nat Med 21, 549–551. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD and Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schwartz SM, Virmani R. and Rosenfeld ME (2000) The good smooth muscle cells in atherosclerosis. Curr Atheroscler Rep 2, 422–429. [DOI] [PubMed] [Google Scholar]
- Sedding DG, B.-D. RC (2005) In Vitro Cultivation of Vascular Smooth Muscle Cells. In: Practical Methods in Cardiovascular Research, (Dhein S. MFW, Delmar M. ed.), pp. 630–639. Springer, Berlin, Heidelberg. [Google Scholar]
- Seeree P, Janvilisri T, Kangsamaksin T, Tohtong R. and Kumkate S. (2019) Downregulation of ABCA1 and ABCG1 transporters by simvastatin in cholangiocarcinoma cells. Oncol Lett 18, 5173–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankman LS, Gomez D, Cherepanova OA et al. (2015) KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 21, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin V, Vickneson K, Kofidis T, Woo CC, Lin XY, Foo R. and Shanahan CM (2020) Role of Vascular Smooth Muscle Cell Plasticity and Interactions in Vessel Wall Inflammation. Front Immunol 11, 599415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatikos A, Dronadula N, Ng P, Palmer D, Knight E, Wacker BK, Tang C, Kim F. and Dichek DA (2019) ABCA1 Overexpression in Endothelial Cells In Vitro Enhances ApoAI-Mediated Cholesterol Efflux and Decreases Inflammation. Hum Gene Ther 30, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatikos A, Knight E, Vojtech L, Bi L, Wacker BK, Tang C. and Dichek DA (2020) Exosome-Mediated Transfer of Anti-miR-33a-5p from Transduced Endothelial Cells Enhances Macrophage and Vascular Smooth Muscle Cell Cholesterol Efflux. Hum Gene Ther 31, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrenyuk Y, Nishi H, Long X. et al. (2015) Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler Thromb Vasc Biol 35, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker BK, Dronadula N, Zhang J. and Dichek DA (2017) Local Vascular Gene Therapy With Apolipoprotein A-I to Promote Regression of Atherosclerosis. Arterioscler Thromb Vasc Biol 37, 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dubland JA, Allahverdian S. et al. (2019) Smooth Muscle Cells Contribute the Majority of Foam Cells in ApoE (Apolipoprotein E)-Deficient Mouse Atherosclerosis. Arterioscler Thromb Vasc Biol 39, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L, Wang N. and Tall AR (2010) Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol 30, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]