Abstract
Targeted protein degradation (TPD) using Proteolysis Targeting Chimeras (PROTACs) and molecular glue degraders has arisen as a powerful therapeutic modality for eliminating disease-causing proteins from cells. PROTACs and molecular glue degraders employ heterobifunctional or monovalent small molecules, respectively, to chemically induce the proximity of target proteins with E3 ubiquitin ligases to ubiquitinate and degrade specific proteins via the proteasome. While TPD is an attractive therapeutic strategy for expanding the druggable proteome, only a relatively small number of E3 ligases out of the >600 E3 ligases encoded by the human genome have been exploited by small molecules for TPD applications. Here, we review the existing E3 ligases that have thus far been successfully exploited for TPD and discuss chemoproteomics-enabled covalent screening strategies for discovering new E3 ligase recruiters. We also provide a chemoproteomic map of reactive cysteines within hundreds of E3 ligases which may represent potential ligandable sites that can be pharmacologically interrogated to uncover additional E3 ligase recruiters.
Introduction
Targeted protein degradation (TPD) has arisen as a powerful chemical biology platform for inducing the degradation of specific proteins in, on, or outside of cells and has made a tremendous impact on drug discovery in both academic and industrial settings 1–3. TPD uses heterobifunctional or monovalent small molecules or biologics to induce the proximity of a component of the cellular degradation machinery with a neo-substrate protein, inducing the degradation of that specific protein. Heterobifunctional degraders, or Proteolysis Targeting Chimeras (PROTACs), consist of a protein-targeting ligand linked to an E3 ubiquitin ligase recruiter to induce the proximity of an E3 ligase with a neo-substrate protein, causing the ubiquitination and elimination of that protein through proteasomal degradation (Fig. 1) 1–3. Molecular glue degraders also bring together an E3 ligase with a neo-substrate protein, but employ monovalent small molecules rather than heterobifunctional small molecules 4. While the genesis of TPD as a potential therapeutic paradigm arose from heterobifunctional or monovalent small molecules that induce the proximity of an E3 ubiquitin ligase with a neosubstrate protein to ubiquitinate and degrade the target protein via the proteasome, next-generation TPD strategies have arisen that exploit 1) the targeted degradation of cell surface and extracellular proteins through the lysosome with Lysosome Targeting Chimeras (LYTACs); 2) the targeted degradation of larger intracellular proteins and cell compartments through autophagy with Autophagy Targeting Chimeras (AUTACs); 3) the targeted degradation of transcription factors with Transcription Factor Targeting Chimeras (TRAFTACs) and Transcription Factor PROTACs (TF-PROTACs); 4) the targeted degradation of bacterial proteins through the recruitment of bacterial proteases with BacPROTACs; and even 5) the targeted degradation of RNA with Ribonuclease Targeting Chimeras (RIBOTACs) 5–9. Induced proximity paradigms are even being extended beyond degradation to include the targeted manipulation of other protein post-translational modifications beyond ubiquitination, including 1) targeted phosphorylation with Phosphorylation Inducing Chimeric Small-Molecules (PHICs); 2) targeted dephosphorylation with Phosphatase Recruiting Chimeras (PHORCs); and 3) targeted deubiquitination with Deubiquitinase Targeting Chimeras (DUBTACs) 10–12. These induced proximity paradigms have garnered significant interest within pharmaceutical companies because of the potential of TPD and next-generation induced proximity paradigms to tackle the “undruggable” proteome - representing most of the proteome which still cannot be therapeutically exploited using classical drug discovery approaches. All these potential therapeutic paradigms are enabled by the discovery of small molecule, peptide, or biologic recruitment handles that, either by themselves or through linkage to protein-targeting ligands, enable the induced proximity of a protein modulator with a neo-substrate protein of interest.
Figure 1. Targeted protein degradation with heterobifunctional PROTACs.
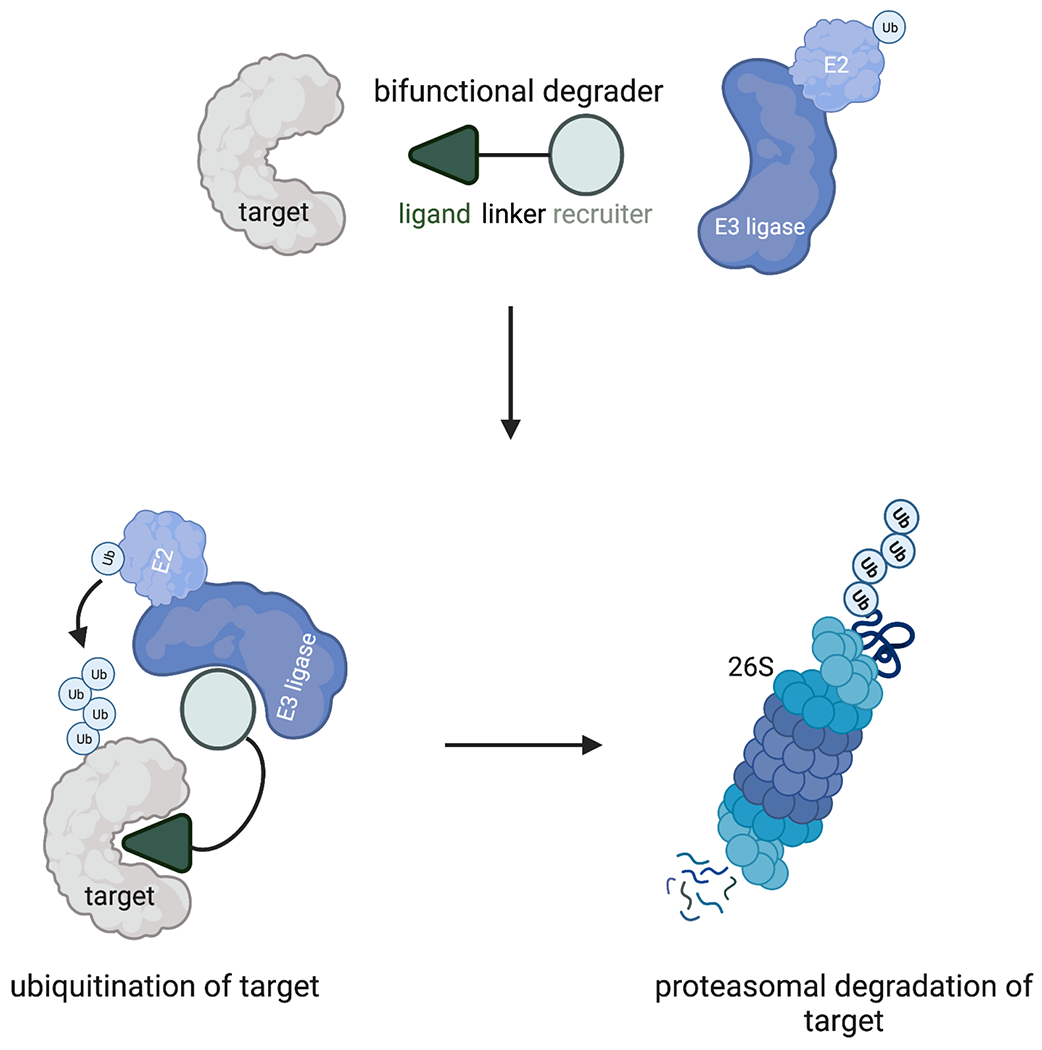
PROTACs consist of a protein-targeting ligand linked to an E3 ubiquitin ligase recruiter via a linker to induce the proximity of an E3 ligase with a neo-substrate target protein to induce ubiquitination and proteasome-mediated degradation of the target protein.
Despite the emergence of many PROTAC and molecular glue degrader drug candidates in the clinic, in clinical development, or entering clinical trials and the development of new induced proximity paradigms beyond TPD, a major bottleneck in using classical PROTACs and molecular glue degraders for TPD is still the paucity of pharmacological tools that exist for the ubiquitin-proteasome system, particularly for E3 ubiquitin ligases. E3 ubiquitin ligases are of particular interest because they are responsible for substrate recognition and specificity in the ubiquitin-proteasome system. E3 ligases are also the most diverse component of the ubiquitin-proteasome system, with over 600 estimated members.13,14 Yet, only a handful of E3 ligase recruiters currently exist with most attention placed on two E3 ligases - cereblon (CRBN) and von Hippel-Lindau (VHL) 15–17. These two E3 ligase recruiters are likely insufficient to degrade any and every protein target of interest with the desired selectivity. Evidence of this include the lack of degradation observed against KRAS G12C with CRBN-based recruiters, but achieved with VHL recruiters 18,19. In a recent beautiful study by Donovan, Ferguson, Sim, Gray, and Fischer et al. where they developed >90 CRBN- and VHL-based kinase PROTACs with various linkers and protein-targeting ligands, the authors impressively showed the degradation of >200 kinases, but there was still a significant number of kinases that were not degraded out of the >500 kinases that were detected 20.
The discovery of new E3 ligase ligands will undoubtedly help in expanding the scope of TPD and the targets that can be degraded by PROTACs and molecular glue degraders. Furthermore, the development of tissue- or cell type- or cell compartment-restricted E3 ligase recruiters may enable the development of degraders that achieve location-specific degradation of targets. Here, we review current E3 ligase recruiters, ligands, and molecular glues that have been successfully used in TPD and discuss chemoproteomic and covalent ligand screening approaches that have been used to discover new E3 ligase recruiters. We also present an aggregated chemoproteomic atlas of reactive cysteines that can potentially be targeted by cysteine-reactive covalent ligands across a large swath of the human E3 ligase family.
1. Ubiquitin-Proteasome System and E3 Ligases
The ubiquitin-proteasome system is a major regulator of protein homeostasis and is responsible for the degradation of a wide range of cellular proteins. Degradation occurs through an enzyme cascade that results in ubiquitination, a covalent post-translational modification. Hershko, Ciechanover, and Rose discovered this cascade in the 1970s while studying the degradation of denatured globin. They showed that degradation was ATP-dependent and proteins were marked for degradation by the covalent addition of multiple molecules of a small protein later identified as ubiquitin21,22. Ubiquitin is activated via the consumption of ATP by an E1 ubiquitin-activating enzyme. The activated ubiquitin is transferred from the E1 enzyme to the E2 ubiquitin conjugation enzyme via a transthiolation reaction. Ubiquitin is then transferred to the substrate by an E3 ubiquitin ligase. The transfer is accomplished via an isopeptide bond between the C-terminal glycine of ubiquitin and a lysine side chain on the substrate. The cycle is repeated to poly-ubiquitinate the substrate, marking it for proteasomal degradation (Fig. 1)23. The system, though complex and regulated, is generally applicable to the diverse proteome.
E3 ligases are classified based on their mechanisms of action. There are two main classes: RING E3 ligases, which recruit E2 ubiquitin conjugates via the RING domain and catalyze the transfer of ubiquitin directly to the substrate, and HECT (Homologous to the E6-AP Carboxyl Terminus) E3 ligases, which transfer ubiquitin to a substrate only after forming a thioester bond with ubiquitin. Within RING E3 ligases, there are 3 subcategories. Cullin-Really Interesting New Gene (RING) ligases (CRLs) are multisubunit complexes where a Cullin protein links an E2-binding subunit and a target-binding subunit.24 Anaphase promoting complex (APC) ligases are also multisubunit complexes that bring E2 proteins and the target protein together without a linker protein. RING-between-RING (RBR) ligases are considered RING E3 ligases because they bind to E2 proteins via their RING domain, but they form a thioester bond with ubiquitin before transferring it to the target protein.
2. E3 Ligase Recruiters
In 2001, the first example of artificial E3 ligase recruitment was shown with the E3 ligase ß-TrCP. A proteolysis targeting chimera (PROTAC) linked a ß-TrCP peptide ligand to a covalent methionine aminopeptidase 2 (MetAP2) ligand, resulting in the degradation of MetAP2 25. A peptide ligand was also used as the recognition sequence for the von Hippel-Lindau (VHL) E3 ligase in the first cell-permeable PROTAC in 2004 26. As interest in E3 ligase recruitment grew, the field shifted away from peptide ligands and toward reversible small-molecule E3 ligase recruiters with the recruitment of the MDM2 E3 ligase by the ligand nutlin in 2008 27. Reversible small-molecule ligands continued to gain recognition with the recruitment of the E3 ligase cIAP in 2010 28. Thalidomide, an immunomodulatory drug (IMiD), was shown to bind to the E3 ligase cereblon (CRBN) in 2010 by Ito, Ando, and Handa 29. In recent years, more covalent small-molecule E3 ligase recruiters have been identified.
2.1. Von Hippel-Lindau (VHL)
The von Hippel-Lindau (VHL) complex E3 ligase consists of VHL, elognins B and C, cullin 2, and RING box protein 1. VHL was one of the first E3 ligases targeted artificially by mimicking the recognition sequence of hypoxia-inducible factor-1 (HIF-1α), the primary target of VHL. In the early 2000s, the central hydroxyproline residue in the ALAPYIP recognition sequence of HIF-1α was determined to be essential for the formation of the HIF-1α/VHL complex via hydrogen bonding between the hydroxyproline of HIF-1α and a hydroxyproline pocket on VHL 30,31. In 2004, Schneekloth and Crews et al. used the ALAPYIP recognition sequence as a VHL recruiter for the first cell-permeable PROTAC, targeting and degrading FK506 binding protein (FKBP12) 26. This set the stage for interfering with proteins on a post-translational level, presenting the opportunity to create chemical knockdowns to study protein function without genetic modification. In 2005, a PROTAC targeting estrogen receptor-α (ERα) was developed that used a shorter peptide recognition sequence to recruit VHL. The shorter, pentapeptide sequence used for VHL recruitment improved cell permeability of the PROTAC. This E2-penta PROTAC successfully degraded ERα and angiogenesis was inhibited as a result of ERα degradation 32. VHL continued to be recruited with peptide recognition sequences resembling the HIF-1α sequence, but as more proteins were targeted, issues with cell permeability and drug-like properties became more apparent 33.
Following the discovery of small molecule recruiters for mouse double minute 2 homolog (MDM2) and cellular inhibitor of apoptosis protein (cIAP), discussed later in this review, a more drug-like small molecule ligand for VHL was identified. In 2012, Buckley, Crews, and Ciulli et al. identified a small molecule ligand that interrupted the interaction between HIF-1α and VHL. By analyzing the crystal structure of the HIF-1α/VHL complex, they were able to identify the key residues for binding and design small-molecule analogues to the key binding motif 34. Between 2012 and 2015, the original small molecule VHL recruiter was optimized to yield a VHL ligand with nanomolar potency 35,36. In 2015, Bondeson and Crews et al. used this optimized VHL recruiter in several PROTACs to specifically degrade ERRa and receptor interacting serine/threonine kinase 2 (RIPK2) in cells and in vivo in mice (Fig. 2) 36. Also in 2015, Zengerle, Chan, and Ciulli et al. designed a PROTAC linking JQ1, a bromo- and extra-terminal (BET) protein inhibitor, to the small-molecule VHL ligand. JQ1 is a non-selective BET inhibitor, but they observed selectivity of the PROTAC for BRD4. At the time, chemical knockdown had only been achieved for all BET family members, but none selectively. A different downstream pharmacological response was observed for BRD4 degradation than non-specific BET inhibition. The PROTAC was successful at low concentrations and the endogenous expression of HIF-1α was not impacted 37. The VHL recruiter has since been used in many PROTACs to degrade a wide range of targets 2,20,38,39.
Figure 2.
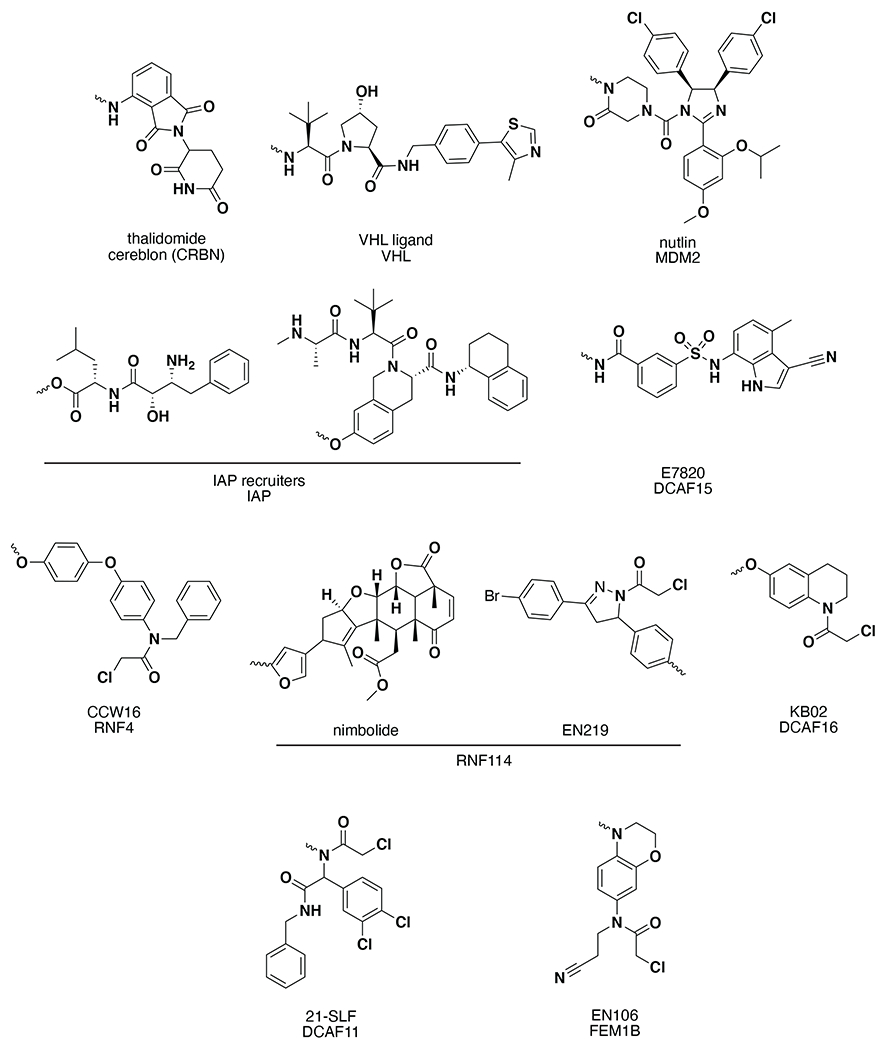
Existing E3 ligase recruiters for targeted protein degradation applications.
More recently, Bond, Chu, and Crews et al. targeted the oncogenic G12C mutant form of Kirsten rat sarcoma viral oncogene homolog (KRAS) with a small-molecule PROTAC recruiting VHL. The Crews lab designed a PROTAC linking MRTX849, a covalent KRASG12C ligand, to a VHL ligand. Degradation of KRASG12C was observed along with downstream suppression of MAPK signaling. This was the first successful PROTAC to target KRASG12C in cancer cells 40. This work follows the unsuccessful degradation of KRASG12C by the Gray lab using cereblon recruiters 41.
2.2. Cereblon (CRBN)
Cereblon (CRBN) is one of the substrate receptors for the Cullin-RING E3 ubiquitin ligase 4 (CRL4) containing DDB1, CUL4, and RBX1. While the endogenous substrates of cereblon are still under investigation, CRBN recognizes neo-substrates upon binding to thalidomide and other immunomodulatory drug (IMiD) analogs by engaging in ternary complexes, subsequently leading to the ubiquitination and proteasome-mediated degradation of these neo-substrates. Thalidomide, a drug originally developed for morning sickness in pregnant women in the 1950’s, was found to exert profound birth defects called phocomelia, hallmarked by shorted limbs 42. CRBN was identified as the primary target of thalidomide responsible for these birth defects by Ito and Handa et al. in 2010 43. The neo-substrate protein responsible for the birth defects associated with thalidomide, Sal-like protein 4 (SALL4), was not uncovered until more recently through concurrent discoveries by Matyskiela and Chamberlain et al. and Donovan and Fischer et al. in 2018 44,45. Once CRBN was identified as the target of thalidomide responsible for its immunomodulatory effects, less toxic IMiDs such as lenalidomide and pomalidomide were developed as cancer drugs to target CRBN as treatments for multiple myeloma 46. A series of studies in 2014 and 2015 provided both structural and biological insights, showing that different thalidomide analogs altered the E3 ligase neo-substrate specificity of CRBN and that the anti-cancer effects of lenalidomide and pomalidomide were through CRBN-mediated degradation of neo-substrate transcription factors Ikaros (IKZF1) and Aiolos (IZKF3) 47–51. Subsequent studies have continued to showcase the diversity of neo-substrate degradation with an expanding scope of IMiD analogs 48,52–55.
In 2015, Winter and Bradner et al. demonstrated that IMiD CRBN recruiters could be linked to protein-targeting ligands to selectively degrade specific proteins in cells (Fig. 2). They showed selective degradation of the transcriptional coactivator BRD4 and FKBP12 by linking IMiDs to their previously developed BET family protein ligand JQ1 and FKBP12 ligand SLF, respectively, in a CRBN-dependent manner 56. Alongside this paper in 2015, Lu and Crews et al. also published on IMiD-based PROTACs for BRD4 57. These results opened up the possibility for a fully synthetic drug-like heterobifunctional degraders that could be used to specifically degrade target proteins in cells, and spurred the development of many IMiD-based PROTACs that have now been exploited against countless protein targets as well as PROTACs that have entered clinical development 2,20,38,39.
2.3. MDM2
In 2008, the Crews lab demonstrated that nutlin, a known mouse double minute 2 homolog (MDM2) E3 ligase ligand that displaces the interaction between MDM2 and the tumor suppressor p53, could be used in a PROTAC to recruit MDM2 to degrade the androgen receptor (AR) (Fig. 2). AR was targeted by a selective androgen receptor modulator (SARM) linked to nutlin by a polyethylene glycol (PEG) linker 27. Using a small-molecule E3 ligase ligand instead of the peptidic E3 ligase ligands that existed at the time helped to increase both the cell permeability and stability of PROTACs while decreasing the molecular weight. The Crews lab found success in recruiting MDM2 with nutlin again in 2019 with the successful degradation of BRD4. Because nutlin inhibits the interactions between MDM2 and p53, MDM2-based PROTACs have the potential to both degrade their target and stabilize p53. The Crews lab demonstrated that their MDM2 PROTAC worked synergistically to exert anti-cancer activity by both degrading BRD4 and stabilizing p53 58.
2.4. cIAP1
The cellular inhibitor of apoptosis protein (cIAP1) degrades caspase proteins and is overexpressed in many cancer cells. In 2008, the Naito lab demonstrated that methyl bestatin (MeBS) bound to cIAP1 and promoted auto-ubiquitylation, facilitating its proteasomal degradation (Fig. 2) 59. This study indicated that cIAP1’s intrinsic ubiquitin ligase activity had the potential to be manipulated for the degradation of other proteins. In 2010, Itoh and Hashimoto designed specific and nongenetic inhibitor-of-apoptosis protein-dependent protein erasers (SNIPERs) that utilized MeBS to recruit cIAP. The original SNIPER contained an ester, which was readily hydrolyzed in the cell and caused off-target degradation 28. Amide-linked SNIPERs were designed with an all-trans retinoic acid (ATRA) ligand to bind the cellular retinoic acid binding protein II (CRABP-II). The selectivity of the SNIPER was improved, and degradation of CRABP-II was observed 60. In 2011, the group demonstrated that the SNIPER technology could be applied to different protein targets by degrading a variety of nuclear receptors (retinoic acid receptor, estrogen receptor-α, and androgen receptor) 61.
Following the facile expansion of SNIPER technology, the Naito lab designed a SNIPER(TACC3) in 2014 to target the spindle regulatory protein transforming acidic coiled-coil-3 (TACC3). Though degradation of TACC3 was observed, it was determined that cIAP1 was not the E3 ligase responsible for degradation. Through mechanistic studies, it was determined that the E3 ligase anaphase-promoting complex/cyclosome (APC/CCDHI) was responsible for mediating TACC3 degradation 62.
In more recent years, cIAP ligands have been improved over the original compounds, as demonstrated by GlaxoSmithKline (GSK) in 2020 with the PROTAC-mediated degradation of receptor-interacting serine/threonine protein kinase 2 (RIPK2) (Fig. 2) 63. This study demonstrated that RIPK2 could be selectively degraded in vivo at nanomolar potency. It is important to note that the RIPK2 PROTAC is more potent than the RIPK2 inhibitor alone, due to the catalytic pharmacological benefits offered by PROTACs 63. These findings demonstrate that PROTACs recruiting a variety of E3 ligases have therapeutic potential.
In 2021, Genentech demonstrated that E3 ubiquitin ligases could degrade themselves in a proteasome-dependent manner using modified X-linked inhibitor of apoptosis protein (XIAP) recruitment ligands 64. This novel degradation modality is composed of an XIAP recruitment ligand linked to a nucleophilic primary amine. The degradation depends on the isopeptide bond formation between the nucleophilic amine and the C-terminus of ubiquitin, leading to the XIAP-promoted ubiquitination of the small molecule recruitment ligand. This complex is then recruited to the proteasome, leading to the degradation of XIAP. Interestingly, the ubiquitylation state of XIAP is not changed, and the degradation depends only on the binding to and E3 ubiquitin ligase function of XIAP. This K-tag approach demonstrates not only the successful recruitment of XIAP, but also the expansion of the field of targeted protein degradation to include the self-degradation of E3 ubiquitin ligases 64.
2.5. RNF4
In 2019, Ward and Nomura discovered a small-molecule cysteine-reactive covalent RING finger protein 4 (RNF4) recruiter using activity-based protein profiling (ABPP)-based covalent ligand screens, to be described in more depth later. RNF4 is an E3 ligase that ubiquitinates SUMOylated proteins for proteasomal degradation 65. Cysteine-reactive covalent ligands were screened against fluorescent cysteine-reactive iodoacetamide probe (IA-rhodamine) labeling of pure human RNF4 in a gel-based ABPP screen, described later in this review. The initial covalent ligand hit TRH 1-23 was found to react with the zinc-coordinating cysteines C132 and C135 within RNF4 without inhibiting RNF4 ubiquitination activity 65. Upon hit optimization, CCW 16 was identified as the most potent compound against RNF4 and was subsequently linked to JQ1 to demonstrate the RNF4-dependent degradation of BRD4 (Fig. 2)65. This study demonstrated the utility of target-based covalent ligand screening to discover new E3 ligase recruiters.
2.6. RNF114
Concurrent with the discovery of covalent RNF4 recruiters, the Neem tree-derived anti-cancer natural product nimbolide was discovered as a covalent RNF114 recruiter 66. In 2019, Spradlin, Maimone, and Nomura et al. discovered that nimbolide reacts preferentially with the intrinsically disordered N-terminal cysteine 8, involved in substrate recognition of the E3 ubiquitin ligase RNF114 using the ABPP-based chemoproteomic approaches discussed later in this review 66. The authors showed that nimbolide exerts its anti-cancer effects by inhibiting RNF114 substrate interactions with tumor suppressors p21 and p57 leading to their accumulation and eventually cell death. More importantly, the authors demonstrated that nimbolide could be used as a covalent RNF114 recruiter for PROTACs in TPD, where they linked nimbolide to JQ1 to show both selective degradation of BRD4 and stabilization of endogenous RNF114 tumor suppressor substrates (Fig. 2) 66. Subsequent studies showed that nimbolide could also be used to degrade the fusion oncogene BCR-ABL in leukemia cancer cells. In this study, the authors demonstrated that RNF114-based degraders showed preferential degradation of BCR-ABL over c-ABL compared to cereblon or VHL-based degraders that showed preferential degradation of c-ABL 67.
To develop a more synthetically tractable RNF114 recruiter, Luo, Spradlin, and Nomura et al. performed a target-based covalent ligand screen using gel-based ABPP approaches against RNF114 to identify a fully synthetic recruiter, EN219, that mimicked the action of nimbolide in preferentially targeting C8 of RNF114 in cells 68. The authors subsequently linked EN219 to JQ1 to demonstrate RNF114-dependent degradation of BRD4 in cancer cells (Fig. 2) 68. While nimbolide was discovered as an RNF114 recruiter through target identification studies using ABPP-based chemoproteomic approaches, this later study showcased how target-based covalent ligand screening can be used to discover new E3 ligase recruiters against specific E3 ligases of interest 68,69.
2.7. DCAF16
Also concurrent with the discovery of covalent RNF4 and RNF114 recruiters, Zhang and Cravatt et al. discovered a novel covalent DDB1 and CUL4 associated factor 16 (DCAF16) E3 ligase recruiter that could be deployed in TPD applications in 2019 using a cellular covalent PROTAC screening strategy coupled with downstream chemoproteomic target identification 70. The authors linked covalent and promiscuous scout ligands to the FKBP12 ligand SLF, and screened for the selective degradation of nuclear FKBP12 that possessed a C-terminal nuclear localization signal. Through this effort, they identified the covalent scout ligand KB02 as a recruiter for TPD applications (Fig. 2). To identify the E3 ligase target of KB02, they performed FKBP12 pulldown proteomic studies to identify ubiquitin-proteasome system proteins that were specifically enriched upon treatment of cells with their KB02-based PROTAC. Through this effort, they identified and subsequently validated DCAF16, a substrate recognition component of the CUL4-DDB1 E3 ligase complex, as the E3 ligase target of KB02 responsible for the degradation activity of their PROTAC. The authors demonstrated that the KB02-based PROTAC was able to degrade its target proteins with low fractional engagement of DCAF16. With a KB02-based PROTAC linked to JQ1, they also demonstrated degradation of BRD4 70.
2.8. DCAF15
Since the mid-2000s, sulfonamides have been utilized in the clinic for cancer treatment due to their antitumor activity 71–73. However, the mechanisms of action of sulfonamide derivatives indisulam, E7820, and chloroquinoxaline sulfonamide were uncharacterized. In 2017, Uehara et al. and Han et al. independently reported that the sulfonamide derivatives form a complex between the coactivator of activating protein-1 and estrogen receptors like RNA-binding protein 39 (RBM39) (also known as CAPERα) and the DCAF15-DDB1-CUL4 complex. The formation of the complex led to the induced ubiquitination and proteasomal degradation of RBM39 74,75. In 2019, three crystal structures of the RBM39-sulfonamide-DCAF15-DDB1-CUL4 complex were reported 76–78. In 2020, Li and Chen et al. reported the first DCAF15-targeting PROTAC based on E7820 (Fig. 2) 79.
2.9. KEAP1
In 2020, Tong, Luo, Maimone, and Nomura et al. demonstrated that targeted degradation of BRD4 could be enabled by exploiting the triterpene derivative bardoxolone methyl (CDDO-Me), a ligand reported to activate the antioxidant NRF2 pathway by targeting the E3 ligase Kelch-like ECH-associated protein 1 (KEAP1) 80. While DCAF16, RNF4, and RNF114 recruiters acted irreversibly towards their respected E3 ligases, bardoxolone was unique as an E3 ligase recruiter due to its presumed covalent reversible interactions with cysteines on KEAP1. Bardoxolone may also interact with targets beyond KEAP1 and it will be of future interest to better understand the mechanisms through which bardoxolone may act as an recruiter for TPD applications.
2.10. DCAF11
In 2021, Zhang and Cravatt et al. discovered another covalent E3 ligase recruiter against the Cullin-RING E3 ligase substrate receptor DCAF11, using a similar approach to their strategy to discover their DCAF16 recruiter 81. In this study, the authors screened a library of PROTACs consisting of the FKBP12 ligand SLF linked to a wider panel of electrophilic ligands to identify compounds that degraded luciferase-conjugated FKBP12 in a luciferase reporter assay. Through this effort, they identified 21-SLF as a covalent PROTAC that degraded luciferase-FKBP12 in 22Rv1 cancer cells, but not in other cancer cell lines tested (Fig. 2). Upon pulldown proteomic studies to identify E3 ligase components that were enriched with FKBP12 and PROTAC treatment, they identified and validated DCAF11 as the E3 ligase responsible for the degradation of 21-SLF. Interestingly, the authors showed that while this PROTAC caused degradation through targeting C460 on DCAF11, in the absence of this cysteine, C443 and C485 also serve as additional engagement sites that support 21-SLF-induced target degradation. Using this DCAF11 recruiter, the authors also demonstrated degradation of the androgen receptor in 22Rv1 cells 81.
2.11. FEM1B
Also in 2021, Henning, Manford, Rape, and Nomura et al. discovered a cysteine-reactive covalent E3 ligase recruiter against the CUL2 E3 ligase FEM1B through a targeted functional screen 82. FEM1B was recently discovered as a critical regulator of the cellular reductive stress response, a cellular environment where there is a persistent depletion of reaction oxygen stress 83. Manford and Rape et al. discovered that FEM1B, under reductive stress, recognizes its substrate FNIP1, leading to FEM1B-dependent ubiquitination and proteasome-mediated degradation of FNIP1 and restoration of mitochondrial activity and redox homeostasis 83. In this study, the authors identified a key cysteine residue, C186, that was critical for substrate recognition. To target this substrate recognition cysteine in FEM1B as a possible FEM1B recruitment site, Henning, Manford, Rape, and Nomura et al. performed a fluorescence polarization screen with a cysteine-reactive covalent ligand library to identify compounds that would disrupt FEM1B interactions with a fluorescently-tagged FNIP1 degron peptide sequence. Through this screen, the authors identified a covalent ligand EN106 that bound to C186 of FEM1B to displace FEM1B-FNIP1 degron interactions (Fig. 2). They subsequently linked this EN106 recruiter to JQ1 to demonstrate selective FEM1B-dependent BRD4 degradation in cells 82. This study highlighted the utility of covalent ligand libraries coupled with functional E3 ligase-substrate degron displacement screens to discover new recruiters against specific E3 ligases of interest.
3. Discovery Strategies for New E3 Ligase Recruiters
Despite the significant advances made in recent years, only a handful of E3 ligase recruiters exist for the >600 E3 ligases that can potentially be exploited for targeted protein degradation or other induced proximity paradigms. As has already been described in the above examples, chemoproteomic platforms and covalent ligand discovery approaches have arisen as powerful strategies for discovering new E3 ligase recruiters, and more broadly for discovering covalent ligands and ligandable sites across the wider proteome. One particularly useful chemoproteomic strategy has been the activity-based protein profiling (ABPP) approach. ABPP uses activity-based or reactivity-based chemical probes to profile proteome-wide reactive, functional, and ligandable sites directly in complex biological systems 84–87.
Weerapana and Cravatt et al. demonstrated in 2010 that broad promiscuous cysteine-reactive probes could be used in complex proteomes using a quantitative proteomic platform, termed isotopic tandem orthogonal proteoslysis-ABPP (isoTOP-ABPP), to identify solvent-accessible cysteines as well as hyperreactive cysteines that were enriched in functional sites or binding pockets due to the local protein microenvironment 88. Since this discovery over 10 years ago, quantitative mass-spectrometry based ABPP platforms have been coupled with covalent ligand screening paradigms targeting to radically expand the scope of proteome-wide ligandability. Starting with discoveries from Wang and Cravatt et al. showing that isoTOP-ABPP could be used in a competitive format to compete lipid electrophiles to enable target identification89 to Backus and Cravatt et al. demonstrating that this platform could be used to identify more drug-like covalent ligands against a broad swatch of ligandable cysteines in the proteome90, there have been an increasing number of studies using chemoproteomics-enabled covalent ligand discovery approaches against cysteines, lysines, and other amino acids to enable ligand discovery against the proteome.69,91–99
As previously described in this review, chemoproteomics-enabled covalent ligand discovery platforms, whether through target-based or through target discovery approaches, have greatly enabled the expansion of E3 ligase recruiters for targeted protein degradation and PROTAC applications, including covalent recruiters against RNF4, DCAF16, RNF114, DCAF11, and FEM1B 65,69,81,82,100. Recently, Henning and Nomura et al. extended this approach to develop a deubiquitinase recruiter against OTUB1 and showed proof-of-concept for Deubiquitinase Targeting Chimeras (DUBTACs) for targeted protein stabilization by stabilizing cystic fibrosis transmembrane conductance regulator (CFTR) 12. As such, isoTOP-ABPP and other chemoproteomics-enabled covalent ligand discovery platforms are powerful approaches for uncovering unique ligandable sites and corresponding ligands to enable TPD, PROTACs, and other induced proximity-based therapeutic modalities.
Our lab, like many others in the TPD field, has been performing many ABPP-based chemoproteomic experiments using reactivity-based chemical probes for various target identification, target engagement, and selectivity profiling studies of covalently-acting small molecules; accumulating large datasets that include many probe-modified peptide data from human proteomes. To better enable the mining of potential reactive cysteines that may exist across the human E3 ligase family of proteins, we have aggregated all of our chemoproteomic data from 455 distinct experiments using the alkyne-functionalized iodoacetamide probe and the isoTOP-ABPP method first reported by Weerapana and Cravatt et al. 88 across various human cell line proteomes. We have quantified the total number of spectral counts for each tryptic peptide identified within each E3 ligase family member across all experiments (Fig. 3A; Table S1). These include data from our research group’s published papers as well as our currently unpublished studies 12,65,68,69,96,97,99,101–109. We show reactive cysteines identified across a representative set of E3 ligases in Table 1.
Figure 3. Using chemoproteomic platforms to identify potential reactive and ligandable cysteines across the human E3 ligase family.
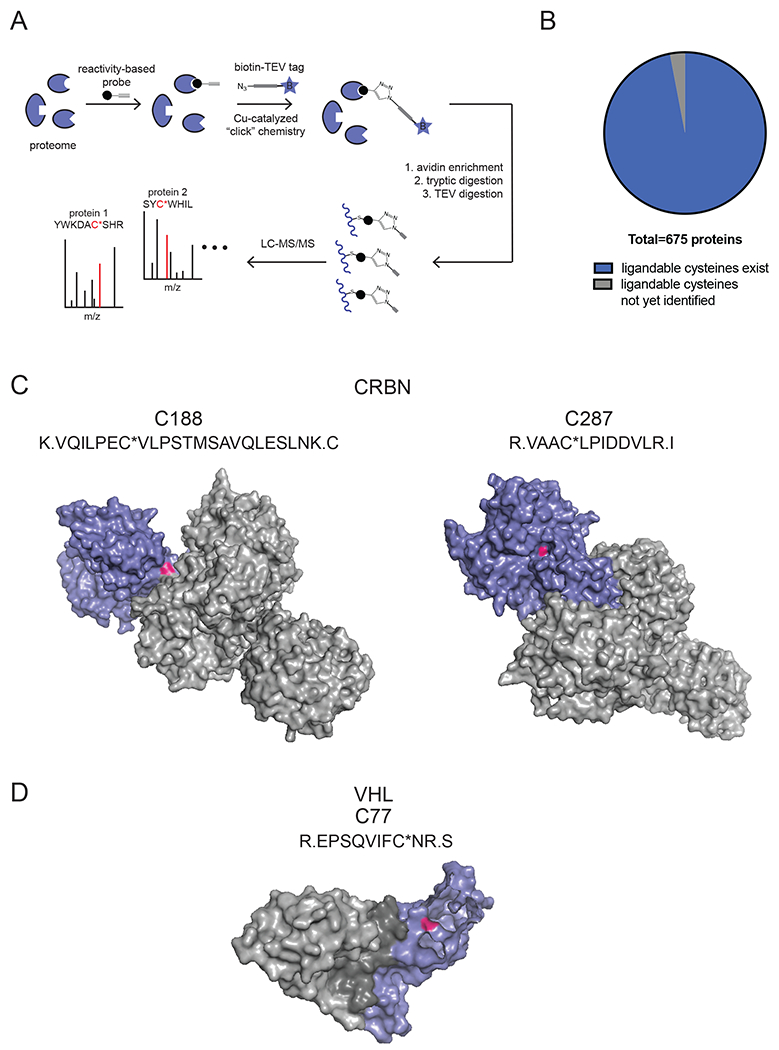
(A) ABPP-based chemoproteomic approaches using reactivity-based probes. Reactivity-based alkyne functionalized probes can be used to label reactive cysteines in complex proteomes, after which a TEV protease-cleavable biotin-azide tag can be appended to probe-labeled proteins using copper-catalyzed azide-alkyne cycloaddition (CuAAC). Probe-modified proteins can be avidinenriched and digested with trypsin, and probe-modified tryptic peptides can be eluted by TEV protease for quantitative proteomic analysis. (B) Aggregating our lab’s chemoproteomic datasets of all probe-modified cysteines from ABPP experiments, we have identified probe-modified cysteines across 97% of human E3 ligases. Data can be found in Supplemental Table 1. (C) Probe-modified tryptic peptides and structure of the reactive cysteines C188 and C287 in CRBN. Shown is the structure of CRBN-DDB1 with CRBN in blue, DDB1 in gray, and C188/C287 in magenta (PDB 4tz4). (D) Probe-modified tryptic peptides and structure of the reactive cysteine C77 in VHL. Shown is the structure of VHL-elongin B/C with VHL in blue, elongin B/C in gray, and C77 in magenta (PDB 1vcb).
Table 1.
Representative list of reactive cysteines within E3 ligases*
| E3 ligase | Uniprot ID | Reactive Cysteine(s) |
|---|---|---|
| RNF114 | Q9Y4L5 | C8**, C110 |
| RNF4 | P78317 | C132/C135** |
| DCAF16 | Q9NXF7 | C177 or C179** |
| DCAF11 | Q8TEB1 | C460** |
| FEM1B | Q9UK73 | C186** |
| CRBN | Q96SW2 | C188***, C287 |
| VHL | P40337 | C77*** |
| RNF2 | C99496 | C72*** |
| RNF14 | Q9UBS8 | C262***, C417 |
| RNF20 | Q5VTR2 | C383, C905, C924 |
| RNF25 | Q96BH1 | C316*** |
| RNF31 | Q96EP0 | C504***, C551 |
| RNF111 | Q6ZNA4 | C968*** |
| RNF113A | O15541 | C15*** |
| RNF123 | Q5XPI4 | C461*** |
| RNF126 | Q9BV68 | C32 |
| RNF128 | Q8TEB7 | C15*** |
| RNF130 | Q86XS8 | C320*** |
| RNF149 | Q8NC42 | C295*** |
| RNF167 | Q9H6Y7 | C271*** |
| RNF180 | Q86T96 | C28*** |
| RNF213 | Q63HN8 | C614*** |
| UBR1 | Q8IWV7 | C180, C1603*** |
| UBR2 | Q8IWV8 | C1360, C1619***, C576 |
| UBR4 | Q5T4S7 | C1358, C2554***, C4049, C2222, C1274 |
| ZNF598 | Q86UK7 | C456*** |
| KCTD21 | Q4G0X4 | C166*** |
| KBTBD7 | Q8WVZ9 | C527/C529*** |
| KLHL20 | Q9Y2M5 | C356*** |
| KLHL24 | Q6TFL4 | C359*** |
| KEAP1 | Q14145 | C23, C288, C196, C319 |
| FBXL18 | Q96ME1 | C468*** |
| FBXL7 | Q9UJT9 | C386/C401*** |
| FBXW8 | Q8N3Y1 | C579*** |
| FBXO10 | Q9UK96 | C430*** |
| FBXO22 | Q8NEZ5 | C228*** |
| FBXO30 | Q8TB52 | C592***, C433, C723/C725 |
| FBXO9 | Q9UK97 | C68*** |
| SKP1 | P63208 | C120, C160 |
| SKP2 | Q13309 | C113***, C240, C205, C345 |
| ASB6 | Q9NWX5 | C153*** |
| ASB9 | Q96DX5 | C216*** |
| RAB40C | Q96S21 | C159*** |
| NOSIP | Q9Y314 | C8*** |
| PRPF19 | Q9UMS4 | C298***, C230, C351 |
| STUB1 | Q9UNE7 | C199***, C48 |
| UBE4A | Q14139 | C79***, C989, C1002 |
| DCAF5 | Q96JK2 | C503*** |
| DCAF7 | P61962 | C61***, C120 |
| DCAF8 | Q5TAQ9 | C297, C272 |
| DCAF13 | Q9NV06 | C190, C215, C120 |
Reactive cysteines annotated are derived from the aggregate chemoproteomic data in Table S1. Listed cysteines show at least 7 aggregate spectral counts for the particular site.
From experimental data from Zhang, Cravatt et al. 2019; Spradlin, Nomura et al. 2019; Ward, Nomura et al. 2019; Zhang, Cravatt et al. 2021; or Henning, Nomura et al. 202165,66,81,82,100.
Represents the dominant probe-modified site over other sites identified in the same E3 ligase from the aggregate chemoproteomic data in Table S1.
Across 675 E3 ubiquitin ligases, substrate receptors, and related components mined, we identified probe-modified cysteines across 653 of these proteins. These sites of probe-labeling represent potential solvent-accessible reactive cysteines that exist across 97% of E3 ligases (Fig. 3B). We have included a Table S1 that includes all probe-modified sites within these 653 proteins identified from our aggregate chemoproteomic data with the individual peptides and sites of modification identified for each E3 ligase, the aggregate spectral counts identified for each site, and the number of experiments the particular probe-modified tryptic peptide has been observed in. There are several caveats to this list of probe-modified sites within human E3 ligases that we point out here before further interpretation of these data below. First, these 675 proteins mined do not represent the total list of all human E3 ligases. There are likely many more E3 ligases that we failed to mine here. Second, while we believe that within these data exist many potential ligandable cysteines within E3 ligases, there are also likely a large fraction of these sites that do not represent true ligandable sites and may just represent surface cysteines that are not part of binding pockets. Third, there are very likely many reactive and ligandable cysteines within E3 ligases that are not represented in this list. This may be for many reasons, including E3 ligases that may 1) be of low abundance; 2) not be expressed in the cell lines profiled to date; 3) show poor ionization of tryptic peptides; 4) show poor reactivity with the iodoacetamide probe; or 5) show destabilization, unfolding, or aggregation upon cell lysis. Fourth, this list only includes probe-modified cysteines and does not include other amino acids that can potentially be interrogated by other reactivity-based probes. These data merely represent an aggregate list of probe-modified cysteines wherein we have quantified the number of times each particular probe-modified tryptic peptide has appeared across our group’s internal chemoproteomic datasets.
To identify potentially interesting cysteines that could be targeted by covalent ligands, we postulated that probe-modified cysteines within E3 ligases that showed higher aggregate spectral counts for a particular cysteine, compared to other sites within the same E3 ligase, may represent more reactive and potentially more ligandable cysteines. Caveats to this premise include cysteines located in regions within a protein sequence that do not yield suitable tryptic peptides with respect to ionization and compatibility with mass spectrometry-based sequencing, and labeling of surface-exposed cysteines that may not be part of binding pockets. However, we conjectured that the aggregate chemoproteomic data would still yield potential ligandable sites within E3 ligases that may be of interest to the TPD community.
Consistent with this premise, if we assess RNF114, a RING E3 ligase for which we have previously identified nimbolide and EN219 as covalent recruiters that can be used in PROTAC applications68,69, we observe nine probe-modified cysteines - C8, C49, C32/C52, C64/C67, C110, C173, and C176. However, among these nine sites, C8 collectively shows 1641 aggregate spectral counts over the next most recognized site C110 with 40 spectral counts (Table S1). Both nimbolide and EN219 react specifically with C8 on RNF114. However, this does not always correlate. For example, in DCAF16 we observe three probe-modified cysteines - C119, C173, and C179; where C119 shows the highest number of aggregate spectral counts. However, Zhang and Cravatt et al. found that their KB02 likely engages with C177 or C179, and not C119. However, C119 may represent another additional site that could be liganded for DCAF16 recruitment. FEM1B and DCAF11 either have not yet appeared in our chemoproteomic profiling experiments or were captured with low aggregate spectral counts.
Nonetheless, we still believe that this dataset likely has many potential entry points into ligand discovery against E3 ligases. For example, in assessing CRBN, an E3 ligase for which IMiDs exist as recruiters but for which a covalent recruiter has not yet been disclosed, we observe four probe-modified cysteines - C188, C287, C326, and C441 - in which C188 and C287 show the highest aggregate spectral counts with 422 and 100, respectively, compared to the other sites (Table S1). Interestingly, C188 sits at the interface between CRBN and DDB1, indicating that targeting this site might either disrupt or stabilize CRBN/DDB1 interactions (Fig. 3C). In contrast, C287 appears to be in an allosteric site within CRBN that sits at the bottom of defined pocket (Fig. 3C). C287 may be a potential ligandable cysteine that can be targeted within CRBN. VHL also possesses a probe-modified cysteine C77 which also appears to be part of a potential binding pocket (Fig. 3D).
An important consideration for using this database of potential ligandable cysteines within E3 ligases to prioritize development of covalent E3 ligase recruiters is the location of the cysteine in the protein. Many E3 ligase recruiters that have been discovered thus far, including for CRBN, VHL, RNF114, DCAF15, cIAP, and FEM1B, target substrate recognition domains within the E3 ligases. Whether an E3 ligase-targeting ligand needs to bind to a substrate recognition site or not to be successfully recruited for TPD remains unclear. Targeting allosteric sites within the E3 ligase that are not directly involved in substrate recognition may still yield successful recruitment. However, if the cysteine rests within a known interaction domain between the E3 ligase and an important functional adapter protein necessary for the function of the E3 ligase complex, these sites would likely not be ideal for recruiter development.
Other important considerations for prioritizing E3 ligases within the database include the cell-type, tissue, or cell compartment expression of the E3 ligase in relation to the target of interest if one aims to achieve location-specific degradation of a protein. Another critical consideration is the functional role of the individual E3 ligases. Many E3 ligases remain poorly characterized as to their endogenous biochemical, cellular, and physiological functions, and the E3 ligases listed in our Table S1 may cause toxicity upon recruitment, may not be involved in ubiquitin conjugation, and may not confer specific ubiquitin linkages that destine a protein for degradation versus the many other roles of ubiquitin or ubiquitin-like proteins 110, Some classes of E3 ligases, such as the RING-in-between RING (RBR) proteins are highly regulated and are autoinhibited and may not be ideal or more complicated for recruitment and TPD 111. The ligandability and the ability to develop potent and selective chemical matter for a particular E3 ligase must be balanced with these aforementioned aspects associated with the biology of each E3 ligase.
Hundreds of additional probe-modified cysteines exist across the >600 human E3 ligases mined here against our lab’s aggregate chemoproteomic data. Many of these sites may represent unique ligandable sites that can be accessed with covalent ligand discovery approaches, and we encourage the ubiquitin proteasome and targeted protein degradation fields to mine this database and deploy covalent ligand screening platforms to expand the arsenal of E3 ligase recruiters that can be exploited in PROTACs and molecular glue degraders.
4. Conclusions and Future Directions
In this review, we have summarized the E3 uibiquitin ligases that have been successfully recruited and manipulated to date and present our aggregate chemoproteomic data of reactive cysteines that have been identified across the human E3 ligase family of over 600 members.
Supplementary Material
Acknowledgement
We thank the members of the Nomura Research Group and Novartis Institutes for BioMedical Research for critical reading of the manuscript. This work was supported by Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies (NB-CPACT) for all listed authors. This work was also supported by the Nomura Research Group and the Mark Foundation for Cancer Research ASPIRE Award.
Footnotes
Competing Financial Interests Statement
DKN is a co-founder, shareholder, and adviser for Frontier Medicines.
References
- (1).Burslem GM; Crews CM Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181 (1), 102–114. 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cromm PM; Crews CM Targeted Protein Degradation: From Chemical Biology to Drug Discovery. Cell Chem. Biol 2017, 24 (9), 1181–1190. 10.1016/j.chembiol.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Nalawansha DA; Crews CM PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol 2020, 27 (8), 998–1014. 10.1016/j.chembiol.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schreiber SL The Rise of Molecular Glues. Cell 2021, 184 (1), 3–9. 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- (5).Banik SM; Pedram K; Wisnovsky S; Ahn G; Riley NM; Bertozzi CR Lysosome-Targeting Chimaeras for Degradation of Extracellular Proteins. Nature 2020, 584 (7820), 291–297. 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Takahashi D; Moriyama J; Nakamura T; Miki E; Takahashi E; Sato A; Akaike T; Itto-Nakama K; Arimoto H AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76 (5), 797–810.e10. 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- (7).Samarasinghe KTG; Jaime-Figueroa S; Burgess M; Nalawansha DA; Dai K; Hu Z; Bebenek A; Holley SA; Crews CM Targeted Degradation of Transcription Factors by TRAFTACs: TRAnscription Factor TArgeting Chimeras. Cell Chem. Biol 2021, 28 (5), 648–661.e5. 10.1016/j.chembiol.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Morreale FE; Kleine S; Leodolter J; Ovchinnikov S; Kley J; Kurzbauer R; Hoi DM; Meinhart A; Hartl M; Haselbach D; Kaiser M; Clausen T BacPROTACs Mediate Targeted Protein Degradation in Bacteria. bioRxiv 2021, 2021.06.09.447781. 10.1101/2021.06.09.447781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu X; Haniff HS; Childs-Disney JL; Shuster A; Aikawa H; Adibekian A; Disney MD Targeted Degradation of the Oncogenic MicroRNA 17-92 Cluster by Structure-Targeting Ligands. J. Am. Chem. Soc 2020, 142 (15), 6970–6982. 10.1021/jacs.9b13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Siriwardena SU; Munkanatta Godage DNP; Shoba VM; Lai S; Shi M; Wu P; Chaudhary SK; Schreiber SL; Choudhary A Phosphorylation-Inducing Chimeric Small Molecules. J. Am. Chem. Soc 2020, 142 (33), 14052–14057. 10.1021/jacs.0c05537. [DOI] [PubMed] [Google Scholar]
- (11).Yamazoe S; Tom J; Fu Y; Wu W; Zeng L; Sun C; Liu Q; Lin J; Lin K; Fairbrother WJ; Staben ST Heterobifunctional Molecules Induce Dephosphorylation of Kinases–A Proof of Concept Study. J. Med. Chem 2020, 63 (6), 2807–2813. 10.1021/acs.jmedchem.9b01167. [DOI] [PubMed] [Google Scholar]
- (12).Henning NJ; Boike L; Spradlin JN; Ward CC; Belcher B; Brittain SM; Hesse M; Dovala D; McGregor LM; McKenna JM; Tallarico JA; Schirle M; Nomura DK Deubiquitinase-Targeting Chimeras for Targeted Protein Stabilization. bioRxiv 2021, 2021.04.30.441959. 10.1101/2021.04.30.441959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Komander D; Rape M The Ubiquitin Code. Annu. Rev. Biochem 2012, 81, 203–229. 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- (14).Chen ZJ; Sun LJ Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol. Cell 2009, 33 (3), 275–286. 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- (15).Jevtić P; Haakonsen DL; Rapé M An E3 Ligase Guide to the Galaxy of Small-Molecule-Induced Protein Degradation. Cell Chem. Biol 2021, S2451-9456(21)00157-4. 10.1016/j.chembiol.2021.04.002. [DOI] [PubMed] [Google Scholar]
- (16).Kannt A; Ðikić I Expanding the Arsenal of E3 Ubiquitin Ligases for Proximity-Induced Protein Degradation. Cell Chem. Biol 2021, S2451-9456(21)00162-8. 10.1016/j.chembiol.2021.04.007. [DOI] [PubMed] [Google Scholar]
- (17).Bond MJ; Crews CM Proteolysis Targeting Chimeras (PROTACs) Come of Age: Entering the Third Decade of Targeted Protein Degradation. RSC Chem. Biol 2021, 2 (3), 725–742. 10.1039/d1cb00011j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bond MJ; Chu L; Nalawansha DA; Li K; Crews CM Targeted Degradation of Oncogenic KRASG12C by VHL-Recruiting PROTACs. ACS Cent. Sci 2020, 6 (8), 1367–1375. 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zeng M; Xiong Y; Safaee N; Nowak RP; Donovan KA; Yuan CJ; Nabet B; Gero TW; Feru F; Li L; Gondi S; Ombelets LJ; Quan C; Jänne PA; Kostic M; Scott DA; Westover KD; Fischer ES; Gray NS Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chem. Biol 2020, 27 (1), 19–31.e6. 10.1016/j.chembiol.2019.12.006. [DOI] [PubMed] [Google Scholar]
- (20).Donovan KA; Ferguson FM; Bushman JW; Eleuteri NA; Bhunia D; Ryu S; Tan L; Shi K; Yue H; Liu X; Dobrovolsky D; Jiang B; Wang J; Hao M; You I; Teng M; Liang Y; Hatcher J; Li Z; Manz TD; Groendyke B; Hu W; Nam Y; Sengupta S; Cho H; Shin I; Agius MP; Ghobrial IM; Ma MW; Che J; Buhrlage SJ; Sim T; Gray NS; Fischer ES Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell 2020, 183 (6), 1714–1731.e10. 10.1016/j.cell.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ciechanover A; Hod Y; Hershko A A Heat-Stable Polypeptide Component of an ATP-Dependent Proteolytic System from Reticulocytes. 1978. Biochem. Biophys. Res. Commun 2012, 425 (3), 565–570. 10.1016/j.bbrc.2012.08.025. [DOI] [PubMed] [Google Scholar]
- (22).Hershko A; Ciechanover A; Rose IA Resolution of the ATP Dependent Proteolytic System from Reticulocytes: A Component That Interacts with ATP. Proc. Natl. Acad. Sci. U. S. A 1979, 76 (7), 3107–3110. 10.1073/pnas.76.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bett JS Proteostasis Regulation by the Ubiquitin System. Essays Biochem. 2016, 60 (2), 143–151. 10.1042/EBC20160001. [DOI] [PubMed] [Google Scholar]
- (24).Mészáros B; Kumar M; Gibson TJ; Uyar B; Dosztányi Z Degrons in Cancer. Sci. Signal 2017, 10 (470). 10.1126/scisignal.aak9982. [DOI] [PubMed] [Google Scholar]
- (25).Sakamoto KM; Kim KB; Kumagai A; Mercurio F; Crews CM; Deshaies RJ Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. U. S. A 2001, 98 (15), 8554–8559. 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Schneekloth JS; Fonseca FN; Koldobskiy M; Mandal A; Deshaies R; Sakamoto K; Crews CM Chemical Genetic Control of Protein Levels: Selective in Vivo Targeted Degradation. J. Am. Chem. Soc 2004, 126 (12), 3748–3754. 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- (27).Smith AR; Pucheault M; Tae HS; Crews CM Targeted Intracellular Protein Degradation Induced by a Small Molecule: En Route to Chemical Proteomics. Bioorg. Med. Chem. Lett 2008, 18 (22), 5904–5908. 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Itoh Y; Ishikawa M; Naito M; Hashimoto Y Protein Knockdown Using Methyl Bestatin-Ligand Hybrid Molecules: Design and Synthesis of Inducers of Ubiquitination-Mediated Degradation of Cellular Retinoic Acid-Binding Proteins. J. Am. Chem. Soc 2010, 132 (16), 5820–5826. 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- (29).Ito T; Ando H; Suzuki T; Ogura T; Hotta K; Imamura Y; Yamaguchi Y; Handa H Identification of a Primary Target of Thalidomide Teratogenicity. Science 2010, 327 (5971), 1345–1350. 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- (30).Epstein ACR; Gleadle JM; McNeill LA; Hewitson KS; O’Rourke J; Mole DR; Mukherji M; Metzen E; Wilson MI; Dhanda A; Tian YM; Masson N; Hamilton DL; Jaakkola P; Barstead R; Hodgkin J; Maxwell PH; Pugh CW; Schofield CJ; Ratcliffe PJC Elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases That Regulate HIF by Prolyl Hydroxylation. Cell 2001, 107 (1), 43–54. 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- (31).Hon WC; Wilson MI; Harlos K; Claridge TDW; Schofield CJ; Pugh CW; Maxwell PH; Ratcliffe PJ; Stuart DI; Jones EY Structural Basis for the Recognition of Hydroxyproline in HIF-1α by PVHL. Nature 2002, 417 (6892), 975–978. 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- (32).Bargagna-Mohan P; Baek SH; Lee H; Kim K; Mohan R Use of PROTACS as Molecular Probes of Angiogenesis. Bioorg. Med. Chem. Lett 2005, 15 (11), 2724–2727. 10.1016/j.bmcl.2005.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang X; Feng S; Fan J; Li X; Wen Q; Luo N New Strategy for Renal Fibrosis: Targeting Smad3 Proteins for Ubiquitination and Degradation. Biochem. Pharmacol 2016, 116, 200–209. 10.1016/j.bcp.2016.07.017. [DOI] [PubMed] [Google Scholar]
- (34).Buckley DL; Van Molle I; Gareiss PC; Tae HS; Michel J; Noblin DJ; Jorgensen WL; Ciulli A; Crews CM Targeting the von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the VHL/HIF-1α Interaction. J. Am. Chem. Soc 2012, 134 (10), 4465–4468. 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Galdeano C; Gadd MS; Soares P; Scaffidi S; Van Molle I; Birced I; Hewitt S; Dias DM; Ciulli A Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities. J. Med. Chem 2014, 57 (20), 8657–8663. 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bondeson DP; Mares A; Smith IED; Ko E; Campos S; Miah AH; Mulholland KE; Routly N; Buckley DL; Gustafson JL; Zinn N; Grandi P; Shimamura S; Bergamini G; Faelth-Savitski M; Bantscheff M; Cox C; Gordon DA; Willard RR; Flanagan JJ; Casillas LN; Votta BJ; Den Besten W; Famm K; Kruidenier L; Carter PS; Harling JD; Churcher I; Crews CM Catalytic in Vivo Protein Knockdown by Small-Molecule PROTACs. Nat. Chem. Biol 2015, 11 (8), 611–617. 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zengerle M; Chan KH; Ciulli A Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol 2015, 10 (8), 1770–1777. 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Pettersson M; Crews CM PROteolysis TArgeting Chimeras (PROTACs) — Past, Present and Future. Drug Discov. Today Technol 2019, 31, 15–27. 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Dale B; Cheng M; Park K-S; Kaniskan HÜ; Xiong Y; Jin J Advancing Targeted Protein Degradation for Cancer Therapy. Nat. Rev. Cancer 2021. 10.1038/s41568-021-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bond MJ; Chu L; Nalawansha DA; Li K; Crews CM Targeted Degradation of Oncogenic KRASG12Cby VHL-Recruiting PROTACs. ACS Cent. Sci 2020. 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zeng M; Xiong Y; Safaee N; Nowak RP; Donovan KA; Yuan CJ; Nabet B; Gero TW; Feru F; Li L; Gondi S; Ombelets LJ; Quan C; Jänne PA; Kostic M; Scott DA; Westover KD; Fischer ES; Gray NS Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chem. Biol 2020, 27 (1), 19–31.e6. 10.1016/j.chembiol.2019.12.006. [DOI] [PubMed] [Google Scholar]
- (42).Ito T; Yamaguchi Y; Handa H Exploiting Ubiquitin Ligase Cereblon as a Target for Small-Molecule Compounds in Medicine and Chemical Biology. Cell Chem. Biol 2021, S2451-9456(21)00205-1. 10.1016/j.chembiol.2021.04.012. [DOI] [PubMed] [Google Scholar]
- (43).Ito T; Ando H; Suzuki T; Ogura T; Hotta K; Imamura Y; Yamaguchi Y; Handa H Identification of a Primary Target of Thalidomide Teratogenicity. Science 2010, 327 (5971), 1345–1350. 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- (44).Matyskiela ME; Couto S; Zheng X; Lu G; Hui J; Stamp K; Drew C; Ren Y; Wang M; Carpenter A; Lee C-W; Clayton T; Fang W; Lu C-C; Riley M; Abdubek P; Blease K; Hartke J; Kumar G; Vessey R; Rolfe M; Hamann LG; Chamberlain PP SALL4 Mediates Teratogenicity as a Thalidomide-Dependent Cereblon Substrate. Nat. Chem. Biol 2018, 14 (10), 981–987. 10.1038/s41589-018-0129-x. [DOI] [PubMed] [Google Scholar]
- (45).Donovan KA; An J; Nowak RP; Yuan JC; Fink EC; Berry BC; Ebert BL; Fischer ES Thalidomide Promotes Degradation of SALL4, a Transcription Factor Implicated in Duane Radial Ray Syndrome. eLife 2018, 7, e38430. 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lopez-Girona A; Mendy D; Ito T; Miller K; Gandhi AK; Kang J; Karasawa S; Carmel G; Jackson P; Abbasian M; Mahmoudi A; Cathers B; Rychak E; Gaidarova S; Chen R; Schafer PH; Handa H; Daniel TO; Evans JF; Chopra R Cereblon Is a Direct Protein Target for Immunomodulatory and Antiproliferative Activities of Lenalidomide and Pomalidomide. Leukemia 2012, 26 (11), 2326–2335. 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Krönke J; Udeshi ND; Narla A; Grauman P; Hurst SN; McConkey M; Svinkina T; Heckl D; Comer E; Li X; Ciarlo C; Hartman E; Munshi N; Schenone M; Schreiber SL; Carr SA; Ebert BL Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells. Science 2014, 343 (6168), 301–305. 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Krönke J; Fink EC; Hollenbach PW; MacBeth KJ; Hurst SN; Udeshi ND; Chamberlain PP; Mani DR; Man HW; Gandhi AK; Svinkina T; Schneider RK; McConkey M; Järås M; Griffiths E; Wetzler M; Bullinger L; Cathers BE; Carr SA; Chopra R; Ebert BL Lenalidomide Induces Ubiquitination and Degradation of CK1α in Del(5q) MDS. Nature 2015, 523 (7559), 183–188. 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lu G; Middleton RE; Sun H; Naniong MV; Ott CJ; Mitsiades CS; Wong KK; Bradner JE; Kaelin WG The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science 2014, 343 (6168), 305–309. 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chamberlain PP; Lopez-Girona A; Miller K; Carmel G; Pagarigan B; Chie-Leon B; Rychak E; Corral LG; Ren YJ; Wang M; Riley M; Delker SL; Ito T; Ando H; Mori T; Hirano Y; Handa H; Hakoshima T; Daniel TO; Cathers BE Structure of the Human Cereblon-DDB1-Lenalidomide Complex Reveals Basis for Responsiveness to Thalidomide Analogs. Nat. Struct. Mol. Biol 2014, 21 (9), 803–809. 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- (51).Fischer ES; Böhm K; Lydeard JR; Yang H; Stadler MB; Cavadini S; Nagel J; Serluca F; Acker V; Lingaraju GM; Tichkule RB; Schebesta M; Forrester WC; Schirle M; Hassiepen U; Ottl J; Hild M; Beckwith REJ; Harper JW; Jenkins JL; Thomä NH Structure of the DDB1-CRBN E3 Ubiquitin Ligase in Complex with Thalidomide. Nature 2014, 512 (7512), 49–53. 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Matyskiela ME; Lu G; Ito T; Pagarigan B; Lu C-C; Miller K; Fang W; Wang N-Y; Nguyen D; Houston J; Carmel G; Tran T; Riley M; Nosaka L; Lander GC; Gaidarova S; Xu S; Ruchelman AL; Handa H; Carmichael J; Daniel TO; Cathers BE; Lopez-Girona A; Chamberlain PP A Novel Cereblon Modulator Recruits GSPT1 to the CRL4(CRBN) Ubiquitin Ligase. Nature 2016, 535 (7611), 252–257. 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- (53).Sievers QL; Petzold G; Bunker RD; Renneville A; Słabicki M; Liddicoat BJ; Abdulrahman W; Mikkelsen T; Ebert BL; Thomä NH Defining the Human C2H2 Zinc Finger Degrome Targeted by Thalidomide Analogs through CRBN. Science 2018, 362 (6414). 10.1126/science.aat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Nishiguchi G; Keramatnia F; Min J; Chang Y; Jonchere B; Das S; Actis M; Price J; Chepyala D; Young B; McGowan K; Slavish PJ; Mayasundari A; Jarusiewicz JA; Yang L; Li Y; Fu X; Garrett SH; Papizan JB; Kodali K; Peng J; Pruett Miller SM; Roussel MF; Mullighan C; Fischer M; Rankovic Z Identification of Potent, Selective, and Orally Bioavailable Small-Molecule GSPT1/2 Degraders from a Focused Library of Cereblon Modulators. J. Med. Chem 2021, 64 (11), 7296–7311. 10.1021/acs.jmedchem.0c01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Powell CE; Du G; Che J; He Z; Donovan KA; Yue H; Wang ES; Nowak RP; Zhang T; Fischer ES; Gray NS Selective Degradation of GSPT1 by Cereblon Modulators Identified via a Focused Combinatorial Library. ACS Chem. Biol 2020, 15 (10), 2722–2730. 10.1021/acschembio.0c00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Winter GE; Buckley DL; Paulk J; Roberts JM; Souza A; Dhe-Paganon S; Bradner JE Phthalimide Conjugation as a Strategy for in Vivo Target Protein Degradation. Science 2015, 348 (6241), 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Lu J; Qian Y; Altieri M; Dong H; Wang J; Raina K; Hines J; Winkler JD; Crew AP; Coleman K; Crews CM Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol 2015, 22 (6), 755–763. 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Hines J; Lartigue S; Dong H; Qian Y; Crews CM MDM2-Recruiting PROTAC Offers Superior, Synergistic Antiproliferative Activity via Simultaneous Degradation of BRD4 and Stabilization of P53. Cancer Res. 2019, 79 (1), 251–262. 10.1158/0008-5472.CAN-18-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Sekine K; Takubo K; Kikuchi R; Nishimoto M; Kitagawa M; Abe F; Nishikawa K; Tsuruo T; Naito M Small Molecules Destabilize CIAP1 by Activating Auto-Ubiquitylation. J. Biol. Chem 2008, 283 (14), 8961–8968. 10.1074/jbc.M709525200. [DOI] [PubMed] [Google Scholar]
- (60).Itoh Y; Ishikawa M; Kitaguchi R; Sato S; Naito M; Hashimoto Y Development of Target Protein-Selective Degradation Inducer for Protein Knockdown. Bioorg. Med. Chem 2011, 19 (10), 3229–3241. 10.1016/j.bmc.2011.03.057. [DOI] [PubMed] [Google Scholar]
- (61).Itoh Y; Kitaguchi R; Ishikawa M; Naito M; Hashimoto Y Design, Synthesis and Biological Evaluation of Nuclear Receptor-Degradation Inducers. Bioorg. Med. Chem 2011, 19 (22), 6768–6778. 10.1016/j.bmc.2011.09.041. [DOI] [PubMed] [Google Scholar]
- (62).Ohoka N; Nagai K; Hattori T; Okuhira K; Shibata N; Cho N; Naito M Cancer Cell Death Induced by Novel Small Molecules Degrading the TACC3 Protein via the Ubiquitin-Proteasome Pathway. Cell Death Dis. 2014, 5 (11), 1–10. 10.1038/cddis.2014.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Mares A; Miah AH; Smith IED; Rackham M; Thawani AR; Cryan J; Haile PA; Votta BJ; Beal AM; Capriotti C; Reilly MA; Fisher DT; Zinn N; Bantscheff M; MacDonald TT; Vossenkamper A; Dace P; Churcher I; Benowitz AB; Watt G; Denyer J; Scott-Stevens P; Harling JD Extended Pharmacodynamic Responses Observed upon PROTAC-Mediated Degradation of RIPK2. Commun. Biol 2020, 3 (1), 140. 10.1038/s42003-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).den Besten W; Verma K; Yamazoe S; Blaquiere N; Phung W; Izrael-Tomasevic A; Mulvihill MM; Helgason E; Prakash S; Goncharov T; Vucic D; Dueber E; Fairbrother WJ; Wertz I; Yu K; Staben ST Primary Amine Tethered Small Molecules Promote the Degradation of X-Linked Inhibitor of Apoptosis Protein. J. Am. Chem. Soc 2021, 143 (28), 10571–10575. 10.1021/jacs.1c05269. [DOI] [PubMed] [Google Scholar]
- (65).Ward CC; Kleinman JI; Brittain SM; Lee PS; Chung CYS; Kim K; Petri Y; Thomas JR; Tallarico JA; McKenna JM; Schirle M; Nomura DK Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. ACS Chem. Biol 2019, 14 (11), 2430–2440. 10.1021/acschembio.8b01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Spradlin JN; Hu X; Ward CC; Brittain SM; Jones MD; Ou L; To M; Proudfoot A; Ornelas E; Woldegiorgis M; Olzmann JA; Bussiere DE; Thomas JR; Tallarico JA; McKenna JM; Schirle M; Maimone TJ; Nomura DK Harnessing the Anti-Cancer Natural Product Nimbolide for Targeted Protein Degradation. Nat. Chem. Biol 2019, 15 (7), 747–755. 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Tong B; Spradlin JN; Novaes LFT; Zhang E; Hu X; Moeller M; Brittain SM; McGregor LM; McKenna JM; Tallarico JA; Schirle M; Maimone TJ; Nomura DK A Nimbolide-Based Kinase Degrader Preferentially Degrades Oncogenic BCR-ABL. ACS Chem. Biol 2020, 15 (7), 1788–1794. 10.1021/acschembio.0c00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Luo M; Spradlin JN; Boike L; Tong B; Brittain SM; McKenna JM; Tallarico JA; Schirle M; Maimone TJ; Nomura DK Chemoproteomics-Enabled Discovery of Covalent RNF114-Based Degraders That Mimic Natural Product Function. Cell Chem. Biol 2021, 28 (4), 559–566.e15. 10.1016/j.chembiol.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Spradlin JN; Hu X; Ward CC; Brittain SM; Jones MD; Ou L; To M; Proudfoot A; Ornelas E; Woldegiorgis M; Olzmann JA; Bussiere DE; Thomas JR; Tallarico JA; McKenna JM; Schirle M; Maimone TJ; Nomura DK Harnessing the Anti-Cancer Natural Product Nimbolide for Targeted Protein Degradation. Nat. Chem. Biol 2019, 15 (7), 747–755. 10.1038/s41589-019-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Zhang X; Crowley VM; Wucherpfennig TG; Dix MM; Cravatt BF Electrophilic PROTACs That Degrade Nuclear Proteins by Engaging DCAF16. Nat. Chem. Biol 2019, 15 (7), 737–746. 10.1038/s41589-019-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Baur M; Gneist M; Owa T; Dittrich C Clinical Complete Long-Term Remission of a Patient with Metastatic Malignant Melanoma under Therapy with Indisulam (E7070). Melanoma Res. 2007, 17 (5), 329–331. 10.1097/CMR.0b013e3282ef4189. [DOI] [PubMed] [Google Scholar]
- (72).Mita M; Kelly KR; Mita A; Ricart AD; Romero O; Tolcher A; Hook L; Okereke C; Krivelevich I; Rossignol DP; Giles FJ; Rowinsky EK; Takimoto C Phase I Study of E7820, an Oral Inhibitor of Integrin α-2 Expression with Antiangiogenic Properties, in Patients with Advanced Malignancies. Clin. Cancer Res 2011, 17 (1), 193–200. 10.1158/1078-0432.CCR-10-0010. [DOI] [PubMed] [Google Scholar]
- (73).Bekaii-Saab TS; Mortazavi A; Hicks LG; Zalupski M; Pelley RJ; Chan KK; Kraut EH A Phase II Study of Chloroquinoxaline Sulfonamide (CQS) in Patients with Metastatic Colorectal Carcinoma (MCRC). Invest. New Drugs 2006, 24 (4), 343–346. 10.1007/s10637-005-4827-3. [DOI] [PubMed] [Google Scholar]
- (74).Uehara T; Minoshima Y; Sagane K; Sugi NH; Mitsuhashi KO; Yamamoto N; Kamiyama H; Takahashi K; Kotake Y; Uesugi M; Yokoi A; Inoue A; Yoshida T; Mabuchi M; Tanaka A; Owa T Selective Degradation of Splicing Factor CAPERα By Anticancer Sulfonamides. Nat. Chem. Biol 2017, 13 (6), 675–680. 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- (75).Han T; Goralski M; Gaskill N; Capota E; Kim J; Ting TC; Xie Y; Williams NS; Nijhawan D Anticancer Sulfonamides Target Splicing by Inducing RBM39 Degradation via Recruitment to DCAF15. Science 2017, 356 (6336). 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- (76).Bussiere DE; Xie L; Srinivas H; Shu W; Burke A; Be C; Zhao J; Godbole A; King D; Karki RG; Hornak V; Xu F; Cobb J; Carte N; Frank AO; Frommlet A; Graff P; Knapp M; Fazal A; Okram B; Jiang S; Michellys PY; Beckwith R; Voshol H; Wiesmann C; Solomon JM; Paulk J Structural Basis of Indisulam-Mediated RBM39 Recruitment to DCAF15 E3 Ligase Complex. Nat. Chem. Biol 2020, 16 (1), 15–23. 10.1038/s41589-019-0411-6. [DOI] [PubMed] [Google Scholar]
- (77).Faust TB; Yoon H; Nowak RP; Donovan KA; Li Z; Cai Q; Eleuteri NA; Zhang T; Gray NS; Fischer ES Structural Complementarity Facilitates E7820-Mediated Degradation of RBM39 by DCAF15. Nat. Chem. Biol 2020, 16 (1), 7–14. 10.1038/s41589-019-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Du X; Volkov OA; Czerwinski RM; Tan HL; Huerta C; Morton ER; Rizzi JP; Wehn PM; Xu R; Nijhawan D; Wallace EM Structural Basis and Kinetic Pathway of RBM39 Recruitment to DCAF15 by a Sulfonamide Molecular Glue E7820. Structure 2019, 27 (11), 1625–1633.e3. 10.1016/j.str.2019.10.005. [DOI] [PubMed] [Google Scholar]
- (79).Li L; Mi D; Pei H; Duan Q; Wang X; Zhou W; Jin J; Li D; Liu M; Chen Y In Vivo Target Protein Degradation Induced by PROTACs Based on E3 Ligase DCAF15. Signal Transduct. Target. Ther 2020, 5 (1), 4–6. 10.1038/s41392-020-00245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Tong B; Luo M; Xie Y; Spradlin JN; Tallarico JA; McKenna JM; Schirle M; Maimone TJ; Nomura DK Bardoxolone Conjugation Enables Targeted Protein Degradation of BRD4. Sci. Rep 2020, 10 (1), 15543. 10.1038/s41598-020-72491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zhang X; Luukkonen LM; Eissler CL; Crowley VM; Yamashita Y; Schafroth MA; Kikuchi S; Weinstein DS; Symons KT; Nordin BE; Rodriguez JL; Wucherpfennig TG; Bauer LG; Dix MM; Stamos D; Kinsella TM; Simon GM; Baltgalvis KA; Cravatt BF DCAF11 Supports Targeted Protein Degradation by Electrophilic Proteolysis-Targeting Chimeras. J. Am. Chem. Soc 2021, 143 (13), 5141–5149. 10.1021/jacs.1c00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Henning NJ; Manford AG; Spradlin JN; Brittain SM; McKenna JM; Tallarico JA; Schirle M; Rape M; Nomura DK Discovery of a Covalent FEM1B Recruiter for Targeted Protein Degradation Applications. bioRxiv 2021, 2021.04.15.439993. 10.1101/2021.04.15.439993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Manford AG; Rodríguez-Pérez F; Shih KY; Shi Z; Berdan CA; Choe M; Titov DV; Nomura DK; Rape M A Cellular Mechanism to Detect and Alleviate Reductive Stress. Cell 2020, 183 (1), 46–61.e21. 10.1016/j.cell.2020.08.034. [DOI] [PubMed] [Google Scholar]
- (84).Nomura DK; Dix MM; Cravatt BF Activity-Based Protein Profiling for Biochemical Pathway Discovery in Cancer. Nat. Rev. Cancer 2010, 10 (9), 630–638. 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Evans MJ; Cravatt BF Mechanism-Based Profiling of Enzyme Families. Chem. Rev 2006, 106 (8), 3279–3301. 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- (86).Moellering RE; Cravatt BF How Chemoproteomics Can Enable Drug Discovery and Development. Chem. Biol 2012, 19 (1), 11–22. 10.1016/j.chembiol.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Spradlin JN; Zhang E; Nomura DK Reimagining Druggability Using Chemoproteomic Platforms. Acc. Chem. Res 2021, 54 (7), 1801–1813. 10.1021/acs.accounts.1c00065. [DOI] [PubMed] [Google Scholar]
- (88).Weerapana E; Wang C; Simon GM; Richter F; Khare S; Dillon MBD; Bachovchin DA; Mowen K; Baker D; Cravatt BF Quantitative Reactivity Profiling Predicts Functional Cysteines in Proteomes. Nature 2010, 468 (7325), 790–795. 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Wang C; Weerapana E; Blewett MM; Cravatt BF A Chemoproteomic Platform to Quantitatively Map Targets of Lipid-Derived Electrophiles. Nat. Methods 2014, 11 (1), 79–85. 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Backus KM; Correia BE; Lum KM; Forli S; Horning BD; González-Páez GE; Chatterjee S; Lanning BR; Teijaro JR; Olson AJ; Wolan DW; Cravatt BF Proteome-Wide Covalent Ligand Discovery in Native Biological Systems. Nature 2016, 534 (7608), 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Hacker SM; Backus KM; Lazear MR; Forli S; Correia BE; Cravatt BF Global Profiling of Lysine Reactivity and Ligandability in the Human Proteome. Nat. Chem 2017, 9 (12), 1181–1190. 10.1038/nchem.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Brulet JW; Borne AL; Yuan K; Libby AH; Hsu K-L Liganding Functional Tyrosine Sites on Proteins Using Sulfur-Triazole Exchange Chemistry. J. Am. Chem. Soc 2020, 142 (18), 8270–8280. 10.1021/jacs.0c00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Kuljanin M; Mitchell DC; Schweppe DK; Gikandi AS; Nusinow DP; Bulloch NJ; Vinogradova EV; Wilson DL; Kool ET; Mancias JD; Cravatt BF; Gygi SP Reimagining High-Throughput Profiling of Reactive Cysteines for Cell-Based Screening of Large Electrophile Libraries. Nat. Biotechnol 2021, 39 (5), 630–641. 10.1038/s41587-020-00778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Bar-Peled L; Kemper EK; Suciu RM; Vinogradova EV; Backus KM; Horning BD; Paul TA; Ichu T-A; Svensson RU; Olucha J; Chang MW; Kok BP; Zhu Z; Ihle NT; Dix MM; Jiang P; Hayward MM; Saez E; Shaw RJ; Cravatt BF Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 2017, 171 (3), 696–709.e23. 10.1016/j.cell.2017.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Vinogradova EV; Zhang X; Remillard D; Lazar DC; Suciu RM; Wang Y; Bianco G; Yamashita Y; Crowley VM; Schafroth MA; Yokoyama M; Konrad DB; Lum KM; Simon GM; Kemper EK; Lazear MR; Yin S; Blewett MM; Dix MM; Nguyen N; Shokhirev MN; Chin EN; Lairson LL; Melillo B; Schreiber SL; Forli S; Teijaro JR; Cravatt BF An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell 2020, 182 (4), 1009–1026.e29. 10.1016/j.cell.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Grossman EA; Ward CC; Spradlin JN; Bateman LA; Huffman TR; Miyamoto DK; Kleinman JI; Nomura DK Covalent Ligand Discovery against Druggable Hotspots Targeted by Anti-Cancer Natural Products. Cell Chem. Biol 2017, 24 (11), 1368–1376.e4. 10.1016/j.chembiol.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Bateman LA; Nguyen TB; Roberts AM; Miyamoto DK; Ku W-M; Huffman TR; Petri Y; Heslin MJ; Contreras CM; Skibola CF; Olzmann JA; Nomura DK Chemoproteomics-Enabled Covalent Ligand Screen Reveals a Cysteine Hotspot in Reticulon 4 That Impairs ER Morphology and Cancer Pathogenicity. Chem. Commun. Camb. Engl 2017, 53 (53), 7234–7237. 10.1039/c7cc01480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Ward CC; Kleinman JI; Nomura DK NHS-Esters As Versatile Reactivity-Based Probes for Mapping Proteome-Wide Ligandable Hotspots. ACS Chem. Biol 2017, 12 (6), 1478–1483. 10.1021/acschembio.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Chung CY-S; Shin HR; Berdan CA; Ford B; Ward CC; Olzmann JA; Zoncu R; Nomura DK Covalent Targeting of the Vacuolar H+-ATPase Activates Autophagy via MTORC1 Inhibition. Nat. Chem. Biol 2019, 15 (8), 776–785. 10.1038/s41589-019-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Zhang X; Crowley VM; Wucherpfennig TG; Dix MM; Cravatt BF Electrophilic PROTACs That Degrade Nuclear Proteins by Engaging DCAF16. Nat. Chem. Biol 2019, 15 (7), 737–746. 10.1038/s41589-019-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Counihan JL; Wiggenhorn AL; Anderson KE; Nomura DK Chemoproteomics-Enabled Covalent Ligand Screening Reveals ALDH3A1 as a Lung Cancer Therapy Target. ACS Chem. Biol 2018, 13 (8), 1970–1977. 10.1021/acschembio.8b00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Counihan JL; Duckering M; Dalvie E; Ku W-M; Bateman LA; Fisher KJ; Nomura DK Chemoproteomic Profiling of Acetanilide Herbicides Reveals Their Role in Inhibiting Fatty Acid Oxidation. ACS Chem. Biol 2017, 12 (3), 635–642. 10.1021/acschembio.6b01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Ford B; Bateman LA; Gutierrez-Palominos L; Park R; Nomura DK Mapping Proteome-Wide Targets of Glyphosate in Mice. Cell Chem. Biol 2017, 24 (2), 133–140. 10.1016/j.chembiol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- (104).Boike L; Cioffi AG; Majewski FC; Co J; Henning NJ; Jones MD; Liu G; McKenna JM; Tallarico JA; Schirle M; Nomura DK Discovery of a Functional Covalent Ligand Targeting an Intrinsically Disordered Cysteine within MYC. Cell Chem. Biol 2021, 28 (1), 4–13.e17. 10.1016/j.chembiol.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Isobe Y; Okumura M; McGregor LM; Brittain SM; Jones MD; Liang X; White R; Forrester W; McKenna JM; Tallarico JA; Schirle M; Maimone TJ; Nomura DK Manumycin Polyketides Act as Molecular Glues between UBR7 and P53. Nat. Chem. Biol 2020, 16 (11), 1189–1198. 10.1038/s41589-020-0557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Cho H; Shen Q; Zhang LH; Okumura M; Kawakami A; Ambrose J; Sigoillot F; Miller HR; Gleim S; Cobos-Correa A; Wang Y; Piechon P; Roma G; Eggimann F; Moore C; Aspesi P; Mapa FA; Burks H; Ross NT; Krastel P; Hild M; Maimone TJ; Fisher DE; Nomura DK; Tallarico JA; Canham SM; Jenkins JL; Forrester WC CYP27A1-Dependent Anti-Melanoma Activity of Limonoid Natural Products Targets Mitochondrial Metabolism. Cell Chem. Biol 2021, S2451-9456(21)00143-4. 10.1016/j.chembiol.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Berdan CA; Ho R; Lehtola HS; To M; Hu X; Huffman TR; Petri Y; Altobelli CR; Demeulenaere SG; Olzmann JA; Maimone TJ; Nomura DK Parthenolide Covalently Targets and Inhibits Focal Adhesion Kinase in Breast Cancer Cells. Cell Chem. Biol 2019, 26 (7), 1027–1035.e22. 10.1016/j.chembiol.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Roberts AM; Miyamoto DK; Huffman TR; Bateman LA; Ives AN; Akopian D; Heslin MJ; Contreras CM; Rape M; Skibola CF; Nomura DK Chemoproteomic Screening of Covalent Ligands Reveals UbA5 As a Novel Pancreatic Cancer Target. ACS Chem. Biol 2017, 12 (4), 899–904. 10.1021/acschembio.7b00020. [DOI] [PubMed] [Google Scholar]
- (109).Roberts LS; Yan P; Bateman LA; Nomura DK Mapping Novel Metabolic Nodes Targeted by Anti-Cancer Drugs That Impair Triple-Negative Breast Cancer Pathogenicity. ACS Chem. Biol 2017, 12 (4), 1133–1140. 10.1021/acschembio.6b01159. [DOI] [PubMed] [Google Scholar]
- (110).Yau R; Rape M The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol 2016, 18 (6), 579–586. 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- (111).Smit JJ; Sixma TK RBR E3-Ligases at Work. EMBO Rep. 2014, 15 (2), 142–154. 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


