Abstract
Genomic and epidemiological surveillance are paramount for the discovery of new viruses with the potential to cross species barriers. Here, we present a new member of the genus Alphavirus found in Trichoprosopon and Wyeomia mosquitoes, tentatively named Pirahy virus (PIRAV). PIRAV was isolated from mosquito pools collected in a rural area of Piraí do Sul, South Brazil. In vitro assays revealed that PIRAV replicates and causes cytopathic effects in vertebrate cell lines such as Vero E6, SH-SY5Y, BHK-21 and UMNSAH/DF-1. Genomic signature analysis supports these results showing a dinucleotide and codon usage balance compatible with several hosts. Phylogenetic analyses placed PIRAV basal to the Venezuelan equine encephalitis complex. Genome analyses, electron microscopy, and biological characterization show findings that may alert for the emergence of a new arbovirus in South America.
Keywords: alphavirus, emerging arboviruses, phylogeny, CpG dinucleotide, codon adaptation, Trichoprosopon, Wyeomyia
1. Introduction
The emergence of a virus is characteristically related both to the evolution and adaptation of viruses and to anthropogenic factors. Unplanned demographic expansion, biodiversity destruction, and global warming directly impact the balance of wild transmission cycles that may impact viral dispersion and new transmission cycles (Gould et al. 2017).
Effective surveillance that enables the discovery of new emerging pathogens before they cause damage is considered the best choice to anticipate and mitigate the effects of emerging infectious diseases (Taylor, Latham, and Woolhouse 2001; King et al. 2006; Jones et al. 2008; Keusch et al. 2010; Day et al. 2012; Allen et al. 2017).
This situation is especially relevant in Brazil, where the circulation of arboviruses from the Flaviviridae and Togaviridae families such as dengue virus, yellow fever virus, Zika virus, West Nile virus, and chikungunya virus (CHIKV) overlaps. Most of them cause similar clinical symptoms at the first stages of infection in humans, impairing differential diagnostic (Carvalho et al. 2019; Barrio-Nuevo et al. 2020; Costa et al. 2021; Brasil. Ministério da Saúde 2021).
Alphaviruses are distributed worldwide and comprise at least 40 described species and a dozen of variants or subtypes of enveloped, single-stranded positive-sense RNA viruses, of which one-third have economic or medical importance (Atkins 2013). Their genome is approximately 11.7 kb in length, with a capped 5′ end and a poly-A tail at the 3′ end, divided into two open reading frames (ORFs) that generate the non-structural proteins (nsP) and the structural proteins (sP) (Strauss and Strauss 1994).
Alphaviruses are geographically divided into two distinct groups, the Old-World and the New World viruses (Weaver et al. 1993). Members of the Old-World alphaviruses, such as CHIKV, Sindbis virus (SINV), Ross River virus (RRV), O’nyong-nyong virus, Barmah Forest virus, and Semliki Forest virus, often cause symptoms like high fever, headache, tiredness, vomiting, diarrhea, aching tendons, joint swelling, muscle pain, or skin rash (Suhrbier, Jaffar-Bandjee, and Gasque 2012). Whereas infections with New World alphaviruses, like Venezuelan equine encephalitis virus (VEEV), Western equine encephalitis viruses (WEEV), Eastern equine encephalitis virus (EEEV), and Everglades virus (EVEV) are associated with encephalitis in humans and domestic animals (Forrester et al. 2012; Griffin 2013). These viruses are maintained in nature by transmission between hematophagous arthropods and susceptible animal hosts, including humans. Additionally, systematic high-throughput analysis has led to the description of an increasing number of non-pathogenic alphaviruses, apparently restricted to insects (Nasar et al. 2012; Hermanns et al. 2017, 2020; Torii et al. 2018; Batovska et al. 2020).
In this study, we describe a new member of the genus Alphavirus, tentatively named Pirahy virus (PIRAV), which was isolated from a pool of Trichoprosopon mosquitoes and also identified in a pool of Wyeomia mosquitoes collected in Piraí do Sul municipality, Brazil, during epidemiological surveillance efforts. PIRAV was isolated in a cell culture model and successfully infected vertebrate cell lines, causing marked cytopathic effects. Phylogenetic analyses, codon adaptation index, dinucleotide usage, electron microscopy, and biological characterization of this new alphavirus are presented and may alert for the potential emergence of a new arbovirus in South America.
2. Results
2.1. Viral screening in mosquitoes and genetic analyses
A total of 4,640 mosquitoes divided into 661 pools were collected at different regions from Paraná State (southern Brazil) for alphavirus and flavivirus molecular prospection. Mosquitoes were morphologically identified as belonging to fifteen distinct genera as follows: Aedeomyia, Aedes, Anopheles, Chagasia, Coquillettidia, Culex, Haemagogus, Limatus, Mansonia, Psorophora, Runchomyia, Sabethes, Shannoniana, Trichoprosopon, and Wyeomyia. Two mosquito pools (codes MS772 and MS773), containing three female mosquitoes each, yielded positive results for alphavirus using a generic RT-PCR protocol (Sánchez-Seco et al. 2001). All specimens were collected on 25 April 2017 at Piraí do Sul, Brazil (24°31’40’’S 49°53’02’’W) (Fig. 1). In addition to the two alphavirus-positive pools, other 73 pools comprising 325 mosquitoes obtained specifically from Piraí do Sul were analyzed and yielded no detectable results for alphavirus (Table S1).
Figure 1.
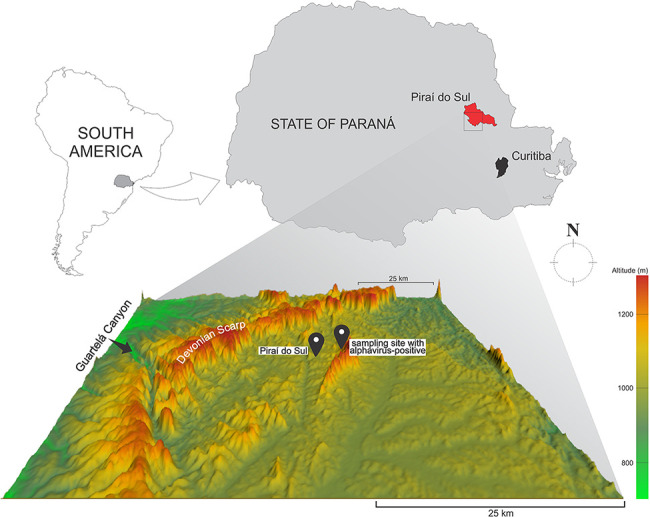
The location of Piraí do Sul municipality, State of Paraná, Brazil, where the mosquito pools MS772 and MS773 were sampled. Below, a relief map showing the Guartelá Canyon and the Devonian Scarp.
The alphavirus cDNA fragments obtained from the MS772 and MS773 pools were amplified using the combined protocols of Sánchez-Seco et al. (Sánchez-Seco et al. 2001) and Hermanns et al. (Hermanns et al. 2017). The amplicons (653 and 1,003 bp in length, both targeting the nsP4 gene were sequenced. Nucleotide alignment with sequences retrieved from GenBank using a BLAST algorithm revealed 75 to 76 per cent of nucleotide identity with Pixuna virus (PIXV) and VEEV, respectively.
The identity of the mosquito species from the MS772 and MS773 pools was confirmed by partial sequencing of the cytochrome C oxidase subunit I (COI) mitochondrial gene. The 689-bp fragment exhibited 94 per cent sequence identity for multiple species of Wyeomyia as the mosquito host in the MS772 pool, corroborating the morphological identification of Wyeomyia limai. The molecular identification of a 696-bp fragment showed over 97 per cent identity for Trichoprosopon pallidoventer as the mosquito host in the MS773 pool.
Viral isolation was successfully achieved from the Trichoprosopon pool (MS773) homogenate inoculated in C6/36 cells. Cytopathic effect (CPE) was observed 5 days post-inoculation. Viral identity was confirmed by indirect immunofluorescence assay (IFA), RT-PCR, and sequencing of the RT-PCR amplicons.
Next-generation sequencing (NGS) of the third passage of the isolated virus yielded a near-complete viral genome in a single contig containing 11,190 nt with a coverage average of 1,756×. Genome organization and amino acid size of each protein are displayed in Fig. 2. The analysis of the near-complete genome sequence of the new isolate showed a 77 per cent identity with VEEV. This result fulfills the criteria of the International Committee on Taxonomy of Viruses for a new species definition: a minimum of a 21 per cent divergence at nucleotide level within an antigenic complex. The nucleotide sequence was deposited in the GenBank database under the accession number OK539813.
Figure 2.
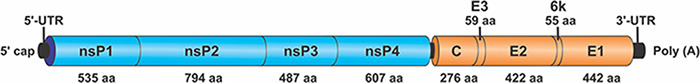
Schematic diagram representing the genome organization of PIRAV.
The phylogenetic relationship of the new virus, tentatively named Pirahy (PIRAV), within the genus Alphavirus was inferred using the maximum likelihood (ML) method. Alignments were analyzed using either the complete genomes or with the nsP and sP genomic regions. Phylogenetic analysis placed PIRAV in the base of the VEEV complex with a bootstrap value of 100 per cent, indicating strong support for these nodes. The same result was observed when the nsP and sP trees were analyzed separately (Fig. 3). The polyprotein sequence of PIRAV shows ≤50 per cent overall amino acid identity compared to species of VEEV complex (Table 1). PIRAV nsP4 and nsP1 proteins have more than 70 and 60 per cent identity to VEEV at the amino acid level, respectively, whereas nsP3 and E2 proteins showed less than 30 per cent identity of amino acid sequences compared to all species of VEEV complex.
Figure 3.
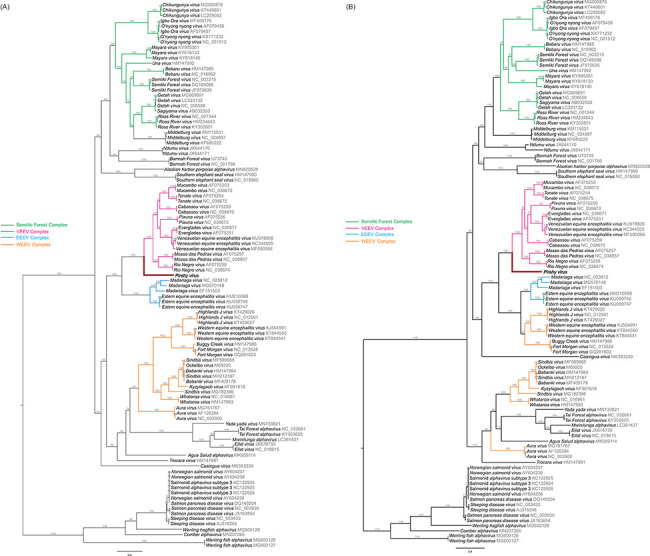
Maximum likelihood analysis of PIRAV incorporated with previously described alphaviruses and their variants. Phylogenetic trees were built based either on the A) structural and B) non-structural genomic regions of alphaviruses. PIRAV branch is highlighted in red and named in bold. GenBank accession numbers are plotted close to all taxa. Bootstrap values are shown above the branches. The scale bar correspond to 0.4 and indicates the number of substitutions per site.
Table 1.
Amino acid sequence identities of individual proteins between PIRAV and closely related alphaviruses of VEEV complex.
| Amino acid sequence identity to PIRAV (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species/proteins | nsP1 | nsP2 | nsP3 | nsP4 | Capsid | E3 | E2 | 6k | E1 | Total |
| Cabassou virus | 63 | 59 | 27 | 71 | 45 | 27 | 27 | 44 | 27 | 49 |
| Everglades virus | 65 | 57 | 21 | 73 | 41 | 30 | 27 | 39 | 24 | 47 |
| Mosso das Pedras virus | 62 | 59 | 16 | 72 | 43 | 31 | 28 | 29 | 24 | 46 |
| Mucambo virus | 65 | 60 | 20 | 74 | 49 | 46 | 28 | 39 | 27 | 50 |
| Pixuna virus | 60 | 56 | 5 | 71 | 47 | 24 | 25 | 25 | 25 | 45 |
| Rio Negro virus | 63 | 59 | 24 | 71 | 48 | 42 | 38 | 24 | 25 | 49 |
| Tonate virus | 62 | 57 | 24 | 73 | 44 | 37 | 27 | 32 | 25 | 48 |
| Venezuelan Equine Encephalitis virus | 65 | 59 | 22 | 73 | 44 | 31 | 29 | 31 | 25 | 49 |
In addition, to detect putative recombination events in the genome of the new alphavirus, seven algorithms in the RDP4 software were performed (Martin et al. 2015). No recombination events were detected in PIRAV using sequence alignments refined by Gblocks software (Castresana 2000).
2.2. PIRAV genomic signatures
To investigate PIRAV adaptability to potential hosts, we performed dinucleotide usage analysis (Lobo et al. 2009), which included alphaviruses that infect humans (CHIKV, VEEV, PIXV), insect-specific alphaviruses (Eilat virus (EILV), Taï forest virus (TAFV), Mwinilunga virus (MWAV)), and several vertebrate (Pan troglodytes, Macaca mulatta, Homo sapiens, Equus caballus, Myotis lucifugus, Rattus norvegicus, Mus musculus, Gallus gallus, Anas platyrhynchos, Danio rerio) and invertebrate (Culex quinquefasciatus, Aedes aegypti, Anopheles gambiae, Drosophila melanogaster) animals. As expected, there is suppression of dinucleotides CG and TA in vertebrates due to the presence of proteins that can recognize those dinucleotides at both DNA and RNA level in these organisms and trigger an immune response (Mansur, Smith, and Ferguson 2014; Takata et al. 2017; Schwerk et al. 2019). Accordingly, Diptera does not show a bias against CG (Bewick et al. 2016). Insect-specific viruses display a dinucleotide profile much like their hosts, while viruses that have to shuttle between hosts (arboviruses) display a mixed dinucleotide bias, with a clear suppression of CG and TA, although not at the same extent as observed in vertebrates. In this analysis, PIRAV groups together with the arboviruses CHIKV and VEEV (Fig. 4A). Normalized codon adaptation index (nCAI) and relative codon deoptimization index (nRCDI) are complementary indexes that can be used to evaluate the relation of virus codon adaptation to its hosts. Both nCAI (Fig. 4B) and nRCDI (Fig. 4C) analysis indicate a match of codon usage to a variety of vertebrate hosts, similar to what is observed for VEEV and notably different from EILV, an insect-specific alphavirus.
Figure 4.
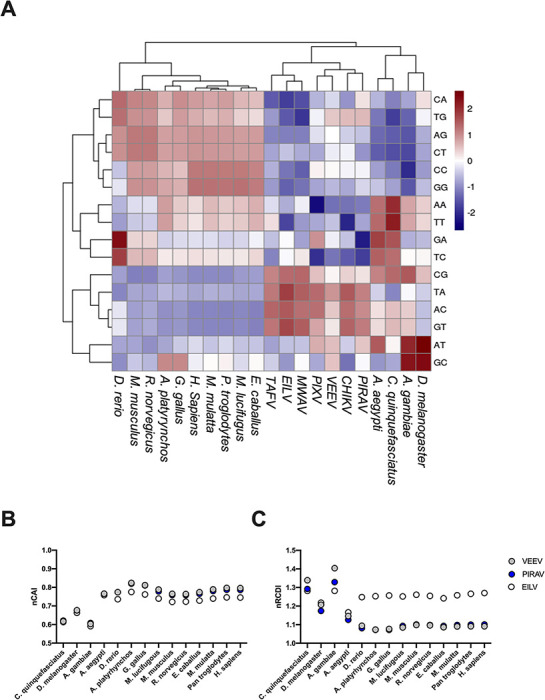
Genomic signatures for host and viral species. (A) Hierarchical cluster analysis of dinucleotide odds ratio values for hosts and viruses included in this study. (B) Normalized CAI and (C) RCDI for VEEV, PIRAV, and EILV in different hosts. VEEV, PIRAV, and EILV are represented in gray, blue, and white, respectively.
Altogether, these genomic signatures support the phylogenetic analysis that places PIRAV at the base of VEEV complex and suggests that this virus may replicate in both vertebrate and invertebrate hosts.
2.3. In vitro characterization of PIRAV
As phylogenetic and genomic signatures analysis suggested the grouping of PIRAV with viruses that are able to infect vertebrates, we evaluated the ability of PIRAV to infect a variety of cells.
Immunofluorescence assays showed that PIRAV was able to replicate in vertebrate cell lines causing cytopathic effects in vitro in a time-dependent manner (Figs 5 and S2). Morphological changes and cell death were observed in human neuroblastoma SH-SY5Y cells infected with PIRAV (Fig. 5A), VEEV (Fig. 5B), and CHIKV (Fig. 5C), when compared to the mock-infected cells (Fig. 5D). PIRAV as well as the human pathogenic VEEV and CHIKV successfully replicated in C6/36 mosquito cells (Fig. 5E), as well as in Vero E6 (Fig. 5F), SH-SY5Y (Fig. 5G), and BHK-21 (Fig. 5H) lineages. Loss of viability was observed for all the above-mentioned cell lines in different degrees, although it was less pronounced in BHK-21 cells infected with PIRAV compared to the other viruses. Interestingly, UMNSAH/DF-1 chicken cells seem to be permissive to VEEV, and to a lesser extent to PIRAV but not to CHIKV (Fig. 5I). ZEM-2S (Danio rerio embryo) cells (Fig. 5J) were refractory to infection by the three alphaviruses tested.
Figure 5.
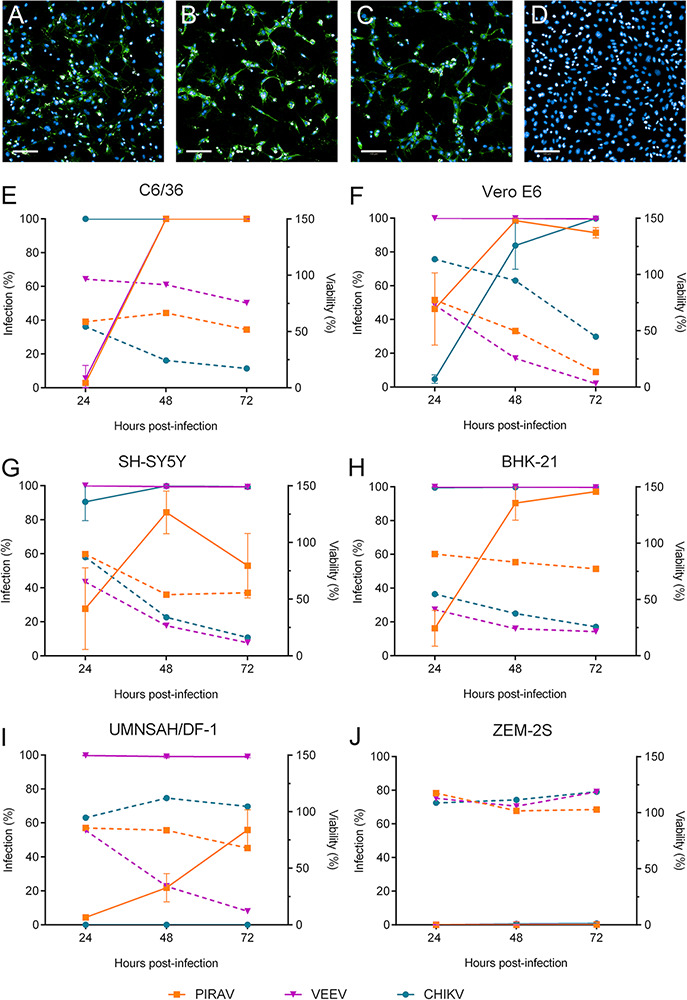
PIRAV infects different cell lines. Immunofluorescence images show the SH-SY5Y cell line 24 h after infection with Pirahy (A), VEEV (B), or CHIKV (C) at an MOI of 1.0 or left uninfected (D). Alphavirus E1 protein is shown in green. Scale bar = 100 µm. Graphs show the time course of PIRAV replication in different cell lineages. Percentages of infection (solid lines) and cell viability (dashed lines) from 24 to 72 h.p.i. are shown for C6/36 (E), Vero E6 (F), SH-SY5Y (G), BHK-21 (H), UMNSAH/DF-1 (I), and ZEM-S2 (J) lineages. Infection was performed at an MOI of 0.1, and results are presented as the mean of duplicate infection (E and J) or as the means from triplicate experiments (F–I). Bars represent the means ± SD.
2.4. PIRAV infection in human peripheral blood mononuclear cells
Once we confirmed the ability of PIRAV to infect and replicate in a variety of vertebrate cells in vitro, including human-derived lineage (SH-SY5Y), we further investigated the susceptibility of human cells to PIRAV infection. Since arboviruses of medical importance have been shown to target blood mononuclear cells during infection and given the crucial role of these cells during immune response, we verified if peripheral blood mononuclear cells (PBMCs) are susceptible to PIRAV infection in vitro.
PBMCs isolated from healthy donors were infected with PIRAV, CHIKV, or VEEV at two different multiplicity of infection (MOI 1 and 10). Infection frequency was analyzed by flow cytometry after 24, 48, and 72 h post-infection (h.p.i.) in each cell population (CD14+, CD4+, CD8+, and CD19+). CD8+ and CD4+ cells were susceptible to PIRAV infection, in addition to CD14+ and CD19+ cells from one donor. PIRAV, CHIKV, and VEEV infection peaked at 48 h.p.i. (Fig. 6). We also analyzed cell death at 24, 48 and 72 h.p.i. using annexin V–7-aminoactinomycin D (7-AAD) staining. While the viability of lymphocytes infected with PIRAV was overall lower than mock cells, it remained relatively stable at all timepoints (Fig. S3). Although statistical significance was not observed for monocytes, infection by PIRAV at MOI 10 seemed to induce progressive cell death in this population. A similar pattern was observed for monocytes infected with CHIKV, as cell viability progressively decreased at each timepoint. Interestingly, VEEV infection induced an increase in cell viability at 48 h.p.i. before decreasing at 72 h.p.i.
Figure 6.
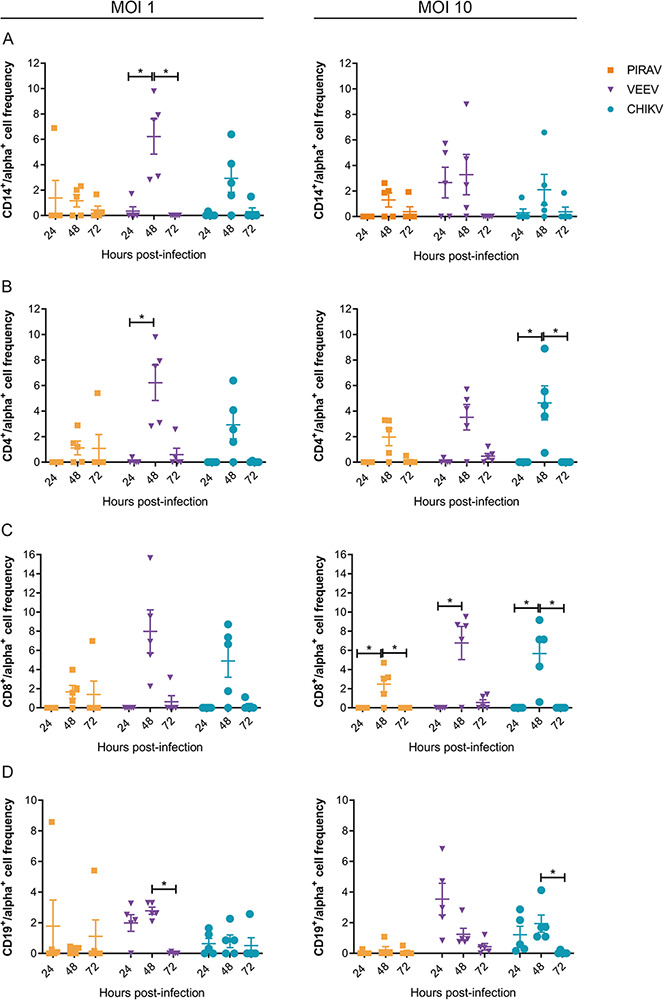
Human PBMCs are susceptible to infection by PIRAV. PBMCs from five healthy donors were infected with PIRAV (orange), VEEV (purple), or CHIKV (blue) at MOIs of 1 and 10. Infection was analyzed by flow cytometry at 24, 48, and 72 h.p.i. by intracellular detection of alphavirus envelope protein. Graphs show frequencies of infection of monocytes (A), CD4+ (B), CD8+ lymphocytes and B lymphocytes, which were normalized by subtracting the fluorescence from mock samples. Bars represent the means of infection ± SD.
2.5. Transmission electron microscopy of PIRAV
Nucleocapsids (NC) and mature virions (MV) were observed by transmission electron microscopy in both C6/36 and Vero E6 cells infected with PIRAV (Fig. 7). MV were observed budding from the plasma membrane (Fig. 7D and H, arrows), while NC were seen in cytoplasm, frequently close to the endoplasmic reticulum (Fig. 7C and G, black arrowheads) or lining up along the cytoplasmic side of vacuolar membranes (Fig. 7C, white arrowhead). Morphometric analysis of PIRAV MV and NC, as well as MV and NC from VEEV (used as a control) in the same host cells (Vero E6 and C6/36), was performed (Table 2). For PIRAV, the sizes of the MV particles ranged from 38 to 58 nm in diameter in Vero E6 cells and 42 to 57 nm in C6/36 cells. NC particle sizes ranged from 16 to 34 nm in diameter in Vero E6 cells, and 21 to 33 nm in C6/36 cells. For VEEV, the sizes of the MV particles ranged from 40 to 66 nm in diameter in Vero E6 cells and 34 to 60 nm in C6/36 cells. NC particle sizes ranged from 14 to 36 nm in diameter in Vero E6 cells and 20 to 33 nm in C6/36 cells (n = 100 for all conditions), which is an expected range for alphaviruses.
Figure 7.
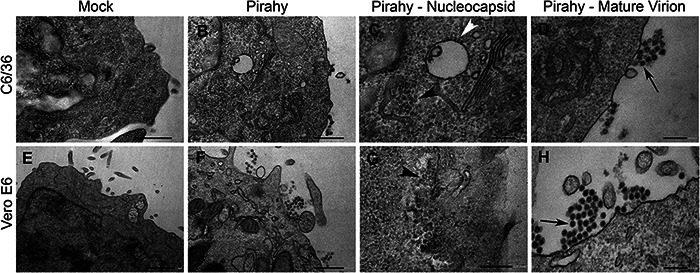
Transmission Electron Micrographs of C6/36- and Vero E6-infected cells. Top row, C6/36 cells. Bottom row, Vero E6 cells. (A and E) Mock cells. (B–D and F–H) PIRAV-infected cells. (C and G) Higher magnification of the infected cell cytoplasm showing NC close to the endoplasmic reticulum (black arrowheads) and lining up along the cytoplasmic side of vacuolar membranes (withe arrowhead). (D and H) Higher magnification of MV close to the plasma membrane (arrows). Scale bars: A, B, and F = 500 nm; C, D, G, and H = 200 nm; E = 1 µm.
Table 2.
Morphometric analysis of PIRAV and VEEV MV and nucleocapsid cores.
| PIRAV (nm ± SD) | VEEV (nm ± SD) | |||
|---|---|---|---|---|
| MV | NC | MV | NC | |
| C6/36 | 49 ± 3 | 28 ± 3 | 47 ± 5 | 26 ± 2 |
| Vero E6 | 51 ± 4 | 26 ± 4 | 51 ± 5 | 22 ± 4 |
Values are expressed as mean diameter (nm) ± standard deviation (SD) (n = 100). MV, mature virion; NC, nucleocapsid core.
3. Discussion
3.1. Viral screening in mosquito pools
Mosquito pools infected with PIRAV were sampled in Piraí do Sul, a city in the State of Paraná, South Brazil. Several dipterous species from the family Culicidae are involved in the transmission of arboviruses and comprise epidemiologically important insects. PIRAV was isolated from Trichoprosopon mosquito pools and also detected in Wyeomyia pools suggesting that these species might be the potential mosquitoes hosts of PIRAV, although further studies are necessary to ascertain its vectorial capacity and dynamics of transmission to vertebrates. Both genera are predominantly forest mosquitoes but can also be found in disturbed areas, present diurnal hematophagic activity, as other Sabethini, and are involved in arboviruses transmission. Anhembi virus (AMBV), Bussuquara virus (BSQV), Ilheus virus, and St. Louis encephalitis virus (SLEV) have been isolated from Trichoprosopon or Wyeomyia species (Downs, Anderson, and Aitken 1956; Galindo and de Rodaniche 1961; Fonseca et al. 1975; Zavortink, Roberts, and Hoch 1983). Generally, viruses from the VEEV complex from which PIRAV has evolutionary proximity are transmitted to vertebrates by the Culex, Psorophora, and Aedes mosquito species (Forrester et al. 2017). However, Trichoprosopon and Wyeomyia species are also related to alphaviruses transmission within the VEEV complex. The alphaviruses Mucambo virus (MUCV) and CABV were isolated from Wyeomyia species (Yuill 1986) and PIXV seems to be naturally found in Trichoprosopon species in Brazil (De Andrade et al. 1964).
3.2. Phylogenetic analysis
The most updated and comprehensive phylogeny of alphaviruses based on whole genomes shows PIRAV placed basally in the clade of VEEV complex with maximum branch support and grouped in the cluster of New World alphaviruses that encompass encephalitic alphaviruses of the Americas.
Although about 50 per cent overall amino acid identity compared to species of VEEV complex, the phylogenetic relationship of PIRAV with the ancestor that originated the VEEV complex reinforces the possibility of PIRAV to infect vertebrate hosts in nature. Not all viruses in the VEEV complex cause pathogenicity in humans, but all have a vertebrate host. Members of the VEEV complex occur in the Neotropical realm that includes part of Florida, where EVEV circulates, usually transmitted by Culex mosquitoes causing neurological disease in humans and horses (Lewis et al. 1974; Calisher et al. 1980). In nature, VEEV uses multiple species of rodents as the main reservoirs, horses as amplification hosts, increasing spillover to humans, and bats as accidental reservoirs and potential dispersing hosts (Guzmán-Terán et al. 2020). MUCV and Mosso das Pedras virus cause flu-like symptoms in humans (Demucha Macias and S’anchez Spindola 1965; Lord 1974), Tonate virus was isolated from humans presenting with mild febrile illnesses (Digoutte and Girault 1976) and Rio Negro virus was supposedly associated with acute febrile illness in humans (Contigiani et al. 1993). Little is known about the viral infection of PIXV and CABV in humans, but both are associated with rodents, equids, bats, and marsupials (Attoui et al. 2007). Although no cases of encephalitis caused by enzootic alphavirus have been recorded as of the time the PIRAV-infected mosquitoes were collected, continuous epidemiological surveillance efforts should be implemented.
Phylogenetics and recombination analyses support the already described ancestral recombinant origin of Highlands J virus, Fort Morgan virus, and WEEV (Hahn et al. 1988), in which the capsid and nsP genes are phylogenetic related to an EEEV-like virus, whereas its envelope glycoprotein genes are more related to a Sindbis-like virus. To address this issue, we performed recombination analyses and no evidence of recombination events in the PIRAV genome were detected. Overall, the nsP tree was highly similar to that observed in complete genome phylogeny and differed only by the position of Una virus and Aura virus (Figs 3 and S1).
3.3. PIRAV genomic signatures
Predicting potential emergence and spread is a key goal in virology, as forecasting these events has implications for vaccine design, drug design, and surveillance of viral pathogens (Dolan, Whitfield, and Andino 2018). Combining analyses, like phylogenetic approaches, NGS, cytosine-phosphate-guanine (CpG) content, and codon adaptation index provide a framework for anticipating potential emergence and allow preparedness (Lobo et al. 2009; Coutinho, Franco, and Lobo 2015; Aguiar et al. 2015; Velazquez-Salinas et al. 2016).
Dinucleotide biases are not sufficient to determine a virus’ host (Di Giallonardo et al. 2017), and PIRAV has not been associated with illness in mammals so far. However, the genomic signatures evaluated here groups PIRAV with well-known human pathogens, such as VEEV and CHIKV, from the same family and puts it in clear contrast with insect-exclusive alphaviruses, such as EILV. Both codon adaptation usage indexes and dinucleotide bias analysis, together with its ability to replicate in mammalian and primary human cells, suggest that PIRAV could be an emergent virus.
3.4. In vitro characterization of PIRAV
To investigate whether PIRAV may infect vertebrate cells, we performed in vitro infection using cell lineages derived from different organisms. Since transmission cycles of encephalitic alphaviruses, such as VEEV and EEEV, are maintained by sylvatic reservoirs like avians and rodents (Go, Balasuriya, and Lee 2014), the UMNSAH/DF-1 cell line from chicken (Gallus gallus) and the BHK-21 fibroblasts from hamster (Mesocricetus auratus) were included in this analysis. It was observed that PIRAV replicates successfully and causes CPE in the arthropod-derived C6/36 line and also in the vertebrate-derived Vero E6 (monkey), SH-SY5Y (human), BHK-21, and UMNSAH/DF-1 cells.
It has been previously shown that laboratory rodents, including hamsters and mice, are susceptible to infection with all the VEE complex viruses (Weaver et al. 2004). Our results showed that PIRAV infects hamster-derived BHK-21 cells at an infection rate similar to VEEV and CHIKV (from the Semliki Forest complex). Interestingly, PIRAV induced less cell death than VEEV or CHIKV in BHK-21 cells, even with a high infection rate (Figs 5 and S2).
PIRAV infects the UMNSAH/DF-1 chicken fibroblast to a lesser extent than VEEV. This cell lineage was also refractory to CHIKV infection. Accordingly, avians are involved in the sylvatic cycle of New World but not of Old-World alphaviruses. Experimental infection of various animal species with CHIKV supports these data, since it revealed that, unlike rodents, avians do not develop viremia nor relevant neutralizing antibodies titers after the challenge (Bowen et al. 2016). In summary, our results indicate that PIRAV has a potential to infect vertebrates, including avian. On the other hand, replication of all the alphaviruses tested in this study failed to efficiently infect zebrafish ZEM-2S cells, regardless of the MOI.
As PIRAV placed basally to the Venezuelan Equine Encephalitis complex, we sought to investigate whether this virus could infect the human neuronal SH-SY5Y cell line. PIRAV replicates in this lineage, leading to morphological alterations and cell death. The infection rate and cell death, however, were lower for PIRAV than for VEEV and CHIKV, even using a higher MOI (Fig. S2). Interestingly, unlike the other viruses tested, we observed an increase in infection of SH-SY5Y cells by PIRAV within 48 h of infection, followed by a decline within 72 h.p.i., while cell viability remained on a plateau between 48 and 72 h.p.i. This effect was less evident when a tenfold higher MOI was used.
3.5. PIRAV infection on human PBMCs
Human PBMCs were shown to be alphavirus targets. In vitro studies have suggested that human monocytes and macrophages are susceptible to CHIKV and RRV infection, respectively (La Linn, Aaskov, and Suhrbier 1996; Her et al. 2010). Similar findings were observed in PBMCs isolated from patients acutely infected with CHIKV, showing active infection of monocytes and, to a lesser extent, of B lymphocytes and plasmacytoid dendritic cells (Her et al. 2010; Michlmayr et al. 2018). Here, we demonstrated that human PBMCs are susceptible to PIRAV infection in vitro. Among populations analyzed, CD4+ and CD8+ were more susceptible to PIRAV, while CD14+ monocytes and CD19+ B lymphocytes seemed to be infected only by CHIKV and VEEV, except by one donor whose CD14+ and CD19+ cells also appeared to be infected by PIRAV. Infection kinetics demonstrated that intracellular viral protein detection peaked at 48 h.p.i. followed by a decrease at 72 h.p.i. Infection frequency decrease could be explained either by inherent controlling of infection by cells and/or by cell death. Indeed, infection at 72 h with CHIKV or VEEV decreases cell viability in monocytes and, to a lesser degree, in lymphocytes, while PIRAV infection slightly reduces cell viability in monocytes at a high MOI. Accordingly, animal models of VEEV infection showed that VEEV replication in lymphoid tissues causes necrosis and lymphocyte apoptosis (Jackson, Sengupta, and Smith 1991).
3.6. Transmission electron microscopy of PIRAV
Alphaviruses’ replication and assembly occur in the cytoplasm and are enveloped by budding at the plasma membrane, where the virions exit the cell. Alphaviruses are enveloped viruses with icosahedral symmetry, forming spherical particles with size of approximately 70 nm diameter (Jose, Snyder, and Kuhn 2009). However, although the sizes of MV and NC from both PIRAV and VEEV were very similar, the average size of MV and NC in Vero E6 and C6/36 cells (Table 2) were smaller than 70 nm. A large range of sizes for MV and NC for PIRAV and VEEV in both host cells were also observed. A variety of diameters and densities of NC were previously observed in BHK cells infected with VEEV, and the authors suggested that these physical characteristics likely reflect differences in capsid protein packing (Lamb et al. 2010).
The measurement of other alphaviruses particle size in ultrathin sections also showed smaller average sizes when compared with the well-known regular 70 nm. The southern elephant seal virus (SESV), from the elephant seal louse L. macrorhini, showed extracellular MV with a diameter of 55 nm (±1 nm) and cytoplasmic NC particles with 29 nm (±2 nm), values very close to what was observed for PIRAV and VEEV in this work. Interestingly, when enveloped spherical particles of SESV obtained from the tissue culture supernatants were measured using negative-contrast electron microscopy, the MV size was improved to 65 nm (±3 nm) (La Linn et al. 2001). This notable difference could be a result of different artifacts induced by regular transmission electron microscopy technique, such as chemical fixation and shrinkage of resin thin sections during imaging with the electron beam. On the other hand, direct observation of negative stained virus particles or even cryo-fixation through flash freeze of samples in its native state can preserve the ultrastructural characteristic of viruses.
4. Conclusions
The description of the new virus, tentatively named PIRAV, might bring contributions to the field of virology in several ways: (1) Phylogenetically, the incorporation of PIRAV sequences could help to elucidate the temporal divergence of the VEEV complex. Times to most recent common ancestors of the VEEV complex were estimated at ca. 149–973 years ago by Forrester et al. (35), a broad confidence limit that could be solved incorporating PIRAV into the analyses. (2) Clinically, it allows the development of new reagents for diagnostic applications, such as monoclonal antibodies. (3) Epidemiologically, potentially emerging viruses are important for strengthening epidemiological surveillance, and may help solve undiagnosed/misdiagnosed cases due to unknown viruses.
5. Material and methods
5.1. Specimen collection
Mosquito collection was performed by the Epidemiological Surveillance group of the Paraná State Health Service between 4 February 2017 to 5 March 2020 as part of the surveillance program for arboviral circulation in the Paraná State, South Brazil. Twenty-one municipalities of the East, Northeast, North, and Northwest regions from the State of Paraná, South Brazil, were evaluated according to the interest of surveillance services, which included Piraí do Sul municipality. Adult mosquitoes, preferably females, were collected using insect aspirators and by active collection techniques between 9:00 a.m. and 10:00 p.m. and were pooled according to morphological identification, sampling location, and date. Morphological identification was performed according to Lane (Lane 1953) and Forattini (Forattini 2002). Alphavirus and flavivirus prospection were performed in our facilities of the Reference Laboratory of Emerging Viruses of the Carlos Chagas Institute/Fiocruz-PR.
5.2. Molecular screening
The screening in the pools of mosquitoes for arboviral identification was performed as described by Tschá et al. (Tschá et al. 2019). Briefly, pools of mosquitoes were mechanically lysed in RNase-free phosphate-buffered saline (PBS) and RNA was extracted using the QIAamp Viral RNA Mini KitTM (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Alphavirus and flavivirus prospection were performed using generic primers and protocol described by Sánchez-Seco et al. (Sánchez-Seco et al. 2001, 2005). The alphavirus-positive pools in this set screening were confirmed using an additional generic RT-PCR protocol (Hermanns et al. 2017). All PCR products were visualized on an 1.5 per cent agarose gel stained with ethidium bromide. PCR products were purified using the High Pure PCR Product Purification Kit (Roche, Mannheim, Germany) before conventional Sanger sequencing for viral identity verification.
5.3. Identification of mosquito species
Fragments of the cytochrome C oxidase subunit I (COI) mitochondrial gene were amplified using genetic markers to confirm the mosquito species identity in the alphavirus-positive pools. The primer sets used were LCO1490 and HCO2198 (Folmer et al. 1994). The genetic material of insects was obtained from mosquito homogenates using a Genomic DNA Extraction Kit (RBC Real GenomicsTM, Banqiao City, Taiwan) according to the manufacturer’s protocol. PCRs were performed using an initial denaturation of 3 min at 95°C followed by 35 cycles: 30 s at 95°C, 30 s at 48°C, and 45 s at 72°C, with a final extension at 72°C for 5 min. PCRs contained 1 × PCR buffer, 2 mM MgCl2, 0.4 mM dNTP, 2.5 U of Taq DNA polymerase, 1 μM of each of the forward and reverse primers, and 1 μL of template DNA in a final volume of 25 μL. PCR amplicon was purified using a High Pure PCR Product Purification Kit (Roche) before conventional Sanger sequencing.
5.4. Viral isolation and titration
PIRAV was isolated from the mosquito lysates that tested positive for alphavirus in the RT-PCR tests. The lysate was diluted in serum-free Leibovitz’s L-15 medium (Gibco, Waltham, USA) and added to C6/36 cells (ATCC® CRL-1660; Manassas, USA). After a 1-h incubation period, additional Leibovitz’s L-15 medium supplemented with 5 per cent fetal bovine serum (FBS), 0.26 per cent tryptose (Sigma-Aldrich, St. Louis, USA), and 25 µg/mL gentamicin (Gibco) was added to the monolayers. Morphological changes were followed daily and viral infection was confirmed by RT-PCR and IFA using an anti-Alphavirus E1 protein (clone 1A4B.6, cat. MAB8754, Merck, Temecula, USA) as primary antibody.
A plaque-forming assay in Vero E6 cells (Sigma-Aldrich, 85020206) was established to titer virus stocks. On the day before the assay, 1 × 105 cells were seeded in 24-well plates and incubated at 37°C with 5 per cent CO2. Titration was preceded by infecting cells with tenfold dilutions of viral supernatants in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 medium (DMEM/F-12—Gibco/Invitrogen) in duplicate. After 1-h incubation at 37°C, the inoculum was replaced with 500 µL of overlay, which was composed of 1.6 per cent carboxymethyl cellulose, and DMEM/F-12 supplemented with 2 per cent FBS, 100 IU/μg/mL penicillin/streptomycin. The plates were incubated at 37°C with 5 per cent CO2 for 7 days. Then, the cells were fixed with 3 per cent paraformaldehyde in PBS and stained with crystal violet.
5.5. Phylogenetics and recombination analyses
Genomic RNA derived from the isolated virus was amplified and sequenced using a combination of the forward generic primers from the protocol of Sánchez-Seco et al. (Sánchez-Seco et al. 2001) and the reverse generic primers from Hermanns et al. (Hermanns et al. 2017) to increase the fragment length and to confirm the isolate identity. The PCR cycling conditions were the same present in Hermanns et al. (Hermanns et al. 2017). The PCR amplicons were purified using a High Pure PCR Product Purification Kit (Roche) before Sanger sequencing.
Posteriorly, the whole-genome sequencing of the isolated virus was performed with the RNA extracted from the supernatant of infected cells, which was precipitated with 7 per cent polyethylene glycol 8000/2.3 per cent sodium chloride prior to RNA purification. Purified RNA was obtained with RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions and converted to cDNA using the High-Capacity RNA-to-cDNATM Kit (Thermo Fisher Scientific). The Nextera®XT kit (Illumina, San Diego, USA) was employed in the library preparation and genome sequencing was performed on the MiSeq platform (Illumina) using a 2 × 250-bp paired-end sequencing strategy. The obtained reads were uploaded to CLC Genomics Workbench v.10.5 (Qiagen) and assembled using the de novo assembly pipeline in the default configuration.
Phylogenetic analyses and recombination assays were performed to investigate the evolutionary relationship of the virus being described here. The consensus sequence generated after genome sequencing was aligned with representative sequences for all alphavirus species, its variants and subtypes described so far. Up to three sequences per virus species were downloaded from GenBank depending upon availability and incorporated in the analysis.
Nucleotide sequences were codon aligned using MACSE v2.03 (Ranwez et al. 2011). To refine the phylogenetic analysis and remove poorly aligned positions with the amount of non-informative sites, the initial aligned sequence dataset was cleaned using Gblocks software v. 0.91b (Castresana 2000). Alignments encompassing the alphavirus nsP and the sP regions were generated and analyzed separately. Furthermore, the full genome sequences were analyzed with concatenated fragments of the two ORFs (results in Fig. S1, Supplementary Material). In addition, the initial codon alignment was used to generate an amino acid identity matrix based on each PIRAV genome-encoded protein against the closest related taxonomic group. Phylogenetic analyses were performed using the ML approach. Prior to the tree reconstruction, the nucleotide substitution model was inferred by JModelTest v.0.1 (Posada 2008) using the corrected Akaike Information Criterion (AICc). ML trees were inferred using IQ-TREE, and branch support was calculated using ultrafast bootstrap approximation algorithm with 1,000 replicates (Nguyen et al. 2015). The phylogenetic trees were rooted to the midpoint and viewed in FigTree v1.4.4 (Rambaut 2019).
The full-length genome alignment before and after the refinement by Gblocks software (Castresana 2000) were screened in order to identify signals of recombination breakpoints in PIRAV sequence using seven detection methods: BootScan (Salminen et al. 1995; Martin et al. 2005), Chimaera (Posada and Crandall 2001), GENECONV (Sawyer 1989; Padidam, Sawyer, and Fauquet 1999), MaxChi (Smith 1992; Posada and Crandall 2001), SiScan (Gibbs, Armstrong, and Gibbs 2000), 3-Seq (Boni, Posada, and Feldman 2007), and the original RDP (Martin and Rybicki 2000) performed triplet-by-triplet using the Recombination Detection Program v.4.16 (RDP4) (Martin et al. 2015). Both BootScan and SiScan were used to check any new signals and to explore for recombination signals detected by all other methods used. The highest acceptable P value cut-off was set to 0.01 and sequences were set as linear. All other parameters were set as default settings.
5.6. Codon analysis
Sequences of all hosts and viruses were retrieved from the National Center for Biotechnology Information in FASTA format. The codon usage table of all hosts was retrieved from the CoCoPUTs database (Alexaki et al. 2019).
Codon adaptation index (CAI) was performed to predict the relative adaptation of the viruses to hosts (Puigbò, Bravo, and Garcia-Vallve 2008). And the Relative Codon Deoptimization Index (RCDI) to evaluate the similarity of the codon usage between virus sequences and hosts (Puigbò, Aragonès, and Garcia-Vallvé 2010). The CAI and RCDI values were computed using the local version of CAIcal-v1.4 (http://genomes.urv.cat/CAIcal/—last accessed 9 September 2021).
5.7. Dinucleotide analysis
Dinucleotide odds ratio is the ratio of the observed and expected frequencies of a dinucleotide in a sequence. This ratio shows patterns of dinucleotide favored by an organism and may indicate selectional and mutational pressures. The dinucleotide odds ratio was calculated using the method shown in (Lobo et al. 2009). Genomic signature analysis was performed using the closest clade to the one of interest which has the better annotated genome.
5.8. Infection of multiple cells lines
The ability of PIRAV to infect arthropod and vertebrate-derived cell lines was evaluated as follows. C6/36 cells derived from Aedes albopictus larvae (ATCC® CRL-1660) and ZEM-2S from Danio rerio embryo fibroblast (ATCC® CRL-2147™) were maintained, respectively, in Leibovitz’s L-15 or LDF (50 per cent L-15, 35 per cent DMEM, and 15 per cent Ham’s F12 media) medium at 28°C. Vero E6 cells from Cercopithecus aethiops kidney (Sigma-Aldrich, 85020206), UMNSAH/DF-1 from Gallus gallus embryo fibroblasts (ATCC® CRL-12203™), BHK-21 from Mesocricetus auratus kidney fibroblasts (ATCC® CCL-10™), and SH-SY5Y from a metastatic neuroblastoma found in human bone marrow (ATCC® CRL-2266™) were grown in DMEM/F-12 medium, supplemented with 5 per cent FBS and 100 IU/ml penicillin (Gibco), 100 µg/ml streptomycin (Gibco), at 37°C with 5 per cent CO2. Cells were seeded in Cellstar® 96-well optical microplates (Greiner Bio-One, Kremsmünster, Austria). After 24 h, cells were infected in duplicate with PIRAV at a MOI of 0.1 and 1.0. For comparison purposes, cells were similarly infected with CHIKV BR/2015/15010 isolate or with a wild type VEEV. Inoculum was removed after 1 h of incubation and replaced with the appropriate media. The culture supernatants were collected and the cells were fixed with cold methanol:acetone (1:1 v/v) at the timepoints 24, 48 and 72 h post-infection (h.p.i.).
Viral replication was assessed by IFA using the primary antibody anti-Alphavirus E1 protein (clone 1A4B.6). It was diluted 1:1000 in blocking buffer (1 per cent bovine serum albumin in PBS) and added to the fixed monolayers for 1 h at 37°C. After the incubation, the cells were washed thrice with washing buffer (0.05 per cent tween 20 in PBS) and further incubated for 1 h with a solution composed of Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen), 3 µM DAPI, Evans blue and blocking buffer. Images were acquired with the Operetta CLS high-content imaging system (PerkinElmer) with a 20× objective. The percentage of infected cells was quantified using the Harmony software (PerkinElmer). Infected cells were those stained with Alexa Fluor 488. The total cell number per image was determined with the DAPI-stained nuclei. Viability of the different groups was calculated compared to mock-infected cells nuclei counting, at the same timepoint.
5.9. Infection of human PBMCs
All donors of the study provided written consent (approved by the Human Ethics Research Committee of Fiocruz under number CAAE: 60643816.6.0000.5248) before participating. Peripheral blood samples of five healthy adult donors ranging from ages 22–30 were collected for isolation of PBMCs by density gradient separation with Ficoll-Paque PLUS (density 1.077 g/mL) (GE Life Science). PMBCs were incubated with PIRAV, CHIKV, and VEEV at MOIs 1 and 10 for 1 h at 37°C and 5 per cent CO2. Uninfected (mock) PBMCs were also cultured as controls. After incubation, the viral inoculum was removed, cells were washed twice with PBS (Lonza) and resuspended in RPMI 1640 medium (Lonza) supplemented with 100 IU/ml penicillin (Gibco), 100 µg/ml streptomycin (Gibco), and 10 per cent FBS. PBMCs were then plated at 5 × 105 cells/well (96-well plates) and maintained at 37°C with 5 per cent CO2 for 24, 48, and 72 h. At each timepoint, culture supernatant was collected and stored at −80°C for viral titration and cells were used for immunophenotyping, intracellular labeling of viral protein and annexin V–7-aminoactinomycin D (7-AAD) staining.
5.10. Immunophenotyping and intracellular labeling
Cells were centrifuged at 3000 rpm for 2 min and recovered in blocking buffer (PBS plus 5 per cent FBS and 1 per cent AB human serum (Lonza)). After 20 min of incubation at room temperature, cells were centrifuged and permeabilized with Cytofix/Cytoperm (BD Biosciences) for 20 min at room temperature. Cells were then washed in Perm/Wash (BD Biosciences) and stained with an anti-alphavirus antibody (clone 1A4B.6) at 1:200 (vol/vol) dilution in Perm/Wash, followed by an Alexa Fluor 488 conjugated anti-mouse IgG antibody (Invitrogen) diluted at 1:400 (vol/vol) in Perm/Wash. Lastly, cells were incubated with a mixture of the following monoclonal antibodies at 1:200 (vol/vol) diluted in blocking buffer: anti-CD4-PE, anti-CD8-PE-Cy5, anti-CD19-PE-Cy7, anti-CD14-BV421 and anti-CD3-APC-Cy7 (BD Biosciences). Cells were washed twice with Perm/Wash between all incubations and were recovered in PBS plus 1.5 per cent paraformaldehyde before flow cytometry analysis on a FACSCanto II cytometer (BD Biosciences).
5.11. Anexin V 7-AAD staining
Cell culture plates were centrifuged at 3000 rpm for 2 min and recovered in binding buffer (BD Biosciences). After 15 min of incubation at room temperature, plates were centrifuged again and cells were incubated for 15 min with annexin V (BD Biosciences) and 7-AAD at 1:100 (vol/vol) dilution in binding buffer. Cells were then washed and recovered in binding buffer before cytometry analysis on a FACSCanto II cytometer (BD Biosciences).
5.12. Transmission electron microscopy
C6/36 and Vero E6 cells were infected with PIRAV or VEEV at a MOI of 0.025 (PIRAV), 0.0025 (VEEV, C6/36) or 0.05 (VEEV, Vero E6) for 72 (C6/36) or 48 h (VeroE6). Mock or infected (PIRAV or VEEV) C6/36 and Vero E6 cells were fixed (2.5 per cent glutaraldehyde, 4 per cent paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.2) at room temperature for 1 h. After washing twice with 0.1 M cacodylate buffer, cells were fixed in 1 per cent OsO4, 0.8 per cent KFe (CN)6 and 5 mM CaCl2 diluted in 0.1 M cacodylate buffer at room temperature for 45 min. Cells were washed twice with 0.1 M cacodylate buffer, dehydrated in increasing concentrations of acetone, and embedded in Poly/Bed 812 resin for 72 h at 60°C. Ultrathin sections (60 nm) were collected in copper grids, stained for 45 min with uranyl acetate and for 5 min with lead citrate. Samples were observed in a JEOL JEM-1400 transmission electron microscope operating at 90 keV.
5.13. Morphometric analysis
Diameters of MV (only MV at the extracellular media were measured) and NC (only NC observed inside the host cell were measured) were determined in transmission electron micrographs of thin sections. Randomly selected cells were imaged in a JEOL JEM-1400 transmission electron microscope equipped with a digital camera (8 megapixels CCD). Profiles of different infected cells were acquired and, in each image, different virus particles and/or NC were measured using ImageJ software (Schneider, Rasband, and Eliceiri 2012). Sample size was 100 (MV and NC) for all conditions.
Supplementary Material
Acknowledgements
To Dr Andrea Cristine Koishi for assistance with the image analysis, Dr Ingrid Larissa Melo de Souza for cell culture support, and Dr Mara Eliza Gasino Joineau for kindly providing us with VEEV. The authors also thank the Program for Technological Development in Tools for Health—PDTIS-FIOCRUZ—for the use of the microscopy and flow cytometry facilities at the Instituto Carlos Chagas/Fiocruz-PR and Coordenação de Vigilância em Saúde e Laboratórios de Referência (CVSLR/Fiocruz) for continuous support. This work was partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq and Fundação Oswaldo Cruz—Fiocruz [grant number PROEP/ICC 442319/2019-3]. C.N.D.S. and D.M. received a CNPq fellowship.
Contributor Information
Marcel Kruchelski Tschá, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Andreia A Suzukawa, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Gabriela Flavia Rodrigues-Luiz, Departamento de Microbiologia, Imunologia e Parasitologia, Centro de Ciências Biológicas (CCB), Universidade Federal de Santa Catarina (UFSC), Av. Prof. Henrique da Silva Fontes 2754, Florianópolis, SC 88040-900, Brazil.
Allan Martins da Silva, Laboratório Central, Secretaria de Estado da Saúde do Paraná, Rua Sebastiana Santana Fraga 1001, São José dos Pinhais, PR 83060-500, Brazil.
Allan Henrique Depieri Cataneo, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Gabriela Mattoso Coelho, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Adão Celestino Ferreira, Núcleo de Entomologia de Foz do Iguaçu, Secretaria de Estado da Saúde do Paraná, R. Santos Dumont 460, Foz do Iguaçu, PR 85851-040, Brazil.
Lia Carolina Soares Medeiros, Laboratório de Biologia Celular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Daniel Mansur, Departamento de Microbiologia, Imunologia e Parasitologia, Centro de Ciências Biológicas (CCB), Universidade Federal de Santa Catarina (UFSC), Av. Prof. Henrique da Silva Fontes 2754, Florianópolis, SC 88040-900, Brazil.
Camila Zanluca, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Claudia N Duarte dos Santos, Laboratório de Virologia Molecular, Instituto Carlos Chagas/Fiocruz-PR, Rua Prof. Algacyr Munhoz Mader 3775, Curitiba, PR 81350-010, Brazil.
Data availability
The PIRAV genome sequence obtained in this study has been deposited in GenBank under accession number OK539813.
Supplementary data
Supplementary data is available at Virus Evolution online.
Funding
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Oswaldo Cruz (Fiocruz).
Conflict of interest:
The authors have declared that no conflict of interest exists.
Author contributions
M.K.T., A.A.S., D.S.M., C.Z., and C.N.D.S. contributed to the study design; A.C.F. collected the moquitoes; A.M.S. performed the morphological identification of the mosquitoes; M.K.T. performed the phylogenetic analysis; A.A.S., A.H.D.C., and G.M.C. performed the cell culture experiments; L.C.A.S.M. performed electron microscopy and analyzed the images; G.F.R.L and D.S.M. performed bioinformatics analyses. M.K.T., A.A.S., G.F.R.L., A.H.D.C., G.M.C., L.C.A.S.M., D.S.M., C.Z., and C.N.D.S. wrote the manuscript. All authors approved the final version of the manuscript.
Etymology
Pirahy virus (PIRAV) derives from the first name of the village that gave rise to the current name of Piraí do Sul municipality, State of Paraná, Brazil.
References
- Aguiar E. R. G. R. et al. (2015) ‘Sequence-independent Characterization of Viruses Based on the Pattern of Viral Small RNAs Produced by the Host’, Nucleic AcidsResearch, 43: 6191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexaki A. et al. (2019) ‘Codon and Codon-Pair Usage Tables (Cocoputs): Facilitating Genetic Variation Analyses and Recombinant Gene Design’, Journal of Molecular Biology, 431: 2434–41. [DOI] [PubMed] [Google Scholar]
- Allen T. et al. (2017) ‘Global Hotspots and Correlates of Emerging Zoonotic Diseases’, Nature Communications, 8: 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins G. J. (2013) ‘The Pathogenesis of Alphaviruses’, ISRNVirology, 2013: 1–22. [Google Scholar]
- Attoui H. et al. (2007) ‘Complete Nucleotide Sequence of Middelburg Virus, Isolated from the Spleen of a Horse with Severe Clinical Disease in Zimbabwe’, Journal of General Virology, 88: 3078–88. [DOI] [PubMed] [Google Scholar]
- Barrio-Nuevo K. M. et al. (2020) ‘Detection of Zika and Dengue Viruses in Wild-caught Mosquitoes Collected during Field Surveillance in an Environmental Protection Area in São Paulo, Brazil’, PLoS One, 15: e0227239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batovska J. et al. (2020) ‘Coding-Complete Genome Sequence of Yada Yada Virus, a Novel Alphavirus Detected in Australian Mosquitoes’, Microbiology Resource Announcements, 9: e01476–19.doi: 10.1128/MRA.01476-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A. J. et al. (2016) ‘Evolution of DNA Methylation across Insects’, Molecular Biology and Evolution, 34: 654–5.doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M. F., Posada D., and Feldman M. W. (2007) ‘An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets’, Genetics, 176: 1035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen R. A. et al. (2016) ‘Viremia in North American Mammals and Birds after Experimental Infection with Chikungunya Viruses’, The American Journal of Tropical Medicine and Hygiene, 94: 504–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil. Ministério da Saúde (2021) ‘Monitoramento dos casos de arboviroses urbanas causados por vírus transmitidos pelo mosquito Aedes (dengue, chikungunya e zika), semanas epidemiológicas 1 a 37, 2021’.
- Calisher C. H. et al. (1980) ‘Everglades Virus Infection in Man, 1975’, Southern Medical Journal, 73: 1548. [DOI] [PubMed] [Google Scholar]
- Carvalho F. R. et al. (2019) ‘Simultaneous Circulation of Arboviruses and Other Congenital Infections in Pregnant Women in Rio de Janeiro, Brazil’, ActaTropica, 192: 49–54. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) ‘Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis’, Molecular Biology and Evolution, 17: 540–52. [DOI] [PubMed] [Google Scholar]
- Contigiani M. et al. (1993) ‘Presencia de anticuerpos contra virus de la encefalitis equina Venezolana subtipo VI en pacientes con enfermedad febril aguda’, Revista Argentina de Microbiología, 25: 244–51. [PubMed] [Google Scholar]
- Costa É. A. et al. (2021) ‘West Nile Virus in Brazil’, Pathogens, 10: 896.doi: 10.3390/pathogens10070896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho T. J. D., Franco G. R., and Lobo F. P. (2015) ‘Homology-Independent Metrics for Comparative Genomics’, Computational and Structural Biotechnology Journal, 13: 352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M. J. et al. (2012) ‘Surveillance of Zoonotic Infectious Disease Transmitted by Small Companion Animals’, Emerging Infectious Diseases, 18.doi: 10.3201/eid1812.120664. [DOI] [Google Scholar]
- De Andrade A. H. P. et al. (1964) ‘The Venezuelan Equine Encephalomyelitis Complex of Group a Arthropod-Borne Viruses, Including Mucambo and Pixuna from the Amazon Region of Brazil’, The American Journal of Tropical Medicine and Hygiene, 13: 723–7. [DOI] [PubMed] [Google Scholar]
- Demucha Macias J., and S’anchez Spindola I. (1965) ‘Two Human Cases of Laboratory Infection with Mucambo Virus’, The American Journal of Tropical Medicine and Hygiene, 14: 475–8. [PubMed] [Google Scholar]
- Di Giallonardo F. et al. (2017) ‘Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species’, Journal of Virology, 91: e02381–16.doi: 10.1128/JVI.02381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digoutte J., and Girault G. (1976) ‘Résultats de l’étude chez la souris du pouvoir protecteur du virus Tonate et de deux souches de virus Cabassou contre la souche neurovirulente Everglades du groupe VEE’, Annals of Microbiology, 127B: 429–37. [PubMed] [Google Scholar]
- Dolan P. T., Whitfield Z. J., and Andino R. (2018) ‘Mapping the Evolutionary Potential of RNA Viruses’, Cell Host &Microbe, 23: 435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs W. G., Anderson C. R., and Aitken T. H. G. (1956) ‘The Isolation of Ilhéus Virus from Wild Caught Forest Mosquitoes in Trinidad’, The American Journal of Tropical Medicine and Hygiene, 5: 621–5. [DOI] [PubMed] [Google Scholar]
- Folmer O. et al. (1994) ‘DNA Primers for Amplification of Mitochondrial Cytochrome C Oxidase Subunit I from Diverse Metazoan Invertebrates’, Molecular Marine Biology and Biotechnology, 3: 294–9. [PubMed] [Google Scholar]
- Fonseca I. E. M. et al. (1975) ‘Bertioga (Guama Group) and Anhembi (Bunyamwera Group), Two New Arboviruses Isolated in São Paulo, Brazil’, The American Journal of Tropical Medicine and Hygiene, 24: 131–4. [DOI] [PubMed] [Google Scholar]
- Forattini O. (2002) Culicidologia Médica: Identificação, biologia, epidemiologia. São Paulo: Editora da Universidade de São Paulo, p. 2. [Google Scholar]
- Forrester N. L. et al. (2012) ‘Genome-Scale Phylogeny of the Alphavirus Genus Suggests a Marine Origin’, Journal of Virology, 86: 2729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- —— et al. (2017) ‘Evolution and Spread of Venezuelan Equine Encephalitis Complex Alphavirus in the Americas’, PLoS Neglected Tropical Diseases, 11: e0005693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo P., and de Rodaniche E. (1961) ‘Isolation of the Virus of Ilhéus Encephalitis from Mosquitoes Captured in Panama’, The American Journal of Tropical Medicine and Hygiene, 10: 393–4. [DOI] [PubMed] [Google Scholar]
- Gibbs M. J., Armstrong J. S., and Gibbs A. J. (2000) ‘Sister-Scanning: A Monte Carlo Procedure for Assessing Signals in Recombinant Sequences’, Bioinformatics, 16: 573–82. [DOI] [PubMed] [Google Scholar]
- Go Y. Y., Balasuriya U. B. R., and Lee C. (2014) ‘Zoonotic Encephalitides Caused by Arboviruses: Transmission and Epidemiology of Alphaviruses and Flaviviruses’, Clinical and ExperimentalVaccineResearch, 3: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E. et al. (2017) ‘Emerging Arboviruses: Why Today?’ OneHealth, 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. E. (2013) ‘Alphaviruses’. In: Knipe, D. M., and Howley, P. M. (eds), Fields Virology. pp. 651–86. Lippincott Williams & Wilkins: Philadelphia. [Google Scholar]
- Guzmán-Terán C. et al. (2020) ‘Venezuelan Equine Encephalitis Virus: The Problem Is Not over for Tropical America’, Annals of Clinical Microbiology and Antimicrobials, 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S. et al. (1988) ‘Western Equine Encephalitis Virus is a Recombinant Virus’, Proceedings of the National Academy of Sciences of the United States of America, 85: 5997–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her Z. et al. (2010) ‘Active Infection of Human Blood Monocytes by Chikungunya Virus Triggers an Innate Immune Response’, The Journal of Immunology, 184: 5903–13. [DOI] [PubMed] [Google Scholar]
- Hermanns K. et al. (2020) ‘Agua Salud Alphavirus Defines a Novel Lineage of Insect-specific Alphaviruses Discovered in the New World’, Journal of General Virology, 101: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- —— et al. (2017) ‘Discovery of a Novel Alphavirus Related to Eilat Virus’, Journal of General Virology, 98: 43–9. [DOI] [PubMed] [Google Scholar]
- Jackson A. C., Sengupta S. K., and Smith J. F. (1991) ‘Pathogenesis of Venezuelan Equine Encephalitis Virus Infection in Mice and Hamsters’, Veterinary Pathology, 28: 410–8. [DOI] [PubMed] [Google Scholar]
- Jones K. E. et al. (2008) ‘Global Trends in Emerging Infectious Diseases’, Nature, 451: 990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose J., Snyder J. E., and Kuhn R. J. (2009) ‘A Structural and Functional Perspective of Alphavirus Replication and Assembly’, FutureMicrobiology, 4: 837–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusch G. T. et al. (2010) Sustaining Global Surveillance and Response to Emerging Zoonotic Diseases. National Academies Press: Washington, DC, pp. 1–340. [PubMed] [Google Scholar]
- King D. A. et al. (2006) ‘Infectious Diseases: Preparing for the Future’, Science, 313: 1392–3. [DOI] [PubMed] [Google Scholar]
- La Linn M., Aaskov J. G., and Suhrbier A. (1996) ‘Antibody-dependent Enhancement and Persistence in Macrophages of an Arbovirus Associated with Arthritis’, Journal of General Virology, 77: 407–11. [DOI] [PubMed] [Google Scholar]
- La Linn M. et al. (2001) ‘Arbovirus of Marine Mammals: A New Alphavirus Isolated from the Elephant Seal Louse, Lepidophthirus Macrorhini’, Journal of Virology, 75: 4103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb K. et al. (2010) ‘Structure of a Venezuelan Equine Encephalitis Virus Assembly Intermediate Isolated from Infected Cells’, Virology, 406: 261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J. (1953) Neotropical Culicidae V.II. Universidade de São Paulo: São Paulo. [Google Scholar]
- Lewis A. L. et al. (1974) ‘Venezuelan Equine Encephalomyelitis in Florida: Endemic Virus Circulation in Native Rodent Populations of Everglades Hammocks’, The American Journal of Tropical Medicine and Hygiene, 23: 513–21. [DOI] [PubMed] [Google Scholar]
- Lobo F. P. et al. (2009) ‘Virus-Host Coevolution: Common Patterns of Nucleotide Motif Usage in Flaviviridae and Their Hosts’, PLoS One, 4: e6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord R. D. (1974) ‘History and Geographic Distribution of Venezuelan Equine Encephalitis’, Bulletin of thePanAmericanHealthOrganization, 8: 100–10. [PubMed] [Google Scholar]
- Mansur D. S., Smith G. L., and Ferguson B. J. (2014) ‘Intracellular Sensing of Viral DNA by the Innate Immune System’, Microbesand Infection, 16: 1002–12. [DOI] [PubMed] [Google Scholar]
- Martin D., and Rybicki E. (2000) ‘RDP: Detection of Recombination Amongst Aligned Sequences’, Bioinformatics, 16: 562–3. [DOI] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, VirusEvolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- —— et al. (2005) ‘A Modified Bootscan Algorithm for Automated Identification of Recombinant Sequences and Recombination Breakpoints’, AIDS Research and Human Retroviruses, 21: 98–102. [DOI] [PubMed] [Google Scholar]
- Michlmayr D. et al. (2018) ‘Comprehensive Innate Immune Profiling of Chikungunya Virus Infection in Pediatric Cases’, Molecular Systems Biology, 14: e7862.doi: 10.15252/msb.20177862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F. et al. (2012) ‘Eilat Virus, a Unique Alphavirus with Host Range Restricted to Insects by RNA Replication’, Proceedings of the National Academy of Sciences, 109: 14622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M., Sawyer S., and Fauquet C. M. (1999) ‘Possible Emergence of New Geminiviruses by Frequent Recombination’, Virology, 265: 218–25. [DOI] [PubMed] [Google Scholar]
- Posada D. (2008) ‘jModelTest: Phylogenetic Model Averaging’, Molecular Biology and Evolution, 25: 1253–6. [DOI] [PubMed] [Google Scholar]
- Posada D., and Crandall K. A. (2001) ‘Evaluation of Methods for Detecting Recombination from DNA Sequences: Computer Simulations’, Proceedings of the National Academy of Sciences, 98: 13757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P., Aragonès L., and Garcia-Vallvé S. (2010) ‘RCDI/eRCDI: A Web-server to Estimate Codon Usage Deoptimization’, BMC Research Notes, 3: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P., Bravo I. G., and Garcia-Vallve S. (2008) ‘CAIcal: A Combined Set of Tools to Assess Codon Usage Adaptation’, BiologyDirect, 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2019), FigTree V1.4: Tree Figure Drawing Tool. GitHub. <https://github.com/rambaut/figtree/releases> accessed 9 Sep 2021.
- Ranwez V. et al. (2011) ‘MACSE: Multiple Alignment of Coding SEquences Accounting for Frameshifts and Stop Codons’, PLoS One, 6: e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M. O. et al. (1995) ‘Identification of Breakpoints in Intergenotypic Recombinants of HIV Type 1 by Bootscanning’, AIDS Research and Human Retroviruses, 11: 1423–5. [DOI] [PubMed] [Google Scholar]
- Sánchez-Seco M. P. et al. (2005) ‘Generic RT-nested-PCR for Detection of Flaviviruses Using Degenerated Primers and Internal Control Followed by Sequencing for Specific Identification’, Journal of VirologicalMethods, 126: 101–9. [DOI] [PubMed] [Google Scholar]
- —— et al. (2001) ‘A Generic nested-RT-PCR Followed by Sequencing for Detection and Identification of Members of the Alphavirus Genus’, Journal of VirologicalMethods, 95: 153–61. [DOI] [PubMed] [Google Scholar]
- Sawyer S. (1989) ‘Statistical Tests for Detecting Gene Conversion’, Molecular Biology and Evolution, 6: 526–38. [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) ‘NIH Image to ImageJ: 25 Years of Image Analysis’, NatureMethods, 9: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk J. et al. (2019) ‘RNA-binding Protein Isoforms ZAP-S and ZAP-L Have Distinct Antiviral and Immune Resolution Functions’, Nature Immunology, 20: 1610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M. (1992) ‘Analyzing the Mosaic Structure of Genes’, Journal of Molecular Evolution, 34: 126–9. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., and Strauss E. G. (1994) ‘The Alphaviruses: Gene Expression, Replication, and Evolution’, Microbiological Reviews, 58: 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhrbier A., Jaffar-Bandjee M.-C., and Gasque P. (2012) ‘Arthritogenic Alphaviruses—an Overview’, Nature ReviewsRheumatology, 8: 420–9. [DOI] [PubMed] [Google Scholar]
- Takata M. A. et al. (2017) ‘CG Dinucleotide Suppression Enables Antiviral Defence Targeting Non-self RNA’, Nature, 550: 124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L. H., Latham S. M., and Woolhouse M. E. J. (2001) ‘Risk Factors for Human Disease Emergence’, Philosophical Transactions of the Royal Society ofLondonSeriesB: Biological Sciences, 356: 983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S. et al. (2018) ‘Discovery of Mwinilunga Alphavirus: A Novel Alphavirus in Culex Mosquitoes in Zambia’, VirusResearch, 250: 31–6. [DOI] [PubMed] [Google Scholar]
- Tschá M. K. et al. (2019) ‘Identification of a Novel Alphavirus Related to the Encephalitis Complexes Circulating in Southern Brazil’, EmergingMicrobes& Infections, 8: 920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L. et al. (2016) ‘Selective Factors Associated with the Evolution of Codon Usage in Natural Populations of Arboviruses’, PLoS One, 11: e0159943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C. et al. (2004) ‘Venezuelan Equine Encephaltis’, Annual Review of Entomology, 49: 141–74. [DOI] [PubMed] [Google Scholar]
- —— et al. (1993) ‘A Comparison of the Nucleotide Sequences of Eastern and Western Equine Encephalomyelitis Viruses with Those of Other Alphaviruses and Related RNA Viruses’, Virology, 197: 375–90. [DOI] [PubMed] [Google Scholar]
- Yuill T. M. (1986) ‘The Ecology of Tropical Arthropod-Borne Viruses’, Annual Review of Ecology and Systematics, 17: 189–219. [Google Scholar]
- Zavortink T. J., Roberts D. R., and Hoch A. L. (1983) ‘Tricoprosopon Digitatum - Morphology, Biology, and Potential Medical Importance’, Mosquito Systematics, 15: 141–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PIRAV genome sequence obtained in this study has been deposited in GenBank under accession number OK539813.


