SUMMARY
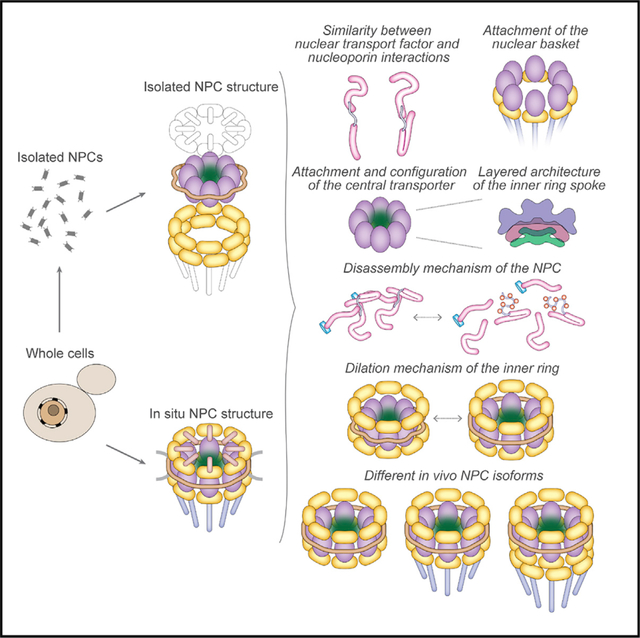
Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub-nanometer resolution to show how flexible connectors tie together different structural and functional layers. These connectors may be targets for phosphorylation and regulated disassembly in cells with an open mitosis. Moreover, some nucleoporin pairs and transport factors have similar interaction motifs, which suggests an evolutionary and mechanistic link between assembly and transport. We provide evidence for three major NPC variants that may foreshadow functional specializations at the nuclear periphery. Cryo-electron tomography extended these studies, providing a model of the in situ NPC with a radially expanded inner ring. Our comprehensive model reveals features of the nuclear basket and central transporter, suggests a role for the lumenal Pom152 ring in restricting dilation, and highlights structural plasticity that may be required for transport.
In brief
A comprehensive model of the yeast NPC reveals an interconnected architecture of the core scaffold and provides an understanding of the isoforms and structural plasticity that may be associated with different functional states.
Graphical Abstract
INTRODUCTION
Nuclear pore complexes (NPCs) are cylindrical assemblies with 8-fold symmetry that form a gateway in the nuclear envelope (NE) for the selective exchange of macromolecules between the cytoplasm and nucleus. These transport machines stabilize a pore in the NE, termed the pore membrane, and are key players in RNA processing and chromatin organization (D’Angelo, 2018; Fernandez-Martinez and Rout, 2021; Kuhn and Capelson, 2019; Ptak et al., 2014). Given this central role, it is not surprising that NPC defects are linked to many diseases and that nuclear transport is a target for therapeutics (Jühlen and Fahrenkrog, 2018; Simon and Rout, 2014). NPCs from the Saccharomyces are smaller (52 MDa) (Kim et al., 2018) than their vertebrate cousins (~109 MDa) (Ori et al., 2013); however, both are composed of about 30 conserved nucleoporins (Nups).
In the yeast NPC, ~550 Nups in multiple copies are arranged in three coaxial rings: an inner ring (IR) with 8 massive subunits (termed spokes) is flanked by cytoplasmic and nuclear outer rings to form the core scaffold. In addition, a Pom152 ring is anchored to the spokes and extends into the NE lumen (Kim et al., 2018). A cylindrical basket is attached to the nuclear outer ring and together with the Nup82 complex on the cytoplasmic side forms a pipeline for RNA processing and export (Fernandez-Martinez et al., 2016; Gaik et al., 2015). The scaffold provides anchor points for an array of Nups with intrinsically disordered Phe-Gly (FG) repeat domains that project inward to form the central transport path (Kim et al., 2018; Yamada et al., 2010); these FG domains provide binding sites for the bidirectional, facilitated diffusion of nuclear transport factors with macromolecular cargos (Knockenhauer and Schwartz, 2016; Paci et al., 2021).
Positional maps and molecular models have been generated for NPCs from yeast, humans, amphibians, and algae using cryo-electron tomography (cryo-ET), single particle electron cryo-microscopy (cryo-EM), X-ray crystallography, and integrative structure determination (Bui et al., 2013; Huang et al., 2020; Kim et al., 2018; Kosinski et al., 2016; Lin et al., 2016; Mosalaganti et al., 2018; von Appen et al., 2015; Zhang et al., 2020). These models have revealed a remarkable structural redundancy and plasticity of the NPC. Thus, the number and composition of the outer rings can vary between species (Bui et al., 2013; Kim et al., 2018; Mosalaganti et al., 2018), and changes in the functional size of the NPC have been observed (Feldherr and Akin, 1994; Mahamid et al., 2016). Moreover, recent 3D maps obtained by cryo-ET of cell cryo-sections from C. reinhardtii, S. cerevisiae, S. pombe, and H. sapiens have revealed a large radial expansion of the inner ring (Mosalaganti et al., 2018; Allegretti et al., 2020; Zimmerli et al., 2021; Schuller et al., 2021).
Even with these steps forward, our understanding of NPC structure and its role in transport remains incomplete. To address this issue, we determined three-dimensional (3D) structures of isolated and in situ yeast NPCs with radially contracted and expanded inner rings. Our comprehensive model provides insights into the structure, assembly, transport mechanism, and evolution of the NPC.
RESULTS
Overview of isolated and in situ yeast NPCs
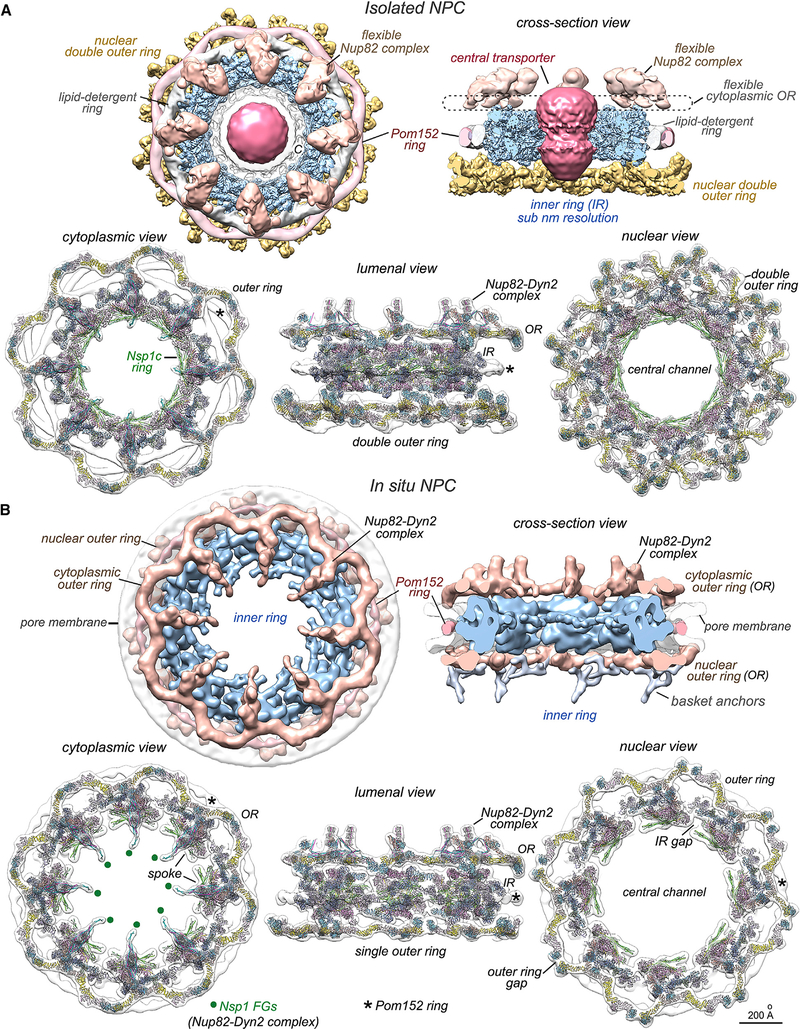
The large size, flexibility, and dynamic properties of the NPC present a challenge to structural studies; thus, a divide-and-conquer approach is required to determine a comprehensive structure. We used cryo-EM and single particle analysis of isolated NPCs to generate a combined 3D map with the inner ring and double outer ring at 7.6 and 11 Å resolution, respectively (Figure 1A, top; Methods S1; Table S1). Cryogenic, focused ion beam milling of yeast cells, cryo-ET, and subtomogram analysis were then used to provide a 3D map of the in situ NPC at 30–40 Å resolution (Figures 1B [top]; Table S1). These density maps reveal the NPC in two different states and highlight important specimen differences. Notably, the isolated NPC lacks a pore membrane while retaining a lipid-detergent ring, has an hourglass-shaped central transporter, and contains a double outer ring on the nuclear side. In addition, the Nup82 complex and cytoplasmic outer ring are blurred due to detergent extraction (Figure 1A [top]). Conversely, the Nup82-Dynein 2 (Dyn2) complex of the in situ NPC is well resolved, and basket anchors are visualized on the single nuclear outer ring (Figure 1B [top right]). However, the central transporter was not imaged clearly with the limited in situ dataset.
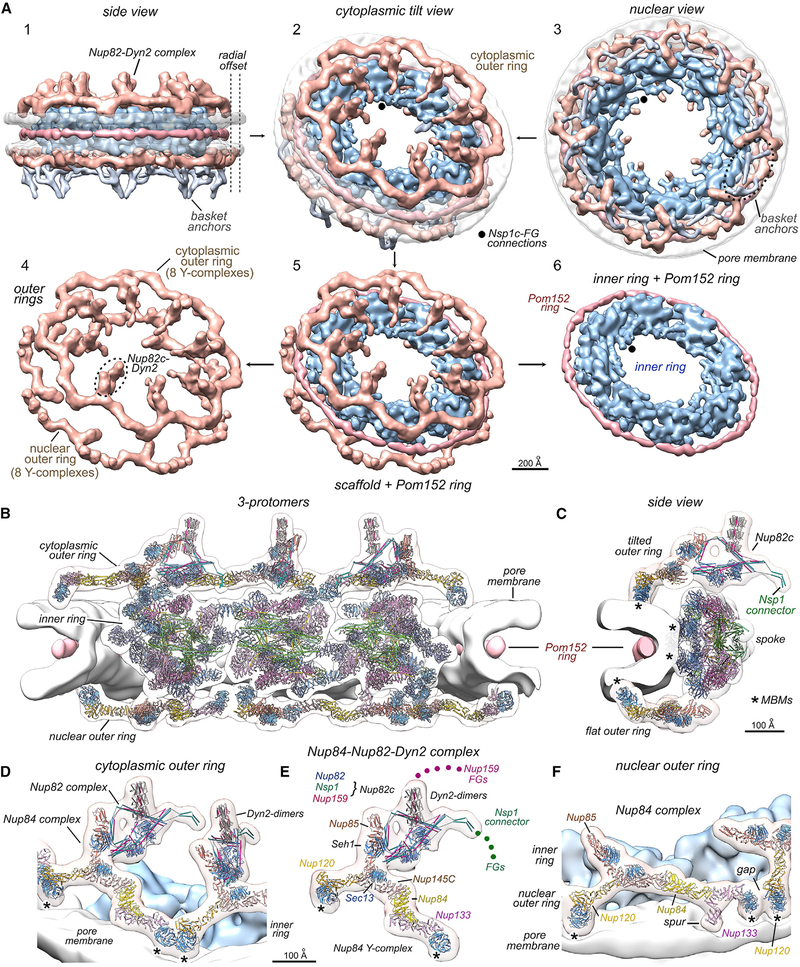
Figure 1. Overview of isolated and in situ yeast NPC structures.
(A) (top) Cytoplasmic and cross-section views of a composite 3D map from the isolated NPC: double nuclear outer ring (gold), inner ring (IR, light blue), Nup82 complex and cytoplasmic outer ring (tan), Pom152 ring (light pink), membrane-detergent ring (white), central transporter (red), and spoke-transporter connections (marked “C”).
(bottom) Three views of molecular models docked in a composite, 3D density map of the isolated NPC. Nup color coding is defined for spokes in Figures 2G and 2H and for Y-complexes in Figure 4D. Only half of the NPC model is shown in each view and the Pom152 ring is marked (*).
(B) (top) Two views of the in situ yeast NPC: outer rings (OR, tan), Pom152 ring (pink), pore membrane (transparent white), and basket anchors (silver).
(bottom) Three views of molecular models docked in the combined 3D density map of the in situ NPC. Positions of Nsp1 FG domain anchor points in the Nup82 complex are indicated (green dots). Scale bar 200 Å.
Crystal structures and homology models of Nups were docked within composite density maps to create molecular structures of the isolated and in situ NPC (Figures 1A and 1B [bottom]). We found that density for the Nup82 complex can be overlayed after aligning the inner rings of the isolated and in situ NPC to bring these maps into register, so we used the cytoplasmic outer ring and Nup82-Dyn 2 complexes from the in situ structure to create a composite, in silico model of the isolated NPC (Figure 1A [bottom]; Video S1). Higher-resolution models of the inner ring and double outer rings from the isolated NPC were then used to create a model of the in situ NPC (Figure 1B [bottom]). A direct comparison reveals a large radial dilation of the in situ inner ring (compare Figures 1A and 1B) (Allegretti et al., 2020; Mosalaganti et al., 2018; Schuller et al., 2021) that is coupled with an expanded membrane pore and Pom152 ring. This dilation creates a central channel that is ~2× larger in volume; this alters the packing density of FG domains and may affect nuclear transport.
Functional domains of the inner ring at sub-nanometer resolution
The inner ring has nearly perfect 2-fold symmetry in the isolated NPC, and a majority of the ~715 ɑ-helices in each spoke were resolved in the single particle structure (Figures 1A and 2A–2D). In total, 28 Nup domains and flexible connectors were modeled (Figures 1A [bottom], S1, S2, S3, S4, and S5) with a root mean square deviation (rmsd) of ~8Å for Cαs in core domains, relative to a previous integrative model (Kim et al., 2018). This improved model is supported by mapping 206 cross-links to the inner ring with 95% satisfaction (Figure S5B). Major insights include the visualization of functional layers, domain rearrangements within the spoke, and the identification of flexible connectors.
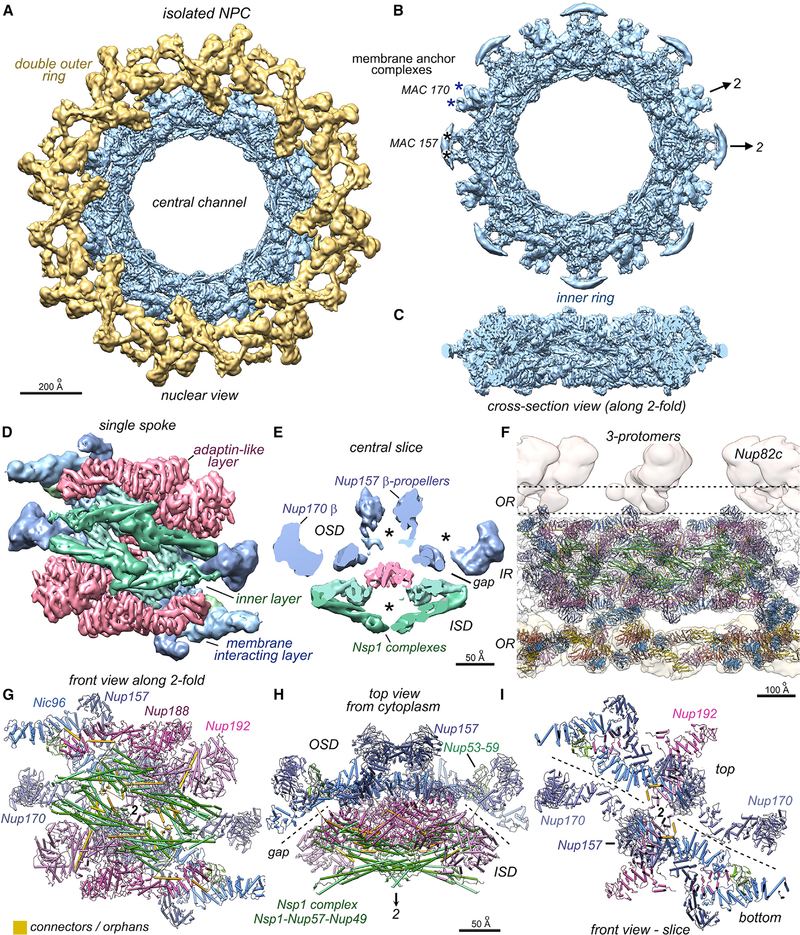
Figure 2. Structure of the isolated yeast NPC.
(A) Composite 3D map of the inner ring and double outer rings viewed from the nuclear side: nuclear, double outer ring (gold), inner ring (IR, light blue).
B. The inner ring is shown with local 2-fold axes and membrane attachment complexes that contain Nup157 pairs, Pom152 and Pom34 (MAC157, black asterisks), and Nup170 pairs with Ndc1 (MAC170; blue asterisks).
(C) A cross-section of the inner ring with ɑ-helical rods throughout.
(D) A single spoke is segmented and viewed along the central 2-fold axis to reveal the three major layers.
(E) A central slice through the spoke with the orientation of Figure 2H shows the gap between inner and outer spoke domains (ISD and OSD, respectively). Cavities are marked with asterisks.
(F) A central section with three docked models for the spokes and double outer ring (oriented as in C). β-propellers in the Y-complexes are highlighted (light blue) and the disordered cytoplasmic outer ring is indicated (dashed lines).
(G) Close-up of a modeled spoke viewed along the 2-fold axis. Connector helices are colored gold.
(H) The spoke contains two domains, as indicated by dashed lines (gap), with distinctive functions. The Nup color palette reflects their distribution within the inner (green), adaptin-like (pink-red), and membrane-interacting (blue) layers.
(I) A thin slab viewed along the central 2-fold axis reveals a vertical separation of the outer spoke domain (dashed lines; also see Figure 3). Relevant scale bars are indicated on the panels.
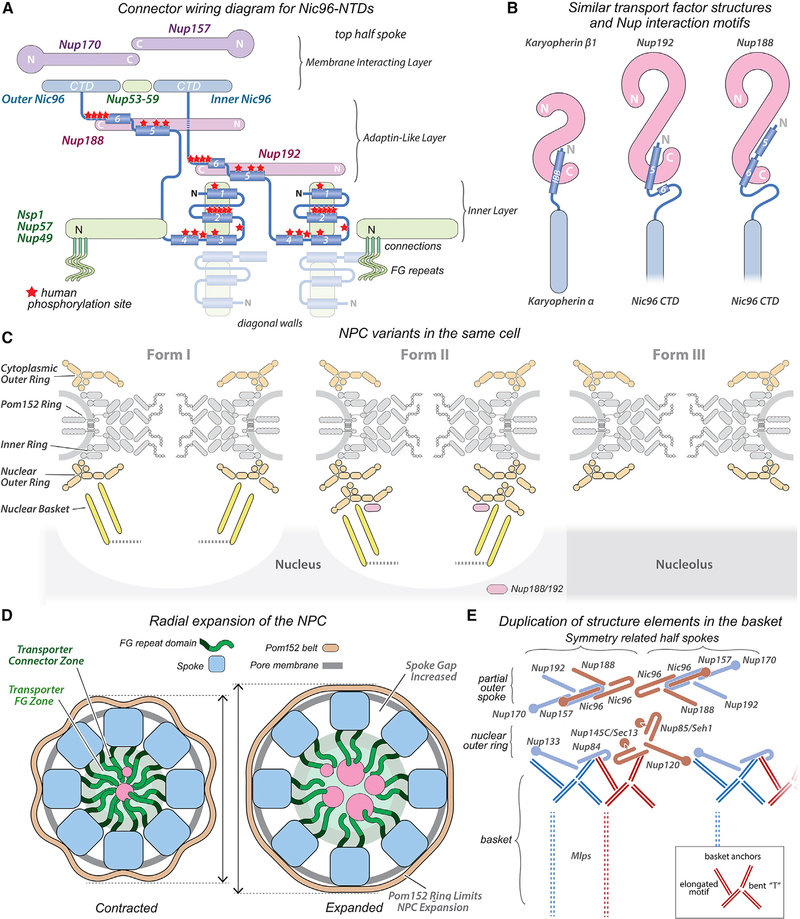
The spoke is comprised of an inner spoke domain (ISD), with two layers and an outer spoke domain (OSD) with a single layer (see gap, Figures 2E and 2H). The inner layer of the ISD contains two pairs of laterally offset Nsp1 heterotrimers (Nsp1, Nup49, and Nup57) related by 2-fold symmetry that face into the central channel (Figures 1A [bottom], 2D–2H, S3), while symmetry-related pairs of Nup188 and Nup192 form an intermediate, adaptin-like layer (Figures 2D and S3B). Both layers bind to an array of “orphan” connectors (Figures 2G and S3). The membrane interaction layer in the OSD contains four Nic96 C-terminal domains (CTDs), which form keystone supports (Kim et al., 2018) for Nup157 and Nup170 (2 copies each), while Nup53-Nup59 heterodimers form bridges between adjacent Nic96 CTDs in the top and bottom half of this layer (Figures S2C and S3A).
This multilayered organization reflects the functional specialization of Nups within the spoke (Figures 2D and S3; Video S2). The inner layer creates a scaffold to deploy FG repeats from the N termini of Nsp1, Nup57, and Nup49 into the transport path. The first 3-helix bundle of each Nsp1 heterotrimer forms an elongated coiled coil (Figures 2G, S2A, and S3D) on the inner face of the spoke, and these features create a ring with an diameter of ~425 Å that defines the central channel (Figure 1A [bottom]) (Kim et al., 2018; Kosinski et al., 2016; Lin et al., 2016). In addition, Nsp1 complexes from the top and bottom half of the spoke each contribute a pair of 3-helix bundles that extend inward to form a diagonal wall, with two walls per spoke (Figures 3A, 3B, and S2A). This also creates a local 2-fold axis for structural elements in each wall.
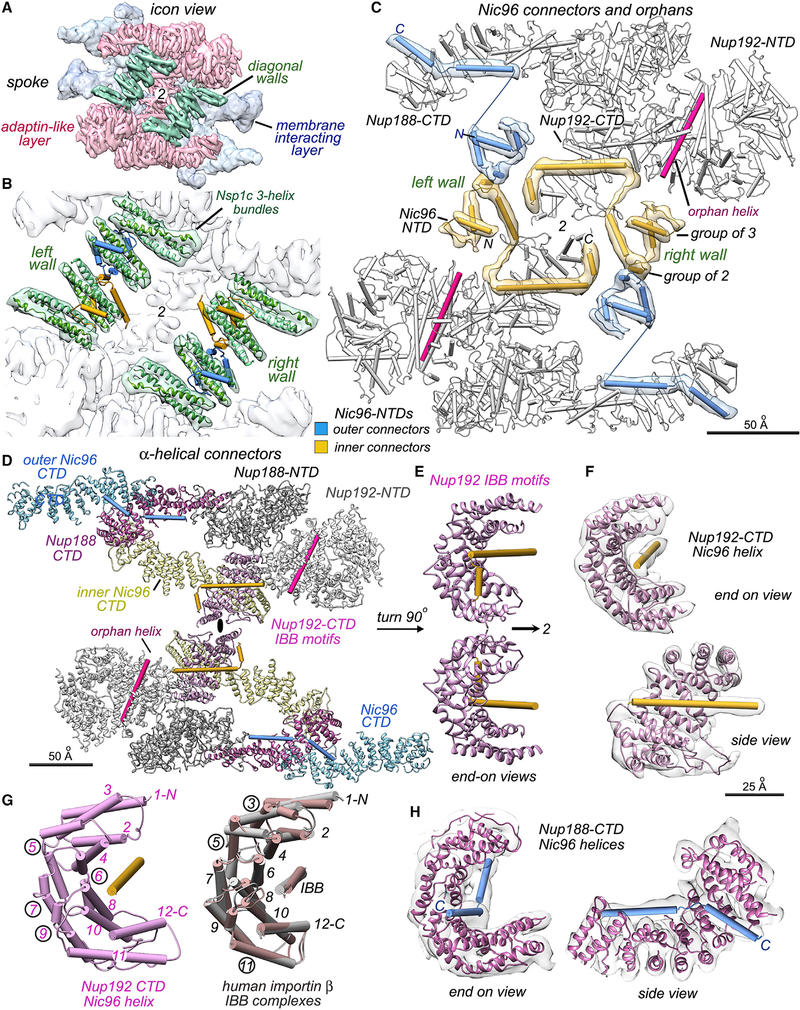
Figure 3. Orphans and interaction motifs for Nic96 NTDs in the spoke.
(A) Nsp1 complex three-helix bundles and Nic96 NTD connectors form two diagonal walls (green iso-surface). Figures 3A–3D are viewed along the central 2-fold axis (marked 2 or a black ellipse).
(B) Nic96 connector ɑ-helices (gold and blue cylinders) form a bridge between opposing half walls (green); other connectors bind to 3-helix bundles from the Nsp1 complex.
(C) Connector helices and strands with overlayed map density are superimposed on Nups 188 and 192 (white cylinders). Each Nic96 NTD forms a group of 3 elements, a group of 2 helices, and a pair of helices that bind to Nup188 or Nup192 CTDs. In addition, two orphan helices (pink) bind to Nup192 NTDs.
(D) Connector interactions are shown with Nups displayed as ribbons. Outer and inner Nic96 CTDs (light blue, light yellow) form keystone supports behind Nup192 and Nup188. Nup192 CTDs form Karyopherin-like interactions with Nic96 connector helices. Nup192 and Nup188 NTDs are shown in white and gray, while their CTDs are color coded.
(E) A pair of Nup192 molecules with IBB-like motifs (from panel 3D) are shown after a 90 degree rotation counterclockwise about a vertical axis.
(F) Two views of the Nup192/IBB-like motif are superimposed with the 3D density map.
(G) Left: the Nup192/IBB-like motif is shown with numbered helices (N- to C-, 1 to 12). Right: overlay of human Importin β co-crystal structures with IBBs of Importin-ɑ (tan, PDB 1QGK) and Snurportin 1 (gray, PDB 3LWW). Numerals of split helices are circled.
(H) Two views of the Nup188 CTD with a pair of connector helices and the overlayed 3D density map. The curved Nup188 tail binds consecutive Nic96 NTD ɑ-helices in a long groove. Scale bars are indicated on the panels.
Nups 192 and 188 in the middle layer contain adaptin-like domains (Figures S2B and S3B) (Flemming et al., 2012; Sampathkumar et al., 2013) that connect the inner layer of Nsp1 complexes to the membrane interacting layer (Figures 2D and S3C). By comparison with previous models, Nup192 and Nup188 CTDs rearrange when they interact with partner Nups and connectors; for example, CTD tails of Nup192 are bent and stacked back-to-back to form a central column in the spoke (Figure S3B, center). Moving outward, the membrane-interacting layer may help shape and stabilize the pore membrane (Figures 2H and S3A) (Kim et al., 2018; Kosinski et al., 2016; Lin et al., 2016); Nups 157 and 170 have amphipathic membrane binding motifs (MBMs) in their N-terminal β-propellers, while Nup53 contains a C-terminal membrane interaction motif (Stuwe et al., 2015). Our data support the idea that NPCs have evolved for flexibility (Beck et al., 2018; Field and Rout, 2019; Kim et al., 2018; Pulupa et al., 2020). Spoke movements may be facilitated by gaps and voids between Nups (asterisks, Figure 2E, dashed lines in Figures 2H and 2I) and by limited edgewise contacts between neighbors.
The role of Nic96 connectors in NPC structure and assembly
Biochemical and structural data have begun to show how flexible connectors tie together folded Nup domains to stabilize the spoke (Fischer et al., 2015; Hampoelz et al., 2019; Kim et al., 2018; Lin et al., 2016; Mosalaganti et al., 2018). In particular, two sequence motifs, known as short linear motifs (SLiMs), have been recognized in the N termini of yeast Nic96 and human Nup93. The first motif interacts with the Nsp1 complex in the inner layer (Chug et al., 2015; Lin et al., 2016; Stuwe et al., 2015), while the second motif binds to Nup188 and Nup192 in the adaptin-like layer (Amlacher et al., 2011; Fischer et al., 2015; Lin et al., 2016).
At the current resolution, orphan densities were revealed in the spoke that could not be accounted for by docked models. The density of these orphans is similar to neighboring Nups which indicates a high occupancy. Three orphans (group of 3) interact with 3-helix bundles in the diagonal walls, and a gap at the center of each wall is filled by 4 orphans arranged in two groups of 2 (Figures 3B, 3C, and S2A). We also identified 3 orphan helices that bind to Nup192 and two orphan helices that bind to Nup188, which are present in two copies per spoke. In total, 30 orphan densities were mapped in the spoke that arise from flexible connectors. To solve the identity and connectivity of these orphans, we applied an integrative threading approach (Saltzberg et al., 2019) to compute Nup identity, copy identifier, and sequence segments for structure elements (SEs) in each orphan (Figure S4; STAR Methods). This resulted in 28 orphan SEs being confidently assigned to contiguous sequences in four Nic96 N-terminal domains (NTDs) (Figure S4E).
In the resulting model, the group of 3 orphans (Figure 3C) is similar to the helix-strand-helix motif in the crystal structure of a Nic96 SLiM bound to the Chaetomium Nsp1 complex (Figure S5F) (Stuwe et al., 2015). Differences between our model and the crystal structure include a repositioning of C-terminal 3-helix bundles in the spoke to form a diagonal wall. Two paired ɑ-helices at the center of each wall (group of 2, Figure 3C) immediately follow the helix-strand-helix motif (Figures 3C and S5A). The final two orphan helices in each Nic96 NTD bind to Nup188 or to Nup192 CTDs; these ɑ-helix pairs contain helices of different lengths and originate from the same sequence in the outer and inner copies of Nic96 (Figures 3C–3E). Thus, the same Nic96 sequence can form different ɑ-helix pairs that bind to Nup188 or Nup192. In total, the N termini of four Nic96 molecules contribute 28 connectors with 24 ɑ-helices and 4 strands per spoke. Two unassigned orphans are bound to Nup192 NTDs (Figures 3C [pink cylinders], S3D, and S4B [SE8]); these long, rod-like connectors may originate from Nup53/59, Nup100, Nup116, or Nup145N.
Collectively, these 28 connectors play important roles in spoke architecture. First, we showed that Nic96 interacts with every other Nup to form a keystone that holds much of this structure together (Kim et al., 2018); here, we find that Nic96 connectors are critical for the recruitment of Nsp1 complexes and tie together the three spoke layers (Figures 3A–3C; Video S2). This network could provide the inner ring with flexibility to sustain expansion-contraction motions during transport. Second, our structure may provide insights into the mechanism by which NPCs assemble and disassemble. Rapid assembly during interphase may be dependent on the ability of flexible connectors to form extensive interactions between Nups in different layers, tying these components together quickly and efficiently into the mature inner ring. S. cerevisiae has a closed mitosis; however, the overall architecture of the spoke is highly conserved (Figure S8) (Zimmerli et al., 2021). Partial mitotic disassembly in Aspergillus (a related fungus) is directly dependent on connector-containing Nups (Nup145N, Nup116, and Nup100 [De Souza et al., 2004]). Importantly, Nic96, Nup188, and Nup192 undergo mitotic disassembly upon activation of NIMA and Cdk1 kinases (Osmani et al., 2006); these Nups are tied together by flexible connectors in our structure. Moreover, homologous regions in human Nups are hyper-phosphorylated at mitosis, including NTD connectors of Nup93, a Nic96 homolog (Figure S5A). This process disrupts the spoke and promotes NPC dis-assembly (Dephoure et al., 2008; Linder et al., 2017).
Nucleoporins and transport factors share common interaction mechanisms
Extended ɑ-helical solenoids of Nup188 and Nup192 are similar to Karyopherin transport factors, which indicates that Nups and Karyopherins may share a common evolutionary origin (Alber et al., 2007b; Devos et al., 2004; Devos et al., 2006; Field et al., 2011; Flemming et al., 2012; Sampathkumar et al., 2013). Here, we show that pairwise interactions are formed by CTD tails of Nup192 and Nup188 with ɑ-helices from Nic96 NTDs.
These data provide insights into how the nuclear localization signal (NLS) recognition system may have arisen, and point to a mechanistic link between spoke assembly and transport. In particular, a long ɑ-helix of Nic96 is cupped by a spiral of ɑ-helices in the Nup192 CTD; this motif resembles the interaction between Karyopherin β and its transport cargos (Figures 3D–3F) (Bhardwaj and Cingolani, 2010; Cingolani et al., 1999). An adjacent, short orphan helix may stabilize this interaction which occurs twice at the spoke center (Figures 3D and 3E). This motif is shown side-by-side with overlayed crystal structures of human Importin β1 with the ɑ-helical, NLS-like importin beta binding (IBB) domain of Importin ɑ (Figure 3G [right, tan], PDB 1QGK) (Cingolani et al., 1999) and with the Snurportin 1 IBB (Figure 3G, gray, PDB 3LWW) (Bhardwaj and Cingolani, 2010). The similar topology of ɑ-helices in the Nup192 CTD and β-Importin structures is indicated by the numbering scheme. The curved tail of Nup188 also binds a short C-terminal helix from the Nic96 NTD in a manner reminiscent of a transport factor-IBB interaction (Figures 3D and 3H, blue cylinders) (Bhardwaj and Cingolani, 2010; Cingolani et al., 1999).
A double outer ring in a subset of yeast NPCs
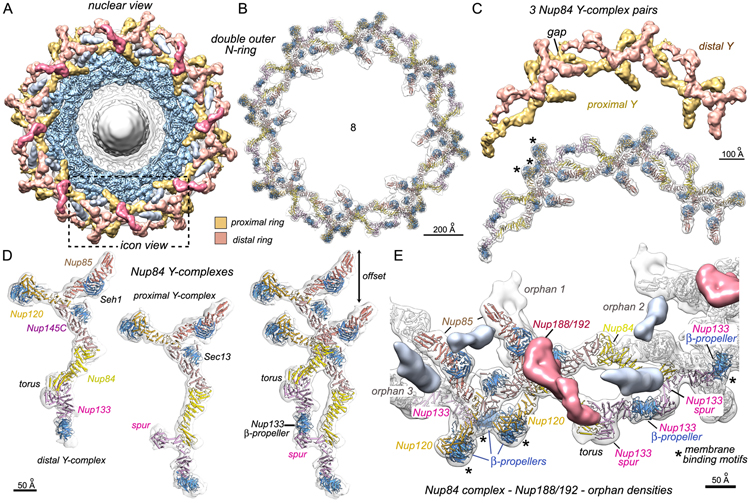
Single outer rings in yeast NPCs are formed by the head-to-tail arrangement of 8 Y-shaped Nup84 complexes (Alber et al., 2007b; Allegretti et al., 2020; Fernandez-Martinez et al., 2012; Kelley et al., 2015; Lutzmann et al., 2002). In isolated NPCs, the single outer rings are disordered (Kim et al., 2018); however, we identified particles with a double outer ring (~20%) and solved the structure at ~11.3 Å resolution (Figures 2A; Methods S1). The double outer ring is present on the nuclear surface (Figures 1A, [top right] and 2F)—as confirmed by quantitative in situ fluorescence (see below in this section). A double outer ring has also been observed on the nuclear face of NPCs in C. reinhardtii and S. pombe (Hampoelz et al., 2019; Zimmerli et al., 2021). However, we now show that at least three NPC variants are present in the yeast nucleus.
We modeled 14 Nupsin two Y-shaped Nup84 complexes, starting with Nup crystal structures (Kelley et al., 2015). The complete model provides a snapshot of Y-complexes in two ring geometries (Figures 4A–4C); the membrane proximal ring is continuous while the distal ring contains a gap between adjacent Y-complexes (Figure 4C, top). The two Y-shaped molecules form typical staggered contacts (Figure 4D, right) (Fernandez-Martinez et al., 2012; Kelley et al., 2015) with distinct tail conformations (Figure 4D [left and middle]). The β-propellers of Nup120 and Nup133 contain amphipathic MBMs; however, only three of the four β-propellers are involved in membrane contacts A Nup133 β-propeller is sandwiched between two Nup120 β-propellers to create a three-point landing at a single site on the membrane surface (Figures 4C and 4E, asterisks). Conversely, a Nup84 domain and the Nup133 spur form a torus-shaped bulge in the tail of the distal Y-complex (Figure 4D, left) that pulls the adjacent β-propeller upward to interact with the Nup133 spur from a Y-complex in the proximal ring (Figures 4D [right] and 4E). Thus, the Nup133 spur domain has two distinct functions in the double outer ring.
Figure 4. A double outer ring in a subset of yeast NPCs.
(A) The isolated yeast NPC with a double outer ring segmented into proximal (gold) and distal (tan) rings. Density for Nup188/192 (red) and two orphans (silver) are shown. A double ring protomer is outlined (dashed box).
(B) Model of Y-complexes in the double outer ring with the overlaid density map.
(C) Top: three pairs of Y-complexes from the 3D density map. A gap is visible between neighboring Y-complexes in the distal ring (tan). Bottom: molecular models are shown in the map with MBMs indicated (*).
(D) Y-complexes from the distal (left) and proximal (center) outer rings are shown, along with a staggered pair (right). A torus-like bulge and Nup133 spur are present in the tails of distal and proximal Y-complexes, respectively.
(E) The pair of Y-complexes outlined in Figure 4A is shown with density for a Nup188/192 molecule (red) and three orphans (numbered 1–3). Orphans, MBMs(*), and a tripod of β-propellers are labeled. Scale bars are indicated on the panels.
We then identified orphan densities in the double outer ring. First, an orphan density that resembles Nup188 or Nup192 runs along the top of the distal Y-complex (Figures 4A and 4E, in red). This unexpected moonlighting by Nup188/192 parallels previous observations of vertebrate homologs (Nup188/Nup205), which are present in both the outer and inner rings (Bui et al., 2013; Huang et al., 2020). In vertebrates, Nup188/205 molecules occupy the inward facing surface of the highly tilted cytoplasmic outer rings (Huang et al., 2020), while in yeast a single copy of either Nup188 or Nup192 sits on the upper surface of the double ring, interacts with the torus of a distal Y-complex (Figure 4E), and may help stabilize this unusual tail conformation. Second, three additional orphans associate with each protomer in the double outer ring. The first orphan interacts with Nup85 and points toward the central channel (Figure 4E, orphan 1), while two orphans (silver) are located between adjacent Y-complexes. (Figure 4E, orphans 2 and 3). The identity of these orphans remains to be determined.
To further characterize the outer rings, we used quantitative in situ fluorescence with one module of Nup84 tagged with a split green fluorescent protein (GFP) to monitor the outer rings, while the remaining module was placed on a reporter localized to either the nucleoplasmic or cytoplasmic side of the NE. The reconstitution of fluorescence signal confirmed that all NPCs have outer rings on their nuclear and cytoplasmic faces (Figures S6A and S6B). In addition, a population of NPCs is located over the dense crescent with low levels of Mlp1, and a population with higher levels of Mlp1 is excluded from this region. Signal distribution curves show that a subset of the “high” Mlp1 NPCs have, on average, more outer ring signal than “low” Mlp1 NPCs (~20%), and the extra signal is on the nuclear side (Figure S6C). Thus, each nucleus contains NPC variants with single and double outer rings on their nucleoplasmic faces, while some lack nuclear baskets and are differentially localized at the nuclear periphery. Overall, the data are consistent with three NPC variants co-existing in yeast cells: (1) the predominant form has two single outer rings that frame the inner ring (Form I); (2) a second form has a single outer ring on the cytoplasmic surface paired with a double outer ring on the nuclear surface (Form II), both with nuclear baskets; and (3) a third NPC variant (Form III) has two single outer rings, lacks the nuclear basket, and is enriched over the nucleolus.
A comprehensive structure of the yeast NPC in situ
The NPC is a dynamic assembly that undergoes conformational changes with a range of effective diameters (Feldherr et al., 1998; Mahamid et al., 2016) and exhibits a large radial expansion in cells (Allegretti et al., 2020; Mosalaganti et al., 2018; Zimmerli et al., 2021). We used cryogenic, focused ion beam milling of cells at mid-log phase, cryo-ET, and subtomogram analysis to generate a 3D density map of the in situ NPC. This map is comparable to a recent structure (Allegretti et al., 2020) (Figures S7A–S7D; Table S1; STAR Methods) and provides additional insights. An overview of the in situ NPC is shown in Figure 5A along with an exploded view of the core scaffold. The canonical in situ NPC has single outer rings on both faces and a radially expanded configuration for the spokes, the pore membrane, and Pom152 ring (Figure 1B). Moreover, there is a striking difference between the outer rings with the diameter of the nuclear outer ring being ~100 Å larger than the cytoplasmic ring (radial offset, Figure 5A).
Figure 5. Structure of the in situ NPC.
(A) Panels 1–3: three views are shown for the in situ yeast NPC: inner ring (blue), cytoplasmic and nucleoplasmic outer rings (tan), Pom152 ring (pink), pore membrane (transparent white), and basket anchors (silver). Connections from Nsp1 complexes to FG domains are indicated with black dots. Cytoplasmic and nuclear outer rings differ in diameter (radial offset, dashed lines). Panels 4–6: the core scaffold with the Pom152 ring is separated into component rings.
(B) Cross-section of the in situ NPC with molecular models for three protomers.
(C) Side view of a nearly complete protomer. β-propellers with MBM contacts are indicated (*).
(D and E) Model for the tilted cytoplasmic outer ring with the Nup82 complex (Nup82c) and Dyn2 dimers. β-propellers with MBMs (*) and N-terminal FG domains from Nsp1 and Nup159 are indicated (colored dots).
(F) Tilted view of the flat nucleoplasmic outer ring with docked models. A gap is present between adjacent Y-complexes and MBM contact sites are indicated (*); the Nup133 spur contacts the membrane. Scale bars are indicated.
We leveraged models for the isolated NPC, the Nup82 complex (Fernandez-Martinez et al., 2016), and a recent study (Allegretti et al., 2020) to build a model of the in situ NPC. The cytoplasmic outer ring is tilted relative to the plane of the NE due to interactions with the Nup82 complex, which overhangs the central channel. Conversely, the outer ring on the nuclear face lies flat on the membrane (Figures 5B and 5C). In the inner ring, 3-helix bundles from opposing Nsp1 complexes extend into the spoke interior to form two diagonal walls that tie the top and bottom halves together (Figure S7E, panels 1 and 2). A cavity is located between the diagonal walls and lies in front of a central column formed by Nup192 CTDs (Figure S7E, panels 2–4), while the membrane interacting layer is resolved at higher radius (Figure S7E, panel 5). A local asymmetry may arise from differential contacts to the outer rings as reflected by the position of Nup188.
The Nup82 complex is comprised of Nup82, Nup159, and Nsp1 and forms a non-canonical dimer that interacts with Nup85 in Y-complexes of the cytoplasmic outer ring (Figures 5C–5E) (Allegretti et al., 2020; Fernandez-Martinez et al., 2016). We modeled three Dyn2 dimers with Nup159 peptides (Gaik et al., 2015; Romes et al., 2012) in a vertical feature of the Nup82 complex; however, their rotational alignment has not been determined. This feature may direct N-terminal FG domains of Nup159 into a region above the central channel along with tethered Dbp5 (an RNA helicase; Allegretti et al., 2020). In addition, linker density from Nsp1 molecules may position FG repeats in the transport path (Figures 5C and 5E). The single, nuclear outer ring contains a gap between Nup133 and Nup120 β-propellers in adjacent Y-complexes (Figure 5F); these gaps increase the outer ring diameter (Figure 5A, panel 1). We also find that the Nup133 spur domain interacts with the pore membrane along with β-propellers of Nup133 and Nup120 (asterisks, Figure 5F). A 3D map of the in situ NPC with a double outer ring was not determined due to the low frequency of this feature. The structure of the in situ NPC is summarized in Video S3.
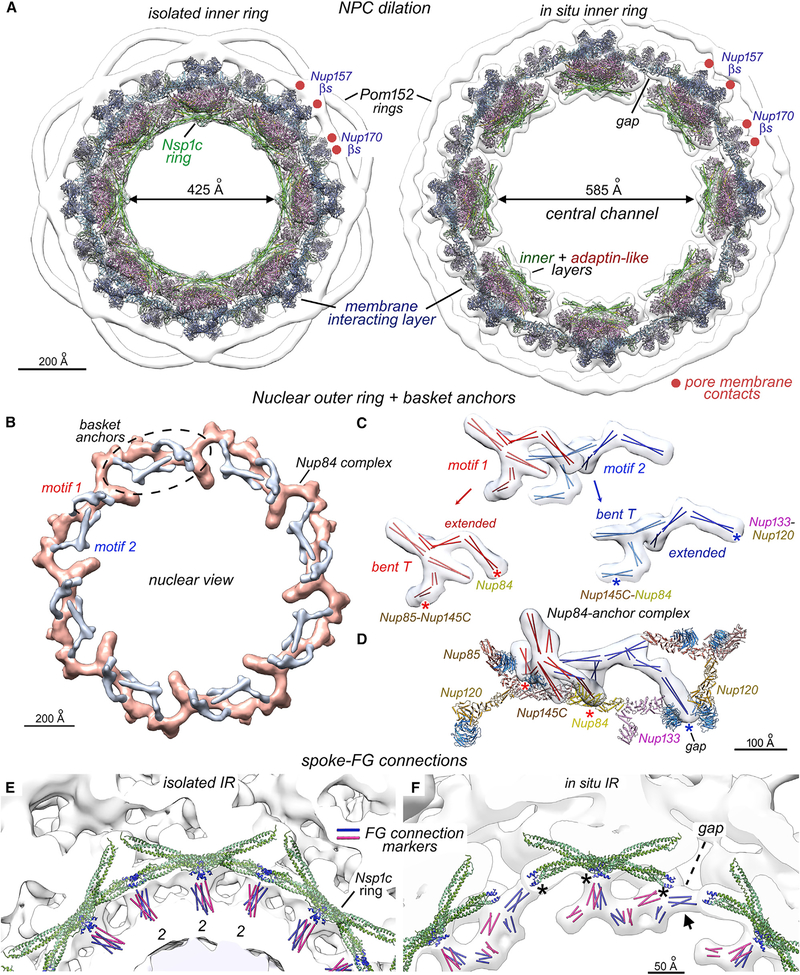
A side-by-side comparison of inner rings from isolated and in situ NPCs reveals the plasticity of this transport machine (Figure 6A). The spoke, and by inference the pore membrane, undergoes a large radial movement (~80 Å) during inner ring dilation (Zimmerli et al., 2021), and the diameter of the central channel is increased from ~425 Å to ~585 Å. This nearly doubles the volume accessible to FG domains, which project into the central channel. Gaps are formed between adjacent spokes (Figure 6A) and lateral contacts are lost, while Nsp1 complexes in each spoke become separated yet remain facing into the central channel. Nup170 molecules from adjacent spokes retain contacts with the pore membrane and with each other during dilation (Figure 6A, Nup170, red dots). In addition, the NTD of Nup192 and a CTD of Nic96 make an inter-spoke contact.
Figure 6. Inner ring dilation, basket anchors, and FG domain connections.
(A) Models for aligned inner rings from the isolated (left) and in situ NPC (right). Radial dilation increases the diameter of the central channel and creates a gap between adjacent spokes. Nup157 and Nup170 membrane attachment sites (red dots) and spoke layers are indicated.
(B) The nuclear outer ring (tan) with basket anchor complexes (silver) is viewed from the nucleoplasm. Two anchor motifs are present for each Y-complex (dashed oval).
(C) Basket anchor complexes have two similar motifs (red and blue); each motif contains a “bent T” and an extended SE that were mapped with 2-helix coiled coils. Contact sites with the Y-complex are indicated (red and blue asterisks).
(D) Tilted view of a Y-complex in the nuclear outer ring with basket anchors.
(E) Extended 3-helix bundles of the Nsp1 complex (green) form a ring that faces the central channel. Connections to the central transporter are mapped with rods (red and blue) and are aligned with N termini (dark blue) of Nsp1 heterotrimers at local 2-fold axes (2).
(F) FG connections of the in situ NPC originate from three points on each spoke (*) that align with the N-termini of Nsp1 complexes (dark blue). Radial expansion creates a gap between adjacent spokes and separates adjacent Nsp1 complexes; a lateral contact is formed between adjacent spokes at the gap (arrow). Scale bars are indicated.
The structural plasticity exhibited by the yeast inner ring may be facilitated by flexible links between the spokes and outer rings, by limited interactions between adjacent spokes, and by a fenestrated architecture that allows small movements of flexible Nups within each spoke. Aligned models of isolated and in situ spokes have an average rmsd of ~9.5 Å for Cɑ backbones after excluding Nup170. Models of the yeast spoke can be docked into cryo-ET maps of human and algal NPCs with isolated and in situ conformations (Figure S8) (Allegretti et al., 2020; Kosinski et al., 2016; Mosalaganti et al., 2018). The quality of this cross-species docking underscores the high level of structural conservation in the spoke, which includes a demarcation into three structural and functional layers, the formation of a central column by Nup192 homologs, and the presence of diagonal walls, which suggests a conserved role for Nic96 and Nup93 connectors.
The conformation of the inner ring is not determined solely by the presence or absence of the pore membrane, as NPCs in isolated NEs have a contracted conformation (Kosinski et al., 2016; Lin et al., 2016). In fact, recent work suggests that nuclear membrane tension may play a role in dilation (Zimmerli et al., 2021). We suggest that a dilated inner ring with an expanded membrane pore and Pom152 ring (Figures 1B and 6A) may represent a “tense state” of in situ NPC, which can relax to a more compact conformation, typified by the inner ring in isolated NPCs (Kim et al., 2018; Kosinski et al., 2016; Lin et al., 2016).
Basket anchor pairs on the nuclear outer ring
A basket extends from the nuclear face of the NPC and this feature has been identified in multiple organisms (Rout and Blobel, 1993; Strambio-de-Castillia et al., 2010). Mlp1 and Mlp2 paralogs are the major basket components in yeast and are predicted to be coiled coil proteins (Niepel et al., 2013; Strambio-de-Castillia et al., 1999). Our map of the in situ NPC revealed a pair of bifurcated densities that associate with each Y-complex in the nuclear outer ring and point toward the interior (Figures 1C (silver), 5A, and 6B). Their extended shape and position suggest that these motifs are anchors for the nuclear basket. We docked 14 two-helix coiled coils into these densities as markers (Figure 6C). The resulting model contains two similar motifs highlighted in red and blue, each with a “bent T” and an “extended” element (Figure 6D). The base of the red T interacts with the Nup85-Nup145C interface while the extended structural element abuts Nup84 (Figure 6D, red asterisks). The blue anchor motif also has two anchor points on the Y-complex but only one is visible in Figure 6D (blue asterisk). In this case, a short stem from the bent T is associated with the Nup145C-Nup84 interface, while the extended element inserts into a gap between Nup133 and Nup120 β-propellers from adjacent Y-complexes. This model provides a rationale for gaps observed in the nuclear outer ring.
The topology of the basket anchors suggests that coiled coils and segments from Mlp1/2, Nup145N and Nup60 (Kim et al., 2018) could contribute to these features. However, the observation of two topologically similar motifs that associate with each Y-complex serves to validate these features and suggests that the recurring theme of laterally offset Nup duplicates or paralog pairs within the NPC may pertain to the basket, as exemplified by Mlp1 and Mlp2 (Alber et al., 2007b; Devos et al., 2004; Devos et al., 2006; Kim et al., 2018).
Membrane interactions of the inner ring
Interactions with the NE are critical for assembly and for maintaining the NPC within the pore membrane aperture (Kosinski et al., 2016; Mosalaganti et al., 2018, Allegretti et al., 2020; Schuller et al., 2021). In the isolated NPC, a lipid-detergent ring encircles the inner ring (Figure 1A) and encompasses partly resolved transmembrane domains (TMDs) of Pom152, Pom34, and Ndc1 (Figures 2B [asterisks], S9A, and S9B [dashed ovals and asterisks]). MBMs in β-propellers of Nup157 and Nup170 (Drin et al., 2007; Kim et al., 2014; Mészáros et al., 2015; Shi et al., 2014) contact the pore membrane, and these interactions are consistent with the protocoatomer hypothesis (Devos et al., 2004; Kim et al., 2018). The density map for the isolated NPC, coupled with peptide cross-link data (Kim et al., 2018), indicate that Nup157 β-propellers interact with Pom152 near the spoke 2-fold axis (MAC157) (Figures 2B and S9B [black asterisks]), while adjacent Nup170 β-propellers interact with Ndc1 dimers (MAC170) (Figure 2B and S9B [blue asterisks]) (Mansfeld et al., 2006; Vollmer et al., 2012). Diagonally arrayed, membrane-interacting layers in adjacent spokes form a nearly continuous band when viewed from the pore membrane (Figure S9C, red dots). Similar contacts are present in the in situ NPC, but with a larger spacing between adjacent layers (Figure S9F, red dots). Nup53-Nup59 dimers are located between neighboring Nic96 CTDs and help tether the inner ring to the pore membrane (Figures 9F [green dots], S9C, and S9E [green asterisks]) (Lin et al., 2016; Kim et al., 2018; Marelli et al., 2001). A local asymmetry at higher radius in the in situ NPC may result from altered membrane interactions, as contacts for Nup170, Nup53-Nup59, and Nup157 are more extensive on the left side of the spoke when viewed from the cytoplasm (see asterisks, Figure S9E). Pom152 and Ndc1 follow the local 2-fold symmetry of the radially contracted inner ring, while these critical interactions may be altered during radial expansion.
Two Pom152 molecules are anchored to each spoke by their NTDs while their CTDs form a lumenal ring in the NPC (Kim et al., 2018) (Figures 1A and 1B). Each Pom152 has 10 immunoglobulin-like repeats (Hao et al., 2018; Upla et al., 2017) that overlap with a molecule from a neighbor to form a lumenal arch, and 8 arches create a sinusoidal ring in the isolated NPC (Figures 1A [inside ring] and S9G). Cross-linking data suggest that Pom34 may associate with the TMD of Pom152 (Kim et al., 2018). The Pom152 ring in the isolated NPC has an outer contour of ~400 Å for one repeat, while dilation expands the sinusoidal ring into a nearly circular profile with a repeat of ~440 Å (Figures 5A and S9G [outside ring]). Integrative models for the Pom152 rings revealed two anti-parallel strands of immunoglobulin-like domains, aligned in a side-by-side manner (Figure S9H). The position, robust construction, and interaction of the Pom152 ring with the spokes suggests that this belt-like feature may help limit the extent of radial expansion by the inner ring and pore membrane (Figure 7D).
Figure 7. Diagrammatic summary of major insights into the yeast NPC.
(A) A wiring diagram for Nic96 connectors that extend from CTDs in the membrane interacting layer to the diagonal walls. Red stars indicate approximate positions of phosphorylation sites in human Nup93: membrane interacting layer (blue-purple), adaptin-like layer (red), and inner layer (green).
(B) The IBB helix of Importin-ɑ/Karyopherin ɑ interacts with Importin-β/Karyopherin β1, and this motif is compared with Nic96 NTD helices bound to Nup192 and Nup188.
(C) Three structural variants of the NPC are present in the same yeast cell.
(D) Inner ring dilation forms gaps between adjacent spokes; this is accompanied by a radial displacement of the pore membrane and expansion of the Pom152 ring.
(E) Duplication within major NPC sub-structures: related Nups are shown in red-brown and light blue. Basket anchor motifs (red and royal blue double lines) are present in two copies per Y-complex. This lateral duplication may encompass the basket itself (vertical dashed lines). (inset) Cartoon of a basket anchor motif with labelled structure elements.
Nsp1 complex connections to FG domains in the central channel
The N termini of Nsp1 complexes in the inner ring extend into the central channel where they account for ~38% of the FG domains in the transport path (Kim et al., 2018; Yamada et al., 2010; Eibauer et al., 2015). Three rods per Nsp1 complex were used—as linear markers—to track these connections, which were resolved at ~20Å resolution (Methods S1). In total, four connecting densities per spoke are present in two groups, with one group of 3 red and 3 blue rods located at the central 2-fold axis and a second group at the boundary between spokes (Figures 1A [top] and 6E). We find that Nsp1 complexes are positioned precisely so that their N-terminal FG repeats may extend through the connections into the transport channel. Our 3D map of the in situ yeast NPC also displays prominent connections, but with a different configuration (Figure 6F). During dilation, each spoke moves radially outward to create a gap between adjacent Nsp1 complexes (Figures 6A, 6E, and 6F [gap]). This causes paired connections from adjacent spokes in the contracted inner ring to split laterally and creates a visible bridge from the N termini of each Nsp1 complex, while connections in the center group remain intact until they bifurcate as they enter the channel. Radial dilation also creates a new lateral contact with FG connections from an adjacent spoke (Figure 6F [arrow]).
We then calculated a global average of the central transporter in the isolated NPC, which is viewed at a higher threshold due to its flexibility and structural heterogeneity (Figure 1A [top]; ~310Å diameter by ~460–500Å in height) (Kim et al., 2018; Akey and Radermacher, 1993; Yang et al., 1998). The central transporter has two cylindrical regions with a less dense central waist, and spoke connections encircle the waist (Figure 6E). The transporter is less ordered near the center and at regions facing the cytoplasm and nucleoplasm (Methods S1). Our data suggest that the transporter is created by densely packed FG domains along with trapped transport factors and cargo complexes (Kim et al., 2018). Central transporter density is also present in a global 3D map of the in situ NPC; however, a larger dataset is required to characterize this feature. Even so, inner ring dilation increases the volume of the central channel by ~2-fold and alters the pattern of connections to Nup FG domains. We surmise that radial expansion may alter the distribution of FG domains to facilitate transport.
DISCUSSION
In this work, a multi-pronged approach has provided a comprehensive picture of the yeast NPC, generating insights into the architectural principles, evolutionary origins, and structure-function relationships of this transport machine.
A layered organization and flexible connectors suggest a mechanism for reversible NPC assembly
We resolved the inner ring at sub-nanometer resolution and identified 28 connectors from Nic96 NTDs that tie together the inner and outer domains of the yeast spoke. Our model supports the idea that the layered organization of Nup folds in the NPC may reflect its evolution. Nups in the outer rings and the membrane-interacting layer share similar folds with COPI and COPII families of vesicle coating proteins, which probably merged to form a proto-NPC (Field and Rout, 2019). β-propellers in these Nups contain MBMs that help anchor the NPC to the pore membrane. Moving inward, the next layer is comprised of two adaptin-like proteins, underscoring the common evolutionary origin of vesicle coating proteins and the NPC. These proteins also share structural homology with Karyopherin transport factors. The innermost layer contains Nsp1 complexes that form two diagonal walls and provide anchor points to position N-terminal FG repeat domains within the central channel (Figures 6E, 6F, and 7A).
A key question is “how can NPCs assemble and disassemble so quickly?” We find that flexible Nic96 connectors tie together the layers of the inner ring (Figure 7A) and may play a crucial role during NPC assembly. Connectors in the Nic96 NTD are targets for reversible phosphorylation in eukaryotes that undergo mitotic dis-assembly and re-assembly, along with SLiMs of Nup53/Nup59, Nup100, Nup116, and Nup145N. (De Souza et al., 2004; Laurell et al., 2011; Linder et al., 2017). We speculate that disassembly may be correlated with a phosphorylation-dependent “untying” of these connectors (see red stars, Figure 7A). Assembly would involve a tying together of Nups, which may occur upon dephosphorylation at the end of telophase or may use newly synthesized Nups during interphase (Kutay et al., 2021). In the simplest model, Nic96 CTDs associate with Nup157 and Nup170 to create the membrane-interacting layer at the pore membrane. This would provide anchor points for Nic96 NTDs during assembly to interconnect the three structural layers of the spoke. These connectors may allow the NPC to accommodate large changes in diameter and adapt to local stresses during transport.
A link between nucleoporin assembly and NLS mediated transport
Our analysis indicates that spoke assembly may be dependent on interaction motifs which are akin to those that occur between transport factors (Figure 7B). The ɑ-helical solenoid of the Nic96 CTD is structurally similar to Karyopherin ɑ, while Nup192 and Nup188 may share a common ancestor with β-Karyopherins (Andersen et al., 2013; Sampathkumar et al., 2013; Stuwe et al., 2014). Interactions of the Nic96 NTD with Nup192, and to a lesser extent with Nup188, resemble the binding of the IBB domain of Karyopherin ɑ and NLSs with Karyopherin β1 (Figure 7B) (Bhardwaj and Cingolani, 2010; Cingolani et al., 1999). These observations provide structural evidence that suggests a common evolutionary origin for the NPC and its soluble transport machinery.
NPCs are modular structures with multiple forms in yeast nuclei
Structural conservation of the inner ring contrasts with an expanding list of NPCs from species with different configurations of the outer rings. These differences are achieved by altering the copy number of Y-complexes, by adding side-specific Nups, and by repurposing Nups for structural roles in the outer rings (Kim et al., 2018; Kosinski et al., 2016; von Appen et al., 2015). In this study, we asked the following: are all NPCs in the same nucleus fundamentally the same? Remarkably, the answer is “no.” When taken together, the data suggest that at least three NPC isoforms coexist in yeast (Figure 7C). The NPC is a modular assembly with a “Lego”-like ability to have major scaffold elements added, reorganized, and/or removed. Differences in the number of outer rings could affect the rigidity of the scaffold; this property may be adapted to the requirements of a particular cell type and organism or even a distinct region at the NE. These structural variants may represent NPCs that are specialized in transporting specific cargos or in performing transcriptional control and directing chromatin organization (Fernandez-Martinez and Rout, 2021; Sumner and Brickner, 2021). In addition, the nuclear double outer ring may provide anchor sites for regulatory factors, interact with the NE to stabilize the pore membrane, and contribute to the functional asymmetry of NPCs.
Dynamic alterations of the NPC may play a central role in transport
The NPC is a dynamic assembly with a range of effective diameters, and recent studies have revealed a large dilation of the inner ring, which creates an expanded central channel (Schuller et al., 2021; Zimmerli et al., 2021). Our analysis provides additional evidence for changes that occur during dilation. Radial movements of the spokes are coupled with a displacement of the pore membrane and alterations in the Pom152 ring (Figure 7D); these changes may reflect the transition from a relaxed to a tense configuration of the inner ring. We also provide insights into membrane protein interactions that anchor the inner ring within the membrane pore. These interactions are mediated by Pom152, Pom34, and Ndc1; they appear to be 2-fold symmetric in the isolated NPC but may become less symmetric during radial dilation. Yeast strains with Pom152 deletions are viable (Wozniak et al., 1994), but even so, the lumenal ring is connected to the spokes and may function like a belt to help limit radial dilation. Some membrane proteins can be transported between the outer and inner membranes through the NPC, but this mechanism remains mysterious (Antonin et al., 2011). The gap between spokes in the dilated inner ring may provide an access point at the pore membrane for this transport process, but Nup170 and Ndc1 molecules may need to be displaced.
Our comprehensive model suggests that local duplication of Nups may play critical roles in NPC assembly and function. At the nuclear periphery, the basket plays a role in targeting and preprocessing of nuclear export complexes including messenger RNA protein complexes. We mapped pairs of basket anchors that extend from the nuclear outer ring. These features appear to follow the pattern of lateral duplication established for the core scaffold, and this theme may extend into the rest of the basket (Figure 7E). Since nuclear baskets are present on NPCs with both single and double outer rings, their anchors must adapt to different ring geometries.
Finally, inner ring dilation is associated with a ~2-fold increase in the volume of the central channel (Figure 7D). Therefore, increased transport levels may be correlated with diameter changes, as observed in Xenopus oocytes (Feldherr et al., 1998) and a recent in situ study of S. pombe NPCs (Zimmerli et al., 2021). Dilation may also help explain changes in transport induced by mechanical forces (Elosegui-Artola et al., 2017; Jacchetti et al., 2021; Kassianidou et al., 2019). We find that the central channel is partitioned into distinct regions including a zone of spoke connections and a central zone of FG repeats. We suggest that the packing density of FG domains may be altered during inner ring dilation, as reflected by changes in spoke connections to these domains; this reorganization may have important consequences for transport. Indeed, fundamental principles that govern dynamic FG domain organization remain to be uncovered in this “undiscovered” country.
Limitations of the study
The current study has created a detailed molecular model of Nups within yeast NPCs, with a precision that is limited by the local resolution and quality of the homology models. Recent advances in structure prediction with deep learning methods promise to provide improved templates for molecular modeling of the NPC (Jumper et al., 2021). In addition, further work is needed to improve the resolution of major ring systems within the NPC and to further characterize connectors, membrane anchor complexes, the Pom152 ring, and the nuclear basket.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael Rout (rout@rockefeller.edu).
Materials availability
Yeast strains and plasmids generated in this study will be distributed without restriction upon request.
Data and code availability
Cryo-EM 3D density maps and associated models have been deposited in the EMDB and RCSB, respectively, with accession codes EMD-24224, EMD-24225, EMD-24232, PDB: 7N85, EMD-24231, PDB: 7N84, EMD-24258 and PDB: 7N9F. Integrative modeling data have been deposited in the PDB-Dev: PDBDEV_00000097. Fluorescence data is available at https://www.stowers.org/research/publications/libpb-1583 and cross-linking data have been deposited at Zenodo (https://zenodo.org/) accession code 5721514.
Software scripts and data for integrative modeling are available at https://github.com/integrativemodeling/NPC_3.0.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Yeast strains and materials
Unless otherwise stated, all S. cerevisiae strains used in this study were grown at 30°C in YPD media (1% yeast extract, 2% bactopeptone, and 2% glucose).
METHOD DETAILS
Immuno-purification of the endogenous S. cerevisiae NPC
Intact S. cerevisiae NPCs were isolated following the method described (Kim et al., 2018). To minimize local domain movements, purified NPCs were mildly cross-linked by the addition of DSS (DiSuccinimidylSuberate, Creative Molecules) to yield a final concentration of 0.5 mM and incubated for 30 min at 25°C with gentle agitation in a shaker (900 rpm). The reaction was then quenched by addition of ammonium bicarbonate to a final concentration of 50 mM. After Cysteine reduction, cross-linked samples were separated by NuPage SDS-PAGE (4%–12%, Invitrogen) for quality control and concentration estimation. Gels were briefly stained by GelCode Blue Stain Reagent (Thermo Fisher Scientific) to enable the visualization of the protein complex. Aliquots of crosslinked NPCs (~0.3 mg/mL) were flash frozen and stored at −80°C in small aliquots in buffer: 20 mM HEPES (pH7.5), 50 mM Potassium acetate, 20 mM NaCl, 2 mM MgCl2, 1 mM DTT, 0.1% Tween 20) supplemented with 10% glycerol. Samples were thawed only once prior to making grids for data collection.
Chemical cross-linking and MS (CX-MS) analysis of affinity-purified yeast NPCs.
NPCs were immuno-purified from Mlp1 tagged S. cerevisiae strains (Kim et al., 2018). After native elution, 1.0 mM disuccinimidyl suberate (DSS) was added and the sample was incubated at 25°C for 40 min with shaking (1,200 rpm). The reaction was quenched by adding a final concentration of 50 mM freshly prepared ammonium bicarbonate and incubating for 20 min with shaking (1,200 rpm) at 25°C. The sample (50 μg) was then concentrated and denatured at 98°C for 5 min in a solubilization buffer (10% solution of 1-dodecyl-3-methylimidazolium chloride (C12-mim-Cl) in 50 mM ammonium bicarbonate, pH 8.0, 100 mM DTT). After denaturation, the sample was centrifuged at 21,130 g for 10 min and the supernatant was transferred to a 100 kDa MWCO ultrafiltration unit (MRCF0R100, Microcon). The sample was quickly spun at 1,000 g for 2 min and washed twice with 50 mM ammonium bicarbonate. After alkylation (50 mM iodoacetamide), the cross-linked NPC in-filter was digested by trypsin and lysC O/N at 37°C. After proteolysis, the sample was recovered by centrifugation and peptides were fractionated into 10–12 fractions by using a stage tip self-packed with basic C18 resins (Dr. Masch GmbH). Fractionated samples were pooled prior to LC/MS analysis.
Desalted cross-link peptides were dissolved in the sample loading buffer (5% Methanol, 0.2% FA), separated with an automated nanoLC device (nLC1200, Thermo Fisher), and analyzed by an Orbitrap Q Exactive HFX (Pharma mode) mass spectrometer (Thermo Fisher) as previously described (Xiang et al., 2020; Xiang et al., 2021). Briefly, peptides were loaded onto an analytical column (C18, 1.6 μm particle size, 100 Å pore size, 75cμmc × c25ccm; IonOpticks) and eluted using a 120-min liquid chromatography gradient. The flow rate was approximately 300 nL/min. The spray voltage was 1.7 kV. The QE HF-X instrument was operated in the data-dependent mode, where the top 10 most abundant ions (mass range 380 – 2,000, charge state c4 – 8) were fragmented by high-energy collisional dissociation (HCD). The target resolution was 120,000 for MS and 15,000 for tandem MS (MS/MS) analyses. The quadrupole isolation window was 1.8cTh; the maximum injection time for MS/MS was set at 200cms.
The raw data were searched with pLink2 (Chen et al., 2019b). An initial MS1 search window of 5 Da was allowed to cover all isotopic peaks of the cross-linked peptides. The data were automatically filtered using a mass accuracy of MS1 ≤ 10 ppm (parts per million) and MS2 ≤ 20 ppm of the theoretical monoisotopic (A0) and other isotopic masses (A+1, A+2, A+3, and A+4) as specified in the software. Other search parameters included cysteine carbamidomethyl as a fixed modification and methionine oxidation as a variable modification. A maximum of two trypsin missed-cleavage sites was allowed. The initial search results were obtained using a default 5% false discovery rate (FDR) expected by the target-decoy search strategy. Spectra were manually verified to improve data quality (Kim et al., 2018; Shi et al., 2014). Cross-linking data were analyzed and plotted with CX-Circos (http://cx-circos.net).
Mapping Nic96 interactions with Nup188/192 by in vitro reconstitution
Full length (FL) Nic96 along with C- and N-terminal truncations were constructed and purified as previously described (Fernandez-Martinez et al., 2012; Kim et al., 2018). Affinity purified and natively eluted full length Nup188 and Nup192 (Fernandez-Martinez et al., 2012) were incubated with the indicated affinity matrix-bound Nic96 complexes in binding buffer (20 mM HEPES-KOH pH 7.4, 50 mM potassium acetate, 20 mM sodium chloride, 2 mM magnesium chloride, 0.1% Tween-20, 1 mM DTT, 1/250 cOmplete ™ protease inhibitor complex (Sigma-Aldrich)) for 1 h at 4°C. After appropriate washes, the bound complexes were resolved by SDS-PAGE and stained with Coomassie R-250.
Quantitative fluorescence imaging
Standard PCR techniques were used to generate strains expressing C-terminally tagged Nup fusion proteins. The outer ring component Nup84 was tagged with the GFP1–10 fragment of split-GFP, and plasmids expressing GFP11 reporters localized to the nucleoplasm (GFP11-mCherry-Pus1) or outer nuclear membrane/ER (GFP11-mCherry-Scs2-TM) were used to reconstitute GFP fluorescence and visualize rings on the nucleoplasmic and cytoplasmic faces of the NPC independently. Nic96-mTurquoise2 was used to detect and segment NPCs during analysis (see below).
Cells were grown in synthetic complete media lacking leucine (SC-leu; 6.7 g Yeast Nitrogen Base without amino acids, 2.0 g SC-Leu supplement mixture (Sunrise Scientific), 2% Glucose, pH 5.8) overnight at 30°C, back diluted and grown to an OD600 of 0.6–0.8 prior to harvesting. Cells were then pelleted for 3 min at 3000 × g and resuspended in 1 mL of 4% paraformaldehyde, 100 mM sucrose and fixed for 15 min at room temperature. Following three washes with 1 mL 1×PBS, the cell pellet was resuspended in 20 μL Prolong Diamond mounting media (Invitrogen) and slides were prepared by applying 3 μL of cells between slides and number 1.5 glass coverslips. Slides were cured overnight at room temperature and stored at 4 degrees until imaging.
Image stacks covering the entire nuclear volume were acquired with sub-Nyquist sampling and 0.126–0.220 nm step sizes on a Leica SP8 confocal microscope equipped with a HC PL APO CS2 100×, 1.40 NA objective. 458 nm, 496 nm and 561 nm lasers were used to excite mTurquosie2, rGFP and mCherry respectively, and emission for the corresponding channels were in the ranges of 463–495 nm, 505–550 nm and 566–660 nm. The pinhole size was set to 0.6 Airy unit to increase resolution, and raw images were automatically processed with default HyVolution settings. Individual nuclei were manually cropped from deconvoluted image stacks and analyzed with custom plugins written in ImageJ (NIH). Downstream analysis and data visualization was performed using R (v. 3.6.1) and RStudio (v. 1.1.442). All original data and custom code used for analysis can be accessed from the Stowers Original Data Repository at https://www.stowers.org/research/publications/libpb-1583.
NPCs were detected in three dimensions using a “track max not mask” approach. Briefly, the brightest voxel in the image was found and a spheroid with a diameter of 12 pixels in x and y and 3 slices in z was masked around that voxel. This process was repeated until no voxels were left unmasked above 20% of the maximum intensity in the image. Next the average intensities of these presumptive NPCs were measured with a spheroid of diameter 8 pixels in x and y and 2.5 slices in z. Average intensity measurements for each image and channel were normalized to the average NPC intensity for the entire image. After normalization, foci with an intensity less than 0.35 in the Nic96-mTurqouise2 channel were removed to avoid poorly detected NPCs and background regions. For analysis of Nup84 levels with respect to Mlp1 intensity, NPCs were classified as either “high” or “low” based on whether the normalized Mlp1-mCherry intensity was above or below the mean Mlp1 intensity (1.0). The ratio of Nup84-rGFP to Nic96-mTurqouise2 was used for measurement purposes as it corrects for potential detection efficiency issues that are shared across fluorescence channels.
Cryo-EM single particle data collection and movie processing
Sample grids were optimized to obtain a range of NPC views with a high loading; thus, particles were adsorbed on a carbon film overlayed onto Quantifoil 400 mesh carbon holey grids (2/2, hole diameter 2.4 uM). Grids were glow discharged followed by an extended floatation with carbon side down on 5 μL drops in a humid chamber. This was followed by 3 stepwise transfers onto 20 μL drops of blotting buffer (NPC buffer without glycerol) supplemented with 0.1% DeoxyBigChaps (wt/wt), while keeping the backside of the grids dry. Each grid was mounted in a Mark III Vitrobot at 10°C and 100% relative humidity, blot buffer was removed with a manual blot from the bottom edge of the grid through a side port in the humid chamber; an appropriate volume of blot buffer was pipetted immediately onto the grid, which was double blotted and plunge frozen in liquid ethane cooled by liquid N2.
Super-resolution movies were collected as LZW compressed tiffs (2 movies per hole, 4015 in total) using SerialEM (Mastronarde, 2005) on a Titan-Krios at 300 kV with a K2 Summit direct electron detector (Cryo-EM Core, University of Massachusetts Medical School). A physical pixel size of 2.66 Å (corrected magnification 37,651) was used to target the 6–9 Å resolution range (~1100 Å diameter by 600 Å height). To check particle distribution and contamination, a script written by Dr. Chen Xu was used to automatically collect test images that covered ~9 holes at 200 um defocus from each candidate grid mesh with the attached CCD. Medium magnification montages were then collected for good meshes at a much lower dose to minimize total exposure. Navigator parameters were set to have 6–7 groups of holes per mesh, with one focus spot per group using an in-house script. For the movies, 40 frames were collected with 0.5 s exposure/frame, a total dose of 40 e/Å2 and a calculated defocus range of −1.5 to −3.8 um with a K2 Summit direct electron detector in super-resolution counting mode (Gatan Inc.) with a Bioquantum GIF and 20eV slit width. Image quality was monitored “on the fly” using a script (written by Dr. Chen Xu), which does local motion correction with IMOD (Kremer et al., 1996) on the imaging computer and creates power-pair jpegs with a motion-corrected micrograph image paired with the fitted power spectrum from CTFFIND4.
After data collection, movies were decompressed, gain corrected and aligned with Motioncor2 (v1.2.3) (Zheng et al., 2017) to produce dose corrected micrographs, which were used to estimate per micrograph contrast transfer function (CTF) parameters with GCTF (Zhang, 2016). Manual triage was done with Motioncor2/GCTF power-pairs using the “eye of Gnome” viewer (eog) and awk scripts to eliminate poor micrographs (3218 remained; ~80%). After parameter optimization, NPCs were picked with Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/) using an image stack of equi-spaced projection views that were calculated from our tomographic model with C8 symmetry ((Kim et al., 2018); EMD-7321) using EMAN2 (e2project3d.py (Tang et al., 2007)). For initial screening, particles were extracted in 310 × 310 boxes with a pixel size of 5.32 Å to remove outliers with 2D and 3D Maximum likelihood-based classification in RELION 2.0 (Scheres; 2012), which resulted in 26049 selected NPCs.
Single particle processing
Spokes and co-axial rings-
To evaluate the overall quality and angular coverage, the dataset was processed initially with particles extracted at 5.32 Å/pixel. Refinements with C8 symmetry and a low pass filtered starting model from tomography (Kim et al., 2018), gave a structure with RELION that was virtually identical to the published map but with many more features (global resolution 24 Å). Subsequent refinements with imposed C8 symmetry used particle images after subtraction of the central transporter density and local soft masks that focused respectively on the inner ring and each of the two distal ring features. This resulted in a composite 3D map with a local resolution range of 14.4 to 30 Å in which features of the double outer ring began to emerge.
For multi-body refinements with density subtraction, we started with particles at 5.32 Å/pixel and after test refinements in RELION 3.0 (Nakane et al., 2018), re-centered NPCs were extracted at 2.66 Å/pixel in 620 × 620 boxes. In brief, we divided the processing into two sequential multi-body cycles to avoid memory limitations due to the requirement for multiple masks, coupled with the large box size of the particles (Methods S1). The first cycle identified the best protomers and was centered on ~1.5× subunits within the full particle in the original box, wherein a subunit is comprised of a spoke and distal ring “features” located on either surface. Following subunit extraction and re-centering within a smaller box, a second multi-body refinement step focused on three regions within the protomer. Refinements in which masks were centered on the spoke 2-fold axis were of noticeably higher quality than those centered on the 2-fold axis that passes between spokes. To start, we expanded particle line entries in the consensus C8 alignment star file for the full NPC, by duplicating every particle line entry 8x with a cyclic permutation of the first Euler angle in 45° increments (relion_particle_symmetry_expand). This 8-fold expanded particle star file and the original image stacks were used in a C1 refinement with a 1.5x spoke mask to bring each protomer into a common alignment.
For the first multi-body refinement step, we created a soft mask that covered the remaining protomers (~6.5x) in the ring-like particle, along with the central transporter using a C8 symmetrized 3D map and then subtracted the density within this mask from the particle stack in each refinement iteration by using a zero alignment flag in the multi-body star file. After refinement, density subtracted image stack files for the entire ~1.5x protomer were written out and used for focused 3D classification with local refinements to eliminate any remaining damaged subunits. A local C1 consensus refinement was then done for the re-centered and density subtracted protomers, which were subjected to a second round of multi-body refinement with three overlapping masks that covered the spoke and each outer ring. We then focused on the spoke and appropriate images were re-extracted and a local 3D classification with a soft mask was run (+/− 9 degrees) to further clean the dataset. Refinements with C1 symmetry and a mask produced a map for the spoke at ~8 Å resolution (local range 7.9–11Å ).
NPCs were imaged on a thin carbon support film that dominates the CTF estimation and this value reflects a particles position along the optical axis relative to true focus. Hence, the median value of the CTF for the particle center is dependent on its orientation on the carbon film. For the NPC, this offset will become more significant at higher resolution. We created a script based on the geometry of the particles and the Euler angles (Liu et al., 2019) to calculate the z-offset (median defocus change) to each NPC center. An ambiguity in sign and thus, the direction of z-change was resolved by carrying out two separate 3D refinements with all per particle CTF values calculated with the same directional offset: the resolution of the final map was used as an indicator of the correct orientation. This resulted in a refined spoke 3D map with an improved local resolution range (corrected 6.6–11 Å), but the median FSC0.143 improved by only 0.1 Å with a larger mask. Per particle CTF corrections improved the best ordered, central 60%–70% of the map, while more peripheral, flexible regions were slightly worse, accounting for the small global change in FSC0.143. This spoke map was 2-fold symmetrized and normalized in EMAN2 (e2proc3d.py–process = normalize.local), which allows densities across the map to be represented more equally at a single threshold.
A similar two-step multi-body approach with particles at 3.99 Å/pixel was used to process the outer ring with a cobra hood shape and revealed a double outer ring of Nup84 Y-complexes. We found that overlapping V-shaped regions from Nup120, Nup85 and Seh1 in two offset Nup84 complexes were combined to create the hood shape in the original 3D maps. In the second round of multi-body refinement, two masks were applied to the image stack with density subtracted transporter; the zero subtraction mask contained the inner ring, membrane density and Nup82 complex density, while the second alignment mask covered ~1.5x subunits in the double outer ring. After re-extraction of centered outer ring particles, 3D classification with tau 8 and local refinement in multiple steps with gradually improved masks identified particles with a double outer ring (~20%) and gave a final 3D map with a global resolution of ~11.3 Å using a generous mask.
Maps from final refinements for the spoke and double outer ring were cloned to create 8 copies and aligned within the best 3D density map of the full NPC in Chimera (Pettersen et al., 2004), rotated appropriately and added together to create complete rings. A visual inspection with docked models in Coot did not reveal anomalous features at subunit boundaries in the reassembled 3D maps at the current resolution. Final 3D maps were masked in EMAN2 and dusted in Chimera to remove noise. The cytoplasmic outer ring could not be improved with a multi-body approach due to its flexibility. The structure determination for the inner ring and double outer rings is summarized in Methods S1 and Table S1.
Central transporter and spoke connections
The central transporter and spoke connections were analyzed separately; an image stack at 5.32 Å/pixel was used for a C8 refinement in RELION. To focus on the transporter, a 3D classification with 8 classes was done with the skip_align option and a mask which included the central transporter and a small region of the surrounding spoke. Two classes with low density were eliminated and the remaining 6 classes were grouped together, auto-refined and post-processed. To focus on connections between the spokes and transporter, a 3D classification with 6 classes was carried out with skip_align and a mask that eliminated the central region of the transporter, both distal rings and part of the membrane remnant. Particles in the two best classes were merged, auto-refined and post-processed with C8 symmetry. Unfortunately, signal for the lumenal Pom152 ring was too delocalized to identify a better ordered subset with 3D focused classification in the isolated NPC.
Modeling the spoke and double outer ring
We started with an integrative model of the yeast spoke (PDBDEV_00000012) that was based on ~25 crystal structures (Kim et al., 2018) and docked 28 Nups within the spoke 3D density map as rigid bodies in Chimera. We then used manual modeling in Coot (Casañal et al., 2020), Chimera (Pettersen et al., 2004), Phenix realspace_refine (Afonine et al., 2018) and molecular dynamics flexible fitting (MDFF; (Trabuco et al., 2008)) to create a model based on a majority of the ~715 ɑ-helices within the spoke. We included flexible connectors from N-termini of four Nic96 molecules and 2 orphan helices to give 26 novel ɑ-helices and 4 strands, along with tail domains with altered conformations from six Nups (two copies each of Nups 192, 188 and 170); the full spoke model contains ~15,600 residues in the ordered domains (~1.69 MDa).. A novel Integrative threading was used to position sequences of Nic96 N-termini within the orphans (see below, (Saltzberg et al., 2019)), which was followed by an additional round of refinements to create a final model (PDB 7N85; EMD-24232). Nup domains in the structure have undergone local movements and re-arrangements with an average Cα rmsd of ~8Å, when compared to an earlier integrative model of the spoke (Kim et al., 2018), with larger changes at Nup contacts and due to rearrangements of the Nsp1 complex. For the double outer ring, 14 Nups were docked into the repeating unit (~1.1 MDa) using yeast homology models based on 5 crystal structures (Kelley et al., 2015) that were split into 22 domains with a total of ~10,000 residues. The improved resolution allowed us to model complete paths for two Y-complexes within the double ring along with a copy of Nup188 and a putative basket anchor in a rod-like orphan density. Initial models were subjected to manual remodeling in Coot, Chimera, and refinements with Phenix realspace_refine (while ensuring that perturbations of the β-propellers did not occur from over-refinement). Final fitting steps used MDFF: a gscale value of 0.6 was used for medium resolution models (7–10Å), while flexible fitting into lower resolution maps used values of 0.15–0.3. Typical restraints were used for protein geometry in refining the double ring model (PDB 7N84, EMD-24231).
Mapping cross-links to the NPC structures
A total of 2,053 DSS and EDC crosslinks were obtained previously for the isolated NPC (Kim et al., 2018). To assess whether the crosslinks agree with the NPC’s structure, these crosslinks were mapped on to the cryo-EM structure by determining the Cα–Cα distance of crosslinked residue pairs. A DSS or EDC crosslink was classified as satisfied by the structure if the corresponding Cα–Cα distance is less than 35 or 30 Å, respectively (Figure S5B–S5E) (Kim et al., 2018).
Initial model for 30 orphan densities within the spoke
At this stage, a total of 30 orphan densities were identified in the spoke that likely arise from N-termini of Nic96 and Nup53/Nup59, functional orthologs of CtNup53 and possibly from other Nups. We identified 20 orphans combined in the two diagonal walls. In addition, 10 orphan helices are associated with two copies Nup192 and two copies of Nup188. Physical, biochemical and structural restraints were used to create an initial model for connectors in the orphan densities. Previous work showed that a large connector is present in the N terminus of CtNup53 which interacts with the NTD of Nup192 (Lin et al., 2016). We observed a long helix associated with the NTD of Nup192 that may be attributed to Nup59 because this protein interacts with Nup170 and Nup192 in pullout experiments and behaves like CtNup53. This left 28 orphans in a spoke that may be contributed by connectors from four Nic96 N termini.
The Nic96 N terminus contains 204 residues, so the sum of orphan structural elements + connector residues in each modeled chain should not exceed this value. Moreover, the distance between diagonal walls is too great to allow a single Nic96 N terminus to be located in both walls. Previous work identified two conserved sequence motifs in the N-termini of Nic96 and its homolog, Nup93 that interact with Nsp1 and p62 complexes, respectively and with the C-terminal tails of Nup188 and Nup192/Nup205 (Lin et al., 2016). The 5 orphans in each diagonal half-wall are present in two groups with linker density visible at a lower threshold for some pairs: a group of three structural elements (two helices and a strand) and a group of two helices. After manual docking of poly-Ala ɑ-helices and strands in Chimera and Coot, we found that the topology of the group of 3 structural elements is similar to the helix-strand-helix motif of a Nic96 SLiM observed in a crystal structure with a Ct-Nsp1 heterotrimer ((Stuwe et al., 2015); PDB 5CWS; Figure S5F). Structure prediction of the Nic96 N terminus also suggested that two helices may be positioned between the group of 3 structural elements and a pair of helices located near the end of the NTD that are bound to Nup192 or Nup188. The final pair of connectors from the innermost copy of Nic96 may interact with a CTD of Nup192 while these connectors from the outer copy of Nic96 probably bind to the Nup188 CTD. Manually docked helices and strands were refined with Phenix realspace_refine (Afonine et al., 2018) and these coordinates were used as input to an integrated threading approach to test all possible structure element orientations and sequences from Nups as connectors within the orphan densities; this analysis did not use the crystal structure of the Nic96 SLiM (Stuwe et al., 2015).
Integrative threading of Nup sequences into spoke orphan structure elements
A novel integrative threading approach (Alber et al., 2007a; Kim et al., 2018; Rout and Sali, 2019; Russel et al., 2012; Saltzberg et al., 2019) was applied to determine the protein, its copy identifier, and the sequence of orphan structure elements (SE) in the inner ring. All possible arrangements derived from the spoke C2-symmetry were considered, but only one resulted in a model that sufficiently satisfied the input data: this option splits the 30 orphan densities into two symmetric subsets of 15 orphan densities each. Integrative modeling proceeded through the standard four stages, as follows:
Stage 1 - Gathering information
The data used for integrative structure modeling is summarized in Table S2, which is available on-line (https://zenodo.org/record/5721514#.YcY9laROlhE), record number 5662389.
Stage 2 - Representing subunits and translating data into spatial restraints
We represent the NPC model using a threading representation. The representation defines the degrees of freedom of the model whose values are fit to input information.
Threading representation.
We first identified a total of 30 orphan densities in the inner-ring cryo-EM density map. These densities most likely correspond to distinct SEs, including ɑ-helices and β strands. A poly-Ala sequence of suitable length was manually fit into each rod-like orphan density, followed by refinement using Phenix realspace_refine as described. Each SE is defined by four degrees of freedom that map it to specific residues in the sequence: 1) Start Residue, defining the first residue in the sequence to which the SE is mapped; 2) Length, the number of residues in the sequence overlapping with the SE; 3) Offset, the index of the first SE residue that is assigned to the sequence; and 4) Polarity, which defines the direction of the mapping between the SE and the sequence (Figure S4C).
Coarse-grained structural representation of the NPC scaffold.
If an atomic structure of a protein segment is present in the isolated NPC structure described above, it was represented by the corresponding rigid body configurations of a fine- and coarse-grained representation of 1- and 10-residue beads, respectively (Table S2, (Kim et al., 2018), references therein).
Scoring function for threading representation.
Alternative models are ranked via a scoring function. The scoring function is a sum of terms, each one of which restrains some aspect of the model based on a subset of input information (e.g., stereochemistry, crosslinks, sequences). We impose three scoring terms that restrain the four degrees of freedom (Saltzberg et al., 2019) for each of the 30 SEs. First, the secondary structure restraint is computed by comparing the secondary structure assignment in the model with the secondary structure propensity predicted from the input sequence. Second, the loop end-to-end distance (i.e., C- to N-termini distance between two sequentially adjacent SEs) restraint relies on the relationship between the model loop length and its end-to-end distance as derived from the CATH 3D structural database. Third, the SE crosslinking restraint corresponds to a harmonic upper-bound on the distance spanned by two cross-linked residues. When one or both crosslinked residue(s) have undefined position(s), we compute the distance from inferred localization volumes.
Stage 3 - Enumeration of threading solutions to produce an ensemble of structures that satisfy the restraints
Sequences from appropriate Nups were threaded through the backbone of the 30 SEs in a spoke. We improved the efficiency of threading by explicitly considering the C8- and C2-symmetries of the inner ring (Figure 2B).
Symmetry: We defined the coordinate system such that the C2-symmetry is imposed simply by cloning a bead in the C2-symmetry unit at (x, y, z) to (x, −y,−z) (equivalent to a rotation of 180° around the x axis). In addition, a second 2-fold axis was considered for sequence threading of the orphan densities (Figure S3C). Based on symmetry, we grouped the SEs into five groups: 1) SEs 1–3 that bind to the Nsp1/Nup49/Nup57 coiled-coils, 2) SEs 4 and 5 that localize between copies of the Nsp1/Nup49/Nup57 coiled-coils, 3) SE 6 that binds to the Nup188 CTD, 4) SE 6 that binds to Nup192 CTD, and 5) SE 7 that binds the Nup192 NTD (Figure S3B). All possible symmetry arrangements derived from the spoke C2-symmetry were considered, such that 30 orphan SE were split into two symmetric subsets of 15 SEs each.
Threading enumeration.
We constructed all possible sequence assignments of a subset of sequences to positions of the SEs by enumerating of the threading degrees of freedom. The subset of sequences included Nic96 (residues 1–205), Nup53 (residues 1–247), Nup59 (residues 1–265), Nup100 (residues 551–815), Nup116 (residues 751–965), and Nup145N (residues 201–458). These segments correspond to regions that have previously been modeled as flexible beads and do not contain FG repeats (Table S2) (Kim et al., 2018). The enumeration begins by assigning a protein identity and copy number to each of the five SE groups. Next, the scoring function is evaluated for all possible assignments of the four SE keys described above. For each SE group, identical keys are applied to all symmetry copies of the group (Figure S4).
Stage 4 - Analyzing and validating the model ensemble
Model validation follows five steps (Sali, 2021; Saltzberg et al., 2021; Viswanath et al., 2017): (i) selection of models for validation; (ii) estimation of sampling precision; (iii) estimation of model precision, (iv) quantification of the match between the model and information used to compute it, and (v) quantification of the match between the model and information not used in the computation. Next, we discuss each one of these validations separately.
Model selection. Models were filtered to find those that sufficiently satisfy the input information; a secondary structure restraint is satisfied if the RaptorXProperties score for the sequence-to-SE mapping is higher than 0.6; a loop end-to-end distance restraint is satisfied if the distance between the termini of two sequential SEs is violated by less than 3 times the standard deviation parameter used to evaluate the restraint; similarly, a crosslink restraint is also satisfied when an evaluated distance is violated by less than 3 standard deviations.
Sampling precision. Because threading solutions were enumerated, no sampling precision needed to be calculated.
Model precision. Model precision (uncertainty) is defined by the variation among the filtered models; the precision of each SE in each cluster was assessed by computing the standard deviation of its starting residue over all models in the cluster.
Information used in modeling. By selection, filtered models satisfy the input information.
Information not used in modeling. Perhaps the most convincing test of a model is by comparing it to information not used in the computation (a generalization of cross-validation). We compared the structure of the Nic96 NTD bound to the Nsp1/Nup49/Nup57 coiled-coil domain to the X-ray structure of the same sub-complex from Chaetomium thermophilum (Stuwe et al., 2015) (Figure S5F). Additionally, the mapping of known Nic96 NTD phosphorylation sites from different species to the integrative threading model rationalized how these site modifications might regulate NPC disassembly and reassembly (Figure S5A), further increasing our confidence in the modeling. Phosphorylation sites were retrieved from the PhosphoSitePlus server (Hornbeck et al., 2015), a manually curated database of experimentally confirmed phosphorylation sites. Finally, the models are consistent with biochemical considerations as described in the initial modeling section (Lin et al., 2016)
Docking the yeast spoke into human and algal NPC 3D density maps
Previous work has shown that the overall architecture of the spoke is conserved based on comparisons of low resolution spoke models that were built with Nup homology models and crystal structures, which were docked into alga/human and yeast/human tomographic 3D density maps (Kim et al., 2018; Mosalaganti et al., 2018). To do a systematic comparison and to validate our new models, we docked yeast spoke models from contracted and expanded inner rings into available 3D maps for the isolated human NPC in nuclear envelopes (EMD-3103; (von Appen et al., 2015)) and the in situ algal NPC, respectively (EMD-4355; (Mosalaganti et al., 2018)). The appropriate yeast spoke model was docked with Chimera (Pettersen et al., 2004) into the 3D density map with a similar conformation for the inner spoke ring. Docked models were then refined within the density maps with MDFF (Trabuco et al., 2008) and a gscale of 0.3 for 150–250 ps with restraints for secondary structure, cispeptides and chirality. The rmsd between the original, manually docked and MDFF refined models was 13.4 Å for the isolated yeast spoke docked into the human NPC 3D density map and ~9.1 Å for the in situ yeast spoke docked into the algal in situ density map (after excluding Nup170 for the latter comparison) (Figure S8). This cross-species docking shows that density maps for human and algal spokes are consistent with many structural features of the yeast spoke.
In situ tomography of S. cerevisiae NPCs- data collection and pre-processing
W303 yeast cells were harvested during log-phase growth and diluted to an 0.8 × 107 cells/mL in YPD media. Five μL of this diluted sample was applied to glow-discharged 200-mesh, Quantifoil R2/1 grids (Electron Microscopy Sciences), excess media was manually blotted from the grid back side (opposite to the carbon substrate where the cells were deposited) and the grid was plunge frozen in a liquid ethane-propane mixture (50/50 volume, Airgas) using a custom-built vitrification device (Max Planck Institute for Biochemistry, Martinsried). Frozen grids were clipped into AutoGrids with a notch to allow milling at shallow grazing angles (Thermo Fisher Scientific). The cryo-FIB milling was performed in an Aquilos DualBeam (Thermo Fisher Scientific). Clusters of a few yeast cells were marked for milling at an angle of ~11–14° (corresponding to stage tilts of 18–21°). Lamellae of ~11–18 μm in width and thicknesses ranging from 120 to 180 nm were prepared using rectangular milling patterns as noted previously (Wagner et al., 2020).
Grids containing lamellae were loaded into a Titan Krios G3 (Thermo Fisher Scientific) operated at 300 kV. First, lamellae were identified in low magnification TEM images and their coordinates were stored. At a nominal magnification of ~2250X or 3600X, the eucentricity at each of the lamellae coordinates was determined and another set of images at a nominal magnification of 8700X was used to generate montages of each lamella in SerialEM (Mastronarde, 2005). The nuclear membrane and NPCs were identified and marked in these montages as targets for data acquisition. Tilt series were acquired on the Krios equipped with a post-column energy filter and a K2 Summit direct detector in counting and dose fractionation modes (Gatan). Typical tilt-series parameters were as follows: tilt range: ~ ± 60°, nominal magnification 42000X, tilt increment 3°(4–5° for some samples), pixel size of 3.43 Å and an effective defocus range of −2 to −11 μm. All image-acquisition was done using SerialEM software (Mastronarde, 2005) (Table S1). Frames of the tilt-series movies were motion-corrected using Motioncorr2 (Zheng et al., 2017). Non-dose weighted and motion-corrected image stacks were imported into EMAN2 for subsequent processing, including CTF estimation, reconstruction, subtomogram, and sub-tilt refinement (Chen et al., 2019a).
Subtomographic analysis of in situ NPCs
Data processing, beginning with collected tilt series, followed the standard subtomogram analysis workflow in EMAN2.9 (Chen et al., 2019a). Briefly, a few tilt series were automatically reconstructed at 4x down-sampling (1k × 1k × 512). Automatic CTF processing was used to check handedness and determine the correct choice of tilt-axis. Once the tilt axis was determined, all tilt series were automatically aligned and reconstructed at 4x down-sampling using e2tomogram.py followed by automatic, tilt-aware CTF determination. NPCs were manually selected from 930 tomograms, avoiding partial NPCs that were cut by the FIB milling, yielding 518 particles. Complete NPCs were re-extracted and reconstructed from the raw tilt series at 2x down-sampling (6.9 Å/pix). The relatively low yield resulted from a compromise between preparing thin lamella to increase data quality and the goal of including complete NPCs, which are comparable in diameter to the optimal lamella thickness.
Numerous alignment runs were completed on whole NPCs and on individual protomers with a variety of different focused masks to identify regions of high variability and to determine the optimal approach for final refinement. In the end, we used a single mask centered on a protomer without additional focused refinements. For the final refinement NPCs were iteratively and globally aligned. The final alignment soft mask covered the spoke ring extending into the membrane and included parts of the cytoplasmic and nucleoplasm rings. Then, 8-fold symmetry was imposed, considering the clear symmetry in the scaffold of the NPC. Based on the alignment, NPCs were mapped back into the tomograms. A few misaligned particles (24) were found to lie outside the plane of the nuclear envelope, and were discarded. During alignment, other particles were placed with the cytoplasm and nucleoplasm inverted; these particles (91, ~18%) were flipped. This was followed by another round of whole-NPC alignment with rotation and translation constrained to 10 pixels and 24 degrees of rotation in all three Euler angles.
Final NPC orientations from this refinement were used to extract 8 protomers from each NPC (C8 symmetry expansion), each in a box large enough to contain all of the density in a 90 degree wedge (one protomer plus two halves), centered on a spoke and extending just past the center of the NPC. These 3656 particles, which were pre-aligned based on the overall NPC alignment, were then iteratively refined using a single alignment soft mask focused on the spoke and outer rings. This protomer alignment was constrained to a maximum of 5 degrees of rotation and 8 pixels of translation per iteration with a classkeep of 0.8 (an EMAN parameter to limit the fraction of particles to include in the average for the next iteration). The final average was performed under a simple spherical mask extending well into the membrane. Per-particle and per-tilt CTF refinement did not yield useful results for the NPC, presumably due to the inherent flexibility and the small number of particles.
Local resolution was estimated using the EMAN2 implementation of mFSC (Penczek, 2020) following the ‘gold standard’ refinements and indicates a range of 30–50 Å in the spoke, outer rings and Pom152 domains. Maps were locally Wiener filtered as part of the mFSC calculation, which computes an FSC curve for every voxel in the map (Figure S7A, S7B). Reliable visual detail (comparison of even/odd maps) matches or exceeds that of EMD-10198 for the yeast NPC, which claims 25 Å resolution. We hypothesize that this difference is due to masking used in the earlier structure, but cannot assess this as even and odd maps were not deposited. Furthermore, we provide a direct comparison of our tomographic 3D map with selected views of EMD-10198 (Figure S7C, S7D). As reported in this work, additional peripheral features are clearly resolved in our 3D map including: connections from Nsp1 complexes in the spoke and from the Nup82 complex, along with orphan densities and basket anchor complexes on the nuclear outer ring, improved Pom152 ring density and spoke contact sites on the pore membrane. These differences may arise from masking steps required to divide the protomer into three overlapping regions for separate alignments and from local filtering and reassembly of the C8 protomer that occurred in previous work (Allegretti et al., 2020), whereas we used a strategy that aligns the entire protomer within a wedge containing two subunits. Alternatively, the amplitudes of Fourier components in this resolution range may be weak, resulting in 3D maps that may not differ significantly in this resolution range. In fact, some combination of these factors may relate to the observed differences in estimated resolution.
Refinement of the Pom152 ring from the in situ NPC
The NPC is an intrinsically flexible assembly, and refinements of one segment may smear out the density of other segments. To refine the Pom152 ring, the 3656 C8 protomers, as described above (pre-aligned based on the overall NPC alignment), were iteratively refined using a soft alignment mask that focused on the double nuclear membrane region. For these iterative refinements, the rotation and translation of the pre-aligned protomer particles were constrained to 8 pixels and 5 degrees of rotation in all three Euler angles. The final average was performed under a simple soft mask that covered the entire asymmetric subunit of the protomer and extended into the membrane regions.
Composite maps and modeling the in situ NPC
The final 3D density map for the in situ NPC was used to create models for spokes in the inner ring and for Nup84 Y-complexes in the outer rings, using methods already described. In this synergistic analysis, the inner and outer rings were modeled as rigid bodies, starting with conformations observed in isolated NPCs, followed by manual rebuilding and a limited MDFF refinement (Trabuco et al., 2008). Additional components were docked manually in Chimera including: the Nup82 complex with 3 pairs of Dyn2 dimers bound to sequential DID repeats of Nup159 (Romes et al., 2012) in the cytoplasmic outer ring, connections from Nsp1 in the Nup82 complex and from Nsp1 complexes in the inner ring that extend into the central channel. The connection models used rods as linear markers and 14 2-helix coiled coil segments were used to model basket anchor densities that emanate from each Y-complex in the nuclear outer ring. Docked models were cloned to form complete rings and used to segment the map by zoning in Chimera with a 20 Å radius to capture additional, nearest neighbor densities and these mini-maps were used for figures. SEGGER (Pintilie et al., 2010) was used to segment the Pom152 ring prior to integrative structure modeling.
To compare maps of the isolated and in situ NPCs segmented inner rings were used for an initial alignment. The inner ring from the isolated NPC was scaled, low pass filtered and boxed in EMAN2.1 to give a comparable resolution with the same pixel and box size as the inner ring in the in situ 3D map. Both inner rings were then aligned for D8 symmetry (which is approximate for the in situ inner ring) with e2symsearch3D.py, and the spoke 2-fold along the x axis. The 8-fold symmetrized, inner ring for the in situ NPC was then aligned to the D8 symmetrized map from this feature to create mutually-centered 3D maps for the two configurations. Outer rings were then placed in the correct orientation relative to their respective, aligned inner ring maps in Chimera.
Integrative modeling of the lumenal Pom152 ring
Integrative modeling of the Pom152 ring in isolated and in situ NPCs proceeded through the standard four stages as described above (Alber et al., 2007a; Kim et al., 2018; Rout and Sali, 2019; Russel et al., 2012). The data used for integrative structure modeling of the Pom152 rings is summarized in Table S2. Each the 10 Pom152 Ig-like domains was represented as a rigid body using a 1-residue-per-bead representation; all other regions were represented as flexible strings of beads (Table S2). We modeled 6 copies of Pom152 spanning 3 spokes. The scoring terms that restrain the spatial degrees of freedom included: first, cross-link restraints based on total of 1425 DSS and EDC. The same set of cross-links was used for modeling the isolated and in situ Pom152 ring because only small conformational changes are expected (Chen and Rappsilber, 2019). Second, the EM density restraint corresponding to a cross-correlation between the Gaussian Mixture Model (GMM) representation of the Pom152 subunits and the GMM representation of the cryo-EM or FIB-milled cryo-ET density maps for the Pom152 ring (i.e., EM maps; Table S2) (Bonomi et al., 2019); third, transmembrane domain restraints that localize the coarse beads in the predicted transmembrane domains (Pom152111–200) within the pore membrane (Kim et al., 2018); fourth, Pom152 peri-nuclear restraints applied to the CTD of Pom152 (residues 201–1337). Finally, excluded volume and sequence connectivity restraints were applied to all components (Kim et al., 2018). Structural models were computed using Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo (MC) algorithm (Shi et al., 2014; Swendsen and Wang, 1986). Each MC step consisted of a series of random transformations (i.e., rotations and/or translations) of the flexible beads and rigid bodies (Table S2). We improved the efficiency of structural sampling by explicitly considering the C8- and C2-symmetries of the inner ring. Analysis and validation of the structural models of the Pom152 rings followed the previously published five steps (Saltzberg et al., 2021) and results from this validation are summarized in (Table S2). The integrative structure modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI) package and our open-source Integrative Modeling Platform (IMP) package (Russel et al., 2012), version 2.8 (https://integrativemodeling.org). Files containing input data, scripts, and output results are available at https://github.com/integrativemodeling/NPC_3.0 and in the worldwide Protein Data Bank (wwPDB) PDB-Dev repository for integrative structures (PDBDEV_00000097).
Figures
All Figure panels of cryo-EM density maps and models were created using Chimera (Pettersen et al., 2004).
Quantitation
Resolution of cryo-EM density maps was estimated with the FSC-0.143 criterion (Rosenthal and Henderson, 2003). Local resolution 3D maps were calculated in RELION 3.0 (Nakane et al., 2018) and with mFSC (Penczek, 2020) in EMAN2 for isolated and in situ NPC volumes, respectively.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
| Rabbit IgG Protein A Purified | Innovative Research | Cat.# IR-RB-GF; RRID:AB_1501660 |
| anti-GFP llama nanobody LAM-16 | (Fridy et al., 2014) | anti-GFP nanobody |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| PreScission protease | GE Healthcare Life Sciences | Cat.# 27–0843–01 |
| Coomassie R250 | MP Biomedicals | Cat.# 190682 |
| GelCode Blue Stain Reagent | Thermo Fisher Scientific | Cat.# 24592 |
| DSS(DiSuccinimidyl Suberate)-H12 | Creative molecules | Cat.# 001S |
| Coomassie Blue R250 | MP Biomedicals | Cat.# 190682 |
| Paraformaldehyde, 16% (Aqueous) | Ted Pella | Cat #18505 |
| Trypsin Sequencing Grade, modified | Roche | Cat.# 11418033001 |
| Iodoacetamide | Sigma | Cat.# I6125–10 g |
| ProLong Glass Antifade Mountant | Invitrogen | P36980 |
| Nupage LDS Sample buffer | Thermo Fisher Scientific | Cat.# NP0007 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Dynabeads M270 Epoxy | Thermo Fisher Scientific | Cat# 143.02D |
|
Deposited data | ||
| Chemical Cross-linking with Mass Spectrometry readout datasets | This paper | Zenodo (https://zenodo.org/) Accession#: 5721514 |
| Original data and custom code used for analysis at Stowers Original Data Repository | This paper | https://www.stowers.org/research/publications/libpb-1583 |
| Low-resolution, multi-part 3D structure of the isolated yeast NPC | This paper | EMD-24225 |
| Double nuclear outer ring of the isolated yeast NPC | This paper | EMD-24231 |
| Fitted model for the double nuclear outer ring of the isolated yeast NPC | This paper | PDB 7N84 |
| Inner ring spoke from the isolated yeast NPC | This paper | EMD-24232 |
| Fitted model for the inner ring spoke from the isolated yeast NPC | This paper | PDB 7N85 |
| Combined 3D structure of the isolated yeast NPC | This paper | EMD-24222 |
| Structure of the in situ yeast NPC | This paper | EMD-24258 |
| Fitted model for the in situ yeast NPC | This paper | PDB 7N9F |
| Integrative models for Pom152 rings | This paper | PDB-Dev PDBDEV_00000097 |
| Files with input data, scripts, and output results for the integrative threading of Nic96 NTDs and modeling | This paper | https://github.com/integrativemodeling/NPC_3.0 |
| Integrative model of yeast NPC | (Kim et al., 2018) | PDB-Dev PDBDEV_00000012 |
| Crystal structure of a Nic96 R1 SLiM bound to the Chaetomium Nsp1 complex | (Stuwe et al., 2015) | PDB 5CWS |
| Crystal structure human Karyopherin/Importin β1 co-crystallized with the α-helical, IBB (Importin beta binding) domain of Karyopherin/Importin α | (Cingolani et al., 1999) | PDB 1QGK, 1QGR |
| Crystal structure human Karyopherin/Importin β1 co-crystallized with the Importin β1-Snurportin 1 IBB | (Bhardwaj and Cingolani, 2010) (Mitrousis et al., 2008) |
PDB 3LWW PDB 2Q5D |
| Tomographic 3D map of human NPC from nuclear envelopes | (von Appen et al., 2015) | EMD-3103 |
| In situ tomographic 3D map of the algal NPC | (Mosalaganti et al., 2018) | EMD-4355 |
| In situ tomographic 3D map of the S. cerevisiae NPC | (Allegretti et al., 2020) | EMD-10198 |
|
| ||
| Experimental models: Saccharomyces cerevisiae strains | ||
|
| ||
| MATα ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100 MLP1 -PPX-ProteinA::HIS5 | (Kim et al., 2018) | Mlp1ppxPrA |
| MATα bar1 ura3–1 his3–11 leu2–3,112 trp1–1 ADE2 LYS2, Nic96-mTurq2-HIS3, Nup84-GFP1–10-URA3MX + pSJ1321 (mCherry-GFP11-PUS1-LEU2) | This study | SLJ13070 |
| MATα bar1 ura3–1 his3–11 leu2–3,112 trp1–1 ADE2 LYS2, Nic96-mTurq2-HIS3, Nup84-GFP1–10-URA3MX+ pSJ1568 (mCherry-GFP11-SCS2-TM-LEU2) | This study | SLJ13072 |
| MATα bar1 ura3–1 his3–11 leu2–3,112 trp1–1 ADE2 LYS2, Nic96-mTurq2-HIS3, Nup84-GFP1–10-URA3MX, Mlp1-mCherry-HYGMX + pSJ1679 (GFP11-Pus1-LEU2) | This study | SLJ13221 |
| MATα bar1 ura3–1 his3–11 leu2–3,112 trp1–1 ADE2 LYS2, Nic96-mTurq2-HIS3, Nup84-GFP1–10-URA3MX, Mlp1-mCherry-HYGMX + pSJ2536 (GFP11-SCS2TM-LEU2) | This study | SLJ13263 |
|
| ||
| Recombinant DNA: List of plasmids used in this study | ||
|
| ||
| pRS315-NOP1pr-GFP11-mCherry-PUS1 | (Smoyer et al., 2016); Addgene #86413 | pSJ1321 |
| pRS315-NOP1pr-GFP11-mCherry-SCS2TM | (Smoyer et al., 2016), Addgene #86416 | pSJ1568 |
| pRS315-NOP1pr-GFP11-PUS1 | (Smoyer et al., 2016), Addgene #86414 | pSJ1679 |
| pRS315-NOP1pr-GFP11-SCS2TM | This study | pSJ2536 |
|
| ||
| Software and algorithms | ||
|
| ||
| pLink | (Yang et al., 2012) | http://pfind.ict.ac.cn/software/pLink/ |
| UCSF Chimera, version 1.13 | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| IMP, version 2.8 | (Russel et al., 2012) | https://integrativemodeling.org/ |
| RELION 3.0 | (Nakane et al., 2018) | https://github.com/3dem/relion |
| MotionCor2 (v1.2.3) | (Zheng et al., 2017)) | https://msg.ucsf.edu/software |
| GCTF | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Gautomatch | ———————— | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Coot (0.8.9.2) | (Casañal et al., 2020) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| NAMD/MDFF Molecular dynamics flexible fitting | (Trabuco et al., 2008) | https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName=NAMDhttps://github.com/ryanmcgreevy/mdff |
| EMAN2 | (Tang et al., 2007) (Chen et al., 2019a) |
https://blake.bcm.edu/emanwiki/EMAN2/Install |
| SerialEM | (Mastronarde, 2005) | https://bio3d.colorado.edu/SerialEM/ |
|
Other | ||
| Mass Spectrometer | Thermo Fisher Scientific | Orbitrap Fusion |
| Mass Spectrometer | Thermo Fisher Scientific | Q Exactive Plus |
| Liquid Chromatograph | Thermo Fisher Scientific | Easy-nLC 1000 |
| Easy-Spray column | Thermo Fisher Scientific | ES800 |
| NuPage 4–12% Bis-Tris Gel 1.0mm x 10 well | Thermo Fisher Scientific | Cat.# NP0321Box |
| Quantifoil holy carbon grids | Quantifoil Micro Tools | R2/2 400 mesh copper |
| Quantifoil holy carbon grids R2/1 200 mesh copper | Electron Microscopy Sciences | Q2100CR1 |
Highlights.
A comprehensive model is presented of the yeast nuclear pore complex (NPC)
Connectors link together different structural and functional layers in the NPC
Multiple structural and functional NPC isoforms co-exist in each cell
Modular construction allows structural plasticity and inner ring dilation of the NPC
ACKNOWLEDGMENTS
We thank members of the Chait, Villa, and Rout laboratories for technical and intellectual support. D.S. is supported by a Damon Runyon Postdoctoral Fellowship (DRG-2364-19). J.M.V. was funded by an Ruth L. Kirschstein NRSA Postdoctoral Fellowship (F32 GM133096). This work was supported by NIH P41 GM109824 (B.T.C., M.P.R., and A.S.), NIH R01 GM112108 and R01 GM117212 (M.P.R.), NIH R01 GM45377 (C.W.A.), NIH P01 GM121203 and NIH R01 GM080139 (S.J.L.), NIH DP2 GM123494 (E.V.), NSF-1818129 (J.F.-M.), the Pew Scholars Program (E.V.), NSF MRI DBI 1920374 (E.V.), and the Stowers Institute for Medical Research (S.L.J.). E.V. is an investigator of the Howard Hughes Medical Institute. We acknowledge the use of the UC San Diego cryo-EM facility, which was built and equipped with funds from the University of California San Diego and an initial gift from Agouron Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.12.015.
REFERENCES
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, and Radermacher M (1993). Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007a). Determining the architectures of macromolecular assemblies. Nature 450, 683–694. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007b). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. [DOI] [PubMed] [Google Scholar]
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turoňová B, Karius K, et al. (2020). In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800. [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, and Hurt E (2011). Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289. [DOI] [PubMed] [Google Scholar]
- Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, and Schwartz TU (2013). Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2, e00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ungricht R, and Kutay U (2011). Traversing the NPC along the pore membrane: targeting of membrane proteins to the INM. Nucleus 2, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Mosalaganti S, and Kosinski J (2018). From the resolution revolution to evolution: structural insights into the evolutionary relationships between vesicle coats and the nuclear pore. Curr. Opin. Struct. Biol. 52, 32–40. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, and Cingolani G (2010). Conformational selection in the recognition of the snurportin importin beta binding domain by importin beta. Biochemistry 49, 5042–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi M, Hanot S, Greenberg CH, Sali A, Nilges M, Vendruscolo M, and Pellarin R (2019). Bayesian Weighing of Electron Cryo-Microscopy Data for Integrative Structural Modeling. Structure 27, 175–188 e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, and Beck M (2013). Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Casañal A, Lohkamp B, and Emsley P (2020). Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 29, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZA, and Rappsilber J (2019). Quantitative cross-linking/mass spectrometry to elucidate structural changes in proteins and their complexes. Nat. Protoc. 14, 171–201. [DOI] [PubMed] [Google Scholar]
- Chen M, Bell JM, Shi X, Sun SY, Wang Z, and Ludtke SJ (2019a). A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Meng JM, Cao Y, Yin JL, Fang RQ, Fan SB, Liu C, Zeng WF, Ding YH, Tan D, et al. (2019b). A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hülsmann BB, Pleiner T, and Görlich D (2015). Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex. Science 350, 106–110. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, and Müller CW (1999). Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA (2018). Nuclear pore complexes as hubs for gene regulation. Nucleus 9, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Hashmi SB, and Osmani SA (2004). Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14, 1973–1984. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, and Gygi SP (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, and Rout MP (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, and Sali A (2006). Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA 103, 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, and Antonny B (2007). A general amphipathic α-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14, 138–146. [DOI] [PubMed] [Google Scholar]
- Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, and Medalia O (2015). Structure and gating of the nuclear pore complex. Nat. Commun. 6, 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, et al. (2017). Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410 e1314. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, and Akin D (1994). Variations in signal-mediated nuclear transport during the cell cycle in BALB/c 3T3 cells. Exp. Cell Res. 215, 206–210. [DOI] [PubMed] [Google Scholar]
- Feldherr C, Akin D, and Moore MS (1998). The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J. Cell Sci. 111, 1889–1896. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, and Rout MP (2021). One Ring to Rule them All? Structural and Functional Diversity in the Nuclear Pore Complex. Trends Biochem. Sci. 46, 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, and Rout MP (2012). Structure-function mapping of a heptameric module in the nuclear pore complex. J. Cell Biol. 196, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, et al. (2016). Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 167, 1215–1228 e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, and Rout MP (2019). Pore timing: the evolutionary origins of the nucleus and nuclear pore complex. F1000Res. 8, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Sali A, and Rout MP (2011). Evolution: On a bender–BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 193, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Teimer R, Amlacher S, Kunze R, and Hurt E (2015). Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol. 22, 774–781. [DOI] [PubMed] [Google Scholar]
- Flemming D, Devos DP, Schwarz J, Amlacher S, Lutzmann M, and Hurt E (2012). Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 177, 99–105. [DOI] [PubMed] [Google Scholar]
- Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, and Rout MP (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaik M, Flemming D, von Appen A, Kastritis P, Mücke N, Fischer J, Stelter P, Ori A, Bui KH, Baßler J, et al. (2015). Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. The Journal of cell biology 208, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B, Andres-Pons A, Kastritis P, and Beck M (2019). Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys. 48, 515–536. [DOI] [PubMed] [Google Scholar]
- Hao Q, Zhang B, Yuan K, Shi H, and Blobel G (2018). Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string. Cell Discov. 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, and Shi Y (2020). Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res. 30, 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacchetti E, Nasehi R, Boeri L, Parodi V, Negro A, Albani D, Osellame R, Cerullo G, Matas JFR, and Raimondi MT (2021). The nuclear import of the transcription factor MyoD is reduced in mesenchymal stem cells grown in a 3D micro-engineered niche. Sci. Rep. 11, 3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühlen R, and Fahrenkrog B (2018). Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease. Histochem. Cell Biol. 150, 593–605. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Kalita J, and Lim RYH (2019). The role of nucleocytoplasmic transport in mechanotransduction. Exp. Cell Res. 377, 86–93. [DOI] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, and Schwartz TU (2015). Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 22, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, et al. (2014). Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol. Cell. Proteomics 13, 2911–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, and Schwartz TU (2016). The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 164, 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. (2016). Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kuhn TM, and Capelson M (2019). Nuclear Pore Proteins in Regulation of Chromatin State. Cells 8, 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Jühlen R, and Antonin W (2021). Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol. 31, 1019–1033. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, and Kutay U (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550. [DOI] [PubMed] [Google Scholar]
- Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. (2016). Architecture of the symmetric core of the nuclear pore. Science 352, aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder MI, Kohler M, Boersema P, Weberruss M, Wandke C, Marino J, Ashiono C, Picotti P, Antonin W, and Kutay U (2017). Mitotic Disassembly of Nuclear Pore Complexes Involves CDK1- and PLK1-Mediated Phosphorylation of Key Interconnecting Nucleoporins. Dev Cell 43, 141–156 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Jih J, Dai X, Bi GQ, and Zhou ZH (2019). Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, and Hurt E (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- Mansfeld J, Güttinger S, Hawryluk-Gara LA, Panté N, Mall M, Galy V, Haselmann U, Mühlhäusser P, Wozniak RW, Mattaj IW, et al. (2006). The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22, 93–103. [DOI] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, and Wozniak RW (2001). A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. Journal of cell biology 153, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mészáros N, Cibulka J, Mendiburo MJ, Romanauska A, Schneider M, and Köhler A (2015). Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev. Cell 33, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrousis G, Olia AS, Walker-Kopp N, and Cingolani GJ (2008). Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol Chem. 283, 7877–7884. [DOI] [PubMed] [Google Scholar]
- Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, and Beck M (2018). In situ architecture of the algal nuclear pore complex. Nat. Commun. 9, 2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kimanius D, Lindahl E, and Scheres SH (2018). Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, and Strambio-De-Castillia C (2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 24, 3920–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. (2013). Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 9, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, and Osmani SA (2006). Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17, 4946–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci G, Caria J, and Lemke EA (2021). Cargo transport through the nuclear pore complex at a glance. J. Cell Sci. 134, jcs247874. [DOI] [PubMed] [Google Scholar]
- Penczek PA (2020). Reliable cryo-EM resolution estimation with modified Fourier shell correlation. IUCrJ 7, 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, and Gossard DC (2010). Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol 170, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak C, Aitchison JD, and Wozniak RW (2014). The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr. Opin. Cell Biol. 28, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulupa J, Prior H, Johnson DS, and Simon SM (2020). Conformation of the nuclear pore in living cells is modulated by transport state. eLife 9, e60654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romes EM, Tripathy A, and Slep KC (2012). Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction. J. Biol. Chem. 287, 15862–15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryo-microscopy. J. Mol. Biol. 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Rout MP, and Blobel G (1993). Isolation of the yeast nuclear pore complex. J. Cell Biol. 123, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, and Sali A (2019). Principles for Integrative Structural Biology Studies. Cell 177, 1384–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velázquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, and Sali A (2012). Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A (2021). From integrative structural biology to cell biology. J. Biol. Chem. 296, 100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzberg DJ, Hepburn M, Pilla KB, Schriemer DC, Lees-Miller SP, Blundell TL, and Sali A (2019). SSEThread: Integrative threading of the DNA-PKcs sequence based on data from chemical cross-linking and hydrogen deuterium exchange. Prog. Biophys. Mol. Biol. 147, 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzberg DJ, Viswanath S, Echeverria I, Chemmama IE, Webb B, and Sali A (2021). Using Integrative Modeling Platform to compute, validate, and archive a model of a protein complex structure. Protein Sci. 30, 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, et al. (2013). Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 21, 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wojtynek M, Mankus D, Tatli M, Kronenberg-Tenga R, Regmi SG, Dip PV, Lytton-Jean AKR, Brignole EJ, Dasso M, et al. (2021). The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman-Duhovny D, Sali A, Rout MP, and Chait BT (2014). Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol. Cell. Proteomics 13, 2927–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, and Rout MP (2014). Cancer and the nuclear pore complex. Adv. Exp. Med. Biol. 773, 285–307. [DOI] [PubMed] [Google Scholar]
- Smoyer CJ, Katta SS, Gardner JM, Stoltz L, McCroskey S, Bradford WD, McClain M, Smith SE, Slaughter BD, Unruh JR, and Jaspersen SL (2016). Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 215, 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, and Rout MP (1999). Proteins connecting the nuclear pore complex with the nuclear interior. Journal of Cell Biology 144, 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Niepel M, and Rout MP (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nature reviews Molecular cell biology 11, 490–501. [DOI] [PubMed] [Google Scholar]
- Stuwe T, Lin DH, Collins LN, Hurt E, and Hoelz A (2014). Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA 111, 2530–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. (2015). Architecture of the fungal nuclear pore inner ring complex. Science 350, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner MC, and Brickner J (2021). The Nuclear Pore Complex as a Transcription Regulator. Cold Spring Harb. Perspect. Biol. 46, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swendsen RH, and Wang JS (1986). Replica Monte Carlo simulation of spin glasses. Phys. Rev. Lett. 57, 2607–2609. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, and Schulten K (2008). Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, et al. (2017). Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure 25, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath S, Chemmama IE, Cimermancic P, and Sali A (2017). Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures. Biophys. J. 113, 2344–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, and Antonin W (2012). Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31, 4072–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. (2015). In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner FR, Watanabe R, Schampers R, Singh D, Persoon H, Schaffer M, Fruhstorfer P, Plitzko J, and Villa E (2020). Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat. Protoc. 15, 2041–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, and Rout MP (1994). POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. Journal of cell biology 125, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, Schneidman-Duhovny D, Zhang C, and Shi Y (2020). Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370, 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Sang Z, Bitton L, Xu J, Liu Y, Schneidman-Duhovny D, and Shi Y (2021). Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst 12, 220–234 e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al. (2010). A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9, 2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, and Akey CW (1998). Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1, 223–234. [DOI] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. (2012). Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906. [DOI] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li S, Zeng C, Huang G, Zhu X, Wang Q, Wang K, Zhou Q, Yan C, Zhang W, et al. (2020). Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res. 30, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, et al. (2021). Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM 3D density maps and associated models have been deposited in the EMDB and RCSB, respectively, with accession codes EMD-24224, EMD-24225, EMD-24232, PDB: 7N85, EMD-24231, PDB: 7N84, EMD-24258 and PDB: 7N9F. Integrative modeling data have been deposited in the PDB-Dev: PDBDEV_00000097. Fluorescence data is available at https://www.stowers.org/research/publications/libpb-1583 and cross-linking data have been deposited at Zenodo (https://zenodo.org/) accession code 5721514.
Software scripts and data for integrative modeling are available at https://github.com/integrativemodeling/NPC_3.0.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.