ABSTRACT
We report the complete genome sequence of Clostridium cadaveris IFB3C5, a strain isolated from the resected tumor of a treatment naive colorectal cancer patient. This genome is comprised of a singular chromosome of approximately 3.63 Mbp in length, contains two plasmids, and has an overall mean GC content of 31.7%.
ANNOUNCEMENT
Clostridium cadaveris, first isolated in 1899 (1), is a rod-shaped, Gram-positive anaerobic bacterium typically present in the human gastrointestinal tract (2, 3). Reported pathogenic associations include equine idiopathic colitis (4), a human abscess (5), bacteremia (6), and chronic osteomyelitis (7). Here, we report the isolation of C. cadaveris IFB3C5, a strain cultivated from the necrotic tissue of a colorectal cancer tumor.
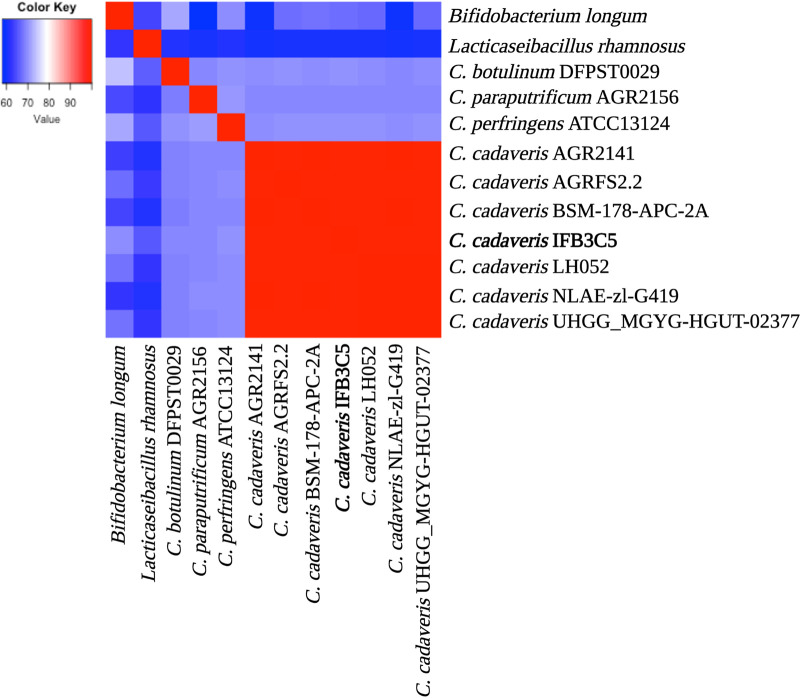
C. cadaveris IFB3C5 was isolated from a cryopreserved colon adenocarcinoma of a 67-year-old treatment-naive female colorectal cancer patient, originally resected in 1989 in Seattle, WA. Classification as C. cadaveris is based on 16S rRNA gene sequencing and average nucleotide identity analysis (Table 1 and Fig. 1). C. cadaveris IFB3C5 was cultured under anaerobic conditions (Oxoid, Thermo Fisher Scientific, USA). High-molecular-weight genomic DNA was extracted using the MasterPure DNA purification kit (Epicentre, Lucigen, USA). Single-molecule real-time sequencing (SMRT-Seq) (8) was carried out on a PacBio Sequel I instrument (Pacific Biosciences, USA). QuBit double-stranded DNA (dsDNA) broad-range (BR) assays (Thermo Fisher Scientific, USA), determined the DNA concentration, and 3 μg of DNA was sheared to an average size of 12 kb using G-tube (Covaris, USA). Libraries were generated using the SMRTbell Express template prep kit 2.0 (Pacific Biosciences), and pooled libraries were size selected via the BluePippin system (Sage Sciences, USA) at a 4-kb minimum threshold. The Pacific Biosciences SMRTAnalysis pipeline version 9.0.0.92188 first processed sequencing reads and then assembled them using Microbial Assembler, which includes an error correction step for chromosomal contiguity and rotation to place the first nucleotide at the chromosomal replication gene, dnaA. Genome assembly showed 21,182 polymerase reads that were further partitioned into 195,640 subreads with an N50 value of 5,553 nucleotides and a total number of subread bases of 787,902,561 with a mean coverage of 212×. Genome assembly resulted in three contigs: a chromosomal sequence of 3,619,347 bp and two putative plasmids of 4,819 bp and 1,618 bp.
TABLE 1.
Publicly available genome assemblies used for ANI analysis
| Species | Strain | Accession no. | Isolation source |
|---|---|---|---|
| C. cadaveris | AGR2141 | GCF_000424205.1 | Rumen microbiome |
| C. cadaveris | BSM-178-APC-2A | GCF_012844035.1 | Pig fecal sample |
| C. cadaveris | AGRFS2.2 | GCF_013390975.1 | Dairy farm |
| C. cadaveris | NLAE-zl-G419 | GCF_900113105.1 | |
| C. cadaveris | LH052 | GCF_900217165.1 | Human preterm infant fecal sample |
| Clostridium paraputrificum | AGR2156 | GCF_000424025.1 | Rumen microbiome |
| Clostridium perfringens | ATCC 13124 | GCF_000013285.1 | |
| Clostridium botulinum | DFPST0029 | GCF_003058345.1 | Contaminated food specimen |
| Bifidobacterium longum | 51A | GCF_004936435.1 | Human fecal sample |
| Lacticaseibacillus rhamnosus | UMB0004 | GCF_002848015.1 | Catheter |
FIG 1.
Heat map of average nucleotide identity (ANI) values. The genome of C. cadaveris IFB3C5 was compared to publicly available genomes of six additional C. cadaveris strains, three different Clostridium species, and two outgroups, i.e., Bifidobacterium longum and Lacticaseibacillus rhamnosus (16) (Table 1), using JSpeciesWS (17). Red indicates a higher ANI value, whereas blue indicates a lower ANI value. C. cadaveris IFB3C5 had an ANI score above 99% against each C. cadaveris strain, scores of 68 to 70% against other species of Clostridium, and scores of 59 to 68% against B. longum and L. rhamnosus outgroups (16). The heat map was generated using the heatmap.2 function from the gplots package on RStudio (version 1.4.1103) (18). The final figure was created on BioRender.
Genome annotation using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (9) identified 3,392 coding sequences, a GC content of 31.7%, and 112 RNAs. Methylome annotation via the Restriction Enzyme Database (REBASE) (10) identified two putative restriction-modification (RM) systems, a type I RM system with the modified bipartite motif ACBN6TCTG and a type II RM system with the modified motif CRAAAAR. For the latter, a similar motif, CAAAAA, influences sporulation in the related organism Clostridioides difficile (11). Detection of RM systems prompted investigation into CRISPR defense systems. CRISPRDetect (12) and CRISPRCasTyper (13) analyses identified a type I-B CRISPR-Cas system with a 58-spacer array.
PlasMapper (14) identified replication-associated genes in both putative plasmids. Putative plasmids showed no significant similarity to each other via BLASTN alignment, supporting the notion that C. cadaveris IFB3C5 carries two distinct plasmids. Antimicrobial resistance gene detection via the Comprehensive Antibiotic Resistance Database (CARD) (15) identified a chromosomal variant in the gyrB gene, which encodes fluoroquinolone resistance.
Currently, seven incomplete C. cadaveris genome assemblies are publicly available. This first complete C. cadaveris genome sequence may therefore advance pangenome analysis of this species, especially in the context of tissue necrosis associated with human disease.
Data availability.
The BioProject accession number for this genome, as well as that for many other human-associated bacterial isolates, is PRJNA549513. The RefSeq assembly accession number is GCF_020911725.1. The genome sequence was deposited in GenBank under the accession number CP076620. The base modification files are available with the GenBank accession and methylome analysis at REBASE under organism 49902 (http://rebase.neb.com/cgi-bin/onumget?49902). and methylome analysis is available at REBASE under organism number 49902.
ACKNOWLEDGMENTS
The research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award number R01 DE027850 (to C.D.J.) and the National Cancer Institute (NCI) under award number R00 CA229984-03 (to S.B.) and by a Washington Research Foundation postdoctoral fellowship (to M.A.Z.-R.).
We thank Rich Roberts and Dana Macelis for REBASE analysis.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Christopher D. Johnston, Email: johnston@fredhutch.org.
Steven R. Gill, University of Rochester School of Medicine and Dentistry
REFERENCES
- 1.Klein E. 1899. Ein Beitrag zur Bakteriologie der Leichenverwesung. Zentralbl Bakteriol Orig 1:278–284. [Google Scholar]
- 2.Willis AT. 2014. Anaerobic bacteriology: clinical and laboratory practice. Butterworth-Heinemann, Oxford, United Kingdom. [Google Scholar]
- 3.Stolk-Engelaar V, Verwiel J, Bongaerts G, Linsen V, Lacquet L, Cox A. 1997. Pleural empyema due to Clostridium difficile and Clostridium cadaveris. Clin Infect Dis 25:160. doi: 10.1086/516893. [DOI] [PubMed] [Google Scholar]
- 4.Staempfli HR, Prescott JF, Brash ML. 1992. Lincomycin-induced severe colitis in ponies: association with Clostridium cadaveris. Can J Vet Res 56:168–169. [PMC free article] [PubMed] [Google Scholar]
- 5.Leung J, Sasson M, Patel SR, Viveiros K. 2009. Clostrium cadaveris intra-peritoneal abscess. Am J Gastroenterol 104:2635–2636. doi: 10.1038/ajg.2009.347. [DOI] [PubMed] [Google Scholar]
- 6.Knight CG, Heitmann PT, McDonald CR. 2021. Clostridium cadaveris bacteraemia with associated superior mesenteric vein thrombus. ANZ J Surg 91:E531–E532. doi: 10.1111/ans.16538. [DOI] [PubMed] [Google Scholar]
- 7.Corrigan RA, Lomas-Cabeza J, Stubbs D, McNally M. 2020. Clostridium cadaveris osteomyelitis: an unusual pathogen which highlights the importance of deep tissue sampling in chronic osteomyelitis. J Bone Jt Infect 5:96–100. doi: 10.7150/jbji.43801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 9.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira PH, Ribis JW, Garrett EM, Trzilova D, Kim A, Sekulovic O, Mead EA, Pak T, Zhu S, Deikus G, Touchon M, Lewis-Sandari M, Beckford C, Zeitouni NE, Altman DR, Webster E, Oussenko I, Bunyavanich S, Aggarwal AK, Bashir A, Patel G, Wallach F, Hamula C, Huprikar S, Schadt EE, Sebra R, van Bakel H, Kasarskis A, Tamayo R, Shen A, Fang G. 2020. Epigenomic characterization of Clostridioides difficile finds a conserved DNA methyltransferase that mediates sporulation and pathogenesis. Nat Microbiol 5:166–180. doi: 10.1038/s41564-019-0613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas A, Staals RH, Morales SE, Fineran PC, Brown CM. 2016. CRISPRDetect: a flexible algorithm to define CRISPR arrays. BMC Genomics 17:356. doi: 10.1186/s12864-016-2627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russel J, Pinilla-Redondo R, Mayo-Muñoz D, Shah SA, Sørensen SJ. 2020. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas loci. CRISPR J 3:462–469. doi: 10.1089/crispr.2020.0059. [DOI] [PubMed] [Google Scholar]
- 14.Dong X, Stothard P, Forsythe IJ, Wishart DS. 2004. PlasMapper: a web server for drawing and auto-annotating plasmid maps. Nucleic Acids Res 32:W660–W664. doi: 10.1093/nar/gkh410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiu R, Caim S, Alcon-Giner C, Belteki G, Clarke P, Pickard D, Dougan G, Hall LJ. 2017. Preterm infant-associated Clostridium tertium, Clostridium cadaveris, and Clostridium paraputrificum strains: genomic and evolutionary insights. Genome Biol Evol 9:2707–2714. doi: 10.1093/gbe/evx210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WH, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2015. gplots: various R programming tools for plotting data. R package version 2.17.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The BioProject accession number for this genome, as well as that for many other human-associated bacterial isolates, is PRJNA549513. The RefSeq assembly accession number is GCF_020911725.1. The genome sequence was deposited in GenBank under the accession number CP076620. The base modification files are available with the GenBank accession and methylome analysis at REBASE under organism 49902 (http://rebase.neb.com/cgi-bin/onumget?49902). and methylome analysis is available at REBASE under organism number 49902.