ABSTRACT
We describe the extended-spectrum β-lactamase blaVEB-3 gene found in an IncA/C plasmid in Aeromonas veronii strain SW3814, which was collected from a freshwater lake in southern California, United States.
ANNOUNCEMENT
Aeromonas bacteria are common in aquatic environments and may serve as reservoirs of antibiotic resistance genes (ARGs) (1–3). These ARGs are often associated with mobile genetic elements (MGEs) (4), which may facilitate their transfer to bacteria of clinical significance (5–7).
Aeromonas veronii strain SW3814 was collected from a freshwater lake in southern California (English Springs Park [33.9951N, 117.756W]). Water was filtered with 0.45-μm filters (GN-6; Pall) and placed on CHROMagar orientation medium (CHROMagar, Paris, France) containing 4 μg/mL cefotaxime (Sigma-Aldrich, St. Louis, MO). A purple isolate, labeled SW3814, was identified as Aeromonas veronii using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker, Billerica, MA). SW3814 was grown (for DNA extraction and tests) overnight in tryptic soy broth (BD Bacto) at 35°C and stored in 25% glycerol at −80°C. Genomic DNA was obtained using a FastPrep homogenizer (MP Biomedicals) with 0.1-mm silica spheres, followed by the DNA extraction method described by Maniatis et al. (8). DNA was quantified with a Qubit fluorimeter (Life Technologies). An Illumina library was prepared with a Nextera DNA Flex library preparation kit, loaded into a 300-cycle high-output flow cell (2 × 150-bp paired-end reads), and run in a MiniSeq instrument with System Suite v2.0.0 (Illumina, San Diego, CA); 2,241,828 Illumina reads were obtained (reads with quality scores of >Q30, 91.1%). Illumina reads were quality filtered using fastp v0.23.1 (9). An Oxford Nanopore Technologies (ONT) (Oxford, UK) library was prepared using SQK-LSK109 and EXP-NBD196 kits, loaded into a FLO-MIN106D flow cell, and run in a MinION ONT device for 75 h. Base calling and quality filtering of ONT reads were conducted using Guppy for GPU v4.5.2; 18,373 ONT reads were obtained (mean size, 12,977 bp; minimum size, 1,000 bp; maximum size, 73,198 bp; reads with quality scores of >Q20, 63.2%). The genome was assembled using Unicycler v0.4.8-beta (10). The genome was annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) with the best-placed reference protein set and GeneMarkS-2+ v5.3 (11–13). The plasmid copy number was obtained with Unicycler depth. Default parameters were used for all software. The Center for Genomic Epidemiology was used to annotate ARGs using ResFinder v2.1 (14). To determine the association of blaVEB-3 with MGEs, a BLASTn search was performed against the NCBI GenBank nucleotide database using the sequences of blaVEB-3 and IS6100. The top six matches with the greatest query coverage were used to create Fig. 1B.
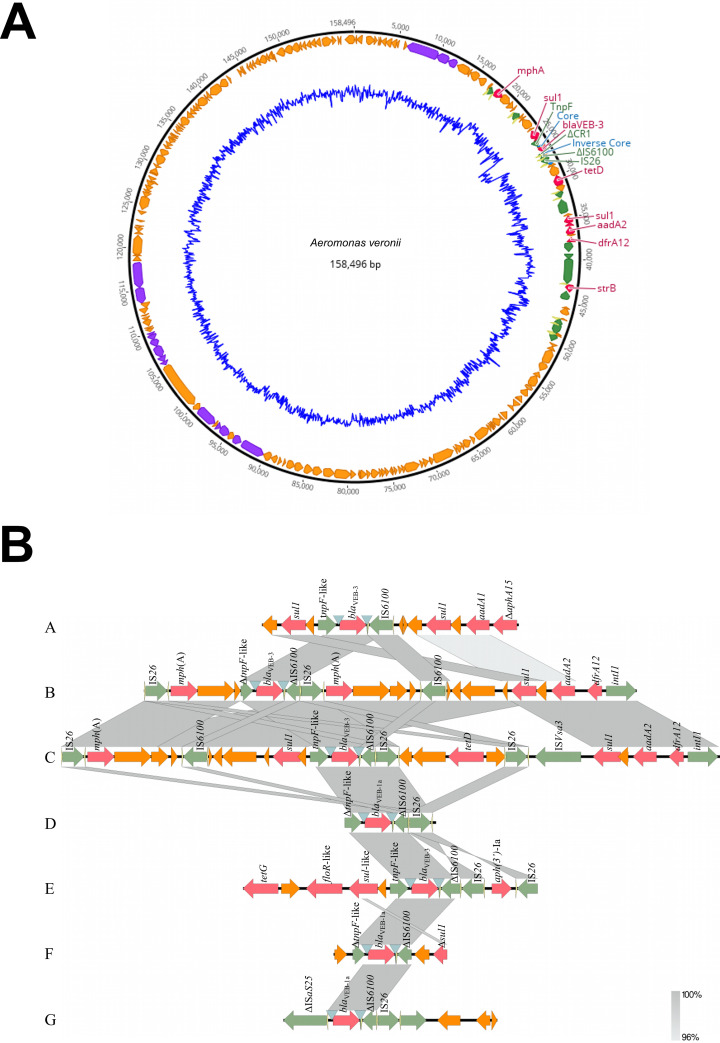
FIG 1.
(A) Genetic map of p158496, displayed in the outermost ring. Open reading frames are represented by arrows in the direction of transcription; they are color coded according to their putative functions, as follows: purple, conjugation machinery; red, ARGs; green, MGEs; yellow, inverted repeats; orange, all other coding sequences. The inner blue ring displays the GC content of the plasmid. Genes of interest are labeled. This figure was created using Geneious v11.1.5. (B) Schematic representation of isolates from the NCBI GenBank database containing the blaVEB gene with regions homologous to those found in p158496. Open reading frames are represented by arrows in the direction of transcription; they are color coded according to their putative functions, as follows: red, ARGs; green, MGEs; yellow, inverted repeats; orange, all other coding sequences. Inverted blue triangles represent the recombination core and inverse core sites. The identity between adjacent sequences is shown as gray shading. Δ indicates the truncation of a gene. Descriptions of isolates A through G are presented in Table 1. This figure was created using EasyFig.
SW3814 has a chromosome of 4,684,058 bp (GC content, 58.4%; Illumina coverage, 53.8×; ONT coverage, 45.8×), an IncA/C plasmid of 158,496 bp with 2.12 plasmid copies per chromosome (GC content, 51.8%), and a 1,739-bp plasmid with 16.92 copies per chromosome (GC content, 56.4%). ARGs found in the IncA/C plasmid were aadA2, blaVEB-3, sul1, tetD, dfrA12, mphA, and strB (Fig. 1A). The plasmid conferred resistance to five classes of antibiotics, classifying the strain as multidrug resistant (15). The blaVEB-3 gene was flanked by MGEs IS6100 and IS26 and a TnpF-like integrase (Fig. 1B). Similar genetic neighborhoods were found in other environmental and clinical bacteria (Fig. 1B and Table 1). The association of the VEB extended-spectrum β-lactamase (ESBL) with MGEs on an IncA/C plasmid highlights the potential for environmental Aeromonas strains to harbor and disseminate ARGs.
TABLE 1.
Descriptions of isolates listed in Fig. 1B
| Isolate | GenBank accession no. | Species | Country | Isolation source | Collection date(s) | Genetic location |
|---|---|---|---|---|---|---|
| A | GQ926879.1 | Acinetobacter pittii | Taiwan | Blood of hospital patient | 1999–2007a | Plasmid |
| B | CP018201.1 | Aeromonas hydrophila | China | Water | 2012 | Chromosome |
| C | CP083462.1 | Aeromonas veronii | USA | Lake in recreational park | 2015 | Plasmid |
| D | HM370390.1 | Aeromonas caviae | France | Seine river | 2009 | Chromosome |
| E | CP006657.1 | Klebsiella pneumonia | China | Blood of hospital patient | 2010 | Plasmid |
| F | HM370392.1 | Aeromonas allosaccharophila | France | Seine river | 2009 | Plasmid |
| G | HM370391.1 | Aeromonas veronii | France | Seine river | 2009 | Plasmid |
Clinical isolates were collected from three hospitals in Taiwan from 1999 to 2007.
Data availability.
The genome sequence data for SW3814 have been deposited in NCBI GenBank under BioProject accession number PRJNA762937, BioSample accession number SAMN21418941, SRA accession number PRJNA762937, and GenBank accession numbers CP083461 (assembled chromosome), CP083462 (plasmid p158496), and CP083463 (plasmid p1739).
ACKNOWLEDGMENTS
This study was supported by NIH grant GM055246, awarded to L.M.-B.
L.M.-B. conceived the main conceptual ideas and methodology, created a platform for data curation, acquired funding, provided resources, helped supervise and provide project administration, validated the results, and wrote the final manuscript. K.G.L. performed antimicrobial susceptibility testing, formal analysis of the sequenced plasmid, interpretation of the results, visualization, and writing of the manuscript.
Contributor Information
Luis Mota-Bravo, Email: lmota@uci.edu.
David Rasko, University of Maryland School of Medicine.
REFERENCES
- 1.Gomes S, Fernandes C, Monteiro S, Cabecinha E, Teixeira A, Varandas S, Saavedra M. 2021. The role of aquatic ecosystems (River Tua, Portugal) as reservoirs of multidrug-resistant Aeromonas spp. Water 13:698. doi: 10.3390/w13050698. [DOI] [Google Scholar]
- 2.Jacobs L, Chenia HY. 2007. Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int J Food Microbiol 114:295–306. doi: 10.1016/j.ijfoodmicro.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Piotrowska M, Popowska M. 2014. The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann Microbiol 64:921–934. doi: 10.1007/s13213-014-0911-2. [DOI] [Google Scholar]
- 4.Piotrowska M, Popowska M. 2015. Insight into the mobilome of Aeromonas strains. Front Microbiol 6:494. doi: 10.3389/fmicb.2015.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Pickup RW. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl Environ Microbiol 66:3883–3890. doi: 10.1128/AEM.66.9.3883-3890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt AS, Bruun MS, Dalsgaard I, Larsen JL. 2001. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl Environ Microbiol 67:5675–5682. doi: 10.1128/AEM.67.12.5675-5682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIntosh D, Cunningham M, Ji B, Fekete FA, Parry EM, Clark SE, Zalinger ZB, Gilg IC, Danner GR, Johnson KA, Beattie M, Ritchie R. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J Antimicrob Chemother 61:1221–1228. doi: 10.1093/jac/dkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maniatis T, Fritsch EF, Sambrook JK. 1982. Molecular cloning: a laboratory manual, p 191–195. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 9.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, O'Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O'Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data for SW3814 have been deposited in NCBI GenBank under BioProject accession number PRJNA762937, BioSample accession number SAMN21418941, SRA accession number PRJNA762937, and GenBank accession numbers CP083461 (assembled chromosome), CP083462 (plasmid p158496), and CP083463 (plasmid p1739).