Abstract
Genome-wide association studies (GWAS) and linkage studies have had limited success in identifying genome-wide significantly linked regions or risk loci for diabetic nephropathy (DN) in individuals with type 1 diabetes (T1D). As GWAS cohorts have grown, they have also included more documented and undocumented familial relationships. Here we computationally inferred and manually curated pedigrees in a study cohort of >6,000 individuals with T1D and their relatives without diabetes. We performed a linkage study for 177 pedigrees consisting of 452 individuals with T1D and their relatives using a genome-wide genotyping array with >300,000 single nucleotide polymorphisms and PSEUDOMARKER software. Analysis resulted in genome-wide significant linkage peaks on eight chromosomal regions from five chromosomes (logarithm of odds score >3.3). The highest peak was localized at the HLA region on chromosome 6p, but whether the peak originated from T1D or DN remained ambiguous. Of other significant peaks, the chromosome 4p22 region was localized on top of ARHGAP24, a gene associated with focal segmental glomerulosclerosis, suggesting this gene may play a role in DN as well. Furthermore, rare variants have been associated with DN and chronic kidney disease near the 4q25 peak, localized on top of CCSER1.
Introduction
Diabetic nephropathy (DN; diabetic kidney disease [DKD]) is a microvascular complication of diabetes that causes progressive decline in kidney function. Approximately one third of individuals with type 1 diabetes (T1D) and half of individuals with type 2 diabetes develop some degree of kidney function impairment (1). For a significant proportion of these individuals, DN eventually leads to severe impairment of kidney function, end-stage renal disease (ESRD), which can only be treated with dialysis or kidney transplantation.
Both genetic and environmental factors play a role in development and progression of the disease. DN segregates in families, with sibling recurrence risk of 2.3. Overall, genetic factors are thought to explain approximately one-third of the DN risk in those with T1D, although after excluding static risk factors (e.g., sex), genetics were shown to explain half of the disease risk (2,3).
Although up to 20 susceptibility genes and loci have been found for DN by recent genome-wide association studies (GWAS) with increasingly large sample sizes, much of the predicted genetic risk of the disease remains unexplained (4,5). GWAS are best powered to detect common variants; however, many common diseases also have rarer variants that affect disease risk at the individual level even more than common variants do (2). Traditional GWAS do not capture rare variants, and whole-exome or -genome sequencing of unrelated individuals would require considerable sample sizes to detect such rare and low-frequency variants. Many of these variants may also be population specific, and using pedigree-based data, linkage methods may serve as a more efficient design for initial variant discovery (6).
Several previous linkage studies have found a genome-wide significant or suggestive linkage peak on chromosome 3q in Finnish and other populations (7). More recently, a significant linkage peak was found on chromosome 22 in Danish, Finnish, and French sibling pairs (8). However, these linkage peaks do not occur in overlapping regions with GWAS findings, and the genetic background of the findings remains largely unclear.
Here we present a genetic linkage study performed in small pedigrees consisting of Finnish individuals with T1D and genotyped with a modern genome-wide genotyping chip that included additional exonic variants on top of common genome-wide variants. Related individuals were extracted from GWAS data, and pedigrees were built computationally based on the identity-by-descent matrix (IBD) of their genetic distances; second- and third-degree relationships were manually assessed and confirmed to construct the pedigrees.
Research Design and Methods
Genotyping and Imputation
In total, 6,152 participants, including individuals with T1D and their relatives, were genotyped on three batches with Illumina HumanCoreExome Bead arrays 12–1.0, 12–1.1, and 24–1.0 (Illumina, San Diego, CA) at the University of Virginia. Genotyping, variant calling, quality control (QC), and imputation have been described earlier (9). Variants were called with the zCall algorithm to optimize calling of rare single nucleotide polymorphisms (SNPs). QC included removal of variants with low genotyping call rate, deviation from Hardy-Weinberg equilibrium, allele frequency difference >20% (minor allele frequency 5%), or allele frequency difference >5% (minor allele frequency >5%) with the 1000 Genomes EUR population. Human reference genome GRCh37 was used for genotype coordinates.
A total of 6,019 individuals passed QC, when samples with genotyping rate <0.95, extreme heterozygosity, sample mix-ups, and genetic outliers were removed. After genotype QC, phased haplotypes (SHAPEIT [version 2r837]) of 316,899 SNPs were used for imputation with 1000 Genomes EUR phase 3 (version 5) via Minimac3/Minimac3-omp (version 1.0.14) (10–12).
Study Cohort
At the Finnish Diabetic Nephropathy (FinnDiane) study visit, each patient underwent a thorough clinical investigation as described earlier (13). Micro- or macroalbuminuria was defined based on urinary albumin excretion rate (AER) in two of three consecutive timed overnight or 24-h urine collections as previously described (13). ESRD was defined as ongoing dialysis or receipt of a kidney transplant. ESRD diagnosis year was verified from hospital records.
Individuals with micro- or macroalbuminuria or ESRD were set as cases, and those with normal AER for at least 15 years were set as controls. We included individuals with T1D with diabetes onset before age 40 years and insulin treatment started within 1 year of diagnosis of diabetes. All participants provided written consent for this study.
Pedigrees
Pedigrees were built based on individuals’ pairwise genetic distances in SNP data. KING software was used to compute pairwise genetic distances (based on proportion of alleles with IBD = 0, IBD = 1, and IBD = 2) between individuals in the genotypic data (14). Pairs of individuals with a third-degree relationship or closer (IBD = 0 < [1 − (1/25/2)]; kinship coefficient > [1/29/2]) were considered for automatic inferring of pedigrees with PRIMUS software (15).
Because IBD = 0 and kinship coefficients may be highly similar between different types of second- and third-degree relative pairs, we used only automatic pedigree construction methods for pedigrees with complete parent-offspring and full-sibling familial relations. We used more distant relation predictions as a basis to search and verify actual relationship types using multiple sources such as population registries, medical records, and questionnaires. Second-degree relationships (representing either grandparent-grandchild pairs, avuncular pairs, or half-siblings) were labeled as avuncular, if the age difference between individuals was >30 years (unlikely to be half-siblings) but <35 years (unlikely to be grandparent-grandchild pairs). Unconfirmed second- and third-degree familial relations were not added to pedigrees, and final pedigree structures were checked using PedCheck software for general structure. PLINK was used to check the Mendelian error rate for SNPs (16,17).
Parametric Two-Point Linkage Analysis
Linkage analysis for autosomal markers was performed with PSEUDOMARKER software for directly genotyped SNPs by using default parameters and a recessive inheritance model, resembling an affected sibling-pairs model for concordant sibling pairs. Frequency of phenocopies was 0, and logarithm of odds (LOD) score of 3.3 was considered the genome-wide significant threshold (18,19). We subsequently ran the linkage analysis on imputed markers for ±1 Mbp from the genome-wide significant linkage peaks, except for the HLA region. Markers were limited to those with imputation quality r2 > 0.75. We also performed separate analysis with pedigrees with only discordant and concordant individuals for DN, along with complex trees that included at least one unaffected individual and two or more affected ones. We conducted an additional linkage analysis using the dominant inheritance model to confirm overlapping peaks in both models.
HLA Allele Prediction and Validation of HLA Allele Imputation
We previously genotyped HLA alleles for DQA1, DQB1, and DRB1 genes for 4,279 FinnDiane study participants (20). Because almost half of the individuals in the pedigrees did not have HLA alleles typed, we imputed HLA alleles using SNP2HLA software based on GWAS data from the HLA region and the Type 1 Diabetes Genome Consortium reference genotype HLA allele panel (21).
In Silico Expression Quantitative Trait Locus and Methylation Quantitative Trait Locus Mapping Using Public Data Sets
We performed an in silico expression quantitative train locus (eQTL) study for markers with genome-wide significant linkage LOD scores for DN by searching two databases with kidney eQTL data: Human Kidney eQTL Atlas and NephQTL (22). We also queried the GTEx database for eQTL changes on various tissues and methylation QTLs (mQTLs) from the mQTLdb database for changes on CpG island methylation in blood (23,24).
In Silico Replication of Linkage Peaks
Of the diabetic nephropathy studies conducted thus far, that by Salem et al. (5) includes the highest number of individuals (n = 19,327). Therefore, to search for further evidence for replication of our linkage peaks, we used the publicly available meta-analysis results of that study (https://t2d.hugeamp.org/datasets.html). On the basis of phenotype similarity with the current study and replication data set, we used the albuminuria-based all DKD phenotype (micro- or macroalbuminuria or ESRD vs. normal AER). Of note, data also include unrelated individuals from the current linkage study.
Data and Resource Availability
The data sets generated and/or analyzed during the current study are not publicly available, because participants’ written consent does not allow data sharing. Data are locally available from the corresponding author on reasonable request.
Results
Individual and Pedigree Characteristics
Of the total 177 pedigrees, more than half (n = 95) consisted of sibling pairs either concordant or discordant for DN. Furthermore, the data set included 40 pedigrees with sibships and more complex relationship structures, resulting in a median pedigree size of three. A total of 263 individuals were affected DN cases (micro- or macroalbuminuria or ESRD), and 120 individuals with T1D had had normal AER for at least 15 years and were assigned as unaffected individuals. Of note, 57 individuals had either unknown DN status or short disease duration, or they were parents without diabetes of included participants; these individuals were included in the families but with an unknown phenotype (Table 1). Nine of the complex pedigrees are described in Supplementary Fig. 1.
Table 1.
Trees and individuals in pedigrees included in linkage analysis
| Pedigree | Trees | Individuals | ||
|---|---|---|---|---|
| Affected | Unaffected | Unknown | ||
| Concordant sibling pair | 40 | 80 | 0 | 0 |
| Discordant sibling pair | 55 | 55 | 55 | 0 |
| Sibship | 9 | 14 | 13 | 5 |
| Parent-offspring pair | 15 | 20 | 9 | 0 |
| Cousin pair | 7 | 10 | 4 | 0 |
| Avuncular pair | 8 | 12 | 3 | 0 |
| Larger tree | 43 | 64 | 44 | 52 |
Data presented as n. Affected indicates n of affected individuals with DN; unaffected, n of individuals with normal AER; unknown, n of individuals with unknown kidney disease status.
There were more men (59.6%) among affected individuals; 52.3% of unaffected individuals were women (Table 2). As expected, affected individuals had higher systolic and diastolic blood pressure and longer diabetes duration. They were also younger and had slightly higher HbA1c.
Table 2.
Clinical characteristics of affected and unaffected individuals in pedigrees
| Affected (n = 255) | Unaffected (n = 128) | P | |
|---|---|---|---|
| Women, % | 40.4 | 52.3 | <0.0001 |
| Age, years | 59.5 ± 11.9 | 57.3 ± 11.0 | NS |
| BMI, kg/m2 | 26.4 ± 4.6 | 25.8 ± 3.8 | NS |
| Age at diabetes onset, years | 12.2 ± 7.5 | 16.0 ± 9.7 | <0.004 |
| Diabetes duration, years | 36.9 ± 10.8 | 30.5 ± 11.4 | <0.0001 |
| HbA1c, % | 8.74 ± 1.67 | 8.23 ± 1.23 | <0.05 |
| Systolic BP, mmHg | 142 ± 21 | 131 ± 17 | <0.0001 |
| Diastolic BP, mmHg | 81 ± 12 | 77 ± 8 | <0.05 |
| Median (IQR) AER, mg | 7.99 (4.56–11.955) | 157.29 (39.2–955) | <0.0001 |
| eGFR, mL/min per 1.73 m2 | 51 ± 19 | 97 ± 20 | <0.0001 |
| Microalbuminuria, n | 78 | — | — |
| Macroalbuminuria, n | 85 | — | — |
| ESRD, n | 92 | — | — |
Data presented as mean ± SD unless otherwise indicated. BP, blood pressure; eGFR, estimated glomerular filtration rate.
Genome-Wide Linkage Analysis
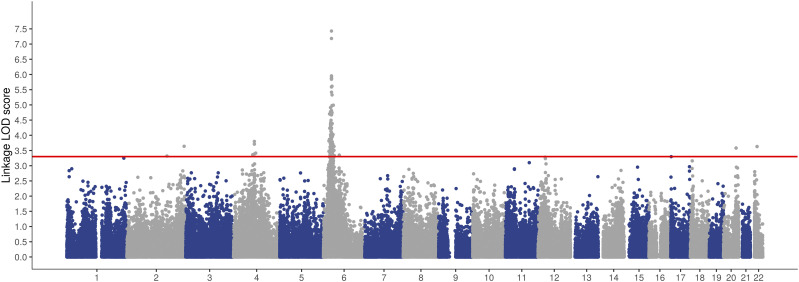
We performed parametric two-point linkage analysis for the 177 pedigrees and exome chip marker data using PSEUDOMARKER software. Analysis resulted in genome-wide significant linkage peaks (LOD score >3.3) on 2q24.3, 2q37.2, 4q21–22, 4q25, 6p21–22, 20q13.2, and 22q12.1 (Fig. 1, Table 3, and Supplementary Fig. 2). The highest linkage peak occurred at chromosome region 6p21–6p22, located at 22.7–45.2 Mb, with LOD scores up to 7.33 and altogether 165 markers on the region exceeding LOD scores ≥3.3 (Supplementary Fig. 3). Multiple linkage peaks on chromosomes 2 and 4 likely originated from separate genetic signals as a result of long distance between regions with significant LOD scores. Linkage analysis was also run for imputed genotypic data for each significant linkage peak (except HLA region), starting 1 Mb before the first genome-wide significant marker and ending 1 Mb after the last significant marker. The imputed marker set resulted in an increased number of markers reaching LOD scores >3.3 but did not yield higher linkage LOD scores for imputed regions (Table 3). Additionally, we identified suggestive linkage peaks with LOD scores ≥3.0 on chromosomes 1q42.3, 11q22.1, 12p11.22, 17p13.3, and 18p11.23 (Table 3).
Figure 1.
Manhattan plot of genome-wide linkage analysis. Plot shows results of genome-wide linkage analysis LOD scores. Analysis showed linkage peaks with genome-wide significant LOD scores >3.3 on chromosomes 2q24.3, 2q37.2, 4q21–22, 4q25, 6p21–22, 20q13.2, and 22q12.1. The highest and widest peak occurs on chromosome 6p HLA region.
Table 3.
Genome-wide significant and suggestive linkage peaks for DN
| Region | Linkage LOD score (imputed)* | n of markers with LOD scores ≥3.3 (imputed)* | Noteworthy nearby genes |
|---|---|---|---|
| Genome-wide significant linkage peaks | |||
| 2q24.3 | 3.31 (3.4) | 1 (5) | GRB14: insulin resistance, fat distribution, BMI (37) |
| 2q37.2 | 3.63 (3.7) | 1 (20) | AGAP1 |
| 4q21.21 | 3.62 | 1 | FRAS1: familial Fraser syndrome, kidney function and development (38,39) |
| 4q22 | 3.80 (3.9) | 2 (93) | ARHGAP24: FSGS (29); PTPN13: DN (3) |
| 4q25 | 3.41 (3.6) | 1 (41) | CCSER1 |
| 6p21–22 | 7.42 | 165 | HLA region |
| 20q13.2 | 3.58 (3.4) | 2 (2) | TSHZ2: renal pelvis development in human and mice (40) |
| 22q12.1 | 3.63 (5.1) | 2 (8) | |
| Suggestive linkage peaks | |||
| 1q42.3 | 3.25 | 1 | NID1: encodes GBM protein nidogen, plausible associations with GBM diseases (41) |
| 11q22.1 | 3.1 | 1 | |
| 12p11.22 | 3.28 | 3 | |
| 17p13.3 | 3.29 | 1 | |
| 18p11.23 | 3.16 | 1 |
FSGS, focal segmental glomerulosclerosis; GBM, glomerular basement membrane.
Linkage LOD scores and n of markers with LOD scores ≥3.3 are given for directly genotyped data and, in parentheses, for imputed data.
Markers with significant linkage LOD scores showed high genotyping quality and heterozygosity (Supplementary Table 1). Additional genome-wide linkage analysis with dominant inheritance showed that peaks on chromosomes 4q25, 6p21–22, and 20q13.2 had genome-wide significant LOD scores in both recessive and dominant models (Supplementary Table 2 and Supplementary Fig. 4). We also performed linkage analysis with individuals with T1D and macroalbuminuria and ESRD as affected and individuals with normal AER as controls. Analysis showed that three of eight significant peaks had LOD scores ≥3.3 as well with macroalbuminuria and ESRD as affected phenotype (Supplementary Table 3).
HLA Haplotype Imputation and HLA Haplotype–Stratified Linkage Analysis
HLA region and in particular MHC class II haplotypes are the most important genetic risk factors for T1D, and a high linkage peak on the HLA region is well documented for T1D. Therefore, we performed a separate linkage analysis for the chromosome 6 linkage region with HLA haplotype stratification, based on imputed HLA alleles and pedigree types. On the basis of comparison within a subset of FinnDiane individuals with directly genotyped HLA alleles, imputation of MHC class II alleles was highly accurate; prediction of HLA-DQA1 matched the genotyped allele with 99.1%, HLA-DQB1 with 98.0%, and HLA-DRB1 with 97.2% accuracy (two or four digits depending on HLA typing accuracy). Allele predictions were combined as haplotypes, and MHC class II haplotype risk scores for T1D were based on a chart by Erlich et al. (25).
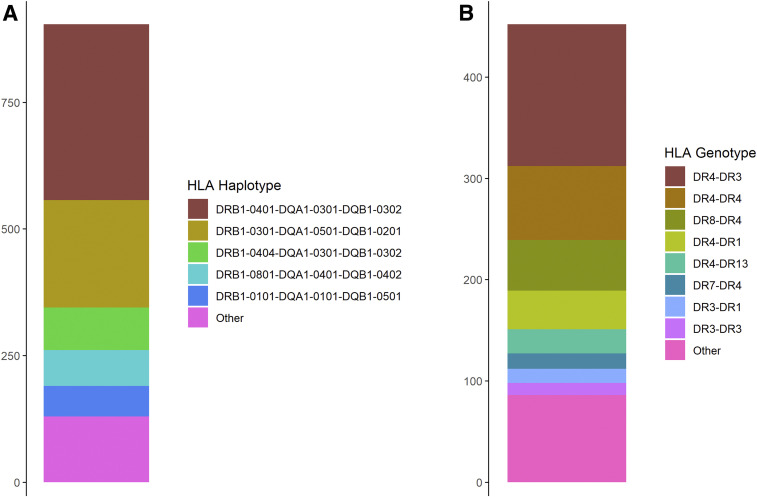
For 89.2% of participants, at least one of two HLA haplotypes was either DRB1*0301-DQA1*0501-DQB1*0201 or DRB1*0401-DQA1*0301-DQB1*0302 (DR4) (Fig. 2). Furthermore, 68.6% of participants had at least one DR4 haplotype, indicating that they had highly similar HLA haplotype–based genetic risk for T1D. In stratified analysis including only individuals with DR4 serotype as cases or controls, linkage remained, with LOD score of 7.43 at 6p22 (33.28 Mb).
Figure 2.
Imputed HLA haplotype and genotype frequencies for patients in pedigrees. HLA alleles were imputed using SNP2HLA software and Type 1 Diabetes Genome Consortium reference panel and then combined into HLA haplotypes and genotypes. A: A majority of haplotypes were high T1D risk DRB1*0301-DQA1*0501-DQB1*0201 (DR3) or DR4 haplotypes. B: Moreover, >80% of individuals had either one or two DR4 haplotypes, which further highlights the homogenic genetic risk for T1D in this cohort.
Subpedigree Type–Based Linkage Analysis
To determine which pedigree type was the leading contributor to linkage signals with LOD scores >3.3, we ran the linkage analysis separately for different pedigree types. Most linkage signals originated from the 57 pedigrees concordant for DN, with fewer signals originating from the 77 discordant pedigrees or 9 larger pedigrees that included at least 2 concordant individuals and 1 discordant member (Supplementary Table 4). Because HLA region is the main T1D genetic risk component, trees concordant for DN could not distinguish between the linkage signals originating from DN and T1D. Outside the HLA region, DN-linked regions did not overlap with markers or regions previously linked or associated with T1D (Supplementary Table 5).
In Silico Replication of Linkage Peaks
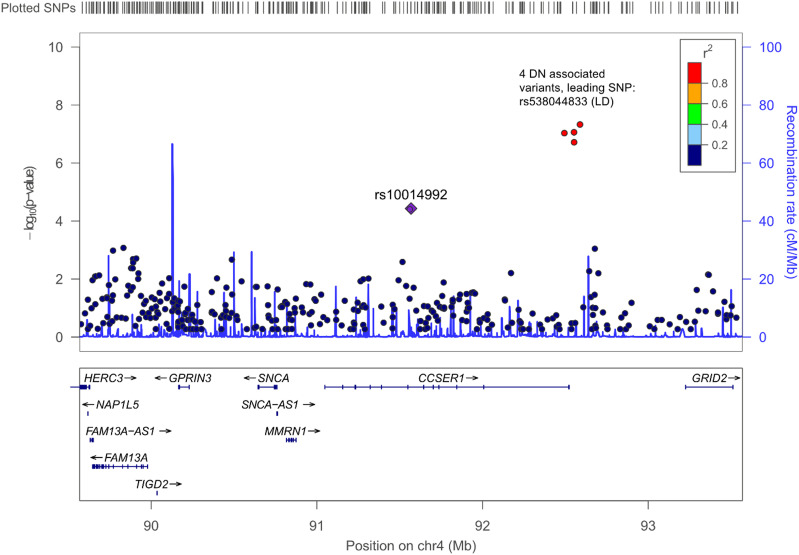
We performed in silico replication for linkage peaks using the newest JDRF Diabetic Nephropathy Collaborative Research Initiative GWAS for DN with 19,327 individuals (5). Of regions with genome-wide significant linkage, we found significant association on the CCSER1 intron on chromosome 4q25 (92.5 Mb away) with lead SNP rs538044833 (P = 2.795 × 10−8). More detailed examination indicated that the association was found in only one cohort (Scottish Diabetes Research Network Type 1 Bioresource; N = 4,689); therefore, this association was not previously reported. The alternative allele of the lead SNP was present in 10 cases and 20 controls, yielding a raw odds ratio of 3.0. Furthermore, suggestive associations were found in the Diabetic Nephropathy Collaborative Research Initiative GWAS at chromosome 2 (163.9 Mb/2q24.3; P = 2.189 × 10−5), chromosome 2 (234.8 Mb/2q37.2; P = 2.619 × 10−5), chromosome 4 (88.1 Mb/4q21.21; P = 6.403 × 10−6), and chromosome 22 (29.1 Mb/22q12.1; P = 5.709 × 10−6) (Fig. 3, Supplementary Fig. 2, and Supplementary Table 6).
Figure 3.
LocusZoom regional association plot of 4q25, showing significant linkage and association signals. The y-axis indicates the −log10 (P values), instead of LOD scores, for both linkage and association results. The purple diamond shows DN linkage P value for the lead SNP in our current linkage study, whereas red circles with rs numbers display association P values from Salem et al. (5). The remaining SNPs are colored according to their r2 correlation with the lead SNP.
In Silico Associations With Gene Expression and DNA Methylation
We studied whether lead variants had eQTL or mQTL associations within the flanking region. Of 11 non–chromosome 6 variants with LOD scores ≥3.3, there were three nominal eQTL associations for either glomerular or tubular allele–specific expression in the NephQTL database: rs10033307-BMP2K (glomerulus; P = 0.022), rs10014992-CCSER1 (tubule; P = 0.011), and rs200655-TSHZ2 (tubule; P = 0.043). Furthermore, five SNPs showed significant eQTL associations in various tissues in the GTEx database (Supplementary Table 7). In the mQTL database, three DN-linked variants at chromosome 4 (85.6–91.5 Mb) showed significant mQTL associations with CpG islands in blood (Supplementary Table 8).
Discussion
Here we performed linkage analysis for DN in 177 computationally inferred and manually curated pedigrees using dense genome-wide genotyping chips combined with analytical methods that allowed analysis across a combination of multiple types of pedigrees. To our knowledge, this is the first genetic linkage study for DN or any diabetic complication performed with a dense marker set. We found evidence of genetic linkage for DN on multiple novel linked regions on 2q24.3, 2q37.2, 4q21–22, 4q25, 6p21–22, 20q13.2, and 22q12.1.
Recent GWAS have yielded only few susceptibility loci for DN, and replication in other populations has had limited success. Of the newly identified linkage peaks, only a few regions overlapped with these GWAS loci. This is not surprising, because pedigree-based linkage studies are expected to find linked regions within families. For example, the best-known susceptibility variants for breast cancer in BRAC1 and BRAC2 genes were originally found with linkage studies, long before GWAS with sample sizes of tens or hundreds of thousands of individuals were possible (26). Regarding diabetic complications, our recent discovery of a CACNB2 association with diabetic retinopathy was initially based on a suggestive linkage signal at chromosome 10p12 (27).
On the basis of gene function, expression in kidneys, and associated kidney diseases, we identified plausible DN genes located under or close to these linkage peaks (Table 3). Aside from the linkage peak at the HLA region on chromosome 6, the chromosome 4q peak was the widest, with markers with significant LOD scores between 79 and 91 Mb. It remains uncertain whether the peak occurs as a result of one or multiple signals, but long distances between markers with significant LOD scores (i.e., rs10033307 at chromosome 4 [79.3 Mb]; rs4129430, rs11097033, and rs1482085 at chromosome 4 [85.9–86.6 Mb]; and rs10014992 chromosome 4 [91.6 Mb]) point toward multiple separate signals (Supplementary Fig. 2C–E). The peak at 91.0–92.5 Mb had a genome-wide significant LOD score with a dominant inheritance model (Supplementary Table 2). These markers with significant linkage LOD scores occurred on top of FRAS1 (79.0–79.5 Mb), ARHGAP24 (86.4–86.9 Mb), and CCSER1 genes (91.0–92.5 Mb), respectively. All these genes have been associated with kidney function or diseases in previous studies (Supplementary Table 9).
We searched for replication of linkage loci in a recent GWAS meta-analysis on DKD and found a genome-wide significant association with CCSER1 intronic variant rs538044833 and DKD close to the 4q25 (91.5 Mb) peak (P = 2.795 × 10−8) (5) (Fig. 3, Supplementary Fig. 2F, and Supplementary Tables 5 and 9). Another rare CCSER1 intronic variant (rs553908921) was also associated with chronic kidney disease (CKD) at genome-wide significant level (P = 2.19 × 10−8) in the recent HUNT study (28). Both associated variants were rare, and in the case of rs538044833, the association was observed only in the Scottish cohort from the meta-analysis. Furthermore, eQTL data suggested that linkage analysis lead SNP rs10014992 was associated with tubular CCSER1 expression. Although additional studies are needed to establish CCSER1 as a DN- and CKD-associated gene, these studies strongly support rare variation in CCSER1 as a risk factor for DN.
The ARHGAP24 gene directly under the linkage peak at 4q22.1 (85.9–86.6 Mb) has been associated with familial focal segmental glomerulosclerosis, a rare familial kidney disease, but not with DN (29) (Supplementary Fig. 1D and Supplementary Table 9). Furthermore, the orthologous Q156R mutation reduces the capability of mouse Arhgap24 to deactivate Rac1 (29). The Salem et al. (5) meta-analysis also showed a suggestive association 1.6 Mb from the peak (Supplementary Fig. 1D and Supplementary Table 6).
Of the SNPs with significant linkage, chromosome 4q variants accounted for the majority of the identified eQTL and mQTL activity (Supplementary Tables 7 and 8). For example, the blood-based mQTL database showed significant mQTL effect with ARHGAP24 intronic variant rs1482085 (LOD score 3.39) and CpG island cg20784207 at birth (P = 3.30 × 10−11) and childhood (P = 3.87 × 10−10) (Supplementary Table 8). Although the associations were not found in kidney tissue, possibly because of a lack of large enough eQTL and mQTL databases for the kidney, blood-based associations could also be considered interesting because of the microvascular nature of DN.
Other plausible DN genes under the linkage peaks included AGAP1 on chromosome 2q37.2, GRB14 at 2q24.3, and TSHZ2 close to 20q13.2, which are associated with insulin resistance, Rac1 activation pathway, and renal development (Supplementary Table 9). For these linkage peaks outside chromosome 4, the Salem et al. (5) meta-analysis showed suggestive associations with DKD for variants on top of or close to regions on 2q24.3, 2q37.2, and 22q12.1 (5) (Supplementary Table 6).
The HLA region has been identified as a leading genetic factor in some kidney diseases such as idiopathic membranous nephropathy (30). For DN, previous studies have yielded contradictory findings on whether genes localized on the HLA region, such as AGER and tumor necrosis factor-α, play a significant role in development of DN. Of note, no significant associations with DN have been found on the HLA region in GWAS.
In this study, the widest chromosome 6 linkage peak with 165 genome-wide significant markers ranged from 22.8 to 45.1 Mb. The region with 13 markers with LOD scores >5 occurred between 33 and 35 Mb (highest with LOD 7.3 at 33.5 Mb). Tumor necrosis factor-α, AGER, and MHC class II genes are located at 31.5–33 Mb. There were also several signals throughout the region, including a peak at 25 Mb occurring on top of the LRRC16A gene, which has been associated with gout (31). Several studies, including our previous linkage analysis, have shown a linkage peak at chromosome 6p21 (8). Because this linkage peak occurred on the HLA region and concordant and discordant sibling pairs showed similar haplotype inheritance patterns, it was presumed that the peak originated from the genetic risk of T1D and not from DN. We HLA stratified the pedigrees by assigning the affected and unaffected phenotypes for individuals with high T1D risk HLA DRB1-0401-DQA1-0301-DQB1-0302 haplotype. This analysis showed increased LOD scores in the HLA region, when compared with the main linkage analysis. We further studied the origin of the signal by running linkage analyses separately for different pedigree types (concordant individuals only, discordant individuals, and complex trees). These analyses showed that pedigrees including only concordant affected individuals were responsible for a majority of the LOD scores (Supplementary Table 4). Because of the superior power of the analysis including affected concordant individuals only compared with the analysis including discordant individuals, along with the relatively small number of complex pedigrees, this may not be surprising. However, because the analysis including concordant individuals only could not distinguish between DN and T1D, the origin of the peak remained ambiguous.
In general, a majority of LOD scores for other DN-linked markers originated from pedigrees including only concordant individuals. However, outside the HLA region, most of these peaks did not co-occur with previously found T1D-associated variants or linked regions (Supplementary Table 5). Because genetic studies of T1D have been more powerful compared with DN studies as a result of larger sample sizes, we did not expect the nonoverlapping linked regions to be novel T1D regions.
Of note, HLA allele imputations performed for the haplotype-stratified linkage analysis were significantly more accurate than previous HLA allele predictions using Illumina 610 K genotyping chip data for 161 Finnish individuals (32). With SNP2HLA and HLA*IMP software and the HapMap2 reference panel, they achieved <20% imputation accuracy for the HLA-DRB1 gene, compared with 99.1% for HLA-DQA1, 98.0% for HLA-DQB1, and 97.2% for HLA-DRB1 in our current study (32). We speculate that the main cause for the drastic difference in imputation accuracy between the studies could be due to the different reference panels and different genotyping platform used (i.e., Illumina CoreExome chip includes more exomic variants compared with most other genotyping chips, which may also improve imputation quality for MHC class II alleles).
Altogether, five suggestive linkage peaks had linkage LOD scores between 3 and 3.3 (Table 3). The chromosome 1q42.3 peak with LOD score of 3.2 is especially notable because it was located between NID1 and GPR137B. Previous studies have suggestively associated NID1 with Goodpasture’s syndrome, also called glomerular basement membrane disease. Nidogen-1, encoded by NID1, is able to bind to other basement membrane proteins such as type IV collagen (33). Rare variants in the COL4A3 gene encoding the α3 subunit of type IV collagen have been associated with Alport syndrome, and recently, a common COL4A3 variant was also associated with DN in a large GWAS involving 19,406 individuals with T1D (5,34). This is one example in which a serious, usually monogenic disease is caused by a deleterious rare variant, whereas the risk of a complex disease affecting the same tissue type or organ is affected by common variants located in the same gene.
Previous studies have found linked regions on multiple chromosomes, including the most recent findings on chromosomes 3q21–25 and 22q11 (7,8). Our linkage analysis did not result in significant linkage peaks with these previously reported ones (Supplementary Table 10 and Supplementary Fig. 2C and H). Nevertheless, the former 3q21–25 region contained three markers with linkage LOD scores >2.5. Likewise, a suggestive linkage signal with LOD score of 2.80 was found at 22q11, which could indicate a lack of power to detect a genome-wide signal in this region rather than lack of replication. Of the 440 individuals in the pedigrees in this study, 175 were also included in our previous study, but most previous linkage studies were conducted using sparse microsatellite platforms, and lower linkage LOD scores could have resulted from platform differences between the SNP microarray and microsatellites (35).
The study included a limited number of pedigrees and a limited number of individuals within the pedigrees. Although this is a common limitation of pedigree-based studies, this study has benefited from the development of modern analysis methods that enable the study of a larger variety of pedigree types together. Supporting association evidence for a chromosome 4q peak is also based on rare variants, and these could be invalidated as more data become available. However, finding regions with low frequency and rare variants with large effect sizes is a main advantage of linkage studies compared with GWAS. We also note that the validity of eQTL associations has met increasing criticism (36). As genotyping and sequencing become more affordable, and with the rise of large nationwide genotyping and sequencing initiatives, studies with larger families, including multiple siblings, cousins, and so on, will become possible.
Because the Finns have well-documented differences in low-frequency and rare variant patterns compared with other European populations, linkage peaks in these Finnish pedigrees do not necessarily exist in pedigrees in other countries. Therefore, replication in other Finnish families would be ideal. However, the FinnDiane study already covers a substantial proportion of Finnish individuals with T1D, and there is no other T1D study cohort with pedigrees and GWAS data available in Finland without significant overlap with the FinnDiane study cohort. Because only a few individuals included in this study have participated in our whole-exome and -genome studies, it is currently not statistically feasible to study variants under the linkage peaks using our sequencing data. However, because more individuals will be sequenced, we expect to be able to pinpoint the variants behind these signals in our future studies.
Our study aimed to identify genetic loci linked to DN in Finnish families including individuals with T1D. Results include eight regions with genome-wide significant linkage signals. Analysis also resulted in peaks on top of or close to genes, such as ARHGAP24, that have previously been associated with kidney disease or kidney function but not yet with DN and a peak on top of CCSER1, where rare variants in two studies have been associated with DN and CKD. Although the most conspicuous peak occurred at the HLA region of chromosome 6, it is likely to have originated at least in part from T1D risk. To pinpoint and confirm linked genes, more studies are needed.
Article Information
Acknowledgments. The authors thank M. Parkkonen, M. Korolainen, A.-R. Salonen, A. Sandelin, and J. Tuomikangas (Folkhälsan Research Center, Helsinki, Finland) for technical assistance. The authors also acknowledge the physicians and nurses at each center taking part in the enrollment and clinical characterization of participants (Supplementary Table 11 provides a list of study centers and investigators involved in the FinnDiane study).
Funding. This study was supported by funding from JDRF (1SRA-2016-333-M-R), Folkhälsan Research Foundation, Wilhelm and Else Stockmann Foundation, Liv och Hälsa Society, Helsinki University Hospital research funds (EVO TYH2018207), Academy of Finland (275614, 299200, and 316664), Novo Nordisk Foundation (OC0013659), and European Foundation for the Study of Diabetes Young Investigator Research Award funds.
Duality of Interest. P.-H.G. has served on advisory boards for AbbVie, Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, Cebix, Eli Lilly, Janssen, Medscape, MSD, Mundipharma, Novartis, Novo Nordisk, and Sanofi; has received lecture honoraria from Astellas, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Elo Water, Genzyme, Medscape, MSD, Mundipharma, Novartis, Novo Nordisk, and Sanofi; and has received investigator-initiated grants from Eli Lilly and Roche. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.H. performed analyses and wrote the manuscript, N.S. supervised analyses and edited the manuscript, E.V. performed quality controls for the genotyping data, C.F. and V.H. helped in constructing pedigrees with distant relatives, J.B.C. helped with meta-analysis replication data, S.J.M. and H.M.C. were responsible for the Scottish cohort of the meta-analysis, and P.-H.G. guided analyses and edited the manuscript. J.H. and P.-H.G. are the guarantors of this work, and as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.13507995.
References
- 1.Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev 2013;93:137–188 [DOI] [PubMed] [Google Scholar]
- 2.Freund MK, Burch KS, Shi H, et al. Phenotype-specific enrichment of Mendelian disorder genes near GWAS regions across 62 complex traits. Am J Hum Genet 2018:103:535–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandholm N, Van Zuydam N, Ahlqvist E, et al.; FinnDiane Study Group; DCCT/EDIC Study Group; GENIE Consortium; SUMMIT Consortium . The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol 2017;28:557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahlström E, Sandholm N. Progress in defining the genetic basis of diabetic complications. Curr Diab Rep 2017;17:80. [DOI] [PubMed] [Google Scholar]
- 5.Salem RM, Todd JN, Sandholm N, et al.; SUMMIT Consortium, DCCT/EDIC Research Group, GENIE Consortium . Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J Am Soc Nephrol 2019;30:2000–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim ET, Würtz P, Havulinna AS, et al.; Sequencing Initiative Suomi (SISu) Project . Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014;10:e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogus JJ, Poznik GD, Pezzolesi MG, et al. High-density single nucleotide polymorphism genome-wide linkage scan for susceptibility genes for diabetic nephropathy in type 1 diabetes: discordant sibpair approach. Diabetes 2008;57:2519–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wessman M, Forsblom C, Kaunisto MA, et al.; FinnDiane Study Group . Novel susceptibility locus at 22q11 for diabetic nephropathy in type 1 diabetes. PLoS One 2011;6:e24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Syreeni A, Sandholm N, Cao J, et al.; DCCT/EDIC Research Group; FinnDiane Study Group . Genetic determinants of glycated hemoglobin in type 1 diabetes. Diabetes 2019;68:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton A, Brooks LD, Durbin RM, et al.; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature 2015;526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods 2011;9:179–181 [DOI] [PubMed] [Google Scholar]
- 13.Syreeni A, El-Osta A, Forsblom C, et al.; FinnDiane Study Group . Genetic examination of SETD7 and SUV39H1/H2 methyltransferases and the risk of diabetes complications in patients with type 1 diabetes. Diabetes 2011;60:3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010;26:2867–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staples J, Qiao D, Cho MH, Silverman EK, Nickerson DA, Below JE; University of Washington Center for Mendelian Genomics . PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am J Hum Genet 2014;95:553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gertz EM, Hiekkalinna T, Le Digabel S, Audet C, Terwilliger JD, Schäffer AA. PSEUDOMARKER 2.0: efficient computation of likelihoods using NOMAD. BMC Bioinformatics 2014;15:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiekkalinna T, Schäffer AA, Lambert B, Norrgrann P, Göring HHH, Terwilliger JD. PSEUDOMARKER: a powerful program for joint linkage and/or linkage disequilibrium analysis on mixtures of singletons and related individuals. Hum Hered 2011;71:256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Söderlund J, Forsblom C, Ilonen J, et al.; FinnDiane Study Group . HLA class II is a factor in cardiovascular morbidity and mortality rates in patients with type 1 diabetes. Diabetologia 2012;55:2963–2969 [DOI] [PubMed] [Google Scholar]
- 21.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillies CE, Putler R, Menon R, et al.; Nephrotic Syndrome Study Network (NEPTUNE) . An eQTL landscape of kidney tissue in human nephrotic syndrome. Am J Hum Genet 2018;103:232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaunt TR, Shihab HA, Hemani G, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol 2016;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonsdale J, Thomas J, Salvatore M, et al.; GTEx Consortium . The genotype-tissue expression (GTEx) project. Nat Genet 2013;45:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erlich H, Valdes AM, Noble J, et al.; Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford D, Easton DF, Stratton M, et al.; The Breast Cancer Linkage Consortium . Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 1998;62:676–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vuori N, Sandholm N, Kumar A, et al.; FinnDiane Study . CACNB2 is a novel susceptibility gene for diabetic retinopathy in type 1 diabetes. Diabetes 2019;68:2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham SE, Nielsen JB, Zawistowski M, et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat Commun 2019;10:1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akilesh S, Suleiman H, Yu H, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest 2011;121:4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanescu HC, Arcos-Burgos M, Medlar A, et al. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 2011;364:616–626 [DOI] [PubMed] [Google Scholar]
- 31.Sakiyama M, Matsuo H, Shimizu S, et al. Common variant of leucine-rich repeat-containing 16A (LRRC16A) gene is associated with gout susceptibility. Hum Cell 2014;27:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlachopoulou E, Lahtela E, Wennerström A, et al. Evaluation of HLA-DRB1 imputation using a Finnish dataset. Tissue Antigens 2014;83:350–355 [DOI] [PubMed] [Google Scholar]
- 33.Chew C, Lennon R. Basement membrane defects in genetic kidney diseases. Front Pediatr 2018;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kashtan CE, Ding J, Garosi G, et al. Alport syndrome: a unified classification of genetic disorders of collagen IV α345: a position paper of the Alport Syndrome Classification Working Group. Kidney Int 2018;93:1045–1051 [DOI] [PubMed] [Google Scholar]
- 35.Osterholm A-M, He B, Pitkaniemi J, et al. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int 2007;71:140–145 [DOI] [PubMed] [Google Scholar]
- 36.Huang QQ, Ritchie SC, Brozynska M, Inouye M. Power, false discovery rate and Winner’s Curse in eQTL studies. Nucleic Acids Res 2018;46:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gondoin A, Hampe C, Eudes R, et al. Identification of insulin-sensitizing molecules acting by disrupting the interaction between the insulin receptor and Grb14. Sci Rep 2017;7:16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGregor L, Makela V, Darling SM, et al. Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat Genet 2003;34:203–208 [DOI] [PubMed] [Google Scholar]
- 39.Pitera JE, Scambler PJ, Woolf AS. Fras1, a basement membrane-associated protein mutated in Fraser syndrome, mediates both the initiation of the mammalian kidney and the integrity of renal glomeruli. Hum Mol Genet 2008;17:3953–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins D, Caubit X, Dimovski A, et al. Analysis of TSHZ2 and TSHZ3 genes in congenital pelvi-ureteric junction obstruction. Nephrol Dial Transplant 2010;25:54–60 [DOI] [PubMed] [Google Scholar]
- 41.Funk SD, Lin M-H, Miner JH. Alport syndrome and Pierson syndrome: diseases of the glomerular basement membrane. Matrix Biol 2018;71–72:250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]