Abstract
Mastitis, inflammation of the mammary gland, is the most prevalent disease in dairy cattle that has a potential impact on profitability and animal welfare. Specifically designed multi-omics studies can be used to prioritize candidate genes and identify biomarkers and the molecular mechanisms underlying mastitis in dairy cattle. Hence, the present study aimed to explore the genetic basis of bovine mastitis by integrating microarray and RNA-Seq data containing healthy and mastitic samples in comparative transcriptome analysis with the results of published genome-wide association studies (GWAS) using a literature mining approach. The integration of different information sources resulted in the identification of 33 common and relevant genes associated with bovine mastitis. Among these, seven genes—CXCR1, HCK, IL1RN, MMP9, S100A9, GRO1, and SOCS3—were identified as the hub genes (highly connected genes) for mastitis susceptibility and resistance, and were subjected to protein-protein interaction (PPI) network and gene regulatory network construction. Gene ontology annotation and enrichment analysis revealed 23, 7, and 4 GO terms related to mastitis in the biological process, molecular function, and cellular component categories, respectively. Moreover, the main metabolic-signalling pathways responsible for the regulation of immune or inflammatory responses were significantly enriched in cytokine–cytokine-receptor interaction, the IL-17 signaling pathway, viral protein interaction with cytokines and cytokine receptors, and the chemokine signaling pathway. Consequently, the identification of these genes, pathways, and their respective functions could contribute to a better understanding of the genetics and mechanisms regulating mastitis and can be considered a starting point for future studies on bovine mastitis.
Keywords: mastitis, transcriptome sequencing, hub genes, multi-omics data, regulatory networks, bovine
1. Introduction
Over the last decade, advances in high-throughput genotyping and sequencing technologies [1], along with progress in developing computational methods [2], have led to a revolution towards a better understanding of the genetic architecture underlying complex traits and diseases, with exceptional depth. To date, several studies have focused on integrating different information sources (“omics” datasets) to create robust insights into complex molecular functional mechanisms by reinforcing complementary evidence from multiple levels [3,4,5]. In this regard, the results of different types of multi-layer studies have been reported, ranging from simple combinations (two different kinds of -omics data) to more comprehensive and computationally demanding ones (multiple kinds of -omics data). Incorporating two layers under a systems biology framework can involve approaches that integrate genomics and transcriptomics [6,7,8], metabolomics and transcriptomics [9,10,11], proteomics and transcriptomics [12,13], and proteomics and metabolomics analyses [14,15] to functionally characterize the interactions at the molecular level for traits of interest in humans and in livestock species.
Bovine mastitis is a common and costly disease, which has a considerable effect on the profitability of the production system, owing to its negative impacts on milk yield, quality, and reproductive performance; early culling; animal welfare issues; and the cost of treatment [16,17,18,19]. The inflammation of the mammary gland occurs in response to infection with pathogenic microorganisms or physiological and metabolic changes [20,21]. Although the heritability of mastitis is low [22], and genetic correlations between mastitis and production traits are unfavorable [23,24], genetic improvement in terms of mastitis resistance is a major breeding goal. It is also known that mastitis is highly genetically correlated with somatic cell count (SCC), which consists of macrophages, lymphocytes, and epithelial cells, and consequently, this can be used as an important indicator of udder health [25]. Furthermore, selection for correlated traits, such as reduced SCC (indicating increased mastitis resistance) could be an interesting alternative, allowing scientists to infer and comprehend the genetic and molecular mechanisms underlying these traits. In other words, the discovery of genomic regions, disease-causing genes, and biomarkers associated with mastitis is of essential importance in improving the diagnosis and treatment of the disease. In the literature, numerous studies have been carried out to identify functional candidate genes associated with mastitis based on genome-wide association studies (GWAS) [26,27,28,29,30] and transcriptome studies [6,16,31,32]. On the other hand, concordance among these studies is low, indicating difficulties in identifying reliable candidate genes for mastitis. New approaches integrating GWAS results with additional sources of information can overcome this challenge. Hence, it is worth investigating the molecular regulatory mechanisms through which mastitis can be developed. Therefore, the objective of this study was to use the integration of previously published RNA-Seq and microarray data with GWAS results to identify and prioritize potential hub genes and create reconstructions of the protein–protein interaction (PPI) and gene regulatory networks, as well as modeling of the three-dimensional hub protein structure involved in pathological processes related to mastitis in dairy cattle.
2. Materials and Methods
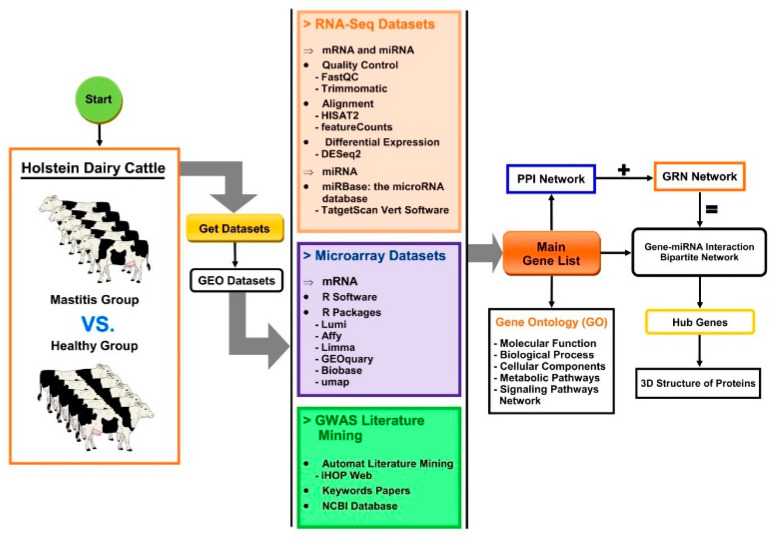
The overall workflow for the data collection and the analysis of relevant genes related to mastitis in dairy cattle is presented in Figure 1.
Figure 1.
Schematic of the workflow used to reconstruct the metabolic pathways of mastitis in dairy cattle. The main gene list was prepared from RNA-Seq and microarray datasets, and literature mining. The protein–protein interaction network (PPI), gene regulatory network (GRN), and interactive bipartite network of gene–miRNA interactions were reconstructed using Cytoscape.
2.1. Data Collection
Collection and evaluation of the available data is the first step in better understanding the reconstruction of molecular networks and the biological basis in terms of the identification of candidate genes, gene regulation, interactions, protein–protein interaction (PPI), and metabolic signaling networks. In this study, the microarray and RNA-sequencing (RNA-Seq) datasets, available in the public repository of the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), were retrieved for samples of mastitis and healthy Bos taurus species. The accession numbers for the RNA-Seq and microarray datasets are shown in Table 1. Six Holstein cows from first to third lactations and days in milk (DIM) ranging from 7 to 236 were included in the GSE131607 dataset. All cows were kept in freestall housing at the University of California–Davis, fed a total mixed ration (TMR), and given ad libitum access to water. Two different samples were taken from each cow after diagnosis using the California Mastitis Test, one sample from the mastitic quarter (n = 6), and the other sample taken diagonally across from the mastitic quarter, which was confirmed as the healthy quarter (n = 6), based on having a somatic cell count (SCC) less than 100,000 cells/mL milk [16]. The GSE15020 and GSE15022 datasets were related to microarray analysis from a study by Mitterhuemer et al. [31]. Fifteen healthy German Holstein Frisian cows in mid-lactation (3 to 6 months postpartum) were included in the study. Quarter milk samples were collected and tested weekly before the trial to ensure that they contained <50,000 somatic cells/mL and were free of mastitis pathogens. The animals were inoculated in one quarter with E. coli and slaughtered after 6 h (n = 5) or 24 h (n = 5) in two different infection methods. Five cows, considered as controls, received no treatment and were slaughtered after 24 h. In total, 89 healthy and 75 diseased German Holstein cows were tested for the GSE93082 dataset. Diseases were distinguished by either systemic (extra-mammary) occurrence or those affecting the mammary gland (mastitis) to account for influences on the milk composition from local inflammatory processes. All cows were examined thoroughly by the dairy herd manager, trained staff, or a veterinarian. Healthy animals (2–4 years old, 1st to 3rd lactation, one animal 4th and one 8th lactation) which had no clinical signs of disease and no abnormalities in the udder or milk, with a reported somatic cell count less than 100,000 cells/mL were chosen as controls. Most of the control samples were taken during early lactation within 10 to 100 days postpartum. Diseased animals were in the 1st to 8th lactation period from 10 to 220 days postpartum. The milk samples of control animals or cows with extra-mammary diseases were collected and tested from one quarter or a composite milk sample (equal volumes from all 4 quarters mixed) [32]. In the GSE75379 dataset, sixteen healthy primiparous Holstein cows were inoculated with live E. coli into one mammary quarter at four to six weeks after parturition. The cows were housed in straw-bedded tie-stalls, where they were individually fed and given free access to water. The animals were fed using a TMR based on corn silage, minerals, and vitamins for ad libitum intake twice daily. Daily feed intake and milk yield at each milking were recorded. Prior to the start of the study period, cows were considered healthy and free of mastitis-causing pathogens based on body temperature, white blood cell count (WBC), California Mastitis Test, glutaraldehyde test, SCC, and bacteriological examinations of milk samples. Control quarters were selected based on bacteriological tests, in which quarter foremilk SCCs were <181,000 cells/mL at 24 h post-intramammary infection (IMI). Biopsy specimens of healthy and diseased udder tissue were performed 24 h post-IMI in infected and non-infected (control) mammary quarters [33].
Table 1.
Summary of the GEO accession numbers for RNA-Seq and microarray data sets.
| No. | Data Type | GEO a Accession | Platforms | Samples (M:H) b | Citation |
|---|---|---|---|---|---|
| 1 | RNA-Seq | GSE131607 | GPL15749 (Illumina HiSeq 2000) | 12 (6:6) | Asselstine et al. [16] |
| 2 | RNA-Seq | GSE75379 | GPL15749 (Illumina HiSeq 2000) | 18 (6:12) | Moyes et al. [33] |
| 3 | Microarray | GSE93082 | GPL2112 ((Bovine) Affymetrix Bovine Genome Array) | 12 (6:6) | Zoldan et al. [32] |
| 4 | Microarray | GSE15020 | GPL2112 ((Bovine) Affymetrix Bovine Genome Array) | 10 (5:5) | Mitterhuemer et al. [31] |
| 5 | Microarray | GSE15022 | GPL2112 ((Bovine) Affymetrix Bovine Genome Array) | 10 (5:5) | Mitterhuemer et al. [31] |
a GEO, Gene Expression Omnibus; b M, number of mastitis samples, and H, number of healthy samples.
2.2. Differential Gene Expression Analysis
Microarray data were pre-processed and normalized using the Lumi package [34] and the GCRMA algorithm (GeneChip Robust Multi-array Averaging) method, implemented in the Affy package in R software, to remove the variance and to prepare the data for further analysis [35]. Gene expression analysis was performed in R/Bioconductor software to screen the significant differential expression genes (DEGs) according to the comparison of the test and control data using the packages Limma [36], GEOquary [37], Biobase [38], and Umap [39].
Concerning RNA-Seq data, the quality of the raw data was assessed using FastQC software (v0.11.9) [40]. Then, based on the results of the raw data quality control, the sequences were edited to remove the adapters, PCR primers, and low-quality reads using Trimmomatic software (v0.38.0) [41]. Alignment sequences and mapping of reads were conducted on the Bos taurus reference genome (http://ftp.ensembl.org/pub/release-103/fasta/bos_taurus/dna/ (accessed on 20 September 2021)) using HISAT2 software (v2.1.0) [42]. For transcript quantification, featureCounts software (v2.0.1) was employed to measure the total raw counts of mapped reads [43]. DESeq2 software (v2.11.40.6) was applied for the measurement of final differences in gene expression [44]. In addition to DEGs, the identification of miRNAs was also performed in the RNA-Seq datasets, simultaneously. Finally, the threshold for statistical significance of the differential expression of each gene was obtained with the criteria of a |log fold-change (FC)| ≥ 2.0 and a false discovery rate (FDR) ≤ 0.05 in accession numbers related to microarray and RNA-Seq datasets. The gene lists from the differential expression related to microarray and RNA-Seq analysis were considered Gene Sets 1 and 2, respectively (Supplementary Materials 1 and 2).
2.3. Literature Mining to Discover Candidate Genes for Mastitis
Extensive literature surveys were performed to search for candidate genes using keywords related to bovine mastitis in the PubMed and Google Scholar databases without time limitation. GWAS studies were selected for further detailed review. Then, iHOP (iHOP literature server, http://www.ihop.net.org/ (accessed on 9 October 2021)) was used, which is a web-based tool that allows the exploration of a network of gene and protein interactions by directly navigating the pool of published scientific literature [45]. Finally, the candidate gene list extracted through literature mining was mentioned as Gene Set 3 (Supplementary Material 3).
2.4. Determination of Main Gene List
To identify the relevant candidate genes related to mastitis, 3 datasets (microarray, RNA-Seq, and GWAS) from the differential expression and GWAS analyses were integrated. Subsequently, genes that were common to the three gene sets were selected as the main gene list for further analysis. The number of shared DE genes between the 3 datasets was analyzed using the R package VennDiagram v1.6.18 [46].
2.5. Functional Enrichment and KEGG Pathway Analysis
Gene ontology (GO) and enrichment analysis were performed using the online programs DAVID [47] (Database for Annotation, Visualization, and Integrated Discovery), PANTHER [48] (Protein ANalysis THrough Evolutionary Relationships), GeneCards (www.genecards.org/ (accessed on 9 October 2021)), g:Profiler [49] (https://biit.cs.ut.ee/gprofiler/gost (accessed on 9 October 2021)), and the STRING database [50] (https://string-db.org (accessed on 9 October 2021)), which are comprehensive web tools that help to explore the biological process (BP), molecular function (MF), and cellular component (CC) of the mined gene set. The pathway enrichment of the identified genes was provided in the Kyoto Encyclopedia of Genes and Genomes (KEGG). Gene Ontology terms with FDR < 0.05 were considered significantly enriched for the identified genes.
2.6. Identification of miRNAs and Target Gene Prediction
The functional annotation of the expressed miRNAs consisted of the functional annotation of their potential target genes. The potentially targeted genes were predicted using miRBase [51] (https://www.mirbase.org/ (accessed on 9 October 2021)) and Targetscan [52]. The predicted target genes were selected and submitted to DAVID, KEGG, Reactome pathways, and the PANTHER database for the enrichment target genes of each miRNA.
2.7. Reconstruction of Omics Multi-Layers Networks
The miRNA–gene bipartite network was reconstructed based on the master gene list and the molecular interactions documented in related papers and in online interaction databases. Protein–protein interaction (PPI) data were abstracted from the Biomolecular Interaction Network Database (BIND), the Database of Interacting Proteins (DIP), the Biological General Repository for Interaction Datasets (BioGRID), and the Mammalian Protein–Protein Interactions Database (MIPS). Finally, PPI network analysis was performed using the STRING database to explore interactions between genes, specifically in Bos taurus species. Each miRNA and target gene was entered into the database and resulting interactions were imported into the networks using Cytoscape software v3.8.2. (National Institute of General Medical Sciences, Bethesda Softworks, Rockville, MD, USA) [53]. Genes and miRNAs in generated networks are represented as nodes and the interactions between these nodes as edges. Furthermore, the metabolic-signaling pathway enrichment of the PPI network was reconstructed using ClueGO v.2.5.5 [54].
2.8. Modeling of Three-Dimensional (3D) Structure of Hub Proteins
The SWISS-MODEL template-based approach [55] (https://www.swissmodel.expasy.org/interactive (accessed on 15 October 2021)) was used to predict the 3D structures of hub proteins using individual FASTA sequences and reference PDB files. The resulting PDB files are enclosed in Supplementary Material 4.
3. Results
3.1. Transcriptome Analysis for Identifying Differentially Expressed Genes (DEGs)
To obtain better insights into the molecular mechanism and the genetic basis of mastitis, we investigated the pattern of transcriptome profiles of mastitis samples versus healthy samples in dairy cattle. The experimental data used for the study were obtained from the GEO database, consisting of microarray and RNA-Seq datasets, as presented in Table 1. The analysis of differentially expressed genes between mastitis and healthy cows was performed based on a fold change > ±2 and a false discovery rate (FDR) < 0.05. The results of the statistical analysis of the microarray datasets showed a total of 564 significant genes from three datasets as follows. The first dataset (GSE93082): 378 DE genes, the second dataset (GSE15020): 177 DE genes, and the third dataset (GSE15022): nine DE genes. Concerning the analysis of RNA-Seq, a total of 774 genes were differentially expressed in the mastitic versus healthy group comparison, of which 442 and 332 genes were detected from accession numbers GSE131607 and GSE75379, respectively. Gene counts and a detailed summary of the alignments for all GEO accession numbers are provided as Gene Sets 1 and 2 for microarray and RNA-Seq analyses, respectively, in Supplementary Materials 1 and 2.
3.2. Identification of miRNAs
Based on analysis of the RNA-Seq dataset with access number GSE75379, we identified eight miRNAs out of the 332 DE genes with functions related to mastitis: bta-mir-339a, bta-mir-24-2, bta-mir-222, bta-mir-27a, bta-mir-146a, bta-mir-23a, bta-mir-142, and bta-mir-223. Considering the threshold of a fold change > ±2 and FDR < 0.05, all differentially expressed miRNAs were overexpressed in the mastitic cows compared to the healthy cows (Table 2).
Table 2.
Information about differentially expressed miRNAs between the mastitis and healthy samples in dairy cattle based on GSE75379.
| miRNA Name | miRNA Region | Fold Change | p-Value | FDR | ||
|---|---|---|---|---|---|---|
| BTA | miRNA Start | miRNA End | ||||
| bta-mir-339a | 25 | 41736134 | 41736211 | 2.0472 | 0.0032 | 0.0490 |
| bta-mir-24-2 | 7 | 11839032 | 11839103 | 2.5302 | 0.0001 | 0.0056 |
| bta-mir-222 | X | 98125920 | 98126030 | 3.2194 | 4.88 × 10−6 | 0.0002 |
| bta-mir-27a | 7 | 11838877 | 11838949 | 3.6647 | 1.13 × 10−8 | 1.16 × 10−6 |
| bta-mir-146a | 7 | 72071548 | 72071646 | 3.9677 | 2.26 × 10−8 | 2.20 × 10−6 |
| bta-mir-23a | 7 | 11838702 | 11838776 | 3.9826 | 8.97 × 10−9 | 9.60 × 10−7 |
| bta-mir-142 | 19 | 9301432 | 9301518 | 4.2675 | 2.69 × 10−13 | 6.83 × 10−11 |
| bta-mir-223 | X | 94562822 | 94562929 | 4.4983 | 3.05 × 10−11 | 5.58 × 10−9 |
3.3. Literature Mining and Identification of Main Gene List
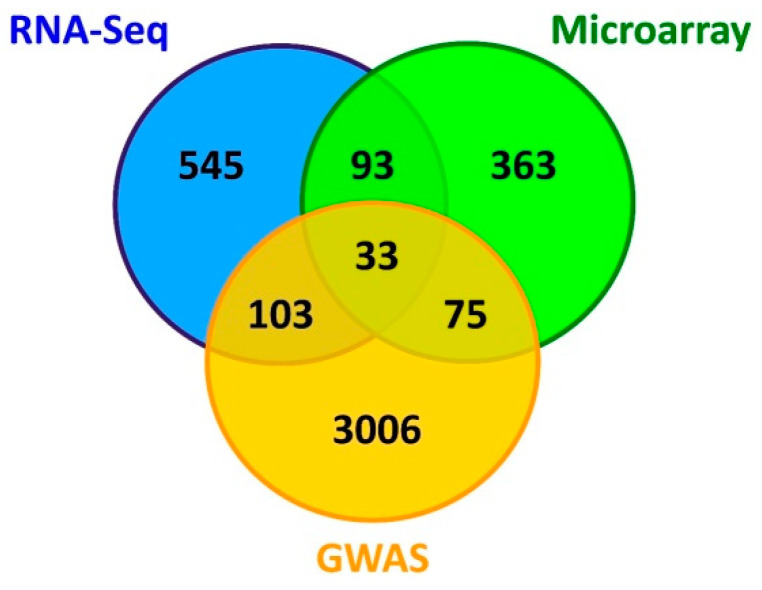
Literature mining was carried out using the iHOP web tool to increase the study’s accuracy and to obtain previous evidence for associations between the identified genes and mastitis in dairy cattle. Based on literature mining, 3217 candidate genes were considered as Gene Set 3 (Supplementary Material 3). The Venn diagram shows the number of genes that are unique or common among the three gene sets for mastitis (Figure 2). Notably, there is a remarkably small overlap between the three datasets. Overall, 33 genes were common in the Gene Sets 1, 2, and 3 relating to microarray, RNA-Seq, and GWAS datasets, respectively, which were named as the main genes involved in mastitis and were considered for subsequent analysis (Table 3). The results of differential expression analysis of 33 common genes based on the RNA-Seq datasets are presented in Table 3. When comparing healthy cows to mastitis cows, of 33 DE genes, nine were underexpressed in the mastitis cows and 24 genes were overexpressed in the mastitis cows, based on their fold-change values (fold change > ±2).
Figure 2.
Venn diagram of significant genes among the three types of dataset, including microarray, RNA-Seq, and GWAS data related to mastitis in dairy cattle.
Table 3.
Summary list of 33 common genes (main genes) in the integrated studies of gene expression and GWAS associated with mastitis in dairy cattle *.
| Gene Symbol | Gene Name | Gene Region | Fold Change | p-Value | FDR | ||
|---|---|---|---|---|---|---|---|
| Chr | Gene Start | Gene End | |||||
| CSN3 | casein kappa | 6 | 85645854 | 85658926 | −4.1767 | 8.29 × 10−10 | 1.55 × 10−6 |
| CSN1S2 | casein alpha-S2 | 6 | 85529905 | 85548556 | −3.9696 | 3.53 × 10−8 | 1.85 × 10−5 |
| CSN2 | casein beta | 6 | 85449164 | 85457744 | −3.7916 | 2.92 × 10−8 | 1.74 × 10−5 |
| RHPN2 | rhophilin Rho GTPase binding protein 2 | 18 | 43404074 | 43474596 | −3.4556 | 1.38 × 10−6 | 0.0002 |
| CSN1S1 | casein alpha s1 | 6 | 85411118 | 85429268 | −3.4530 | 2.94 × 10−7 | 7.41 × 10−5 |
| LALBA | lactalbumin alpha | 5 | 31183432 | 31213145 | −3.1099 | 1.23 × 10−5 | 0.0009 |
| ACSS2 | acyl-CoA synthetase short chain family member 2 | 13 | 64186743 | 64233568 | −2.6729 | 4.02 × 10−10 | 1.05 × 10−6 |
| RHOU | ras homolog family member U | 28 | 697339 | 706882 | −2.4960 | 0.0001 | 0.0047 |
| KRT7 | keratin 7 | 5 | 27674854 | 27689030 | −2.3804 | 0.0003 | 0.0110 |
| SGK1 | serum/glucocorticoid regulated kinase 1 | 9 | 72305979 | 72418535 | 2.0107 | 1.85 × 10−14 | 5.70 × 10−12 |
| TRIB1 | tribbles pseudokinase 1 | 14 | 14779050 | 14787206 | 2.0650 | 4.36 × 10−5 | 0.0024 |
| LYST | lysosomal trafficking regulator | 28 | 8379173 | 8523114 | 2.1066 | 0.0009 | 0.0212 |
| VAV1 | vav guanine nucleotide exchange factor 1 | 7 | 17664498 | 17728163 | 2.1138 | 3.31 × 10−5 | 0.0013 |
| GRO1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 6 | 89072611 | 89075133 | 2.2062 | 0.0003 | 0.0105 |
| F5 | coagulation factor V | 16 | 37159073 | 37238306 | 2.2729 | 3.94 × 10−10 | 6.01 × 10−8 |
| SERPINE1 | serpin family E member 1 | 25 | 35596139 | 35617193 | 2.3498 | 0.0002 | 0.0065 |
| BASP1 | brain abundant membrane attached signal protein 1 | 20 | 55908762 | 55964145 | 2.4174 | 3.40 × 10−8 | 3.14 × 10−6 |
| CD40 | CD40 molecule | 13 | 74842191 | 74853116 | 2.4243 | 1.20 × 10−5 | 0.0005 |
| TNFRSF6B | TNF receptor superfamily member 6b | 13 | 54054302 | 54055810 | 2.4416 | 0.0001 | 0.0035 |
| SLC16A3 | solute carrier family 16 member 3 | 19 | 50634317 | 50642204 | 2.4821 | 8.56 × 10−6 | 0.0004 |
| CXCR1 | chemokine (C-X-C motif) receptor 1 | 2 | 106215131 | 106219158 | 2.5332 | 0.0002 | 0.0065 |
| SOCS3 | suppressor of cytokine signaling 3 | 19 | 53840159 | 53840858 | 2.5589 | 0.0001 | 0.0047 |
| CCDC88B | coiled-coil domain containing 88B | 29 | 42630756 | 42645750 | 2.6392 | 4.24 × 10−8 | 3.81 × 10−6 |
| TNFAIP6 | TNF alpha induced protein 6 | 2 | 44747145 | 44764214 | 2.7306 | 0.0001 | 0.0040 |
| S100A9 | S100 calcium binding protein A9 | 3 | 17115128 | 17117984 | 2.9459 | 5.39 × 10−6 | 0.0005 |
| PSTPIP2 | proline-serine-threonine phosphatase interacting protein 2 | 24 | 45737786 | 45832060 | 2.9685 | 3.43 × 10−10 | 1.05 × 10−6 |
| ALOX5AP | arachidonate 5-lipoxygenase activating protein | 12 | 30108987 | 30138259 | 3.2299 | 1.64 × 10−11 | 3.14 × 10−9 |
| CCL19 | C-C motif chemokine ligand 19 | 8 | 76054024 | 76055932 | 3.5366 | 4.73 × 10−7 | 3.31 × 10−5 |
| MMP9 | matrix metallopeptidase 9 | 13 | 74746976 | 74754303 | 3.5921 | 8.64 × 10−8 | 7.35 × 10−6 |
| HCK | HCK proto-onco, Src family tyrosine kinase | 13 | 61563070 | 61608503 | 4.2225 | 3.23 × 10−18 | 1.69 × 10−15 |
| S100A12 | S100 calcium binding protein A12 | 3 | 17102722 | 17104173 | 4.4133 | 4.22 × 10−11 | 7.47 × 10−9 |
| S100A8 | S100 calcium binding protein A8 | 3 | 17085577 | 17086827 | 4.7179 | 6.81 × 10−12 | 1.36 × 10−9 |
| IL1RN | interleukin 1 receptor antagonist | 11 | 46815591 | 46837831 | 4.9613 | 8.55 × 10−16 | 3.36 × 10−13 |
* Information on common differentially expressed genes between the mastitis and healthy samples in dairy cattle provided based on RNA-Seq datasets.
3.4. Functional Annotation and Pathway Enrichment Analysis
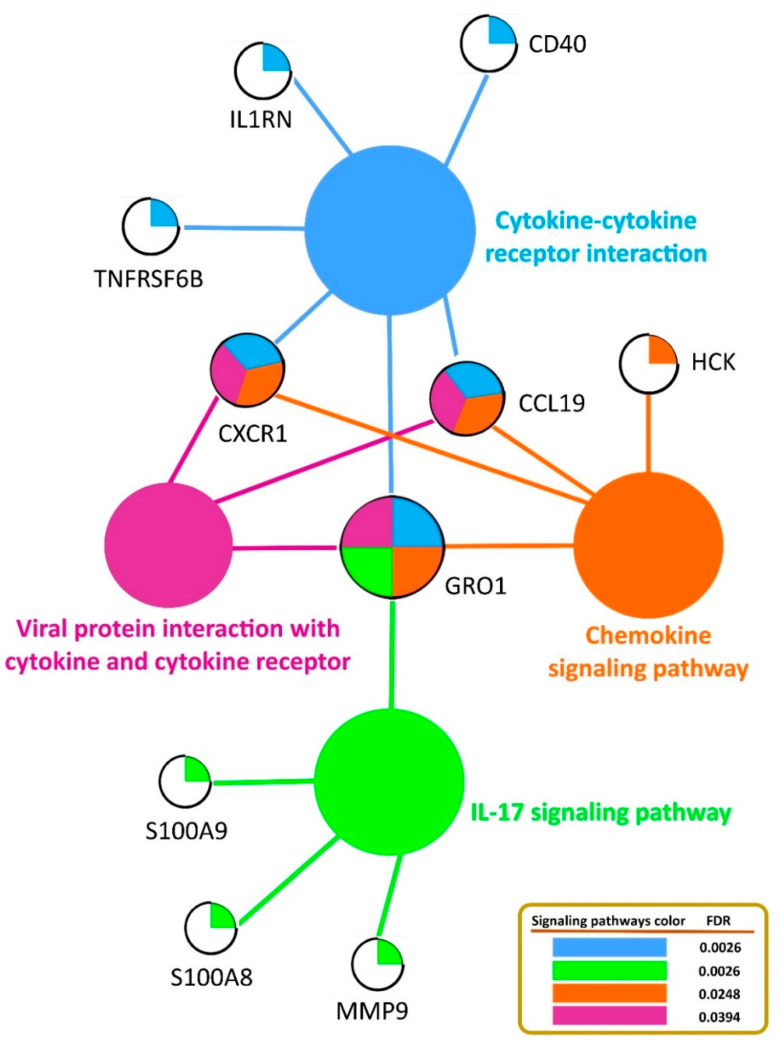
The functional annotation of GO terms was performed based on the biological process (BP), molecular function (MF), and cellular component (CC) to identify the biological meaning and the systematic features of the list of 33 DE genes, using the DAVID, PANTHER, and g:Profiler databases. Twenty-three biological processes were identified, such as response to stimulus, defense response, response to stress, immune response, cellular process, and biological regulation, which were the most significant ones associated with mastitis. The identified DEGs were significantly involved in the seven following functions: antioxidant activity, binding, protein binding, zymogen binding, arachidonic acid binding, Toll-like receptor 4 binding, and RAGE receptor binding for molecular function. Regarding cellular components, four GO terms, including extracellular region, Golgi lumen, extracellular space, and cellular anatomical entity, were identified (Table 4). In addition, KEGG pathway analysis revealed that the identified DE genes involved in mastitis were enriched in cytokine–cytokine-receptor interaction, the IL-17 signaling pathway, viral protein interaction with cytokines and cytokine receptors, and the chemokine signaling pathway (Figure 3).
Table 4.
Top significant gene ontology (GO) terms enriched using genes associated with mastitis in dairy cattle.
| Category | Term_ID | Term | Count | FDR | Genes |
|---|---|---|---|---|---|
| BP 1_DIRECT | GO:0050896 | Response to stimulus | 20 | 3.26 × 10−9 | CSN2, RHPN2, SGK1, CSN1S2, LALBA, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, LYST, F5, RHOU, IL1RN, MMP9, CD40, CSN3, GRO1, S100A12, CXCR1 |
| BP_DIRECT | GO:0032570 | Response to progesterone | 5 | 5.21 × 10−8 | CSN2, CSN1S2, LALBA, CSN1S1, CSN3 |
| BP_DIRECT | GO:0032355 | Response to estradiol | 5 | 3.34 × 10−7 | CSN2, CSN1S2, LALBA, CSN1S1, CSN3 |
| BP_DIRECT | GO:0006952 | Defense response | 9 | 3.93 × 10−6 | CSN1S2, LALBA, S100A9, S100A8, LYST, IL1RN, CD40, GRO1, S100A12 |
| BP_DIRECT | GO:0006950 | Response to stress | 12 | 7.04 × 10−6 | CSN2, CSN1S2, LALBA, S100A9, S100A8, LYST, F5, IL1RN, MMP9, CD40, GRO1, S100A12 |
| BP_DIRECT | GO:0006955 | Immune response | 8 | 2.06 × 10−5 | S100A9, S100A8, LYST, IL1RN, CD40, GRO1, S100A12, CXCR1 |
| BP_DIRECT | GO:0051716 | Cellular response to stimulus | 14 | 2.06 × 10−5 | CSN2, RHPN2, SGK1, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, RHOU, IL1RN, MMP9, CD40, GRO1, CXCR1 |
| BP_DIRECT | GO:0030593 | Neutrophil chemotaxis | 4 | 6.46 × 10−5 | S100A9, S100A8, GRO1, CXCR1 |
| BP_DIRECT | GO:0098542 | Defense response to other organisms | 7 | 6.46 × 10−5 | CSN1S2, LALBA, S100A9, S100A8, LYST, CD40, S100A12 |
| BP_DIRECT | GO:0006954 | Inflammatory response | 6 | 6.92 × 10−5 | S100A9, S100A8, IL1RN, CD40, GRO1, S100A12 |
| BP_DIRECT | GO:0033993 | Response to lipids | 6 | 9.95 × 10−5 | CSN2, CSN1S2, LALBA, CSN1S1, CSN3, GRO1 |
| BP_DIRECT | GO:0065007 | Biological regulation | 17 | 0.00029 | CSN2, RHPN2, SGK1, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, SERPINE1, F5, RHOU, IL1RN, MMP9, CD40, CSN3, GRO1, CXCR1 |
| BP_DIRECT | GO:0052548 | Regulation of endopeptidase activity | 5 | 0.00063 | CSN2, S100A9, S100A8, SERPINE1, MMP9 |
| BP_DIRECT | GO:0045087 | Innate immune response | 5 | 0.0028 | S100A9, S100A8, LYST, CD40, S100A12 |
| BP_DIRECT | GO:0023051 | Regulation of signaling | 8 | 0.0041 | CSN2, S100A9, CSN1S1, SOCS3, S100A8, IL1RN, MMP9, CD40 |
| BP_DIRECT | GO:0042981 | Regulation of apoptotic process | 6 | 0.0041 | S100A9, CSN1S1, SOCS3, S100A8, MMP9, CD40 |
| BP_DIRECT | GO:0050727 | Regulation of inflammatory response | 4 | 0.0046 | CSN2, S100A9, SOCS3, S100A8 |
| BP_DIRECT | GO:0070488 | Neutrophil aggregation | 2 | 0.0046 | S100A9, S100A8 |
| BP_DIRECT | GO:0002523 | Leukocyte migration involved in inflammatory response | 2 | 0.01 | S100A9, S100A8 |
| BP_DIRECT | GO:0009987 | Cellular process | 18 | 0.0104 | CSN2, RHPN2, SGK1, LALBA, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, SERPINE1, LYST, RHOU, IL1RN, MMP9, CD40, GRO1, S100A12, CXCR1 |
| BP_DIRECT | GO:0032268 | Regulation of cellular protein metabolic process | 7 | 0.0221 | CSN2, S100A9, SOCS3, S100A8, SERPINE1, MMP9, CD40 |
| BP_DIRECT | GO:0050793 | Regulation of developmental process | 6 | 0.0276 | CSN2, CSN1S1, SOCS3, RHOU, MMP9, CD40 |
| BP_DIRECT | GO:0071345 | Cellular response to cytokine stimulus | 4 | 0.0302 | SOCS3, CD40, GRO1, CXCR1 |
| MF 2_DIRECT | GO:0016209 | Antioxidant activity | 5 | 2.82 × 10−5 | CSN2, S100A9, CSN1S1, S100A8, ALOX5AP |
| MF_DIRECT | GO:0005488 | Binding | 19 | 0.00034 | CSN2, SGK1, CSN1S2, LALBA, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, SERPINE1, F5, RHOU, IL1RN, MMP9, CD40, CSN3, GRO1, S100A12, CXCR1 |
| MF_DIRECT | GO:0005515 | Protein binding | 14 | 0.00034 | CSN2, CSN1S2, LALBA, S100A9, SOCS3, S100A8, SERPINE1, RHOU, IL1RN, MMP9, CD40, CSN3, GRO1, CXCR1 |
| MF_DIRECT | GO:0035375 | Zymogen binding | 3 | 0.00034 | CSN1S2, SERPINE1, CSN3 |
| MF_DIRECT | GO:0050544 | Arachidonic acid binding | 3 | 0.00034 | S100A9, S100A8, ALOX5AP |
| MF_DIRECT | GO:0035662 | Toll-like receptor 4 binding | 2 | 0.007 | S100A9, S100A8 |
| MF_DIRECT | GO:0050786 | RAGE receptor binding | 2 | 0.033 | S100A9, S100A8 |
| CC 3_DIRECT | GO:0005576 | Extracellular region | 13 | 1.74 × 10−7 | CSN2, CSN1S2, LALBA, S100A9, CSN1S1, S100A8, SERPINE1, F5, IL1RN, MMP9, CSN3, GRO1, S100A12 |
| CC_DIRECT | GO:0005796 | Golgi lumen | 4 | 1.38 × 10−6 | CSN2, CSN1S2, CSN1S1, CSN3 |
| CC_DIRECT | GO:0005615 | Extracellular space | 10 | 2.57 × 10−6 | CSN2, CSN1S2, LALBA, CSN1S1, S100A8, SERPINE1, IL1RN, MMP9, CSN3, GRO1 |
| CC_DIRECT | GO:0110165 | Cellular anatomical entity | 22 | 0.00069 | CSN2, RHPN2, SGK1, CSN1S2, LALBA, S100A9, CSN1S1, SOCS3, S100A8, ALOX5AP, SERPINE1, KRT7, LYST, F5, RHOU, IL1RN, MMP9, CD40, CSN3, GRO1, S100A12, CXCR1 |
1 BP, biological process; 2 MF, molecular function; 3 CC, cellular components.
Figure 3.
The KEGG pathway-based network analysis of significant genes related to mastitis in dairy cattle.
3.5. PPI Network and Identification of Hub Genes
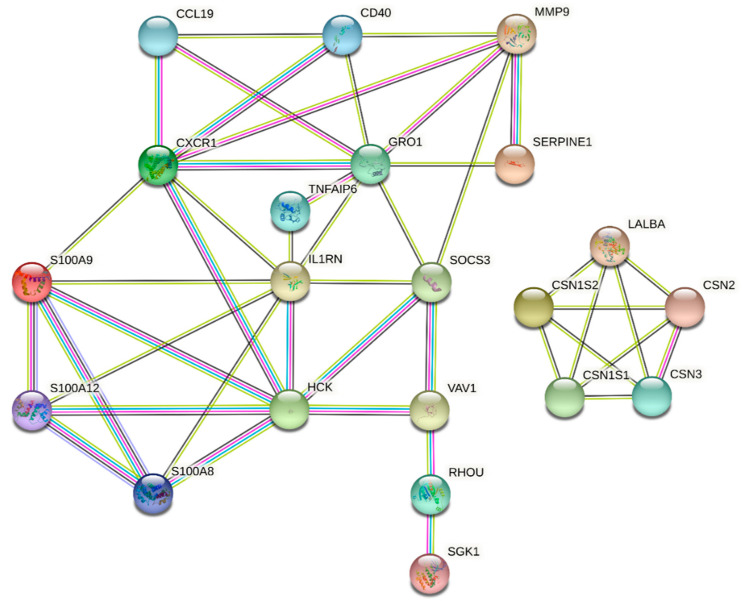
Protein–protein interaction (PPI) networks for up- and downregulated genes were reconstructed with the STRING database, which indicated the physical connection between two or more protein molecules related to biochemical functions (Figure 4). Twenty-one nodes with 45 connections (edges) were represented in the PPI network, as presented in Figure 4. Moreover, we considered hub genes based on their higher-degree connectivity values in the PPI network. A total of seven hub genes, including MMP9, HCK, GRO1, SOCS3, CXCR1, IL1RN, and S100A9, were identified, all of which were overexpressed genes. All the hub proteins identified are protein-coding genes. The functional enrichment analysis demonstrated that hub genes were involved in the majority of molecular functions and biological processes (Table 4).
Figure 4.
Protein–protein interaction (PPI) network analysis of common differentially expressed genes associated with mastitis in dairy cattle.
3.6. Prediction of miRNA-Target Genes and Gene Regulatory Network Reconstruction
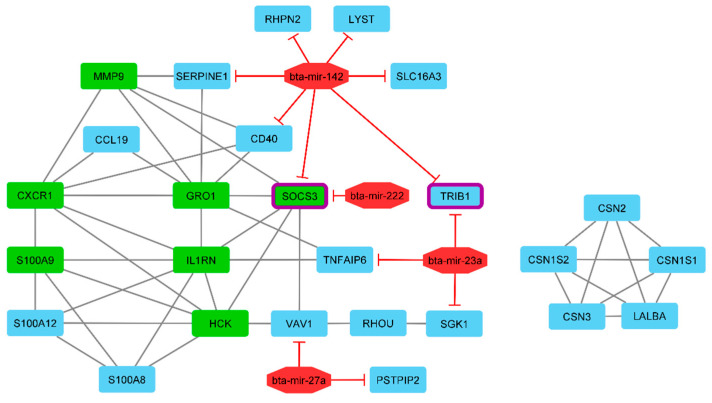
We also aimed to determine whether the expression of miRNAs was associated with that of the 33 DE genes in the mastitic and healthy cows. Among the DE miRNAs, bta-mir-222, bta-mir-27a, bta-mir-23a, and bta-mir-142 suppressed 11 of the identified DE genes as targets of the selected miRNAs. A target gene search using TargetScan demonstrated that bta-mir-142 has seven target genes, namely, RHPN2, LYST, SERPINE1, CD40, SOCS3, TRIB1, and SLC16A3, followed by bta-mir-23a having three target genes, SGK1, TNFAIP6, and TRIB1. In addition, bta-mir-27a suppressed the PSTPIP2 and VAV1 genes, and bta-mir-222 suppressed the SOCS3 gene. The TRIB1 and SOCS3 genes displayed the highest suppression by miRNAs. The identified target genes, associated with their miRNAs, are visualized in Figure 5. For constructing the gene regulatory network, we compiled a list of DE genes and miRNAs (as nodes) involved in mastitis based on literature mining and PPI resources. Briefly, miRNA–gene bipartite networks are commonly represented in an undirected graph format, with nodes representing miRNAs or genes and edges corresponding to interactions (genes–genes and miRNAs–targeted genes). In this network, we identified 30 nodes (26 genes and four miRNAs), with 57 edges interacting with it (Figure 5).
Figure 5.
Interactive bipartite network (gene–miRNA) which demonstrates the regulatory effect on mastitis in dairy cattle. In this network, the quadrilateral points represent genes, and the octagonal points represent miRNAs. Regarding miRNAs and target genes, the edges indicate the suppressive role of miRNAs. The edges also represent the gene–gene interactions. The green quadrilateral nodes represent the hub genes. The quadrilateral nodes that have purple around them are the genes showing the highest suppression by miRNAs.
3.7. Three-Dimensional Modeling of Hub Proteins
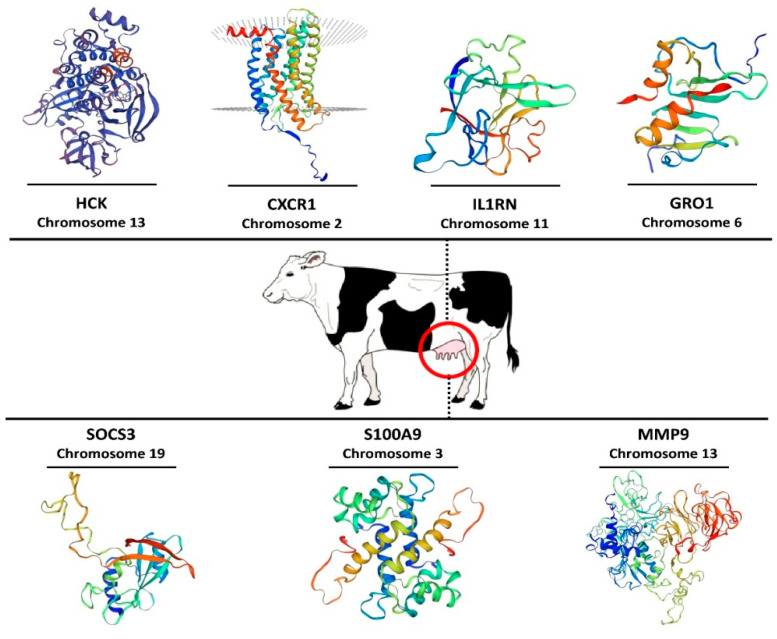
In the present study, we also modeled the 3-dimensional protein structure of the seven hub genes identified in the PPI network that had the most interaction with other genes involved in the network (Figure 6). 3D modeling revealed that the predicted structures of these seven hub proteins were significantly different from each other. Four hub proteins (MMP9, HCK, CXCR1, and S100A9) had the greatest structural complexity compared to the three other proteins.
Figure 6.
Modeling of three-dimensional protein structure for genes with the most interaction (hub genes) in the interactive bipartite network (gene–miRNA) according to the SWISS-MODEL repository.
4. Discussion
Mastitis is a complex trait and is prominent among health-related traits in the cattle industry, exerting a severe impact on profitability and animal welfare. The identification of functional candidate genes and molecular mechanisms involved in mastitis is required, given the persistence of the disease on dairy farms. Moreover, understanding the interplay between molecular and cellular components, with each component interacting at different levels that are entangled in several biological pathways, is important. Hence, the present study provides a general framework to investigate and integrate different sources of transcriptome data and previous results from GWAS studies to identify the genetic basis and key pathways associated with bovine mastitis. Numerous studies have demonstrated that the integration of multiple layers of omics data is a powerful strategy to increase the efficiency and accuracy of candidate gene and biomarker discovery, detecting molecular and biochemical interactions, and the relationships between biological variables in different species [4,7,8,56,57,58]. In this study, the integrative analysis of multiple datasets resulted in prioritizing 33 DE genes as the main gene list, of which nine genes were downregulated and 24 genes were upregulated in the mastitic cows compared with the healthy cows based on their FC values. A list of main detected genes related to mastitis is provided in Table 3, with their main functions (biological process, molecular function, and cellular component) listed in Table 4. Among them, CSN2, CSN3, CSN1S1, CSN1S2, RHPN2, LALBA, ACSS2, RHOU, and KRT7 genes were underexpressed in the mastitic cows, mostly located on chromosomes 6. The most important overexpressed genes in cows with mastitis were GRO1, CXCR1, SOCS3, S100A9, MMP9, HCK, and IL1RN, which were hub genes (highly connected genes) involved in mastitis in this study. The casein cluster is composed of four genes; β-casein (CSN2), κ-casein (CSN3), αs1-casein (CSN1S1), and αs2-casein (CSN1S2), which encode approximately 80% of the protein content of bovine milk [59], and the whey protein gene (LALBA) was downregulated in inflamed mammary glands. LALBA encodes α-lactalbumin and is essential for lactose synthesis, which plays an important role in milk production as an osmotic regulator of milk secretion [60]. A possible explanation for the lower expression levels of these genes could be that the protein content in mastitic milk would decrease due to an antagonistic genetic relationship between mastitis and protein yield [24]. In previous studies, it was also demonstrated that all five genes were observed in enhanced abundance in the mammary glands of lactating dairy cows [61], dairy sheep [62], and lactating dairy goats [63]. Interestingly, as presented in Table 4, these genes were found in a majority of enriched pathways, suggesting possible key regulatory roles for them. Other noteworthy genes (RHPN2, ACSS2, RHOU, and KRT7) showing lower expression in cows with mastitis have critical roles in biological pathways, cellular process, fatty acid synthesis, and metabolism. For instance, ACSS2 (acyl-CoA synthetase short-chain family member 2) is well known to affect mastitis resistance in dairy cows and plays a role in the activation of acetate for de novo fatty acid synthesis [64]. Similarly to our results, Chen et al. [65] reported lower expression for ACSS2 and RHPN2 genes in response to the intramammary infection caused by two different pathogens (Escherichia coli and Streptococcus uberis) in dairy cows. As presented in Table 3, in the mastitis cows, 24 genes were more highly expressed, of which seven genes were considered as hub genes involved in significantly enriched biological processes and KEGG pathways. Subsequently, the PPI networks and gene regulatory networks were constructed based on these hub genes, which showed significant connectivity and which could shed light on the post-transcriptional regulation of gene expression by the identified miRNAs. Furthermore, the functional enrichment analysis resulted in four significant KEGG pathways associated with mastitis, which comprised six hub genes, i.e., GRO1, CXCR1, S100A9, MMP9, HCK, and IL1RN, as presented in Figure 3. Among these genes, GRO1 and CXCR1 were observed in four and three pathways, respectively.
GRO1 (melanoma growth stimulating activity, alpha) also known as CXCL1, is a protein-encoding gene and plays an important role in inflammation and immune defense due to the modulation of leukocyte infiltration [66], which has been previously proposed as a biomarker and therapeutic target in mastitis [67]. This gene is also involved in the metabolic pathways of cytokine–cytokine-receptor interaction, the IL-17 signaling pathway, the chemokine signaling pathway, and viral protein interaction with cytokines and cytokine receptors. In the case of the cytokine–cytokine-receptor interaction pathway, other genes, such as CD40, IL1RN, CXCR1, TNFRSF6B, and CCL19, were found to be involved in mastitis defense or immune response, as all these genes were upregulated in the mastitic cows based on their FC values. The significant role of cytokines in the immune response to infectious agents is well known because they are soluble extracellular proteins or glycoproteins that are crucial intercellular regulators and mobilizers of cells engaged in innate as well as adaptive inflammatory host defenses, cell growth, differentiation, cell death, and cell development and repair processes. It was previously reported that cytokines can participate in activation of the host defense mechanisms during mastitis [68,69]. The CXCR1 and CCL19 genes were also enriched in two other pathways of the chemokine signaling pathway and in viral protein interaction with cytokines and cytokine receptors. CXCR1 (chemokine (C-X-C motif) receptor 1), identified as a hub gene, is a protein-encoding gene for major pro-inflammatory cytokine receptors [70] that is introduced as a potential genetic marker for resistance to mastitis in dairy cows [71,72]. In our study, the gene CXCR1 was involved in seven GO terms—response to stimulus, immune response, cellular response to stimulus, neutrophil chemotaxis, biological regulation, cellular process, and cellular response to cytokine stimulus for biological processes (Table 4). In addition, earlier studies have reported that a non-synonymous mutation, c.365C > T, located in exon II of the CXCR1 gene is associated with susceptibility to mastitis in different breeds of cattle [73,74]. The viral protein interaction with cytokines and the cytokine receptor pathway is an immune system pathway which has a key role in the inflammatory responses to infection and may activate or inhibit cytokine signaling and possibly affect different aspects of immunity. Furthermore, the S100A8, S100A9, and MMP9 genes have been recognized as components of the IL-17 signaling pathway. This pathway plays crucial roles in both acute and chronic inflammatory responses. In fact, the interleukin 17 (IL-17) family, as a subset of cytokines, signals via their correspondent receptors and activates downstream pathways that include NF-kappaB, MAPKs, and C/EBPs to induce the expression of antimicrobial peptides, cytokines, and chemokines. S100A9 and MMP9, which were identified as hub genes and which were upregulated in the mastitic cows, play key roles in the regulation of immune response and inflammatory pathways [65,66].
SOCS3 (suppressor of cytokine signalling 3) was another hub gene identified with higher expression levels, which encodes an intracellular inhibitor of cytokine signaling and has a crucial role in the initial steps of the recognition of a pathogen-associated molecular pattern (PAMP) in the innate immune cells [75]. Furthermore, in the regulatory network, SOCS3 is suppressed by bta-mir-142 and bta-mir-222. These two miRNAs showed upregulation and their target gene, SOCS3, showed the lower expression than them in mastitic cows. We characterized bta-mir-222, bta-mir-27a, bta-mir-23a, and bta-mir-142 as the major miRNAs which play a prominent role in regulating this network of genes and these were upregulated in the mastitic cows. The gene regulatory network showed that the greatest target genes (seven genes) were suppressed by bta-mir-142. There is evidence that miRNAs play a critical role in the regulation of inflammation and immune function during infection with mastitis in dairy cattle [76,77,78,79].
Modeling of the 3D protein structure of hub proteins can be an invaluable aid in order to better understand the details of a particular protein because studies of protein structure and function are becoming a promising approach in the field of bioinformatics. Functional characterization of a protein is often facilitated by its 3D structure. Hence, it is necessary that a 3D structure is determined in examining the proteins’ function at the molecular level. Sequence identity, as a measure of the expected accuracy of a model represented, >30% indicates a relatively good predictor of the model [80]. When sequence identity drops below 30%, the main problem becomes the identification of related templates and their alignment with the sequence to be modeled. Based on our results, among the seven hub proteins, HCK, CXCR1, S100A9, and MMP9 showed the highest structural complexity, with sequence identities of 94.6%, 75.4%, 69.9%, and 47.5%, respectively, whereas the proteins of SOCS3, IL1RN, and GRO1 had the lowest structural complexity with sequence identities of 92.6%, 80.1%, and 72.5%, respectively (Figure 6). Consequently, these findings demonstrate the relevance of integrating results from transcriptomic and functional analyses for a better understanding of the function of important genes and molecular mechanisms responsible for mastitis development.
5. Conclusions
The integration of multi-omics data resulted in the identification of 33 common and relevant genes associated with bovine mastitis. Among these, seven genes (CXCR1, HCK, IL1RN, MMP9, S100A9, GRO1, and SOCS3) were identified as the hub genes and these can be explored as potential candidate genes for mastitis susceptibility and resistance. Functional annotation and enrichment analysis identified 23, 7, and 4 GO terms related to mastitis in the biological process, molecular function, and cellular component categories, respectively. We identified eight differentially expressed miRNAs, of which four suppressed 11 of the identified genes as their targets. Furthermore, the reconstruction of the regulatory network of genes associated with their miRNAs sheds light on the post-transcriptional regulation of this network. Therefore, this study provides a general framework to investigate and incorporate multiple layers of omics data from high-throughput technologies or available pathway annotation databases, which has led to the elucidation of molecular networks, the cellular and molecular-level features, and the genetic and biological basis of mastitis in dairy cattle.
Acknowledgments
The authors wish to express their thanks to the Gene Expression Omnibus (GEO) open repository database for providing a platform for the availability of molecular data to move forward to more detailed conclusions and interoperations of earlier outcomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cimb44010023/s1, Supplementary Material 1: The gene list from the differential expression related to microarray datasets analyses; Supplementary Material 2: The gene list from the differential expression related to RNA-Seq datasets analyses; Supplementary Material 3: The candidate gene list extracted from GWAS results using literature mining; Supplementary Material 4: Predicted structures of the hub proteins in PDB format.
Author Contributions
Conceptualization, M.N., F.G. and S.Z.; methodology, M.N. and F.G.; formal analysis, M.N. and F.G.; resources, D.L.; data curation, F.G., S.Z. and Z.M.; investigation, N.P., S.G., M.H.B., M.R.B. and M.A.H.; writing—original draft preparation, M.N.; writing—review and editing, N.P., M.N.P., H.W.B., K.-S.M. and D.L.; supervision, D.L. and M.N.; project administration, M.N., D.L. and S.Z.; funding acquisition, D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Animal Science Research Institute of Iran (ASRI) with project (21313094-990767).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matukumalli L.K., Lawley C.T., Schnabel R.D., Taylor J.F., Allan M.F., Heaton M.P., O’Connell J., Moore S.S., Smith T.P., Sonstegard T.S. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE. 2009;4:e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meuwissen T.H., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fondi M., Liò P. Multi-omics and metabolic modelling pipelines: Challenges and tools for systems microbiology. Microbiol. Res. 2015;171:52–64. doi: 10.1016/j.micres.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghafouri F., Bahrami A., Sadeghi M., Miraei-Ashtiani S.R., Bakherad M., Barkema H.W., Larose S. Omics multi-layers networks provide novel mechanistic and functional insights into fat storage and lipid metabolism in poultry. Front. Genet. 2021;12:646297. doi: 10.3389/fgene.2021.646297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang L., Sahana G., Su G., Yu Y., Zhang S., Lund M.S., Sørensen P. Integrating sequence-based GWAS and RNA-Seq provides novel insights into the genetic basis of mastitis and milk production in dairy cattle. Sci. Rep. 2017;7:45560. doi: 10.1038/srep45560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gòdia M., Reverter A., González-Prendes R., Ramayo-Caldas Y., Castelló A., Rodríguez-Gil J.-E., Sánchez A., Clop A. A systems biology framework integrating GWAS and RNA-seq to shed light on the molecular basis of sperm quality in swine. Genet. Sel. Evol. 2020;52:72. doi: 10.1186/s12711-020-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramayo-Caldas Y., Mármol-Sánchez E., Ballester M., Sánchez J.P., González-Prendes R., Amills M., Quintanilla R. Integrating genome-wide co-association and gene expression to identify putative regulators and predictors of feed efficiency in pigs. Genet. Sel. Evol. 2019;51:1–17. doi: 10.1186/s12711-019-0490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kromer J.O., Sorgenfrei O., Klopprogge K., Heinzle E., Wittmann C. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J. Bacteriol. 2004;186:1769–1784. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sana T.R., Fischer S., Wohlgemuth G., Katrekar A., Jung K.-H., Ronald P.C., Fiehn O. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics. 2010;6:451–465. doi: 10.1007/s11306-010-0218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., Tschaplinski T.J., Engle N.L., Carroll S.L., Martin S.L., Davison B.H., Palumbo A.V., Rodriguez M., Brown S.D. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genom. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S., Chen L., Te R., Qiao J., Wang J., Zhang W. Complementary iTRAQ proteomics and RNA-seq transcriptomics reveal multiple levels of regulation in response to nitrogen starvation in Synechocystis sp. PCC 6803. Mol. BioSyst. 2013;9:2565–2574. doi: 10.1039/c3mb70188c. [DOI] [PubMed] [Google Scholar]
- 13.Kühl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A., Filipovska A., Larsson N.-G. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife. 2017;6:e30952. doi: 10.7554/eLife.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu F., Cheng V.W., Wu Y., Tang Y., Weiner J.H., Li L. Comparative proteomic and metabolomic analysis of Staphylococcus warneri SG1 cultured in the presence and absence of butanol. J. Proteome Res. 2013;12:4478–4489. doi: 10.1021/pr400533m. [DOI] [PubMed] [Google Scholar]
- 15.Ma Q., Zhou J., Zhang W., Meng X., Sun J., Yuan Y.-J. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PLoS ONE. 2011;6:e26108. doi: 10.1371/journal.pone.0026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asselstine V., Miglior F., Suarez-Vega A., Fonseca P., Mallard B., Karrow N., Islas-Trejo A., Medrano J., Cánovas A. Genetic mechanisms regulating the host response during mastitis. J. Dairy Sci. 2019;102:9043–9059. doi: 10.3168/jds.2019-16504. [DOI] [PubMed] [Google Scholar]
- 17.Cai Z., Guldbrandtsen B., Lund M.S., Sahana G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genom. 2018;19:656. doi: 10.1186/s12864-018-5050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar N., Manimaran A., Kumaresan A., Jeyakumar S., Sreela L., Mooventhan P., Sivaram M. Mastitis effects on reproductive performance in dairy cattle: A review. Trop. Anim. Health Prod. 2017;49:663–673. doi: 10.1007/s11250-017-1253-4. [DOI] [PubMed] [Google Scholar]
- 19.Yang F., Chen F., Li L., Yan L., Badri T., Lv C., Yu D., Zhang M., Jang X., Li J. Three novel players: PTK2B, SYK, and TNFRSF21 were identified to be involved in the regulation of bovine mastitis susceptibility via GWAS and post-transcriptional analysis. Front. Immunol. 2019;10:1579. doi: 10.3389/fimmu.2019.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakhtiarizadeh M.R., Mirzaei S., Norouzi M., Sheybani N., Vafaei Sadi M.S. Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach. Front. Genet. 2020;11:722. doi: 10.3389/fgene.2020.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oviedo-Boyso J., Valdez-Alarcón J.J., Cajero-Juárez M., Ochoa-Zarzosa A., López-Meza J.E., Bravo-Patino A., Baizabal-Aguirre V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J. Infect. 2007;54:399–409. doi: 10.1016/j.jinf.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Heringstad B., Klemetsdal G., Ruane J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000;64:95–106. doi: 10.1016/S0301-6226(99)00128-1. [DOI] [Google Scholar]
- 23.Halasa T., Huijps K., Østerås O., Hogeveen H. Economic effects of bovine mastitis and mastitis management: A review. Veter. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- 24.Heringstad B., Chang Y., Gianola D., Klemetsdal G. Genetic association between susceptibility to clinical mastitis and protein yield in Norwegian dairy cattle. J. Dairy Sci. 2005;88:1509–1514. doi: 10.3168/jds.S0022-0302(05)72819-8. [DOI] [PubMed] [Google Scholar]
- 25.Li N., Richoux R., Boutinaud M., Martin P., Gagnaire V. Role of somatic cells on dairy processes and products: A review. Dairy Sci. Technol. 2014;94:517–538. doi: 10.1007/s13594-014-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meredith B., Lynn D., Berry D., Kearney F., Bradley D., Finlay E., Fahey A. A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front. Genet. 2013;4:229. doi: 10.3389/fgene.2013.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sodeland M., Kent M., Olsen H., Opsal M., Svendsen M., Sehested E., Hayes B., Lien S. Quantitative trait loci for clinical mastitis on chromosomes 2, 6, 14 and 20 in Norwegian Red cattle. Anim. Genet. 2011;42:457–465. doi: 10.1111/j.1365-2052.2010.02165.x. [DOI] [PubMed] [Google Scholar]
- 28.Wagner P., Yin T., Brügemann K., Engel P., Weimann C., Schlez K., König S. Genome-Wide Associations for Microscopic Differential Somatic Cell Count and Specific Mastitis Pathogens in Holstein Cows in Compost-Bedded Pack and Cubicle Farming Systems. Animals. 2021;11:1839. doi: 10.3390/ani11061839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Ma P., Liu J., Zhang Q., Zhang Y., Ding X., Jiang L., Wang Y., Zhang Y., Sun D. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015;16:111. doi: 10.1186/s12863-015-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welderufael B., Løvendahl P., De Koning D.-J., Janss L.L., Fikse W. Genome-wide association study for susceptibility to and recoverability from mastitis in Danish Holstein cows. Front. Genet. 2018;9:141. doi: 10.3389/fgene.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitterhuemer S., Petzl W., Krebs S., Mehne D., Klanner A., Wolf E., Zerbe H., Blum H. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genom. 2010;11:138. doi: 10.1186/1471-2164-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoldan K., Schneider J., Moellmer T., Fueldner C., Knauer J., Fuerll M., Starke A., Specht M., Reiche K., Hackermueller J. Discovery and validation of immunological biomarkers in milk for health monitoring of dairy cows-results from a multiomics approach. J. Adv. Dairy Res. 2017;5:2. [Google Scholar]
- 33.Moyes K., Sørensen P., Bionaz M. The impact of intramammary Escherichia coli challenge on liver and mammary transcriptome and cross-talk in dairy cows during early lactation using RNAseq. PLoS ONE. 2016;11:e0157480. doi: 10.1371/journal.pone.0157480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du P., Kibbe W.A., Lin S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 35.Gautier L., Cope L., Bolstad B., Irizarry R. Affy-Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis S., Meltzer P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 38.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInnes L., Healy J., Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv. 20181802.03426 [Google Scholar]
- 40.Andrews S. FastQC: A quality control tool for high throughput sequence data. Retrieved May. 2010;17:2018. [Google Scholar]
- 41.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez J.M., Hoffmann R., Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H., Boutros P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. Swiss-Model: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Backman M., Flenkenthaler F., Blutke A., Dahlhoff M., Ländström E., Renner S., Philippou-Massier J., Krebs S., Rathkolb B., Prehn C. Multi-omics insights into functional alterations of the liver in insulin-deficient diabetes mellitus. Mol. Metab. 2019;26:30–44. doi: 10.1016/j.molmet.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dao M.C., Sokolovska N., Brazeilles R., Affeldt S., Pelloux V., Prifti E., Chilloux J., Verger E.O., Kayser B.D., Aron-Wisnewsky J. A data integration multi-omics approach to study calorie restriction-induced changes in insulin sensitivity. Front. Physiol. 2019;9:1958. doi: 10.3389/fphys.2018.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee B., Zhang S., Poleksic A., Xie L. Heterogeneous multi-layered network model for omics data integration and analysis. Front. Genet. 2020;10:1381. doi: 10.3389/fgene.2019.01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrell Jr H., Jimenez-Flores R., Bleck G., Brown E., Butler J., Creamer L., Hicks C., Hollar C., Ng-Kwai-Hang K., Swaisgood H. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004;87:1641–1674. doi: 10.3168/jds.S0022-0302(04)73319-6. [DOI] [PubMed] [Google Scholar]
- 60.Zidi A., Casas E., Amills M., Jordana J., Carrizosa J., Urrutia B., Serradilla J.M. Genetic variation at the caprine lactalbumin, alpha (LALBA) gene and its association with milk lactose concentration. Anim. Genet. 2014;45:612–613. doi: 10.1111/age.12152. [DOI] [PubMed] [Google Scholar]
- 61.Seo M., Lee H.-J., Kim K., Caetano-Anolles K., Jeong J.Y., Park S., Oh Y.K., Cho S., Kim H. Characterizing milk production related genes in Holstein using RNA-seq. Asian-Australas. J. Anim. Sci. 2016;29:343. doi: 10.5713/ajas.15.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García-Gámez E., Gutiérrez-Gil B., Sahana G., Sánchez J.-P., Bayón Y., Arranz J.-J. GWA analysis for milk production traits in dairy sheep and genetic support for a QTN influencing milk protein percentage in the LALBA gene. PLoS ONE. 2012;7:e47782. doi: 10.1371/journal.pone.0047782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi H., Zhu J., Luo J., Cao W., Shi H., Yao D., Li J., Sun Y., Xu H., Yu K. Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats. Funct. Integr. Genom. 2015;15:309–321. doi: 10.1007/s10142-014-0420-1. [DOI] [PubMed] [Google Scholar]
- 64.Bionaz M., Loor J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008;9:366. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X., Cheng Z., Zhang S., Werling D., Wathes D.C. Combining genome wide association studies and differential gene expression data analyses identifies candidate genes affecting mastitis caused by two different pathogens in the dairy cow. Open J. Anim. Sci. 2015;5:358–393. doi: 10.4236/ojas.2015.54040. [DOI] [Google Scholar]
- 66.Sharifi S., Pakdel A., Ebrahimi M., Reecy J.M., Fazeli Farsani S., Ebrahimie E. Integration of machine learning and meta-analysis identifies the transcriptomic bio-signature of mastitis disease in cattle. PLoS ONE. 2018;13:e0191227. doi: 10.1371/journal.pone.0191227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnzon C.-F., Artursson K., Söderlund R., Guss B., Rönnberg E., Pejler G. Mastitis pathogens with high virulence in a mouse model produce a distinct cytokine profile in vivo. Front. Immunol. 2016;7:368. doi: 10.3389/fimmu.2016.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aderem A., Ulevitch R.J. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 69.Taraktsoglou M., Szalabska U., Magee D.A., Browne J.A., Sweeney T., Gormley E., MacHugh D.E. Transcriptional profiling of immune genes in bovine monocyte-derived macrophages exposed to bacterial antigens. Vet. Immunol. Immunopathol. 2011;140:130–139. doi: 10.1016/j.vetimm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 70.Dinarello C.A. Proinflammatory cytokines. Chest. 2000;118:503–508. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 71.Lahouassa H., Rainard P., Caraty A., Riollet C. Identification and characterization of a new interleukin-8 receptor in bovine species. Mol. Immunol. 2008;45:1153–1164. doi: 10.1016/j.molimm.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 72.Mao Y., Zhu X., Li R., Chen D., Xin S., Zhu Y., Liao X., Wang X., Zhang H., Yang Z. Methylation analysis of CXCR1 in mammary gland tissue of cows with mastitis induced by Staphylococcus aureus. Genet. Mol. Res. 2015;14:12606–12615. doi: 10.4238/2015.October.19.4. [DOI] [PubMed] [Google Scholar]
- 73.Pokorska J., Dusza M., Kułaj D., Żukowski K., Makulska J. Single nucleotide polymorphisms in the CXCR1 gene and its association with clinical mastitis incidence in Polish Holstein-Friesian cows. Genet. Mol. Res. 2016;15:gmr15017573. doi: 10.4238/gmr.15027247. [DOI] [PubMed] [Google Scholar]
- 74.Zhou L., Wang H., Ju Z., Zhang Y., Huang J., Qi C., Hou M., An L., Zhong J., Wang C. Association of novel single nucleotide polymorphisms of the CXCR1 gene with the milk performance traits of Chinese native cattle. Genet. Mol. Res. 2013;12:2725–2739. doi: 10.4238/2013.July.30.10. [DOI] [PubMed] [Google Scholar]
- 75.Strillacci M.G., Frigo E., Schiavini F., Samoré A.B., Canavesi F., Vevey M., Cozzi M.C., Soller M., Lipkin E., Bagnato A. Genome-wide association study for somatic cell score in Valdostana Red Pied cattle breed using pooled DNA. BMC Genet. 2014;15:106. doi: 10.1186/s12863-014-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Den Breems N.Y., Nguyen L.K., Kulasiri D. Integrated signaling pathway and gene expression regulatory model to dissect dynamics of Escherichia coli challenged mammary epithelial cells. Biosystems. 2014;126:27–40. doi: 10.1016/j.biosystems.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 77.Ju Z., Jiang Q., Liu G., Wang X., Luo G., Zhang Y., Zhang J., Zhong J., Huang J. Solexa sequencing and custom micro RNA chip reveal repertoire of micro RNA s in mammary gland of bovine suffering from natural infectious mastitis. Anim. Genet. 2018;49:3–18. doi: 10.1111/age.12628. [DOI] [PubMed] [Google Scholar]
- 78.Lawless N., Foroushani A.B., McCabe M.S., O’Farrelly C., Lynn D.J. Next generation sequencing reveals the expression of a unique miRNA profile in response to a gram-positive bacterial infection. PLoS ONE. 2013;8:e57543. doi: 10.1371/journal.pone.0057543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L., Huang J., Zhang X., Ju Z., Qi C., Zhang Y., Li Q., Wang C., Miao W., Zhong J. One SNP in the 3′-UTR of HMGB1 gene affects the binding of target bta-miR-223 and is involved in mastitis in dairy cattle. Immunogenetics. 2012;64:817–824. doi: 10.1007/s00251-012-0641-1. [DOI] [PubMed] [Google Scholar]
- 80.Fiser A. Template-based protein structure modeling. Comput. Biol. 2010;373:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Materials.