Abstract
CRISPR-Cas13 systems have been developed for precise RNA editing, and can potentially be used therapeutically when temporary changes are desirable or DNA editing is challenging. We identified and characterized an ultra-small family of Cas13b - Cas13bt - that can mediate mammalian transcript knockdown. We engineered compact variants of REPAIR and RESCUE RNA editors by functionalizing Cas13bt with adenosine and cytosine deaminase domains, and demonstrated packaging of the editors within a single AAV.
RNA-targeting CRISPR-Cas13 systems have been harnessed for a variety of applications1, including programmable RNA editing2,3. RNA editing is a promising therapeutic strategy that allows for installation of temporary, non-heritable edits. However, therapeutic delivery of Cas13-based RNA editing systems remains challenging, in part because the size of Cas13-based RNA editors developed so far exceed the packaging capacity of adeno-associated virus (AAV), the most widely used viral vector for gene delivery4,5.
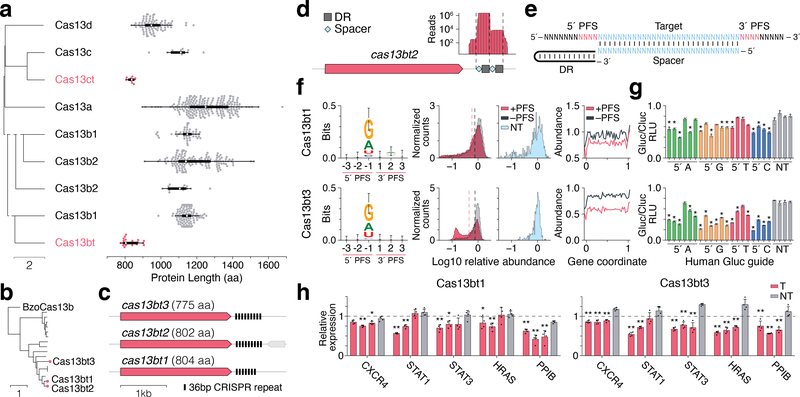
To overcome this limitation, we performed an iterative HMM profile search of Cas13s in prokaryotic and viral genomes and metagenomes, identifying 5843 candidates. Phylogenetic analysis revealed novel groups of ultra-compact Cas13 proteins that form distinct branches within the Cas13b and Cas13c subtypes (Fig. 1a, b, Supplementary Fig. 1a, b), hereafter referred to as Cas13bt and Cas13ct, respectively. Unlike other CRISPR-Cas13b systems6, the genomic loci encoding the Cas13bt subfamily lack any accessory genes (Fig. 1c). Relative to BzoCas13b (1224 aa), the smallest Cas13bts (775–804 aa) have 26 large (>5aa) deletions that total 408 aa (Supplementary Fig. 1c). As Cas13b’s are more active than Cas13c’s in mammalian systems and support programmable RNA editing2, we focused our analysis on a group of 16 ultra-compact Cas13bt’s (Cas13bt1 to Cas13bt16).
Figure 1 |. Cas13bt is a functional family of small Cas nucleases.
(A) UPGMA dendrogram and protein size distribution of Cas13 variants (new subfamilies in red). Box plots show mean (white circle), lower and upper quartiles (thick line), and 1.5 times the interquartile range (thin line). From top to bottom n = 87, 12, 10, 223, , 49, 152, 16, 123, 22.
(B) Phylogenetic tree of Cas13bt proteins (experimentally studied proteins in red).
(C) Cas13bt locus organization.
(D) Cas13bt2 crRNA expression from E. coli heterologous expression.
(E) Schematic of PFS orientation relative to target sequence.
(F) E. coli essential gene screen shows Cas13bt1 (top) and Cas13bt3 (bottom) mediate interference with a weak 5´ D (A/G/T) PFS. Left: Weblogo of PFS preference based on top 1% of most depleted sequences, top n = 304, bottom n = 313). Total height corresponds to mean entropy, error bars as in ref. 15. Middle: Histograms of the distribution of fold depletion (log10 relative abundance) of both targeting and non-targeting spacers. Dashed lines represent the mean log10 relative abundance. Right: line plots showing relative abundance in final library of spacers targeting regions across normalized positions in the target transcript.
(G) Evaluation of Cas13bt1 and Cas13bt3 for knockdown of luciferase reporter in HEK293FT cells. All values were normalized to a transfection control with crRNA alone. Data are presented as mean ± SD, n = 4. T: targeting crRNA, NT: non-targeting crRNA, RLU: relative luminescence units. *P < 0.05 via two-tailed t-test incorporating Bonferroni correction.
(H) qPCR measurement of Cas13bt1- and Cas13bt3-mediated knockdown of endogenous transcripts in HEK293FT cells (mean ± SD; n = 4 biological replicates, each average of 4 technical replicates), normalized to a transfection control with crRNA alone via ddCt correction. Statistical significance was assessed using two-tailed t-tests compared to the negative control condition for each gene, respectively, *P < 0.05, **P < 0.01.
See Table S11 for p-values where relevant. SD: standard deviation.
To experimentally characterize Cas13bt, we first identified the required CRISPR RNA (crRNA) components. We transformed E. coli with a plasmid containing one of the Cas13bt loci, Cas13bt2, with its CRISPR array truncated to two direct repeats (DRs) and performed small RNA sequencing to determine the configuration of the mature crRNA. In agreement with previously characterized Cas13b proteins, we found that the crRNA of Cas13bt2 also has a 3´ DR (Fig. 1d). To determine if Cas13bt proteins are also capable of mediating crRNA-guided RNA targeting, we performed an RNA interference screen using a library of crRNAs that were programmed to target essential gene transcripts in E. coli (Supplementary Fig. 2a)6. Two of the three tested members of the Cas13bt subfamily, Cas13bt1 and Cas13bt3, mediated depletion of targeting spacers in E. coli (Fig. 1f, Supplementary Fig. 2b–d). Analysis of the sequence flanking sites targeted by depleted crRNAs revealed that both Cas13bt1 and Cas13bt3 have a permissive 5´ D (A/G/T) protospacer flanking sequence (PFS) preference (Fig. 1f and Supplementary Fig. 3). Additionally, crRNAs targeting the 5´ UTR and beginning of the coding sequence (CDS) were more depleted (Fig. 1f).
Cas13s have previously been reported to exhibit collateral RNA cleavage activity upon crRNA-guided binding of their ssRNA target7. In vitro evaluation of Cas13bt3 showed that Cas13bt also performs target-specific collateral RNA cleavage, and that this collateral activity is mediated by the HEPN domains (Supplementary Fig. 4b). This target-specific collateral activity may render this subfamily amenable for use in diagnostic platforms such as SHERLOCK8.
To evaluate the efficacy of Cas13bt-mediated knockdown of RNA in human cells, we tested Cas13bt1 and Cas13bt3 using a set of 20 crRNAs targeting a Gaussia luciferase (Gluc) mRNA. We found that both proteins promoted crRNA-guided Gluc knockdown in HEK293FT cells (Fig. 1g). Catalytically inactivating the HEPN domains of Cas13bt1 and Cas13bt3 abolished their RNA knockdown activity (Supplementary Fig. 4c). Further, we found that the PFS preference detected in E. coli was not required for crRNA-guided RNA knockdown in HEK293FT cells, similar to other Cas13’s (Fig. 1g)2. Both Cas13bt1 and Cas13bt3 also mediated crRNA-guided knockdown of endogenous transcripts in HEK293FT cells (Fig. 1h, Supplementary Fig. 5).
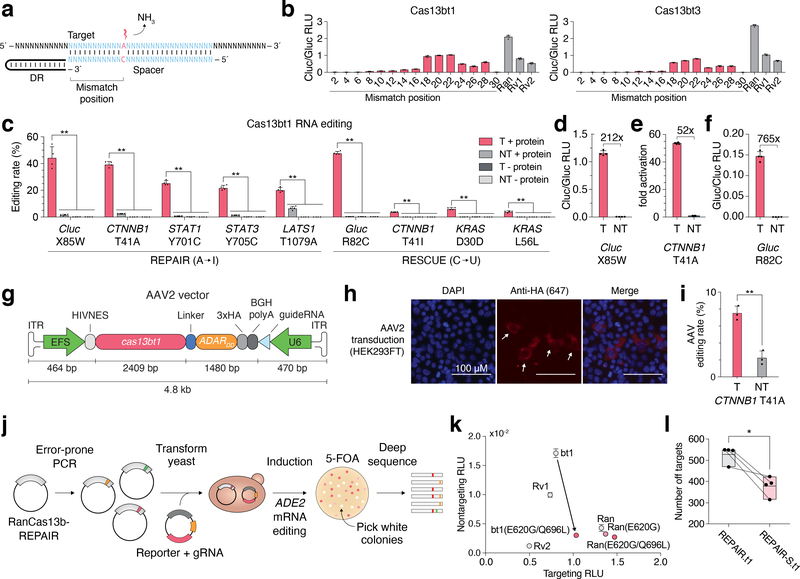
To develop Cas13bt’s for RNA editing, we fused catalytically inactive versions of Cas13bt1 and Cas13bt3 with a hyperactive mutant of the RNA adenosine deaminase ADAR2 catalytic domain (ADAR2dd(E188Q)) to construct REPAIR.t1 and REPAIR.t3, respectively. We evaluated the ability of these programmable adenosine deaminases to revert a Trp85Stop mutation in the Cypridina luciferase (Cluc) mRNA by introducing a specific A-to-I mutation. Targeted RNA editing is achieved when ADAR2 deaminates a specific mismatched adenosine on the target RNA within the RNA duplex formed between the target RNA and the crRNA spacer (Fig. 2a)2,9. Previous studies have shown that the location of the mismatched target adenosine within the target RNA-crRNA duplex can dramatically affect the RNA editing efficiency. To establish optimal parameters for crRNA design, we tested a range of mismatch positions within the target RNA-crRNA duplex2,3. We found that for the crRNA we used to target Cluc, both REPAIR.t1 and REPAIR.t3 showed optimal editing when the mismatched target adenosine is positioned within 18–22 nt of the 5´ end of the target site. Editing efficiency was comparable to the previously described REPAIRv1 and v2 systems, consisting of dPspCas13b fused to either ADAR2dd(E488Q) or ADAR2(E488Q/T375G) respectively, and lower than dRanCas13b fused to ADAR2dd(E488Q) (Fig. 2b)2,3.
Figure 2 |. RNA editing with Cas13bt.
(A) Schematic of crRNA design for RNA editing. Mismatch at target adenosine shown in red. Mismatch position is measured from the 5´ end of the target.
(B) Evaluation of RNA editing of a W85X Cypridina luciferase reporter in HEK293FT cells. Data are presented as mean ± SD, n = 4 for REPAIR.t1 and n = 3 for REPAIR.t3. Rv1: REPAIRv1; Rv2: REPAIRv22; Ran: RanCas13b-REPAIR3.
(C) Quantification of RNA editing by REPAIR.t1 and RESCUE.t1 by deep sequencing. Statistical significance for interaction of +/– protein and targeting versus non-targeting crRNA was assessed by 2-way ANOVA followed by post-hoc analysis using Games-Howell pairwise comparison. **P < 0.01. For this panel and panels D-F, values show mean ± SD, n = 4, and T: targeting crRNA, NT: Non-targeting crRNA.
(D) Quantification of Cluc restoration by REPAIR.t1.
(E) Quantification of beta-catenin reporter activation of REPAIR.t1.
(F) Quantification of Gluc restoration by RESCUE.t1.
(G) Schematic of AAV2 vector. ITR: inverted terminal repeat; EFS: short Ef1-alpha promoter.
(H) Representative immunofluorescence staining of HEK293FT cells transduced with AAV2 encoding REPAIR.t1 (n = 2).
(I) Quantification of RNA editing of CTNNB1 in HEK293FT cells by AAV2-mediated expression of REPAIR.t1. T: targeting crRNA, NT: Non-targeting crRNA. Data are presented as mean ± SD, n = 3, Statistical significance was assessed with a two-tailed t-test, **P < 0.01.
(J) Schematic of a directed evolution approach for engineering specific ADAR2dd variants.
(K) Evaluation of specificity-enhancing ADAR2dd mutants applied to REPAIR.t1 targeting the W85X mutation in a Cypridina luciferase reporter. bt1: REPAIR.t1; other construct abbreviations are as in (B). Data are presented as mean ± standard deviation of non-targeting RLU, n = 3.
(L) Quantitative comparison of off-target editing between REPAIR.t1 variants. REPAIR-S: ADAR2dd(E488Q/E620G/Q696L); REPAIR: ADAR2dd(E488Q). Data are presented as mean with top and bottom boundaries denoting minimum and maximum, n = 4. Statistical significance was assessed using a two-tailed paired t-test, *P < 0.05. Paired samples are shown by connected lines between the two conditions.
See Table S11 for exact p-values where relevant. SD: standard deviation.
We additionally constructed cytosine RNA editors with an evolved ADAR2dd capable of cytidine deamination3 (RESCUE.t1 and RESCUE.t3) and directed both editors to reporter and endogenous transcripts in HEK293FT cells. We found that these fusion proteins were capable of mediating C-to-U editing of all targets (Fig. 2c–f and Supplementary Fig. 6) and can edit when the mismatch is positioned between 14–28 nt of the 5´ end of the target site.
To demonstrate the ability of Cas13bt-REPAIR to edit functionally relevant targets, we targeted RNA regions corresponding to previously characterized phosphorylation sites3. In particular, we attempted to alter activation of the Wnt/beta-catenin pathway by editing Thr41 codon of CTNNB1, a site known to promote degradation of beta-catenin when phosphorylated10. We found that REPAIR.t1 achieved 40% editing at this site, converting the codon to alanine (A) and leading to a 52-fold increase in beta-catenin signaling as measured by a beta-catenin driven (TCF/LEF) luciferase reporter (see Methods), which may be relevant for applications such as promoting regeneration after acute liver failure11,12 (Fig. 2c, e). REPAIR.t1 was also able to efficiently edit sites corresponding to phosphorylated residues in the STAT1, STAT3, and LATS1 transcripts as measured by targeted RNA sequencing (Fig. 2c).
To assess the potential of using Cas13bt fusion proteins in a single viral vector, we packaged REPAIR.t1 along with a crRNA expression cassette in a recombinant AAV2 vector and delivered the system to HEK293FT cells (Fig. 2g). Immunofluorescence staining demonstrated that REPAIR.t1 delivered by AAV was expressed and localized to the cytoplasm with the help of a nuclear export signal (Fig. 2h, Supplementary Fig. 7). RNA sequencing showed site specific A-to-I editing of 7.5%±0.8% at the CTNNB1 Thr41 site following enrichment for transduced cells (see Methods) (Fig. 2i). This demonstrates the feasibility of using a single AAV genome for delivery. However, as editing rates achieved using AAV2 delivery is lower than plasmid DNA transfection, further optimization would likely be needed to boost the editing efficiency, such as targeting the editing machineries to specific subcellular compartments to achieve the optimal editing rate for specific target transcripts. When attempting in vivo delivery to specific tissues, AAV serotypes should be chosen to maximize delivery and expression efficiency, for example AAV8 for liver13 and PHP.eB for brain in C57BL/6 mice14.
Finally, we quantified the transcriptome-wide specificity of REPAIR.t1 and found the number of off-target edits introduced by this system was comparable to REPAIRv1 (Supplementary Fig. 8). Although off-target edits in RNA are transient, they may lead to deleterious protein products which may cause adverse effects. To improve the specificity of REPAIR.t1, we used a yeast-based directed evolution approach to identify two promising mutations in ADAR2dd (E620G and Q696L) (Fig. 2j, Supplementary Figs. 9,10) that reduce promiscuous deamination activity. We incorporated these two mutations in REPAIR.t1 and found that the number of off-target edits decreased without reducing the on-target activity (Fig. 2k, l, Supplementary Fig. 8). With additional mutagenesis and optimization, it may be possible to even further reduce off-target activity for off-target sensitive applications.
The small size of Cas13bt proteins provides new opportunities for programmable RNA modulation, particularly in vivo. We have shown here that Cas13bt1 and 3 can be used to generate compact REPAIR and RESCUE constructs compatible with delivery by AAV, thereby further advancing the development of programmable RNA editing technologies.
METHODS
Data curation and search pipeline
The bioinformatics analysis in this study were performed on contigs from public sequence databases including National Center for Biotechnology Information’s prokaryotic and virus databases (http://ncbi.nlm.nih.gov) and Whole Genome Shotgun database (https://www.ncbi.nlm.nih.gov/genbank/wgs/), MG-RAST (https://www.mg-rast.org/)16, and the Department of Energy Joint Genome Institute’s online resources (http://jgi.doe.gov)17,18, totaling 3.17 trillion bp. All open reading frames larger than 80 aa were annotated resulting in 10 billion putative proteins for further analysis. Previously developed Cas13 profiles19 were used to identify Cas13 family proteins with HMMER3.220 using a minimum bit-score threshold of 25. A group of small (~800aa) but divergent Cas13b’s were identified and used to generate a new profile for a second HMMER search with the same settings to retrieve additional members of this subfamily. In total, 5843 Cas13 loci were identified.
Phylogenetic analysis
For phylogenetic analysis and classification, the 5843 candidate genes were clustered using MMseqs2 with a minimum sequence identity of 50% and minimum coverage of 70%21,22. Proteins within each cluster were clustered at 90% identity and 80% minimum coverage for redundancy reduction. Each redundancy-reduced cluster was aligned using MAFFT23 with default parameters. Proteins identified as truncated or partial on the basis of partial match to a larger protein in an alignment, and/or clusters entirely composed of partial/truncated proteins were removed from the analysis.
The aligned redundancy reduced clusters were converted into HHsuite profiles using all columns with less than 50% gaps, and each of these profiles was searched against each other with profile-profile alignment using HHsearch24. The resulting pairwise bit-scores between clusters, sij, where i,j denote clusters i and j, respectively, were used to construct a classification dendrogram. First, the asymmetric bit-scores were symmetrized by setting sij = (sij + sji)/2. Then, pseudo-distances were calculated by setting dij = - (log sij - log min(sii , sjj))/2 to generate a distance matrix25. A UGPMA dendrogram for classification was constructed using these distances. Branches and the subtrees of the dendrogram were contracted without modifying their topology to highlight known subtypes and subgroups within each subtype. Lengths in amino acids (aa) of the redundancy reduced proteins from each subtree were used to generate protein size distributions.
Non-redundancy reduced Cas13bt proteins, along with one Cas13b protein sequence (BzoCas13b) as an outgroup were aligned using MAFFT. A phylogenetic tree was constructed using FastTree26, and the tree was rooted using the outgroup.
Design and cloning of bacterial expression plasmid constructs
All cloning in this study was performed using chemically competent Stbl3 E. coli (NEB) unless otherwise noted. All PCR for cloning was performed using 2X Phusion Flash High-Fidelity Master Mix (Thermo Fisher) unless otherwise noted.
The Cas13bt2 full locus was synthesized and cloned into the BamHI site of pACYC184 by GenScript.
To clone bacterial expression plasmids for the PFS screen, Cas13bt protein coding sequences were human codon optimized using GeneArt GeneOptimizer (Thermo Fisher) and synthesized by GenScript into a pcDNA3.1(+) backbone. Genes were amplified by PCR to add a pLac promoter and cloned into a pBR322 backbone (NEB) digested with EcoRV (Thermo Fisher) by Gibson assembly.
crRNA expression cassettes for each DR corresponding to each Cas13bt of interest were synthesized by IDT, amplified by PCR, and cloned into a pACYC184 backbone digested with EcoRV and BamHI (Thermo Fisher) by Gibson assembly. All primers are listed in Supplementary Table 4 and final constructs in Supplementary Table 10.
Design and cloning of mammalian expression plasmid constructs
Mammalian crRNA expression cassettes were amplified from pC0048 (Addgene plasmid # 103854; http://n2t.net/addgene:103854; RRID:Addgene_103854)2 using primers to add the DR for each Cas13bt ortholog of interest and cloned into pC0048 digested with LguI and KpnI (Thermo Fisher) using Gibson assembly.
Mammalian protein expression cassettes were cloned by amplifying previously mentioned synthesized Cas13bt genes by PCR and cloning into pC0053 (Addgene plasmid # 103869; http://n2t.net/addgene:103869; RRID:Addgene_103869)2 digested with HindIII and NotI (Thermo Fisher), either alone or with ADAR2dd(E488Q) amplified from pC0053 for REPAIR constructs and pC0078 (Addgene plasmid # 130661; http://n2t.net/addgene:130661; RRID:Addgene_130661)3 for RESCUE constructs. Site directed mutagenesis was used to create catalytically inactivated Cas13bt’s. ADAR2 mutants derived from directed evolution screens were cloned by introduction of mutations via PCR primers. All primers are listed in Supplementary Table 4 and final constructs in Supplementary Table 10.
crRNA spacers were cloned into expression backbones by Golden Gate assembly as previously described27. Spacer sequences are listed in Supplementary Tables 6 and 8.
Bacterial RNA sequencing
Bacterial RNA sequencing was performed as previously described28. Briefly, 5 mL overnight cultures of a Stbl3 E. coli colony transformed with a plasmid containing the locus of interest was spun down and resuspended in 1 mL of TRI Reagent (Zymo Research). After a 5-minute room temperature incubation, 250 uL of 0.5 mm Zirconia beads was added and the Trizol resuspension was vortexed vigorously for 30s to 1 min. 200 uL chloroform was added, samples were inverted gently, incubated at room temperature for 3 minutes, and then spun down at 12000xg for 5 min at 4°C. Following centrifugation, the aqueous fraction was used as input to the Qiagen miRNeasy kit, as per the manufacturer’s instructions.
Purified RNA was treated with DNase I (NEB), purified again using RNA Clean & Concentrate-25 (Zymo Research), and treated with T4 polynucleotide kinase (PNK) (NEB). PNK-treated RNA was again purified using RNA Clean & Concentrate-25 (Zymo Research), and ribosomal RNA was removed using the Ribominus Transcriptome Isolation Kit (Yeast and Bacteria) (Thermo Fisher). Samples were subsequently treated with RNA 5´ polyphosphatase (Epicentre) and purified again using an RNA Clean & Concentrate-5 kit (Zymo Research). Purified RNA was used as input to the NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB). Library preparation was performed as per the manufacturer’s instructions, except with a final PCR of 20 cycles. Libraries were quantified by qPCR using the KAPA Library Quantification Kit for Illumina (Roche) on a StepOnePlus Real-Time PCR System (Thermo Fisher) and sequenced on an Illumina NextSeq. Reads were mapped using BWA.
E. coli essential gene PFS screen
Libraries were designed and cloned as previously described6. Briefly, the library of spacers was cloned into each Cas13bt pJ23119-spacer-DR backbone containing a chloramphenicol resistance gene using Golden Gate assembly with a 5:1 ratio of spacer library to pre-digested backbone with 210 cycles. Libraries were transformed into Endura Electrocompetent Cells (Lucigen) by electroporation and plated over five 22.7cmx22.7cm chloramphenicol LB agar plates. 12 hours after plating, libraries were scraped from plates and DNA was extracted using the Macherey-Nagel Nucleobond Xtra Maxiprep Kit (Macherey-Nagel).
200 ng of library plasmid and 200 ng Cas13bt gene plasmid containing an ampicillin resistance gene were transformed into 100 uL of Endura Electrocompetent Cells (Lucigen) by electroporation as per the manufacturer’s protocol and plated across four 22.7cmx22.7cm ampicillin/chloramphenicol LB agar plates per biological replicate, with three biological replicates per condition. 10–12 hours post-transformation, libraries of transformants were scraped from the plates and DNA was extracted using the Macherey-Nagel Nucleobond Xtra Maxiprep Kit (Macherey-Nagel). Libraries were prepared from extracted DNA for next generation sequencing using primers in Supplementary Table 5 with NEBNext High-Fidelity 2X PCR Master Mix (NEB) and sequenced on an Illumina NextSeq.
Spacers with 100 or more total reads across all replicates in the control sample were retained for analysis. Average spacer abundance (fraction of reads relative to all reads) was calculated as the average abundance of each spacer across all replicates. Relative abundance was calculated as the ratio of the spacer abundance in the experimental condition relative to the spacer abundance in the input library. To account for off-target cleavage activity, a two-component Gaussian mixture distribution was fit to the log10 relative abundance distribution of non-targeting (negative control) spacers, and the Gaussian component with the higher mean, m, was used as the null distribution as it contains fewer non-targeting guides with off-target activity. log10 abundances for all spacers in the Cas13bt library were normalized by subtracting off m (see Supplementary Fig. 2b). Significantly depleted spacers were selected as those with normalized log10 relative abundances below −5σ, where σ is the standard deviation of the null distribution. Weblogos were then generated (https://weblogo.berkeley.edu/logo.cgi)15 using all significantly depleted spacers and separately using the top 1% most depleted spacers of all spacers.
To analyze gene position preference, all spacers were searched against their respective CDS targets in E. coli using the genome NC_010473.1 to identify their coordinates relative to the respective CDS. Gene coordinates were normalized by the CDS length. Bins of normalized gene coordinates from −0.1 to 1.1 were generated with a step size of 0.025. For each bin, and each CDS, the average relative abundance of spacers with start positions that fall in the bin was computed. Then the overall abundance for the bin was taken to be the average relative abundance for each CDS in the bin averaged across all CDSs, excluding CDSs with no spacers in the given bin. This analysis was performed using Python and is available via Github.
Mammalian cell culture and transfection
Mammalian cell culture experiments were performed in the HEK293FT line (Thermo Fisher catalog #R70007) grown in Dulbecco’s Modified Eagle Medium with high glucose, sodium pyruvate, and GlutaMAX (Thermo Fisher), additionally supplemented with 1× penicillin–streptomycin (Thermo Fisher), 10 mM HEPES (Thermo Fisher), and 10% fetal bovine serum (VWR Seradigm). All cells were maintained at confluency below 80%.
All transfections were performed with Lipofectamine 2000 (Thermo Fisher) in 96-well plates unless otherwise noted. Cells were plated at approximately 20,000 cells/well 16–20 hours prior to transfection to ensure 90% confluency at the time of transfection. For each well on the plate, transfection plasmids were combined with OptiMEM I Reduced Serum Medium (Thermo Fisher) to a total of 25 μl. Separately, 23 μl of OptiMEM was combined with 2 μl of Lipofectamine 2000. Plasmid and Lipofectamine solutions were then combined and pipetted onto cells.
Mammalian RNA knockdown assays
HEK293FT cells were transfected as described with 75 ng of a plasmid encoding either a Cas13bt ortholog or GFP expressed from a CMV promoter, 150 ng of a plasmid encoding a crRNA expressed from a human U6 promoter and, where relevant, 45 ng of reporter plasmid. After 48 h, RNA was harvested as described previously27 with 2x the amount of recommended DNase and a 20 minute lysis step. RNA expression was measured by qPCR using commercially available TaqMan probes (Thermo Fisher) (Supplementary Table 7) on a LightCycler 480 II (Roche) with GAPDH as an endogenous internal control in 5 uL multiplexed reactions. Probes and primer sets were generally selected to amplify across the Cas13 target site so as to minimize detection of cleaved transcripts. Each of the 4 biological replicate is the average of 4 technical qPCR replicates, and relative expression was calculated using the ddCt method29 with a negative control condition consisting of the corresponding crRNA expression plasmid co-transfected with the GFP expression plasmid rather than a Cas13bt expression plasmid. Negative control values are the average of 4 biological replicates. Statistical significance was assessed using a two-tailed t-test.
For luciferase reporter assays, media was aspirated from cells and Cypridina and Gaussia luciferase activity in the media was measured as relative luminescence units (RLU) using Gaussia and Cypridina Luciferase Assay Kits (Targeting Systems) with an injection protocol on a Biotek Synergy Neo 2 (Agilent). Each experimental luciferase measurement was normalized to the appropriate control luciferase measurement (i.e., if Cypridina luciferase was targeted, the Gaussia luciferase measurement was used as the control value and vice versa). For knockdown assays, normalized luciferase values were then again normalized to an average normalized luciferase measurement of 4 biological replicates of a negative control condition consisting of the corresponding crRNA expression plasmid co-transfected with a GFP expression plasmid rather than a Cas13 expression plasmid. Error bars were calculated in GraphPad Prism 7 and represent the standard deviation of the luciferase values normalized to negative control transfection, n = 4. Statistical significance was assessed by pairwise comparison of each targeting condition with each of the 4 non-targeting conditions using a two-tailed T-test. The maximum p-value was taken from each of the 4 pairwise comparisons for each targeting condition, and significance was assessed by comparison using an experiment-wide critical value of α=0.05 with Bonferroni correction using n = 4.
Mammalian RNA editing assays
HEK293FT cells were transfected as described with 150 ng of plasmid encoding a dCas13b ortholog-ADAR2dd(E488Q) fusion expressed from a CMV promoter, 300 ng of a plasmid encoding a crRNA expressed from a human U6 promoter and, where relevant, 45 ng of a reporter plasmid. After 48 h, RNA was harvested as described above and reverse transcription was performed as described27 using gene-specific primers for the relevant target transcript (Supplementary Table 9). cDNA was used as input for library preparation of next-generation sequencing libraries (Supplementary Table 5) using NEBNext High-Fidelity 2X PCR Master Mix (NEB), and amplicons were sequenced on an Illumina MiSeq. Editing was quantified by counting the number of reads at which the expected edited position in the amplicon was called as a G (for A-to-I editing) or T (for C-to-U editing) and dividing by the total number of reads in the sample using Python (available via Github). Unless otherwise noted, all reported data is the average of 4 biological replicates.
For AAV-transduced cells, 250 uL of AAV2 was mixed with 750 uL media, and media on HEK293FT cells was replaced with AAV-containing media solutions in 12-well plates. After approximately 40 hours, RNA was harvested as previously described30. Briefly, cells were dissociated from plates with TrypLE (Thermo Fisher) to a single cell suspension and fixed with fixation buffer (4% paraformaldehyde, 0.1% saponin, 1:100 SuperaseIn RNAse inhibitor (Invitrogen) in phosphate buffered saline (PBS)) for 30 min at 4°C. Cells were then stained with anti-HA primary antibodies (Roche Anti-HA High Affinity 3F10 rat IgG and Cell Signaling Technologies Anti-HA C29F4 rabbit mAb) at 1:100 dilution each in staining buffer (0.1% saponin, 1% BSA, 1:100 SuperaseIn RNAse inhibitor in PBS), followed by staining with secondary antibodies (AlexaFluor 488 Goat anti-rabbit and AlexaFluor 555 Goat anti-rat, Invitrogen) at 1:2000 dilution in staining buffer, each for 30 min at 4°C. Cells were then sorted by FACS for HA-positive cells and de-crosslinked in RecoverAll digestion buffer with proteinase K (Invitrogen) for 3 hours at 50°C. RNA was extracted using a QuickRNA microprep kit (Zymo Research) and libraries were prepared as described above.
Luciferase reporter assays for RNA editing were performed as described above, with the modification that normalized luciferase values were not normalized to a GFP control condition. For CTNNB1 targeting, we used a previously reported engineered luciferase reporter generated by replacing the EF1alpha promoter driving Gaussia luciferase expression in our dual luciferase reporter plasmid with a promoter derived either from an M50 Super 8X TOPFlash (TOP) or M51 Super 8X FOPFlash (FOP) reporter3. M50 Super 8x TOPFlash (Addgene plasmid # 12456; http://n2t.net/addgene:12456; RRID:Addgene_12456) and M51 Super 8x FOPFlash (TOPFlash mutant) (Addgene plasmid # 12457; http://n2t.net/addgene:12457; RRID:Addgene_12457) were gifts from Randall Moon31. 8x TOPFlash and FOPFlash reporters contain 8 beta catenin binding sites either intact (TOP) or scrambled (FOP); the TOPFlash reporter thus provides a metric of beta-catenin activation when compared to background as measured by the FOPFlash reporter with otherwise similar conditions. Luciferase activity was measured for these custom dual luciferase reporters for each protein/crRNA condition and normalized as described for a dual luciferase reporter. Fold activation was calculated by taking the ratio of the average TOP measurement and dividing by average FOP measurement, and error was calculated by a standard error propagation formula.
Optimal spacers for all target sites tested were determined by tiling spacers across the site of interest, varying the distance of the mismatch from the DR from 14 bp to 28 bp in intervals of 2 bp.
Statistical significance between +/– protein and targeting/non-targeting crRNA conditions was assessed using two-way ANOVA followed by post-hoc analysis using Games-Howell pairwise comparison.
RNA editing specificity
HEK293FT cells were transfected as described for mammalian RNA editing assays. After 48 h, RNA was harvested using a QIAGEN RNeasy Plus 96 kit as per the manufacturer’s protocol. The mRNA fraction was enriched using an NEBNext Poly(A) Magnetic Isolation Module (NEB). Libraries were prepared using an NEBNExt Ultra II Directional RNA library prep kit (NEB) as per the manufacturer’s protocol and sequenced on an Illumina NextSeq. Each sample was sequenced with an average read depth of 8 million reads per sample and randomly downsampled to 5 million reads per sample. Data was analyzed using a previously described custom pipeline on the FireCloud computational framework and downstream analysis was performed using a previously described custom Python script2,3. Any significant edits found in eGFP-transfected conditions were considered to be SNPs or artifacts of the transfection and filtered out. An additional layer of filtering for known SNP positions was performed using the Kaviar method for identifying SNPs32.
AAV production
Recombinant AAV2 vectors packaging base editing and GFP control cassettes described above were generated using triple plasmid transfection in HEK293T cells as described previously33. Briefly, 60 ug Adenoviral helper genes (Addgene plasmid #112867; http://n2t.net/addgene:112867; RRID:Addgene_112867, a gift from James M. Wilson), 50 ug AAV2 Rep/AAV2 Cap (Addgene plasmid # 104963; http://n2t.net/addgene:104963; RRID:Addgene_104963, a gift from Melina Fan), and 30 ug ITR-flanked transgene were transfected into five 15 cm plates of HEK293T cells. Media and cells were harvested 5 days post transfection. Media was then combined with PEG8000 in 2.5M NaCl at a 4:1 ratio, incubated overnight at 4°C, and centrifuged at 3000xg for 30 min at 4°C to pellet the precipitate. Cell pellets were sonicated to lyse cells and release viral particles. Combined sonicated cell pellets and media precipitate were digested with DNAse for 1 hour at 37°C before iodixanol gradient ultracentrifugation. Iodixanol fractions of 17%, 25%, 40%, and 60% were prepared as previously described, and viral preparations were loaded on top of the gradient34. Following 20 hours of ultracentrifugation at 28000 RPM, gradients were fractionated, and peak fractions at the 40%/60% interface were subject to buffer exchange and concentration with Pierce Protein Concentration Columns according to manufacturer instructions (Thermo Fisher).
For determination of viral titers, viral preparation samples were treated with DNAse (10mg/mL) to remove unencapsidated viral genomes. Following inactivation of DNAse with 0.5 M EDTA, samples were digested with Proteinase K to release encapsidated viral genomes. Viral DNA was then measured against a vector standard (Addgene #37825-AAV2) by quantitative PCR with primers against the ITRs (5´-AACATGCTACGCAGAGAGGGAGTGG-3´ and 5´-CATGAGACAAGGAACCCCTAGTGATGGAG-3´) using a Roche Lightcycler 480 II (Roche).
DATA AVAILABILITY
Deep sequencing data from whole transcriptome sequencing was deposited as a BioProject under Project ID PRJNA641934. Data from main figures are available in the Supplementary Information section.
CODE AVAILABILITY
All Python scripts used for data analysis are available in a GitHub repository found here: https://github.com/fengzhanglab/Cas13bt-analysis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Eloe-Fadrosh for generously sharing JGI data from study Ga0114919; D.L. Valentine and J. Tarn for generously sharing JGI data from study Ga0180434; E. Puccio for assistance with AAV production; F.E. Demircioglu for assistance with protein purification; members of the Zhang lab for advice and discussions, R. Macrae for discussion and editing of the manuscript, and R. Belliveau for technical support. S.K. is supported by a National Science Foundation Graduate Research Fellowship. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201), HHMI, the Open Philanthropy Project, The G. Harold and Leila Y. Mathers, Bill and Melinda Gates, and Edward Mallinckrodt, Jr. Foundations, the Poitras Center for Psychiatric Disorders Research at MIT, the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT, the Yang-Tan Center for Molecular Therapeutics, and by Lisa Yang, the Phillips family, and J. and P. Poitras.
Footnotes
COMPETING INTERESTS
F.Z. is a co-founder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences. S. K, H. A and F.Z are co-inventors on US provisional patent application 62/905,645 relating to the Cas proteins described in this manuscript. The remaining authors declare no competing interests.
REFERENCES
- 1.Terns MP CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol. Cell 72, 404–412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox DBT et al. RNA editing with CRISPR-Cas13. Science (80-. ). 1027, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abudayyeh OO et al. A cytosine deaminase for programmable single-base RNA editing. Science (80-. ). 365, 382–386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong JY, Fan PD & Frizzel RA Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther 7, 2101–12 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, Yang H & Colosi P Effect of genome size on AAV vector packaging. Mol. Ther 18, 80–86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smargon AA et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. 5573, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gootenberg JS et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science (80-. ). 360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews MM et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol 23, 426–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonald BT, Tamai K & He X Wnt/β-Catenin Signaling: Components, Mechanisms, and Diseases. Dev. Cell 17, 9–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte U et al. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am. J. Pathol 175, 1056–1065 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhushan B et al. Pro-regenerative signaling after acetaminophen-induced acute liver injury in mice identified using a novel incremental dose model. Am. J. Pathol 184, 3013–3025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattenhorn LM et al. Adeno-Associated Virus Gene Therapy for Liver Disease. Hum. Gene Ther 27, 947–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 16.Keegan KP, Glass EM & Meyer F MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol. Biol 1399, 207–233 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Nordberg H et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42, 26–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen IMA et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shmakov SA, Makarova KS, Wolf YI, Severinov KV & Koonin EV Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. U. S. A 115, E5307–E5316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy SR Accelerated profile HMM searches. PLoS Comput. Biol 7, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinegger M & Söding J MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Steinegger M & Söding J Clustering huge protein sequence sets in linear time. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinegger M et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 473, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova KS et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol 18, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS & Arkin AP FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joung J et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zetsche B et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD & Livak KJ Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Hrvatin S, Deng F, O’Donnell CW, Gifford DK & Melton DA MARIS: Method for analyzing RNA following intracellular sorting. PLoS One 9, 1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veeman MT, Slusarski DC, Kaykas A, Louie SH & Moon RT Zebrafish Prickle, a Modulator of NoncanonicalWnt/Fz Signaling, Regulates Gastrulation Movements. Curr. Biol 13, 680–685 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Glusman G, Caballero J, Mauldin DE, Hood L & Roach JC Kaviar: An accessible system for testing SNV novelty. Bioinformatics 27, 3216–3217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen S, Troupes AN, Pulicherla N & Asokan A Multiple Roles for Sialylated Glycans in Determining the Cardiopulmonary Tropism of Adeno-Associated Virus 4. J. Virol 87, 13206–13213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grieger JC, Choi VW & Samulski RJ Production and characterization of adeno-associated viral vectors. Nat. Protoc 1, 1412–1428 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing data from whole transcriptome sequencing was deposited as a BioProject under Project ID PRJNA641934. Data from main figures are available in the Supplementary Information section.