Brief Summary
Humans harbor a large quantity of microbes in the intestinal tract and have evolved symbiotic relationships with many of them. Several specific bacterial pathobionts have been associated with liver disease pathogenesis. Although bacteriophages (phages) and eukaryotic viruses (collectively known as “the virome”) outnumber bacteria and fungi in the intestine, little is known about the intestinal virome in patients with liver disease. Here, we will summarize changes in the fecal virome associated with fatty liver diseases and cirrhosis. As natural predators of bacteria, phages can precisely edit the bacterial microbiota. We will describe the potential of phages to target specific bacterial pathobionts as a novel treatment approach for liver disease and describe challenges to clinical applications.
Keywords: mycobiome, virome, phageome, cirrhosis, gut-liver-axis
1. Introduction
Work over the last few decades have increasingly shed light on the myriad of ways in which the different microbial communities that colonize our guts are influencing human health and disease risk [1]. These microbial communities are composed of bacteria, fungi, viruses, and archaea that together encode over a hundred-fold more genes than the human genome [2]. The composition of the our gut microbiome is significantly influenced even from birth by our surrounding environment and these encoded genes in turn have the potential for both benefit and harm [3]. Most of our understanding of how the gut microbiota affects human disease has been focused on bacteria, but with new advances in metagenomic methods, viruses are beginning to receive more attention.
The human intestinal virome is made up of about 90% bacteriophages (phages or prokaryotic viruses) and 10% eukaryotic viruses [4]. Eukaryotic DNA and RNA viruses include plant and mammalian viruses and can have intestinal cells as their host. Some eukaryotic viruses can affect human health by causing disease, like the well-known enteric pathogens Norovirus, Rotavirus, and Enterovirus, while others are not pathogenic. Plant viruses are likely derived from the diet [5]. On the other hand, the phageome consists of approximately 1015 bacteriophages and is largely composed of the order Caudovirales (family Siphoviridae, Myoviridae, and Podoviridae) and family Microviridae [6]. In healthy subjects, the intestinal viral microbiome exhibits a high level of interpersonal heterogeneity with relative intrapersonal stability [7, 8]. However, changes in the virome community can be seen with changes in lifestyle such as diet and with different disease states [8, 9]. Deep sequencing of the intestinal viral microbiome in healthy individuals suggests that there is a small core group of phages shared among a majority of people, with a wider range of more rare phages that are unique to individuals [10]. Understanding how the composition of the intestinal viral microbiome differs amongst individuals with different disease states will help elucidate the mechanism by which the viral microbiome influences disease. Already, differences in the viral microbiome have been implicated in the pathogenesis of obesity, type 2 diabetes, colon cancer, inflammatory bowel disease, and more.
Here, we will review current literature focused on the human intestinal virome in liver disease. An estimated 1.5 billion people have chronic liver disease worldwide and an estimated 1.2 million cirrhotics will die per year, making it one of the leading causes of death globally [11]. Our existing strategies for reversing or preventing progression of liver disease are limited and often liver transplantation is the only therapy available to patients once they progress to end-stage liver disease. In recent years, we have improved our understanding of how the intestinal microbiome contributes to liver disease and with that, there is increased interest in targeting the intestinal microbiome to treat liver disease. Hence, we will also review the use of phage therapy in gastrointestinal and liver disease and summarize the key bacteria of interest in various liver diseases that may serve as potential targets for phage therapy in the future.
2. Intestinal virome in patients with liver disease
Both eukaryotic viruses and bacteriophages have been implicated in liver disease pathogenesis. Among the eukaryotic viruses are the known pathogenic and hepatotropic viruses, hepatitis A (Picornaviridae family) and E (Hepeviridae family) viruses, which can be transmitted by the oral-fecal route and detected in stool. Both HAV and HEV exist in two forms, a non-enveloped form comprised of a capsid surrounding the RNA genome and a quasi-enveloped form that is masquerading in a layer of host cell membrane [12]. The non-enveloped form, which is found in the stool and saliva of infected individuals, can survive harsh conditions such as transit through the gastrointestinal tract and cross the intestinal barrier into the blood via mechanisms not yet well understood. Recent work suggests that once in the blood, HAV harnesses endosomal gangliosides to infect hepatocytes and Kupffer cells [13], where it replicates and exits back into the bloodstream in its quasi-enveloped form, which camouflages its antigenic proteins from neutralizing antibodies [14]. Other known eukaryotic viruses that can be found in the intestinal virome and cause liver injury include Epstein-Barr virus (EBV), Cytomegalovirus (CMV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [15–17].
Recent work by Jiang et al. investigating the intestinal virome in patients with alcoholic hepatitis and alcohol use disorder demonstrated that variations in intestinal viral taxa are associated with disease severity and mortality [18]. Comparing the intestinal viromes of patients with alcoholic hepatitis, alcohol use disorder, and controls, fecal samples from patients with alcohol use disorder had significantly higher viral diversity and richness compared to controls, and this was generally correlated with lower bacterial diversity. In alcoholic hepatitis patients, Escherichia-, Enterobacteria-, and Enterococcus phages were overrepresented compared to controls, while Parabacteroides phages were underrepresented. Further, increased abundance of Staphylococcus phages and Citrobacter phages were associated with higher disease severity.
Aside from differences in phage composition, fecal samples from patients with alcoholic hepatitis also contained significantly more mammalian viruses, such as those from the Parvoviridae and Herpesviridae families, than controls. Herpesviridae was only present in fecal samples from alcoholic hepatitis patients, with most of the assigned reads attributed to EBV [18]. It is unclear why EBV is only detected in the guts of patients with alcoholic hepatitis, though a possible hypothesis is suppression of immunosurveillance in these patients. Alternatively, EBV reactivation might predispose the development of hepatitis in alcoholic patients. Notably, a study of the intestinal virome in nonalcoholic fatty liver disease (NAFLD) did not observe increased proportions of mammalian viruses in these patients as compared to controls [19]. Further studies are needed to confirm and characterize the intestinal mammalian virus population in patients with alcohol-associated liver disease as this may shed light on its pathogenesis.
Another difference noted between NAFLD and alcohol-associated liver disease is that patients with NAFLD and fibrosis had significantly lower intestinal viral diversity and proportionately fewer phages compared to controls [19]. Incorporating fecal viral diversity with clinical data into a model to non-invasively predict histologic fibrosis severity significantly improved its diagnostic accuracy compared with clinical data alone. Additionally, the abundance of several Lactococcus and Leuconostoc phages were inversely correlated with severity of liver fibrosis, whereas the abundance of Lactobacillus phage was positively correlated with severity of liver fibrosis. Though the abundance of some phages were inversely correlated with their respective bacterial hosts, the viral diversity did not correlate with bacterial diversity. It is difficult to draw conclusions regarding how liver disease affects the phage/bacteria relationship with data from a single timepoint.
One study evaluated phage/bacteria interactions across two timepoints in compensated cirrhotics before and after 8 weeks of treatment with rifaximin [20]. This study found a significant reduction in the genus-level richness of the bacterial but not viral population after rifaximin use. Decreased complexity of bacterial-phage interactions was also seen after rifaximin, with complete collapse of bacterial-phage interactions seen in phages directed against pathobionts such as Streptococcus, Pseudomonas, and Enterobacteriaceae spp. These changes are most likely secondary to the direct impact of rifaximin on the bacterial population, and it is unclear how much cirrhosis contributed to these dynamics. Cross-sectional analysis revealed that phage-bacterial correlation network complexity was highest in controls, lowest in cirrhotics taking only lactulose, and improved in cirrhotics taking both lactulose and rifaximin [20]. A notable technical difference between these studies is that this study performed metagenomic sequencing of fecal DNA whereas the prior two studies used filtration techniques to isolate RNA- and DNA-containing viral particles from stool, followed by metagenomic sequencing.
Research on the intestinal virome is in its infancy and a causative link between changes in the phageome and disease has not been established. It remains to be seen whether changes in the virome are drivers of disease, or whether they are the result of disease. Future longitudinal studies are required to confirm virome changes in independent cohorts of patients, and to test the stability of the fecal virome and its correlation with liver disease severity over time. The analysis of the virome depends on metagenomic sequencing, methods for virome research have not been standardized, and only a small fraction of all sequences can be assigned to known viral taxa in public databanks. Improvements in bioinformatic analysis will lead to a better understanding of phage-bacteria interaction dynamics. This will allow us to answer the question whether changes in phages are drivers for bacterial dysbiosis or vice versa.
3. Phage biology and their therapeutic potential
3.1. Bacteriophages - natural predators of bacteria.
Phages are viruses that infect bacteria and are considered to be the most numerous group of viruses on the planet with an estimated 1031 total phage particles [21]. Shortly after the discovery of phages by Frederick Twort in 1915 [22], they were used to treat bacterial hemorrhagic dysentery. Phage therapy is the practice of using preparations of infectious phages to treat bacterial infections, which has the advantage over antibiotics of targeting specific bacterial species or strains while self-replicating and spreading to infect additional target bacterial cells. Phage therapy became very popular throughout the world to treat a wide range of diseases caused by both Gram-positive and Gram-negative pathogens, such as Staphylococcus, Streptococcus, Vibrio, Klebsiella, Enterobacter, Shigella, Escherichia, Pseudomonas and Providencia to name a few [23]. Commercial production of phage cocktails was initiated in France by what would later become L’Oreal [24], followed by the Eliava Institute of Bacteriophage, Microbiology and Virology (EIBMV) (Tbilisi, Georgia) and the Hirszfeld Institute of Immunology and Experimental Therapy (HIIET) (Wroclaw, Poland). In the US, pharmaceutical giant Eli Lilly (Indianapolis, IN) produced seven phage cocktails [24].
After the discovery and use of antibiotics, phage therapy fell out of favor in many western countries, particularly the US. Much of the concern regarding the efficacy of phages as a therapeutic stemmed from reproducibility issues where the same successful cocktail of phages used on one patient did not work for all patients. This was presumably due to the narrow host range of the selected phages. Another problem was inflammatory responses to the phage cocktail [25]. This was more likely due to contamination of the lysate by bacterial endo and exotoxins used to grow the phages in production rather than an immune response to the phages. Likewise, there was concern that the rapid lysis of cells by phage-encoded lytic enzymes can cause septic shock; however, this argument also applies to bactericidal antibiotics.
Phages come in all shapes and sizes with genomes consisting of either double-stranded or single-stranded DNA or RNA, which were originally used to define them into 21 morphotypes before nucleic acid sequencing technologies were used for taxonomic classification [26, 27]. Phages can be tailed, polyhedral, filamentous, pleomorphic, enveloped or not, but it is the double-stranded DNA tailed phages, in the order Caudovirales, that are the most common and are the phages commonly used for therapeutic purposes. In addition to morphotype and nucleic acid type, there are two main lifestyle categories of phages, lytic (a.k.a., virulent, obligately lytic) and temperate. Lytic phages can replicate only via the lytic life cycle that ends with the destruction of the infected bacterial cell and release of progeny phages (Figure 2A) while temperate phages are capable of choosing between the lytic and lysogenic life cycles (Figure 2B). The latter includes the integration of phage DNA into the bacterial chromosome and its passive replication [28–30]. Temperate phages show reduced lytic abilities, may incorporate and transfer (i.e. transduce) bacterial DNA, including drug resistance and pathogenicity genes, and can convert a bacterium into a “lysogen” (i.e., bacterial cell with a viral genome integrated into the bacterial chromosome) that becomes immune to superinfection by the same phage or related phages. Therefore, temperate phages have historically not been used as therapeutics [31]. However, temperate phages can be modified to become obligately lytic [32, 33] and since temperate phages are commonly found in bacteria, their modification could expand the arsenal of therapeutic phages, at least for some pathogens with limited numbers of or a lack of lytic phages [34].
Figure 2. Bacteriophage life cycles.
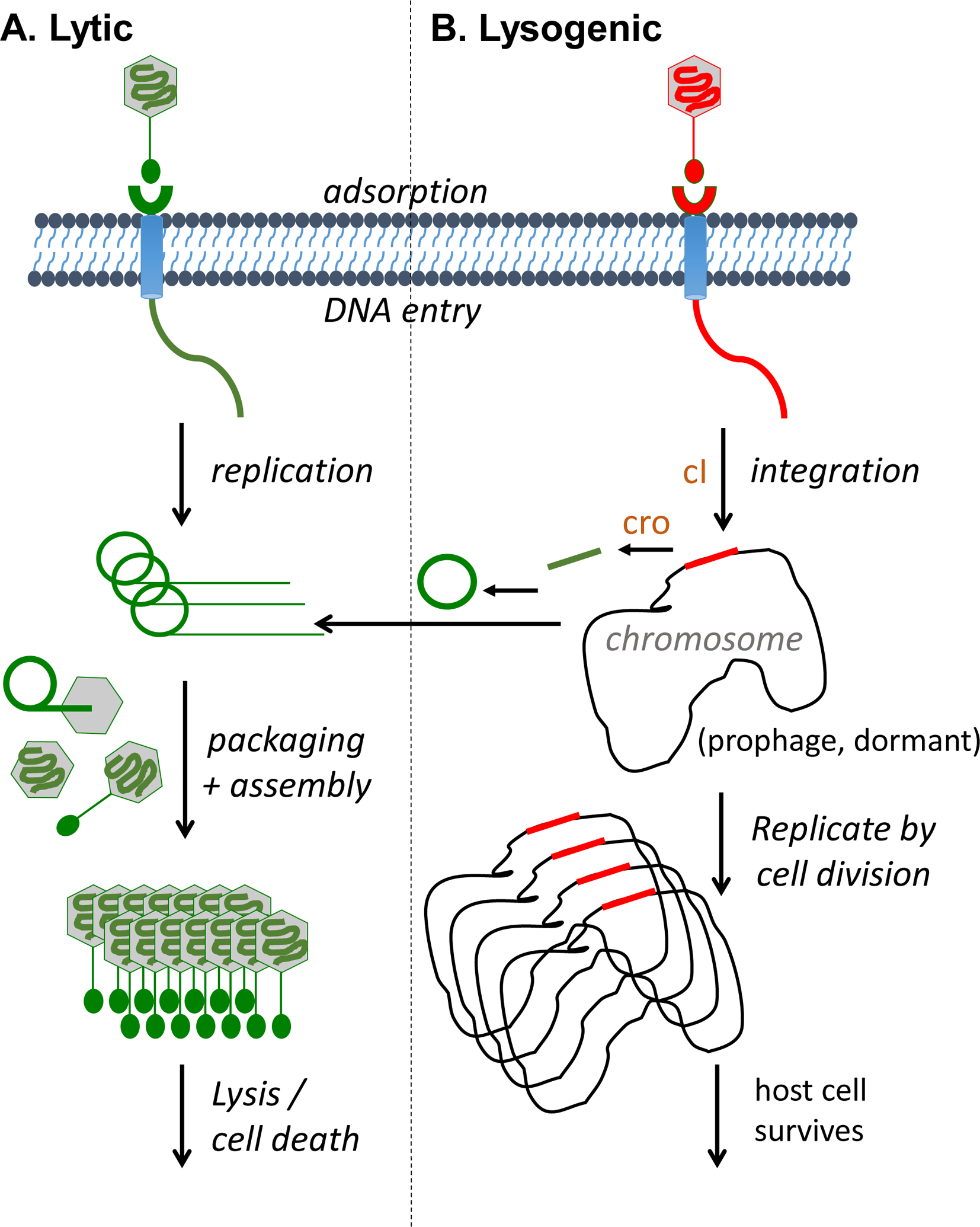
Depicted are the two main types of tailed phage life cycles, lytic (A) and lysogenic (B). Lytic growth consists of replicating the genome, expressing structural proteins, packaging the genome into particles, assembling the mature virions, and ending in lysis and cell death of the host bacterium. In contrast, lysogenic growth is a storage state whereby the phage integrates its genome into the bacterial host chromosome surviving by vertical transmission through host cell division. Lysogenic phages can also undergo lytic growth under certain conditions. Proteins related to the repressors cI and cro (brown color) are responsible for switching from lysogenic to lytic development.
Although the host range of a phage tends to be quite narrow (e.g., strain/serotype- or species-specific) [35], there are lytic phages that can infect more distantly related bacteria [36]. Host specificity is largely determined by receptor-binding proteins (RBPs), a.k.a. anti-receptors [37–43]. These phage-encoded proteins enable high affinity binding of phage virions to receptors located on the outer surface of bacterial cells such as lipopolysaccharide (LPS), lipoteichoic acid, capsular polysaccharide, flagella and pili [44]. Swapping domains or altering the sequence of specific regions of RBPs has resulted in altered host range specificity to enable the phage to attach to different strains or different bacterial genera [45–48]. Phages typically encode a single RBP, but can encode more than one, resulting in polyvalency (i.e., the ability to bind to more than one receptor and potentially more than one host organism/strain) [49].
3.2. Resurgence of phage therapy
The widespread overprescribing of antibiotics by physicians coupled with the overuse of antibiotics in the livestock industry are two key factors that are thought to have led to the global spread of antibiotic-resistant bacteria [50, 51]. This poses a serious public health problem since there are few, or in some cases, no drugs available to treat life-threatening bacterial infections. Antibiotics are not entirely safe either, as they can cause allergic reactions and severe side effects, including organ damage and a clearing of the normal commensal gut microbiota, leaving the gut vulnerable to secondary infections by opportunistic pathogens such as Clostridioides difficile [52]. Because phages are host specific rather than broad spectrum like many antibiotics, phage therapy has the potential to have fewer off-target effects on beneficial bacterial microbiome species. Phages are now being used to treat livestock infections [53–55], to prevent food spoilage [56–61], in human compassionate use cases, and in clinical trials (see below).
3.3. Phage therapy in gastrointestinal diseases
Over the last two decades, several clinical trials have been performed with T4-like phages (Table 1). Oral administration of phage cocktails is considered to be safe in both healthy adults and children, with only occasional side effects independent of phage dosage [62–65]. To determine the safety and efficacy of phage therapy on treating gastrointestinal infections, a T4-like coliphage cocktail was orally given to children hospitalized with acute diarrhea for four days. Non-bacterial causes of diarrhea were not ruled out. No adverse events were reported, suggesting the overall safety of the phage cocktail [66]. However, substantial intestinal replication of phages was not observed, and phage treatment did not show improvement over placebo control in quantitative diarrhea parameters, such as stool output and frequency [66]. This could be explained by the fact that only half of the patients actually harbored Escherichia coli (E. coli) strains susceptible to the administered phages, and E. coli only represented 5% of the total fecal bacteria. Overall, this trial confirmed the safety of phage treatments in children with diarrhea. Though the trial failed to show efficacy, this could potentially be explained by low E. coli abundance in the stool samples or symptoms caused by a non-bacterial source such as viral gastroenteritis.
Table 1:
Overview of recent studies of phage-related treatment in gastrointestinal diseases (English literature only)
| Type of study | Phage (target) | Dose and method | Subject | Result and conclusion | Reference |
|---|---|---|---|---|---|
| Clinical trial | Phage T4 (E. coli) | 105 PFU/ml, dose A, 103 PFU/ml, dose B, Oral administration | 15 healthy individuals | Safe, but E. coli abundance not changed | Bruttin [62] |
| Clinical trial | T4-like Phages (E. coli) | 3×109 PFU/ml, dose A, 3×107 PFU/ml, dose B, Oral administration | 15 healthy individuals | Safe, gut microbiota profile not affected | Sarker [63] |
| Clinical trial | Commercial phage cocktail ColiProteus (E. coli) | 20ml for adults, 10ml for children, and 10-fold dilution Oral administration | 5 healthy adults, 10 healthy children | Overall safe, with occasional reported side effect independent of dosage | McCallin [110] |
| Clinical trial | T4-like Phages or ColiProteus (E. coli) | 108 or 106 PFU for older children (T4-like phages), 107 or 105 PFU for younger children (T4-like phages), 5×108 or 109 PFU for all (ColiProteus), Oral administration |
20 older children, 20 younger children | Both cocktails are safe | Sarker [65] |
| Clinical trial | T4-like Phages or ColiProteus (E. coli) | 3.6×108 PFU (T4-like phages), 1.4×109 PFU (ColiProteus), Oral administration | 120 children with diarrhea | Safe, but lack of efficacy | Sarker [111] |
| Clinical case report | Phage cocktail (A. baumannii) | 5×109 PFU Intracavitary and Intravenous | 68-year-old male with necrotizing pancreatitis complicated by pancreatic pseudocyst | Patient completely recovered | Schooley [67] |
| Clinical trial | Phage cocktail PreforPro (E. coli) | One 15mg capsule, Oral administration | 32 healthy individuals with mild to moderate gastrointestinal distress | Safe and tolerable, but no difference from placebo | Gindin [112] |
| Clinical trial | Phage cocktail PreforPro (E. coli), together with probiotics Bifidobacterium animalis subspecies lactis strain BL04 | One 15mg capsule, Oral administration | 68 healthy individuals with mild to moderate gastrointestinal distress | Safe and tolerable, but no compelling evidence of efficacy | Grubb [113] |
| Preclinical study | Phage cocktail (adherent-invasive E. coli) | 3×107 PFU Oral administration | Wild-type mice colonized with 108 CFU of adherent-invasive E. coli, Dextran sodium sulfate-induced colitis | Fecal E. coli level decreased, dextran sodium sulfate-induced colitis ameliorated | Galtier [70] |
| Preclinical study | Phage cocktail (E. faecalis) | 1010 PFU Oral administration | Gnotobiotic mice colonized with stool samples from cytolysin-positive patients with alcoholic hepatitis, Ethanol-induced liver disease | Fecal E. faecalis level decreased, ethanol-induced liver disease ameliorated | Duan [87] |
One successful case was reported in 2016, in which a 68-year-old male patient was suffering from necrotizing pancreatitis complicated by a pancreatic pseudocyst infected with multi-drug-resistant Acinetobacter baumannii [67]. Phages were applied by intracavitary and intravenous routes, and the patient completely recovered after five months [67]. Although this is only a case report, the obvious clinical improvements suggest that phage therapy might be useful for treating bacterial infections, and especially those caused by multidrug-resistant bacteria.
Phage therapy may also be a promising way to precisely edit the gut microbiota. Two randomized, placebo-controlled trials have been performed to determine the safety and efficacy of phages in adults suffering mild to moderate gastrointestinal distress (e.g., gas, bloating, diarrhea, constipation, etc) (NCT03269617; NCT04511221). Over the 28-day study, oral administration of the coliphage cocktail was considered to be safe and tolerated. Patients experienced a similar reduction in gastrointestinal symptom severity during both the treatment and placebo periods, suggesting that the phage therapy was ineffective, but there was also no evidence that patients’ initial symptoms were secondary to overgrowth of the bacteria targeted by the phages administered. Future studies can evaluate phage therapy efficacy by documenting the interactions of phages in these cocktails with the specifically targeted bacterial strains obtained from the subjects.
In addition to these clinical trials, there are also some preclinical with encouraging results (Table 1). Adherent-invasive E. coli (AIEC) have been shown to be involved in the pathogenesis of inflammatory bowel diseases [68, 69]. Administration of a phage cocktail against these E. coli strains reduced intestinal AIEC colonization in transgenic mice expressing human AIEC receptor [70]. Furthermore, wild-type mice colonized with AIEC were protected from dextran sodium sulfate-induced colitis upon phage treatment, with less E. coli in feces, as well as in ileal and colonic sections [70]. To evaluate the ability of phage cocktail in targeting AIEC strains in patients, ileal biopsies from patients with Crohn’s disease were spiked with an AIEC strain. Active phage replication was detected 5 hours and 24 hours after phage administration, confirming the killing potential of phages in such environment [70]. A phase 1/2a placebo-controlled clinical trial was therefore initiated, to assess the safety and efficacy of the phage cocktails in patients with inactive Crohn’s Disease (NCT03808103).
4. Potential for phage therapy in liver disease
4.1. Known bacterial pathobionts driving liver disease as potential targets for phage therapy
Our existing knowledge of the taxonomic differences in the bacterial microbiota of patients with liver disease can help guide further investigation into potential targets for phage therapy. In the following subsections, we summarize fecal bacteria changes in selected human liver diseases (Table 2).
Table 2:
Bacterial genera and species with known correlations to different etiologies of liver disease
| Increased | Decreased | |
|---|---|---|
| Nonalcoholic fatty liver disease | ||
| Alcohol-associated liver disease | ||
| Autoimmune hepatitis |
|
|
| Primary sclerosing cholangitis | ||
| Cirrhosis |
4.1.1. Non-alcoholic fatty liver disease (NAFLD)
NAFLD is a spectrum of disease beginning with excessive fat deposition in the liver in the absence of significant alcohol use that can progress to liver inflammation, known as non-alcoholic steatohepatitis (NASH), and eventually fibrosis [71]. Several studies have found a decreased fecal abundance of Faecalibacterium and specifically Faecalibacterium prausnitzii in both obese and non-obese NASH patients [72–74]. Ruminococcus was enriched in obese NASH patients in one study [75], but reduced in other studies of obese [73] and non-obese NASH patients [74]. Ruminococcus obeum was specifically found to be reduced in NAFLD patients in one study [76]. Advanced fibrosis secondary to NASH is associated with an overall decrease in intestinal bacterial diversity and an increase in the relative abundance of Gram-negative bacteria such as Bacteroides and Escherichia [76–80]. Although no causative role of these bacterial strains for steatohepatitis has been demonstrated in preclinical models, Yuan et al. demonstrated that an ethanol-producing Klebsiella pneumoniae (K. pneumoniae) strain was present in 60% of a Chinese cohort of NAFLD patients. Introduction of this strain into mice induced steatohepatitis [81].
4.1.2. Alcohol-associated liver disease
Heavy alcohol use leads to a spectrum of liver disease beginning with steatosis, which can be reversible, but can progress to steatohepatitis and fibrosis in susceptible patients [82]. Studies of the intestinal bacterial microbiome in patients with alcohol-associated have revealed enrichment of Enterobacteriaceae [83, 84] and a reduction of Lactobacillus [84], Bacteroidetes [83, 85], and Akkermansia [86]. A recent study by Duan et al. demonstrated that patients with alcoholic hepatitis have an increased relative abundance of Enterococcus faecalis (E. faecalis) and specifically a strain that secrets the exotoxin cytolysin. Presence of cytolysin-secreting E. faecalis correlated with the severity of liver disease and with mortality in patients with alcoholic hepatitis, and oral administration of cytolysin-positive E. faecalis promotes ethanol-induced liver injury in mice [87].
4.1.3. Autoimmune hepatitis
Autoimmune hepatitis is a chronic inflammatory liver disease with poorly understood pathogenesis, though genetic susceptibility and loss of tolerance against liver antigens are proposed mechanisms [88]. Patients with autoimmune hepatitis have an overrepresentation of potential pathobionts, including Veillonella species such as Veillonella dispar, in their fecal microbiomes [89]. Translocation of Enterococcus gallinarum (E. gallinarum) to the liver triggered an autoimmune response in mice genetically predisposed to autoimmunity. Subsequent antibiotic treatment prevented the formation of pathogenic autoantibodies and T cells, thus improving mortality [90]. E. gallinarum DNA was detected in the livers of most of patients with autoimmune hepatitis but in none of the healthy control livers [90].
4.1.4. Primary sclerosing cholangitis (PSC)
Primary sclerosing cholangitis (PSC) is a cholestatic liver disease characterized by inflammation of the bile ducts leading to stricturing and sclerosis and eventually progressive biliary fibrosis and cirrhosis. Several recent studies compared the fecal bacterial microbiota of patients with PSC and healthy controls [91]. Patients with PSC are consistently shown to have lower bacterial microbiome diversity than healthy controls. Additionally, Veillonella has been consistently shown by many studies to be enriched in the stool of PSC patients compared to healthy controls [92–97]. Enterococcus, Streptococcus and Lactobacillus are also frequently enriched in PSC patients, whereas there is a relative depletion of short chain fatty acid (SCFA)-producing Firmicutes, such as Faecalibacterium and Coprococcus. Germ-free mice inoculated with fecal matter from patients with PSC were more susceptible to hepatobiliary injury by diethyldithiocarbamate and harbored K. pneumoniae, Proteus mirabilis, and E. gallinarum in their mesenteric lymph nodes [98]. Further, specific K. pneumoniae strains could induce pore formation on human intestinal epithelial organoids, suggesting that increased bacterial translocation could be a potential mechanism of increased susceptibility to hepatobiliary injury [98].
4.1.5. Cirrhosis
Patients with cirrhosis have decreased proportions of beneficial, autochthonous taxa, such as Lachnospiraceae and Ruminococcaceae and instead have an overrepresentation of potentially pathogenic bacteria such as Enterobacteriaceae, Staphylococcaceae, and Enterococcaceae, whose abundance correlates with disease progression and endotoxemia [85, 99, 100]. Another study found higher relative abundance of bacteria normally associated with oral flora in the intestinal microbiome of cirrhotics, as well as increased Veillonella and Streptococcus species as compared to controls [2]. Changes in intestinal bacterial microbiome composition have also been correlated with severity of liver disease. The ratio of autochthonous taxa such as Ruminococcaceae, Lachnospiraceae, and Clostridiales to non-autochthonous ones such as Enterobacteriaceae and Bacteroidaceae was much higher in healthy individuals than cirrhotics and decreased with higher MELD score and degree of hepatic decompensation [101]. Moreover, increased relative abundance of pathogenic bacteria was associated with the development of complications such as hepatic encephalopathy.
Cirrhotic patients not only have increased relative abundance of pathogenic bacteria in their intestinal bacterial microbiomes, they also have increased risk of bacterial translocation, a process which bacteria migrate from the intestinal lumen to extraintestinal sites [102]. Aerobic Gram-negative bacteria, such as E. coli, K. pneumoniae, Pseudomonas aeruginosa, and other Enterobacteriaceae, translocate much more readily than anaerobic bacteria [103]. Notably, these species have also been implicated in decompensation of cirrhosis [104]. Increasingly, studies are comparing cirrhotic patients based on etiology of cirrhosis and while many intestinal bacterial communities are shared across the spectrum of liver disease etiologies, differences in the patterns of dysbiosis across the different etiologies of cirrhosis may provide a better understanding of the mechanisms underlying these associations.
4.2. Preclinical phage utilization in liver disease
Although no clinical trial using phage therapy for patients with liver disease has been published, two preclinical studies used phage therapy to treat liver disease. Duan et al. demonstrated that intestinal levels of E. faecalis are significantly increased in patients with alcoholic hepatitis [87]. Furthermore, the presence of a specific strain of E. faecalis that produces the bacterial exotoxin cytolysin correlates with increased severity of disease and mortality in patients with alcoholic hepatitis. Transplantation of feces from cytolysin-positive patients with alcoholic hepatitis worsened ethanol-induced liver disease in gnotobiotic mice, whereas treatment of these mice with specific phages targeting cytolytic E. faecalis by oral gavage, reversed the exacerbation of liver disease. No improvement in liver disease was seen in the gnotobiotic mice treated with phages targeting non-cytolytic E. faecalis. This preclinical study demonstrates the utility of targeting specific species of the intestinal bacterial microbiome to modify disease progression.
Another study showed that selective elimination of the ethanol-producing K. pneumoniae strain using phages prior to fecal transplantation into mice prevented development of diet-induced steatohepatitis [81]. These studies are good examples of how elimination of pathobionts by phages can improve liver disease in mouse models.
5. Conclusion and future directions
Recent advances in the field of microbiota research have identified a few bacterial strains that correlate with liver disease in patients and that are causatively linked to disease pathogenesis, as targets for therapy. Despite the renewed interest in phage therapy, there are many roadblocks preventing phage therapy from being the standard of care. One major roadblock is the narrow host range of phages, which limits wide therapeutic utility and the use of the same phages in different patients. One possibility is to use a cocktail of multiple phages. Limited host range can also be addressed through natural or engineered alterations in phage-encoded receptor-binding proteins, capable of targeting different hosts [45–47, 49, 105–107]. To avoid using phages with undesirable off-targets (i.e., to commensal bacteria), experiments should include analysis of effects of any potential therapeutic phage on the composition of the microbiome. Phage host range will never be as broad as standard of care broad-spectrum antibiotics. In addition, phages can be made to bind multiple receptors (i.e., polyvalent), thereby extending the host range of a single virion [46]. By targeting more than one receptor, we avoid many of the obstacles that bacteria have evolved to prevent phage adsorption (e.g., mutations to or physically blocking of receptors with extracellular polysaccharides). Blocking of receptors by biofilms can be avoided by expressing an extracellular polysaccharide-degrading enzyme in the phage [108].
Other obstacles to widespread use of phage therapeutics have to do with pharmacokinetics and pharmacodynamics within the human body. For example, some phages administered systemically can be cleared rapidly from circulation [109]. It is therefore critical to determine the dose of phages being administered and their clearance from the site where they are applied to. This will allow that phages will be present at the specific site long enough to lyse bacteria.
One of the crucial factors to the success of phage therapy is to screen the patients for the presence and sufficient abundance of the targeted bacteria in the intestine and to test the susceptibility of the target bacteria against the phages. Combining this personalized treatment approach with the precise execution by phages, phage-based therapies could become powerful new drugs to treat many diseases including liver diseases.
Figure 1. Intestinal virome.
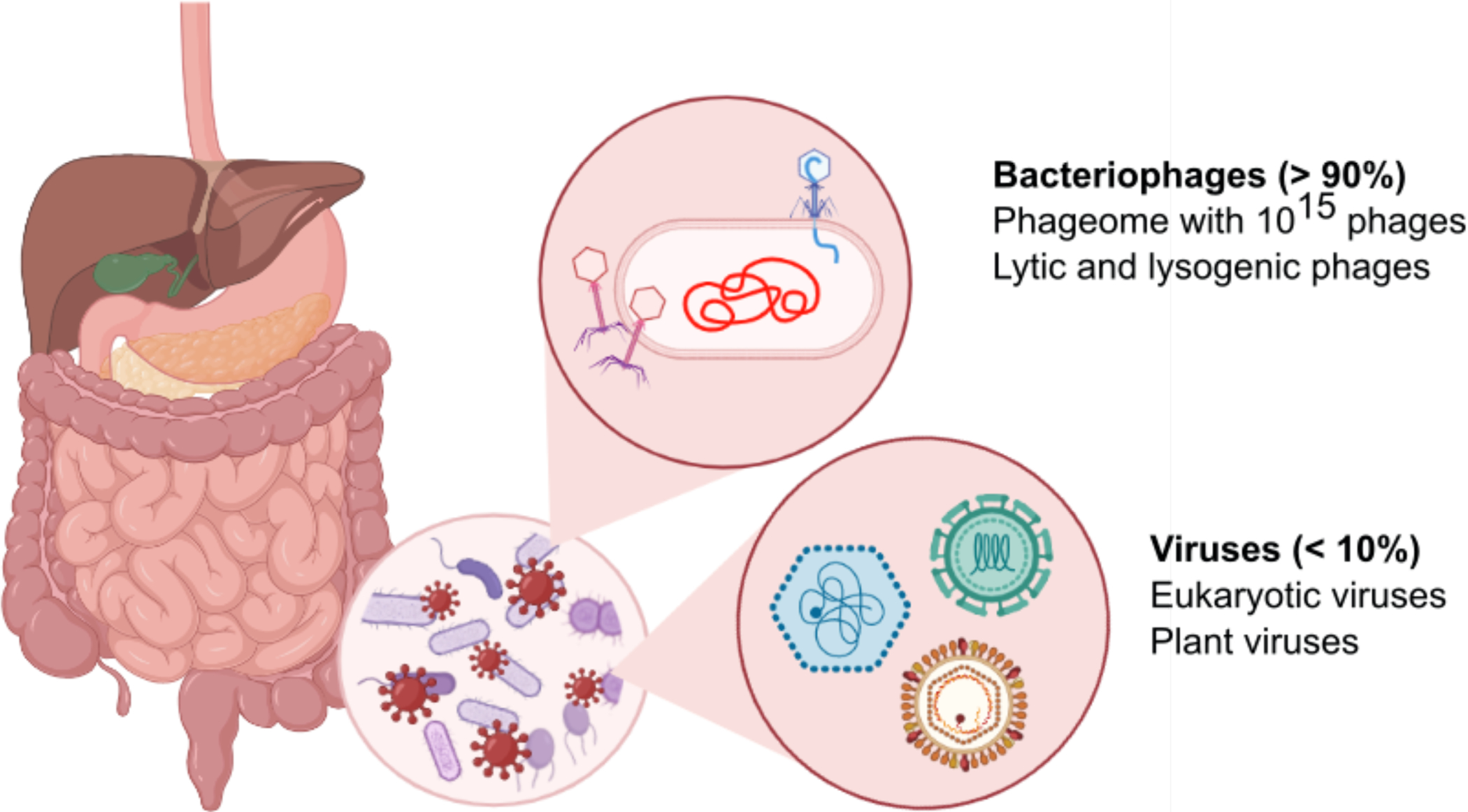
The intestine and liver are intimately connected and communicate via the portal vein and the common bile duct. The intestinal microbiota contains bacteria, fungi, archaea and viruses. Viruses in the intestinal virome are predominantly phages (also called phageome), but also contain some eukaryotic viruses. Lytic phages can lyse bacteria and contribute to changes in the bacterial microbiota. Graphic illustration was created in Biorender.com.
Key Points.
Over 90% of the human intestinal virome is composed of bacteriophages, with eukaryotic plant and mammalian viruses making up the remaining fraction.
Patients with liver disease exhibit differences in both the diversity and composition of their intestinal viromes compared to healthy control, though the impact of these differences on the bacterial microbiome needs more study.
Use of phage therapy for the treatment of multi-drug resistant infections shows promise in case reports, and several clinical trials involving phage therapy are underway.
Our knowledge of taxonomic differences in the bacterial microbiomes of patients with liver disease specific to etiology can help inform potential targets for phage therapy.
Preclinical data suggest that selective targeting of bacterial strains such as E. faecalis or K. pneumoniae by phage therapy can modify liver disease progression, such as in alcoholic hepatitis or steatohepatitis respectively.
Acknowledgements:
This review was supported in part by NIH grants R01 AA24726, R01 AA020703, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and a Biocodex Microbiota Foundation Grant (to B.S.) and services provided by NIH centers P30 DK120515 and P50 AA011999. C.H. is supported by T32 DK007202.
Conflicts of interest:
B.S. has been consulting for Ferring Research Institute, HOST Therabiomics, Intercept Pharmaceuticals, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda. B.S.’s institution UC San Diego has received research support from Axial Biotherapeutics, BiomX, CymaBay Therapeutics, NGM Biopharmaceuticals, Prodigy Biotech and Synlogic Operating Company. B.S. is founder of Nterica Bio. UC San Diego has filed several patents with B.S. as inventor related to this work.
Bibliography
- [1].Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016;375:2369–2379. [DOI] [PubMed] [Google Scholar]
- [2].Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- [3].Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. [DOI] [PubMed] [Google Scholar]
- [4].Li Y, Handley SA, Baldridge MT. The dark side of the gut: Virome-host interactions in intestinal homeostasis and disease. J Exp Med 2021;218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 2006;4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rowan-Nash AD, Korry BJ, Mylonakis E, Belenky P. Cross-Domain and Viral Interactions in the Microbiome. Microbiol Mol Biol Rev 2019;83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010;466:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 2011;21:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci U S A 2016;113:10400–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moon AM, Singal AG, Tapper EB. Contemporary Epidemiology of Chronic Liver Disease and Cirrhosis. Clin Gastroenterol Hepatol 2020;18:2650–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. Naked Viruses That Aren’t Always Naked: Quasi-Enveloped Agents of Acute Hepatitis. Annu Rev Virol 2014;1:539–560. [DOI] [PubMed] [Google Scholar]
- [13].Das A, Barrientos R, Shiota T, Madigan V, Misumi I, McKnight KL, et al. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat Microbiol 2020;5:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Webb GW, Kelly S, Dalton HR. Hepatitis A and Hepatitis E: Clinical and Epidemiological Features, Diagnosis, Treatment, and Prevention. Clin Microbiol Newsl 2020;42:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Markin RS. Manifestations of Epstein-Barr virus-associated disorders in liver. Liver 1994;14:1–13. [DOI] [PubMed] [Google Scholar]
- [16].Vento S, Cainelli F. Is there a role for viruses in triggering autoimmune hepatitis? Autoimmun Rev 2004;3:61–69. [DOI] [PubMed] [Google Scholar]
- [17].Adams DH, Hubscher SG. Systemic viral infections and collateral damage in the liver. Am J Pathol 2006;168:1057–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jiang L, Lang S, Duan Y, Zhang X, Gao B, Chopyk J, et al. Intestinal Virome in Patients With Alcoholic Hepatitis. Hepatology 2020;72:2182–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, et al. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 2020;159:1839–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bajaj JS, Sikaroodi M, Shamsaddini A, Henseler Z, Santiago-Rodriguez T, Acharya C, et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 2021;70:1162–1173. [DOI] [PubMed] [Google Scholar]
- [21].Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A 1999;96:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Twort FW. An investigation on the nature of ulta-microscopic viruses. Lancet 1915;2:1241–1243. [Google Scholar]
- [23].Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage 2011;1:66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sulakvelidze A, Alavidze Z, Morris JG Jr. Bacteriophage therapy. Antimicrob Agents Chemother 2001;45:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Adhya S, Merril CR, Biswas B. Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harb Perspect Med 2014;4:a012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ackermann HW. Frequency of morphological phage descriptions in the year 2000. Brief review. Arch Virol 2001;146:843–857. [DOI] [PubMed] [Google Scholar]
- [27].Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Echols H Developmental pathways for the temperate phage: lysis vs lysogeny. Annu Rev Genet 1972;6:157–190. [DOI] [PubMed] [Google Scholar]
- [29].Young R Bacteriophage lysis: mechanism and regulation. Microbiol Rev 1992;56:430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hobbs Z, Abedon ST. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett 2016;363. [DOI] [PubMed] [Google Scholar]
- [31].Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol 2010;11:2–14. [DOI] [PubMed] [Google Scholar]
- [32].Zhang H, Fouts DE, DePew J, Stevens RH. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology (Reading) 2013;159:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc Natl Acad Sci U S A 2018;115:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol 2015;23:185–191. [DOI] [PubMed] [Google Scholar]
- [35].Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev 2004;28:127–181. [DOI] [PubMed] [Google Scholar]
- [36].Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, et al. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol 1998;64:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang J, Hofnung M, Charbit A. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J Bacteriol 2000;182:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, et al. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J Bacteriol 2006;188:2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brondsted L. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901–1 and Tuc2009. J Bacteriol 2006;188:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, et al. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol Microbiol 2012;83:289–303. [DOI] [PubMed] [Google Scholar]
- [41].Garcia-Doval C, van Raaij MJ. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc Natl Acad Sci U S A 2012;109:9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tetart F, Repoila F, Monod C, Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol 1996;258:726–731. [DOI] [PubMed] [Google Scholar]
- [43].Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 2005;115:101–107. [DOI] [PubMed] [Google Scholar]
- [44].Lindberg AA. Bacteriophage receptors. Annu Rev Microbiol 1973;27:205–241. [DOI] [PubMed] [Google Scholar]
- [45].Ando H, Lemire S, Pires DP, Lu TK. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst 2015;1:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dunne M, Rupf B, Tala M, Qabrati X, Ernst P, Shen Y, et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep 2019;29:1336–1350 e1334. [DOI] [PubMed] [Google Scholar]
- [47].Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MT, et al. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019;179:459–469 e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Latka A, Lemire S, Grimon D, Dams D, Maciejewska B, Lu T, et al. Engineering the Modular Receptor-Binding Proteins of Klebsiella Phages Switches Their Capsule Serotype Specificity. mBio 2021;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Scholl D, Rogers S, Adhya S, Merril CR. Bacteriophage K1–5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J Virol 2001;75:2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 2009;157:2893–2902. [DOI] [PubMed] [Google Scholar]
- [51].Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 2015;109:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med 2002;346:334–339. [DOI] [PubMed] [Google Scholar]
- [53].Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM. Alternatives to antibiotics: utilization of bacteriophage to treat colibacillosis and prevent foodborne pathogens. Poult Sci 2005;84:655–659. [DOI] [PubMed] [Google Scholar]
- [54].Callaway TR, Edrington TS, Brabban AD, Anderson RC, Rossman ML, Engler MJ, et al. Bacteriophage isolated from feedlot cattle can reduce Escherichia coli O157:H7 populations in ruminant gastrointestinal tracts. Foodborne Pathog Dis 2008;5:183–191. [DOI] [PubMed] [Google Scholar]
- [55].Wall SK, Zhang J, Rostagno MH, Ebner PD. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl Environ Microbiol 2010;76:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl Environ Microbiol 2009;75:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Anany H, Chen W, Pelton R, Griffiths MW. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in meat by using phages immobilized on modified cellulose membranes. Appl Environ Microbiol 2011;77:6379–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Guenther S, Loessner MJ. Bacteriophage biocontrol of Listeria monocytogenes on soft ripened white mold and red-smear cheeses. Bacteriophage 2011;1:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Patel J, Sharma M, Millner P, Calaway T, Singh M. Inactivation of Escherichia coli O157:H7 attached to spinach harvester blade using bacteriophage. Foodborne Pathog Dis 2011;8:541–546. [DOI] [PubMed] [Google Scholar]
- [60].Guenther S, Herzig O, Fieseler L, Klumpp J, Loessner MJ. Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int J Food Microbiol 2012;154:66–72. [DOI] [PubMed] [Google Scholar]
- [61].Hooton SP, Atterbury RJ, Connerton IF. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int J Food Microbiol 2011;151:157–163. [DOI] [PubMed] [Google Scholar]
- [62].Bruttin A, Brüssow H. Human Volunteers Receiving Escherichia coli Phage T4 Orally: a Safety Test of Phage Therapy. Antimicrobial Agents and Chemotherapy 2005;49:2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sarker SA, McCallin S, Barretto C, Berger B, Pittet A-C, Sultana S, et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 2012;434:222–232. [DOI] [PubMed] [Google Scholar]
- [64].McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, et al. Safety analysis of a Russian phage cocktail: From MetaGenomic analysis to oral application in healthy human subjects. Virology 2013;443:187–196. [DOI] [PubMed] [Google Scholar]
- [65].Sarker SA, Berger B, Deng Y, Kieser S, Foata F, Moine D, et al. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environmental microbiology 2017;19:237–250. [DOI] [PubMed] [Google Scholar]
- [66].Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, Charton F, et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016;4:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrobial Agents and Chemotherapy 2017;61:e00954–00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflammatory Bowel Diseases 2007;13:1277–1283. [DOI] [PubMed] [Google Scholar]
- [69].Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 2018;67:574–587. [DOI] [PubMed] [Google Scholar]
- [70].Galtier M, Sordi LD, Sivignon A, de Vallée A, Maura D, Neut C, et al. Bacteriophages Targeting Adherent Invasive Escherichia coli Strains as a Promising New Treatment for Crohn’s Disease. Journal of Crohn’s and Colitis 2017;11:840–847. [DOI] [PubMed] [Google Scholar]
- [71].Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA 2015;313:2263–2273. [DOI] [PubMed] [Google Scholar]
- [72].Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One 2013;8:e62885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer SE, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Scientific Reports 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Duarte SMB, Stefano JT, Miele L, Ponziani FR, Souza-Basqueira M, Okada L, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr Metab Cardiovasc Dis 2018;28:369–384. [DOI] [PubMed] [Google Scholar]
- [75].Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017;65:451–464. [DOI] [PubMed] [Google Scholar]
- [76].Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 2017;25:1054–1062 e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Caussy C, Hsu C, Lo MT, Liu A, Bettencourt R, Ajmera VH, et al. Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology 2018;68:918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019;69:107–120. [DOI] [PubMed] [Google Scholar]
- [80].Shen F, Zheng RD, Sun XQ, Ding WJ, Wang XY, Fan JG. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int 2017;16:375–381. [DOI] [PubMed] [Google Scholar]
- [81].Yuan J, Chen C, Cui J, Lu J, Yan C, Wei X, et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab 2019;30:675–688 e677. [DOI] [PubMed] [Google Scholar]
- [82].Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol 2004;34:9–19. [DOI] [PubMed] [Google Scholar]
- [83].Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol 2012;302:G966–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tuomisto S, Pessi T, Collin P, Vuento R, Aittoniemi J, Karhunen PJ. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol 2014;14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011;54:562–572. [DOI] [PubMed] [Google Scholar]
- [86].Grander C, Adolph TE, Wieser V, Lowe P, Wrzosek L, Gyongyosi B, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 2018;67:891–901. [DOI] [PubMed] [Google Scholar]
- [87].Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019;575:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mieli-Vergani G, Vergani D, Czaja AJ, Manns MP, Krawitt EL, Vierling JM, et al. Autoimmune hepatitis. Nat Rev Dis Primers 2018;4:18017. [DOI] [PubMed] [Google Scholar]
- [89].Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020;69:569–577. [DOI] [PubMed] [Google Scholar]
- [90].Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018;359:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Little R, Wine E, Kamath BM, Griffiths AM, Ricciuto A. Gut microbiome in primary sclerosing cholangitis: A review. World J Gastroenterol 2020;26:2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ruhlemann M, Liwinski T, Heinsen FA, Bang C, Zenouzi R, Kummen M, et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther 2019;50:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ruhlemann MC, Heinsen FA, Zenouzi R, Lieb W, Franke A, Schramm C. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 2017;66:753–754. [DOI] [PubMed] [Google Scholar]
- [94].Kummen M, Holm K, Anmarkrud JA, Nygard S, Vesterhus M, Hoivik ML, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017;66:611–619. [DOI] [PubMed] [Google Scholar]
- [95].Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 2017;23:4548–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016;65:1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020;69:92–102. [DOI] [PubMed] [Google Scholar]
- [98].Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 2019;4:492–503. [DOI] [PubMed] [Google Scholar]
- [99].Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol 2012;303:G675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol 2012;302:G168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wiest R, Rath HC. Gastrointestinal disorders of the critically ill. Bacterial translocation in the gut. Best Pract Res Clin Gastroenterol 2003;17:397–425. [DOI] [PubMed] [Google Scholar]
- [103].Steffen EK, Berg RD, Deitch EA. Comparison of translocation rates of various indigenous bacteria from the gastrointestinal tract to the mesenteric lymph node. J Infect Dis 1988;157:1032–1038. [DOI] [PubMed] [Google Scholar]
- [104].Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, et al. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J Clin Microbiol 2010;48:2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Tetart F, Repoila F, Monod C, Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol 1996;258:726–731. [DOI] [PubMed] [Google Scholar]
- [106].Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 2005;115:101–107. [DOI] [PubMed] [Google Scholar]
- [107].Duplessis M, Moineau S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol 2001;41:325–336. [DOI] [PubMed] [Google Scholar]
- [108].Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A 2007;104:11197–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Merril CR, Scholl D, Adhya SL. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2003;2:489–497. [DOI] [PubMed] [Google Scholar]
- [110].McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology 2013;443:187–196. [DOI] [PubMed] [Google Scholar]
- [111].Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, Charton F, et al. Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 2016;4:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gindin M, Febvre HP, Rao S, Wallace TC, Weir TL. PHAGE Study: Bacteriophages as Novel Prebiotics. ClinicalTrials.gov (NCT03269617). 2017.
- [113].Grubb DS, Wrigley SD, Freedman KE, Wei Y, Vazquez AR, Trotter RE, et al. PHAGE-2 Study: Supplemental Bacteriophages Extend Bifidobacterium animalis subsp. lactis BL04 Benefits on Gut Health and Microbiota in Healthy Adults. Nutrients 2020;12:2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2013;11:868–875 e861–863. [DOI] [PubMed] [Google Scholar]
- [115].Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 2013;57:601–609. [DOI] [PubMed] [Google Scholar]
- [116].Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, et al. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci Rep 2016;6:32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A 2014;111:E4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lang S, Fairfied B, Gao B, Duan Y, Zhang X, Fouts DE, et al. Changes in the fecal bacterial microbiota associated with disease severity in alcoholic hepatitis patients. Gut Microbes 2020;12:1785251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Elsherbiny NM, Rammadan M, Hassan EA, Ali ME, El-Rehim ASA, Abbas WA, et al. Autoimmune Hepatitis: Shifts in Gut Microbiota and Metabolic Pathways among Egyptian Patients. Microorganisms 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020;69:92–102. [DOI] [PubMed] [Google Scholar]
- [121].Chen Y, Ji F, Guo J, Shi D, Fang D, Li L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep 2016;6:34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun 2019;10:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Bajaj JS, Betrapally NS, Gillevet PM. Decompensated cirrhosis and microbiome interpretation. Nature 2015;525:E1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab 2020;32:901. [DOI] [PMC free article] [PubMed] [Google Scholar]


