Abstract
Several morphologically dissimilar ascomycete fungi including Schizosaccharomyces, Taphrina, Saitoella, Pneumocystis, and Neolecta have been grouped into the taxon Taphrinomycotina (Archiascomycota or Archiascomycotina), originally based on rRNA phylogeny. These analyses lack statistically significant support for the monophyly of this grouping, and although confirmed by more recent multigene analyses, this topology is contradicted by mitochondrial phylogenies. To resolve this inconsistency, we have assembled phylogenomic mitochondrial and nuclear data sets from four distantly related taphrinomycotina taxa: Schizosaccharomyces pombe, Pneumocystis carinii, Saitoella complicata, and Taphrina deformans. Our phylogenomic analyses based on nuclear data (113 proteins) conclusively support the monophyly of Taphrinomycotina, diverging as a sister group to Saccharomycotina + Pezizomycotina. However, despite the improved taxon sampling, Taphrinomycotina continue to be paraphyletic with the mitochondrial data set (13 proteins): Schizosaccharomyces species associate with budding yeasts (Saccharomycotina) and the other Taphrinomycotina group as a sister group to Saccharomycotina + Pezizomycotina. Yet, as Schizosaccharomyces and Saccharomycotina species are fast evolving, the mitochondrial phylogeny may be influenced by a long-branch attraction (LBA) artifact. After removal of fast-evolving sequence positions from the mitochondrial data set, we recover the monophyly of Taphrinomycotina. Our combined results suggest that Taphrinomycotina is a legitimate taxon, that this group of species diverges as a sister group to Saccharomycotina + Pezizomycotina, and that phylogenetic positioning of yeasts and fission yeasts with mitochondrial data is plagued by a strong LBA artifact.
Keywords: Fungi, Taphrinomycotina, Schizosaccharomyces, Pneumocystis, phylogeny, mitochondria, long-branch attraction artifact
Introduction
Ascomycota are currently subdivided into three major taxa (Hibbett et al. 2007): Saccharomycotina (Hemiascomycota; budding yeasts), Pezizomycotina (Euascomycota; for the most part filamentous fungi, e.g., Neurospora), and Taphrinomycotina (Archiascomycota). The taxon Taphrinomycotina was initially created based on rRNA phylogeny (Nishida and Sugiyama 1993), regrouping diverse fungal species of previously uncertain taxonomic affiliation: 1) Schizosaccharomyces species (fission yeasts; previously considered to be highly divergent members of the budding yeast lineage), 2) Taphrina (several fungal plant pathogens), 3) the anamorphic yeast-like Saitoella, a suspected ascomycete or basidiomycete, and 4) Neolecta irregularis, a fungus with filamentous cell growth that forms complex fruiting bodies (unique within this group of organisms). Yet, the statistical support for this grouping with rRNA data is well below standards (for details, see Leigh et al. 2003). Addition of potential taphrinomycotina taxa, for instance more Schizosaccharomyces species or Pneumocystis carinii (a unicellular lung pathogen [Edman et al. 1988] that like Schizosaccharomyces divides by binary fission), has not improved the outcome. Evidently, resolving this question requires substantially more than just rRNA data.
Several multigene analyses have more recently been conducted to overcome the apparent problems with inferring fungal relationships. These analyses differ in their choice of genes. First, data sets with six or fewer nuclear genes also produce conflicting phylogenies. For instance in an early overview paper (Baldauf et al. 2000), Schizosaccharomyces, the only Taphrinomycotina included in this analysis, groups with Saccharomycotina, although without significant support. This topology is recovered by a more recent analysis (Diezmann et al. 2004) but contradicted by others (James et al. 2006; Liu et al. 2006; Spatafora et al. 2006; Sugiyama et al. 2006) that find high bootstrap support for Taphrinomycotina as a monophyletic grouping as a sister group to Saccharomycotina + Pezizomycotina. Yet, rigorous statistical testing (e.g., by applying the approximate unbiased [AU] test [Shimodaira 2002]) has not been performed in these cases, and because most sequence information was obtained by polymerase chain reaction, less genomic sequence information was available to exclude potentially misleading gene paralogs. Additional reasons why analyses with small data sets are more likely misled by phylogenetic artifacts are discussed elsewhere (Delsuc, Brinkmann, and Philippe 2005). Finally, in two of these analyses (James et al. 2006; Spatafora et al. 2006), both rRNA and protein sequences were used in the same data set, which implies the use of mixed-model analyses, complicating rigorous statistical AU testing. The applied Bayesian analyses are known to largely overestimate confidence when using real data as these evolve in much more complex ways than implemented in current models (Erixon et al. 2003; Taylor and Piel 2004; Mar et al. 2005). In turn, when applying the AU test to alternative analyses that are restricted to the nucleotide level, the risk of error due to compositional bias (rRNA vs. protein gene sequences) increases.
In phylogenomic analyses that utilize the maximum amount of discrete sequence data, Schizosaccharomyces pombe consistently diverges as a sister group to Saccharomycotina + Pezizomycotina with significant support (e.g., Philippe et al. 2004; Fitzpatrick et al. 2006; Robbertse et al. 2006; Dutilh et al. 2007). Yet, the question of Taphrinomycotina monophyly remains open as genome-size data sets are not available for other taphrinomycotina lineages. Finally, mitochondrial data sets with 13 proteins and 3 Schizosaccharomyces species consistently support a grouping of fission yeasts with Saccharomycotina (Bullerwell et al. 2003; Leigh et al. 2003). Obviously, the use of multigene data sets is insufficient to tackle the given phylogenetic question without paying close attention to potential phylogenetic artifacts (Delsuc, Brinkmann, and Philippe 2005). In the analyses of mitochondrial data (Bullerwell et al. 2003; Leigh et al. 2003), the authors suggest that the grouping of Saccharomycotina and Schizosaccharomyces may be due to a long-branch attraction (LBA) artifact, which causes clustering of fast-evolving lineages irrespective of their true evolutionary relationships. A common strategy to overcome this artifact involves the complete elimination of fast-evolving species; yet in the mitochondrial data set, all Schizosaccharomyces and budding yeast species are fast evolving. Other less radical options include the exclusion of fast-evolving sequence positions (Brinkmann and Philippe 1999) or the use of more realistic evolutionary models, for example, the CAT model (Lartillot et al. 2007). Evidently, such improvements at the analytical level should be combined with improved taxon sampling, with particular emphasis on the addition of slowly evolving species. Finally, congruence with analyses with alternative data sets (e.g., nuclear vs. mitochondrial) is an indication that results are accurate.
In the present study, we take advantage of new data provided by both nuclear and mitochondrial genome projects for all key taphrinomycotina species except Neolecta, which unfortunately has not yet been grown in culture. We compare two large data sets, one with 113 nuclear and another with 13 mitochondrial proteins, and conclude that Taphrinomycotina is indeed a monophyletic group diverging as a sister group to Saccharomycotina + Pezizomycotina.
Materials and Methods
Construction of cDNA Libraries and Expressed Sequence Tag Sequencing
Saitoella complicata (NRLL Y-17804) and Taphrina deformans (NRRL T-857) cDNA libraries were constructed from strains grown on glycerol medium, following recently published protocols (Rodriguez-Ezpeleta et al., forthcoming). Plasmids were purified using the QIAprep 96 Turbo Miniprep Kit (Qiagen, Valencia, CA), sequencing reactions were performed with the ABI Prism BigDyeTM Terminators version 3.0/3.1 (PerkinElmer, Wellesley, MA), and a total of 3,840 S. complicata and 3,919 T. deformans expressed sequence tags (ESTs) were sequenced on an MJ BaseStation. Trace files were imported into the TBestDB database (http://tbestdb.bcm.umontreal.ca/searches/login.php) (O'Brien et al. 2007) for automated processing, including assembly as well as automated gene annotation by AutoFact (Koski et al. 2005). Pneumocystis carinii sequences were obtained from the Pneumocystis Genome Project (http://pgp.cchmc.org).
Mitochondrial Sequencing
Saitoella complicata and T. deformans were grown with vigorous shaking in liquid medium (1% yeast extract and 3% glycerol). The harvested cells were disrupted by manual shaking with glass beads, and mitochondrial DNA (mtDNA) was isolated following a whole cell lysate protocol (Lang and Burger 2007) and sequenced using a random procedure (Burger et al. 2007).
Data Set Construction
The nuclear data set was assembled by adding EST and genomic sequences from GenBank to a previously published alignment (Rodriguez-Ezpeleta et al. 2007). Paralogous proteins were identified and removed from the alignment as described (Roure et al. 2007). Gblocks (Castresana 2000) (default parameters) was used to extract unambiguously aligned regions. The inclusion of some missing data allowed us to add more genes and species. From originally 174 proteins, 113 were selected to minimize the degree of missing data in phylogenetic analysis. The final alignment has a total number of 29,387 amino acid positions and 54 species. The average proportion of missing data is 25% per species. The proportion of missing data for each species is listed in supplementary table S1 (Supplementary Material online).
The mitochondrial protein alignment includes our new T. deformans and S. complicata sequences as well as sequences retrieved from public data repositories (GenBank and the Pneumocystis Genome Project). Sequences of 13 mitochondrial proteins (cox1, 2, 3, cob, atp6, 9 and nad1, 2, 3, 4, 4L, 5, 6) were selected for phylogenetic analysis. An application developed in-house (mams) was used for automatic protein alignment with Muscle (Edgar 2004), removal of ambiguous regions with Gblocks (Castresana 2000), and concatenation. The final data set contains 2,596 amino acid positions with missing data only in Schizosaccharomyces species and in Saccharomyces cerevisiae (46.2% missing positions for those species), which both lost all nad genes, coding for subunits of complex I of the respiratory chain.
Phylogenetic Analysis of the Nuclear Data Set
Phylogenetic analyses were performed at the amino acid level. The concatenated nuclear protein data sets were analyzed either by maximum likelihood (ML) or Bayesian inference (BI) methods. Three ML programs, Treefinder (Jobb et al. 2004), PhyML (Guindon and Gascuel 2003), and RAxML (Stamatakis 2006) were used with the Whelan and Goldman (WAG) + gamma model with four categories. In case of BI methods, we used either MrBayes (Ronquist and Huelsenbeck 2003; WAG + gamma model, 500,000 generations, first 100,000 generations removed as burn-in, analysis repeated three times with identical results) or PhyloBayes (version 2) (Lartillot and Philippe 2004; CAT model, 3,000 cycles, first 1,000 cycles removed as burn-in, analysis repeated three times with identical results). The reliability of internal branches was either evaluated based on 100 (ML) bootstrap replicates or on posterior probabilities (PPs).
Likelihood tests of competing tree topologies were also performed. An exhaustive set of 945 topologies was generated by constraining trusted internal branches (monophyly of Saccharomycotina, Pezizomycotina, Ascomycota, Basidiomycota, and the grouping of Zygomycota and Chytridiomycota), leaving the four Taphrinomycotina unconstrained within Ascomycota. The sitewise likelihood values for each topology were estimated using Tree-Puzzle (Schmidt et al. 2002), and P values for each topology were calculated with CONSEL (Shimodaira and Hasegawa 2001).
Phylogenetic Analysis of the Mitochondrial Data Set with the Slow–Fast Method
LBA artifacts may possibly be overcome by elimination of fast-evolving sequence positions with the slow–fast (SF) method (Brinkmann and Philippe 1999). Briefly, the data set is split into monophyletic groups, and the number of substitutions for each position in each group is estimated using a maximum parsimony criterion with PAUP* (Swofford 2000). These numbers are summed over all groups of the data set, providing an estimate of the variability for each position. A number of data sets (in the current analysis, 14) are then constructed with an increasing fraction of fast-evolving sequence positions.
Trees and bootstrap support (100 replicates) for the subdata sets were estimated with RAxML, as Treefinder and PhyML were often trapped in local optima with these relatively small data sets.
Results
Phylogenetic Analysis of the Nuclear Data Set
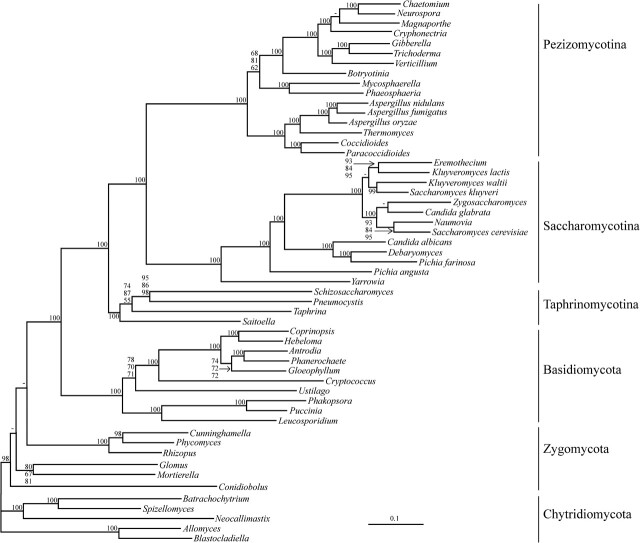
Our nuclear data set contains 113 orthologous proteins (29,387 amino acid positions) from 54 fungal species, including 33 Ascomycota and representatives of the three other major fungal groups (Basidiomycota, Zygomycota, and Chytridiomycota). In the phylogenetic tree shown in figure 1, the monophyly of Ascomycota, Saccharomycotina, Pezizomycotina, and Basidiomycota are recovered with significant support by both ML and BI methods. In addition, Taphrinomycotina form a significantly supported monophyletic group (>99% bootstrap proportion [BP] and PP 1.0). The grouping of S. pombe with P. carinii receives 95% support using Treefinder, 86% with RAxML, and 98% with PhyML; the branching order of S. complicata and T. deformans remains unresolved.
FIG. 1.—
Phylogeny based on nucleus-encoded protein sequences. This tree was inferred from 113 nucleus-encoded proteins (29,387 amino acid positions), with three ML (Treefinder, PhyML, and RAxML) and two BI (MrBayes and PhyloBayes) methods, either using the WAG + Gamma (four categories) model or the CAT model (PhyloBayes). The PP using MrBayes and PhyloBayes are 1.0 for all branches, except for the one that groups Taphrina and Saitoella (PP 0.6). Numbers at internal branches represent support values obtained with 100 bootstrap replicates on the concatenated data set with Treefinder/RAxML/PhyML. When all support values are identical, only one is indicated.
Data sets including ESTs usually contain a fraction of missing data, amounting for S. complicata and T. deformans to 66.8% and 56.8%, respectively. The data set contains 113 proteins, but only one single protein contains sequences from all 54 species (rpl4B). To test the potential influence of missing data, we reduced the data set to the most complete 76 proteins, thereby decreasing missing positions for these two species to 43.0% and 39.9%, respectively. The inferred tree topologies remain the same, and support values for the monophyly of Taphrinomycotina are only moderately effected, decreasing or decreasing slightly depending on the analysis method (ML inferences, BP > 95%; MrBayes, PP 1.0; and PhyloBayes, PP 0.99; supplementary fig. S1, Supplementary Material online).
Likelihood Test of Competing Topologies
Using the original complete nuclear data set, both ML and BI approaches yield identical, well-supported tree topologies. To assess the level of confidence with a strict, alternative approach, we performed likelihood-based tests of competing tree topologies with CONSEL (Shimodaira and Hasegawa 2001), with the complete data set (113 proteins). The corresponding 10 top-ranking topologies according to AU test P values are shown in table 1. All scenarios in which Taphrinomycotina are paraphyletic are rejected with confidence (P < 0.01), thus confirming the monophyly of Taphrinomycotina as well as their position as a sister group to Saccharomycotina + Pezizomycotina.
Table 1.
Likelihood Tests of Alternative Tree Topologies
| Rank | Tree Topology | Taphrinomycotina | ΔlnLa | AU Test |
| 1 | Best tree (fig. 1) | Monophyletic | −14.4 | 0.869 |
| 2 | ((T.d, S.c), (S.p, P.c)) | Monophyletic | 14.4 | 0.297 |
| 3 | (S.c, (P.c, (T.d, S.p))) | Monophyletic | 27.6 | 0.131 |
| 4 | ((T.d, S.p), (P.c, S.c)) | Monophyletic | 45.1 | 0.032 |
| 5 | (P.c, (S.c, (S.p, T.d))) | Monophyletic | 50.2 | 0.011 |
| 6 | (T.d, (S.c, (S.p, P.c)), (Sacch,Pezi)) | Paraphyletic | 163.1 | 0.007 |
| 7 | ((S.p, (T.d, P.c)), ((S.c, Sacch), Pezi)) | Paraphyletic | 525.6 | 0.007 |
| 8 | ((T.d, S.c), (P.c, (S.p, (Sacch, Pezi)))) | Paraphyletic | 243.0 | 0.005 |
| 9 | (S.p, ((S.c, T.d), (P.c, (Sacch, Pezi)))) | Paraphyletic | 265.6 | 0.004 |
| 10 | ((S.p, P.c), ((S.c, T.d), (Sacch, Pezi))) | Paraphyletic | 99.2 | 0.004 |
NOTE.—A total of 945 topologies were generated by constraining well-supported internal branches (monophyly of Saccharomycotina, Pezizomycotina, Ascomycota, Basidiomycota, and Zygomycota plus Chytridiomycota as outgroup), leaving the four Taphrinomycotina unconstrained within Ascomycota. Table 1 lists the P values of the 10 top-ranking topologies based on the AU test (data model as in fig 1). The following abbreviations are used: P.c: Pneumocystis carinii; S.c: Saitoella complicata; S.p: Schizosaccharomyces pombe; T.d: Taphrina deformans; Sacch: Saccharomycotina; and Pezi: Pezizomycotina. In the five best topologies, Taphrinomycotina are monophyletic. All other topologies in which they are paraphyletic are rejected at a significance level less than 0.01.
Log likelihood difference.
Phylogenetic Analysis of Mitochondrial Data Sets
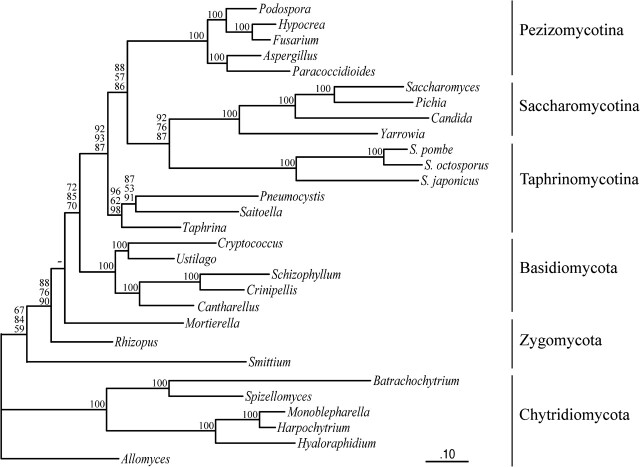
The mitochondrial data set contains 2,596 amino acid positions from 13 well-conserved mitochondrial proteins, including 29 species from the four major fungal lineages. In ML analyses, the newly added Taphrinomycotina (T. deformans, P. carinii, and S. complicata) group as a sister group to Saccharomycotina + Pezizomycotina (fig. 2), and as in previously published analyses (Bullerwell et al. 2003; Leigh et al. 2003; Pramateftaki et al. 2006), S. pombe, Schizosaccharomyces japonicus, and Schizosaccharomyces octosporus group with Saccharomycotina. Yet, due to the addition of the new Taphrinomycotina species, the support for the grouping of fission yeasts with budding yeasts is noticeably lower (fig. 2; 92% with Treefinder, 76% with RAxML, and 87% with PhyML). In our experience, the heuristic search of RAxML is most effective in avoiding local minima; thus, the 76% confidence level of RaxML in this analysis is the most reliable. BI analyses (using MrBayes and PhyloBayes) inferred the same topology as ML approaches, with more than PP 0.99 for all internal branches except the one leading to Chytridiomycota (PP 0.64).
FIG. 2.—
Phylogeny based on concatenated proteins encoded by mtDNA. The sequences of 13 proteins (cox1, cox2, cox3, cob, atp6, atp9 and nad1, nad2, nad3, nad4, nad4L, nad5, nad6) were concatenated (2,596 amino acid positions). For details on inference methods, see figure 1. The BI with MrBayes has PPs of at least 0.99, except for the internal branch that groups Allomyces and other chytrids (PP 0.64).
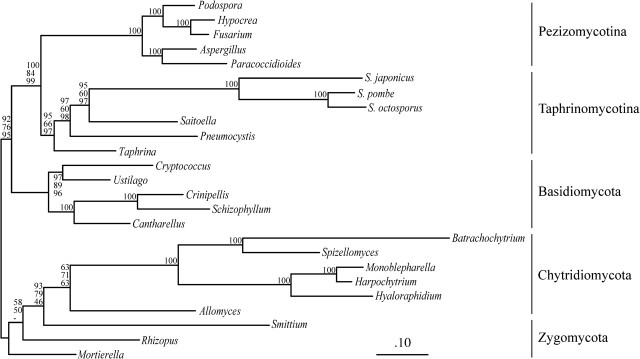
As Schizosaccharomyces and Saccharomycotina species have relatively long branches, they are suspected to group due to an LBA artifact. If this interpretation is correct, removing Saccharomycotina is expected to relocate the Schizosaccharomyces to its correct position. Indeed, instead of grouping with Pezizomycotina, the three Schizosaccharomyces group with other Taphrinomycotina after Saccharomycotina are removed. The monophyly of Taphrinomycotina receives varying support (Treefinder, BP 95%; RAxML, 66%; PhyML, 97%; and MrBayes, PP 1.0; fig. 3).
FIG. 3.—
Phylogenetic analysis of mitochondrial data set after removing Saccharomycotina. All Saccharomycotina were removed from the complete mitochondrial data set. The analyses were performed as in figure 1. The Schizosaccharomyces group with other Taphrinomycotina with various BPs among three ML methods (TreeFinder: 95%, RAxML: 66%, and PhyML: 97%), the PP of BI using MrBayes is 1.0.
We further explored the use of a fast-evolving fungal outgroup, which was expected to draw Schizosaccharomyces away from Saccharomycotina, toward the outgroup. To test this prediction, we reduced the original mitochondrial data set to 19 species, including all 15 Ascomycota plus four (fast evolving) Chytridiomycota. Indeed, analyses of this data set with ML and BI methods position Schizosaccharomyces closer to the fungal divergence point (supplementary fig. S2, Supplementary Material online), although with marginal support (Treefinder, BP 71%; RAxML, 57%; PhyML, 86%; and MrBayes, PP 0.95).
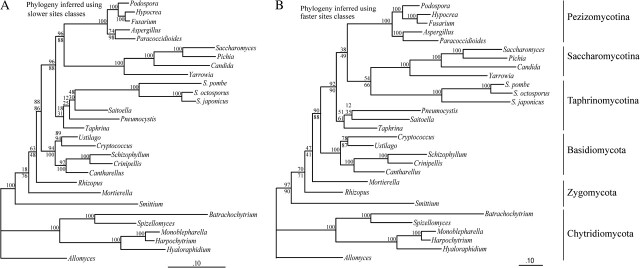
Finally, we analyzed the mitochondrial data set with the SF method, which is designed to reduce the effect of LBA by selecting slowly evolving positions, thus increasing the ratio of phylogenetic signal to noise (Delsuc, Brinkmann, and Philippe 2005). A series of data matrices containing increasing fractions of fast-evolving positions were analyzed with both ML and BI methods (fig. 4). In the data sets with the most slowly evolving sites and most reliable phylogenetic information (S2–S5; only results from S3 and S5 are shown in fig. 4A ; for more details, see supplementary fig. S3, Supplementary Material online), the Schizosaccharomyces lineage grouped together with other Taphrinomycotina. Yet, although there was good support (BP of 96% or 88%) to reject a grouping of Saccharomycotina plus Taphrinomycotina, there was not significant BP support to the monophyly of Taphrinomycotina, due to the small size of the remaining data sets (S2 contains only 1,023 amino acid positions, S3: 1,223; S4: 1,436; and S5: 1,638). In fact, addition of further fast-evolving positions to S5 resulted in decrease of support, as expected in a classical case of LBA. Finally, as more fast-evolving positions were included (the S6–S14 data sets), Schizosaccharomyces grouped with Saccharomycotina, and the BP for this incorrect topology increased (the result from S7 and S9 is shown in fig. 4B ). The evolution of BP supports for all S data sets is shown in supplementary figure S3 (Supplementary Material online).
FIG. 4.—
Impact of fast-evolving positions on the inferred phylogeny from proteins encoded by mtDNA. The SF method was used to generate a series of 14 data sets (S0, S1, S2, …, S14) with an increasing fraction of fast-evolving sequence positions. The phylogenies were inferred using RAxML on these data sets (WAG + gamma with four categories). Results with S3 and S5 are shown in (A) and with S7 and S9 in (B). Numbers at internal branches represent BP obtained with 100 bootstrap replicates, which are in the order S3, S5 (A) and S7, S9 (B) from top. When all bootstrap values are >95%, only one value is presented.
Discussion
The Nuclear Data Set Significantly Supports the Monophyly of Taphrinomycotina
Two previous analyses of five or six genes supported monophyletic Taphrinomycotina (James et al. 2006; Spatafora et al. 2006); however, data sets with few genes are often misled by stochastic error. Our phylogenetic analysis is first in using a large number (113) of nucleus-encoded proteins from most key taphrinomycotina species and concludes with high confidence that Taphrinomycotina is monophyletic. Some authors have claimed that missing data (in our case, due to partial EST sequencing) may result in unstable tree topologies (Anderson 2001; Sanderson et al. 2003). Yet, consistent with other work (Wiens 2003; Philippe et al. 2004), our ML analysis did not confirm this claim. Our explanation is that the effect of missing data is negligible when using large data sets with a strong phylogenetic signal. The comparison of alternative topologies with the AU test confirmed the monophyly of Taphrinomycotina with high confidence (P < 0.01), although the relationships among Taphrinomycotina remain to be resolved. Additional data from the ongoing S. octosporus and S. japonicus genome projects are expected to improve tree resolution, and complete genome sequences from Taphrina and Saitoella (slowly evolving taphrinomycotina genomes that we expect to be more gene rich and more typical for Taphrinomycotina than those of Schizosaccharomyces) are required for confident inference of their phylogenetic position. Finally, EST or genome sequencing will be required to confirm that N. irregularis belongs in Taphrinomycotina.
The Mitochondrial Tree Topology Is Sensitive to Phylogenetic Artifacts
Mitochondrion-encoded protein data have been successfully used to resolve a large variety of phylogenetic questions, in some cases, predicting for the first time deep relationships with high confidence (e.g., Lang et al. 2002). Yet, mitochondrial genes tend to have a high A + T sequence bias that contributes to phylogenetic artifacts, particularly in lineages with elevated evolutionary rates. For instance, in a previous analysis that includes three Schizosaccharomyces species, Schizosaccharomyces plus Saccharomycotina group with strong support (BP 95%), although an alternative (likely correct) position of Schizosaccharomyces as sister group to Saccharomycotina + Pezizomycotina was not rejected by an AU test (Bullerwell et al. 2003). In this study, amino acid instead of nucleotide sequences have been used to decrease the effect of A + T bias. Yet, after inclusion of further complete mitochondrial data from three slowly evolving Taphrinomycotina (S. complicata, T. deformans, and P. carinii), the position of Schizosaccharomyces does not change, although the bootstrap support for this topology decreases to 76% (fig. 2). This result is consistent with the suggestion that adding more sequences (particularly from slowly evolving species) usually helps to reduce the effect of LBA (for a review, see Delsuc, Brinkmann, and Philippe 2005).
We have further tested whether Schizosaccharomyces mitochondrial sequences contain little phylogenetic signal and a strong tendency for LBA by inferring a phylogeny with a distant fungal outgroup composed of four fast-evolving Chytridiomycota. In this case, Schizosaccharomyces changes its position, away from Saccharomycotina toward the outgroup, apparently due to LBA with the distantly related Chytridiomycota. When Saccharomycotina are removed from the original data set, Taphrinomycotina become monophyletic, though without significant support. Finally, positional sorting with the SF method confirms our interpretation. Only the slowest evolving positions (S2–S5 data matrix) are able to recover the tree topology inferred with the nuclear data set, although only with marginal statistical support. Our analyses strongly suggest that the grouping of Schizosaccharomyces with Saccharomycotina in trees based on mitochondrial data is due to an LBA artifact.
Limitations of Mitochondrial Sequence Data in Phylogenetic Analysis
A limitation of mitochondrial genome data is their small data size compared with nuclear genomes. The most popular mitochondrial data set contains only 13 proteins, including some that are rather small (atp9, nad4L) and others that are fast evolving (nad2, nad6) and are therefore of limited value for the inference of deep phylogenies. To expand these data sets, mitochondrial genes that were transferred to the nucleus might be added. Yet, because the A + T content and other evolutionary constraints are different in mitochondrial and nuclear genomes, evolutionary models and inference methods might have to be adapted.
Conclusion
The current analysis ends a long-standing controversy on the phylogenetic position of Schizosaccharomyces species: We conclude that they are part of Taphrinomycotina, branching as a sister group to Saccharomycotina + Pezizomycotina. Yet, the phylogenetic identity of Neolecta, another putative representative of this group, remains to be assessed by phylogenomic analysis.
Supplementary Material
Acknowledgments
Salary and interaction support from the Canadian Institute for Advanced Research (B.F.L.), the Canadian Institute of Health Research (B.F.L.), the “Bourses d'Excellence biT” (CIHR; Y.L.) is acknowledged, and the “Programa de Formación de Investigadores del Departamento de Educación, Universidades e Investigación” (Government of Basque Country; N.R.-E.). J.W.L. is supported by a Student Research Award from the Nova Scotia Health Research Foundation. We thank Lise Forget and Tom Sesterhenn for help in mtDNA sequencing and sequence interpretation.
References
- Anderson JS. The phylogenetic trunk: maximal inclusion of taxa with missing data in an analysis of the lepospondyli (Vertebrata, Tetrapoda) Syst Biol. 2001;50:170–193. doi: 10.1080/10635150119889. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Forget L, Lang BF. Evolution of monoblepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res. 2003;31:1614–1623. doi: 10.1093/nar/gkg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Lavrov DV, Forget L, Lang BF. Sequencing complete mitochondrial and plastid genomes. Nat Protoc. 2007;2:603–614. doi: 10.1038/nprot.2007.59. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Diezmann S, Cox CJ, Schonian G, Vilgalys RJ, Mitchell TG. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol. 2004;42:5624–5635. doi: 10.1128/JCM.42.12.5624-5635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, van Noort V, van der Heijden RT, Boekhout T, Snel B, Huynen MA. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23:815–824. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC, Kovacs JA, Masur H, Santi DV, Elwood HJ, Sogin ML. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol. 2003;52:665–673. doi: 10.1080/10635150390235485. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, et al. (67 co-authors) A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. (70 co-authors) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koski LB, Gray MW, Lang BF, Burger G. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Burger G. Purification of mitochondrial and plastid DNA. Nat Protoc. 2007;2:652–660. doi: 10.1038/nprot.2007.58. [DOI] [PubMed] [Google Scholar]
- Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Leigh J, Seif E, Rodriguez N, Jacob Y, Lang BF. Fungal evolution meets fungal genomics. In: Arora DK, editor. Handbook of Fungal Biotechnology. New York: Marcel Dekker Inc; 2003. pp. 145–161. [Google Scholar]
- Liu YJ, Hodson MC, Hall BD. Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes. BMC Evol Biol. 2006;6:74. doi: 10.1186/1471-2148-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar JC, Harlow TJ, Ragan MA. Bayesian and maximum likelihood phylogenetic analyses of protein sequence data under relative branch-length differences and model violation. BMC Evol Biol. 2005;5:8. doi: 10.1186/1471-2148-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Sugiyama J. Phylogenetic relationships among Taphrina, Saitoella, and other higher fungi. Mol Biol Evol. 1993;10:431–436. doi: 10.1093/oxfordjournals.molbev.a040012. [DOI] [PubMed] [Google Scholar]
- O'Brien EA, Koski LB, Zhang Y, Yang L, Wang E, Gray MW, Burger G, Lang BF. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35:D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Pramateftaki PV, Kouvelis VN, Lanaridis P, Typas MA. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 2006;6:77–90. doi: 10.1111/j.1567-1364.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- Robbertse B, Reeves JB, Schoch CL, Spatafora JW. A phylogenomic analysis of the Ascomycota. Fungal Genet Biol. 2006;43:715–725. doi: 10.1016/j.fgb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Teijeiro S, Forget L, Burger G, Lang BF Forthcoming. Totowa (NJ): Humana press Inc; Generation of cDNA libraries: protists and fungi. In: P J, editor. Methods in molecular biology: expressed sequence tags. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 2007;7(Suppl 1):S2. doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Driskell AC, Ree RH, Eulenstein O, Langley S. Obtaining maximal concatenated phylogenetic data sets from large sequence databases. Mol Biol Evol. 2003;20:1036–1042. doi: 10.1093/molbev/msg115. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, et al. (33 co-authors) A five-gene phylogeny of Pezizomycotina. Mycologia. 2006;98:1018–1028. doi: 10.3852/mycologia.98.6.1018. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sugiyama J, Hosaka K, Suh SO. Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia. 2006;98:996–1005. doi: 10.3852/mycologia.98.6.996. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony and other methods. Sunderland (MA): Sinauer; 2000. [Google Scholar]
- Taylor DJ, Piel WH. An assessment of accuracy, error, and conflict with support values from genome-scale phylogenetic data. Mol Biol Evol. 2004;21:1534–1537. doi: 10.1093/molbev/msh156. [DOI] [PubMed] [Google Scholar]
- Wiens JJ. Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol. 2003;52:528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.