SUMMARY
Aim of this study was to reconstruct the phylogeography of variola virus (VARV) in the XX century, using 47 VARV whole genome sequences available in public databases, through two different methods for ancestral character reconstruction: a frequently used Bayesian framework and a fast maximum-likelihood (ML) based method.
The substitution rate of the whole VARV genome was estimated to be between 6.7×10−6 and 1.1×10−5 substitutions/site/year. Both ML and Bayesian methods gave similar trees topology, showing two distinct monophyletic groups: one (known as P1) including the great part of variola major and the second (P2) including West African and American (variola minor) isolates and close evolutionary rate estimations, between 6.73×10−6 and 1.1×10−5 for the whole genome.
The phylogeographical reconstruction of P1 suggested that the common ancestor of the variola major circulating in the Old World between the 1940s and the 1970s most probably originated in the Far East in the first decades of the XX century, and then spread to Indian subcontinent in the 1920s. India represented a center of further spread of VARV to eastern Africa in the 1940s and to the Middle East in the 1960s.
The phylogeographic scenario obtained by the maximum-likelihood based method was congruent with that obtained by Bayesian framework, but the analysis was faster indicating the usefulness of this method in the analyses of large viral genomes.
Our results may help to explain the controversial reconstructions of the history of VARV obtained using long or short timescale for calibration.
Keywords: Variola virus, Poxvirus, evolution
INTRODUCTION
Smallpox was once a severe infectious disease caused by the variola virus (VARV) whose outbreaks across the world were a major cause of mortality. It was responsible for hundreds of millions of deaths as it not only negatively affected populations’ growth in the Old World, but also contributed to the crises affecting established civilizations in the New World when their populations were decimated after they came into contact with Europeans [1].
However, in 1980, it was officially certified that the intensified Smallpox Eradication Programme (SEP) approved by WHO in 1966 for eliminating endemic disease in Africa, Asia, and South America, and preventing its return to Europe, had ensured the global elimination of a disease for the first time in human history. The last natural case of smallpox was recorded in Somalia in 1977 [2–4]. Two principal variants of VARV infections were variola major, which was associated with an overall case fatality rates (CFR) of around 20%, and variola minor (also called alastrim), which was common in western Africa and the New World in the late XIX century but had lower CFR of <1%. There was no credible description of smallpox in the Americas or sub-Saharan Africa before the westward exploration of the XV century exported the disease to their aboriginal populations [5].
Although it has been claimed that cases of smallpox dating back thousands of years were observed in Egypt, China and India, the timescale of the emergence of variola virus and its evolution are unclear because there are significant gaps in historical medical records and controversies concerning the ancient or more recent origin of various forms of smallpox [5].
Several Authors have reconstructed the timescale of the evolution of VARV on a phylogenetic basis analyzing the few genomes available in public databases. The findings of phylogenetic studies suggest that the origin of smallpox went back about 3000–4000 YA, while the divergence between variola minor (P2) and variola major (P1) have been dated to 700–1000 YA [6, 7]. A study based on the use of viral ancient DNA obtained from a Lithuanian mummy of the XVII century, suggested a more recent time (about 250 YA) for the divergence between PI and P2 [8, 9]. A more recent analysis based on ancient VARV DNA obtained from the remains of individuals who lived in North Europe in the Viking Age allowed to backdate the origin of VARV between 600 and 1050 CE [10].
In a previous study based on hemagglutinin sequences, we described the evolutionary history of the entire Orthopoxvirus (OPV) genus: it was estimated that the root of the VARV clade dated back to 720 YA (corresponding to about 1300 AD) and that variola major and minor diverged about 500 YA (corresponding to about XV–XVI century) [1]. The main limitation of this study was that it was based on the analysis of partial conserved regions of the viral genome.
Despite the presence of several studies on the evolutionary history of Poxviruses and VARVs, specific studies on the phylogeography of the virus causing the last human epidemics are still scarce. Poxviridae have some of the largest genomes among vertebrate viruses (between 130 and 375 kb in length) and this may represent a limitation in the use of particularly time-consuming approaches for the phylogeographical analysis such as the classical Bayesian approach. One possible solution is represented by the use of maximum-likelihood methods allowing the use of complex models on large viral datasets for the ancestral character reconstruction (ACR) in a calendar timescale. Particularly, a recent developed fast and simple approach based on maximum likelihood which uses decision-theory concepts to associate each node of a tree to a set of likely states, implemented in PastML software, it has been recently proposed as a valid alternative to very time-consuming approaches for the reconstruction of phylogeography [11].
The aims of this study were to reconstruct on a spatial and temporal scale the phylogeography of VARV during the XX century, analyzing all the whole VARV genomes present in public databases (which include viral strains isolated between years 1940s and 1970s) and to assess the correlations between the phylogeographical reconstruction and the historical data available. Moreover, in this study we evaluated the employment of different methods for ACR comparing the commonly used Bayesian framework with a faster and simpler likelihood-based method.
MATERIALS AND METHODS
Sequence datasets
A total of 47 whole genome (WG) sequences of VARV with known sampling time and location, were retrieved from public databases (Genbank at: http://www.ncbi.nlm.nih.gov/genbank/).
The VARV sequences came from Ethiopia (n=2), Somalia (n=3), Tanzania (n=1), Sudan (n=2), South Africa (ZA), Botswana (n=2), Sierra Leone (n=1), Nigeria (n=1), Ghana (n=1), Benin (n=1), Bangladesh (n=4), Nepal (n=1), India (n=4), Germany (n=1), the United Kingdom (n=4), Afghanistan (n=1), Iran (n=1), Kuwait (n=1), Pakistan (n=1), Syria(n=1), Yugoslavia (n=1), Japan (n=3), China (n=1), Korea (n=1), Indonesia (n=2), Mexico (n=1) and Brazil (n=1).
Phylogenetic analysis by ML and Bayesian methods
Root-to-tip regression analysis was performed to investigate the temporal signal of the dataset by using TempEst [12].
The maximum likelihood tree of the data set was estimated by IQ-TREE v. 1.6.12 (http://www.iqtree.org/), using a Kimura’s three parameter model with unequal base frequencies and proportion of invariable sites (K3Pu+F+I) model selected by ModelFinder implemented in the program [13, 14]. Branch supports were obtained by ultrafast bootstrapping approximation with 1,000 parametric replicates [15]. Maximum Likelihood dating was obtained by the least square dating method (LSD2) integrated in IQ-TREE with 100 replicates to obtain confidence intervals in tMRCA estimates [16].
Given the lack of a representative number of variola minor sequences (P2), a subset of 40 strains exclusively containing P1 sequences (thus excluding alastrim) was used for the Bayesian phylogeographical analysis.
A general time reversible model (GTR) was used for analysis.
A Bayesian Markov Chain Monte Carlo (MCMC) method implemented in BEAST 1.8.4 was used to estimate evolutionary rates and the time of the Most Recent Common Ancestor (tMRCA) [17]. The coalescent priors of a constant population size, exponential growth, logistic growth, and a piecewise-constant Bayesian skyline plot (BSP), were tested under strict clock conditions as previously described [1, 18]. The chains were run for 30 million generations and sampled every 3,000 steps until reaching convergence which was assessed on the basis of the effective sampling size (ESS>200) after a 10% burn-in using Tracer software v.1.6 (http://tree.bio.ed.ac.uk/software/tracer/). Uncertainty in the estimates was indicated by 95% highest probability density (HPD) intervals, and the best fitting models were selected using a Bayes Factor (BF) with marginal likelihoods implemented in BEAST.
The strength of the evidence against the null hypothesis (H0) was evaluated as follows: 2lnBF <2 no evidence; 2–6 weak evidence; 6–10 strong evidence, and >10 very strong evidence [19]. A negative 2lnBF indicates evidence in favor of H0. A less restrictive Bayesian skyline plot was used as the coalescent prior. Only values of >6 were considered significant. BF calculations were made using Tracer v.1.6.
The maximum clade credibility (MCC) tree was then selected from the posterior tree distribution after excluding a 10% burn-in using the TreeAnnotator program v.1.8.4 included in the BEAST package, and visualized using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The tMRCA estimates were expressed as the mean number of years and 95%HPD years before the most recent sampling date, corresponding to 1977.
Phylogeographical analysis by ML and Bayesian methods
Each taxon was assigned to eight discrete groups on the basis of their sampling location: Africa (eastern Africa/EA, n=8; central Africa/CA, n=2; southern Africa/SA, n=4 and western Africa/WA=4); India (IN, n=13), the Middle East (ME, n=6), the Far East (FE, n=7) and America (AM=3). In the case of the strains isolated in Europe, where smallpox was already eradicated in the 1940s, we used the probable place of origin of the infection rather than the sampling location. PastML20 (https://pastml.pasteur.fr/) was used to reconstruct the ancestral geographic states along the ML dated tree. A maximum likelihood-based method (marginal posterior probabilities approximation-MPPA) and the F81-like model were used. The PartML generated tree was visualized and edited using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The Bayesian phylogeographical analysis of the subset of 40 P1 sequence was made using Beast 1.8.4 by assigning the isolates to six groups (the same groups described above, but without America and West Africa). The analyses were made using the continuous-time Markov chain (CTMC) process over discrete sampling locations [20] and a Bayesian stochastic search variable selection (BSSVS) approach in order to allow diffusion rates to be zero with a positive prior probability.
In order to estimate the direction of dispersion, the sequence data were fitted for two discretetrait models: a symmetrical model (which assumes non-zero rates of change between each pair of discrete states are equal) and the asymmetrical model, which assumes that the non-zero rates of change between each pair of discrete states are different. Comparison of the posterior and prior odds that the individual rates are non-zero provides a formal BF for testing the significance of the linkage between locations. Rates yielding a BF >3 were considered significant [20].
MCMC chains of the two datasets were run for 30 million generations and sampled every 3,000 generations until reaching convergence which was assessed on the basis of the effective sampling size (ESS ≥200) after a 10% burn-in by using Tracer software v.1.6. The final MCC tree was visualized using FigTree v. 1.4.2, which is freely available on the web (http://tree.bio.ed.ac.uk/software/figtree/), and the most probable location at each node was highlighted by labeling the branches with different colors.
In order to visualize diffusion rates over time, it is possible to convert the location-annotated MCC tree to a keyhole mark-up language (KML) file that is suitable for viewing with geo-referencing software using the SPREAD program (available at http://www.kuleuven.ac.be/aidslab/phylogeography/SPREAD.html). The migration routes indicated by the tree were visualized using Google Earth in order to provide a spatial projection (http://earth.google.com).
RESULTS
Root-to-tip regression analysis
Root-to-tip regression analysis (Figure 1) of the temporal signal revealed a strong association between genetic distances and sampling days (a correlation coefficient of 0.87 and a coefficient of determination (R2) of 0.76 with a slope (rate) of 1.05×10−5 substitutions/site/year, s/s/y).
Figure 1.
Root to Tip analyses of VARV isolates.
Evolutionary rates and time of the most recent common ancestor (tMRCA) estimations
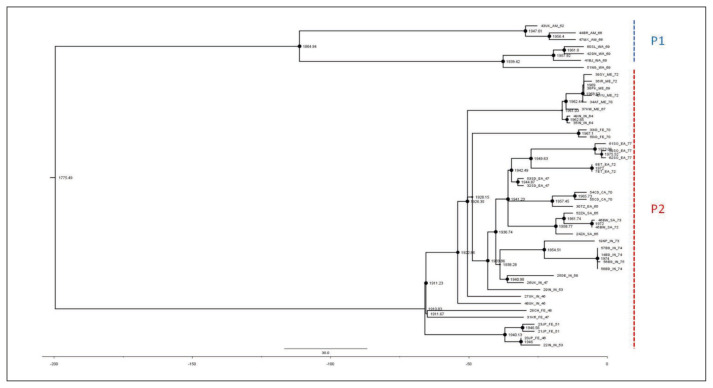
The ML dated tree of the whole genomes (Figure 2) showed two main significant clades with high statistical support (bootstrap=100 and posterior probability, pp=1) corresponding to the P1 and P2 lineages. The P1 lineage included isolates from all over the Old World (Africa, Asia and Europe) whereas the P2 lineage only included strains from America (alastrim) and western Africa. The evolutionary rate estimated by ML method resulted in a mean 8.4×10−6 s/s/y (95%CI, Confidential Interval: 6.73×10−6–1.1×10−5) and a mean tMRCA for the root of the tree corresponding to 1775 (95%CI: 1711–1825), while the mean tMRCAs of the P1 and P2 clades were 1911 (95%CI: 1898–1921) and 1865 (95%CI: 1819–1892), respectively.
Figure 2.
Dated ML tree, including all VARV whole genome sequences available (P1 = VARV major; P2 = VARV minor, alastrim). Node sizes are proportional to bootstrap support-max size = 100%). The scale axis below of the tree shows the years before the last sampling time, corresponding to 1977.
Bayesian analysis showed a topology of the tree identical to that obtained by ML and similar evolutionary rates (mean 9.41×10−6 s/s/y −95%HPD, Highest Posterior Density: 8.3×10−6–1.1×10−5) and tMRCA estimates (Table 1).
Table 1.
tMRCA estimates by ML and Bayesian methods.
| Maximum Likelihood | Bayesian | |||||
|---|---|---|---|---|---|---|
| Node | tMRCA a | 95% CIL b | 95% CIH c | tMRCA | 95% HPDL c | 95% HPDH d |
| Root | 1775.5 | 1711 | 1817 | 1794 | 1770 | 1816 |
| P1e | 1911 | 1898 | 1921 | 1912 | 1906 | 1919 |
| P2f | 1864 | 1819 | 1892 | 1874 | 1859 | 1888 |
tMRCA, time of the Most Recent Common Ancestor;
95% CI, 95% Confidential interval: Low limit;
95% CI, 95% Confidential interval-High limit;
95% HPDL, Highest Posterior Density Low Interval;
95% HPDH, Highest Posterior Density High Interval.
Phylogeographical reconstruction
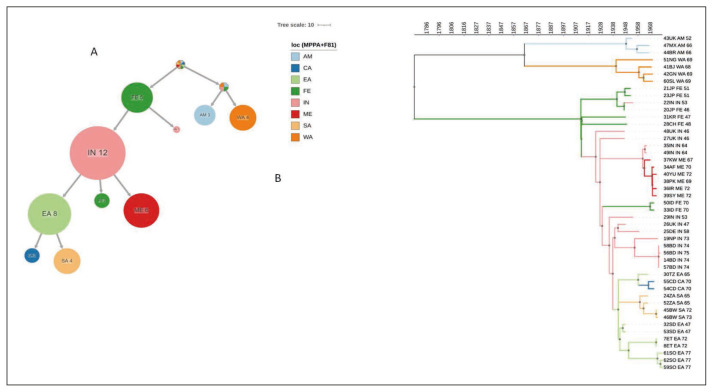
The maximum-likelihood based ACR using PastML (Figure 3A) showed that the location of the tree root was unresolved, as well as that of the P2 clade. On the contrary, the root of the P1 clade was resolved to Far East from where the epidemic spread to India, through at least two independent introductions: one forming a large cluster including 12 sequences and the second including only one sample. India acted as a centre of further spread to East Africa, the Middle East and the Far East. From East Africa, the epidemic further spread to South and Central Africa.
Figure 3.
Ancestral reconstruction of VARV phylogeography by PastML, using MPPA with F81-like model. (a) compressed visualtization. (b) full tree. Different colors correspond to different geographical areas (see legend. Loc = location). Calendar years are reported on the axis. CA = central Africa; EA = eastern Africa; FE = Far East; IN = India; ME = Middle East; SA = southern Africa, WA = western Africa, AM = America.
The phylogeographic tree timeline (Figure 3B) showed that the epidemic reached India in the first decades of 1900s (between 1911, the root of P1 clade and 1922, the first node most probably located in India), spread to East Africa in early 1940s and to the Middle East in the 1960s. From East Africa the epidemic spread to South Africa between late 1950s and 1960s and to Central Africa in 1960s.
Given the uncertainty of the P2 root and the scarce number of isolates of this strain, the Bayesian phylogeographical analysis was limited to the P1 clade.
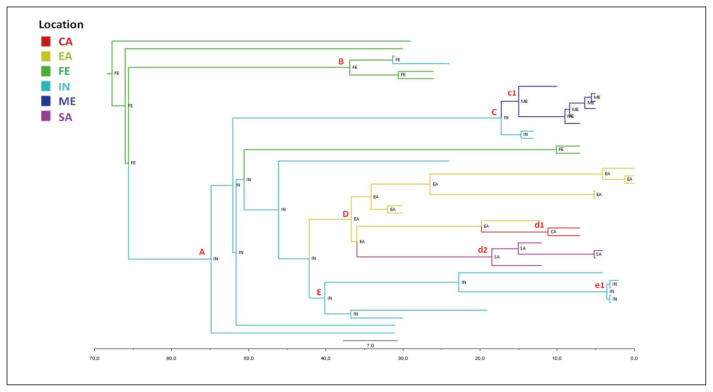
The maximum credibility tree shown in Figure 4 indicated the Far East as the most probable location of P1 clade root with a state posterior probability (spp=1).
Figure 4.
Bayesian phylogeographical tree of P1 genomes. Branches are coloured on the basis of the most probable location of the descendent nodes that is indicated. The colour legend is shown in the panel (bottom left). (CA = central Africa; EA = eastern Africa; FE = Far East; IN = India; ME = Middle East; SA = southern Africa). The scale axis below the tree shows the years before the last sampling time (1977).
The tree showed several nested clades (Table 2), the largest of which (node A) have India as a most probable place of origin (spp=1), as well as the deepest nodes of the clade (nodes C and E). The terminal subclades included isolates from homogeneous geographic areas segregating significantly and showing different MRCA-locations (node c1: Middle East, node d1: Central Africa, node d2: South Africa, node D: East Africa, and node B: Far East).
Table 2.
MRCA-locations and tMRCA estimations in different clades.
| Clade | Location | spp a | tMRCA b | 95% HPDL c | 95% HPDH d | Year | 95% HPDL | 95% HPDH |
|---|---|---|---|---|---|---|---|---|
| Root | FE | 1 | 67.8 | 61 | 76.11 | 1909.2 | 1901 | 1916 |
| A | IN | 1 | 54.9 | 49.5 | 60.9 | 1922.1 | 1916.1 | 1927.5 |
| B | FE | 1 | 36.9 | 34.1 | 40 | 1940.1 | 1937 | 1942.9 |
| C | IN | 1 | 17.2 | 15.2 | 19.4 | 1959.8 | 1957.6 | 1961.8 |
| c1 | ME | 0.99 | 15 | 12.8 | 17.4 | 1962 | 1959.6 | 1964.2 |
| D | EA | 1 | 36.7 | 33.8 | 40 | 1940.3 | 1937 | 1943.2 |
| d1 | CA | 1 | 11.2 | 9.3 | 13.2 | 1965.8 | 1963.8 | 1967.7 |
| d2 | SA | 1 | 18.5 | 15.7 | 21.2 | 1958.5 | 1955.8 | 1961.3 |
| E | IN | 0.96 | 40.1 | 36.3 | 44.4 | 1936.9 | 1932.6 | 1940.7 |
| e1 | IN* | 1 | 3.5 | 3.1 | 4.1 | 1973.5 | 1972.9 | 1973.9 |
spp, state posterior probability;
tMRCA, time of the Most Recent Common Ancestor;
95% HPDL, Highest Posterior Density Low Interval;
95% HPDH, Highest Posterior Density High Interval;
Bangladesh;
FE: Far East, IN: India, ME: Middle East, EA: eastern Africa, CA: central Africa, SA: southern Africa.
The Bayesian symmetrical and asymmetrical phylogeographic diffusion models indicated five highly supported linkages from eastern to southern Africa (Bayes Factor, BF=26.4) and central Africa (BF=64.5), from the Far East to India (BF=427.5), and from India to the Middle East (BF=99.6) and eastern Africa (BF=34.5).
Phylogeographical fluxes (Figure 5) indicated that the tMRCA of the P1 clade existed in the Far East in the first decade of 1900s (mean 1909 – 95%HPD: 1901–1916) and moved to India in 1920s (mean 1922 – 95%HPD: 1916–1927) from where it arrived in eastern Africa in 1940s (mean 1940 – 95%HPD: 1937–1943) from which the virus spread to all the continent, reaching southern Africa in the 1950s (mean 1958 – 95%HPD: 1956–1961) and central Africa in the 1960s (mean 1966 – 95%HPD: 1964–1968). Finally the virus reached the Middle East starting always from India in the 1960s (mean 1962 – 95%HPD: 1960–1964).
Figure 5.
Significant non-zero migration rates of VARV P1 worldwide supported by a Bayes Factor >4. The migrations were calculated using SPREAD program. The dates in which the virus entered the area and the direction of fluxes are shown (CA = central Africa; EA = eastern Africa; FE = Far East; IN = India; ME = Middle East; SA = southern Africa).
DISCUSSION
Several authors have attempted to reconstruct the evolutionary history of the human smallpox virus by using time scaled phylogeny based on sequences/genomes sampled in the XX century, or based on ancient DNA from mummies, obtaining contrasting results on the origin of VARV [6, 8, 10, 21, 22].
A recent work published by us aiming at reconstructing the evolutionary history of the entire Orthopoxvirus genus, led to an estimated tMRCA of about 10,000 YA for the entire animal tree root and a mean tMRCA of 720 YA for the VARV species [1].
This preliminary analysis was based on a single gene (B7R encoding for the viral protein hemagglutinin) and on an historical/anthropological calibration that assumed the post-Colombian divergence of the South American and western Africa variola minor clades, and indicated a mean evolutionary rate of about 6×10−6 substitutions/site/year.
In the present study, we aimed to reconstruct a possible phylogeographic scenario of the VARVs circulating in the XX century immediately before its eradication employing a short timescale to calibrate the molecular clock by using “heterochronous” VARV sequences with known sampling dates available in public databases. Since all these isolates were obtained in a short interval of time, between the 1940s and 1970s, they represent only a very small fraction of a long evolutionary history.
The newly estimated mean evolutionary rate on the entire viral genomes is between 6.73×10−6 and 1.1×10−5 sub/site/year, including in the lower bound the value previously estimated on hemagglutinin gene. Under these conditions, we estimate that the divergence of P1 and P2 occurred in the XVIII century, and that the time of radiation of P1 and P2 was respectively the first decade of 1900s and the second half of 1800s. Comparable results have recently been obtained by Duggan et al., who studied an ancient strain of VARV obtained from a Lithuanian mummy [8].
These results, indicating an apparently recent history of VARV, may be due to the disappearance of basal lineages after vaccination and more probably are reporting the history of the extant lineages. This explanation was also suggested by the studies based on the few available ancient sequences showing that they are always basal to all the XX century sequences. In particular, the recent identification of a now-extinct sister clade from northern-European human remains dating back to the Viking age, allowed the origin of the VARV to be dated back to about 1700 YA [10]. The oldest sequences in our analysis were isolated in the 1940s, when endemic lineages had already been eliminated in a number of countries, including Europe.
The long branch connecting P1 and P2 in the complete VARV phylogeny suggest the existence of major bottlenecks in both clades during the XIX and XX centuries, probably because the increase in global smallpox vaccination led to the extinction of various lineages, as proposed by Duggan et al [8]: for example, the last major outbreaks of variola major in England and Wales occurred in 1902–1905 [2, 23]. However, the deleterious effect of the First World War on the health conditions of civilians led to large smallpox epidemics affecting tens of thousands of people in Europe, and these contrasting phenomena may explain the phylogenetic bottleneck observed [2].
Although smallpox had been eliminated from almost all European countries by the late 1930s, isolated incidents occurred during and after the Second World War in Europe causing small outbreaks due to imported cases from colonial possessions or neighboring countries in which VARV was still endemic [2, 24]. Nevertheless, the original European strain was already extinct in that period and for this reason we decided to use the most probable place of origin of the infection rather than the sampling location for the sequences from Europe.
Our phylogeographical reconstruction of the P1 variola major clade suggests that the common ancestor of the strains circulating in the Old World between the 1940s and the 1970s originated in the Far East in the first two decades of the XX century, and then spread to Indian subcontinent in the 1920s.
Smallpox was highly endemic in the Far East in the first decades of the XX century, and caused mainly seasonal epidemics [2].
The migration of the virus to the Indian sub-continent in the 1920s may have been due to the close relationships between former British colonies, (particularly between British India and Hong Kong), before and during the First World War. Although the introduction of smallpox vaccinations in British India in the late XIX century led to a decrease in the number of deaths, smallpox remained endemic even after the partition of British India in 1947. Particularly, a major epidemic in Indian port cities in the 1930s required the restriction of regional maritime trading. With the sole exception of Sri Lanka, smallpox was definitively eliminated from the Indian subcontinent only after the 1970s [2].
In our reconstruction, India acted as an important center of further spreading of VARV to East Africa in the 1940s and to the Middle East in the ‘60s.
During the second World War several British Indian divisions were engaged in the East African campaign possibly justifying the exportation of Asian smallpox to East Africa.
Although smallpox vaccinations were introduced in eastern Africa in the 1940s, the disease remained endemic until the 1970s, and Somalia suffered the very last variola major epidemic in 1977. In our phylogeographic scenario, the virus spread to other African countries, in the central and southern part of the continent between 1950s and 1960s, in particular in the former British colonies where important communities of South Asian descendent people were living [2].
Finally, Muslim pilgrimages to the holy cities and the presence of migrant workers from endemic areas in the Gulf States contributed to the resurgence of smallpox in Iran, Iraq and Syria in the early 1960s, justifying our observation of a flux of virus from India to the Middle East in the ‘60s [2, 25].
A second aim of this study was the comparison between different approaches for ACR: a commonly used, highly flexible Bayesian approach and a faster and simpler maximum-likelihood based approach [11, 17]. The ACR obtained were largely congruent both in time and space, however, the Bayesian approach took days to reconstruct the phylogeography on 40 P1 genomes, while maximum-likelihood approach allowed us to obtain the closely similar results only in a few hours using all the 47 genomes.
The main limitation of this study is the small number of complete genomes analyzed, only 47 compared to an infection that during the twentieth century caused millions of cases on almost all continents.
Nonetheless, this limitation is common to all studies that have attempted a phylogenetic-based reconstruction of the history of the smallpox virus, including those that rely on ancient DNA that are based on sporadic sampling of frequently incomplete sequences. Nevertheless, all these studies, including ours, can make an important contribution, together with historical and paleoanthropological studies, to the reconstruction of the evolutionary and epidemiological history of an infection that has influenced important phases of human history.
Moreover, our results may help to explain the controversies concerning the phylogenetic reconstruction using different time scales and represent the first application of a new maximum-likelihood approach to the reconstruction of a hypothetical ancestral scenario of VARV.
Footnotes
Data availability statement
All the sequences used in this work are available in Genbank at: http://ncbi.nlm.nih.gov/genbank/.
Conflict of interest disclosure
The authors declare no conflict of interest.
Author contributions
AL, AB, GZ, and MG conceived and designed the study. AB, AL, CDV, GZ, MC and RM made the phylogenetic analyses. AL, AB, GZ, and MG wrote the first draft of the manuscript. All of the authors contributed to revising the manuscript, and read and approved the submitted version.
Funding statement
This research received no external funding.
REFERENCES
- 1.Zehender G, Lai A, Veo C, Bergna A, Ciccozzi M, Galli M. Bayesian reconstruction of the evolutionary history and cross-species transition of variola virus and orthopoxviruses. J Med Virol. 2018;90:1134–41. doi: 10.1002/jmv.25055. [DOI] [PubMed] [Google Scholar]
- 2.Fenner F. Smallpox and its eradication. World Health Organization; 1988. [Google Scholar]
- 3.Sánchez-Sampedro L, Perdiguero B, Mejías-Pérez E, García-Arriaza J, Di Pilato M, Esteban M. The evolution of poxvirus vaccines. Viruses. 2015;7:1726–803. doi: 10.3390/v7041726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shchelkunova GA, Shchelkunov SN. 40 Years without Smallpox. Acta Naturae. 2017;9:4–12. [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Nat Acad Sci. 2007;104:15787–92. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes AL, Irausquin S, Friedman R. The evolutionary biology of poxviruses. Infect Genet Evol. 2010;10:50–9. doi: 10.1016/j.meegid.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babkin IV, Babkina IN. A retrospective study of the orthopoxvirus molecular evolution. Infect Genet Evol. 2012;12:1597–604. doi: 10.1016/j.meegid.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Duggan AT, Perdomo MF, Piombino-Mascali D, et al. 17(th) Century Variola Virus reveals the recent history of Smallpox. Curr Biol. 2016;26:3407–12. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biagini P, Thèves C, Balaresque P, et al. Variola virus in a 300-year-old Siberian mummy. N Engl J Med. 2012;367:2057–9. doi: 10.1056/NEJMc1208124. [DOI] [PubMed] [Google Scholar]
- 10.Mühlemann B, Vinner L, Margaryan A, et al. Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science. 2020:369. doi: 10.1126/science.aaw8977. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol. 2019;36:2069–85. doi: 10.1093/molbev/msz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UF Boot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.To TH, Jung M, Lycett S, Gascuel O. Fast dating using least-squares criteria and algorithms. Syst Biol. 2016;65:82–97. doi: 10.1093/sysbio/syv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 19.Kass RE, Raftery AE. Bayes factors. J Am Stat Ass. 1995;90:773–95. [Google Scholar]
- 20.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babkin IV, Babkina IN. The origin of the variola virus. Viruses. 2015;7:1100–12. doi: 10.3390/v7031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biagini P, Theves C, Balaresque P, et al. Variola virus in a 300-year-old Siberian mummy. New Engl J Med. 2012;367:2057–9. doi: 10.1056/NEJMc1208124. [DOI] [PubMed] [Google Scholar]
- 23.Geddes AM. The history of smallpox. Clin Dermatol. 2006;24:152–7. doi: 10.1016/j.clindermatol.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Millward G. Vaccinating Britain: Mass vaccination and the public since the Second World War. Manchester UK: © Gareth Millward; 2019. p. 2019. [PubMed] [Google Scholar]
- 25.Ristanovic E, Gligic A, Atanasievska S, Protic-Djokic V, Jovanovic D, Radunovic M. Smallpox as an actual biothreat: lessons learned from its outbreak in ex-Yugoslavia in 1972. Ann Ist Super Sanita. 2016;52:587–97. doi: 10.4415/ANN_16_04_21. [DOI] [PubMed] [Google Scholar]