Abstract
Background
The SARS-CoV-2 omicron (B.1.1.529) variant, which was first identified in November, 2021, spread rapidly in many countries, with a spike protein highly diverged from previously known variants, and raised concerns that this variant might evade neutralising antibody responses. We therefore aimed to characterise the sensitivity of the omicron variant to neutralisation.
Methods
For this cross-sectional study, we cloned the sequence encoding the omicron spike protein from a diagnostic sample to establish an omicron pseudotyped virus neutralisation assay. We quantified the neutralising antibody ID50 (the reciprocal dilution that produces 50% inhibition) against the omicron spike protein, and the fold-change in ID50 relative to the spike of wild-type SARS-CoV-2 (ie, the pandemic founder variant), for one convalescent reference plasma pool (WHO International Standard for anti-SARS-CoV-2 immunoglobulin [20/136]), three reference serum pools from vaccinated individuals, and two cohorts from Stockholm, Sweden: one comprising previously infected hospital workers (17 sampled in November, 2021, after vaccine rollout and nine in June or July, 2020, before vaccination) and one comprising serum from 40 randomly sampled blood donors donated during week 48 (Nov 29–Dec 5) of 2021. Furthermore, we assessed the neutralisation of omicron by five clinically relevant monoclonal antibodies (mAbs).
Findings
Neutralising antibody responses in reference sample pools sampled shortly after infection or vaccination were substantially less potent against the omicron variant than against wild-type SARS-CoV-2 (seven-fold to 42-fold reduction in ID50 titres). Similarly, for sera obtained before vaccination in 2020 from a cohort of convalescent hospital workers, neutralisation of the omicron variant was low to undetectable (all ID50 titres <20). However, in serum samples obtained in 2021 from two cohorts in Stockholm, substantial cross-neutralisation of the omicron variant was observed. Sera from 17 hospital workers after infection and subsequent vaccination had a reduction in average potency of only five-fold relative to wild-type SARS-CoV-2 (geometric mean ID50 titre 495 vs 105), and two donors had no reduction in potency. A similar pattern was observed in randomly sampled blood donors (n=40), who had an eight-fold reduction in average potency against the omicron variant compared with wild-type SARS-CoV-2 (geometric mean ID50 titre 369 vs 45). We found that the omicron variant was resistant to neutralisation (50% inhibitory concentration [IC50] >10 μg/mL) by mAbs casirivimab (REGN-10933), imdevimab (REGN-10987), etesevimab (Ly-CoV016), and bamlanivimab (Ly-CoV555), which form part of antibody combinations used in the clinic to treat COVID-19. However, S309, the parent of sotrovimab, retained most of its activity, with only an approximately two-fold reduction in potency against the omicron variant compared with ancestral D614G SARS-CoV-2 (IC50 0·1–0·2 μg/mL).
Interpretation
These data highlight the extensive, but incomplete, evasion of neutralising antibody responses by the omicron variant, and suggest that boosting with licensed vaccines might be sufficient to raise neutralising antibody titres to protective levels.
Funding
European Union Horizon 2020 research and innovation programme, European and Developing Countries Clinical Trials Partnership, SciLifeLab, and the Erling-Persson Foundation.
Introduction
The SARS-CoV-2 omicron variant (B.1.1.529) has rapidly replaced the highly transmissible delta variant (B.1.617.2) in many countries.1 Compared with the original SARS-CoV-2 virus, the archetypical omicron (BA.1) variant harbours two deletions, one insertion, and 30 amino acid differences in the viral spike protein, including many mutations known or predicted to confer resistance to neutralising antibodies. However, their combined effect, and the phenotypic effects of a number of novel omicron mutations, were unknown.
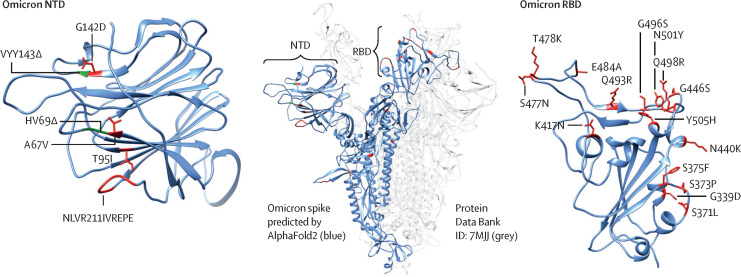
The deletions and insertions in the viral spike protein of the omicron variant are located within the N-terminal domain, a known target of neutralising antibodies,2 and the receptor binding domain, which exhibits 15 non-synonymous mutations, many of which cluster in and around the angiotensin-converting enzyme 2 (ACE2) receptor binding motif (figure 1 ). Mutations at amino acid positions 484, 417, and 501 are common to multiple variants of concern, and these three mutations alone (but E484K instead of E484A in the omicron variant) explain the majority of resistance exhibited by the beta (B.1.351) variant,3 which has no other receptor binding domain mutations. Deep mutational scanning data suggest that E484A and K417N, in addition to G446S and Q493R (which are not present in other variants of concern) are the largest contributors to the resistance profile of the omicron variant.4
Figure 1.
Omicron spike mutations
Changes in the NTD (left) and RBD (right) that have potential immunological significance are labelled. Residues on either side of a deletion are shown in green, and point mutations and insertions are shown in red. Changes are visualised on a model of an omicron spike protomer.12 NTD=N-terminal domain. RBD=receptor binding domain.
Research in context.
Evidence before this study
Towards the end of 2021, the novel SARS-CoV-2 omicron (B.1.1.529) variant rapidly replaced the highly transmissible delta (B.1.617.2) variant in many countries. Sequencing showed that the omicron variant was extensively diverged from all other previously known lineages and harboured a number of mutations in the viral spike protein, including many mutations known or predicted to confer resistance to neutralising antibodies. However, the combined effect of these mutations, and the phenotypic effects of a number of novel omicron mutations, were unknown, and no experimental data were available on the resistance of omicron to neutralising antibodies at the onset of this study. We searched PubMed from database inception to Dec 19, 2021, without language restrictions, for articles using the search terms: “((SARS-CoV-2) AND ((Neutralisation) OR (Neutralisation)) AND ((Omicron) OR (B.1.1.529)))”. Our search yielded 17 articles, of which four were directly relevant. Two preprints evaluated neutralisation of live omicron isolates for recipients of the BNT162b2 mRNA vaccine (Pfizer–BioNTech), reporting an approximate 36-fold to 40-fold reduction in sensitivity relative to ancestral SARS-CoV-2 isolates. Two preprints reported resistance of omicron spike pseudotyped viruses to neutralisation by serum from convalescent and vaccinated individuals (BNT162b2, mRNA-1273 [Moderna], and Ad26.COV2.S [Johnson & Johnson] vaccine recipients), demonstrating low or undetectable titres against the omicron variant after infection or primary vaccination but substantial cross-neutralisation of omicron after an additional mRNA vaccine dose. An additional preprint not identified in the literature search reported on the neutralisation of the omicron variant by a panel of monoclonal antibodies (mAbs), showing that the omicron variant evaded neutralisation by approximately 85% of mAbs tested.
Added value of this study
This study provides in-vitro data on the sensitivity of the omicron variant to antibody-mediated neutralisation and quantifies the loss of neutralisation potency by vaccine and convalescent serum standards, as well as by five mAbs incorporated in licensed antibody therapies. Furthermore, we assessed neutralising activity against the omicron variant in sera from two Swedish cross-sectional cohorts, providing insight into population-level immunity against the omicron variant in Sweden at present. The initial serum neutralisation results from this study were disseminated just 13 days after the omicron variant was first reported to WHO.
Implications of all the available evidence
In agreement with several studies done in parallel, we identified that most mAbs incorporated into therapeutics licensed for human use have little or no neutralising activity against the omicron variant, which is likely to undermine their efficacy. One mAb, S309 (the parent of sotrovimab), retained potency against omicron, suggesting that sotrovimab might retain clinical utility. Furthermore, the results of this study and others have shown that the omicron variant displays profound escape from neutralising antibodies in serum samples obtained after infection or vaccination, which is likely to underpin the reductions in vaccine effectiveness observed in real-world settings. However, in a real-world cohort of blood donors from Stockholm (Sweden), reduced but detectable cross-neutralisation of the omicron variant was evident, suggesting that loss of protection at the population level might be less substantial in certain groups. We showed that although little or no cross-neutralisation of the omicron variant was identified in the sera of convalescent individuals, individuals who had been infected before being vaccinated had considerably higher neutralising potency against the omicron variant, highlighting the benefit of vaccination in individuals who have been previously infected. This finding is in line with other studies that emphasised the benefits of an additional vaccine dose in broadening neutralising antibody responses, including against the omicron variant.
Such substantial antigenic drift might undermine protection afforded by currently licensed vaccines and monoclonal antibodies (mAbs) used in the clinic. We therefore aimed to characterise, using a pseudotyped virus assay, the sensitivity of the omicron variant to neutralisation by relevant monoclonal antibodies, pooled serum from vaccine recipients and convalescent individuals, serum samples from previously infected and previously infected-then-vaccinated hospital workers, and serum from a random sample of blood donors.
Methods
Serum samples
For this cross-sectional study, we studied two cohorts. The first cohort comprised serum samples from 40 blood donors (anonymised and therefore with unknown exposure and vaccination status), donated during week 48 of 2021 (Nov 29–Dec 5), in Stockholm, Sweden. The second cohort comprised serum samples from previously infected hospital workers at the Karolinska University Hospital (Stockholm, Sweden; as described previously5) who were confirmed to be SARS-CoV-2 positive by PCR in April or May, 2020. 17 previously infected hospital workers had serum sampled in November, 2021, after vaccine rollout, and nine had serum sampled in June or July 2020, before vaccination (defined as convalescent samples hereafter). Informed consent was obtained from all participants included in the hospital worker and convalescent cohorts as part of an ethics approval (decision number 2020-01620, with amendments 2020-02881, 2020-05630, and 2021-04377) from the Swedish Ethical Review Authority. The blood donor cohort and the omicron-positive sample from which the spike was cloned were anonymised and thus not subject to ethical approvals, as per advisory statement 2020–01807 from the Swedish Ethical Review Authority.
Plasma and serum pooled standards were also studied, including the first WHO International Standard for anti-SARS-CoV-2 immunoglobulin (20/136), pooled from convalescent patients in 2020. Additionally, pooled serum standards from vaccinated individuals (including BEI Resources NRH-17727, NRH-17846, and NRH-20012)6 were provided by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health. For the three pools, serum was isolated approximately 3 months after completion of the primary vaccination series (mean 69 days [SD 14·9] for recipients of the BNT162b2 mRNA vaccine [Pfizer–BioNTech], 102 days [13·3] for recipients of the mRNA-1273 [Moderna] vaccine, and 83 days [19·0] for recipients of the Ad26.COV2.S [Johnson & Johnson] vaccine) from six donors with no history of infection who were aged between 18 and 55 years (mean age 44·2 years [SD 15·1] for BNT162b2 recipients, 52·2 years [16·0] for mRNA-1273 recipients, and 36·8 years [12·5] for Ad26.COV2.S recipients).7 The interval between first and second vaccine doses was approximately 3 weeks for BNT162b2 recipients and 4 weeks for mRNA-1273 recipients, with no second dose given for Ad26.COV2.S recipients.
mAb production
We produced in-house versions of clinically relevant mAbs: casirivimab (REGN-10933),7 imdevimab (REGN-10987),7 bamlanivimab (LY-CoV555),8 etesevimab (LY-CoV016),8 and S3099 (from which sotrovimab was derived). mAbs were expressed in Expi293 cells and purified using protein G Agarose columns (appendix pp 3–4).
Pseudovirus neutralisation assay
For use in the pseudovirus neutralisation assay, an omicron variant spike protein was molecularly cloned from an anonymised diagnostic sample, suspected to contain omicron due to S-gene target failure, which was subsequently confirmed by sequencing. The approach and primers used for the construction of the omicron spike expression plasmid are described in the appendix (pp 2–3, 5). The resulting spike plasmid had an amino acid sequence identical to the omicron consensus, using native codons from amino acids 43–1000, and was codon optimised outside of this region. Plasmids encoding the spikes from the B.1 (D614G), mu (B.1.621), and delta variants were obtained from the G2P-UK National Virology consortium.10
HEK293T cells (CRL-3216; ATCC, Manassas, VA, USA) and HEK293T-ACE2 cells (stably expressing human ACE2) were cultured in Dulbecco's Modified Eagle Medium (high glucose, with sodium pyruvate) supplemented with 10% fetal calf serum, 100 units per mL penicillin and 100 μg/mL streptomycin. Cultures were maintained in a humidified 37°C incubator (5% CO2).
The pseudovirus neutralisation assay was done as previously described.11 Spike-pseudotyped lentivirus particles were generated by the co-transfection of HEK293T cells with a relevant spike plasmid, an HIV gag-pol packaging plasmid (8455; Addgene, Watertown, MA, USA), and a lentiviral transfer plasmid encoding firefly luciferase (170674; Addgene) using polyethylenimine.
Neutralisation was assessed in HEK293T-ACE2 cells. Briefly, pseudoviruses sufficient to produce approximately 30 000 relative light units were incubated with serial three-fold dilutions of serum for 60 min at 37°C in a black-walled 96-well plate. 10 000 HEK293T-ACE2 cells were then added to each well, and plates were incubated for 48 h. Luminescence was measured using Bright-Glo Luciferase Assay System (Promega, Madison, WI, USA) on a GloMax Navigator Microplate Luminometer (Promega). Neutralisation was calculated relative to the average of eight control wells infected in the absence of serum. All fold-changes reported use titres from neutralisation assays run in parallel.
Statistical analysis
Individual ID50 values for each sample against each variant were calculated in GraphPad Prism (version 9.0) by fitting a four-parameter logistic curve to neutralisation by serial three-fold dilutions of serum. For comparisons of titres across variants, we used non-parametric Wilcoxon matched-pairs tests. To assess whether fold reductions were normally distributed, we used a Shapiro-Wilk test in Prism. We assayed all available samples and no sample size calculations were performed.
The omicron spike protein used for the visualisation (figure 1) was modelled in AlphaFold2 (appendix p 4). We created a list of amino acid positions mutated in the omicron variant that were potentially relevant for mAb escape on the basis of whether, for each reported mAb, they were proximal to the mAb–receptor binding domain interface, and we cross-referenced this list with deep mutational scanning13 data against that specific antibody (appendix p 4).
Role of the funding source
The funders had no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or the decision to submit for publication.
Results
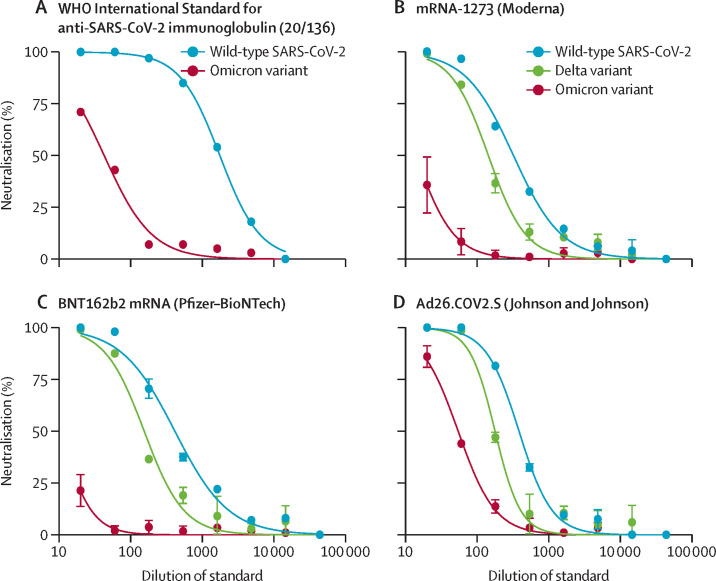
The WHO International Standard Immunoglobulin (20/136) had approximately 40-fold lower ID50 titres against omicron compared with wild-type SARS-CoV-2 (from 1765 [0·6 IU/ml] to 43 [23·4 IU/ml]; figure 2A ), indicating substantial resistance to antibodies elicited by infection with early variants of SARS-CoV-2.
Figure 2.
Neutralisation of the SARS-CoV-2 omicron (B.1.1.529) variant by reference reagents
Neutralisation of the omicron variant and wild-type SARS-CoV-2 by the first WHO International Standard for anti-SARS-CoV-2 immunoglobulin from convalescent individuals (A), and neutralisation of the omicron variant, delta (B.1.617.2) variant, and wild-type SARS-CoV-2 by pooled sera standards from recipients of the mRNA-1273 vaccine (Moderna; B), BNT162b2 mRNA vaccine (Pfizer-BioNTech; C), and the Ad26.Cov2.S vaccine (Johnson & Johnson; D). Error bars show SD around the mean. The WHO International Standard for anti-SARS-CoV-2 immunoglobulin was assayed only once per variant due to reagent limitations.
We assessed neutralisation of the omicron variant, delta variant, and wild-type SARS-CoV-2 by pooled serum standards6 from recipients of BNT162b2 (Pfizer-BioNTech), mRNA-1273 (Moderna), and Ad26.COV2.S (Johnson & Johnson) vaccines. We found that neutralisation of omicron was substantially reduced (by seven-fold to >40-fold), across the vaccine standard serum pools (figure 2B–D; appendix p 9).
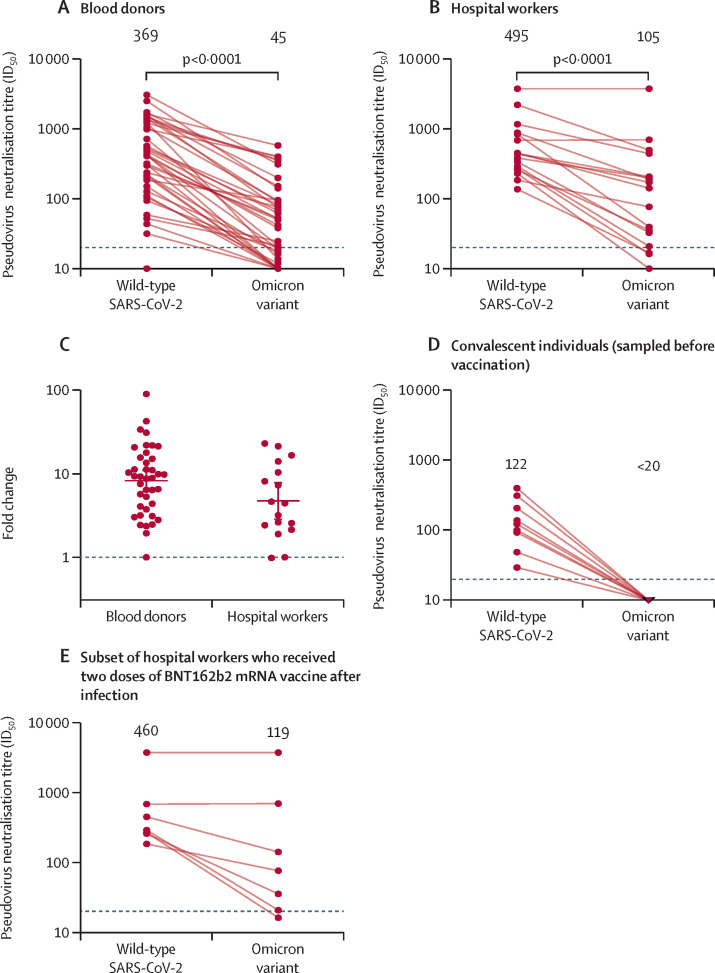
While neutralising activity in serum sampled shortly following vaccination provides critical information about the antibody responses elicited and boosted by vaccines, immunity at the population level and real-world vaccine protection incorporates not just vaccination but a variety of previous and subsequent exposures, as well as waning14, 15 of the responses to these. Therefore, to provide a snapshot of immunity at the population level before the introduction of omicron, we assessed neutralisation by sera from two cohorts from Stockholm, Sweden. Geometric mean neutralising ID50 titres for the blood donors were approximately eight-fold lower against the omicron variant than against wild-type virus (figure 3A ). However, the reduction in neutralising activity was heterogeneous, with some sera nearly 90-fold less potent and others having no significant reduction in potency, indicating the presence of cross-neutralising antibodies in a subset of donors. Similarly, geometric mean ID50 titres from hospital workers were around five-fold lower against the omicron variant than against wild-type virus (figure 3B), and considerable inter-individual variation was observed (figure 3C; table 1 ). Fold changes in neutralising ID50 titres in both cohorts were consistent with a log-normal distribution (appendix p 10), and the potency with which a sample neutralised the omicron variant correlated with its ability to cross-neutralise other variants (appendix p 8).
Figure 3.
Neutralisation of the omicron (B.1.1.529) variant by serum samples
Paired pseudovirus neutralisation titres against wild-type SARS-CoV-2 and the omicron variant in the blood donor cohort (n=40; A) and hospital worker cohort (n=17; B), and comparison of the fold reduction in titres between the blood donor and hospital worker cohorts (C). Neutralisation titres against wild-type SARS-CoV-2 and the omicron variant for samples obtained from previously infected hospital workers before vaccination (n=9; D) and for the subset of the hospital workers who received two doses of the BNT162b2 mRNA vaccine (n=7; E). Numbers above the graphs are geometric mean titres. Dotted lines in parts A, B, D, and E show the lowest dilution tested in the assay; the dotted line in part C indicates no difference in geometric mean titre. Error bars in part C show the geometric mean and 95% CI. ID50=reciprocal serum dilution that produces 50% inhibition.
Table 1.
ID50 titres against omicron and wild-type SARS-CoV-2 in hospital workers infected with SARS-CoV-2 in early 2020
| Wild-type SARS-CoV-2 ID50 | Omicron ID50 | Fold change in ID50 (wild-type vs omicron) | Self-reported vaccine history* | |
|---|---|---|---|---|
| Individual A | 840 | 43 | 20 | Unknown |
| Individual B | 442 | 213 | 2 | ChAdOx1 and BNT162b2 (4 months) |
| Individual C | 686 | 734 | 1 | Two doses of BNT162b2 (4 months) |
| Individual D | 185 | 79 | 2 | Two doses of BNT162b2 (8 months) |
| Individual E | 879 | 214 | 4 | Two doses of ChAdOx1 (5 months) |
| Individual F | 230 | 10 | 23 | Unknown |
| Individual G | 137 | 17 | 8 | ChAdOx1 and BNT162b2 (4 months) |
| Individual H | 259 | 36 | 7 | Two doses of BNT162b2 (8 months) |
| Individual I | 440 | 179 | 2 | Two doses of ChAdOx1 (4 months) |
| Individual J | 2202 | 549 | 4 | Unknown |
| Individual K | 382 | 207 | 2 | Two doses of ChAdOx1 (5 months) |
| Individual L | 451 | 155 | 3 | Two doses of BNT162b2 (5 months) |
| Individual M | 338 | 36 | 9 | Unknown |
| Individual N | 269 | 16 | 17 | Two doses of BNT162b2 (8 months) |
| Individual O | 3748 | 4053 | 1 | Two doses of BNT162b2 (2 months) |
| Individual P | 291 | 22 | 13 | Two doses of BNT162b2 (4 months) |
| Individual Q | 1166 | 493 | 2 | Unknown |
Age of individuals ranged from 28 to 74 years (median 54 years [IQR 41–62]). ID50=reciprocal serum dilution that produces 50% inhibition.
Calendar months between most recent immunisation and day of serum sampling are shown in parentheses.
Historical samples obtained from nine hospital workers after confirmed SARS-CoV-2 infection, but before vaccination, showed a near-complete loss of neutralising activity against the omicron variant (figure 3D). However, for seven hospital workers who received two doses of the BNT162b2 vaccine after infection, robust cross-neutralisation of the omicron variant was evident in a number of individuals (figure 3E), highlighting the improvement in the neutralisation of the omicron variant afforded by vaccination in previously infected individuals.
We assessed the sensitivity of the omicron variant to neutralisation by several clinically relevant mAbs that have previously been licensed or authorised for human use. Casirivimab (REGN-10933), imdevimab (REGN-10987), etesevimab (Ly-CoV016), and bamlanivimab (Ly-CoV555) did not neutralise the omicron variant at the highest concentration tested (10 μg/mL; table 2 ). However, the parent antibody of sotrovimab, S309, maintained its activity, with only a two-fold reduction in potency against the omicron variant compared with ancestral B.1 (D614G) virus (table 2), which is likely to be attributable to the location of the epitope that sotrovimab binds to being outside of the highly mutated receptor binding motif.
Table 2.
Neutralising potency of clinically relevant monoclonal antibodies against the SARS-CoV-2 omicron (B.1.1.529) variant
| D614G IC50 (μg/mL) | Omicron IC50 (μg/mL) | Fold change in IC50 (omicron vs D614G) | Mutations* | |
|---|---|---|---|---|
| Casirivimab (REGN-10933)7 | 0·009 | >10 | >1100 | 417†, 484†, 493†, 477, 478 |
| Imdevimab (REGN-10987)7 | 0·008 | >10 | >1200 | 440†, 446† |
| Bamlanivimab (LY-CoV555)8 | 0·007 | >10 | >1400 | 484†, 493†, 478 |
| Etesevimab (LY-CoV16)8 | 0·04 | >10 | >270 | 417†, 493, 501, 505, 477 |
| Sotrovimab (S309)9 | 0·1 | 0·2 | 2 | 339 |
IC50=50% inhibitory concentration.
Mutated amino acid positions modelled on the omicron receptor binding domain, proximal to the antibody interface, are listed.
Functional evidence (from deep mutational scanning data13) for an effect on antibody binding of mutations at that amino acid position.
Discussion
Neutralising antibodies are a mechanistic correlate of SARS-CoV-2 vaccine protection.16 Although other components of the immune system contribute to protection from severe disease, the significant reduction in neutralisation sensitivity observed in this study is likely to translate into a reduction in vaccine-mediated protection against infection. This hypothesis is supported by the rapid spread of the omicron variant in countries with high vaccine coverage.17
We found that there was a marked reduction in neutralisation potency against the omicron variant for serum pools from convalescent donors and recently vaccinated individuals, and from individual convalescent donors sampled soon after initial infection. This finding is consistent with several other contemporaneous studies.18, 19, 20 However, sera from a high-exposure cohort5 of hospital workers who had been infected, then vaccinated, achieved substantial cross-neutralisation of the omicron variant, which correlates with their ability to cross-neutralise other variants. This finding suggests that responses to the SARS-CoV-2 spike protein broaden with increasing antigenic exposure, which has been shown for other variants in the context of both previous infections21 and three doses of vaccination.22
Serum samples from a cohort of blood donors in Stockholm also showed substantial cross-neutralisation of the omicron variant. On average, the fold reduction in neutralising antibody titres against the omicron variant was only marginally greater than that of the previously infected, then vaccinated, hospital worker cohort. Such cross-neutralisation in a real-world cohort would not have been predicted from the responses observed soon after vaccination.18 At the time of sampling, most individuals in Sweden had only received two vaccine doses, thus this breadth of antibody response might be explained by the frequency of exposure to SARS-CoV-2 before, or following, vaccination in Stockholm. At the time of anonymous blood donor sampling, 78% of adults (≥18 years) had received two vaccine doses.23 Testing recommendations in Sweden make it difficult to directly estimate the proportion of the population who had been infected with SARS-CoV-2. In early 2021, before mass vaccination, the seroprevalence of anti-SARS-CoV-2 antibodies in blood donors in Stockholm was estimated at around 20%,24 and there have subsequently (before sampling of the current blood donor cohort) been two waves of SARS-CoV-2 infection, dominated by the highly transmissible alpha (B.1.1.7) and delta variants. Previous infection rates are thus expected to have been high in the blood donor cohort, which could contribute to cross-neutralising responses. Alternatively, systematic differences might exist in cross-neutralisation for samples obtained immediately after a second immunisation compared with those sampled later. Affinity maturation of antibody lineages over the course of months after SARS-CoV-2 infection enabled the cross-neutralisation of heterologous sarbecoviruses and SARS-CoV-2 variants of concern,25 which has also been demonstrated for the omicron variant.26
From a global health perspective, the marked reduction in neutralisation of the omicron variant by serum obtained from previously infected, but unvaccinated individuals raises the question as to whether such individuals can be considered immune. Vaccine effectiveness afforded by two doses seems to be significantly reduced against infection with the omicron variant.27 Furthermore, the cross-neutralising antibody responses in individuals who had been infected then vaccinated in the hospital worker cohort indicate that vaccination of previously infected individuals is of considerable value.
mAbs represent important treatment and prophylactic options for certain categories of patients, and can significantly reduce morbidity in those otherwise at risk for severe COVID-19.28 Considering the complete resistance of the omicron variant to several clinically relevant mAbs, now supported by several studies,20, 29 treatment options should be informed by rapid SARS-CoV-2 genotyping in regions where the omicron variant and other variants are co-circulating. This finding also highlights the need for rapid diversification of our clinical mAb portfolio to protect against unpredictable reduction in potency against future variants and for the further development of small-molecule antivirals against more conserved targets. There is also a need to rapidly screen variants for their sensitivity to clinical antibody therapeutics.
Methodologically, the current standard practice for generating pseudovirus spike expression plasmids for novel variants relies on site-directed mutagenesis when only a small number of mutations differ from an existing plasmid construct or gene synthesis to generate entire spike genes. Exceptional urgency is demanded by the emergence of a rapidly spreading novel variant with a large number of spike mutations. Molecular cloning from a diagnostic sample allowed us to circumvent gene synthesis delays and share pseudovirus neutralisation data just 8 days after receipt of the diagnostic samples suspected to contain the omicron variant and 13 days after the variant was first reported to WHO. One risk associated with this approach is that if the expression of a non-codon-optimised spike protein is too low, pseudovirus entry into target cells might be too inefficient to accurately quantify neutralisation. For this reason, our cloning strategy retained as much of the codon-optimised backbone as possible, especially in the C-terminal region of the spike, which is not mutated in the omicron variant. It is not clear whether such a strategy would universally succeed with all variants; therefore, a dual approach that attempts gene synthesis and direct cloning (when samples are available) would mitigate this risk.
Limitations of this study include the absence of vaccination and infection information for the anonymous blood donors and the heterogeneity in vaccination histories of the hospital workers. As a result, the drivers of the observed variability in the ability to cross-neutralise the omicron variant could not be assessed. Furthermore, although we evaluated serum pools post-vaccination with BNT162b2, mRNA-1273, and Ad26.COV2.S vaccines, each of these were pools of serum from only six individuals, and a single individual with cross-neutralising antibodies could influence the apparent cross-neutralisation for the entire pool. Nevertheless, the use of these standardised reagents enables future efforts to calibrate vaccine titres, across assays and variants. Additionally, we have not yet assessed the relative sensitivity of other members of the omicron clade, including BA.2, nor of emerging sublineages with additional mutations that might have a substantial impact on neutralising antibodies.
Ultimately, long-term protection against SARS-CoV-2, including antigenic variants that will arise, might require updated vaccines or vaccines that elicit more broadly cross-neutralising antibodies. Until such vaccines are available, our data from two different cohorts suggest that there is incomplete loss of neutralisation against the omicron variant. It has previously been shown with other variants that a third dose of an unmodified vaccine might have a broadening effect on the antibody response.23 Even in the absence of a broadening effect, in many donors, the magnitude of reduction in neutralisation observed against the omicron variant suggests that it might be possible to boost antibody titres into a protective range using currently licensed vaccines. Indeed, emerging evidence suggests that a third dose of licensed vaccines enhances protection against symptomatic infection, hospitalisation, and death associated with the omicron variant.30
Data sharing
All relevant data for the hospital worker cohort is included in table 1. Individual-level neutralisation data for the blood donor and convalescent cohorts will be available without restriction upon reasonable request, upon publication. Requests should be directed to the corresponding author (benjamin.murrell@ki.se).
Declaration of interests
STR is a cofounder of and held shares in deepCDR Biologics, which has been acquired by Alloy Therapeutics. DJS, GBKH, and BM have intellectual property rights associated with antibodies that neutralise the omicron variant. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
We thank the G2P-UK National Virology consortium funded by the UK Medical Research Council and UK Research and Innovation (grant MR/W005611/1) and the Barclay Lab at Imperial College London for providing B.1, B.1.351, B.1.617.2, and B.1.621 spike-encoding plasmids. We thank Penny Moore and The National Institute For Communicable Diseases Of South Africa for providing a B.1.351 spike plasmid, which was generated using funding from the South African Medical Research Council. The pCMV-dR8.2 dvpr plasmid was a gift from Bob Weinberg (Addgene plasmid 8455; Addgene, Watertown, MA, USA). The pBOBI-FLuc plasmid was a gift from David Nemazee (Addgene plasmid 170674; Addgene). We acknowledge all staff at the Department of Clinical Microbiology, Karolinska University Hospital (Stockholm, Sweden), involved in SARS-CoV-2 routine diagnostics, S-gene screening, and sequencing. We thank Klara Lenart and Karin Loré for providing the WHO International Standard for anti-SARS-CoV-2 immunoglobulin. We thank Matthew Hall and Jack Stapleton for providing the vaccine standards (pooled human serum samples for the Pfizer-BioNTech vaccine [BEI resources NRH-17727], Moderna vaccine [BEI resources NRH-17846], and Johnson & Johnson vaccine [BEI resources NRH-20012]). This study was supported, in part, by funding from the European Union Horizon 2020 research and innovation programme (grant number 101003653 [CoroNAb]) to GBKH, STR, and BM; the European and Developing Countries Clinical Trials Partnership programme supported by the European Union (grant number RIA2020EF-3030-RADIATES) to BM; SciLifeLab (VC-2021-0033) to BM and JA; and the Erling-Persson Foundation to BM and GBKH.
Contributors
DJS, DPM, GBKH, JA, and BM conceptualised the study. DJS and BM did the formal analysis. DJS, CK, XCD, and RD conducted the assays. AP, DJS, and BM designed the methodology. DJS and BM were responsible for the figures and tables. RAE, STR, JD, GBKH, and JA provided resources. DJS, GBKH, STR, JA, and BM oversaw the study. DJS and BM wrote the initial draft. DJS, DPM, GBKH, JA, and BM reviewed and edited the manuscript. DJS and BM have accessed and verified all of the data. DJS, GBKH, JA, and BM were responsible for the decision to submit the manuscript for publication.
Supplementary Material
References
- 1.Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 omicron variant in southern Africa. Nature. 2022 doi: 10.1038/s41586-022-04411-y. published online Jan 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCallum M, De Marco A, Lempp FA, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332. doi: 10.1016/j.cell.2021.03.028. 47.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 4.Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463. doi: 10.1016/j.chom.2021.02.003. 76.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elfström KM, Blomqvist J, Nilsson P, et al. Differences in risk for SARS-CoV-2 infection among healthcare workers. Prev Med Rep. 2021;24 doi: 10.1016/j.pmedr.2021.101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang J, Katz L, Winokur PL, et al. Establishment of human post-vaccination SARS-CoV-2 standard reference sera. medRxiv. 2022 doi: 10.1101/2022.01.24.22269773. published online Jan 25, 2022. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlieb RL, Nirula A, Chen P, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto D, Park Y-J, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 10.Newman J, Thakur N, Peacock TP, et al. Neutralising antibody activity against SARS-CoV-2 variants, including omicron, in an elderly cohort vaccinated with BNT162b2. bioRxiv. 2021 doi: 10.1101/2021.12.23.21268293. published online Dec 24. (preprint). [DOI] [Google Scholar]
- 11.Sheward DJ, Mandolesi M, Urgard E, Kim C. Beta RBD boost broadens antibody-mediated protection against SARS-CoV-2 variants in animal models. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirdita M, Ovchinnikov S, Steinegger M. ColabFold—making protein folding accessible to all. [DOI] [PMC free article] [PubMed]
- 13.Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin EG, Lustig Y, Cohen C, et al. Waning immune humoral response to BNT162b2 COVID-19 vaccine over 6 months. N Engl J Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seow J, Graham C, Merrick B, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 17.UK Health Security Agency Technical briefing 32. SARS-CoV-2 variants of concern and variants under investigation in England. Dec 17, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042046/Technical_Briefing_32.pdf
- 18.Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021 doi: 10.1038/s41586-021-04387-1. published online Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. bioRxiv. 2021 doi: 10.1101/2021.12.07.21267432. published online Dec 8. (preprint). [DOI] [Google Scholar]
- 20.Planas D, Saunders N, Maes P, et al. Considerable escape of SARS-CoV-2 omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. published online Dec 23. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Muecksch F, Schaefer-Babajew D, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi A, Koch M, Wu K, et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 2021;27:2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folkhälsomyndigheten Statistik för vaccination mot COVID-19. https://www.folkhalsomyndigheten.se/folkhalsorapportering-statistik/statistikdatabaser-och-visualisering/vaccinationsstatistik/statistik-for-vaccination-mot-covid-19/
- 24.Castro Dopico X, Muschiol S, Christian M, et al. Seropositivity in blood donors and pregnant women during the first year of SARS-CoV-2 transmission in Stockholm, Sweden. J Intern Med. 2021;290:666–676. doi: 10.1111/joim.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muecksch F, Weisblum Y, Barnes CO, et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853. doi: 10.1016/j.immuni.2021.07.008. 68.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheward DJ, Pushparaj P, Das H, et al. Structural basis of omicron neutralization by affinity-matured public antibodies. bioRxiv. 2022 doi: 10.1101/2022.01.03.474825. published online Jan 4. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. 2021 doi: 10.1056/NEJMc2119270. published online Dec 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinreich DM, Sivapalasingam S, Norton T, et al. REGEN-COV antibody combination and outcomes in outpatients with COVID-19. N Engl J Med. 2021;385:e81. doi: 10.1056/NEJMoa2108163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-04385-3. published online Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UK Health Security Agency COVID-19 vaccine surveillance report (week 4) Jan 27, 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050721/Vaccine-surveillance-report-week-4.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data for the hospital worker cohort is included in table 1. Individual-level neutralisation data for the blood donor and convalescent cohorts will be available without restriction upon reasonable request, upon publication. Requests should be directed to the corresponding author (benjamin.murrell@ki.se).