Abstract
Macroautophagy/autophagy is a conserved catabolic recycling pathway in which cytoplasmic components are sequestered, degraded, and recycled to survive various stress conditions. Autophagy dysregulation has been observed and linked with the development and progression of several pathologies, including cardiovascular diseases, the leading cause of death in the developed world. In this review, we aim to provide a broad understanding of the different molecular factors that govern autophagy regulation and how these mechanisms are involved in the development of specific cardiovascular pathologies, including ischemic and reperfusion injury, myocardial infarction, cardiac hypertrophy, cardiac remodelling, and heart failure.
Keywords: Autophagosome, Cardiomyocyte, Heart, Lysosome, Vascular
1. Introduction
Autophagy is an evolutionarily conserved catabolic recycling pathway in which different cellular components ranging from protein aggregates to entire organelles are targeted for degradation to promote cell survival under different types of stress.1 Autophagy induction has been observed under a diverse range of physiological and pathological conditions, including hypoxia,2 endoplasmic reticulum (ER) stress,3,4 oxidative stress and particularly nutrient starvation. All these stimuli are also involved in cardiovascular development, metabolism and disease; thus, autophagy regulation is a relevant subject for cardiovascular pathology.
2. Macroautophagy, microautophagy, and chaperone-mediated autophagy
Autophagy is mainly divided into three main branches, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), each of which has been described in detail elsewhere.5,6 Whereas some studies have shown the importance of microautophagy and CMA in the heart,7 the main volume of research points to macroautophagy as the main autophagic branch regulating both the physiological and pathological mechanisms involved with the cardiovascular system. Thus, for the remainder of this review, macroautophagy will be referred to as autophagy. In brief, during macroautophagy, cytoplasmic components targeted for degradation are enclosed by a rapidly expanding cup-shaped membrane called the phagophore. Expansion and maturation of the phagophore lead to the sequestration of the cargo inside double-membrane vesicles (autophagosomes) that ultimately fuse with the vacuole (in yeast and plants) or lysosome (in metazoans). Once fused together, the sequestered cargo is degraded by resident vacuolar/lysosomal hydrolases, generating macromolecules that can be then transported back into the cytosol to be recycled by the cell (Figure 1).1
Figure 1.
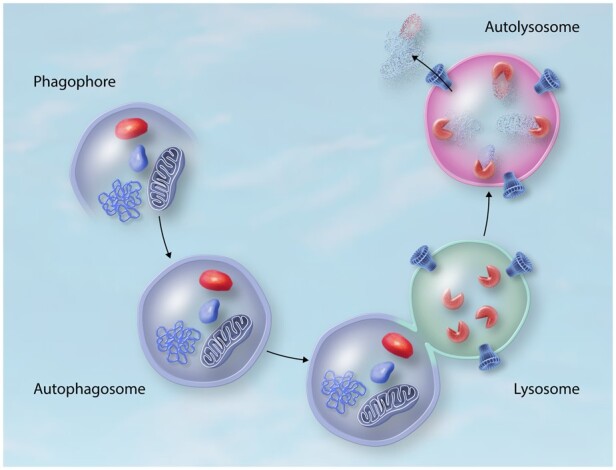
During macroautophagy different cytoplasmic components are enclosed by the expanding phagophore. Maturation of the phagophore results in the sequestration of the cargo inside double-membrane autophagosomes. Fusion between autophagosomes and lysosomes results in the formation of an autolysosome, where the sequestered cargo is degraded by lysosomal hydrolases. Macromolecules obtained from cargo degradation are transported back into the cytosol to be reutilized.
3. Autophagy induction
Autophagy activation is a tightly regulated process that depends on the transcriptional,8 post-transcriptional,8,9 and post-translational8 regulation of several autophagy-related (ATG) genes and their corresponding proteins. In nutrient-rich conditions, autophagy is usually kept at low levels; however, during nutrient starvation, autophagy activity is highly upregulated.8 Thus, autophagy regulation by nutrient availability depends on several crucial cellular energy sensors such as MTORC1, AKT/PKB, AMPK, and PRKA/PKA.
Activation of the ULK Ser/Thr kinase complex is one of the early events required for autophagy induction. The ULK complex is formed by the catalytic subunits ULK1/2,10,11 the regulatory scaffold protein ATG13,12,13 RB1CC1/FIP200,14 and the stabilizing protein ATG101.15 As part of the initiation step of the autophagy pathway, the ULK complex phosphorylates downstream ATG proteins leading to their activation and recruitment. Activation of the ULK complex during nutrient starvation conditions leads to ULK1 auto-phosphorylation13,14 at Thr18016 and Ser104717 which stabilizes its catalytic activity and in turn leads to the phosphorylation of several targets including BECN1 at Ser1418 and Ser30,19 ATG14 at Ser29,20 AMBRA1 at Ser465 and Ser635,19 ATG16L1 at Ser278,21 RB1CC122 at Ser943, Ser986 and Ser1323,19 ATG1322 at Ser31823 and Ser389,19 and ATG101 at Ser11 and Ser203.19 In nutrient-rich conditions, the ULK complex activity is inhibited by MTORC1-dependent phosphorylation of both ULK1 at Ser75724 and ATG13 at multiple sites.10,12,22 MTORC1 is a major sensor of the amino acid availability inside the cell, being activated by high amino acid levels through binding to an RRAG GTPase dimer25 and the small GTPase RHEB.26 Thus, MTORC1 is a major autophagy suppressor integrating nutrient and energy signalling into autophagy regulation. Furthermore, MTORC1 signalling has also been implicated in the transcriptional regulation of autophagy. During nutrient-rich conditions, MTORC1 phosphorylates the transcription factor TFEB at Ser211,27 preventing its nuclear translocation and the transcription of several lysosomal28 and ATG genes, including UVRAG, WIPI, MAP1LC3B, SQSTM1, VPS11, VPS18, and ATG9B.29
Closely related to MTORC1 signalling, the AKT/PKB pathway responds to growth factors to inhibit autophagy.30 Activation of INSR (insulin receptor) and IGF1R (insulin like growth factor 1 receptor) triggers the activation of AKT, which in turn phosphorylates the GTPase activating protein TSC2 at Ser393 and Thr1462, preventing RHEB inhibition and leading to autophagy suppression by MTORC1.31 Furthermore, AKT directly activates MTORC1 by phosphorylation at Ser2448.32 AKT activation depends on the formation of phosphatidylinositol(3,4,5)trisphosphate (PtdInsP3) in the plasma membrane; the phosphoinositide phosphatase PTEN is thus able to induce autophagy by dephosphorylating PtdInsP3 and downregulating AKT signalling.33 Autophagy is also directly inhibited by AKT, which can directly phosphorylate BECN1 at Ser295 and possibly Ser234, as well as ULK1 at Ser774.16,34 Similar to MTORC1, AKT can also regulate autophagy at the transcriptional level by phosphorylating the FOXO family of transcription factors, specifically FOXO1 and FOXO3, preventing their translocation to the nucleus and inhibiting the transcription of multiple autophagy genes such as ATG4, ATG12, BECN1, BNIP3, LC3, PIK3C3/VPS34, ULK1, and ULK2.35–37
Whereas MTORC1 prevents autophagy activation, AMPK has been linked with autophagy induction.24 AMPK is a heterotrimeric Ser/Thr kinase complex that can sense the energy status of the cell by binding AMP and ADP.38,39 A decrease in the ATP:AMP ratio inside the cell results in more AMP binding to AMPK, which in turn leads to the phosphorylation of PRKAA/AMPK α-subunit at Thr172 by STK11/LBK1, resulting in AMPK activation.40–42 Once activated, AMPK seeks to restore energy homeostasis inside the cell by upregulating catabolic pathways that can generate ATP and downregulating anabolic processes that consume energy. In this regard, AMPK plays both a direct and indirect role in autophagy regulation. Upon activation, AMPK can directly induce autophagy by phosphorylating ULK1 at Ser55524,43 and BECN1 at Ser93 and Ser96.44 Additionally, AMPK can also indirectly activate autophagy by preventing MTORC1-dependent inhibition of ULK1.26,45,46 Interestingly, ULK1-dependent phosphorylation of all three subunits of AMPK has been proposed as a negative feedback loop to terminate autophagy induction.47,48 Recently, a kinase substrate screen discovered that the cyclin-dependent kinase CCNY–CDK16 complex is a novel AMPK phosphorylation target involved in AMPK-mediated autophagy induction.49
PRKA/PKA is a cyclic AMP-dependent Ser/Thr protein kinase that phosphorylates LC3, preventing its recruitment to phagophores and inhibiting autophagy.50 PRKA/PKA has also been implicated in regulating vascular network formation in endothelial cells by phosphorylating ATG16L1. PRKA/PKA-dependent phosphorylation of ATG16L1 leads to its degradation, reducing autophagy, a mechanism that could be involved in regulating the stabilization of nascent vascular endothelium.51
Whereas nutrient depletion constitutes the main stimulus for autophagy induction in some organisms, other types of cellular stress are also involved in autophagy regulation. These additional types of stress, relevant components of the autophagy machinery, and mechanism include the following: (i) hypoxia involving HIF1A, BNIP3, and BNIP3L, which can lead to BCL2-BECN1 dissociation;52,53 MTORC1 inhibition, resulting in BECN1, and ATG14 phosphorylation.54 (ii) ER stress can induce autophagy through several mechanisms involving ERN1/IRE1α and MAPK/JNK driving BCL2-BECN1 dissociation;55–57 EIF2AK3/PERK and EIF2A/eIF2α leading to ATF4-dependent translational activation of ATG genes;58–64 ATL3, CCPG1, RETREG1, RTN3, SEC62, and TEX264 receptor-dependent reticulophagy.65,66 (iii) Oxidative stress leading to inactivation of MTORC1 and AKT, along with AMPK-dependent autophagy activation.67–69
4. Membrane nucleation
Generation and assembly of the phagophore require membrane nucleation by the class III phosphatidylinositol 3-kinase (PtdIns3K). This lipid kinase protein complex catalyzes the formation of phosphatidylinositol-3-phosphate (PtdIns3P), which serves as a recruitment factor for PtdIns3P-binding proteins such as those of the WIPI/WDR45 family.70 The PtdIns3K complex is formed by several core proteins, including the catalytic subunit PIK3C3/VPS34, the regulatory subunit PIK3R4/VPS15 and BECN1.71 Whereas these three proteins constitute the core of the PtdIns3K complex, depending on the other interacting partners, at least three other PtdIns3K subcomplexes involved in autophagy have been described.72,73 The PtdIns3K complex I is formed by the three core proteins, in addition to the BECN1-binding proteins ATG14 and AMBRA1, and the ATG14-binding protein NRBF2.74–76 This complex positively regulates autophagy by promoting recruitment of the PtdIns3K complex to the phagophore and inducing the generation of PtsIns3P. In this regard, ATG14 is directly associated with the ability of the PtdIns3K complex I to translocate to the phagophore;74 AMBRA1 improves BECN1 and PIK3C3 interaction and catalytic activity;76 and NRBF2 modulates PIK3C3 activity by promoting complex I assembly.75 The PtdIns3K complex II is formed by the above-mentioned PtdIns3K core proteins in addition to UVRAG.77 Although both ATG14 and UVRAG stabilize and directly interact with BECN1, their binding is mutually exclusive, generating two protein complexes with different autophagic functions73 (Figure 2). Furthermore, UVRAG binding partners RUBCN/RUBICON and SH3GLB1/Bif-1 further differentiate the PtdIns3K complex II into two subpopulations with different functions. Whereas the PtdIns3K complex in which UVRAG binds SH3GLB1 stimulates autophagy by promoting the autophagosome maturation step in which autophagosomes and lysosomes fuse,78,79 UVRAG binding to RUBCN inhibits autophagy.73 UVRAG can be negatively regulated by MTORC1-mediated phosphorylation, which increases UVRAG and RUBCN interaction.80
Figure 2.
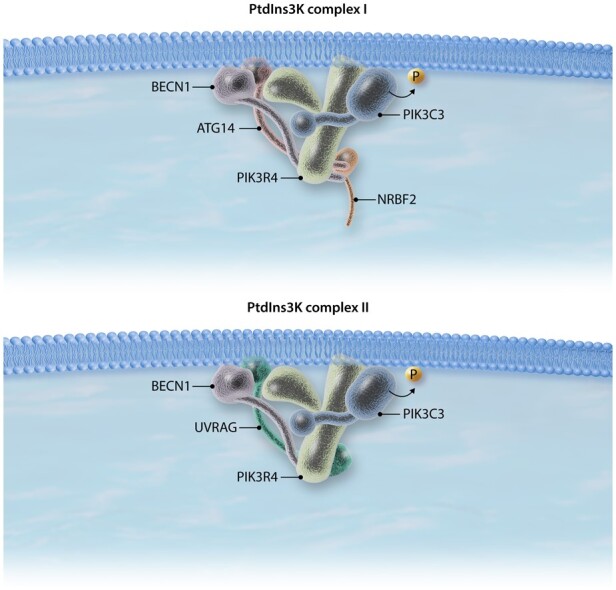
The proposed structure of the PtdIns3K complex I (top) and PtdIns3K complex II (bottom). Note that not all PtdIns3K complex I and complex II factors, including RUBCN and SH3GLB1, are shown.
5. Autophagosome membrane source
The membrane source from which the phagophore and subsequently the autophagosome are formed have long been a source of debate. Different studies have indicated that the initial phagophore membrane could originate from the ER,81trans-Golgi,82 mitochondria,83 endosomes,84 or the plasma membrane.85 ATG9A is the only essential transmembrane core autophagy protein, and it plays a key role in phagophore expansion,82 in a process regulated by the adaptor complexes AP1, AP2,86,87 and AP4.88 In turn, AP1/2 binding is regulated by the phosphorylation of the ATG9A N terminus by SRC during nutrient-rich conditions, and ULK1 during stress.86 Recently, the ATG9A structure has been solved, providing insight into its ability to bend and bind highly curved membranes, consistent with its role as a membrane transporter.89 Furthermore, the ATG9A structure also revealed a possible lipid scramblase activity, which could control autophagosome size.90 The ATG9A C terminus is responsible for binding ATG2A,89 which works as a funnel, tethering the ER membrane to the growing phagophore and mediating lipid transfer from the ER.91,92
In addition to ATG9A-containing vesicles, the ER-Golgi intermediate compartment (ERGIC) has also been proposed as another possible membrane source for phagophore formation.93 During nutrient-rich conditions coat protein complex II (COPII) vesicles travel from the ER to the Golgi as part of the secretory pathway. However, when nutrients are depleted and the secretory pathway is inhibited, COPII vesicles are repurposed for phagophore formation. During this process, the PtdIns3K complex I is recruited to the ERGIC, leading to the budding of specialized COPII vesicles that serve as a platform for LC3 lipidation, an essential step in phagophore expansion and later autophagosome maturation.93,94 This process is regulated by ULK1, which during nutrient starvation phosphorylates the COPII essential protein subunit SEC23B, preventing its proteasomal degradation and leading to SEC23B accumulation and the generation of autophagic COPII vesicles from the ERGIC.95
Consistent with the idea that the ER is a major membrane source for autophagosome formation, different microscopy and 3D tomography studies have highlighted the connection between ER cisternae and pha-gophore-autophagosome membranes.96,97 A PtdIns3P-enriched ER-subdomain (the omegasome) is linked to autophagosome biogenesis, providing a dynamic platform for recruiting several ATG proteins.98 Omegasome formation is dependent on PtdIns3K complex I ER localization, mediated by ATG14;99 subsequent PtdIns3P generation recruits ZFYVE1/DFCP198 and WIPI/WDR45 family proteins.70 WIPI2B is required for LC3 lipidation.70 This process is regulated by WIPI2 recruitment and direct binding to ATG16L1, which together with ATG12 and ATG5 forms the ATG12–ATG5-ATG16L1 complex, required for efficient LC3 lipidation and phagophore expansion.100 Other WIPI family members have also been implicated in autophagy regulation. WDR45/WIPI4 forms a complex with ATG2A, allowing the latter to tether PtdIns3P-containing vesicles to non-Ptdns3P-containing membranes.101,102 Also, the ULK1 complex associates with omegasomes in their early stages in a PtdIns3P-dependent manner.103
Multiple studies have indicated that autophagosomes can form at mitochondria-associated ER membranes (MAM), specific sites where the ER and mitochondria are in close proximity to one another.83,104 During starvation conditions, ATG14 is recruited to the MAM by the ER-resident SNARE protein STX17, which is required for complete autophagosome formation.104 Furthermore, disrupting ER-mitochondria contacts by depleting the tethering proteins MFN2 or PACS1 impairs ATG14 recruitment and starvation-induced autophagy.83,104 ATG2A also localizes to the MAM by binding the outer mitochondria membrane protein complex formed by TOMM70–TOMM40, which in turn is required for pha-gophore expansion and autophagic flux.105 Interestingly, the omegasome marker ZFYVE1 is also enriched in the MAM during starvation conditions, suggesting a unified system in which omegasomes and ER-mitochondria contacts are adjacent to each other.104
Taken together, an overall model has started to emerge in which the ER and ER-associated membranes serve as the main platform and membrane source for phagophore formation. Membranes from other organelles are very likely to contribute to autophagosome formation by generating membrane contact sites with the ER. Electron microscopy and electron tomography studies have shown phagophores making contact sites with membranes from late endosomes, Golgi, and mitochondria, sometimes even simultaneously.106 Therefore, it is reasonable to think of the ER as an expanding membranous system across the cell, interconnecting with different organelles that supply the phagophore with their membranes (Figure 3).
Figure 3.
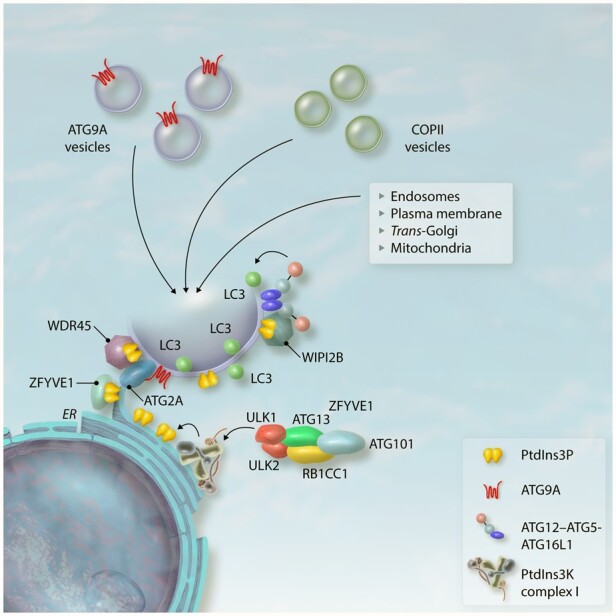
Translocation of the ULK complex to the ER leads to the phosphorylation and activation of the PtdIns3K complex I, which generates PtdIns3P-rich ER subdomains known as omegasomes. ZFYVE1, WDR45/WIPI4, and WIPI2B are recruited to omegasomes by binding PtdIns3P. WIPI4 binds ATG2A, which tethers the ER to the growing phagophore and transports lipids from one side to the other. ATG9A binds ATG2A, and the former moves lipids from one side of the membrane leaflet to the other, expanding the phagophore. WIPI2B recruits the ATG12–ATG5-ATG16L1 complex, inducing the lipidation of LC3 at the phagophore. Different sources provide membranes for phagophore expansion.
6. Expansion of the phagophore membrane
Two interconnected ubiquitin-like conjugation systems are involved in expanding the phagophore membrane, the ATG12–ATG5-ATG16L1 complex and the lipidation of the LC3/GABARAP family (Figure 4). The mechanism involved in the conjugation of LC3/GABARAP to phosphatidylethanolamine (PE), involving the action of the ATG12–ATG5-ATG16L1 complex as an E3 ligase has been described in great detail elsewhere.107–109 Binding between ATG16L1 and WIPI2 may control the targeting of the ATG12–ATG5-ATG16L1 complex and in turn the site of LC3/GABARAP lipidation.100 ATG16L1 is also able to interact with the ULK1 complex subunit RB1CC1/FIP200, which plays a role in the recruitment of the ATG12–ATG5-ATG16L1 complex to the phagophore.110 Recently, ATG16L1 has been proposed to bind directly to membranes;111 thus, it seems that the localization of ATG12–ATG5-ATG16L1 depends on multiple factors, including protein and lipid interactions.
Figure 4.
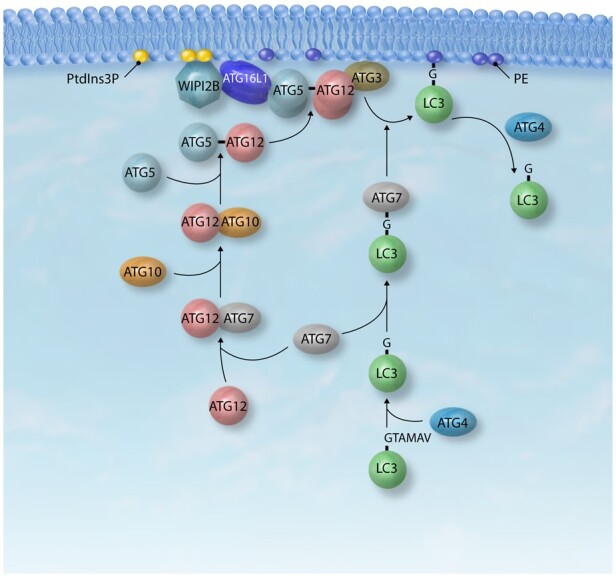
The ubiquitin-like proteins ATG12 and the LC3/GABARAP family go through activation by ATG7 and conjugation by ATG10 and ATG3, respectively. Covalent binding between ATG12–ATG5 leads to forming a dimeric ATG12–ATG5-ATG16L1 complex, which is recruited to the phagophore by WIPI2B, promoting the ligation of LC3 to PE. LC3-II/LC3–PE can be deconjugated by ATG4, the same protein involved in LC3 initial processing. GTAMAV, C-terminal amino acid residues; PE, phosphatidylethanolamine.
Beyond their role in the expansion and maturation of autophagosomes, when covalently bound to PE, the LC3/GABARAP family serve as interaction platforms for a diverse range of different proteins, physically linking them to the phagophore membrane. Among these LC3/GABARAP-binding proteins are various autophagy receptors that play an important role in selective autophagy, tethering the cargo targeted for degradation to the growing phagophore. These receptors include SQSTM1/p62, OPTN, TOLLIP, and NBR1, which participate in the autophagic degradation of various types of ubiquitinated cargo; BNIP3L and FUNDC1 which have been linked to mitophagy;112 RETREG1, CCPG1, SEC62, RTN3, and TEX264, which have been connected to reticulophagy;65 and CALCOCO2 that has been reported to induce the selective autophagic degradation of intracellular pathogens.113 Binding between these receptor proteins and LC3/GABARAP is generally mediated by the LC3-interacting region (LIR), a distinct amino acid sequence closely resembling Trp-X-X-Leu/Ile.109,114 A specific type of LIR termed GABARAP-interacting motif has been described, which resembles the sequence Trp/Phe-Val/Ile-X-V and shows a higher binding affinity for the GABARAP subfamily.115 Besides the autophagy receptor proteins, several other core components of the autophagy machinery such as ULK1, ATG13, RB1CC1,116 BECN1, ATG14,117 ATG4,118 and ATG7119 have been proposed to possess LIR sequences that allow them to interact with LC3/GABARAP and be recruited to the phagophore. Even though most of the research involving LC3/GABARAP and their interacting partners has been focused on the interface between the LIR and its corresponding docking site on the various LC3/GABARAP proteins, a recent publication has reported a novel alternative binding site in this family that binds ubiquitin-interacting motifs, opening the door for the discovery of new LC3/GABARAP-interacting partners and mechanisms of autophagy regulation.120
7. Lysosomal fusion and cargo degradation
In the last stages of autophagy, complete autophagosomes must fuse with lysosomes to release their cargo for degradation. This process requires a spatial approach between the two organelles that allow for membrane tethering and fusion to occur. Mature autophagosome trafficking is mediated by microtubules that serve as tracks, allowing the transport of complete autophagosomes to the perinuclear region, where lysosomes are usually located during starvation.121,122 Once autophagosomes and lysosomes are sufficiently close, tethering and fusion are triggered by multiple factors, including RAB7 and the HOPS complex,123,124 STX17, SNAP29, VAMP7, and VAMP8.125–127 Specificity of the lysosome-autophagosome fusion process is provided by several different adaptor proteins; for example, the RAB7 effector protein PLEKHM1 directly interacts with the HOPS complex and LC3/GABARAP through an LIR, providing a bridge between autophagosomes and the different members of the tethering complex.128 Once fusion is completed, the inner autophagosomal membrane is degraded inside the newly formed autolysosome, where lipases and acid hydrolases including cathepsins CTSB, CTSD, and CTSL degrade the cargo, generating essential molecules such as amino acids that can be transported back to the cytosol to be recycled by the cell. Recent studies have identified the lysosomal multi-spanning transmembrane protein SLC38A9 as a central amino acid transporter responsible for recycling amino acids from the lysosomal lumen to the cytosol.129–131 SLC38A9 senses Arg, which plays a modulating role in the lysosomal efflux of Gln and Leu and other essential amino acids to the cytosol.132,133 SLC38A9-dependent amino acid efflux is sensed by MTORC1,129–133 which is activated, generating a negative feedback loop that finally inhibits autophagy.
8. Cardiac autophagy
Autophagy has a vital role in the normal and diseased heart.134–136 Basally, cardiomyocytes display beneficial autophagy to degrade misfolded proteins and damaged organelles.134,137 Moreover, autophagy is required for normal cardiac development.138 Knockdown or knockout of essential autophagy genes, including Atg5, results in defects in cardiac morphogenesis, notably in valve development and chamber septation.139 However, this process is altered during metabolic stresses (e.g. diabetes, lipotoxicity), ischemia/reperfusion (I/R) injury, myocardial infarction (MI), cardiac hypertrophy, cardiac remodelling, and heart failure (HF).134–136 Because our previous review covered cardiovascular autophagy up to 2014,134 we focus mainly on the new findings described since 2015. The main discoveries in the cardiovascular area are described in Table 1.
Table 1.
Timeline describing principal findings in cardiovascular autophagy
| Year | Description | |
|---|---|---|
| Heart | 1976 | First description of autophagy in cardiomyocytes140 |
| 2000 | First description of autophagy in hearts associating accumulation of autophagic vacuoles and cardiomyopathy in LAMP2-deficient mice141 | |
| 2001 | Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy142 | |
| 2003 | Myocytes die by multiple mechanisms in failing human hearts143 | |
| 2007 | Cardiac autophagy is a maladaptive response to haemodynamic stress | |
| 2007 | Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMPK and BECN1 in mediating autophagy144 | |
| 2007 | The role of autophagy in cardiomyocytes in the basal state and in response to haemodynamic stress145 | |
| 2008 | Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy | |
| 2009 | Markers of autophagy are downregulated in failing human heart after mechanical unloading146 | |
| 2010 | Deacetylation of FOXO by SIRT1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes147 | |
| 2012 | Autophagy proteins LC3B, ATG5, and ATG12 participate in quality control after mitochondrial damage and influence lifespan148 | |
| 2012 | First description of autophagy in cardiac fibroblast assessing the role of ADRB2/beta(2)-adrenergic receptor in the regulation of cardiac fibroblast autophagy and collagen degradation149 | |
| 2012 | Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure150 | |
| 2012 | Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury151 | |
| 2013 | Mechanical unloading activates FOXO3 to trigger BNIP3-dependent cardiomyocyte atrophy152 | |
| 2013 | PINK1-phosphorylated MFN2 is a PRKN receptor for culling damaged mitochondria153 | |
| 2013 | Autophagy regulates endothelial cell processing, maturation, and secretion of VWF154 | |
| 2013 | STK4/MST1 inhibits autophagy by promoting the interaction between BECN1 and BCL2155 | |
| 2014 | Autophagy regulates vascular endothelial cell NOS3 and EDN1 expression induced by laminar shear stress in an ex vivo perfused system156 | |
| 2015 | PRKN-mediated mitophagy directs perinatal cardiac metabolic maturation in mice157 | |
| 2014 | PRKN-independent mitophagy requires DNM1L and maintains the integrity of mammalian heart and brain158 | |
| 2015 | Endogenous DNM1L mediates mitochondrial autophagy and protects the heart against energy stress159 | |
| 2015 | Interdependence of PRKN-mediated mitophagy and mitochondrial fission in adult mouse hearts160 | |
| 2015 | SIRT7 contributes to myocardial tissue repair by maintaining the TGFB signaling pathway161 | |
| 2016 | DNM1L-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure162 | |
| 2016 | Restoration of autophagy in endothelial cells from patients with diabetes mellitus improves nitric oxide signaling163 | |
| 2016 | Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification164 | |
| 2016 | Cardioprotection and lifespan extension by the natural polyamine spermidine165 | |
| 2017 | Endothelial-specific deletion of Atg7 attenuates arterial thrombosis in mice166 | |
| 2017 | Endothelial cell autophagy maintains shear stress-induced nitric oxide generation via glycolysis-dependent purinergic signaling to endothelial NOS3167 | |
| 2017 | Autophagy is required for endothelial cell alignment and atheroprotection under physiological blood flow168 | |
| 2018 | Endothelial autophagic flux hampers atherosclerotic lesion development169 | |
| 2018 | BECN1-dependent autophagy protects the heart during sepsis170 | |
| 2018 | TLR4 contributes to a myofibroblast phenotype in cardiac fibroblasts and is associated with autophagy after myocardial infarction in a mouse model171 | |
| 2019 | Mitophagy is essential for maintaining cardiac function during high fat diet-induced diabetic cardiomyopathy172 | |
| 2020 | Downregulation of BECN1 promotes direct cardiac reprogramming173 | |
| Blood vessels | 1997 | First description of autophagy in vascular endothelial cells and cardiac endothelial cells obtained from mouse hearts after long-term exposure to non-metabolizable sugars174 |
| 2001 | Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells175 | |
| 2006 | First description of autophagy in VSMC by evaluating the effects of IGF1 and TNF in the regulation of autophagy through MAPK/JNK and AKT pathways in human atherosclerotic vascular smooth cells176 | |
| 2011 | BECN1 deficiency is associated with increased hypoxia-induced angiogenesis177 | |
| 2015 | Defective autophagy in VSMCs accelerates senescence and promotes neointima formation and atherogenesis178 | |
| 2018 | Impaired mitochondrial respiration in human carotid plaque atherosclerosis: a potential role for PINK1 in VSMC energetics179 | |
| 2019 | Altered mitochondrial quality control in ATG7-deficient VSMCs promotes enhanced apoptosis and is linked to unstable atherosclerotic plaque phenotype180 | |
| 2019 | VSMC plasticity and autophagy in dissecting aortic aneurysms181 | |
| 2020 | Defective autophagy in vascular smooth muscle cells increases passive stiffness of the mouse aortic vessel wall182 | |
| 2019 | FOXO1 inhibits autophagosome-lysosome fusion leading to endothelial autophagic-apoptosis in diabetes183 | |
| 2019 | Endothelial-specific deficiency of ATG5 attenuates ischemia-related angiogenesis184 | |
| 2020 | Mitophagy contributes to endothelial adaptation to simulated microgravity185 | |
| 2020 | Defective autophagy in vascular smooth muscle cells alters vascular reactivity of the mouse femoral artery186 | |
| 2020 | Endothelial autophagy deficiency induces IL6-dependent endothelial mesenchymal transition and organ fibrosis187 | |
| 2020 | Laminar flow inhibits the STK3–STK4 (HIPPO)–YAP1 pathway via autophagy and SIRT1-mediated deacetylation against atherosclerosis188 | |
| 2020 | Ischemia induces autophagy of endothelial cells and stimulates angiogenic effects in a hindlimb ischemia mouse model189 |
9. Mitochondrial dysfunction and mitophagy
Because of constant beating, the heart is an organ that requires a continuous high supply of ATP. The heart produces and utilizes ∼6 kg ATP per day.190 Therefore, mitochondrial metabolism and function are essential for cardiac homeostasis.191 Mitophagy, the selective autophagic degradation of mitochondria, is an important mediator of mitochondrial quality control in cardiac myocytes. Removing dysfunctional mitochondria through mitophagy is essential for maintaining cardiomyocyte energetic and metabolic requirements.191 In developing hearts, the MFN2–PINK1–PRKN pathway, which poly-ubiquitinates damaged mitochondria to promote mitophagy, changes the cardiac metabolism by redirecting the mitochondrial substrate preference to fatty acids.157 Through this mechanism, foetal cardiomyocyte mitochondria are removed by PRKN-mediated mitophagy and replaced by mature adult mitochondria.157 Cardiac mitophagy is also observed during cardiac ischemia by a complex consisting of ULK1, RAB9, RIPK1, and DNM1L.192 Downregulation of mitophagy mediates the development of mitochondrial dysfunction and HF.162 In this model, an isoform shift from PRKAA2/AMPKα2 to PRKAA1/AMPKα1 is observed in mouse hearts and in heart samples from HF patients.193 BAG3, a co-chaperone of HSPA/HSP70, is also recruited to depolarized mitochondria, together with PRKN to promote mitophagy,194 in part by regulating translation of LC3B.195 Suppression of BAG3 in cardiac myocytes reduces autophagy flux and mitophagy.194 Conversely, administration in the heart of BAG3 using an adeno-associated virus improves left ventricular ejection fraction (LVEF) after MI.196 Restoration of PRKAA2 activates the PINK1–PRKN–SQSTM1 pathway that increases cardiac mitophagy associated with the improvement in mitochondrial function, removal of damaged mitochondria, decrease in reactive oxygen species (ROS) production, and cardiomyocyte apoptosis.193
Single-nucleotide polymorphisms in the PRKN are linked to blood pressure in Nigerian and Korean families, suggesting an association between mitophagy and hypertension.197 Moreover, spermidine supplementation in the diet of salt-sensitive rats enhances cardiac mitophagy and delays the development of hypertensive heart disease.198 Also, in a unilateral renovascular hypertension pig model, valsartan, a well-known antihypertensive drug, reduces left ventricular hypertrophy and increases mitophagy and mitochondrial biogenesis.198 These data suggest that the increase of mitochondrial turnover decreases hypertensive-dependent cardiac remodelling.
10. Ischemia/reperfusion
Myocardial ischemia is one of the main causes of sudden cardiac death worldwide, and activation of autophagy protects cardiomyocytes from I/R injury.134 During ischemia, autophagy is triggered as a pro-survival mechanism in response to nutrient and oxygen deprivation.199 However, during reperfusion, autophagy has beneficial or detrimental effects depending on the experimental model and whether it involves a BECN1- or an AMPK-MTOR-dependent activation of auto-phagy.144,199,200 More recent findings indicate that induction of autophagy by simultaneous administration of PT1, a specific activator of AMPK, and 3HOI-BA-01, a potent inhibitor of MTOR, reduces cardiomyocyte death triggered by simulated I/R. In vivo administration of PT1 or 3HOI-BA-01 in a murine I/R injury model stimulates autophagy and reduces infarct size.201 Likewise, cardiomyocyte-specific autophagy disruption with a conditional atg7 knockout leads to myofibrillar disarray and severe contractile dysfunction and worsens the I/R injury with cardiac hypertrophy and severe cardiac fibrosis.202 Autosis, a form of cell death due to excessive activation of autophagy, is induced in cardiomyocytes exposed to I/R.203 This autosis is associated with RUBCN upregulation, autophagic flux attenuation, and autophagosome accumulation.203 Furthermore, inhibition of excessive I/R-induced autophagy by trimetazidine protects the rat hearts from I/R-induced HF.204
11. Cardiac fibrosis
The differentiation of cardiac fibroblast to myofibroblast is one of the most important features of fibrosis.205 Autophagy is implicated in cardiac fibrosis because autophagy inhibition blocks fibroblast-to-myofibroblast phenoconversion and inhibits myofibroblast cell migration and contractility.206 Moreover, TGFB1 treatment of human atrial fibroblasts induces autophagy and enhances the fibrogenic response, suggesting an association between the myofibroblast phenotype and autophagy.207 In streptozotocin-induced diabetic rats, alterations in autophagy correlate with cardiac fibrosis, indicating a potential synergistic role for autophagy in diabetic cardiac fibrosis.208 Moreover, in vitro and in vivo studies show that inhibition of PARP1 partially decreases autophagy, abrogates cardiac fibrosis, and significantly improves cardiac function post-MI.209 In a mouse model of lipopolysaccharide-induced sepsis, cardiac-specific overexpression of BECN1 promotes autophagy, improves cardiac function, and alleviates inflammation and fibrosis.170
12. Diabetic cardiomyopathy
Diabetic cardiomyopathy is a cardiovascular disease characterized by morphological, functional, and metabolic changes in the heart produced as a complication of type 2 diabetes mellitus (T2DM).210 Excessive autophagy is observed in the right atrial appendages collected from diabetic and non-diabetic patients.211 Similarly, animal models of T2DM show increased autophagosomes in the heart that is further increased upon chloroquine injection. Importantly, in vitro genetic deletion of Becn1 in adult rat cardiomyocytes exposed to high glucose markedly inhibits autophagy and triggers apoptosis suggesting a pathological role of autophagy in the T2DM heart.211 Moreover, excessive autophagy in T2DM induces HF aggravation after MI through defective mitophagy, associated with excessive inflammasome activation, secretion of IL18, and cell death.212 Prevention of inflammation by NFKB blockade with pyrrolidine dithiocarbamate reduces oxidative stress and improves mitochondrial integrity, thus restoring cardiac function in T2DM.213 In hearts from obese db/db T2DM mice, LC3 lipidation, SQSTM1, and phosphorylated MTOR levels are increased, but CTSD level is decreased, and very few lysosomes are detected, despite the abundance of autophagic vacuoles. In these hearts, AMPK activity and ATP content are decreased. These findings suggest that in this T2DM model, the autophagic flux is blocked at the final step.214 In high-fat diet-induced obese mice, a failure to upregulate LC3 lipidation or to clear SQSTM1 in the heart after fasting are observed, although mRNA for Lc3b and Sqstm1 are appropriately upregulated. These data suggest that hearts of diet-induced obese mice also exhibit impaired autophagy.215
In hearts from streptozotocin-induced type 1 diabetic mice (T1DM), diastolic dysfunction is observed,214,216 although autophagic activity is increased, as evidenced by increases in LC3-II and CTSD, and decreased SQSTM1, and by the abundance of autophagic vacuoles and lysosomes.214 The increase of blood glucose level in T1DM induces mitochondrial damage by O-GlcNAcylation of many electron transport chain subunits and other mitochondrial proteins.217 Inhibition of autophagic flux by chloroquine decreases autolysosomes, cardiomyocyte apoptosis, and cardiac fibrosis, but increases both Lc3 and Sqstm1 expression.218 Surprisingly, mitophagy is inhibited in hearts from T1DM mice.219 Moreover, there is substantial attenuation of cardiac damage in mice deficient for BECN1 and ATG16L1 as evidenced by an improvement in cardiac function along with decreased levels of oxidative stress, interstitial fibrosis, and cardiomyocyte apoptosis. In contrast, diabetes-induced cardiac injury is aggravated by BECN1 overexpression.220 In the same way, administration of 1,25-dihydroxyvitamin-D3 and resveratrol improves diabetic cardiomyopathy by restoring the impaired cardiac autophagy in streptozotocin-induced diabetic rats.221,222
13. Heart failure
HF is a clinical syndrome due to a structural and/or functional cardiac abnormality, which results in a reduction in cardiac output and/or elevated intracardiac pressure.223 HF affects patients ranging from those with normal (≥50%) LVEF, known as HF with preserved EF (HFpEF), to those with reduced (<40%) LVEF, known as HF with reduced EF (HFrEF).223 All these patients display signs of HF with evidence of abnormal cardiac structure and function.224 Epidemiological studies have shown that the incidence of HFrEF and HFpEF are almost equally distributed, but the incidence of HFpEF is increasing with time.225 The role of autophagy is well described in HFrEF;134 however, its role in HFpEF is poorly defined. Recently, a gene expression analysis performed on heart biopsies obtained from HFrEF, HFpEF, and control patients revealed that autophagy genes are strikingly downregulated in HFpEF patients.226
Initial findings indicated that basal constitutive cardiac-specific autophagy impairment, obtained by a tamoxifen-induced deletion of Atg5, shows disorganized sarcomere structure, mitochondrial misalignment and aggregation in cardiac myocytes, and cardiac hypertrophy, left ventricular dilatation, and contractile dysfunction, indicating the occurrence of HFrEF.145 HFrEF progression can be prevented by rapamycin.227 In fact, activation of basal autophagy by rapamycin, improves diastolic function in old mice beginning at 2–4 weeks and progress throughout 10-week treatment. Autophagy was transiently induced during the first week of treatment associated with mitochondrial biogenesis, suggesting the replacement of damaged mitochondria.228 In a rat model of HFrEF induced by MI, histological and echocardiographic measurements showed that rapamycin treatment improves myocardial function and inhibits cardiac remodelling at 8 weeks post-MI mechanism involving inhibition of the MTORC1 and ER stress pathways.229 In the same model, the increase of MMP9 (matrix metallopeptidase 9) inhibits cardiomyocyte and cardiac fibroblast autophagy in the peri-infarct site, whereas ablation of MMP9 increases cardiac autophagy.230 Moreover, in a model of HF induced by pressure overload, nitric oxide and natriuretic peptides induce cardioprotection by activation of autophagy through a PRKG1–TSC2–RHEB-dependent inhibition of MTORC1.231 In patients with dilated cardiomyopathy, autophagic vacuoles in cardiomyocytes are associated with a better HFrEF prognosis, suggesting autophagy could be involved in the prevention of myocardial deterioration.232 In patients with idiopathic dilated cardiomyopathy, mechanical unloading with a left ventricular assist device decreases markers of autophagy BECN1, ATG12–ATG5, and LC3-II.146 Because the mechanical unloading decreases the energy demand of the failing heart these results suggest that autophagy is activated as an adaptive mechanism. Moreover, myocardial CTSD, a major lysosomal protease induced by MI, protects against cardiac dysfunction and remodelling, which is in part through promoting cardiac myocyte autophagic flux.233
Conversely, autophagy, assessed by anti-LC3-II staining and vacuole formation, is extensively detected in myocardial samples from patients with ischemic cardiomyopathy and idiopathic dilated cardiomyopathy, with the occurrence of necroptosis and oncosis exceeding that of apoptosis.234 In a mouse model of MI, necroptosis is triggered by autophagy flux dysfunction, and this cell death process contributes to loss of cardiomyocytes, adverse ventricular remodelling, and HFrEF.235 Moreover, persistent autophagy triggered by upregulation of TLR3 expression and signalling induced by MI, promotes HFrEF and lethality.236 Likewise, in a model of pressure overload in mice induced by TAC, left ventricular wall thickness is increased at 1 week, associated with a decrease in autophagy.237 At 9 weeks, this model develops HFrEF characterized by a further increase in left ventricular wall thickness, with the intensification of cardiomyocyte autophagy and apoptosis.237 In this model, activation of AMPK by injecting AICAR improves cardiac function by attenuating autophagy.238 These results suggest that in HFrEF, autophagy can be beneficial or detrimental, depending on the context of the disease.
14. Cardiac ageing and regeneration
Cardiac ageing is characterized by fibrosis, hypertrophy, dysfunctional mitochondria, and misfolded protein accumulation, and impaired autophagy.239 Inhibition of autophagy exacerbates cardiac dysfunction accompanied by the accumulation of dysfunctional organelles and misfolded proteins.240 Reduction of autophagy during ageing correlates with the hypermethylation of Atg5 and Lc3b gene promoter regions in macrophages from aged mice associated with reduced gene expression;241 treatment with Trdmt1/Dnmt2 siRNA or a methyltransferase inhibitor restores Atg5 and Lc3 expression.241 Interestingly, a homozygous BECN1F121A knock-in mouse, a mutation that decreases the interaction of BECN1 with the negative regulator BCL2, has increased basal autophagy in several organs, including the heart.242 These mice show an increased life span with decreased cardiac and renal pathological changes and spontaneous tumorigenesis.242
Conversely, cardiac regeneration is considered a promising tool to treat cardiac diseases. Zebrafish, but not adult mammals, are capable of cardiac tissue regeneration. In an injury model of ventricular apex resection, a tightly regulated autophagic response is observed during the early stages of the regeneration process. Inhibition of autophagy by rapamycin impairs cardiac regeneration.243 These findings clearly show the double-edged sword effect that autophagy has in cardiac homeostasis.
15. Autophagy in the vascular system
The vascular tissue is mainly constituted by vascular smooth muscle cells (VSMCs) and endothelial cells. VSMCs exhibits several phenotypes in response to endogenous and environmental factors contributing to the genesis and progression of vascular diseases.244 Comprehensive overviews of the role of autophagy in VSMCs were previously and extensively described.134,135
16. VSMC phenotype
Several studies have shown that autophagy controls VSMC differentiation and dedifferentiation. Dedifferentiated VSMCs are characterized by increased cell migration, proliferation, extracellular matrix protein synthesis, secretion, and decreased contractile protein content.244 This phenotype is responsible for vessel formation and reparation. However, when it is dysregulated, it is responsible for vascular diseases (Figure 5).244 Inhibition of autophagy with 3-methyladenine delays the differentiation of epicardial progenitor cells into coronary VSMCs while its activation with rapamycin induces an early transient differentiation of epicardial progenitor cells and enhanced migration.245 Moreover, GDF11, an inhibitor of cardiac hypertrophy in mice,246 promotes carotid VSMC differentiation and prevents cell dedifferentiation induced by autophagosome accumulation triggered by western diet, lysosome function deficiency, and inflammation.247
Figure 5.
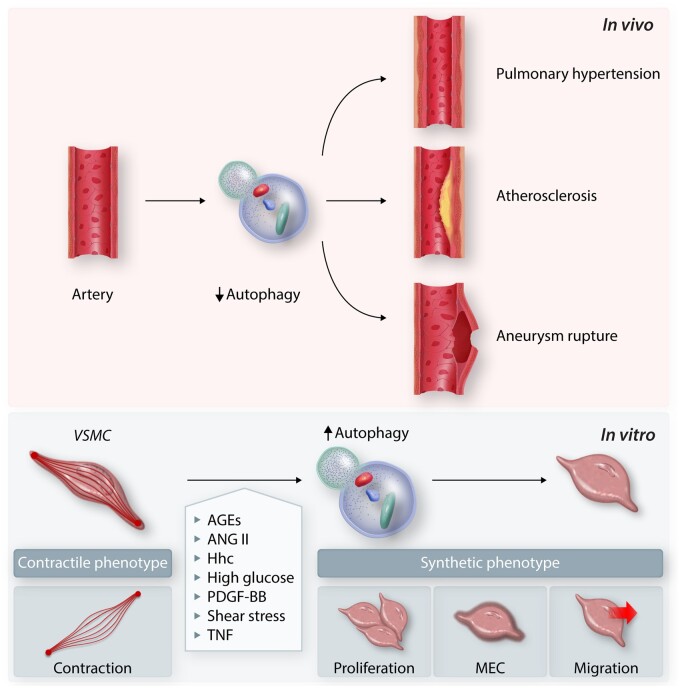
Role of autophagy in the development of vascular diseases using in vivo models and VSMC phenotypic change using in vitro models. AGES, advanced glycation end products; Ang II, angiotensin II; Hhc, hyperhomocysteinemia; PDGF-BB, platelet-derived growth factor-BB; TNF, tumor necrosis factor; VSMC, vascular smooth muscle cell.
Conversely, angiotensin II (Ang II), through an AGTR1–RHOA kinase-dependent signalling pathway, activates autophagy in VSMCs by a mechanism involving the increase of BECN1, PIK3C3, ATG12–ATG5, ATG4, and ATG7 protein levels and BECN1 phosphorylation, suggesting activation of phagophore initiation and expansion.248 Ang II regulates contractile protein content in VSMCs through an autophagy-dependent mechanism.248 Inhibition of autophagy by SEPTIN4,249 and cortistatin, a neuropeptide highly homologous to somatostatin,250 inhibits Ang II-induced VSMC proliferation. Similarly, an AMPK–MTORC1 signalling-dependent induction of autophagy in VSMCs is described for SPARC. SPARC upregulates LC3-II, BECN1, and ATG5, triggering an autophagy-dependent VSMC dedifferentiation.251 Regulation of cell migration by autophagy via the degradation of KAT2A is also described for VSMC.252 Therefore, VSMC dedifferentiation processes, i.e. proliferation, migration, contractile, and extracellular matrix protein synthesis, are highly regulated by autophagy.
17. Endothelial function
The endothelium is a critical regulator of vascular health and function. Shear stress generated by the blood flow is one of the most important stimuli that control endothelial function.253 Steady laminar shear stress (5 or 15 dynes/cm2) induces autophagy in human and rabbit endothelial cells, associated with the induction of endothelial NOS3/eNOS (nitric oxide synthase 3) and inhibition of expression of the vasoconstrictor EDN1/ET-1 (endothelin 1).156 The mechanism involves the deacetylation of ATG5 and ATG7 in a SIRT1-dependent manner.254 In contrast, oscillatory or low levels of shear stress reduce autophagy, uncouple NOS3, and trigger proatherogenic responses.168,254,255 From a translational point of view, using primary endothelial cells collected from the radial artery of men, showed that 60 min of rhythmic handgrip exercise activates autophagy, increases NOS3 phosphorylation, and increases nitric oxide and generation.256 In diabetic patients, autophagic flux is reduced, which impairs NOS3 activation.163 These data suggest that steady laminar shear stress induces autophagy with a beneficial vasodilatory effect, and diabetes disrupts these mechanisms. Moreover, disturbed blood flow promotes atherosclerosis, whereas laminar flow has a protective action by preventing endothelial apoptosis, senescence, and inflammation.169 The mechanism involves activation of endothelial autophagy that triggers SIRT1 expression that inhibits STK3/MST2–STK4/MST1 (HIPPO)-YAP1 signalling that interrupts atherosclerotic plaque formation.188
18. Vascular remodelling
Pathological vascular remodelling is characterized by the narrowing of the vessel lumen, mobilization of VSMC to the intima layer, exacerbated extracellular matrix production (fibrosis) and infiltration by immune cells.257 Dedifferentiated VSMCs have been extensively described in the development and progression of atherosclerosis, hypertension, restenosis, and neointimal formation.258,259 Various rapamycin analogues (rapalogs) have been used in drug-eluting stents for treating arterial restenosis. Two major rapalogs, everolimus, and biolimus strongly inhibit neointimal hyperplasia.260 However, everolimus exerts cytotoxicity by increasing ROS and reducing energy metabolism. In contrast, biolimus preferentially induces autophagy by activating ULK1. The implantation of biolimus-eluting stent reduces endothelial loss and inflammation in porcine coronary arteries.260
VSMCs with specific genetic atg7 deletion show a reduction in serum-induced cell growth and cell proliferation rate and an increase in cell death.261 Furthermore, an atg7 conditional knockout mouse crossed with Apoe (apolipoprotein E)-deficient mouse shows a reduction in medial cellularity and an increase in TUNEL-positive cells in the descending aorta. This mouse also shows a reduced survival rate due to aortic rupture associated with increased markers of atherosclerosis.261 Using another VSMC-specific atg7 knockout mouse continuously infused with Ang II (1.5 mg/kg/day) also exhibits increased mortality associated with cardiac rupture, MI, end-organ damage, venous distention, pleural effusion, and abdominal aortic dissections.262 Deletion of Atg5 in VSMCs increases cell death susceptibility, enhances ER stress activation, promotes VSMC inflammation, and increases the incidence of fatal rupture of aortic aneurysm (Figure 5).181 TFEB is downregulated in aortic aneurysm lesions, and TFEB deficiency increases VSMC apoptosis and promotes abdominal aortic aneurysm formation in mice.263 In contrast, TFEB activation has the opposite effect.263 Additionally, metformin preserves aortic elastin and collagen and reduces aortic cell apoptosis abdominal aortic aneurysms in patients and rat models. The mechanism involves the inactivation of the PI3K–AKT–MTORC1 pathway and the decrease of mRNA and protein levels of LC3B and BECN1.264 Inhibition of vascular inflammation by using BP-1-102, a novel potent STAT3 inhibitor, decreases abdominal aortic aneurysm by preserving autophagy.265 These data suggest that the fine-tuning of autophagy flux is required to maintain aorta wall integrity.
19. Atherosclerosis
VSMC dedifferentiation is a key step in the development of atherosclerotic plaque. Therefore, disruption of autophagy has important consequences on atherosclerosis genesis and progression. Besides macrophages, VSMCs are also an important source of foam cells in atherosclerosis. In a high-fat diet-fed apoe knockout mice model of atherosclerosis, activation of the P2RY12 receptor inhibits autophagy in VSMCs, which decreases cholesterol efflux and promotes VSMC-derived foam cell formation.266
Atherosclerotic plaque instability predisposes to the occurrence of cardiovascular events such as MI and stroke. A mouse model of plaque instability in VSMC-specific atg7 knockout mice crossed with apoe knockout mice, shows that defective autophagy in VSMCs increases plaque instability and the risk of rupture (Figure 5).267 Conversely, enhancement of autophagy in VSMCs or macrophages by using trehalose or by overexpression of TFEB exerts athero-protective effects, reducing plaque formation with a reduction of inflammation.268 Moreover, in the context of proatherogenic stimuli, the deficiency of autophagy induces inflammation in coronary artery VSMCs by promoting NLRP3 inflammasome formation and activation.269 These data suggest that autophagy is also a critical process in maintaining immune homeostasis during atherosclerosis.
20. Pulmonary hypertension
Pulmonary hypertension (PH) is a progressive disease characterized by excessive proliferation of pulmonary arterial vascular smooth muscle cells (PASMC).270 More than 70% of familial PH and 20% of idiopathic PH patients carry heterozygous mutations in BMPR2.271 BMPR2 is degraded by autophagy in pulmonary artery endothelial cells and PASMC.271 Mutations in BMPR2 are sufficient to trigger an increased autophagic flux.271 Similarly, pulmonary microvascular endothelial cells from end-stage idiopathic PH patients present an elevated autophagic flux.271 Along these lines, inhibition of autophagy with chloroquine272 or paclitaxel273 blocks PASMC proliferation (Figure 5).
21. Autophagy as a pharmacological target in cardiovascular diseases and current limitations
Despite several pharmacological agents and regimens that modulate autophagy, very few clinical trials have evaluated their effects in cardiovascular diseases. Of these, only exercise, trehalose, and hydroxychloroquine have been evaluated (Table 2). Another important current limitation in the field is the lack of cell/organ-targeted delivery of exogenous autophagy regulators for cardiac or vascular repair. We are still far from having an effective autophagy therapy. However, insights obtained from such works will provide inspiration for future clinical trials to assess the autophagic response for therapeutic gain. Careful consideration must be kept in mind because of the dual role of autophagy in cytoprotection and cell death.
Table 2.
Clinical trials involving autophagy in the cardiovascular system
| Name of the study | Study type | Description | Clinical trial identifier |
|---|---|---|---|
| MUcociliary Clearance IN Stroke (MUCINS) | Observational | Evaluation of autophagy in respiratory tissue (nasal, tracheal, and bronchial) | NCT03884166 |
| Transforming Growth Factor Beta Signalling in the Development of Muscle Weakness in Pulmonary Arterial Hypertension | Observational | Determination of the contribution of atrophy and autophagy to muscle wasting in PAH | NCT01847716 |
| Intermittent Pneumatic Compression With and Without Exercise to Improve Functioning in Peripheral Artery Disease (INTERCEDE) | Interventional | Evaluation among PAD participants whether intermittent pneumatic compression therapy combined with exercise improves autophagy (LC3, LAMP2, PRKN) at 6-month follow-up, compared to exercise alone | NCT03871075 |
| Autophagy and Venous Endothelial Function | Observational | Evaluation of endothelial function in venous samples from patients with venous insufficiency before and after treatment with autophagy enhancer spermidine | NCT04138134 |
| Autophagy Maintains Vascular Function Through a Novel Glycolysis-linked Pathway Regulating eNOS | Interventional | Evaluation of BECN1, ATG3, ATG5, ATG7, LAMP1, LAMP2, and SQSTM1 after rhythmic handgrip exercise or chronic exercise training | NCT04200560 |
| Autophagy, Oxidative Stress, and Hippo Signalling in Human Aortic Aneurysm | Observational | Comparison between the levels of STK3–STK4 (HIPPO) signalling and autophagy markers observed in aortic aneurysms and the levels assessed in the adjacent non-aneurysmatic aortic portions | NCT03211000 |
| Effects of Trehalose and Polyphenols in Vasculopathic Patients | Interventional | Change of autophagy (LC3) in PAD patients after mixed supplementation of trehalose and polyphenols | NCT04061070 |
| Effect of Hydroxychloroquine on Atrial Fibrillation Recurrence | Interventional | Recurrence rate of atrial fibrillation after radiofrequency catheter ablation, anticoagulant therapy, and hydrochloroquine treatment (200 mg, bidpo) | NCT03592823 |
| Potential Impact of Neuroimmune and Autophagic Alterations on the Progression and Severity of Human Atherosclerotic Process | Observational | Characterization of the autophagic process and correlation with neural modulations of the stability/instability plaque | NCT03922698 |
PAD, peripheral arterial disease; PAH, pulmonary arterial hypertension.
22. Conclusions
As autophagy remains an important factor in maintaining cardiovascular homeostasis, further research is required to pinpoint novel pharmacological targets that will allow us to harness the benefits of controlling autophagy levels to treat different cardiac pathologies.
Funding
This work was supported by grants from the National Institutes of Health (GM131919 to D.J.K.), the Agencia Nacional de Investigacion y Desarrollo (ANID), Chile: Fondo Nacional de Desarrollo Científico y Tecnológico/FONDECYT (1180157 to M.C. and 120049 to S.L.), and Fund for Financing Research Centers in Priority Areas/FONDAP (15130011 to M.C. and S.L.).
Conflict of interest: none declared.
References
- 1. Lahiri V, Hawkins WD, Klionsky DJ.. Watch what you (self-) eat: autophagic mechanisms that modulate metabolism. Cell Metab 2019;29:803–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mazure NM, Pouyssegur J.. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol 2010;22:177–180. [DOI] [PubMed] [Google Scholar]
- 3. Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S.. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell 2018;44:217–232.e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Solda T, D'Antuono R, Raimondi A, Jung M, Melnyk A, Schorr S, Schreiber A, Simonelli L, Varani L, Wilson-Zbinden C, Zerbe O, Hofmann K, Peter M, Quadroni M, Zimmermann R, Molinari M.. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 2016;18:1173–1184. [DOI] [PubMed] [Google Scholar]
- 5. Cuervo AM, , Wong E.. Chaperone-mediated autophagy: roles in disease and aging. Cell Res 2014;24:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schuck S. Microautophagy- distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 2020;133:jcs246322.doi: 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- 7. Pedrozo Z, Torrealba N, Fernandez C, Gatica D, Toro B, Quiroga C, Rodriguez AE, Sanchez G, Gillette TG, Hill JA, Donoso P, Lavandero S.. Cardiomyocyte ryanodine receptor degradation by chaperone-mediated autophagy. Cardiovasc Res 2013;98:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feng Y, Yao Z, Klionsky DJ.. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 2015;25:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gatica D, Hu G, Liu X, Zhang N, Williamson PR, Klionsky DJ.. The Pat1-Lsm complex stabilizes ATG mRNA during nitrogen starvation-induced autophagy. Mol Cell 2019;73:314–324.e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganley IG, Lam Du H, Wang J, Ding X, Chen S, Jiang X.. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009;284:12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan EY, Kir S, Tooze SA.. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007;282:25464–25474. [DOI] [PubMed] [Google Scholar]
- 12. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N.. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009;20:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan EY, Longatti A, McKnight NC, Tooze SA.. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 2009;29:157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N.. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008;181:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N.. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009;5:973–979. [DOI] [PubMed] [Google Scholar]
- 16. Bach M, Larance M, James DE, Ramm G.. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J 2011;440:283–291. [DOI] [PubMed] [Google Scholar]
- 17. Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J.. Mapping the phosphorylation sites of Ulk1. J Proteome Res 2009;8:5253–5263. [DOI] [PubMed] [Google Scholar]
- 18. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL.. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ.. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell 2015;59:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, Voytas D, Kim DH.. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 2016;12:547–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian W, Alsaadi R, Guo Z, Kalinina A, Carrier M, Tremblay ME, Lacoste B, Lagace D, Russell RC.. An antibody for analysis of autophagy induction. Nat Methods 2020;17:232–239. [DOI] [PubMed] [Google Scholar]
- 22. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH.. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009;20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, Steeves MA, Yang CY, Prater SM, Kim DH, Thompson CB, Youle RJ, Ney PA, Cleveland JL, Kundu M.. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell 2011;43:572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J, Kundu M, Viollet B, Guan KL.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM.. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoki K, Li Y, Xu T, Guan KL.. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003;17:1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martina JA, Chen Y, Gucek M, Puertollano R.. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012;8:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A.. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473–477. [DOI] [PubMed] [Google Scholar]
- 29. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A.. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K.. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 2008;183:101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC.. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 2002;10:151–162. [DOI] [PubMed] [Google Scholar]
- 32. Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR.. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999;344(Pt 2):427–431. [PMC free article] [PubMed] [Google Scholar]
- 33. Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E.. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 2001;276:35243–35246. [DOI] [PubMed] [Google Scholar]
- 34. Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B.. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012;338:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sengupta A, Molkentin JD, Yutzey KE.. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem 2009;284:28319–28331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M.. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007;6:458–471. [DOI] [PubMed] [Google Scholar]
- 37. Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL.. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472–483. [DOI] [PubMed] [Google Scholar]
- 38. Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG.. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest 2004;113:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheung PC, Salt IP, Davies SP, Hardie DG, Carling D.. Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem J 2000;346(Pt 3):659–669. [PMC free article] [PubMed] [Google Scholar]
- 40. Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG.. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2003;2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D.. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 2003;13:2004–2008. [DOI] [PubMed] [Google Scholar]
- 42. Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC.. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 2004;101:3329–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ.. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011;331:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL.. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013;152:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inoki K, Zhu T, Guan KL.. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003;115:577–590. [DOI] [PubMed] [Google Scholar]
- 46. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ.. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008;30:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B.. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 2011;7:696–706. [DOI] [PubMed] [Google Scholar]
- 48. Holczer M, Hajdú B, Lőrincz T, Szarka A, Bánhegyi G, Kapuy O.. Fine-tuning of AMPK-ULK1-mTORC1 regulatory triangle is crucial for autophagy oscillation. Sci Rep 2020;10:17803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dohmen M, Krieg S, Agalaridis G, Zhu X, Shehata SN, Pfeiffenberger E, Amelang J, Butepage M, Buerova E, Pfaff CM, Chanda D, Geley S, Preisinger C, Sakamoto K, Luscher B, Neumann D, Vervoorts J.. AMPK-dependent activation of the Cyclin Y/CDK16 complex controls autophagy. Nat Commun 2020;11:1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cherra SJ 3rd, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT.. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol 2010;190:533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao X, Nedvetsky P, Stanchi F, Vion AC, Popp O, Zuhlke K, Dittmar G, Klussmann E, Gerhardt H.. Endothelial PKA activity regulates angiogenesis by limiting autophagy through phosphorylation of ATG16L1. Elife 2019;8:e46380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, Mazure NM.. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009;29:2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF.. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 2007;27:6229–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, Higgins L, Kim DH.. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 2018;14:584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K.. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006;26:9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D.. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000;287:664–666. [DOI] [PubMed] [Google Scholar]
- 57. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B.. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008;30:678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D.. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099–1108. [DOI] [PubMed] [Google Scholar]
- 59. Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T.. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007;14:230–239. [DOI] [PubMed] [Google Scholar]
- 60. Wang J, Li Y, Duan J, Yang M, Yu Y, Feng L, Yang X, Zhou X, Zhao Z, Sun Z.. Silica nanoparticles induce autophagosome accumulation via activation of the EIF2AK3 and ATF6 UPR pathways in hepatocytes. Autophagy 2018;14:1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A.. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013;41:7683–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL.. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010;29:4424–4435. [DOI] [PubMed] [Google Scholar]
- 63. Ranjitha HB, Ammanathan V, Guleria N, Hosamani M, Sreenivasa BP, Dhanesh VV, Santhoshkumar R, Sagar BKC, Mishra BP, Singh RK, Sanyal A, Manjithaya R, Basagoudanavar SH.. Foot-and-mouth disease virus induces PERK-mediated autophagy to suppress the antiviral interferon response. J Cell Sci 2020;134:jcs240622. [DOI] [PubMed] [Google Scholar]
- 64. Luhr M, Torgersen ML, Szalai P, Hashim A, Brech A, Staerk J, Engedal N.. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem 2019;294:8197–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grumati P, Dikic I, Stolz A.. ER-phagy at a glance. J Cell Sci 2018;131:jcs217364. [DOI] [PubMed] [Google Scholar]
- 66. Wilkinson S. ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS J 2019;286:2645–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin DS, Huang YW, Ho CS, Hung PL, Hsu MH, Wang TJ, Wu TY, Lee TH, Huang ZD, Chang PC, Chiang MF.. Oxidative insults and mitochondrial DNA mutation promote enhanced autophagy and mitophagy compromising cell viability in pluripotent cell model of mitochondrial disease. Cells 2019;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y, Azad MB, Gibson SB.. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 2009;16:1040–1052. [DOI] [PubMed] [Google Scholar]
- 69. Chen L, Xu B, Liu L, Luo Y, Yin J, Zhou H, Chen W, Shen T, Han X, Huang S.. Hydrogen peroxide inhibits mTOR signaling by activation of AMPKalpha leading to apoptosis of neuronal cells. Lab Invest 2010;90:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA.. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010;6:506–522. [DOI] [PubMed] [Google Scholar]
- 71. Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T.. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001;2:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Itakura E, Kishi C, Inoue K, Mizushima N.. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008;19:5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T.. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009;11:385–396. [DOI] [PubMed] [Google Scholar]
- 74. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q.. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008;105:19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, Li M, Yue Z.. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun 2014;5:3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F.. Ambra1 regulates autophagy and development of the nervous system. Nature 2007;447:1121–1125. [DOI] [PubMed] [Google Scholar]
- 77. Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU.. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006;8:688–699. [DOI] [PubMed] [Google Scholar]
- 78. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG.. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007;9:1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU.. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008;10:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH.. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 2015;57:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A.. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009;11:1433–1437. [DOI] [PubMed] [Google Scholar]
- 82. Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA.. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006;119:3888–3900. [DOI] [PubMed] [Google Scholar]
- 83. Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J.. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010;141:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC.. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013;154:1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC.. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010;12:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, Luo Q, Feng D, Zhu Y, Chen Q.. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res 2017;27:184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Imai K, Hao F, Fujita N, Tsuji Y, Oe Y, Araki Y, Hamasaki M, Noda T, Yoshimori T.. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci 2016;129:3781–3791. [DOI] [PubMed] [Google Scholar]
- 88. Mattera R, Park SY, De Pace R, Guardia CM, Bonifacino JS.. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc Natl Acad Sci USA 2017;114:E10697–E10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guardia CM, Tan XF, Lian T, Rana MS, Zhou W, Christenson ET, Lowry AJ, Faraldo-Gomez JD, Bonifacino JS, Jiang J, Banerjee A.. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep 2020;31:107837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T.. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 2020;27:1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Maeda S, Otomo C, Otomo T.. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 2019;8:e45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ.. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 2019;218:1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ge L, Melville D, Zhang M, Schekman R.. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2013;2:e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ge L, Zhang M, Schekman R.. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 2014;3:e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jeong YT, Simoneschi D, Keegan S, Melville D, Adler NS, Saraf A, Florens L, Washburn MP, Cavasotto CN, Fenyo D, Cuervo AM, Rossi M, Pagano M.. The ULK1-FBXW5-SEC23B nexus controls autophagy. Elife 2018;7:e42253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL.. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009;5:1180–1185. [DOI] [PubMed] [Google Scholar]
- 97. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A.. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy 2010;6:301–303. [DOI] [PubMed] [Google Scholar]
- 98. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT.. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008;182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T.. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 2010;190:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA.. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 2014;55:238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Otomo T, Chowdhury S, Lander GC.. The rod-shaped ATG2A-WIPI4 complex tethers membranes in vitro. Contact (Thousand Oaks) 2018;1:251525641881993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chowdhury S, Otomo C, Leitner A, Ohashi K, Aebersold R, Lander GC, Otomo T.. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc Natl Acad Sci USA 2018;115:E9792–E9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT.. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 2013;126:5224–5238. [DOI] [PubMed] [Google Scholar]
- 104. Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T.. Autophagosomes form at ER-mitochondria contact sites. Nature 2013;495:389–393. [DOI] [PubMed] [Google Scholar]
- 105. Tang Z, Takahashi Y, He H, Hattori T, Chen C, Liang X, Chen H, Young MM, Wang HG.. TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep 2019;28:1744–1757.e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Biazik J, Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL.. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 2015;11:439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mizushima N, Sugita H, Yoshimori T, Ohsumi Y.. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 1998;273:33889–33892. [DOI] [PubMed] [Google Scholar]
- 108. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T.. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F.. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 2008;13:1211–1218. [DOI] [PubMed] [Google Scholar]
- 110. Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N.. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 2013;14:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lystad AH, Carlsson SR, de la Ballina LR, Kauffman KJ, Nag S, Yoshimori T, Melia TJ, Simonsen A.. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat Cell Biol 2019;21:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gatica D, Lahiri V, Klionsky DJ.. Cargo recognition and degradation by selective autophagy. Nat Cell Biol 2018;20:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F.. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009;10:1215–1221. [DOI] [PubMed] [Google Scholar]
- 114. Klionsky DJ, Schulman BA.. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 2014;21:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, Lohr F, Wakatsuki S, Dotsch V, Dikic I, Dobson RC, McEwan DG.. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep 2017;18:1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T.. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 2012;287:39275–39290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Birgisdottir AB, Mouilleron S, Bhujabal Z, Wirth M, Sjottem E, Evjen G, Zhang W, Lee R, O'Reilly N, Tooze SA, Lamark T, Johansen T.. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 2019;15:1333–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Skytte Rasmussen M, Mouilleron S, Kumar Shrestha B, Wirth M, Lee R, Bowitz Larsen K, Abudu Princely Y, O'Reilly N, Sjøttem E, Tooze SA, Lamark T, Johansen T.. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy 2017;13:834–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, Ohsumi Y, Inagaki F.. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell 2011;44:462–475. [DOI] [PubMed] [Google Scholar]
- 120. Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD.. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 2019;177:766–781.e724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kimura S, Noda T, Yoshimori T.. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 2008;33:109–122. [DOI] [PubMed] [Google Scholar]
- 122. Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z.. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006;281:36303–36316. [DOI] [PubMed] [Google Scholar]
- 123. Gutierrez MG, Munafo DB, Beron W, Colombo MI.. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004;117:2687–2697. [DOI] [PubMed] [Google Scholar]
- 124. Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL.. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004;117:4837–4848. [DOI] [PubMed] [Google Scholar]
- 125. Tian X, Zheng P, Zhou C, Wang X, Ma H, Ma W, Zhou X, Teng J, Chen J.. DIPK2A promotes STX17- and VAMP7-mediated autophagosome-lysosome fusion by binding to VAMP7B. Autophagy 2020;16:797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Saleeb RS, Kavanagh DM, Dun AR, Dalgarno PA, Duncan RR.. A VPS33A-binding motif on syntaxin 17 controls autophagy completion in mammalian cells. J Biol Chem 2019;294:4188–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N.. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 2014;25:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I.. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 2015;57:39–54. [DOI] [PubMed] [Google Scholar]
- 129. Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, Straub C, Kim C, Park J, Sabatini BL, Sabatini DM.. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015;347:188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G.. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015;519:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jung J, Genau HM, Behrends C.. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol 2015;35:2479–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, Sabatini DM.. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 2017;171:642–654.e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Scalise M, Galluccio M, Pochini L, Cosco J, Trotta M, Rebsamen M, Superti-Furga G, Indiveri C.. Insights into the transport side of the human SLC38A9 transceptor. Biochim Biophys Acta Biomembr 2019;1861:1558–1567. [DOI] [PubMed] [Google Scholar]
- 134. Gatica D, Chiong M, Lavandero S, Klionsky DJ.. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res 2015;116:456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lavandero S, Chiong M, Rothermel BA, Hill JA.. Autophagy in cardiovascular biology. J Clin Invest 2015;125:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lavandero S, Troncoso R, Rothermel BA, Martinet W, Sadoshima J, Hill JA.. Cardiovascular autophagy: concepts, controversies, and perspectives. Autophagy 2013;9:1455–1466. [DOI] [PubMed] [Google Scholar]
- 137. Morales PE, Arias-Duran C, Avalos-Guajardo Y, Aedo G, Verdejo HE, Parra V, Lavandero S.. Emerging role of mitophagy in cardiovascular physiology and pathology. Mol Aspects Med 2020;71:100822. [DOI] [PubMed] [Google Scholar]
- 138. Zhang J, Liu J, Huang Y, Chang JY, Liu L, McKeehan WL, Martin JF, Wang F.. FRS2alpha-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res 2012;110:e29–e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lee E, Koo Y, Ng A, Wei Y, Luby-Phelps K, Juraszek A, Xavier RJ, Cleaver O, Levine B, Amatruda JF.. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy 2014;10:572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sybers HD, Ingwall J, DeLuca M.. Autophagy in cardiac myocytes. Recent Adv Stud Cardiac Struct Metab 1976;12:453–463. [PubMed] [Google Scholar]
- 141. Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P.. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 2000;406:902–906. [DOI] [PubMed] [Google Scholar]
- 142. Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H.. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J 2001;65:965–968. [DOI] [PubMed] [Google Scholar]
- 143. Kostin S, Pool L, ElsäSser A, Hein S, Drexler HCA, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, KlöVekorn W-P, Schaper J, Myocytes die by multiple mechanisms in failing human hearts. Circ Res 2003;92:715–724. [DOI] [PubMed] [Google Scholar]
- 144. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J.. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007;100:914–922. [DOI] [PubMed] [Google Scholar]
- 145. Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K.. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007;13:619–624. [DOI] [PubMed] [Google Scholar]
- 146. Kassiotis C, Ballal K, Wellnitz K, Vela D, Gong M, Salazar R, Frazier OH, Taegtmeyer H.. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation 2009;120:S191–S197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J.. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 2010;107:1470–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mai S, Muster B, Bereiter-Hahn J, Jendrach M.. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy 2012;8:47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Aránguiz-Urroz P, Canales J, Copaja M, Troncoso R, Vicencio JM, Carrillo C, Lara H, Lavandero S, Díaz-Araya G.. Beta(2)-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim Biophys Acta 2011;1812:23–31. [DOI] [PubMed] [Google Scholar]
- 150. Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K.. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 2012;485:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A.. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation 2012;125:3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cao DJ, Jiang N, Blagg A, Johnstone JL, Gondalia R, Oh M, Luo X, Yang KC, Shelton JM, Rothermel BA, Gillette TG, Dorn GW, Hill JA.. Mechanical unloading activates FoxO3 to trigger Bnip3-dependent cardiomyocyte atrophy. J Am Heart Assoc 2013;2:e000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Chen Y, Dorn GW 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013;340:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA, Wu XS, Rovira II, Fergusson MM, Weigert R, Connelly PS, Daniels MP, Komatsu M, Cao L, Finkel T.. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med 2013;19:1281–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, Lim DS, Isobe M, Sadoshima J.. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013;19:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Guo F, Li X, Peng J, Tang Y, Yang Q, Liu L, Wang Z, Jiang Z, Xiao M, Ni C, Chen R, Wei D, Wang GX.. Autophagy regulates vascular endothelial cell eNOS and ET-1 expression induced by laminar shear stress in an ex vivo perfused system. Ann Biomed Eng 2014;42:1978–1988. [DOI] [PubMed] [Google Scholar]
- 157. Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW 2nd. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 2015;350:aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, Gabrielson K, Kass DA, Iijima M, Sesaki H.. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 2014;33:2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, Abdellatif M, Sadoshima J.. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res 2015;116:264–278. [DOI] [PubMed] [Google Scholar]
- 160. Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, Dorn GW 2nd. Interdependence of Parkin-mediated mitophagy and mitochondrial fission in adult mouse hearts. Circ Res 2015;117:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Araki S, Izumiya Y, Rokutanda T, Ianni A, Hanatani S, Kimura Y, Onoue Y, Senokuchi T, Yoshizawa T, Yasuda O, Koitabashi N, Kurabayashi M, Braun T, Bober E, Yamagata K, Ogawa H.. Sirt7 contributes to myocardial tissue repair by maintaining transforming growth factor-beta signaling pathway. Circulation 2015;132:1081–1093. [DOI] [PubMed] [Google Scholar]
- 162. Shirakabe A, Zhai P, Ikeda Y, Saito T, Maejima Y, Hsu CP, Nomura M, Egashira K, Levine B, Sadoshima J.. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation 2016;133:1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Fetterman JL, Holbrook M, Flint N, Feng B, Breton-Romero R, Linder EA, Berk BD, Duess MA, Farb MG, Gokce N, Shirihai OS, Hamburg NM, Vita JA.. Restoration of autophagy in endothelial cells from patients with diabetes mellitus improves nitric oxide signaling. Atherosclerosis 2016;247:207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Li DL, Wang ZV, Ding G, Tan W, Luo X, Criollo A, Xie M, Jiang N, May H, Kyrychenko V, Schneider JW, Gillette TG, Hill JA.. Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification. Circulation 2016;133:1668–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Büttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Mühlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F.. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med 2016;22:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yau JW, Singh KK, Hou Y, Lei X, Ramadan A, Quan A, Teoh H, Kuebler WM, Al-Omran M, Yanagawa B, Ni H, Verma S.. Endothelial-specific deletion of autophagy-related 7 (ATG7) attenuates arterial thrombosis in mice. J Thorac Cardiovasc Surg 2017;154:978–988.e971. [DOI] [PubMed] [Google Scholar]
- 167. Bharath LP, Cho JM, Park SK, Ruan T, Li Y, Mueller R, Bean T, Reese V, Richardson RS, Cai J, Sargsyan A, Pires K, Anandh Babu PV, Boudina S, Graham TE, Symons JD.. Endothelial cell autophagy maintains shear stress-induced nitric oxide generation via glycolysis-dependent purinergic signaling to endothelial nitric oxide synthase. Arterioscler Thromb Vasc Biol 2017;37:1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Vion AC, Kheloufi M, Hammoutene A, Poisson J, Lasselin J, Devue C, Pic I, Dupont N, Busse J, Stark K, Lafaurie-Janvore J, Barakat AI, Loyer X, Souyri M, Viollet B, Julia P, Tedgui A, Codogno P, Boulanger CM, Rautou PE.. Autophagy is required for endothelial cell alignment and atheroprotection under physiological blood flow. Proc Natl Acad Sci USA 2017;114:E8675–E8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Kheloufi M, Vion AC, Hammoutene A, Poisson J, Lasselin J, Devue C, Pic I, Dupont N, Busse J, Stark K, Lafaurie-Janvore J, Barakat AI, Loyer X, Souyri M, Viollet B, Julia P, Tedgui A, Codogno P, Boulanger CM, Rautou PE.. Endothelial autophagic flux hampers atherosclerotic lesion development. Autophagy 2018;14:173–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sun Y, Yao X, Zhang QJ, Zhu M, Liu ZP, Ci B, Xie Y, Carlson D, Rothermel BA, Sun Y, Levine B, Hill JA, Wolf SE, Minei JP, Zang QS.. Beclin-1-dependent autophagy protects the heart during sepsis. Circulation 2018;138:2247–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yang R, Song Z, Wu S, Wei Z, Xu Y, Shen X.. Toll-like receptor 4 contributes to a myofibroblast phenotype in cardiac fibroblasts and is associated with autophagy after myocardial infarction in a mouse model. Atherosclerosis 2018;279:23–31. [DOI] [PubMed] [Google Scholar]
- 172. Tong M, Saito T, Zhai P, Oka SI, Mizushima W, Nakamura M, Ikeda S, Shirakabe A, Sadoshima J.. Mitophagy is essential for maintaining cardiac function during high fat diet-induced diabetic cardiomyopathy. Circ Res 2019;124:1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Wang L, Ma H, Huang P, Xie Y, Near D, Wang H, Xu J, Yang Y, Xu Y, Garbutt T, Zhou Y, Liu Z, Yin C, Bressan M, Taylor JM, Liu J, Qian L.. Down-regulation of Beclin1 promotes direct cardiac reprogramming. Sci Transl Med 2020;12:eaay7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wildenthal K, Dees JH, Buja LM.. Cardiac lysosomal derangements in mouse heart after long-term exposure to nonmetabolizable sugars. Circ Res 1977;40:26–35. [DOI] [PubMed] [Google Scholar]
- 175. Dorn BR, Dunn WA Jr, Progulske-Fox A.. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect Immun 2001;69:5698–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Jia G, Cheng G, Gangahar DM, Agrawal DK.. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol 2006;84:448–454. [DOI] [PubMed] [Google Scholar]
- 177. Lee SJ, Kim HP, Jin Y, Choi AM, Ryter SW.. Beclin 1 deficiency is associated with increased hypoxia-induced angiogenesis. Autophagy 2011;7:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Grootaert MO, da Costa Martins PA, Bitsch N, Pintelon I, De Meyer GR, Martinet W, Schrijvers DM.. Defective autophagy in vascular smooth muscle cells accelerates senescence and promotes neointima formation and atherogenesis. Autophagy 2015;11:2014–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Docherty CK, Carswell A, Friel E, Mercer JR.. Impaired mitochondrial respiration in human carotid plaque atherosclerosis: a potential role for Pink1 in vascular smooth muscle cell energetics. Atherosclerosis 2018;268:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nahapetyan H, Moulis M, Grousset E, Faccini J, Grazide MH, Mucher E, Elbaz M, Martinet W, Vindis C.. Altered mitochondrial quality control in Atg7-deficient VSMCs promotes enhanced apoptosis and is linked to unstable atherosclerotic plaque phenotype. Cell Death Dis 2019;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Clement M, Chappell J, Raffort J, Lareyre F, Vandestienne M, Taylor AL, Finigan A, Harrison J, Bennett MR, Bruneval P, Taleb S, Jorgensen HF, Mallat Z.. Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler Thromb Vasc Biol 2019;39:1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. De Munck DG, Leloup AJA, De Meyer GRY, Martinet W, Fransen P.. Defective autophagy in vascular smooth muscle cells increases passive stiffness of the mouse aortic vessel wall. Pflugers Arch 2020;472:1031–1040. [DOI] [PubMed] [Google Scholar]
- 183. Zhang H, Ge S, He K, Zhao X, Wu Y, Shao Y, Wu X.. FoxO1 inhibits autophagosome-lysosome fusion leading to endothelial autophagic-apoptosis in diabetes. Cardiovasc Res 2019;115:2008–2020. [DOI] [PubMed] [Google Scholar]
- 184. Sprott D, Poitz DM, Korovina I, Ziogas A, Phieler J, Chatzigeorgiou A, Mitroulis I, Deussen A, Chavakis T, Klotzsche-von Ameln A.. Endothelial-specific deficiency of ATG5 (autophagy protein 5) attenuates ischemia-related angiogenesis. Arterioscler Thromb Vasc Biol 2019;39:1137–1148. [DOI] [PubMed] [Google Scholar]
- 185. Locatelli L, Cazzaniga A, De Palma C, Castiglioni S, Maier JAM.. Mitophagy contributes to endothelial adaptation to simulated microgravity. FASEB J 2020;34:1833–1845. [DOI] [PubMed] [Google Scholar]
- 186. De Munck DG, De Moudt S, Roth L, De Meyer GRY, Martinet W, Fransen P.. Defective autophagy in vascular smooth muscle cells alters vascular reactivity of the mouse femoral artery. Front Physiol 2020;11:548943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Takagaki Y, Lee SM, Dongqing Z, Kitada M, Kanasaki K, Koya D.. Endothelial autophagy deficiency induces IL6-dependent endothelial mesenchymal transition and organ fibrosis. Autophagy 2020;16:1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Yuan P, Hu Q, He X, Long Y, Song X, Wu F, He Y, Zhou X.. Laminar flow inhibits the Hippo/YAP pathway via autophagy and SIRT1-mediated deacetylation against atherosclerosis. Cell Death Dis 2020;11:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Jeong IH, Bae WY, Choi JS, Jeong JW.. Ischemia induces autophagy of endothelial cells and stimulates angiogenic effects in a hindlimb ischemia mouse model. Cell Death Dis 2020;11:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Taegtmeyer H. Energy metabolism of the heart: from basic concepts to clinical applications. Curr Probl Cardiol 1994;19:59–113. [DOI] [PubMed] [Google Scholar]
- 191. Lopez-Crisosto C, Pennanen C, Vasquez-Trincado C, Morales PE, Bravo-Sagua R, Quest AFG, Chiong M, Lavandero S.. Sarcoplasmic reticulum-mitochondria communication in cardiovascular pathophysiology. Nat Rev Cardiol 2017;14:342–360. [DOI] [PubMed] [Google Scholar]
- 192. Saito T, Nah J, Oka SI, Mukai R, Monden Y, Maejima Y, Ikeda Y, Sciarretta S, Liu T, Li H, Baljinnyam E, Fraidenraich D, Fritzky L, Zhai P, Ichinose S, Isobe M, Hsu CP, Kundu M, Sadoshima J.. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest 2019;129:802–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Wang B, Nie J, Wu L, Hu Y, Wen Z, Dong L, Zou MH, Chen C, Wang DW.. AMPKalpha2 protects against the development of heart failure by enhancing mitophagy via PINK1 phosphorylation. Circ Res 2018;122:712–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Tahrir FG, Knezevic T, Gupta MK, Gordon J, Cheung JY, Feldman AM, Khalili K.. Evidence for the role of BAG3 in mitochondrial quality control in cardiomyocytes. J Cell Physiol 2017;232:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Rodriguez AE, Lopez-Crisosto C, Pena-Oyarzun D, Salas D, Parra V, Quiroga C, Morawe T, Chiong M, Behl C, Lavandero S.. BAG3 regulates total MAP1LC3B protein levels through a translational but not transcriptional mechanism. Autophagy 2016;12:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Knezevic T, Myers VD, Su F, Wang J, Song J, Zhang XQ, Gao E, Gao G, Muniswamy M, Gupta MK, Gordon J, Weiner KN, Rabinowitz J, Ramsey FV, Tilley DG, Khalili K, Cheung JY, Feldman AM.. Adeno-associated virus serotype 9-driven expression of BAG3 improves left ventricular function in murine hearts with left ventricular dysfunction secondary to a myocardial infarction. JACC Basic Transl Sci 2016;1:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Jin HS, Hong KW, Kim BY, Kim J, Yoo YH, Oh B, Jeong SY.. Replicated association between genetic variation in the PARK2 gene and blood pressure. Clin Chim Acta 2011;412:1673–1677. [DOI] [PubMed] [Google Scholar]
- 198. Eisenberg T, Abdellatif M, Zimmermann A, Schroeder S, Pendl T, Harger A, Stekovic S, Schipke J, Magnes C, Schmidt A, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Carmona-Gutierrez D, Pietrocola F, Pieber TR, Sigrist SJ, Linke WA, Muhlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F.. Dietary spermidine for lowering high blood pressure. Autophagy 2017;13:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A.. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol 2006;40:846–852. [DOI] [PubMed] [Google Scholar]
- 200. Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A.. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy 2012;8:1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Huang L, Dai K, Chen M, Zhou W, Wang X, Chen J, Zhou W.. The AMPK agonist PT1 and mTOR inhibitor 3HOI-BA-01 protect cardiomyocytes after ischemia through induction of autophagy. J Cardiovasc Pharmacol Ther 2016;21:70–81. [DOI] [PubMed] [Google Scholar]
- 202. Li S, Liu C, Gu L, Wang L, Shang Y, Liu Q, Wan J, Shi J, Wang F, Xu Z, Ji G, Li W.. Autophagy protects cardiomyocytes from the myocardial ischaemia-reperfusion injury through the clearance of CLP36. Open Biol 2016;6:160177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Nah J, Zhai P, Huang CY, Fernandez AF, Mareedu S, Levine B, Sadoshima J.. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest 2020;130:2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Wu S, Chang G, Gao L, Jiang D, Wang L, Li G, Luo X, Qin S, Guo X, Zhang D.. Trimetazidine protects against myocardial ischemia/reperfusion injury by inhibiting excessive autophagy. J Mol Med (Berl) 2018;96:791–806. [DOI] [PubMed] [Google Scholar]
- 205. Norambuena-Soto I, Núñez-Soto C, Sanhueza-Olivares F, Cancino-Arenas N, Mondaca-Ruff D, Vivar R, Díaz-Araya G, Mellado R, Chiong M.. Transforming growth factor-beta and Forkhead box O transcription factors as cardiac fibroblast regulators. Biosci Trends 2017;11:154–162. [DOI] [PubMed] [Google Scholar]
- 206. Gupta SS, Zeglinski MR, Rattan SG, Landry NM, Ghavami S, Wigle JT, Klonisch T, Halayko AJ, Dixon IM.. Inhibition of autophagy inhibits the conversion of cardiac fibroblasts to cardiac myofibroblasts. Oncotarget 2016;7:78516–78531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH, Chen S, Klonisch T, Halayko AJ, Ambrose E, Singal R, Dixon IM.. Autophagy is a regulator of TGF-beta1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis 2015;6:e1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Gao H, Yang Q, Dong R, Hou F, Wu Y.. Sequential changes in autophagy in diabetic cardiac fibrosis. Mol Med Rep 2016;13:327–332. [DOI] [PubMed] [Google Scholar]
- 209. Wang H, Yang X, Yang Q, Gong L, Xu H, Wu Z.. PARP-1 inhibition attenuates cardiac fibrosis induced by myocardial infarction through regulating autophagy. Biochem Biophys Res Commun 2018;503:1625–1632. [DOI] [PubMed] [Google Scholar]
- 210. Westermeier F, Riquelme JA, Pavez M, Garrido V, Diaz A, Verdejo HE, Castro PF, Garcia L, Lavandero S.. New molecular insights of insulin in diabetic cardiomyopathy. Front Physiol 2016;7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Munasinghe PE, Riu F, Dixit P, Edamatsu M, Saxena P, Hamer NS, Galvin IF, Bunton RW, Lequeux S, Jones G, Lamberts RR, Emanueli C, Madeddu P, Katare R.. Type-2 diabetes increases autophagy in the human heart through promotion of Beclin-1 mediated pathway. Int J Cardiol 2016;202:13–20. [DOI] [PubMed] [Google Scholar]
- 212. Durga Devi T, Babu M, Mäkinen P, Kaikkonen MU, Heinaniemi M, Laakso H, Ylä-Herttuala E, Rieppo L, Liimatainen T, Naumenko N, Tavi P, Ylä-Herttuala S.. Aggravated postinfarct heart failure in type 2 diabetes is associated with impaired mitophagy and exaggerated inflammasome activation. Am J Pathol 2017;187:2659–2673. [DOI] [PubMed] [Google Scholar]
- 213. Mariappan N, Elks CM, Sriramula S, Guggilam A, Liu Z, Borkhsenious O, Francis J.. NF-kappaB-induced oxidative stress contributes to mitochondrial and cardiac dysfunction in type II diabetes. Cardiovasc Res 2010;85:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Kanamori H, Takemura G, Goto K, Tsujimoto A, Mikami A, Ogino A, Watanabe T, Morishita K, Okada H, Kawasaki M, Seishima M, Minatoguchi S.. Autophagic adaptations in diabetic cardiomyopathy differ between type 1 and type 2 diabetes. Autophagy 2015;11:1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Andres AM, Kooren JA, Parker SJ, Tucker KC, Ravindran N, Ito BR, Huang C, Venkatraman V, Van Eyk JE, Gottlieb RA, Mentzer RM Jr. Discordant signaling and autophagy response to fasting in hearts of obese mice: implications for ischemia tolerance. Am J Physiol Heart Circ Physiol 2016;311:H219–H228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Chandramouli C, Reichelt ME, Curl CL, Varma U, Bienvenu LA, Koutsifeli P, Raaijmakers AJA, De Blasio MJ, Qin CX, Jenkins AJ, Ritchie RH, Mellor KM, Delbridge LMD.. Diastolic dysfunction is more apparent in STZ-induced diabetic female mice, despite less pronounced hyperglycemia. Sci Rep 2018;8:2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wende AR, Schell JC, Ha CM, Pepin ME, Khalimonchuk O, Schwertz H, Pereira RO, Brahma MK, Tuinei J, Contreras-Ferrat A, Wang L, Andrizzi CA, Olsen CD, Bradley WE, Dell'Italia LJ, Dillmann WH, Litwin SE, Abel ED.. Maintaining myocardial glucose utilization in diabetic cardiomyopathy accelerates mitochondrial dysfunction. Diabetes 2020;69:2094–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Yuan X, Xiao YC, Zhang GP, Hou N, Wu XQ, Chen WL, Luo JD, Zhang GS.. Chloroquine improves left ventricle diastolic function in streptozotocin-induced diabetic mice. Drug Des Devel Ther 2016;10:2729–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Kobayashi S, Patel J, Zhao F, Huang Y, Kobayashi T, Liang Q.. Novel dual-fluorescent mitophagy reporter reveals a reduced mitophagy flux in type 1 diabetic mouse heart. J Am Osteopath Assoc 2020;120:446–455. [DOI] [PubMed] [Google Scholar]
- 220. Xu X, Kobayashi S, Chen K, Timm D, Volden P, Huang Y, Gulick J, Yue Z, Robbins J, Epstein PN, Liang Q.. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem 2013;288:18077–18092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Wei H, Qu H, Wang H, Ji B, Ding Y, Liu D, Duan Y, Liang H, Peng C, Xiao X, Deng H.. 1,25-Dihydroxyvitamin-D3 prevents the development of diabetic cardiomyopathy in type 1 diabetic rats by enhancing autophagy via inhibiting the beta-catenin/TCF4/GSK-3beta/mTOR pathway. J Steroid Biochem Mol Biol 2017;168:71–90. [DOI] [PubMed] [Google Scholar]
- 222. Wang B, Yang Q, Sun YY, Xing YF, Wang YB, Lu XT, Bai WW, Liu XQ, Zhao YX.. Resveratrol-enhanced autophagic flux ameliorates myocardial oxidative stress injury in diabetic mice. J Cell Mol Med 2014;18:1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, Gonzalez-Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P; ESC Scientific Document Group. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129–2200. [DOI] [PubMed] [Google Scholar]
- 224. Redfield MM. Heart failure with preserved ejection fraction. N Engl J Med 2016;375:1868–1877. [DOI] [PubMed] [Google Scholar]
- 225. Gladden JD, Linke WA, Redfield MM.. Heart failure with preserved ejection fraction. Pflugers Arch 2014;466:1037–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Hahn VS, Knutsdottir H, Luo X, Bedi K, Margulies KB, Haldar SM, Stolina M, Yin J, Khakoo AY, Vaishnav J, Bader JS, Kass DA, Sharma K.. Myocardial gene expression signatures in human heart failure with preserved ejection fraction. Circulation 2021; 143:120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Zhang C, Liu A, Su G, Chen Y.. Effect of rapamycin on the level of autophagy in rats with early heart failure. J Cell Biochem 2019;120:4065–4070. [DOI] [PubMed] [Google Scholar]
- 228. Chiao YA, Kolwicz SC, Basisty N, Gagnidze A, Zhang J, Gu H, Djukovic D, Beyer RP, Raftery D, MacCoss M, Tian R, Rabinovitch PS.. Rapamycin transiently induces mitochondrial remodeling to reprogram energy metabolism in old hearts. Aging (Albany NY) 2016;8:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Gao G, Chen W, Yan M, Liu J, Luo H, Wang C, Yang P.. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int J Mol Med 2020;45:195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Nandi SS, Katsurada K, Sharma NM, Anderson DR, Mahata SK, Patel KP.. MMP9 inhibition increases autophagic flux in chronic heart failure. Am J Physiol Heart Circ Physiol 2020;319:H1414–H1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Ranek MJ, Kokkonen-Simon KM, Chen A, Dunkerly-Eyring BL, Vera MP, Oeing CU, Patel CH, Nakamura T, Zhu G, Bedja D, Sasaki M, Holewinski RJ, Van Eyk JE, Powell JD, Lee DI, Kass DA.. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature 2019;566:264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Saito T, Asai K, Sato S, Hayashi M, Adachi A, Sasaki Y, Takano H, Mizuno K, Shimizu W.. Autophagic vacuoles in cardiomyocytes of dilated cardiomyopathy with initially decompensated heart failure predict improved prognosis. Autophagy 2016;12:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Wu P, Yuan X, Li F, Zhang J, Zhu W, Wei M, Li J, Wang X.. Myocardial upregulation of cathepsin D by ischemic heart disease promotes autophagic flux and protects against cardiac remodeling and heart failure. Circ Heart Fail 2017;10:e004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Corsetti G, Chen-Scarabelli C, Romano C, Pasini E, Dioguardi FS, Onorati F, Knight R, Patel H, Saravolatz L, Faggian G, Scarabelli TM.. Autophagy and oncosis/necroptosis are enhanced in cardiomyocytes from heart failure patients. Med Sci Monit Basic Res 2019;25:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Zhang H, Yin Y, Liu Y, Zou G, Huang H, Qian P, Zhang G, Zhang J.. Necroptosis mediated by impaired autophagy flux contributes to adverse ventricular remodeling after myocardial infarction. Biochem Pharmacol 2020;175:113915. [DOI] [PubMed] [Google Scholar]
- 236. Gao T, Zhang SP, Wang JF, Liu L, Wang Y, Cao ZY, Hu QK, Yuan WJ, Lin L.. TLR3 contributes to persistent autophagy and heart failure in mice after myocardial infarction. J Cell Mol Med 2018;22:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Li B, Chi RF, Qin FZ, Guo XF.. Distinct changes of myocyte autophagy during myocardial hypertrophy and heart failure: association with oxidative stress. Exp Physiol 2016;101:1050–1063. [DOI] [PubMed] [Google Scholar]
- 238. Li Y, Wang Y, Zou M, Chen C, Chen Y, Xue R, Dong Y, Liu C.. AMPK blunts chronic heart failure by inhibiting autophagy. Biosci Rep 2018;38:BSR20170982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J.. Aging and autophagy in the heart. Circ Res 2016;118:1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, Oka T, Tamai T, Oyabu J, Murakawa T, Nishida K, Shimizu T, Hori M, Komuro I, Takuji Shirasawa TS, Mizushima N, Otsu K.. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010;6:600–606. [DOI] [PubMed] [Google Scholar]
- 241. Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, Assani K, Kopp B, Marsh C, Dakhlallah D, Amer AO.. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics 2016;11:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Fernandez AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, Marciano DK, Schiattarella GG, Bhagat G, Moe OW, Hu MC, Levine B.. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018;558:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Chavez MN, Morales RA, Lopez-Crisosto C, Roa JC, Allende ML, Lavandero S.. Autophagy activation in zebrafish heart regeneration. Sci Rep 2020;10:2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Chiong M, Cartes-Saavedra B, Norambuena-Soto I, Mondaca-Ruff D, Morales PE, García-Miguel M, Mellado R.. Mitochondrial metabolism and the control of vascular smooth muscle cell proliferation. Front Cell Dev Biol 2014;2:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Liu Y, Liu B, Li Y, Li Y, Du J, Deng S, Jing X, She Q.. Autophagy is involved in the differentiation of epicardial progenitor cells into vascular smooth muscle cells in mice. Exp Cell Res 2019;375:60–71. [DOI] [PubMed] [Google Scholar]
- 246. Garrido-Moreno V, Díaz-Vegas A, López-Crisosto C, Troncoso MF, Navarro-Marquez M, García L, Estrada M, Cifuentes M, Lavandero S.. GDF-11 prevents cardiomyocyte hypertrophy by maintaining the sarcoplasmic reticulum-mitochondria communication. Pharmacol Res 2019;146:104273. [DOI] [PubMed] [Google Scholar]
- 247. Yuan X, Bhat OM, Lohner H, Li N, Zhang Y, Li PL.. Inhibitory effects of growth differentiation factor 11 on autophagy deficiency-induced dedifferentiation of arterial smooth muscle cells. Am J Physiol Heart Circ Physiol 2019;316:H345–H356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Mondaca-Ruff D, Riquelme JA, Quiroga C, Norambuena-Soto I, Sanhueza-Olivares F, Villar-Fincheira P, Hernandez-Diaz T, Cancino-Arenas N, San Martin A, Garcia L, Lavandero S, Chiong M.. Angiotensin II-regulated autophagy is required for vascular smooth muscle cell hypertrophy. Front Pharmacol 2018;9:1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Wang N, Xu F, Lu S, Zhang N, Sun Y.. Septin4 as an autophagy modulator regulates Angiotensin-II mediated VSMCs proliferation and migration. Biochem Biophys Res Commun 2020;525:272–279. [DOI] [PubMed] [Google Scholar]
- 250. Wang Y, Zhang X, Chen W, Gao L, Li J, Song T, Chi J, Zhang X, Shi Z, Dong Y, Yin X, Liu Y.. Cortistatin ameliorates Ang II-induced proliferation of vascular smooth muscle cells by inhibiting autophagy through SSTR3 and SSTR5. Life Sci 2020;253:117726. [DOI] [PubMed] [Google Scholar]
- 251. Li T, Tan X, Zhu S, Zhong W, Huang B, Sun J, Li F, Wang Y.. SPARC induces phenotypic modulation of human brain vascular smooth muscle cells via AMPK/mTOR-mediated autophagy. Neurosci Lett 2019;712:134485. [DOI] [PubMed] [Google Scholar]
- 252. Ouyang C, Mu J, Lu Q, Li J, Zhu H, Wang Q, Zou MH, Xie Z.. Autophagic degradation of KAT2A/GCN5 promotes directional migration of vascular smooth muscle cells by reducing TUBA/alpha-tubulin acetylation. Autophagy 2019;16:1753–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Chatterjee S. Endothelial mechanotransduction, redox signaling and the regulation of vascular inflammatory pathways. Front Physiol 2018;9:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Liu J, Bi X, Chen T, Zhang Q, Wang SX, Chiu JJ, Liu GS, Zhang Y, Bu P, Jiang F.. Shear stress regulates endothelial cell autophagy via redox regulation and Sirt1 expression. Cell Death Dis 2015;6:e1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Zhang JX, Qu XL, Chu P, Xie DJ, Zhu LL, Chao YL, Li L, Zhang JJ, Chen SL.. Low shear stress induces vascular eNOS uncoupling via autophagy-mediated eNOS phosphorylation. Biochim Biophys Acta Mol Cell Res 2018;1865:709–720. [DOI] [PubMed] [Google Scholar]
- 256. Park SK, La Salle DT, Cerbie J, Cho JM, Bledsoe A, Nelson A, Morgan DE, Richardson RS, Shiu YT, Boudina S, Trinity JD, Symons JD.. Elevated arterial shear rate increases indexes of endothelial cell autophagy and nitric oxide synthase activation in humans. Am J Physiol Heart Circ Physiol 2019;316:H106–H112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Renna NF, de Las Heras N, Miatello RM.. Pathophysiology of vascular remodeling in hypertension. Int J Hypertens 2013;2013:808353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Campbell GR, Campbell JH.. Smooth muscle phenotypic changes in arterial wall homeostasis: implications for the pathogenesis of atherosclerosis. Exp Mol Pathol 1985;42:139–162. [DOI] [PubMed] [Google Scholar]
- 259. Cecchettini A, Rocchiccioli S, Boccardi C, Citti L.. Vascular smooth-muscle-cell activation: proteomics point of view. Int Rev Cell Mol Biol 2011;288:43–99. [DOI] [PubMed] [Google Scholar]
- 260. Kim Y, Park JK, Seo JH, Ryu HS, Lim KS, Jeong MH, Kang DH, Kang SW.. A rapamycin derivative, biolimus, preferentially activates autophagy in vascular smooth muscle cells. Sci Rep 2018;8:16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Osonoi Y, Mita T, Azuma K, Nakajima K, Masuyama A, Goto H, Nishida Y, Miyatsuka T, Fujitani Y, Koike M, Mitsumata M, Watada H.. Defective autophagy in vascular smooth muscle cells enhances cell death and atherosclerosis. Autophagy 2018;14:1991–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Ramadan A, Singh KK, Quan A, Plant PJ, Al-Omran M, Teoh H, Verma S.. Loss of vascular smooth muscle cell autophagy exacerbates angiotensin II-associated aortic remodeling. J Vasc Surg 2018;68:859–871. [DOI] [PubMed] [Google Scholar]
- 263. Lu H, Sun J, Liang W, Chang Z, Rom O, Zhao Y, Zhao G, Xiong W, Wang H, Zhu T, Guo Y, Chang L, Garcia-Barrio MT, Zhang J, Chen YE, Fan Y.. Cyclodextrin prevents abdominal aortic aneurysm via activation of vascular smooth muscle cell transcription factor EB. Circulation 2020;142:483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Wang Z, Guo J, Han X, Xue M, Wang W, Mi L, Sheng Y, Ma C, Wu J, Wu X.. Metformin represses the pathophysiology of AAA by suppressing the activation of PI3K/AKT/mTOR/autophagy pathway in ApoE(-/-) mice. Cell Biosci 2019;9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Wu QY, Cheng Z, Zhou YZ, Zhao Y, Li JM, Zhou XM, Peng HL, Zhang GS, Liao XB, Fu XM.. A novel STAT3 inhibitor attenuates angiotensin II-induced abdominal aortic aneurysm progression in mice through modulating vascular inflammation and autophagy. Cell Death Dis 2020;11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Pi S, Mao L, Chen J, Shi H, Liu Y, Guo X, Li Y, Zhou L, He H, Yu C, Liu J, Dang Y, Xia Y, He Q, Li JH, Hu Y, Miao Y, Yue Y, Hu B.. The P2RY12 receptor promotes VSMC-derived foam cell formation by inhibiting autophagy in advanced atherosclerosis. Autophagy 2020;17:980–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Masuyama A, Mita T, Azuma K, Osonoi Y, Nakajima K, Goto H, Nishida Y, Miyatsuka T, Mitsumata M, Watada H.. Defective autophagy in vascular smooth muscle cells enhances atherosclerotic plaque instability. Biochem Biophys Res Commun 2018;505:1141–1147. [DOI] [PubMed] [Google Scholar]
- 268. Evans TD, Jeong SJ, Zhang X, Sergin I, Razani B.. TFEB and trehalose drive the macrophage autophagy-lysosome system to protect against atherosclerosis. Autophagy 2018;14:724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Yuan X, Bhat OM, Meng N, Lohner H, Li PL.. Protective role of autophagy in Nlrp3 inflammasome activation and medial thickening of mouse coronary arteries. Am J Pathol 2018;188:2948–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Chen R, Jiang M, Li B, Zhong W, Wang Z, Yuan W, Yan J.. The role of autophagy in pulmonary hypertension: a double-edge sword. Apoptosis 2018;23:459–469. [DOI] [PubMed] [Google Scholar]
- 271. Gomez-Puerto MC, van Zuijen I, Huang CJ, Szulcek R, Pan X, van Dinther MA, Kurakula K, Wiesmeijer CC, Goumans M-J, Bogaard H-J, Morrell NW, Rana AA, Ten Dijke P.. Autophagy contributes to BMP type 2 receptor degradation and development of pulmonary arterial hypertension. J Pathol 2019;249:356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Guo L, Li Y, Tian Y, Gong S, Chen X, Peng T, Wang A, Jiang Z.. eIF2alpha promotes vascular remodeling via autophagy in monocrotaline-induced pulmonary arterial hypertension rats. Drug Des Devel Ther 2019;13:2799–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Feng W, Wang J, Yan X, Zhai C, Shi W, Wang Q, Zhang Q, Li M.. Paclitaxel alleviates monocrotaline-induced pulmonary arterial hypertension via inhibition of FoxO1-mediated autophagy. Naunyn Schmiedebergs Arch Pharmacol 2019;392:605–613. [DOI] [PubMed] [Google Scholar]