Abstract
The genetic relatedness of French isolates of Streptococcus pneumoniae highly resistant to amoxicillin (MIC, ≥4 μg/ml, equal to or exceeding those of penicillin) was investigated by molecular fingerprinting. The results suggest that high-level resistance to amoxicillin has emerged within preexisting penicillin-resistant clones.
In France, penicillin resistance rates in Streptococcus pneumoniae of up to 42.8% were reported in 1996 (7). Penicillin-resistant S. pneumoniae isolates show increased resistance to other β-lactam antibiotics. However, MICs of amoxicillin and extended-spectrum cephalosporins (cefotaxime and ceftriaxone) are generally about equal to or two to four times less than the MIC of benzylpenicillin (17). Thus, these drugs are currently recommended for the treatment of pneumococcal infections. Recently, pneumococci with high-level resistance to amoxicillin (amoxicillin MICs, ≥4 μg/ml) and/or extended-spectrum cephalosporins (cefotaxime MICs, ≥4 μg/ml) have been identified in France. The emergence of such strains is of considerable concern, because it further limits the available options for the therapy of severe pneumococcal infections.
For this report, we analyzed 29 clinical isolates of S. pneumoniae for which the amoxicillin MICs were ≥4 μg/ml (SPA4) recovered from 29 patients in different cities widely separated geographically across France by rRNA gene restriction pattern analysis to determine their overall genetic relatedness and examined DNA fingerprinting of the pbp1a, pbp2b, and pbp2x genes to determine the relatedness of the pbp genes. For comparison, 11 isolates with various levels of susceptibility to amoxicillin (MICs, <4 μg/ml) representative of the strains currently recovered in France were also studied: 3 French derivatives of the serotype 23F penicillin-resistant pandemic clone (4), 1 member of the serotype 9V/14 penicillin-resistant clone (4), 1 member of the serotype 6B penicillin-resistant clone, and 6 penicillin-intermediate strains (serotypes 23F, 14, and 19F). In addition, two Spanish penicillin-resistant clones (serotype 23F and 6B) and the penicillin-susceptible reference strain R6 were included in the study. These isolates and their relevant properties are listed in Table 1. Penicillin, amoxicillin, cefotaxime, and ceftriaxone MICs were determined by the dilution method on Mueller-Hinton agar supplemented with 5% sheep blood according to the National Committee for Clinical Laboratory Standards (NCCLS) guidelines (16). Serotyping was performed by coagglutination with antiserum-coated latex particles.
TABLE 1.
Phenotypic and genotypic characteristics and origin of the pneumococcal isolates in this study
| Isolate | MIC (μg/ml)
|
Serotype | Isolation
|
RFLP pbp gene pattern
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillin G | Amoxicillin | Cefotaxime | Ceftriaxone | Location | Yr | Sourcea | pbp1a | pbp2b | pbp2x | ||
| R6 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | NT | United States | 1946 | Unknown | X | XIV | XVII |
| 11758 | 0.25 | ≤0.03 | ≤0.03 | ≤0.03 | 23F | France | 1989 | CSF | X | XIX | XIII |
| 16120b | 1 | 0.5 | 1 | 0.5 | 14 | France | 1991 | CSF | I | I | III |
| 73211 | 2 | 4 | 1 | 1 | 14 | France | 1997 | AOM | I | V | VIII |
| 73295 | 2 | 4 | 1 | 1 | 14 | France | 1997 | AOM | I | IV | VII |
| 73510 | 4 | 8 | 1 | 0.5 | 14 | France | 1997 | AOM | IX | XI | VI |
| 73575 | 2 | 8 | 4 | 2 | 14 | France | 1997 | AOM | I | VII | II |
| 73012 | 2 | 4 | 1 | 0.5 | 14 | France | 1997 | NP | I | V | VIII |
| 73394 | 4 | 8 | 1 | 1 | 14 | France | 1997 | BLD | I | V | VIII |
| 73529 | 2 | 4 | 1 | 1 | 14 | France | 1997 | BLD | I | V | VIII |
| 73234 | 8 | 8 | 8 | 2 | 14 | France | 1997 | AOM | IV | XVII | I |
| 72830 | 4 | 8 | 4 | 1 | 14 | France | 1997 | SPU | VI | IV | VI |
| 73084 | 4 | 8 | 1 | 0.5 | 19F | France | 1997 | BAL | IX | II | VI |
| 17904c | 1 | 0.25 | 0.25 | 0.25 | 6B | Spain | 1993 | Unknown | I | XII | X |
| 72734 | 2 | 4 | 0.5 | 0.5 | 6B | France | 1997 | Unknown | I | III | VI |
| 72853 | 2 | 4 | 0.5 | 0.5 | 6B | France | 1997 | BLD | I | III | VI |
| 73042 | 2 | 4 | 1 | 0.5 | 6B | France | 1997 | AOM | I | VII | VII |
| 73171 | 1 | 4 | 0.5 | 0.5 | 6B | France | 1997 | SPU | I | III | VI |
| 73202 | 2 | 4 | 0.5 | 0.5 | 6B | France | 1997 | BLD | I | III | VI |
| 73328 | 2 | 4 | 1 | 0.5 | 6B | France | 1997 | SPU | I | VI | VI |
| 73333 | 2 | 4 | 1 | 0.5 | 6B | France | 1997 | BLD | I | XI | XI |
| 17977 | 0.5 | 0.25 | 0.25 | 0.5 | 6B | France | 1991 | CSF | I | XV | X |
| 60702d | 1 | 1 | 1 | 0.5 | 6B | France | 1996 | CSF | I | XIII | X |
| 11551e | 2 | 2 | 2 | 1 | 23F | France | 1989 | CSF | I | I | I |
| 65127 | 4 | 8 | 2 | 1 | 23F | France | 1996 | SPU | I | IX | I |
| 72521 | 2 | 8 | 2 | 1 | 23F | France | 1997 | BAL | I | IX | I |
| 456f | 2 | 2 | 2 | 1 | 23F | Spain | 1984 | Unknown | I | I | I |
| 73437 | 4 | 8 | 1 | 1 | 23F | France | 1997 | CUNJ | I | IX | XII |
| 72493 | 4 | 8 | 1 | 1 | 23F | France | 1997 | BAL | I | II | V |
| 72782 | 2 | 8 | 1 | 0.5 | 23F | France | 1997 | NP | I | IX | I |
| 72997 | 2 | 8 | 1 | 0.5 | 23F | France | 1997 | NP | VI | IX | I |
| 73311 | 8 | 8 | 8 | 1 | 23F | France | 1997 | NP | II | IX | IX |
| 9481e | 2 | 1 | 1 | 0.5 | 23F | France | 1988 | CSF | I | I | I |
| 73543 | 2 | 8 | 1 | 1 | 23F | France | 1997 | BAL | I | VIII | I |
| 43458e | 2 | 2 | 2 | 1 | 23F | France | 1994 | CSF | I | I | I |
| 72636 | 2 | 8 | 1 | 0.5 | 23F | France | 1997 | SPU | I | III | IV |
| 16294 | 0.125 | ≤0.03 | ≤0.03 | ≤0.03 | 19F | France | 1991 | CSF | XII | XVI | XVIII |
| 8966 | 0.125 | ≤0.03 | 0.06 | ≤0.03 | 14 | France | 1988 | CSF | VII | XVI | XVI |
| 71969 | 4 | 8 | 4 | 2 | 23F | France | 1997 | AOM | V | IV | I |
| 72487 | 4 | 8 | 4 | 2 | 23F | France | 1997 | NP | VIII | IV | I |
| 7473 | 0.125 | ≤0.03 | 0.12 | 0.06 | 23F | France | 1987 | CSF | X | XVIII | XIV |
| 9566 | 0.5 | ≤0.03 | 0.12 | 0.06 | 23F | France | 1988 | CSF | XI | XVIII | XV |
| 73058 | 4 | 8 | 1 | 1 | 19F | France | 1997 | CUNJ | III | X | I |
CSF, cerebrospinal fluid; SPU, sputum; AOM, acute otitis media; NP, nasopharyngeal swab; BAL, bronchoalveolar lavage; CUNJ, cunjunctival pus; BLD, blood.
Isolate representative of the serotype 9V/14 penicillin-resistant French clone.
Isolate representative of the serotype 6B penicillin-resistant Spanish clone.
Isolate representative of the serotype 6B penicillin-resistant French clone.
Isolates representative of the French derivatives of the pandemic 23F multiresistant Spanish clone.
Isolate representative of the pandemic serotype 23F multiresistant Spanish clone.
DNA ribotyping was performed as previously described (1) after digestion with HindIII and EcoRI (Boehringer Mannheim, Mannheim, Germany). pbp1a, pbp2b, and pbp2x were PCR amplified from chromosomal DNA by using the primers previously described (2, 6, 14). HinfI and DdeI-StyI were used for fingerprinting pbp1a, DdeI and AluI were used for pbp2b, and DdeI and HinfI were used for pbp2x. After digestion of the amplified fragments (pbp1a, nucleotides 786 to 3194; pbp2b, nucleotides 812 to 2317; pbp2x, nucleotides 478 to 2533), electrophoresis was performed on 6% polyacrylamide gels which were stained with ethidium bromide, and the DNA bands were visualized with a UV transilluminator. All patterns were analyzed with Bio-Gene V.96 computer software (Vilber Lourmat, Marne-la-Vallée, France). For each DNA fingerprint, patterns obtained with the two enzymes were combined. These combined patterns were used for the similarity estimation and cluster analysis, in which the similarity among strains was estimated by the Dice comparison, and the clustering of strains was determined by the unweighted pair group method.
The serotypes of and MICs for the clinical isolates studied are presented in Table 1. The 29 SPA4 isolates belong to only four serotypes: 23F (n = 11), 6B (n = 7), 14 (n = 9), and 19F (n = 2). The MICs of amoxicillin for all of these isolates were equal to or 1 to 2 dilutions higher than the MICs of penicillin. As demonstrated by the cefotaxime MICs, eight of these isolates were resistant to the extended-spectrum cephalosporins according to the NCCLS criteria, and the cefotaxime MICs for six of them were ≥4 μg/ml. Cefotaxime MICs were equal to or 1 dilution higher than penicillin MICs for seven isolates. Interestingly, ceftriaxone MICs, which are generally equal to or 1 dilution less than cefotaxime MICs (9), were 2 or 4 dilutions less than cefotaxime MICs for four isolates (71969, 72830, 73234, and 73311).
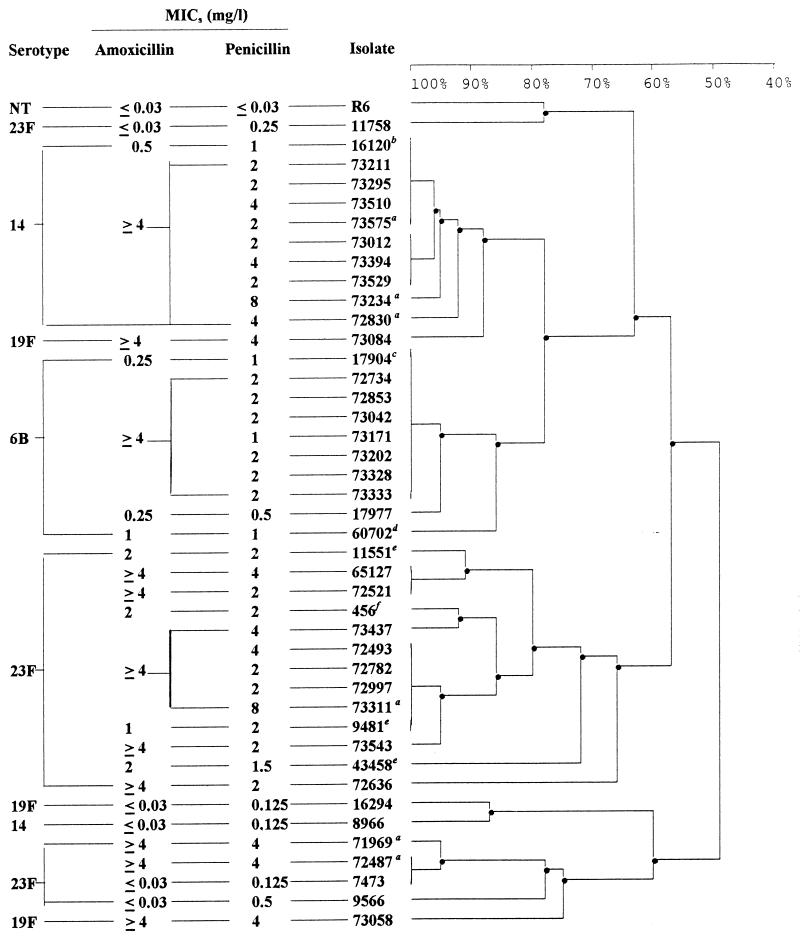
Digestion with HindIII generated 18 different patterns among the 43 isolates and 10 patterns among the 29 SPA4 isolates. Digestion with EcoRI generated 15 different patterns among the 43 isolates and 10 patterns among the 29 SPA4 isolates. Combination of the results obtained with the two enzymes generated 24 ribotypes among the 43 isolates and 14 ribotypes among the 29 SPA4 isolates. A dendrogram constructed from the cluster analysis shows estimates of the genetic relationships among the 24 ribotypes (Fig. 1). Among the 11 serotype 23F SPA4 isolates, 9 were genotypically indistinguishable or closely related (>70% homology) to the serotype 23F French and Spanish clones. The remaining two serotype 23F isolates exhibited slightly different patterns which placed them near the 23F intermediate strains. The serotype 14 SPA4 isolates were indistinguishable or closely related (>90% homology) to the serotype 9V/14 resistant French clone previously described (4). The seven serotype 6B SPA4 isolates were indistinguishable from the serotype 6B Spanish clone and closely related to the current French penicillin-resistant 6B strain.
FIG. 1.
Dendrogram depicting genetic distance and overall relatedness of pneumococcal isolates on the basis of rrn RFLP data. a, isolates for which cefotaxime MICs were ≥4 μg/ml. See Table 1 for the other characteristics of the isolates. (b, c, d, e, and f are representative of the serotype 9V/14 penicillin-resistant French clone, the serotype 6B penicillin-resistant Spanish clone, the serotype 6B penicillin-resistant French clone, the French derivatives of the pandemic 23F multiresistant Spanish clone, and the pandemic serotype 23F multiresistant Spanish clone, respectively.)
By using the combination of the results of the restriction fragment length polymorphism (RFLP) obtained with the two enzymes for each pbp gene, 12 different RFLP patterns were found for pbp1a, 19 were found for pbp2b, and 18 were found for pbp2x. All clinical isolates gave pbp2b and pbp2x gene patterns that differed from those of the penicillin-susceptible strain R6. Only two intermediate-resistant isolates (7473 and 11758) gave the same pbp1a gene pattern as R6. Eight different RFLP pbp1a patterns were found among the SPA4 isolates. One of them was unique to 20 isolates and was indistinguishable from that of 8 comparator isolates (Table 1). The SPA4 isolates gave 11 different pbp2b patterns that were all different from those of the isolates for which amoxicillin MICs were <2 μg/ml. pbp fingerprinting revealed 10 different patterns for pbp2x, all but 1 being distinct from those of the comparator isolates (Table 1). The six isolates for which cefotaxime MICs were ≥4 μg/ml exhibited six different pbp1a gene patterns and four different pbp2b and pbp2x gene patterns. With the combination of the ribotyping and pbp RFLP data, the following SPA4 isolates were indistinguishable: 65127 and 72521 (serotype 23F); 72734, 72853, 73171, and 73202 (serotype 6B); and 73012, 73394, and 73529 (serotype 14).
In France, the penicillin-resistant S. pneumoniae isolates (penicillin MICs, >1 μg/ml) belong to a limited number of serogroups (23, 9, 19, 6, and 14, representing 91.6% of all penicillin-resistant isolates) (7). The amoxicillin MICs for all of these isolates were <4 μg/ml. Pneumococci for which amoxicillin MICs were ≥4 μg/ml have recently been detected in the United States (3). During 1997, 29 SPA4 clinical isolates from 29 patients and belonging to four different serotypes were isolated across France, pointing to the dissemination of new clones. Among them, the cefotaxime MICs for six isolates belonging to two serotypes (23F and 14) were ≥4 μg/ml. Only a few isolates showing a high level of resistance to extended-spectrum cephalosporins (MICs, ≥4 μg/ml) have previously been reported in Spain, the United States, and the United Kingdom (2, 11, 13, 15). The spread of such strains would compromise the use of these antibiotics in serious infections like meningitis (5, 12).
Based on the results of our overall genomic DNA analysis, SPA4 isolates are indistinguishable from or closely related to isolates of the predominant resistant clones of the same serotype currently found in France. This suggests that high-level resistance to amoxicillin has emerged de novo within preexisting clones. Four distinct lineages of SPA4 isolates were distinguished. The first group was highly related by serotyping and ribotyping to the pandemic multiresistant Spanish serotype 23F clone. A second lineage was related to the serotype 23F intermediate strains. Another was derived from the serotype 9V/14 resistant French clone, and the last was derived from the Spanish resistant clone 6B, which has spread to Iceland and the United Kingdom (18). Thus, for each SPA4 lineage, we identified a related amoxicillin-susceptible group. Inside each lineage, great heterogeneity was observed in the pbp patterns, pointing to multiplicity in the sequence modifications. Only pbp2b patterns were always different from those of the comparator isolates.
High-level resistance to penicillin was previously associated with low-level resistance to amoxicillin and extended-spectrum cephalosporins. Indeed, treatment of infections due to penicillin-resistant pneumococci has required a change from penicillin to amoxicillin, cefotaxime, or ceftriaxone (10). Thus, amoxicillin and extended-spectrum cephalosporins have been used increasingly in France (8). The simultaneous occurrence of high-level resistance to amoxicillin in several pneumococcal clones in France suggests that amoxicillin-resistant pneumococci are likely to be recovered with increasing frequency, given the strong selective pressure resulting from extensive use of β-lactams for the treatment of pneumococcal infections in France. DNA sequencing of pbp genes (especially pbp2b) is warranted to determine the genetic modifications responsible for the high level of resistance to amoxicillin.
REFERENCES
- 1.Bingen E, Denamur E, Lambert-Zechowsky N, Aujard Y, Brahimi N, Geslin P, Elion J. Analysis of DNA restriction fragment length polymorphism extends the evidence for breast milk transmission in Streptococcus agalactiae late-onset neonatal infection. J Infect Dis. 1992;165:569–573. doi: 10.1093/infdis/165.3.569. [DOI] [PubMed] [Google Scholar]
- 2.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:255–260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 3.Doern G V, Brueggemann A, Holley H P, Jr, Rauch A M. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients in the United States during the winter months of 1994 to 1995: results of a 30-center national surveillance study. Antimicrob Agents Chemother. 1996;40:1208–1213. doi: 10.1128/aac.40.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doit C, Denamur E, Picard B, Geslin P, Elion J, Bingen E. Mechanisms of spread of penicillin resistance in Streptococcus pneumoniae strains causing meningitis in children in France. J Infect Dis. 1996;174:520–528. doi: 10.1093/infdis/174.3.520. [DOI] [PubMed] [Google Scholar]
- 5.Doit C P, Bonacorsi S P, Fremaux A J, Sissia G, Cohen R, Geslin P L, Bingen E H. In vitro killing activities of antibiotics at clinically achievable concentrations in cerebrospinal fluid against penicillin-resistant Streptococcus pneumoniae isolated from children with meningitis. Antimicrob Agents Chemother. 1994;38:2655–2659. doi: 10.1128/aac.38.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 7.Geslin P. Rapport d’activité année 1996. Créteil, France: Centre National de Référence du Pneumocoque; 1996. [Google Scholar]
- 8.Guillemot D, Maison P, Carbon C, Balkau B, Vauzelle-Kervroëdan F, Sermet C, Bouvenot G, Eschwège E. Trends in antimicrobial drug use in the community—France, 1981–1992. J Infect Dis. 1998;177:492–497. doi: 10.1086/517384. [DOI] [PubMed] [Google Scholar]
- 9.Hass D W, Stratton C W, Griffin J P, Weeks L, Alls S C. Diminished activity of ceftizoxime in comparison to cefotaxime and ceftriaxone against Streptococcus pneumoniae. Clin Infect Dis. 1995;20:671–676. doi: 10.1093/clinids/20.3.671. [DOI] [PubMed] [Google Scholar]
- 10.Jackson M A, Mithal Y, Robins-Browne R M, Gaspar M N, Koornhof H J. Relatively penicillin-resistant pneumococcal infections in pediatric patients. Pediatr Infect Dis. 1984;3:129–132. doi: 10.1097/00006454-198403000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Johnson A P, Livermore D M, Woodford N, Quoraishi A, Freeman R. High level β-lactam resistance in strains of Streptococcus pneumoniae isolated in the UK. J Antimicrob Chemother. 1998;42:115–116. doi: 10.1093/jac/42.1.115. [DOI] [PubMed] [Google Scholar]
- 12.Klugman K P, Friedland I R. Antibiotic-resistant pneumococci in pediatric disease. Microb Drug Resist. 1995;1:5–8. doi: 10.1089/mdr.1995.1.5. [DOI] [PubMed] [Google Scholar]
- 13.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 15.Munoz R, Downson C G, Daniels M, Coffey T J, Martin C, Hackenbeck R, Spratt B G. Genetic of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2477. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial suceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1994. [Google Scholar]
- 17.Pankuch G A, Jacobs M R, Appelbaum P C. Comparative activity of ampicillin, amoxycillin, amoxycillin/clavulanate and cefotaxime against 189 penicillin-susceptible and -resistant pneumococci. J Antimicrob Chemother. 1995;35:883–888. doi: 10.1093/jac/35.6.883. [DOI] [PubMed] [Google Scholar]
- 18.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]