Abstract
In silico molecular docking studies, in vitro toxicity and in silico predictions on the biological activity profile, pharmacokinetic properties, drug–likeness, ADMET (absorption, distribution, metabolism, excretion, and toxicity) physicochemical pharmacokinetic data, and target proteins and toxicity predictions were performed on six copper(II) complexes with the non–steroidal anti–inflammatory drugs ibuprofen, loxoprofen, fenoprofen and clonixin as ligands, in order to investigate the ability of these complexes to interact with the key therapeutic target proteins of SARS–CoV–2 (Severe Acute Respiratory Syndrome Coronavirus 2) 3C–like cysteine main protease (3CLpro/Mpro), viral papain–like protease (PLpro), RNA–dependent RNA polymerase (RdRp), and non–structural proteins (Nsps) Nsp16–Nsp10 2′–O–methyltransferase complex, and their capacity to act as antiviral agents, contributing thus to understanding the role they can play in the context of coronavirus 2019 (COVID–19) pandemic. Cytotoxic activity against five human cancer and normal cell lines were also evaluated.
Keywords: SARS–CoV–2 target proteins, 3C–like cysteine main protease, Papain–like protease, RNA–dependent RNA polymerase, Nsp16–Nsp10 2′–O–methyltransferase complex, in silico predictive tools
Abbreviations: 2′OMTase, 2′–O–methyltransferase; 3CLpro/Mpro, 3C–like cysteine main protease; A549, lung adenocarcinoma; ADMET, absorption, distribution, metabolism, excretion, and toxicity; BBB, blood–brain barrier; BL2, Blocking loop 2; CNR1, cannabinoid receptor 1; CNS, central nervous system; COVID–19, coronavirus disease 2019; DDI, drug–drug interactions; DPP4, Dipeptidyl Peptidase 4; E, envelope glycoprotein; FDA, U.S. Food and Drug Administration; GPCR, G–protein–coupled receptors ligand; Hb, hydrogen bond; Hclon, clonixin; Hfeno, fenoprofen; HIA, human intestinal absorption; Hibu, ibuprofen; Himi, imidazole; Hloxo, loxoprofen; Hph, hydrophobic; M, membrane glycoprotein; MCF7, breast adenocarcinoma; MD, Molecular Dynamic; MDCK, Madin–Darby Canine Kidney; MERS–CoV, Middle East Respiratory Syndrome coronavirus; Mpro, main protease; MRC5, normal human lung; N, nucleocapsid; neoc, neocuproine, 2,9–dimethyl–1,10–phenanthroline; NiRAN, nidovirus–specific RdRp–associated nucleotidyltransferase; NSAID(s), non–steroidal anti–inflammatory drug(s); Nsps, non–structural proteins; OVCAR–3, human ovarian cancer; P, polar; PASS, Prediction of Activity Spectra for Substances; PC–3, prostate adenocarcinoma; PDB, Protein Data Bank; PLpro, PLpro, PLpro, papain–like protease; py, pyridine; PPB, plasma protein binding; RdRp, RNA–dependent RNA polymerase; RMSF, root–mean square fluctuation; Ro5, Lipinski's Rule of Five; SAH, S–adenosylhomocysteine; SAM, S–adenosyl methionine; SARS–CoV–2, Severe acute respiratory syndrome coronavirus 2; TGI, total growth inhibition; TPSA, Topological Polar Surface Area; U5G, Boceprevil; WDI, World Drug Index; WHO, World Health organization.
Graphical abstract
Copper(II) complexes with non-steroidal anti-inflammatory drugs as ligands may act as antiviral agents against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS–CoV–2), interfering with 3C–like cysteine main protease, papain-like protease, RNA–dependent RNA–polymerase, and non–structural proteins (Nsps) Nsp16–Nsp10 2′–O–methyltransferase, preventing in its replication.
1. Introduction
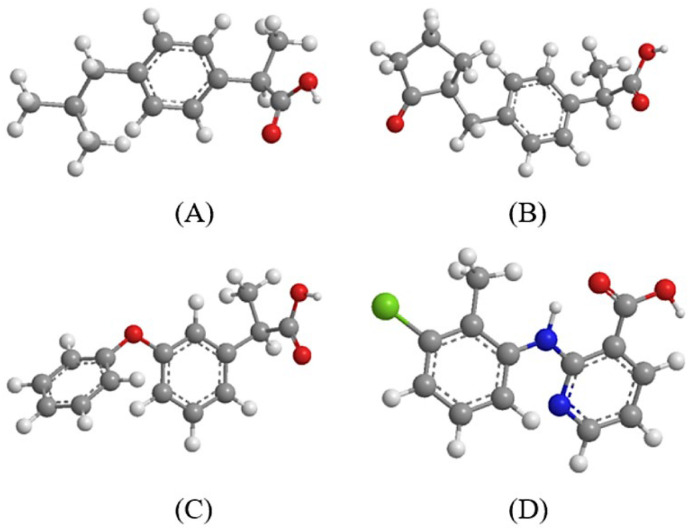
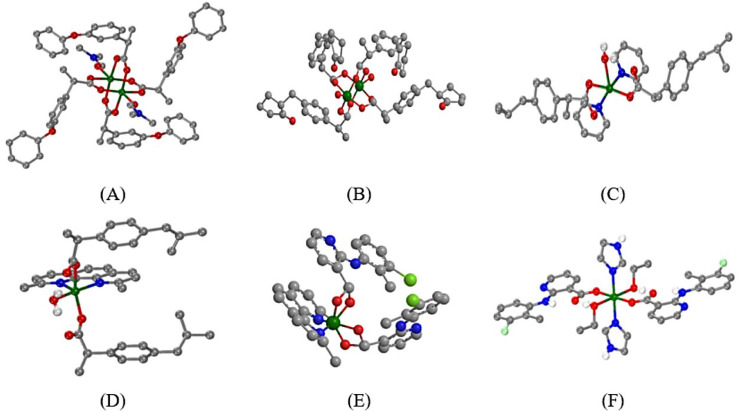
The synthesis, characterization and biological activity of six copper(II) complexes with the non–steroidal anti–inflammatory drugs (NSAIDs) ibuprofen (Hibu), loxoprofen (Hloxo), fenoprofen (Hfeno), and clonixin (Hclon) as ligands (Fig. 1 ) in the absence or presence of nitrogen–donors 2,9–dimethyl–1,10–phenanthroline (neocuproine, neoc), imidazole (Himi) or pyridine (py) as co–ligands has been recently reported. In this recent report, six Cu(II)–NSAID complexes, namely [Cu2(feno–O,O′)4(DMF)2] (1), [Cu2(loxo–O,O′)4(H2O)2] (2), [Cu(ibu–O)2(py)2(Η2Ο)] (3), [Cu(ibu–O)2(neoc)(H2Ο)] (4), [Cu(clon–O,O′)2(neoc)] (5) and [Cu(clon–O)2(Himi)2(EtOH)2] (6) (molecular structures depicted in Fig. 2 ), were characterized by IR and UV–vis spectroscopy, and by single–crystal X–ray crystallography. In addition, the interaction of the complexes with calf–thymus DNA and serum albumins as well as their free radical scavenging activity were examined by in vitro experiments and in silico calculations [1].
Fig. 1.
(A) ibuprofen, (B) loxoprofen, (C) fenoprofen, (D) clonixin. (C: grey, O: red, H: white, N: blue, Cl: light green.)
Fig. 2.
(A) – (F) Structures of complexes 1–6, respectively (adapted from [1]). (C: grey, O: red, H: white, N: blue, Cu: dark green, Cl: light green.)
In continuation of this research, in silico studies were carried out in an attempt to explore the ability of the complexes to act as potent SARS–CoV–2 (Severe Acute Respiratory Syndrome Coronavirus 2) antiviral compounds, and to elucidate a possible mechanism of action. SARS–CoV–2 contains a single–stranded positive–RNA genome of approximately 29,800 bases which encodes four structural proteins – spike (S) glycoprotein, small envelope (E) glycoprotein, membrane (M) glycoprotein, and nucleocapsid (N) – and sixteen non–structural proteins (Nsp1–Nsp16) that play important roles in virus transcription and replication [2].
The spike protein is the focus of ongoing efforts to develop a vaccine against SARS–CoV–2 [3]. Focusing on the binding of a drug to only a single target may restrict the therapeutic prospect of a potential new drug. It may be more effective to consider attacking several points in the virus, which are responsible for its life cycle or which affect the host's immune response of the host.
Among the non–structural proteins (Nsps) in SARS–CoV–2, Mpro/Nsp5 [4], PLpro/Nsp3 [5], RdRp/Nsp12 [6], and helicase/NTPase (Nsp13) [7] are considered essential to the viral cycle [8]. Therefore, these have been targeted for docking and/or for the development of small–molecule inhibitors due to their clear biological functions [9]. In this context, complexes 1–6 were used for evaluating potential antiviral activity against SARS–CoV–2 via interaction with significant coronavirus protein targets 3C–like cysteine main protease (3CLpro/Mpro), viral papain–like protease (PLpro), RNA–dependent RNA polymerase (RdRp), and Nsp16–Nsp10 2′–O–methyltransferase complex.
Some metal–based drugs have shown remarkable effects in combating virus replication and inhibiting a few key viral enzymes in the SARS–CoV–2 virus [10,11]. In a recent study supported by protein–protein interactions between SARS–CoV–2 and human proteins, a large number of metalloproteins have been identified as metallo–binding interactors of SARS–CoV–2 proteins [12]. The majority of these metalloproteins interact with the accessory protein SARS–CoV–2 orf8, the structural membrane protein SARS–CoV–2 M and the non–structural proteins SARS–CoV–2 Nsp12 and Nsp13. In a review article, Cirri et al [10] explored potential metallo–therapeutics for COVID–19 concluding that metallodrugs might offer an excellent opportunity to develop new antiviral drugs against COVID–19. Metal complexes as potential antiviral agents against SARS–CoV–2 target proteins have been discussed in a minireview by Karges and Cohen in which they proposed mechanisms of action [13]. A series of gold complexes functionalized with N–heterocyclic carbene or alkynyl ligands as well as Auranofin, an antirheumatic agent, have been studied as inhibitors of ACE2 [14]. These metal complexes are also known inhibitors of the PLpro of SARS–CoV–2, presenting a possible multimodal mechanism of action. Another recent study evaluated, through molecular docking, the interactions of 4′–acetamidechalcones with enzymatic and structural targets of SARS–CoV–2 and with the host's human angiotensin–converting enzyme 2 (hACE2) such as the spike protein, the main protease (Mpro), and the Nsp16–Nsp10 heterodimer methyltransferase [15]. Furthermore, computational studies of selected transition metal complexes as potential drug candidates against the SARS–CoV–2 virus [16,17] reveal them to be potential inhibitors of spike protein, main protease (Mpro), and RNA–dependent RNA polymerase (RdRp) enzymes. Another category of metal complexes, Re(I) tricarbonyl complexes, were also found to demonstrate coordinate covalent enzymatic inhibition of 3CLpro/Mpro [18]. In addition, a new class of noble metal complexes has been evaluated as potential SARS–CoV–2 antiviral agents [19]. These findings demonstrate that two pentamethylcyclopentadienyl (Cp*) rhodium piano–stool complexes have direct virucidal activity against SARS–CoV–2. A molecular docking study on binuclear Co(II), Ni(II), Cu(II), and Zn(II) diimine Schiff base complexes targeting SARS–CoV–2 3CLpro/Mpro revealed that the Ni(II) complex showed the best binding capacity with Mpro [20].
On the contrary, a study of Grifagni et al [21] with a zinc ion bound to coronavirus Mpro rather than as part of a coordination compound investigates the zinc binding affinity and its ability to inhibit the SARS–CoV–2 Mpro. The study shows that zinc ion is coordinated to catalytic dyad Cys145/His41 and two water molecules completing a tetrahedral geometry (SARS–CoV–2 Mpro structures deposited at the Protein Data Bank (PDB) with ID codes: 7NXH and 7NWX, respectively). The zinc binding site has been also compared with that of other non–viral cysteine–proteases reported to bind zinc, such as cathepsin S both in the metal–bound and metal–free state, reporting similar to those of SARS–CoV–2 Mpro, structural features. Interestingly, cathepsin S is predicted in our Target Prediction approach adopted through Swiss Target Prediction method to be bound with high possibility to complex 5. The same is valid for cathepsins D and E as target proteins predicted to be bound to complex 1. The affinity of the protein for the zinc ion, although high, appears to be not sufficient to inactivate SARS–CoV–2 Mpro, as almost all the intracellular zinc ion is bound to other proteins with similar or higher affinity. A more proper approach in the design of potent and more selective inhibitors of SARS–CoV–2 Mpro should be the development of metallodrugs with metal ions, coordinated to suitable ligands capable of interacting with additional sites on the protein surface. This may provide a significant increase in binding affinity. To that end, in the present study we are exploring the potential antiviral activity of copper(II) complexes in the context of binding to NSPs of SARS–CoV–2 with the employment of in silico molecular docking calculations.
Additionally, in vitro toxicity and a variety of computational tools were employed to predict the complete biological activity profile of complexes 1–6. Predictive tools include general pharmacological potential, prediction of target proteins affected by the compounds, toxicity predictions, and calculation of ADMET (absorption, distribution, metabolism, excretion, and cytotoxicity) parameters, pharmacokinetic properties, drug–like nature and medicinal chemistry friendliness of the compounds.
2. Experimental
2.1. In silico computational methods
2.1.1. In silico molecular docking calculations
A series of in silico studies were employed in order to predict the potential antiviral activity of the complexes. We adopted molecular docking calculations on various target proteins related to SARS–CoV–2 virus, including proteins and enzymes involved in various stages of viral infection of the host cell, playing a functional role in the viral life cycle and its replication and transcription. Details concerning the in silico molecular docking procedures are given in the Supporting information file (Section S1).
2.1.2. Molecular pharmacokinetic properties, drug–likeness and toxicity predictions
A chemo–informatics analysis of complexes 1–6 was carried out in order to calculate some physicochemical parameters, such as octanol/water partition coefficient (LogP) (as a measure of their lipophilicity), Topological Polar Surface Area (TPSA), molecular volume, hydrogen bonding capacity, and molecular weight (MW), which play a vital role in generation and determination of the bioactivity and drug–likeness of the complexes. Additionally, pharmacokinetic properties and toxicity prediction studies were adopted. Details concerning the experimental procedures used are found in the Supplementary data file (Section S3 and S4).
2.2. In vitro toxicity studies
The in vitro toxicity activity of complex 3 was evaluated against four well–established human cancer cell lines: ovarian cancer (OVCAR–3) cells, breast adenocarcinoma (MCF7) cells, prostate adenocarcinoma (PC–3) cells, lung adenocarcinoma (A549) cells, and normal human lung (MRC5) cells and cytostatic (growth inhibition: IC50, TGI) and cytotoxic/cytocidal (IC50) activity were determined. Detailed procedures regarding the in vitro study of the biological activity of the complex are given in the Supporting Information (Section S2).
3. Results and discussion
3.1. In silico molecular docking studies on SARS–CoV–2 target proteins
In silico molecular docking calculations were employed to evaluate the ability of complexes 1–6 and the NSAIDs Hfeno, Hloxo, Hibu, and Hclon to bind to a number of target proteins related to SARS–CoV–2 viral infection including: (i) 3C–like cysteine main protease (3CLpro/Mpro) (PDB IDs: 6LU7, 7C6S, and 6XHM), (ii) papain–like protease (PLpro) (PDB ID: 6W9C), (iii) RNA–dependent RNA polymerase (RdRp) (PDB ID: 6M71), and (iv) Nsp16–Nsp10 2′–O–methyltransferase complex (PDB ID: 6W4H). The best–scored pose of docked compounds in each target macromolecule was selected for evaluation of binding interactions. Binding free energy for each pose was also determined and the pose with the lowest free binding energy was selected for further visualization studies. The computed binding energies for the best docking poses of the compounds on these target proteins are shown in Table S1.
3.1.1. 3C–like cysteine main protease
SARS–CoV–2, similar to many other single–stranded RNA viruses, employs a chymotrypsin–like protease (3CL main protease, or 3CLpro/Mpro) to enable the production of non–structural proteins essential for viral replication [22]. 3CLpro/Mpro(Nsp5) has a pivotal role in the proteolytic activation of the SARS–CoV–2 spike at S1/S2 site. Mpro by autocleavage between Nsp4 and Nsp6 [23], removes the N– and C–terminal regions in order to produce mature enzymes. Mpro functions as a cysteine protease cleaving polyproteins at 11 positions to release functional units Nsp6–Nsp16 for virus replication and packaging within the host cells [24].
Detailed investigation of the structure and catalytic mechanism of Mpro has revealed it as an attractive target for anti–coronavirus drug development since it plays a key–role in viral transcription and replication, and no human proteases are known with the same substrate specificity [25], thus making it an attractive and promising therapeutic drug target for SARS–CoV–2 and discovery of novel antiviral protease inhibitor drugs [26]. If an individual is already infected with the virus, then a drug that binds to the protease and stops it from creating mature viral proteins could be administered and finally viral replication could be slowed. At the time this work was completed, no protease inhibitors targeting SARS–CoV 3CLpro/Mpro had been approved by U.S. Food and Drug Administration (FDA), despite significant research effort during the past fifteen years [27].
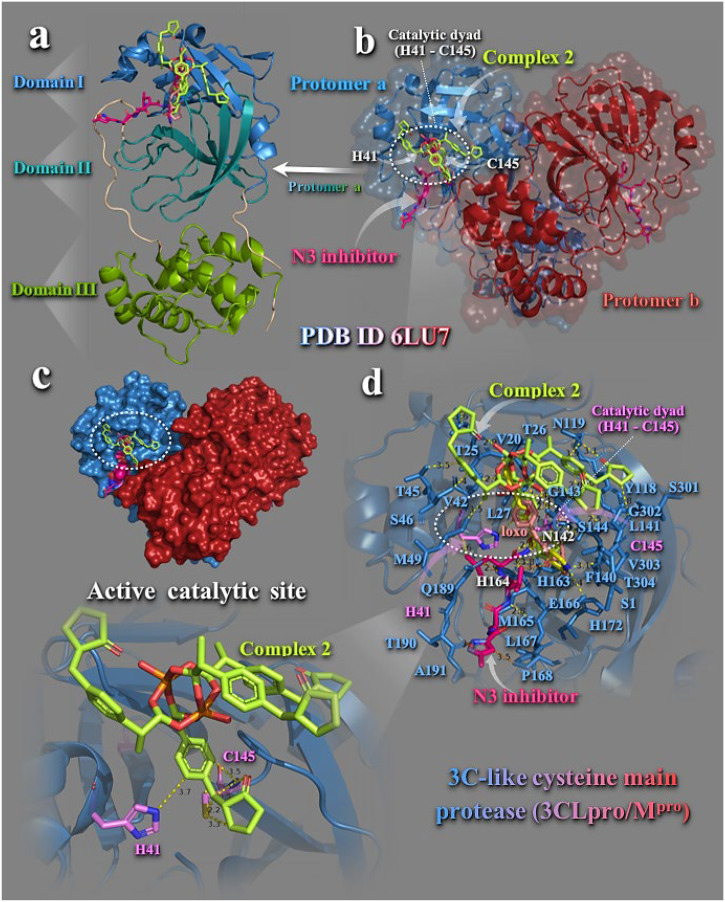
Structural and functional role of 3CLpro/Mpro is reported in the Supporting Information (Section S1.1). Mpro, unlike other chymotrypsin–like enzymes and many Ser or Cys hydrolases, has a Cys-His catalytic dyad instead of a canonical Ser(Cys)-His-Asp(Glu) triad, with the substrate–binding site located in a cleft between domains I and II [28], and domain III is responsible for the enzyme dimerization, enabling the active form of the macromolecule [29]. In the active catalytic site, which constitutes another binding site of the protein, the important residues taking place in the mechanism of action are cysteine C145 and histidine H41 (this is why Mpro is a cystine protease). Nucleophilic C145 attacks the amide bond of a peptide and H41 electron rich nitrogen atom grabs the cysteine proton leaving it with enough negative charge to attack the peptide bond [30]. This type of enzyme is excellent target for irreversible inhibition.
Binding energies for the best–docking pose of complexes 1–6 and Hfeno, Hloxo, Hibu, and Hclon on 3CLpro/Mpro (PBD entry code 6LU7) are cited in Table S1. Best–bound compound (with higher binding capacity and affinity for the protein) was revealed to be complex 2. Complexes 1, 2, 4, and 5 were found to possess better binding affinity than their corresponding NSAIDs, except 3 and 6, for which Hibu and Hclon, respectively, exhibited better binding capacity. The docking pose of best–bound complex 2 (with the lowest binding energy) is illustrated in Fig. 3 , along with its NSAID Hloxo on the crystal structure of the enzyme, superimposed with its inhibitor N3. Complex 2 was shown to bind at the periphery of the substrate-binding region which is located between domains I and II and in the binding site of N3 inhibitor.
Fig. 3.
Docking pose orientation of best–bound complex 2 on the symmetric dimer unit of SARS-CoV-2 3CLpro/Mpro (PBD: 6LU7). Target enzyme illustrating its two subunits (protomers a and b) is depicted either in cartoon–colored by chain (protomers a and b in sky blue and firebrick red, respectively) with additional depiction of a semitransparent surface for the unit colored according to protomer colors (b), or in opaque surface of the homodimer of Mpro colored by chain according to b (c). Structural analysis, along with the binding pose of complex 2 superimposed with the co–crystallized protease inhibitor N3 in the protomer a of the enzyme structure is depicted in (a) illustrating the domains I (residues 8–101), II (residues 102–184), and III (residues 201–303) of the asymmetric unit in cartoon representation colored in sky blue, deep teal, and split pea green, respectively, while loop regions 1–7, 185–200, and 305–306 are shown in wheat color. A close–up view of the binding interaction architecture of 2 in the catalytic active site of the enzyme (protomer a in sky blue cartoon with additional depiction of selected contacting residues flanking the catalytic active cavity rendered in stick model and colored according to cartoon) is shown in (d). The catalytic dyad (H41–C145) is also indicated in dashed white ovals (b, d).Catalytic residues H41 and C145 are depicted in stick model and colored according to atom type in violet C atoms. Complex 2, its NSAID Hloxo, and N3 inhibitor are rendered in stick model and colored according to atom type in limon, salmon, and hot pink C atoms, respectively. Binding contacts of 2 and N3 are shown as dotted yellow and orange lines, respectively. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu: orange and O: red. The final structure was ray–traced with depth cue in the ray–tracing rendering of the cartoon and illustrated with the aid of PyMol Molecular Graphics Systems. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Complex 2 is anchored in a binding cavity of Domain I of protomer a, at the ridge of the antiparallel β–barrel structure (Fig. 3a). Complex 2 adopts a binding position in a way that one of the four loxo wings of 2 is inserted into the hydrophobic S2 subunit consisting of the side chains of H41, M49, F140, L141, N142, G143, H163, T24, T25, M49, T45, Y54, N119, Y118, and M165 residues, at the same place occupied by the Ν3 inhibitor backbone build structure. As it is illustrated in Fig. 3d, complex 2 is stabilized in its binding pocket with one loxo arm placed between histidine H41 and cysteine C145 residues constituting the catalytic dyad (Fig. 3b, d). Binding contacts of 2 with the critical to catalytic activity residues H41 and C145 involve π–π stacking interactions of the aromatic imidazole side chain of histidine with the phenyl ring of loxo moiety of 2 and π–alkyl hydrophobic type contact of C145/CB with the aromatic phe ring of loxo moiety (3.6 Å). Additional binding contacts of 2 are reported in the Supporting Information (Section S1.1). Common contacts of 2 with the inhibitor N3 revealed to be H41, C145, N142, T24, H163, T26, and L141 residues. Residues S46 and M49 of the small helix (residues 45–50) are shown to stabilize one of complex's 2 loxo wings inside the binding pocket. This region of the enzyme has also been identified by molecular dynamics (MD) simulation, as one of the most malleable sites (showing the largest root–mean square fluctuations (RMSF)), possibly able to accommodate various chemical groups [31]. Furthermore, E166 and P168 residues of β–hairpin loop (residues 166–170) are also used for the stabilization of 2 and N3 in the binding cavity.
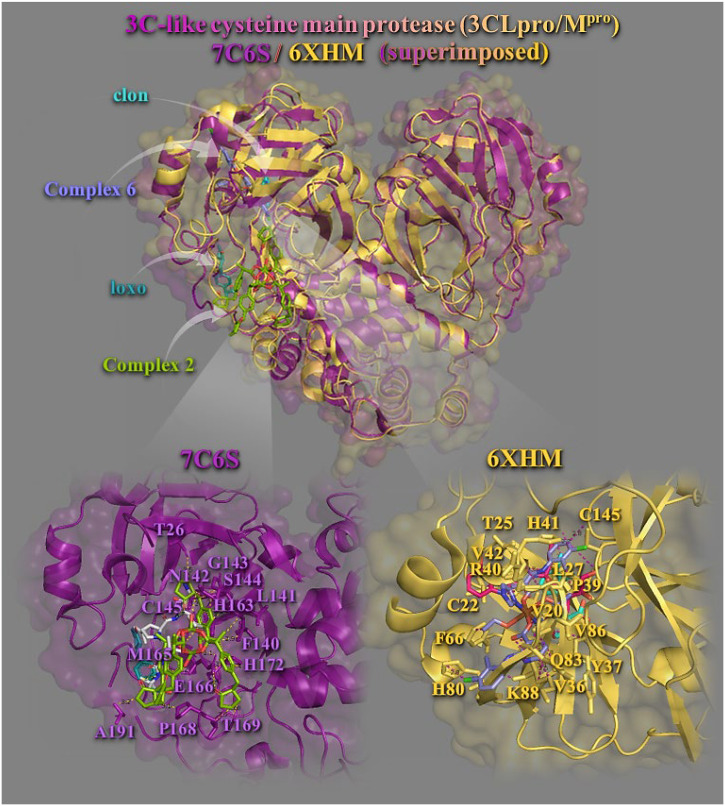
Binding energies for the best docking pose of 1–6 and the NSAIDs on 3CLpro/Mpro (PBD entry codes: 7C6S and 6XHM) are given in Table S1. Best–bound complexes on 7C6S and 6XHM 3CLpro/Mpro proteases revealed to be 2 (for 7C6S) and 6 (for 6XHM). All complexes (with the exception of 2 in 6XHM protein) were found to possess better binding affinity than the corresponding NSAIDs.
Best binding pose of each complex is depicted in Fig. 4 . Both complexes 2 and 6 are docked in a binding pocket at the same place occupied by the co–crystallized inhibitors (boceprevil, U5G) in 7C6S and V2M in 6XHM, respectively (Fig. 4, upper panel). Stabilization of complex 2 may be attributed to π–π stacking, hydrogen bond (Hb), hydrophobic (Hph), π–alkyl, polar (P), π–polar and π–anion electrostatic contacts involving the following residues: glutamate E166 (Hb, P, and π–anion), histidine H163 (π–π), asparagine N142 (Hb), leucine L141 (π–polar), glycine G143 (π–polar), serine S144 (π–polar and P), alanine A191 (P), threonine T26 (P), cysteine C145 (P), phenylalanine F140 (P), T169 (P and Hph), S1 (π–alkyl), H172 (π–alkyl), methionine M165 (π–alkyl), and proline P168 (Hph) (Fig. 4, lower panel). Most interacting residues were found to be common with those of loxo and boceprevil inhibitor. The binding of complex 6 in its pocket was achieved with the formation of hydrogen bond (Hb), hydrophobic (Hph), π–alkyl, polar (P), π–polar and π–cation electrostatic contacts involving the following residues: glutamine Q83 (Hb), lysine K88 (Hb), H41 (Hb), valine V42 (Hb), tyrosine Y37 (Hb), H80 (P), V36 (π–polar), C145 (π–polar), L87 (P), T25 (π–polar), K88 (π–cation and π–alkyl), L27 (π–alkyl), V86 (π–alkyl), C22 (π–alkyl), V20 (π–alkyl), arginine R40 (π–alkyl), F66 (π–alkyl), and P39 (Hph). Most interacting residues found to be common with those of clon and V2M inhibitor. Since complex 6 bears an imidazole ligand, it is interesting to notice the therapeutic potentials of a new series of imidazole derivatives against Mpro and RdRp target proteins of SARS–CoV–2 [32]. The compounds displayed various levels of binding affinities for the SARS–CoV–2 drug targets. Bis–imidazole C2 scored highest against all the targets, with its aromatic rings including the two imidazole groups contributing to the binding.
Fig. 4.
Molecular docking of complexes 2 and 6, and their NSAIDs Hloxo and Hclon, as well as the protease inhibitors boceprevil (U5G) of 7C6S and V2M of 6XHM, on two SARS-COV-2 3CLpro/Mpro enzymes. The final alignment of structures derived by the superimposition of 7C6S and 6XHM proteases. The structures were aligned by PyMol Molecular Graphics System. The overlay of the two resolved structures of 3CLpro/Mpro 7C6S and 6XHM are illustrated in cartoon mode and colored in deep purple and yellow–orange, respectively, with additional depiction of a semitransparent surface colored according to cartoon representation. All compounds are rendered in stick model and colored according to atom type in split pea green (2), slate blue (6), deep teal (Hloxo), cyan (Hclon), white (boceprevil), and hot pink (V2M), C atoms. A close–up view of the binding interaction architecture of 2 and 6 in their binding pockets superimposed with Hloxo and Hclon, respectively, with additional depiction of selected contacting residues rendered in stick model and colored according to cartoon, are shown at the lower panel. Binding contacts are shown as dotted yellow and deep purple lines. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu: split pea green (in 2), brown (in 6), Cl: green, O: red, and N: blue. The final structure was ray–traced with depth cue in the ray–tracing rendering of the cartoon and illustrated with the aid of PyMol Molecular Graphics Systems. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Although copper(II) complexes interact potently with Mpro specifically in close proximity to the critical to catalytic activity residues H41 and C145, yet they do not make covalent contacts with them in order to possess irreversible inhibition, as, for example, the antineoplastic drug carmofur does [33]. For inhibitors, the covalent linkage is critical to maintain its antiviral activity [34]. Furthermore, no copper(II) complexes were able to inhibit the dimerization of 3CLpro/Mpro influencing its functional protease activity (are not docked at the responsible for the enzyme dimerization Domain III).
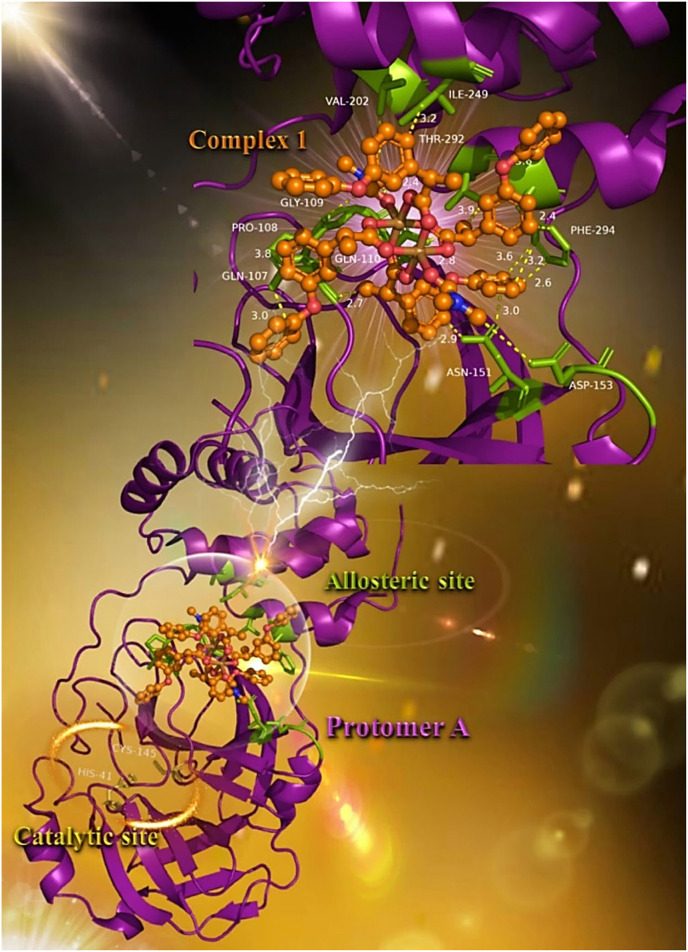
In a recent study of Günther et al [35] performing a large–scale X–ray crystallographic screen of SARS–CoV–2 main protease (3CLpro/Mpro) against two drug–repurposing libraries, beyond the catalytic binding site harboring the catalytic dyad comprised of Cys (C145) and His (H41) in its active site, additionally, two allosteric binding sites were identified, representing attractive targets for drug development against SARS–CoV–2. One of the allosteric inhibitors, AT7519, revealed to bind to an allosteric binding site formed by a deep groove between the catalytic domains and the dimerization domain of the enzyme. In silico docking experiments indicated the binding pocket comprising residues Gln (Q107), Pro (P108), Gly (G109), Gln (Q110), Asn (N151), Asp (D153), Val (V202), Ile (I249), Phe (F294), Thr (T292), Arg (R298), and Tyr (Y154).
It is of interest to notice that one of the studied compounds, complex 1, seems to be anchored at the same place occupied by AT7519, in a deep cleft between the catalytic and dimerization domain of the enzyme (Fig. 5 ). The docking procedure indicates the stabilization of complex 1 in this allosteric binding site with almost all of the binding contacts of AT7519, with the exception of Arg (R298) and Tyr (Y154). Binding interactions of 1 with residues of the allosteric site are reported in the Supporting Information (Section S1.1).
Fig. 5.
Docking pose orientation of best–bound complex 1 on protomer a of SARS-CoV-2 3CLpro/Mpro (PBD: 6LU7). Target enzyme is depicted in cartoon colored in deep purple. A close–up view of the binding interaction architecture of 1 in the allosteric binding site of the enzyme in a deep cleft between the catalytic and dimerization domain of the enzyme with additional depiction of selected contacting residues rendered in stick model and colored in split pea green is shown in the upper panel of the molecular structure. Binding residues of complex 1 in the allosteric binding pocket are Gln (Q107), Pro (P108), Gly (G109), Gln (Q110), Asn (N151), Asp (D153), Val (V202), Ile (I249), Phe (F294), and Thr (T292). The two critical residues of the catalytic site (H41–C145) are also indicated in yellow–orange sticks (lower panel). Complex 1 is rendered in ball–and–stick model and colored according to atom type in orange. Binding contacts are shown as dotted yellow lines. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu: brown, O: red, and N: blue. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics Systems. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Oral antiviral candidate drug Paxlovid™ (a combination of Pfizer's investigational antiviral PF–07321332 and a low dose of ritonavir, an antiretroviral medication traditionally used to treat HIV by inhibiting cytochrome P450 and slowing the metabolism of protease inhibitors), a specifically SARS–CoV–2 3CL protease inhibitor designed by Pfizer, was found to reduce the risk of hospitalization or death by 89% compared to placebo in non–hospitalized high–risk adults with COVID–19. Paxlovid is going to be approved by the FDA and is currently under phase II/III EPIC–HR (Evaluation of Protease Inhibition for COVID–19 in High–Risk Patients), II/III EPIC–SR (Evaluation of Protease Inhibition for COVID–19 in Standard–Risk Patients), and EPIC–PEP (Evaluation of Protease Inhibition for COVID–19 in Post–Exposure Prophylaxis) studies. The trial will finish in early 2022. PF–00835231 has been found to bind covalently to the protease's active site, inactivating the enzyme. The crystal structure of Mpro in complex with PF–073213327SI9 at 1.6 Å resolution (PDB ID: 7VH8) has recently resolved and deposited (2021–09–21) [36]. Additionally, crystal structure with PDB ID: 7SI9, has also been resolved and deposited (2021−10−12). The binding pocket of PF–07321332 is found in the active site of Mpro, at the same place where complex 2 is also bound, and is stabilized forming a covalent bond to C145. It is intriguing the fact that almost all the binding residues of PF–073213327SI9 are common with those of complex 2 (catalytic dyad H41/C145, M49, F140, L141, N142, G143, S144, H163, M165, E166, L167, P168, H172, Q189, and T190). New binding residues of PF–073213327SI9 revealed to be Y54, D187, R188, and Q192.
Other SARS–CoV–2 Mpro–targeting compounds like Pfizer's compound, are also in development such as PBI–0451 (by Pardes, oral in phase I), S–217622 (by Shionogi, oral in phase III), EDP–235 (by Entana, oral in phase I planned), and 13b–K (by University of Lubeck, inhaled, preclinical), most of which mimic the peptides cleaved by Mpro, covalently binding to cysteine residues in the active site to inhibit the enzyme.
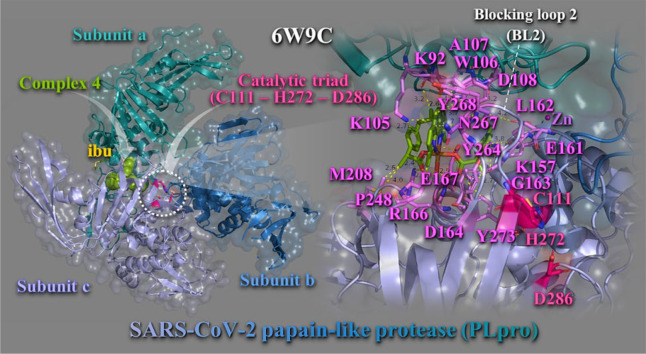
3.1.2. Papain–like protease (PLpro)
Another attractive antiviral target for inhibiting SARS–CoV–2 virus is papain–like protease (PLpro) non–structural protein 3 (Nsp3), an essential component of the replicase–transcriptase complex which cleaves pp1a and pp1ab viral polypeptides [37]. Inhibition of this viral protease results in impaired viral replication in host cells. Along with protease activity, the PLpro possesses deubiquitinating activity, which is important in immune regulation. PLpro is a cysteine protease with a classical Cys-His-Asp catalytic triad (Cys112, His273, Asp287), which cleaves the viral polyprotein, releasing Nsp1, Nsp2, and Nsp3 [38].
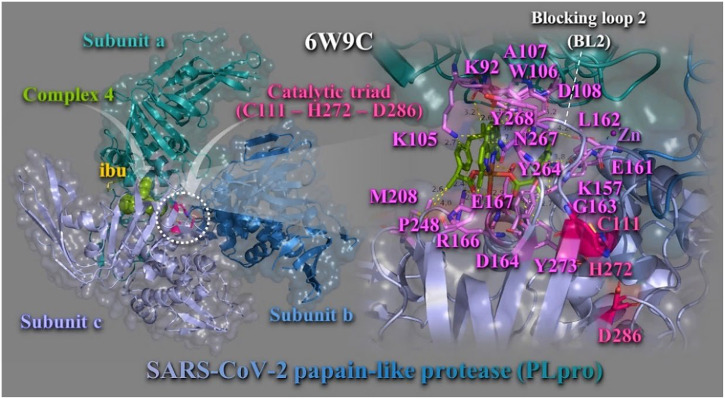
Binding energies for the best–docking pose of complexes 1–6 and the NSAIDs on SARS-CoV-2 PLpro (PBD ascension number: 6W9C) are given in Table S1. Best bound compound (with higher binding capacity and affinity for the protein) was revealed to be complex 4. Complexes 3 and 4 were found to possess better binding affinity than Hibu and 2 better than Hloxo, while 1 and 5, 6 were found to exhibit lower binding capacity than Hfeno and Hclon, respectively. The binding of best docking scored pose of complex 4 (with the lowest binding energy), superimposed with its NSAID Hibu, is illustrated in Fig. 6 . Complex 4 is shown to be anchored in the active site of the enzyme that contains a classic catalytic triad composed of Cys-His-Asp, where Cys acts as nucleophile (Fig. 6, left panel). Complex 4 is stabilized adjacent to Blocking loop 2 (BL2) in the Palm Domain and in close proximity to catalytic triad C111–H272–D286. The molecular docking results demonstrated that the binding residues responsible for the stabilization of complex 4 in the binding site include lysine K105 (π–cation), tryptophane W106 (π–π T–shape with neoc moiety aromatic rings, π–alkyl, and π–polar), tyrosine Y264 (π–polar), asparagine N267 (π–polar), Y273 (π–polar), Y268 (π–π stacking with neoc moiety rings), arginine R166 (hydrogen bond), lysine K157 (polar), alanine A107 (π–alkyl), glutamate E161 (polar), aspartate D164 (hydrogen bond), glycine G163 (π–polar), proline P248 (hydrophobic), glutamate Q167 (π–polar), D108 (polar), leucine L162 (π–polar), K92 (polar), and methionine M208 (polar and hydrophobic) (Fig. 6, right panel). These binding residues are part of subunits A and C. Residues of subunit B do not make any contact with 4. Residues denoted in italics are located in palm domain, underscored in finger domain, and the rest in the thumb domain of the enzyme. N267 and Y268 residues are part of the BL2 region of the PLpro. Common residues responsible also for the binding of Hibu stabilization in the structure are Y264, R166, G163, and L162.
Fig. 6.
Docking pose orientation of best bound complex 4 and Hibu (rendered in sphere and stick model and colored by atom type in split pea green and yellow–orange C atoms, respectively) on the crystal structure of SARS–CoV–2 PLpro (PBD: 6W9C). Target protein is illustrated in cartoon colored by deep teal (subunit A), sky blue (subunit B), and light blue (subunit C), with additional depiction of a semitransparent surface colored according to cartoon representation and indication of the catalytic triad in dashed white oval. A close–up view of the binding interaction architecture of 4 in its binding pocket superimposed with Hibu and additional depiction of selected contacting residues rendered in stick model and colored according to atom type in violet C atoms, are shown at the right panel. Complex 4 and Hibu are found to be stabilized in close proximity to catalytic triad (C111, H272, and D286) colored by atom type in hot pink C atoms. Zn ion is shown as purple sphere. Binding contacts are shown as dotted yellow lines. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu: orange, O: red, and N: blue. The final structure was ray–traced with depth cue in the ray–tracing rendering of the cartoon and illustrated with the aid of PyMol Molecular Graphics Systems. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The binding contact residue Y268 of complex 4 was also frequently involved in intermolecular bonds with a number of dietary compounds including (−)–epigallocatechin gallate and cyanidin–3–O–glucoside. Additionally, in the 6w9c structure of SARS–CoV–2 PLpro, 3 k and GRL–0617 naphthalene–based inhibitors formed hydrogen bond and salt bridge contacts with Y264 (the first) and hydrogen bonds with G266 and N267 (the second) [39]. A series of natural phytochemical compounds such as oleanolic acid, ursolic acid, 3b–acetoxyolean–12–en–27–oic acid, and isovitexin of Vitex negundo, showed inhibitory activity against PLpro of SARS–CoV–2 [40]. The PLpro–ursolic acid complex shows that it stabilizes the protein through conventional hydrogen bonding at amino acid residues Asp108 and coheres at Ala107, Pro248, and Tyr264 with Alkyl, π–Alkyl interaction. Ligand 3b–acetoxyolean–12–en–27–oic acid forms the conventional hydrogen bond and alkyl bond with residues Gly160 and Leu162, respectively. The isovitexin stabilizes the structure through Pi–anion and amide Pi–stacked interaction with the residues Glu161 and Gly160. In another study, organoselenium compound ebselen proved to be potent PLpro inhibitor. Ebselen binding with 6W9C structure of PLpro identified critical binding interaction that are common with the binding contacts of complex 4, such as K105, W106, Y268, L289, and A288.
3.1.3. RNA–dependent RNA–polymerase (RdRp) (Nsp12–Nsp7–Nsp8 complex)
The replication of SARS–CoV–2 genome is dominated by a replication/transcription complex which contains several subunits. The complex is composed of viral Nsps and the core of the complex is the multimeric protein RNA–dependent RNA polymerase (RdRp) in Nsp12 [41]. RdRp is an essential enzyme encoded in the genomes of all RNA–containing viruses with no DNA stage, i.e. RNA viruses, including SARS–CoV–2. Inhibition of RdRp activity is blocking replication and prevents viral transcription [42]. RdRp catalyzes SARS–CoV–2 RNA replication from an RNA template and, hence, is an obvious target for antiviral drug design. The function of the Nsp12 requires accessory factors, including one Nsp7 and two Nsp8. More specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. RdRps are responsible for transcription and replication of viral RNAs [43].
RdRps can be used as drug targets for viral pathogens as their function is not necessary for eukaryotic survival. Currently, the US FDA has approved remdesivir inhibitor of SARS-CoV-2 RdRp. Remdesivir is a nucleotide analog which inhibits the function of RdRp by covalently binding to and interrupting termination of the nascent RNA through early or delayed termination or preventing further elongation of the RNA polynucleotide [42]. Structural and functional role of RdRp is reported in the Supporting Information (Section S1.2).
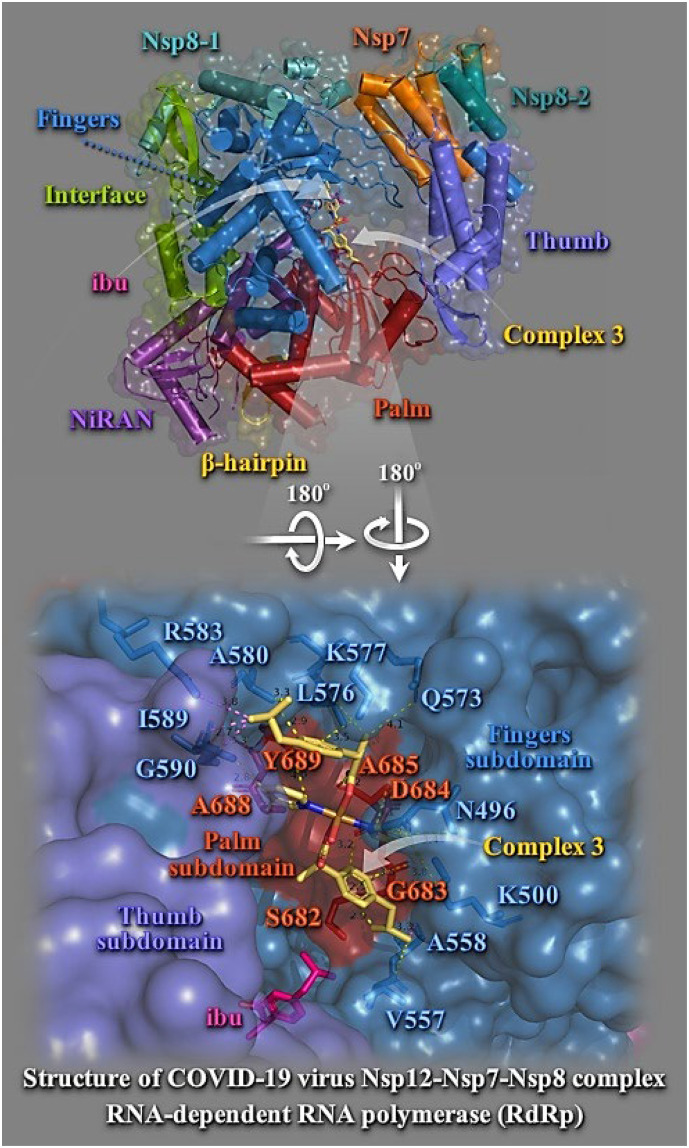
Binding energies for the best docking pose of complexes 1–6 and the NSAIDs on SARS-CoV-2 RdRp (PBD ascension number: 6M71) are summarized in Table S1. Best–bound compound (with higher binding capacity and affinity for the protein) was revealed to be complex 5. All complexes were found to possess better binding affinity than the corresponding NSAIDs. Best binding pose of each complex is depicted in Fig. 7 . Schematic diagram for the domain organization of the RdRp complex, containing Nsp12, Nsp7, and two copies of Nsp8 (Nsp8–1 and Nsp8–2), showing important domains in RdRp is illustrated in Fig. 7. The N–terminal portion of Nsp12 contains a β–hairpin (residues V31 to K68) and a unique nidovirus–specific RdRp–associated nucleotidyltransferase (NiRAN, residues V69 to R249) extension domain comprising of six helices with three stranded β sheets at the N–terminus. The β–hairpin is sandwiched by the palm subdomain in RdRp core and NiRAN forming close contacts to stabilize the overall structure [44]. The C–terminal catalytic domain of Nsp12 (residues L366 to F932) adopts a canonical cupped right–handed configuration of all viral RdRp composed of the Fingers, Palm, and Thumb subdomains, connecting to NiRAN through an interface subdomain (residues A250 to R365). The core protein looks like a cupped right hand which is further sub–divided into subdomain including Finger domain (L366 to A581 and K621–G679), Palm domain (T582 to P620 and T680 to Q815), and Thumb domain (H816 to Q932) [45]. Nevertheless, complex 5 was found to be positioned in a higher and lower binding energy pocket surrounded by NiRAN and Palm subdomains in one, and NiRAN, Fingers and Interface subdomains, in the other, are far from the active catalytic site and may exert its effects through altered interaction with other components of the replication–transcription complex or with the RNA template Nsp12 (NiRAN, interface, fingers, palm and thumb) (Fig. 7, upper panel). Complex 3, although found to be docked to the protein in higher binding energy, is positioned in a pocket formed between the loop connecting fingers domain with palm domain, in one side, and the lower sheet of the β–sheet cluster, in the other, in close proximity to remdesivir binding site, near the catalytic site. Hibu is located at this second part of the pocket. Complex 3 is stabilized at the vicinity of the active catalytic site, nevertheless not being able to make contacts with its critical residues. Binding contacts of 3 include stable interactions with the residues: K500, N496, V557, A558, Q573, L576, K577, A580, R583, I589, G590, S682, G683, D684, A685, A688, and Y689. Binding residues common with remdesivir are K500, Q573, S682, G683, D684, A685, A688, and Y689 (Fig. 7, lower panel). Complexes 6 and 1, bound to the protein via higher binding energy than 3, were found to make common contacts with remdesivir with the inclusion of D760, T680, A688, T687, and S682 residues (the first), and T680, N691, and D760 residues (the second). It is interesting that 1 is bound to the protein with D760, one of the catalytic site residues of motif C.
Fig. 7.
(Upper panel) Docking pose orientation of best bound complex 3 and Hibu (rendered in stick model and colored by atom type in yellow–orange and hot pink C atoms, respectively) on the crystal structure of SARS–CoV–2 RdRp (PBD: 6M71). Target protein is illustrated in cartoon with α–helices shown as cylinders, β–strands as arrows, and loops as strands, colored according to structure characterization of subdomain components of the RdRp complex as follows: β–hairpin, yellow–orange; NiRAN, violet purple; Interface, split pea green; Fingers, sky blue; Palm, firebrick red; Thumb, slate blue; Nsp7, orange; Nsp8–1, teal; and Nsp8–2, deep teal, with additional depiction of a semitransparent surface colored according to cartoon representation. (Lower panel) A close–up view of the binding interaction architecture of 3 in its binding pocket superimposed with Hibu illustrated as semitransparent surface with additional depiction of selected contacting residues rendered in stick model and colored according to atom type in sky blue and firebrick red corresponding to Fingers and Palm, respectively. Binding contacts are shown as dotted yellow and violet lines. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu: orange, and O: red. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics Systems.
3.1.4. 2′–O–methyltransferase (2′OMTase) (Nsp16–Nsp10 complex)
2′–O–methyltransferase (2′OMTase), a Nsp16 non–structural protein, is a methyltransferase enzyme that mediates mRNA cap 2’–O–ribose methylation to the 5′–cap structure of viral mRNAs, and forms a heterodimer together with its allosteric activator Nsp10 [46]. The role Nsp16 plays in the virus's life cycle is to perform the final step of RNA cap synthesis. Capping of the 5′–end of mRNA stabilizes it, preventing degradation by the host cell, and helps to reduce an innate immune response. The N7–methyl guanosine cap is a prerequisite for binding of Nsp16, therefore plays an essential role in viral mRNAs cap methylation and effective RNA translation, which is essential to evade the host immune system [47]. Viruses that infect eukaryotic organisms generally modify the 5′–cap of viral RNAs to mimic the hosts mRNA structure. In this manner, the virus protects itself from degradation by 5′–3′ exoribonucleases, ensures efficient translation and helps to prevent recognition by the host immune system. SARS–CoV–2 encodes one S–adenosyl–methionine (SAM)–dependent methyltransferase (a 2′OMTase also known as the nonstructural protein 16, Nsp16) [48]. Inhibition of the Nsp16 2′OMTase may represent a promising therapeutic target for the identification of new anti–SARS–CoV–2 drugs to treat COVID–19. The functional role of 2′OMTase is reported in the Supporting Information (Section S1.3).
Binding energies for the best docking pose of complexes 1–6 and the NSAIDs on SARS–CoV–2 2′OMTase (Nsp16–Nsp10 heterodimer complex) (PBD: 6W4H) are given in Table S1. Best–bound compound (with higher binding capacity and affinity for the protein) was revealed to be complex 5. All complexes were found to possess better binding affinity than the respective NSAIDs with the exception of complex 2.
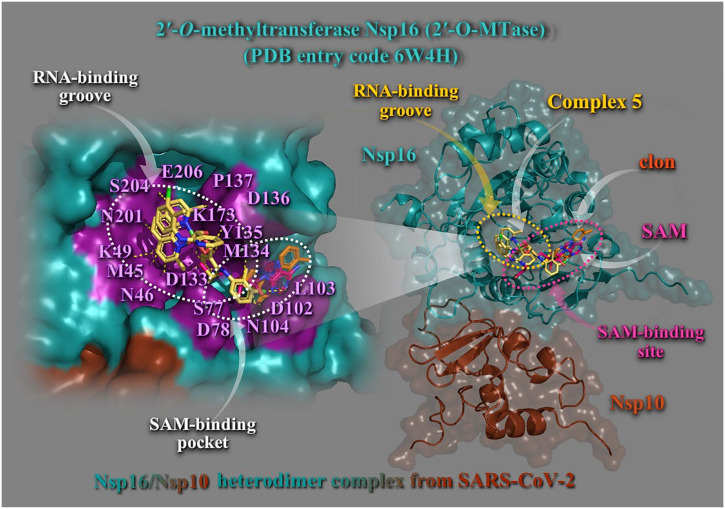
Complex 5 binds at the same region with both SAM and the RNA molecule binding to a groove proximal to the SAM binding pocket (Fig. 8 ). Complex 5 seems to occupy the SAM binding site acting as antagonist for SAM binding. Complex 5 is positioned in a binding cavity sharing both SAM and RNA binding pockets formed by helices αΖ, αΕ, η5/αΗ, αD, η4, αΑ, and η1, as well as β–sheets β1 to β8 (Fig. 8, right panel). Binding interactions stabilizing 5 in its binding cavity include: π–polar with neoc moiety (asparagine N46, N104, N201, serine S204, and glutamate E206), π–cation with neoc moiety (lysine K49), π–π stacking and π–alkyl with neoc (tyrosine Y135), π–alkyl with neoc (methionine M45), and polar (leucine L103, Y135) (Fig. 8, left panel). Binding interactions of Hclon include: K173 (π–cation), D133 (π–anion), Y135 (π–π stacking, π–alkyl, and π–polar), proline P137 (π–alkyl), S77 (π–polar), aspartate D102 (π–anion), D136 (π–polar), D78 (π–anion), and M134 (π–alkyl). Binding residues common with SAM are indicated in bold face. Additional binding residues stabilizing SAM in its binding pocket include: D117, Y50, glycine G74, alanine A75, G76, histidine H72, G84, cysteine C118, and P83. Common contacts of 5 with the residues of the catalytic active site are K173, E206, K49, and D133 (underscored). Cap binding residues coordinating m7GpppA involve Y33, S205, threonine T175, Y135, K140, and H177. The guanosine ring of m7GpppA stacked with tyrosine Y135 which revealed to be also a binding contact of 5, while the phosphate groups of m7GpppA were mostly stabilized by side–chain atoms of Y33, Y135, K140, T175, S204, and S205. It is interesting that serine S204 was found also to interact with 5. D133 binding contact of complex 5 is also involved in the RNA–binding.
Fig. 8.
(Right panel) Docking pose orientation of best–bound complex 5 and Hclon (rendered in stick model and colored by atom type in yellow–orange and orange C atoms, respectively) on the crystal structure of SARS–CoV–2 2′OMTase (Nsp16–Nsp10 heterodimer complex illustrated as cartoon colored in deep teal and chocolate, respectively with additional depiction of semitransparent surface) superimposed with SAM (rendered in stick colored in hot pink C atoms) in the small unit crystal form (PDB ID: 6W4H). SAM–binding site and RNA–binding groove are also indicated in dashed yellow and hot pink ovals, respectively. (Left panel) A close–up view of the ligand binding architecture mapping of 5 in its binding pocket superimposed with Hclon and SAM, as well as the residues implicated in SAM and RNA binding pockets (highlighted in deep purple) onto the SARS–CoV–2 Nsp16 structure (opaque surface in deep teal) showing the relative positions of the SAM and RNA binding regions. Binding contacts are shown as dotted yellow lines. Hydrogen atoms are omitted from all molecules for clarity. Heteroatom color–code: Cu and Cl: green, N: blue, and O: red. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics Systems.
Notably, L103, D78, D133, D102, and Y135 binding contacts of 5 were found also to stabilize the S–adenosylhomocysteine (SAH) analog sinefungin [34] in the SAM cleft. From the above it may be deduced that complex 5 may antagonize the natural ligand SAM in Nsp16 and thus could be a potent inhibitor of Nsp16, to limit viral replication in infected cells, favoring viral clearance.
3.2. In vitro toxicity studies
The growth inhibition/cytostatic and cytocidal/cytotoxic effects induced by complex 3 against human ovarian adenocarcinoma (OVCAR–3) cells, breast adenocarcinoma (MCF7) cells, prostate adenocarcinoma (PC–3) cells, lung adenocarcinoma (A549) cells, and human normal lung (MRC–5) cells, are presented in Table S2 and reveal that the administered complex caused a dose–dependent inhibition of cell proliferation. The dose–effect curves for all treated cell lines are illustrated in Fig. S1 demonstrating the different chemosensitivity of the cell lines to the studied complex. The order of cell line chemosensitivity for the complex was found to be: OVCAR–3 > A549 > MCF–7 > PC3 ≈ MRC5. Complex 3 exhibited a moderate cytostatic and cytotoxic effect against ovarian and lung cancer cell lines, displaying IC50 values below the threshold of 100 μM. Examining the cytotoxic potency of 3 against cell lines, it was revealed to be similar for OVCAR–3 and A549, exhibiting IC50 values of 66.0 μM and 72.0 μM, respectively. More sensitive to the administration of 3 were demonstrated to be OVCAR–3 cells. Lower toxicity of 3 was revealed for MCF–7 cells, while the rest cell lines (PC3 and MRC–5) were proved to be more resistant to the administration of the complex. Human lung–derived MRC5 cells are highly susceptible to the infection of various human coronaviruses, including HCoV–OC43, HCoV–229E and Middle East respiratory syndrome coronavirus (MERS–CoV) [49]. Furthermore, it is of interest that, since it is well known that ACE2 acts as an entry receptor for SARS–CoV–2, Uemura et al [50] have engineered MRC5 cells to express ACE2 and serve as a model system for the discovery of antivirals targeting SARS–CoV–2. Their results revealed that SARS–CoV–2 replicates efficiently in MRC5/ACE2 cells identifying them as a virus–susceptible human cells, concluding that the MRC5/ACE2 cells will be important in developing specific anti–viral therapeutics and will assist in vaccine development to combat SARS–CoV–2 infections. With the employment of the MRC5/ACE2 cells the researchers managed to evaluate antiviral activities of a number of small molecules, including remdesivir. To this context, the study of in vitro cell toxicity of complex 3 against MRC5 cells would be of some significance despite that it was not used the engineered MRC5/ACE2 cells.
3.3. Molecular pharmacokinetic properties, drug–likeness, target proteins and toxicity predictions
The molecular physicochemical pharmacokinetics properties of complexes 1–6 related to Lipinski's Rule of Five(Ro5) with the employment of Molinspiration property engine to analyze the drug–like properties of the compounds are reported in Table S3. Better approximation to Ro5 was revealed for complexes 3–5 (better bioavailability) and worst for 1, 2, and 6. It has been reported that compounds which are violating two or more of the Lipinski's rule parameters may create problems in bioavailability [51].
The obtained values of drug–likeness score revealed that complexes 1 and 2 presented the lower drug–likeness score (Table S3). Table S3 shows complexes 3 and 4 comply with the Ro5 criteria, violating only the molar mass criterion and marginally the milogP for 4. However, still this discordance is considered to be acceptable for good drug–likeness behavior, as only the violation of more than 2 criteria would render a molecule as a non–orally available drug [52]. Theoretical miLogP values of compounds 3 and 6 and marginally 4, were found to be below 5, suggesting, according to Ro5, good permeability across the cell membrane. According to Veber's Rule complex 5 exhibits TPSA values lower than 140 Å2 and number of rotatable bonds 10 and is thus predicted to have good oral bioavailability according to Veber's Rule. Complexes 1, 3, and 4 comply only with TPSA calculated values. The Ghose filter is also fulfilled for complexes 3–6 (natoms restricted between 20 and 70: 44, 48, 53, and 53, respectively), and marginally for complex 2 (natoms = 76), with only one exception for complex 1 which violates this rule exhibiting natoms = 84.
The probable biological activity profiles for complexes 1–6 and the determination of drug–likeness score of compounds through Molinspiration bioactivity score prediction (G–protein–coupled receptors ligands (GPCR), kinase inhibitors, ion channel modulators, nuclear receptors) are depicted in Table S4. The larger the bioactivity score has higher probability of the specific molecule to be active. The obtained drug–likeness score values from Table S4 reveal that complexes 3 and 4 exhibit bioactivity score higher than −0.50, indicating moderate activity for these compounds (for both compounds considerable protease inhibitor activity was revealed since they displayed bioactivity scores higher than 0.00). On the contrary, complexes 1 and 2 found to possess the lowest bioactivity scores.
The results regarding the drug–likeness parameter predictions and physicochemical characteristics of complexes 1–3, and 5, 6, based on PreADMET application are shown in Table S5. All compounds are computed not qualified to CMC like rule and found to violate Lead like, Rule of Five, and WDI (World Drug Index) like rules, whilst it observed no violation (suitable) for MDDR–like rule. The MDDR–like rule describes a molecule as drug–like or non–drug–like on the basis of the number of rings, rigid bonds, and rotatable bonds. The PreADMET results suggested that all compounds have good drug–like properties, at least for one rule, as they adhered to the MDDR–like rule (did not report any violations regarding the MDDR rule descriptors). However, they did not obey the rest rules. The WDI standard is a measure based on compounds that have molecular properties within the 90% upper limit found in the WDI (World Drug Index). Regarding the WDI–like rule, the compound does not display properties similar to this library.
ADMET prediction [53] is very essential to assess the behavior of compounds that can be used as pharmaceuticals. The ADMET pharmacokinetic data of complexes 1–3, and 5, 6, based on PreADMET application, are reported in Table S6. The ADMET prediction method involving Madin–Darby Canine Kidney cell permeability (MDCK), Caco–2 cell permeability, Human Intestinal Absorption (HIA), skin permeability, and blood–brain barrier (BBB) penetration, was also employed for these compounds by PreADMET, with the results summarized in Table S6. The results suggested that for BBB, all compounds were found to exhibit high absorption displaying values enough higher than the threshold 2.0 (CBrain/CBlood > 2.0), while only complex 6 exhibiting a value of 0.21 displayed middle absorption (0.1–2.0). The penetration of the blood brain barrier is critical in the pharmaceutical field [54], because compounds showing penetration to BBB can be used as central nervous system (CNS) drugs (one category being the NSAID drugs). Thus, all compounds classified as active in the CNS and may cause collateral effects. The latter is also supported in terms of TPSA (Table S3) with a value of 166.22 Å2 (values greater than 140 Å2 exhibit poor permeability).The Caco–2 cell permeability (PCaco–2) (nm/s) and permeability across MDCK cell permeability (PMDCK) (nm/s) cell models have been used as a reliable in vitro model for the prediction of oral drug absorption, with the Caco–2 cells derived from human epithelial colon adenocarcinoma having various routes of drug transport through the intestinal epithelium [55]. All compounds showed medium permeability to Caco–2 cells with the values ranging between 10 and 100 nm/s (Table S6). In accordance with Irvine et al [56], PMDCK (nm/s) system cells can be used as tool for rapid screening permeability. The test compounds presented low permeability in the PMDCK (<25) cell system with permeability values ranging from 0.0338 to 0.0434. ADMET results showed that complexes 1–3, and 5, 6 are well–absorbed by human intestinal [57] with HIA values ranging between 70 and 100% defined for well absorption. Intestinal absorption is defined as a percentage absorbed rather than as a ratio of concentrations, with ADMET predicting the HIA after oral administration. The prediction of HIA can identify the potential candidate drug, as well as predict its route of administration and efficacy [58]. A well–absorbed compound is one that is absorbed at least 90% into the bloodstream in humans. The analysis of PreADMET identified that complexes 1–3, 5 and 6 displayed HIA values of 98.07%, 100.00%, 99.05%, 98.29%, and 94.96%, respectively, being good candidates for oral drug since they are able to be well absorbed by the intestine. Complex 6 showed the lowest absorption equal to 94.96%, whereas complex 2 showed the highest value of HIA equal to 100.00%, as shown in Table S6. The ADMET prediction results of these compounds revealed also an excellent percentage of plasma protein binding (PPB) (100%), with the exception of complex 6 which exhibited PPB value of 62.43%, leading to weak binding (values <90% represent weak binding, and values >90% represent strong binding). For aqueous solubility of compounds 1–5, the predicted values found to be extremely low, indicating very good absorption, with the exception of complex 6. The CYP enzymes, particularly isoforms 1A2, 2C9, 2C19, 2D6 and 3A4, were responsible for about 90% of oxidative metabolic reactions [59]. The more CYP isoforms a given molecule inhibits, the more likely it will be involved in drug–drug interactions (DDI) with many other drugs [60]. Complexes 1–3, 5 and 6 showed to be either inhibitor, substrate or no activity on various CYP isoforms. Drugs that inhibit CYP2C19 and CYP2C9 enzyme activity tend to increase plasma concentrations and, in some cases, adverse effects may occur [61]. According to preADMET, among the screened compounds, only complex 2 observed to inhibit these two CYP isoforms, while the rest compounds predicted to be non–inhibitors. The results revealed that complexes 1–3, and 5 have no CYP2D6 inhibitory activity, which is responsible for the metabolism of many drugs and toxic chemicals.CYP2D6 is involved in the metabolism of drugs such as antiarrhythmics, adrenoceptor antagonists and tricyclic antidepressants [62]. Only complex 6 was found to possess inhibitory activity against CYP2D6. Cytochrome CYP3A4 is an enzyme responsible for the oxidation of small organic molecules (xenobiotics), such as toxins or drugs, so that they can be removed from the body, found mainly in the liver and intestine [63]. According to the preADMET analysis, all complexes found to act on the inhibition of CYP3A4, and at the same time to be substrate of CYP3A4. The MDCK evaluation, useful for the rapid screening of cell membrane permeability, has demonstrated that the compounds have low MDCK cell permeability (values less than 25), i.e. they have low diffusivity in cell membranes. Pgp glycoprotein is an efflux pump found in cells of the small intestine, blood–brain barrier, hepatocytes and proximal tubule of the kidney, serving as a protective function for the body against xenobiotics. Intestinal absorption, biliary excretion and urinary excretion of Pgp substrates can, therefore, be altered by the inhibition or induction of Pgp [64]. According to the preADMET analysis, complexes 1–3, and 5 can act as an inhibitor of Pgp. This factor must be taken into account for their administration in order not to generate bioaccumulation in either animal models or in humans. In contrast, complex 6 does not act as Pgp inhibitor. The solubility of complexes (with the exception of 3 and 6) in pure water as well as in a buffered solution is extremely low. Therefore, for their use as drugs, there is the need to use a pharmaceutical system that makes water solubility feasible. The permeability of the skin is another important parameter since it allows the prediction of the rate of skin permeability, a crucial parameter for the transdermal delivery of medications and for the risk assessment of all chemicals that come into contact with the skin accidentally [65]. Good absorption via the skin is documented for complexes 2 and 3 (higher than −3 threshold value).
Physicochemical descriptors and predicted ADMET parameters, pharmacokinetic properties and drug–likeness nature of complex 5 with the aid of SwissADMET predictions are reported in Table S7. The bioavailability radar of complex 5 is depicted in Fig. S1. From the predicted descriptors it is deduced that complex 5 bears moderate lipophilicity, high human intestinal absorption and ability to act as a P–glycoprotein substrate. Furthermore, it is not shown to be able to pass the BBB, nor to inhibit various CYP metabolism enzymes. Nevertheless, due to its bulk size and number of aromatic rings it seems to violet the adopted rule tests (Lipinski, Ghose, Egan and Muegge, violating from one to three parameters including molecular weight, number of atoms, and logP), with the exception of Veber test for which it possesses drug likeness properties.
Target Prediction approach was adopted through Swiss Target Prediction method. The target proteins predicted with higher possibility to be bound to studied complexes derived from Swiss Target Prediction are reported in Table S8. Some of the proteins depicted in Table S8 with higher possibility to act as targets for complexes 1–6 are renin, apoptosis regulator Bcl–2, cathepsins D and E, dipeptidyl peptidase 4 (DPP4), and cannabinoid receptor 1 (CNR1). Details concerning selected predicted targets are found in the Supplementary data file (Section S1.3). It is of interest that among the most possible predicted targets for complexes 3 and 4 is DPP4. This target, which is identical to adenosine deaminase complexing protein–2, and to the T–cell activation antigen CD26, is an intrinsic type II transmembrane glycoprotein and a serine exopeptidase that cleaves X–proline dipeptides from the N–terminus of polypeptides. DPP4 is highly involved in glucose and insulin metabolism, as well as in immune regulation. This protein was shown to be a functional receptor for human coronavirus MERS–CoV, and protein modeling suggests that it may play a similar role with SARS–CoV–2 [66].
The prediction of compound toxicities is an important part of the drug design development process. Computational toxicity estimations are not only faster than the determination of toxic doses in animals, but can also help to reduce the amount of animal experiments. The rat acute toxicity of the complexes predicted by the in silico tool GUSAR on the basis of PASS (Prediction of Activity Spectra for Substances) technology via four types of administration (intraperitoneal, intravenous, oral, and subcutaneous), are shown in Table S9. Predictions of LD50 values of rat acute toxicity, based on the structural formula of the tested compounds, are reported along with the acute rodent toxicity classification of the compounds. Higher toxicities are documented for complexes 1 and 2 for all routes of administration. According to route of administration complexes 1–6 were estimated to have moderate, slight, and practically non–toxic behavior based on the calculated LD50 values. Furthermore, toxicity profile data parameters of complexes 1–6 based on preADMET application are displayed in Table S10. In vitro values of acute algae (algae_at) and daphnia (daphnia_at) toxicity were predicted to be within the acceptable range with extremely low values. By performing preADMET analysis, it was possible to verify whether the studied complexes are able to inhibit hERG that may cause prolongation of the QT interval (electrocardiographic parameter representing the duration of electrical systole – cardiac contraction), resulting in potentially fatal ventricular tachyarrhythmia called Torsade de Pointes [67]. All complexes displayed medium risk for hERG gene coding for the α–subunit of potassium ion channel in humans. In this case, the administration of these compounds should be cautious in cases of cardiopathies. Additionally, all compounds predicted to be mutagenic according to Ames test. According to the results of Table S10, the toxicity prediction of complexes 1–6 on Carcino Mouse and Carcino Rat models revealed no carcinogenic activity for mice and rats. Prediction of acute Daphnia (fish) toxicity also showed extremely low levels of toxicity for all compounds. The same is valid for small fishes, such as minnow (Pimephales promelas) and medaka (Oryzias latipes) which are used as toxicology research models worldwide, considering the maximum toxic concentration acceptable up to 0.01 mg/L. All compounds displayed practically no toxicity on these organism models (values range in the level of e–7 to e–13). Using preADMET, the compound did not predict to have mutagenic activity, being negative to TA1535_10RLI, TA100_NA, and TA100_10RLI strains. However, mutagenic activity (positive values) was observed only for strain TA1535_NA.
Further toxicity evaluation of complexes 2 and 5 were adopted with the OSIRIS Property Explorer. For complex 5, the prediction revealed no potential toxicity risks of tumorigenicity, irritating effects, mutagenicity, and reproductive effects (predicted via toxicity risks of Property Explorer applet). For complex 2, irritating risk due to its high–risk for irritating effects cyclopentanone fragments was predicted. Medium–risk toxicity risk of reproductive effects was indicated by both cyclopentanone ring fragments and main loxo fragment ligand. Finally, no potential toxicity risks of tumorigenicity and mutagenicity effects were documented.
In general, the studied compounds qualified for drug–likeliness and ADMET showing satisfactory results and can be further selected to study the interactions between the drug target by molecular docking and interaction studies.
4. Conclusions
Molecular docking procedure revealed the ability of the studied copper(II) complexes to interfere with critical targets of SARS–CoV–2. The study has demonstrated that these compounds could potentially bind to 3C–like cysteine main protease, papain–like protease, RNA–dependent RNA–polymerase, and Nsp16–Nsp10 2′–O–methyltransferase complex target proteins of SARS–CoV–2 involved in the progression of COVID–19, highlighting their role in the replication of the virus. Additionally, in vitro toxicity and a variety of computational tools were employed to predict the complete biological activity profile, the pharmacokinetic properties, the drug–like nature, and possible toxicity of the compounds, distinguishing the risky ones. Liable scoring functions for the compounds according to their ADMET properties predicted by in silico models.
Further experimental studies are needed to thoroughly elucidate the underling mechanism of antiviral activity and to explore possible clinical effect of the compounds and the role they can play in COVID–19 as antiviral therapeutic agents with the employment of molecular dynamic simulations, construction of pharmacophore models, and quantum chemical studies to understand structural features essential for the activity.
Author statement
- Dr. Elena Geromichalou, Dr. Panagiotis Dalezis, Mr. Georgios Malis (MSc), and Dr. George D. Geromichalos conceptualized, designed and conducted the project.
- Prof. Dimitrios T. Trafalis and Prof. George Psomas supervised the project.
All authors provided critical comments on manuscript.
Declaration of Competing Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jinorgbio.2022.111805.
Appendix A. Supplementary data
Supplementary material
References
- 1.Malis G., Geromichalou E., Geromichalos G.D., Hatzidimitriou A.G., Psomas G. J. Inorg. Biochem. 2021;224 doi: 10.1016/j.jinorgbio.2021.111563. [DOI] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebedeva N.S., Gubarev Y.A., Mamardashvili G.M., Zaitceva S.V., Zdanovich S.A., Malyasova A.S., Romanenko J.V., Koifman M.O., Koifman O.I. Sci. Rep. 2021;11:19481. doi: 10.1038/s41598-021-99072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang-Show H. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 5.Osipiuk J., Azizi S., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R., Kim Y., Lisnyak V., Maki S., Nicolaescu V., Taylor C., Tesar C., Zhang Y., Zhou Z., Randall G., Michalska K., Snyder S., Dickinson B., Joachimiak A. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X., Peng R., Peng Q., Wang M., Xu Y., Liu S., Tian X., Deng H., Tong Y., Hu X., Zhong J., Wang P., Qi J., Gao G., Shi Y. Cell Rep. 2020;31 [Google Scholar]
- 7.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P., Maruthi K., Eng E., Vatandaslar H., Chait B., Kapoor T., Darst S., Campbell E. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beeraka N.M., Sadhu S.P., Madhunapantula S.V., Pragada R.R., Svistunov A., Nikolenko V.N., Mikhaleva L., Aliev G. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.552925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ullrich S., Nitsche C. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirri D., Pratesi A., Marzo T., Messori L. Expert Opin. Drug Discovery. 2021;16:39–46. doi: 10.1080/17460441.2020.1819236. [DOI] [PubMed] [Google Scholar]
- 11.de Paiva R.E.F., Marçal Neto A., Santos I.A., Jardim A.C.G., Corbi P.P., Bergamini F.R.G. Dalton Trans. 2020;49:16004–16033. doi: 10.1039/d0dt02478c. [DOI] [PubMed] [Google Scholar]
- 12.Chasapis C.T., Georgiopoulou A.K., Perlepes S.P., Bjørklund G., Peana M. J. Inorg. Biochem. 2021;219 doi: 10.1016/j.jinorgbio.2021.111423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karges J., Cohen S.M. ChemBioChem. 2021;22:2600–2607. doi: 10.1002/cbic.202100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil-Moles M., Basu U., Büssing R., Hoffmeister H., Türck S., Varchmin A., Ott I. Chem. Eur. J. 2020;26:15140–15144. doi: 10.1002/chem.202004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida-Neto F.W., Matos M.G.C., Marinho E.M., Marinho M.M., de Menezes R.R.P.P.B., Sampaio T.L., Bandeira P.N., Fernandes C.F.C., Teixeira A.M.R., Marinho E.S., De Lima-Neto P., dos Santos H.S. Biochem. Biophys. Res. Commun. 2021;537:71–77. doi: 10.1016/j.bbrc.2020.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal M., Musib D., Zade A.J., Chowdhury N., Roy M. ChemistrySelect. 2021;6:7429–7435. doi: 10.1002/slct.202101852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynak P., Dulal M., Mithun R. New J. Chem. 2021;45:1924–1933. [Google Scholar]
- 18.Karges J., Kalaj M., Gembicky M., Cohen S.M. Angew. Chem. Int. Ed. Eng. 2021;60:10716–10723. doi: 10.1002/anie.202016768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuong C., DuChane C.M., Webb E.M., Rai P., Marano J.M., Bernier C.M., Merola J.S., Weger-Lucarelli J. Viruses. 2021;13:980. doi: 10.3390/v13060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Refat M.S., Gaber A., Alsanie W.F., Kobeasy M.I., Zakaria R., Alam K. Open Chem. 2021;19:772–784. [Google Scholar]
- 21.Grifagni D., Calderone V., Giuntini S., Cantini F., Fragai M., Banci L. Chem. Commun. 2021;57:7910–7913. doi: 10.1039/d1cc02956h. [DOI] [PubMed] [Google Scholar]
- 22.Muramatsu T., Takemoto C., Kim Y., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngwa W., Kumar R., Thompson D., Lyerly W., Moore R., Reid T., Lowe H., Toyang N. Molecules. 2020;25:2707. doi: 10.3390/molecules25112707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellavite P., Donzelli A. Antioxidants (Basel) 2020;9:742. doi: 10.3390/antiox9080742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ionescu M.I. Protein J. 2020;39:600–618. doi: 10.1007/s10930-020-09933-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai W., Zhang B., Jiang X., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L., Xu Y., Yang H., Liu H. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo S., Kim S., Shin D.H., Kim M. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boopathi S., Poma A.B., Kolandaivel P. J. Biomol. Struct. Dyn. 2020:1–10. [Google Scholar]
- 30.Chou K., Wei D., Zhong W. Biochem. Biophys. Res. Commun. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson T.O., Adegboyega A.E., Iwaloye O., Eseola O.A., Plass W., Afolabi B., Rotimi D., Ahmed E.I., Albrakati A., Batiha G.E., Adeyemi O.S. J. Pharmacol. Sci. 2021;147:62–71. doi: 10.1016/j.jphs.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu X. Yang, Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Rao H. Yang Z. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 34.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J.F. Sci. Signal. 2020;13:eabe1202. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Günther S., Reinke P., Fernández-Garcia Y., Lieske J., Lane T.J., Ginn H.M., Koua G., Ehrt C., Ewert W.…Meents A. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Fang C., Zhang Q., Zhang R., Zhao X., Duan Y., Wang H., Zhu Y., Feng L., Zhao J., Shao M., Yang X., Zhang L., Peng C., Yang K., Ma D., Rao Z., Yang H. 2022. Protein Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Acta Pharm. Sin. B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jahan R., Paul A., Bondhon T., Hasan A., Jannat K., Mahboob T., Nissapatorn V., Pereira M., Wiart C., Wilairatana P. Nat. Prod. Commun. 2021;16:1–13. [Google Scholar]
- 39.Pitsillou E., Liang J., Ververis K., Lim K.W., Hung A., Karagiannis T.C. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.623971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitra D., Verma D., Mahakur B., Kamboj A., Srivastava R., Gupta S., Pandey A., Arora B., Pant K., Panneerselvam P., Ghosh A., Barik D.P., Mohapatra P.K.D. J. Biomol. Struct. Dyn. 2021:1–22. doi: 10.1080/07391102.2021.1873185. [DOI] [PubMed] [Google Scholar]
- 41.Kirchdoerfer R.N., Ward A.B. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S., Fulbabu S.K.M., Sonawane A., Kar P., Sadhukhan S. J. Biomol. Struct. Dyn. 2021;39:6249–6264. doi: 10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elfiky A.A. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald S.M. Wiley Interdiscip. Rev. RNA. 2013;4:351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menachery V.D., Debbink K., Baric R.S. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H.R., Muth D., Kint J., Forlenza M., Müller M.A., Drosten C., Thiel V., Trybala E. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uemura K., Sasaki M., Sanaki T., Toba S., Takahashi Y., Orba Y., Hall W.W., Maenaka K., Sawa H., Sato A. Sci. Rep. 2021;11:5376. doi: 10.1038/s41598-021-84882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok S.W.F., Zeng W., Niu Y., Coghi P., Wu Y., Sin W.M., Ng S.I., Gordillo-Martínez F., Gao J.Y., Law B.Y.K., Liu L., Yao X., Wong V.K.W. Front. Pharmacol. 2018;9:710. doi: 10.3389/fphar.2018.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jadhav P.B., Yadav A.R., Gore M.G. Int. J. Pharm. Bio Sci. 2015;6:142–154. [Google Scholar]
- 53.Reddy G.D., Kumar K.N.V.P., Duganath N., Divya R.A.K. Int. J. Drug Dev. Res. 2012;4:268–282. [Google Scholar]
- 54.Ajay G.W., Bemis M.A. Murcko. J. Med. Chem. 1999;42:4942–4951. doi: 10.1021/jm990017w. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita S., Furubayashi T., Kataoka M., Sakane T., Sezaki H., Tokuda H. Eur. J. Pharmacol. 2000;10:195–204. doi: 10.1016/s0928-0987(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 56.Irvine J.D., Takahashi L., Lockhart K., Cheong J., Tolan J.W., Selick H.E., Grove J.R. J. Pharm. Sci. 1999;88:28–33. doi: 10.1021/js9803205. [DOI] [PubMed] [Google Scholar]
- 57.Egan W.J., Lauri G. Adv. Drug Deliv. Rev. 2002;54:273–289. doi: 10.1016/s0169-409x(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 58.Saurav K., Zhang W., Saha S., Zhang H., Li S., Zhang Q., Wu Z., Zhang G., Zhu Y., Verma G. Interdiscip. Sci. 2014;6:187–196. doi: 10.1007/s12539-013-0200-y. [DOI] [PubMed] [Google Scholar]
- 59.Williams J.A., Hyland R., Jones B.C., Smith D.A., Hurst S., Goosen T.C., Peterkin V., Koup J.R., Ball S.E. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 60.Cheng F., Yu Y., Zhou Y., Shen Z., Xiao W., Liu G., Li W., Lee P.W., Tang Y. J. Chem. Inf. Model. 2001;51:2482–2495. doi: 10.1021/ci200317s. [DOI] [PubMed] [Google Scholar]
- 61.van Booven D., Marsh S., McLeod H., Carrillo M.W., Sangkuhl K., Klein T.E., Altman R.B. Pharmacogenet. Genomics. 2010;20:277–281. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertilsson L., Dahl M., Dal’en P., Al-Shurbaji A. Br. J. Clin. Pharmacol. 2002;53:111–122. doi: 10.1046/j.0306-5251.2001.01548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai D., Tang J., Rose R., Hodgson E., Bienstock R.J., Mohrenweiser H.W., Goldstein J.A. J. Pharmacol. Exp. Ther. 2001;299:825–831. [PubMed] [Google Scholar]
- 64.Wessler J.D., Grip L.T., Mendell J., Giugliano R.P. J. Am. Coll. Cardiol. 2013;61:2495–2502. doi: 10.1016/j.jacc.2013.02.058. [DOI] [PubMed] [Google Scholar]
- 65.Singh S., Singh J. Med. Res. Rev. 1993;13:569–621. doi: 10.1002/med.2610130504. [DOI] [PubMed] [Google Scholar]
- 66.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danker T., Moller C. Front. Pharmacol. 2014;5:203. doi: 10.3389/fphar.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material