Abstract
Whole-genome sequencing (WGS) data of a bacterial strain IITK SM2 isolated from an aquifer located in the middle Indo-Gangetic plain is reported here, along with its physiological, morphological, biochemical, and redox-transformation characteristics in the presence of dissolved arsenic (As). The aquifer exhibits oxidizing conditions relative to As speciation. Analyses based on 16S rRNA and recN sequences indicate that IITK SM2 was clustered with C. youngae NCTC 13708T and C. pasteuri NCTC UMH17T. However, WGS analyses using the digital DNA-DNA hybridization and Rapid Annotations using Subsystems Technology suggest that IITK SM2 belongs to a strain of C. youngae. This strain can effectively reduce As(V) to As(III) but cannot oxidize As(III) to As(V). It exhibited high resistance to As(V) [32,000 mg L−1] and As(III) [1,100 mg L−1], along with certain other heavy metals typically found in contaminated groundwater. WGS analysis also indicates the presence of As-metabolizing genes such as arsC, arsB, arsA, arsD, arsR, and arsH in this strain. Although these genes have been identified in several As(V)-reducers, the clustering of these genes in the forms of arsACBADR, arsCBRH, and an independent arsC gene has not been observed in any other Citrobacter species or other selected As(V)-reducing strains of Enterobacteriaceae family. Moreover, there were differences in the number of genes corresponding to membrane transporters, virulence and defense, motility, protein metabolism, phages, prophages, and transposable elements in IITK SM2 when compared to other strains. This genomic dataset will facilitate subsequent molecular and biochemical analyses of strain IITK SM2 to identify the reasons for high arsenic resistance in Citrobacter youngae and understand its role in As mobilization in middle Indo-Gangetic plain aquifers.
1. Introduction
The Citrobacter genus, which belongs to the Enterobacteriaceae family, was first described in 1932 [1]. Till now, 18 species of Citrobacter have been identified [2, 3] from varied sources like soil, water, sewage, feces, and intestinal gut of animals and humans [4, 5]. Members of Citrobacter are enteric Gram-negative and rod-shaped coliform bacteria with 1.0 × 2.0–6.0 μm in size [6]. Some strains of Citrobacter are opportunistic pathogens and can cause infections in immunocompromised patients [7]. It is often reported that Citrobacter species are the cause of meningitis in infants [8]. Among the different species of Citrobacter, C. youngae causes inflammation in the peritoneum, the membrane that covers the inner wall of the abdomen [9]. Furthermore, different strains of Citrobacter are known to be resistant to several heavy metals, including arsenic (As; [10, 11]).
Arsenic (As) is a geogenic metalloid contaminant that has affected the health of animals and humans [12–15]. Sustained consumption of As-polluted water [>10 μg/L, WHO permissible limit [16]] can cause acute to chronic health problems in humans [13, 15]. Depending on its electronic configuration, As can exhibit multiple oxidation states: +V [arsenates; As(V)], +III [arsenites; As(III)], 0, and –III [15, 17]. In nature, +V and +III are the predominant oxidation states of As [14]. These arsenic species can exist in inorganic (iAsV/III) and methylated (mAsV/III) forms [18]. In mammalian systems, As(III) is identified as more toxic than As(V) [19]. Monomethylated arsenous acid [MMA(III)] is considered to be the most toxic of all forms of As(V) and As(III) [20, 21]. However, MMA(III) and other mAs forms, such as monomethylated arsenic acid [MMA(V)] and dimethylated As species, DMA(III), and DMA(V), are generally not detected in food or water. At times they are found in urine or its by-products [22, 23].
The iAsV/III forms are the major forms of As present in groundwater [15]. In aquifers, As redox transformations can be affected by site-specific conditions such as the prevalent pH, redox potential (EH), co-ions, and the presence of labile organic matter [12, 14]. Although the source of arsenic pollution in groundwater is mainly inorganic [24–26], involvement of the indigenous microbial population in As-redox transformations and mobilization cannot be ignored if abundant labile organic matter was present [18, 27]. These microbes can metabolize arsenic due to the presence of As-metabolizing genes in their system [18, 19]. These As-metabolizing organisms are classified as (a) As(V) reducers [28, 29] and/or (b) As(III) oxidizers [30, 31]. The presence of these organisms in groundwater could indicate microbially mediated arsenic transformation and resultant speciation [27]. Specifically, As(V) reducers have the potential to control As concentrations in the groundwater due to their capacity to bioreduce host minerals of As(V) [32]. The bacterial genera known for their efficient reduction of As(V) to As(III) are Sulfurospirillum, Bacillus, Wolinella, Clostridium, Staphylococcus, Desulfomicrobium, and Citrobacter [33–35].
In this study, a strain of Citrobacter youngae IITK SM2 (hereafter referred to as strain “IITK SM2”) was isolated from middle Indo-Gangetic plain (IGP) groundwater in India, at conditions that were oxidizing with respect to arsenic speciation. The objectives of the study were as follows: (i) identification of major genes, including As-resistant genes, present in this strain through whole-genome sequencing (WGS); (i) determination of physiological, morphological, biochemical, and redox-transformation characteristics of this isolate in the presence of dissolved arsenic; and (iii) identification of features in the arsenic-operon system contained in this strain relative to other organisms. To the best of our knowledge, this is the first strain of Citrobacter youngae which shows effective As(V) reduction to As(III). This information will be helpful in the identification of the As(V) metabolizing enzymes and other proteins involved in the influx and efflux of As relative to this bacteria.
2. Materials and Methods
2.1. Study Site, Sample Collection, and Analysis
Groundwater was sampled from a previously identified As-polluted aquifer in Baikunthpur, Uttar Pradesh, India (26°33′47.3″N and 80°15′18.5″E), situated in the Indo-Gangetic plains [[36]; Figure S1 of the supporting information]. Two different groundwaters, As-polluted and As-unpolluted, were sampled from this site (Figure S1). For each sample, pH, temperature (°C), conductivity (μS cm−1), and redox potential (EH; V) were measured at the site using a portable multiparameter meter (Thermo Orion Star A329) and suitable electrodes. Three sets of water samples were collected from the polluted aquifer. Two sets were filtered using 0.2 μm nylon syringe filters (Cole-Parmer). One of these filtered samples was immediately acidified using 1% (v/v) trace-metal grade HNO3 for the determination of dissolved total arsenic (AsT) and other elements using inductively coupled plasma mass spectrometry (ICP-MS). The other set of filtered samples was left unacidified and was collected without headspace in 15 mL centrifuge tubes. This filtered-unacidified sample set was utilized for dissolved carbon analysis using total organic carbon (TOC) analyzer, and for the measurement of dissolved inorganic As(V) and As(III) using ion chromatography coupled with ICP-MS (IC-ICP-MS). Due to the reported inaccuracies in measured redox potential values using electrode [37–41], EH was also estimated using measured As(V) and As(III) concentrations using the Nernst equation for As-polluted groundwater [42] (Section S1 of the Supporting Information).
For bacterial culturing, unfiltered groundwater samples were collected in sterilized tubes without headspace and were sealed with Parafilm. All these samples were placed in ice gel packs and transported within 1 h to the laboratory. Subsequently, the unfiltered samples were transferred to predefined As-amended agar plates inside a laminar hood.
2.2. Chemicals
All solutions and buffers were prepared in ultrapure water (Milli-Q, resistivity > 18.2 MΩ − cm) and were either filtered using 0.2 μm nylon syringe filters or autoclaved at 121°C for 20 min before use. For preparing 100 mg mL−1 stock solutions of As(V) and As(III), Na2HAsO4·7H2O and NaAsO2, respectively, were used. For qualitative As redox transformation test, a stock solution of 1 M AgNO3 was prepared prior to use and was stored in dark at 4°C. Chemicals used in this study, and their manufacturers and purities, are detailed in Table S1 of the Supporting Information.
2.3. Analytical Techniques
Elemental concentrations, including total dissolved arsenic (AsT), were measured using inductively coupled plasma mass spectrometry (ICP-MS; Thermo iCAP-Qc), with germanium as an internal standard for As measurement. All standards and samples were analyzed in a 1% HNO3 matrix. Dissolved As(V) and As(III) concentrations were measured using ion chromatography coupled with ICP-MS (IC-ICP-MS; Thermo Scientific iCAP Q with the Thermo Scientific Dionex ICS-5000 IC). Analytes were eluted in 100 mM (NH4)2CO3 over IonPac AS7 analytical column. Dissolved total carbon (TC) and inorganic carbon (DIC) concentrations were measured with total organic carbon analyzer (TOC-L; Shimadzu TNM-L ROHS), and dissolved TOC was estimated from TC and DIC (TOC = TC–DIC). The optical density of cultures was monitored by the measurement of absorbance at 600 nm (OD600; Biospectrometer; Eppendorf). The method detection limits of various techniques are listed in Table S2 of the Supporting Information.
Morphology of the bacterial strain was determined microscopically. The Gram-staining test followed by optical microscopy was performed at 100x magnification (Quasmo; Ecostar-plus). For higher magnification and better resolution, tungsten scanning electron microscopy with associated energy dispersive X-ray spectroscopy (W-SEM-EDX; JEOL JSM 6010 LA) was used. Before SEM-EDX analysis, samples were gold-coated at 7-10 nm.
2.4. Isolation of Arsenic-Resistant Bacteria
To isolate As-resistant bacteria, 100 μL of As-polluted groundwater was added to each of the As-amended Lysogeny agar plates (0-400 mg L−1) and spread using sterilized glass beads in a laminar flow hood. These plates were incubated at 37°C for 24 h, and sixteen distinct colonies were isolated upon visual identification. However, only two isolates grew in 400 mg L−1 each of As(III)- and As(V)-containing Lysogeny agar plates. Of these isolates, the strain (IITK SM2) which showed more efficient growth in the presence of arsenic was selected for WGS. Strain IITK SM2 was inoculated in Lysogeny broth (LB) and diluted several times by streaking the culture on variables As-supplemented (0-10,000 mg L−1) Lysogeny agar plates until single colonies were obtained. For further analyses, these cultures were preserved in 12–15% glycerol solution at -80°C.
2.5. Morphological, Physiological, and Biochemical Characterization of Strain IITK SM2
Experiments were performed with strain IITK SM2 to determine its bacterial group, optimum growth conditions, and response of bacterial growth to specific biochemical tests. Experimental details of these tests are provided in Section S2 of the Supporting Information.
2.5.1. Effect of Arsenic on the Growth: Kinetics and Morphology
The growth profile of IITK SM2 was studied in the absence and presence of arsenic [As(V) or As(III)]. Before starting this experiment, it was confirmed that dissolved As was not present in the background media. Initially, a single colony of this isolate from Lysogeny agar plate was inoculated at 30°C (optimum temperature) and 120 rpm in arsenic-free minimum salt media (MSM; Table S3 of the Supporting Information), supplemented with 10 mM glucose as the only C source. Optical absorbance at 600 nm (OD600) of this culture was regularly recorded (BioSpectrometer® basic; Eppendorf) until an OD600 of ~0.1 was reached. Thereafter, ~1% of the bacterial suspension was individually transferred to set-up different cultures which contained 0 mM, 1.33 mM, 3.33 mM, 6.67 mM, 10 mM, 13.33 mM, and 20 mM of either As(V) or As(III) in 10 mM glucose-supplemented MSM. These cultures were incubated at 30°C and 120 rpm. After 8 h of transfer, the OD600 was measured for all the cultures. This experiment was performed three times in triplicate. Among all the concentrations investigated, the maximum biomass was obtained at 10 mM of As(V) dosage (Figure S2 of the Supporting Information). Consequently, 10 mM of As was chosen as the optimum dosage for comparative growth profile study in the presence of either 10 mM As(V) or 10 mM As(III) along with the As-free control. Doubling times for all these three cultures [10 mM As(V), 10 mM As(III), and 0 mM As] were calculated from respective growth curves by considering two log-unit increase in bacterial population. For these three cultures, morphological characterization of bacterial cells was performed using SEM. Details of sample preparation for SEM analysis are discussed in the section S3 of the Supporting Information.
2.5.2. Resistance to As and Other Heavy Metals Typically Present in Contaminated Groundwater
Minimum inhibitory concentration (MIC) tests were performed to evaluate the resistance of strain IITK SM2 to As(III) and As(V), along with certain heavy metals found in contaminated groundwaters like Fe(III), Cr(VI), Mn(II), Ba(II), and Zn(II).The MIC was defined as the lowest metal concentration at which no bacterial growth was observed [43]. The detailed procedure used for determining MIC is discussed in the section S4 of the Supporting Information.
2.6. Redox Transformation of Arsenic
The capability of strain IITK SM2 to transform As(V) to As(III) or vice versa was qualitatively estimated by the formation of colored precipitates upon addition of silver nitrate, per a slightly modified procedure from what is detailed in a previous study [44]. This procedure is discussed briefly in Section S5 of the Supporting Information. Again, MSM supplemented with 10 mM glucose was used as the culturing media without any background As(V) and As(III). Arsenic was either added as As(V) or As(III) to obtain final concentrations as 0, 50, 100, and 500 mg L−1 of each form of arsenic. Also, systems without bacterial cultures were initiated as controls with the same set of As concentrations (0-500 mg L−1). For confirmation of dissolved arsenic concentrations, AsT was measured using ICP-MS, and As(V) and As(III) were measured using IC-ICP-MS before and after the addition of strain IITK SM2.
2.7. Molecular Characterization
To identify the genus and species of strain IITK SM2, 16SrRNA and whole-genome sequencing were performed. The methods used for the isolation of genomic DNA and for 16S rRNA sequencing are detailed in Section S6 of the Supporting Information.
2.7.1. Whole-Genome Sequencing (WGS)
Libraries were constructed in alignment with microbial WGS with the recommendations of Nextera™ DNA flex library preparation kit from Illumina Inc. To determine the mean fragment size, libraries were loaded and analyzed on a high-sensitivity D1000 ScreenTape. The Illumina libraries were diluted to 4 nM pooled, spiked with 5% PhiX, premade Illumina library, and loaded onto a Miseq v2 kit. The sequencing was performed for 2 x 150 cycles. The raw data obtained from Illumina MiSeq were recorded as FASTQ files. The adapter-free reads were obtained using an adapter trimming plugin. The quality check of the reads was done using FastQC v0.11.3 [45]. High-quality reads were obtained using Trimmomatic v0.39 [46]. De novo assembly and scaffolding were done using SPAdes v3.14.1 [47], where filtered reads were assembled without biasing the assembly to any known genome. The quality check was performed using QUAST v5.0.2 [48]. For assembly of the genome, only contigs with more than 500 bp were considered. The g-DNA sequences were assembled into 30 different contigs. The assembly was annotated using National Center for Biotechnology Information, Prokaryotic Genome Annotation Pipeline [[49]; NCBI-PGAP v2020-09-24 build4894)]. The g-DNA of the strain was mapped to reference organisms—Citrobacter freundii FDAARGOS 549 (GenBank accession number NZ_CP033744.1) and Citrobacter youngae NCTC 13708 (GenBank accession number NZ_UFWE01000006.1) using Bowtie 2 [50].
2.7.2. Sequence Comparison of IITK SM2 with Different Strains
To identify whether the isolate belongs to a new species or not, type (strain) genome server (TYGS) using formula 2 was applied for the computation of digital DNA-DNA hybridization (dDDH) value [51]. Furthermore, subsystem features in strain IITK SM2 were also compared with features of different Citrobacter species using rapid annotations with subsystem technology [RAST, [52]. The different types of strains used for these analyses are detailed in Table S4 of the Supporting Information.
Based on the coding sequences (CDs), orthologous gene clusters were determined by comparing the genome of isolate IITK SM2 with genomes of closely related strains. These strains were chosen based on dDDH values. For this clustering, the OrthoVenn web server was used [53]. Default parameters such as e-value cut-off of 10−5and inflation value of 1.5 were used to compare protein similarity and to generate orthologous clusters, respectively.
Through the WGS, arsenic-resistant genes were also identified in the g-DNA of this strain. The arrangement of these genes, or ars operon in IITK SM2, was compared with genetic arrangement of different species of Citrobacter and selected strains of Enterobacteriaceae (Table S4 of the Supporting Information).
2.7.3. Phylogenetic Analysis
The evolutionary relationship of strain IITK SM2 was determined by the comparison of its 16S rRNA sequence with sequences of closely related bacterial species. Furthermore, to accurately differentiate among different species, the highly conserved recN gene sequence was used. For these analyses, sequences of related bacterial strains were downloaded from the NCBI database using basic local alignment search tool [BLAST, [54]]. Multiple alignments of protein sequences of As(V)-reductase gene arsC identified in strain IITK SM2 were performed using CLUSTAL_W [55]. Also, to understand the evolutionary history of these genes, arsC sequences of our isolate were compared with sequences of bacterial species from different genera than of strain IITK SM2. Accession numbers of different strains used for these analyses are detailed in Table S4 of the Supporting Information.
The bootstrap method was used for testing the phylogeny with 1000 replicates [56]. Phylogenetic trees of different sequences mentioned above were prepared using the neighbor-joining [NJ] method [57]. For computing the evolutionary distances, the p-distance method was used [58] and the units were reported in base differences per site. MEGA X was used for conducting these evolutionary analyses [59].
3. Results and Discussion
3.1. Geochemistry of As-Polluted Aquifer
The As-polluted groundwater sampled from the study site exhibited oxidizing conditions with respect to arsenic speciation as suggested by the measured dissolved arsenic, co-solutes, and calculated redox potential (EHm~112 mV). On an average, ~70 μg L−1 of total dissolved arsenic (AsT) was recorded, of which the dissolved inorganic As(V) and dissolved inorganic As(III) concentrations were~43 μg L−1 and~22 μg L−1, respectively. Methylated forms of arsenic were not measured in groundwater because it is well documented in the literature that simple methylated forms such as MMA(III), DMA(V), MMA(V), and DMA(V) are usually not detected in food or water [15, 22, 23, 60, 61] but are detected in urine or its by-products [62–64]. Furthermore, the sum of these dissolved inorganic As(V) and As(III) was within the 10% of AsT, which also suggests that methylated forms of arsenic, if present, were in negligible quantity in the groundwater of our interest (Table 1). Higher concentration of As(V) as compared to As(III) supports the prevalence of oxidising conditions with respect to arsenic speciation. However, the presence of significant concentration of the reduced form of arsenic [~22 μg L−1 as As(III)] in such oxidizing aquifers indicated a potential role of As(V)-reducing organisms. Furthermore, high average DIC (~1296 mg L−1) and TOC (~40 mg L−1) concentrations as compared to unpolluted aquifer (DIC~186 mg L−1 and TOC~11 mg L−1) suggest the potential role of microbial activity in this groundwater. Other water quality parameters are detailed in Table 1.
Table 1.
Geochemical parameters of groundwater samples collected from the study site in Kanpur, India.
| Parameter | Units | As-polluted groundwater | As-unpolluted groundwater |
|---|---|---|---|
| (i) General water quality parameters | |||
| pH | — | 7.11 | 7.32 |
| Measured redox potential (EHm) | mV | 248 | 269 |
| Calculated redox potential (EHc) | mV | 112 | n.a. |
| Temperature | °C | 25.9 | 28.2 |
| Conductivity | μS cm−1 | 4021 | 1023 |
| Alkalinity | mg L−1 as CaCO3 | 600 | 550 |
| Estimated depth of the aquifer | m | 12.2 | 30 |
| Total organic carbon | mg L−1 as C | 40 | 11 |
| Dissolved inorganic carbon | mg L−1 as C | 1296 | 186 |
|
| |||
| (ii) Elemental concentrations | |||
| Total arsenic (AsT) | μg L−1 | 70 | 2 |
| As(V) | 43 | bdl | |
| As(III) | 22 | bdl | |
| Mn | 1671 | 16 | |
| U | 27 | 22 | |
| Al | 2 | bdl | |
| Cr | 6 | bdl | |
| Fe | 106 | 838 | |
| Ni | 3 | 10 | |
| Cu | 9 | 2 | |
| Zn | 23 | 259 | |
| Ba | 569 | 83 | |
| V | bdl | bdl | |
| Co | 1 | bdl | |
| Se | 10 | bdl | |
| Mo | 92 | bdl | |
|
| |||
| Ca | mg L−1 | 101.5 | 69 |
| Na | 638.2 | 206 | |
| Mg | 88.9 | 48 | |
| K | 604.7 | 16 | |
| P | 4.7 | bdl | |
bdl: below detection limit. n.a.: not applicable.
One of the most widely accepted mechanism of arsenic mobilization in groundwater is reductive dissolution of iron (oxy)hydroxide. [FeOOH(s)] [24, 25, 65–73]. However, this mechanism is more prevalent under reducing conditions [66]. For the oxidizing groundwater from where the strain IITK SM2 was isolated as in this study, As(V) might still be released from reductive dissolution of FeOOH but the presence of As(III) in such aquifers hints towards the microbially mediated As(V)-reduction to As(III). However, a detailed and careful study would be required for identifying the role and resultant mechanisms of such microbes in arsenic speciation in oxidizing aquifers. This would require systematic investigation and comparison of indigenous microbial population from both As-free and As-polluted groundwaters.
3.2. Characterization of Bacterial Strain IITK SM2
3.2.1. Classification of the Isolate
The isolate was rod-shaped Gram-negative, catalase-positive, and motile bacteria, which exhibited a negative starch hydrolysis test. Strain IITK SM2 can grow over pH 4–10, with an optimum pH of 7.25. Furthermore, the isolate grew over 15–45°C, with an optimum growth at 30°C at pH 7.25 (Figure S3 of the Supporting Information). The strain could tolerate NaCl up to 6% (w/v) and indicated an optimal growth at 1.5% (w/v) of the salt. After incubation for 2 d in an anaerobic chamber, colonies grown in Lysogeny agar plates were circular, opaque, and yellow, which suggested that the isolate was a facultative anaerobe. Furthermore, strain IITK SM2 showed resistance to ampicillin (100 μg L−1) and hygromycin (50 μg L−1), but growth was not observed in the presence of kanamycin (50 μg L−1), or chloramphenicol (25 μg L−1), or ciprofloxacin (20 μg L−1), or gentamycin (10 μg L−1), or streptomycin (50 μg L−1).
The strain IITK SM2 belongs to genus Citrobacter, as determined by 16S rRNA sequencing, and is closely related to Citrobacter murliniae CDC 2970-59T (98.7%), Citrobacter freundii ATCC 8090T (98.3%), Citrobacter werkmanii CDC 0876-58T (98.2%), and Citrobacter youngae GTC 1314T (97.8%). Furthermore, a distance-based phylogenetic tree construction revealed that the strain was clustered with Citrobacter freundii ATCC 8090 (Figure 1).
Figure 1.
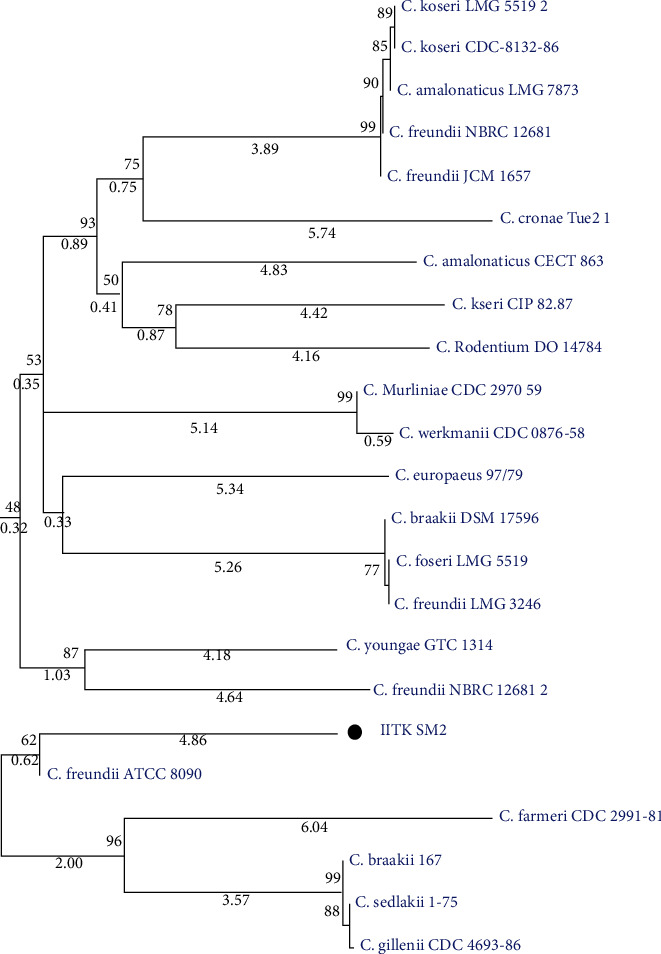
Phylogenetic position of strain IITK SM2 relative to other strains of genus Citrobacter based on 16S rRNA sequences. The neighbor-joining method was used for tree construction [57]. Percentage bootstrap values corresponding to 1000 replicates are shown next to the branches in “bold”. Branch lengths are shown in “narrow italics” below each branch. The tree was drawn to scale.
3.2.2. Resistance to Heavy Metals
Among the metal species considered in this study, IITK SM2 showed resistance to As(V), As(III), Fe(III), Cr(VI), Mn(II), Ba(II), and Zn(II) up to certain levels (Table 2). Of these species, the maximum resistance was observed for As. The minimum inhibitory concentrations (MIC) of As(V) and As(III) were 32,000 mg L−1 (427 mM) and 1100 mg L−1 (~14.7 mM), respectively. To the best of our knowledge, such high MICs of As(III) and As(V) have not been reported for any other Citrobacter species till date. A comparison of MIC of Gram-negative rod-shaped arsenate reducers is made in Table S5 of the Supporting Information. Furthermore, the MIC for Fe(III), Mn(II), Cr(VI), Ba(II), and Zn(II) were estimated to be 17.9 mM, 91.0 mM, 0.2 mM, 14.7 mM, and 3.8 mM, respectively (Table 2).
Table 2.
Characterization of Citrobacter youngae IITK SM2 isolated from As-polluted groundwater of Baikunthpur, India.
| Characteristics | Strain IITK SM2 |
|---|---|
| (a) General parameters | |
| Gram staining test | Gram-negative |
| Shape | Rod |
| Cell size (μm) | 0.3–0.6 × 2.2–3.4 |
| Pigmentation (in Lysogeny agar plate) | Yellow |
|
| |
| (b) Growth conditions | |
| pH range | 4–10 |
| Optimum pH | 7.25 |
| Temperature range (°C) | 15-45 |
| Optimum temperature (°C) | 30 |
| NaCl tolerance (%; w/v) | 0-6 |
| Optimum NaCl tolerance (%; w/v) | 1.5 |
|
| |
| (c) Minimum inhibitory concentration | |
| Dissolved As(III) | 1,100 mg L−1 (~14.7 mM) |
| Dissolved As(V) | 32,000 mg L−1 (~427.1 mM) |
| Dissolved Fe(III) | 1,000 mg L−1 (~17.9 mM) |
| Dissolved Mn(II) | 5,000 mg L−1 (~91.0 mM) |
| Dissolved Cr(VI) | 10 mg L−1 (~0.2 mM) |
| Dissolved Ba(II) | 1100 mg L−1 (~14.7 mM) |
| Dissolved Zn(II) | 250 mg L−1 (~3.8 mM) |
|
| |
| (d) Antibiotic resistanceα | |
| Ampicillin | 100 μg L−1 |
| Hygromycin | 50 μg L−1 |
|
| |
| (e) Other specific parameters | |
| Utilization of D-glucose | + |
| Catalase | + |
| Motility | + |
| Starch hydrolysis | — |
| G + C content (%) | 51.7 |
| Arsenic transformation capability | Reduces As(V) to As(III) |
α No resistance to kanamycin (50 μg L−1), chloramphenicol (25 μg L−1), ciprofloxacin (20 μg L−1), gentamycin (10 μg L−1), and streptomycin (50 μg L−1) was observed.
3.2.3. Impact of Arsenic on Growth and Morphology
The growth of IITK SM2 was found to be more in the presence of arsenic as compared to As-free condition in minimum salt media (MSM) supplemented with glucose (Figures S2 and 2a). The lag phase of this strain varied with the type of As (III versus V, Figure S2) stress provided. For the comparative growth profile study investigated for 0 mM As, 10 mM As(V) and 10 mM As(III) conditions, the shortest lag phase was observed in the absence of any As (8 h), followed by increasing lag phases in the presence of 10 mM As(III) (32 h) and 10 mM As(V) (48 h) (Figure 2(a)). Logarithmic growth was observed between 8-24 h, 32-60 h, and 56-96 h for As-free, 10 mM As(V)-containing and 10 mM As(III)-containing conditions, respectively. The maximum biomass was observed in the presence of As(V) followed by As(III) and As-free conditions (Figure 2(a)). The growth rate (k) and doubling time (DT) observed in the absence of As was 0.08 h−1 and 8.9 h, respectively. In As-stressed conditions, DT increased to 10.5 h and 17.4 h in the systems containing 10 mM As(V) [k = 0.07 h−1] and 10 mM As(III) [k = 0.04 h−1], respectively. Although the addition of dissolved As(V) and As(III) retarded the growth rate, no inhibitory effect of arsenic on the growth of this strain was observed at 10 mM of As dosage. On the contrary, higher biomass was obtained in As-stressed conditions suggesting that the isolate metabolizes As and obtains energy for its growth [74, 75]. Furthermore, more growth in the presence of As(V) as compared to As(III) indicated that strain IITK SM2 possibly had a mechanism to effectively metabolize As(V) relative to As(III).
Figure 2.
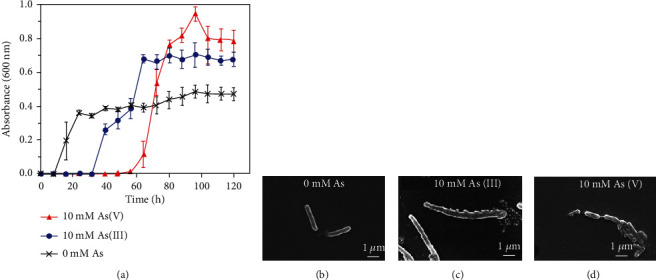
Effect of dissolved As(V) and As(III) on the (a) growth and (b, c, and d) morphology of strain IITK SM2. Bacteria were inoculated in 10 mM glucose-containing minimum salt media (MSM) and supplemented with either 10 mM As(V), or 10 mM As(III), or no As. Error bars correspond to standard deviations of the means from triplicate experiments. No background As(V) and As(III) was detected in MSM.
The IITK SM2 strain was rod-shaped as confirmed by SEM analysis (Figures 2(b)–2(d)). The average length of a bacterial cell in the absence of dissolved As was 2.8 ± 0.6 μm (Figure 2(b)). However, in the presence of As(III), the length of bacteria increased to 5.6 ± 2.2 μm, which suggested that As(III) induced stress condition to this isolate (Figure 2(c)) that resulted in filamentation [76]. This elongation indicated that cell division might be affected due to As(III) stress. However, in the presence of As(V), this strain was found to be clustered together, with no significant change in the cell size (2.0 ± 0.6 μm) as compared to that of the As-free condition (Figure 2(d)).
3.2.4. Redox Transformation of Arsenic by IITK SM2
Qualitative silver nitrate assay indicated that strain IITK SM2 was an As(V) reducer (Figure 3). The formation of yellow- and brown-colored precipitates was observed with standard salts of As(III) [As(III)s; Figure 3a] and As(V) [As(V)s; Figure 3c], respectively, possibly due to precipitation of Ag3AsIIIO3(s) and Ag3AsVO4(s) [44]. However, in the presence of strain IITK SM2, the yellow color in precipitated solids was retained under As(III)-stressed conditions [As(III)I; Figure 3(b)], but brown precipitates were not observed for As(V) conditions [As(V)I; Figure 3(d)]. In fact, the precipitated solids were increasingly yellow with increasing As(V) concentrations. Further, measurements of AsT by ICP-MS, and of As(V) and As(III) by IC-ICP-MS, before and after the reaction with IITK SM2, confirmed these qualitative results. A complete reduction of As(V) to As(III) was observed in As(V)-amended conditions, whereas no change was observed in the conditions initially containing As(III) (Table S6 of the Supporting Information). Overall, results indicate that the isolate can mediate As(V) reduction to As(III), but not vice-versa. The presence of arsenate reducers, like C. youngae IITK SM2, in the middle Gangetic plain groundwater, could be the reason for significant concentrations of dissolved As(III) in oxidizing conditions for arsenic speciation.
Figure 3.

Redox transformation capability of strain IITK SM2 qualitatively confirmed with silver nitrate test. Isolates were cultured in minimum salt media, and 10 mM glucose was used as the carbon source. Color precipitates were obtained by adding Tris-HCl (0.1 M; pH = 7.4) and AgNO3. White precipitates were developed in the absence of any arsenic due to the formation of AgCl. Yellow [Ag3AsO3(s)] and brown precipitates [Ag3AsO4(s)] represent the presence of As(III) and As(V), respectively. Whereas As(III)s and As(V)s represent systems without any bacterial inoculation, The As(III)I and As(V)I represent systems with bacterial inoculation [1% (v/v); OD~ 1]. No background As(V) and As(III) was detected in MSM.
3.3. Genes Identified in Arsenic Metabolism of IITK SM2
3.3.1. Whole-Genome Sequencing (WGS) and Comparison with Other Organisms
To identify the species of IITKSM2, WGS was performed. The mean fragment size of PCR-enriched library of g-DNA of the strain was found to be 564 bp with a concentration of 14.1 ng μL−1. In total, 2,672,974 bp raw reads were obtained for g-DNA, whereas total reads that survived after trimming and filtering were 1,778,915. The final genome size was found to be 4,857,938 bp with a guanine-cytosine (GC) content of 51.70% with N50 value of 438,827 bp. The respective assembly length was ~4.8 Mbp. The NCBI-PGAP annotation showed that the g-DNA contained a total of 4725 genes, which were associated with 4553 coding sequences (CDs) and 66 tRNA, 6 rRNA, and 9 noncoding RNA (ncRNA) sequences. Besides, 91 pseudogenes were also present.
Distance-based phylogeny developed by considering recN sequence indicated that IITK SM2 was clustered with C. youngae NCTC 13708T and C. pasteuri NCTC UMH17T (Figure S4 of the Supporting Information). However, the draft sequence of g-DNA of the strain showed the maximum average nucleotide identity (ANI) of 86.7% with Citrobacter youngae NCTC 13708T, followed by 62.43% ANI with Citrobacter freundii FDAARGOS 549T. Moreover, DNA-DNA hybridization (dDDH) analysis suggested that the intergenomic distance of our isolate was the closest to C. youngae NCTC 13708T (85.8%), followed by C. youngae CCUG 30791T (83.8%). As the proposed cut-off for species delineation is 70% [77, 78], these distances confirm that IITK SM2 belongs to a strain of C. youngae (Table 3). Furthermore, the difference in GC content between IITK SM2 and type strains of C. youngae was ≤0.1%, which confirms that the isolate is a strain of C. youngae.
Table 3.
Intergenomic distance of isolate “IITK SM2” with different strains of Citrobacter using digital DNA-DNA hybridization (dDDH) analysis. Recommended formula 2α was used for estimating the intergenomic distance. Accession numbers of strains used in this analysis are detailed in Table S4 of the Supporting Information.
| Type of strain | dDDH value (%) | GC content difference (%) |
|---|---|---|
| Citrobacter youngae NCTC 13708 | 85.8 | 0.10 |
| Citrobacter youngae 30791 | 83.8 | 0.09 |
| Citrobacter pasteurii CIP 55.13 | 60.7 | 0.10 |
| Citrobacter freundii FDAARGOS 549 | 42.0 | 0.03 |
| Citrobacter freundii NBRC 12681 | 41.9 | 0.04 |
| Citrobacter portucalensis A60 | 39.2 | 0.31 |
| Citrobacter europaeus 97/79 | 39.0 | 0.18 |
| Citrobacter braakii ATCC 51113 | 38.9 | 0.20 |
| Citrobacter cronae Tue2-1 | 36.7 | 0.71 |
| Citrobacter werkmanii NBRC 105721 | 36.4 | 0.37 |
| Citrobacter tructae SNU WT2 | 32.9 | 0.25 |
| Citrobacter koseri NCTC 10786 | 26.4 | 2.10 |
| Citrobacter amalonaticus NCTC 10805 | 25.2 | 1.73 |
| Citrobacter rodentium NBRC 105723 | 24.7 | 2.94 |
| Citrobacter sedlakii NBRC 105722 | 24.4 | 3.01 |
α [51].
Although IITK SM2 belongs to C. youngae, it is different from other C. youngae strains as suggested by the comparison of subsystem features and gene clustering of different Citrobacter species. The coverage of subsystem features in IITK SM2 and comparison of counts of each feature in different Citrobacter species using Rapid Annotations using Subsystems Technology (RAST) server suggested that IITK SM2 was different from the other type strains of C. youngae (Figure 4 and Table 4). Subsystem features grouped under virulence, disease, defense (F3), phages, prophages, transposable elements, plasmids (F7), and membrane transporter (F8) were much higher in strain IITK SM2 as compared to other type strains of C. youngae [NCTC 13708T and CCUG 30791T; Table 4]. On the contrary, much lesser feature counts of motility and chemotaxis (F14) and protein metabolism (F12) were observed in IITK SM2. Furthermore, a Venn diagram of protein clustering of strain IITK SM2 with closely related Citrobacter strains, NCTC 13708, CCUG 30791, and FDAARGOS 549, indicated a total of 4653 protein clusters (Figure 4(b)). Of these, 4590 orthologous clusters that contained at least two strains and 3686 were single-copy clusters. Although IITK SM2 shared 3718 orthologous protein clusters with the other three strains, maximum clusters (4184) were shared with CCUG 30791. Most of the unique orthologous clusters identified in IITK SM2 represented proteins with unknown functions. However, two protein clusters were identified as the IS11595 family transposase and metal binding proteins. These analyses confirmed that our isolate is a novel strain of Citrobacter youngae and was named Citrobacter youngae IITK SM2.
Figure 4.
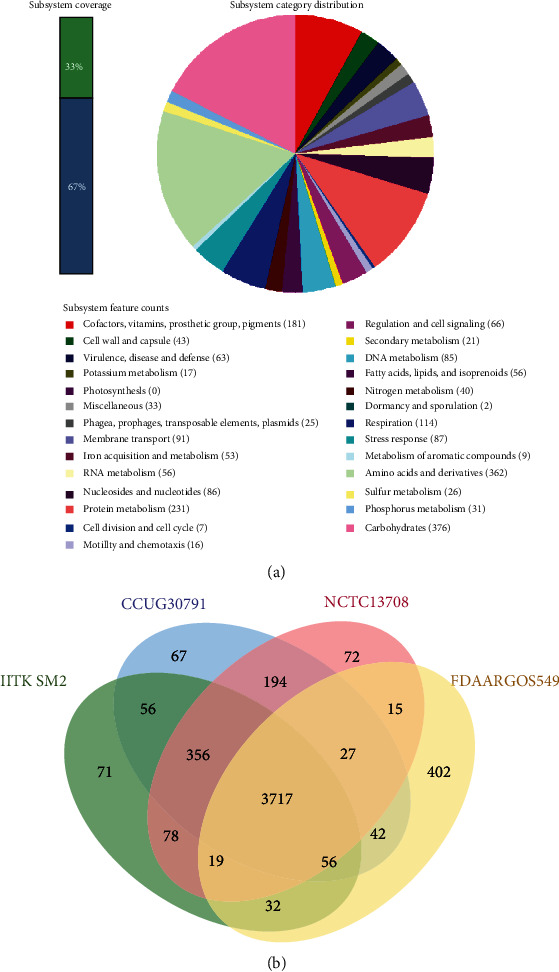
Distribution of subsystem features (a) and orthologous protein clusters (b) in the genome of Citrobacter youngae strain IITK SM2 of subsystem features was performed using the rapid annotations using subsystems technology (RAST) server, where RASTtk annotation scheme was used [57]. The Venn diagram of the clustering of proteins based on the coding sequences (CDs) was constructed using the whole genome sequences of isolate IITK SM2 and of strains of Citrobacter youngae (CCUG 30791 and NCTC 13708) and Citrobacter freundii (FDAARGOS 549).
Table 4.
Comparison of subsystem features in different Citrobacter species using rapid annotations using subsystems technology (RAST) server. The annotation scheme used was RASTtk [57]. Accession numbers of different type strains are mentioned in Table S4 of the Supporting Information.
| S.no. | Parameters | Number of parameters/features | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C. youngae IITK SM2a | C. youngae NCTC 13708 | C. youngae CCUG 30791 | C. Freundii FDAARGOS 549 | C. Sedlakii NBRC 105722 | C. Cronae Tue2-1 | C. Tructae SNU WT2 | C. Braakii ATCC 51113 | C. Rodentium NBRC 105729 | ||
| General parameters | ||||||||||
| G1 | Total number of features | 4725 | 4855 | 4872 | 4856 | 4507 | 5608 | 4686 | 5717 | 5297 |
| G2 | Number of coding sequences | 4644 | 4747 | 4789 | 4747 | 4426 | 5514 | 4578 | 5619 | 5297 |
| G3 | Number of RNAs | 81 | 108 | 83 | 109 | 81 | 94 | 108 | 98 | 70 |
| G4 | GC content | 51.7 | 51.8 | 51.8 | 51.7 | 54.7 | 52.7 | 51.9 | 51.9 | 54.6 |
|
| ||||||||||
| Subsystem feature | ||||||||||
| F1 | Cofactors, vitamins, prosthetic groups, and pigments | 181 | 179 | 180 | 182 | 175 | 200 | 179 | 183 | 180 |
| F2 | Cell wall and capsule | 43 | 44 | 44 | 43 | 41 | 44 | 46 | 47 | 42 |
| F3 | Virulence, disease, and defense | 63 | 53 | 55 | 53 | 50 | 74 | 52 | 59 | 39 |
| F4 | Potassium metabolism | 17 | 17 | 17 | 17 | 15 | 17 | 17 | 17 | 14 |
| F5 | Photosynthesis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F6 | Miscellaneous | 33 | 33 | 33 | 35 | 28 | 34 | 35 | 35 | 19 |
| F7 | Phages, prophages, transposable elements, and plasmids | 25 | 7 | 10 | 14 | 5 | 42 | 15 | 26 | 13 |
| F8 | Membrane transport | 91 | 70 | 72 | 74 | 77 | 86 | 67 | 106 | 79 |
| F9 | Iron acquisition and metabolism | 53 | 53 | 54 | 46 | 27 | 37 | 39 | 42 | 26 |
| F10 | RNA metabolism | 56 | 57 | 57 | 57 | 55 | 59 | 57 | 58 | 60 |
| F11 | Nucleosides and nucleotides | 86 | 80 | 80 | 82 | 79 | 87 | 78 | 81 | 84 |
| F12 | Protein metabolism | 231 | 253 | 236 | 262 | 255 | 250 | 210 | 247 | 198 |
| F13 | Cell division and cell cycle | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| F14 | Motility and chemotaxis | 16 | 101 | 97 | 15 | 101 | 98 | 96 | 100 | 89 |
| F15 | Regulation and cell signaling | 66 | 65 | 68 | 61 | 54 | 88 | 63 | 68 | 51 |
| F16 | Secondary metabolism | 21 | 21 | 21 | 23 | 4 | 21 | 21 | 22 | 5 |
| F17 | DNA metabolism | 85 | 78 | 78 | 74 | 83 | 103 | 74 | 102 | 89 |
| F18 | Fatty acids, lipids, and isoprenoids | 56 | 55 | 61 | 54 | 58 | 58 | 56 | 56 | 52 |
| F19 | Nitrogen metabolism | 40 | 40 | 40 | 51 | 45 | 50 | 40 | 40 | 46 |
| F20 | Dormancy and sporulation | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| F21 | Respiration | 114 | 111 | 114 | 119 | 106 | 135 | 116 | 117 | 103 |
| F22 | Stress response | 87 | 86 | 86 | 96 | 93 | 100 | 89 | 93 | 94 |
| F23 | Metabolism of aromatic compounds | 9 | 9 | 9 | 20 | 35 | 17 | 6 | 13 | 34 |
| F24 | Amino acids and derivatives | 362 | 364 | 368 | 385 | 331 | 399 | 349 | 369 | 336 |
| F25 | Sulfur metabolism | 26 | 26 | 28 | 23 | 22 | 23 | 21 | 21 | 26 |
| F26 | Phosphorus metabolism | 31 | 31 | 31 | 31 | 29 | 31 | 30 | 31 | 30 |
| F27 | Carbohydrates | 376 | 387 | 378 | 411 | 367 | 425 | 387 | 394 | 396 |
a This study.
3.3.2. Presence of Arsenic-Resistant Genes
From the draft genome of isolate, Citrobacter youngae IITK SM2 As-resistant genes were identified (Figure 5(a)). These genes belongs to the ars operon system [33, 35, 79]. Genes corresponding to arsC, arsB, arsA, arsD, arsR, and arsH were present in the g-DNA of this isolate. These genes have specific functions. The arsenate reductase arsC, which encodes a protein of smaller molecular weight (13-15 kDa), belongs to the thioredoxin superfamily and mediates the reduction of As(V) to As(III) in the cytoplasm [79, 80]. The ars operon further contains an efflux pump (arsB) specific to arsenic, which encodes arsenite permease and extrudes As(III) out of the cell [81]. Resistance to As(V) and As(III) is provided by the expressions of arsC and arsB genes, which are controlled by a transcriptional repressor, arsR [80, 82]. The presence of these As-resistant genes could be the possible reason of such high MICs of As(V) and As(III) observed in strain IITK SM2. In addition to these genes, As-operon also contains arsD and arsA. It is known that arsD exhibits a weak As(III)-responsive transcriptional repressor activity [83], and the gene arsA provides higher resistance to elevated levels of As(III) by encoding intracellular ATPase, which forms a dimer with arsB [79].
Figure 5.
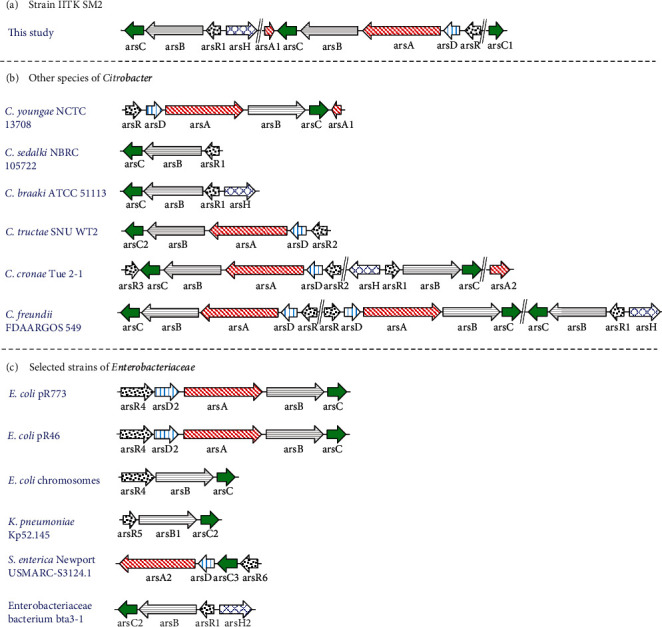
Comparison of arsenic-resistant genes identified in strain IITK SM2 with genes of other reference organisms: arsenic operon system in (a) strain IITK SM2, (b) other Citrobacter species, and (c) selected strains of Enterobacteriaceae a genus other than Citrobacter. The whole-genome sequence of each strain was considered for representing this schematic. Numbers appended after the name of some As-resistant genes indicate the difference in nucleotides. The gene bank or accession number of Citrobacter species used in this analysis is shown in Table S4 of the Supporting Information.
The arsenic-resistant gene arsH is a NADPH-dependent flavin mononucleotide reductase [84] and was also identified in the g-DNA of IITK SM2. It was reported that the presence of an arsH gene increased resistance to inorganic As species (iAsV/III) in some bacteria [85, 86]. However, some studies proved that neither overexpression nor mutation of arsH protein provided resistance to inorganic As in Thiobacillus ferrooxidans [87] and cyanobacterium Synechocystis sp. PCC 6803 [88]. The exact function of the arsH gene remains unclear. Recently, a study showed that arsH detoxified organoarsenic compounds like MMA(III) and aromatic arsenic species by their oxidation to MMA(V) [89]. These As-resistant genes were arranged in three distinct ways, which contained (1) arsCBRH, (2) arsACBADR, and (3) an independent arsC gene in strain IITK SM2 (Figure 5(a)). It is possible that the presence of these genes might have a role in high arsenic resistance exhibited by IITK SM2.
3.3.3. Comparison of As-Resistant Genes in Strain IITK SM2 with Genes in Reference Organisms
The distribution of ars genes identified in the isolate differed from other type strains of Enterobacteriaceae and especially of Citrobacter species (Figure 5). C. youngae NCTC 13708 was the closest to IITK SM2, which contained the unique six gene operon (arsACBADR) but was regulated in the opposite direction compared to IITK SM2 (Figure 5(b)). Such a sextet gene cluster was neither observed for any other Citrobacter species nor for any mentioned pioneer strains of the Enterobacteriaceae family. Besides this, operon IITK SM2 contained the arsCBRH operon and an independent arsC gene (Figure 5(a)), which were absent in NCTC 13708. These differences suggest that our isolate could be even more effective in arsenic resistance than NCTC 13708. Although arsCBRH operon was present in other Citrobacter species, such as C. braakii ATCC 51113T, C. freundii FDAARGOS 549T, and strain bta3-1T, the independent arsC gene was not identified in any of the chosen reference strains (Figures 5(b) and 5(c)). Earlier studies have referred to such proteins as “fusion proteins,” which if functional could provide evolutionary advantage in sensing and/or detoxifying As(III) in the environment [90, 91]. Other reference chromosomes or plasmids mostly contain the five-gene arsCBADR operon identified in C. tructae SNU WT2T, C. freundii FDAARGOS 549T and E. coli (R773 and R46), or the three-gene arsRBC operon present in C. sedalki NBRC 105722T, K. pneumoniae Kp52.145T, and E. coli chromosomes. An exception in the arrangement of As metabolizing genes in C. cronae Tue2-1T was observed, where two transacting repressors were identified (arsRCBDAR). These arsCBADR and arsRBC were not identified in strain IITK SM2. Even though a dDDH value of ~84% suggested that the other strain of C. youngae, CCUG 30791, was closer to IITK SM2, no As-metabolizing genes were identified in this type strain. The identified unique arrangement of ars genes in this isolate was consistent with the possibility that IITK SM2 is a novel As-resistant strain of C. youngae.
3.3.4. Presence of Three Arsenate-Reductase (arsC) Genes
The isolate IITK SM2 contained three different arsC genes designated as arsC1, arsC2, and arsC3 (Figure 6). The numbers of amino acids in these arsenate reductases were 119, 141, and 141, respectively. The alignment of their protein sequences suggested that 41 amino acids were completely conserved in these three genes (Figure 6(a)). Furthermore, the protein sequences of arsC2 and arsC3 showed ~83% similarity. The phylogenetic tree based on NJ method suggested that the smaller arsenate-reductase, arsC1, clustered with S. enterica (GAS71778.1), whereas arsC2 and arsC3 were not clustered with any arsC's of the selected bacterial strains. However, arsC2 and arsC3 were clustered together (Figure 6(b)). This analysis suggested that the smaller arsenate reductase in strain IITK SM2 might have evolved from Salmonella-type strains, whereas the larger arsC's might be native of Citrobacter. The presence of these arsenate reductases along with arsB, arsA, arsD, arsR, and arsH in strain IITK SM2 could be responsible for its high arsenic resistance and for As(V) reduction to As(III) (Figure S5). Detailed mechanism and kinetics of microbially mediated arsenate reduction by this isolate would help to understand the role of IITK SM2 on arsenic speciation in groundwater.
Figure 6.
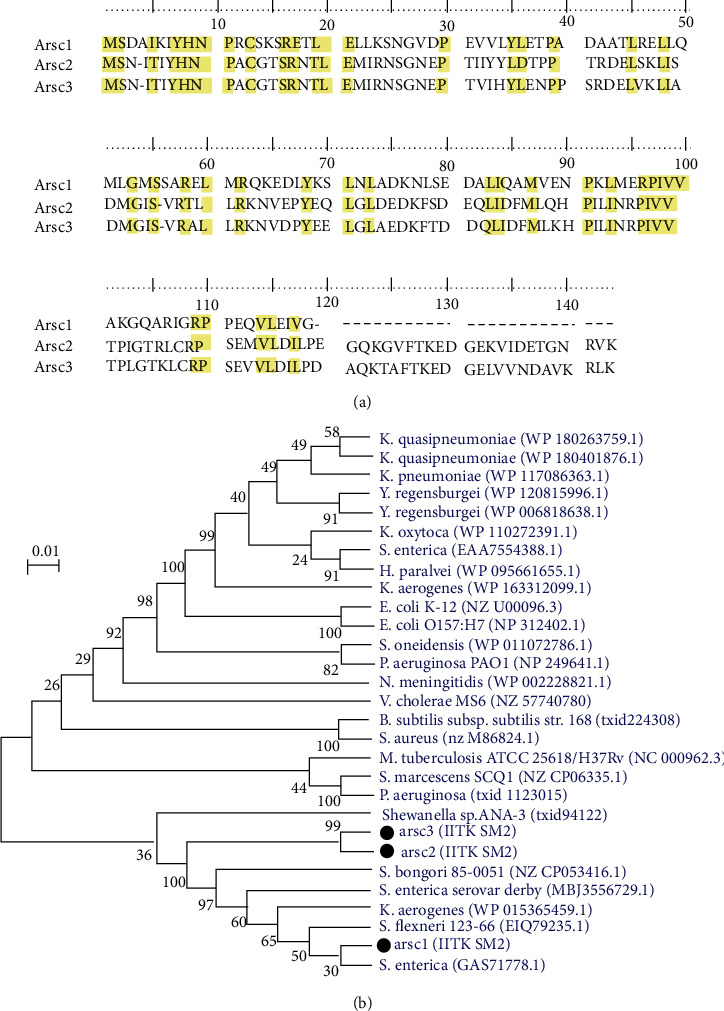
Multiple alignment (a) and phylogeny (b) of arsenate-reductase gene (arsC) sequences identified in the strain Citrobacter youngae IITK SM2. Multiple alignments of arsC sequences (arsC1, arsC2, and arsC3) of IITK SM2 were performed using CLUSTAL_W. Nucleotides conserved in all the three arsC genes are highlighted in yellow. The neighbor-joining method [57] was used for developing distance-based phylogeny of arsC genes. For this phylogeny, arsC genes in strain IITK SM2 were compared with other As(V)-reducing organisms (excluding strains of Citrobacter). Percentage bootstrap values corresponding to 1000 replicates are shown next to the branches. The tree shown in (b) was drawn to scale.
4. Conclusions
The Gram-negative, rod-shaped facultative anaerobe bacterial strain IITK SM2 could survive under high concentrations of dissolved As and could reduce As(V) to As(III). Apart from dissolved As, this isolate also showed resistance to some of the other heavy metals found in groundwater, such as Fe(III), Mn(II), Zn(II), Ba(II), and Cr(VI). Enhanced bacterial growth was observed in the presence of dissolved As(III) and As(V), but the former was more toxic to the cells than the latter. The IITK SM2 is a novel strain of Citrobacter youngae and is different from other strains in terms of a number of critical subsystem features related to membrane transporters, virulence and defense, motility, protein metabolism, and phages, prophages, and transposable elements. The presence of As-metabolizing genes such as arsC, arsB, arsA, arsD, arsR, and arsH were identified in the genome of this strain. A unique clustering of As-resistant genes was also found in the g-DNA of IITK SM2 as (1) arsCBRH, (2) arsACBADR, and (3) an independent arsC gene, which was not observed in any other Citrobacter species and selected strains of Enterobacteriaceae family. Furthermore, two different varieties of arsC genes were identified, where one As(V)-reductase gene might have evolved from Salmonella and the other may have been native to Citrobacter. The information presented in this study could contribute to the mechanistic understanding of the biogeochemical processes that control elevated arsenic prevalence in groundwater, which could help in developing long term in situ mobilization techniques for As-remediation.
Acknowledgments
We acknowledge Mibiome Therapeutics LLP and Macrogen genomics, India, for sequencing and analysis of the genome. We thank MHRD for providing teaching assistantships to Prem Anand Murugan and Hariharan Vedi Chinnasamy for their doctoral studies. We acknowledge the central facility of the Center for Environmental Sciences and Engineering and Department of Biological Sciences and Bioengineering for various facilities provided to carry out this work. Advanced Centre for Materials science (ACMS) at IITK is acknowledged for Scanning Electron Microscope (SEM) facility. We thank all the members of Environmental Geochemistry laboratory, and Microbial Adaptation and Chromatin Remodeling Laboratory at IITK for helping in miscellaneous purpose, and especially, Chitral Chatterjee (Ph.D. student) is acknowledged for providing useful suggestions in the manuscript. This work has been supported by the Department of Science and Technology, Government of India, [Grant No. DST/TM/WTI/2K15/109(G)] awarded to Dr. Abhas Singh under the Water Technology Initiative. We thank DBT-IYBA for the fellowship provided to Dr. Saravanan Matheshwaran. The draft genome sequencing was funded through a research grant of Prime Minister's Research Fellowship provided to Akshat Verma by the Ministry of Human Resource and Development (MHRD).
Abbreviations
- As:
Arsenic
- As(III):
Arsenite
- As(V):
Arsenate
- IGP:
Indo-Gangetic plain
- g-DNA:
Genomic DNA
- WGS:
Whole-genome sequencing
- LB:
Lysogeny broth
- GC:
Guanine-cytosine
- MSM:
Minimum salt media
- MIC:
Minimum inhibitory concentration
- MHB:
Mueller Hinton broth
- SEM:
Scanning electron microscopy
- ICP-MS:
Inductively coupled plasma mass spectroscopy
- IC-ICP-MS:
Ion chromatography coupled with inductively coupled plasma mass spectroscopy
- NCBI:
National Center for Biotechnology Information
- BLAST:
Basic local alignment search tool
- TOC:
Total organic carbon
- DIC:
Dissolved inorganic carbon
- dDDH:
Digital DNA-DNA hybridization
- TYGS:
Type (strain) genome server
- NJ:
Neighbor joining
- PGAP:
Prokaryotic genome annotation pipeline
- OM:
Organic matter
- MMA(III):
Monomethylated arsenous acid
- MMA(V):
Monomethylated arsenic acid
- RAST:
Rapid annotations using subsystems technology.
Contributor Information
Abhas Singh, Email: abhas@iitk.ac.in.
Saravanan Matheshwaran, Email: saran@iitk.ac.in.
Data Availability
The GenBank accession number for the 16S rRNA sequence is MZ477215. The accession number of the whole-genome shotgun project of Citrobacter youngae IITK SM2 registered at DDBJ/ENA/GenBank is JAGIYN000000000. In this study, the version described is JAGIYN000000000.1. Other data are included within the manuscript.
Conflicts of Interest
The authors declare that no competing financial interests exist, and this work has been carried out in compliance with ethical standards.
Authors' Contributions
Akshat Verma, Abhas Singh, and Saravanan Matheshwaran contributed to the conceptualization; Akshat Verma, Prem Anand Murugan, and Hariharan Vedi Chinnasamy contributed to the methodology; Akshat Verma and Saravanan Matheshwaran contributed to the software; Akshat Verma, Abhas Singh, and Saravanan Matheshwaran contributed to the validation; Akshat Verma, Hariharan Vedi Chinnasamy, and Saravanan Matheshwaran contributed to the formal analysis; Akshat Verma, Saravanan Matheshwaran, and Abhas Singh contributed to the investigation; Saravanan Matheshwaran, Abhas Singh, Akshat Verma, and Hariharan Vedi Chinnasamy contributed to the resources; Akshat Verma, Prem Anand Murugan, and Saravanan Matheshwaran contributed to the data curation; Akshat Verma contributed to the writing–original draft preparation; Saravanan Matheshwaran and Abhas Singh contributed to the writing–review and editing; and Akshat Verma and Prem Anand Murugan contributed to the visualization of the study. Saravanan Matheshwaran and Abhas Singh supervised the study. Saravanan Matheshwaran and Abhas Singh were responsible for project administration of the study. Saravanan Matheshwaran, Abhas Singh, and Akshat Verma were responsible for the funding acquisition for the study.
Supplementary Materials
A Supporting Information file of 24 pages containing six sections, five figures, and six tables is available online. This file contains details of sample collections, analysis techniques, experimental procedure, phylogenetic analysis based on recN sequence, map of sampling location, composition of media used, results from biochemical experiments, details of chemicals and media used, method detection limits of various techniques used, and comparison of strain IITK SM2 with other arsenate-reducers identified in the literature.
References
- 1.Werkman C. H., Gillen G. F. Bacteria producing trimethylene glycol. Journal of Bacteriology . 1932;23(2):167–182. doi: 10.1128/jb.23.2.167-182.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeolu M., Alnajar S., Naushad S., Gupta R. S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales Ord. Nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. Nov., Pectobacteriaceae fam. Nov., Yersiniaceae fam. Nov., Hafniaceae fam. Nov., Morganellaceae fam. Nov., and Budviciaceae fam. Nov. International Journal of Systematic and Evolutionary Microbiology . 2016;66(12):5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 3.Wang J. T., Chang S. C. Citrobacter species. 2016. http://www.antimicrobe.org/b93.asp .
- 4.Schloissnig S., Arumugam M., Sunagawa S., et al. Genomic variation landscape of the human gut microbiome. Nature . 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J. T., Chang S. C., Chen Y. C., Luh K. T. Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods. Journal of Microbiology, Immunology and Infection . 2000;33(4):258–262. [PubMed] [Google Scholar]
- 6.Rogers L., Power K., Gaora P. O., Fanning S. Escherichia coli and other Enterobacteriaceae: occurrence and detection. In: Caballero B., Finglas P. M., Toldrá F., editors. Encyclopedia of Food and Health . Oxford, UK: Academic Press; 2016. pp. 545–551. [Google Scholar]
- 7.Whalen J. G., Mully T. W., English J. C. Spontaneous Citrobacter freundii infection in an immunocompetent patient. Archives of Dermatology . 2007;143(1):115–126. doi: 10.1001/archderm.143.1.124. [DOI] [PubMed] [Google Scholar]
- 8.Crawford S. E., Daum R. S. Bacterial pneumonia, lung abscess, and empyema. Pediatric Respiratory Medicine . 2008;2:501–553. [Google Scholar]
- 9.Chen K. J., Chen T. J., Sue Y. M. Citrobacter youngae and Pantoea agglomerans peritonitis in a peritoneal dialysis patient. Peritoneal Dialysis International . 2013;33(3):336–337. doi: 10.3747/pdi.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matyar F. Antibiotic and heavy metal resistance in bacteria isolated from the eastern Mediterranean Sea coast. Bulletin of Environmental Contamination and Toxicology . 2012;89(3):551–556. doi: 10.1007/s00128-012-0726-4. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Huang N., Shao J., Hu M., Zhao Y., Huo M. Coupling heavy metal resistance and oxygen flexibility for bioremoval of copper ions by newly isolated _Citrobacter freundii_ JPG1. Journal of Environmental Management . 2018;226:194–200. doi: 10.1016/j.jenvman.2018.08.042. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson J. F., Gavis J. A review of the arsenic cycle in natural waters. Water Research . 1972;6(11):1259–1274. doi: 10.1016/0043-1354(72)90052-8. [DOI] [Google Scholar]
- 13.International Agency for Research on Cancer. A review of human carcinogens: metals, arsenic, fibres and dusts. International Agency for Research on Cancer: Monographs on the Evaluation of Carcinogenic Risks to Humans . 2012;100 [PMC free article] [PubMed] [Google Scholar]
- 14.Smedley P. L., Kinniburgh D. G. A review of the source, behaviour and distribution of arsenic in natural waters. Applied Geochemistry . 2002;17(5):517–568. doi: 10.1016/S0883-2927(02)00018-5. [DOI] [Google Scholar]
- 15.Bissen M., Frimmel F. H. Arsenic—a review. Part I: occurrence, toxicity, speciation, mobility. Acta Hydrochimica et Hydrobiologica . 2003;31(1):9–18. doi: 10.1002/aheh.200390025. [DOI] [Google Scholar]
- 16.WHO. Guidelines for drinking-water quality: fourth edition incorporating the first addendum . Geneva, Switzerland: World Health Organization; 2017. [PubMed] [Google Scholar]
- 17.Subcommittee on Arsenic in Drinking Water, National Research Council (US) Arsenic in Drinking Water . Washington, DC, USA: Joseph Henry Press; 1999. [Google Scholar]
- 18.Oremland R. S., Stolz J. F. Arsenic, microbes and contaminated aquifers. Trends in Microbiology . 2005;13(2):45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Rosen B. P. Families of arsenic transporters. Trends in Microbiology . 1999;7(5):207–212. doi: 10.1016/S0966-842X(99)01494-8. [DOI] [PubMed] [Google Scholar]
- 20.Pace C., Banerjee T. D., Welch B., Khalili R., Dagda R. K., Angermann J. Monomethylarsonous acid, but not inorganic arsenic, is a mitochondria-specific toxicant in vascular smooth muscle cells. Toxicology In Vitro . 2016;35:188–201. doi: 10.1016/j.tiv.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Styblo M., del Razo L., Vega L., et al. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Archives of Toxicology . 2000;74(6):289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 22.Braman R. S., Foreback C. C. Methylated forms of arsenic in the environment. Science . 1973;182(4118):1247–1249. doi: 10.1126/science.182.4118.1247. [DOI] [PubMed] [Google Scholar]
- 23.Sharma V. K., Sohn M. Aquatic arsenic: toxicity, speciation, transformations, and remediation. Environment International . 2009;35(4):743–759. doi: 10.1016/j.envint.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan V. S., Nickson R. T., Chauhan D., Iyengar L., Sankararamakrishnan N. Ground water geochemistry of Ballia district, Uttar Pradesh, India and mechanism of arsenic release. Chemosphere . 2009;75(1):83–91. doi: 10.1016/j.chemosphere.2008.11.065. [DOI] [PubMed] [Google Scholar]
- 25.Nickson R., McArthur J., Burgess W., Ahmed K. M., Ravenscroft P., Rahman M. Arsenic poisoning of Bangladesh groundwater. Nature . 1998;395(6700):p. 338. doi: 10.1038/26387. [DOI] [PubMed] [Google Scholar]
- 26.Nickson R. T., McArthur J. M., Ravenscroft P., Burgess W. G., Ahmed K. M. Mechanism of arsenic release to groundwater, Bangladesh and West Bengal. Applied Geochemistry . 2000;15(4):403–413. doi: 10.1016/S0883-2927(99)00086-4. [DOI] [Google Scholar]
- 27.Oremland R. S., Stolz J. F. The ecology of arsenic. Science . 2003;300(5621):939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 28.Ahmann D., Roberts A. L., Krumholz L. R., Morel F. M. Microbe grows by reducing arsenic. Nature . 1994;371(6500):p. 750. doi: 10.1038/371750a0. [DOI] [PubMed] [Google Scholar]
- 29.Laverman A. M., Blum J. S., Schaefer J. K., Phillips E., Lovley D. R., Oremland R. S. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Applied and Environmental Microbiology . 1995;61(10):3556–3561. doi: 10.1128/aem.61.10.3556-3561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green H. H. Description of a bacterium which oxidizes arsenite to arsenate, and of one which reduces arsenate to arsenite, isolated from a cattle-dipping Etank. South African Journal of Science . 1918;14:465–467. [Google Scholar]
- 31.Oremland R. S., Hoeft S. E., Santini J. M., Bano N., Hollibaugh R. A., Hollibaugh J. T. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Applied and Environmental Microbiology . 2002;68(10):4795–4802. doi: 10.1128/AEM.68.10.4795-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborne T. H., McArthur J. M., Sikdar P. K., Santini J. M. Isolation of an arsenate-respiring bacterium from a redox front in an arsenic-polluted aquifer in West Bengal, Bengal Basin. Environmental Science & Technology . 2015;49(7):4193–4199. doi: 10.1021/es504707x. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay R., Rosen B., Phung L. T., Silver S. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiology Reviews . 2002;26(3):311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 34.Silver S., Phung L. T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Applied and Environmental Microbiology . 2005;71(2):599–608. doi: 10.1128/AEM.71.2.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver S., Phung L. T., Rosen B. P. Arsenic metabolism: resistance, reduction, and oxidation. In: Frankenberger W. T., editor. Environmental Chemistry of Arsenics . New York: Marcel Dekker; 2001. pp. 247–272. [Google Scholar]
- 36.Nilling J. J., Verma A., Singh A. Relative kinetics of precipitation and adsorption of arsenic(V) in systems with dissolved iron(II) Barcelona Spain: Goldschmidt Abstracts; 2020. [Google Scholar]
- 37.Lovley D. R., Goodwin S. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochimica et Cosmochimica Acta . 1988;52(12):2993–3003. doi: 10.1016/0016-7037(88)90163-9. [DOI] [Google Scholar]
- 38.Lindberg R. D., Runnells D. D. Ground water redox reactions: an analysis of equilibrium state applied to Eh measurements and geochemical modeling. Science . 1984;225(4665):925–927. doi: 10.1126/science.225.4665.925. [DOI] [PubMed] [Google Scholar]
- 39.Berner R. A. A new geochemical classification of sedimentary environments. Journal of Sedimentary Research . 1981;51(2):359–365. doi: 10.1306/212F7C7F-2B24-11D7-8648000102C1865D. [DOI] [Google Scholar]
- 40.Thorstenson D. C. The concept of electron activity and its relation to redox potentials in aqueous geochemical systems . Virginia U.S.A: US Department of the Interior, Geological Survey; 1984. [Google Scholar]
- 41.Stumm W., Morgan J. Aquatic Chemistry: An Introduction Emphasizing Chemical Equilibria in Natural Waters . New York: John Wiley and Sons; 1981. [Google Scholar]
- 42.Benjamin M. M. Water Chemistry . Waveland Press; 2014. [Google Scholar]
- 43.Andrews J. M. Determination of minimum inhibitory concentrations. Journal of Antimicrobial Chemotherapy . 2001;48(Supplement 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 44.Simeonova D. D., Lièvremont D., Lagarde F., Muller D. A. E., Groudeva V. I., Lett M. C. Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiology Letters . 2004;237(2):249–253. doi: 10.1111/j.1574-6968.2004.tb09703.x. [DOI] [PubMed] [Google Scholar]
- 45.Andrews S. FastQC: a quality control tool for high throughput sequence data . Cambridge, United Kingdom: Babraham Bioinformatics; 2010. [Google Scholar]
- 46.Bolger A. M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics . 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A., Nurk S., Antipov D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology . 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics . 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatusova T., DiCuccio M., Badretdin A., et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Research . 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods . 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff J. P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature Communications . 2019;10(1):p. 2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aziz R. K., Bartels D., Best A. A., et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics . 2008;9(1):1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Coleman-Derr D., Chen G., Gu Y. Q. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research . 2015;43(W1):W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson D. A., Cavanaugh M., Clark K., et al. GenBank. Nucleic Acids Research . 2012;41(D1):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows Interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research . 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution . 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 57.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution . 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 58.Zuckerkandl E., Pauling L. Evolving Genes and Proteins . Academic Press; 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
- 59.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution . 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.EFSA. Panel on contaminants in the food chain (CONTAM) Scientific Opinion on Arsenic in Food EFSA Journal . 2009;7:1351–1355. [Google Scholar]
- 61.Yousefsani B. S., Pourahmad J., Hosseinzadeh H. The mechanism of protective effect of crocin against liver mitochondrial toxicity caused by arsenic III. Toxicology Mechanisms and Methods . 2018;28(2):105–114. doi: 10.1080/15376516.2017.1368054. [DOI] [PubMed] [Google Scholar]
- 62.Bhattacharyya R., Chatterjee D., Nath B., Jana J., Jacks G., Vahter M. High arsenic groundwater: mobilization, metabolism and mitigation–an overview in the Bengal Delta Plain. Molecular and Cellular Biochemistry . 2003;253(1):347–355. doi: 10.1023/A:1026001024578. [DOI] [PubMed] [Google Scholar]
- 63.Sodhi K. K., Kumar M., Agrawal P. K., Singh D. K. Perspectives on arsenic toxicity, carcinogenicity and its systemic remediation strategies. Environmental Technology & Innovation . 2019;16, article 100462 doi: 10.1016/j.eti.2019.100462. [DOI] [Google Scholar]
- 64.Spayd S. E., Robson M. G., Xie R., Buckley B. T. Importance of arsenic speciation in populations exposed to arsenic in drinking water. Human and Ecological Risk Assessment: An International Journal . 2012;18(6):1271–1291. doi: 10.1080/10807039.2012.722824. [DOI] [Google Scholar]
- 65.Acharyya S. K., Chakraborty P., Lahiri S., Raymahashay B. C., Guha S., Bhowmik A. Arsenic poisoning in the Ganges delta. Nature . 1999;401(6753):p. 545. doi: 10.1038/44052. [DOI] [PubMed] [Google Scholar]
- 66.Harvey C. F., Swartz C. H., Badruzzaman A. B. M., et al. Arsenic mobility and groundwater extraction in Bangladesh. Science . 2002;298(5598):1602–1606. doi: 10.1126/science.1076978. [DOI] [PubMed] [Google Scholar]
- 67.Fendorf S., Michael H. A., Geen A. V. Spatial and temporal variations of groundwater arsenic in south and Southeast Asia. Science . 2010;328(5982):1123–1127. doi: 10.1126/science.1172974. [DOI] [PubMed] [Google Scholar]
- 68.Postma D., Trang P. T., Sø H. U., et al. A model for the evolution in water chemistry of an arsenic contaminated aquifer over the last 6000 years, Red River floodplain, Vietnam. Geochimica et Cosmochimica Acta . 2016;195:277–292. doi: 10.1016/j.gca.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Islam F. S., Gault A. G., Boothman C., et al. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature . 2004;430(6995):68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- 70.Bhattacharya P., Claesson M., Bundschuh J., et al. Distribution and mobility of arsenic in the Río Dulce alluvial aquifers in Santiago del Estero Province, Argentina. Science of the Total Environment . 2006;358(1):97–120. doi: 10.1016/j.scitotenv.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 71.Bhowmick S., Nath B., Halder D., et al. Arsenic mobilization in the aquifers of three physiographic settings of West Bengal, India: understanding geogenic and anthropogenic influences. Journal of Hazardous Materials . 2013;262:915–923. doi: 10.1016/j.jhazmat.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 72.Chauhan V. S., Yunus M., Sankararamakrishnan N. Geochemistry and mobilization of arsenic in Shuklaganj area of Kanpur–Unnao district, Uttar Pradesh, India. Environmental Monitoring and Assessment . 2012;184(8):4889–4901. doi: 10.1007/s10661-011-2310-5. [DOI] [PubMed] [Google Scholar]
- 73.Kocar B. D., Polizzotto M. L., Benner S. G., et al. Integrated biogeochemical and hydrologic processes driving arsenic release from shallow sediments to groundwaters of the Mekong delta. Applied Geochemistry . 2008;23(11):3059–3071. doi: 10.1016/j.apgeochem.2008.06.026. [DOI] [Google Scholar]
- 74.Anderson C. R., Cook G. M. Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Current Microbiology . 2004;48(5):341–347. doi: 10.1007/s00284-003-4205-3. [DOI] [PubMed] [Google Scholar]
- 75.Guo H., Liu Z., Ding S., Hao C., Xiu W., Hou W. Arsenate reduction and mobilization in the presence of indigenous aerobic bacteria obtained from high arsenic aquifers of the Hetao basin, Inner Mongolia. Environmental Pollution . 2015;203:50–59. doi: 10.1016/j.envpol.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 76.Fulladosa E., Murat J. C., Villaescusa I. Effect of cadmium(II), Chromium(VI), and arsenic(V) on long-term viability- and growth-inhibition assays using Vibrio fischeri marine bacteria. Archives of Environmental Contamination and Toxicology . 2005;49(3):299–306. doi: 10.1007/s00244-004-0170-5. [DOI] [PubMed] [Google Scholar]
- 77.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics . 2016;32(6):929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences . 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosen B. P. Biochemistry of arsenic detoxification. FEBS Letters . 2002;529(1):86–92. doi: 10.1016/S0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 80.Stolz J. F., Basu P., Santini J. M., Oremland R. S. Arsenic and selenium in microbial metabolism. Annual Review of Microbiology . 2006;60(1):107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 81.Meng Y. L., Liu Z., Rosen B. P. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. Journal of Biological Chemistry . 2004;279(18):18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- 82.Silver S., Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiological Reviews . 1992;56(1):195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y., Rosen B. P. Metalloregulatory properties of the ArsD repressor. Journal of Biological Chemistry . 1997;272(22):14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- 84.Ye J., Yang H. C., Rosen B. P., Bhattacharjee H. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Letters . 2007;581(21):3996–4000. doi: 10.1016/j.febslet.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neyt C., Iriarte M., Thi V. H., Cornelis G. R. Virulence and arsenic resistance in yersiniae. Journal of Bacteriology . 1997;179(3):612–619. doi: 10.1128/jb.179.3.612-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryan D., Colleran E. Arsenical resistance in the IncHI2 plasmids. Plasmid . 2002;47(3):234–240. doi: 10.1016/S0147-619X(02)00012-4. [DOI] [PubMed] [Google Scholar]
- 87.Butcher B. G., Deane S. M., Rawlings D. E. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Applied and Environmental Microbiology . 2000;66(5):1826–1833. doi: 10.1128/AEM.66.5.1826-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.López-Maury L., Florencio F. J., Reyes J. C. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. Journal of Bacteriology . 2003;185(18):5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen J., Bhattacharjee H., Rosen B. P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Molecular Microbiology . 2015;96(5):1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ben Fekih I., Zhang C., Li Y. P., et al. Distribution of arsenic resistance genes in prokaryotes. Frontiers in Microbiology . 2018;9(2473) doi: 10.3389/fmicb.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu B., Song J., Beitz E. Novel channel enzyme fusion proteins confer arsenate resistance. Journal of Biological Chemistry . 2010;285(51):40081–40087. doi: 10.1074/jbc.M110.184457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Supporting Information file of 24 pages containing six sections, five figures, and six tables is available online. This file contains details of sample collections, analysis techniques, experimental procedure, phylogenetic analysis based on recN sequence, map of sampling location, composition of media used, results from biochemical experiments, details of chemicals and media used, method detection limits of various techniques used, and comparison of strain IITK SM2 with other arsenate-reducers identified in the literature.
Data Availability Statement
The GenBank accession number for the 16S rRNA sequence is MZ477215. The accession number of the whole-genome shotgun project of Citrobacter youngae IITK SM2 registered at DDBJ/ENA/GenBank is JAGIYN000000000. In this study, the version described is JAGIYN000000000.1. Other data are included within the manuscript.


