Abstract
Neutral sphingomyelinases have an important role in generation of ceramide and phosphorylcholine from sphingomyelins which then act as secondary messengers in various signaling pathways of the cellular machinery. They function ubiquitously with a predominant role in the central nervous system. Neutral sphingomyelinase type 3, encoded by SMPD4 gene has recently been reported to cause a severe autosomal recessive neurodevelopmental disorder with congenital arthrogryposis and microcephaly. We report a 22-month-old girl having characteristic features of neurodevelopmental delay, prenatal onset growth failure, arthrogryposis, microcephaly and brain anomalies including severe hypomyelination, simplified gyral pattern and hypoplasia of corpus callosum and brainstem. Additionally, she was noted to have nystagmus and visual impairment secondary to macular dystrophy and retinal pigment epithelial stippling at posterior pole. Copy number variant analysis from trio whole exome sequencing (ES) enabled identification of a homozygous 11kb deletion encompassing exons 18 −20 of SMPD 4 gene, confirming the diagnosis of SMPD4-related disorder in her.
Keywords: SMPD4, neurodevelopmental disorder, arthrogryposis, growth failure, microcephaly, hypomyelination, brain anomalies, visual impairment
Introduction
Sphingomyelin (SM) is the most abundant sphingolipid, expressed ubiquitously with high expression in the CNS. It is one of the major components of plasma membrane, maintaining cellular integrity [1]. SM metabolism is tightly regulated by sphingomyelin synthases (SMSs) and sphingomyelinases (SMases). Hydrolysis of SM to ceramide and phosphorylcholine is performed by SMases, of which there are three types: (i) Acid sphingomyelinase (a-SMase) encoded by SMPD1, having two isoforms: lysosomal l-SMase and secretory s-SMase isoform; (ii) Four neutral sphingomyelinases (n-SMase 1–4), encoded by SMPD2–5 genes; and (iii) Alkaline SMase (alk-SMase) encoded by ENPP7, secreted in the intestine where it digests the exogenous sphingomyelins [2].
Recently, bi-allelic pathogenic variants in SMPD4, have been identified to cause a severe autosomal recessive neurodevelopmental disorder with arthrogryposis, microcephaly and structural brain anomalies [3, 4]. Here, we report a child with similar clinical findings harboring a homozygous partial deletion of SMPD4.
Case report
We ascertained a 22-months-old female with chief complaints of profound global developmental delay, severe failure to thrive and congenital contractures of multiple joints. The proband was born at term as small for gestational age (birth weight- 2 kg; −2.5 SD), with feeble cry. Multiple contractures of joints of both upper and lower limbs were noted. She had feeding difficulty, requiring nasogastric feeds till 3 months of age. She did not achieve any motor or cognitive developmental milestones. She had visual inattention and no speech development. Her feeding continued to be poor, with only liquid diet fed by a spoon. There was no history of seizures or any other abnormal movements.
In family, a small for gestation baby girl (Fig 1A III.1) was born to parents earlier who was noted to have contractures of multiple joints at birth, a feeble cry, feeding difficulty, frequent vomiting and severe failure to thrive. She died at 5 months of age owing to respiratory difficulty.
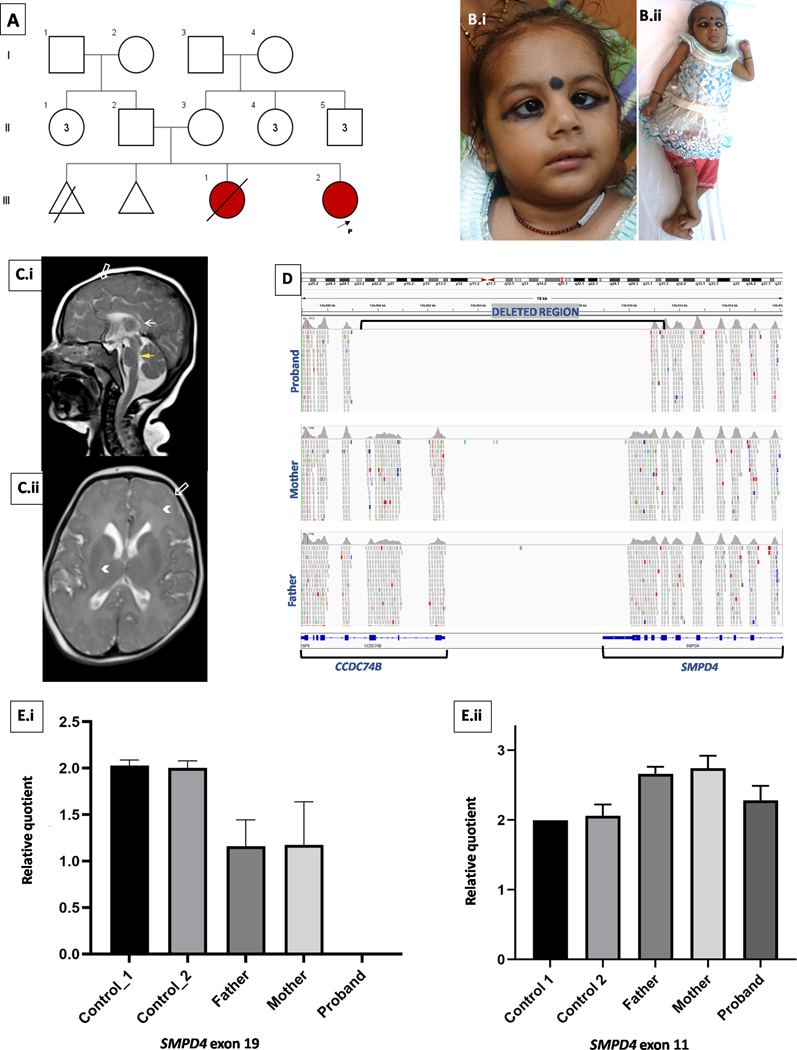
Fig 1.
Pedigree of the family with proband marked with arrow, III2 (A). Clinical photographs of the proband showing strabismus, epicanthic folds (B.i.) hands and ankle contractures (B.ii). Neuroimaging of brain at 1 year 10 months showed hypoplasia of the corpus callosum and pons (yellow/white arrow, C.i), simplified gyral pattern (open arrow, C.i & Cii) and T2W hyperintensities of cerebral white matter suggesting delayed myelination (arrow heads, C.ii). Integrated genomic viewer snapshot of exome sequencing data shows contiguous gene deletion of SMPD4 and CCDC74B in homozygous state in proband and heterozygous state in parents (D). Quantitative PCR using genomic DNA showed no amplification of SMPD4 exon 19 in proband, her parents showed half the amplification as compared to controls corresponding to one copy number (E.i). No significant difference in amplification was seen in SMPD4 exon 11 between the control sample, parents and proband (E.ii).
On examination at 22-months-age, the proband appeared alert and comfortable. Her weight was 3.6 kg (−7.47 SD), length 63.4 cm (−6.8 SD) and occipito-frontal head circumference was 36 cm (−7.8 SD) [5]. She displayed dysmorphic features viz. bilateral epicanthic folds, deep set eyes, short and anteverted nose, bilateral convergent squint, nystagmus, flexion deformities of limbs (cortical thumb, dorsiflexion of feet bilaterally) (Fig 1B). Bilateral knees had fixed extension deformity. There was no visual response to stimuli. Generalized hypotonia, preserved deep tendon reflexes, and abnormal tonic movements of upper limbs were noted, along with hand stereotypy viz. frequent mouthing. Eye evaluation showed macular dystrophy, & retinal pigment epithelial stippling at posterior pole in both eyes. There was bilateral nystagmus and convergent squint with clear anterior segment and lens bilaterally. Investigations including complete blood count, liver and renal function tests, thyroid function and MS/MS analysis of acyl carnitines and amino acids, lactate and ammonia were normal. MRI brain performed at 22-months-age revealed global hypomyelination (complete absence of white matter myelination in brain including posterior limb of internal capsule and cerebellar peduncles), simplified gyral pattern, hypoplasia of corpus callosum and brain stem (Fig. 1C). The proband succumbed to her illness at 25-months-age.
Methods
Exome sequencing & Quantitative PCR
Parent child trio ES was performed on genomic DNA using exome research panel, Integrated DNA Technologies (Coralville, Iowa, United States) followed by massively parallel sequencing on Illumina NextSeq Platform (Illumina, San Diego, CA, USA). The raw reads were aligned against GRCh37/hg19 genome assembly. cn.MOPS [6] exome depth [7] and XHMM [8] tools were used to call CNVs in the proband and our in-house exome sequencing dataset of 1069 unrelated individuals followed by manual inspection of variants of interest using integrated genomic viewer. Region of homozygosity analysis was performed using AutoMap. [9] Exome based pairwise kinship analysis was performed to check for relatedness between the parents using KING.11 with kinship co-efficient cut-off of 0.0884. Quantitative PCR (qPCR) was on genomic DNA of the proband, clinically normal parents and two unrelated healthy controls as previously described [10]. Detailed methodology is provided in ssupplementary section.
Results
ES analysis rendered no disease-causing single nucleotide variants in concordance with observed phenotype. CNV analysis within the exome detected approximately 11 Kb homozygous deletion (chr2:130899707–130911023) spanning exons 18–20 of SMPD4 (NM_017951.5) and exons 1–3 of CCDC74B (NM_207310.4) in the proband (Fig. 1D). This variant was not observed in gnomAD and our-inhouse ES dataset of 1069 unrelated individuals. This variant does not lie in a region of homozygosity in the proband. Pairwise kinship analysis from parental ES data revealed a kinship co-efficient of 0.0182 indicating unrelatedness. Upon qPCR, no amplification of exon 19 of SMPD4 was detected in the proband, suggesting homozygous deletion and values of amplicons in her parents corresponded to one copy number indicative of heterozygous deletion (Fig. 1E). Values of amplicons of exon 11 of SMPD4 in proband and her parents were comparable to that of diploid unrelated control sample (Fig. 1F).
Discussion
The proband showed remarkable similarities with the 33 previously reported cases from 13 families [3,4]. Similar to published cases, the proband had neurodevelopmental delay, microcephaly and congenital arthrogryposis. Additionally, she had visual impairment with macular dystrophy which are novel findings, not reported in the previous cases. There was severe postnatal growth failure in the proband, with weight and length being 7 and 6 standard deviations below the mean for age, a fact not highlighted in previous cases. Even though IUGR was a prominent feature in majority in Magini’s cohort, postnatal growth failure was significant only in one family. Our patient also did not manifest seizures, cardiac involvement or diabetes. There is a wide spectrum of the disorder, and a genotype-phenotype correlation. Magini et al mentioned the cardiomyopathy and diabetes were seen only in the longer surviving, thus mildly affected patients, carrying milder missense variants. Seizures were also noted in 59% of cases (Table 1).
Table 1.
Clinical and Neuroradiological features observed in proband in comparison with those reported in literature [3, 4]
| Clinical and Neuroradiological features | Number and percentages of assessed cases reported in literature [3,4] | Present study |
|---|---|---|
| Growth Parameters and Survival | ||
| IUGR | 15/22 (68%) | Yes |
| Primary microcephaly | 17/24 (70.8%) | Yes |
| Postnatal growth failure | 3/14 (21.3%) | Yes |
| Postnatal death (less than 12 months) | 7/19 (36.8%) | No (Died at 25 months) |
|
| ||
| Clinical features | ||
| Facial dysmorphism | 15/15 (100%) | Yes |
| Congenital arthrogryposis | 20/23 (87%) | Yes |
| Developmental delay | 7/7 (100%) | Yes |
| Seizures | 10/17 (58.8%) | No |
| Hypotonic | 4/15 (26.7%) | Yes |
| Hypertonic | 9/15 (60%) | No |
| Persistent respiratory distress | 11/18 (61.1%) | No |
| Congenital heart defect | 12/21 (57.1%) | NK* |
| Diabetes mellitus | 2/19 10.5% | NK |
| Visual impairment (macular dystrophy) | NK | Yes |
|
| ||
| Neuro-imaging (MRI Brain) | ||
| Simplified gyral pattern | 12/18 66.7% | Yes |
| Thin corpus callosum | 12/23 52.2% | Yes |
| Hypomyelination | 7/19 36.8% | Yes |
| Cerebellar hypoplasia | 12/23 52.2% | No |
| Brainstem hypoplasia | 3/20 15% | Yes |
NK= Not known
Neuroimaging findings of SMPD4 related disorder include simplified gyral pattern, hypoplasia and severe hypomyelination. There was near absence of myelination in the proband, features indicating an insult to the early developing brain, starting from intra-uterine life itself. The timing coincides with activity of the neutral sphingomyelinases, particularly n-SMase3 whose function has been demonstrated to be its highest in the early stages of brain development [10].
SMPD4, encodes for n-SMase 3 having a protein-protein interaction domain and a C-terminal transmembrane domain [2]. Loss of function of SMPD4 induces ER stress, autophagy, impaired sphingolipids homeostasis, cell cycle dysregulation [3]. This may partially explain the severe growth failure as well as microcephaly in the SMPD4 related disorder. Individuals with homozygous missense variants had milder phenotype compared to individuals with truncating variants in SMPD4 [3]. The severe phenotype observed in the proband is in concordance with previously reported individuals with loss-of-function SMPD4 variants.[3]
The homozygous deletion observed in the proband was inherited from her parents. It was neither observed in gnomAD, nor in our in-house dataset of 1069 unrelated individuals. Though the presence of a completely identical and rare deletion in the parents suggests the origin of this variant from a common ancestor, parents were clinically documented to be non-consanguineous. Also, pairwise kinship analysis from parental ES data revealed a kinship co-efficient of 0.0182 indicating unrelatedness in them. Further, the variant does not lie in a region of homozygosity in the proband. Hence, there is insufficient evidence to support this variant as a recurrent, founder or that arising from a recent ancestor at present.
In conclusion, we present a rare autosomal recessive disorder due to homozygous partial SMPD4 deletion causing a unique constellation of severe growth and neurodevelopmental delay, arthrogryposis, microcephaly and brain anomalies. Visual impairment was a novel finding, expanding the phenotype.
Supplementary Material
Acknowledgements
We thank the proband and her family for participating in this study. A part of this work was funded by National Institutes of Health, USA under the project titled ‘Genetic Diagnosis of Neurodevelopmental Disorders in India’ Grant ID - R01 HD093570 01 A1 (PI). The trio-WES was funded by Medgenome Laboratories Ltd. The authors also acknowledge the contributions from Dr Renu Saxena, head of Molecular Genetics laboratory and Dr I C Verma and Dr Ratna D Puri, senior clinical geneticists at Sir Ganga Ram Hospital, for significant intellectual discussions.
Footnotes
Data Availability
The data pertaining to this work will be made available upon a reasonable request to the corresponding author.
Conflict of interest
All the authors declare that they have no conflict of interest.
References
- 1.Piccinini M, Scandroglio F, Prioni S, Buccinnà B, Loberto N, Aureli M, et al. Deregulated sphingolipid metabolism and membrane organization in neurodegenerative disorders. Mol Neurobiol. 2010;41:314–40. doi: 10.1007/s12035-009-8096-6. [DOI] [PubMed] [Google Scholar]
- 2.Bienias K, Fiedorowicz A, Sadowska A, Prokopiuk S, Car H. Regulation of sphingomyelin metabolism. Pharmacol Rep. 2016;68:570–81. doi: 10.1016/j.pharep.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Magini P, Smits DJ, Vandervore L, Schot R, Columbaro M, Kasteleijn E, et al. Loss of SMPD4 Causes a Developmental Disorder Characterized by Microcephaly and Congenital Arthrogryposis. Am J Hum Genet. 2019;105:689–705. doi: 10.1016/j.ajhg.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravenscroft G, Clayton JS, Faiz F, Sivadorai P, Milnes D, Cincotta R, et al. Neurogenetic fetal akinesia and arthrogryposis: genetics, expanding genotype-phenotypes and functional genomics. J Med Genet. 2020:jmedgenet-2020–106901. doi: 10.1136/jmedgenet-2020-106901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Multicentre Growth Reference Study Group. WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr Suppl. 2006;450:76–85. [DOI] [PubMed] [Google Scholar]
- 6.Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic acids research. 2012;40(9):e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, Kumararatne D, Doffinger R, Nejentsev S. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012. Nov 1;28(21):2747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromer M, Purcell SM. Using XHMM Software to Detect Copy Number Variation in Whole-Exome Sequencing Data. Curr Protoc Hum Genet. 2014. Apr 24;81:7.23.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinodoz M, Peter VG. AutoMap is a high performance homozygosity mapping tool using next-generation sequencing data. 2021;12(1):518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somashekar PH, Upadhyai P, Narayanan DL, Kamath N, Bajaj S, Girisha KM, et al. Phenotypic diversity and genetic complexity of PAX3-related Waardenburg syndrome. Am J Med Genet A. 2020;182:2951–2958. doi: 10.1002/ajmg.a.61893. [DOI] [PubMed] [Google Scholar]
- 11.Svennerholm L, Boström K, Fredman P, Månsson JE, Rosengren B, Rynmark BM. Human brain gangliosides: developmental changes from early fetal stage to advanced age. Biochim Biophys Acta. 1989;1005:109–17. doi: 10.1016/0005-2760(89)90175-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Zheng T, Wang T, Li Q, Wang F, Liang X, Chen J, Teng J. CCDC74A/B are K-fiber crosslinkers required for chromosomal alignment. BMC Biol. 2019. Sep 14;17(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.