Abstract
Nonhuman primates and especially rhesus macaques (Macaca mulatta) have been indispensable animal models for studies of various aspects of neurobiology, developmental psychology, and other aspects of neuroscience. While remarkable progress has been made in our understanding of influences on atypical human social behavior, such as that observed in autism spectrum disorders (ASD), many significant questions remain. Improved understanding of the relationships among variation in specific genes and variation in expressed social behavior in a nonhuman primate would benefit efforts to investigate risk factors, developmental mechanisms, and potential therapies for behavioral disorders including ASD. To study genetic influences on key aspects of social behavior and interactions—individual competence and/or motivation for specific aspects of social behavior—we quantified individual variation in social interactions among juvenile rhesus macaques using both a standard macaque ethogram and a macaque-relevant modification of the human Social Responsiveness Scale. Our analyses demonstrate that various aspects of juvenile social behavior exhibit significant genetic heritability, with estimated quantitative genetic effects similar to that described for ASD in human children. We also performed exome sequencing and analyzed variants in 143 genes previously suggested to influence risk for human ASD. We find preliminary evidence for genetic association between specific variants and both individual behaviors and multi-behavioral factor scores. To our knowledge, this is the first demonstration that spontaneous social behaviors performed by free-ranging juvenile rhesus macaques display significant genetic heritability and then to use exome sequencing data to examine potential macaque genetic associations in genes associated with human ASD.
Keywords: animal, autism spectrum disorder, developmental, disease models, macaca mulatta, neurobiology, psychology, social behavior, whole exome sequencing
Lay Summary
In order to better understand both the genetic mechanisms that cause and possible therapies for treating autism spectrum disorders, it will be valuable to understand how genetic differences among nonhuman primates can influence brain function and resulting differences in social behavior. Rhesus macaques exhibit many complex social behaviors and are amenable to various kinds of genetic analysis, making them a highly relevant animal model. We watched more than 200 rhesus macaques in a naturalistic environment and used two different measures to identify differences among the animals in their expression of various social behaviors. We then used quantitative analyses of the differences among the animals and found that some behaviors were strongly influenced by genetic variation and may be linked to specific DNA sequence variants, just as has been demonstrated in humans.
INTRODUCTION
Primates, both humans and nonhuman species, form strong social bonds that provide support, protection and access to resources that are critical for survival. Social and communication deficits, such as those present in human autism spectrum disorder (ASD), hinder the individual from functioning appropriately in a community. Given that the prevalence of ASD is estimated at 1 in 59 children in the US (Maenner et al., Maenner et al., 2020) and that early diagnosis and intervention can change the trajectory of the disorder (Pickles et al., 2016; Wetherby et al., 2018), significant research is focused on identifying early markers for screening/diagnosis. ASD is a developmental disorder with high heritability and nearly equal contributions of genetics and environment to the overall risk in a population (Sandin et al., 2017). Multiple genome sequencing studies in humans have identified sets of genes exhibiting variants with elevated frequencies in ASD relative to controls, although the majority of these variants each contribute <0.1%–1% of risk (Satterstrom et al., 2020). Very few of these genes have clear loss of function mutations that support their role in ASD, and together the many de novo mutations now identified collectively explain <5% of overall liability (Robinson et al., 2016). The emerging understanding implicates a network of interacting proteins that can be perturbed in multiple ways to interface with the environment and lead to different trajectories for ASD (Chahrour et al., 2016); the specific constellation of causal factors may be different for almost every individual with ASD.
Despite the important mechanistic information on neurodevelopmental processes provided to date by genetic rodent models of ASD (Ryan et al., 2018), only a few studies have examined those processes in nonhuman primate (NHP) models with social behaviors and brain anatomy, function and developmental trajectories that more closely resemble our species (Bauman & Schumann, 2018; Zhu et al., 2018). Recent studies have begun to develop transgenic NHP models of ASD-related behavioral deficits, focusing on mutations in the SHANK3 and MECP2 genes (Z. Liu et al., 2016) and to explore early social markers that predict low sociability in macaques (Kovacs-Balint et al., 2019; Matheson & Bernstein, 2000; Sclafani et al., 2016). However, NHP species exhibit substantial levels of spontaneous individual variation in social behavior, without need for genetic engineering that disrupts normal behavioral development. A number of studies have investigated variation in behaviors related to social interactions through the experimental manipulation of environmental circumstances (Johnson et al., 2015; Kalin & Shelton, 2003; Rogers, 2018) (Brent & Veira, 2002; Fairbanks, 2001; Hopkins et al., 2014; Kinnally et al., 2008). Several of these studies have documented significant genetic heritability of behavioral responses to conspecifics or other similar challenges (Fairbanks et al., 2004; Fawcett et al., 2014; Johnson et al., 2015; Rogers, 2018; Rogers et al., 2013). Thus, although transgenic studies will likely be quite valuable, a different strategy likely to generate useful insight into both the neurobehavioral mechanisms and circuits that underly social competence and social motivation, as well as helping to identify specific genes that influence this inter-individual variation, is to investigate the functional impact of naturally occurring spontaneous genetic variants identified among primates living in social groups. The value of analyzing variability in sociality within naturalistic social groups of macaques has been recognized by various researchers, but investigation of genetic contributions is just beginning (Beisner et al., 2020; Myers et al., 2021; Talbot et al., 2020; Talbot et al., 2021; Warren et al., 2020).
Although genetic heritability of other cognitive, behavioral reactivity/anxiety and related neural phenotypes has been reported in macaques and other NHPs (D. Hopkins, William, Russell, & Schaeffer, 2014; Rogers, 2018) and mutual eye gaze has been recently shown to be moderately heritable in chimpanzees (W. D. Hopkins et al., 2020) little is known about the heritability of spontaneous social behaviors and their associations with naturally-occurring genetic variants among macaques housed in complex social groups. A 2018 study on SNVs in the macaque orthologs of OXTR and AVPR1A/B, specific genes regulating human social behavior, estimated modest contributions to single behaviors observed in a free-ranging colony (Madlon-Kay et al., 2018). The purpose of the present study was to examine the heritability of social behavioral phenotypes in a juvenile cohort of rhesus monkeys from the Yerkes National Primate Research Center breeding colony, and to identify naturally occurring genetic variants segregating in these animals. The longer-term goal is to develop a more complete understanding of gene-phenotype relationships in nonhuman primates that can help to illuminate gene-phenotype relationships in humans relevant to individual variation in complex human sociality. Such information from nonhuman primates will, we believe, help to reveal genetic mechanisms relevant to some aspects of ASD. To our knowledge, ours is the first study using both traditional, macaque-based behavioral measurements and an adapted human behavioral scale to demonstrate robust, significant genetic heritability of individual variation in rhesus macaque social behavior using observations of naturally-occurring social behaviors among macaques living in complex, undisturbed groups. To accomplish this, we collected behavioral observations on more than 200 juvenile rhesus macaques of known pedigree while they were housed in species-typical social groups. In addition, we begin to identify potentially functional genetic variation by performing whole exome sequencing (WES) in the same animals characterized for spontaneous social behavior.
To document behavioral variation among this cohort of juvenile rhesus macaques, we used two quantitative behavioral measures: traditional primate ethograms (Altmann, 2006; Herman et al., 2003; Mccormack et al., 2015; Raper et al., 2014) and an adapted human measure of atypical social behavior, the juvenile macaque Social Responsiveness Scale (jmSRS, [Kovacs Balint et al., 2021]), consisting of 14 scored items. We then used exploratory factor analysis to identify intrinsic factors, their loadings and associations between jmSRS and ethogram behavioral factors. In parallel, we conducted exome sequencing using DNA from the individuals with behavioral data and performed quantitative trait association analysis to begin investigating specific single nucleotide variants (SNVs) that may be associated with the behavioral phenotypes and factors. Although atypical social development is found in several human developmental disorders (a few examples being ASD, attention deficit hyperactivity disorder, William’s syndrome, and schizophrenia) (Greene et al., 1997; Mier & Kirsch, 2015; Royston et al., 2019) we focused our genetic studies on genes that have been associated with ASD specifically because (1) social deficits are one of the main symptoms associated with ASD, (2) the SRS tool used to investigate atypical social behaviors in our juvenile group was first developed as a rapid measure of atypical social development in ASD and (3) to limit the number of genes to be included in our analyses given the relatively small sample size. Our results show high heritability in social behaviors among juvenile macaques living in semi-naturalistic social housing, without need for experimental manipulation to reveal differences. The levels of heritability are similar to levels of social behaviors in humans. We also provide preliminary but suggestive evidence of specific variants in ASD-associated genes in humans that may influence macaque social behaviors.
METHODS
Subjects and housing
A total of 212 juvenile rhesus monkeys (M. mulatta, 98 females, 114 males) maintained at the Yerkes National Primate Research Center Field Station (Lawrenceville, GA) were studied between 12–18 months of age. Subjects lived in their natal social groups consisting of 55–130 adult females with their subadult, juvenile, and infant offspring and two to four adult males. This housing preserves opportunity for critical species-specific, complex social experiences for these study subjects. The groups were housed in outdoor compounds with access to indoor climate-controlled housing areas. Animals were fed a standard commercial low-fat, high-fiber diet (Purina Mills International, LabDiets, St. Louis, MO) ad libitum in the morning and afternoon, supplemented each day with seasonal fruits or vegetables, and water was freely available. All of the procedures described here were performed in accordance with the Animal Welfare Act and the U.S. Department of Health and Human Services “Guide for the Care and Use of Laboratory Animals” and approved by the Emory University Institutional Animal Care and Use Committee.
Behavioral data collection
Behavioral observations were performed on 211 macaques; one animal was excluded due to assignment to a different study. Thirty-minute behavioral observations were collected on four different days (approximately 2 h total observation), using an adaptation of a well-established rhesus monkey ethogram based on published methods (Altmann, 2006; Herman et al., 2003; Mccormack et al., 2015; Raper et al., 2014). Four trained observers with an inter-rater reliability of Cohen’s k = 0.83 collected all the behavioral data in this study.
Nineteen separate behaviors were collected to measure social affiliative behaviors (such as time spent in proximity, contact and grooming other animals), social play, time spent alone, aggression, submission, and nonsocial behaviors such as anxiety (see operational definition of behaviors in Table 1; for detailed description of the behavioral data collection methods, see [Kovacs Balint et al., 2021]).
TABLE 1.
Behaviors collected with our ethogram during focal observations and their operational definitions
| Behavior | Coded as | Operational definitions |
|---|---|---|
|
| ||
| Contact | Duration | Majority of body is touching another animal. |
| Following | Duration | Persistent trailing of another animal; both moving simultaneously, within arm’s reach. |
| Grooming | Duration | One animal combing through the hair of another, usually with hands, but can be with mouth. |
| Proximity | Duration | Subject is within arm’s length to another animal. Not scored for animals passing by or when animals are in motion. |
| Solitary play | Duration | Does not involve a partner, vigorous play by oneself. Manipulate/tactile an object (e.g., rock, poop, cage), climb or swing for at least 3 s. |
| Aggression | Frequency | Hostile interaction between two animals, with or without physical contact between the animals (e.g., slap or grab and attack, bite, physical threat, open mouth, barking, lunge). |
| Anxiety | Frequency | Sum of scratching, yawning, body-shaking and self-directed behaviors (e.g., self-exploring, self-grooming). |
| Breaking proximity | Frequency | Leaving behind another animal (beyond arm’s reach), end of being in proximity (within arm’s length) with another animal. |
| Display | Frequency | Shaking or bouncing vigorously to convey dominance status. |
| Eye-gaze | Frequency | Making direct eye contact with another animal, if prolonged, 1 additional occurrence coded for every 3 s. |
| Grimace | Frequency | Open mouth wide to show teeth with a closed jaw to another animal, pulling back of lips to display teeth. |
| Groom soliciting | Frequency | Posture to solicit grooming from another animal, can groom solicit from more than one animal simultaneously. |
| Scream | Frequency | High pitch, high intensity screech or loud chirp. |
| Sex-related behavior | Frequency | Sum of hip touch (subject places two hands on hip of the other animal) and mount (subject’s feet are clasped on the outside of the ankles of the other animal) behaviors. |
| Sitting alone | Frequency | Sitting out of proximity (arm’s length) to other animals. |
| Social play | Frequency | Playing with another animal, including wrestle, play chase and playing with tail. |
| Submissive behavior | Frequency | Sum of lip-smacking (opening and closing lips) and withdrawing (avoiding or pulling away from another animal) behaviors. |
| Touch | Frequency | Subject approaches and touches another animal, distinct from grooming; if prolonged, one additional occurrence coded for every 3 s. |
Juvenile macaque social responsiveness scale
The jmSRS is a novel screening tool adapted and validated by our group for juvenile macaques, composed of 14 items that measure global dimensions of typical and atypical social behaviors, as well as stereotypic and odd behaviors of relevance to ASD (Kovacs Balint et al., 2021). The jmSRS was adapted from the adult macaque SRS (mSRS; [Feczko et al., 2016]), which was originally adapted from the human SRS (Constantino et al., 2003). After the fourth behavioral observation was collected for each subject, social responsiveness was rated by the observers for each of the 14 items in the jmSRS instrument (Table 2) using a 5-point Likert scale (1 = not true or 0%, 2 = sometimes true or 25%, 3 = often true or 50%, 4 = almost always true or 75%, and 5 = always true or 100%).
TABLE 2.
Items of the jmSRS instrument. R: Reverse-coded items. Modified from Kovacs-Balint et al., 2021, with permission
| Item 1. Seems self-confident when interacting with others. R |
| Item 2. Would rather be alone than with others. |
| Item 3. Behaves in ways that seem strange or bizarre for others of comparable age/rank/gender categories. |
| Item 4. Is not well coordinated in physical activities. |
| Item 5. Responds appropriately to other monkeys’ vocalizations and facial expressions. R |
| Item 6. Avoids eye contact or has unusual eye contact. |
| Item 7. Plays appropriately with peers. R |
| Item 8. Avoids social interactions with others. |
| Item 9. Is socially awkward. |
| Item 10. Has a restricted or unusually narrow range of interests. |
| Item 11. Has repetitive, odd behaviors such as hand flapping, rocking/swaying, tumbling or spinning. |
| Item 12. Is too tense in social situations, for example, walks stiffly, stiffens or freezes when others approach. |
| Item 13. Stares or gazes off into space. |
| Item 14. Manifests species and status-typical reaction to loss of a valued resource. R |
Behavioral data analysis
Behavioral observations
To begin, the frequency or duration (where applicable) of each observed social behavior was calculated, then divided by the total observation time for the given subject (in hours) to produce behavioral rates/hour (for frequencies) or percent time (for durations). Z-scores were then calculated from both frequency and duration measures per hour to rescale the different data types to the same scale.
Exploratory factor analysis (EFA) was performed on the whole set of behaviors using IBM SPSS Statistics (IBM Corporation, Armonk, NY) to examine how the observed behaviors were interrelated, and for data reduction purposes. We used “principal axis factoring” as the extraction method, and the “latent root criterion” (eigenvalues greater than one) to determine the number of factors to extract. Varimax orthogonal rotation was used to achieve more meaningful factor solutions. Behavioral factor scores were then calculated individually, using “factor score method” (multiplying the occurrence of the observed behavior by the factor weights, called factor loadings).
Juvenile social rank was also measured, which at this age is based on their mothers’ matrilineal social rank. Matrilineal social rank was assessed from aggression and submission behaviors exhibited during dyadic agonistic interactions during group observations with high rank defined as the top 33% most dominant, low rank as the lowest 33%, and middle rank as those in between.
Juvenile macaque SRS
Using the 14 items in the jmSRS scored for all the juveniles by our observers, and after items #1, 5, 7, and 14 were reverse-coded to match the rest of items in directionality (i.e., so that the higher scores represented greater social impairment), EFA was performed using IBM SPSS Statistics to determine the overall factor structure of the jmSRS items. We used the “percentage of variance” criterion, which determined the number of factors to extract, and Promax rotation method to achieve simpler and theoretically more meaningful factor solutions. Factor scores were then calculated for each subject individually, using the factor score method. For the detailed description of the adaptation and validation of the jmSRS, see (Kovacs Balint et al., 2021).
Genomics
Sequencing and variant calling
Rhesus macaque DNA samples (n = 208, due to sample collection difficulties and one case of low DNA-yield) were enriched for exome sequences using the Rhexome v2 capture reagent (Caskey et al., 2019) and sequenced using the Illumina NovaSeq system. The 150 bp paired end reads were aligned to the rhesus Mmul_8.0.1 (rheMac8) reference genome assembly (https://www.ncbi.nlm.nih.gov/assembly/GCF_000772875.2/) using BWA mem with an average on target sequence depth of 66.8× across the samples. Picard MarkDuplicates version 1.105 (http://broadinstitute.github.io/picard/) was used to identify and mark duplicate reads. The GATK v3.3.0 best practices pipeline (Depristo et al., 2011; Mckenna et al., 2010) was used to identify single nucleotide variants (SNVs) and small insertions/deletions (indels). Variant Effect Predictor software (VEP) (Mclaren et al., 2016) was used to annotate variants based on merged Ensembl and RefSeq gene models.
Heritability
The software package Sequential Oligogenic Linkage Analysis Routines (SOLAR) version 8.1.1 (Almasy & Blangero, 1998) was used to estimate heritability (h2r) and heritability p values for all behavioral phenotypes. SOLAR implements a variance components approach to estimating additive genetic (narrow sense) heritability. Briefly, if individual variation in any given phenotype is influenced by genetic differences among those individuals, then the expectation is that pairwise phenotypic differences among individuals will be negatively correlated with pairwise kinship. That is, the more closely related any two individuals are, the smaller should be the quantitative difference between them in phenotype. We tested for significant genetic heritability in social behavioral phenotypes by testing across all possible pairs of study animals for statistically significant negative correlation between pairwise phenotypic differences and pairwise kinship. Prior to heritability analyses, the behavioral phenotypes were normalized using the SOLAR rank-based inverse-normal transformation (i-normal) routine. The normalized behavioral phenotypes and known pedigree relationships among subject animals were used as inputs to the polygenic model of inheritance. Heritability was estimated across all animals in two ways: using just sex as a covariate and also using both sex and social dominance rank as covariates. Among the juveniles analyzed using SOLAR, individuals had an average of 0.03 full-sibs and 9.3 half-sibs in the cohort.
Genetic association
Genetic variants were analyzed for quantitative trait association with each behavioral phenotype using the software packages Factored Spectrally Transformed Linear Mixed Models (FaST-LMM) version 0.2.24 (Lippert et al., 2011) and Genome-wide Efficient Mixed Model Association algorithm (GEMMA) version 0.98 (X. Zhou & Stephens, 2012). Both of these software packages implement a linear mixed model that takes into account potential relatedness among samples; we chose these due to the known genealogical relatedness among the animals tested. We used the FaST-LMM single_snp function that performs a single SNV association analysis using cross validation over the chromosomes and restricted maximum likelihood (REML). For GEMMA, a standardized relatedness matrix was generated, and this matrix was used to perform a univariate LMM association analysis. GEMMA also applies a filter that removes SNVs with a minor allele frequency (MAF) < 0.01 from the analysis. Quantitative trait association analyses were performed based on different measures of behavior: (1) individual behaviors based on a well-established ethogram, (2) behavioral EFA (or BFA) performed on the individual behaviors for data reduction and (3) EFA performed on the jmSRS for data reduction. Association analyses were performed across all animals using both sex and social dominance rank as covariates. Although we identified SNV data from the full exome, we only analyzed SNVs from a limited set of 143 genes of interest (see below and Table S1). Variants of interest were further examined by lifting over (Hinrichs, 2006) the rhesus positions to the orthologous human (GRCh38) positions and retrieving CADD version 1.5 PHRED scores that predict the functional impact of variants (Rentzsch et al., 2018).
Given our relatively small sample size, association testing analyses were limited to a set of 143 highconfidence genes (Table S1). These genes were selected because they were either confidence level 1 (high confidence) or 2 (strong candidate) in the SFARI Gene database (Larsen et al., 2016), meaning they have been implicated in multiple ASD genetic reports; they appeared on the list of ASD-related genes reported in the large scale exome sequencing study from the Autism Sequencing Consortium (Satterstrom et al., 2020); they have been suggested to be relevant to macaque social behavior; or a combination of these elements.
RESULTS
Behavioral observations
Behavioral EFA (BFA) was performed on 15 individual behaviors, for data reduction purposes. “Scream” behavior was removed from the analysis due to low occurrence rates (scored for less than 25% of the subjects during the 2 h. of observation). “Grooming duration” and “following duration” were removed during model refinement due to lack of significant loading on any one factor. Final EFA of the observed behaviors revealed that six components had eigenvalues greater than 1.0, and these components accounted for 20.62% (BFA factor 1: BFA-1), 12.23% (BFA-2), 10.33% (BFA-3), 8.27% (BFA-4), 7.50% (BFA-5) and 6.69% (BFA-6) of the variance, respectively (the first six factors explained 65.63% of the variance). Factor loadings are presented in Table 3; distributions of the factor scores calculated for each animal based on the six EFA factors are shown in Figure S1.
TABLE 3.
Factor loadings of the behavioral exploratory factor analysis (BFA)
| Factor | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| BFA-1 | BFA-2 | BFA-3 | BFA-4 | BFA-5 | BFA-6 | |
|
| ||||||
| Submissive behavior | 0.755 | −0.065 | 0.116 | −0.048 | −0.017 | −0.177 |
| Grimace | 0.548 | 0.018 | −0.142 | 0.062 | 0.136 | −0.027 |
| Touch | 0.547 | 0.070 | 0.281 | 0.114 | −0.131 | 0.233 |
| Anxiety | 0.522 | −0.027 | 0.245 | 0.357 * | −0.122 | −0.093 |
| Leave behind | 0.293 | 0.650 | 0.220 | 0.195 | −0.274 | −0.120 |
| Proximity (duration) | −0.163 | 0.651 | −0.097 | 0.051 | −0.042 | −0.096 |
| Social play | −0.028 | −0.372 | 0.131 | 0.115 | −0.085 | −0.24 |
| Aggression | 0.122 | 0.066 | 0.576 | 0.017 | −0.131 | 0.071 |
| Display | −0.068 | −0.108 | 0.482 | 0.319 * | −0.074 | −0.055 |
| Sex-related behaviors | 0.042 | −0.084 | 0.322 | 0.014 | 0.144 | −0.018 |
| Eye-gaze | 0.222 | 0.153 | 0.365 * | 0.541 | −0.056 | 0.004 |
| Groom soliciting | 0.119 | 0.256 | 0.265 | 0.63 | 0.006 | −0.043 |
| Solitary play (duration) | 0.005 | −0.067 | −0.06 | 0.355 | −0.004 | −0.014 |
| Sitting alone | 0.007 | −0.068 | −0.004 | −0.037 | 0.887 | −0.072 |
| Contact (duration) | −0.103 | 0.116 | 0.043 | −0.044 | −0.08 | 0.772 |
| Eigenvalue | 3.093 | 1.834 | 1.549 | 1.240 | 1.124 | 1.003 |
| % of variance | 20.621 | 12.227 | 10.329 | 8.269 | 7.497 | 6.688 |
Note: Bold black letters indicate high factor loadings,
indicates cross-loading behaviors. Eigenvalues and percent of variance explained of each factor are shown at the bottom of the table.
Behaviors with high-factor loadings on BFA-1 (submissive behavior, touch, grimace, and anxiety) represent anxiety, fear, and behaviors related to low social rank. Behaviors loading on BFA-2 (leave behind, proximity = positive loading-; social play = negative loading-) are related to keeping and breaking proximity with others and juvenile social play. BFA-3 includes sex- and dominance-related behaviors (aggression, display, hip-touch/mount) and eye-gaze. Behaviors with high factor-loadings on BFA-4 include behaviors that also load positively in BFA-3 (eye-gaze, display) and BFA-1 (anxiety) but combined with high groom soliciting and solitary play. BFA-5 and BFA-6 had only one behavior with high loading each (sitting alone and contact, respectively); thus, they could not be formally considered as separate behavioral factors and we instead examined the associations of the individual behaviors with SNVs.
Juvenile macaque SRS
The jmSRS measures individual differences in social responsiveness among juvenile macaques. EFA revealed that together the first four components (Table 4) explained 66.56% of the data, and these components accounted for 39.84% (jmSRS factor 1: jmSRS-1), 9.95% (jmSRS-2), 9.01% (jmSRS-3), and 7.76% (jmSRS-4) of the variance, respectively. Each item had a high (above 0.4) factor loading on at least one factor, and only two items (“Behaves in ways that seems strange or bizarre for others of comparable age/rank/gender categories.” and “Is too tense in social situations, e.g., walks stiffly, stiffens or freezes when others approach.”) showed significant loadings on more than one factor (Table 4). For more details on the adaptation and validation of the jmSRS for juvenile macaques, see (Kovacs Balint et al., 2021).
TABLE 4.
Factor structure of the jmSRS
| Factor | ||||
|---|---|---|---|---|
|
| ||||
| jmSRS-1 | jmSRS-2 | jmSRS-3 | jmSRS-4 | |
|
| ||||
| Item 1. Seems self-confident when interacting with others. R | 0.585 | −0.084 | 0.265 | 0.038 |
| Item 2. Would rather be alone than with others. | 0.752 | 0.045 | 0.121 | −0.028 |
| Item 7. Plays appropriately with peers. R | 0.561 | 0.077 | 0.021 | 0.184 |
| Item 8. Avoids social interactions with others. | 0.926 | 0.055 | 0.019 | −0.257 |
| Item 10. Has a restricted or unusually narrow range of interests. | 0.505 | 0.117 | 0.092 | 0.206 |
| Item 14. Manifests species and status-typical reaction to loss of a valued resource. R | 0.538 * | −0.021 | −0.426 | 0.206 |
| Item 5. Responds appropriately to other monkeys’ vocalizations and facial expressions. R | 0.124 | 0.411 | −0.196 | 0.166 |
| Item 6. Avoids eye contact or has unusual eye contact. | −0.115 | 0.853 | −0.096 | 0.033 |
| Item 9. Is socially awkward. | 0.275 | 0.617 | −0.034 | −0.124 |
| Item 12. Is too tense in social situations, for example, walks siffly, stiffens or freezes when others approach. | 0.182 | 0.688 | 0.088 | −0.207 |
| Item 4. Is not well coordinated in physical activities. | 0.181 | −0.264 | 0.556 | 0.186 |
| Item 13. Stares or gazes off into space. | −0.002 | 0.204 | 0.596 | 0.062 |
| Item 3. Behaves in ways that seem strange or bizarre for others of comparable age/rank/gender categories. | −0.044 | 0.408 | 0.195 | 0.538 * |
| Item 11. Has repetitive, odd behaviors such as hand flapping, rocking/swaying, tumbling or spinning. | −0.028 | −0.123 | 0.134 | 0.590 |
| Eigenvalue | 5.577 | 1.394 | 1.261 | 1.086 |
| % of variance | 39.834 | 9.954 | 9.011 | 7.757 |
Note: Items with high-factor loadings are listed in bold. R: Reverse-coded items;
: Significant cross-loadi arengs. Eigenvalues anpercent of variance explained of each factor shown at the bottom of the table.
Correlations between the four jmSRS factors (jmSRS-1, jmSRS-2, jmSRS-3, jmSRS-4) and the BFA factors generated for the behavioral observations are shown in Table 5. Validity of the jmSRS factors was demonstrated by the significant positive correlations found between: (1) BFA-1 (anxiety, fear and social subordinate behaviors) and jmSRS-1 (poor social interactions), jmSRS-3 (poor physical motor coordination, staring off into space and atypical status-related reactions to loss of resources), and jmSRS-4 (repetitive and odd behaviors); (2) BFA-2 (leave behind, proximity -positive loading- and social playnegative loading-) and jmSRS-3; (3) BFA-3 (dominance-related behaviors and eye-gaze) and jmSRS-1, −3 and −4; (4) BFA-4 (solitary play, dominance and anxiety behaviors) and jmSRS-1-3; Significant negative correlations were found between BFA5 and jmSRS-3 and −4.
TABLE 5.
Correlations between the jmSRS and the BFA factor scores
| Behavioral Factors | ||||||||
|---|---|---|---|---|---|---|---|---|
| BFA-1 | BFA-2 | BFA-3 | BFA-4 | BFA-5 | BFA-6 | |||
|
| ||||||||
| jmSRS FACTORS | jmSRS-1 | Spearman’s rho | 0.228** | 0.105 | 0.198** | 0.184** | −0.017 | −0.057 |
| p value | 0.001 | 0.129 | 0.004 | 0.007 | 0.805 | 0.407 | ||
| jmSRS-2 | Spearman’s rho | 0.090 | 0.117 | 0.038 | 0.158* | 0.026 | 0.029 | |
| p value | 0.191 | 0.091 | 0.587 | 0.022 | 0.705 | 0.677 | ||
| jmSRS-3 | Spearman’s rho | 0.300** | 0.206** | 0.270** | 0.294** | −0.158* | −0.066 | |
| p value | 9.0 × 10−6 | 0.003 | 7.1 × 10−5 | 1.4 × 10−5 | 0.022 | 0.338 | ||
| jmSRS-4 | Spearman’s rho | 0.165* | 0.104 | 0.204** | 0.124 | −0.167* | −0.060 | |
| p value | 0.017 | 0.132 | 0.003 | 0.071 | 0.015 | 0.388 | ||
Note:
p < 0.05;
p < 0.01.
Heritability
Using the SOLAR software package (Almasy & Blangero, 1998), we estimated heritability of social behaviors across the macaque pedigree. We found 20 aggregate or single behaviors with significant (p < 0.05) heritability (Table 6) and note that this level of genetic effect is similar to estimates for ASD in humans. Given the matriarchal and strongly hierarchical nature of macaque social structures, and the potential sex effects, we used sex and dominance rank as covariates in the heritability estimates. The resulting heritability estimates changed little based on the use of any of these covariates, suggesting that the estimates are robust. However, our relatively small sample size does produce substantial standard errors that must be considered when interpreting the heritability estimates. Furthermore, our social rank tertile classification is likely not granular enough to detect subtle effects of social dominance or to fully control for the effects of those non-genetic factors. Nevertheless, we did detect significant rank effects on several phenotypes each: for sex, these were BFA-1, BFA-2, contact, display, leave behind, and social play; for rank, the behaviors/factors were BFA-1, display, follow duration, and leave behind.
TABLE 6.
Social behaviors with significant heritability (p < 0.05). Heritability was calculated using SOLAR after rank-based inverse-normal transformation (inormal). Sex and rank were used as covariates
| Behavior | Heritability (h2 r) | h2r Std err | h2r p-value | Sex significant | Rank significant |
|---|---|---|---|---|---|
|
| |||||
| BFA-1 | 0.43 | 0.20 | 6.00E-03 | Yes | Yes |
| BFA-2 | 0.65 | 0.20 | 2.11E-05 | Yes | No |
| BFA-3 | 0.58 | 0.19 | 1.13E-04 | No | No |
| BFA-4 | 0.84 | 0.19 | 4.00E-07 | No | No |
| jmSRS-1 | 0.57 | 0.20 | 7.64E-04 | No | No |
| jmSRS-2 | 0.44 | 0.24 | 2.44E-02 | No | No |
| jmSRS-3 | 0.49 | 0.22 | 3.68E-03 | No | No |
| jmSRS-4 | 0.39 | 0.18 | 3.51E-03 | No | No |
| ZANXIETY | 0.50 | 0.22 | 3.84E-03 | No | No |
| ZCONTACT | 0.51 | 0.24 | 5.20E-03 | Yes | No |
| ZDISPLAY | 0.27 | 0.18 | 2.24E-02 | Yes | Yes |
| ZFOLLOW_DUR | 0.69 | 0.26 | 1.18E-03 | No | Yes |
| ZGROOM_SOLICIT | 0.80 | 0.17 | 3.17E-08 | No | No |
| ZLEAVE_BEHIND | 0.54 | 0.21 | 9.78E-04 | Yes | Yes |
| ZSOCIAL_PLAY | 0.36 | 0.28 | 3.14E-02 | Yes | No |
| ZSOLITARY_PLAY_DUR | 0.86 | 0.28 | 9.26E-05 | No | No |
| ZSIT_ALONE | 0.74 | 0.31 | 3.30E-03 | No | No |
| ZTOUCH | 0.29 | 0.20 | 4.13E-02 | Yes | No |
Phenotyping versus genotyping data
Exome sequencing and variant calling identified a total of 193,509 SNVs and 8255 indels across the 208 individuals. This includes 2439 SNVs and 169 indels in the 143 genes of particular interest. Among the sequenced individuals there was an average of 17903.67 homozygous alternate and 37008.33 heterozygous sites exome wide and 141.16 homozygous alternate and 262.17 heterozygous sites in our genes of interest. Association analyses of aggregate behavioral EFA and jmSRS factor scores along with the individual behaviors identified a set of four notable SNVs detected by the two association software packages FaST-LMM and GEMMA at a significance of p < 7 × 10−7 (Table 7 and Table S2). This is a consensus significance threshold for exome studies for SNVs with a minor allele frequency (MAF) > = 0.01 (Fadista et al., 2016) consistent with the GEMMA MAF filter that removes SNVs with a MAF < 0.01. Lowering the shared threshold to a significance of p < 1 × 106, as suggested for some human exome sequencing studies (Fadista et al., 2016), identified a linked set of three more SNVs in SMARCA4; we consider these to have only a suggestive association with the jmSRS-3 factor, which measures poor motor coordination, staring off into space and atypical status-related reactions to loss of resources (Table S2 and Figure S2 for individual animals with variants). FAST-LMM alone identified 48 SNVs associated at the p < 7 × 10−7 level with behavioral traits of interest. However, since 18 of these were SNVs found only in one individual, we did not consider these associations to be robust and meaningful. GEMMA alone identified three more SNVs associated at the p < 7 × 10−7 level with behavioral traits of interest, in the genes MAGEL2 and SHANK2 for the factor jmSRS-3 (poor motor coordination, staring off into space and atypical reactions to loss of resources) and in the gene CNTNAP2 for jmSRS-4 (repetitive and odd behaviors) (Table S2 and Figure S2 for individual animals with variants).
TABLE 7.
Significant (p < 7 × 10 −7) associations between specific genetic variants and behavioral factors, in both GEMMA and FAST-LMM results
| Behavior | Chromosome: Position | Ref/alt | Ref/alt Allele count | Gene | Consequence | FastLMM p | Gemma Wald p | CADD PHRED score |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| jmSRS-3 | 3:60389877 | T/C | 6/410 | ABCA13 | missense variant p.Gln3931Arg |
6.54E-07 | 1.61E-08 | 15.30 |
| jmSRS-3 | 5:139396723 | G/A | 409/7 | NAA15 | synonymous variant p. Thr744Thr | 2.36E-08 | 7.39E-09 | 5.386 |
| ZAGGRESSION | 7:28322629 | G/T | 405/11 | LEO1 | synonymous variant p.Arg514Arg | 1.08E-07 | 7.17E-08 | 22.60 |
| ZGROOMING_DUR | 9:287974 | G/C | 411/5 | DIP2C | synonymous variant p.Thr938Thr | 4.25E-07 | 4.26E-07 | 15.11 |
The significantly associated SNVs (Table 7) are missense or synonymous variants. Three SNVs had high CADD PHRED scores: 15.3 in ABCA13, 22.6 in LEO1 and 15.11 in DIP2C. A CADD PHRED score of 10 predicts a SNV is among the 10% most functional changes in the human genome while a CADD PHRED score of 20 indicates the change is among the 1% most functional changes (Rentzsch et al., 2018). The remaining SNV, in NAA15, has a relatively low CADD PHRED score of 5.386, so it may be in linkage with other causative variants or alterations (although it is still possible this variant has an effect itself). Two of the SNVs were associated with the behavioral factor jmSRS-3, again associated with poor motor coordination, staring off into space and atypical reactions to loss of resources. Two individual behaviors had significantly associated SNVs: aggression and grooming duration.
We also examined indels. There were no indels identified as associated with phenotype at p < 7 × 10−7 by both FaST-LMM and GEMMA in our genes of interest.
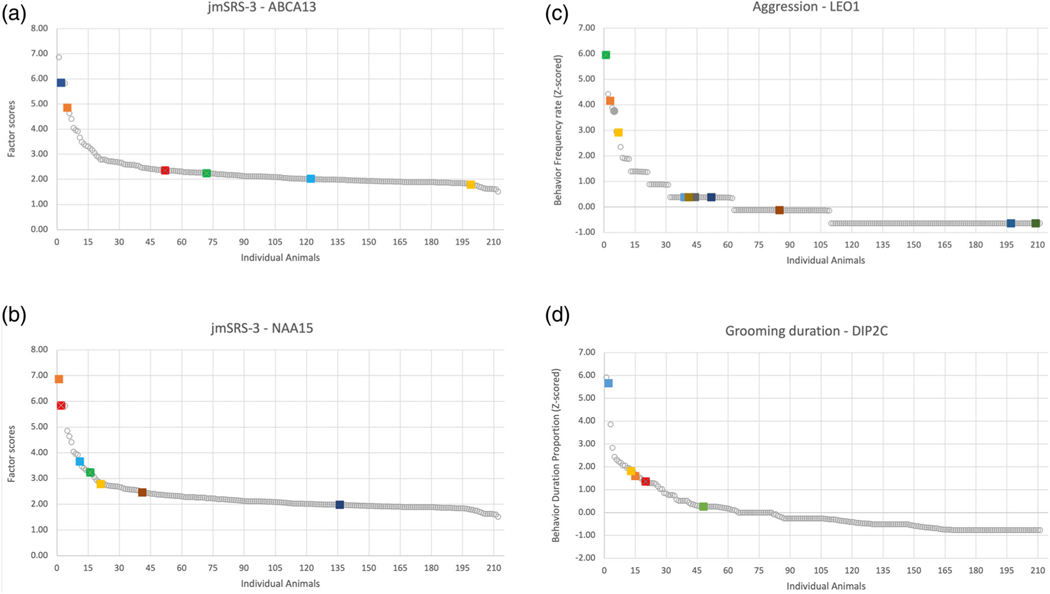
To further explore the association results, we went back to the specific animals carrying the alternate allele for these associated variants. We found that the carrier animals are largely skewed toward the tail of the behavioral distributions. Figure 1A highlights the jmSRS-3 scores of the six animals with the alternate T allele in ABCA13 at position 3:60389877 (note that our data suggest the Mmul 8.0.1 (rheMac8) reference genome contains a low-frequency allele T as the “reference,” since this population largely contains the supposed “alternate” allele C in this position. The nucleotide at this position has been changed to C in Mmul_10). Similarly, Figure 1B–D show the animals with the alternate alleles in the genes NAA15, LEO1, and DIP2C respectively.
FIGURE 1.
Individual animal scores on specific behaviors or factors. (a) Population distribution of scores on the jmSRS-3, where animals with the alternate T allele in ABCA13 at position 3:60389877 are highlighted. (b) Population distribution of scores on the jmSRS-3, where animals with the alternate A allele at position 5:139396723 for NAA15 allele are highlighted. (c) Population distribution of behavioral frequency rate for aggression where animals with the alternate T allele in LEO1, position 7:28322629 are highlighted. (d) Population distribution of behavior duration proportion for grooming duration, where animals with the C allele in DIP2C, position 9:287974 are highlighted
DISCUSSION
NHPs, and especially rhesus macaques, have long been critical to research concerning the neurobiology, physiology and psychosocial development of complex behaviors. Neuroscientists have used studies of rhesus macaques to investigate maternal–infant attachment and human attachment theory (Bowlby, 1951), the effect of early social environment and adversity on infant development and many other topics including more clinically focused analyses such as drug or alcohol abuse (Wakeford et al., 2018). Studies of temperament, anxiety-related behaviors and other aspects of personality have generally used experimental paradigms in which animals are challenged with carefully designed environmental or social situations in order to elicit variation in behavioral responses (Beisner et al., 2020; Fairbanks et al., 2004; Fawcett et al., 2014; Johnson et al., 2015; Kalin & Shelton, 2003; Rogers, 2018; Rogers et al., 2013). Although these challenge paradigms can be highly informative in eliciting significant inter-individual variation, there is also significant value in identifying the source of natural behavioral variation among individuals within a NHP species, and assessing the factors (environmental, genetic, epigenetic or otherwise) that underlie this behavioral variation. Similarly, transgenic models are being developed for multiple behavioral conditions, such as the SHANK3 models generated by Zhou et al. (2019); while these models recapitulate some aspects of atypical social behavior, they have multiple drawbacks, including time and expense; the presence of more complex/larger mutations not naturally found in human or macaque populations; and the combination of the phenotypes of multiple conditions, such as ASD and Phelan-McDermid syndrome in the SHANK3 model specifically. It was a desire to identify spontaneous natural variation in social behaviors and quantify extreme social phenotypes that are potentially relevant to modeling ASD-like traits in rhesus macaques that motivated the present study.
Autism spectrum disorder is often cited as one of or perhaps the most highly heritable behavioral condition in humans, with recent estimates of heritability (h2) as high as 80% in large population studies (Bai et al., 2019). Still, debate has surrounded whether the genetic drivers related to ASD are affecting the entire phenotype or act on separate specific domains of ASD symptoms (Shuster et al., 2014), while simultaneously acknowledging the potential contribution of environment (Cheroni et al., 2020). The latter interpretation of the heritability of subdomains is consistent with our findings of moderate to high heritability scores for the social behaviors measured both through traditional NHP ethograms and adapted human SRS measures. We are not assuming that the macaques spontaneously display variation that parallels ASD in full, but rather we focus on genetic analyses of specific elements of the behavioral repertoire of macaques with the goal of understanding genotype–phenotype relationships in an animal model closely related to humans with its own complex system of social signals, behaviors and patterns of interaction. Previous work using SOLAR to measure heritability of six factors derived from the autism diagnostic interview-Revised (ADI-R), another commonly used measure of social behavior in clinical autism diagnosis, found h2 from 0.32 to 0.69 in a group of 618 families from the Autism Genome Project Phase I (X.-Q. Liu et al., 2011). A 2019 update of this study with more samples and more conservative methods reported h2 ranging from 0.21 for repetitive behavior to 0.53 for social interaction components of the ADI-R (Yousaf et al., 2020). In our BFA measures, we also obtained substantial heritability estimates for measures primarily focused on social interactions (or lack thereof), including h2 of 0.84 +/− 0.19 for BFA-4, which is driven by a combination of juvenile social behaviors including groom soliciting and eye gaze, and to a lesser degree display, anxiety, and solitary play duration. BFA-4 is positively and significantly correlated with three of the four jmSRS factors, each of them displaying h2 in a range of 0.44–0.57. As in juvenile humans, developing macaques are strongly attracted to direct gazing into the eyes of others very early in life, which increases as social experiences accrue (Paukner et al., 2018). However, as the animals with low social dominance rank mature, they start avoiding direct eye gaze during agonistic interactions because it is a threatening social behavior. The high heritability of eye gaze seen in a human twin study, with monozygotic twin-twin concordance of 0.91 compared to dizygotic concordance of 0.35 (Constantino et al., 2017), further suggests that BFA-4’s strong heritability may be driven in part by this trait. However, unlike some previous macaque studies of eye gaze, the inclusion of eye-gaze behavior in this factor did not render significant covariance with sex or rank (Paukner et al., 2018).
Among the individual behaviors, we saw the highest heritability levels for groom soliciting, with an h2 of 0.80 ± 0.17 (Table 6); sitting alone, with an h2 of 0.79 ± 0.21; and solitary play duration, with an h2 of 0.86 ± 0.28. Impaired social interaction is one of the diagnostic criteria for ASD in humans, and it is therefore interesting that these behaviors related to social interaction (or lack thereof) show the highest heritability in our macaque population. None of these three behaviors’ heritability was affected by sex or social rank. However, the heritability of several other behaviors did covary with sex, suggesting that the behavior/factor’s heritability differs between males and females: BFA-1 (anxiety, fear and behaviors related to low social rank), BFA-2 (keeping and breaking proximity with others and social play), contact, display, leave behind and social play. The heritability of behaviors where we observed significant covariance with rank, suggesting they differ between dominant and subordinate animals, were BFA-1, display, follow duration, and leave behind.
The high point estimates of heritability detected in this study should be interpreted with caution, as they could be partially explained by our small sample size. Nevertheless, the p-values assigned to the heritability estimates indicate that there is strong evidence for genetic influence on these phenotypes, while the substantial standard errors indicate our study is unable to generate a precise estimate of the degree of genetic effect. In parallel, our tertile classification of social dominance rank may not be granular enough to capture some of the more subtle effects of rhesus macaque hierarchical social structure. Madlon-Kay et al. studied adult social behaviors in a group of free-ranging macaques with a similar sample size, and tested for narrow-sense heritabilities for the single behaviors of receiving/giving grooming, passive contact, approach, and forms of aggression (Madlon-Kay et al., 2018). Our estimated heritabilities for juveniles are significantly higher than theirs for adults, which are generally not statistically significant. Instead, our estimates using juvenile subjects are more consistent with heritability estimates reported for human children, reported to be approximately 80% for ASD overall in a multinational cohort study (Bai et al., 2019).
In order to focus our genetic variant association analyses, we only analyzed a list of 143 genes with prior link to ASD or social behavior in macaques (Table S1). We also chose a conservative threshold for significance, based on human exome sequencing studies. These choices should be kept in mind when interpreting the resulting associations. Most variants that show associations with behaviors map to the jmSRS-3 factor, which includes items related to staring off into space, atypical reactivity to loss of preferred resources based on one’s social rank and poor physical motor coordination. These behaviors are typically more common in nursery-reared animals (Harlow, 1969; Nelson & Winslow, 2009; Roy, 1981), but our study population lived in a semi free-ranging social environment where abnormal and repetitive behaviors are not commonly observed. Therefore, we feel confident that the association observed is likely significant and biobehaviorally relevant, although due to the sample size should be considered preliminary. We do note the contrast between the heritability results of the jmSRS factors and the association findings; for example, the jmSRS-1 factor, where several important social behaviors load (typical and atypical), such as avoiding social interactions and playing appropriately with peers, has a heritability of 0.57 (+/− 0.20) but did not yield significant genetic associations in our study. We suggest this is likely due to the complexity of behaviors loaded into jmSRS-1, in contrast to the two behaviors loaded onto jmSRS-3. We also note that our study included exome sequencing only, and therefore would not capture either copy number variants or regulatory variants outside the exome method used. Finally, we would note that it’s possible many variants of smaller effect contribute to jmSRS-1 and this could produce an observed effect of high heritability but no variants associated at the significance levels we set. This would be consistent with the emerging model of common variants adding small but additive risk in human ASD (Klei et al., 2021).
The first of two genetic variants significantly associated with the jmSRS-3 factor is in the gene ABCA13, which has been previously reported to associate with atypical behavior in both macaques and Drosophila, including impaired social interactions (Ueoka et al., 2018; Yoshida et al., 2016). The gene encodes a transmembrane transporter in ATP-binding cassette family and is likely involved in transporting lipid molecules in the central nervous system and other tissues (Ueoka et al., 2018) In the SFARI Gene database, where genes associated with ASD are divided into four categories based on the strength of evidence for that association (Larsen et al., 2016), ABCA13 falls in the lowest category, “suggestive evidence.” Rare genetic variants in ABCA13 have been reported in both de novo and familial cases of ASD but are fairly rare compared to other genetic variants. The variant we found, 3:60389877, is listed as a reference T allele in the Mmul_8.0.1 genome assembly we used, but the large majority of our study animals carry the C allele. Given the multiple lines of evidence suggesting a role for ABCA13 in social behavior, we suggest this gene is worthy of further study in nonhuman primates.
The second and most strongly associated genetic variant with the jmSRS-3 factor is in the gene NAA15, part of a complex involved in N-alpha-acetylation, a protein modification essential for cell function. This gene is classified as the highest confidence for association with ASD by SFARI Gene. Human missense variants in NAA15 have been associated with multiple neurodevelopmental phenotypes, including ASD and intellectual disability (ID), and missense mutations in the paralogue of NAA15 generated locomotion defects in C. elegans (Wong et al., 2019). Our jmSRS-3 factor, of course, contains the behavior “poor motor coordination.” The macaque polymorphism is a synonymous variant in threonine also seen in humans (gnomAD) and other macaques (mGAP database) at very low frequencies and does not have any obvious effects on the expressed protein, in keeping with its low CADD score. It may be in linkage with another variant, or the variant may have more significant effects in macaques than in humans; we view both as important questions for follow-up studies.
Only one genetic variant (in LEO1) was significantly associated with aggression in this pilot sample. This variant has been reported in the mGAP database and in the human dbSNP database, but not in the database of human clinically associated variants, ClinVar. Intragroup aggression, especially in juvenile animals, tends to occur at a relatively low levels and intensity in free-ranging conditions where groups are stable and well-integrated. Under such conditions, the majority of agonistic interactions consist of gestures of dominance and submission related to social hierarchy, such as non-contact aggression (threats, chases), withdrawals, fear-grimaces and scream vocalizations. However, higher intensity contact aggression is also observed when tensions escalate (slaps, grabs, bites) (Bernstein & Williams, 1983). Aggressive behaviors may also occur during fights among males over access to sexually receptive females or during stressful situations such as introducing an unfamiliar animal to the group (see (Hall, 1964)). LEO1 is a component of the RNA polymerase II complex. Relatively few mutations in and around the gene have been associated with ASD, primarily in the promoter region (Brandler et al., 2018). These mutations have only been described through largescale sequencing studies, and the current literature does not describe any human LEO1 mutation carriers in sufficient detail to know if aggression was part of any phenotypic spectrum. The high CADD score associated with the variant we observed suggests a potential functional effect despite it being a synonymous change.
Finally, one genetic variant (DlP2C) was significantly associated with grooming duration. Social grooming is a very salient affiliative behavior in rhesus macaques and animals spend a high percentage of their time engaged in it (Wooddell et al., 2017). Grooming in primates serves two primary adaptive functions: a hygienic function to remove ticks, insects, dirt, and other materials from the fur and importantly, a social purpose allowing animals to bond and build or reinforce social relationships (Barton, 1985). This synonymous variant is predicted to appear in multiple isoforms of DIP2C. It is reported at 1% frequency of current samples in mGAP while humans have a G > A SNV reported at the same position, not the G > C SNV we observe in the macaques. The DIP2C protein is predicted to bind transcription factors, particularly in the nervous system and has been classified as a strong candidate for involvement in ASD based on multiple rare loss of function variants [https://gene.sfari.org/database/human-gene/DIP2C].
Variants in three genes were significantly associated with jmSRS-3 behaviors in the GEMMA program but only reached our suggestive association threshold in FAST-LMM (individual data points of variant animals shown in Figure S2). Of particular interest to us is the associated variant in SHANK2, a missense G to A change resulting in amino acid substitution from Ala or Thr, affecting multiple isoforms of the protein. Mutations in the SHANK2 protein are clearly associated with ASD and other behavioral disorders; this is consistent with their role as scaffolding proteins at excitatory synapses (Caumes et al., 2020). Mice with loss-of-function mutations in SHANK2 show increases in repetitive behaviors and altered social behavior (Zaslavsky et al., 2019). Therefore, it is possible that the nature of the missense change observed in our macaques could produce similar repetitive behavior or alterations in sociality. This gene is part of the same family as SHANK3, which has become an interesting NHP transgenic model for ASD in both macaque and marmoset studies (Y. Zhou et al., 2019). Again, the SHANK3 mutations introduced by CRISPR-Cas9 experiments into macaques were not SNVs but were a mosaicism of indels leading to either deleted protein segments or frameshifts and loss of function of the protein, which have never been previously reported in macaque populations and therefore could be expected to yield more disruptive phenotypes than those we would see in our naturalistic population. Altogether, this evidence underscores the value of expanding analyses of naturally occurring macaque genetic variants in SHANK2 and their associations with social behavior collected in complex naturalistic social settings.
Of the variants exhibiting suggestive association in analyses employing both software programs, SMARCA4 is associated with factor jmSRS-3 through multiple variants. The gene encodes a chromatin-remodeling factor required for synapse development (Zhang et al., 2015) and is considered a strong candidate for ASD association [https://gene.sfari.org/database/human-gene/SMARCA4]. In humans, there has been particular interest in postzygotic mosaic mutations in SMARCA4 (Lim et al., 2017). Putative gain of function or dominant negative missense mutations lead to Coffin-Siris syndrome, a rare genetic disorder that includes intellectual disability and delayed development of speech and motor skills (Dsouza et al., 2019). Heterogeneity of resulting phenotype across patients is thought to be related to specific variants and frequencies within any given patient (Mitrakos et al., 2020). The C to T variant identified in our sample has not previously been reported in macaques but is seen in very low frequencies in gnomAD and assigned “likely-benign” clinical significance in ClinVar. All are synonymous changes in the wobble position of an alanine codon (https://www.ncbi.nlm.nih.gov/snp/rs200087760). The second variant associated with jmSRS-3 in SMARCA4 is also a synonymous variant, this time in a glutamine codon. It also has not been reported previously in macaques [mGAP] but is not reported thus far in ClinVar and occurs at very low frequencies in the ExAC database of humans (https://www.ncbi.nlm.nih.gov/snp/rs200665075).
It is important to recognize that the small sample size of this study limits our power to identify gene-phenotype associations. Therefore, failure in this study to find association between any given gene and our behavioral phenotypes should not be taken as evidence that variation in that gene has no effect on the behaviors studied. Additional larger studies are warranted. In addition, the particular cohort of macaques studied here may not be segregating for functional variation in a specific gene, but mutations in that gene may be segregating in other macaque populations, and analyses of those populations might reveal associations that cannot be detected in this cohort.
This exploratory study has yielded some enticing leads that warrant further study. First, we identified extreme social phenotypes including age- and species-atypical behaviors in this population of 200+ male and female juvenile macaques using a novel screening tool (the jmSRS) adapted from the SRS used for ASD diagnosis in children. The human SRS shows high sensitivity and validity based on comparisons with more traditional behavioral measures (ethogram-based), and a similar approach has also been used by other groups studying rhesus macaques (Talbot et al., 2020; Talbot et al., 2021). Second, we found strong evidence for significant additive genetic heritability of patterns of social behavior (both play/affiliative behaviors and solitary behaviors) and non-social behaviors (e.g., anxiety) using multiple behavioral measures in macaques. These results support further use of these measures to investigate individual variation in NHP social behavior and the exploration of various factors (genetics, age, sex, social dominance, etc.) and their interactions. Improved understanding of mechanisms that drive primate behavioral variation will help to delineate the biological underpinnings of social development and social deficits in children, despite the recognition that NHP do not fully manifest the characteristics of any human clinical syndromes. Third, we found that several behavioral phenotypes exhibit effects of sex and/or social dominance rank in addition to also showing significant heritability. Finally, we also found significant associations between particular behavioral phenotypes and individual rhesus macaque SNVs in genes previously found to be associated with human ASD. These genephenotype associations should be explored in larger studies in order to provide robust replication. However, we can conclude there is strong evidence for heritability and preliminary evidence for specific genomic variants that are associated with variance in social behavior among macaques. To our knowledge, this is the first study to demonstrate significant genetic heritability of typical and atypical naturally occurring social behaviors among juvenile macaques living in complex species-typical social groups.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Jennifer Whitley, Josh Bailey, Manuel Bautista and Jessica Johnson, members of the Yerkes National Primate Research Center -YNPRC- Field Station Phenotyping group (PhenX: Identification of Unique Phenotypes at the YNPRC Breeding Colony), and Colony Management for collection of social rank behavioral data; Leonard Howell for the conceptualization and support of the PhenX initiative; Rebecca Herman for developing the data extraction/error checking programs and Hasse Walum and Melinda Higgins for statistical guidance. We also wish to thank Donna Muzny, Kim Walker and Harsha Doddapeneni of the Human Genome Sequencing Center for their assistance in generating the whole exome sequencing data. This study was supported by the National Institutes of Health: Intramural research program of the National Human Genome Research Institute (CG), P50 MH100029 from the National Institute of Mental Health (JB and MMS), P51 OD011132 Office of Research Infrastructure Programs (YNPRC Base Grant = JB, MMS, JR, ZKB), the Yerkes Pilot Research Project Program funded by the YNPRC Base Grant (OD P51OD011132–56, ZKB); and by an Emory University Research Committee (URC) Interdisciplinary Award (00085818, MMS and CG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The YNPRC is fully accredited by AAALAC, International.
Funding information
Emory University, Grant/Award Number: Emory University Research Committee (URC) Interdis; National Human Genome Research Institute, Grant/Award Number: Intramural research program; National Institute of Mental Health, Grant/Award Number: P50 MH100029; National Institutes of Health, Grant/Award Number: P51 OD011132
Footnotes
CONFLICT OF INTERESTS
The authors declare no competing interests.
DATA AVAILABILITY STATEMENT
Exome sequence datasets have been deposited in the sequence read archive (SRA) under BioProject PRJNA732910, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA732910.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- Almasy L, & Blangero J. (1998). Multipoint quantitative-trait linkage analysis in general pedigrees. The American Journal of Human Genetics, 62(5), 1198–1211. 10.1086/301844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann SA (2006). A field study of the sociobiology of rhesus monkeys, Macaca mulatta. Annals of the New York Academy of Sciences, 102(2), 338–435. 10.1111/j.1749-6632.1962.tb13650.x [DOI] [PubMed] [Google Scholar]
- Bai D, Yip B, Windham GC, Sourander A, Francis R, Yoffe R, Glasson E, Mahjani B, Suominen A, Leonard H, Gissler M, Buxbaum JD, Wong K, Schendel D, Kodesh A, Breshnahan M, Levine SZ, Parner ET, Hansen SN, Hultman C, … Sandin S. (2019). Association of Genetic and Environmental Factors with Autism in a 5-country cohort. JAMA Psychiatry, 76(10), 1035–1043. 10.1001/jamapsychiatry.2019.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton R. (1985). Grooming site preferences in primates and their functional implications. International Journal of Primatology, 6(5), 519–532. 10.1007/BF02735574 [DOI] [Google Scholar]
- Bauman MD, & Schumann CM (2018). Advances in nonhuman primate models of autism: Integrating neuroscience and behavior. Experimental Neurology, 299, 252–265. 10.1016/j.expneurol.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisner B, Braun N, Posfai M, Vandeleest J, D’Souza R, & Mccowan B. (2020). A multiplex centrality metric for complex social networks: Sex, social status, and family structure predict multiplex centrality in rhesus macaques. PeerJ, 8, e8712. 10.7717/peerj.8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein IS, & Williams LE (1983). Ontogenetic changes and the stability of rhesus monkey dominance relationships. Behavioural Processes, 8(4), 379–392. 10.1016/0376-6357(83)90025-6 [DOI] [PubMed] [Google Scholar]
- Bowlby J. (1951). Maternal care and mental health. Bulletin of the World Health Organization, 3(3), 355–533. [PMC free article] [PubMed] [Google Scholar]
- Brandler WM, Antaki D, Gujral M, Kleiber ML, Whitney J, Maile MS, Hong O, Chapman TR, Tan S, Tandon P, Pang T, Tang SC, Vaux KK, Yang Y, Harrington E, Juul S, Turner DJ, Thiruvahindrapuram B, Kaur G, Wang Z, … Sebat J. (2018). Paternally inherited cis-regulatory structural variants are associated with autism. Science, 360(6386), 327–331. 10.1126/science.aan2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent L, & Veira Y. (2002). Social behavior of captive Indochinese and insular long-tailed macaques (Macaca fascicularis) following transfer to a new facility. International Journal of Primatology, 23(1), 147–159. [Google Scholar]
- Caskey JR, Wiseman RW, Karl JA, Baker DA, Lee T, Maddox RJ, Raveendran M, Harris RA, Hu J, Muzny DM, Rogers J, & O’Connor DH (2019). MHC genotyping from rhesus macaque exome sequences. Immunogenetics, 71(8–9), 531–544. 10.1007/s00251-019-01125-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumes R, Smol T, Thuillier C, Balerdi M, Lestienne-Roche C, Manouvrier-Hanu S, & Ghoumid J. (2020). Phenotypic spectrum of SHANK2-related neurodevelopmental disorder. European Journal of Medical Genetics, 63, 104072. 10.1016/j.ejmg.2020.104072 [DOI] [PubMed] [Google Scholar]
- Chahrour M, O’Roak BJ, Santini E, Samaco RC, Kleiman RJ, & Manzini MC (2016). Current perspectives in autism Spectrum disorder: From genes to therapy. Journal of Neuroscience, 36, 11402–11410. 10.1523/JNEUROSCI.2335-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroni C, Caporale N, & Testa G. (2020). Autism spectrum disorder at the crossroad between genes and environment: Contributions, convergences, and interactions in ASD developmental pathophysiology. Molecular Autism, 11, 69. 10.1186/s13229-020-00370-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, Metzger LM, Shoushtari CS, Splinter R, & Reich W. (2003). Validation of a brief quantitative measure of autistic traits: Comparison of the social responsiveness scale with the autism diagnostic interview-revised. Journal of Autism and Developmental Disorders, 33, 427–433. 10.1023/A:1025014929212 [DOI] [PubMed] [Google Scholar]
- Constantino JN, Kennon-McGill S, Weichselbaum C, Marrus N, Haider A, Glowinski AL, Gillespie S, Klaiman C, Klin A, & Jones W. (2017). Infant viewing of social scenes is under genetic control and is atypical in autism. Nature, 547(7663), 340–344. 10.1038/nature22999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, & Daly MJ (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43(5), 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza NR, Zimmermann MT, & Geddes GC (2019). A case of coffin–Siris syndrome with severe congenital heart disease and a novel SMARCA4 variant. Molecular Case Studies, 5(3), a003962. 10.1101/mcs.a003962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Manning AK, Florez JC, & Groop L. (2016). The (in) famous GWAS P-value threshold revisited and updated for low-frequency variants. European Journal of Human Genetics, 24, 1202–1205. 10.1038/ejhg.2015.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks LA (2001). Individual differences in response to a stranger: Social impulsivity as a dimension of temperament in vervet monkeys. Journal of Comparative Psychology, 115, 22–28. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, Newman TK, Bailey JN, Jorgensen MJ, Breidenthal SE, Ophoff RA, Comuzzie AG, Martin LJ, & Rogers J. (2004). Genetic contributions to social impulsivity and aggressiveness in vervet monkeys. Biological Psychiatry, 55(6), 642–647. 10.1016/j.biopsych.2003.12.005 [DOI] [PubMed] [Google Scholar]
- Fawcett GL, Dettmer AM, Kay D, Raveendran M, Higley JD, Ryan ND, Cameron JL, & Rogers J. (2014). Quantitative genetics of response to novelty and other stimuli by infant rhesus macaques (Macaca mulatta) across three behavioral assessments. International Journal of Primatology, 35(1), 325–339. 10.1007/s10764-014-9750-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko EJ, Bliss-Moreau E, Walum H, Pruett JR, & Parr LA (2016). The macaque social responsiveness scale (mSRS): A rapid screening tool for assessing variability in the social responsiveness of rhesus monkeys (Macaca mulatta). PLoS One, 11, e0145956. 10.1371/journal.pone.0145956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene RW, Biederman J, Faraone SV, Sienna M, & GarciaJetton J. (1997). Adolescent outcome of boys with attentiondeficit/hyperactivity disorder and social disability: Results from a 4-year longitudinal follow-up study. Journal of Consulting and Clinical Psychology, 65(5), 758–767. 10.1037/0022006X.65.5.758 [DOI] [PubMed] [Google Scholar]
- Hall KRL (1964). Aggression in monkey and ape societies. In Carthy JD & Ebling FJ (Eds.), The natural history of aggression (pp. 51–64). Academic Press. [Google Scholar]
- Harlow HF (1969). Effects of various mother-infant relationships on rhesus monkey behaviour. In Determinants of infant behaviour (Vol. 4). Methuen. [Google Scholar]
- Herman RA, Measday MA, & Wallen K. (2003). Sex differences in interest in infants in juvenile rhesus monkeys: Relationship to prenatal androgen. Hormones and Behavior, 43(5), 573–583. 10.1016/s0018-506x(03)00067-9 [DOI] [PubMed] [Google Scholar]
- Hinrichs AS (2006). The UCSC genome browser database: Update 2006. Nucleic Acids Research, 34(90001), D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins D, William R, Jamie L, & Schaeffer J. (2014). Chimpanzee intelligence is heritable. Current Biology, 24(14), 1649–1652. 10.1016/j.cub.2014.05.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins WD, Keebaugh AC, Reamer LA, Schaeffer J, Schapiro SJ, & Young LJ (2014). Genetic influences on receptive joint attention in chimpanzees (pan troglodytes). Scientific Reports, 4, 3774. 10.1038/srep03774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins WD, Mulholland MM, Reamer LA, Mareno MC, & Schapiro SJ (2020). The role of early social rearing, neurological, and genetic factors on individual differences in mutual eye gaze among captive chimpanzees. Scientific Reports, 10(1), 7412. 10.1038/s41598-020-64051-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Z, Brent L, Alvarenga JC, Comuzzie AG, Shelledy W, Ramirez S, Cox L, Mahaney MC, Huang YY, Mann JJ, Kaplan JR, & Rogers J. (2015). Genetic influences on response to novel objects and dimensions of personality in Papio baboons. Behavior Genetics, 45(2), 215–227. 10.1007/s10519-0149702-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin NH, & Shelton SE (2003). Nonhuman primate models to study anxiety, emotion regulation, and psychopathology. Annals of the New York Academy of Sciences, 1008(1), 189–200. 10.1196/annals.1301.021 [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Whiteman HJ, Mason WA, Mendoza SP, & Capitanio JP (2008). Dimensions of response to novelty are associated with social engagement and aggression in adult male rhesus macaques (Macaca mulatta). Journal of Comparative Psychology, 122(2), 195–203. [DOI] [PubMed] [Google Scholar]
- Klei L, McClain LL, Mahjani B, Panayidou K, De Rubeis S, Grahnat A-CS, Karlsson G, Lu Y, Melhem N, Xu X, Reichenberg A, Sandin S, Hultman CM, Buxbaum JD, Roeder K, & Devlin B. (2021). How rare and common risk variation jointly affect liability for autism spectrum disorder. Molecular Autism, 12(1), 1–13. 10.1186/s13229-021-00466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs Balint Z, Raper J, Michopoulos V, Howell LH, Gunter C, Bachevalier J, & Sanchez MM (2021). Validation of the social responsiveness scale (SRS) to screen for atypical social behaviors in juvenile macaques. PLoS One, 16(5), e0235946. 10.1371/journal.pone.0235946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs-Balint Z, Feczko E, Pincus M, Earl E, Miranda-Dominguez O, Howell B, Morin E, Maltbie E, Li L, Steele J, Styner M, Bachevalier J, Fair D, & Sanchez M. (2019). Early developmental trajectories of functional connectivity along the visual pathways in rhesus monkeys. Cerebral Cortex, 29(8), 3514–3526. 10.1093/cercor/bhy222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen E, Menashe I, Ziats MN, Pereanu W, Packer A, & Banerjee-Basu S. (2016). A systematic variant annotation approach for ranking genes associated with autism spectrum disorders. Molecular. Autism, 7(1), 4. 10.1186/s13229-016-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D’Gama AM, Kim SN, Hill RS, Goldberg AP, Poultney C, Minshew NJ, Kushima I, Aleksic B, Ozaki N, Parellada M, Arango C, Penzol MJ, Carracedo A, Kolevzon A, … Walsh CA (2017). Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nature Neuroscience, 20, 1217–1224. 10.1038/nn.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, & Heckerman D. (2011). FaST linear mixed models for genomewide association studies. Nature Methods, 8, 833–835. 10.1038/nmeth.1681 [DOI] [PubMed] [Google Scholar]
- Liu XQ, Georgiades S, Duku E, Thompson A, Devlin B, Cook EH, Wijsman EM, Paterson AD, & Szatmari P. (2011). Identification of genetic loci underlying the phenotypic constructs of autism Spectrum disorders. Journal of the American Academy of Child & Adolescent Psychiatry, 50, 687–696.e613. 10.1016/j.jaac.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, Wang Y, Zhang CC, Nie YH, Chen ZF, Bian WJ, Zhang L, Xiao J, Lu B, Zhang YF, Zhang XD, Sang X, Wu JJ, Xu X, Xiong ZQ, … Qiu Z. (2016). Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MECP2. Nature, 530, 98–102. 10.1038/nature16533 [DOI] [PubMed] [Google Scholar]
- Madlon-Kay S, Montague MJ, Brent L, Ellis S, Zhong B, Snyder-Mackler N, Horvath JE, Skene J, & Platt ML (2018). Weak effects of common genetic variation in oxytocin and vasopressin receptor genes on rhesus macaque social behavior. In. American Journal of Primatology, 80, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Shaw KA, Baio J, Washington A, Patrick M, DiRienzo M, Christensen DL, Wiggins LD, Pettygrove S, Andrews JG, Lopez M, Hudson A, Baroud T, Schwenk Y, White T, Rosenberg CR, Lee LC, Harrington RA, Huston M, … Dietz PM (2020). Prevalence of autism spectrum disorder among children aged 8years-autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveillance Summaries, 69, 1–12. 10.15585/MMWR.SS6904A1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson MD, & Bernstein IS (2000). Grooming, social bonding, and agonistic aiding in rhesus monkeys. American Journal of Primatology, 51(3), 177–186. 10.1002/1098-2345(200007)51:3177::aid-ajp23.0.co;2-k [DOI] [PubMed] [Google Scholar]
- McCormack K, Howell BR, Guzman D, Villongco C, Pears K, Kim H, Gunnar MR, & Sanchez MM (2015). The development of an instrument to measure global dimensions of maternal care in rhesus macaques (Macaca mulatta). American Journal of Primatology, 77(1), 20–33. 10.1002/ajp.22307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, & DePristo MA (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, & Cunningham F. (2016). The Ensembl variant effect predictor. Genome Biology, 17(1), 122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mier D, & Kirsch P. (2015). Social-cognitive deficits in schizophrenia. In Social behavior from rodents to humans (pp. 397–409). Springer International Publishing. [Google Scholar]
- Mitrakos A, Lazaros L, Pantou A, Mavrou A, Kanavakis E, & Tzetis M. (2020). Coffin-Siris syndrome 4-related Spectrum in a Young woman caused by a heterozygous SMARCA4 deletion detected by high-resolution aCGH. Molecular Syndromology, 11(3), 141–145. 10.1159/000508563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AK, Talbot CF, Del Rosso LA, Maness AC, Simmons S, Garner JP, Capitanio JP, & Parker KJ (2021). Assessment of medical morbidities in a rhesus monkey model of naturally occurring low sociality. Autism Research, 14, 1332–1346. 10.1002/aur.2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EE, & Winslow JT (2009). Non-human primates: Model animals for developmental psychopathology. Neuropsychopharmacology, 34(1), 90–105. 10.1038/npp.2008.150 [DOI] [PubMed] [Google Scholar]
- Paukner A, Slonecker EM, Murphy AM, Wooddell LJ, & Dettmer AM (2018). Sex and rank affect how infant rhesus macaques look at faces. Developmental Psychobiology, 60(2), 187–193. 10.1002/dev.21579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles A, Le Couteur A, Leadbitter K, Salomone E, Cole-Fletcher R, Tobin H, Gammer I, Lowry J, Vamvakas G, Byford S, Aldred C, Slonims V, McConachie H, Howlin P, Parr JR, Charman T, & Green J. (2016). Parent-mediated social communication therapy for young children with autism (PACT): Long-term follow-up of a randomised controlled trial. The Lancet, 6736, 2501–2509. 10.1016/S0140-6736(16)31229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper J, Stephens SBZ, Henry A, Villarreal T, Bachevalier J, Wallen K, & Sanchez MM (2014). Neonatal amygdala lesions Lead to increased activity of brain CRF systems and hypothalamic-pituitary-adrenal Axis of juvenile rhesus monkeys. Journal of Neuroscience, 34(34), 11452–11460. 10.1523/jneurosci.0269-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, & Kircher M. (2018). CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research, 45, 846–860. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, Maller J, Samocha KE, Sanders SJ, Ripke S, Martin J, Hollegaard MV, Werge T, Hougaard DM, iPSYCH-SSI-Broad Autism Group, Neale BM, Evans DM, Skuse D, Mortensen PB, Børglum AD, … Daly MJ (2016). Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nature Genetics, 48, 552–555. 10.1038/ng.3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. (2018). The behavioral genetics of nonhuman primates: Status and prospects. American Journal of Physical Anthropology, 165, 23–36. 10.1002/ajpa.23384 [DOI] [PubMed] [Google Scholar]
- Rogers J, Raveendran M, Fawcett GL, Fox AS, Shelton SE, Oler JA, Cheverud J, Muzny DM, Gibbs RA, Davidson RJ, & Kalin NH (2013). CRHR1 genotypes, neural circuits and the diathesis for anxiety and depression. Molecular Psychiatry, 18(6), 700–707. 10.1038/mp.2012.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MA (1981). Abnormal behaviors in nursery-reared squirrel monkeys (Saimiri sciureus). American Journal of Primatology, 1(1), 35–42. 10.1002/ajp.1350010105 [DOI] [PubMed] [Google Scholar]
- Royston R, Waite J, & Howlin P. (2019). Williams syndrome. Current Opinion in Psychiatry, 32(2), 60–66. 10.1097/yco.0000000000000477 [DOI] [PubMed] [Google Scholar]
- Ryan AM, Berman RF, & Bauman MD (2018). Bridging the species gap in translational research for neurodevelopmental disorders. Neurobiology of Learning and Memory, 165, 106950. 10.1016/j.nlm.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, & Reichenberg A. (2017). The heritability of autism Spectrum disorder. JAMA, 318, 1182–1184. 10.1001/jama.2017.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, Stevens C, Reichert J, Mulhern MS, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, … Buxbaum JD (2020). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell, 1–17, 568–584.e23. 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani V, Del Rosso LA, Seil SK, Calonder LA, Madrid JE, Bone KJ, Sherr EH, Garner JP, Capitanio JP, & Parker KJ (2016). Early predictors of impaired social functioning in male rhesus macaques (Macaca mulatta). PLoS One, 11(10), e0165401. 10.1371/journal.pone.0165401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J, Perry A, Bebko J, & Toplak ME (2014). Review of factor analytic studies examining symptoms of autism spectrum disorders. Journal of Autism and Developmental Disorders, 44, 90–110. 10.1007/s10803-013-1854-3 [DOI] [PubMed] [Google Scholar]
- Talbot CF, Garner JP, Maness AC, Mccowan B, Capitanio JP, & Parker KJ (2020). A psychometrically robust screening tool to rapidly identify socially impaired monkeys in the general population. Autism Research, 13(9), 1465–1475. 10.1002/aur.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot CF, Maness AC, Capitanio JP, & Parker KJ (2021). The factor structure of the macaque social responsiveness scale-revised predicts social behavior and personality dimensions. American Journal of Primatology, 83, e23234. 10.1002/ajp.23234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueoka I, Kawashima H, Konishi A, Aoki M, Tanaka R, Yoshida H, Maeda T, Ozaki M, & Yamaguchi M. (2018). Novel Drosophila model for psychiatric disorders including autism spectrum disorder by targeting of ATP-binding cassette protein a. Experimental Neurology, 300, 51–59. 10.1016/j.expneurol.2017.10.027 [DOI] [PubMed] [Google Scholar]
- Wakeford AGP, Morin EL, Bramlett SN, Howell LL, & Sanchez MM (2018). A review of nonhuman primate models of early life stress and adolescent drug abuse. Neurobiology of Stress, 9, 188–198. 10.1016/j.ynstr.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Harris RA, Haukness M, Fiddes IT, Murali SC, Fernandes J, Dishuck PC, Storer JM, Raveendran M, Hillier LW, Porubsky D, Mao Y, Gordon D, Vollger MR, Lewis AP, Munson KM, DeVogelaere E, Armstrong J, Diekhans M, Walker JA, … Eichler EE (2020). Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science, 370. 10.1126/science.abc6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherby AM, Woods J, Guthrie W, Delehanty A, Brown JA, Morgan L, Holland RD, Schatschneider C, & Lord C. (2018). Changing developmental trajectories of toddlers with autism Spectrum disorder: Strategies for bridging research to community practice. Journal of Speech, Language, and Hearing Research, 61(11), 2615–2628. 10.1044/2018_jslhr-l-rsaut-18-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WR, Brugman KI, Maher S, Oh JY, Howe K, Kato M, & Sternberg PW (2019). Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Human Molecular Genetics, 28, 2271–2281. 10.1093/hmg/ddz051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooddell LJ, Kaburu SSK, Murphy AM, Suomi SJ, & Dettmer AM (2017). Rank acquisition in rhesus macaque yearlings following permanent maternal separation: The importance of the social and physical environment. Developmental Psychobiology, 59(7), 863–875. 10.1002/dev.21555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Go Y, Kushima I, Toyoda A, Fujiyama A, Imai H, Saito N, Iriki A, Ozaki N, & Isoda M. (2016). Singleneuron and genetic correlates of autistic behavior in macaque. Science Advances, 2, e1600558. 10.1126/sciadv.1600558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf A, Waltes R, Haslinger D, Klauck SM, Duketis E, Sachse M, Voran A, Biscaldi M, Schulte-Rüther M, Cichon S, Nöthen M, Ackermann J, Koch I, Freitag CM, & Chiocchetti AG (2020). Quantitative genome-wide association study of six phenotypic subdomains identifies novel genome-wide significant variants in autism spectrum disorder. Translational. Psychiatry, 10(1). 10.1038/s41398020-00906-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavsky K, Zhang WB, McCready FP, Rodrigues DC, Deneault E, Loo C, Zhao M, Ross PJ, El Hajjar J, Romm A, Thompson T, Piekna A, Wei W, Wang Z, Khattak S, Mufteev M, Pasceri P, Scherer SW, Salter MW, & Ellis J. (2019). SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nature Neuroscience, 22, 556–564. 10.1038/s41593-019-0365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cao M, Chang CW, Wang C, Shi X, Zhan X, Birnbaum SG, Bezprozvanny I, Huber KM, & Wu JI (2015). Autism-associated chromatin regulator Brg1/SMARCA4 is required for synapse development and MEF2-mediated synapse remodeling. Molecular and Cellular Biology, 36, MCB.00534–00515. 10.1128/MCB.00534-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, & Stephens M. (2012). Genome-wide efficient mixed-model analysis for association studies. Nature Genetics, 44(7), 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Sharma J, Ke Q, Landman R, Yuan J, Chen H, Hayden DS, Fisher JW 3rd, Jiang M, Menegas W, Aida T, Yan T, Zou Y, Xu D, Parmar S, Hyman JB, Fanucci-Kiss A, Meisner O, Wang D, Huang Y, … Yang S. (2019). Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature, 570(7761), 326–331. 10.1038/s41586-019-1278-0 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Sousa A, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrandez-Peral L, Gulden FO, Yang M, Miller DJ, Marques-Bonet T, Imamura Kawasawa Y, Zhao H, & Sestan N. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science, 362, eaat8077. 10.1126/science.aat8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.