Abstract
Benign prostatic hyperplasia (BPH) is a progressive expansion of peri-urethral prostate tissue common in aging men. Patients with enlarged prostates are treated with 5-alpha reductase inhibitors (5ARIs) to shrink prostate volume by blocking the conversion of testosterone to dihydrotestosterone (DHT). A reduction in DHT levels can elicit atrophy and apoptosis of prostate secretory luminal cells, which results in a favorable clinical response characterized by improved lower urinary tract symptoms. However, the histologic response to 5ARI treatment is often heterogeneous across prostate acini and lower urinary tract symptoms can persist to require surgical intervention. We used two spatial profiling approaches to characterize gene expression changes across histologically normal and atrophied regions in prostates from 5ARI-treated men. Objective transcriptomic profiling using the Visium spatial gene expression platform showed that 5ARI-induced atrophy of prostate luminal cells correlated with reduced androgen receptor signaling and increased expression of urethral club cell genes including LTF, PIGR, OLFM4, SCGB1A1 and SCGB3A1. Prostate luminal cells within atrophied acini adapted to decreased DHT conditions by increasing NF-κB signaling and anti-apoptotic BCL2 expression, which may explain their survival. Using GeoMx digital spatial profiling with a probe set to assess ~18,000 RNA targets, we confirmed that atrophied acini expressing SCGB3A1 displayed higher levels of club cell markers compared to histologically normal acini with NKX3-1 expression. In addition, club-like cells within regions of 5ARI-induced atrophy closely resembled true club cells from the prostatic urethra. A comparison of histologically normal regions from 5ARI-treated men and histologically normal regions from untreated men revealed few transcriptional differences. Taken together, our results describe a heterogeneous response to 5ARI treatment where cells in atrophied acini undergo an adaptation from a prostate secretory luminal to a club cell-like state in response to 5ARI treatment.
Keywords: benign prostatic hyperplasia, luminal epithelia, club cell, spatial transcriptomics, 5 alpha reductase inhibitor
Introduction
Benign prostatic hyperplasia (BPH) is a pleomorphic expansion of stromal and glandular tissue that is commonly treated with 5-alpha reductase inhibitors (5ARIs) such as finasteride or dutasteride [1]. Finasteride can reduce prostate volume by ~19% after one year of treatment through a reduction in prostatic dihydrotestosterone (DHT) levels, resulting in apoptosis of secretory prostate luminal cells and acinar shrinkage [2]. Unlike androgen deprivation therapies (ADT) such as enzalutamide, 5ARI treatment leads to a heterogeneous response with involution of some acini, but not others [1]. This heterogeneity likely contributes to incomplete clinical response to 5ARI treatment, which may necessitate surgery to alleviate obstructive urinary symptoms [3]. Understanding the mechanisms by which some prostate luminal cells survive extended periods of 5ARI treatment could lead to improved therapeutic outcomes by more accurately predicting which patients will achieve a clinical response.
BPH occurs predominantly in the prostate transition zone [4]. Distal prostate acini drain secretions into the prostatic urethra through a proximal ductal network in the transition zone. We discovered that the human prostatic urethra is populated by club and hillock epithelial cell types that extend into the proximal ducts of the transition zone [5]. Recently, single cell RNA-sequencing of the mouse and human prostate demonstrated that prostate luminal cells that survive androgen deprivation undergo a transcriptional shift to a state resembling a urethral cell type [6].
To gain a better understanding of the molecular signatures associated with the heterogeneous response to 5ARI treatment, we used two different spatial transcriptomics platforms. Using the Visium platform, we found that 5ARI-induced glandular atrophy was associated with a gradual transition of prostate luminal cells to a club-like identity. Atrophied acini were characterized by high NF-κB and anti-apoptosis signaling that are normally restricted to the prostatic urethra. Using GeoMx digital spatial profiling, we compared prostate luminal (NKX3-1+) and club (SCGB3A1+) regions from 5ARI-treated and untreated men. SCGB3A1+ atrophied regions from 5ARI-treated men displayed a similar transcript profile to true club cells from the urethra. Prostate luminal cells from acini that did not undergo atrophy after 5ARI treatment displayed a similar profile to normal prostate luminal cells from untreated men. Results from both platforms describe how prostate luminal cells adopt a club-like state following 5ARI treatment while some acini remain unaffected.
Materials and methods
Human tissues
BPH specimens were obtained from patients undergoing simple prostatectomy at UT Southwestern Medical Center. Normal prostate specimens were obtained from organ donors whose families were consented at the Southwest Transplant Alliance under IRB STU 112014-033 (supplementary material, Table S1). Fresh human tissue samples were dissected into portions for digestion of fresh tissue, fixation in 10% formalin, fixation in OCT embedding medium or flash freezing in liquid nitrogen.
Mass spectrometry
Human prostate samples were analyzed using a previously published [7] high pressure liquid chromatographic assay with tandem mass spectrometric detection (LC-MS/MS) for testosterone (T), dihydrotestosterone (DHT), finasteride and dutasteride. Two separate runs were performed on different cohorts of BPH patient samples (supplementary material, Table S2).
Visium spatial transcriptomics
Spatial transcriptomics was performed using the Visium platform from 10x Genomics (Pleasanton, CA, USA). Cryosections from OCT-embedded untreated and 5ARI-treated prostate samples were placed on Visium spatial slides. mRNA bound by printed capture oligos with spatial barcodes on the slide was converted to cDNA. cDNA was transferred from the slide for library preparation and subsequent sequencing using a NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA). Data were merged using Seurat [8] and cell type identities from our normal prostate reference dataset [9] were assigned to the Visium capture dots. Supplementary tables contain sequencing metrics and gene lists for custom scores (supplementary material, Table S3 and Table S4 respectively).
Morphometric analysis
Epithelial acini underlying Visium capture dots categorized as prostate luminal, intermediate luminal, intermediate club-like and club-like were manually outlined in ImageJ [10]. Morphometric analysis was carried out by calculating area and circularity of the outlined regions using inbuilt ImageJ functions.
Single cell RNA-sequencing
Prostate tissue from an 18-year-old organ donor was dissected into urethra/proximal and distal prostate tissue and digested as previously described [5]. Single cells were loaded onto the 10x Genomics chromium controller. Single cell data were analyzed as described previously [9]. Cell types were identified using Seurat’s label transfer method [11] with the same reference used for the Visium data [9]. Sequencing metrics can be found in supplementary material, Table S3.
Tissue immunofluorescence staining
Immunofluorescence staining on paraffin or frozen sections was performed as described previously [12]. Primary and secondary antibody information is presented in supplementary material, Table S5.
RNA in situ hybridization
RNA in situ hybridization was performed using 5-micron paraffin sections using a RNAscope Multiplex fluorescence V2 assay kit (Advanced Cell Diagnostics, Newark, CA, USA) following manufacturer’s instructions. RNAscope probe information is presented in supplementary material, Table S5.
Primary cell culture
Prostate tissue from organ donors was digested into single cells as previously described [5] and cultured in WIT-P media (Rockland Immunochemicals, Pottstown, PA, USA) to promote expansion of epithelial cells. Cells were pretreated with 25 μM BMS-345541 (Sigma Aldrich, St. Louis, MO, USA) or DMSO for 1 h. Cells were stimulated with 10 ng/ml TNF-alpha (Thermo Fisher Scientific, Waltham, MA, USA) for 24 h to induce NF-κB activation. RT-qPCR reactions were set up using iTaq Universal SYBR Green Supermix (Bio-Rad Inc., Hercules, CA, USA). Primer information is provided in supplementary material, Table S5.
GeoMX whole transcriptome assays
Spatial profiling using the GeoMx Whole Transcriptome Atlas RNA assay was performed as described previously [13] with modifications. 5-micron thick paraffin sections of prostate tissue were labeled with RNAscope probes to detect NKX3-1 and SCGB3A1 using the Multiplex Fluorescent V2 kit (Advanced Cell Diagnostics). Sections were hybridized overnight with the GeoMx human whole transcriptome RNA assay probe set (NanoString Technologies, Seattle, WA, USA) and nuclei were labeled with Syto13. Slide scanning, ROI selection, segmentation and barcode collection were performed using the GeoMx Digital Spatial Profiler (NanoString Technologies). DNA barcode libraries were prepared according to manufacturer instructions and sequenced on Nextseq 550 (Illumina). Data analysis was performed using the GeoMx data analysis suite.
For detailed protocols see Supplementary materials and methods.
Results
Visium spatial transcriptomics reveals increased transcriptional heterogeneity of prostate epithelia from 5ARI-treated men
Prostate tissue from BPH patients undergoing simple prostatectomy were subjected to mass spectrometry to validate altered hormone levels in 5ARI-treated patients (supplementary material, Table S2). BPH patients with detectable drug levels by mass spectrometry (5ARI-treated) had reduced DHT levels compared to treatment-naïve (untreated) patients (supplementary material, Figure S1A). Testosterone concentrations were significantly increased in 5ARI-treated patients (supplementary material, Figure S1B).
Epithelial atrophy does not occur uniformly across the entire prostate in 5ARI-treated patients [1]. To characterize how atrophied acini differ transcriptionally from normal acini, we performed Visium spatial transcriptomics on prostate specimens from four 5ARI-treated men and two untreated men. The Visium platform produces a transcriptomic profile of intact tissue sections by capturing mRNA on an underlying grid of equidistant dots, each containing approximately 5 or 6 cells. Using the Seurat label transfer function [11], the transcript signature of each capture dot was assigned a probability score for each of 11 discrete cell types described in our previously published human prostate reference dataset [9]. The cell type corresponding to the highest probability score was assigned as the identity of the capture dot (Figure 1A).
Figure 1. Increased transcriptional heterogeneity of prostate epithelial cells in 5ARI-treated men.
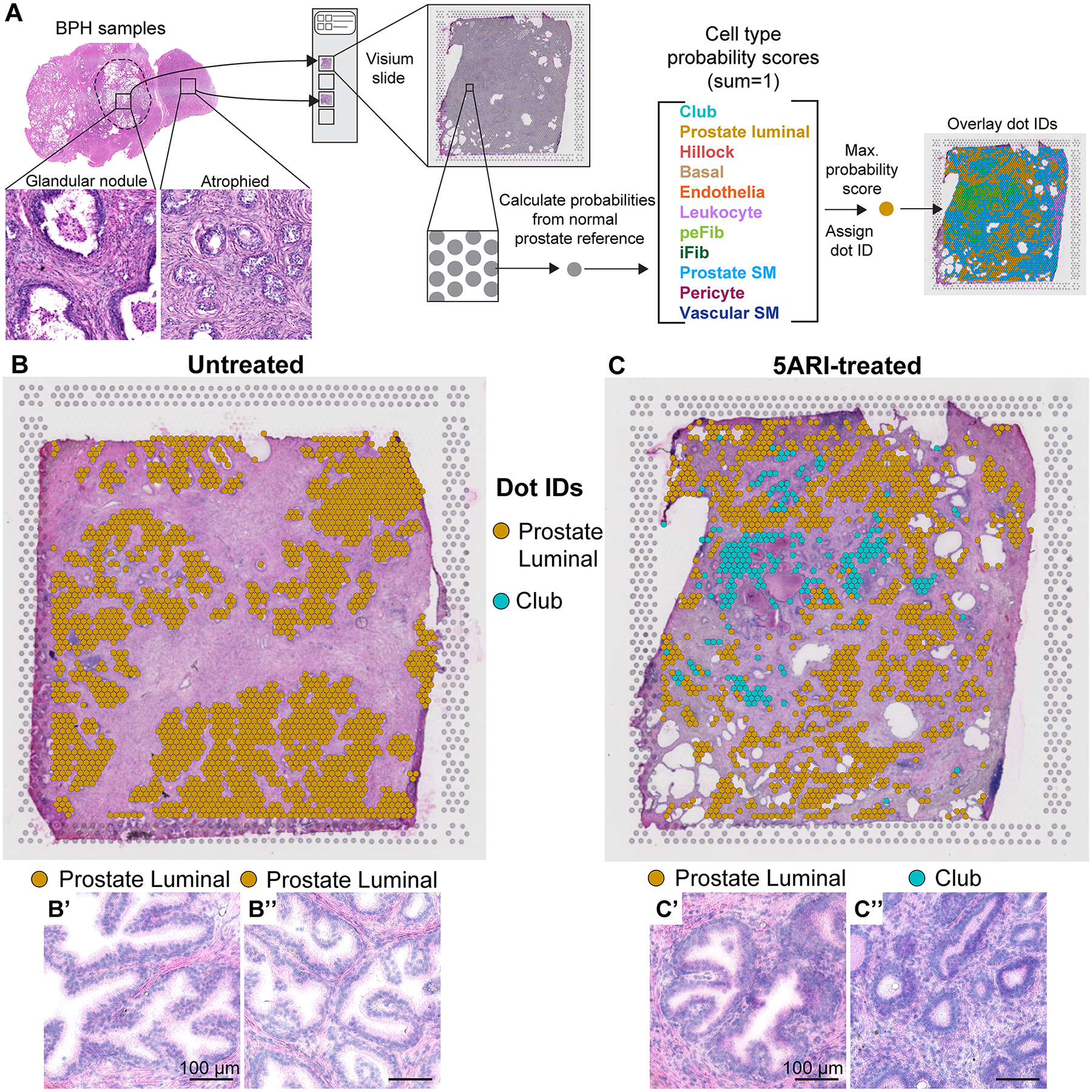
(A) Cryosections from untreated and 5ARI-treated prostates were placed on Visium spatial gene expression slides. Transcriptional data from individual capture dots were compared to normal prostate single cell RNA-sequencing dataset Joseph et al 2021 [9] to obtain identities for each capture dot. Cell type probability scores for 11 cell types in the prostate were calculated which together sum to 1. The cell type associated with the highest probability score value was assigned as the identity of the capture dot. Capture dot identities were overlaid on the image of the original section stained with hematoxylin and eosin. Dots identified as prostate luminal and club were overlaid on slide scans of prostates from (B) untreated and (C) 5ARI-treated men. Magnified regions from (B) underlying dots identified as prostate luminal are shown in B’,B”. Magnified regions from (C) underlying dots identified as prostate luminal and club are shown in C’ and C” respectively. Scale bar, 100 μm. Abbreviations: peFib-Peri epithelial fibroblast, iFib-Interstitial fibroblast, SM-Smooth muscle.
Glandular epithelial regions in untreated men were composed entirely of capture dots labeled as “prostate luminal” (Figure 1B, supplementary material, Figure S2). In contrast, prostate tissue from 5ARI-treated men displayed epithelial heterogeneity with capture dots in some regions identifying as “club” (Figure 1C, supplementary material, Figure S2). Notably, epithelial structures underlying capture dots identified as “prostate luminal” had normal glandular morphology with infolded lumens while acini underlying regions identified as “club” were smaller in size and resembled atrophy associated with androgen deprivation (Figure 1B’,B”–C’,C”) [14].
Prostate luminal cells exhibit a spectrum of club cell gene expression in 5ARI-treated men
We showed previously that club cells are largely confined to the prostatic urethra and proximal ducts in non-diseased young adult men [5]. Cells with club gene profiles are increased in abundance in aged BPH patients [12]. Based on previous evidence from ADT-treated men [6], we set out to test whether small atrophied acini displaying club cell signatures in 5ARI-treated men could represent a survival adaption of prostate luminal cells to a low DHT environment. Compared to untreated men with acini containing only prostate luminal cells (Figure 1B), 5ARI-treated men displayed a mix of epithelia identified as club and prostate luminal (Figure 1C).
We wanted to determine whether capture dots identified as prostate luminal or club were distinct groups, or whether a spectrum of luminal and club cell states existed in 5ARI-treated men. Accordingly, we plotted a spatial heatmap of club scores and prostate luminal scores (Figure 2A–D, supplementary material, Figure S3) onto prostate tissue sections from untreated and 5ARI-treated men. Regions with increased club scores were observed in the 5ARI-treated group but absent in the untreated group. In addition, regions with high prostate luminal scores and high club scores appeared to be mutually exclusive (Figure 2A–D, supplementary material, Figure S3).
Figure 2. Spectrum of prostate luminal and club scores in 5ARI-treated men.
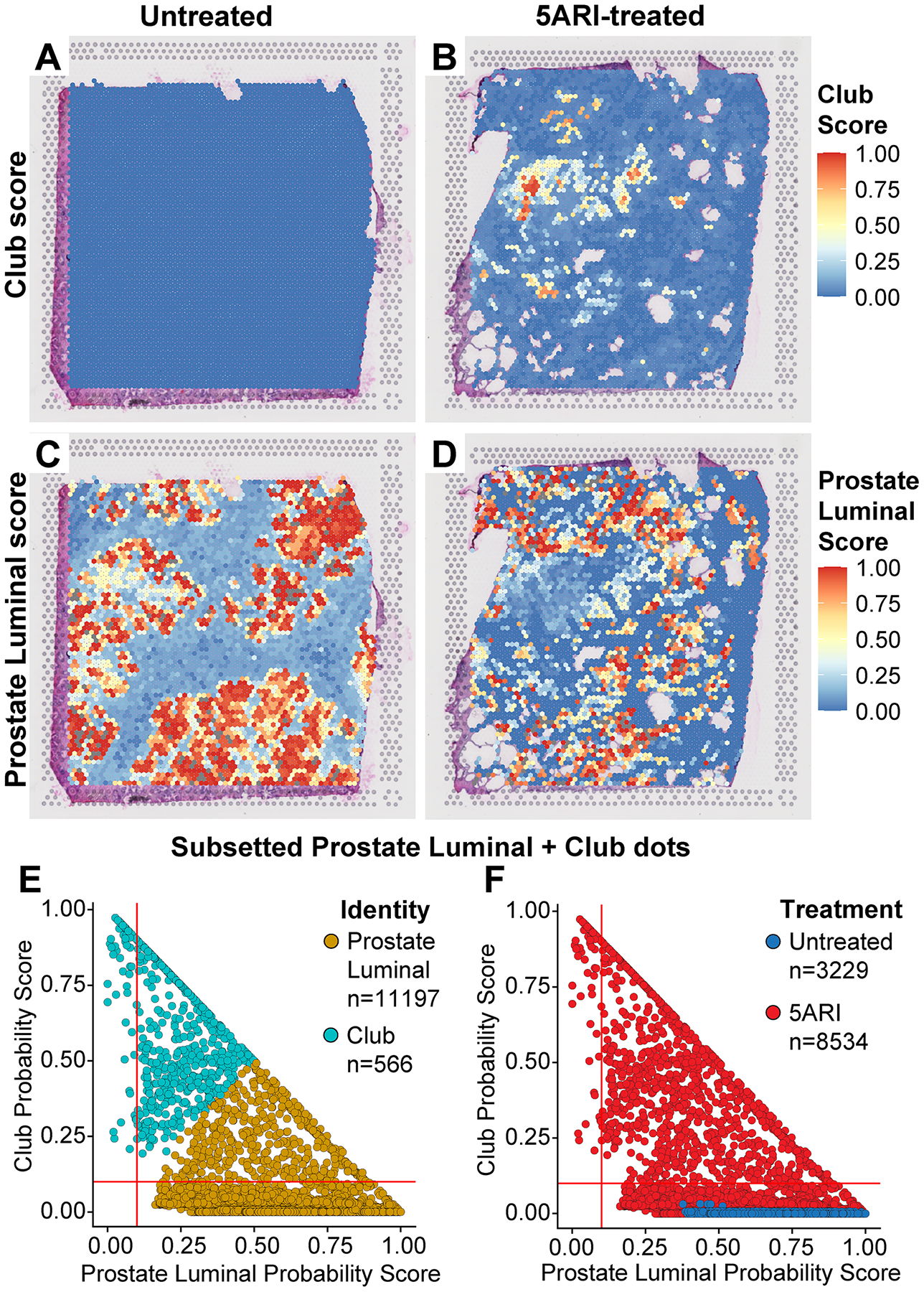
Club score values are plotted as a spatial heatmap on prostates from (A) untreated and (B) 5ARI-treated BPH patients. Prostate luminal score values are plotted as a spatial heatmap on (C) untreated and (D) 5ARI-treated BPH samples. Dots identified as prostate luminal and club were subsetted. Prostate luminal and club score values of these dots are plotted against each other. Dots are shaded by (E) cell identity and (F) treatment status.
To visualize the distribution of prostate luminal and club scores, we subsetted capture dots that identified as prostate luminal or club from all analyzed sections. Club and prostate luminal probability scores of these capture dots were plotted against each other (Figure 2E,F). The results show a continuum of club and prostate luminal scores between the two groups. This distribution was only observed in the 5ARI-treated group and not in the untreated group. Notably, we did not observe a spectrum of prostate luminal to hillock scores after 5ARI treatment, suggesting a specific luminal to club-like adaptation (Figure S4A,B). To test whether basal cells show differentiation to a club-like state, we plotted scores of dots identifying as basal and club. Basal dots did not show an increase in club score in 5ARI prostates indicating that a basal to club transition is unlikely (supplementary material, Figure S4C–D).
In the young normal prostate, club cells are spatially and transcriptionally distinct from prostate luminal cells, localizing to the urethral epithelium and proximal ducts. To confirm that a spectrum of club and prostate luminal cell states does not exist in normal prostate, we performed scRNA-seq on a young organ donor specimen that was dissected into urethra/proximal ducts and distal prostate. We subsetted single cells identified as club and prostate luminal based on our reference scRNA-seq dataset [9] and plotted their club and prostate luminal scores. As expected for two distinct populations, we observed a clear separation of prostate luminal and club cells from the new young donor specimen (supplementary material, Figure S4E). Additionally, these data corroborate our previous finding [5] that urethral club cells are only found in the urethra and proximal region of young healthy prostates (supplementary material, Figure S4F). Given that club cells are very rarely observed away from proximal prostatic ducts, our data suggest that prostate luminal cells in 5ARI-treated men are being skewed towards a club-like transcriptional profile rather than a dramatic expansion of urethral club cells.
Prostate luminal to club-like transition is correlated with morphological response to 5ARI
The morphological response to androgen deprivation therapies includes a spectrum of acinar atrophy and involution, cytoplasmic clearing, nuclear and nucleolar shrinkage [14]. To determine whether the spectrum of prostate luminal and club scores in 5ARI-treated patients correlated with the spectrum of morphological changes, we defined four bins: prostate luminal (<0.1 club score), intermediate prostate luminal (>0.1 club score), intermediate club-like (>0.1 luminal score), and club-like (<0.1 luminal score) (Figure 3A).
Figure 3. Club like gene expression is associated with morphological atrophy in 5ARI-treated men.

Dots identified as prostate luminal and club were subsetted. (A) Prostate luminal dots were categorized into ‘prostate luminal’ (<0.1 club score) and ‘intermediate luminal’ (>0.1 club score). Club dots were classified into ‘club-like’ (<0.1 prostate luminal score) and ‘intermediate club-like’ (>0.1 prostate luminal score). The four categories were overlaid on images from (B) untreated and (C) 5ARI-treated BPH prostates. Images of magnified regions underlying (D) untreated prostate luminal dots, (E) 5ARI-treated prostate luminal dots, (F) 5ARI-treated intermediate luminal dots, (G) 5ARI-treated intermediate club-like dots and (H) 5ARI-treated club-like dots. Morphometric analysis was performed on acini from all 5 categories. Plots showing (I) acinus area, (J) acinus circularity (values from 0–1, 1 is a perfect circle), (K) epithelial cell heights, (L) lumen area, (M) lumen circularity and (N) cell layer thickness. Scale bar, 100 μm. Asterisks indicate significant p-values (* p<0.05, **<0.01, ***<0.001, ****<0.0001). Abbreviations: Int-Intermediate, PrLE-Prostate luminal epithelium, IntLE-Intermediate luminal epithelium, IntClub-Intermediate club.
These four categories were mapped onto prostate sections from untreated and 5ARI-treated patients that had undergone Visium profiling (Figure 3B,C and supplementary material, Figure S5). In untreated men, acini underlying prostate luminal dots had characteristic infolding and tall, ‘fluffy’ secretory cells. In 5ARI-treated patients, secretory acini underlying prostate luminal dots had typical infolding. However, prostate acinus size and shape gradually changed in the intermediate prostate luminal, intermediate club-like and club-like cell categories (Figure 3D–H).
We quantified these changes by performing morphometric analysis on prostate acini from the four transcript bins. The largest acini were observed underlying prostate luminal dots from untreated patients. Acini underlying prostate luminal dots from 5ARI-treated patients had reduced area compared to the untreated group. Acini underlying intermediate luminal, intermediate club-like and club-like dots had a further decrease in area (Figure 3I). In contrast, circularity (1=circle) was lowest in acini from untreated patients, which can be attributed to high levels of infolding. Acini from 5ARI-treated patients had higher circularity with the highest value observed in acini underlying club-like capture dots (Figure 3J). This corresponds to the lack of infolding and smooth circular lumens in acini underlying club-like dots. Likely due to reduced accumulation of secretions, we observed a decrease in cell heights in glandular acini from 5ARI-treated patients compared to untreated (Figure 3K). Changes in lumen area and circularity matched the decrease in acinar area and circularity observed in 5ARI-treated patients (Figure 3L,M). A slight increase in cell layer thickness was observed in atrophied acini from 5ARI-treated patients, which often had 3 or 4 cell layer thickness compared to the 2-cell layer thickness in prostate acini from untreated patients (Figure 3N).
Progressive increase in club gene expression and decreased androgen signaling in prostate luminal cells from 5ARI-treated men.
We assessed gene expression changes across prostate luminal, intermediate prostate luminal, and intermediate club-like and club-like categories. Looking at a set of 25 prostate luminal DEGs from our previously published single cell data [5], we saw a gradual decrease in expression of several androgen-dependent secretory genes including MSMB, KLK3, ACPP, KLK2 and KLK4 between the prostate luminal to club-like categories. In addition, expression of prostate-specific transcription factor NKX3-1 also followed a similar pattern (Figure 4A–C and supplementary material, Figure S6A and Table S6). Expression of these androgen-dependent genes was highest in prostate luminal dots from untreated men with a gradual reduction in expression from prostate luminal to club-like categories in 5ARI-treated men.
Figure 4. Progressive increase in club gene expression in situ in prostate luminal cells from 5ARI-treated men.
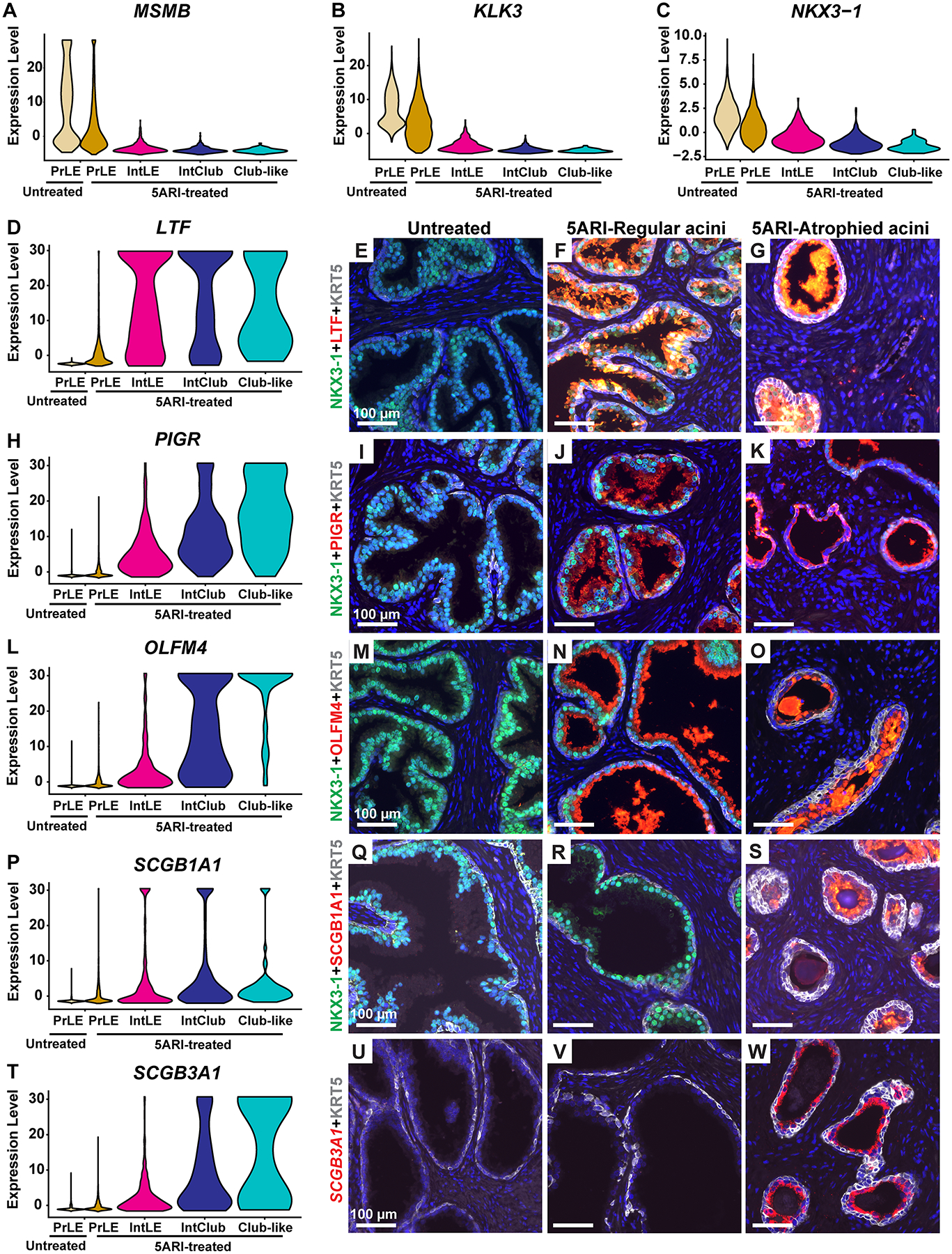
Violin plot depicting expression of the androgen receptor target genes (A) MSMB, (B) KLK3 and (C) NKX3-1 in prostate luminal, intermediate luminal, intermediate club-like and club-like dots. (D) Violin plot for LTF expression. Immunostaining with antibodies against LTF (in red), KRT5 (in white) and NKX3-1 (in green) in (E) untreated prostates, (F) regular acini and (G) atrophic acini from 5ARI-treated prostates. (H) Violin plot for PIGR expression. Immunostaining with antibodies against PIGR (in red), KRT5 (in white) and NKX3-1 (in green) in (I) untreated prostates, (J) regular acini and (K) atrophic acini from 5ARI-treated prostates. (L) Violin plot for OLFM4 expression. Immunostaining with antibodies against OLFM4 (in red), KRT5 (in white) and NKX3-1 (in green) in (M) untreated prostates, (N) regular acini and (O) atrophic acini from 5ARI-treated prostates. (P) Violin plot for SCGB1A1 expression. Immunostaining with antibodies against SCGB1A1 (in red), KRT5 (in white) and NKX3-1 (in green) in (Q) untreated prostates, (R) regular acini and (S) atrophic acini from 5ARI-treated prostates. (T) Violin plot for SCGB3A1 expression. In situ hybridization with probes against SCGB3A1 (in red) and immunostaining with antibodies against KRT5 (in white) in (U) untreated prostates, (V) regular acini and (W) atrophic acini from 5ARI-treated prostates. DAPI staining is in blue. Scale bars, 100 μm. Abbreviations: PrLE-Prostate luminal epithelium, IntLE-Intermediate luminal epithelium, IntClub-Intermediate club.
We observed a reciprocal increase in club cell genes including SCGB3A1, LCN2, PIGR, OLFM4, LTF and SCGB1A1 between the prostate luminal to club-like categories. Expression of most club cell genes was negligible in prostate luminal cells in untreated men with a gradual increase from prostate luminal to club-like categories in 5ARI-treated men (supplementary material, Figure S6B and Table S6).
We validated the overall increase in club cell gene expression in atrophied regions by performing immunofluorescence staining for club cell markers LTF, PIGR, OLFM4, SCGB1A1 and SCGB3A1 in prostate tissue from untreated and 5ARI-treated men. Prostate samples from untreated men showed little to no expression of club cell markers, whereas expression of each of these markers was increased in 5ARI-treated men. Strikingly, we could observe co-labeling of cells for NKX3-1 and club cell markers suggesting that these cells are prostate luminal cells in which expression of club cell markers are being induced. Our staining also validated the decrease in NKX3-1 expression in small atrophied acini containing club-like cells compared to larger acini from both untreated and 5ARI-treated men (Figure 4D–W). Images with individual channels isolated are provided in supplementary material, Figures S7–S11.
Considering the reduced expression of several androgen-regulated genes after 5ARI treatment, we investigated gene and protein expression of androgen receptor (AR) across prostate luminal and club-like categories. AR gene expression was highest in the prostate luminal category and showed a modest decline in intermediate luminal, intermediate club-like and club-like categories (supplementary material, Figure S12A). We observed nuclear AR expression in prostate luminal cells from untreated men. Nuclear AR was reduced in acini expressing the club cell marker LTF in 5ARI-treated men (supplementary material, Figure S12B–D). In 5ARI-treated prostates, we also observed a modest increase in expression of Trop2/TACSTD2 and PSCA, which are well studied in the context of androgen resistance (supplementary material, Figure S12E–F).
NF-κB and anti-apoptotic signaling are increased in atrophied regions from 5ARI-treated men
Compared to prostate luminal dots, club-like dots from 5ARI treated patients show an increase in immune and inflammatory pathways including those involved in antigen presentation, chemokine signaling and toll-like receptor signaling (supplementary material, Table S7). The NF-κB pathway is a key pro-inflammatory signaling pathway. Several studies have shown that AR and NF-κB signaling are mutually antagonistic [15,16]. To determine whether NF-κB signaling was increased in prostates of 5ARI-treated men, we extracted a list of NF-κB target genes (supplementary material, Table S4) [17] to create a combined NF-κB target score for each transcriptional bin. In 5ARI-treated men, we observed a gradual increase in NF-κB target score from prostate luminal to club-like categories (Figure 5A). This was corroborated by increased in situ nuclear localization of phospho-RELA/p65, a subunit of the NF-κB complex, in small atrophied acini from 5ARI-treated men compared to normal acini with large lumens (Figure 5B–D).
Figure 5. NF-κB and anti-apoptotic signaling is increased in atrophied regions from 5ARI-treated men.
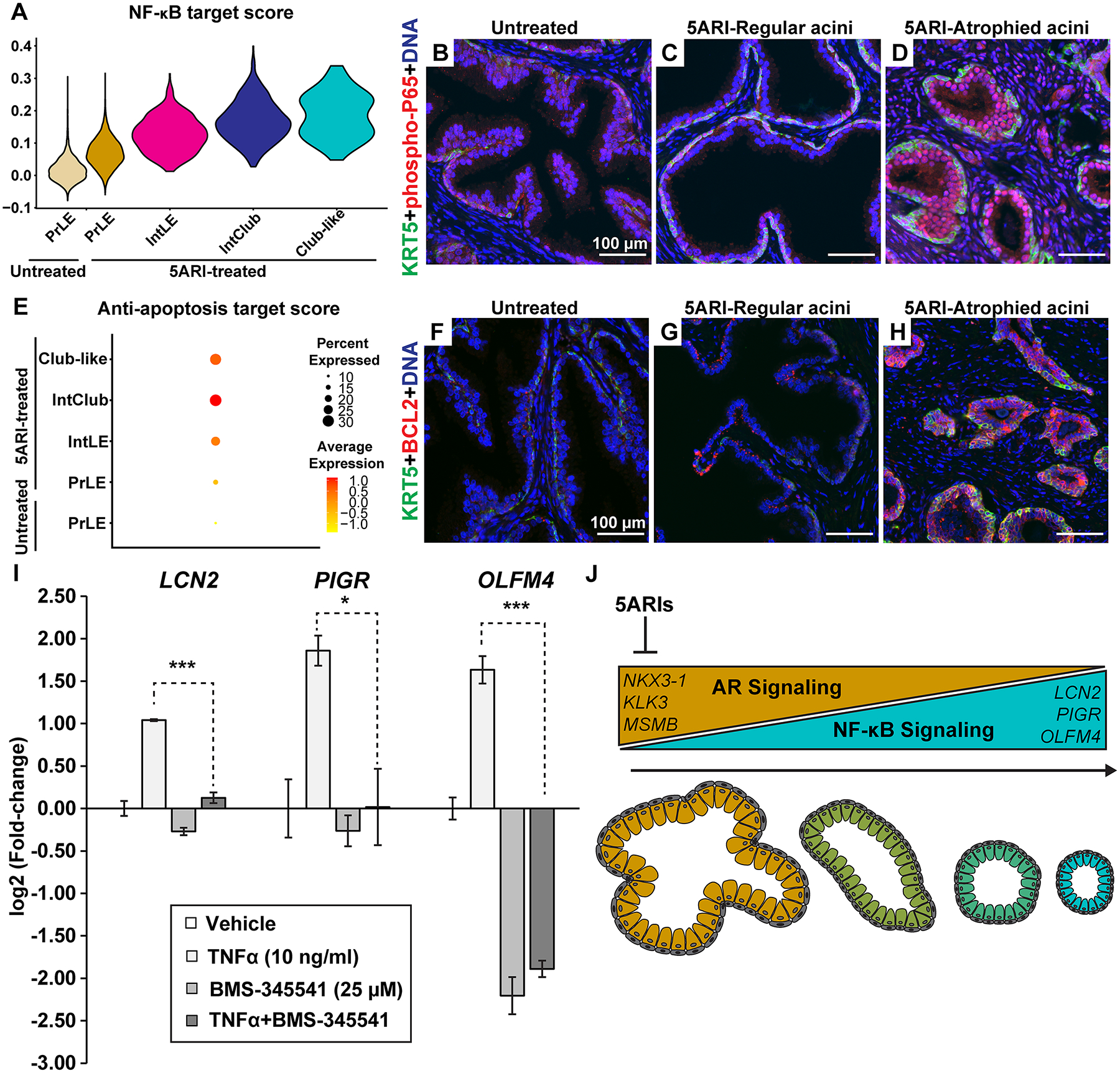
(A) Violin plot depicting NF-κB target gene scores in prostate luminal, intermediate luminal, intermediate club-like and club-like dots. Immunostaining with antibodies against phospho-P65 (in red) and KRT5 (in green) in (B) untreated prostates, (C) regular acini and (D) atrophic acini from 5ARI-treated prostates. (E) Violin plot depicting anti-apoptotic target gene scores in prostate luminal, intermediate luminal, intermediate club-like and club-like dots. Immunostaining with antibodies against BCL2 (in red) and KRT5 (in green) in (F) untreated prostates, (G) regular acini and (H) atrophic acini from 5ARI-treated prostates. (I) Primary prostate epithelial cells treated with TNFα in combination with the NF-κB pathway inhibitor BMS-345541. Results representative of experiments conducted on primary prostate cells derived from at least 2 separate patients. Asterisks represent significant p values (* p<0.05, *** p<0.001). (J) Schematic showing decrease in AR signaling and a reciprocal increase in NF-κB signaling after 5ARI treatment along with a transition from in-folded prostate acini to small atrophied acini. DAPI staining is in blue. Scale bars, 100 μm. Abbreviations: PrLE-Prostate luminal epithelium, IntLE-Intermediate luminal epithelium, IntClub-Intermediate club.
NF-κB mediated activation of anti-apoptotic signaling genes such as BCL2 [18] could be driving survival of atrophied acini in 5ARI-treated men. Anti-apoptotic gene expression, measured as a custom score based on a list of anti-apoptotic genes [19] (supplementary material, Table S4), was increased in the club categories from 5ARI-treated men compared to prostate luminal categories (Figure 5E). Furthermore, increased BCL2 immunoreactivity was observed in atrophied acini from 5ARI-treated men (Figure 5F–H). Similar to regions of atrophy, club cells from the prostatic urethra of a young organ donor had a higher baseline NF-κB score compared to prostate luminal cells (supplementary material, Figure S13A). Urethral cells displayed increased expression of nuclear phospho-RELA/p65 and the NF-κB target gene BCL2 compared to surrounding prostate tissue (supplementary material, Figure S13B–E).
Next, we tested whether increased NF-κB signaling could drive the expression of club genes in primary prostate cells. Several club genes [5] including LCN2 [20], PIGR [21], and OLFM4 [22] are targets of the NF-κB pathway. Treatment with the NF-κB agonist TNFα [23] upregulated expression of LCN2, PIGR and OLFM4 mRNA and this increase was abrogated by pre-treatment and incubation with the NF-κB pathway inhibitor BMS-345541 [24] (Figure 5I). Taken together, our results show that a decrease in AR signaling is accompanied by an increase in NF-κB signaling in atrophied acini (Figure 5J).
Spatial profiling of segmented cell types in prostatic urethra and BPH confirms transcript similarity between urethral club cells and club-like atrophy
To further validate epithelial heterogeneity occurring in 5ARI-treated patients over larger regions including nodular growths, we performed digital spatial profiling on tissue sections hybridized with the GeoMx whole transcriptome atlas RNA probe set (~18,000 RNA probes). Prostate tissue sections from untreated and 5ARI-treated men (n=3 patients/group) were hybridized with fluorescent probes to SCGB3A1 and NKX3-1 to distinguish club cells and prostate luminal cells, respectively (Figure 6A,B, supplementary material, Figure S14). From the untreated group, regions of interest (ROIs) were selected from the urethral epithelium to capture SCGB3A1 expressing urethral club cells (Figure 6A’). ROIs were also selected from NKX3-1-expressing glandular regions representing prostate luminal cells from untreated patients (Figure 6A”). From the 5ARI-treated group, atrophied regions within the prostate with SCGB3A1 labeling were selected to represent club-like regions (Figure 6B’). Atrophied regions had notably reduced NKX3-1 labeling with some residual expression. Histologically normal regions from 5ARI-treated patients that did not undergo atrophy and retained NKX3-1 expression were selected as prostate luminal areas (Figure 6B”). Segmented areas of interest based on SCGB3A1 or NKX3-1 labeling were illuminated with UV light by the GeoMx Digital spatial profiler to photocleave barcoded DNA tags on RNA target probes. Library preparation, sequencing and downstream analyses provided normalized counts of RNA targets within each segmented ROI.
Figure 6. Spatial profiling of segmented cell types in prostatic urethra and BPH confirms transcriptional similarity between urethral club cells and club-like atrophy.

Paraffin tissue sections labeled with probes to NKX3-1 (in green) and SCGB3A1 (in red) were hybridized with the GeoMx whole transcriptome atlas RNA probe set. (A) Representative images of scanned slides showing ROI selection and cell marker segmentation in prostate sections from (A) untreated and (B) 5ARI-treated BPH patients. Images are representative of n=3 slides/group. Magnified regions show representative ROIs and segmentation of (A’) urethral club cells (SCGB3A1+) and (A”) histologically normal prostate luminal regions (NKX3-1+) from untreated men. Representative ROIs and segmentation of (B’) atrophied club-like regions (SCGB3A1+) and (B”) histologically normal prostate luminal regions (NKX3-1+) from 5ARI-treated men. (C) UMAP plot of 4 segmented categories. An intermediate prostate luminal category from 5ARI-treated patients is circled by dotted lines. (D–I) Images of segmented ROIs labeled on the UMAP plot in panel (C) representing the spectrum of normal to atrophied prostate acini. (J) Differential expression analysis was performed comparing club-like and PrLE categories from 5ARI-treated patients (n=18 ROIs/group from 3 patients). Top 25 genes sorted by log2 fold-change that were up and down in the club-like category (FDR<0.05) were used for unsupervised clustering of all ROIs from 5ARI and untreated patients. Results are depicted in a heatmap of normalized target counts. (K) DEGs in urethral club versus prostate luminal segments from untreated men. (L) DEGs in club-like cells versus prostate luminal segments from 5ARI-treated men. (M) Common DEGs between (K) and (L). (N) DEGs between urethral club segments in untreated men versus club-like segments in 5ARI-treated men. (O) DEGs between prostate luminal segments in untreated versus 5ARI-treated men. (P) Volcano plot of DEGs in urethral club versus prostate luminal segments from untreated men. (Q) Volcano plot of DEGs in prostate luminal cells versus club-like segments from 5ARI-treated men. Syto13 staining for DNA is in blue. Gray scale bar, 2.5 mm. White scale bar, 100 μm. Abbreviations: PrLE-Prostate Luminal, Ur-Urethral.
On a UMAP plot, untreated urethral club and prostate luminal segments clustered furthest apart while 5ARI-treated club-like and prostate luminal segments were closer in proximity (Figure 6C). A small group of 5ARI-treated prostate luminal segments were categorized as an intermediate prostate luminal group due to their proximity to the club-like segments on the UMAP plot (Figure 6C, circled). Images of representative segments from each of the four groups and the intermediate prostate luminal group showed a change in prostate glandular morphology from histologically normal to atrophied (Figure 6D–I and supplementary material, Figure S15A–E). The small group of intermediate prostate luminal segments from 5ARI-treated patients, although retaining NKX3-1 expression, had characteristics of atrophy including smoother lumens and smaller lumen sizes (Figure 6F and supplementary material, Figure S15B–D). The intermediate group showed a modest increase in expression of key club cells genes [5] and decreased expression of NKX3-1 compared to the remaining prostate luminal segments from the 5ARI-treated group (supplementary material, Figure S15F–M). This corroborates our previously described Visium spatial transcriptomics findings that indicated a transition from prostate luminal to an atrophied phenotype accompanied by an increase in club gene expression.
Differential expression analysis of the GeoMx data confirmed increased expression of club cell genes (SCGB3A1, SCGB1A1, LTF, PIGR and OLFM4) and decreased androgen regulated genes (NKX3-1, MSMB and KLK3) in atrophied regions (club-like) compared to histologically normal regions (PrLE) from 5ARI-treated patients (n=3 patients) (Figure 6J,L,Q and supplementary material, Table S8). Unsupervised clustering of all segments based on the top 25 significant DEGs (up and down) showed that NKX3-1+ and SCGB3A1+ segments clustered apart irrespective of treatment status (Figure 6J). Comparison of urethral club segments (Ur Club) and prostate luminal (PrLE) segments from untreated patients confirmed differential expression of previously described club cell genes [5] including SCGB3A1, LCN2 and PIGR (Figure 6K,P and supplementary material, Table S8). Comparison of untreated urethral club to prostate luminal segments yielded 4,774 significant DEGs (False discovery rate, FDR<0.05) (Figure 6K,P and supplementary material, Table S8) while the comparison of 5ARI-treated club-like to prostate luminal segments showed 3090 DEGs (FDR<0.05) (Figure 6L,Q and supplementary material, Table S8). This represented an overlap of 2,111 shared DEGs between the two comparisons (Figure 6M). In contrast, the comparison of club-like atrophied regions from 5ARI-treated patients and urethral club cells from untreated patients revealed only 66 DEGs (Figure 6N and supplementary material, Figure S16A and Table S8). These results demonstrate that the high degree of transcript similarity between regions of 5ARI-induced club-like atrophy and urethral club cells.
While the histologic response to 5ARIs is strongly associated with a prostate luminal to club cell transition, we noted that some nodules in 5ARI-treated patients were resistant to atrophy and club-like differentiation. Response to 5ARI treatment is known to be heterogeneous, but nodules have not been identified as uniquely resistant [1]. Our spatial profiling results demonstrate high concordance between phenotypically normal NKX3-1+ luminal epithelia from 5ARI-treated versus untreated men with only 38 DEGs between the 2 groups (Figure 6O and supplementary material, Figure S16B and Table S8). Because only minimal molecular differences can be detected between histologically normal prostate acini from untreated and 5ARI-treated men, it is possible that the ~2-fold increase in testosterone in the prostates of 5ARI-treated men (Figure S1B) is capable of maintaining prostate luminal identity in some glands. Other causes of the heterogeneous response to 5ARIs should be explored further to improve clinical responses.
Discussion
5-alpha reductase inhibitors (5ARIs) are used to shrink prostate volume and reduce obstructive symptoms in BPH patients [25]. To characterize the heterogeneous histologic response to 5ARIs [1] molecularly, we used two different spatial transcriptomics approaches. The progressive morphological atrophy of prostate acini that is attributed to 5ARI treatment was correlated with high expression of club cell markers including SCGB1A1, SCGB3A1, LTF, PIGR and OLFM4. Considering the low abundance of normal club cells in 5ARI-naïve BPH specimens, the emergence of a club-like state is likely an adaptation of prostate luminal cells to androgen deprivation and not the survival and expansion of pre-existing club cells. Luminal cells with a club-like signature were also observed in benign cells from prostate cancer patients treated with androgen deprivation therapy [6], although it is not yet clear whether prostate cancer cells undergo a club cell transition during androgen deprivation.
After castration in mice, surviving luminal epithelia of the distal prostate also acquire urethral or ‘L2’ cell markers such as Tacstd2 and/or Trop2 [6,12]. This has caused some confusion regarding the putative role of Sca-1+/Trop2+/Psca+/Krt4+ proximal urethral luminal epithelia as progenitors for Nkx3-1+ distal prostate luminal cells. Although proximal urethral luminal cells survive better in 3D spheroid culture conditions, lineage tracing of this cell type through castration and regeneration does not support their role as a progenitor for the distal prostate [6,26]. Our results confirm that the histologic response of prostate glands to androgen deprivation through 5ARI treatment is correlated with a transcriptional adaptation of prostate luminal epithelia to a club-like identity. Moreover, it is possible that club-like cells in atrophied regions could regrow prostate tissue after cessation of 5ARI treatment [27].
In addition to androgen deprivation, age-related androgen decline and chronic inflammation could potentially induce a similar luminal to club adaptation. Serum testosterone levels decrease in men with age by ~2–3% annually [28] and androgen deprivation is known to drive prostate inflammation [29]. Inflammation is positively correlated with a loss of luminal AR expression [30] and BPH patients with higher baseline inflammation display greater prostate volume [31]. A prostate luminal to club adaptation could also be occurring in normal aging as well. Aging is accompanied by increased abundance of urethral-like cells in the mouse and human prostate [32], but further studies on the effects of aging and decreasing androgen levels on luminal to club transition are needed.
Finally, glandular nodules containing histologically normal NKX3-1+ luminal epithelia can be seen adjacent to regions of atrophy in the same patient. A transcriptional comparison of NKX3-1+ luminal epithelia from histologically normal glands inside 5ARI-resistant nodules versus histologically normal glands in untreated men revealed minimal differences (Figure 6O, supplementary material, Figure S16B). Although AR signaling could be driven by the high levels of testosterone resulting from 5ARI treatment (supplementary material, Figure S1B), this does not explain how adjacent regions in the same tissue undergo atrophy. It is possible that there is a unique stromal composition within non-responding nodules compared to the regions that undergo atrophy. In future studies, understanding the stroma within regions of atrophy and within non-responding nodules could help decipher mechanisms of 5ARI treatment resistance. In addition, it should be determined whether a certain nodular size is predictive of resistance to 5ARI treatment, which could reduce unnecessary treatment and improve clinical outcomes.
Supplementary Material
Figure S1. Reduced DHT levels and increased testosterone levels in 5ARI-treated men compared to untreated
Figure S2. Replicate samples show increased transcript heterogeneity of prostate epithelium from 5ARI-treated men
Figure S3. Replicate samples show increased club cell signature in 5ARI-treated prostates
Figure S4. A spectrum of scores does not exist between prostate luminal and hillock cells from 5ARI-treated prostates or prostate luminal and urethral club cells from young normal prostate
Figure S5. Spatial plots of prostate luminal and club categories from replicate samples
Figure S6. Dot plots of prostate luminal and club cell genes across categories
Figure S7. LTF and NKX3-1 immunofluorescence staining of untreated and 5ARI treated BPH prostates
Figure S8. PIGR and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S9. OLFM4 and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S10. SCGB1A1 and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S11. Fluorescence staining to detect SCGB3A1 mRNA and NKX3-1 protein in untreated and 5ARI-treated BPH prostates
Figure S12. Changes in AR, TACSTD2 and PSCA expression in 5ARI-treated prostates
Figure S13. Increased NF-κB signaling and BCL2 expression in urethral club cells compared to prostate luminal cells
Figure S14. ROI selection and segmentation of replicate samples for GeoMx spatial profiling
Figure S15. Intermediate prostate luminal regions in 5ARI-treated samples
Figure S16. Comparison of club (SCGB3A1+) and prostate luminal (NKX3-1+) segments from untreated and 5ARI-treated patients
Table S1. Sample information
Table S2. Mass spectrometry results
Table S3. Visium sequencing metrics
Table S4. Lists for NF-κB and anti-apoptotic gene score
Table S5. Key resources table
Table S7. Pathway analysis 5ARI treated prostate luminal versus club-like dots
Table S8. GeoMx annotations and differentially expressed genes
Table S6. Visium differentially expressed genes
Acknowledgments
We thank the families of organ donors at the Southwest Transplant Alliance for their commitment to basic science research. We acknowledge the assistance of the UT Southwestern Tissue Resource (5P30CA142543), DNA Microarray core and the Whole Brain Microscopy Core. Financial support came from NIH R01 DK115477 and U54DK104310 (DWS), AUA research scholar award 659333 (DBJ); and the generous donations of the Smith, Penson and Harris families to the UT Southwestern Department of Urology (CGR). We acknowledge the assistance of NCI P30CA016056 to the Roswell Park Comprehensive Cancer Center for support of the Bioanalytical, Metabolomics, and Pharmacokinetics Shared Resource.
Footnotes
No conflicts of interest were declared
SUPPLEMENTARY MATERIAL ONLINE
References 32–35 are cited only in supplementary materials
Data availability statement
Visium and single cell RNA-sequencing data have been deposited in the Gene Expresion Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) (GSE178934 and GSE179312).
References
- 1.Strand DW, Costa DN, Francis F, et al. Targeting phenotypic heterogeneity in benign prostatic hyperplasia. Differentiation; research in biological diversity 2017; 96: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gormley GJ, Stoner E, Bruskewitz RC, et al. The effect of finasteride in men with benign prostatic hyperplasia. The Finasteride Study Group. The New England journal of medicine 1992; 327: 1185–1191. [DOI] [PubMed] [Google Scholar]
- 3.McConnell JD, Roehrborn CG, Bautista OM, et al. The long-term effect of doxazosin, finasteride, and combination therapy on the clinical progression of benign prostatic hyperplasia. The New England journal of medicine 2003; 349: 2387–2398. [DOI] [PubMed] [Google Scholar]
- 4.McNeal JE. Origin and evolution of benign prostatic enlargement. Investigative urology 1978; 15: 340–345. [PubMed] [Google Scholar]
- 5.Henry GH, Malewska A, Joseph DB, et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell reports 2018; 25: 3530–3542.e3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karthaus WR, Hofree M, Choi D, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science (New York, NY 2020; 368: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilton JH, Titus MA, Efstathiou E, et al. Androgenic biomarker prof|ling in human matrices and cell culture samples using high throughput, electrospray tandem mass spectrometry. The Prostate 2014; 74: 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021; 184: 3573–3587.e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph DB, Henry GH, Malewska A, et al. Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions. J Pathol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019; 177: 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph DB, Henry GH, Malewska A, et al. Urethral luminal epithelia are castration-insensitive cells of the proximal prostate. The Prostate 2020; 80: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 2020; 38: 586–599. [DOI] [PubMed] [Google Scholar]
- 14.Bostwick DG, Qian J, Civantos F, et al. Does finasteride alter the pathology of the prostate and cancer grading? Clin Prostate Cancer 2004; 2: 228–235. [DOI] [PubMed] [Google Scholar]
- 15.Ko S, Shi L, Kim S, et al. Interplay of nuclear factor-kappaB and B-myb in the negative regulation of androgen receptor expression by tumor necrosis factor alpha. Mol Endocrinol 2008; 22: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelius T, Filleur S, Yemelyanov A, et al. Androgen receptor targets NFkappaB and TSP1 to suppress prostate tumor growth in vivo. Int J Cancer 2007; 121: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 1999; 18: 6853–6866. [DOI] [PubMed] [Google Scholar]
- 18.Catz SD, Johnson JL. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene 2001; 20: 7342–7351. [DOI] [PubMed] [Google Scholar]
- 19.Jourdan M, Reme T, Goldschmidt H, et al. Gene expression of anti- and pro-apoptotic proteins in malignant and normal plasma cells. Br J Haematol 2009; 145: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iannetti A, Pacifico F, Acquaviva R, et al. The neutrophil gelatinase-associated lipocalin (NGAL), a NF-kappaB-regulated gene, is a survival factor for thyroid neoplastic cells. Proc Natl Acad Sci U S A 2008; 105: 14058–14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno ME, Frantz AL, Rogier EW, et al. Regulation of the polymeric immunoglobulin receptor by the classical and alternative NF-κB pathways in intestinal epithelial cells. Mucosal Immunol 2011; 4: 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KK, Park KS, Song SB, et al. Up regulation of GW112 Gene by NF kappaB promotes an antiapoptotic property in gastric cancer cells. Mol Carcinog 2010; 49: 259–270. [DOI] [PubMed] [Google Scholar]
- 23.Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci U S A 1989; 86: 2336–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke JR, Pattoli MA, Gregor KR, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem 2003; 278: 1450–1456. [DOI] [PubMed] [Google Scholar]
- 25.Marberger M Drug Insight: 5α-reductase inhibitors for the treatment of benign prostatic hyperplasia. Nat Clin Pract Urol 2006; 3: 495–503. [DOI] [PubMed] [Google Scholar]
- 26.Kwon OJ, Choi JM, Zhang L, et al. The Sca-1(+) and Sca-1(−) mouse prostatic luminal cell lineages are independently sustained. Stem cells (Dayton, Ohio) 2020; 38: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim W, Jung JH, Kang TW, et al. Clinical effects of discontinuing 5-alpha reductase inhibitor in patients with benign prostatic hyperplasia. Korean journal of urology 2014; 55: 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman HA, Longcope C, Derby CA, et al. Age trends in the level of serum testosterone and other hormones in middle-aged men: longitudinal results from the Massachusetts male aging study. J Clin Endocrinol Metab 2002; 87: 589–598. [DOI] [PubMed] [Google Scholar]
- 29.Sorrentino C, Musiani P, Pompa P, et al. Androgen deprivation boosts prostatic infiltration of cytotoxic and regulatory T lymphocytes and has no effect on disease-free survival in prostate cancer patients. Clin Cancer Res 2011; 17: 1571–1581. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Kwon OJ, Henry G, et al. Non-Cell-Autonomous Regulation of Prostate Epithelial Homeostasis by Androgen Receptor. Mol Cell 2016; 63: 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert G, Descazeaud A, Nicolaiew N, et al. Inflammation in benign prostatic hyperplasia: a 282 patients’ immunohistochemical analysis. The Prostate 2009; 69: 1774–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowell PD, Fox JJ, Hashimoto T, et al. Expansion of Luminal Progenitor Cells in the Aging Mouse and Human Prostate. Cell reports 2019; 28: 1499–1510 e1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaari G, Bolen CR, Thakar J, et al. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res 2013; 41: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA Science 2014; 343: 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Reduced DHT levels and increased testosterone levels in 5ARI-treated men compared to untreated
Figure S2. Replicate samples show increased transcript heterogeneity of prostate epithelium from 5ARI-treated men
Figure S3. Replicate samples show increased club cell signature in 5ARI-treated prostates
Figure S4. A spectrum of scores does not exist between prostate luminal and hillock cells from 5ARI-treated prostates or prostate luminal and urethral club cells from young normal prostate
Figure S5. Spatial plots of prostate luminal and club categories from replicate samples
Figure S6. Dot plots of prostate luminal and club cell genes across categories
Figure S7. LTF and NKX3-1 immunofluorescence staining of untreated and 5ARI treated BPH prostates
Figure S8. PIGR and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S9. OLFM4 and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S10. SCGB1A1 and NKX3-1 immunofluorescence staining of untreated and 5ARI-treated BPH prostates
Figure S11. Fluorescence staining to detect SCGB3A1 mRNA and NKX3-1 protein in untreated and 5ARI-treated BPH prostates
Figure S12. Changes in AR, TACSTD2 and PSCA expression in 5ARI-treated prostates
Figure S13. Increased NF-κB signaling and BCL2 expression in urethral club cells compared to prostate luminal cells
Figure S14. ROI selection and segmentation of replicate samples for GeoMx spatial profiling
Figure S15. Intermediate prostate luminal regions in 5ARI-treated samples
Figure S16. Comparison of club (SCGB3A1+) and prostate luminal (NKX3-1+) segments from untreated and 5ARI-treated patients
Table S1. Sample information
Table S2. Mass spectrometry results
Table S3. Visium sequencing metrics
Table S4. Lists for NF-κB and anti-apoptotic gene score
Table S5. Key resources table
Table S7. Pathway analysis 5ARI treated prostate luminal versus club-like dots
Table S8. GeoMx annotations and differentially expressed genes
Table S6. Visium differentially expressed genes
Data Availability Statement
Visium and single cell RNA-sequencing data have been deposited in the Gene Expresion Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) (GSE178934 and GSE179312).


