Abstract
Background
MicroRNAs (miRNAs) function as potential diagnostic biomarkers in various cancers. This study aimed to evaluate the roles of miR‐205‐5p in lung cancer progression and diagnosis.
Materials and Methods
MiR‐205‐5p was detected by quantitative real‐time PCR. The effect of miR‐205‐5p on cell proliferation and metastasis was estimated by MTT and flow cytometry. The expression of TP53INP1 and related genes was analyzed by immunoblotting. The diagnostic value of miR‐205‐5p was analyzed using receiver operating characteristic (ROC) curve analysis, sensitivity, and specificity.
Results
The miR‐205‐5p was increased in lung cancer tissues. MiR‐205‐5p mimics were promoted but its inhibitor suppressed cell proliferation and metastasis compared with control treatment in vitro and in vivo. By regulating the 3′ untranslated region, miR‐205‐5p could negatively regulate TP53INP1 expression, which further inhibited the expression of RB1 and P21, but increased that of cyclinD1. Moreover, the serum miR‐205‐5p levels of patients with lung cancer were significantly higher than those of normal controls, and they were correlated with patients' gender, drinking status, and clinical stage. The area under the ROC curve of serum miR‐205‐5p in the diagnosis of non‐small‐cell lung cancer was 0.8250, respectively. The finding supported its possession of high diagnostic efficiency for lung cancer.
Conclusions
MiR‐205‐5p promoted lung cancer cell proliferation and metastasis by negatively regulating the novel target TP53INP1, which further affected the expression of P21, RB1, and cyclin D1. Serum miR‐205‐5p is a novel and valuable biomarker for lung cancer diagnosis.
Keywords: biomarker, lung cancer, miR‐205‐5p, TP53INP1, tumorigenesis
MiR‐205‐5p was upregulated in lung cancer and promoted lung cancer cell proliferation by negatively regulating a novel target TP53INP1, which further affected the expression of P21, RB1, and cyclin D1. A high level of serum miR‐205‐5p is a valuable biomarker for the diagnosis of patients with lung cancer.

INTRODUCTION
Lung cancer is the carcinogenesis of lung tissue cells, and it seriously threatens public health as its incidence and mortality are the highest among malignant tumors worldwide. 1 In 2017, the China Cancer Center reported that the number of new cases of lung cancer was 1.2 million per year, and 80% of lung cancer is non‐small‐cell lung cancer (NSCLC). 1 , 2 , 3 The early symptoms of patients with lung cancer are not obvious, and the detection of lung cancer is often in middle or late stage. Therefore, early diagnosis is the key measure to improve the survival rate and reduce mortality. The development of lung cancer is closely related not only to environmental factors but also genetic factors. The limitations of application in early clinical screening are due to the lack of satisfactory sensitivity and specificity of tumor markers, therefore establishing higher specificity and sensitivity of markers is important for lung cancer diagnosis and therapy.
MicroRNAs (miRNAs), as endogenous and non‐coding small RNAs, 18–22 nucleotides in length, play the crucial roles of oncogenes and suppressive genes in tumor progression by regulating the targeted gene. 4 , 5 , 6 , 7 The expression of miRNAs is specific in different tissues and cells due to their different functions, which are involved in differentiation, development, and cell cycle. 8 MiRNA dysregulation could induce many cases of cancer, and understanding the functions of miRNAs in tumors is not only helpful to explain the occurrence and development of tumors, but also beneficial for patients to obtain accurate treatment.
MiR‐205‐5p has been reported to play crucial roles in tumorigenesis of lung cancer. 9 , 10 , 11 , 12 , 13 The expression of miR‐205‐5p in lung squamous cell carcinoma (LUSC) and small‐cell lung cancer (SCLC) tissues was reported to increase significantly compared with that in adjacent tissues. 10 In serum, significantly higher miR‐205‐5p relative expression was observed in patients with NSCLC than in the control group. 11 Ulivi et al. studied the prognostic value of miRNAs in 182 patients with lung cancer, and found that miR‐205‐5p was significantly associated with survival of patients with LUSC. 12 Although the abovementioned studies showed that miR‐205‐5p levels were increased in lung cancer tissues, the detailed mechanism of miR‐205‐5p in tumorigenesis and its role as a diagnostic biomarker for lung cancer needed to be further studied. Here, miR‐205‐5p promoted lung cancer cell proliferation and metastasis and its targeted gene. Serum miR‐205‐5p levels were also measured to explore the roles of miR‐205‐5p in the diagnosis of lung cancer.
METHODS
Patients and serum samples
Seventy‐five patients with lung cancer were recruited from Dongying People's Hospital, Affiliated Teaching Hospital of Binzhou Medical University, Shandong Province, China. All patients were pathologically diagnosed with lung cancer for the first time. None of them received any radiotherapy or chemotherapy, or had undergone surgical treatment. Sixty‐two healthy volunteers were enrolled as healthy controls without any physical illness or tumor during the medical checkup in this hospital. The study protocol was approved by the Medical Ethics Committee of Binzhou Medical University and written informed consent was obtained from all subjects before the study.
Sample preparation and miRNA extraction
Blood samples were collected from each sample between 7:00 a.m. and 8:00 a.m. The blood samples were centrifuged at 3000 rpm for 30 min at room temperature to separate serum. The serum samples were then stored at −80°C. MiRNAs were extracted from 200 μL of serum with a miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacture's instructions. In brief, 200 μL of serum sample was treated with 1 mL of QIAzol solution (Qiagen). MiRNAs were extracted after adding 1.6 × 108 copies of miRNeasy Serum/Plasma Spike‐In Control (Qiagen). They were then eluted in 14 μL of RNase‐free water, and quality was verified using an Eppendorf Bio Spectrometer (Eppendorf). The miRNA specimens were stored at −80°C.
Cell culture
Human lung cancer A549 and H1975 cell lines were purchased from the Shanghai Institute of Cell Biology, China. The cells were cultured in dulbecco's modification of eagle's medium (DMEM) medium with 10% fetal bovine serum, then cultured at 37°C with 5% CO2 in a humid incubator.
Reverse transcription and qRT‐PCR
MiRNAs were added with poly (A) tail by Poly (A) polymerase (Ambion). Reverse transcription was performed by SuperTag Polymerase (TaKaRa Biotechnology). Then, miR‐205‐5p levels were detected using the StepOnePlus Real‐Time PCR System (Thermo Fisher Scientific) with a QuantiTect SYBR Green RT‐PCR Kit (Qiagen). The primers used to amplify miR‐205‐5p (GenePharm Co., Ltd) were forward, 5′‐TCCTTCATTCCACCGGAGTC‐3′ and reverse, 5′‐AACATGTACAGTCCATGGATG‐3′. The PCR cycles incubation at 95°C for 10 min, followed by 40 cycles of 95°C 15 s and 60°C for 1 min. MiR‐39 was used as a control, 5′‐AGCTGATTTCGTCTTGGTAATA‐3′. All reactions were performed in triplicate. The results were represented as cycle threshold (CT) and calculated using the 2−ΔΔCT method. 14
Cell viability assays
The transfected cells were seeded into 96‐well plates at a density of 1 × 104 cells/well. A 3‐(4,5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyltetrazolium bromide solution (20 μL of 5 mg/mL MTT; Sigma) was added to the cultures (for a total volume of 200 μL) and incubated for 4 h at 37°C. DMSO was then added and absorbance at 570 nm was measured.
CCK8 assay
A549 and H1975 cells were seeded into 96‐well plates with 1 × 104 cells/well. After miRNA or antisense oligonucleotides (ASO) treatment for 24, 48 or 72 h, 10 μL of CCK8 (Biosharp) was added to each well. The plates were incubated in the incubator for 1–4 h. The absorbance was measured by one ELISA reader (Multiskan FC, Thermo Fisher Scientific) at 450 nm.
Transwell migration assay
Cell migration was performed using Corning Costar Transwell chambers with 8 μm pore size membranes (Sigma). After treatment, A549 and H1975 cells incubated in a 500‐μL serum‐free medium were seeded into the upper chamber. Then, 600 μL of 1640 medium supplemented with 10% calf serum was added to the lower chamber. After 24 h of treatment, lower chamber cells were stained with 1% crystal violet (Sigma) in 2% ethanol for 20 min. The stained cells were counted under a microscope (DM6000B, Leica). The experiment assay was analyzed in triplicate.
Luciferase reporter assay
The primer sequence for the wild‐type TP53‐INP1‐3′ untranslated region (UTR) was forward, 5′‐TGTCAAAGTGAGTTAGAAGT‐3′; reverse, 5′‐AAGTAAGTAGATAGGGATGC‐3′. For the luciferase assay, Lipofectamine 2000 was used to co‐transfect A549 cells with the miR‐205‐5p and pGL3‐tp53‐inp1 luciferase vectors. The firefly luciferase activity was measured 24 h after transfection, and the results were normalized against β‐gal. Each sample was assayed in triplicate.
Apoptosis assay
Apoptosis assay was performed 48 h after transfection by using Annexin V‐FITC/PI in accordance with the manufacture's protocols (KeyGEN Biotech. Co. Ltd), as described in a previous study. 15
Immunoblotting
Total protein was extracted from the transfected cells by using RIPA lysis buffer (Beyotime) according to the manufacturer's instructions. The blots were incubated with primary antibodies at 4°C. The following antibody dilutions were used: anti‐TP53INP1 (1:1000; Bioss), anti‐P21 (1:1000; Proteintech Group), anti‐RB1 (1:1000; Bioworld Technology), anti‐cylin D1 (1:1500; Bioworld Technology), and anti‐GAPDH (1:10000; Bioworld Technology). After incubation with HRP‐conjugated secondary antibodies was performed, the protein bands were visualized using an enhanced chemiluminescence reagent (Beyotime Biotechnology).
Cancer cell xenografts
A549 cells treated with miR‐205‐5p, ASO‐205, or controls were harvested after transfection. In total, 2 × 106 A549 cells were injected subcutaneously into the backs of BALB/C‐nu nude mice aged 6–8 weeks (purchased from HFK Bio‐Technology). The primary tumors were estimated daily by a caliper. One month later, the mice were euthanized by intraperitoneally injecting a barbiturate.
Statistical analyses
The correlation of serum miR‐205‐5p levels with the clinical features of lung cancer, such as age, gender, smoking history, alcohol history, histology, TNM (tumor, node, metastasis) stage, and pathological type, were analyzed using SPSS 22.0 software (IBM Corp.). Quantitative data were tested for normality and homogeneity. Normal data were presented as mean ± standard deviation. Non‐normal data were presented as median and quartiles. Categorical data were expressed as percentages. Intergroup difference was compared using Student's t‐test for normal data, the Wilcoxon rank‐sum test for non‐normal data, and the chi‐square (χ 2 ) test for categorical data. Non‐normal data was compared using the Mann–Whitney U test. The receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value of serum miR‐205‐5p for differentiation between patients with lung cancer and healthy subjects. A value of p < 0.05 was considered statistically significant.
RESULTS
MiR‐205‐5p levels were increased in lung cancer tissues
MiRNAs, as small nonprotein coding RNAs, act as tumor suppressors and oncogenes through negative gene regulation in various cancers. 16 Although miR‐205‐5p plays important roles in tumorigenesis of human cancers, it can act either as an oncogene or as tumor‐suppressive gene in a certain microenvironment. 17 For further investigation of its roles in lung cancer, miRNA microarray datasets were download from the NCBI Gene Expression Omnibus (GEO) through accession code GSE27705 (https://www.ncbi.nlm.nih.gov). The expression of 33 miRNAs, including miR‐205‐5p, increased, but that of 30 miRNAs decreased in NSCLC tissues (n = 20) compared with control tissues (n = 10) (Figure 1a,b). Data from the cancer genome atlas (TCGA) further proved that miR‐205‐5p expression was significantly increased in lung adenocarcinoma tissues (n = 447) (Figure 1c) and lung squamous‐cell carcinoma tissues (n = 336) compared with that in normal controls (n = 44, p < 0.01) (Figure 1d). These results indicate that miR‐205‐5p may promote the progression of lung cancer.
FIGURE 1.
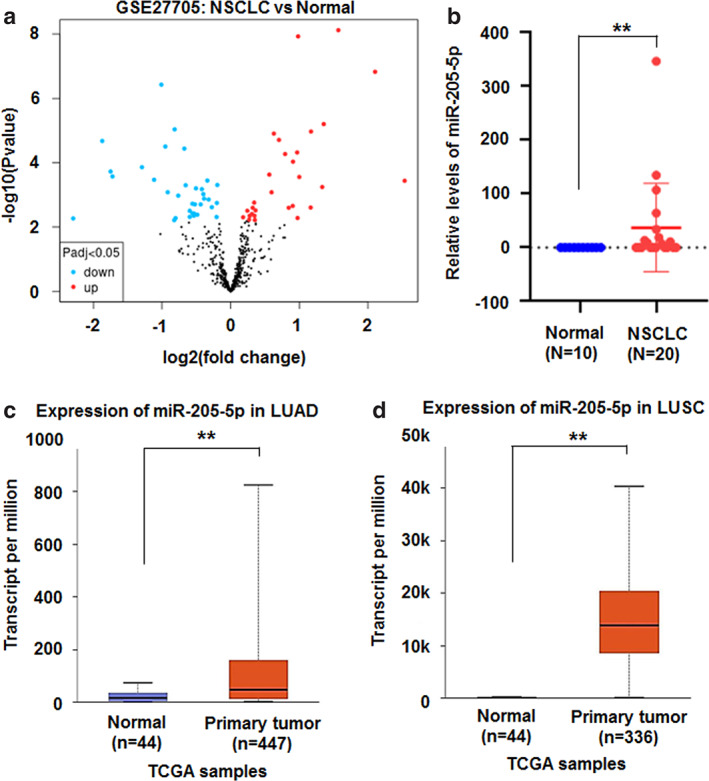
Higher miR‐205‐5p levels in lung cancer tissues. (a) Volcano plot of miRNA datasets GSE27705. Red, upregulated miRNAs; blue, downregulated miRNAs. (b) miR‐205‐5p expression was increased in non‐small‐cell lung cancer (n = 20) compared with normal tissues (n = 10) from GSE27705. Data were shown as median (interquartile range), **p < 0.01, Mann–Whitney U test. (c, d) miR‐205‐5p levels increased in lung adenocarcinoma (LUAD, n = 447) and lung squamous cell carcinoma (LUSC, n = 336) compared with normal controls (n = 44) from TCGA, respectively. Data were shown as median (interquartile range), **p < 0.01. TCGA, the cancer genome atlas
MiR‐205‐5p promotes the growth and metastasis of lung cancer cells
Lung cancer cell lines A549 and H1975 were transfected with miR‐205‐5p mimics or ASO‐205‐5p (miR‐205 inhibitor) to further investigate the role of miR‐205‐5p in lung cancer progression. Compared with scrambled controls, miR‐205‐5p was significantly overexpressed and promoted cell proliferation in miR‐205‐5p mimic‐treated A549 and H1975 cells compared with scrambled control treatment (Figure 2a–d). CCK8 assay further showed that miR‐205‐5p overexpression promoted d A549 and H1975 cell proliferation (Figure 2e,f). Cell migration assay demonstrated that miR‐205‐5p mimics also increased the metastasis of A549 and H1975 cells compared with scrambled control (Figure 2g,h). The apoptotic number of A549 or H1975 cells did not obviously change in the miR‐205‐5p mimic‐treated cultures (Figure 2i,j).
FIGURE 2.
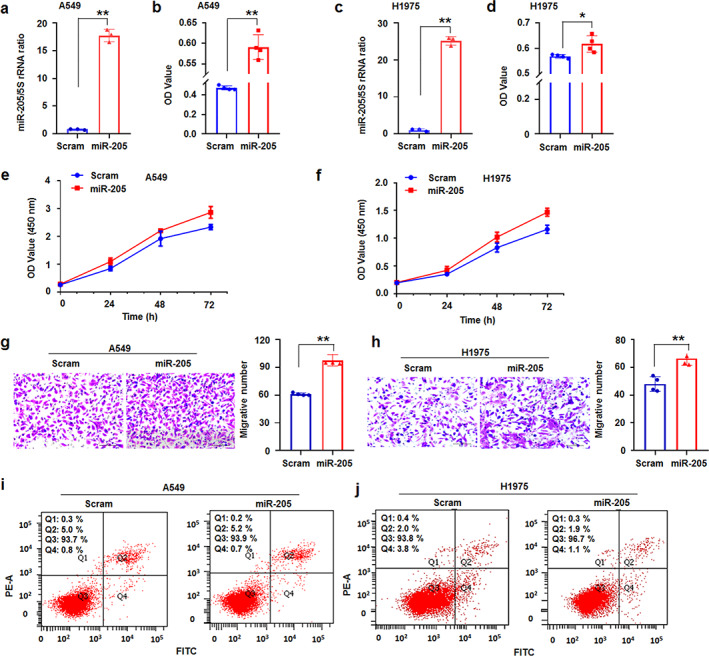
MiR‐205‐5p promoted lung cancer cell progression. (a) qRT‐PCR showed that miR‐205‐5p was upregulated in miR‐205‐5p‐treated A549 cells. (b) MTT assay illustrated that miR‐205‐5p promoted A549 cell proliferation. (c) qRT‐PCR showed that miR‐205‐5p was overexpressed in miRNA ‐treated H1975 cells. (d) miR‐205‐5p promoted H1975 cell proliferation. (e, f) CCK8 assay showed that miR‐205‐5p overexpression promoted A549 and H1975 cell proliferation, respectively. (g, h) miR‐205‐5p promoted the metastasis of A549 and H1975 cells, respectively. (I, j) miR‐205‐5p does not increase theA549 and H1975 cell apoptosis, respectively. Data expressed as mean ± SD for triplicate experiments. *p < 0.05, **p < 0.01; Student's t‐test
However, ASO‐205‐5p (miR‐205 inhibitor) treatment inhibited A549 and H1975 cell proliferation compared with control treatment (Figure 3a–d). CCK8 assay also showed that ASO‐205‐5p suppressed A549 and H1975 cell growth (Figure 3e,f). ASO‐miR‐205‐5p significantly inhibited cell metastasis (Figure 3g,h) and induced cell apoptosis (Figure 4i,j) of A549 or H1975 cells compared with control treatment. The cell experiments supported that miR‐205‐5p promoted lung cancer progression.
FIGURE 3.
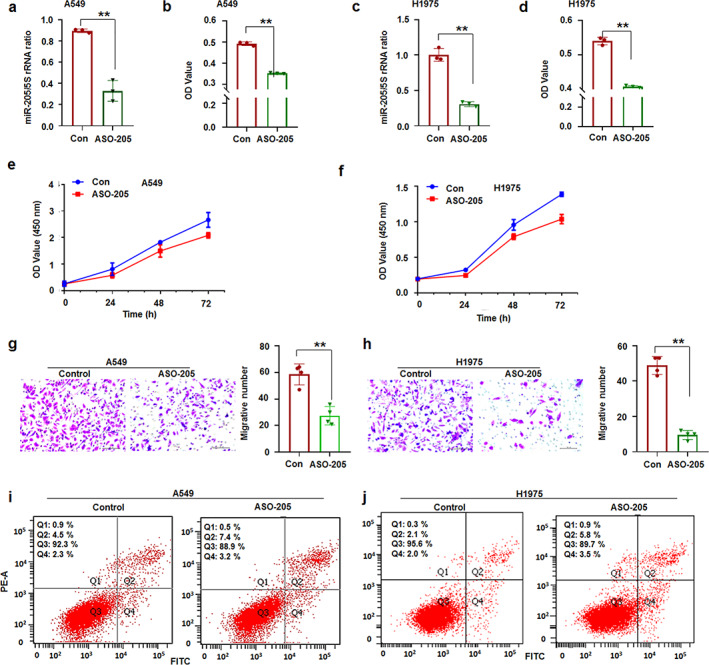
ASO‐205‐5p prevented lung cancer cell progression. (a) qRT‐PCR analysis. ASO‐205 inhibitor suppressed miR‐205‐5p levels in A549 cells. (b) MTT assay. ASO‐205 suppressed A549 cell proliferation. (c) qRT‐PCR analysis. miR‐205‐5p was decreased in ASO‐205‐treated H1975 cells. (d) MTT assay. ASO‐205 suppressed H1975 cell proliferation. (e,f) CCK8 assay. ASO‐205‐5p suppressed A549 and H1975 cell growth, respectively. (g, h) ASO‐205 suppressed the metastasis of A549 and H1975 cells, respectively. (i, j) ASO‐205 increased the number of apoptotic A549 and H1975 cells, respectively. Data expressed as mean ± SD for triplicate experiments. **p < 0.01; Student's t‐test
FIGURE 4.
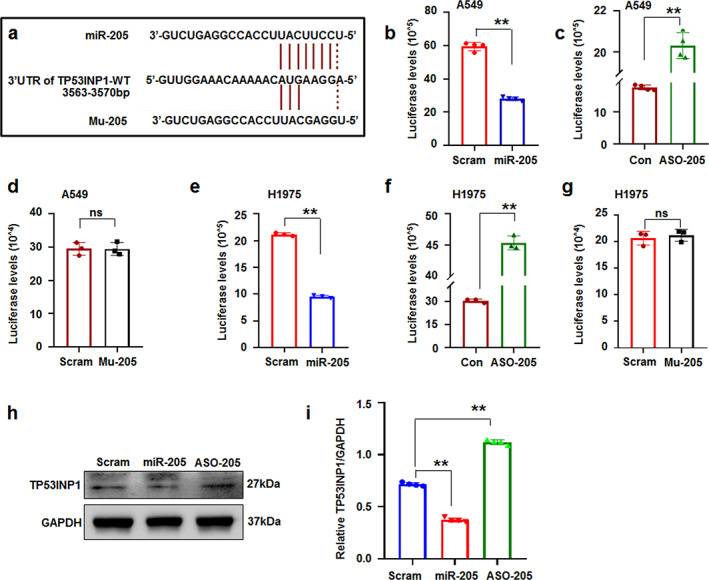
TP53INP1 is a novel target of miR‐205. (a) Diagram of the miR‐205‐5p putative binding sites alignment with TP53INP1‐UTR. (b) MiR‐205‐5p reduced luciferase levels in pc‐luci‐TP53INP1‐UTR‐treated A549 cells. (c) ASO‐205 raised luciferase levels in pc‐luci‐TP53INP1‐UTR‐treated A549 cells. (d) Mutant miRNA could not affect luciferase levels in pc‐luci‐TP53INP1‐UTR‐treated A549 cells. (e–g) MiR‐205‐5p decreased, ASO‐205 increased, and mutant miRNA has no effect on luciferase expression in pc‐luci‐TP53INP1‐UTR‐treated H1975 cells, respectively. (h, i) Immunoblotting analysis and relative TP53INP1/GAPDH ratio. MiR‐205‐5p decreased TP53INP1 levels. Data expressed as mean ± SD for triplicate experiments. **p < 0.01; Student's t‐test or ANOVA
MiR‐205‐5p targets and negatively regulates TP53INP1‐related genes
MiRNAs could directly regulate gene expression via the 3′‐UTR of its targeted gene. 17 Though miR‐205‐5p could enhance the process of lung cancer, 18 its detailed roles in lung cancer remain unclear. To further investigate the mechanism of miR‐205‐5p in regulating cancer cell proliferation, the target genes of miR‐205‐5p were predicted online (http://www.targetscan.org/index.html) and the results showed that the 3′‐UTR of tumor protein p53 inducible nuclear protein 1 (TP53INP1) has the targeted sites of miR‐205‐5p (Figure 4a). Then, the full‐length wild‐type TP53INP1 3′‐UTR was cloned and inserted into the downstream of luciferase reporter to construct pcDNA‐luci‐TP53INP1‐3′UTR. Luciferase reporter assay demonstrated that miR‐205‐5p suppressed luciferase expression in miR‐205‐5p and TP53INP1‐3′UTR‐transfected A549 cells compared with scrambled control treatment (Figure 4b). MiR‐205‐5p inhibitor (ASO‐205) increased the luciferase levels compared with that in control‐treated cultures (Figure 4c). Mutant miR‐205‐5p (Mu‐205) could not increase the luciferase levels in A549 cells (Figure 4d). Similar luciferase changes were demonstrated in miR‐205‐5p or inhibitor or mutant miRNA‐transfected H1975 cells (Figure 4e–g), indicating that miR‐205‐5p could regulate luciferase expression via TP53INP1 3′‐UTR.
Immunoblotting revealed that the TP53INP1 protein levels in miR‐205‐5p‐treated cells significantly decreased and levels in ASO‐205‐treated cultures increased compared with that in control treatment (Figure 4h,i). The results further supported that miR‐205‐5p could regulate TP53INP1 protein levels.
As a nuclear protein, TP53INP1 can bind to p53 and then regulates p53 transcriptional activity on P21 promoters. 19 Furthermore, TP53INP1 induces G1 arrest and increases p53‐mediated apoptosis, which could influence cell cycle progression by regulating cyclin D1 and RB1. 20 , 21 As TP53INP1 is a downstream target of miR‐205‐5p, it can regulate the expression of TP53INP1‐related genes, such as RB1, P21, and cyclin D1. The results of the present study showed that overexpression of miR‐205‐5p inhibited the expression of RB1 and P21, but increased that of cyclinD1. ASO‐205 upregulated the expression of RB1 and P21 but decreased that of cyclinD1 (Figure 5a,b). Therefore, miR‐205‐5p could regulate the expression of TP53INP1 and its related genes, which may be related to the oncogenic role of miR‐205‐5p in promoting lung cancer progression.
FIGURE 5.
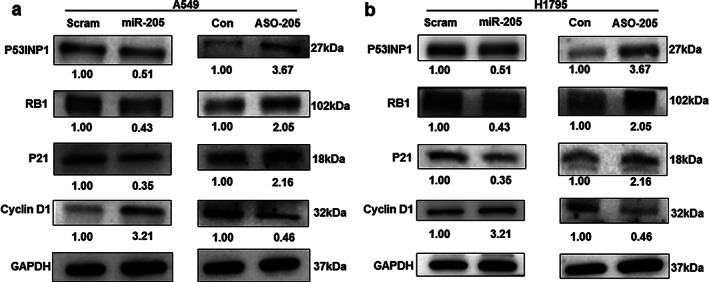
miR‐205‐5p regulated the expression of TP53INP1‐related proteins. (a, b) Immunoblotting analysis. miR‐205‐5p downregulated RB1 and p21 but upregulated cyclin D1 expression, while ASO‐205 increased RB1 and p21 but reduced cyclin D1 levels in A549 and H1975 cells, respectively
miR‐205‐5p promoted lung cancer growth in xenograts
To further study the roles of miR‐205‐5p in lung cancer in vivo, A549 cells treated with miR‐205‐5p or control were hypodermically injected to produce xenografts into nude mice as previously described. 15 Every 2 days, the volumes of the xenografts and the weights of mice were measured. The mice were sacrificed at 1 month after treatment. MiR‐205‐5p could promote tumor proliferation both in weights and volumes compared with control groups (Figure 6a–c). As expected, miR‐205‐5p was overexpressed but TP53INP1 was reduced in miR‐205‐5p‐treated xenografts compared with the control groups (Figure 6d,e). However, miR‐205‐5p inhibitor suppressed tumor proliferation in weights and volumes (Figure 6f–h). MiR‐205‐5p also decreased and TP53INP1 increased in ASO‐205‐treated xenografts compared with the control groups (Figure 6i,j). The results demonstrate that miR‐205‐5p plays an oncogenic role in lung cancer growth in vivo.
FIGURE 6.
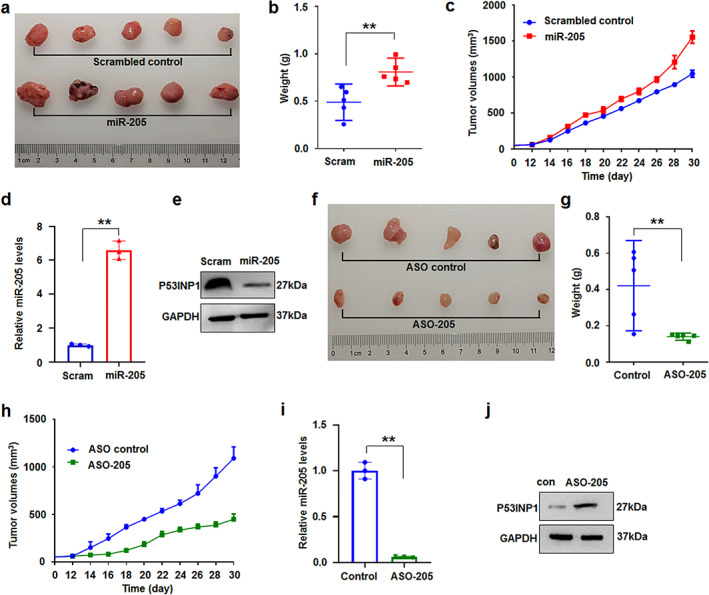
miR‐205‐5p regulated cell proliferation in vivo. (a–c) Analysis of miR‐205‐5p‐treated‐xenograft tumors in vivo (n = 5). Quantitative data of tumor weight (b) and dynamically detected volume change (c) of xenografts were increased in miR‐205‐5p‐treated xenografts. Data were expressed as median (interquartile range), **p < 0.01; Mann–Whitney U. (d, e) miR‐205‐5p levels were increased by qRT‐PCR analysis and TP53INP1 protein was reduced by immunoblotting in miR‐205‐5p‐treated‐xenografts, respectively. Data expressed as mean ± SD for triplicate experiments. **p < 0.01. (f–h) Quantitative data of tumor weight (g) was reduced and dynamically detected volume was inhibited (h) in ASO‐205‐treated‐xenograft tumors in vivo (n = 5). Data were expressed as median (interquartile range), ** p < 0.01; Mann–Whitney U. (i, j) miR‐205‐5p levels were decreased by qRT‐PCR analysis and TP53INP1 was upregulated in ASO‐205‐treated xenografts, respectively. Data expressed as mean ± SD for triplicate experiments. **p < 0.01
Higher levels of miR‐205‐5p in serum of patients with lung cancer
MiRNAs are frequently dysregulated in lung cancer, and they could be novel diagnostic and prognostic biomarkers for treatment of lung cancer. 2 A total of 75 patients with lung cancer and 62 normal controls were recruited into the present study to investigate the role of miR‐205‐5p in lung cancer diagnosis. The demographic and clinicopathological characteristics of the participants are presented in Table 1. No significant differences in gender (p = 0.816), age (p = 0.691), smoking history (p = 0.067), alcohol history (p = 0.197), and histology (p = 0.080) were found between patients with lung cancer and healthy controls.
TABLE 1.
The demographic and clinicopathological characteristics of patients with lung cancer and healthy controls
| Patients/n (%) | Controls/n (%) (%) | Test Statisticsvalue. | p value | |
|---|---|---|---|---|
| Total | 75 | 62 | ||
| Gender | ||||
| Male | 39 (52.0) | 31 (50.0) | χ 2 = 0.054 | 0.816 |
| Female | 36 (48.0) | 31 (50.0) | ||
| Age (mean ± SD, years) | 61.99 ± 9.91 | 62.68 ± 10.17 | t = 0.398 | 0.691 |
| Smoking history | ||||
| Yes | 25 (33.3) | 12 (19.4) | χ 2 = 3.364 | 0.067 |
| No | 50 (66.7) | 50 (80.6) | ||
| Alcohol history | ||||
| Yes | 13 (17.3) | 6 (9.7) | χ 2 = 1.665 | 0.197 |
| No | 62 (82.7) | 56 (90.3) | ||
| Pathological type | ||||
| Adenocarcinomas | 42 (56.0) | |||
| Squamous cell carcinomas | 24 (32.0) | |||
| Small‐cell lung cancer | 9 (12.0) | |||
Because the case number of SCLC is small (n = 9), this study focused on NSCLC. The results showed that the serum miR‐205‐5p levels examined in patients with NSCLC (n = 66), including 42 lung adenocarcinoma (LUAD) and 24 lung squamous cell carcinoma (LUSC), were markedly higher than those in healthy controls (n = 62) (p < 0.01; Figure 7a and Table 2.).
FIGURE 7.
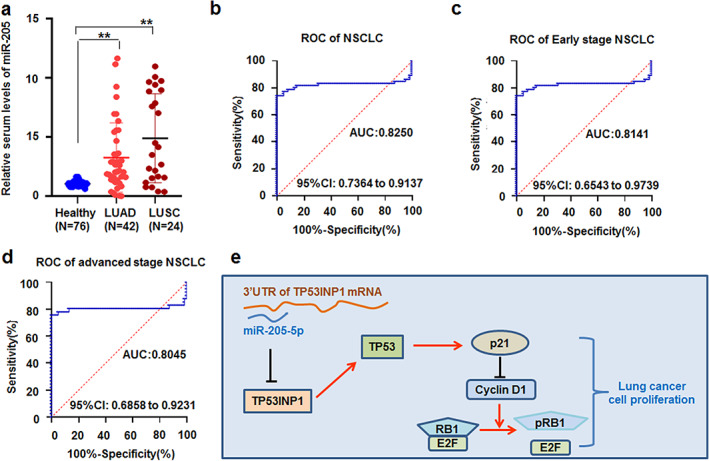
miR‐205‐5p levels in the serum of patients with lung cancer. (a) The levels of miR‐205‐5p increased in serum between patients with lung adenocarcinoma and lung squamous cell carcinoma compared with healthy controls. ** p < 0.001, Kruskal‐Wallis H test. (b) The receiver operating characteristic (ROC) curve of serum miR‐205‐5p was analyzed in patients with non‐small‐cell lung cancer (NSCLC). (c, d) The ROC curve of serum miR‐205‐5p was estimated in early and advanced TNM stage NSCLC patients, respectively. (e) Proposed model by which miR‐205‐5p regulates lung cancer cell proliferation by TP53INP1‐related genes
TABLE 2.
Association between plasma miR‐205‐5p levels and clinicopathological characteristics in patients with NSCLC
| Group | n | Median | (P 25, P 75) | p value a | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 33 | 3.479 g | (1.639, 8.840) | 0.045 | |
| Female | 33 | 2.033 | (1.372,3.620) | ||
| Age | |||||
| ≥62 | 37 | 2.055 | (1.179, 6.350) | 0.361 | |
| <62 | 29 | 2.991 | (1.650, 5.581) | ||
| Smoking history | |||||
| Yes | 21 | 5.419 | (1.650,8.841) | 0.036 | |
| No | 45 | 2.157 | (1.162,4.316) | ||
| Alcohol history | |||||
| Yes | 14 | 6.183 | (1.896,9.024) | 0.047 | |
| No | 52 | 2.155 | (1.324,4.535) | ||
| Pathological type | |||||
| Adenocarcinomas | 42 | 2.101 | (1.377,4.403) | ||
| Squamous cell carcinomas | 24 | 3.810 | (1.559,8.868) | 0.131 | |
| TNM stage | |||||
| I–II | 19 | 2.607 | (1.235,4.956) | ||
| III–IV | 41 | 2.907 | (1.615,7.020) | 0.364 | |
| Unclear | 6 | 1.628 | (1.581,1.937) | ||
Wilcoxon rank sum exact test or Kruskal–Wallis rank sum test.
Risk factors and clinical value of serum miR‐205‐5p in NSCLC
The association between serum miR‐205‐5p levels and the clinicopathological characteristics of patients with NSCLC was analyzed to evaluate the risk factors and the clinical value of miR‐205. MiR‐205‐5p levels were significantly higher in male patients than in female patients (median 3.479 vs. 2.033, p = 0.045). The serum miR‐205‐5p level in patients with NSCLC who smoke was significantly higher than that in patients who are nonsmokers (median 5.419 vs. 2.157, p = 0.036). The serum miR‐205‐5p level in patients with lung cancer who drink alcohol was significantly higher than that in nonalcoholic patients (median 6.183 vs. 2.155, p = 0.047). However, the results showed no significant association between miR‐205‐5p and age (p = 0.361), pathological type (p = 0.131), and TNM stage (p = 0.364), as shown in Table 2.
Serum miR‐205‐5p levels is a potential diagnostic marker for NSCLC
ROC curve analysis was performed to evaluate the diagnostic power of serum miR‐205‐5p in discriminating patients with lung cancer from healthy controls. According to the ROC curve analysis, the cut‐off value was defined as 1.498, with the highest specificity and sensitivity. The results showed that serum miR‐205‐5p levels may be a valuable biomarker for NSCLC patients with an area under the ROC curve (AUC) of 0.8250 (95% confidence interval [CI] 0.7364–0.9137; Figure 7b). To further analyze the roles of serum miR‐205‐5p levels in the analysis of early and advanced NSCLC by TNM stage, the results showed that the AUC of serum miR‐205‐5p level for patients with NSCLC at early stage (I + II) and advanced stage (III + IV) are 0.8141 (95% CI 0.6543–0.9739; Figure 7c) and 0.8045 (95% CI 0.6858–0.9231; Figure 7d), respectively.
DISCUSSION
Thus far, lung cancer has not been treated as a uniform specific treatment regimen because the pathogenesis has not yet been completely elucidated. 22 Accumulating evidence has broadened the awareness of the mechanisms of tumor and treatment methods. 23 , 24 Cancer‐related miRNAs play important roles in lung cancer occurrence and development. The expression of miRNAs is useful to classify human cancers, distinguish tumor subtypes, and correlate with prognosis. In the present study, the miR‐205‐5p levels in lung cancer tissues increased. MiR‐205‐5p promoted lung cancer cell proliferation and metastasis by negatively regulating a novel target TP53INP1, which further affected the expression of P21, RB1, and cyclin D1 (Figure 7e). A high level of serum miR‐205‐5p is also a valuable biomarker for patients with lung cancer.
MiR‐205‐5p is related to various tumors, such as endometrial cancer, urinary bladder cancer, prostate cancer, gastric cancer, breast cancer, lung cancer, and colon cancer. 25 , 26 , 27 , 28 , 29 However, the roles of miR‐205‐5p in cancer remain controversial. The miR‐205‐5p levels in endometrial cancer tissues and cell lines increased compared with those in controls. MiR‐205‐5p could affect the progression of endometrial cancer by regulating PTEN expression. 25 In a study of urinary bladder and prostate cancer, miR‐205‐5p was upregulated in these cancer samples, and it was used to differentiate benign prostatic hyperplasia from malignant cases. 26 The miR‐205‐5p levels in breast cancer stem cells also increased, which regulated the tumorigenic properties of breast cancer cells. MiR‐205‐5p knock‐down in breast cancer suppressed tumor proliferation and metastasis. 27 However, Zhang et al. demonstrated that miR‐205‐5p expression was low in gastric cancer cells and miR‐205‐5p upregulation significantly impaired cancer cell proliferation. Xenografts of gastric cancer revealed that miR‐205‐5p inhibited tumor proliferation by suppressing neovascularization. 28
MiR‐205‐5p played oncogenic or tumor‐suppressive roles in the tumorigenesis of human cancers. 17 In the present study, further investigation of the roles of miR‐205‐5p in lung cancer through miRNA datasets from GEO and TCGA showed that miR‐205‐5p in NSCLC tissues increased compared with that in control tissue. These results were supported by previous studies 11 , 30 that showed that miR‐205 was upregulated in NSCLC tissues. These data verified the oncogenic roles of miR‐205‐5p in the tumorigenesis of NSCLC.
Growing evidence shows that miR‐205‐5p has a potential for diagnosis of lung cancer. 11 , 31 , 32 Significantly higher miR‐205‐5p levels were observed in the serum of patients with NSCLC compared with the control group. 11 A study of serum exosomal miRNAs demonstrated that miR‐205‐5p was more abundant in serum exosomes from patients with NSCLC. 31 Upregulation of miR‐205‐5p also increased the risk of lung cancer and the diagnostic AUC accuracy of miR‐205‐5p for NSCLC was 0.825, indicating that it may be a promising biomarker for the early diagnosis of lung cancer. 32 In this study, the efficacy of miR‐205‐5p in the diagnosis of lung cancer was estimated. The levels of serum miR‐205‐5p were found to be significantly increased in patients with lung cancer compared with those in normal controls, and a high level of miR‐205‐5p expression was correlated with male gender and alcohol history. Subgroup analyses further showed that the expression of serum miR‐205 in LUAD, LUSC, and SCLC was significantly higher than in normal controls. These results indicate that miR‐205 may be a diagnostic indicator of lung cancer.
MiRNAs maintain cell homeostasis by negatively regulating their relative gene. 16 The targets of miR‐205‐5p were predicted by Targetscan, and the results suggest that the 3′‐UTR of tumor suppressor TP53INP1 is a target of miR‐205‐5p. Luciferase reporter assays and immunoblotting further proved that miR‐205‐5p could negatively regulate TP53INP1 expression, indicating that TP53INP1 is a novel target of miR‐205‐5p.
TP53INP1 is a tumor suppressor gene, and its overexpression leads to cell cycle arrest (G1 phase) and p53‐dependent or independent apoptosis. 33 TP53INP1 interacts with p53 and two kinases, homeodomain‐interacting protein kinase‐2 and protein kinase Cδ. TP53INP1 also resulted in decreased P21 levels in p53 wide‐type cell lines. 34 In addition, p53 could further regulate cyclin D1, 20 which activated CDK4/CDK6 to influence cell cycle by RB1. 21 Thus, the RB1, P21, and cyclin D1 levels in miR‐205‐5p‐treated lung cancer cells were analyzed in the present study, and the results showed that miR‐205‐5p could decrease the expression of TP53INP1, RB1, and P21 and increase that of cyclin D1, indicating that the function of miR‐205 in lung cancer is related to regulating TP53INP1, RB1, P21, and cyclin D1.
In summary, this study demonstrated that miR‐205‐5p could promote lung cancer cell proliferation and migration by downregulating TP53INP1‐related genes. The findings also indicate that the serum levels of miR‐205‐5p are useful in the diagnosis of patients with lung cancer.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
University and written informed consents were obtained from all subjects before the study.
AUTHOR CONTRIBUTIONS
S.Y.X. and G.B.S. designed the study, performed data analysis, and revised the paper. J.J.Y. collected clinical samples. Y.L.Z. and J.X.Z. performed cell culture, miRNA detection, cell proliferation, and luciferase assay. Y.B.W. and J.F.P. performed immunoblotting and apoptosis assay. C.J.F., M.H.H. and R.W. produced xenografts in vivo. P.Y.W. performed data analysis. All authors approved the final manuscript.
ETHICS STATEMENT
This study protocol was approved by the Medical Ethics Committee of Binzhou Medical.
ACKNOWLEDGMENTS
We acknowledge professional editing support from ShineWrite.com (service@shinewrite.com) in editing the English text of a draft of this manuscript. The present study was supported by the National Natural Science Foundation of China (No.81772281, 31371321), the Shandong Science and Technology Committee (No. ZR2019MH022, ZR2020KH015), the Yantai Science and Technology Committee (2018XSCC051), the Education Department of Shandong Province (2019KJK014), and the Shandong Province Taishan Scholar Project (no. ts201712067).
Zhao Y‐L, Zhang J‐X, Yang J‐J, Wei Y‐B, Peng J‐F, Fu C‐J, et al. MiR‐205‐5p promotes lung cancer progression and is valuable for the diagnosis of lung cancer. Thorac Cancer. 2022;13:832–843. 10.1111/1759-7714.14331
Yu‐Long Zhao and Jia‐Xiang Zhang contributed equally to this work.
Funding information National Natural Science Foundation of China, Grant/Award Number: 81772281
Contributor Information
Guang‐Bin Sun, Email: 793283846@qq.com.
Shu‐Yang Xie, Email: shuyangxie@aliyun.com.
DATA AVAILABILITY STATEMENT
Source data for figures and Supporting Information figures are provided within the online content of this paper.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Du X, Zhang J, Wang J, Lin X, Ding F. Role of miRNA in lung cancer‐potential biomarkers and therapies. Curr Pharm Des. 2018;23(39):5997–6010. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4. O'Bryan S, Dong S, Mathis JM, Alahari SK. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer. 2017;72:1–11. [DOI] [PubMed] [Google Scholar]
- 5. Honardoost M, Rad S. Triangle of AKT2, miRNA, and tumorigenesis in different cancers. Appl Biochem Biotechnol. 2018;185(2):524–40. [DOI] [PubMed] [Google Scholar]
- 6. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. [DOI] [PubMed] [Google Scholar]
- 7. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10(7):396–404. [DOI] [PubMed] [Google Scholar]
- 8. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. [DOI] [PubMed] [Google Scholar]
- 9. Huang W, Hu J, Yang DW, Fan XT, Jin Y, Hou YY, et al. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med. 2012;186(11):1160–7. [DOI] [PubMed] [Google Scholar]
- 10. Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR‐205‐5p and promoting VEGFA expression. Cancer Biomark. 2019;25(1):101–13. [DOI] [PubMed] [Google Scholar]
- 11. Jiang M, Zhang P, Hu G, Xiao Z, Xu F, Zhong T, et al. Relative expressions of miR‐205‐5p, miR‐205‐3p, and miR‐21 in tissues and serum of non‐small cell lung cancer patients. Mol Cell Biochem. 2013;383(1–2):67–75. [DOI] [PubMed] [Google Scholar]
- 12. Ulivi P, Petracci E, Marisi G, Baglivo S, Chiari R, Billi M, et al. Prognostic role of circulating miRNAs in early‐stage non‐small cell lung cancer. J Clin Med. 2019;8(2):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Farrell HE, Bowman RV, Fong KM, Yang IA. Plasma extracellular vesicle miRNAs can identify lung cancer, current smoking status, and stable COPD. Int J Mol Sci. 2021;22(11):5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C[T]) method. Methods (San Diego, Calif). 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q, Yan YF, Lv Q, Li YJ, Wang RR, Sun GB, et al. miR‐4293 upregulates lncRNA WFDC21P by suppressing mRNA‐decapping enzyme 2 to promote lung carcinoma proliferation. Cell Death Dis. 2021;12(8):735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. [DOI] [PubMed] [Google Scholar]
- 17. Ferrari E, Gandellini P. Unveiling the ups and downs of miR‐205 in physiology and cancer: transcriptional and post‐transcriptional mechanisms. Cell Death Dis. 2020;11(11):980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. [DOI] [PubMed] [Google Scholar]
- 19. Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y, et al. miR‐205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non‐small cell lung cancer. Cancer Res. 2013;73(17):5402–15. [DOI] [PubMed] [Google Scholar]
- 20. Chen JH, Tsou TC, Chiu IM, Chou CC. Proliferation inhibition, DNA damage, and cell‐cycle arrest of human astrocytoma cells after acrylamide exposure. Chem Res Toxicol. 2010;23(9):1449–58. [DOI] [PubMed] [Google Scholar]
- 21. Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–72. [DOI] [PubMed] [Google Scholar]
- 22. Ren F, Chen J. Diagnosis and treatment strategies of multiple primary lung cancer: a systematic review. Drug Eval. 2017;14(16):31–8. [Google Scholar]
- 23. Boeri M, Sestini S, Fortunato O, Verri C, Suatoni P, Pastorino U, et al. Recent advances of microRNA‐based molecular diagnostics to reduce false‐positive lung cancer imaging. Expert Rev Mol Diagn. 2015;15(6):801–13. [DOI] [PubMed] [Google Scholar]
- 24. Powrozek T, Mlak R, Dziedzic M, Malecka‐Massalska T, Sagan D. Analysis of primary‐miRNA‐3662 and its mature form may improve detection of the lung adenocarcinoma. J Cancer Res Clin Oncol. 2017;143(10):1941–6. [DOI] [PubMed] [Google Scholar]
- 25.Xin W, Zhao S, Han X, Zhao P, Yu H, Gao X, et al. lncRNA LA16c‐313D11.11 modulates the development of endometrial cancer by binding to and inhibiting microRNA‐205‐5p function and indirectly increasing PTEN activity. Int J Oncol 2020;57(1):355–363. [DOI] [PubMed] [Google Scholar]
- 26. Ghorbanmehr N, Gharbi S, Korsching E, Tavallaei M, Einollahi B, Mowla SJ. miR‐21‐5p, miR‐141‐3p, and miR‐205‐5p levels in urine‐promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019;79(1):88–95. [DOI] [PubMed] [Google Scholar]
- 27. De Cola A, Lamolinara A, Lanuti P, Rossi C, Iezzi M, Marchisio M, et al. MiR‐205‐5p inhibition by locked nucleic acids impairs metastatic potential of breast cancer cells. Cell Death Dis. 2018;9(8):821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Zhang J, Pang X, Chen Z, Zhang Z, Lei L, et al. MiR‐205‐5p suppresses angiogenesis in gastric cancer by downregulating the expression of VEGFA and FGF1. Exp Cell Res. 2021;404(2):112579. [DOI] [PubMed] [Google Scholar]
- 29. Gulei D, Magdo L, Jurj A, Raduly L, Cojocneanu‐Petric R, Moldovan A, et al. The silent healer: miR‐205‐5p up‐regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up‐regulating E‐cadherin expression. Cell Death Dis. 2018;9(2):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA‐21 and microRNA‐205 overexpression in non‐small cell lung cancer by quantitative real‐time RT‐PCR. Clin Chem. 2008;54(10):1696–704. [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Jiang X, Li J, Wang J, Binang H, Shi S, et al. Serum exosomal miR‐1269a serves as a diagnostic marker and plays an oncogenic role in non‐small cell lung cancer. Thorac Cancer. 2020;11(12):3436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Sui J, Shen X, Li C, Yao W, Hong W, et al. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of lung cancer. Oncol Rep. 2017;37(6):3543–53. [DOI] [PubMed] [Google Scholar]
- 33. Shahbazi J, Lock R, Liu T. Tumor protein 53‐induced nuclear protein 1 enhances p53 function and represses tumorigenesis. Front Genet. 2013;4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bell E, Lunec J, Tweddle DA. Cell cycle regulation targets of MYCN identified by gene expression microarrays. Cell Cycle. 2007;6(10):1249–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data for figures and Supporting Information figures are provided within the online content of this paper.


