Abstract
Scope
Lack of biomarkers is a challenge for the accurate assessment of protein intake and interpretation of observational study data. The study aims to identify biomarkers of a protein‐rich dietary pattern.
Methods and Results
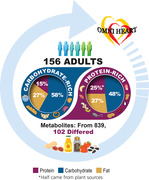
The Optimal Macronutrient Intake Trial to Prevent Heart Disease (OmniHeart) trial is a randomized cross‐over feeding study which tested three dietary patterns with varied macronutrient content (carbohydrate‐rich; protein‐rich with about half from plant sources; and unsaturated fat‐rich). In 156 adults, differences in log‐transformed plasma metabolite levels at the end of the protein‐ and carbohydrate‐rich diet periods using paired t‐tests is examined. Partial least‐squares discriminant analysis is used to identify a set of metabolites which are influential in discriminating between the protein‐rich versus carbohydrate‐rich dietary patterns. Of 839 known metabolites, 102 metabolites differ significantly between the protein‐rich and the carbohydrate‐rich dietary patterns after Bonferroni correction, the majority of which are lipids (n = 35), amino acids (n = 27), and xenobiotics (n = 24). Metabolites which are the most influential in discriminating between the protein‐rich and the carbohydrate‐rich dietary patterns represent plant protein intake, food or beverage intake, and preparation methods.
Conclusions
The study identifies many plasma metabolites associated with the protein‐rich dietary pattern. If replicated, these metabolites may be used to assess level of adherence to a similar dietary pattern.
Keywords: biomarkers, dietary pattern, feeding study, macronutrient profiles, metabolomics, protein
Lack of biomarkers is a challenge for the accurate assessment of protein intake. The present study aims to identify biomarkers of a protein‐rich dietary pattern relative to a carbohydrate‐rich dietary pattern. The study finds that metabolites which represent the protein‐rich dietary pattern include biomarkers of plant protein intake, food or beverage intake, and preparation methods.
1. Introduction
Diet plays a central role in the etiology of chronic diseases. Whether protein intake relative to carbohydrate intake is associated with chronic diseases has been a major interest in nutrition research. However, lack of biomarkers is a challenge for the accurate assessment of protein intake and interpretation of observational study data. Typically, in observational studies, protein intake is assessed using a food frequency questionnaire, 24‐h dietary recalls, or diet records. These methods have limitations (e.g., recall bias, social desirability bias, underreporting) and are subject to systematic errors (e.g., inaccuracies in nutrition databases used to analyze dietary intake).[ 1 , 2 ] Objective biomarkers which are not influenced by these errors can contribute to accurate estimation of protein intake. For protein intake, 24‐h urea nitrogen is considered the gold standard, but it can suffer from incompleteness, and is burdensome for participants to collect all urine output over a 24‐h period. Furthermore, multiple 24‐h urine collections may be needed to approximate usual protein intake.[ 3 , 4 ] Therefore, collection of 24‐h urea nitrogen may be difficult in large epidemiological studies. Novel biomarkers of protein intake, which are not subject to systematic errors from self‐reported diets and are less burdensome for study participants are needed.
Controlled feeding studies provide an excellent opportunity to discover biomarkers of dietary intake. All participants in controlled feeding studies are provided with foods that are developed from standardized menus, which reduce variability in dietary intake across participants, and ensure intake of similar types of foods and preparation methods. The Optimal Macronutrient Intake Trial to Prevent Heart Disease (OmniHeart) was a controlled feeding study which tested whether three healthful diets that varied in macronutrient composition improved cardiovascular risk factors.[ 5 ] The trial found that, compared to the carbohydrate‐rich dietary pattern, the protein‐rich dietary pattern improved cardiovascular risk factors [e.g., reduced systolic blood pressure (SBP) and diastolic blood pressure (DBP) and reduced low‐density lipoprotein (LDL), high‐density lipoprotein (HDL), total cholesterol, non‐high‐density lipoprotein, and triglycerides].[ 5 ]
Untargeted metabolomic profiling, an approach which detects many small molecules in biospecimens, can help identify metabolites which may be used as surrogate measures of protein intake. Thus, in the present study, we aimed to identify biomarkers of the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern in the OmniHeart trial.
2. Results
More than half of the trial participants was men and African American (Table 1 ). More than 40% were college graduates, current alcohol consumers, and had obesity. A majority of the trial participants were never smokers. Nearly 20% of the trial participants had blood pressure levels in the hypertensive range. These baseline characteristics were nearly identical for the original trial participants (n = 164) and our analytic sample (n = 156).
Table 1.
Baseline characteristics of participants in the OmniHeart trial
| Characteristic a) | Trial participants (N = 164) | Analytic sample (N = 156) |
|---|---|---|
| Age, y | 53.1 (10.8) | 53.0 (10.6) |
| Women, n (%) | 73 (44.5) | 70 (44.9) |
| African American, n (%) | 90 (54.9) | 85 (54.5) |
| Income, n (%) | ||
| <$30000 | 52 (31.7) | 49 (31.4) |
| $30000–$59999 | 60 (36.6) | 57 (36.5) |
| ≥$60000 | 45 (27.4) | 43 (27.6) |
| Unknown or refused | 7 (4.3) | 7 (4.5) |
| Education, n (%) | ||
| High school graduate or less | 33 (20.1) | 32 (20.5) |
| Some college | 56 (34.1) | 53 (34.0) |
| College graduate | 75 (45.7) | 71 (45.5) |
| Smoking, n (%) | ||
| Current smoker | 18 (11.0) | 18 (11.5) |
| Former smoker | 46 (28.0) | 42 (26.9) |
| Never smoker | 100 (61.0) | 96 (61.5) |
| Current alcohol consumer, n (%) | 73 (44.5) | 69 (44.2) |
| Total energy intake, kcal | 2315 (1174) | 2316 (1187) |
| BMI, kg/m2 | 30.3 (6.1) | 30.0 (5.8) |
| BMI category, n (%) | ||
| Not overweight or obese | 34 (20.7) | 34 (21.8) |
| Overweight | 55 (33.5) | 52 (33.3) |
| Obese | 75 (45.7) | 70 (44.9) |
| SBP, mm Hg | 131 (9) | 131 (9) |
| DBP, mm Hg | 77 (8) | 77 (8) |
| Hypertensive status b) | 32 (20) | 30 (19) |
Values are n (%) for categorical variables and mean (standard deviation) for continuous variables;
Hypertensive status was defined as SBP ≥140 mmHg or DBP ≥90 mmHg.
BMI indicates body mass index; DBP, diastolic blood pressure; SBP, systolic blood pressure.
Of the 839 known metabolites, 102 metabolites (12%) differed significantly between the protein‐rich dietary pattern and the carbohydrate‐rich dietary pattern after Bonferroni correction (Table 2 ). The most common metabolite categories represented were lipids (n = 35, 34%), amino acids (n = 27, 26%), and xenobiotics (n = 24, 24%) (Figure 1 ). The proportion of metabolites that were amino acids (26% vs 23%), cofactors and vitamins (6% vs 3%), and xenobiotics (24% vs 13%) was higher among the subset of metabolites that were statistically significantly different between the protein‐rich dietary pattern and carbohydrate‐rich dietary pattern relative to the distribution of these metabolite categories in the data set.
Table 2.
Metabolites (N = 102) significantly associated with the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern in the OmniHeart trial
| Metabolite | Superpathway | Subpathway | Mean differencea) | p‐value |
|---|---|---|---|---|
| Hydroxyasparagine | Amino acid | Alanine and aspartate metabolism | –0.0690 | 1.85 × 10−7 |
| Creatine | Amino acid | Creatine metabolism | 0.1189 | 2.23 × 10−6 |
| Pyroglutamine* | Amino acid | Glutamate metabolism | −0.1294 | 4.88 × 10−5 |
| N‐acetylglycine | Amino acid | Glycine, serine and threonine metabolism | 0.1827 | 3.62 × 10−7 |
| Glycine | Amino acid | Glycine, serine and threonine metabolism | –0.0548 | 2.35 × 10−5 |
| Betaine | Amino acid | Glycine, serine and threonine metabolism | –0.0514 | 1.72 × 10−5 |
| Guanidinosuccinate | Amino acid | Guanidino and acetamido metabolism | 0.3052 | 4.47 × 10−12 |
| 1‐methyl‐5‐imidazoleacetate | Amino acid | Histidine metabolism | 0.2676 | 2.00 × 10−6 |
| hydantoin‐5‐propionate | Amino acid | Histidine metabolism | 0.1899 | 2.05 × 10−5 |
| 1‐ribosyl‐imidazoleacetate* | Amino acid | Histidine metabolism | −0.0670 | 4.87 × 10−5 |
| Tiglyl carnitine (C5) | Amino acid | Leucine, isoleucine and valine metabolism | 0.3195 | 1.93 × 10−16 |
| Isobutyrylcarnitine (C4) | Amino acid | Leucine, isoleucine and valine metabolism | 0.2009 | 2.44 × 10−8 |
| 3‐hydroxyisobutyrate | Amino acid | Leucine, isoleucine and valine metabolism | 0.1937 | 2.42 × 10−6 |
| 3‐methylglutaconate | Amino acid | Leucine, isoleucine and valine metabolism | 0.1570 | 8.21 × 10−8 |
| N,N,N‐trimethyl‐5‐aminovalerate | Amino acid | Lysine metabolism | −0.1228 | 4.34 × 10−12 |
| S‐methylcysteine | Amino acid | Methionine, cysteine, SAM and taurine metabolism | −0.1868 | 4.44 × 10−7 |
| Phenylacetate | Amino acid | Phenylalanine metabolism | 0.2406 | 3.27 × 10−5 |
| 2‐hydroxyphenylacetate | Amino acid | Phenylalanine metabolism | 0.2030 | 9.48 × 10−7 |
| Tryptophan betaine | Amino acid | Tryptophan metabolism | 0.5422 | 2.91 × 10−25 |
| Indolepropionate | Amino acid | Tryptophan metabolism | −0.3145 | 1.24 × 10−8 |
| Xanthurenate | Amino acid | Tryptophan metabolism | 0.2351 | 7.98 × 10−6 |
| N‐formylanthranilic acid | Amino acid | Tryptophan metabolism | 0.2310 | 2.06 × 10−7 |
| 3‐indoxyl sulfate | Amino acid | Tryptophan metabolism | 0.1916 | 2.92 × 10−5 |
| 6‐bromotryptophan | Amino acid | Tryptophan metabolism | −0.0944 | 9.46 × 10−9 |
| 3‐methoxytyrosine | Amino acid | Tyrosine metabolism | −0.0975 | 2.54 × 10−5 |
| Urea | Amino acid | Urea cycle; arginine and proline metabolism | 0.2094 | 1.19 × 10−19 |
| Argininate* | Amino acid | Urea cycle; arginine and proline metabolism | 0.2048 | 6.57 × 10−7 |
| Glycerate | Carbohydrate | Glycolysis, gluconeogenesis, and pyruvate metabolism | −0.0781 | 5.50 × 10−7 |
| Ribulonate/xylulonate/lyxonate* | Carbohydrate | Pentose metabolism | –0.1664 | 3.71 × 10−9 |
| Arabonate/xylonate | Carbohydrate | Pentose Metabolism | –0.1121 | 5.32 × 10−5 |
| Oxalate (ethanedioate) | Cofactors and vitamins | Ascorbate and aldarate metabolism | –0.1075 | 1.20 × 10−8 |
| N1‐Methyl‐2‐pyridone‐5‐carboxamide | Cofactors and vitamins | Nicotinate and nicotinamide metabolism | 0.1493 | 9.76 × 10−6 |
| Gamma‐CEHC glucuronide* | Cofactors and vitamins | Tocopherol metabolism | −0.3241 | 5.09 × 10−7 |
| Gamma‐CEHC | Cofactors and vitamins | Tocopherol metabolism | −0.3051 | 6.59 × 10−13 |
| delta‐CEHC | Cofactors and vitamins | Tocopherol metabolism | –0.2690 | 2.23 × 10−6 |
| Pyridoxate | Cofactors and vitamins | Vitamin B6 metabolism | 0.1542 | 8.98 × 10−7 |
| Succinylcarnitine (C4) | Energy | TCA Cycle | −0.0982 | 2.93 × 10−5 |
| 5alpha‐androstan‐3alpha,17beta‐diol monosulfate (1) | Lipid | Androgenic steroids | −0.1710 | 3.28 × 10−6 |
| Androstenediol (3beta,17beta) monosulfate (2) | Lipid | Androgenic steroids | −0.1573 | 1.43 × 10−9 |
| 5alpha‐androstan‐3beta,17beta‐diol disulfate | Lipid | Androgenic steroids | −0.1544 | 4.85 × 10−6 |
| Androsterone sulfate | Lipid | Androgenic steroids | −0.1344 | 1.86 × 10−7 |
| Androstenediol (3beta,17beta) disulfate (1) | Lipid | Androgenic steroids | −0.1300 | 2.47 × 10−7 |
| Androstenediol (3alpha, 17alpha) monosulfate (3) | Lipid | Androgenic steroids | −0.1002 | 2.21 × 10−5 |
| N‐stearoyl‐sphingosine (d18:1/18:0)* | Lipid | Ceramides | −0.1253 | 2.47 × 10−7 |
| Ceramide (d18:2/24:1, d18:1/24:2)* | Lipid | Ceramides | −0.1212 | 2.00 × 10−5 |
| 3,4‐methyleneheptanoylcarnitine | Lipid | Fatty acid metabolism(Acyl Carnitine) | −0.3080 | 8.90 × 10−8 |
| Lignoceroylcarnitine (C24)* | Lipid | Fatty acid Metabolism(Acyl Carnitine) | 0.1494 | 1.31 × 10−6 |
| Picolinoylglycine | Lipid | Fatty acid metabolism(Acyl Glycine) | 0.2212 | 4.83 × 10−9 |
| 16‐hydroxypalmitate | Lipid | Fatty acid, monohydroxy | −0.1073 | 5.88 × 10−5 |
| Glycosyl‐N‐(2‐hydroxynervonoyl)‐sphingosine (d18:1/24:1(2OH))* | Lipid | Hexosylceramides (HCER) | −0.3066 | 8.55 × 10−8 |
| Glycosyl‐N‐nervonoyl‐sphingosine (d18:1/24:1)* | Lipid | Hexosylceramides (HCER) | –0.1456 | 5.65 × 10−10 |
| Glycosyl ceramide (d18:2/24:1, d18:1/24:2)* | Lipid | Hexosylceramides (HCER) | −0.1416 | 2.96 × 10−8 |
| Glycosyl‐N‐stearoyl‐sphingosine (d18:1/18:0) | Lipid | Hexosylceramides (HCER) | −0.1408 | 4.08 × 10−8 |
| Glycosyl‐N‐palmitoyl‐sphingosine (d18:1/16:0) | Lipid | Hexosylceramides (HCER) | −0.0670 | 2.62 × 10−5 |
| 1‐linolenoyl‐GPC (18:3)* | Lipid | Lysophospholipid | −0.2207 | 5.66 × 10−9 |
| 1‐linoleoyl‐GPE (18:2)* | Lipid | Lysophospholipid | −0.1425 | 5.28 × 10−6 |
| 1‐stearoyl‐2‐docosahexaenoyl‐GPC (18:0/22:6) | Lipid | Phosphatidylcholine (PC) | −0.1268 | 1.72 × 10−12 |
| 1‐stearoyl‐2‐oleoyl‐GPC (18:0/18:1) | Lipid | Phosphatidylcholine (PC) | −0.1148 | 2.86 × 10−7 |
| 1‐palmitoyl‐2‐stearoyl‐GPC (16:0/18:0) | Lipid | Phosphatidylcholine (PC) | −0.0751 | 2.81 × 10−6 |
| 1‐palmitoyl‐2‐oleoyl‐GPC (16:0/18:1) | Lipid | Phosphatidylcholine (PC) | −0.0648 | 5.03 × 10−5 |
| 1‐palmitoyl‐2‐arachidonoyl‐GPI (16:0/20:4)* | Lipid | Phosphatidylinositol (PI) | −0.1552 | 7.81 × 10−6 |
| 1‐palmitoyl‐2‐linoleoyl‐GPI (16:0/18:2) | Lipid | Phosphatidylinositol (PI) | −0.1434 | 4.46 × 10−6 |
| 1‐(1‐enyl‐palmitoyl)‐2‐arachidonoyl‐GPE (P‐16:0/20:4)* | Lipid | Plasmalogen | 0.1394 | 4.66 × 10−8 |
| 1‐(1‐enyl‐stearoyl)‐2‐arachidonoyl‐GPE (P‐18:0/20:4)* | Lipid | Plasmalogen | 0.0980 | 1.19 × 10−5 |
| 1‐(1‐enyl‐palmitoyl)‐2‐arachidonoyl‐GPC (P‐16:0/20:4)* | Lipid | Plasmalogen | 0.0901 | 3.29 × 10−8 |
| Sphingomyelin (d18:2/14:0, d18:1/14:1)* | Lipid | Sphingomyelins | −0.0971 | 1.04 × 10−7 |
| Sphingomyelin (d18:1/25:0, d19:0/24:1, d20:1/23:0, d19:1/24:0)* | Lipid | Sphingomyelins | 0.0873 | 2.02 × 10−6 |
| Sphingomyelin (d18:2/23:1)* | Lipid | Sphingomyelins | −0.0859 | 7.32 × 10−6 |
| Sphingomyelin (d17:1/14:0, d16:1/15:0)* | Lipid | Sphingomyelins | −0.0802 | 3.04 × 10−5 |
| Sphingomyelin (d18:2/24:2)* | Lipid | Sphingomyelins | −0.0753 | 2.77 × 10−6 |
| Stearoyl sphingomyelin (d18:1/18:0) | Lipid | Sphingomyelins | −0.0665 | 1.24 × 10−5 |
| Sphingomyelin (d18:2/24:1, d18:1/24:2)* | Lipid | Sphingomyelins | −0.0664 | 3.51 × 10−6 |
| Uracil | Nucleotide | Pyrimidine metabolism, uracil containing | 0.2314 | 1.22 × 10−7 |
| Uridine | Nucleotide | Pyrimidine metabolism, uracil containing | 0.0832 | 3.88 × 10−6 |
| Glycine conjugate of C10H14O2 (1)* | Partially characterized molecules | Partially characterized molecules | −0.2161 | 1.96 × 10−5 |
| Metabolonic lactone sulfate | Partially characterized molecules | Partially characterized molecules | −0.2154 | 4.37 × 10−18 |
| Phenylacetylglutamate | Peptide | Acetylated peptides | 0.3346 | 3.26 × 10−5 |
| Phenylacetylglutamine | Peptide | Acetylated peptides | 0.2313 | 1.07 × 10−5 |
| 1H‐indole‐7‐acetic acid | Xenobiotics | Bacterial/fungal | 0.4521 | 1.46 × 10−6 |
| 4‐ethylphenyl sulfate | Xenobiotics | Benzoate metabolism | 1.2830 | 8.94 × 10−22 |
| 4‐acetylphenyl sulfate | Xenobiotics | Benzoate metabolism | 0.6736 | 1.63 × 10−12 |
| Hippurate | Xenobiotics | Benzoate metabolism | −0.5706 | 4.07 × 10−11 |
| 4‐vinylphenol sulfate | Xenobiotics | Benzoate metabolism | 0.4873 | 2.29 × 10−7 |
| Guaiacol sulfate | Xenobiotics | Benzoate metabolism | −0.3593 | 5.86 × 10−8 |
| Catechol sulfate | Xenobiotics | Benzoate metabolism | −0.3333 | 3.54 × 10−8 |
| Benzoylcarnitine* | Xenobiotics | Chemical | −0.6127 | 6.89 × 10−16 |
| Indoleacetylcarnitine* | Xenobiotics | Chemical | 0.3876 | 6.43 × 10−9 |
| 6‐hydroxyindole sulfate | Xenobiotics | Chemical | 0.2068 | 3.26 × 10−5 |
| Quinate | Xenobiotics | Food component/Plant | −1.0307 | 2.35 × 10−15 |
| Piperine | Xenobiotics | Food component/Plant | 0.8829 | 8.53 × 10−14 |
| Sulfate of piperine metabolite C18H21NO3 (1)* | Xenobiotics | Food component/Plant | 0.7956 | 1.90 × 10−19 |
| Sulfate of piperine metabolite C16H19NO3 (3)* | Xenobiotics | Food component/Plant | 0.7655 | 9.00 × 10−15 |
| Sulfate of piperine metabolite C16H19NO3 (2)* | Xenobiotics | Food component/Plant | 0.7431 | 1.48 × 10−15 |
| Sulfate of piperine metabolite C18H21NO3 (3)* | Xenobiotics | Food component/Plant | 0.6537 | 1.33 × 10−18 |
| Glucuronide of piperine metabolite C17H21NO3 (4)* | Xenobiotics | Food component/Plant | 0.6100 | 3.52 × 10−10 |
| Glucuronide of piperine metabolite C17H21NO3 (3)* | Xenobiotics | Food component/Plant | 0.5843 | 4.53 × 10−10 |
| Genistein sulfate* | Xenobiotics | Food component/Plant | 0.5504 | 3.38 × 10−8 |
| Glucuronide of piperine metabolite C17H21NO3 (5)* | Xenobiotics | Food component/Plant | 0.4446 | 6.10 × 10−8 |
| Pyrraline | Xenobiotics | Food component/Plant | 0.3915 | 5.20 × 10−7 |
| Stachydrine | Xenobiotics | Food component/Plant | −0.3181 | 5.99 × 10−11 |
| 3,4‐methyleneheptanoate | Xenobiotics | Food component/Plant | −0.3098 | 2.89 × 10−5 |
| homostachydrine* | Xenobiotics | Food component/Plant | −0.2137 | 1.22 × 10−8 |
Mean difference represents differences in plasma levels comparing the protein‐rich dietary pattern versus the carbohydrate‐rich dietary pattern at the end of the 6‐week intervention. Positive mean difference indicates that the metabolite was higher after the protein‐rich relative to the carbohydrate‐rich dietary patterns. Negative mean difference indicates that the metabolite was higher after the carbohydrate‐rich relative to the protein‐rich dietary patterns. Only metabolites which passed the Bonferroni threshold are presented (0.05/839 = 5.95 × 10−5). p value is derived from paired t‐test.
Asterisk indicates metabolites not officially confirmed (tier 2 identification).
Figure 1.
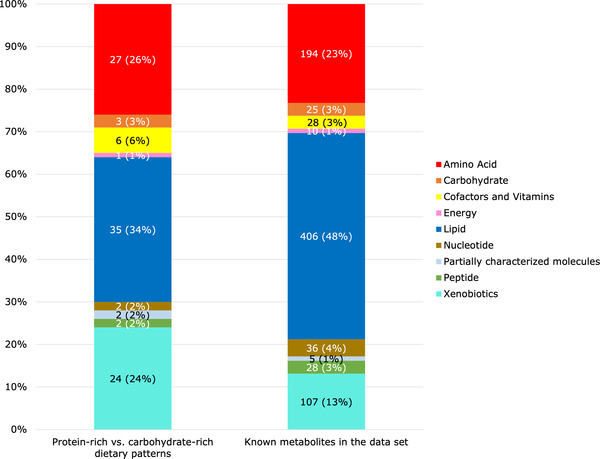
Metabolite categories (n (%)) significantly associated with the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern in the OmniHeart trial. “Known metabolites in the data set” indicates the distribution of all 839 plasma metabolites detected in the OmniHeart trial participants. For example, out of 102 metabolites which differed significantly between the protein‐rich versus carbohydrate‐rich dietary patterns, 24 (24%) metabolites were xenobiotics. Out of 839 known metabolites in the data set, 107 (13%) metabolites were xenobiotics. Considering the distribution of metabolite categories in the data set, xenobiotics (24% vs 13%) and amino acids (26% vs 23%) were overrepresented when we compared the protein‐rich versus carbohydrate‐rich dietary patterns.
Metabolites with strong positive mean differences (>0.5; indicating higher metabolite levels for protein vs carbohydrate) were mostly xenobiotics representing food components/plants (piperine, sulfate of piperine metabolite, glucuronide of pipierine metabolite), metabolites involved in benzoate metabolism (4‐ethylphenyl sulfate, 4‐acetylphenyl sulfate), and an amino acid (tryptophan betaine) (Figure 2 ). Metabolites with weak to moderate positive mean differences (>0 and <0.5) were creatine and essential amino acids related to the metabolism of leucine, isoleucine, valine, phenylalanine, histidine, and tryptophan. Metabolites with weak to moderate negative mean differences (<0 and >–0.5) were mostly lipids in the androgenic steroids, ceramides, hexosylceramides, phosphatidylcholine, phosphatidylinositol, and sphingomyelin pathways. Metabolites with strong negative mean differences (<‐0.5) included three xenobiotics (quinate, benzoylcarnitine, and hippurate).
Figure 2.
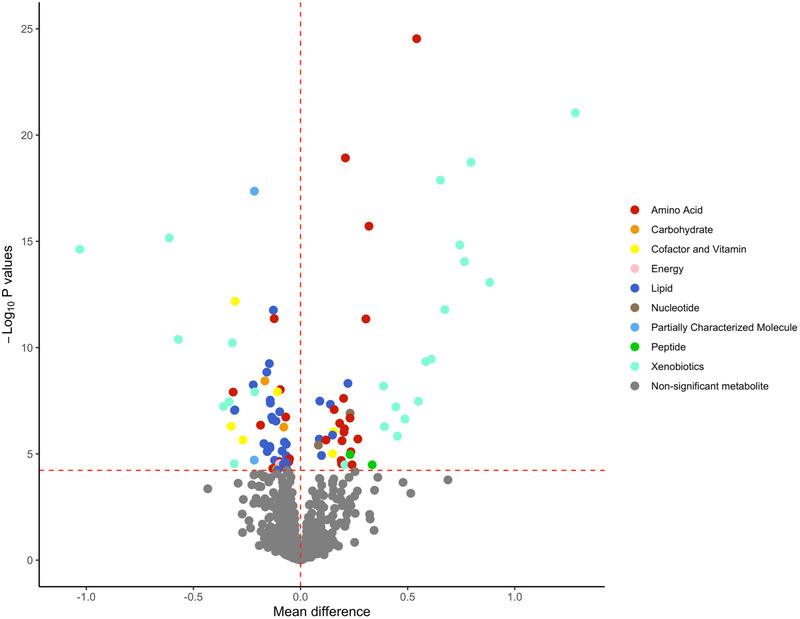
Volcano plot of mean differences and p values for the association between individual plasma metabolites and protein‐rich dietary pattern versus carbohydrate‐rich dietary pattern. The red horizontal dashed line represents the Bonferroni‐adjusted threshold (0.05/839 = 5.95 × 10−5) and the red vertical dashed line is set at mean difference of 0. Metabolites located to the right of the vertical dashed line indicate that the plasma levels of these metabolites were higher after the protein‐rich relative to the carbohydrate‐rich dietary patterns. Metabolites located to the left of the vertical dashed line indicate that the plasma levels of these metabolites were higher after the carbohydrate‐rich relative to the protein‐rich dietary patterns.
Metabolites significantly different between the two diets were involved in a total of 95 subpathways (Table S1, Supporting Information). Of these 95 pathways, six pathways (hexosylceramides, food component/plant, tocopherol metabolism, androgenic steroids, tryptophan metabolism, and benzoate metabolism) were overrepresented (Fisher's exact test p < 0.05) (Table 3 ). Of these pathways, hexosylceramindes had the smallest p value (p = 1.30 × 10−4).
Table 3.
Pathway overrepresentation analysis of metabolites significantly different between the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern
| Superpathway | Subpathways | Significant metabolites | Total metabolites | Fisher's exact p‐value |
|---|---|---|---|---|
| Lipid | Hexosylceramides (HCER) | 5 | 6 | 1.30 × 10−4 |
| Xenobiotics | Food component/Plant | 14 | 49 | 0.001 |
| Cofactors and vitamins | Tocopherol metabolism | 3 | 5 | 0.01 |
| Lipid | Androgenic steroids | 6 | 19 | 0.02 |
| Amino acid | Tryptophan metabolism | 6 | 20 | 0.03 |
| Xenobiotics | Benzoate metabolism | 6 | 22 | 0.04 |
Using partial least‐squares discriminant analysis(PLS‐DA), the first and second components explained 17.1% and 14.3% of the variance, respectively. The 10 metabolites which were influential in discriminating the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern were all xenobiotics: 4‐ethylphenyl sulfate, quinate, piperine, 5 sulfate or glucuronide piperine metabolites, 4‐acetylphenyl sulfate, and benzoylcarnitine (Table 4 ).
Table 4.
Top 10 metabolites influential in distinguishing protein‐rich dietary pattern versus carbohydrate‐rich dietary pattern in the OmniHeart trial (N = 156)
| Metabolite a) | Superpathway | Subpathway | VIP | C‐statistics | |
|---|---|---|---|---|---|
| Testing sample (N = 78) | Validation sample (N = 78) | ||||
| 4‐ethylphenyl sulfate | Xenobiotics | Benzoate metabolism | 3.74 | 0.867 | 0.894 |
| Quinate | Xenobiotics | Food component/Plant | 3.00 | 0.834 | 0.868 |
| Piperine | Xenobiotics | Food component/Plant | 2.57 | 0.826 | 0.829 |
| Sulfate of piperine metabolite C18H21NO3 (1)* | Xenobiotics | Food component/Plant | 2.32 | 0.877 | 0.876 |
| Sulfate of piperine metabolite C16H19NO3 (3)* | Xenobiotics | Food component/Plant | 2.23 | 0.862 | 0.784 |
| Sulfate of piperine metabolite C16H19NO3 (2)* | Xenobiotics | Food component/Plant | 2.16 | 0.863 | 0.805 |
| 4‐acetylphenyl sulfate | Xenobiotics | Benzoate metabolism | 1.96 | 0.849 | 0.741 |
| Sulfate of piperine metabolite C18H21NO3 (3)* | Xenobiotics | Food component/Plant | 1.90 | 0.872 | 0.861 |
| Benzoylcarnitine* | Xenobiotics | Chemical | 1.78 | 0.870 | 0.823 |
| Glucuronide of piperine metabolite C17H21NO3 (4)* | Xenobiotics | Food component/Plant | 1.78 | 0.776 | 0.754 |
| All 10 metabolites | 0.996 | 0.989 | |||
Variable importance in projection (VIP) scores were calculated from partial least‐squares discriminant analysis (PLS‐DA). C‐statistics were calculated using conditional logistic regression with protein‐rich dietary pattern as the response variable and each of the individual metabolites as the exposure variable. “All 10 metabolites” refers to the panel of 10 metabolites in this table.
Asterisk indicates metabolites not officially confirmed (tier 2 identification).
These 10 metabolites were predictive of the protein‐rich dietary pattern individually (testing sample range in C‐statistics = 0.776–0.877; validation sample range in C‐statistics = 0.741–0.894) and collectively (testing sample C‐statistic = 0.996, validation sample C‐statistic = 0.989) (Table 4).
When we examined the top 15 metabolites, we observed four additional xenobiotics and an amino acid which were influential in discriminating between the protein‐rich dietary pattern and the carbohydrate‐rich dietary pattern: glucuronide of piperine metabolite, hippurate, genistein sulfate, tryptophan betaine, and 4‐vinylphenol sulfate. Among the top 15 metabolites, 4‐ethylphenyl sulfate, and 4‐acetylphenyl sulfate were highly correlated with each other ( = 0.74) (Figure S2, Supporting Information). Similarly, all piperine metabolites were highly correlated with each other ( > 0.7).
Out of 249 unknown metabolites, 48 metabolites were significantly different between the protein‐rich dietary pattern and the carbohydrate‐rich dietary pattern, 21 of which were higher after the protein diet intervention and 27 of which were lower after the protein diet intervention compared to the carbohydrate diet intervention (Table S2, Supporting Information). Unknown metabolites with the smallest p values were positively associated with the protein‐rich dietary pattern (e.g., p < 1 × 10−25 for X–11847, X–11299, X–11483, and X–11858).
3. Discussion
An untargeted metabolomic platform identified 102 metabolites that were significantly different between the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern, the majority of which were lipids, amino acids, and xenobiotics. We found that compounds involved in the metabolism of hexosylceramides were strongly associated with the protein‐rich dietary pattern. Prior studies using data from trials have examined changes in metabolite levels associated with amount of protein intake,[ 6 , 7 ] level of glycemic load,[ 8 , 9 ] hypocaloric diets which differed in fat, glycemic index, or carbohydrate,[ 10 ] and specific dietary patterns (e.g., Mediterranean diet, prudent diet, Western‐style diet, habitual diet).[ 11 , 12 , 13 ] To our knowledge, the present study is the first to identify a set of plasma metabolites which represent a protein‐rich dietary pattern compared to a carbohydrate‐rich dietary pattern in the context of a healthy diet.
Broadly, the top 10 metabolites associated with the protein‐rich dietary pattern represented 1) metabolites reflective of plant protein intake which are produced or converted through gut microbial activity, 2) metabolites directly derived from foods and beverages (quinate, hippurate, and catechol sulfate), and 3) metabolites which may reflect preparation methods or consumption habit, e.g., use of spices (piperine and several piperine metabolites). In the present study, 4‐ethylphenyl sulfate was highly predictive of the protein‐rich dietary pattern, and 4‐acetylphenyl sulfate, 4‐vinylphenol sulfate, and genistein sulfate were among the top 15 metabolites which were representative of the protein‐rich dietary pattern compared to the carbohydrate‐rich dietary pattern. Levels of all of these metabolites were higher in the protein‐rich dietary pattern compared to the carbohydrate‐rich dietary pattern. 4‐ethylpheyl sulfate is a gut flora metabolite, which has previously been associated with consumption of soy products (tofu, soy milk).[ 14 , 15 , 16 ] To a variable extent, isoflavoids in soy products are converted into phytoestrogens such as genistein by the gut microbiota.[ 17 ] 4‐vinylphenol sulfate, which was previously associated with peanut or total nut intake,[ 14 , 15 ] is believed to be a polyphenolic gut metabolite, although the origin of this metabolite is less well‐established than 4‐ethylphenyl sulfate. 4‐acetylphenyl sulfate is a novel metabolite that has not been associated with dietary patterns or dietary composition in prior studies. Given the high correlation between this metabolite and 4‐ethylphenyl sulfate, and modest correlation with 4‐vinylphenol sulfate, the higher levels of 4‐acetylphenyl sulfate may have been due to the plant protein intake. These metabolites are consistent with prior findings that higher protein intake influences the gut microbiota.[ 18 ]
In our study, plasma levels of quinate, hippurate, and catechol sulfate were lower after the protein diet intervention, which may be due to lower intake of fruits and juices. It has been suggested that quinate, hippurate, and catechol sulfate are metabolites of chlorogenic acid bacterial metabolism[ 16 , 19 ] or metabolites that are directly derived from foods or beverages.[ 10 , 19 ] For instance, chlorogenic acid is found in coffee beans and fruits such as peaches, pears, apples, and prunes.[ 20 ] Piperine and several piperine metabolites were among the top 10 metabolites which represented the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern. Piperine is found in black pepper.[ 21 ] In our study, piperine and piperine metabolites were all positively associated with the protein‐rich dietary pattern. Given that black pepper was allowed, this may primarily reflect consumption habits of the participants (e.g., addition of black pepper to foods which may have been used to enhance flavoring).
Our results were comparable to prior metabolomics studies of protein intake. In our study, levels of creatinine, urea, and many metabolites involved in the metabolism of essential amino acids were different between the protein‐rich and carbohydrate‐rich dietary patterns. Our findings for creatinine and urea overlapped with results from a controlled feeding study of elderly men who were randomly assigned to receive higher protein intake.[ 7 ] Findings on creatinine, urea, and uridine also replicated in another controlled feeding study which used nuclear magnetic resonance spectroscopy to identify biomarkers of macronutrient intake.[ 13 ] Further, we replicated other metabolites (creatinine, urea, phenylacetate, hydantoin‐5‐propionate, 3‐indoxyl sulfate, isobutyrylcarnitine, tiglylcarnitine) which were reported as candidate biomarkers of higher protein intake in a study of individuals with chronic kidney disease.[ 6 ] Metabolites which were involved in the metabolism of essential amino acids may be biomarkers of protein intake, given that essential amino acids can only be supplied from the diet. Importantly, tryptophan betaine was positively associated with the protein‐rich dietary pattern in our study and was one of the top 15 metabolites which were representative of the protein‐rich dietary pattern compared to the carbohydrate‐rich dietary pattern. Tryptophan betaine has been reported as a biomarker of plant proteins such as chickpeas and lentils,[ 22 ] nuts,[ 23 , 24 ] and healthy dietary patterns, such as the Dietary Approaches to Stop Hypertension (DASH) diet in prior studies.[ 25 , 26 , 27 ] The consistency of some of our findings with other protein metabolome studies is encouraging, but replication is necessary to confirm our proposed panel of metabolites, which are intended to represent healthy dietary patterns with higher protein and lower carbohydrate composition.
In our study, levels of most lipids were lower on the protein‐rich dietary pattern relative to the carbohydrate‐rich dietary pattern. In pathway analysis, hexosylcermides (in the lipid superpathway) were overrepresented. Plasma levels of lipids are affected by dietary carbohydrate intake. Carbohydrate‐rich diets increase de novo lipogenesis and production of certain phosphatidylcholines.[ 28 ] Thus, it is biologically plausible that levels of lipids were lower after the protein‐rich dietary pattern compared to the carbohydrate‐rich dietary pattern. For hexosylceramides, only one study reported an association with macronutrient intake. In free living Chinese adults in Singapore, plasma hexosylceramides were negatively associated with dietary protein intake, assessed using a food frequency questionnaire.[ 29 ] In the same study, hexosylceramides were negatively associated with LDL, HDL, and total cholesterol.[ 30 ] Hexosylceramides are formed by glycosylation of ceramides. Ceramides are bioactive lipids which play important roles in cellular signaling, and accumulation of these lipids is associated with increased atherosclerosis and cardiovascular disease risk.[ 31 ] Circulating levels of ceramides are thought to be influenced by dietary intake as well. In the Prevención con Dieta Mediterránea (PREDIMED) trial, individuals with higher baseline ceramide concentrations who were assigned to the two Mediterranean diet groups (either supplemented with nuts or extra virgin olive oil) had a similar risk of cardiovascular disease as those with lower baseline ceramide concentrations.[ 32 ] In contrast, individuals with higher baseline ceramide concentrations who were assigned to the control diet had elevated risk of incident cardiovascular disease.[ 32 ] The mechanism through which dietary protein and carbohydrate affects plasma levels of ceramides and hexosylceramides remains unclear and requires further investigation. However, our study suggests that hexosylceramides may represent a pathway through which higher protein, within the framework of a healthy dietary pattern, is associated with cardiovascular risk factors. In the future, examining the associations between candidate diet biomarkers from our study and cardiovascular risk factors may provide useful insights on the underlying metabolic pathway.
The present study has several strengths. We used data from a controlled feeding study. Participants received foods and had a high level of adherence, which increases confidence that the differences in metabolite levels observed represents the true biological effect of differences in macronutrient intake. The OmniHeart trial was a cross‐over study, in which participants served as their own control. This design minimized confounding by age, sex, and race/ethnicity. Further, the trial held other factors, most notably weight, constant throughout the duration of the study. Stabilizing weight removes the potentially confounding effect of weight change on metabolites. Additionally, the underlying study population included women and minority groups, thus these results are likely generalizable to the general US population.
Limitations should be noted. The feeding period, while long for a feeding study, was relatively short (6 weeks per diet period). It is unclear if these metabolites are representative of longer‐term intake of a protein‐rich healthy dietary pattern. However, we replicated many metabolites from observational studies, which assessed usual consumption of dietary patterns or food items. There was a lower level of confidence (tier 2 identification) for the identification of several of the top 10 and top 15 metabolites representative of the protein diet. It would be worthwhile to confirm the identity of these metabolites with greater confidence as well as the unknown metabolites that were statistically significantly associated with the protein‐rich dietary pattern and assessing if these metabolites replicate as biomarkers of healthy dietary patterns in future studies. Further, testing prediction in an independent sample and studying the effect of diets on the gut microbiome would be informative. Biospecimens from the OmniHeart trial were in storage for more than 10 years. Degradation of metabolites may have occurred, but it would be expected to be non‐differential across dietary patterns. Lastly, the OmniHeart trial was not originally designed to test differences in biomarkers of macronutrient content, which may in part explain why we found metabolites of specific foods and beverages to be significantly different between two diet interventions.
In conclusion, using data from the OmniHeart trial, we identified many metabolites (most of which were lipids, amino acids, and xenobiotics) that were significantly different between the protein‐rich dietary pattern and the carbohydrate‐rich dietary pattern. The top 10 metabolites which discriminated the two dietary patterns represented plant protein intake, metabolites in foods (e.g., fruits) and beverages (e.g., coffee, juices), and metabolites in spices (e.g., black pepper). If replicated, these metabolites may be used to assess adherence to a similar dietary pattern as the OmniHeart protein‐rich dietary pattern in observational studies and clinical trials.
4. Experimental Section
Study Design and Study Population
The OmniHeart trial was a randomized, three‐period crossover, controlled feeding study conducted in two clinical centers in the US (Baltimore, Maryland and Boston, Massachusetts) ( Clinical Trial Registration: https://clinicaltrials.gov/ct2/show/NCT00051350; Unique identifier: registered as NCT00051350). The trial evaluated the effects of three dietary patterns on blood pressure and serum lipids (registered as NCT00051350 at clinicaltrails.gov).[ 5 ] These three dietary patterns were different in macronutrient intake, varying in 1) carbohydrate, 2) protein (about half from plant sources), and 3) unsaturated fat. Details on the study design had been reported.[ 5 , 33 ] Briefly, healthy adults (≥30 years of age) with SBP 120–159 mmHg and DBP 80–99 mmHg were eligible. Individuals with chronic conditions such as diabetes or cardiovascular disease, elevated LDL (>5.70 mmol L−1), elevated fasting triglycerides (>8.48 mmol L−1), and elevated body weight (>159 kg), and those taking blood pressure‐lowering or lipid‐lowering medication were excluded. Institutional review boards at both clinical centers approved the study. Procedures were followed in accordance with the ethical standards of the institutional review boards. Participants provided written documentation of informed consent. The present study was approved by the Johns Hopkins IRB‐X Committee (CR00037373/NA_00 069360).
For the present metabolomic study, plasma specimens stored in the National Health, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC) was used.[ 34 ] Blood specimens were collected at the end of each dietary intervention. For the present study, the interest was to compare the plasma metabolome of the protein‐rich dietary pattern and the carbohydrate‐rich dietary pattern. Of the 164 participants of the OmniHeart trial, the study excluded three participants with no stored specimens and five participants with incomplete metabolomics data (i.e., metabolomics data not available for both the carbohydrate and protein intervention periods) (Figure S1, Supporting Information). The analytic sample of the present metabolomics study was 156 participants.
Dietary Exposures
After a 6‐day run‐in period, participants were randomly assigned to one of six sequences of the three dietary patterns. Each feeding period lasted 6 weeks with a wash‐out period of 2–4 weeks between each diet intervention (Figure S1, Supporting Information). Details on study diets had been reported in prior publications.[ 5 , 33 ] Briefly, the dietary pattern rich in carbohydrates was similar to the DASH diet, with 58% carbohydrate, 15% protein, and 27% fat (6% saturated fat; 13% monounsaturated fat; 8% polyunsaturated fat) from total energy intake. Carbohydrate intake in the OmniHeart carbohydrate diet was slightly higher (58%) than the DASH diet (55%). The protein‐rich dietary pattern had 48% carbohydrate, 25% protein, and 27% fat from total energy intake, with the same composition of total, saturated, monounsaturated, and polyunsaturated fat as the carbohydrate‐rich dietary pattern. In the protein‐rich dietary pattern, plant proteins (legumes, grains, nuts, and seeds) comprised about half of the total protein intake. Other protein sources, such as meat (beef, pork, ham), poultry, fish, low‐fat dairy product, and egg product substitute were higher for the protein‐rich dietary pattern compared to the carbohydrate‐rich dietary pattern. The protein‐rich dietary pattern was also higher in soy products (7.3 g day−1 in the protein diet vs 0.5 g day−1 in the carbohydrate diet at 2100 kcal day−1), but lower in full‐fat dairy products (0.2 serving per day in the protein diet vs 0.7 serving per day in the carbohydrate diet). The protein‐rich dietary pattern was lower in fruits and juices (3.8 serving per day in the protein‐rich vs 6.6 serving per day in the carbohydrate‐rich dietary patterns), but higher in vegetables (5.4 serving per day in the protein‐rich vs 4.4 serving per day in the carbohydrate‐rich dietary patterns). Fiber, potassium, magnesium, and calcium intake was similar in all diets, and sodium level was set to the intermediate level in the DASH‐Sodium diet (2300 mg day−1 at 2100 kcal day−1) for both diets.
All foods were prepared in the research kitchens using standardized recipes and consistent food brands. Participants received all meals, and ate one meal at the respective clinical center on weekdays and received other foods to take home. Throughout the study period, participants were instructed to consume only the foods they received from the study. Adherence to the diet was measured by observing how participants ate their foods onsite and checking participants’ daily food diaries. In daily diaries, participants recorded if they missed any foods or ate any foods that were not on the diet. Based on this information, the trial found that adherence to the diets was high.[ 5 ] Participants reported consuming all study foods and not eating non‐study foods on >95% of person‐days on all diet periods. Participants’ weight was measured every weekday and was kept constant by adjusting total energy intake. Participants were asked to drink no more than three caffeinated beverages per day and to maintain their habitual level of physical activity and alcohol consumption throughout the trial. The instructions for having coffee, tea, or alcohol did not vary by diets. Participants were allowed to add certain spices (e.g., black pepper, cayenne pepper, lemon pepper seasoning, dried Italian seasoning, curry powder, onion powder, garlic powder, an all‐purpose seasoning packet) to their foods.
Metabolomic Profiling
Untargeted metabolomic profiling was conducted by Metabolon (Durham, North Carolina) using plasma specimens collected after fasting for 8–12 h. Samples were divided into five fractions. Two fractions were analyzed using two reverse phase ultra‐high performance liquid chromatography‐mass spectrometry (UPLC‐MS/MS) with a positive ion mode electrospray ionization (ESI), one analyzed using UPLC‐MS/MS with a negative ion mode ESI, another one analyzed using hydrophilic interaction UPLC‐MS/MS with a negative ion mode ESI, and the remaining fraction was reserved for back‐up. A Waters ACQUITY liquid chromatographer and a ThermoFisher Scientific Q‐Exactive high resolution spectrometer with a heated ESI source and ThermoFisher Scientific Orbitrap mass analyzer was used for liquid chromatography‐mass spectrometry. After data were processed, peaks were identified using Metabolon's in‐house software, and matched to an extensive chemical library which had information on purified standard compounds. All detected metabolites were either a “tier 1” or “tier 2” identifications. To be considered a “tier 1” identification, at least two orthogonal measurements (e.g., accurate mass, retention time, fragmentation pattern) matched to an authentic reference.[ 35 , 36 ] We indicated metabolites with a lower level of identification confidence (e.g., “tier 2”) in a footnote.
All biospecimens were de‐identified, and laboratory technicians were blinded to assigned diet and any other characteristics of the participants. Samples were analyzed in a single batch and in random order. Twelve blind duplicates (six pairs) were included for quality control. In blind duplicates, 77% of metabolites had correlations ≥0.8 and 93% metabolites had coefficients of variation <20%, indicating high validity of the metabolomic profiling.
A total of 1243 metabolites were identified. The study excluded 109 metabolites with >80% missing values across samples. For the remaining 1134 metabolites, missing values were imputed with the minimum detectable level for the specific metabolite. These metabolites were then rescaled to a median of 1 by dividing by the batch‐specific median and log‐transformed (loge). The study further excluded 46 metabolites with variance on log scale <0.01 or missing variance. Outliers were capped at five standard deviations (SDs). After this data cleaning processing, 1088 metabolites remained, of which 839 metabolites were known (named) and 249 metabolites were unknown (unnamed).
Statistical Analysis
The baseline characteristics of the trial participants (n = 164) and the analytic sample (n = 156) using means (standard deviations) for continuous variables and proportions for categorical variables were examined.
In this study, paired t‐tests on log‐transformed known metabolites was used to assess differences in metabolite levels, comparing the protein‐rich dietary pattern to the carbohydrate‐rich dietary pattern (reference). To account for multiple testing, the Bonferroni approach (0.05/839 metabolites = 5.95 × 10−5) was used. The study considered the analyses of known metabolites as the primary analyses.
Next, for known metabolites, a pathway analysis using subpathway information provided by Metabolon was conducted. To evaluate whether the observed number of metabolites significant at the Bonferroni threshold was different than the expected number of metabolites, Fisher's exact test was used.
Then, PLS‐DA) to identify the top 10 plasma metabolites which were influential in discriminating between the protein‐rich and carbohydrate‐rich dietary patterns was used. This method was used to narrow down the significant metabolites to a reasonable number so that they may be more feasible to be assessed as biomarkers of the protein‐rich dietary pattern. The top 10 metabolites were selected using the metabolites with the highest variable importance in projection (VIP) scores. This PLS‐DA model was validated using random permutation testing with 2000 iterations and found that the risk of overfitting was low (p < 0.05). C‐statistics was calculated to assess whether these metabolites predict the protein‐rich dietary pattern relative to carbohydrate‐rich dietary pattern. For the analyses on C‐statistics, conditional logistic regression models with the protein‐rich dietary pattern as the outcome and added each of the top 10 metabolites one at a time, then all of the 10 metabolites simultaneously was used. C‐statistics was calculated first in a randomly selected sample of half of the study participants (testing sample, n = 78) and then in the remaining sample (validation sample, n = 78).
As a secondary analysis, the top 15 influential metabolites to expand on potential plasma biomarkers of the protein‐rich dietary pattern and to confirm previously proposed biomarkers of protein intake, and assessed correlations between these metabolites was examined. The correlation coefficients between the top 15 metabolites was calculated. Lastly, changes in levels of unknown metabolites using paired t‐tests was examined and used the Bonferroni approach (0.05/249 unknown metabolites = 2.01 × 10−4) to account for multiple testing. All analyses were conducted using Stata version 15 (StataCorp, College Station, TX, USA) and R version 4.1.0 (R Foundation for Statistical Computing, Vienna, Austria).
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
H.K. drafted the manuscript and conducted the statistical analysis. A.H.L., K.W., K.E.W., E.R.M, III, J.C., and L.J.A. contributed to data interpretation and critical revision of the manuscript. C.M.R. was involved in all aspects of the study from study design to analysis to critical revision of the manuscript. All authors read and approved the final manuscript.
Supporting information
Supporting information
Acknowledgements
C.M.R. was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; K01 DK107782, R03 DK128386) and from the National Heart, Lung, and Blood Institute (NHLBI; R01 HL153178). The funding agencies had no role in study design, data collection, analysis, drafting of the manuscript, and the decision to submit this manuscript for publication. The authors thank the staff and participants of the OmniHeart trial.
Kim H., Lichtenstein A. H., White K., Wong K. E., Miller E. R., Coresh J., Appel L. J., Rebholz C. M., Plasma Metabolites Associated with a Protein‐Rich Dietary Pattern: Results from the OmniHeart Trial. Mol. Nutr. Food Res. 2022, 66, 2100890. 10.1002/mnfr.202100890
Data Availability Statement
Original data and codebook of the OmniHeart Trial are available upon request pending application and approval from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinating Center: https://biolincc.nhlbi.nih.gov/studies/omniheart/?q=Omni Heart. The remaining metabolomics data that support the results of the present study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Kipnis V., Midthune D., Freedman L., Bingham S., Day N. E., Riboli E., Ferrari P., Carroll R. J., Public Health Nutr. 2002, 5, 915. [DOI] [PubMed] [Google Scholar]
- 2. Freedman L. S., Schatzkin A., Midthune D., Kipnis V., J. Natl. Cancer Inst. 2011, 103, 1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bingham S. A., J. Nutr. 2003, 133, 921S. [DOI] [PubMed] [Google Scholar]
- 4. Hedrick V. E., Dietrich A. M., Estabrooks P. A., Savla J., Serrano E., Davy B. M., Nutr J. 2012, 11, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Appel L. J., Sacks F. M., Carey V. J., Obarzanek E., Swain J. F., Miller E. R., Conlin P. R., Erlinger T. P., Rosner B. A., Laranjo N. M., Charleston J., McCarron P., Bishop L. M., for the OmniHeart Collaborative Research Group , J. Am. Med. Assoc. 2005, 294, 2455. [DOI] [PubMed] [Google Scholar]
- 6. Rebholz C. M., Zheng Z., Grams M. E., Appel L. J., Sarnak M. J., Inker L. A., Levey A. S., Coresh J., Am. J. Clin. Nutr. 2019, 109, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durainayagam B., Mitchell C. J., Milan A. M., Zeng N., Sharma P., Mitchell S. M., Ramzan F., Knowles S. O., Sjödin A., Wagner K.‐H., Roy N. C., Fraser K., Cameron‐Smith D., Front. Nutr. 2019, 6, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barton S., Navarro S. L., Buas M. F., S Y., Gu H., Djukovic D., Raftery D., Kratz M., Neuhouser M. L., Lampe J. W., Food Funct. 2015, 6, 2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Navarro S. L., Tarkhan A., Shojaie A., Randolph T. W., Gu H., Djukovic D., Osterbauer K. J., Hullar M. A., Kratz M., Neuhouser M. L., Lampe P. D., Raftery D., Lampe J. W., Am. J. Clin. Nutr. 2019, 110, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esko T., Hirschhorn J. N., Feldman H. A., Hsu Y.‐H. H., Deik A. A., Clish C. B., Ebbeling C. B., Ludwig D. S., Am. J. Clin. Nutr. 2017, 105, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wellington N., Shanmuganathan M., de Souza R. J., Zulyniak M. A., Azab S., Bloomfield J., Mell A., Ly R., Desai D., Anand S. S., Britz‐McKibbin P., Nutrients 2019, 11, 2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vázquez‐Fresno R., Llorach R., Urpi‐Sarda M., Lupianez‐Barbero A., Estruch R., Corella D., Fitó M., Arós F., Ruiz‐Canela M., Salas‐Salvadó J., Andres‐Lacueva C., J. Proteome Res. 2015, 14, 531. [DOI] [PubMed] [Google Scholar]
- 13. Zheng C., Gowda G. A. N., Raftery D., Neuhouser M. L., Tinker L. F., Prentice R. L., Beresford S. A. A., Zhang Y., Bettcher L., Pepin R., Djukovic D., Gu H., Barding G. A., Song X., Lampe J. W., Eur. J. Nutr. 2021, 60, 4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y., Hodge R. A., Stevens V. L., Hartman T. J., McCullough M. L., Metabolites 2020, 10, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guertin K. A., Moore S. C., Sampson J. N., Huang W.‐Y., Xiao Q., Stolzenberg‐Solomon R. Z., Sinha R., Cross A. J., Am. J. Clin. Nutr. 2014, 100, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu G. D., Compher C., Chen E. Z., Smith S. A., Shah R. D., Bittinger K., Chehoud C., Albenberg L. G., Nessel L., Gilroy E., Star J., Weljie A. M., Flint H. J., Metz D. C., Bennett M. J., Li H., Bushman F. D., Lewis J. D., Gut 2016, 65, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Correia M. S. P., Jain A., Alotaibi W., Young Tie Yang P., Rodriguez‐Mateos A., Globisch D., Free Radic. Biol. Med. 2020, 160, 745. [DOI] [PubMed] [Google Scholar]
- 18. Beaumont M., Portune K. J., Steuer N., Lan A., Cerrudo V., Audebert M., Dumont F., Mancano G., Khodorova N., Andriamihaja M., Airinei G., Tomé D., Benamouzig R., Davila A.‐M., Claus S. P., Sanz Y., Blachier F., Am. J. Clin. Nutr. 2017, 106, 1005. [DOI] [PubMed] [Google Scholar]
- 19. Pallister T., Jackson M. A., Martin T. C., Zierer J., Jennings A., Mohney R. P., MacGregor A., Steves C. J., Cassidy A., Spector T. D., Menni C., Sci. Rep. 2017, 7, 13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wishart D. S., Feunang Y. D., Marcu A., Guo A. C., Liang K., Vázquez‐Fresno R., Sajed T., Johnson D., Li C., Karu N., Sayeeda Z., Lo E., Assempour N., Berjanskii M., Singhal S., Arndt D., Liang Y., Badran H., Grant J., Serra‐Cayuela A., Liu Y., Mandal R., Neveu V., Pon A., Knox C., Wilson M., Manach C., Scalbert A., Nucleic Acids Res. 2018, 46, D608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derosa G., Maffioli P., Sahebkar A., Adv Exp Med Biol 2016, 928, 173. [DOI] [PubMed] [Google Scholar]
- 22. Garcia‐Aloy M., Ulaszewska M., Franceschi P., Estruel‐Amades S., Weinert C. H., Tor‐Roca A., Urpi‐Sarda M., Mattivi F., Andres‐Lacueva C., Mol. Nutr. Food Res. 2020, 64, 1901137. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y., Gapstur S. M., Carter B. D., Hartman T. J., Stevens V. L., Gaudet M. M., McCullough M. L., J. Nutr. 2018, 148, 932. [DOI] [PubMed] [Google Scholar]
- 24. Playdon M. C., Sampson J. N., Cross A. J., Sinha R., Guertin K. A., Moy K. A., Rothman N., Irwin M. L., Mayne S. T., Stolzenberg‐Solomon R., Moore S. C., Am. J. Clin. Nutr. 2016, 104, 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rebholz C. M., Lichtenstein A. H., Zheng Z., Appel L. J., Coresh J., Am. J. Clin. Nutr. 2018, 108, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim H., Rebholz C. M., Curr. Atheroscler. Rep. 2021, 23, 26. [DOI] [PubMed] [Google Scholar]
- 27. Playdon M. C., Moore S. C., Derkach A., Reedy J., Subar A. F., Sampson J. N., Albanes D., Gu F., Kontto J., Lassale C., Liao L. M., Männistö S., Mondul A. M., Weinstein S. J., Irwin M. L., Mayne S. T., Stolzenberg‐Solomon R., Am. J. Clin. Nutr. 2017, 105, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inoue M., Senoo N., Sato T., Nishimura Y., Nakagawa T., Miyoshi N., Goda T., Morita A., Miura S., J. Nutr. Biochem. 2017, 50, 83. [DOI] [PubMed] [Google Scholar]
- 29. Seah J. Y. H., Chew W. S., Torta F., Khoo C. M., Wenk M. R., Herr D. R., Tai E. S., van Dam R. M., Metabolites 93, 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chew W. S., Torta F., Ji S., Choi H., Begum H., Sim X., Khoo C. M., Khoo E. Y. H., Ong W.‐Y., Van Dam R. M., Wenk M. R., Tai E. S., Herr D. R., JCI Insight 2019, 5, e126925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaurasia B., Summers S. A., Trends Endocrinol. Metab. 2015, 26, 538. [DOI] [PubMed] [Google Scholar]
- 32. Wang D. D., Toledo E., Hruby A., Rosner B. A., Willett W. C., Sun Q., Razquin C., Zheng Y., Ruiz‐Canela M., Guasch‐Ferré M., Corella D., Gómez‐Gracia E., Fiol M., Estruch R., Ros E., Lapetra J., Fito M., Aros F., Serra‐Majem L., Lee C.‐H., Clish C. B., Liang L., Salas‐Salvadó J., Martínez‐González M. A., Hu F. B., Circulation 2017, 135, 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swain J. F., McCarron P. B., Hamilton E. F., Sacks F. M., Appel L. J., J. Am. Diet Assoc. 2008, 108, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giffen C. A., Carroll L. E., Adams J. T., Brennan S. P., Coady S. A., Wagner E. L., Biopreserv. Biobank 2015, 13, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schrimpe‐Rutledge A. C., Codreanu S. G., Sherrod S. D., McLean J. A., J. Am. Soc. Mass Spectrom. 2016, 27, 1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W.‐M., Fiehn O., Goodacre R., Griffin J. L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A. N., Lindon J. C., Marriott P., Nicholls A. W., Reily M. D., Thaden J. J., Viant M. R., Metabolomics 2007, 3, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
Original data and codebook of the OmniHeart Trial are available upon request pending application and approval from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinating Center: https://biolincc.nhlbi.nih.gov/studies/omniheart/?q=Omni Heart. The remaining metabolomics data that support the results of the present study are available from the corresponding author upon reasonable request.


