Abstract
The polymerase (pol) coding sequence was determined for 40 independent clinical cytomegalovirus isolates sensitive to ganciclovir and foscarnet. Sequence alignments showed >98% interstrain homology and amino acid variation in only 4% of the 1,237 codons. Almost all variation occurred outside of conserved functional domains where resistance mutations have been identified.
The technical difficulty and time-consuming nature of cell culture-based susceptibility assays have resulted in a need for rapid genotypic assays for cytomegalovirus (CMV) drug resistance (9). Development of such assays will depend on knowledge about the relative frequency of various resistance-related mutations and the extent of interstrain variation that occurs among drug-sensitive clinical isolates.
The CMV DNA polymerase (pol; UL54) (reviewed in reference 1) and the UL97 phosphotransferase are two viral genes where certain mutations have been linked to drug resistance (9). Ganciclovir resistance results from mutations in UL97 (4), pol (11), or both (13). All known foscarnet and cidofovir resistance mutations map to the pol gene, and cidofovir-resistant pol mutants are generally cross-resistant to ganciclovir (7, 8). It is important to distinguish resistance mutations from natural interstrain sequence variation. Some of the observed amino acid changes in the pol genes of resistant clinical isolates (5, 6, 11, 13) have subsequently been validated as resistance markers by transfer of mutations to laboratory strains of CMV, but others have not. We performed a multicenter sequencing study to assess the range of interstrain variation in the pol gene of drug-sensitive clinical isolates and to compare the loci of variation with resistance mutations that have been identified to date.
Clinical CMV isolates for this study were collected at four geographically separate institutions from 40 unrelated subjects who had received organ transplants or had human immunodeficiency virus infection. Some patients had received prior anti-CMV therapy. Susceptibility to ganciclovir and foscarnet was determined by plaque reduction in fibroblast cell culture. The 50% inhibitory drug concentration (IC50) was determined, and an isolate was considered sensitive if the ganciclovir IC50 was 6 μM or less and the foscarnet IC50 was 400 μM or less (10). For the 40 isolates selected, the ganciclovir IC50 ranged from 0.8 to 5.8 μM and the foscarnet IC50 ranged from 60 to 280 μM. All isolates were therefore considered sensitive to both drugs.
The entire 3.7-kb CMV pol coding sequence of all isolates was amplified by PCR from DNA extracts of infected cell cultures. A full-length PCR product was obtainable by using the GeneAmp XL PCR kit (PE Applied Biosystems, Branchburg, N.J.) and the following primers which flank the pol coding region (forward, 5′-GTCAGCCTCTCACGGTCCGCTAT-3′; reverse, 5′-CTCAGTCTCAGCAGCATCATCAC-3′). The PCR products were sequenced without cloning, by using primers internal to the amplified segment (11) that produced overlapping coverage of the pol coding sequence. Sequencing reactions were performed by dideoxy chain termination chemistry, with an automated sequencer (ABI 373 or 377) and fluorescent dideoxy terminators (AmpliTaq FS cycle sequencing kit; PE Biosystems, Foster City, Calif.), or by manual methods with the Thermo Sequenase 33P terminator cycle sequencing kit (Amersham, Arlington Heights, Ill.).
Derived sequences of each isolate were aligned with the strain AD169 reference sequence (EMBL accession no. X17403) by using a sequence text editor. Codons which differed from strain AD169 were assessed as to validity of the raw sequence data, and repeat sequencing reactions were performed to recheck any ambiguity. Amino acid differences were tabulated, graphed, and compared with previously published pol resistance mutations. Sequence homology calculations at the nucleotide and amino acid levels were also performed to compare each isolate with strain AD169.
We found >98% interstrain homology at both the nucleotide and the amino acid level. In comparison to the AD169 sequence, the number of nucleotide changes within the 3,729-bp pol coding sequence for each strain ranges from 25 to 63. The average number of nucleotide changes per strain is 46. There are a total of 282 variant nucleotides, of which >80% produce silent mutations. Among the 1,237 codons in the pol coding sequence, each isolate has nine or fewer amino acid changes compared to the AD169 sequence. A change from the AD169 sequence was observed at 47 codons (Table 1). This represents 4% of the total codons in pol. There are only four codons (655, 685, 897, and 1122) where 10 or more isolates have an amino acid configuration different from the most common one. In most of the rest of the codons that show variation, only one or two isolates show any change. Changes that are not amino acid substitutions include a codon insertion at 885 and codon deletions at 1151 and 1156 (Table 1).
TABLE 1.
Pol amino acid differences of clinical isolates from strain AD169
| Codon | Change | No. of isolates | Codon | Change | No. of isolates | |
|---|---|---|---|---|---|---|
| 15 | A15T | 1 | 870 | D870H | 2 | |
| 24 | S24L | 2 | 873 | V873L | 1 | |
| 32 | S32P | 1 | 874 | G874R | 6 | |
| 95 | V95E | 1 | 885 | A885T | 37 | |
| 142 | H142Y | 1 | 885 | A885S | 1 | |
| 143 | G143S | 1 | 885 | Insertion of T | 1 | |
| 347 | G347D | 1 | 887 | P887S | 2 | |
| 355 | V355A | 4 | 890 | L890F | 4 | |
| 464 | S464F | 1 | 897 | S897L | 11 | |
| 515 | D515G | 1 | 898 | N898D | 35 | |
| 626 | A626V | 1 | 899 | E899K | 1 | |
| 628 | P628L | 1 | 953 | V953A | 2 | |
| 631 | A631G | 1 | 1020 | L1020I | 4 | |
| 640 | M640R | 1 | 1116 | N1116H | 7 | |
| 655 | S655L | 20 | 1122 | A1122T | 23 | |
| 663 | S663N | 1 | 1133 | G1133S | 2 | |
| 669 | F669L | 3 | 1146 | S1146N | 1 | |
| 676 | S676G | 2 | 1147 | N1147S | 3 | |
| 678 | G278S | 1 | 1149 | R1149T | 3 | |
| 685 | N685S | 25 | 1151 | Deletion of 1151 | 1 | |
| 688 | A688V | 3 | 1153 | P1153S | 1 | |
| 693 | A693T | 1 | 1156 | Deletion of 1156 | 1 | |
| 697 | Q697H | 1 | 1162 | P1162L | 1 | |
| 868 | Q868R | 1 | 1235 | S1235T | 1 |
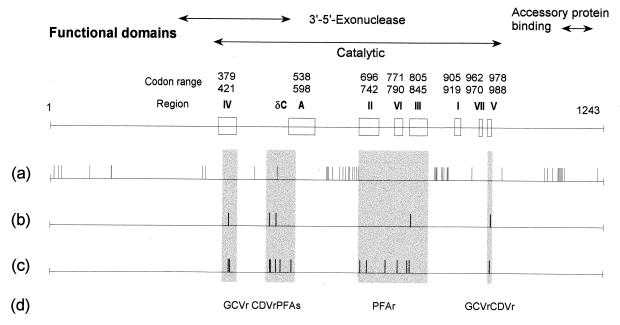
Codons with amino acid changes are strongly clustered outside the previously defined conserved catalytic sequences of pol (12) (Fig. 1). The greatest concentration of variant codons is at 626 to 697, 868 to 899, and 1116 to 1162. Within the conserved regions I to VII and A (Fig. 1), variation occurs only at codon 697 (on the fringe of domain II). In the area between codon 492 and region A (also known as delta-region C) (7, 13), which includes some known resistance mutations (Fig. 1), one isolate has an amino acid change at codon 515. There is no overlap between the amino acid variation observed in these drug-sensitive isolates and the published pol drug resistance mutations that have been proven by marker transfer experiments. The list of codons where a single mutation confers a resistance phenotype upon transfer to strain AD169 or Towne includes 408, 412, 501, 513, 522, 545, 700, 715, 781, 802, 809, and 987 (2, 5–7, 12, 14).
FIG. 1.
CMV DNA polymerase mutation map. Shown at top are the conserved functional domains and their codon ranges (boxed). Loci of amino acid changes are mapped below. (a) Codons showing variation in drug-sensitive isolates in this study; (b) codons mapped to drug resistance in laboratory strains (8, 12, 14); (c) codons mapped to drug resistance in clinical isolates (2, 5–7, 9, 11, 13); (d) drug resistance phenotypes associated with drug-resistant mutants according to region. Regions known to be associated with drug resistance are shaded. GCV, ganciclovir; CDV, cidofovir; PFA, foscarnet.
This study of drug-sensitive CMV clinical isolates demonstrates the very high level of sequence conservation of the DNA polymerase coding region, the strong clustering of the amino acid changes outside the functional conserved domains, and the absence of overlap between baseline sequence variation and the mutations that have been shown to be related to drug resistance. The absence of significant sequence divergence in pol is in contrast to the envelope glycoprotein gB gene which is directly adjacent to pol. In that gene, grouped variation results in sequence divergence of up to 12% among isolates, about 10-fold greater than that seen here in pol (3), and the variation affects functional domains and epitopes.
Increasing information is available concerning the pol mutations in clinical isolates that are markers of drug resistance (5–7) (Fig. 1). Dual resistance to ganciclovir and cidofovir has been mapped to pol region IV (codons 379 to 421), codon 987 in region V, and a region between codons 501 and 545 (5, 7, 8, 12, 13). The only baseline variation observed in these codon ranges is in the latter area (at codon 515). Foscarnet resistance has been mapped to codons 700, 715, 756, 781, 802, 809, and 821, involving regions II, III, and VI (codon range 696 to 845). Mutations in this region sometimes confer low-grade cross-resistance to ganciclovir and other drugs (5–8). No baseline amino acid variation was observed in this region except at codon 697. An amino acid change at codon 676 was previously reported in association with a ganciclovir-cidofovir-resistant isolate (13). The same change was found in two sensitive isolates in this study and has been found by marker transfer to be unrelated to resistance to ganciclovir and cidofovir (7).
Based on current knowledge, genotypic assays for resistance should focus on the pol codon ranges 379 to 421, 492 to 545, 696 to 845, and 978 to 988. The sequence database obtained from this study should be useful for optimization of these genotypic assays.
Nucleotide sequence accession numbers.
Sequences determined in this study have been deposited in GenBank under accession no. AF133589 through AF133628.
Acknowledgments
We thank Matthew Shiveley, Ernest Winkfield, Julia Clark, and Joe Zhou for excellent technical assistance and W. Lawrence Drew and Richard C. Miner for providing nine CMV isolates and their sensitivity data.
This work was supported by subcontracts to the participating laboratories from Social and Scientific Systems based on an NIH/NIAID Cooperative Agreement (AI38858). Sequences obtained at the University of Colorado Health Sciences Center were provided by the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility, which is supported by the NIH/NCI Cancer Core Support Grant (CA46934). Identification of pol resistance mutations was supported by VA research funds and NIH/NIAID grant AI39938 to S.C. C.S.C. was supported by NIH grants AI41690 and U01 AI27659.
REFERENCES
- 1.Anders D G, McCue L A. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology. 1996;39:378–388. doi: 10.1159/000150508. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti F, Underwood M R, Stanat S C, Biron K K, Chou S, Sarasini A, Silini E, Gerna G. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J Virol. 1996;70:1390–1395. doi: 10.1128/jvi.70.3.1390-1395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S, Dennison K M. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229–1234. doi: 10.1093/infdis/163.6.1229. [DOI] [PubMed] [Google Scholar]
- 4.Chou S, Guentzel S, Michels K R, Miner R C, Drew W L. Frequency of UL97 phosphotransferase mutations related to ganciclovir resistance in clinical cytomegalovirus isolates. J Infect Dis. 1995;172:239–242. doi: 10.1093/infdis/172.1.239. [DOI] [PubMed] [Google Scholar]
- 5.Chou S, Marousek G, Guentzel S, et al. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 6.Chou S, Marousek G, Parenti D M, et al. Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J Infect Dis. 1998;178:526–530. doi: 10.1086/515648. [DOI] [PubMed] [Google Scholar]
- 7.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cihlar T, Fuller M D, Mulato A S, Cherrington J M. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology. 1998;248:382–393. doi: 10.1006/viro.1998.9299. [DOI] [PubMed] [Google Scholar]
- 9.Crumpacker C S, editor. Drug resistance in cytomegalovirus: current knowledge and implications for patient management. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12(Suppl. 1):S1–S22. [PubMed] [Google Scholar]
- 10.Drew W L, Miner R, Saleh E. Antiviral susceptibility testing of cytomegalovirus: criteria for detecting resistance to antivirals. Clin Diagn Virol. 1993;1:179–185. doi: 10.1016/0928-0197(93)90012-t. [DOI] [PubMed] [Google Scholar]
- 11.Erice A, Gil-Roda C, Perez J L, et al. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immuno-compromised patients. J Infect Dis. 1997;175:1087–1092. doi: 10.1086/516446. [DOI] [PubMed] [Google Scholar]
- 12.Lurain N S, Thompson K D, Holmes E W, Read G S. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J Virol. 1992;66:7146–7152. doi: 10.1128/jvi.66.12.7146-7152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan V, Biron K K, Talarico C, Stanat S C, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]