Abstract
The term autophagy encompasses different pathways that route cytoplasmic material to lysosomes for degradation and include macroautophagy, chaperone-mediated autophagy and microautophagy. Since these pathways are crucial for degradation of aggregate-prone proteins and dysfunctional organelles like mitochondria, they help to maintain cellular homeostasis. As post-mitotic neurons cannot dilute unwanted protein and organelle accumulation by cell division, the nervous system is particularly dependent on autophagic pathways. This dependence may be a vulnerability as people age and these processes become less effective in the brain. Here we will review how the different autophagic pathways may protect against neurodegeneration, giving examples of both polygenic and monogenic diseases. We have considered how autophagy may have roles in normal CNS functions and the relationships between these degradative pathways and different types of programmed cell death. Finally, we will provide an overview of recently described strategies for upregulating autophagic pathways for therapeutic purposes.
Graphical Abstract
The term autophagy encompasses different pathways enabling lysosomal degradation of cytoplasmic material, like macroautophagy, chaperone-mediated autophagy and microautophagy. Fleming et al. review how different autophagic pathways protect against neurodegeneration, and consider recently described therapeutic strategies exploiting autophagic upregulation.
A. Overview of different autophagy pathways
The term autophagy describes a set of mechanistically distinct processes that diverse cells use to deliver cytoplasmic contents to lysosomes for degradation that includes macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy. In this review, we will briefly describe these pathways, how they impact neurological functions in health and disease and the current pharmacological efforts to boost autophagic activities as therapeutic strategies in neurodegenerative diseases.
Macroautophagy core pathway
The first morphologically distinct structure in macroautophagy is the cup-shaped, double-membraned phagophore, whose edges extend and fuse to become an autophagosome (Fig. 1). Autophagosome biogenesis is complex and involves multiple proteins and lipids from various membrane sources, including the endoplasmic reticulum (ER), ER/mitochondria contact sites (MAM), ER exit sites, recycling endosomes, Golgi and plasma membrane (Axe et al., 2008; Ge and Schekman, 2014; Puri et al., 2013).
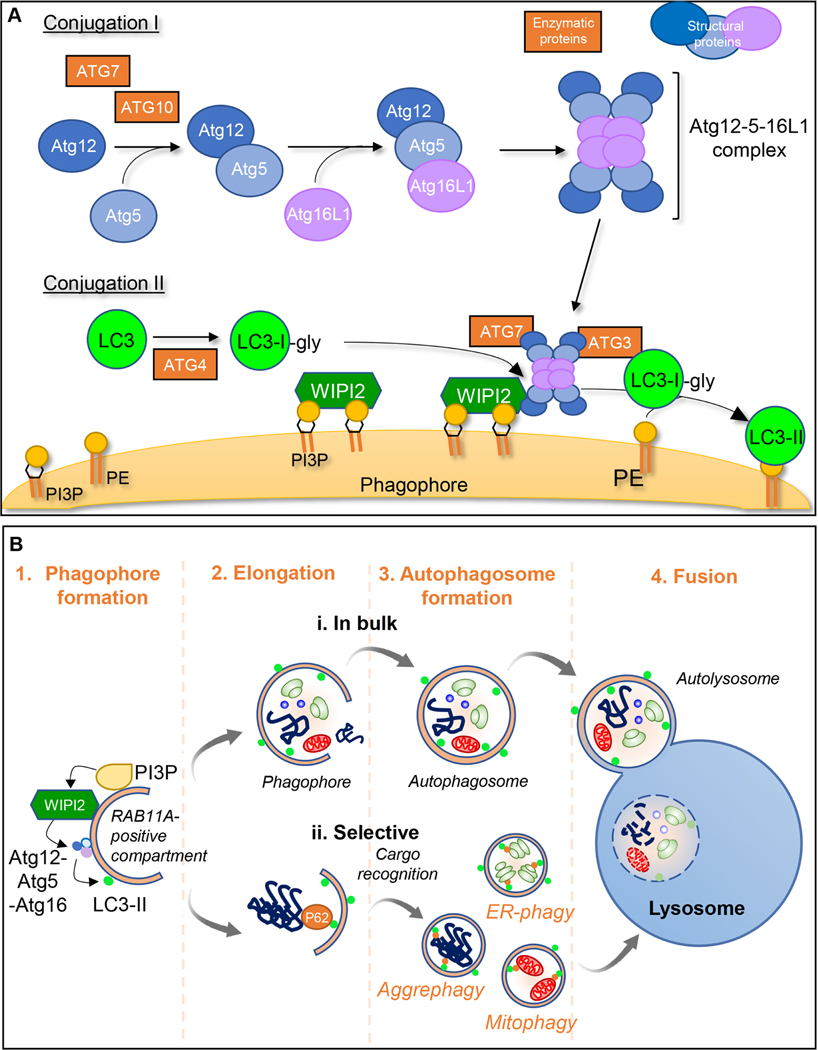
Figure 1. Schematic representation of macroautophagy:
Cell components to be degraded are engulfed in a double membraned structure called the phagophore, the edges of which elongate and close to form autophagosomes. These ultimately fuse with the lysosomal membrane for cargo degradation. A. Early steps in macroautophagy involve 2 ubiquitin-like conjugation cascades. Conjugation I leads to the formation of the Atg5-Atg12 conjugate mediated by ATG7 (E1-like) and ATG10 (E2-like). This then forms a complex with ATG16L1. During Conjugation II, ATG4 cleaves the C-terminus of LC3 generating LC3-I whose C-terminal glycine can be conjugated to phosphatidylethanolamine (PE) by ATG7 (E1-like), ATG3 (E2-like) and ATG5-ATG12/ATG16L1 (E3-like) (generating lipidated LC3 (LC3-II)). The other ATG8 family members (GABARAPs) use the same machinery to enable their conjugation to PE as LC3 proteins. The sites of LC3 conjugation to membranes are determined by the ATG5-ATG12-ATG16L1 complex, which localizes to the surface of the forming phagophore by interacting with WIPI2. WIPI2 is recruited to these membranes by binding both to phosphatidylinositol 3-phosphate (PI3P) and RAB11A. B. During autophagosome formation, the phagophore double membrane elongates and fuses to form a double-membraned vesicle termed the autophagosome. Cargo within autophagosomes can be trapped in a (i) bulk or (ii) selective manner by autophagy cargo receptors, such as P62, leading to the selective autophagy of specific substrates. Completion of vesicle closure to engulf regions of cytoplasm and organelles or to engulf specific cargoes such as aggregates (aggrephagy), mitochondria (mitophagy) or ribosomes (ribophagy) is followed by release from the recycling endosome-RAB11A platform to which the LC3 conjugates. Finally, the autophagosome outer membrane fuses with the lysosomal membrane and cargo is released for complete degradation in the lysosomal lumen.
The molecular components of macroautophagy were originally described in yeast, where genetic screens identified more than 40 ATG (autophagy related) proteins, most of them with mammalian orthologues, that regulate macroautophagy at its different stages (Mizushima et al., 2011). A key event in the formation of phagophores is the conjugation of members of the ubiquitin-like ATG8 family (Fig. 1), including the LC3 and GABARAP (gamma-aminobutyric acid receptor-associated protein) subfamilies, to phosphatidylethanolamine in precursor membranes. This event occurs on RAB11A-positive recycling endosomes in a wide range of cells, including primary neurons (Puri et al., 2018). Thereafter, the autophagosomes are closed by ESCRT machinery (Takahashi et al., 2018) then released in a step mediated by DNM2 (Puri et al., 2020).
It is likely that there is lipid transfer to growing autophagosomes via the ATG2-WIPI4 complex (Maeda et al., 2019) and that some lipid remodelling occurs on nascent autophagosomes mediated by the scramblase function of ATG9 that assists membrane expansion (Matoba et al., 2020). Most of the cell biology and biochemistry of mammalian autophagy has been studied in fibroblasts and cancer cell lines. While the pathway conservation across species suggests that the core biology will be essentially similar in all cells, there may be neuron- or glial-specific adaptions.
Selective Macroautophagy (mitophagy, ER-phagy, pexophagy, aggrephagy)
Macroautophagy degrades long-lived, aggregate-prone proteins, protein complexes, and dysfunctional or damaged organelles. Substrate degradation can be facilitated by machinery that enables their preferential sequestration via adaptor proteins that bridge components of the substrate (generally surface proteins which are ubiquitinated) and elements of the nascent autophagosome, typically LC3 (Fig. 1). These forms of selective autophagy include mitophagy (mitochondria), pexophagy (peroxisomes), ribophagy (ribosomes), ER-phagy (ER) and aggrephagy (aggregate-prone proteins), among others.
Mitophagy
Mitophagy exists in several forms, depending on the mechanism of recruitment of the phagophore membrane to mitochondria. The most well-studied form relies on the phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase (PTEN)-induced putative kinase 1 (PINK1) and the ubiquitin ligase Parkin. After mitochondrial damage, PINK1 levels, normally low, increase and PINK1 auto-phosphorylates to recruit cytosolic Parkin to the mitochondrial membrane, triggering mitophagy (Matsuda, 2016). In PINK1-Parkin-dependent mitophagy, autophagy receptors such as optineurin (OPTN), NDP52 (nuclear domain 10 protein 52) and TAX1BP1 (Trans-activating transcriptional regulatory protein of HTLV-1 (TAX1) binding protein 1) recruit the autophagosome biogenesis machinery to mitochondria. (Geisler et al., 2010; Narendra et al., 2010). Neuronal PINK1 or Parkin deletions in mice cause only mild phenotypes, highlighting the existence of alternative mitophagy processes, including receptor-mediated (NIX/BNIP3L), ubiquitin-mediated (MUL1-dependent) and lipid-mediated (through exposure of the inner membrane mitochondrial lipid cardiolipin) pathways (Evans and Holzbaur, 2020).
ER-phagy (reticulophagy)
Constitutive ER turnover is required to maintain proper cellular function. Autophagy contributes to ER remodelling via ER-phagy or reticulophagy. The ER-phagy adaptor FAM134B (family with sequence similarity 134, member B), interacts directly with LC3 or GABARAP and cooperates with Reticulon 3 (RTN3- RHD-containing protein) on the ER membrane to trigger the recruitment of the specific area of the ER that need to be degraded inside the autophagosome (Grumati et al., 2017). Other ER-phagy receptors, include cell-cycle progression gene 1 (CCPG1), Sec62, Atlastin-3 (ATL3) and testis expressed 264 (TEX264) (Chino et al., 2019; Fumagalli et al., 2016). The existence of multiple receptors suggests a redundancy or selectivity for types of ER. FAM134B and RTN3L are involved in starvation-induced ER-phagy, while CCPG1 is induced during ER stress and Sec62 participates in the recovery of ER stress (recovER-phagy). Mutations in FAM134B and ATL3 have been linked to peripheral neuropathy, but the other ER-receptors have not been studied yet in the nervous system.
Pexophagy
Damaged or excess peroxisomes are degraded through autophagy via pexophagy, where ubiquitination of PEX3 or PMP34 on the cytosolic face of peroxisomes enables recognition by autophagy adaptors P62 or NBR1, which target peroxisomes for autophagic degradation (Jo et al., 2020).
Aggrephagy
Most neurodegenerative diseases manifest with the accumulation of aggregates in vulnerable cell populations in the brain. Selective degradation of protein aggregates is called aggrephagy. Misfolded, aggregate-prone proteins are ubiquitinated, recognized and linked to the autophagy machinery by adaptors like P62, NBR1, Tollip (Cue5 in yeast), OPTN (Shen et al., 2017) and TAX1BP1 (Sarraf et al., 2020).
Lysophagy
Damaged lysosomes are degraded by selective autophagy via lysophagy, which protects cells from lysosomal cell death. Lysophagy appears to be ubiquitination-dependent and involves various canonical autophagy receptors, like TAX1BP1 (Eapen et al., 2021) and TBK1 (Eapen et al., 2021), a protein mutated in various diseases, including forms of ALS.
Major regulators
Macroautophagy can be stimulated by stresses including nutrient depletion, growth factor deprivation, oxidative stress and protein aggregation (Menzies et al., 2017). Nutrient starvation inhibits mTORC1 (mechanistic target of rapamycin complex 1), a highly conserved negative regulator of autophagy (Saxton and Sabatini, 2017). In nutrient-rich conditions, mTORC1 interacts and phosphorylates ULK1, to inhibit autophagy. Upon starvation, mTORC1 sites on ULK1 are dephosphorylated and ULK1 dissociates from mTORC1, thereby activating ULK1 kinase activity (Hosokawa et al., 2009). In mammals, mTORC1 activity is stimulated by growth factors through inhibition of the tuberous sclerosis complex (TSC) 1 and 2 (Dibble and Manning, 2013). Amino acids signal to mTORC1 through Rag GTPases independently of the TSC complex (Sancak et al., 2008). Recently, leucine was shown to signal to mTORC1 in most cells, including neurons and glia, via its metabolite Acetyl-coenzyme A (Ac-CoA), which stimulates acetylation of raptor and subsequent activation of mTORC1 and inhibition of autophagy (Son et al., 2020). Autophagy can also be induced by low cellular energy levels/low glucose (increased AMP/ATP) sensed by AMP-activated protein kinase (AMPK). In glucose-deprived cells, AMPK activates ULK1, which then phosphorylates and activates the lipid kinase PIKfyve (FYVE finger-containing phosphoinositide kinase), thereby increasing synthesis of the phosphatidylinositol 5-phosphate PI(5)P (Karabiyik et al., 2021). PI(5)P upregulates autophagosome synthesis and induces autophagic flux in the absence of PI(3)P in a VPS34-independent manner (Vicinanza et al., 2015). Another key regulator of autophagy and lysosomal biogenesis is transcription factor EB (TFEB) (Settembre et al., 2013).
Non autophagic role of the ATG protein conjugation system
Some ATGs have autophagy-independent functions. In LC3-associated phagocytosis (LAP), ATGs contribute to immune regulation and inflammatory responses, particularly in phagocytic cells. In LAP, LC3 is conjugated to phagosome single membranes (LAPosomes) that directly fuse to lysosomes (Galluzzi and Green, 2019). LAP is independent on the ULK1/FIP200/ATG13/ATG101 macroautophagy complex and is unresponsive to starvation or intracellular stress (Heckmann et al., 2017). Toll-like receptors (TLRs) and immunoglobulin (Ig) receptors participate in cargo recognition to activate LAP. The recognition of opsonized foreign particles leads to the recruitment of LAP regulatory machinery to the phagosome and the engulfed substrates are degraded following fusion with lysosomes (Heckmann et al., 2019).
LC3 can also be conjugated to RAB5-, clathrin-positive endosomes that contain amyloid-β. This new function, called LANDO (LC3-associated endocytosis), requires ATGs, such as LC3, Rubicon and ATG5, but not FIP200. LANDO was described in microglia, where it enables removal of amyloid-β and ameliorates pathology in a murine model of Alzheimer’s Disease (AD) (Heckmann et al., 2019).
Non-canonical macroautophagy
Macroautophagy-like processes can also occur in the absence of some key ATG proteins, in what is called non-canonical macroautophagy. Some cell types lacking Atg5 or Atg7, essential for canonical macroautophagy, still perform macroautophagy in response to specific stressors (Nishida et al., 2009). Autophagosome formation in this case is independent of the ubiquitin-like protein systems ATG5-ATG12, ATG7-ATG8, ATG16 and ATG9, but requires ULK1 and PI3K-Beclin 1-VPS34 complexes (Nishida et al., 2009). ATG5-ATG7-independent autophagosomes originate from the trans-Golgi membrane in a RAB9-dependent manner (Honda et al., 2020). Non-canonical macroautophagy has been shown to contribute to mitochondrial clearance during erythroid maturation or iPSC cell differentiation and to the degradation of proinsulin granules in glucose-deprived β-cells (Honda et al., 2020). Neuronal non-canonical autophagy is required for the degradation of ceruloplasmin to prevent iron deposition (Yamaguchi et al., 2020).
Chaperone-mediated autophagy
In CMA, substrate proteins are translocated across the lysosomal membrane, where they are targeted following the recognition of a KFERQ-like targeting motif in their sequences by the heat shock cognate 71kDa protein (HSC70) (Fig. 2). The HSC70/substrate complex binds the cytosolic tail of the lysosome-associated membrane protein type 2A (LAMP-2A) (Cuervo and Dice, 1996), triggering its multimerization into a translocation complex. After unfolding, the substrate is internalized into the lysosomal lumen assisted by a lysosome resident HSC70, and the LAMP-2A complex is disassembled (Kaushik and Cuervo, 2018). LAMP-2A levels and its assembly/disassembly in the lysosomal membrane are rate-limiting for CMA.
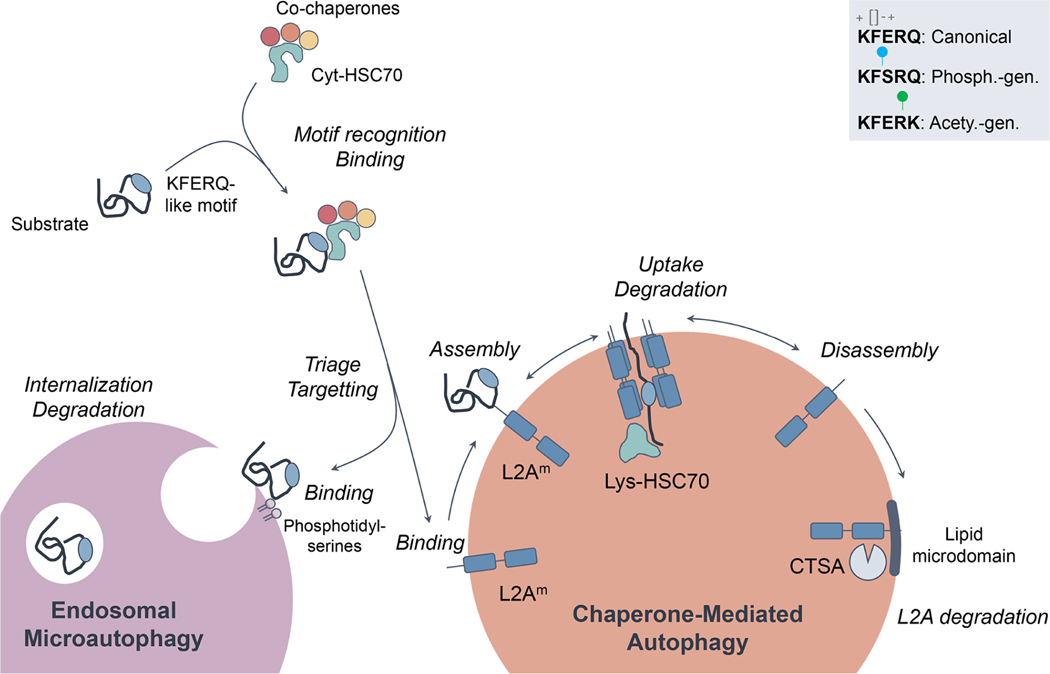
Figure 2. Schematic representation of chaperone-mediated autophagy (CMA) and endosomal microautophagy (eMI).
The first steps for cargo recognition are shared by CMA and eMI and are mediated by the binding of HSC70 to a targeting motif in the protein sequence biochemically related to the pentapeptide KFERQ. The inset (top right) highlights the chemical requirements for KFERQ motifs and the post-translational modifications such as phosphorylation or acetylation that can generate motifs by providing the missing charges. This motif is necessary and sufficient for CMA, whereas it is necessary but insufficient for eMI. Additional, as yet unknown, mediators are required for eMI targeting. HSC70 binds to the surface of late endosomes via phosphoserine and triggers assembly of the ESCRT machinery for internalization of substrate into intraluminal vesicles. In CMA, binding of the HSC70/substrate complex to LAMP2A at the lysosomal membrane triggers its multimerization to form a translocation complex that mediates the internalization of the substrate protein into the lumen for degradation. LAMP2A is actively disassembled from the complex to initiate a new cycle of binding/internalization. LAMP2A also mobilizes laterally to incorporate into lipid microdomains for its own degradation triggered by cathepsin A (CTSA). Phosph. gen.: phosphorylation generated. Acetyl. gen: Acetylation generated.
HSC70 is the only chaperone required for CMA cargo recognition (Chiang et al., 1989), while HSP90, HSP40, the HSP70-HSP90 organizing protein (Hop), the HSP70-interacting protein (Hip), and the BCL2-associated athanogene-1 protein (BAG-1), facilitate substrate unfolding, enhance substrate/LAMP2A binding and stabilize LAMP2A during its multimerization (Kaushik and Cuervo, 2018). In all cell types studied to date, LAMP2A translocation complex stability is regulated by the monomeric form of the intermediate filament protein glial fibrillary (GFAP) and the elongation factor-1 α (EF1α) in a GTP-dependent manner (Bandyopadhyay et al., 2010), and by lysosomal AKT (Arias et al., 2015). Phosphorylation of lysosomal membrane AKT by mTORC2 is inhibitory for CMA, whereas its dephosphorylation by the PHLPP1 kinase-phosphatase that binds to lysosomes in a Rac-1 dependent manner, leads to maximal CMA activation (Arias et al., 2015). The CMA target of AKT is GFAP, also recently shown to be phosphorylated by class I PI3K with a similar inhibitory effect on CMA (Endicott et al., 2020). CMA is also transcriptionally upregulated by NFAT, NRF2 and TPD52, whereas signalling through retinoic acid receptor alpha (RARα) and growth hormone signalling transcriptionally repress CMA (Anguiano et al., 2013).
The KFERQ-like motif is necessary and sufficient for CMA targeting, but because binding to HSC70 is dependent on the chemical properties of the motif (i.e. charge and polarity) (Kaushik and Cuervo, 2018), multiple amino acid combinations can build a functional (canonical) KFERQ-like motif. Post-translational modifications (PTMs), such as phosphorylation or acetylation (Fig. 2), can also generate functional motifs (by adding the missing charges) or disrupt a motif (i.e. through ubiquitination of the residue’s lysine). Around 46% of the proteome contains canonical motifs and an additional 30% can be generated via PTMs (Kirchner et al., 2019), but CMA degradation of those proteins only occurs when their motif becomes accessible to the chaperone (i.e. by protein conformational changes or dissociation of protein-protein interactions).
Microautophagy and endosomal microautophagy
Microautophagy involves lysosomal degradation of cellular components via membrane invaginations in compartments of the endolysosomal system (Fig. 3). Beside proteins, microautophagy can also degrade organelles in yeast such as mitochondria, lipid droplets, ER, peroxisomes and even nuclear fragments (Schuck, 2020); although thus far, only micro-ER-phagy has been identified in mammalian systems (Loi et al., 2019).
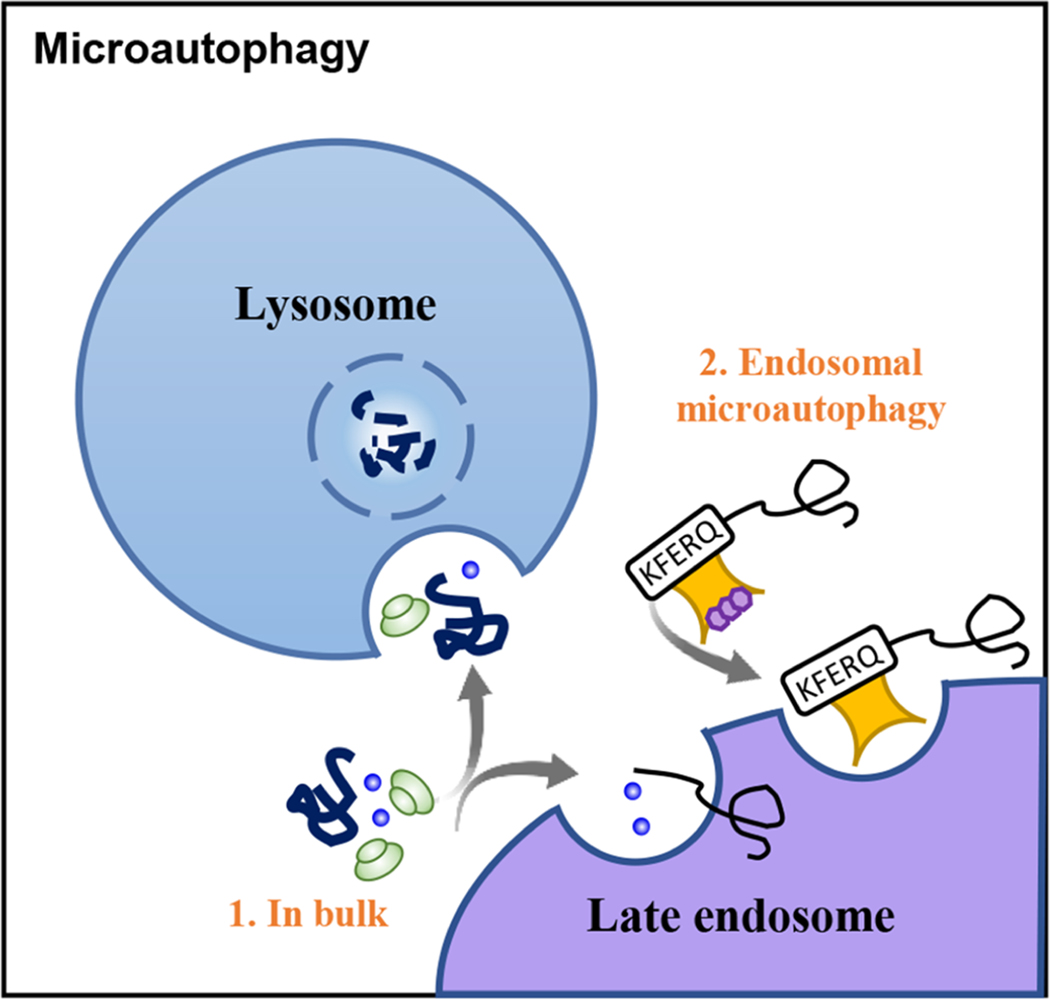
Figure 3. Schematic representation of microautophagy:
Bulk of proteins and cells components such as organelles (1. In bulk) can be integrated into lysosomes and late endosomes directly through invaginations at the lysosomal membrane. Cytosolic proteins targeted by Hsc70 can be also selectively degraded by its internalization into late endosome invaginations in a process known as Endosomal microautophagy (2).
Mammalian microautophagy pathways all target their cargo to late endosomes/multivesicular bodies (LE/MVBs) (Krause and Cuervo, 2021). The term endosomal microautophagy (eMI) refers to the degradation of cytosolic proteins in LE/MVBs and can be further sub-classified based on the selectivity of the cargo and the molecular machinery involved. The first type of eMI identified requires, as in CMA, recognition of a KFERQ-like motif by HSC70 (Fig. 2) (Kirchner et al., 2019; Sahu et al., 2011). HSC70 binds to phosphatidylserine in LE/MVB membranes (Morozova et al., 2016), triggering substrate internalization into the lumen via membrane invaginations that form in an ESCRT-dependent manner. Cargo degradation protein can occur in the LE/MVB compartment itself, although the bulk of degradation occurs after LE/MVB-lysosome fusion. An additional type of mammalian eMI, independent of HSC70 but dependent on some of the ESCRT proteins involved in eMI, has been identified as part of the early response to amino acid starvation (Mejlvang et al., 2018). This pathway degrades several macroautophagy receptors, possibly regulating the switch between selective and in bulk macroautophagy.
Little is known about the regulation and functional implications of eMI in mammals. In Drosophila, eMI is upregulated in response to oxidative and genotoxic stress (Mesquita et al., 2020) and to nutrient deprivation (Jacomin et al., 2021; Mukherjee et al., 2016). Basal eMI has been shown to contribute to local turnover of proteins in Drosophila neuromuscular junction synapses and its blockage leads to impaired neurotransmission (Uytterhoeven et al., 2015).
Autophagic crosstalk
The ubiquitin-proteasome system (UPS), mostly responsible for degradation of soluble and short-lived proteins, shares some key players with autophagy, like ubiquitin. This small protein can modulate the degradation rates of proteins or organelles through the UPS or macroautophagy (Komander and Rape, 2012). Moreover, ubiquitination of the core macroautophagic machinery serves as an important regulatory mechanism (reviewed in (Yin et al., 2020). The macroautophagy receptor, P62, has also been reported to escort ubiquitinated proteins for proteasomal degradation (Seibenhener et al., 2004), depending on its oligomerization state (Lu et al., 2017).
Accumulating evidence suggests a compensatory balance between these two degradation pathways, whereby UPS inhibition upregulates macroautophagic proteins by mechanisms still not fully understood (reviewed in (Sun-Wang et al., 2020). Proteasome inhibition enhances nuclear translocation of TFEB (Li et al., 2019a), and other transcription factors that induce autophagy, like Nrf1 and Nrf2 (nuclear factor erythroid-2-like 1 and 2) (Pajares et al., 2016; Sha et al., 2018). The proteasome itself can be a substrate of macroautophagy under stress conditions in the process called proteaphagy (Cohen-Kaplan et al., 2016; Cuervo et al., 1995). Although macroautophagy-deficient cells appear to accumulate proteasomes due to decreased proteaphagy and increased expression of the proteasome subunits (Wang et al., 2013), degradation of proteasome substrates seems to be compromised in these cells, without changes in proteasome activity (Korolchuk et al., 2009).
Different types of autophagy, although not redundant, are capable of compensating for each other. Early studies revealed that mouse fibroblasts with defective macroautophagy displayed elevated constitutive CMA activity. Compensatory activation of macroautophagy and the UPS occurs in a variety of non-neuronal tissues in CMA-deficient mouse models (mouse fibroblasts, liver, T cells) (Kaushik and Cuervo, 2018). However, this compensation is not universal as it was not observed, for example, in the retina or in hematopoietic stem cells (Dong et al., 2021; Rodriguez-Muela et al., 2013). Likewise, compensatory activation of macroautophagy was not observed in CMA-defective neurons, leading to an early phenotype of proteostasis failure (already evident at 6 months) (Bourdenx et al., 2021). In fact, a direct comparison of insoluble fractions isolated from the brains of CMA- (Lamp2A−/−) and macroautophagy-deficient (Atg7−/−) mice highlighted that these two pathways handle different portions of the proteome in neurons (only ~50% of the aggregated proteins were similar). Understanding the mechanisms behind reciprocal macroautophagy and CMA compensation in non-neuronal cells could identify novel therapeutic targets to upregulate these pathways in neurons in neurodegenerative conditions.
B. Roles of autophagic pathways in the healthy nervous system
Maintaining neuronal function
Neurons are post-mitotic, long-lived cells that require robust protein and organelle quality control mechanisms. Neuronal loss of ATGs results in accumulation of ubiquitin-positive protein aggregates, axonal swellings and neuronal degeneration, demonstrating that basal macroautophagy is critically important for neuronal health (Menzies et al., 2017). Similarly, loss of the CMA receptor LAMP2A disrupts neuronal proteostasis and function (Bourdenx et al., 2021). While impaired macroautophagy and CMA accelerate the accumulation of aggregate-prone proteins (a hallmark of most neurodegenerative diseases), they likely protect neuronal health via several additional mechanisms.
Neurons need to maintain a healthy pool of mitochondria, as they have high energy demands. The strong genetic connection between mitophagy and neurodegeneration emphasizes the importance of this pathway in neurons. However, the role of neuronal mitophagy in non-pathological conditions is unclear. Mice deficient for proteins involved in selective mitophagy, such as P62, PINK1 and Parkin, do not show accumulation of defective mitochondria or neuronal loss (Martinez-Vicente, 2017). In contrast, PINK1 knock-out rats or PINK1/Parkin deficient flies display mitochondrial abnormalities and neurodegeneration (Martinez-Vicente, 2017). The mitochondrial defects in the PINK1/Parkin depleted models may be independent of mitophagy. For example, PARIS (ZNF746), a substrate of PINK1-mediated phosphorylation and Parkin-mediated ubiquitination, accusmulates following Parkin deletion and leads to repression of the peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α), a master coregulator of mitochondrial function, biogenesis, and mitochondrial oxidative stress management (Pirooznia et al., 2020; Shin et al., 2011; Stevens et al., 2015).
Mice with neuronal loss of WIPI4, FIP200 and ATG9A, involved in early stages of autophagosome formation, or WIPI3, involved in alternative macroautophagy, show accumulation of damaged mitochondria (Liang et al., 2010; Yamaguchi et al., 2020; Yamaguchi et al., 2018; Zhao et al., 2015). Furthermore, neurons may have upregulated other mitochondria quality control pathways, such as mitochondrial-derived vesicles (MDVs) that carry oxidised proteins to lysosomes for degradation, thereby saving the entire organelle from clearance (Misgeld and Schwarz, 2017). Hence, bulk macroautophagy or MDVs may provide a quality control mechanism that is constantly active in non-pathologic conditions, whereas mitophagy is trigged upon mild or severe mitochondrial stress (Misgeld and Schwarz, 2017; Yao et al., 2020).
Synapses are dynamic structures highly dependent on controlled turnover of their components. Autophagy is emerging as a crucial player in synapse formation, pruning and plasticity (Birdsall and Waites, 2019; Stavoe and Holzbaur, 2019). Loss-of-ATG7 studies revealed that autophagy is required in motor neurons for presynaptic terminal formation, innervation of the neuromuscular junction (Rudnick et al., 2017), and in dopaminergic neurons for synaptic vesicle degradation regulated by active zone proteins, like Bassoon and Piccolo (Hernandez et al., 2012; Okerlund et al., 2017). Subsequent studies in mice lacking neuronal ATG5 revealed defective selective degradation of tubular ER in axons and increased excitatory neurotransmission through excessive calcium release from ER stores (Kuijpers et al., 2021). The relative contribution of the failure to degrade synaptic vesicles versus calcium storage dysregulation to the observed enhanced neurotransmission requires future investigation. At the postsynaptic terminal, macroautophagy is required for spine pruning, as cortical neurons lacking ATG7 have an excessive number of dendritic spines (Tang et al., 2014). Macroautophagy also drives the degradation of the scaffold proteins PSD95, PICK1 and SHANK3, and of AMPA receptors, leading to synapse destabilization (Compans et al., 2021; Nikoletopoulou et al., 2017; Shehata et al., 2012), and enables retrograde transport of brain-derived neurotrophic factor (BDNF) (Kononenko et al., 2017), a key molecule involved in neuronal differentiation, survival and synaptic plasticity.
The role of macroautophagy in synaptic plasticity has not been fully elucidated. Acute Beclin 1 knockdown revealed the need for macroautophagy induction to enhance activity-dependent plasticity changes in hippocampal neurons after chemical long-term potentiation (LTP) (Glatigny et al., 2019). Similarly, mice deficient in neuronal WIPI4 show attenuated hippocampal LTP (Zhao et al., 2015). In contrast, suppression of macroautophagy is required for BDNF-induced LTP (Nikoletopoulou et al., 2017). During long-term depression, endocytic sorting and autophagy induction regulate the degradation of AMPA receptors (Ehlers, 2000; Shehata et al., 2012). Further studies are required to understand how macroautophagy regulates synaptic strength.
CMA plays an important role in maintenance of the neuronal proteome with higher propensity for aggregation. Acute silencing of LAMP2A in dopaminergic neurons leads to accumulation of α-Synuclein and ubiquitinated proteins, neurodegeneration, and ultimately behavioural impairments (Xilouri et al., 2016). Selective blockage of CMA in excitatory neurons leads to a shift in the solubility of the metastable proteome (proteins close to their solubility limit) (Bourdenx et al., 2021). Entrapment of CMA substrate proteins within aggregates leads to loss of their function and defects in cellular metabolism, endocytosis, and cytoskeleton organization (Bourdenx et al., 2021).
The tight communication between neurons and glia suggests that glial macroautophagy may have profound non-cell-autonomous effects on neuronal function and health. Astrocytes promote neuronal health and survival by providing nutrients, controlling the uptake of neurotransmitters and ions and protecting neurons upon injury. Astrocyte macroautophagy is emerging as an important pathway to support neuronal health by regulating the degradation of misfolded proteins (Janen et al., 2010; Tang et al., 2008) and damaged mitochondria from degenerating neurons (Davis et al., 2014; Morales et al., 2020).
Oligodendrocytes and Schwann cells are the myelin-producing cells of the central and peripheral nervous systems, respectively. Studies using mice lacking ATG5 in oligodendrocyte progenitor cells (OPC) demonstrated that macroautophagy is essential for OPC survival, maturation and proper myelination (Bankston et al., 2019). Similarly, Schwann cell-specific removal of ATG7 caused abaxonal accumulation of excess cytoplasm and organelles, as well as abnormal myelination (Jang et al., 2015), demonstrating that macroautophagy is essential for proper myelination and insulation of axons.
Microglia are the resident immune cells of the brain that constantly sense the neural environment and clear debris to maintain homeostasis (Nimmerjahn et al., 2005). Amyloid-β-induced neuroinflammation was aggravated upon microglial-specific deletion of ATG7, resulting in excessive neuronal damage (Cho et al., 2014), transition of microglia to a proinflammatory status, defects in lipid homeostasis and elevated tau spreading (Xu et al., 2021). Similarly, loss of microglial ATG5 resulted in enhanced neuroinflammation and neurodegeneration in the striatum of conditional knock-out mice (Cheng et al., 2020), suggesting that microglial macroautophagy plays a protective role against aberrant microglial activation. Microglia have also been shown to be involved in synaptic pruning (Paolicelli et al., 2011). Neurons co-cultured with ATG7-deficient microglia showed increased synaptic markers and dendritic spine density, as well as in immature dendritic filopodia (Kim et al., 2017). However, ATG7 is also involved in LAP in peripheral macrophages (Heckmann et al., 2017), therefore further studies are required to determine whether deficiencies in macroautophagy or LAP are responsible for these disease changes. Interestingly, microglia from mice expressing human α-Synuclein in neurons engulf and sequester α-Synuclein by autophagosomes for degradation (Choi et al., 2020). Together these studies show the importance of functional macroautophagy in glia for neuronal function and viability.
Less is known about the function of CMA in glia. Neuronal CMA blockage is associated with astrocyte enlargement and microglial activation, most probably because of neuronal dysfunction (Bourdenx et al., 2021; Xilouri et al., 2016). Mice with full-body CMA blockage did not show overt astrogliosis or microglial activation (Bourdenx et al., 2021) but a functional study is lacking.
Autophagic processes and cell death pathways
Although removal of excessive neurons is important for the development of the nervous system, aberrant neuron death is one of the principal causes of neurological disorders. The cell death pathways that have been reported to interact with autophagy in the nervous system are discussed below.
Necrosis
Necrosis, characterised by plasma membrane rupture can be either regulated and genetically controlled, or unregulated (passive). Multiple types of regulated necrosis have been identified to cause neuronal cell death, including necroptosis, ferroptosis, pyroptosis and parthanatos (Fricker et al., 2018). Necroptosis is mediated by activities of receptor interacting protein kinase 1 (RIPK1), RIPK3, and mixed lineage kinase domain-like pseudokinase (MLKL) and has been reported to regulate axon degeneration induced by glutamate excitotoxicity and contribute to pathologies in conditions like ALS and AD (Degterev et al., 2019). P62 interacts with RIPK1 on early autophagosome structures to form the necrosome and induce necroptosis in response to TNF-related apoptosis inducing ligand (TRAIL) in mouse prostate cells. Knockdown of P62 switches the cell death to TRAIL-induced apoptosis by blocking the formation of necrosome (Goodall et al., 2016).
Ferroptosis
Ferroptosis is an iron-dependent form of cell death which occurs when the phospholipid hydroperoxide (PLOOH) and lipid radicals overwhelm their scavenging systems. Agents that inhibit ferroptosis improve neuronal survival in multiple neurodegeneration models (Zheng and Conrad, 2020), and ferroptosis is involved in glial to neuron conversion after traumatic brain injury (Gascon et al., 2016). Macroautophagy promotes ferroptosis through ferritinophagy (Yang et al., 2019), since free iron is released from ferritin-bound iron via the lysosome. CMA also promotes ferroptosis by degrading glutathione peroxidase 4 (GPX4), which catalyses the reduction of lipid peroxides and prevents ferroptosis (Wu et al., 2019).
Apoptosis
BCL-family proteins stimulate or inhibit macroautophagy and apoptosis, and these activities are at the centre of the mutual regulation of these processes. Macroautophagy both positively and negatively regulates apoptosis through degradation of BCL-family proteins and their regulators (Fricker et al., 2018).
In neurons, BAX is the dominant executor of intrinsic apoptosis. P53 upregulated modulator of apoptosis (PUMA, also called BBC3) directly translocates BAX onto mitochondria in neuronal cells undergoing oxidative stress, thereby inducing apoptosis, a phenomenon that was rescued by PUMA knockout (Steckley et al., 2007). In HeLa cells, autophagy maintains low PUMA mRNA levels through an unknown mechanism (Thorburn et al., 2014). It is unclear whether similar regulation of PUMA exists in neuronal cells.
The extrinsic apoptosis pathway contributes to pathogenesis in neurological disorders. Neuronal specific deletion of caspase-8 renders neurons resistant to TNF-α ligation-induced apoptosis in vitro and increases neuron survival with reduced caspase-3 activation after acute brain injury (Krajewska et al., 2011). The induction of apoptosis in primary cortical neurons by the neurotoxin 6-hydroxydopamine (6-OHDA) was rescued by knockdown of ATG5 or macroautophagy inhibitor 3-methyladenine (Chung et al., 2018).
The relation between CMA and apoptosis has been mostly studied in cancer cells, where CMA prevents apoptosis through degradation of cyclin D1, PUMA or HMGB1, but facilitates immunogenic apoptosis by mediating surface exposure of calreticulin (reviewed in (Arias and Cuervo, 2020)). In untransformed cells, inhibition of CMA increases susceptibility to stressors and leads to apoptosis but the molecular mechanisms remain poorly understood.
Pyroptosis
Pyroptosis is a form of cell death seen in many common neurological diseases, like AD and PD, that manifests with inflammasome-mediated release of caspase 1 from affected cells. Macroautophagy activation protects against this form of cell death in mouse models via diverse pathways, including degradation of a key pyroptosis mediator NLRP3 (Wu et al., 2021). In this way, autophagy may also buffer neuroinflammation, a prevalent process in different neurodegenerative conditions.
Autophagic cell death and Autosis
Usually, macroautophagy promotes cell survival following stress/nutrient limitation by recycling cellular components and providing energy. However, in ischemia/hypoxia and stroke, macroautophagy has been reported to lead to cell death, termed autophagic cell death (ACD). The main morphology of ACD is accumulation of vacuoles with increased macroautophagy flux, and is rescued by knockdown or inhibition of autophagic proteins (Galluzzi and Green, 2019). However, much of the literature in this domain has caveats, including the use of imprecise chemical tools to manipulate autophagy, and difficulties measuring autophagic flux versus steady-state levels of autophagosomes in vivo. For example, autophagic flux can be impaired by defects downstream of autophagosome formation, which lead autophagosome accumulation. Furthermore, a reduction in cell death in such scenarios in macroautophagy knockout cells means that autophagy is required for the death process but not that autophagy is causing the cell death. To robustly support ACD, one needs to show a reduction of cell death when the flux is normalised, and not when it is ablated. Furthermore, some macroautophagy genes may have roles in cell death unconnected with their autophagic functions.
Interestingly, in all the ACD cases reported, inhibition of lysosome function fails to rescue cell death, suggesting that ACD may be an autophagy-independent pathway under the control of autophagy machinery. Moreover, no common upstream initiator-signalling pathway has been found (Galluzzi and Green, 2019).
Role of Different Autophagy Pathways in Enabling Neuronal Functions
Autophagy plays a role in the structural reorganisation of neuronal circuits via axonal growth, dendritic spine formation and pruning, synaptic assembly and vesicle turnover (Fleming and Rubinsztein, 2020; Kulkarni and Maday, 2018; Lieberman and Sulzer, 2019). These autophagy-driven structural changes have a major impact on neuronal function.
Learning and memory
In hippocampal neurons, macroautophagy is upregulated during learning and memory consolidation. Macroautophagy inhibition in the hippocampus of young mice affects their performance in different behavioural tests, indicating a deficit in the formation of novel memories (Glatigny et al., 2019). The mTORC1 complex is known to be over-activated in humans with fragile X syndrome (Hoeffer et al., 2012). In mouse models, this hyper-activation decreases macroautophagy, increases dendritic spine density with aberrant morphology and leads to exaggerated LTD in hippocampal neurons associated with impaired novel object recognition (Yan et al., 2018).
Since hippocampal macroautophagy declines with age, promoting macroautophagy has been investigated as a mechanism to improve memory in aged animals. Injection of plasma from young mice into older mice improves their memory in a macroautophagy-dependent fashion (Glatigny et al., 2019), likely via osteocalcin, a blood-brain barrier penetrant, bone-derived circulating molecule previously demonstrated to act as a hormonal regulator of hippocampal memory (Obri et al., 2018). However, upregulation of macroautophagy may not be a panacea for all memory deficits. For example, in primary neuron cultures, treatment with AICAR, a cell-permeable nucleoside used to induce AMPK hyper-activation, led to macroautophagy-dependent loss of pre- and postsynaptic markers and a decline in neuronal network function (Domise et al., 2019), suggesting that elevated macroautophagy may be as damaging as reduced autophagy and that maintaining the balance of autophagic flux is crucial.
Whole-body CMA-deficient mice have both memory and motor-coordination dysfunction, while blockage of CMA only in excitatory neurons resulted mostly in impaired short-term memory (Bourdenx et al., 2021).
Sleep and circadian rhythm
Many neurodegenerative disorders are associated with altered or poor sleep. Chronic sleep deprivation increases amyloid-β and tau in interstitial fluid and is associated with increased pathology in mouse models (Holth et al., 2019). Daily sleep–wake and feeding–fasting cycles are coupled to the central circadian clock in the suprachiasmatic nucleus. Circadian oscillations involve transcriptional circuits and non-translational controls, such as phosphorylation, which synchronize sleep/wake cycles, food intake and cellular bioenergetics (Reddy and Rey, 2014). Food intake occurs during wake cycles and autophagy is elevated during fasting (sleep). Recent work has shown that TFEB and TFE3 control the rhythmic induction of their transcriptional target genes involved in macroautophagy and lysosomal biogenesis during the light phase (sleep). Liver or muscle-specific knockouts of both TFEB and TFE3 in mice results in loss of the diurnal macroautophagy cycle. TFEB and TFE3 were shown to directly regulate the expression of Rev-erbα (Nr1d1), a transcriptional repressor component of the core clock machinery also involved in the regulation of whole-body metabolism and macroautophagy (Pastore et al., 2019).
A dual interplay between circadian rhythms and CMA has also been recently reported whereby CMA displays central and peripheral circadian activity and, at the same time, contributes to the regulation of circadian cycling through degradation of components of the clock machinery (Juste et al., 2021). Disruption of CMA in vivo and the subsequent impaired degradation by CMA of the positive elements Bmal1 and Clock and the regulatory element Rev-erbα, leads to temporal shifts and amplitude changes of the clock-dependent transcriptional program and fragmented circadian patterns. In contrast to the modulatory effect of TFEB on circadian cycling, the CMA-dependent regulation of the clock is nutrient-independent.
Suggested links with psychiatric diseases
Recent studies have provided some support for the hypothesis that altered neuronal macroautophagy may contribute to depression, bipolar disorder and schizophrenia. Autophagy-inducing drugs acting via independent pathways have antidepressant-like properties in mice (Kara et al., 2018; Kara et al., 2013), and numerous clinically prescribed anti-depressants with diverse pharmacological activities enhance macroautophagy (reviewed in Gassen and Rein, 2019). Although there has been little human clinical research to support these findings, therapeutic concentrations of paroxetine and amitriptyline increase expression of macroautophagy components (Beclin1, phosphor-AKT, LC3II) in blood cells of patients and these changes predict clinical improvement (Gassen et al., 2014). Similarly, in a small study in patients with major depressive disorder, serum levels of Beclin 1 were higher in responders to selective serotonin reuptake inhibitors than non-responders (He et al., 2019).
A smaller but growing body literature provides some support for a link between autophagy and schizophrenia. When mutated, Disrupted-in-Schizophrenia 1 (DISC1) confers susceptibility to psychiatric illness. DISC1 contains an LC3-interacting region (LIR) motif and, when over-expressed, enhances mitophagy (Wang et al., 2019b). In post-mortem samples, transcriptomic analysis revealed over-representation of genes that regulate altered neuronal macroautophagy with down-regulation of expression observed in cortical and hippocampal areas in schizophrenic patients (Ryskalin et al., 2018). In addition, exome sequencing identified four rare ULK1 variants significantly associated with schizophrenia in a case-control study (Al Eissa et al., 2018).
Ageing
Ageing is a major risk factor for human neurodegenerative disease. Autophagy appears to decline with age in many organisms (Hansen et al., 2008). Age-dependent decreases have been reported for macroautophagy gene transcripts in brains from Drosophila (Simonsen et al., 2008) and humans (Lipinski et al., 2010), in mouse retinas (Rodriguez-Muela et al., 2013), and in autophagy proteins in mouse hypothalamus and hippocampus (Glatigny et al., 2019; Kaushik et al., 2012). Changes in lysosomal abundance with age have been extensively documented in multiple peripheral tissues in different experimental models (reviewed in (Nixon, 2020), but expansion of these compartments with age is not a universal feature, since recent studies in C. elegans have revealed age-dependent decreases in lysosome and autophagosome abundance in tissues, such as intestines, muscles and neurons (Chang et al., 2017). This decrease in abundance could explain the reported decreased macroautophagic activity in aged C. elegans (Wilhelm et al., 2017) and it is consistent with the decline in autophagy flux and autophagosome biogenesis observed in mouse brains (Park, 2021). The progressive decline in macroautophagy with age can predispose to toxic protein and organelle accumulation in neurons and compromise neuronal health. Indeed, impairing autophagy reduces lifespan in C. elegans, Drosophila and mice, while induction of macroautophagy extends longevity in these organisms (Hansen et al., 2018).
CMA activity decreases with age in most organs and tissues, predominantly due to reduced stability of LAMP2A at the lysosomal membrane of old organisms (Kaushik et al., 2021). In peripheral tissues, reduced CMA with age contributes to loss of proteostasis, metabolic derangements, immune senescence and loss of stemness, all considered hallmarks of aging. Conversely, genetic restoration of the LAMP2A defect in old organisms has proven sufficient to prevent proteotoxicity, improve the organismal response to stress and restore organ function (Kaushik et al., 2021). Although changes in CMA activity with age in neurons are still poorly characterized, elevated levels of neuronal nitric oxide synthase, previously associated with neuronal aging, have demonstrated sufficient to reduce neuronal LAMP2A levels (Valek et al., 2019).
Most studies on autophagy in the aging brain have followed changes on steady-state markers due to the difficulties of measuring autophagy flux in live organisms. The development of mouse models constitutively expressing fluorescent probes to follow bulk macroautophagy (Lopez et al., 2018), selective macroautophagy (mitophagy, (Sun et al., 2015), or CMA (Dong et al., 2020) opens up the possibility of tracking changes in the dynamics of these processes with age. For example, studies in a mitophagy reporter mouse have demonstrated reduced mitophagy flux in aged hippocampal pyramidal neurons (Sun et al., 2015).
C. Autophagy pathways and neurodegenerative diseases
Mutations in genes encoding proteins involved at all steps in the macroautophagy pathway are implicated in different neurodegenerative diseases. Often, one mutation can affect macroautophagy at multiple stages. This section describes defects associated with selected important examples, while Table 1 summarises a more comprehensive, although not exhaustive, list of autophagy-related genes associated with neurodegenerative disorders and the autophagic disruption caused by some common neurodegenerative disease genes is summarised in Fig. 4. We have not reviewed all neurodegenerative disease genes causing autophagy defects in the text, as extensive reviews have been published previously (Menzies et al., 2017; Stamatakou et al., 2020).
Table 1:
Mutations in neurodegeneration-related genes that affect macroautophagy
| Gene/ Protein | Neurodegenerative disease | Stage of the autophagy pathway were a protein acts | Cellular phenotype upon mutation or loss of function of a protein |
|---|---|---|---|
| Core macroautophagy | |||
| ATG5/ Autophagy-related 5 | Hereditary childhood ataxia | Early phagophore formation and elongation | Decreased autophagosome formation caused by weak binding of ATG5 to ATG12 (Kim et al., 2016) |
| WIPI2/ WD repeat domain phosphoinositide-interacting protein 2 | Severe cognitive impairment | Early phagophore formation and elongation | Decreased autophagosome formation caused by reduced binding of WIPI2b to ATG16L1, as well as ATG5–12 (Jelani et al., 2019) |
| WDR45/ WD repeat domain phosphoinositide interacting protein 4 (WIPI4) | Beta-propeller protein-associated neurodegeneration (BPAN, NBIA, SENDA) | Early phagophore formation and elongation/ Autophagosome-lysosome fusion | Impairment in autophagy flux and an accumulation of LC3-positive autophagosome membranes (Saitsu et al., 2013) and ubiquitinpositive aggregates (Zhao et al., 2015); impaired formation of fusion machinery (Ji et al., 2021) |
| Other genes that impact autophagy | |||
| ATXN3/ Ataxin-3 | Spinocerebellar ataxia type 3 (SCA3) | Early phagophore formation | Impaired initiation in starvation-induced autophagy (Ashkenazi et al., 2017) |
| GJB1/ gap junction protein connexin 32 | Charcot-Marie-Tooth type 1 (CMT1) | Autophagosome formation | Reduced autophagosome formation (Bejarano et al., 2014) |
| VPS35 / Vacuolar protein sorting-associated protein 35 | Parkinson’s disease (PD) | Autophagosome formation and elongation | Abnormal trafficking of mATG9 and autophagy impairment (Zavodszky et al., 2014) |
| EPM2A, EPM2B/ Laforin, Malin | Lafora disease | Autophagosome formation and elongation | Decreased LC3 lipidation and increase in p62 (Aguado et al., 2010); defective regulation of PI3KC3 complex; decreased PI(3)P levels (Sanchez-Martin et al., 2020) |
| TECPR2 / Tectonin beta-propeller repeat containing 2 TECPR2 | Spastic paraplegia type 49 (SPG49) | Phagophore membrane elongation | Decrease in LC3 lipidation and levels of LC3 and WIPI2 (Oz-Levi et al., 2012; Stadel et al., 2015) |
| AP4S1/ AP-4 complex subunit sigma-1 | Hereditary spastic paraplegia (SPG47, SPG52) | Phagophore membrane elongation | Deficiency causes mis-sorting of mATG9 (Davies et al., 2018) |
| PICALM/ Phosphatidylinositol-Binding Clathrin Assembly Protein | Alzheimer’s disease (AD) | Autophagosome formation/ maturation | Autophagosome formation and maturation dysfunction; impaired APP cargo recognition (Moreau et al., 2014; Tian et al., 2013) |
| C9ORF72 / Hexanucleotide-repeat expansions in a non-coding region of chromosome 9 open reading frame 72 | Amyotrophic lateral sclerosis (ALS); frontotemporal dementia (FTD) | Autophagosome formation/ maturation | Impaired autophagy flux (Farg et al., 2014); impaired trafficking of the ULK1 initiation complex to the phagophore (Webster et al., 2016) |
| SNCA/ α-synuclein | Parkinson’s disease (PD) | Autophagosome formation/ maturation | Impaired autophagosome transport (Tanik et al., 2013; Volpicelli-Daley et al., 2014); abnormal mATG9 trafficking (Winslow et al., 2010); disturbed TFEB-mediated lysosomal biogenesis (Decressac et al., 2013) |
| VCP/ Valosin-containing protein VCP | Inclusion body myopathy with earlyonset Paget disease and frontotemporal dementia (IBMPFD), Amyotrophic lateral sclerosis (ALS), Charcot-Marie-Tooth type 2 (CMT2), tauopathies | Autophagosome formation / maturation | Loss of ATPase activity causes impaired autophagosome formation (Hill et al., 2021); impaired autophagosome maturation (Ju et al., 2009; Tresse et al., 2010); recruited to damaged lysosomes to facilitate lysophagy (Papadopoulos et al., 2020) |
| SPG11/ spatacsin | Hereditary spastic paraplegia (SPG11) | Autophagosome maturation | Impaired autophagic lysosome reformation (Chang et al., 2014; Vantaggiato et al., 2019) |
| ZFYVE26 / spastizin (SPG15) | Hereditary spastic paraplegia (SPG15) | Autophagosome maturation | Disrupts interaction with Beclin 1 which impairs autophagosome maturation (Vantaggiato et al., 2013) and lysosomal biogenesis (Chang et al., 2014; Vantaggiato et al., 2019) |
| RAB7A / Ras-associated protein Rab-7a | Charcot-Marie-Tooth type 2 (CMT2) | Autophagosome maturation | Impaired autophagosome-lysosome fusion (Ganley et al., 2011) |
| ALS2/ ALSIN | Amyotrophic lateral sclerosis (ALS) | Autophagosome maturation | Impaired autophagosome fusion with endosomes through its regulation of RAB5 (Ravikumar et al., 2008) |
| UBQLN2 / Ubiquilin 2 | Amyotrophic lateral sclerosis (ALS) | Autophagosome formation/ maturation/ Lysosomal function | Impaired autophagosome maturation (N’Diaye et al., 2009); impaired LC3 lipidation (Rothenberg et al., 2010); impaired autophagic flux caused by lysosomal defect (Şentürk et al., 2019; Wu et al., 2020); decrease in the levels of autophagic proteins (Chen et al., 2018) |
| HTT / Huntingtin | Huntington’s disease | Autophagosome formation/ Autophagosome-lysosome fusion/ Cargo recognition | Impaired cargo recognition (Martinez-Vicente et al., 2010; Ochaba et al., 2014; Rui et al., 2015); decreased autophagosome transport (Wong and Holzbaur, 2014b); impaired starvation-induced autophagy initiation (Ashkenazi et al., 2017) |
| CHMP2B/ Charged multivesicular body protein 2b | Amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD) | Autophagosome-lysosome fusion Required for eMI | Impaired autolysosome formation (Filimonenkoet al., 2007; Lee et al., 2007) |
| MAPT/ microtubule-associated protein tau | Alzheimer’s disease (AD), tauopathies | Autophagosome-lysosome fusion | Dysfunction of the retrograde axonal transport of autophagosomes (Butzlaff et al., 2015; Majid et al., 2014) |
| SNX14/ Sorting nexin-14 | Spinocerebellar Ataxia Type 20 (SCAR20) | Autophagosome-lysosome fusion | Impaired autophagosome clearance (Akizu et al., 2015); impaired lysosomes (Bryant et al., 2018) |
| DCTN1/ Dynactin | Amyotrophic lateral sclerosis (ALS) | Autophagosome-lysosome fusion | Impaired autophagosome-lysosome fusion (Ravikumar et al., 2005) |
| PSEN1/ Presenilin 1 | Alzheimer’s disease (AD) | Lysosomal function | Impaired autophagosome-lysosome fusion and defective lysosomal acidification (Chong et al., 2018; Leeet al., 2010; Zhang et al., 2012) |
| GBA/ Lysosomal acid glucosylceramidase | Parkinson’s disease (PD) | Lysosomal function | Impaired autophagy caused by lysosomal dysfunction (Murphyet al., 2014); impaired removal of dysfunctional mitochondria by mitophagy (Li et al., 2019b) |
| ATP13A2/ Polyaminetransporting ATPase 13A2 | Parkinson’s disease (PD) | Lysosomal function | Impaired lysosome acidification (Bento et al., 2016; Dehay et al., 2012) |
| SYT11/ synaptotagmin 11 | Parkinson’s disease (PD) | Lysosomal function | Lysosomal dysfunction ((Bentoet al., 2016) |
| GRN/ Progranulin | Frontotemporal lobar degeneration (FTLD) | Lysosomal function | Impaired lysosomal function (Logan et al., 2021);deficiency in neurons increases autophagy flux and causes abnormally enlarged lysosomes (Elia et al., 2019) |
| FIG4/ Polyphosphoinositide phosphatase FIG4 | Charcot-Marie-Tooth disease 4J (CMT4J), Amyotrophic lateral sclerosis 11 (ALS11), Yunis-Varon syndrome (YVS) | Lysosomal function | Impaired production of PI(3,5)P(2), impaired endo-lysosomes, accumulation of p62 (Ferguson et al., 2009) |
| VPS13D/ Vacuolar protein sorting 13D | ataxia and/or spastic paraplegia | ? | Accumulation of damaged mitochondria (Anding et al., 2018) |
| PEX13/ Peroxisomal membrane protein PEX13 | Human peroxisome biogenesis disorders (PBDs) | ? | Impaired mitophagy without disruption of general autophagy (Lee et al., 2017) |
| Macroautophagy adaptors | |||
| SQSTM1/ Sequestosome-1 (p62) | Amyotrophic lateral sclerosis (ALS), Frontotemporal lobar degeneration (FTLD) | Cargo recognition | Impaired recruitment into autophagosomes (Goode et al., 2016); impaired cargo recognition and protein clearance (Gal et al., 2009) |
| OPTN/ Optineurin | Amyotrophic lateral sclerosis (ALS) | Cargo recognition/ Autophagosome formation and maturation | Impaired cargo recognition and protein clearance (Shenet al., 2015); impaired ATG5-ATG12-ATG16L recruitment to phagophore and decreased formation (Bansal et al., 2018; Song et al., 2018); impaired autophagosome trafficking to lysosomes (Sundaramoorthy et al., 2015; Tumbarello et al., 2012) |
| TBK1/TANK-binding kinase 1 | Amyotrophic lateral sclerosis (ALS) | Promotes efficient cargo recognition by OPTN and regulates other autophagic proteins | Impaired mitophagy (Moore and Holzbaur, 2016; Pilli et al., 2012; Richteret al., 2016) |
| RETREG1/ Reticulophagy regulator 1 (FAM134B) | Hereditary sensory and autonomic neuropathy type II (HSAN II) | ER-phagy receptor | Impaired ER-phagy though decrease binding to autophagy modifiers LC3 and GABARAP (Bhaskaraet al., 2019; Khaminets et al., 2015) |
| ATL1 and ATL3/ Atlastin 1 and 3 | hereditary spastic paraplegia (HSP), hereditary sensory and autonomic neuropathies (HSAN) | ER-phagy receptor / autophagy initiation at ER | Depletion inhibits ER-phagy, acts downstream of FAM134B during ER-phagy (Liang et al., 2018); impaired association with GABARAP and impaired ER-phagy (Chenet al., 2019); impaired recruitment of ULK1 complex to ER-specific site of autophagosome formation (Liu et al., 2021) |
| PINK1/ Serine/threonine-protein kinase PINK1 | Parkinson’s disease (PD) | Sensor of mitochondrial stress/ Promotes efficient mitophagy | Impaired mitophagy (reviewed in (Ge et al., 2020) |
| PRKN/ E3 ubiquitin-protein ligase PARKIN | Parkinson’s disease (PD) | Amplifies damage mitochondria detection signal/ Promotes efficient mitophagy | Impaired mitophagy (reviewed in(Geet al., 2020) |
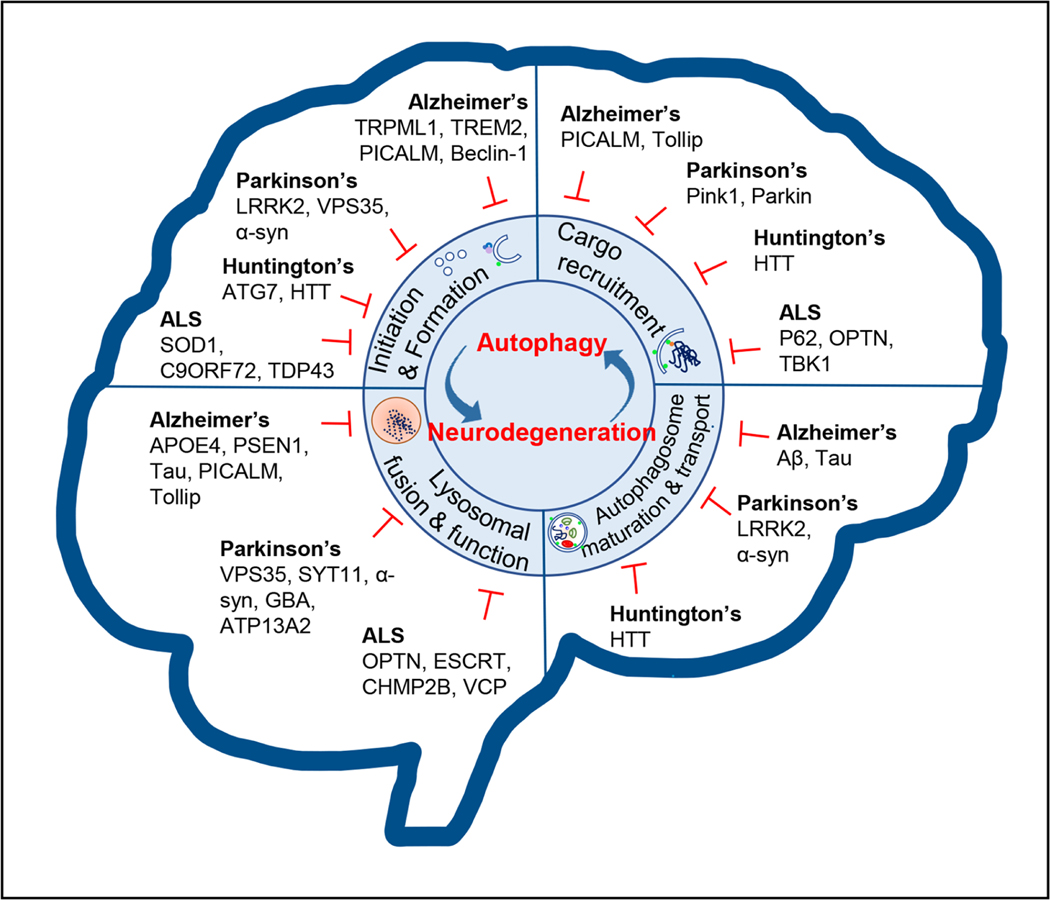
Figure 4. Overview of the role of macroautophagy in the nervous system in health and neurodegeneration.
Autophagy is fundamental to sustain the homoeostasis and function of the CNS. Perturbations in the macroautophagy pathway at different stages have been observed during neurodegeneration and distinct disease-associated genes are also key contributors to macroautophagy dysfunction. Defective autophagy compromises protein clearance and organelle turnover, leading to the accumulation of toxic proteins and damaged cellular components that finally alter neuronal function and induce neuronal loss.
Mutations in core autophagy genes
Mutations in core macroautophagy genes have been identified in relatively few human neurodegenerative diseases. This may be because core macroautophagy components are vital for cellular homeostasis and may cause early lethality if mutated. A recent study identified recessive mutations in ATG7 in 12 patients from 5 families with a range of neurodevelopmental disorders affecting the cerebellum and corpus callosum with additional muscle, and endocrine involvement as well as facial dysmorphism. While fibroblasts from one family had no obviously detectable LC3-II, those from other families showed LC3-II conjugation, indicative of functional autophagy. Furthermore, in the family where no LC3-II was detected, autophagosomes were evident in muscle biopsies (Collier et al., 2021). Hence, further analysis is needed to determine whether the mutations result in a complete loss-of-function and whether there is compensation from non-canonical autophagy. Homozygotes with a mutation in ATG5 (E122D) manifest with childhood ataxia and cells derived from these patients display decreased autophagic flux and reduced conjugation of ATG12 to ATG5 (Kim et al., 2016). X-linked dominant mutations in another core autophagy gene, WDR45 (encoding protein WIPI4) cause human β-propeller protein-associated neurodegeneration (BPAN) mainly in females (Saitsu et al., 2013) and in patient-derived lymphoblastoid cells, decreased stability of WIPI4 and accumulation of aberrant early autophagic structures were observed. An autosomal recessive missense mutation in WIPI2 results in severe syndromic cognitive impairment and loss of brain volume (Jelani et al., 2019).
Mutations in genes involved in early stages of autophagosome formation
Several neurodegenerative disease-causing mutations have been identified in genes involved in membrane trafficking events required for autophagosome biogenesis. A mutation in VPS35 (D620N), a core retromer component, has been described in Parkinson’s disease (PD) patients and in subsequent studies was shown to cause ATG9 mislocalization (Zavodszky et al., 2014; Zimprich et al., 2011). Similarly, an ATG9 trafficking defect was observed in cells derived from patients with early-onset progressive spastic paraplegia (SPG47 & SPG52) which are deficient for AP-4 pathway function (Davies et al., 2018).
Mutations in genes involved in substrate recognition and selective autophagy
Mutations in the autophagy receptor P62 have been identified in cases of familial and sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) (reviewed in (Deng et al., 2017), leading to disrupted degradation of SOD1 and TDP-43 (Gal et al., 2009).
Mutations in FAM134B (also called RETREG1), an ER-phagy receptor, cause hereditary sensory and autonomic neuropathy type II (HSAN II) and compromise ER-phagy, leading to ER expansion (Bhaskara et al., 2019; Khaminets et al., 2015). Mutations in ATL3 that cause hereditary sensory and autonomic neuropathy type I (HSAN I) impair starvation-induced ER-phagy through direct disruption of the association of ATL3 with GABARAP (Chen et al., 2019).
OPTN mutations inhibit its ability to recruit LC3 to damaged mitochondria and to induce mitophagy (Shen et al., 2015; Wong and Holzbaur, 2014a), and ALS-associated mutations in this kinase reduce binding of TBK1 to OPTN therefore decreasing mitophagy (Richter et al., 2016).
Mutations in genes involved in autophagosome trafficking, maturation and fusion with lysosomes
Autophagosomes generated in axons rely on retrograde transport to fuse with perinuclear lysosomes. Therefore, neurons are particularly vulnerable to disruption in trafficking. Dynein–dynactin is a molecular motor required for fast retrograde transport of autophagosomes, organelles, RNAs and proteins along microtubules and is the target of mutations causing axonal Charcot-Marie-Tooth hereditary neuropathy type 2 (CMT2) and ALS (Puls et al., 2003). Defects in trafficking have also been associated with disease-causing mutations in α-Synuclein (PD), C9ORF72 (ALS) and with hyperphosphorylated tau (seen in tauopathies, see section D).
Fusion events required for autophagosome maturation involve many components of the endocytic pathway, such as the small GTPase RAB protein family. Mutations in RAB7, encoding the late endosomal RAB7A protein, cause Charcot–Marie–Tooth type 2B disease (Verhoeven et al., 2003). Similarly, mutations in a RAB5 regulatory protein Alsin, can result in recessive ALS (Yang et al., 2001). ALS can be also caused by mutations in CHMP2B, one of the subunits of the ESCRT-III complex, which impair autophagosome maturation (Filimonenko et al., 2007; Lee et al., 2007). Disruption to the ESCRT machinery may also interfere with eMI by disrupting proper LE/MVB biogenesis.
Mutations in genes involved in lysosomal function
Macroautophagy completion requires lysosomal digestion of autophagic cargo and subsequent release of recycled metabolites. The degradative capacity of lysosomes depends on low pH, which is mediated by vacuolar ATPase (vATPase). Mutations in genes associated with familial forms of neurodegeneration, such as PSEN1 (AD) (Lee et al., 2010), α-Synuclein (Decressac et al., 2013) and ATP13A2 (PD) (Bento et al., 2016; Dehay et al., 2012) affect lysosomal pH. Following autolysosome fusion and cargo degradation, lysosomes undergo a process called reformation. SPG11 and SPG15, encoded by genes mutated in hereditary spastic paraplegia, have been implicated in this process and loss of SPG11 or SPG15 result in the depletion of lysosomes capable of fusing with maturing autophagosomes (Vantaggiato et al., 2019).
Mutations in genes involved in CMA
Loss-of-function mutations in LAMP2 cause Danon disease that manifests with cardiomyopathy, myopathy, variable mental retardation and with progressive retinal degeneration (Cenacchi et al., 2020). Most mutations in this disease occur in the part of the gene common to all spliced LAMP2 protein variants. Since only the LAMP2A splice variant of this gene is required for CMA, whereas the other variants LAMP2B and LAMP2C contribute to macroautophagy and lysosomal degradation of DNA and RNA, respectively, the mutations will cause defects in multiple types of autophagy. Interestingly, expression of each of the LAMP2 proteins has been shown to be differentially affected in PD patients brains, with the earlier changes occurring in LAMP2A (Sala et al., 2016). However, further studies on the mechanism that regulate splicing of this gene are needed to determine whether these differences are linked to specific gene mutations. Further studies are also needed for the heterozygous variant in the LAMP2 gene promoter that significantly reduces LAMP2 transcription in a PD patient (Sala et al., 2016). Mutations in different proteins related to PD and frontotemporal dementia associated with reduced CMA activity are summarized in Table 2.
Table 2.
Mutations in neurodegeneration-related genes with impact on CMA
| Gene | Mutation/s | Associated disease | Effect on CMA | References |
|---|---|---|---|---|
| SNCA | A53T A30P | PD | Blocks CMA uptake | (Cuervo et al., 2004) |
| LRRK2 | G2019S R1441C D1994A | PD | Reduces LAMP2A stability and blocks CMA uptake | (Orenstein et al., 2013) |
| VPS35 | D620N | PD | Reduces LAMP2A stability | (Tang et al., 2015) |
| UCH-L1 | I93M | PD | Blocks CMA substrates degradation | (Kabuta et al., 2008) |
| LAMP2 | LAMP24127A>C | PD | Reduces LAMP2A transcription | (Pang et al., 2012) |
| MAPT | P301L | FTD | Blocks CMA degradation | (Caballeroet al., 2018) |
D. Polygenic diseases
Tauopathies and AD
Tauopathies are characterized by the intracellular accumulation and aggregation of tau into paired helical filaments and neurofibrillary tangles and include frontotemporal dementias (FTDs), AD, progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD).
Multiple studies demonstrate the roles of different types of autophagy in the degradation of tau. Tau colocalises with LC3 and P62 in post-mortem brains of familial AD, CBD, and PSP patients and the expression of P62 is reduced in AD patients (Liu et al., 2017). Tau associates with the autophagy cargo receptors NDP52 and OPTN, which also colocalize with neurofibrillary tangles and dystrophic neurites in AD patients (Osawa et al., 2011; Xu et al., 2019). Clearance of tau variants associated with different tauopathies occurs via different autophagic routes (Caballero et al., 2018). For example, phosphorylation of tau inhibits its degradation by macroautophagy and results in tau clearance via an endolysosomal pathway dependent on ESCRT complex and the small GTPase RAB35 (Vaz-Silva et al., 2018). About 50% of tau is degraded by macroautophagy and the other 50% is degraded by non-macroautophagy pathways (CMA and eMI) (Caballero et al., 2021). Interestingly, tau acetylation, a pathological post-translational modification, and pathogenic mutations, such as the FTD-related mutation A152T, favor tau degradation by macroautophagy, whereas the P301L mutation reduces overall tau degradation (Caballero et al., 2018).
Compromised removal by CMA of pathogenic forms of tau, including both mutant and post translationally modified, is further aggravated by the toxic effect that these proteins exert on this process. In vivo, expression of pathogenic tau protein is sufficient to induce neuronal- (but not astrocytic) specific inhibition of CMA (Bourdenx et al., 2021). Although direct evaluation of CMA activity in human brain in a cell-type specific manner is not currently possible, analysis of the transcriptional expression of the components of the CMA network in the brain of AD patients using single-nuclei RNAseq demonstrated a neuronal specific transcriptional inhibition of CMA (Bourdenx et al., 2021) that correlated with the Braak stages of tau pathology. The reduction of CMA score was higher in excitatory than in inhibitory neurons, thus corresponding well with the higher vulnerability to tau pathology of excitatory neurons.
The mechanisms behind tau’s toxicity on CMA are under investigation, but at least for acetylated and P301L tau, translocation of these proteins through the LAMP2A multimeric complex is compromised due to loss of their pH-dependent interaction with luminal HSC70 (Caballero et al., 2021). Persistent occupancy of the CMA translocation complex by pathogenic tau blocks degradation of other CMA substrates. Interestingly, this dependence on lysosomal pH for uptake through CMA, so far only described for tau, explains tau’s highly efficient degradation by this pathway (Caballero et al., 2021) and highlights that the reported impaired lysosomal acidification in some forms of familial AD could also lead to reduced tau degradation by CMA (Wolfe et al., 2013).
Tau can also be a substrate of KFERQ-selective eMI, although, as is the case for CMA, pathogenic tau mutations and posttranslational modifications inhibit eMI activity at different stages (i.e. substrate binding to LE/MVB, internalization or degradation in the lumen) (Caballero et al., 2018). As different types of autophagy can clear tau, this enables re-routing of this aggregate-prone protein from one autophagy type to another when one of these routes is compromised. For example, acetylated tau can be re-routed to eMI for degradation when CMA is inhibited (Caballero et al., 2021). This re-routing of acetylated tau to eMI can lead to its extracellular release in exosomes through fusion of LE/MVBs with the plasma membrane (Caballero et al., 2021). This may provide a mechanism to remove toxic products, such as acetylated tau, from neurons when they are unable to be degraded (Perez et al., 2019) but may also contribute to disease progression through extracellular release of pathogenic tau (Caballero et al., 2021). The factors that determine whether the eMI re-routed protein is degraded in LE/MVB or released extracellularly remain unknown.
AD, the most common tauopathy and neurodegenerative disorder, is characterized by the accumulation of intracellular tau tangles, and extracellular amyloid-β deposits (Aβ plaques). Macroautophagy has been implicated in the production of amyloid-β within the autophagic vesicles and its secretion to the extracellular matrix, contributing to plaque formation in vivo in mice and Drosophila models of amyloid toxicity (Yu et al., 2004). Levels of Beclin 1 are decreased in patients with AD at early stages (Lachance et al., 2019), whereas Beclin 1 overexpression reduced amyloid aggregation in a mouse model of AD (Pickford et al., 2008). Microglial Beclin 1 modulates the inflammatory response in Becn1+/−/APPPS1 mice (Houtman et al., 2019) and microglial amyloid-β removal and phagocytosis is significantly impaired in Beclin 1+/− mice (Lucin et al., 2013). Amyloid-β proteins also induce the accumulation of deficient mitochondria, driven by the depletion of cytosolic Parkin and PINK1 accumulation, resulting in defective mitophagy (Cummins et al., 2019; Fang et al., 2019). TREM2, an inflammation-linked risk factor for late-onset AD (Jonsson et al., 2013), is also associated with mTOR pathway dysregulation in AD, where there is also downregulation of the Beclin 1-PI3K and ULK1/2 complexes (Lachance et al., 2019).
Brains from AD patients show a selective loss of nuclear TFEB in the hippocampus, negatively correlating with the severity of the neuropathology and reduced expression of TFEB target genes is also observed in AD patient fibroblasts and iPSC-derived human AD neurons (reviewed in (Cortes and La Spada, 2019).
Familial AD and genetic risk factors
Mutations in the genes encoding amyloid precursor protein (APP) and presenilin (PSEN) cause autosomal dominant early-onset forms of AD (reviewed in Raybould and Sims, 2021). PSEN1 mutations increase lysosomal pH by deregulating the maturation of lysosomal v-ATPase and consequently reduce autophagic cargo degradation (Nixon, 2013). The APOE4 allele, involved lipid trafficking and metabolism, is the most common AD genetic risk factor (Bu, 2009). APOE4 carriers have more pronounced reductions in LC3, P62 and LAMP1 compared to APOE3 carriers (Parcon et al., 2018). In astrocytes from mice expressing human APOE4 variant, autophagosome formation and cargo degradation were defective (Simonovitch et al., 2016). Levels of phosphatidylinositol biphosphates, essential for autophagic function, are reduced in post-mortem human brain tissues of APOE4 carriers (Dall’Armi et al., 2013) and in primary neurons and astrocytes derived from knock-in mice expressing human APOE4 (Zhu et al., 2015). APOE4 may also affect the expression of P62, LAMP2 and MAP1LC3B by competing with TFEB for the binding to the CLEAR domains in their promoters (Parcon et al., 2018).
The impairment of macroautophagy in AD may also be a consequence of variants in genes with a functional role in macroautophagy, such as SORL1, PICALM and PLD3 (see Table 1 for wildtype function). Reduced levels of SORL1, which regulates protein trafficking between the trans-Golgi network, are observed in iPSC-derived neurons from AD patients (Hung et al., 2021). Phosphatidylinositol Binding Clathrin Assembly Protein (PICALM) identified in a GWAS study in a locus associated with AD risk, is abnormally cleaved in AD brains and its levels inversely correlate with LC3-II and Beclin 1 levels. In addition, alternative splicing of the protein correlates with tau aggregation and Braak stages (Raj et al., 2018). PICALM regulates endocytosis of critical SNAREs involved in both autophagosome biogenesis and degradation (Moreau et al., 2014).
The link between macroautophagy and AD is further supported by several genetic studies that associate autophagy-related genes with AD. Pathway enrichment algorithms analysing the data from three GWAS studies found an enrichment in autophagic and endolysosomal genes associated with genetic variants that increase risk of AD (Gao et al., 2018).
Gradual loss of CMA with age may also become a risk factor for AD. Thus, CMA blockage in a mouse model of AD accelerated tau phosphorylation and aggregation, tau propagation, and amyloid-β extracellular deposition (Bourdenx et al., 2021; Caballero et al., 2021). CMA deficiency increases similarity between the proteomes of brains from mouse model of disease and AD patients, thus mimicking part of the disease usually missing in conventional models (Bourdenx et al., 2021).
Although much interest has been focused on extracellular fragments (amyloid-β 40 and 42), the C-terminal fragment (CTF) of APP (C99 – originating from β-secretase cleavage) induces autophagy-lysosome impairments independently of amyloid-β accumulation (Lauritzen et al., 2016). Interestingly, APP CTFs contain a KFERQ motif (763KFFEQ768) that could be utilized for its degradation through CMA or eMI (Park et al., 2016a). Consistent with that notion, CMA blockage in a genetic AD mouse model caused CTF accumulation (Bourdenx et al., 2021).
Parkinson’s disease (PD)
PD is characterized by the loss of dopaminergic neurons in the substantia nigra, the presence of intraneuronal inclusions (Lewy bodies, LBs) in neuronal soma and neurites enriched with filamentous forms of α-Synuclein (Spillantini et al., 1997).
α-Synuclein has been proposed to play roles in synaptic function, its levels are a major determinant of PD severity, and multiplications of the SNCA (α-Synuclein -encoding) locus cause autosomal dominant forms of PD. α-Synuclein can be cleared by the proteasome, macroautophagy and CMA (Cuervo et al., 2004; Webb et al., 2003). α-Synuclein accumulation compromises autophagic flux, as the presence of inclusion bodies containing α-Synuclein impairs autophagosome maturation and fusion with lysosomes (Tanik et al., 2013). Indeed, disruptions in the macroautophagy machinery appear to be central to PD pathogenesis, with different PD-causing mutations affecting various stages of the autophagy itinerary (Karabiyik et al., 2017).
α-Synuclein overexpression in mammalian cells and transgenic mice compromises autophagosome biogenesis by inhibiting the GTPase RAB1 leading to mislocalisation of ATG9 (Winslow et al., 2010). The expression of mutant α-synuclein (A53T) leads to an accumulation of mitochondria-containing autophagosomes in PC12 cells (Stefanis et al., 2001), and to increased autophagosomal engulfment of healthy, polarized mitochondria in primary neurons, causing an abnormal clearance of functional mitochondria and therefore, a bioenergetic deficit (Choubey et al., 2011). In addition, results from Drosophila suggest that α-Synuclein expression impairs the autophagic flux in ageing adult neurons by disrupting the F-actin cytoskeleton (Sarkar et al., 2021).
Although CMA contributes to clearance of α-Synuclein in primary cultured cells and in vivo (Cuervo et al., 2004), pathogenic A53T and A30P mutant α-Synuclein proteins are still targeted by HSC70 to lysosomes, but their abnormally enhanced interaction with LAMP2A and their oligomerization at the lysosomal membrane disrupt the CMA translocation complex and block CMA of other substrate proteins (Cuervo et al., 2004). CMA malfunction is not restricted to familial PD, as post-translational modifications on α-Synuclein can also compromise its CMA degradation (Martinez-Vicente et al., 2008). While phosphorylation and covalent oligomerization mask the KFERQ-like motif in α-Synuclein (Kirchner et al., 2019) and disrupt its targeting to lysosomes, dopamine oxidation leads to formation of α-Synuclein-dopamine adducts that are still targeted to lysosomes where they disrupt CMA through similar mechanisms as the α-synuclein mutants (Martinez-Vicente et al., 2008).
Alterations in CMA-related proteins have been reported in the brains of familial and idiopathic PD patients. Reduced levels of LAMP2A and HSC70 in the substantia nigra and amygdala of PD brains occur early in the disease and correlate with α-Synuclein accumulation (Scrivo et al., 2018). Reduced mRNA levels for both proteins suggest that downregulated transcription, in addition to the physical blockage of CMA, may both contribute to compromised CMA in PD. This reduced CMA in the PD brain has a negative impact in neuronal survival, in part, by reducing degradation of the transcription factor MEF2D, normally degraded by CMA. Accumulation of the inactive form of MEF2D prevents its protective function in these cells (Yang et al., 2009). The relation between PD and eMI has not been directly analysed, but a recent study showing that a mutated form of α-synuclein accumulates in the LE/MVB and is secreted in exosomes (Stykel et al., 2021), opens the possibility of a rerouting of α-Synuclein from CMA to eMI, similar to the one described for tau upon CMA blockage (Caballero et al., 2021).
Increasing evidence suggests that other mutations causing familial PD also affect autophagic function. In an autosomal dominant form of PD, a mutation (D620N) in the vacuolar protein sorting 35 (VPS35), a component of the retromer complex, impairs macroautophagy (see Table One). Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are responsible for most autosomal dominant PD cases, increase phosphorylation of different RAB GTPases (Alessi and Sammler, 2018) and disrupt endosome-to-lysosome trafficking. While the most common mutation (G2019S) has been associated with a reduction of macroautophagy flux (Henry et al., 2015; Wallings et al., 2019), LRRK2 deletion in neurons increases both macroautophagy (through the regulation of p62 phosphorylation) and lysosomal degradation (Park et al., 2016b; Wallings et al., 2019). Contrary to this proposed inhibitory role of LRRK2 in neurons, overexpression of LRRK2 in microglial cells increased macroautophagy flux, while LRRK2 silencing reduced degradation (Wallings et al., 2019). Furthermore, the effect of LRRK2 may also differ depending on the type of autophagy. Thus, the LRRK2 G2019S mutant was also suggested to induce mitophagy in multiple studies by causing calcium imbalance, resulting in mitochondrial depolarization (Cherra et al., 2013).
Heterozygous mutations in the gene encoding the lysosomal enzyme glucocerebrosidase 1 (GBA1) are amongst the most common genetic risk factors for PD. iPSC-derived neurons from PD patients with GBA1 mutations showed increased α-Synuclein levels, macroautophagy and lysosomal defects, including impaired fusion between autophagosomes and lysosomes (Schöndorf et al., 2014), further corroborated in neuroblastoma cells with nonsense mutations in GBA1 (Bae et al., 2015) and in GBA1-deficient cells (Magalhaes et al., 2016).
Mutations in ATP13A2/PARK9, which encodes a transmembrane lysosomal P-type ATPase polyamine transporter, cause familial Kufor-Rakeb syndrome with early-onset Parkinsonism and impairs lysosomal function (Dehay et al., 2012). Loss of ATP13A2 causes α-Synuclein accumulation in mammalian cells and in C. elegans (Karabiyik et al., 2017) and in vitro, depletion leads to a decrease in the levels of synaptotagmin 11 (SYT11), another PD-associated gene (Bento et al., 2016). Overexpression of SYT11 rescues the macroautophagy defects observed in ATP13A2-depleted cells, implying that the two proteins lie within the same pathway (Wang et al., 2019a). ATP13A2 also recruits the histone deacetylase 6 (HDAC6) to lysosomes to deacetylate a protein implicated in autophagosome-lysosome fusion, cortactin, resulting in increased fusion and autophagy flux (Borsche et al., 2020).
Autosomal recessive mutations in PTEN-induced kinase 1 (PINK1 or PARK6) and Parkin (PARK2), key regulators of a form of mitophagy, have been discussed above. Low CMA activity in PD has also been linked to alteration of mitochondria dynamics. Failure to degrade PARK7/DJ1 through CMA leads to its accumulation, disruption of mitochondrial homeostasis and cell death (Wang et al., 2016). Similarly, MARCHF5, an E3 ubiquitin ligase required for mitochondrial fission, is a CMA substrate that upon CMA blockage gets stabilized, increasing mitochondrial fission and altering mitochondrial homeostasis. Overexpression of LAMP2A in a 6-OHDA PD mouse model was sufficient to rescue its phenotype (Nie et al., 2020).
The reported interaction between UCH-L1 with LAMP2A, HSC70 and HSP90 is abnormally enhanced in the case of the PD-related UCH-L1 I93M mutant and results in reduced degradation of proteins, including α-Synuclein via CMA (Scrivo et al., 2018). Similarly, although a fraction of cellular LRRK2 undergoes degradation via CMA, mutant forms of LRRK2 block the formation of the LAMP2A translocation complex and degradation of other proteins through CMA (Orenstein et al., 2013). In other instances, reduced CMA by PD-related pathogenic proteins is a consequence of a more general effect in the lysosomal compartment. This occurs with VPS35, whose deficiency or mutant form D620N reduces lysosomal levels of LAMP2A through its inefficient retrieval from the Golgi (Scrivo et al., 2018). Reduced enzymatic activity of GBA1 reduces lysosomal levels of LAMP2A (Murphy et al., 2014), likely by inducing changes in the lipid composition of the lysosomal membrane associated with increased levels of cathepsin A, the enzyme that triggers LAMP2A degradation in lysosomes (Kaushik and Cuervo, 2018). A similar destabilizing effect on lysosomal LAMP2A has been described for the loss of function mutants of PARKK7/DJ1, although the mechanism behind this effect remains unknown (Xu et al., 2017).
ALS
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease characterised by upper and lower motor neurons (MNs) degeneration. While 90% of all ALS cases are sporadic (sALS), around 10% are due to a genetic mutation. To date, more than 20 genes have been associated with ALS (Mathis et al., 2019), including C9ORF72, TAR DNA binding protein (TARDBP/TDP-43), P62 (SQSTM1), TANK binding kinase 1 (TBK1), and optineurin (OPTN), whose mutations can result in autophagy and mitophagy defects, and the accumulation of protein inclusions in familial ALS (fALS) patients.
Hexanucleotide repeat expansions (HREs) in the 5’ non-coding sequence of the C9ORF72 gene have been identified as the most common cause of fALS and frontotemporal dementia due to loss-of-function or gain-of-function through three possible mechanisms: (1) haploinsufficiency, (2) repeat associated non-AUG (RAN) translations creating toxic dipeptide repeat (DPR) proteins and (3) sequestration of RNA-binding proteins (RNPs) through generation of RNA foci involving the C9ORF72 HRE RNA (Wen et al., 2017). While the first mechanism results from loss-of-function, the latter two mechanisms are considered toxic gain-of-function.
Although protein levels of C9ORF72 are reduced by 10–75% in post-mortem tissues, a significant reduction of C9ORF72 expression is not observed in patient-iPSC-derived MNs (Renton et al., 2011). LC3-II levels are decreased in C9ALS- patient-iPSC-derived neurons but silencing of C9ORF72 enhances autophagy via inhibition of mTOR activity and elevation of TFEB levels (Ugolino et al., 2016). C9ORF72 forms a stable complex with two proteins, the Smith-Magenis syndrome chromosome region, candidate 8 (SMCR8) and the WD repeat-containing protein 41 (WDR41). C9ORF72-SMCR8 recruits ULK1 complexes on phagophores in a RAB1A-dependent manner and deficiency of C9ORF72 causes a decrease in ULK1 protein, compromising the initiation of autophagy (Pang and Hu, 2021). The sense repeat RNA of C9ORF72 results in the accumulation of nuclear and cytoplasmic RNA foci, the latter resulting in the formation of stress granules (SG) (Mizielinska et al., 2013). Wild-type C9ORF72 lowers SGs via autophagic degradation by cooperating with P62 (Chitiprolu et al., 2018). Overall, a reduction of autophagic activity caused by C9ORF72 loss-of-function results in the accumulation of TDP-43, DPR, and SGs which would otherwise be eliminated by autophagy.
The accumulation of TDP-43 is frequently seen in ALS and is also mutated in some cases. Knockdown of TDP-43 impacts autophagy at multiple steps, such as downregulation of ATG7 and reduction of dynactin subunit 1 expression resulting in an overall inhibition of autolysosome formation and impaired autophagic flux (Xia et al., 2016). Mutations of OPTN and TBK1 observed in ALS patients prevent ubiquitin binding to OPTN and OPTN binding to TBK1, respectively, compromising mitophagy (Evans and Holzbaur, 2019).
Although few studies have investigated the status of other autophagic pathways in ALS, growing evidence supports possible involvement of CMA failure on ALS disease progression. Inhibition of CMA results in the accumulation of the truncated form of TDP-43 leading to TDP-43 aggregation (Scrivo et al., 2018). Furthermore, TDP-43 aggregates (but not overexpressed soluble TDP-43) promote transcriptional upregulation of LAMP2A and HSC70, which has been interpreted as a possible early protective mechanism against TPD-43 mediated proteotoxicity. However, as TPD-43 aggregation persists, the associated lysosomal damage may likely lead to CMA failure (Scrivo et al., 2018). Interestingly, intrabody-mediated targeting of misfolded TDP-43 protein to CMA has proven effective in reducing TDP-43 aggregates and decreasing neurotoxicity (Tamaki et al., 2018).
CMA has also shown to contribute to degradation of the ubiquitin-like protein, ubiquilin-2, which is mutated in forms of ALS and ALS-like dementia. Physiological clearance by CMA of ubiquilin-2, a known regulator of macroautophagy, has been proposed to contribute to the crosstalk between the two autophagic pathways (Rothenberg et al., 2010). However, expression of mutant UBQLN2P497H in neuronal cells is associated with reduced expression of LAMP2A in the ventral horn of the spinal cord and its accumulation in ubiquilin-2-positive inclusions (Chen et al., 2018).
A still poorly explored change in ALS, but with the potential of linking this condition with CMA and KFERQ-selective eMI, is the sequestration of HSC70 mRNA into inclusions resulting from overexpression of either TDP-43 or C9orf72 (Coyne et al., 2017). Lower levels of HSC70 protein have also been noted in peripheral blood mononuclear cells from sporadic ALS patients. and silencing of HSC70 in neuroblastoma cell line is sufficient to precipitate formation of TDP-43 aggregates (Arosio et al., 2020). Although the multiplicity of intracellular functions of HSC70 precludes the ability to exclusively attribute these changes in proteostasis to defective CMA or eMI, these findings should generate future interest in exploring the status of both pathways in ALS.
E. Therapeutic Strategies
A number of drug-like small molecules and genetic tools upregulate macroautophagy and ameliorate pathologies in cellular and animal models of different neurodegenerative diseases (reviewed in (Lee et al., 2018; Li et al., 2014; Menzies et al., 2017). Here, we will focus on more recent developments and also consider other types of autophagy.
Macroautophagy targeting
L-type Ca2+ channel blockers, such as felodipine, enhance macroautophagy in mouse neurons (Siddiqi et al., 2019). Felodipine has good brain penetration, enhances macroautophagy flux in the brain and was found to reduce mutant huntingtin aggregates a Huntington’s disease (HD) mouse model and to ameliorate motor phenotypes. Felodipine administered to mimic the plasma concentration of humans taking the drug for hypertension, reduced levels of insoluble A53T α-Synuclein, and ameliorated neurodegeneration and motor phenotypes in a PD mouse model (Siddiqi et al., 2019).
Lonafarnib, a brain-penetrant farnesyltransferase-inhibitor reduced levels of pathogenic tau and ameliorated brain shrinkage and behavioural abnormalities in a tauopathy mouse model, likely through inhibition of the GTP-binding protein Rhes (Hernandez et al., 2019).
Genetic deletion of the mGluR5 receptor reverses disease pathology in Q111 mHtt knockin mice (HD model) (Ribeiro et al., 2014). CTEP, a well-characterised, potent, CNS penetrant mGluR5 negative allosteric modulator, facilitated autophagy-dependent clearance of the huntingtin aggregates and improved motor deficits and cognitive impairment in an HD mouse model (Abd-Elrahman and Ferguson, 2019).
The D2/D3 dopamine receptor (DR) agonist pramipexole promotes macroautophagy in cell culture by enhancing Beclin 1 transcription (Wang et al., 2015), and ameliorates disease in the R6/1 HD mouse model where it increased LC3-II and decreased p62 in the striatum (Luis-Ravelo et al., 2018).
PPARα agonists, gemfibrozil and Wy14643, which induce macroautophagy in cellular models via PPARα, reduce soluble amyloid-β levels in the cortex and hippocampus of an AD mouse model, induce macroautophagy and improve behavioural deficits (Luo et al., 2020).
Rapid progress has been made developing small molecules such as molecular glues, that stabilise protein-protein interactions (Schreiber, 2021) and bifunctional molecules, like PRoteolysis TArgeting Chimeras (PROTACs) that degrade proteins by forcing their interaction with a ubiquitin E3 ligase that ubiquitinates the target enabling its subsequent proteasomal clearance (Paiva and Crews, 2019). Similar approaches have been developed for macroautophagy. Molecules dubbed AuTophagosome-TEthering Compounds (ATTECs) bind both LC3 and mHTT to target the mutant protein to phagophores for macroautophagic degradation (Li et al., 2019c) (Fig. 5). Such ATTECs can lower levels of mutant but not wild-type huntingtin, and ameliorate disease in an HD mouse model (Li et al., 2019c).
Figure 5. Autophagy tethering compounds (ATTEC) and the autophagy-targeting chimera (AUTAC) system.
A) ATTEC molecules tether the protein of interest to the autophagosomes by direct binding to the protein of interest and to LC3. A proof-of-concept study using the mutant HTT protein (mHTT) demonstrated that these compounds can degrade mHTT both in cells and in vivo in animal models and demonstrated targeting of mHTT to autophagosomes for subsequent degradation without influencing autophagy activity per se. B) AUTAC technology has a similar design to the PROTAC technology and both use ubiquitination to target proteins for degradation. The AUTAC molecule contains a degradation tag (a guanine derivative called FBnG) which induces K63 polyubiquitination and a ligand which binds to the target protein to provide target specificity. The resulting K63 ubiquitination targets the labelled protein for degradation via macroautophagy.
Chimeric molecules called AUTACs (AUtophagy TArgeting Chimera) (Takahashi et al., 2019) utilize endogenous tagging by S-guanylation of cysteine residues by 8-nitro-cGMP tag to trigger ubiquitination and then macroautophagic clearance. AUTAC molecules consist of Cys-S-p-fluorobenzylguanine (dubbed FBnG), a mimetic of Cys-S-cGMP with improved physicochemical properties, attached to a target-specific ligand via a polyethylene glycol (PEG) linker (Fig. 5). The approach was extended to mitophagy of fragmented mitochondria through the generation of AUTAC4 in which the FBnG is attached to a warhead binding to the mitochondrial translocator protein (TSPO). AUTAC4 promoted mitophagy, reduced apoptosis and maintained levels of intracellular ATP when mitochondrial fragmentation was induced.
Clinical trials of autophagy-enhancing drugs for neurodegeneration are relatively rare. Much of the effort to-date has focused on repurposing and the next phase will likely see the development of specific autophagy-enhancing drugs through focussed medicinal chemistry on well-validated targets. Such effort within the pharmaceutical and biotech sectors should increase the chance of projects progressing from preclinical to clinical studies.
Therapeutic targeting through CMA
The recently described broad role of CMA in neuronal proteostasis and degradation of pathogenic proteins, and the growing evidence of reduced CMA activity in PD, FTD and AD, has made CMA an attractive target to prevent neuronal proteotoxicity and neurodegeneration.
Genetic modulation of CMA in neurodegeneration models has primarily focused on LAMP2A. Lentiviral-based LAMP2A overexpression in rats substantia nigra reduced human α-Synuclein aggregation and protected dopaminergic neurons from degeneration (Xilouri et al., 2013).
Since retinoic acid receptor α (RARα) negatively regulates CMA, activation of this pathway can be attained with retinoic acid derivatives (Anguiano et al., 2013). Extensive medicinal chemistry optimized one of these derivatives (CA77.1), which improved behaviour and tau and Aβ-related pathology in FTD mouse models (Bourdenx et al., 2021). Although interventions that improve lysosomal function should also indirectly enhance CMA, this possibility has been rarely investigated. For example, lactulose has been proposed to improve Aβ pathology through macroautophagy and CMA, but CMA activity was not measured (Lee et al., 2021). Similarly, inhibition of HDAC6 with tubastatin A was reported to reduce PD-related pathology through CMA activation (Francelle et al., 2020). Although CMA activity was not directly measured, the fact that the intervention increased LAMP2A levels justifies future studies.
PROTAC-like approaches have also been attempted for CMA. A fusion molecule containing the KFERQ-like motif and the polyglutamine binding peptide 1, which interacts with polyglutamine chains, was effective in targeting mutant huntingtin for degradation via CMA (Bauer et al., 2010). Intra-striatal delivery of this fusion molecule reduced pathology in HD models and improved the disease phenotype (Bauer et al., 2010). KFERQ-containing peptides designed to bind Aβ oligomers and promote degradation through CMA have been effective (Dou et al., 2020), although this strategy may not involve CMA, as Aβ oligomers are usually extracellular and CMA requires protein unfolding before internalization. It is still possible that KFERQ-tagging targets instead these proteins to LE/MVB via eMI.
F. Conclusions and important future questions
To maximise our understanding of these areas of biology and increase the probabilities of contributing to therapeutic strategies, there are several domains that deserve further exploration. From a basic science perspective, it will be important to appreciate how different autophagic pathways, the UPS and unconventional secretion communicate and influence each other. One challenge is to devise approaches different from knockout systems (e.g. ATG-null cells) or pharmacological inhibitors, as these can affect other systems or induce compensatory effects. One approach that may contribute to such analyses is autophagosome and lysosomal content proteomic profiling (Le Guerroue et al., 2017; Schneider et al., 2015).
We need a better understanding of the roles of autophagic pathways in different glia. This will expand understanding of glial physiology and disease relevance, for example, for neuroinflammatory-like. Furthermore, growing evidence of non-cell autonomous control of neuronal autophagy by glia that could be utilized as a therapeutic target. For example, chemical enhancement of CMA in PD astrocytes has proven protective in vitro against a-Synuclein-mediated toxicity in neurons (di Domenico et al., 2019).
Further studies testing links between autophagy and psychiatric disease and conditions like autism will be important for understanding their etiologies, therapeutic potential and, by extension, may also shed light on processes underpinning the very challenging behavioural features in common neurodegenerative diseases.
Harnessing autophagy-stimulating strategies has real potential for many neurodegenerative diseases, but translational success may depend on the identification of human biomarkers that reflect brain autophagic activities. Dynamic metabolite tracing (metabolic fluxomics) and multiplex quantitative proteomics including PTMs (Wesseling et al., 2020) and more sensitive and specific tools for in vivo imaging of human brains of disease-causing autophagic substrates, like tau, α-Synuclein and huntingtin (van Waarde et al., 2021) may help reveal the molecular signatures of defects in autophagy. Early treatment, ideally in presymptomatic individuals, may be necessary, as disease-protein lowering later in the disease course is not nearly as effective as early removal (Rubinsztein and Orr, 2016). In this respect, trial strategies and approval processes should evolve to enable development of preventive agents administered to healthy/presymptomatic people, analogous to the use of statins in heart disease.
Enhancing autophagosome formation, although effective in disease animal models, may have risk in conditions when autophagosome clearance is defective. This may be resolved by careful in vivo studies aiming to mimic the physiological/disease scenario in a model organism. In advanced states of disease, when multiple autophagy steps and pathways may be compromised, it would be worth considering combinatorial interventions aiming at restoring flux through multiple autophagic pathways, since even partial restoration of each of them may have an additive beneficial effect.
The coordinate functioning of autophagy with the UPS and chaperones also makes an attractive combination of chemical regulators of these components with autophagy enhancing interventions.
Consideration should also be given to the changes on autophagy in aging and the higher probabilities of comorbidities at advanced ages that may contribute to disease (i.e. diabetes and AD). Consequently, it will be important to consider the effects of therapeutic agents not only on the neurodegenerative disease itself, but also on comorbidities and on healthy older people.
Enhancing autophagic pathways to treat diseases or delay symptom onset has the added benefit that constitutive upregulation in animals is generally associated with longevity and other health benefits (e.g. (Fernandez et al., 2018; Pyo et al., 2013). Furthermore, the autophagy-inducing agent may not need to engage its target all of the time like an antihypertensive drug but may have efficacy if administered in a pulsatile fashion e.g. every few days, which would reduce liabilities of the agent, increasing the feasibility of this approach.
ACKNOWLEDGEMENTS
This work was supported by UK Dementia Research Institute (funded by the MRC, Alzheimer’s Research UK and the Alzheimer’s Society), Alzheimer’s Research UK (ARUK-2022DDI-CAM, ARUK-TC2020-1 and ARUK-PG2018C-001), The Tau Consortium, Cambridge Centre for Parkinson-Plus, the National Institutes of Health grants AG054108, AG031782, AG021904, NS100717 and the generous support of the Rainwater Charitable Foundation, The JPB Foundation, The Backus Foundation, The Glenn Foundation and R&R Belfer and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care’. AMS is supported by a Ramon Areces postdoctoral fellowship and GJK by a T32 GM007491 and a T32 GM007288. MB is supported by an ‘Allocation Jeune Chercheur’ from Fondation Alzheimer (France).
Footnotes
CONFLICTS OF INTEREST
D.C.R. is a consultant for Aladdin Healthcare Technologies SE, Drishti Discoveries, Abbvie, PAQ Therapeutics, MindRank AI and Nido Biosciences. A.M.C. is a cofounder and scientific advisor for Life Biosciences, consults for Generian Pharmaceuticals and Cognition Therapeutics and has US patent US9512092. None of the other authors have any potential competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd-Elrahman KS, and Ferguson SSG (2019). Modulation of mTOR and CREB pathways following mGluR5 blockade contribute to improved Huntington’s pathology in zQ175 mice. Mol Brain 12, 35. 10.1186/s13041-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Córdoba SR, Knecht E, and Rubinsztein DC (2010). Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Human molecular genetics 19, 2867–2876. 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N, Cantagrel V, Zaki MS, Al-Gazali L, Wang X, Rosti RO, Dikoglu E, Gelot AB, Rosti B, Vaux KK, et al. (2015). Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat Genet 47, 528–534. 10.1038/ng.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Eissa MM, Fiorentino A, Sharp SI, O’Brien NL, Wolfe K, Giaroli G, Curtis D, Bass NJ, and McQuillin A. (2018). Exome sequence analysis and follow up genotyping implicates rare ULK1 variants to be involved in susceptibility to schizophrenia. Ann Hum Genet 82, 88–92. 10.1111/ahg.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, and Sammler E. (2018). LRRK2 kinase in Parkinson’s disease. Science 360, 36–37. 10.1126/science.aar5683. [DOI] [PubMed] [Google Scholar]
- Anding AL, Wang C, Chang TK, Sliter DA, Powers CM, Hofmann K, Youle RJ, and Baehrecke EH (2018). Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance. Current biology: CB 28, 287–295.e286. 10.1016/j.cub.2017.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, and Cuervo AM (2013). Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol 9, 374–382. 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E, and Cuervo AM (2020). Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol Metab 31, 53–66. 10.1016/j.tem.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E, Koga H, Diaz A, Mocholi E, Patel B, and Cuervo AM (2015). Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell 59, 270–284. 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio A, Cristofani R, Pansarasa O, Crippa V, Riva C, Sirtori R, Rodriguez-Menendez V, Riva N, Gerardi F, Lunetta C, et al. (2020). HSC70 expression is reduced in lymphomonocytes of sporadic ALS patients and contributes to TDP-43 accumulation. Amyotroph Lateral Scler Frontotemporal Degener 21, 51–62. 10.1080/21678421.2019.1672749. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, Squitieri F, Hardenberg MC, Imarisio S, Menzies FM, and Rubinsztein DC (2017). Polyglutamine tracts regulate autophagy. Autophagy 13, 1613–1614. 10.1080/15548627.2017.1336278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, and Ktistakis NT (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701. 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae EJ, Yang NY, Lee C, Lee HJ, Kim S, Sardi SP, and Lee SJ (2015). Loss of glucocerebrosidase 1 activity causes lysosomal dysfunction and α-synuclein aggregation. Experimental and Molecular Medicine 47. 10.1038/emm.2014.128. [DOI] [Google Scholar]
- Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, and Cuervo AM (2010). Identification of regulators of chaperone-mediated autophagy. Mol Cell 39, 535–547. 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankston AN, Forston MD, Howard RM, Andres KR, Smith AE, Ohri SS, Bates ML, Bunge MB, and Whittemore SR (2019). Autophagy is essential for oligodendrocyte differentiation, survival, and proper myelination. Glia 67, 1745–1759. 10.1002/glia.23646. [DOI] [PubMed] [Google Scholar]
- Bansal M, Moharir SC, Sailasree SP, Sirohi K, Sudhakar C, Sarathi DP, Lakshmi BJ, Buono M, Kumar S, and Swarup G. (2018). Optineurin promotes autophagosome formation by recruiting the autophagy-related Atg12–5-16L1 complex to phagophores containing the Wipi2 protein. J Biol Chem 293, 132–147. 10.1074/jbc.M117.801944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, and Nukina N. (2010). Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol 28, 256–263. nbt.1608 [pii] [DOI] [PubMed] [Google Scholar]
- Bejarano E, Yuste A, Patel B, Stout RF, Spray DC, and Cuervo AM (2014). Connexins modulate autophagosome biogenesis. Nat Cell Biol 16, 401–414. 10.1038/ncb2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Ashkenazi A, Jimenez-sanchez M, and Rubinsztein DC (2016). The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nature Communications 7, 1–16. 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, Kudryashev M, Dikic I, and Hummer G. (2019). Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun 10, 2370. 10.1038/s41467-019-10345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsall V, and Waites CL (2019). Autophagy at the synapse. Neuroscience letters 697, 24–28. 10.1016/j.neulet.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsche M, König IR, Delcambre S, Petrucci S, Balck A, Bruggemann N, Zimprich A, Wasner K, Pereira SL, Avenali M, et al. (2020). Mitochondrial damage-associated inflammation highlights biomarkers in PRKN/PINK1 parkinsonism. Brain 143, 3041–3051. 10.1093/brain/awaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx M, Martin-Segura A, Scrivo A, Rodriguez-Navarro JA, Kaushik S, Tasset I, Diaz A, Storm NJ, Xin Q, Juste YR, et al. (2021). Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 184, 2696–2714 e2625. 10.1016/j.cell.2021.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Liu Y, Datta S, Hariri H, Seda M, Anderson G, Peskett E, Demetriou C, Sousa S, Jenkins D, et al. (2018). SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Human molecular genetics 27, 1927–1940. 10.1093/hmg/ddy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu GJ (2009). Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 10, 333–344. 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzlaff M, Hannan SB, Karsten P, Lenz S, Ng J, Voßfeldt H, Prüßing K, Pflanz R, Schulz JB, Rasse T, and Voigt A. (2015). Impaired retrograde transport by the Dynein/Dynactin complex contributes to Tau-induced toxicity. Human molecular genetics 24, 3623–3637. 10.1093/hmg/ddv107. [DOI] [PubMed] [Google Scholar]
- Caballero B, Bourdenx M, Luengo E, Diaz A, Sohn PD, Chen X, Wang C, Juste YR, Wegmann S, Patel B, et al. (2021). Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat Commun 12, 2238. 10.1038/s41467-021-22501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero B, Wang YP, Diaz A, Tasset I, Juste YR, Stiller B, Mandelkow EM, Mandelkow E, and Cuervo AM (2018). Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 17. ARTN e12692. 10.1111/acel.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenacchi G, Papa V, Pegoraro V, Marozzo R, Fanin M, and Angelini C. (2020). Review: Danon disease: Review of natural history and recent advances. Neuropathol Appl Neurobiol 46, 303–322. 10.1111/nan.12587. [DOI] [PubMed] [Google Scholar]
- Chang J, Lee S, and Blackstone C. (2014). Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest 124, 5249–5262. 10.1172/JCI77598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Kumsta C, Hellman AB, Adams LM, and Hansen M. (2017). Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife 6, 1–23. 10.7554/eLife.18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xiao Y, Chai P, Zheng P, Teng J, and Chen J. (2019). ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Current biology : CB 29, 846–855.e846. 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Chen T, Huang B, Shi X, Gao L, and Huang C. (2018). Mutant UBQLN2(P497H) in motor neurons leads to ALS-like phenotypes and defective autophagy in rats. Acta Neuropathol Commun 6, 122. 10.1186/s40478-018-0627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Liao Y, Dong Y, Hu H, Yang N, Kong X, Li S, Li X, Guo J, Qin L, et al. (2020). Microglial autophagy defect causes parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy 16, 2193–2205. 10.1080/15548627.2020.1719723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ 3rd, Steer E, Gusdon AM, Kiselyov K, and Chu CT (2013). Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol 182, 474–484. 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HL, Terlecky SR, Plant CP, and Dice JF (1989). A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385. 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Chino H, Hatta T, Natsume T, and Mizushima N. (2019). Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell 74, 909–921 e906. 10.1016/j.molcel.2019.03.033. [DOI] [PubMed] [Google Scholar]
- Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, Palidwor G, Barry FA, Zinman L, Keith J, et al. (2018). A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 9, 2794. 10.1038/s41467-018-05273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, and Yoon SY (2014). Autophagy in microglia degrades extracellular beta-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 10, 1761–1775. 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Zhang Y, Seegobin SP, Pruvost M, Wang Q, Purtell K, Zhang B, and Yue Z. (2020). Microglia clear neuron-released alpha-synuclein via selective autophagy and prevent neurodegeneration. Nat Commun 11, 1386. 10.1038/s41467-020-15119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong CM, Ke M, Tan Y, Huang Z, Zhang K, Ai N, Ge W, Qin D, Lu JH, and Su H. (2018). Presenilin 1 deficiency suppresses autophagy in human neural stem cells through reducing gamma-secretase-independent ERK/CREB signaling. Cell Death Dis 9, 879. 10.1038/s41419-018-0945-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, Zharkovsky A, and Kaasik A. (2011). Mutant A53T α-Synuclein induces neuronal death by increasing mitochondrial autophagy. Journal of Biological Chemistry 286, 10814–10824. 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Lee J, Jung S, Lee Y, Cho JW, and Oh YJ (2018). Dysregulated autophagy contributes to caspase-dependent neuronal apoptosis. Cell Death Dis 9, 1189. 10.1038/s41419-018-1229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, and Ciechanover A. (2016). p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A 113, E7490–E7499. 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier JJ, Guissart C, Olahova M, Sasorith S, Piron-Prunier F, Suomi F, Zhang D, Martinez-Lopez N, Leboucq N, Bahr A, et al. (2021). Developmental Consequences of Defective ATG7-Mediated Autophagy in Humans. N Engl J Med 384, 2406–2417. 10.1056/NEJMoa1915722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans B, Camus C, Kallergi E, Sposini S, Martineau M, Butler C, Kechkar A, Klaassen RV, Retailleau N, Sejnowski TJ, et al. (2021). NMDAR-dependent long-term depression is associated with increased short term plasticity through autophagy mediated loss of PSD-95. Nat Commun 12, 2849. 10.1038/s41467-021-23133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, and La Spada AR (2019). TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: Molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiol Dis 122, 83–93. 10.1016/j.nbd.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Lorenzini I, Chou CC, Torvund M, Rogers RS, Starr A, Zaepfel BL, Levy J, Johannesmeyer J, Schwartz JC, et al. (2017). Post-transcriptional Inhibition of Hsc70–4/HSPA8 Expression Leads to Synaptic Vesicle Cycling Defects in Multiple Models of ALS. Cell Rep 21, 110–125. 10.1016/j.celrep.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, and Dice JF (1996). A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503. 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, and Knecht E. (1995). Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem 227, 792–800. 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, and Sulzer D. (2004). Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295. 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Cummins N, Tweedie A, Zuryn S, Bertran-Gonzalez J, and Gotz J. (2019). Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J 38. 10.15252/embj.201899360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Armi C, Devereaux KA, and Di Paolo G. (2013). The Role of Lipids in the Control of Autophagy. Current Biology 23, R33–R45. 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AK, Itzhak DN, Edgar JR, Archuleta TL, Hirst J, Jackson LP, Robinson MS, and Borner GHH (2018). AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat Commun 9, 3958. 10.1038/s41467-018-06172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CH, Kim KY, Bushong EA, Mills EA, Boassa D, Shih T, Kinebuchi M, Phan S, Zhou Y, Bihlmeyer NA, et al. (2014). Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci U S A 111, 9633–9638. 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, and Björklund A. (2013). TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A 110, E1817–1826. 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Ofengeim D, and Yuan J. (2019). Targeting RIPK1 for the treatment of human diseases. Proc Natl Acad Sci U S A 116, 9714–9722. 10.1073/pnas.1901179116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, Vital A, Vila M, Klein C, and Bezard E. (2012). Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci U S A 109, 9611–9616. 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Purtell K, Lachance V, Wold MS, Chen S, and Yue Z. (2017). Autophagy Receptors and Neurodegenerative Diseases. Trends Cell Biol 27, 491–504. 10.1016/j.tcb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- di Domenico A, Carola G, Calatayud C, Pons-Espinal M, Munoz JP, Richaud-Patin Y, Fernandez-Carasa I, Gut M, Faella A, Parameswaran J, et al. (2019). Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Reports 12, 213–229. 10.1016/j.stemcr.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, and Manning BD (2013). Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 15, 555–564. 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domise M, Sauve F, Didier S, Caillerez R, Begard S, Carrier S, Colin M, Marinangeli C, Buee L, and Vingtdeux V. (2019). Neuronal AMP-activated protein kinase hyper-activation induces synaptic loss by an autophagy-mediated process. Cell Death Dis 10, 221. 10.1038/s41419-019-1464-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Aguirre-Hernandez C, Scrivo A, Eliscovich C, Arias E, Bravo-Cordero JJ, and Cuervo AM (2020). Monitoring spatiotemporal changes in chaperone-mediated autophagy in vivo. Nat Commun 11, 645. 10.1038/s41467-019-14164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Wang Q, Kao YR, Diaz A, Tasset I, Kaushik S, Thiruthuvanathan V, Zintiridou A, Nieves E, Dzieciatkowska M, et al. (2021). Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 591, 117–123. 10.1038/s41586-020-03129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J, Su P, Xu C, Wen Z, Mao Z, and Li W. (2020). Targeting Hsc70-based autophagy to eliminate amyloid beta oligomers. Biochem Biophys Res Commun 524, 923–928. 10.1016/j.bbrc.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen VV, Swarup S, Hoyer MJ, Paulo JA, and Harper JW (2021). Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy. Elife 10. 10.7554/eLife.72328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD (2000). Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28, 511–525. 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Elia LP, Mason AR, Alijagic A, and Finkbeiner S. (2019). Genetic Regulation of Neuronal Progranulin Reveals a Critical Role for the Autophagy-Lysosome Pathway. The Journal of neuroscience: the official journal of the Society for Neuroscience 39, 3332–3344. 10.1523/JNEUROSCI.3498-17.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott SJ, Ziemba ZJ, Beckmann LJ, Boynton DN, and Miller RA (2020). Inhibition of class I PI3K enhances chaperone-mediated autophagy. J Cell Biol 219. 10.1083/jcb.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CS, and Holzbaur ELF (2019). Autophagy and mitophagy in ALS. Neurobiol Dis 122, 35–40. 10.1016/j.nbd.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CS, and Holzbaur ELF (2020). Quality Control in Neurons: Mitophagy and Other Selective Autophagy Mechanisms. J Mol Biol 432, 240–260. 10.1016/j.jmb.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou YJ, Palikaras K, Adriaanse BA, Kerr JS, Yang BM, Lautrup S, Hasan-Olive MM, Caponio D, Dan XL, et al. (2019). Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci 22, 401-+. 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, et al. (2014). C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Human molecular genetics 23, 3579–3595. 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CJ, Lenk GM, and Meisler MH (2009). Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Human molecular genetics 18, 4868–4878. 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, et al. (2018). Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140. 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, and Simonsen A. (2007). Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179, 485–500. 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, and Rubinsztein DC (2020). Autophagy in Neuronal Development and Plasticity. Trends Neurosci 43, 767–779. 10.1016/j.tins.2020.07.003. [DOI] [PubMed] [Google Scholar]
- Francelle L, Outeiro TF, and Rappold GA (2020). Inhibition of HDAC6 activity protects dopaminergic neurons from alpha-synuclein toxicity. Sci Rep 10, 6064. 10.1038/s41598-020-62678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M, Tolkovsky AM, Borutaite V, Coleman M, and Brown GC (2018). Neuronal Cell Death. Physiol Rev 98, 813–880. 10.1152/physrev.00011.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Solda T, et al. (2016). Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18, 1173–1184. 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- Gal J, Ström AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, and Zhu H. (2009). Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem 111, 1062–1073. 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, and Green DR (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177, 1682–1699. 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, and Jiang X. (2011). Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42, 731–743. 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Casey AE, Sargeant TJ, and Makinen VP (2018). Genetic variation within endolysosomal system is associated with late-onset Alzheimer’s disease. Brain 141, 2711–2720. 10.1093/brain/awy197. [DOI] [PubMed] [Google Scholar]
- Gascon S, Murenu E, Masserdotti G, Ortega F, Russo GL, Petrik D, Deshpande A, Heinrich C, Karow M, Robertson SP, et al. (2016). Identification and Successful Negotiation of a Metabolic Checkpoint in Direct Neuronal Reprogramming. Cell Stem Cell 18, 396–409. 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A, Kirmeier T, Kollmannsberger L, Wagner KV, Dedic N, et al. (2014). Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med 11, e1001755. 10.1371/journal.pmed.1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen NC, and Rein T. (2019). Is There a Role of Autophagy in Depression and Antidepressant Action? Front Psychiatry 10, 337. 10.3389/fpsyt.2019.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, and Schekman R. (2014). The ER-Golgi intermediate compartment feeds the phagophore membrane. Autophagy 10, 170–172. 10.4161/auto.26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Dawson VL, and Dawson TM (2020). PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol Neurodegener 15, 20. 10.1186/s13024-020-00367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, and Springer W. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12, 119–131. 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, Shanley MR, Boudarene N, Rousseaud A, Friedman AK, et al. (2019). Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Current biology: CB 29, 435–448 e438. 10.1016/j.cub.2018.12.021. [DOI] [PubMed] [Google Scholar]
- Goodall ML, Fitzwalter BE, Zahedi S, Wu M, Rodriguez D, Mulcahy-Levy JM, Green DR, Morgan M, Cramer SD, and Thorburn A. (2016). The Autophagy Machinery Controls Cell Death Switching between Apoptosis and Necroptosis. Dev Cell 37, 337–349. 10.1016/j.devcel.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode A, Butler K, Long J, Cavey J, Scott D, Shaw B, Sollenberger J, Gell C, Johansen T, Oldham NJ, et al. (2016). Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy 12, 1094–1104. 10.1080/15548627.2016.1170257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, and Kenyon C. (2008). A Role for Autophagy in the Extension of Lifespan by Dietary Restriction in C. elegans. 4. 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Rubinsztein DC, and Walker DW (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 19, 579–593. 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Zeng D, Xu F, Zhang J, Zhao N, Wang Q, Shi J, Lin Z, Yu W, and Li H. (2019). Baseline Serum Levels of Beclin-1, but Not Inflammatory Factors, May Predict Antidepressant Treatment Response in Chinese Han Patients With MDD: A Preliminary Study. Front Psychiatry 10, 378. 10.3389/fpsyt.2019.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Boada-Romero E, Cunha LD, Magne J, and Green DR (2017). LC3-Associated Phagocytosis and Inflammation. J Mol Biol 429, 3561–3576. 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, and Green DR (2019). LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551 e514. 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AG, Aghamohammadzadeh S, Samaroo H, Chen Y, Mou K, Needle E, and Hirst WD (2015). Pathogenic LRRK2 mutations, through increased kinase activity, produce enlarged lysosomes with reduced degradative capacity and increase ATP13A2 expression. Human molecular genetics 24, 6013–6028. 10.1093/hmg/ddv314. [DOI] [PubMed] [Google Scholar]
- Hernandez D, Torres CA, Setlik W, Cebrian C, Mosharov EV, Tang G, Cheng HC, Kholodilov N, Yarygina O, Burke RE, et al. (2012). Regulation of presynaptic neurotransmission by macroautophagy. Neuron 74, 277–284. 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez I, Luna G, Rauch JN, Reis SA, Giroux M, Karch CM, Boctor D, Sibih YE, Storm NJ, Diaz A, et al. (2019). A farnesyltransferase inhibitor activates lysosomes and reduces tau pathology in mice with tauopathy. Sci Transl Med 11. 10.1126/scitranslmed.aat3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Wrobel L, Ashkenazi A, Fernandez-Estevez M, Tan K, Bürli RW, and Rubinsztein DC (2021). VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nat Chem Biol. 10.1038/s41589-020-00726-x. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Sanchez E, Hagerman RJ, Mu Y, Nguyen DV, Wong H, Whelan AM, Zukin RS, Klann E, and Tassone F. (2012). Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav 11, 332–341. 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holth JK, Fritschi SK, Wang C, Pedersen NP, Cirrito JR, Mahan TE, Finn MB, Manis M, Geerling JC, Fuller PM, et al. (2019). The sleep-wake cycle regulates brain interstitial fluid tau in mice and CSF tau in humans. Science 363, 880–884. 10.1126/science.aav2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Arakawa S, Yamaguchi H, Torii S, Tajima Sakurai H, Tsujioka M, Murohashi M, and Shimizu S. (2020). Association Between Atg5-independent Alternative Autophagy and Neurodegenerative Diseases. J Mol Biol 432, 2622–2632. 10.1016/j.jmb.2020.01.016. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20, 1981–1991. 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtman J, Freitag K, Gimber N, Schmoranzer J, Heppner FL, and Jendrach M. (2019). Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. Embo Journal 38. ARTN e99430. 10.15252/embj.201899430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomin AC, Gohel R, Hussain Z, Varga A, Maruzs T, Eddison M, Sica M, Jain A, Moffat KG, Johansen T, et al. (2021). Degradation of arouser by endosomal microautophagy is essential for adaptation to starvation in Drosophila. Life Sci Alliance 4. 10.26508/lsa.202000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janen SB, Chaachouay H, and Richter-Landsberg C. (2010). Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia 58, 1766–1774. 10.1002/glia.21047. [DOI] [PubMed] [Google Scholar]
- Jang SY, Shin YK, Park SY, Park JY, Rha SH, Kim JK, Lee HJ, and Park HT (2015). Autophagy is involved in the reduction of myelinating Schwann cell cytoplasm during myelin maturation of the peripheral nerve. PLoS One 10, e0116624. 10.1371/journal.pone.0116624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelani M, Dooley HC, Gubas A, Mohamoud HSA, Khan MTM, Ali Z, Kang C, Rahim F, Jan A, Vadgama N, et al. (2019). A mutation in the major autophagy gene, WIPI2, associated with global developmental abnormalities. Brain 142, 1242–1254. 10.1093/brain/awz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Zhao H, Chen D, Zhang H, and Zhao YG (2021). β-propeller proteins WDR45 and WDR45B regulate autophagosome maturation into autolysosomes in neural cells. Current biology : CB 31, 1666–1677.e1666. 10.1016/j.cub.2021.01.081. [DOI] [PubMed] [Google Scholar]
- Jo DS, Park NY, and Cho DH (2020). Peroxisome quality control and dysregulated lipid metabolism in neurodegenerative diseases. Exp Mol Med 52, 1486–1495. 10.1038/s12276-020-00503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. (2013). Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. New Engl J Med 368, 107–116. 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, and Weihl CC (2009). Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 187, 875–888. 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste YR, Kaushik S, Bourdenx M, Aflakpui R, Bandyopadhyay S, Garcia F, Diaz A, Lindenau K, Tu V, Krause G, et al. (2021). Reciprocal regulation of chaperone-mediated autophagy and the circadian clock. Nat Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuta T, Furuta A, Aoki S, Furuta K, and Wada K. (2008). Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem 283, 23731–23738. 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara NZ, Flaisher-Grinberg S, Anderson GW, Agam G, and Einat H. (2018). Mood-stabilizing effects of rapamycin and its analog temsirolimus: relevance to autophagy. Behav Pharmacol 29, 379–384. 10.1097/FBP.0000000000000334. [DOI] [PubMed] [Google Scholar]
- Kara NZ, Toker L, Agam G, Anderson GW, Belmaker RH, and Einat H. (2013). Trehalose induced antidepressant-like effects and autophagy enhancement in mice. Psychopharmacology (Berl) 229, 367–375. 10.1007/s00213-013-3119-4. [DOI] [PubMed] [Google Scholar]
- Karabiyik C, Lee MJ, and Rubinsztein DC (2017). Autophagy impairment in Parkinson’s disease. Essays in Biochemistry 61. 10.1042/EBC20170023. [DOI] [PubMed] [Google Scholar]
- Karabiyik C, Vicinanza M, Son SM, and Rubinsztein DC (2021). Glucose starvation induces autophagy via ULK1-mediated activation of PIKfyve in an AMPK-dependent manner. Dev Cell. 10.1016/j.devcel.2021.05.010. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, Sahu S, Schwartz GJ, Pessin JE, and Singh R. (2012). Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Reports 13, 258–265. 10.1038/embor.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2018). The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19, 365–381. 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Tasset I, Arias E, Pampliega O, Wong E, Martinez-Vicente M, and Cuervo AM (2021). Autophagy and the hallmarks of aging. Ageing Res Rev 72, 101468. 10.1016/j.arr.2021.101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Cho MH, Shim WH, Kim JK, Jeon EY, Kim DH, and Yoon SY (2017). Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Molecular psychiatry 22, 1576–1584. 10.1038/mp.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, et al. (2016). Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5. 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner P, Bourdenx M, Madrigal-Matute J, Tiano S, Diaz A, Bartholdy BA, Will B, and Cuervo AM (2019). Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol 17, e3000301. 10.1371/journal.pbio.3000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, and Rape M. (2012). The ubiquitin code. Annu Rev Biochem 81, 203–229. 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kononenko NL, Classen GA, Kuijpers M, Puchkov D, Maritzen T, Tempes A, Malik AR, Skalecka A, Bera S, Jaworski J, and Haucke V. (2017). Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat Commun 8, 14819. 10.1038/ncomms14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, and Rubinsztein DC (2009). Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33, 517–527. 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, You Z, Rong J, Kress C, Huang X, Yang J, Kyoda T, Leyva R, Banares S, Hu Y, et al. (2011). Neuronal deletion of caspase 8 protects against brain injury in mouse models of controlled cortical impact and kainic acid-induced excitotoxicity. PLoS One 6, e24341. 10.1371/journal.pone.0024341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GJ, and Cuervo AM (2021). Assessment of mammalian endosomal microautophagy. Methods Cell Biol 164, 167–185. 10.1016/bs.mcb.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, Kochlamazashvili G, Stumpf A, Puchkov D, Swaminathan A, Lucht MT, Krause E, Maritzen T, Schmitz D, and Haucke V. (2021). Neuronal Autophagy Regulates Presynaptic Neurotransmission by Controlling the Axonal Endoplasmic Reticulum. Neuron 109, 299–313 e299. 10.1016/j.neuron.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni VV, and Maday S. (2018). Compartment-specific dynamics and functions of autophagy in neurons. Dev Neurobiol 78, 298–310. 10.1002/dneu.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance V, Wang Q, Sweet E, Choi I, Cai CZ, Zhuang XX, Zhang Y, Jiang JL, Blitzer RD, Bozdagi-Gunal O, et al. (2019). Autophagy protein NRBF2 has reduced expression in Alzheimer’s brains and modulates memory and amyloid-beta homeostasis in mice. Mol Neurodegener 14, 43. 10.1186/s13024-019-0342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen I, Pardossi-Piquard R, Bourgeois A, Pagnotta S, Biferi MG, Barkats M, Lacor P, Klein W, Bauer C, and Checler F. (2016). Intraneuronal aggregation of the beta-CTF fragment of APP (C99) induces Abeta-independent lysosomal-autophagic pathology. Acta Neuropathol 132, 257–276. 10.1007/s00401-016-1577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guerroue F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, and Behrends C. (2017). Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell 68, 786–796 e786. 10.1016/j.molcel.2017.10.029. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Yoon YS, and Lee SJ (2018). Mechanism of neuroprotection by trehalose: controversy surrounding autophagy induction. Cell Death Dis 9, 712. 10.1038/s41419-018-0749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, and Gao FB (2007). ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Current biology : CB 17, 1561–1567. 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. (2010). Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158. 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Sumpter R, Zou Z, Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI, and Levine B. (2017). Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep 18, 48–60. 10.15252/embr.201642443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Lai DM, Huang HJ, Lee-Chen GJ, Chang CH, Hsieh-Li HM, and Lee GC (2021). Prebiotic Lactulose Ameliorates the Cognitive Deficit in Alzheimer’s Disease Mouse Model through Macroautophagy and Chaperone-Mediated Autophagy Pathways. J Agric Food Chem 69, 2422–2437. 10.1021/acs.jafc.0c07327. [DOI] [PubMed] [Google Scholar]
- Li C, Wang X, Li X, Qiu K, Jiao F, Liu Y, Kong Q, and Wu Y. (2019a). Proteasome Inhibition Activates Autophagy-Lysosome Pathway Associated With TFEB Dephosphorylation and Nuclear Translocation. Front Cell Dev Biol 7, 170. 10.3389/fcell.2019.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ham A, Ma TC, Kuo SH, Kanter E, Kim D, Ko HS, Quan Y, Sardi SP, Li A, et al. (2019b). Mitochondrial dysfunction and mitophagy defect triggered by heterozygous GBA mutations. Autophagy 15, 113–130. 10.1080/15548627.2018.1509818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kim SG, and Blenis J. (2014). Rapamycin: one drug, many effects. Cell metabolism 19, 373–379. 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang C, Wang Z, Zhu C, Li J, Sha T, Ma L, Gao C, Yang Y, Sun Y, et al. (2019c). Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 575, 203–209. 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, and Guan JL (2010). Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285, 3499–3509. 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, and Corn JE (2018). Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol 217, 3354–3367. 10.1083/jcb.201804185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman OJ, and Sulzer D. (2019). The Synaptic Autophagy Cycle. J Mol Biol. 10.1016/j.jmb.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, and Yuan J. (2010). Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 107, 14164–14169. 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dai C, Fan Y, Guo B, Ren K, Sun T, and Wang W. (2017). From autophagy to mitophagy: the roles of P62 in neurodegenerative diseases. J Bioenerg Biomembr 49, 413–422. 10.1007/s10863-017-9727-7. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhao H, Zhao YG, Hu J, and Zhang H. (2021). Atlastin 2/3 regulate ER targeting of the ULK1 complex to initiate autophagy. J Cell Biol 220. 10.1083/jcb.202012091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan T, Simon MJ, Rana A, Cherf GM, Srivastava A, Davis SS, Low RLY, Chiu CL, Fang M, Huang F, et al. (2021). Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell 184, 4651–4668 e4625. 10.1016/j.cell.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi M, Raimondi A, Morone D, and Molinari M. (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun 10, 5058. 10.1038/s41467-019-12991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A, Fleming A, and Rubinsztein DC (2018). Seeing is believing: methods to monitor vertebrate autophagy in vivo. Open Biol 8. 10.1098/rsob.180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, den Brave F, and Jentsch S. (2017). Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol 19, 732–739. 10.1038/ncb3531. [DOI] [PubMed] [Google Scholar]
- Lucin KM, O’Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E, and Wyss-Coray T. (2013). Microglial Beclin 1 Regulates Retromer Trafficking and Phagocytosis and Is Impaired in Alzheimer’s Disease. Neuron 79, 873–886. 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis-Ravelo D, Estevez-Silva H, Barroso-Chinea P, Afonso-Oramas D, Salas-Hernandez J, Rodriguez-Nunez J, Acevedo-Arozena A, Marcellino D, and Gonzalez-Hernandez T. (2018). Pramipexole reduces soluble mutant huntingtin and protects striatal neurons through dopamine D3 receptors in a genetic model of Huntington’s disease. Exp Neurol 299, 137–147. 10.1016/j.expneurol.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Luo R, Su LY, Li G, Yang J, Liu Q, Yang LX, Zhang DF, Zhou H, Xu M, Fan Y, et al. (2020). Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 16, 52–69. 10.1080/15548627.2019.1596488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Otomo C, and Otomo T. (2019). The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8. 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes J, Gegg ME, Migdalska-Richards A, Doherty MK, Whitfield PD, and Schapira AHV (2016). Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: Relevance to Parkinson disease. Human molecular genetics 25, 3432–3445. 10.1093/hmg/ddw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid T, Ali YO, Venkitaramani DV, Jang MK, Lu HC, and Pautler RG (2014). In vivo axonal transport deficits in a mouse model of fronto-temporal dementia. Neuroimage Clin 4, 711–717. 10.1016/j.nicl.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M. (2017). Neuronal Mitophagy in Neurodegenerative Diseases. Front Mol Neurosci 10, 64. 10.3389/fnmol.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, et al. (2008). Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest 118, 777–788. 10.1172/jci32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, and Cuervo AM (2010). Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 13, 567–576. 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al. (2020). Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27, 1185–1193. 10.1038/s41594-020-00518-w. [DOI] [PubMed] [Google Scholar]
- Matsuda N. (2016). Phospho-ubiquitin: upending the PINK-Parkin-ubiquitin cascade. J Biochem 159, 379–385. 10.1093/jb/mvv125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Olsvik H, Svenning S, Bruun JA, Abudu YP, Larsen KB, Brech A, Hansen TE, Brenne H, Hansen T, et al. (2018). Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol 217, 3640–3655. 10.1083/jcb.201711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Fullgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, et al. (2017). Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 93, 1015–1034. 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- Mesquita A, Glenn J, and Jenny A. (2020). Differential activation of eMI by distinct forms of cellular stress. Autophagy, 1–13. 10.1080/15548627.2020.1783833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld T, and Schwarz TL (2017). Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron 96, 651–666. 10.1016/j.neuron.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, and Isaacs AM (2013). C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol 126, 845–857. 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, and Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132. 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Moore AS, and Holzbaur EL (2016). Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A 113, E3349–3358. 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales I, Sanchez A, Puertas-Avendano R, Rodriguez-Sabate C, Perez-Barreto A, and Rodriguez M. (2020). Neuroglial transmitophagy and Parkinson’s disease. Glia 68, 2277–2299. 10.1002/glia.23839. [DOI] [PubMed] [Google Scholar]
- Moreau K, Fleming A, Imarisio S, Lopez Ramirez A, Mercer JL, Jimenez-Sanchez M, Bento CF, Puri C, Zavodszky E, Siddiqi F, et al. (2014). PICALM modulates autophagy activity and tau accumulation. Nat Commun 5, 4998. 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova K, Clement CC, Kaushik S, Stiller B, Arias E, Ahmad A, Rauch JN, Chatterjee V, Melis C, Scharf B, et al. (2016). Structural and Biological Interaction of hsc-70 Protein with Phosphatidylserine in Endosomal Microautophagy. J Biol Chem 291, 18096–18106. 10.1074/jbc.M116.736744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Patel B, Koga H, Cuervo AM, and Jenny A. (2016). Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984–1999. 10.1080/15548627.2016.1208887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KE, Gysbers AM, Abbott SK, Tayebi N, Kim WS, Sidransky E, Cooper A, Garner B, and Halliday GM (2014). Reduced glucocerebrosidase is associated with increased α-synuclein in sporadic Parkinson’s disease. Brain 137, 834–848. 10.1093/brain/awt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, and Brown EJ (2009). PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10, 173–179. 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, and Youle RJ (2010). p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106. 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie T, Tao K, Zhu L, Huang L, Hu S, Yang R, Xu P, Mao Z, and Yang Q. (2020). Chaperone-mediated autophagy controls the turnover of E3 ubiquitin ligase MARCHF5 and regulates mitochondrial dynamics. Autophagy, 1–16. 10.1080/15548627.2020.1848128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Sidiropoulou K, Kallergi E, Dalezios Y, and Tavernarakis N. (2017). Modulation of Autophagy by BDNF Underlies Synaptic Plasticity. Cell metabolism 26, 230–242 e235. 10.1016/j.cmet.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, and Helmchen F. (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, and Shimizu S. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658. 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Nixon RA (2013). The role of autophagy in neurodegenerative disease. Nat Med 19, 983–997. 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Nixon RA (2020). The aging lysosome: An essential catalyst for late-onset neurodegenerative diseases. Biochim Biophys Acta Proteins Proteom 1868, 140443. 10.1016/j.bbapap.2020.140443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obri A, Khrimian L, Karsenty G, and Oury F. (2018). Osteocalcin in the brain: from embryonic development to age-related decline in cognition. Nat Rev Endocrinol 14, 174–182. 10.1038/nrendo.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL, Yeung SY, Humbert S, et al. (2014). Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A 111, 16889–16894. 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerlund ND, Schneider K, Leal-Ortiz S, Montenegro-Venegas C, Kim SA, Garner LC, Waites CL, Gundelfinger ED, Reimer RJ, and Garner CC (2017). Bassoon Controls Presynaptic Autophagy through Atg5. Neuron 93, 897–913 e897. 10.1016/j.neuron.2017.01.026. [DOI] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, et al. (2013). Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 16, 394–406. 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, and Okamoto K. (2011). Optineurin in neurodegenerative diseases. Neuropathology 31, 569–574. 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- Oz-Levi D, Ben-Zeev B, Ruzzo EK, Hitomi Y, Gelman A, Pelak K, Anikster Y, Reznik-Wolf H, Bar-Joseph I, Olender T, et al. (2012). Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet 91, 1065–1072. 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva SL, and Crews CM (2019). Targeted protein degradation: elements of PROTAC design. Curr Opin Chem Biol 50, 111–119. 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jiménez-Moreno N, García-Yagüe Á, Escoll M, de Ceballos ML, Van Leuven F, Rábano A, Yamamoto M, Rojo AI, and Cuadrado A. (2016). Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12, 1902–1916. 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Chen D, Zhang A, Qin X, and Yan B. (2012). Genetic analysis of the LAMP-2 gene promoter in patients with sporadic Parkinson’s disease. Neuroscience letters 526, 63–67. 10.1016/j.neulet.2012.07.044. [DOI] [PubMed] [Google Scholar]
- Pang W, and Hu F. (2021). Cellular and physiological functions of C9ORF72 and implications for ALS/FTD. J Neurochem 157, 334–350. 10.1111/jnc.15255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C, Kravic B, and Meyer H. (2020). Repair or Lysophagy: Dealing with Damaged Lysosomes. J Mol Biol 432, 231–239. 10.1016/j.jmb.2019.08.010. [DOI] [PubMed] [Google Scholar]
- Parcon PA, Balasubramaniam M, Ayyadevara S, Jones RA, Liu L, Reis RJS, Barger SW, Mrak RE, and Griffin WST (2018). Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement 14, 230–242. 10.1016/j.jalz.2017.07.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim DH, and Yoon SY (2016a). Regulation of amyloid precursor protein processing by its KFERQ motif. BMB Rep 49, 337–342. 10.5483/bmbrep.2016.49.6.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Han S, Choi I, Kim B, Park SP, Joe EH, and Suh YH (2016b). Interplay between Leucine-Rich Repeat Kinase 2 (LRRK2) and p62/SQSTM-1 in Selective Autophagy. PLoS One 11, e0163029. 10.1371/journal.pone.0163029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Frake RA, Karabiyik C, Son SM, Siddiqi FH, Bento CF, Sterk P, Vicinanza M, Pavel M, Rubinsztein DC (2021). Vinexin contributes to autophagic decline in brain ageing across species. Cell Death and Differentiation (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N, Vainshtein A, Herz NJ, Huynh T, Brunetti L, Klisch TJ, Mutarelli M, Annunziata P, Kinouchi K, Brunetti-Pierri N, et al. (2019). Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. EMBO J 38. 10.15252/embj.2018101347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Avila J, and Hernandez F. (2019). Propagation of Tau via Extracellular Vesicles. Front Neurosci 13, 698. 10.3389/fnins.2019.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, and Wyss-Coray T. (2008). The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 118, 2190–2199. 10.1172/Jci33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. (2012). TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234. 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia SK, Yuan C, Khan MR, Karuppagounder SS, Wang L, Xiong Y, Kang SU, Lee Y, Dawson VL, and Dawson TM (2020). PARIS induced defects in mitochondrial biogenesis drive dopamine neuron loss under conditions of parkin or PINK1 deficiency. Mol Neurodegener 15, 17. 10.1186/s13024-020-00363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, et al. (2003). Mutant dynactin in motor neuron disease. Nat Genet 33, 455–456. 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Puri C, Manni MM, Vicinanza M, Hilcenko C, Zhu Y, Runwal G, Stamatakou E, Menzies FM, Mamchaoui K, Bitoun M, and Rubinsztein DC (2020). A DNM2 Centronuclear Myopathy Mutation Reveals a Link between Recycling Endosome Scission and Autophagy. Dev Cell 53, 154–168 e156. 10.1016/j.devcel.2020.03.018. [DOI] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, and Rubinsztein DC (2013). Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299. 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, Bento CF, Renna M, Menzies FM, and Rubinsztein DC (2018). The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev Cell 45, 114–131 e118. 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, and Jung YK (2013). Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nature Communications 4, 1–9. 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Li YI, Wong G, Humphrey J, Wang M, Ramdhani S, Wang YC, Ng B, Gupta I, Haroutunian V, et al. (2018). Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet 50, 1584–1592. 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, and Rubinsztein DC (2005). Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 37, 771–776. 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, and Rubinsztein DC (2008). Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 121, 1649–1660. 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, and Rey G. (2014). Metabolic and nontranscriptional circadian clocks: eukaryotes. Annu Rev Biochem 83, 165–189. 10.1146/annurev-biochem-060713-035623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro FM, Devries RA, Hamilton A, Guimaraes IM, Cregan SP, Pires RG, and Ferguson SS (2014). Metabotropic glutamate receptor 5 knockout promotes motor and biochemical alterations in a mouse model of Huntington’s disease. Human molecular genetics 23, 2030–2042. 10.1093/hmg/ddt598. [DOI] [PubMed] [Google Scholar]
- Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, et al. (2016). Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 113, 4039–4044. 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, and Boya P. (2013). Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 12, 478–488. 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, and Monteiro MJ (2010). Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Human molecular genetics 19, 3219–3232. 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, and Orr HT (2016). Diminishing return for mechanistic therapeutics with neurodegenerative disease duration?: There may be a point in the course of a neurodegenerative condition where therapeutics targeting disease-causing mechanisms are futile. Bioessays 38, 977–980. 10.1002/bies.201600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick ND, Griffey CJ, Guarnieri P, Gerbino V, Wang X, Piersaint JA, Tapia JC, Rich MM, and Maniatis T. (2017). Distinct roles for motor neuron autophagy early and late in the SOD1(G93A) mouse model of ALS. Proc Natl Acad Sci U S A 114, E8294–E8303. 10.1073/pnas.1704294114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. (2015). Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol 17, 262–275. 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskalin L, Limanaqi F, Frati A, Busceti CL, and Fornai F. (2018). mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders. Int J Mol Sci 19. 10.3390/ijms19082226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, and Santambrogio L. (2011). Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20, 131–139. 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, et al. (2013). De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45, 445–449, 449e441. 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- Sala G, Marinig D, Arosio A, and Ferrarese C. (2016). Role of Chaperone-Mediated Autophagy Dysfunctions in the Pathogenesis of Parkinson’s Disease. Front Mol Neurosci 9, 157. 10.3389/fnmol.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin P, Lahuerta M, Viana R, Knecht E, and Sanz P. (2020). Regulation of the autophagic PI3KC3 complex by laforin/malin E3-ubiquitin ligase, two proteins involved in Lafora disease. Biochim Biophys Acta Mol Cell Res 1867, 118613. 10.1016/j.bbamcr.2019.118613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Olsen AL, Sygnecka K, Lohr KM, and Feany MB (2021). Α-Synuclein Impairs Autophagosome Maturation Through Abnormal Actin Stabilization. PLoS Genetics 17, 1–26. 10.1371/JOURNAL.PGEN.1009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Shah HV, Kanfer G, Pickrell AM, Holtzclaw LA, Ward ME, and Youle RJ (2020). Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. Mol Cell 80, 779–795 e710. 10.1016/j.molcel.2020.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, Villarroya F, Santambrogio L, and Cuervo AM (2015). Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264. 10.1111/acel.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöndorf DC, Aureli M, Mcallister FE, Hindley CJ, Mayer F, Scho DC, Schmid B, Sardi SP, Valsecchi M, Hoffmann S, et al. (2014). iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun 5, 1–17. 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- Schreiber SL (2021). The Rise of Molecular Glues. Cell 184, 3–9. 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- Schuck S. (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133. 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- Scrivo A, Bourdenx M, Pampliega O, and Cuervo AM (2018). Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol 17, 802–815. 10.1016/S1474-4422(18)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, and Wooten MW (2004). Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24, 8055–8068. 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şentürk M, Lin G, Zuo Z, Mao D, Watson E, Mikos AG, and Bellen HJ (2019). Ubiquilins regulate autophagic flux through mTOR signalling and lysosomal acidification. Nat Cell Biol 21, 384–396. 10.1038/s41556-019-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, and Ballabio A. (2013). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296. 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Schnell HM, Ruoff K, and Goldberg A. (2018). Rapid induction of p62 and GABARAPL1 upon proteasome inhibition promotes survival before autophagy activation. J Cell Biol 217, 1757–1776. 10.1083/jcb.201708168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, and Inokuchi K. (2012). Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. The Journal of neuroscience: the official journal of the Society for Neuroscience 32, 10413–10422. 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Li HY, Chen GC, Chern Y, and Tu PH (2015). Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 11, 685–700. 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Li Y, Gasparski AN, Abeliovich H, and Greenberg ML (2017). Cardiolipin Regulates Mitophagy through the Protein Kinase C Pathway. J Biol Chem 292, 2916–2923. 10.1074/jbc.M116.753574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, and Dawson TM (2011). PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell 144, 689–702. 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi FH, Menzies FM, Lopez A, Stamatakou E, Karabiyik C, Ureshino R, Ricketts T, Jimenez-Sanchez M, Esteban MA, Lai LX, et al. (2019). Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nature Communications 10. ARTN 1817. 10.1038/s41467-019-09494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovitch S, Schmukler E, Bespalko A, Iram T, Frenkel D, Holtzman DM, Masliah E, Michaelson DM, and Pinkas-Kramarski R. (2016). Impaired Autophagy in APOE4 Astrocytes. J Alzheimers Dis 51, 915–927. 10.3233/Jad-151101. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, and Finley KD (2008). Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Son SM, Park SJ, Stamatakou E, Vicinanza M, Menzies FM, and Rubinsztein DC (2020). Leucine regulates autophagy via acetylation of the mTORC1 component raptor. Nat Commun 11, 3148. 10.1038/s41467-020-16886-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GJ, Jeon H, Seo M, Jo M, and Suk K. (2018). Interaction between optineurin and Rab1a regulates autophagosome formation in neuroblastoma cells. J Neurosci Res 96, 407–415. 10.1002/jnr.24143. [DOI] [PubMed] [Google Scholar]
- Spillantini M, Schmidt M, Lee V, Trojanowski J, Jakes R, and Goedert M. (1997). α-Synuclein in Lewy bodies. Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- Stadel D, Millarte V, Tillmann KD, Huber J, Tamin-Yecheskel BC, Akutsu M, Demishtein A, Ben-Zeev B, Anikster Y, Perez F, et al. (2015). TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export. Mol Cell 60, 89–104. 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Stamatakou E, Wróbel L, Hill SM, Puri C, Son SM, Fujimaki M, Zhu Y, Siddiqi F, Fernandez-Estevez M, Manni MM, et al. (2020). Mendelian neurodegenerative disease genes involved in autophagy. Cell Discov 6, 24. 10.1038/s41421-020-0158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, and Holzbaur ELF (2019). Autophagy in Neurons. Annu Rev Cell Dev Biol 35, 477–500. 10.1146/annurev-cellbio-100818-125242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckley D, Karajgikar M, Dale LB, Fuerth B, Swan P, Drummond-Main C, Poulter MO, Ferguson SS, Strasser A, and Cregan SP (2007). Puma is a dominant regulator of oxidative stress induced Bax activation and neuronal apoptosis. The Journal of neuroscience: the official journal of the Society for Neuroscience 27, 12989–12999. 10.1523/JNEUROSCI.3400-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanis L, Larsen KE, Rideout HJ, Sulzer D, and Greene LA (2001). Expression of A53T mutant but not wild-type α-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. Journal of Neuroscience 21, 9549–9560. 10.1523/jneurosci.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DA, Lee Y, Kang HC, Lee BD, Lee YI, Bower A, Jiang H, Kang SU, Andrabi SA, Dawson VL, et al. (2015). Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc Natl Acad Sci U S A 112, 11696–11701. 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stykel MG, Humphries KM, Kamski-Hennekam E, Buchner-Duby B, Porte-Trachsel N, Ryan T, Coackley CL, Bamm VV, Harauz G, and Ryan SD (2021). alpha-Synuclein mutation impairs processing of endomembrane compartments and promotes exocytosis and seeding of alpha-synuclein pathology. Cell Rep 35, 109099. 10.1016/j.celrep.2021.109099. [DOI] [PubMed] [Google Scholar]
- Sun-Wang JL, Ivanova S, and Zorzano A. (2020). The dialogue between the ubiquitin-proteasome system and autophagy: Implications in ageing. Ageing Res Rev 64, 101203. 10.1016/j.arr.2020.101203. [DOI] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, and Finkel T. (2015). Measuring In Vivo Mitophagy. Mol Cell 60, 685–696. 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy V, Walker AK, Tan V, Fifita JA, Mccann EP, Williams KL, Blair IP, Guillemin GJ, Farg MA, and Atkin JD (2015). Defects in optineurin- and myosin VI-mediated cellular trafficking in amyotrophic lateral sclerosis. Human molecular genetics 24, 3830–3846. 10.1093/hmg/ddv126. [DOI] [PubMed] [Google Scholar]
- Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, Akaike T, Itto-Nakama K, and Arimoto H. (2019). AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol Cell 76, 797–810 e710. 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, He H, Tang Z, Hattori T, Liu Y, Young MM, Serfass JM, Chen L, Gebru M, Chen C, et al. (2018). An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9, 2855. 10.1038/s41467-018-05254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki Y, Shodai A, Morimura T, Hikiami R, Minamiyama S, Ayaki T, Tooyama I, Furukawa Y, Takahashi R, and Urushitani M. (2018). Elimination of TDP-43 inclusions linked to amyotrophic lateral sclerosis by a misfolding-specific intrabody with dual proteolytic signals. Sci Rep 8, 6030. 10.1038/s41598-018-24463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F-L, Erion JR, Tian Y, Liu W, Yin D-M, Ye J, Tang B, Mei L, and Xiong W-C (2015). VPS35 in Dopamine Neurons Is Required for Endosome-to-Golgi Retrieval of Lamp2a, a Receptor of Chaperone-Mediated Autophagy That Is Critical for α-Synuclein Degradation and Prevention of Pathogenesis of Parkinson’s Disease. The Journal of neuroscience 35, 10613–10628. 10.1523/JNEUROSCI.0042-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, et al. (2014). Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131–1143. 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Yue Z, Talloczy Z, Hagemann T, Cho W, Messing A, Sulzer DL, and Goldman JE (2008). Autophagy induced by Alexander disease-mutant GFAP accumulation is regulated by p38/MAPK and mTOR signaling pathways. Human molecular genetics 17, 1540–1555. 10.1093/hmg/ddn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanik SA, Schultheiss CE, Volpicelli-Daley LA, Brunden KR, and Lee VM (2013). Lewy body-like α-synuclein aggregates resist degradation and impair macroautophagy. J Biol Chem 288, 15194–15210. 10.1074/jbc.M113.457408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn J, Andrysik Z, Staskiewicz L, Gump J, Maycotte P, Oberst A, Green DR, Espinosa JM, and Thorburn A. (2014). Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep 7, 45–52. 10.1016/j.celrep.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Chang JC, Fan EY, Flajolet M, and Greengard P. (2013). Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci U S A 110, 17071–17076. 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, and Taylor JP (2010). VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6, 217–227. 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, and Buss F. (2012). Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol 14, 1024–1035. 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, and Wang J. (2016). Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet 12, e1006443. 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytterhoeven V, Lauwers E, Maes I, Miskiewicz K, Melo MN, Swerts J, Kuenen S, Wittocx R, Corthout N, Marrink SJ, et al. (2015). Hsc70–4 Deforms Membranes to Promote Synaptic Protein Turnover by Endosomal Microautophagy. Neuron 88, 735–748. 10.1016/j.neuron.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Valek L, Heidler J, Scheving R, Wittig I, and Tegeder I. (2019). Nitric oxide contributes to protein homeostasis by S-nitrosylations of the chaperone HSPA8 and the ubiquitin ligase UBE2D. Redox Biol 20, 217–235. 10.1016/j.redox.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waarde A, Marcolini S, de Deyn PP, and Dierckx R. (2021). PET Agents in Dementia: An Overview. Semin Nucl Med 51, 196–229. 10.1053/j.semnuclmed.2020.12.008. [DOI] [PubMed] [Google Scholar]
- Vantaggiato C, Crimella C, Airoldi G, Polishchuk R, Bonato S, Brighina E, Scarlato M, Musumeci O, Toscano A, Martinuzzi A, et al. (2013). Defective autophagy in spastizin mutated patients with hereditary spastic paraparesis type 15. Brain 136, 3119–3139. 10.1093/brain/awt227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantaggiato C, Panzeri E, Castelli M, Citterio A, Arnoldi A, Santorelli FM, Liguori R, Scarlato M, Musumeci O, Toscano A, et al. (2019). ZFYVE26/SPASTIZIN and SPG11/SPATACSIN mutations in hereditary spastic paraplegia types AR-SPG15 and AR-SPG11 have different effects on autophagy and endocytosis. Autophagy 15, 34–57. 10.1080/15548627.2018.1507438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Silva J, Gomes P, Jin Q, Zhu M, Zhuravleva V, Quintremil S, Meira T, Silva J, Dioli C, Soares-Cunha C, et al. (2018). Endolysosomal degradation of Tau and its role in glucocorticoid-driven hippocampal malfunction. EMBO J 37. 10.15252/embj.201899084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, et al. (2003). Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet 72, 722–727. 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, and Rubinsztein DC (2015). PI(5)P regulates autophagosome biogenesis. Mol Cell 57, 219–234. 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Gamble KL, Schultheiss CE, Riddle DM, West AB, and Lee VM (2014). Formation of α-synuclein Lewy neurite-like aggregates in axons impedes the transport of distinct endosomes. Mol Biol Cell 25, 4010–4023. 10.1091/mbc.E14-02-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallings R, Connor-Robson N, and Wade-Martins R. (2019). LRRK2 interacts with the vacuolar-type H+-ATPase pump a1 subunit to regulate lysosomal function. Human molecular genetics 28, 2696–2710. 10.1093/hmg/ddz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Cai Z, Tao K, Zeng W, Lu F, Yang R, Feng D, Gao G, and Yang Q. (2016). Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy 12, 1215–1228. 10.1080/15548627.2016.1179401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Cao YL, Li Q, Yang YP, Jin M, Chen D, Wang F, Wang GH, Qin ZH, Hu LF, and Liu CF (2015). A pivotal role of FOS-mediated BECN1/Beclin 1 upregulation in dopamine D2 and D3 receptor agonist-induced autophagy activation. Autophagy 11, 2057–2073. 10.1080/15548627.2015.1100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tan J, Chen T, Han H, Tian R, Tan Y, Wu Y, Cui J, Chen F, Li J, et al. (2019a). ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome–lysosome fusion. Journal of Cell Biology 218, 267–284. 10.1083/jcb.201804165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Yu J, Wong SH, Cheng AS, Chan FK, Ng SS, Cho CH, Sung JJ, and Wu WK (2013). A novel crosstalk between two major protein degradation systems: regulation of proteasomal activity by autophagy. Autophagy 9, 1500–1508. 10.4161/auto.25573. [DOI] [PubMed] [Google Scholar]
- Wang ZT, Lu MH, Zhang Y, Ji WL, Lei L, Wang W, Fang LP, Wang LW, Yu F, Wang J, et al. (2019b). Disrupted-in-schizophrenia-1 protects synaptic plasticity in a transgenic mouse model of Alzheimer’s disease as a mitophagy receptor. Aging Cell 18, e12860. 10.1111/acel.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, and Rubinsztein DC (2003). Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278, 25009–25013. 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, et al. (2016). The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 35, 1656–1676. 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Westergard T, Pasinelli P, and Trotti D. (2017). Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene. Neuroscience letters 636, 16–26. 10.1016/j.neulet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P, Fatou B, Guise AJ, Cheng L, Takeda S, et al. (2020). Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer’s Disease. Cell 183, 1699–1713 e1613. 10.1016/j.cell.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm T, Byrne J, Medina R, Kolundzic E, Geisinger J, Hajduskova M, Tursun B, and Richly H. (2017). Neuronal inhibition of the autophagy nucleation complex extends lifespan in post-reproductive C. elegans. Genes & development 31, 1561–1572. 10.1101/135178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, et al. (2010). α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. Journal of Cell Biology 190, 1023–1037. 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, and Nixon RA (2013). Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci 37, 1949–1961. 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, and Holzbaur EL (2014a). Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111, E4439–4448. 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, and Holzbaur EL (2014b). The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. The Journal of neuroscience: the official journal of the Society for Neuroscience 34, 1293–1305. 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AG, Zhou XG, Qiao G, Yu L, Tang Y, Yan L, Qiu WQ, Pan R, Yu CL, Law BY, et al. (2021). Targeting microglial autophagic degradation in NLRP3 inflammasome-mediated neurodegenerative diseases. Ageing Res Rev 65, 101202. 10.1016/j.arr.2020.101202. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Cai A, Greenslade JE, Higgins NR, Fan C, Le NTT, Tatman M, Whiteley AM, Prado MA, Dieriks BV, et al. (2020). ALS/FTD mutations in UBQLN2 impede autophagy by reducing autophagosome acidification through loss of function. Proc Natl Acad Sci U S A 117, 15230–15241. 10.1073/pnas.1917371117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, Shan B, Pan H, and Yuan J. (2019). Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A 116, 2996–3005. 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, Ren H, Chen D, Han J, Ying Z, and Wang G. (2016). TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. EMBO J 35, 121–142. 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xilouri M, Brekk OR, Landeck N, Pitychoutis PM, Papasilekas T, Papadopoulou-Daifoti Z, Kirik D, and Stefanis L. (2013). Boosting chaperone-mediated autophagy in vivo mitigates alpha-synuclein-induced neurodegeneration. Brain 136, 2130–2146. 10.1093/brain/awt131. [DOI] [PubMed] [Google Scholar]
- Xilouri M, Brekk OR, Polissidis A, Chrysanthou-Piterou M, Kloukina I, and Stefanis L. (2016). Impairment of chaperone-mediated autophagy induces dopaminergic neurodegeneration in rats. Autophagy 12, 2230–2247. 10.1080/15548627.2016.1214777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CY, Kang WY, Chen YM, Jiang TF, Zhang J, Zhang LN, Ding JQ, Liu J, and Chen SD (2017). DJ-1 Inhibits α-Synuclein Aggregation by Regulating Chaperone-Mediated Autophagy. Front Aging Neurosci 9, 308. 10.3389/fnagi.2017.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Propson NE, Du S, Xiong W, and Zheng H. (2021). Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2023418118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang S, and Zheng H. (2019). The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau). Autophagy 15, 583–598. 10.1080/15548627.2018.1532258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Honda S, Torii S, Shimizu K, Katoh K, Miyake K, Miyake N, Fujikake N, Sakurai HT, Arakawa S, and Shimizu S. (2020). Wipi3 is essential for alternative autophagy and its loss causes neurodegeneration. Nat Commun 11, 5311. 10.1038/s41467-020-18892-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J, Suzuki C, Nanao T, Kakuta S, Ozawa K, Tanida I, Saitoh T, Sunabori T, Komatsu M, Tanaka K, et al. (2018). Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy 14, 764–777. 10.1080/15548627.2017.1314897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Porch MW, Court-Vazquez B, Bennett MVL, and Zukin RS (2018). Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc Natl Acad Sci U S A 115, E9707–E9716. 10.1073/pnas.1808247115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen P, Liu J, Zhu S, Kroemer G, Klionsky DJ, Lotze MT, Zeh HJ, Kang R, and Tang D. (2019). Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv 5, eaaw2238. 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, and Mao Z. (2009). Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124–127. 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, et al. (2001). The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29, 160–165. 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- Yao PJ, Eren E, Petralia RS, Gu JW, Wang YX, and Kapogiannis D. (2020). Mitochondrial Protrusions in Neuronal Cells. iScience 23, 101514. 10.1016/j.isci.2020.101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Popelka H, Lei Y, Yang Y, and Klionsky DJ (2020). The Roles of Ubiquitin in Mediating Autophagy. Cells 9. 10.3390/cells9092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Kumar A, Peterhoff C, Shapiro Kulnane L, Uchiyama Y, Lamb BT, Cuervo AM, and Nixon RA (2004). Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide over-production and localization in Alzheimer’s disease. Int J Biochem Cell Biol 36, 2531–2540. 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, and Rubinsztein DC (2014). Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun 5, 1–16. 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Garbett K, Veeraraghavalu K, Wilburn B, Gilmore R, Mirnics K, and Sisodia SS (2012). A Role for Presenilins in Autophagy Revisited: Normal Acidification of Lysosomes in Cells Lacking PSEN1 and PSEN2. Journal of Neuroscience 32, 8633–8648. 10.1523/Jneurosci.0556-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Sun L, Miao G, Ji C, Zhao H, Sun H, Miao L, Yoshii SR, Mizushima N, Wang X, and Zhang H. (2015). The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 11, 881–890. 10.1080/15548627.2015.1047127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, and Conrad M. (2020). The Metabolic Underpinnings of Ferroptosis. Cell metabolism 32, 920–937. 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhong MH, Elder GA, Sano M, Holtzman DM, Gandy S, Cardozo C, Haroutunian V, Robakis NK, and Cai DM (2015). Phospholipid dysregulation contributes to ApoE4-associated cognitive deficits in Alzheimer’s disease pathogenesis. P Natl Acad Sci USA 112, 11965–11970. 10.1073/pnas.1510011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. (2011). A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet 89, 168–175. 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]