Abstract
Introduction
The Coronavirus Disease 2019 (COVID-19) pandemic caused by Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2) emerged in late December 2019. Considering the important role of gut microbiota in maturation, regulation, and induction of the immune system and subsequent inflammatory processes, it seems that evaluating the composition of gut microbiota in COVID-19 patients compared with healthy individuals may have potential value as a diagnostic and/or prognostic biomarker for the disease. Also, therapeutic interventions affecting gut microbial flora may open new horizons in the treatment of COVID-19 patients and accelerating their recovery.
Methods
A systematic search was conducted for relevant studies published from December 2019 to December 2021 using Pubmed/Medline, Embase, and Scopus. Articles containing the following keywords in titles or abstracts were selected: “SARS-CoV-2” or “COVID-19” or “Coronavirus Disease 19” and “gastrointestinal microbes” or “dysbiosis” or “gut microbiota” or “gut bacteria” or “gut microbes” or “gastrointestinal microbiota”.
Results
Out of 1,668 studies, 22 articles fulfilled the inclusion criteria and a total of 1,255 confirmed COVID-19 patients were examined. All included studies showed a significant association between COVID-19 and gut microbiota dysbiosis. The most alteration in bacterial composition of COVID-19 patients was depletion in genera Ruminococcus, Alistipes, Eubacterium, Bifidobacterium, Faecalibacterium, Roseburia, Fusicathenibacter, and Blautia and enrichment of Eggerthella, Bacteroides, Actinomyces, Clostridium, Streptococcus, Rothia, and Collinsella. Also, some gut microbiome alterations were associated with COVID-19 severity and poor prognosis including the increment of Bacteroides, Parabacteroides, Clostridium, Bifidobacterium, Ruminococcus, Campylobacter, Rothia, Corynebacterium, Megasphaera, Enterococcus, and Aspergillus spp. and the decrement of Roseburia, Eubacterium, Lachnospira, Faecalibacterium, and the Firmicutes/Bacteroidetes ratio.
Conclusion
Our study showed a significant change of gut microbiome composition in COVID-19 patients compared with healthy individuals. This great extent of impact has proposed the gut microbiota as a potential diagnostic, prognostic, and therapeutic strategy for COVID-19. There is much evidence about this issue, and it is expected to be increased in near future.
Keywords: COVID-19, SARS-CoV-2, gastrointestinal microbiome, dysbiosis, prognosis, diagnosis, gut microbiota, therapeutic
Introduction
A pandemic caused by Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2) emerged in late December 2019 (Zhu et al., 2020). The World Health Organization (WHO) named the consequent disease as Coronavirus Disease 2019 (COVID-19) and declared it as a global emergency due to the serious public health effects (Jamshidi et al., 2021). According to the report of the WHO, until February 1, 2022, there have been about 376 million confirmed cases and about 5.6 million deaths due to COVID-19 around the world.
The angiotensin-converting enzyme 2 (ACE2) receptor is a known SARS-CoV-2 receptor for entering host cells (Li et al., 2003; Zhou et al., 2020). This receptor is detected in various cells of the body such as the respiratory, digestive, renal, and skin epithelium, suggesting that each of these organs could be a potential target for the virus (Jamshidi et al., 2021; Xue et al., 2021). Moreover, virus RNA and viral particles have been identified in the fecal sample of COVID-19 patients, which may indicate the possibility of virus replication and activity in the human intestine (Gu et al., 2020a; Lamers et al., 2020; Xu et al., 2020).
Gut microbiota plays a well-known role in regulating immune system responses (Donaldson et al., 2016;; Schirmer et al., 2016). Recent studies indicate the role of gut dysbiosis in the pathogenesis of various diseases such as inflammatory bowel disease, type 1 and type 2 diabetes, and celiac disease, as well as chronic respiratory diseases such as asthma, COPD, and cystic fibrosis (Jamshidi et al., 2019; Enaud et al., 2020).
Bacteria in the human intestinal flora appear to affect the respiratory system and lungs (especially the lung microbiota) by producing metabolites, endotoxins, cytokines, and intestinal hormones reaching the bloodstream, which is called the gut–lung axis (Budden et al., 2017; Dang and Marsland, 2019; Zhang et al., 2020).
On the other hand, there is evidence of the role of gut dysbiosis in the severity and prognosis of bacterial (e.g., Streptococcus pneumonia, Klebsiella pneumonia, Pseudomonas aeruginosa, Mycobacterium tuberculosis) and viral (e.g., H1N1 influenza) respiratory infectious diseases in animal models (Ichinohe et al., 2011; Fagundes et al., 2012; Fox et al., 2012; Brown et al., 2017). The use of broad-spectrum antibiotics that target the gut microbiota has led to a poor prognosis in mouse models with infectious lung diseases (Enaud et al., 2020).
Considering the important role of gut microbiota in maturation, regulation, and induction of the immune system and subsequent inflammatory processes, it seems that evaluating the composition of gut microbiota in COVID-19 patients compared with healthy individuals may have potential value as a diagnostic and/or prognostic biomarker of the disease. Also, therapeutic interventions affecting gut microbial flora may open new horizons in the treatment of COVID-19 patients and accelerating their recovery.
Methods
This review conforms to the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) statement (Moher et al., 2009).
Search Strategy and Selection Criteria
To investigate the diagnostic, prognostic, and therapeutic role of the gut microbiota composition in COVID-19, a systematic search was conducted for relevant studies published from December 2019 to December 2021 using Pubmed/Medline, Embase, and Scopus.
Articles containing the following keywords in titles or abstracts were selected: “SARS-CoV-2” or “COVID-19” or “Coronavirus Disease 19” and “gastrointestinal microbes” or “dysbiosis” or “gut microbiota” or “gut bacteria” or “gut microbes” or “gastrointestinal microbiota”. Only studies included if they contained data about the gut microbiota composition in COVID-19 patients. There were no language restrictions. Review articles, duplicate publications, letters, commentary, animal studies, and articles with no relevant data were excluded from the analysis. Two authors (MA and FV) independently screened the articles by title and abstract. Full-text screening was conducted by two other authors independently (AT and YF). In each step, contrarieties were discussed with a third reviewer (PJ).
Data Extraction
A data extraction form designed by two authors (PJ and MJN) and, finally, selected data were extracted from the full text of eligible publications by PJ, YF, AT, MA, and FV. The following data were extracted for further analysis: first author’s name, year of publication, country where the study was executed, type of study, study population, mean age, gender, COVID-19 severity of the cases, comorbidity(ies), microbiota analysis technique, intestinal microbiota alterations, biochemical and immunological alterations, and studied value of gut microbiota alterations in COVID-19. The data were jointly reconciled, and disagreements were discussed and resolved by review authors (PJ, MJN).
Quality Assessment
The critical appraisal checklist for case reports provided by the Joanna Briggs Institute (JBI) was used to perform a quality assessment of the studies (Institute, 2021).
Results
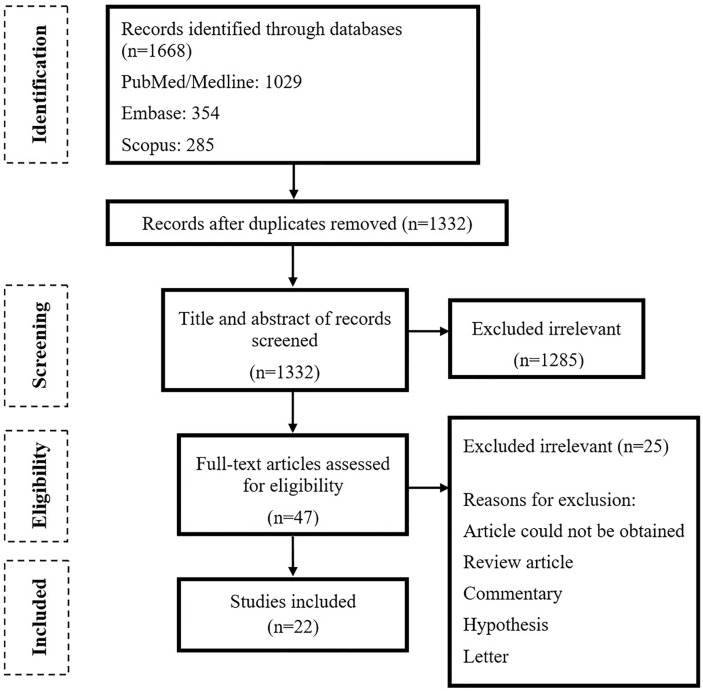
As shown in Figure 1 , the primary search resulted in 1,668 relevant articles, of which 47 articles were selected after title and abstract screening. Following the full-text screening, 22 articles fulfilled the inclusion criteria. Most of the studies were case–control (n = 9) followed by cohort (n = 9), clinical trial (n = 2), and cross-sectional (n = 2) studies. Fifteen of the studies were executed in China, 2 in Italy and 1 in UK, Portugal, India, Egypt, and Korea ( Table 1 ).
Figure 1.
PRISMA flowchart of study selection for inclusion in the systematic review.
Table 1.
Characteristics of the included studies.
| Authors | Year | Country | Type of study | Study population | Gut microbiota analysis technique |
|---|---|---|---|---|---|
| Gu et al. (2020b) | 2020 | China | Cross-sectional | 30 COVID-19, 24 H1N1, 30 HC a | 16S rRNA sequencing |
| d’Ettorre et al. (2020) | 2020 | Italy | Clinical trial | 70 COVID-19 (case: 28, control: 42) b | NM |
| Tang et al. (2020) | 2020 | China | Cohort | 57 COVID-19 (20 non-severe, 19 severe, 18 critical) | q- PCR |
| Zuo et al. (2020b) | 2020 | China | Case–control | 30 cases (COVID-19), 39 control (30 HC a and 9 CAP c ) | Shotgun metagenomic sequencing |
| Zuo et al. (2020b) | 2020 | China | Case–control | 15 cases (COVID-19), 21 control (15 HC a and 6 CAP c ) | Shotgun metagenomic sequencing and RT-PCR |
| Zuo et al. (2021a) | 2020 | China | Cohort | 15 COVID-19 | Shotgun metagenomic sequencing |
| Cao et al. (2021) | 2021 | China | Cohort | 13 COVID-19, 5 HC a | cDNA sequencing, bacteriome sequencing, metagenomic sequencing |
| Liu et al. (2021) | 2021 | China | Clinical trial | 11 COVID-19 | 16s rRNA sequencing |
| Lv et al. (2021b) | 2021 | China | Cohort | 67 COVID-19, 35 H1N1, 48 HC a | q-PCR on DNA extract of fecal samples |
| Lv et al. (2021a) | 2021 | China | Cohort | 56 COVID-19, 47 HC a | NM |
| Yeoh et al. (2021) | 2021 | China | Cohort | 100 COVID-19, 78 non-COVID-19 | Shotgun sequencing of DNA extracted from stools |
| Prasad et al. (2021) | 2021 | UK | Cohort | 30 COVID-19, 16 HC a | 16S rRNA sequencing, metatranscriptomic analysis (plasma samples) |
| He et al. (2021) | 2021 | China | Case–control | 13 cases (COVID-19), 21 control (HC a ) | Metaproteomics |
| Zhou et al. (2021) | 2021 | China | Case–control | 15 cases (recovered COVID-19 patients), 14 control (HC a ) | 16S rRNA sequencing |
| Moreira-Rosário et al. (2021) | 2021 | Portugal | Cross-sectional | 115 COVID-19 (19 mild, 37 moderate, 58 severe) | 16S rRNA sequencing |
| Wu et al. (2021) | 2021 | China | Case–control | 53 cases (COVID-19), 76 control (HC a ) | 16S rRNA sequencing |
| Gaibani et al. (2021) | 2021 | Italy | Case–control | STUDY1: 69 COVID-19, 69 HC
a
STUDY2: 69 COVID-19, 16 non-COVID-19 ICU admitted control |
16S rRNA sequencing |
| Kim et al. (2021) | 2021 | Korea | Case–control | 12 cases (COVID-19), 36 control (HC a ) | 16S rRNA sequencing |
| Zuo et al. (2021b) | 2021 | China | Case–control | 98 cases (COVID-19), 78 control (HC a ) | Shotgun metagenomic sequencing |
| Khan et al. (2021) | 2021 | India | Case–control | 30 cases (COVID-19), 10 control(HC a ) | 16S rRNA sequencing |
| Hegazy et al. (2021) | 2021 | Egypt | Cohort | 200 COVID-19 (122 mild, 78 moderate) | NM |
| Wang et al. (2021b) | 2021 | China | Cohort | 156 COVID-19 (98 mild and moderate, 58 severe and critical) d | NM |
aHealthy control subjects.
b42 patients received hydroxychloroquine, antibiotics, and tocilizumab, alone or in combination, and 28 patients received the same therapy added with oral bacteriotherapy, using a multistrain formulation.
cCommunity-acquired pneumonia.
dThe efficacy of probiotic treatment has been studied only in 16 severe and critical COVID-19 patients (treatment group = 10, control group = 6).
A total of 1,255 confirmed COVID-19 patients were examined in 22 included articles ( Table 1 ). Among the COVID-19 patients whose comorbidity was mentioned by the authors, the most common ones were hypertension (32.8%) and diabetes mellitus (17.8%). Other reported comorbidities were chronic respiratory disease (6.9%), cardiovascular disease (3%), immunosuppression (2.9%), dyslipidemia (2.3%), thrombotic events (2%), and renal impairment (1.6%). See Table 2 .
Table 2.
Characteristics of the study population.
| Authors | Age (Mean) | Gender | COVID-19 Severity | Comorbidity |
|---|---|---|---|---|
| Gu et al. (2020b) | 52.3 years | 49M, 35F | 15 non-severe, 15 severe | HTN (H1N1: 5/24, COVID-19: 9/30) |
| d’Ettorre et al. (2020) | 59.75 years | 41M, 29F | 70 severe: stage III a | None |
| Tang et al. (2020) | 64.17 years | 29M, 28F | 20 non-severe, 19 severe, 18 critical b | HTN: 7 in non-severe, 8 in severe, 12 in critical DM: 3 in non-severe, 0 in severe, 6 in critical |
| Zuo et al. (2020b) | 43 years | 36M, 33F | NM | 11 in COVID-19, 9 in CAP, 0 in healthy…not specified |
| Zuo et al. (2020a) | 51.25 years | 20M, 16F | NM | 6 in COVID-19, 6 in CAP, 0 in healthy…not specified |
| Zuo et al. (2021a) | 53 years | 7M, 8F | 11 moderate to severe, 2 critical (ICU) | HTN: 4, DM: 2, hyperlipidemia: 4, obesity: 1, chronic hepatitis B: 1, renal impairment: 1, duodenal ulcer: 1, left subclavian artery occlusion: 1 |
| Cao et al. (2021) | 48 years | 6M, 7F | 3 severe, 7 moderate, 3 mild | HTN:4, hyperthyroidism:1, gallstone:1, arthritis:1 |
| Liu et al. (2021) | 49.8 years | 6M, 5F | 10 non-severe, 1 severe | None |
| Lv et al. (2021b) | 52 years | 92M, 58F | 36 severe | HTN: 16, diabetes: 10, CVD: 4, liver diseases: 2 |
| Lv et al. (2021a) | 54 years | 57M, 46F | 26 mild, 30 severe | HTN: 18, DM: 7 |
| Yeoh et al. (2021) | 41 years | 86M, 92F | 47 mild, 45 moderate, 5 severe, 3 critical | HTN: 11, hyperlipidemia: 4, DM: 2, CVD: 2, allergic disorders: 7, HIV: 3, asthma: 2 |
| Prasad et al. (2021) | 52 years | 23M, 23F | 11 mild, 17 moderate, 2 severe | DM: 10, thrombotic events: 15 |
| He et al. (2021) | 37.9 years | 22M, 12F | 7 mild, 5 moderate, 1 severe | DM: 1, sinusitis: 1,rhinitis:1 |
| Zhou et al. (2021) | 33.1 years | 8M, 21F | NM | HTN: 2 |
| Moreira-Rosário et al. (2021) | 68 years | 73M, 42F | 19 mild, 37 moderate, 58 severe | DM: 45, HTN: 67, chronic respiratory disease: 21, immunosuppression: 11, hematological-oncological disease: 9 |
| Wu et al. (2021) | 48.5 years | 82M, 46F c | 30 non-severe, 20 Severe | NM |
| Gaibani et al. (2021) | 73 years | 38M, 31F d | NM | HTN: 44, DM: 12, CVD: 5, immunosuppression: 7, CKD:11 |
| Kim et al. (2021) | 26 years | 8M, 4F d | 12 asymptomatic or mild | NM |
| Zuo et al. (2021b) | 33 years | 85M, 91F | 3 asymptomatic, 53 mild, 34 moderate, 5 severe, 3 critical | 55… not specified |
| Khan et al. (2021) | 51.8 years | NM | NM | NM |
| Hegazy et al. (2021) | 41 years | 94M,106 F | 122 mild, 78 moderate | DM: 30, HTN: 34, chronic lung disease: 15, chronic liver disease: 2, CVD:9 |
| Wang et al. (2021b) | 48.5 years | 95M, 61F | 98 mild and moderate, 58 severe and critical | DM: 18, HTN: 31, CVD:15, COPD:12 |
NM, not mentioned; HTN, hypertension; DM, diabetes mellitus; CVD, cardiovascular disease; CKD, chronic kidney disease, COPD, chronic obstructive pulmonary disease.
aAccording to the syndromic classification proposed by the Italian Society of Anesthesia and Resuscitation (SIAARTI).
bRespiratory failure requiring mechanical ventilation, shock, or other organ failure requiring ICU care.
cThere was one missing data among COVID-19 group.
dOnly the data of the COVID-19 group was available.
Ten studies assessed gut microbiota composition alteration by fecal samples; one study used plasma samples, and it was not mentioned by the rest. The most commonly used techniques in these studies for detection and assessment of gut microbiota were 16s rRNA sequencing and shotgun metagenomic sequencing analysis ( Table 1 ).
Gut Microbiome Dysbiosis of COVID-19 Patients
All included studies showed a significant association between COVID-19 and gut microbiota dysbiosis ( Table 3 ). The most alteration in the bacterial composition of COVID-19 patients was depletion in genera Ruminococcus, Alistipes, Eubacterium, Bifidobacterium, Faecalibacterium, Roseburia, Fusicathenibacter, and Blautia and enrichment of Eggerthella, Bacteroides, Actinomyces, Clostridium, Streptococcus, Rotia, and Collinsella. Details are shown in Table 4 .
Table 3.
Association between gut microbiota and COVID-19.
| Authors | Type of study | Studied value | Association between gut microbiota and COVID-19 |
|---|---|---|---|
| Gu et al. (2020b) | Cross-sectional | Diagnostic | Yes |
| d’Ettorre et al. (2020) | Clinical trial | Therapeutic | Yes |
| Tang et al. (2020) | Cohort | Prognostic and diagnostic | Yes |
| Zuo et al. (2020b) | Case–control | None | Yes |
| Zuo et al. (2020a) | Case–control | Prognostic and therapeutic | Yes |
| Zuo et al. (2021a) | Cohort | Prognostic | Yes |
| Cao et al. (2021) | Cohort | Diagnostic and prognostic | Yes |
| Liu et al. (2021) | Clinical trial | Therapeutic (postinfection recovery) | Yes |
| Lv et al. (2021b) | Cohort | Diagnostic | Yes |
| Lv et al. (2021a) | Cohort | Diagnostic and prognostic | Yes |
| Yeoh et al. (2021) | Cohort | Prognostic | Yes |
| Prasad et al. (2021) | Cohort | Diagnostic and prognostic | Yes |
| He et al. (2021) | Case–control | None | Yes |
| Zhou et al. (2021) | Case–control | None | Yes |
| Moreira-Rosário et al. (2021) | Cross-sectional | Prognostic | Yes |
| Wu et al. (2021) | Case–control | Diagnostic and prognostic | Yes |
| Gaibani et al. (2021) | Case–control | Prognostic | Yes |
| Kim et al. (2021) | Case–control | Diagnostic (postinfection recovery) | Yes |
| Zuo et al. (2021b) | Case–control | Diagnostic and prognostic | Yes |
| Khan et al. (2021) | Case–control | Prognostic | Yes |
| Hegazy et al. (2021) | Cohort | Prognostic | Yes |
| Wang et al. (2021b) | Cohort | Therapeutic | Yes |
Table 4.
Gut microbiota alterations.
| Authors | Intestinal microbial alternations |
|---|---|
| Gu et al. (2020b) |
COVID-19 and H1N1 vs. HC
a
: Microbial diversity↓, Streptococcus spp.↑, Escherichia-Shigella spp.↑ H1N1 vs. COVID-19 and HC: phylum (Actinobacteria↓, Firmicutes↓), class (Actinobacteria↓, Erysipelotrichia↓, Clostridia↓), family (Lachnospiraceae↓, Ruminococcaceae↓), Blautia spp.↓, Agathobacter spp.↓, Anaerostipes spp.↓, Fusicatenibacter spp.↓, Eubacterium hallii group↓, unclassified Lachnospiraceae↓, Dorea spp.↓, Faecalibacterium spp.↓, Ruminococcus-2 spp.↓ COVID-19 vs. HC: Ruminococcaceae UCG-013↓, Roseburia spp.↓, Lachnospiraceae family↓(Fusicatenibacter spp.↓, Anaerostipes spp.↓, Agathobacter spp.↓, unclassified Lachnospiraceae↓, Eubacteriumhallii group↓), Streptococcus spp.↑↑ COVID-19 vs. H1N1: Streptococcus spp. ↑↑, Prevotella spp.↓, Ezakiella spp.↓, Murdochiella spp.↓, Porphyromonas spp.↓ COVID-19 dominated by: Streptococcus spp., Rothia spp., Veillonella spp., Erysipelato clostridium spp., Actinomyces spp. HC dominated by: Bifidobacterium spp., Romboutsia spp., Faecalibacterium spp., Fusicatenibacter spp., Eubacterium hallii group, Blautia spp., Collinsella spp. H1N1 dominated by: Enterococcus spp., Prevotella spp., Finegoldia spp., Peptoniphilus spp.Richness, diversity, and structure of the gut microbiota were not significantly different between general and severe COVID-19 patients. |
| d’Ettorre et al. (2020) | The formulation administered in this study contained: Streptococcus thermophilus DSM 32345, Lactobacillus acidophilus DSM 32241, Lactobacillus helveticus DSM 32242, Lactobacillus paracasei DSM 32243, Lactiplantibacillus plantarum DSM 32244, Lactobacillus brevis DSM 27961, Bifidobacterium lactis DSM 32246, Bifidobacterium lactis DSM 32247. |
| Tang et al. (2020) | Lactobacillus spp.↓, Bifidobacterium spp.↓, Faecalibacterium prausnitzii↓, Clostridium butyricum↓, Clostridium leptum↓, Eubacterium rectale↓, Enterobacteriaceae ↓, Bacteroides spp.↓ bEnterococcus spp.↑ (It was increased with disease severity) |
| Zuo et al. (2020b) | Candida albicans↑, Aspergillus flavus↑, Aspergillus niger↑ |
| Zuo et al. (2020a) |
Antibiotic negative COVID-19 group:
Clostridium hathewayi↑, Actinomyces viscosus↑, Bacteroides nordii↑ Antibiotic positive COVID-19 group: Faecalibacterium prausnitzii↓, Lachnospiraceae bacterium↓, Eubacterium rectale↓, Ruminococcus obeum↓, Dorea formicigenerans↓ |
| Zuo et al. (2021a) |
Enriched in fecal samples in high infectivity:
Collinsella aerrofaciens↑, Morganella morganii↑, Streptococcus infantis↑ Enriched in fecal samples in low to non infectivity: Parabacteroides merdae↑, Bacteroides stercoris↑, Alistipes onderdonkii ↑, Lachnospiraceae bacterium↑ |
| Cao et al. (2021) |
COVID-19 vs. HC:Bacteria:
Ruminococcus gnavus↑, Eggerthella spp.↑, Coprobacillus spp.↑, Lachnospiraceae bacterium 2_1_58FAA↑, Clostridium ramosum↑, Eggerthella lenta↑, Lachnospiraceae bacterium 1_4_56FAA↑, Alistipes_sp_AP11↓, Roseburia intestinalis↓, Burkholderiales bacterium 1_1_47↓, Eubacterium_hallii↓, Parasutterella_excrementihominis↓, Alistipes indistinctus↓, Coprobacter fastidiosus↓, Eubacterium eligens↓, Bacterioidales bacterium ph8↓, Bacterioides salyersiae↓, Odoribacter splanchnicus↓, Alistipes shahii↓, Ruminococcus bromii↓, Bacteroides massiliensis↓ Virome: Inviridae↑, Microviridae↑, virgaviridae↑ Antibiotic positive vs. antibiotic negative COVID-19 patients:Bacteria: Subdoligranulum↓, Roseburia inulinivorans↓, Roseburia hominis↓, Parasutterella excrementihominis↓, Lachnospiraceae bacterium 2_1_46FAA↓, Faecalibacterium prausnitzii↓,Dorea formicigenerans↓, Coprococcus catus↓, Collinsella aerofaciens↓, Bacteroides vulgatus↓,Veillonella parvula↑, Coprobacillus spp.↑, Clostridium ramosum↑ Virome: No virus was identified as a differential species. Effect of disease severity on gut microbiome: Virome: Severe cases: Fourteen Microviridaephages, one Inoviridae phage, one Podoviridae phage and one unclassified virus↑ Mild cases: No viral community increased Bacteria: Severe cases: Corynebacterium durum↑, Rothia mucilaginosa↑, Enterococcus faecium↑, Campylobacter gracilis↑, Corynebacterium spp.↑, Enterococcus spp.↑, Rothia spp.↑, Megasphaera spp.↑, Campylobacter spp.↑, Eubacterium spp.↓ Mild cases: Eubacterium rectale↑ |
| Liu et al. (2021) | After intervention(FMT): Proteobacteria ↓, Actinobacteria ↑, Bifidobacterium spp.↑, Faecalibacterium spp.↑, Collinsella spp.↑ |
| Lv et al. (2021b) | COVID-19 vs. others: Candida glabrata↓, Candida parapsilosis↓, Five unclassified species separately belonging to Helotiales↓, Pleosporales↓, Sordariales↓, Microscypha spp.↓,Emericellopsis spp.↓, Cystobasidium spp.↑, An unclassified species of Exidiaceae↓, Trebouxiadecolorans↓, An unclassified species belonging to the kingdom Chromista↓ |
| Lv et al. (2021a) | COVID-19 vs. others: Ruminococcaceae↓, Eubacteriumhallii group↓, Family XIII AD3011 group↓, Anaerostipes spp.↓, Fusicatenibacter spp.↓, Roseburia spp.↓, Faecalibacterium spp.↓, Ruminococcus spp. 5139BFAA↓, Aspergillus rugulosus↓, Aspergillus tritici↓,Penicillium spp.↓, Penicillium citrinum ↓, Actinomyces spp.↑, Sphingomonas spp.↑, Rothia spp.↑, Actinomyces odontolyticus↑, Streptococcus parasanguinis↑, Aspergillus penicillioide↓ |
| Yeoh et al. (2021) | Ruminococcus gnavus↑, Bacteroides dorei↑, Ruminococcus torques↑, Bacteroides vulgates↑, Bacteroides ovatus↑, Bacteroides caccae↑,Akkermansia muciniphila↑, Bifidobacterium adolescentis↑, Eubacterium rectal↑, Ruminococcus bromii↑, Subdoligranulum unclassified↑, Bifidobacterium pseudocatenulatum↑, Faecalibacterium prausnitzii↑, Collinsella aerofaciens↑, Ruminococcus obeum↑, Dorea longicatena↑, Coprococcus comes↑, Dorea formicigenerans↑ |
| Prasad et al. (2021) | Plasma samples: Proteobacteria↑, Firmicutes ↑, Actinobacteria ↑, Acinetobacter spp.↑, Nitrospirillum spp.↑, Cupriavidus spp.↑, Pseudomonas spp.↑, Aquabacterium spp.↑, Burkholderia spp.↑, Caballeronia spp.↑, Paraburkholderia spp.↑, Bravibacterium spp.↑, Sphingomonas spp.↑, Staphylococcus spp.↓, Lactobacillus spp.↓ |
| He et al. (2021) | Ruminococcus gnavus↓, Lachnospiraceae↓, Tyzzerella spp.↓, Blautia spp.↓, Eubacterium spp.↓, Peptostreptococcaceae↓,Butyrivibrio spp.↓, Ruminococcus spp.↓, Lachnoclostridium spp.↓, Bacteroides uniformis↑, Bacteoides graminisolvens↑, Bacteroides coprophilus↑ |
| Zhou et al. (2021) | Phylum (Actinobacteria↑), Family (Lachnospiraceae↓, Desulfovibrionaceae↓), Faecalibacterium spp.↓, Roseburia spp.↓, Fusicatenibacter spp.↓, Ruminococcus spp.↓, Clostridium XVIII↓, Dorea spp.↓, Butyricicoccus spp.↓, Romboutsia spp.↓, Intestinimonas spp.↓, Bilophila spp.↓, Escherichia spp.↑, Flavonifractor spp.↑, Intestinibacter spp.↑, Intestinibacter bartlettii↑, Clostridium aldenense↑, Clostridium bolteae↑, Flavonifractor plautii↑, Clostridium ramosum↑, Faecalibacterium prausnitzii↓, Roseburia inulinivorans↓, Fusicatenibacter saccharivorans↓, Ruminococcus bromii↓, Blautia faecis↓, Butyricicoccus pullicaecorum↓, Intestinimonas butyriciproducens↓ |
| Moreira-Rosário et al. (2021) | Severe cases: Proteobacteria↑, Firmicutes/Bacteroidetes ratio↓, Roseburia spp.↓, Lachnospira spp.↓ |
| Wu et al. (2021) |
Blautia spp.↓, Coprococcus spp.↓, Collinsella spp.↓, Streptococcus spp.↑, Weissella spp.↑, Enterococcus spp.↑, Rothia spp.↑, Lactobacillus spp.↑, Actinomyces spp.↑, Granulicatella spp.↑, Bacteroides caccae↓, Bacteroides coprophilus↓, Blautia obeum↓, Clostridium colinum↓, Clostridium citroniae↑, Bifidobacterium longum↑, Rothia mucilaginosa↑ Associations between gut microbiota disturbance and SARS-CoV-2 viral loads: Prevotella copri and Eubacterium dolichum were positively correlated and Streptococcus anginosus, Dialister spp., Alistipes spp., Ruminococcus spp., Clostridium citroniae, Bifidobacterium spp., Haemophilus spp., and Haemophilus parainfluenzae were negatively correlated with the viral load of SARS-CoV-2. |
| Gaibani et al. (2021) |
Study1
c
: Enterococcaceae↑, Coriobacteriaceae↑, Lactobacillaceae↑, Veillonellaceae↑, Porphyromonadaceae↑, Staphylococcaceae↑, Bacteroidaceae↓, Lachnospiraceae↓, Ruminococcaceae↓,Prevotellaceae↓, Clostridiaceae↓ Study2 c : Enterococcus spp.↑, Klebsiella spp.↓, Ruminococcus spp.↑ |
| Kim et al. (2021) | Escherichia spp.↑, Citrobacter spp.↑, Collinsella spp.↑, Bifidobacterium spp.↑, Bacteroides spp.↓, Butyricimonas spp.↓, Odoribacter spp.↓ |
| Zuo et al. (2021b) | Pepper Mild Mottle Virus (PMMoV)↓, Eukaryotic viruses particularly environment-derived eukaryotic viruses with unknown host↑, Streptococcus phage↑, Escherichia phage↑, Homavirus↑, Lactococcus phage↑, Ralstonia phage↑, Solumvirus↑, Microcystis phage↑ Severe cases: plant-derived RNA virus↓, Pepper Chlorotic Spot Virus (PCSV)↓, Myxococcus phage↓, Rheinheimera phage↓, Microcystis virus↓, Bacteroides phage↓, Murmansk poxvirus↓, Saudi moumouvirus↓, Sphaerotilus phage↓, Tomelloso virus↓, Ruegeria phage↓ |
| Khan et al. (2021) | Firmicutes↓, Bacteroidetes↑, Proteobacteria↑, Actinobacteria↑Severe cases: Bacteroides plebeius↓, Faecalibacterium prausnitzii↓, Roseburia faecis↓, Bifidobacterium spp.↑, Bacteroides caccae↑, Bacteroides ovatus↑, Bacteroides fragilis↑, Ruminococcus gnavus↑, Clostridium bolteae↑, Clostridium citroniae↑, Clostridium hathewayi↑, Parabacteroides distasonis↑ |
| Hegazy et al. (2021) | The formulation used in this study was a yogurt containing Bifidobacterium spp. and Lactobacillus spp. |
| Wang et al. (2021b) | Probiotic administration protocol: Bifidobacterium lactobacillus triplex live tablet; each tablet contained no less than 0.5×107 CFU of live Bifidobacterium longum; live Lactobacillus bulgaricus and Streptococcus thermophilus that was not lower than 0.5×106 CFU, 4 pieces at a time, 3 times a day. |
aHealthy control.
bBacteroides spp. was decreased in all groups, but the decrease was within the lower normal limits. There was no significant difference between the groups.
cStudy1: comparison between COVID-19 patients and healthy controls, Study2: comparison between COVID-19 patients and non-COVID-19 ICU admitted control.
Three articles surveyed the gut mycobiota alterations, and different results have been reported for different species of the same genus. About the Candida spp., an increase in Candida albicans and a decrease in Candida glabrata and Candida parapsilosis were mentioned. In regard to Aspergillus spp., enrichment of Aspergillus flavus and Aspergillus niger and a depletion of Aspergillus rugulosus, Aspergillus tritici, and Aspergillus penicillioides were reported. Also, one study indicated a reduction in seven unclassified species belonging to order Helotiales, Pleosporales, and Sordariales, family Exidiaceae, and genera Microscypha and Emericellopsis in COVID-19 patients ( Table 4 ).
According to all gut microbiota changes that were mentioned in the reviewed articles, a decrease in phyla Firmicutes and Bacteroidetes and an increase in phylum Actinobacteria among COVID-19 patients were inferred.
Association Between Gut Microbiota Composition and COVID-19 Severity
A few studies indicated the role of gut microbiome in COVID-19 severity ( Table 4 ). In severe COVID-19 cases, Bacteroides spp., Parabacteroides spp., Clostridium spp., Bifidobacterium spp., Ruminococcus spp., Campylobacter spp., Rothia spp., Corynebacterium spp., Megasphaera spp., Enterococcus spp., and Aspergillus spp. were increased and Roseburia spp., Eubacterium spp., Lachnospira spp., Faecalibacterium spp., and Firmicutes/Bacteroidetes ratio were decreased significantly. In subjects with mild disease the observed significant change was in the enrichment of Eubacterium spp.
The alteration of the gut virome composition in severe COVID-19 cases was mentioned in two studies. In severe cases, fourteen Microviridae phages, one Inoviridae phage, one Podoviridae phage, and one unclassified virus were increased and plant-derived RNA virus, pepper chlorotic spot virus (PCSV), Myxococcus phage, Rheinheimera phage, Microcystis virus, Bacteroides phage, Murmansk poxvirus, Saudi moumouvirus, Sphaerotilus phage, Tomelloso virus, and Ruegeria phage were decreased significantly. See Table 4 .
One study evaluated the associations between gut microbiota disturbance and SARS-CoV-2 viral loads and revealed that Prevotella copri and Eubacterium dolichum were positively correlated and Streptococcus anginosus, Dialister spp., Alistipes spp., Ruminococcus spp., Clostridium citroniae, Bifidobacterium spp., Haemophilus spp., and Haemophilus parainfluenzae were negatively correlated with the viral load of SARS-CoV-2.
Biochemical and Immunologic Modifications in Relation to Gut Microbiota Alternations in COVID-19 Patients
In most studies, compared with healthy controls, COVID-19 patients had significantly higher levels of interleukin (IL)-2, IL-4, IL-6, IL-10, tumor necrosis factor (TNF)-α, and C-reactive protein (CRP) and lower lymphocyte counts.
According to one study, a positive correlation between Bifidobacterium spp. and prothrombin time (PT) and lactate dehydrogenase (LDH) was shown. Also, a negative correlation was reported between Atopobium spp. and D-dimer, Bacteroides spp. and LDH and creatine kinase (CK) level, Clostridium butyricum, and CRP and neutrophil count, and Faecalibacterium prausnitzii and CRP in critical COVID-19 patients. One study showed a specific relation between some genus of gut microbiota and immunological and biochemical modifications in critical and severe COVID-19 patients. In severe patients, Faecalibacterium prausnitzii and Clostridium leptum had a positive correlation with neutrophil count as well as Eubacterium rectale with IL-6 and Enterobacteriaceae with AST.
Another study indicated the specific relation of some species of gut microbiome and immune cells as the following: Bacteroides ovatus, Lachnospiraceae bacterium, and Eubacterium ventriosum had a positive correlation with CD4 and CD8 lymphocytes and other T-cells, in contrast to Bifidobacterium animalis and Escherichia spp. On the other hand, Faecalibacterium prausnitzii had a positive correlation with NK cells and Coprobacillus spp., Clostridium ramosum, and Clostridium symbiosum had a negative correlation with them.
Studied Value of Gut Microbiome in COVID-19
All of the included studies showed a correlation between intestinal microbiota and COVID-19, and they studied the correlation in different aspects as in the following.
Four studies suggested that microbiota could have therapeutic properties with reducing gastrointestinal (GI) symptoms. Streptococcus, Lactobacillus, and Bifidobacterium were the most common bacterial genera interventions used so far. Nine articles demonstrated intestinal microbiota modifications in infected cases with COVID-19 in which two of them confirmed the value of specified gut microbiota as a diagnostic tool and one of them studied gut microbiota changes during recovery time. Lachnospiraceae are a large family including Fusicathenibacter, Eubacterium hallii group, and Roseburia, and the Ruminococcaceae family including Faecalibacterium prausnitzii and Ruminococcus as well as Clostridium spp., Bacteroides spp., Lactobacillus spp., Rothia spp., Actinomyces spp., Lactobacillus spp., and Streptococcus spp. were the most common bacteria with diagnostic value. Thirteen studies demonstrated a relationship between gut microbiota changes and the intensity and prognosis of COVID-19. Eubacterium, Faecalibacterium, Ruminococcus, Bacteroides, Clostridium, Lactobacillus, Bifidobacterium, and Roseburia were the most notable bacterial genera with prognostic values. Details are shown in Table 3 .
Discussion
The interaction between gut microbiota and viral respiratory diseases such as COVID-19 is a complex, bilateral, and dynamic association. The current study emphasizes the role of the gut–lung axis (GLA) in the pathogenesis of COVID-19. One of the important aspects of GLA is the impact of gut microbiota on the supply and maintenance of the lung immune system, and its correlation with respiratory diseases and infections (He et al., 2017; Dang and Marsland, 2019; Ahmadi Badi et al., 2021; Allali et al., 2021). The gut and lung microbiome are closely related in health or disease conditions (Dickson and Huffnagle, 2015; Enaud et al., 2020; Ahmadi Badi et al., 2021). SARS-CoV-2 may cause a dysbiosis in the lung microbiota to increase the population of inflammatory bacteria such as Klebsiella oxytoca and Rothia mucilaginosa which is associated with acute respiratory distress syndrome (Aktaş and Aslim, 2020; Han et al., 2020; van der Lelie and Taghavi, 2020; Battaglini et al., 2021; Chattopadhyay and Shankar, 2021). The high levels of inflammatory cytokine productions during SARS-CoV-2 invasion interfere with gut mucosal integrity and increasing risk of bacterial disposition to the bloodstream (Chattopadhyay and Shankar, 2021; Prasad et al., 2021).
Based on our findings, gut microbiota in patients with SARS-CoV-2 is significantly affected, possibly due to systemic inflammatory response. Although the underlying mechanism for the observed dysbiosis is unclear, it might happen via downregulation of ACE2 expression that alleviates the intestinal absorption of tryptophan leading to decreased secretion of antimicrobial peptides and changes the composition of gut microbiota (He et al., 2020). In a healthy individual, intact bacteria and their fragments or metabolites such as des-amino-tyrosine and short-chain fatty acids (SCFAs) pass across the intestinal barrier via the mesenteric lymphatic system, reach the lung, and activate the innate immune system by the production of cytokines like type 1 interferon (IFN1) (Antunes et al., 2019). In the current study, Faecalibacterium prausnitzii and Clostridium leptum have a positive correlation with neutrophil counts in COVID-19 patients and a negative correlation with Clostridium butyricum. In addition to the innate immunity, gut microbiota improves the function of CD8+ T-cell effectors (Trompette et al., 2018). There is also evidence that few bacterial species such as Bacteroides ovatus, Lachnospiraceae bacterium 5_1_63FAA, and Eubacterium ventriosum have an anti-inflammatory property in CD4+ and CD8+ T cells (Cao et al., 2021). Natural killer (NK) cells and B cells are also affected by gut microbiota; Coprobacillus spp., Clostridium ramosum, and Clostridium symbiosum are negatively associated with NK cell activity and Bacteroides uniformis, Faecalibacterium prausnitzii, and Subdoligranulum are positively correlated with B cells (Cao et al., 2021).
We found that the dysbiosis of gut microbiota may be a determinant factor in the clinical severity of COVID-19. Increase of the dominant Enterococcus and reduction of Ruminococcaceae and Lachnospiraceae are reported in severe cases of COVID-19 who were admitted to the medical intensive unit (MICU) (Gaibani et al., 2021).
Diversity of gut microbiota reduces in patients with COVID-19, which is associated with pro-inflammatory reaction and increased risk of opportunistic infections. GI symptoms of COVID-19 are reported to be strongly affected by gut microbial composition (Din et al., 2021; Katz-Agranov and Zandman-Goddard, 2021). The expression of the ACE-2 receptor on the surface of small intestine epithelial cells has been significantly associated with the extent of GI symptoms and fecal viral shedding during the course of disease (D’Amico et al., 2020; Patel et al., 2020; Wu et al., 2020; Katz-Agranov and Zandman-Goddard, 2021; Suárez-Fariñas et al., 2021). An increased expression of ACE-2 receptor in COVID-19 may occur due to a dominance of Coprobacillus in gut microbiota (Geva-Zatorsky et al., 2017; Enaud et al., 2020; Zuo et al., 2020a; Massip Copiz, 2021; Rajput et al., 2021; Walton et al., 2021). It is also noteworthy to imply that the expression of ACE-2 on luminal cells may be a determinant factor in microbial composition as the ACE-2 receptor plays some role in amino-acid absorption (Harmer et al., 2002; Dang and Marsland, 2019).
Eggerthella is another genus of bacteria which significantly increases in patients with COVID-19 (d’Ettorre et al., 2020; Cao et al., 2021). Eggerthella may induce colitis via abnormal activation of Th17 in patients with inflammatory diseases. It can interfere with gut integrity and make the patient more susceptible to pathogen invasion including SARS-CoV-2 (Alexander et al., 2019; Katz-Agranov and Zandman-Goddard, 2021).
There is a dominancy of genus Clostridium in patients with COVID-19 (Gu et al., 2020b; Zuo et al., 2020a; Cao et al., 2021; Yamamoto et al., 2021). The increase in Clostridium ramosum and Clostridium hathewayi is associated with the disease severity which may be a risk factor of acute portal vein thrombosis (Zuo et al., 2020; Rokkam et al., 2021). Clostridium difficile may complicate COVID-19 and worsen the GI symptoms (Ferreira et al., 2020; Oba et al., 2020; Sandhu et al., 2020; Khanna and Kraft, 2021). Two butyrate-producing members of this genus, Clostridium butyricum and Clostridium leptum, are decreased in patients with COVID-19 (Tang et al., 2020).
Streptococcus is another important bacterial genus which increases in COVID-19 (d’Ettorre et al., 2020; Donati Zeppa et al., 2020; Gu et al., 2020b; Zuo et al., 2021a). The abundance of Streptococcus is also an indicator of the extent of opportunistic bacterial invasion (Weiser et al., 2018; Tao et al., 2020). Streptococcus abundance is associated with more expressions of IL-18, TNF-α, and IFN-γ and other inflammatory cytokines worsening clinical outcomes (Tao et al., 2020; van der Lelie and Taghavi, 2020; Chhibber-Goel et al., 2021). An altered gut integrity affected by dysbiosis and inflammatory cytokines seems to be the main cause of increase in Streptococcus abundance in COVID-19 (Donati Zeppa et al., 2020). Streptococcus also affects the lung microbiome with proinflammatory activity (Kyo et al., 2019; Yamamoto et al., 2021).
Genus Rothia dominancy increases in patients with COVID-19 (Gu et al., 2020b; Lv et al., 2021a). This genus seems to be associated with inflammatory lung injuries (Han et al., 2020; Chattopadhyay and Shankar, 2021).
The genus Collinsella is an opportunistic pathogenic genus which is widely found in the gut of patients with COVID-19 especially in severe cases and higher infectivity status (Chhibber-Goel et al., 2021; Liu et al., 2021; Massip Copiz, 2021; Rajput et al., 2021; Zuo et al., 2021a). Collinsella aerofaciens abundance altered with the gut mucosal integrity and production of inflammatory mediators such as IL-17, CXCL1, and CXCL5 from the luminal cells (Kalinkovich and Livshits, 2019).
The alteration of genus Parabacteroides in COVID-19 is an area of controversy (Tang et al., 2020; Tao et al., 2020; Chhibber-Goel et al., 2021; Zuo et al., 2021a; Yeoh et al., 2021). It has been suggested that higher levels of Parabacteroides in microbiota are associated with better gut mucosal integrity (Venegas et al., 2019; Tang et al., 2020; Chhibber-Goel et al., 2021).
Ruminococcus species such as Ruminococcus gnavus and Ruminococcus torques increase (Cao et al., 2021; Yeoh et al., 2021), and species including Ruminococcus bromii, Ruminococcus obeum, and Ruminococcus sp. 5139BFAA are reduced in patients with COVID-19 (Zuo et al., 2020a; Cao et al., 2021; Lv et al., 2021). Ruminococcus gnavus and Ruminococcus torques are known as proinflammatory bacteria which previously have been shown to be associated with proinflammatory status and higher production of inflammatory mediators (Matsuoka and Kanai, 2015; Hall et al., 2017; Henke et al., 2019; Yeoh et al., 2021). Reduction of Ruminococcus obeum seems to be secondary to the wide use/misuse of antibiotics in the management of COVID-19 patients (Cyprian et al., 2021). Among Alistipes genera, Alistipes_sp_AP11, Alistipes indistinctus, and Alistipes shahii are reduced and Alistipes onderdonkii increased in COVID-19 (Zuo et al., 2020a; Cao et al., 2021). Alistipes onderdonkii is one of the most important sources of short-chain fatty acid (SCFA) production in the gut that helps to the gut homeostasis (Venegas et al., 2019; Tang et al., 2020). Alistipes is also important in preservation of the gut immunity via being involved in tryptophan synthesis pathways (Gao et al., 2018).
Bacteroides alteration is reported in COVID-19, especially in critically ill patients (Tang et al., 2020; Cao et al., 2021; Chattopadhyay and Shankar, 2021; Yeoh et al., 2021; Zuo et al., 2021a). Bacteroides are the most critical commensal bacterial genera in gut whose alterations are associated with several conditions affecting human health and disease (Ley et al., 2006; Turnbaugh et al., 2006; Larsen et al., 2010; Claesson et al., 2012; Yu et al., 2015; Boursier et al., 2016; Salazar et al., 2017; Belizário et al., 2018; O’Toole and Jeffery, 2018; Yildiz et al., 2018; Antosca et al., 2019; Rahayu et al., 2019; Crovesy et al., 2020; Juárez-Fernández et al., 2021). Bacteroides also have immunomodulatory effects, which is mainly mediated by alterations in production of polysaccharide A, IL-6, IL-7, IL-10, dendritic cells, and CD4+ and CD8+ T cells (Abt et al., 2012; Zhang et al., 2018; Jia et al., 2018; Ramakrishna et al., 2019; Alvarez et al., 2020; Gautier et al., 2021). Bacteroides are also associated with reduction of the expression of the ACE-2 receptor (Chattopadhyay and Shankar, 2021). Use of antibiotics seems to result in increase of Bacteroides caccae in a COVID-19 patient. In antibiotic-naive COVID-19 patients, Bacteroides nordii are more common (Chattopadhyay and Shankar, 2021). On the other hand, species such as Bacteroides massiliensis, Bacteroides dorei, Bacteroides thetaiotaomicron, and Bacteroides ovatus decrease in SARS-CoV-2 infection (Zuo et al., 2020a; Cao et al., 2021). Bacteroides dorei itself is a controversial bacterium with both evidence of increase and decrease in COVID-19 patients. This species is associated with IL-6 and IL-8 and downregulation of the ACE-2 receptor (Yoshida et al., 2018; Yeoh et al., 2021).
Bifidobacterium, a major bacterial genus in the gut, reduces by SARS-CoV-2 (Gu et al., 2020b). In patients who received fecal microbial transplant (FMT), a re-expansion of Bifidobacterium in their gut was demonstrated (Liu et al., 2021). Bifidobacterium spp. are well known for their immunomodulatory effects especially on Th17 and in the amelioration of inflammatory process (de Vrese et al., 2005; Wang et al., 2011; Groeger et al., 2013; Jungersen et al., 2014; King et al., 2014; Han et al., 2016; Schiavi et al., 2016; Bozkurt et al., 2019; Bozkurt and Kara, 2020; Tian et al., 2020; Arenas-Padilla et al., 2021; Chen and Vitetta, 2021; Hong et al., 2021; Milner et al., 2021). Bifidobacterium may be considered as a supplemental therapeutic agent for controlling cytokine storm and inflammation in patients with COVID-19 (Bozkurt and Quigley, 2020; Bozkurt and Quigley, 2020a; Schett et al., 2020; Bhushan et al., 2021 Gautier et al., 2021; Mohseni et al., 2021).
Faecalibacterium decreases in COVID-19 and has been related to the severity of disease (Zuo et al., 2020; Gu et al., 2020; Tang et al., 2020; Yamamoto et al., 2021; Lv et al., 2021). Faecalibacterium is a butyrate-producing genus which positively impacts on intestinal mucosal integrity and is also known to have anti-inflammatory effects (Alameddine et al., 2019; Zuo et al., 2020a; Chhibber-Goel et al., 2021). Fecal transplantation significantly increases the abundance of Faecalibacterium in discharged patients with COVID-19 and has been shown to improve the inflammation states (Sokol et al., 2008; van den Munckhof et al., 2018; Venegas et al., 2019; Liu et al., 2021).
Lachnospiraceae, which is known as SCFA-producing bacteria, decreases in patients with COVID-19 (d’Ettorre et al., 2020; Gu et al., 2020b; Zuo et al., 2020a; Cao et al., 2021; Gaibani et al., 2021; Gautier et al., 2021; Zuo et al., 2021a). It may be attributed to common use of azithromycin and other antibiotics in the management of COVID-19 (Segal et al., 2020).
Genus Roseburia is another commensal gut microbiota which decreases in patients with COVID-19 and other viral diseases such as influenza (Wang et al., 2017; Gu et al., 2020b; Cao et al., 2021; Lv et al., 2021a). SCFAs produced by Roseburia maintain mucosal integrity in healthy adults via modulation of inflammatory mediators especially IL-10 (Koh et al., 2016; Zheng et al., 2017; Haak et al., 2018; Gautier et al., 2021). It has been shown previously that butyrate may preserve lung integrity from cytokine-induced injuries in influenza (Chakraborty et al., 2017; Dang and Marsland, 2019). If we assume that it is true in COVID-19, lower levels of Roseburia result in lower levels of butyrate and consequently more extensive lung injuries due to inflammatory processes.
Eubacterium is a genus with immunomodulatory effects which significantly decrease in gut microbiota of patients with COVID-19 (d’Ettorre et al., 2020; Zuo et al., 2020a; Gu et al., 2020b; Cao et al., 2021; Chattopadhyay and Shankar, 2021; Gautier et al., 2021; Lv et al., 2021; Yeoh et al., 2021). Wide use of antibiotics is considered to be associated with reduction of this genus (Zuo et al., 2020a). This genus similar to Roseburia spp. produces butyrate and modulates inflammation in inflammation-mediated injuries (Koh et al., 2016; Zheng et al., 2017; Haak et al., 2018; Gautier et al., 2021).
Fusicatenibacter is another bacterial genus that reduced during the course of COVID-19 (Gu et al., 2020; Chattopadhyay and Shankar, 2021; Lv et al., 2021a). Fusicatenibacter alteration is a very sensitive biomarker during COVID-19. It is proposed to be a diagnostic tool for COVID-19 (Gu et al., 2020b; Segal et al., 2020; Cyprian et al., 2021; Howell et al., 2021). This genus is also negatively correlated with CRP and procalcitonin levels in patients with COVID-19 (Gu et al., 2020b).
Other members of the gut microbiota are viruses and fungi. Although pathogenic gut viruses are known for more than a century, the term “gut virome” is recently introduced (Reyes et al., 2012). Most of the gut virome consists of bacteriophages that can explain the fact that the virome structure is related to gut bacterial composition in both healthy and COVID-19 people (Minot et al., 2011; Cao et al., 2021). There is a bidirectional relationship between gut virome and infectious diseases; bacteriophages have a significant role in protecting against bacterial infections (Wilks and Golovkina, 2012). Gut virome composition might be affected during COVID-19 (Cao et al., 2021). There are limited data on the alterations of commensal viral and fungal populations in the gut during COVID-19 infection.
Lv et al. showed a strong correlation between altered fungal gut microbiome and inflammatory blood biomarkers (Lv et al., 2021b). Further studies focusing on viral and fungal alterations during the COVID-19 are desired.
The gut microbiota alteration in COVID-19 patients should be considered as a dynamic process (d’Ettorre et al., 2020; Liu et al., 2021; Hussain et al., 2021; Zuo et al., 2020a). To the date of revising this manuscript (January 2022), several registered clinical trials are in progress and the results are not provided yet; however, growing evidence supports the effectiveness of microbiota modulatory actions on fastening the recovery of patients with COVID-19 (Chen and Vitetta, 2021; Hussain et al., 2021; Wang et al., 2021a).
Limitations and Suggestions
A few studies have documented the comorbidities of subjects. However, almost all the studies have missed the impact of comorbidities on gut microbiota alterations in COVID-19 patients compared with the healthy control. It has been shown that gut microbiota may change in patients with hypertension, cardiovascular diseases, diabetes mellitus, hyperlipidemia, and thrombotic events (Huynh, 2020; Avery et al., 2021; Kyriakidou et al., 2021; Mineshita et al., 2021). We strongly propose to investigate the effects of underlying comorbidities in gut microbial composition in patients with COVID-19.
Due to the variations in data analysis techniques such as 16S rRNA sequencing, qPCR, and metagenome sequencing, there was a challenge to compare the bacterial taxa across studies. Since there was a fair diversity in geographical distribution of the current studies (most of them are from China), we cannot ignore the effect of diet and genetic predisposing factors like HLA in gut microbiome compositions. Future studies in different countries are required in this regard.
It is important to mention that different levels of p-value significance were reported in reviewed articles; however, in this study we used only statistically significant findings from the included studies.
Further studies with a larger study population, including the range of patients from mild to severe symptoms, involving the patients who are managed out patiently, focusing on the effectiveness of gut microbiota-targeted therapies for prevention and improvement of COVID-19 patients’ symptoms are desired to light up this topic.
Conclusion
Our study showed a significant alteration of gut microbiome composition in patients with COVID-19 compared to healthy individuals. This great extent of impact has proposed the gut microbiota as a potential diagnostic, prognostic, and potentially therapeutic strategy for COVID-19.
Author Contributions
MN and PJ designed the study. MN, YF, AT, PJ, MA, and FV performed the search and data extraction and wrote the first draft of the manuscript. LS, AHSB, and MM reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study was supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran.
References
- Abt M. C., Osborne L. C., Monticelli L. A., Doering T. A., Alenghat T., Sonnenberg G. F., et al. (2012). Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity 37 (1), 158–170. doi: 10.1016/j.immuni.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi Badi S., Tarashi S., Fateh A., Rohani P., Masotti A., Siadat S. D. (2021). From the Role of Microbiota in Gut-Lung Axis to SARS-CoV-2 Pathogenesis. Mediators Inflamm. 2021, 6611222. doi: 10.1155/2021/6611222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktaş B., Aslim B. (2020). Gut-Lung Axis and Dysbiosis in COVID-19. Turkish J. Biol. 44 (SI-1), 265–272. doi: 10.3906/biy-2005-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alameddine J., Godefroy E., Papargyris L., Sarrabayrouse G., Tabiasco J., Bridonneau C., et al. (2019). Faecalibacterium Prausnitzii Skews Human DC to Prime IL10-Producing T Cells Through TLR2/6/JNK Signaling and IL-10, IL-27, CD39, and IDO-1 Induction. Front. Immunol. 10, 143. doi: 10.3389/fimmu.2019.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M., Ang Q. Y., Turnbaugh P. J. (2019). A Diet-Dependent Enzyme From the Human Gut Microbiome Promotes Th17 Accumulation and Colitis. bioRxiv Preprint Server Biol., 766899. doi: 10.1101/766899 [DOI] [Google Scholar]
- Allali I., Bakri Y., Amzazi S., Ghazal H. (2021). Gut-Lung Axis in COVID-19. Interdiscip. Perspect. Infect. Dis. 2021, 6655380. doi: 10.1155/2021/6655380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C. A., Jones M. B., Hambor J., Cobb B. A. (2020). Characterization of Polysaccharide A Response Reveals Interferon Responsive Gene Signature and Immunomodulatory Marker Expression. Front. Immunol. 11 (2677). doi: 10.3389/fimmu.2020.556813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosca K. M., Chernikova D. A., Price C. E., Ruoff K. L., Li K., Guill M. F., et al. (2019). Altered Stool Microbiota of Infants With Cystic Fibrosis Shows a Reduction in Genera Associated With Immune Programming From Birth. J. Bacteriol. 201 (16), e00274–e00219. doi: 10.1128/JB.00274-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes K. H., Fachi J. L., de Paula R., da Silva E. F., Pral L. P., Dos Santos A., et al. (2019). Microbiota-Derived Acetate Protects Against Respiratory Syncytial Virus Infection Through a GPR43-Type 1 Interferon Response. Nat. Commun. 10 (1), 3273. doi: 10.1038/s41467-019-11152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Padilla M., González-Rascón A., Hernández-Mendoza A., Calderón de la Barca A. M., Hernández J., Mata-Haro V. (2021). Immunomodulation by Bifidobacterium Animalis Subsp. Lactis Bb12: Integrative Analysis of miRNA Expression and TLR2 Pathway-Related Target Proteins in Swine Monocytes. Probiot. Antimicrob. Proteins 20, 1–13. doi: 10.1007/s12602-021-09816-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery E. G., Bartolomaeus H., Maifeld A., Marko L., Wiig H., Wilck N., et al. (2021). The Gut Microbiome in Hypertension: Recent Advances and Future Perspectives. Circ. Res. 128 (7), 934–950. doi: 10.1161/CIRCRESAHA.121.318065 [DOI] [PubMed] [Google Scholar]
- Battaglini D., Robba C., Fedele A., Trancǎ S., Sukkar S. G., Di Pilato V., et al. (2021). The Role of Dysbiosis in Critically Ill Patients With COVID-19 and Acute Respiratory Distress Syndrome. Front. Med. 8, 671714. doi: 10.3389/fmed.2021.671714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belizário J. E., Faintuch J., Garay-Malpartida M. (2018). Gut Microbiome Dysbiosis and Immunometabolism: New Frontiers for Treatment of Metabolic Diseases. Mediators Inflamm. 2018, 2037838. doi: 10.1155/2018/2037838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan I., Sharma M., Mehta M., Badyal S., Sharma V., Sharma I., et al. (2021). Bioactive Compounds and Probiotics–a Ray of Hope in COVID-19 Management. Food Sci. Hum. Wellness 10 (2), 131–140. doi: 10.1016/j.fshw.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F., et al. (2016). The Severity of Nonalcoholic Fatty Liver Disease is Associated With Gut Dysbiosis and Shift in the Metabolic Function of the Gut Microbiota. Hepatology 63 (3), 764–775. doi: 10.1002/hep.28356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt H. S., Kara B. (2020). A New Treatment for Ulcerative Colitis: Intracolonic Bifidobacterium and Xyloglucan Application. Eur. J. Inflamm. 18, 2058739220942626. doi: 10.1177/2058739220942626 [DOI] [Google Scholar]
- Bozkurt H. S., Quigley E. M. (2020. a). The Probiotic Bifidobacterium in the Management of Coronavirus: A Theoretical Basis. Int. J. Immunopathol. Pharmacol. 34, 2058738420961304. doi: 10.1177/2058738420961304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt H., Quigley E. (2020. b). Probiotic Bifidobacteria Against the COVİD-19: A New Way Out! Int. J. Immunopathol. Pharmacol. 34. doi: 10.1177/2058738420961304 [DOI] [Google Scholar]
- Bozkurt H. S., Quigley E. M., Kara B. (2019). Bifidobacterium Animalis Subspecies Lactis Engineered to Produce Mycosporin-Like Amino Acids in Colorectal Cancer Prevention. SAGE Open Med. 7, 2050312119825784. doi: 10.1177/2050312119825784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. L., Sequeira R. P., Clarke T. B. (2017). The Microbiota Protects Against Respiratory Infection via GM-CSF Signaling. Nat. Commun. 8 (1), 1512. doi: 10.1038/s41467-017-01803-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden K. F., Gellatly S. L., Wood D. L., Cooper M. A., Morrison M., Hugenholtz P., et al. (2017). Emerging Pathogenic Links Between Microbiota and the Gut-Lung Axis. Nat. Rev. Microbiol. 15 (1), 55–63. doi: 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- Cao J., Wang C., Zhang Y., Lei G., Xu K., Zhao N., et al. (2021). Integrated Gut Virome and Bacteriome Dynamics in COVID-19 Patients. Gut Microbes 13 (1), 1887722. doi: 10.1080/19490976.2021.1887722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K., Raundhal M., Chen B. B., Morse C., Tyurina Y. Y., Khare A., et al. (2017). The Mito-DAMP Cardiolipin Blocks IL-10 Production Causing Persistent Inflammation During Bacterial Pneumonia. Nat. Commun. 8, 13944. doi: 10.1038/ncomms13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay I., Shankar E. M. (2021). SARS-CoV-2-Indigenous Microbiota Nexus: Does Gut Microbiota Contribute to Inflammation and Disease Severity in COVID-19? Front. Cell. Infect. Microbiol. 11 (96). doi: 10.3389/fcimb.2021.590874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Vitetta L. (2021). Modulation of Gut Microbiota for the Prevention and Treatment of COVID-19. J. Clin. Med. 10 (13), 2903. doi: 10.3390/jcm10132903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber-Goel J., Gopinathan S., Sharma A. (2021). Interplay Between Severities of COVID-19 and the Gut Microbiome: Implications of Bacterial Co-Infections? Gut Pathogens 13 (1), 14. doi: 10.1186/s13099-021-00407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J., Jeffery I. B., Conde S., Power S. E., O’Connor E. M., Cusack S., et al. (2012). Gut Microbiota Composition Correlates With Diet and Health in the Elderly. Nature 488 (7410), 178–184. doi: 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Crovesy L., Masterson D., Rosado E. L. (2020). Profile of the Gut Microbiota of Adults With Obesity: A Systematic Review. Eur. J. Clin. Nutr. 74 (9), 1251–1262. doi: 10.1038/s41430-020-0607-6 [DOI] [PubMed] [Google Scholar]
- Cyprian F., Sohail M. U., Abdelhafez I., Salman S., Attique Z., Kamareddine L., et al. (2021). SARS-CoV-2 and Immune-Microbiome Interactions: Lessons From Respiratory Viral Infections. Int. J. Infect. Dis. 105, 540–550. doi: 10.1016/j.ijid.2021.02.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico F., Baumgart D. C., Danese S., Peyrin-Biroulet L. (2020). Diarrhea During COVID-19 Infection: Pathogenesis, Epidemiology, Prevention, and Management. Clin. Gastroenterol. Hepatol. 18 (8), 1663–1672. doi: 10.1016/j.cgh.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang A. T., Marsland B. J. (2019). Microbes, Metabolites, and the Gut-Lung Axis. Mucosal Immunol. 12 (4), 843–850. doi: 10.1038/s41385-019-0160-6 [DOI] [PubMed] [Google Scholar]
- d’Ettorre G., Ceccarelli G., Marazzato M., Campagna G., Pinacchio C., Alessandri F., et al. (2020). Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 7, 389. doi: 10.3389/fmed.2020.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrese M., Winkler P., Rautenberg P., Harder T., Noah C., Laue C., et al. (2005). Effect of Lactobacillus Gasseri PA 16/8, Bifidobacterium Longum SP 07/3, B. Bifidum MF 20/5 on Common Cold Episodes: A Double Blind, Randomized, Controlled Trial. Clin. Nutr. (Edinburgh Scotland) 24 (4), 481–491. doi: 10.1016/j.clnu.2005.02.006 [DOI] [PubMed] [Google Scholar]
- Dickson R. P., Huffnagle G. B. (2015). The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PloS Pathogens 11 (7), e1004923. doi: 10.1371/journal.ppat.1004923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din A. U., Mazhar M., Waseem M., Ahmad W., Bibi A., Hassan A., et al. (2021). SARS-CoV-2 Microbiome Dysbiosis Linked Disorders and Possible Probiotics Role. Biomed. Pharmacother. = Biomed. Pharmacother. 133, 110947. doi: 10.1016/j.biopha.2020.110947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G. P., Lee S. M., Mazmanian S. K. (2016). Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 14 (1), 20–32. doi: 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati Zeppa S., Agostini D., Piccoli G., Stocchi V., Sestili P. (2020). Gut Microbiota Status in COVID-19: An Unrecognized Player? Front. Cell. Infect. Microbiol. 10 (742). doi: 10.3389/fcimb.2020.576551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enaud R., Prevel R., Ciarlo E., Beaufils F., Wieërs G., Guery B., et al. (2020). The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 10 (9). doi: 10.3389/fcimb.2020.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes C. T., Amaral F. A., Vieira A. T., Soares A. C., Pinho V., Nicoli J. R., et al. (2012). Transient TLR Activation Restores Inflammatory Response and Ability to Control Pulmonary Bacterial Infection in Germfree Mice. J. Immunol. (Baltimore Md 1950) 188 (3), 1411–1420. doi: 10.4049/jimmunol.1101682 [DOI] [PubMed] [Google Scholar]
- Ferreira E., Penna B., Yates E. A. (2020). Should We Be Worried About Clostridioides Difficile During the SARS-CoV2 Pandemic? Front. Microbiol. 11 (2292). doi: 10.3389/fmicb.2020.581343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. C., McConnell K. W., Yoseph B. P., Breed E., Liang Z., Clark A. T., et al. (2012). The Endogenous Bacteria Alter Gut Epithelial Apoptosis and Decrease Mortality Following Pseudomonas Aeruginosa Pneumonia. Shock (Augusta Ga) 38 (5), 508–514. doi: 10.1097/SHK.0b013e31826e47e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P., D’Amico F., Bartoletti M., Lombardo D., Rampelli S., Fornaro G., et al. (2021). The Gut Microbiota of Critically Ill Patients With COVID-19. Front. Cell. Infect. Microbiol. 11 (554). doi: 10.3389/fcimb.2021.670424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., et al. (2018). Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 8, 13. doi: 10.3389/fcimb.2018.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T., David-Le Gall S., Sweidan A., Tamanai-Shacoori Z., Jolivet-Gougeon A., Loréal O., et al. (2021). Next-Generation Probiotics and Their Metabolites in COVID-19. Microorganisms 9 (5), 941. doi: 10.3390/microorganisms9050941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T. G., Ortiz-Lopez A., et al. (2017). Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168 (5), 928–943.e11. doi: 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger D., O’Mahony L., Murphy E. F., Bourke J. F., Dinan T. G., Kiely B., et al. (2013). Bifidobacterium Infantis 35624 Modulates Host Inflammatory Processes Beyond the Gut. Gut Microbes 4 (4), 325–339. doi: 10.4161/gmic.25487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. (2020. a). COVID-19: Gastrointestinal Manifestations and Potential Fecal-Oral Transmission. Gastroenterology 158 (6), 1518–1519. doi: 10.1053/j.gastro.2020.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., et al. (2020. b). Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 71 (10), 2669–2678. doi: 10.1093/cid/ciaa709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak B. W., Littmann E. R., Chaubard J. L., Pickard A. J., Fontana E., Adhi F., et al. (2018). Impact of Gut Colonization With Butyrate-Producing Microbiota on Respiratory Viral Infection Following Allo-HCT. Blood 131 (26), 2978–2986. doi: 10.1182/blood-2018-01-828996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. B., Yassour M., Sauk J., Garner A., Jiang X., Arthur T., et al. (2017). A Novel Ruminococcus Gnavus Clade Enriched in Inflammatory Bowel Disease Patients. Genome Med. 9 (1), 103. doi: 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Ding Z., Shi H., Qian W., Hou X., Lin R. (2016). The Role of Probiotics in Lipopolysaccharide-Induced Autophagy in Intestinal Epithelial Cells. Cell. Physiol. Biochem. 38 (6), 2464–2478. doi: 10.1159/000445597 [DOI] [PubMed] [Google Scholar]
- Han Y., Jia Z., Shi J., Wang W., He K. (2020). The Active Lung Microbiota Landscape of COVID-19 Patients. medRxiv. doi: 10.1101/2020.08.20.20144014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K. L. (2002). Quantitative mRNA Expression Profiling of ACE 2, a Novel Homologue of Angiotensin Converting Enzyme. FEBS Lett. 532 (1-2), 107–110. doi: 10.1016/S0014-5793(02)03640-2 [DOI] [PubMed] [Google Scholar]
- Hegazy M., Ahmed Ashoush O., Tharwat Hegazy M., Wahba M., Lithy R. M., Abdel-Hamid H. M., et al. (2021). Beyond Probiotic Legend: ESSAP Gut Microbiota Health Score to Delineate SARS-COV-2 Infection Severity. Br. J. Nutr. 7, 1–10. doi: 10.1017/S0007114521001926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke M. T., Kenny D. J., Cassilly C. D., Vlamakis H., Xavier R. J., Clardy J. (2019). Ruminococcus Gnavus, a Member of the Human Gut Microbiome Associated With Crohn’s Disease, Produces an Inflammatory Polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 116 (26), 12672–12677. doi: 10.1073/pnas.1904099116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wang J., Li F., Shi Y. (2020). Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front. Microbiol. 11, 1302. doi: 10.3389/fmicb.2020.01302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wen Q., Yao F., Xu D., Huang Y., Wang J. (2017). Gut-Lung Axis: The Microbial Contributions and Clinical Implications. Crit. Rev. Microbiol. 43 (1), 81–95. doi: 10.1080/1040841X.2016.1176988 [DOI] [PubMed] [Google Scholar]
- He F., Zhang T., Xue K., Fang Z., Jiang G., Huang S., et al. (2021). Fecal Multi-Omics Analysis Reveals Diverse Molecular Alterations of Gut Ecosystem in COVID-19 Patients. Analyt. Chim. Acta 1180, 338881. doi: 10.1016/j.aca.2021.338881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong N., Ku S., Yuk K., Johnston T. V., Ji G. E., Park M. S. (2021). Production of Biologically Active Human Interleukin-10 by Bifidobacterium Bifidum BGN4. Microb. Cell Fact. 20 (1), 16. doi: 10.1186/s12934-020-01505-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M. C., Green R., McGill A. R., Dutta R., Mohapatra S., Mohapatra S. S. (2021). SARS-CoV-2-Induced Gut Microbiome Dysbiosis: Implications for Colorectal Cancer. Cancers 13 (11), 2676. doi: 10.3390/cancers13112676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I., Cher G. L. Y., Abid M. A., Abid M. B. (2021). Role of Gut Microbiome in COVID-19: An Insight Into Pathogenesis and Therapeutic Potential. Front. Immunol. 12. doi: 10.3389/fimmu.2021.765965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K. (2020). Novel Gut Microbiota-Derived Metabolite Promotes Platelet Thrombosis via Adrenergic Receptor Signalling. Nat. Rev. Cardiol. 17 (5), 265. doi: 10.1038/s41569-020-0367-y [DOI] [PubMed] [Google Scholar]
- Ichinohe T., Pang I. K., Kumamoto Y., Peaper D. R., Ho J. H., Murray T. S., et al. (2011). Microbiota Regulates Immune Defense Against Respiratory Tract Influenza A Virus Infection. Proc. Natl. Acad. Sci. U. S. A. 108 (13), 5354–5359. doi: 10.1073/pnas.1019378108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi P., Hajikhani B., Mirsaeidi M., Vahidnezhad H., Dadashi M., Nasiri M. J. (2021). Skin Manifestations in COVID-19 Patients: Are They Indicators for Disease Severity? A Systematic Review. Front. Med. 8, 634208. doi: 10.3389/fmed.2021.634208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi P., Hasanzadeh S., Tahvildari A., Farsi Y., Arbabi M., Mota J. F., et al. (2019). Is There Any Association Between Gut Microbiota and Type 1 Diabetes? A Systematic Review. Gut Pathog. 11 (1), 49. doi: 10.1186/s13099-019-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Xie G., Jia W. (2018). Bile Acid–Microbiota Crosstalk in Gastrointestinal Inflammation and Carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15 (2), 111–128. doi: 10.1038/nrgastro.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanna Briggs Institute . (2021). Critical Appraisal Tools (South Australia: Joanna Briggs Institute: University of Adelaide; ). Available at: https://jbi.global/critical-appraisal-tools. [Google Scholar]
- Juárez-Fernández M., Porras D., García-Mediavilla M. V., Román-Sagüillo S., González-Gallego J., Nistal E., et al. (2021). Aging, Gut Microbiota and Metabolic Diseases: Management Through Physical Exercise and Nutritional Interventions. Nutrients 13 (1), 16. doi: 10.3390/nu13010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungersen M., Wind A., Johansen E., Christensen J. E., Stuer-Lauridsen B., Eskesen D. (2014). The Science Behind the Probiotic Strain Bifidobacterium Animalis Subsp. lactis BB-12(®). Microorganisms 2 (2), 92–110. doi: 10.3390/microorganisms2020092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinkovich A., Livshits G. (2019). A Cross Talk Between Dysbiosis and Gut-Associated Immune System Governs the Development of Inflammatory Arthropathies. Semin. Arthritis Rheum. 49 (3), 474–484. doi: 10.1016/j.semarthrit.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Katz-Agranov N., Zandman-Goddard G. (2021). Autoimmunity and COVID-19 - The Microbiotal Connection. Autoimmun. Rev. 20 (8), 102865. doi: 10.1016/j.autrev.2021.102865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Mathew B. J., Gupta P., Garg G., Khadanga S., Vyas A. K., et al. (2021). Gut Dysbiosis and IL-21 Response in Patients With Severe COVID-19. Microorganisms 9 (6), 1292. doi: 10.3390/microorganisms9061292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S., Kraft C. S. (2021). The Interplay of SARS-CoV-2 and Clostridioides Difficile Infection. Future Microbiol. 16 (6), 439–443. doi: 10.2217/fmb-2020-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-N., Joo E.-J., Lee C.-W., Ahn K.-S., Kim H.-L., Park D.-I., et al. (2021). Reversion of Gut Microbiota During the Recovery Phase in Patients With Asymptomatic or Mild COVID-19: Longitudinal Study. Microorganisms 9 (6), 1237. doi: 10.3390/microorganisms9061237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S., Glanville J., Sanders M. E., Fitzgerald A., Varley D. (2014). Effectiveness of Probiotics on the Duration of Illness in Healthy Children and Adults Who Develop Common Acute Respiratory Infectious Conditions: A Systematic Review and Meta-Analysis. Br. J. Nutr. 112 (1), 41–54. doi: 10.1017/S0007114514000075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165 (6), 1332–1345. doi: 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- Kyo M., Nishioka K., Nakaya T., Kida Y., Tanabe Y., Ohshimo S., et al. (2019). Unique Patterns of Lower Respiratory Tract Microbiota are Associated With Inflammation and Hospital Mortality in Acute Respiratory Distress Syndrome. Respir. Res. 20 (1), 246. doi: 10.1186/s12931-019-1203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidou A., Koufakis T., Goulis D. G., Vasilopoulos Y., Zebekakis P., Kotsa K. (2021). Pharmacogenetics of the Glucagon-Like Peptide-1 Receptor Agonist Liraglutide: A Step Towards Personalized Type 2 Diabetes Management. Curr. Pharm. Des. 27 (8), 1025–1034. doi: 10.2174/1381612826666201203145654 [DOI] [PubMed] [Google Scholar]
- Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., et al. (2020). SARS-CoV-2 Productively Infects Human Gut Enterocytes. Sci. (New York NY) 369 (6499), 50–54. doi: 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Vogensen F. K., van den Berg F. W., Nielsen D. S., Andreasen A. S., Pedersen B. K., et al. (2010). Gut Microbiota in Human Adults With Type 2 Diabetes Differs From non-Diabetic Adults. PloS One 5 (2), e9085. doi: 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Human Gut Microbes Associated With Obesity. Nature 444 (7122), 1022–1023. doi: 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., et al. (2003). Angiotensin-Converting Enzyme 2 is a Functional Receptor for the SARS Coronavirus. Nature 426 (6965), 450–454. doi: 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Ye S., Zhu X., He X., Wang S., Li Y., et al. (2021). Gastrointestinal Disturbance and Effect of Fecal Microbiota Transplantation in Discharged COVID-19 Patients. J. Med. Case Rep. 15 (1), 60. doi: 10.1186/s13256-020-02583-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Gu S., Jiang H., Yan R., Chen Y., Chen Y., et al. (2021. b). Gut Mycobiota Alterations in Patients With COVID-19 and H1N1 Infections and Their Associations With Clinical Features. Commun. Biol. 4 (1), 480. doi: 10.1038/s42003-021-02036-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Jiang H., Chen Y., Gu S., Xia J., Zhang H., et al. (2021. a). The Faecal Metabolome in COVID-19 Patients is Altered and Associated With Clinical Features and Gut Microbes. Analyt. Chim. Acta 1152, 338267. doi: 10.1016/j.aca.2021.338267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massip Copiz M. M. (2021). The Gut Microbiota in Patients With COVID-19 and Obesity. FASEB J. 35 (S1). doi: 10.1096/fasebj.2021.35.S1.05159 [DOI] [Google Scholar]
- Matsuoka K., Kanai T. (Eds.) (2015). The Gut Microbiota and Inflammatory Bowel Disease. Seminars in Immunopathology (Springer; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner E., Stevens B., An M., Lam V., Ainsworth M., Dihle P., et al. (2021). Utilizing Probiotics for the Prevention and Treatment of Gastrointestinal Diseases. Front. Microbiol. 12, 689958. doi: 10.3389/fmicb.2021.689958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineshita Y., Sasaki H., H-k K., Shibata S. (2021). Relationship Between Fasting and Postprandial Glucose Levels and the Gut Microbiota, 24 September 2021, PREPRINT (Version 1). available at Research Square [https://doi.org/10.21203/rs.3.rs-917351/v1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S., Sinha R., Chen J., Li H., Keilbaugh S. A., Wu G. D., et al. (2011). The Human Gut Virome: Inter-Individual Variation and Dynamic Response to Diet. Genome Res. 21 (10), 1616–1625. doi: 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Ann. Internal Med. 151 (4), 264–269. doi: 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- Mohseni A. H., Casolaro V., Bermúdez-Humarán L. G., Keyvani H., Taghinezhad S. S. (2021). Modulation of the PI3K/Akt/mTOR Signaling Pathway by Probiotics as a Fruitful Target for Orchestrating the Immune Response. Gut Microbes 13 (1), 1–17. doi: 10.1080/19490976.2021.1886844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Rosário A., Marques C., Pinheiro H., Araújo J. R., Ribeiro P., Rocha R., et al. (2021). Gut Microbiota Diversity and C-Reactive Protein Are Predictors of Disease Severity in COVID-19 Patients. Front. Microbiol. 12, 705020. doi: 10.3389/fmicb.2021.705020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba J., Silva C. A., Toma R. K., Carvalho W. B., Delgado A. F. (2020). COVID-19 and Coinfection With Clostridioides (Clostridium) Difficile in an Infant With Gastrointestinal Manifestation. Einstein (Sao Paulo Brazil) 18, eRC6048. doi: 10.31744/einstein_journal/2020RC6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole P. W., Jeffery I. B. (2018). Microbiome-Health Interactions in Older People. Cell. Mol. Life Sci. 75 (1), 119–128. doi: 10.1007/s00018-017-2673-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K. P., Patel P. A., Vunnam R. R., Hewlett A. T., Jain R., Jing R., et al. (2020). Gastrointestinal, Hepatobiliary, and Pancreatic Manifestations of COVID-19. J. Clin. Virol. 128, 104386. doi: 10.1016/j.jcv.2020.104386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Patton M. J., Floyd J. L., Vieira C. P., Fortmann S., DuPont M., et al. (2021). Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. bioRxiv Preprint Server Biol. 33851159. doi: 10.1101/2021.04.06.438634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayu E. S., Utami T., Mariyatun M., Hasan P. N., Kamil R. Z., Setyawan R. H., et al. (2019). Gut Microbiota Profile in Healthy Indonesians. World J. Gastroenterol. 25 (12), 1478–1491. doi: 10.3748/wjg.v25.i12.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput S., Paliwal D., Naithani M., Kothari A., Meena K., Rana S. (2021). COVID-19 and Gut Microbiota: A Potential Connection. Indian J. Clin. Biochem. 36 (3), 1–12. doi: 10.1007/s12291-020-00948-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna C., Kujawski M., Chu H., Li L., Mazmanian S. K., Cantin E. M. (2019). Bacteroides Fragilis Polysaccharide A Induces IL-10 Secreting B and T Cells That Prevent Viral Encephalitis. Nat. Commun. 10 (1), 2153. doi: 10.1038/s41467-019-09884-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Semenkovich N. P., Whiteson K., Rohwer F., Gordon J. I. (2012). Going Viral: Next-Generation Sequencing Applied to Phage Populations in the Human Gut. Nat. Rev. Microbiol. 10 (9), 607–617. doi: 10.1038/nrmicro2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokkam V. R. P., Kutti G., Sridharan, Vegunta R., Vegunta R., Boregowda U., Mohan B. P. (2021). Clostridium Difficile and COVID-19: Novel Risk Factors for Acute Portal Vein Thrombosis. Case Rep. Vasc. Med. 2021, 8832638. doi: 10.1155/2021/8832638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar N., Valdés-Varela L., González S., Gueimonde M., de Los Reyes-Gavilán C. G. (2017). Nutrition and the Gut Microbiome in the Elderly. Gut Microbes 8 (2), 82–97. doi: 10.1080/19490976.2016.1256525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu A., Tillotson G., Polistico J., Salimnia H., Cranis M., Moshos J., et al. (2020). Clostridioides Difficile in COVID-19 Patients, Detroit, Michigan, USA, March-April 2020. Emerg. Infect. Dis. 26 (9), 2272–2274. doi: 10.3201/eid2609.202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G., Sticherling M., Neurath M. F. (2020). COVID-19: Risk for Cytokine Targeting in Chronic Inflammatory Diseases? Nat. Rev. Immunol. 20 (5), 271–272. doi: 10.1038/s41577-020-0312-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi E., Gleinser M., Molloy E., Groeger D., Frei R., Ferstl R., et al. (2016). The Surface-Associated Exopolysaccharide of Bifidobacterium Longum 35624 Plays an Essential Role in Dampening Host Proinflammatory Responses and Repressing Local TH17 Responses. Appl. Environ. Microbiol. 82 (24), 7185–7196. doi: 10.1128/AEM.02238-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S. P., Vlamakis H., Jaeger M., Oosting M., Franzosa E. A., et al. (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167 (4), 1125–1136.e8. doi: 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal J. P., Mak J. W. Y., Mullish B. H., Alexander J. L., Ng S. C., Marchesi J. R. (2020). The Gut Microbiome: An Under-Recognised Contributor to the COVID-19 Pandemic? Ther. Adv. Gastroenterol. 13, 1756284820974914. doi: 10.1177/1756284820974914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L. G., Gratadoux J.-J., et al. (2008). Faecalibacterium Prausnitzii is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. 105 (43), 16731–16736. doi: 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M., Tokuyama M., Wei G., Huang R., Livanos A., Jha D., et al. (2021). Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps With the Pathogenesis of SARS-CoV-2-Related Disease. Gastroenterology 160 (1), 287–301.e20. doi: 10.1053/j.gastro.2020.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gu S., Gong Y., Li B., Lu H., Li Q., et al. (2020). Clinical Significance of the Correlation Between Changes in the Major Intestinal Bacteria Species and COVID-19 Severity. Eng. (Beijing China) 6 (10), 1178–1184. doi: 10.1016/j.eng.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Zhang G., Wang X., Guo M., Zeng W., Xu Z., et al. (2020). Analysis of the Intestinal Microbiota in COVID-19 Patients and its Correlation With the Inflammatory Factor IL-18. Med. Microecol. 5, 100023. doi: 10.1016/j.medmic.2020.100023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Zhao Y., Yang Y., Wang T., Jin S., Guo J., et al. (2020). The Protective Role of Short-Chain Fatty Acids Acting as Signal Molecules in Chemotherapy- or Radiation-Induced Intestinal Inflammation. Am. J. Cancer Res. 10 (11), 3508–3531. [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E. S., Pattaroni C., Lopez-Mejia I. C., Riva E., Pernot J., et al. (2018). Dietary Fiber Confers Protection Against Flu by Shaping Ly6c(-) Patrolling Monocyte Hematopoiesis and CD8(+) T Cell Metabolism. Immunity 48 (5), 992–1005.e8. doi: 10.1016/j.immuni.2018.04.022 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature 444 (7122), 1027–1031. doi: 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- van den Munckhof I. C., Kurilshikov A., ter Horst R., Riksen N. P., Joosten L., Zhernakova A., et al. (2018). Role of Gut Microbiota in Chronic Low-Grade Inflammation as Potential Driver for Atherosclerotic Cardiovascular Disease: A Systematic Review of Human Studies. Obes. Rev. 19 (12), 1719–1734. doi: 10.1111/obr.12750 [DOI] [PubMed] [Google Scholar]
- van der Lelie D., Taghavi S. (2020). COVID-19 and the Gut Microbiome: More Than a Gut Feeling. Msystems 5 (4), e00453–e00420. doi: 10.1128/mSystems.00453-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas D., Marjorie K., Landskron G., González M., Quera R., Dijkstra G., et al. (2019). Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 10, 277. doi: 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton G. E., Gibson G. R., Hunter K. A. (2021). Mechanisms Linking the Human Gut Microbiome to Prophylactic and Treatment Strategies for COVID-19. Br. J. Nutr. 126 (2), 219–227. doi: 10.1017/S0007114520003980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang J., Cheng Y., Liu X., Huang Y. (2011). Secreted Factors From Bifidobacterium Animalis Subsp. Lactis Inhibit NF-κb-Mediated Interleukin-8 Gene Expression in Caco-2 Cells. Appl. Environ. Microbiol. 77 (22), 8171–8174. doi: 10.1128/AEM.06145-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang H., Sun Y., Ren Z., Zhu W., Li A., et al. (2021. a). Potential Associations Between Microbiome and COVID-19. Front. Med. 8. doi: 10.3389/fmed.2021.785496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang Y., Lu C., Qiu L., Song X., Jia H., et al. (2021. b). The Efficacy of Probiotics in Patients With Severe COVID-19. Ann. Palliat. Med. 10 (12), 12374–12380. doi: 10.21037/apm-21-3373 [DOI] [PubMed] [Google Scholar]
- Wang B., Yao M., Lv L., Ling Z., Li L. (2017). The Human Microbiota in Health and Disease. Engineering 3 (1), 71–82. doi: 10.1016/J.ENG.2017.01.008 [DOI] [Google Scholar]
- Weiser J. N., Ferreira D. M., Paton J. C. (2018). Streptococcus Pneumoniae: Transmission, Colonization and Invasion. Nat. Rev. Microbiol. 16 (6), 355–367. doi: 10.1038/s41579-018-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks J., Golovkina T. (2012). Influence of Microbiota on Viral Infections. PloS Pathogens 8 (5), e1002681. doi: 10.1371/journal.ppat.1002681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Cheng X., Jiang G., Tang H., Ming S., Tang L., et al. (2021). Altered Oral and Gut Microbiota and its Association With SARS-CoV-2 Viral Load in COVID-19 Patients During Hospitalization. NPJ Biofilms Microbiomes 7 (1), 61. doi: 10.1038/s41522-021-00232-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. (2020). Prolonged Presence of SARS-CoV-2 Viral RNA in Faecal Samples. Lancet Gastroenterol. Hepatol. 5 (5), 434–435. doi: 10.1016/S2468-1253(20)30083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Mi Z., Wang Z., Pang Z., Liu H., Zhang F. (2021). High Expression of ACE2 on Keratinocytes Reveals Skin as a Potential Target for SARS-CoV-2. J. Invest. Dermatol. 141 (1), 206–209.e1. doi: 10.1016/j.jid.2020.05.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., et al. (2020). Characteristics of Pediatric SARS-CoV-2 Infection and Potential Evidence for Persistent Fecal Viral Shedding. Nat. Med. 26 (4), 502–505. doi: 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Saito M., Tamura A., Prawisuda D., Mizutani T., Yotsuyanagi H. (2021). The Human Microbiome and COVID-19: A Systematic Review. PloS One 16 (6), e0253293. doi: 10.1371/journal.pone.0253293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y. K., Zuo T., Lui G. C., Zhang F., Liu Q., Li A. Y., et al. (2021). Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients With COVID-19. Gut 70 (4), 698–706. doi: 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz S., Mazel-Sanchez B., Kandasamy M., Manicassamy B., Schmolke M. (2018). Influenza A Virus Infection Impacts Systemic Microbiota Dynamics and Causes Quantitative Enteric Dysbiosis. Microbiome 6 (1), 9. doi: 10.1186/s40168-017-0386-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Emoto T., Yamashita T., Watanabe H., Hayashi T., Tabata T., et al. (2018). Bacteroides Vulgatus and Bacteroides Dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 138 (22), 2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714 [DOI] [PubMed] [Google Scholar]
- Yu B., Dai C. -Q., Chen J., Deng L., Wu X. -I., Wu S., et al. (2015). Dysbiosis of Gut Microbiota Induced the Disorder of Helper T Cells in Influenza Virus-Infected Mice. Hum. Vaccines Immunother. 11 (5), 1140–1146. doi: 10.1080/21645515.2015.1009805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li S., Wang N., Tan H. Y., Zhang Z., Feng Y. (2020). The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 11, 301. doi: 10.3389/fmicb.2020.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhu B., Xu J., Liu Y., Qiu E., Li Z., et al. (2018). Bacteroides Fragilis Protects Against Antibiotic-Associated Diarrhea in Rats by Modulating Intestinal Defenses. Front. Immunol. 9 (1040). doi: 10.3389/fimmu.2018.01040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Kelly C. J., Battista K. D., Schaefer R., Lanis J. M., Alexeev E. E., et al. (2017). Microbial-Derived Butyrate Promotes Epithelial Barrier Function Through IL-10 Receptor-Dependent Repression of Claudin-2. J. Immunol. (Baltimore Md 1950) 199 (8), 2976–2984. doi: 10.4049/jimmunol.1700105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., et al. (2020). A Pneumonia Outbreak Associated With a New Coronavirus of Probable Bat Origin. Nature 579 (7798), 270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang J., Zhang D., Ma W. L., Wang X. (2021). Linking the Gut Microbiota to Persistent Symptoms in Survivors of COVID-19 After Discharge. J. Microbiol. (Seoul Korea) 59 (10), 941–948. doi: 10.1007/s12275-021-1206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A Novel Coronavirus From Patients With Pneumonia in China, 2019. New Engl. J. Med. 382 (8), 727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Liu Q., Zhang F., Lui G. C.-Y., Tso E. Y., Yeoh Y. K., et al. (2021. a). Depicting SARS-CoV-2 Faecal Viral Activity in Association With Gut Microbiota Composition in Patients With COVID-19. Gut 70 (2), 276–284. doi: 10.1136/gutjnl-2020-322294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Liu Q., Zhang F., Yeoh Y. K., Wan Y., Zhan H., et al. (2021). Temporal Landscape of Human Gut RNA and DNA Virome in SARS-CoV-2 Infection and Severity. Microbiome 9 (1), 91. doi: 10.1186/s40168-021-01008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhang F., Lui G. C. Y., Yeoh Y. K., Li A. Y. L., Zhan H., et al. (2020. a). Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 159 (3), 944–955.e8. doi: 10.1053/j.gastro.2020.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhan H., Zhang F., Liu Q., Tso E. Y. K., Lui G. C. Y., et al. (2020. b). Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization Until Discharge. Gastroenterology 159 (4), 1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]