Abstract
Lung cancer screening detects early-stage cancers, but also a large number of benign nodules. Molecular markers can help in the lung cancer screening process by refining inclusion criteria or guiding the management of indeterminate pulmonary nodules. In this study, we developed a diagnostic model based on the quantification in plasma of complement-derived fragment C4c, cytokeratin fragment 21–1 (CYFRA 21–1) and C-reactive protein (CRP). The model was first validated in two independent cohorts, and showed a good diagnostic performance across a range of lung tumor types, emphasizing its high specificity and positive predictive value. We next tested its utility in two clinically relevant contexts: assessment of lung cancer risk and nodule malignancy. The scores derived from the model were associated with a significantly higher risk of having lung cancer in asymptomatic individuals enrolled in a computed tomography (CT)-screening program (OR = 1.89; 95% CI = 1.20 –2.97). Our model also served to discriminate between benign and malignant pulmonary nodules (AUC: 0.86; 95% CI = 0.80 –0.92) with very good specificity (92%). Moreover, the model performed better in combination with clinical factors, and may be used to reclassify patients with intermediate-risk indeterminate pulmonary nodules into patients who require a more aggressive work-up. In conclusion, we propose a new diagnostic biomarker panel that may dictate which incidental or screening-detected pulmonary nodules require a more active work-up.
INTRODUCTION
Lung cancer is the most commonly diagnosed cancer worldwide and the leading cause of cancer death.1 Lung tumors are mostly diagnosed at metastatic stage, where the 5-year survival rate is less than 5%.2 In contrast, the 5-year survival rate of patients with localized lung cancer is close to 60%,2 which evidences the importance of detecting lung cancer at early stages. Screening of subjects at risk with low-dose computed tomography (LDCT) has long been proposed as a strategy to increase the relative frequency of early-stage diagnosis.3 Randomized controlled trials, performed in the United States and Europe, have demonstrated that lung cancer screening with LDCT is associated with a marked diagnostic stage shift towards lung tumors detected at early stages (mostly stage I) with the subsequence reduction of mortality rates from lung cancer.4–6 These results have elicited a notable excitement with the prospects of implementation of CT-based lung cancer screening at the population level.7 In fact, screening of lung cancer in high risk individuals is currently recommended by the U.S. Preventive Services Task Force and other professional societies based on the inclusion criteria in the National Lung Screening Trial-NLST.8 There are two major challenges to improve the cost/benefit ratio of the population-based implementation of lung cancer screening. First, the refinement of risk models and selection criteria to reduce the numbers of individuals unnecessarily undergoing screening. Second, the optimization of the management of patients with indeterminate nodules and of decision-making algorithms regarding follow-up interventions. Contributions to any of these 2 challenges would substantially help in the implementation and sustainability of screening protocols.9 To this aim, the use of molecular biomarkers, derived from the tumor itself or from the host reaction to its presence, offers great potential. These biomarkers may complement image-based screening in different ways. On the one hand, biomarkers associated with genetic predisposition or risk of lung cancer presence in asymptomatic individuals may allow refinement of screening selection criteria.10 Molecular makers could be added to clinical variables already included in risk models. On the other hand, a molecular marker may aid in decision-making protocols in the case of individuals presenting indeterminate pulmonary nodules (IPNs). Almost one fourth of screened individuals present IPNs, but only 4% of them are malignant.4 Actual procedures for IPN management often lead to unnecessary follow-up CTs, or even invasive procedures.11 Molecular markers may help to differentiate patients with malignant IPNs from the large number of subjects with benign nodules. Extensive research efforts are ongoing to evaluate the applicability of molecular markers in these 2 clinical contexts.12
Several experimental observations suggest that the complement system, an essential element of innate immunity, is activated in patients with cancer.13 We previously demonstrated that the classical complement pathway is activated by lung cancer cells.14 C4d, a split product derived from complement activation, was elevated in biological fluids from patients with lung cancer and was associated with poorer prognosis.14 We also showed that plasma C4d levels were associated with an increased risk of lung cancer in asymptomatic individuals from a screening population cohort.14 Patients with malignant IPNs presented significantly higher plasma levels of C4d than those with benign nodules.15 C4d plasma levels were also increased in head and neck cancer patients,16 and may be a prognostic factor in malignant pleural mesothelioma.17
C4d is a breakdown product generated from complement C4 during activation of the classical pathway of complement. Upon C4 fragmentation, C4d remains covalently attached to the plasma membrane whereas C4c, another proteolytic fragment derived from C4, is released to the extracellular milieu.18 Therefore, C4c may be more readily detectable in extracellular fluids, such as plasma. Still, the potential use of C4c as a diagnostic marker for lung cancer has not been evaluated yet.
The aim of the present study was to assess the utility of C4c plasma levels as a diagnostic maker for lung cancer. Moreover, we searched for a combination with other plasma proteins that would improve the diagnostic performance of the test. We describe the development of a diagnostic model based on the quantification of C4c in plasma, along with two cancer-associated proteins, cytokeratin fragment 21–1 (CYFRA 21–1) and C-reactive protein (CRP). The scores derived from this model were associated with a significantly higher risk of having lung cancer in asymptomatic individuals enrolled in a CT-screening program, as well as with the capacity to identify a subpopulation of patients with malignant IPNs that may require a more aggressive work-up.
MATERIAL AND METHODS
Clinical samples.
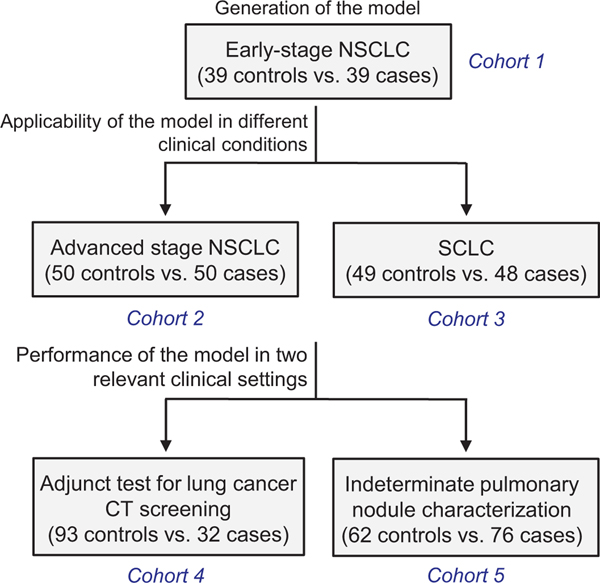
The study included plasma samples from 5 case-control series of lung cancer patients and control individuals. These series and the purpose of their evaluation are summarized in Fig 1. The cohort in which the diagnostic model was developed consisted of plasma samples from 39 patients with early stage non-small cell lung cancer (NSCLC) and 39 control subjects (herein denoted as discovery cohort). The levels of C4d and C4d-containing fragments were previously determined in these samples.14,19 The validation of the model was carried out in 2 sets of samples, one consisting of 50 NSCLC patients at advanced stages and 50 control individuals, and the other of 48 SCLC patients and 49 control individuals. The clinical utility of the model was tested in 2 nested case-control series: a set of 125 subjects enrolled in the International Early Lung Cancer Action Program (I-ELCAP), which included 32 individuals diagnosed with lung cancer and 93 individuals with no evidence of cancer at the completion of the screening protocol, and a set of 138 patients presenting IPNs discovered by chest CT, where 76 of them were diagnosed as lung cancers and the remaining 62 as nonmalignant lesions. All samples were collected between 2003 and 2018 and retrieved from the biorepository of the Clinica Universidad de Navarra, except for the IPN cohort, which was collected prospectively between 2003 and 2011 at the Vanderbilt University Medical Center with the intention to identify diagnostic biomarkers able to differentiate benign from malignant cases. Lung tumors were classified according to the WHO 2004 classification and the International System for Staging Lung Cancer.20 All study protocols were approved by the Institutional Research Ethics Committee, and all patients gave informed consent.
Fig 1.
Schematic representation of the study design. The goal of each analysis and the number of samples per cohort are shown.
Biomarker measurements.
C4c plasma levels were evaluated using an enzyme-linked immunosorbent assay as previously described.21 This assay has been reported to be highly specific, with no cross-reactivity with uncleaved C4 or other proteolytic fragments derived from C4.21 Results were calculated as relative values to those found in a reference sample and expressed as arbitrary units (AU). C4d-containing fragments from C4-activation and C4d were determined as previously described.14,19 Luminex technology and the Human Circulating Cancer Biomarkers Magnetic Bead Panel 1 (HCCBP1MAG-58K, Millipore) were used to evaluate the expression of a set of cancer-related markers in plasma samples. A Cobas analyzer (Roche Diagnostics) was used for the determination of CRP, CYFRA 21–1, IL-6, and prolactin. Determinations were performed blindly.
Statistical analyses.
STARD (Standards for Reporting of Diagnostic Accuracy Studies) guidelines were followed for the presentation of the manuscript.22 The model was defined using the discovery cohort and maintained unaltered throughout the study. No optimal size estimation was performed. Cohort size was based on availability. Normal distribution of the data was tested by the Shapiro-Wilk test. Differences between two or more groups were determined using the Mann-Whitney U test or the Kruskal-Wallis test, respectively. Marker values were expressed as median (25th to 75th percentiles). Logistic regression was used to generate the integrated models. The performance of the individual markers and the models was evaluated by using standard measurements of diagnostic accuracy, including the area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, positive predictive value, negative predictive value, positive likelihood ratio, and negative likelihood ratio. The goodness of fit of the competing diagnostic models was evaluated by comparing their likelihood ratios using the likelihood-ratio test. Differences in performance were also assessed by comparing AUCs using the rocgold command in STATA, which performs tests of equality of ROC area against a “gold standard” ROC curve. Conditional logistic regression was used to estimate odds ratios and 95% CI for lung cancer risk.
Survival curves were generated using the Kaplan-Meier method, and statistically significant differences were analyzed with the log rank test. Confidence intervals for the diagnostic parameters were calculated using the MedCalc statistical software. All other statistical analyses were performed with STATA/IC 12.1. Two-sided P values less than 0.05 were considered statistically significant.
RESULTS
Performance of C4c as a marker for lung cancer diagnosis.
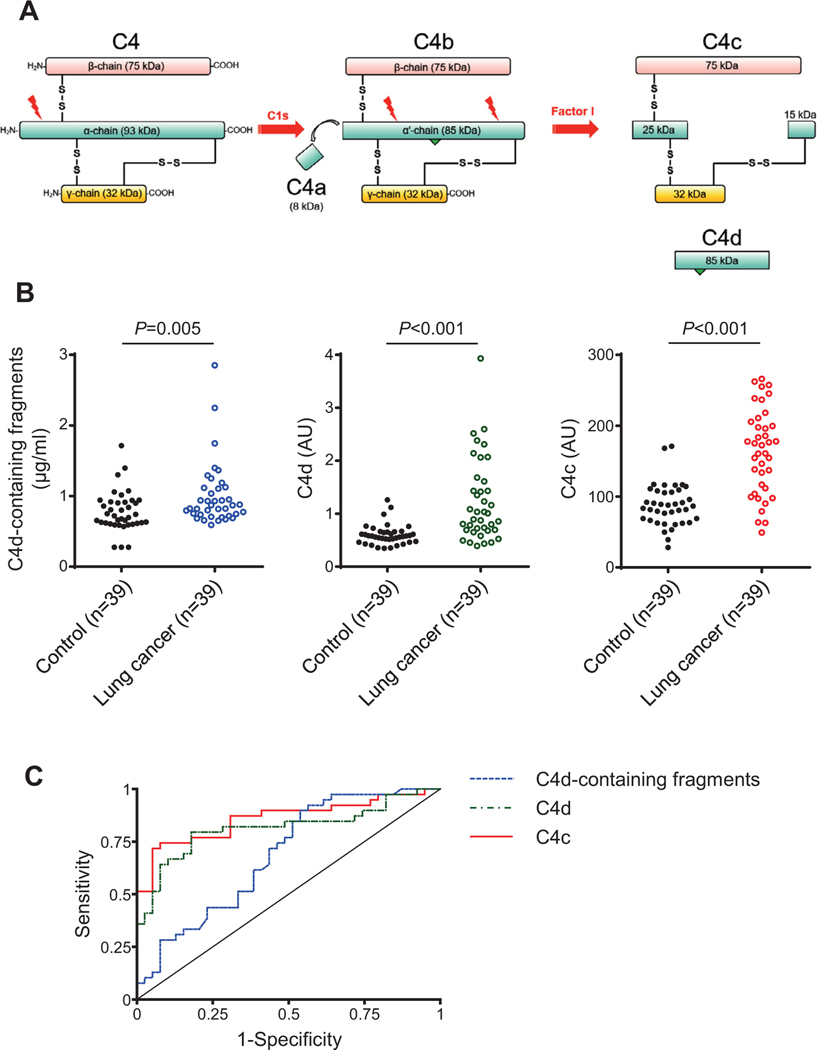
We have previously proposed the use of proteolytic fragments of complement C4 as a diagnostic marker from lung cancer.14,19 Detection was based on the quantification of C4d, a breakdown product generated from complement C4 upon activation of the classical pathway of complement (Fig 2A). In this study, we sought to generate a clinically useful model for lung cancer diagnosis based on the detection of either C4-derived fragments containing C4d (which include C4b, iC4b, and C4d; herein referred as C4d-containing fragments), only C4d, or the soluble fragment C4c. We initially used a series of plasma samples from 39 patients with early stage non-small cell lung cancer (NSCLC) and 39 control subjects matched by age, sex and smoking status. In these samples, C4d-containing fragments14 or C4d19 had previously been determined.
Fig 2.
Performance of C4-derived fragments as diagnostic markers for lung cancer. A) Scheme representing the proteolytic formation of C4-derived fragments upon activation of the classical pathway of complement. C4a and C4c are soluble fragments, whereas C4b and C4d remain covalently attached to the target membrane (the location of the membrane-binding site is indicated by a green triangle). C4b, C4d, and iC4d (an intermediate fragment not shown in the figure) are designated in this manuscript as C4d-containing fragments derived from C4 activation. B) Quantification of C4d-containing fragments, C4d and C4c plasma levels in patients diagnosed with early-stage NSCLC (n = 39) and matched control subjects (n = 39). The P value was calculated using the 2-sided Mann-Whitney U test. AU: Arbitrary units. C) ROC curves obtained from the plasma levels of the three markers.
In the present study, we quantified C4c and compared that with previous quantifications. Samples from lung cancer patients showed significantly higher levels of C4d-containing fragments, C4d or C4c than those from control individuals (Fig 2B). Plasma levels in cases vs controls were: 0.87 (0.74–1.12) vs 0.72 (0.61–0.92) μg/ml for C4d-containing fragments (P = 0.005); 1.01 (0.69–1.61) vs 0.58 (0.48–0.66) arbitrary units (AU) for C4d (P < 0.001); and 173 (112–206) vs 87 (66–108) AU for C4c (P < 0.001). The areas under the ROC curves (AUCs) were 0.69 (95% CI = 0.57–0.81), 0.81 (95% CI = 0.71–0.92) and 0.86 (95% CI = 0.77–0.95), respectively (Fig 2C). Pairwise comparisons using the test for equality of ROC curves showed a better performance for C4c (Table 1). The use of C4c as a diagnostic marker resulted in a high specificity and positive predictive value (Table 2). A significant association was found between C4c levels and malignant nodule size (P = 0.019), whereas no association was observed with other epidemiological or clinical characteristics, such as sex, age, smoking status, cancer histology or stage (Supplementary Table 1). Supplementary Tables 2 and 3 summarize the associations found between the clinicopathological characteristics of the patients and C4d and C4d-containing fragments, respectively. Based on these results, we conclude that C4c is a better marker than other C4-derived fragments to discriminate lung cancer patients from control individuals.
Table 1.
Plasma levels of C4d-containing fragments, C4d and C4c in samples from control subjects (n = 39) and early-stage lung cancer patients (n = 39), and their corresponding areas under the ROC curve (AUC)
| Biomarker* | Control subjects | Cancer patients | P value† | AUC | P value‡ |
|---|---|---|---|---|---|
| C4d-containing fragments (μg/ml) | 0.72(0.61–0.92) | 0.87 (0.74–1.12) | 0.005 | 0.69 (0.57–0.81) | |
| C4d (AU) | 0.58 (0.48–0.66) | 1.01 (0.69–1.61) | <0.001 | 0.81 (0.71–0.92) | 0.088 |
| C4c (idem) | 87 (66–108) | 173(112–206) | <0.001 | 0.86 (0.77–0.95) | 0.011 |
Biomarker levels are expressed as median (25th to 75th percentiles).
Two-sided Mann-Whitney U test.
Test for equality of AUCs using the AUC for C4d-containing fragments as reference.
Table 2.
Diagnostic performance of the determination of the three complement C4-derived fragments in plasma samples from early-stage lung cancer
| Diagnostic parameters* | C4d-containing fragments | C4d | C4c |
|---|---|---|---|
| Sensitivity (%) | 92 (79–98) | 79(64–91) | 72 (55–85) |
| Specificity (%) | 44 (28–60) | 82 (66–92) | 95 (83–99) |
| Accuracy (%) | 68 (56–78) | 81 (70–89) | 83 (73–91) |
| Positive predictive value (%) | 62 (55–69) | 82 (69–90) | 93 (78–98) |
| Negative predictive value (%) | 85 (64–95) | 80 (68–88) | 77 (67–85) |
| Positive likelihood ratio | 1.64(1.22–2.19) | 4.43 (2.22–8.83) | 14.00 (3.58–54.78) |
| Negative likelihood ratio | 0.18(0.06–0.55) | 0.25(0.13–0.47) | 0.30(0.18–0.49) |
Diagnostic parameters were calculated using the cut-off that provides the maximum Youden index. 95% CIs are also shown.
Generation of a C4c-containing diagnostic model.
We aimed to improve the performance of our test by complementing the diagnostic capacity of C4c with previously described cancer markers. In the discovery series in which we had measured C4c, we determined the plasma levels of the 25 following protein markers: AFP, PSA, CA 15–3, CA 19–9, MIF, TRAIL, leptin, IL-6, sFasL, CEA, CA 125, IL-8, HGF, sFas, TNFα, prolactin, SCF, CYFRA 21–1, OPN, FGF2, bHCG, HE4, TGFα, VEGF, and CRP. All of them were determined by Luminex technology, except for CRP, which was evaluated using a Cobas analyzer. We found significantly higher levels of IL-6 (P = 0.022), prolactin (P = 0.003), CYFRA 21–1 (P < 0.001), and CRP (P = 0.004) in the group of lung cancer patients as compared with the control group (Supplementary Fig 1 and Supplementary Table 4). The differences in the plasma levels of IL-6, prolactin, and CYFRA 21–1 were validated in a Cobas analyzer (Supplementary Fig 2). In one control sample, CYFRA 21–1 could not be determined.
We developed regression models to combine the diagnostic capacity provided by C4c, IL-6, prolactin, CYFRA 21–1, and CRP. Univariate analyses performed for each of the markers alone showed that their levels were associated with malignancy, except for IL-6, which was excluded from subsequent analyses (Supplementary Table 5). Next, logistic regression multivariate models were generated in which we evaluated the capacity of prolactin, CYFRA 21–1, CRP and their combinations to add diagnostic value to C4c (Supplementary Table 6). The information provided from both the univariate and multivariate analyses led us to select C4c, CYFRA 21–1, and CRP for the integrative model, excluding prolactin in favor of parsimony. Finally, we assessed the particular contribution of C4c to the classifier by comparing the performance of the three-protein model with that of a model based only on CYFRA 21– 1 and CRP levels. As shown in Supplementary Table 7, the model performed significantly better when C4c was present (P < 0.001), confirming the relevance of this marker in the final model.
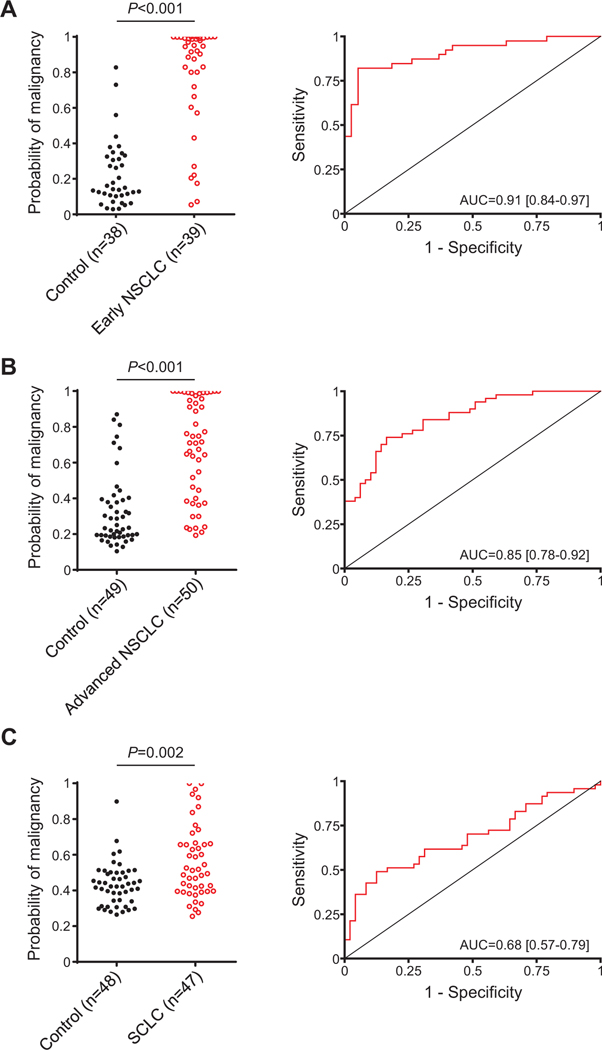
Using the model based on the quantification of C4c, CYFRA 21–1, and CRP, we calculated a malignancy probability score in each control individual and each patient. As shown in Fig 3A, the malignancy probability scores were significantly higher in cancer patients than in control individuals (P < 0.001), which yielded an AUC of 0.91 (95% CI = 0.84 –0.97). The combined diagnostic model showed a strong positive predictive value (94%) and positive likelihood ratio (15.60), as expected from its high specificity (95%). The associations between the predictive probabilities of malignancy and demographics of patients and controls are shown in Supplementary Table 8. The combined marker performed better for the diagnosis of squamous cell carcinomas (P = 0.012) and nodules larger than 2.5 cm (P = 0.003). Interestingly, the malignant probability scores determined by the model were also significantly associated with the outcome of the patients (P = 0.020), suggesting that the model may have prognostic value. No other significant associations were found.
Fig 3.
Generation and validation of the diagnostic model. A) Probabilities of malignancy, calculated using the regression model generated from the quantification of C4c, CYFRA 21–1 and CRP, in plasma samples from control individuals (n = 38) and patients (n = 39) with early NSCLC (stages I and II). B) Probabilities of malignancy in control individuals (n = 49) and patients with advanced NSCLC (n = 50). C) Probabilities of malignancy in control subjects (n = 48) and patients with SCLC (n = 47). The ROC curves obtained from all these probabilities and the AUCs are also shown. P values were calculated using the 2-sided Mann-Whitney U test.
Validation of the diagnostic model.
The capacity of the model to diagnose lung cancer was validated in 2 independent case-control cohorts. The first set consisted of 50 NSCLC patients at advanced stages and 50 control individuals matched by age, sex, and smoking history. The predictive probabilities of the model in these 2 groups were calculated using the regression model defined in the previous section (of note, in one control, CYFRA 21–1 could not be determined). As shown in Fig 3B, lung cancer patients showed significantly higher malignant probability scores that matched control individuals (P < 0.001). The area under the ROC curve was 0.85 (95% CI = 0.78 – 0.92). Associations between the predictive malignant probabilities and the clinicopathological characteristics of the patients are shown in Supplementary Table 9.
We also tested the efficacy of the protein combination panel in a set of plasma samples from SCLC patients (n = 48) and control individuals (n = 49). SCLC is a histology subtype of lung cancer that accounts for around 15% of lung cancer cases. The predictive probabilities of the model were calculated from the levels of C4c, CYFRA 21–1, and CRP using the same formula used previously (of note, C4c could not be determined in 1 control individual, and CRP in 1 patient). Although the diagnostic classifier was less accurate than in the 2 NSCLC cohorts, and in this case C4c did not add diagnostic value to the model (data not shown), the probabilities of malignancy predicted by the model were still significantly higher in the cancer group (P = 0.002), and the model showed a good specificity and positive predictive value (Fig 3C). No significant associations were found between demographics and the predictive probabilities of malignancy (Supplementary Table 10).
The diagnostic performance of the three-protein model in the discovery series and the two validation series is summarized in Table 3. From these analyses, we concluded that our combined model was able to discriminate between lung cancer patients and control individuals with high specificity, prompting us to test its performance in clinically relevant settings.
Table 3.
Summary of the diagnostic performance of the model based on the determination of C4c, CYFRA 21–1, and CRP in the different series of plasma samples used in this study*
| Early NSCLC | Advanced NSCLC | SCLC | Screening | Indeterminate nodules | |
|---|---|---|---|---|---|
| Number of controls | 38 | 49 | 48 | 93 | 62 |
| Number of cases | 39 | 50 | 47 | 30 | 76 |
| Sensitivity (%) | 82 (66–92) | 74 (60–85) | 49 (34–64) | 73 (54–88) | 70 (58–80) |
| Specificity (%) | 95 (82–99) | 84 (70–93) | 88 (75–95) | 70 (60–79) | 92 (82–97) |
| Accuracy (%) | 88 (87–98) | 79 (69–86) | 68 (58–78) | 71 (62–79) | 80 (72–86) |
| Positive predictive value (%) | 94 (80–98) | 82 (71–90) | 80 (63–90) | 44 (35–53) | 91 (82–96) |
| Negative predictive value (%) | 84(72–91) | 76 (66–84) | 63 (56–70) | 89 (82–94) | 71 (64–78) |
| Positive likelihood ratio | 15.60(4.01–60.55) | 4.53 (2.35–8.72) | 3.91 (1.75–8.74) | 2.44(1.67–3.55) | 8.64 (3.68–20.30) |
| Negative likelihood ratio | 0.19(0.10–0.37) | 0.31 (0.19–0.50) | 0.58 (0.43–0.79) | 0.38(0.21–0.70) | 0.33 (0.23–0.47) |
Diagnostic parameters were calculated using the cut-off that provides the maximum Youden index in each series. 95% CIs are also shown.
Application of the diagnostic model for the assessment of lung cancer risk in asymptomatic individuals undergoing lung cancer screening.
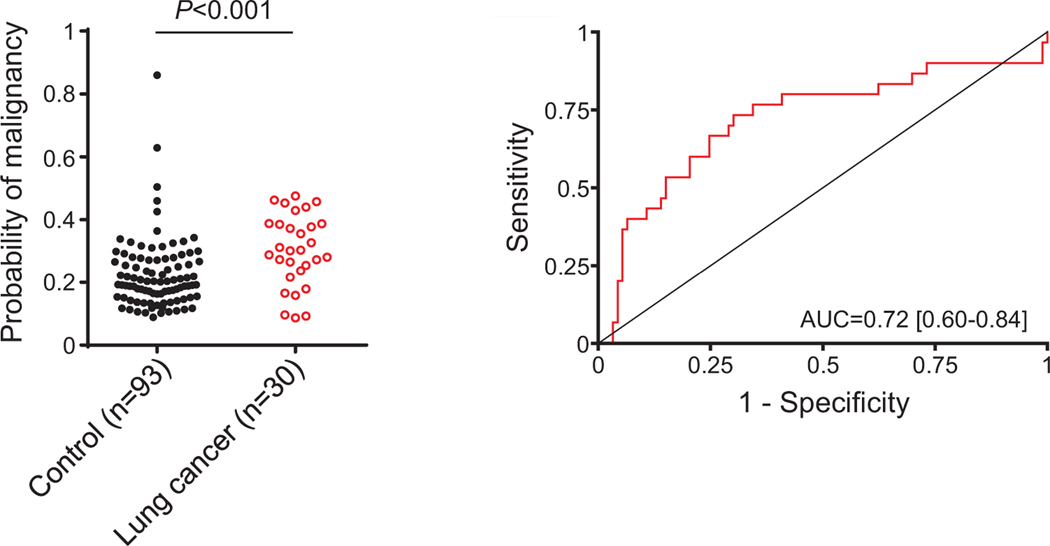
To test the clinical utility of the new diagnostic model, we first assessed its capacity to discriminate between asymptomatic individuals who were or were not diagnosed with lung cancer in the context of a CT-screening program performed at the Clinica Universidad de Navarra.23 We used a cohort of 125 subjects enrolled in the International Early Lung Cancer Detection Program (I-ELCAP). The cohort included 32 individuals diagnosed with lung cancer in the context of the CT screening, and 93 individuals with no evidence of cancer at the completion of the screening protocol. Samples from lung cancer patients were collected at diagnosis for most cases, and no more than 2 months prior to diagnosis for any of them. Scores derived from the regression model based on C4c, CYFRA 21–1 and CRP plasma levels were significantly associated with lung cancer risk (OR = 1.89; 95% CI = 1.20 –2.97). Of note, CRP could not be analyzed in two cancer patients. The statistically significant differences in the probability scores and the area of the ROC curve are shown in Fig 4. Probabilities were higher in individuals with more than 35 pack-years history of smoking, whereas no association was found with sex, age, histology, lesion size, or stage (Supplementary Table 11). The diagnostic yield of the model in this clinical setting is summarized in Table 3. We also evaluated the performance of the model using all lung cancer cases but only those control individuals with pulmonary nodules detected by CT (n = 54). The association with lung cancer risk remained significant (OR = 3.60; 95% CI = 1.61–8.07; Supplementary Fig 3).
Fig 4.
Performance of the protein diagnostic model in the assessment of lung cancer risk in asymptomatic individuals. Probabilities of malignancy were calculated using the regression model generated from the quantification of C4c, CYFRA 21–1 and CRP in plasma samples from asymptomatic individuals who were or were not diagnosed with lung cancer in the context of a CT-screening program (n = 30 and n = 93, respectively). The ROC curve and the AUC obtained from these probabilities are also shown. The P value was calculated using the two-sided Mann-Whitney U test.
In addition, we evaluated the performance of the assay in the 36 control subjects and 13 cancer patients included in the screening cohort who did not meet the 55–74 years old, and at least 30 pack-years NLST selection criteria. The clinicopathological characteristics of these patients are shown in Supplementary Table 12. Higher lung cancer risk (OR = 2.31; 95% CI = 1.15 – 4.67) and predicted probabilities (P = 0.018) were still found in cancer patients than in control subjects. In fact, no differences in the probability scores were found between lung cancer patients who did or did not meet NLST selection criteria (P = 0.155).
In summary, in this part of the study, we provide evidence supporting that our diagnostic model may help in the identification of asymptomatic individuals at high risk of having lung cancer, including those who fall outside the NLST eligibility criteria.
Application of the diagnostic model for the evaluation of indeterminate pulmonary nodules found incidentally.
We finally evaluated the performance of the protein model in the diagnosis of patients presenting with IPNs. We used plasma samples obtained from 138 patients presenting IPNs discovered by chest CT at Vanderbilt University Medical Center. Lung nodules were defined as rounded opacities completely surrounded by lung parenchyma. Seventy-six of these nodules were diagnosed as lung cancers by pathological examination, whereas the remaining 62 were diagnosed as nonmalignant lesions. Demographics of these patients, stratified by diagnosis, are shown in Supplementary Table 13. Diagnosis of patients with nonmalignant nodules included chronic obstructive pulmonary disease, emphysema, inflammatory disease, granulomatous lesions, or hamartomas.
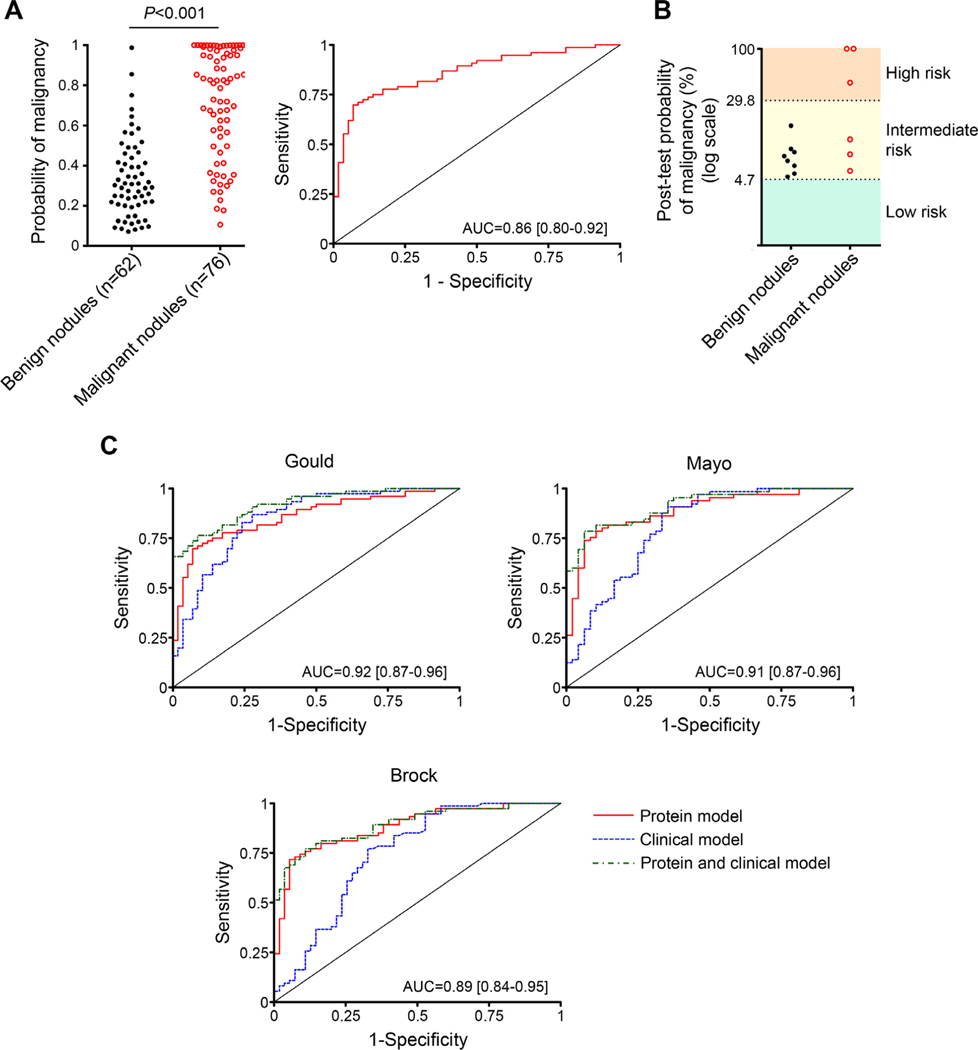
The predicted probabilities of the classifier, calculated from the plasma levels of C4c, CYFRA 21–1, and CRP as indicated above, were compared with the final diagnosis, and a ROC curve was constructed (Fig 5A). The area under the curve was 0.86 (95% CI = 0.80 –0.92). The diagnostic performance of the model is summarized in Table 3. The model showed high specificity (92%), suggesting its utility to rule in the disease. Associations between the predicted probabilities and characteristics of patients and lesions are shown in Supplementary Table 13. No association was found between the probability scores and sex, age, smoking history, histology or stage (although there was a tendency towards higher scores in extensive SCLC as compared with limited disease). Interestingly, larger nodules were significantly associated with higher malignancy probability scores in patients with malignant nodules (P < 0.001), but not in individuals with benign lesions (P = 0.972). A high malignancy score was also significantly associated with worse prognosis (P < 0.001; Supplementary Fig 4).
Fig 5.
Performance of the protein model in the diagnosis of patients with indeterminate pulmonary nodules. A) Malignancy probability scores calculated by the regression model generated from the quantification of C4c, CYFRA 21–1, and CRP in plasma samples from individuals with benign and malignant pulmonary nodules (n = 62 and n = 76, respectively). The P value was determined using the 2-sided Mann-Whitney U test. The ROC curve and AUC obtained from these probabilities is also shown. B) Post-test probabilities of malignancy of individuals with intermediate-risk pulmonary nodules (9–15 mm in diameter) after the application of the diagnostic model. Pre-test probability was stablished as 4.7% based on reported data from more than 20,000 CT-screened individuals.24 In three patients, post-test probabilities were higher than the 29.8% prevalence of lung cancer reported for individuals with nodules higher than 15 mm. C) ROC curves for the performance of the three-protein model, alone or in combination with the Gould, Mayo, or Brock clinical classifiers in patients with benign and malignant pulmonary nodules (58 and 76 patients, respectively, for the Gould model; 48 and 65 patients, respectively, for the Mayo model; and 55 and 74 patients, respectively, for the Brock model). The AUCs of the combined models are shown.
At present, the management of a patient with a CT-detected IPN is mainly guided by the size of the lesion. Nodules lower than 5 mm in diameter generally require no further assessment, whereas if the nodule is 15 mm or larger, this is highly suggestive of malignancy and a biopsy is recommended. Less consensus exists about the action required for intermediate-sized lesions (which may include a follow-up CT, PET scan, bronchoscopy or biopsy sampling). We speculated that the application of our highly specific diagnostic model might increase the probability of malignancy in intermediate-sized IPNs to the next actionable level. To test this hypothesis, patients with intermediate-risk nodules (9 15 mm in diameter) were selected from the Vanderbilt cohort, and their probability of malignancy was determined before and after the application of our diagnostic model (pretest and post-test probabilities, respectively). Four individuals of the benign group and six individuals of the malignant group had intermediate-risk nodules. The prevalence of lung cancer in CT-screened individuals with nodules ranging from 9 to 15 mm has been previously reported as 4.7%.24 Therefore, we used this value as the pre-test probability of malignancy for the intermediate risk nodules. Post-test probability of malignancy in each individual was derived from the pretest probability and the positive likelihood ratio calculated by the three-protein model. Post-test probabilities of malignancy markedly increased in 3 out of the 6 cancer patients with intermediate-risk nodules (Fig 5B). Interestingly, in those three patients, the post-test probability was higher than 29.8%, the probability of malignancy (prevalence) reported for individuals with nodules higher than 15 mm.24 Therefore, in half of the intermediate-risk patients, the application of our diagnostic model resulted in their re-classification as patients in the high-risk group, which may require a more aggressive follow-up (Table 4). The probabilities of malignancy in the benign group were not substantially modified (Fig 5B). Although limited by the number of cases, this analysis suggests that patients with intermediate-risk indeterminate pulmonary nodules may be reclassified as patients with high-risk nodules who may require a more aggressive work-up.
Table 4.
Pre-test and post-test probabilities of malignancy after applying the diagnostic model to Vanderbilt’s patients with an intermediate risk of malignancy based on nodule size (9–15 mm)
| Patient | Sex | Smoking status | Nodule size (mm) | Cancer histology | Pretest probability | Post-test probability |
|---|---|---|---|---|---|---|
| 13 | Female | Former | 11 | ADC | 4.7% | 100% |
| 14 | Male | Current | 11 | SCLC | 4.7% | 100% |
| 16 | Male | Former | 12.5 | SCC | 4.7% | 5.7% |
| 19 | Female | Former | 13.5 | SCLC | 4.7% | 12% |
| 21 | Female | Former | 14 | ADC | 4.7% | 45.2% |
| 25 | Male | Former | 14.5 | ADC | 4.7% | 8.5% |
Finally, we assessed the capacity of our diagnostic classifier to improve the prediction of malignancy in patients with IPNs upon the accuracy provided by clinical factors. We compared our protein model with 3 different clinical models: the Gould model,25,26 based on age, smoking history, nodule diameter and time since quitting smoking, the Mayo model,27 based on age, smoking history, previous extrathoracic cancer and nodule diameter and spiculation, and the parsimonious version of the Brock model,28 based on sex and nodule size, location, and spiculation. The available clinical information and the selection criteria for the models allowed us to apply the Gould model to 134 patients of the Vanderbilt series (58 benign cases and all 76 malignant cases), the Mayo model to 113 patients (48 benign cases and 65 malignant cases), and the Brock model to 129 patients (55 benign cases and 74 malignant cases). The predictive probability scores of the clinical models generated ROC curves with an AUC of 0.85 (95% CI = 0.78 –0.92) for the Gould model, 0.81 (95% CI = 0.73 –0.89) for the Mayo model, and 0.75 (95% CI = 0.65 –0.84) for the Brock model. No statistical differences were observed between the AUCs generated by the protein model and those generated by the Gould or the Mayo models (P = 0.793 and P = 0.111, respectively), whereas the protein model performed significantly better than the Brock model (P = 0.008). Interestingly, as shown in Fig 5C, the models obtained by the combination of our protein model with any of the three clinical models performed significantly better as predictors of malignancy than any of the clinical models alone (P = 0.006 vs the Gould model, P = 0.014 vs the Mayo model, and P = 0.002 vs the Brock model). The diagnostic performances of the three clinical models, alone or in combination with the protein model, are summarized in Supplementary Table 14. The combination of the protein and clinical models showed a remarkably high specificity and positive predictive value. From these analyses, we conclude that the proposed diagnostic signature can be combined with models based on clinical features to identify more accurately pulmonary nodules with high risk of malignancy.
DISCUSSION
We describe here the development of a diagnostic model based on the plasma quantification of the complement fragment C4c and 2 cancer-associated proteins, CYFRA 21–1, and CRP. The model identified those asymptomatic individuals enrolled in a CT-screening trial with higher risk of having lung cancer, and served to discriminate between benign and malignant pulmonary nodules.
Moving lung cancer diagnostic molecular markers from discovery to clinical application has proved to be challenging.12 Some prospective clinical trials have shown promising results in the assessment of indeterminate pulmonary nodules (IPNs). The PANOPTIC study evaluated the use of a protein classifier to identify benign lung nodules in patients with a pretest probability of cancer lower than 50%.29 This classifier is based on the quantification of 2 plasma proteins, LG3BP and C163A (normalization protein) in combination with 5 clinical factors. On the other hand, in the AEGIS 1/2 trials, a 23-gene classifier based on gene expression changes in bronchial airway epithelium was able to show an improvement of the diagnostic performance of bronchoscopy among patients with intermediate-risk nodules.30 In these patients pretest probabilities as high as 60% gave posttest probabilities lower than 10%. Both classifiers achieved high sensitivity and negative predictive value in distinguishing benign from malignant nodules, suggesting that, if used in clinical practice, unnecessary invasive procedures could be reduced. On the other hand, the specificities and positive predictive values of these classifiers were modest and not suitable to “rule in” a diagnosis of cancer. In the context of lung cancer screening, where disease prevalence is low, a test with high specific and positive predictive value may be of greater clinical utility to recommend for a closer follow-up or even for an invasive procedure. The diagnostic model that we have developed has high specificity and positive predictive value across different cohorts of lung cancer patients and high-risk individuals, which may help in the diagnostic management of lung cancer patients.
Our model is defined by the combination of complement C4c with 2 other protein markers, CYFRA 21–1 and CRP. Previous studies have demonstrated that complement C4-derived fragments are significantly elevated in biological fluids from lung cancer patients, and may be of use for diagnosis or prognosis.14,15,19 C4-derived fragments are generated from C4 upon activation of the classical pathway of the complement system, a central humoral component of innate immunity. C4 is cleaved into C4b, which binds to the target cell, and C4a, a soluble factor. Surface-bound C4b can be subsequently proteolyzed and inactivated by factor I into the final breakdown products C4d and C4c.18 Unlike C4d, the diagnostic performance of C4c had not been evaluated yet. In the present study, we show the high specificity and positive predictive value of C4c testing. Even more, the diagnostic performance of C4c was enhanced by the concurrent determination of the two tumor markers CYFRA 21–1 and CRP, resulting in a multivariate diagnostic model with a remarkable capacity to discriminate between plasma samples from controls and lung cancer patients (specificity ranging from 84 to 95).
CYFRA 21–1 is a circulating cytokeratin-19 fragment that shows a high diagnostic capacity for the detection of NSCLC.31–33 High circulating levels of this marker have also been associated with unfavorable prognosis.34,35 On the other hand, CRP is a systemic marker of chronic inflammation. CRP levels have been associated with cancer incident and survival.36 Cancer-free individuals with high levels of CRP show a significantly higher risk of developing lung cancer.37,38 Elevated blood levels of CRP have also been associated with poor survival in lung cancer patients.39 Patients with either NSCLC or SCLC showed higher levels of CRP in circulation than patients with a variety of inflammatory benign lung diseases.40 Nevertheless, the diagnostic capacity of CRP is controversial, possible due to its broad association with inflammation, which limits its use as a stand-alone diagnostic biomarker.
Our model has several strengths. By combining the plasma levels of three proteins into a single score, the model performs better than each marker alone. The model is simple, has been validated in independent series, shows high specificity, performs similarly well across a range of tumor sizes, types and stages, and has been successfully tested in 2 clinically relevant contexts: assessment of lung cancer risk and nodule malignancy probability.
Annual CT screening is recommended for individuals at highest risk for developing lung cancer based on the NLST criteria of age and smoking pack-years. However, 60% –70% of lung cancer cases in U.S. or Canada occur in individuals who fall outside of the NLST criteria.41,42 The predicted lung cancer risk obtained with our model was independent of age and tobacco exposure, showing similar risk probabilities in patients who did or did not meet NLST selection criteria. A blood-based 4-protein risk prediction model, which included CYFRA 21–1, was recently integrated with a smoking-based model to refine screening eligibility beyond NLST criteria.43 In pre-diagnostic samples from lung cancer patients and matched controls, this predictive model yielded a sensitivity of 42% and a specificity of 95%, performing better than the smoking model alone.43 Our protein model performed similarly when tested in patients enrolled in a CT-screening program. The integration of our model with any of the several models that use clinical and imaging features44 may result in an improved method to identify those individuals who will benefit from CT screening.
The vast majority of IPNs, detected either incidentally or in the context of CT-screening protocols, are ultimately benign.45 Current predictive tools to discriminate benign from malignant nodules are suboptimal, and the development of novel approaches able to discriminate benign versus malignant IPNs is an unmet clinical need.11 More particularly, individuals with an intermediate pretest probability pose a diagnostic challenge. A variety of models have been proposed to assess the risk of cancer in patients with IPNs.46—50 A serum proteomic signature distinguished subjects with lung cancer from matched controls with an overall accuracy of 73%, a sensitivity of 58% and a specificity of 86%.46 The application of this proteomic signature to two cohorts of patients and controls with lung nodules resulted in an AUC of 0.64.47 Recently, the diagnostic performance of a radiomic model based on eight imaging features has been validated using a dataset of incidentally identified lung nodules, showing a sensitivity of 92%, a specificity of 62%, a positive predictive value of 74%, and a negative predictive value of 87%.50 The performance of our model in individuals presenting with lung nodules is comparable to the performance of these models, standing out by its high positive predictive value. Based on this feature, our plasma protein model was useful in reclassifying patients with indeterminate-risk nodules into patients with high-risk nodules, without affecting the classification of benign nodules. Although this result should be evaluated with caution, due to the limited number of cases studied, it suggests that a high probability score may be a criterion to recommend for a more active surveillance or evaluation in the subgroup of patients classified as having intermediate-risk nodules. Our results also suggest that the integration of our model with other predictors, such as radiological or clinical features, may result in an even more accurate diagnostic model.
We acknowledge some limitations to this study. The model was derived from a limited selection of circulating proteins. Other complement- and cancer-related markers could also be useful for the model. All analyses were conducted using retrospective samples. Some analyses, such as the reclassification of intermediate-risk IPNs, were carried out with a limited number of cases. Prospective studies with a larger number of intended-use samples are required for validation. Further standardization and calibration of the analytical tests is also necessary, especially for the C4c assay. The need of a proper standardization is reflected in the diverse probabilities found in the controls from the different cohorts studied. Stratified analyses of the performance of the model in association with variables such as smoking status, histology subtype, stage or the presence of inflammatory conditions related to classical complement activation are also warranted.
In conclusion, we have built and validated a multivariate logistic regression model, based on the quantification of C4c, CYFRA 21–1 and CRP. This model has high specificity and positive predictive value, which may be of clinical use to refine risk prior to screening or to help in the identification of those patients with IPNs who require closer examination.
Supplementary Material
AT A GLANCE COMMENTARY.
Ajona, et al
Background
Recent results from randomized controlled trials have rekindled the interest for the implementation of lung cancer screening. However, better diagnostic models are needed to reduce the numbers of unnecessarily screenings, and to optimize the management of patients with indeterminate nodules.
Translational Significance
We developed and validated a molecular diagnostic model able to identify asymptomatic individuals enrolled in a CT-screening program at higher risk of having lung cancer. The model was also able to discriminate which pulmonary nodules may require a more active work-up. This model may improve inclusion criteria and management in the context of lung cancer screening.
ACKNOWLEDGMENTS
The authors thank the STARD Group for helping us in identifying essential items for reporting diagnostic accuracy studies, Yaseelan Palarasah for helping us in the initial determinations of C4c, and Jackeline Agorreta and María José Pajares for their contribution in the collection of clinical samples and data.
Funding: This study was supported by Foundation for Applied Medical Research (FIMA), Centro de Investigación Biomédica en Red Cáncer (CIBER-ONC), Fundación Científica de la Asociación Española Contra el Cáncer, Fundación Ramón Areces, Juan Serrano, Instituto de Salud Carlos III-EU FEDER “Una manera de hacer Europa” (PI17/00411, PI19/00098, PI20/00419), Amadix, and National Cancer Institute (NCI CA152662).
Abbreviations:
- AU
arbitrary units
- AUC
area under the curve
- CRP
C-reactive protein
- CT
= computer tomography
- CYFRA 21–1
cytokeratin fragment 21.1
- IPN
indeterminate pulmonary nodules
- NLST
National Lung Screening Trial
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- ROC
receiver operating characteristic
Footnotes
SUPPLEMENTARY MATERIALS
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.trsl.2021.02.009.
Conflicts of interest: All authors have read the journal’s policy on disclosure of potential conflicts of interest and have disclosed any financial or personal relationship with organizations that could potentially be perceived as influencing the described research. Daniel Ajona, Luis M Montuenga and Ruben Pio are inventors in two patent applications licensed to Amadix that are related to the use of complement-derived fragments in the diagnosis of lung cancer. Ruben Pio has received consultant fees and research funding from Amadix. Ana Martin and Rosa Perez are employees of Amadix.
All authors have read the journal’s authorship agreement and the manuscript has been reviewed by and approved by all named authors.
REFERENCES
- 1.Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol 2019;5:1749–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 3.International Early Lung Cancer Action Program Investigators, Henschke CI, Yankelevitz DF, et al. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–71. [DOI] [PubMed] [Google Scholar]
- 4.National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365: 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastorino U, Silva M, Sestini S, et al. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: new confirmation of lung cancer screening efficacy. Ann Oncol 2019;30:1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med 2020;382:503–13. [DOI] [PubMed] [Google Scholar]
- 7.Duffy SW, Field JK. Mortality reduction with low-dose CT screening for lung cancer. N Engl J Med 2020;382:572–3. [DOI] [PubMed] [Google Scholar]
- 8.Moyer VA. U.S. Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:330–8. [DOI] [PubMed] [Google Scholar]
- 9.Shankar A, Saini D, Dubey A, et al. Feasibility of lung cancer screening in developing countries: challenges, opportunities and way forward. Transl Lung Cancer Res 2019;8:S106–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atwater T, Massion PP. Biomarkers of risk to develop lung cancer in the new screening era. Ann Transl Med 2016;4:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massion PP, Walker RC. Indeterminate pulmonary nodules: risk for having or for developing lung cancer? Cancer Prev Res 2014;7:1173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seijo LM, Peled N, Ajona D, et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol 2019;14:343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pio R, Corrales L, Lambris JD. The role of complement in tumor growth. Adv Exp Med Biol 2014;772:229–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ajona D, Pajares MJ, Corrales L, et al. Investigation of complement activation product C4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst 2013;105:1385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajona D, Razquin C, Pastor MD, et al. Elevated levels of the complement activation product C4d in bronchial fluids for the diagnosis of lung cancer. PLoS One 2015;10:e0119878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajona D, Pajares MJ, Chiara MD, et al. Complement activation product C4d in oral and oropharyngeal squamous cell carcinoma. Oral Dis 2015;21:899–904. [DOI] [PubMed] [Google Scholar]
- 17.Klikovits T, Stockhammer P, Laszlo V, et al. Circulating complement component 4d (C4d) correlates with tumor volume, chemotherapeutic response and survival in patients with malignant pleural mesothelioma. Sci Rep 2017;7:16456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law SKA, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci 1997;6:263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajona D, Okrój M, Pajares MJ, et al. Complement C4d-specific antibodies for the diagnosis of lung cancer. Oncotarget 2017;9:6346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Travis WD, Brambilla E, Muller-Hermelink HK, CC Harris CC. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press; 2004. [Google Scholar]
- 21.Pilely K, Skjoedt M-O, Nielsen C, et al. A specific assay for quantification of human C4c by use of an anti-C4c monoclonal antibody. J Immunol Methods 2014;405:87–96. [DOI] [PubMed] [Google Scholar]
- 22.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Salcedo P, Berto J, De-Torres JPJP, et al. Lung cancer screening: fourteen year experience of the Pamplona Early Detection Program (P-IELCAP). Arch Bronconeumol 2014;51:169–76. [DOI] [PubMed] [Google Scholar]
- 24.Henschke CI, Yip R, Yankelevitz DF, Smith JP. International Early Lung Cancer Action Program Investigators. Definition of a positive test result in computed tomography screening for lung cancer: a cohort study. Ann Intern Med 2013;158:246–52. [DOI] [PubMed] [Google Scholar]
- 25.Schultz EM, Sanders GD, Trotter PR, et al. Validation of two models to estimate the probability of malignancy in patients with solitary pulmonary nodules. Thorax 2008;63:335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gould MK, Ananth L, Barnett PG. Veterans Affairs SNAP Cooperative Study Group. A clinical model to estimate the pre-test probability of lung cancer in patients with solitary pulmonary nodules. Chest 2007;131:383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules: application to small radiologically indeterminate nodules. Arch Intern Med 1997;157:849–55. [PubMed] [Google Scholar]
- 28.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369:910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvestri GA, Tanner NT, Kearney P, et al. Assessment of plasma proteomics biomarker’s ability to distinguish benign from malignant lung nodules: results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) trial. Chest 2018;154:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A Silvestri G A Vachani, Whitney D, et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med 2015;373:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina R, Agusti C, Filella X, et al. Study of a new tumor marker, CYFRA 21–1, in malignant and nonmalignant diseases. Tumor Biol 1994;15:318–25. [DOI] [PubMed] [Google Scholar]
- 32.Molina R, Marrades RM, Augé JM, et al. Assessment of a combined panel of six serum tumor markers for lung cancer. Am J Respir Crit Care Med 2016;193:427–37. [DOI] [PubMed] [Google Scholar]
- 33.Kammer MN, Kussrow AK, Webster RL, et al. Compensated interferometry measures of CYFRA 21–1 improve diagnosis of lung cancer. ACS Comb Sci 2019;21:465–72. [DOI] [PubMed] [Google Scholar]
- 34.Pujol J-L, Molinier O, Ebert W, et al. CYFRA 21–1 is a prognostic determinant in non-small-cell lung cancer: results of a meta-analysis in 2063 patients. Br J Cancer 2004;90: 2097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21–1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol 2003;24:209–18. [DOI] [PubMed] [Google Scholar]
- 36.Chaturvedi AK, Caporaso NE, Katki HA, et al. C-reactive protein and risk of lung cancer. J Clin Oncol 2010;28:2719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allin KH, Bojesen SE, Nordestgaard BG. Baseline C-reactive protein is associated with incident cancer and survival in patients with cancer. J Clin Oncol 2009;27:2217–24. [DOI] [PubMed] [Google Scholar]
- 38.Shiels MS, Katki HA, Hildesheim A, et al. Circulating inflammation markers, risk of lung cancer, and utility for risk stratification. J Natl Cancer Inst 2015;107:djv199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastorino U, Morelli D, Leuzzi G, et al. Baseline and postoperative C-reactive protein levels predict mortality in operable lung cancer. Eur J Cancer 2017;79:90–7. [DOI] [PubMed] [Google Scholar]
- 40.Oremek GM, Sauer-Eppel H, Bruzdziak TH. Value of tumour and inflammatory markers in lung cancer. Anticancer Res 2007;27:1911–5. [PubMed] [Google Scholar]
- 41.Pinsky PF, Berg CD. Applying the National Lung Screening Trial eligibility criteria to the US population: what percent of the population and of incident lung cancers would be covered? J Med Screen 2012;19:154–6. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Midthun DE, Wampfler JA, et al. Trends in the proportion of patients with lung cancer meeting screening criteria. JAMA 2015;313:853–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guida F, Sun N, Bantis LE, et al. Assessment of lung cancer risk on the basis of a biomarker panel of circulating proteins. JAMA Oncol 2018;4:e182078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tammemägi MC. Selecting lung cancer screenees using risk prediction models-where do we go from here. Transl Lung Cancer Res 2018;7:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gould MK, Tang T, Liu I-LA, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med 2015;192:1208–14. [DOI] [PubMed] [Google Scholar]
- 46.Yildiz PB, Shyr Y, Rahman JSM, et al. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol 2007;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pecot CV, Li M, Zhang XJ, et al. Added value of a serum proteomic signature in the diagnostic evaluation of lung nodules. Cancer Epidemiol Biomarkers Prev 2012;21:786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massion PP, Healey GF, Peek LJ, et al. Autoantibody signature enhances the positive predictive power of computed tomography and nodule-based risk models for detection of lung cancer. J Thorac Oncol 2017;12:578–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massion PP, Antic S, Ather S, et al. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med 2020;202:241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maldonado F, Varghese C, Rajagopalan S, et al. Validation of the BRODERS classifier (Benign versus aggressive nODule Evaluation using Radiomic Stratification), a novel high-resolution computed tomography-based radiomic classifier for indeterminate pulmonary nodules,. Eur Respir J 2020;Dec. 10:Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.