Significance
Deep profiling of the plasma proteome at scale has been a challenge for traditional approaches. We achieve superior performance across the dimensions of precision, depth, and throughput using a panel of surface-functionalized superparamagnetic nanoparticles in comparison to conventional workflows for deep proteomics interrogation. Our automated workflow leverages competitive nanoparticle–protein binding equilibria that quantitatively compress the large dynamic range of proteomes to an accessible scale. Using machine learning, we dissect the contribution of individual physicochemical properties of nanoparticles to the composition of protein coronas. Our results suggest that nanoparticle functionalization can be tailored to protein sets. This work demonstrates the feasibility of deep, precise, unbiased plasma proteomics at a scale compatible with large-scale genomics enabling multiomic studies.
Keywords: proteomics, nano–bio interaction, nanoparticle, mass spectrometry, machine learning
Abstract
Deep interrogation of plasma proteins on a large scale is a challenge due to the number and concentration of proteins, which span a dynamic range of over 10 orders of magnitude. Current plasma proteomics workflows employ labor-intensive protocols combining abundant protein depletion and sample fractionation. We previously demonstrated the superiority of multinanoparticle (multi-NP) coronas for interrogating the plasma proteome in terms of proteome depth compared to simple workflows. Here we show the superior depth and precision of a multi-NP workflow compared to conventional deep workflows evaluating multiple gradients and search engines as well as data-dependent and data-independent acquisition. We link the physicochemical properties and surface functionalization of NPs to their differential protein selectivity, a key feature in NP panel profiling performance. We find that individual proteins and protein classes are differentially attracted by specific surface properties, opening avenues to design multi-NP panels for deep interrogation of complex biological samples.
Nanoparticles (NPs) have expanding utility in many fields, including drug therapy, where they are being utilized as targeting and delivery vehicles (1–3). Upon contact with biofluids like blood plasma, a thin biomolecule layer termed the corona forms specifically and reproducibly on the surface of NPs and is composed of proteins, metabolites, and nucleic acids (4, 5). Recent investigations of nano–bio interactions have sought to improve the targeting abilities of nanomaterials and reduce their toxicity by leveraging the predictable process of corona formation (1, 6, 7).
Corona composition is driven by the complex interplay between the physicochemical properties of the NP surface and biomolecules including proteins (8–12). For instance, cross-linked N-isopropylacrylamide and butylacrylamide polymer NPs and graphene nanoflakes are initially bound to high-abundance molecules (e.g., serum albumin) but subsequently at equilibrium are replaced by lower-abundance, higher–binding affinity apolipoproteins AI, All, AIV, and E (13, 14). Depending on the NP type, the number and quantity of proteins in the protein corona change as a function of protein concentrations in serum/plasma (4). Understanding what drives the formation of the protein corona will not only improve the efficacy of nanomedicine (e.g., for drug delivery) but also enable the use of NP coronas to interrogate the complex proteomes of biofluids for basic science and biomarker discovery.
In genomics, scalable whole-genome and transcriptome sequencing workflows developed over the past 2 decades have advanced our understanding of basic biology and translational medicine through untargeted, hypothesis-free data generation approaches such as genome-wide association studies and colocalization analyses. However, even though the proteome is downstream from the genome and transcriptome, and therefore in principle more directly connected to cellular and organismal state and phenotype, hypothesis-free data generation approaches, in particular for deep plasma proteomics, have fallen behind the pace of genomics, primarily because of the large number of protein variants present in biofluids, their highly diverse structures, their wide-ranging concentrations, and the lack of suitable molecular processing tools. Despite a molecular inventory of blood plasma comprising tens of thousands of different proteins and metabolites, which offers a detailed profile of the current status of an organism, there have been relatively few tests developed from this information over the past century of effort (15). The dynamic range of protein concentrations in plasma exceeds 10 orders of magnitude, with only a few highly abundant proteins making up most of the protein mass, presenting a challenge to the identification of novel low-abundance protein biomarkers (15–18). Over the years, complex and resource-intensive workflows have been developed to address these challenges, but their complexity has prevented application to studies with large numbers of samples and large patient cohorts (19).
To enable broad, deep, fast, and unbiased proteomics, we recently demonstrated the utility of the nano–bio interface for plasma profiling in a study identifying candidate biomarkers for early nonsmall cell lung cancer (NSCLC) (20). In that work we showed that proteins have a reproducible and specific binding affinity to different NPs. This attribute facilitates compression of the large dynamic range of the plasma proteome by exploiting the competitive binding equilibria, effectively normalizing the protein concentration by NP binding affinity. We previously showed that the properties of the protein–NP interface allow differential interrogation of complex plasma proteomics samples with superior performance compared to simple workflows based on neat and depleted plasma (20). This facilitated the discovery of NSCLC protein signatures comprising a novel combination of both known and unknown protein biomarkers. Our findings prompted us to compare our multi-NP workflow to the common, labor-intensive high-pH peptide fractionation of depleted plasma as well as to a commercially available deep proteomics workflow. Furthermore, here we investigated the mechanisms by which different NPs differentially interrogate the proteome of complex biological samples, thus enabling broad protein profiling with a multi-NP panel.
High-pH fractionation exploits the hydrophobicity of peptides to reduce the complexity of a sample prior to liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) injection by spreading the peptides across multiple fractions. Those fractions can then be recombined to optimize use of the LC-MS/MS gradient. Although this approach achieves significantly deeper measurement of the measured proteome compared to simpler methods (e.g., neat or depleted plasma), it also increases overall sample acquisition time, effectively reducing practical study size; it may also increase variation by requiring additional processing steps (21). Here we compared NPs to high-pH fractionation and found that our NP workflow provides better performance in terms of precision and depth. To dissect mechanisms that contribute to protein corona formation on NPs, we modeled protein abundance in the corona as a function of NP physicochemical properties including charge and decoration with functional groups. Connecting the physicochemical makeup of NPs with the specific distribution pattern of proteins across NP coronas provides an avenue for rational NP designs tailored to further improve the depth and breadth of proteomic interrogations.
Results
Comparing a Multi-NP Workflow to Conventional Deep Workflows.
A common workflow for deep plasma proteomic profiling is to fractionate peptides according to their hydrophobicity prior to LC-MS/MS. We compared a panel of five NPs to a high-pH fractionation of plasma depleted for high-abundance proteins (deep fractionation) in which 19 fractions were skip-concatenated to 9 final fractions, depleted plasma (without any fractionation), and neat plasma (Fig. 1A) using 30-min gradients and data-independent acquisition (DIA). To facilitate comparison, we used a common pooled plasma sample, the same MS instruments and gradients, and the same downstream processing of acquired MS data. Using the five-NP panel, we identified over 1,700 protein groups across three assay replicates, which translates to 2.5×, 4.4×, and 5.5× more protein groups compared to deep fractionation, depleted, and neat plasma, respectively, for the same gradient (Fig. 1B). The four workflows (Fig. 1B) were processed in independent analysis batches, which was necessary to compare the independent identification performance by each workflow. Since multiple deep workflow variants exist, and data-dependent acquisition (DDA) has been reported to benefit from fractionation more than DIA (23), we performed additional comparisons using the same pooled plasma and longer gradients with DDA as well as a commercially available deep DDA LC-MS/MS pipeline. While the absolute number of detected proteins varied significantly depending on gradient lengths and MS instrumentation, our five-NP workflow achieved an overall superior performance across all conditions. Comparing the precision of peptide quantification between different workflows, five NPs, depleted, and neat plasma yielded median coefficient of variation (CV) <20% (16.9, 19.8, and 8.7%, respectively), while deep fractionation yielded 2× higher median CV (34.2%) compared to the five-NP workflow (Fig. 1C). The neat plasma workflow has the lowest CV across workflows because most of the proteins identified in neat plasma are abundant and easier to measure.
Fig. 1.
DIA workflow comparison. Comparing a five-NP workflow (red) to a 19-concatenated-into-9 high-pH fractionation of depleted plasma strategy (blue), a plasma depletion strategy (green), and neat plasma (purple). (A) Step-by-step comparison of five-NP, high-pH fractionation, depleted, and neat plasma workflows. (B) Median number of protein groups identified by each workflow. Error bars denote SDs of assay replicates. The top dash depicts the number of identified proteins in any of the samples, and the lower dash represents the number of identified proteins in three out of three assay replicates (defined as the complete features). For this comparison, samples belonging to the respective workflows were processed as independent Spectronaut runs. When processed together, the median numbers of protein groups were 1,615, 862, 461, and 375 for five-NP, high-pH, depleted, and neat plasma, respectively. (C) CV of median-normalized peptide intensities filtered for three out of three identifications across assay replicates. Median CV is depicted on each plot. (D) Dynamic range of identified proteins matched with normalized protein intensities from a plasma protein database (22). Protein groups were filtered for complete features. Median log10 intensity of complete features is shown on each boxplot, and the outliers are removed. (E) Percent coverage of the plasma protein database in each workflow (Top) and relative coverage of plasma protein database by the five-NP method to high-pH fractionation (Bottom) over negative protein log10 intensities. The 95% interval is shown in gray. Protein groups were filtered for complete features. (F) UpSet plot showing the protein group overlap between the five-NP and high-pH workflows. Protein groups are filtered for complete features. The workflows included in each bar are shown as colored labels. (G) Comparison of number of proteins with specified functional annotations covered exclusively with five-NP (red), exclusively with high-pH workflow (blue), or both (overlapped). Protein groups were filtered for complete features. The Venn diagrams are proportional to the number of protein groups. Workflows were processed together using Spectronaut for all analyses except for A, in which each workflow was processed separately. All proteins and peptides were conservatively filtered at 1% protein and peptide FDR.
To determine the dynamic range covered by each workflow, the identified proteins were mapped to previously reported deep plasma proteome data (22) and their respective normalized intensities. The panel of five NPs covers more proteins at lower intensity than alternative workflows (Fig. 1C), extending nearly throughout the database’s entire dynamic range, with about 10× greater median depth compared to the conventional deep workflows using the same 30-min gradient and DIA. Moreover, comparing complete identified features (those detected in all three assay replicates), five NPs detected more complete features than the other methods, especially at lower intensity levels (Fig. 1D). To further investigate how each workflow covers the database, we examined the percent coverage at each intensity range, ranking from high- to low-abundance proteins. Deep fractionation covers 18% more high-abundance proteins (top 50% intensity) than the five NPs, while the five NPs produce up to 10× higher coverage than deep fractionation across the lowest two orders of magnitude, capturing 62% more proteins at the lower 50% intensity levels compared to the alternative deep workflow (Fig. 1E). Intriguingly, the proteome coverage for five NPs stays stable within the low-abundance range, supporting the utility of NPs for compression and sampling across the entire dynamic range.
We examined the overlap between identified protein groups in different workflows (Fig. 1F). Out of the 1,706 identified protein groups across the three assay replicates using five NPs, 900 are uniquely identified by the five NPs, 184 are common between all methods, and 169 are common only between NPs and deep fractionation. Comparing five NPs and deep fractionation, the latter contributes only 172 unique protein groups, compared to 979 identified by the five NPs. We next grouped proteins uniquely and commonly identified by five NPs and the deep workflow based on their functional annotations (Fig. 1G). Compared to deep fractionation, five NPs cover up to 4× more proteins annotated in UniProt keywords as putatively phosphorylated (2.8×), glycosylated (1.1×), acetylated (3.3×), and methylated (4×) as well as other functionally relevant classes, including secreted (1.2×) proteins and lipoproteins (2.6×) (Fig. 1G).
To show that the performance differences between workflows are independent from the processing software, we reprocessed the data with a recently published neural network-based proteomics search engine, Data-Independent Acquisition Neural Network [DIA-NN (24)], in library-free mode (SI Appendix, Fig. S1). This analysis identified over 3,000 protein groups across three replicates of the five-NP panel (SI Appendix, Fig. S1A). Compared to alternative data processing workflows, the five-NP workflow has a consistently superior performance in terms of protein group coverage, quantification reproducibility, and dynamic range coverage (SI Appendix, Fig. S1).
Next, we compared the automated five-NP workflow to the above-described deep fractionation workflow using DDA with the injection-to-injection time of 2 h for five-NP samples and 4 h for deep fractionated samples (SI Appendix, Fig. S2). Moreover, to evaluate our own implementation of a conventional deep workflow, we asked a commercial proteomics service facility to perform its deepest plasma proteome workflow on the same plasma pool as an external reference. The outsourced workflow entailed depletion of the 12 most abundant plasma proteins followed by high-pH fractionation into 96 fractions that were skip-concatenated to 12 final fractions, which were then analyzed by DDA over a total of 48 h (2 h for each sample). Using either the same gradient length (outsourced) or longer gradient length (in-house) for deep fractionation workflows, five NPs maintained considerably higher protein group coverage with higher precision across a larger dynamic range (SI Appendix, Fig. S2).
Differential Interrogation of the Plasma Proteome Using NPs.
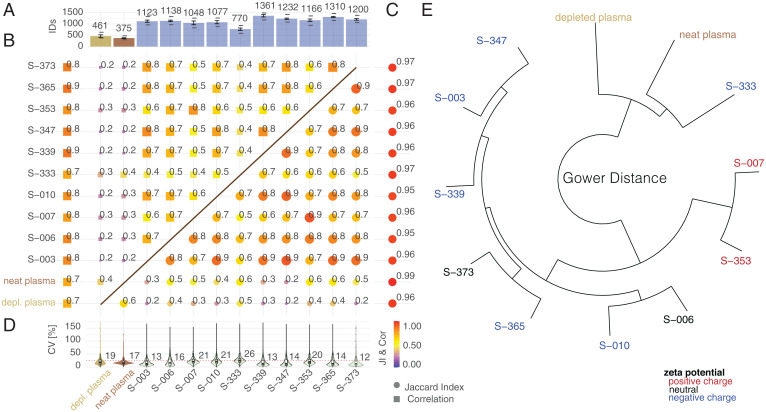
To gain a deeper understanding of the superior performance of the multi-NP panel, we evaluated the individual performance of an extended group of 10 NPs. Compared to neat and depleted plasma, each NP yielded significantly more protein groups with consistently greater depth, ranging from 205 (SP-333) to 363% (SP-339) (neat plasma; Fig. 2A). To determine the extent of overlap between proteins identified by different NPs, as well as both neat and depleted plasma, we calculated the Jaccard index (JI), which is the ratio of the size of the intersection of two sets to the size of the union of the sets. This measure is particularly useful when combining different NPs with the goal of increasing proteome coverage and is informative on the degree of exclusivity and redundancy (though redundancy is not necessarily undesirable, because quantification across multiple NPs could increase precision). NPs exhibited consistent identification patterns in assay replicates with distinct populations of proteins identified across NPs (Fig. 2B).
Fig. 2.
Interrogating the human plasma proteome with a multi-NP panel. (A) Median number of protein groups identified (1% protein, 1% peptide FDR for neat and depleted plasma, with the respective NPs shown as bar plots. Error bars denote SDs of protein IDs in assay replicates. The lower dash represents the number of identified proteins in three out of three assay replicates (complete features). The top dash depicts the number of proteins identified in any of the samples. (B) The top left triangular area shows the JI indicating degree of overlapping identifications as the mean JI across assay replicates (left column) and comparing individual NPs, depleted, and neat plasma (filtering for proteins quantified in three out of three assay replicates). Color and box size are scaled by the magnitude of JI. (C) The bottom right triangular area shows the Pearson correlation coefficient (r) indicating correlation of median normalized log10 intensities as the mean r across assay replicates (right column) and comparing individual NPs and neat plasma (filtering for proteins quantified in three out of three assay replicates, using median normalized intensity comparing pairwise common detections). Color and circle size are scaled by the magnitude of r. (D) Assay precision (CV) calculated for proteins quantified in three out of three assay replicates. Protein intensities were median normalized. Inner boxplots report the 25 (lower hinge), 50, and 75% quantiles (upper hinge). Whiskers indicate observations equal to or outside hinge ± 1.5 × interquartile range (IQR). Outliers (beyond 1.5 × IQR) are not plotted. Violin plots display all data points. (E) Gower distance mapped as distance tree for median protein intensities (filtered for three out of three identifications in assay replicates).
Confidence and precision of quantification can be increased by measuring proteins across multiple NPs. To estimate the degree to which NPs differentially enrich and deplete commonly detected proteins, we calculated correlation coefficients between protein coronas for the same plasma. The correlation of NPs to neat as well as depleted plasma ranges between 0.3 and 0.6, consistent with the particles’ ability to differentially enrich and deplete subsets of the proteome and compress dynamic range (20). Across NPs within our workflow, proteins are more closely correlated with coefficients between 0.5 and 0.9 (Fig. 2C), indicating similarities in protein corona composition across individual NPs. Assay triplicates exhibited high reproducibility (Fig. 2D), as measured by the mean correlation coefficient (close to 1) and the CV (median CVs below 25% for all NPs, with the exception of SP-333 with CV 25.7%).
To further map similarity across all NPs as well as neat and depleted plasma, we calculated the Gower distance (Fig. 2E), which combines qualitative and quantitative similarities to determine sample relations. We observed clustering that coincides with negative and positive charge for NPs SP-347, SP-003, and SP-339 and NPs SP-353 and SP-007, respectively. However, some NPs that cluster together (like SP-365 and SP-373) do not strictly follow that rule (Fig. 2E), suggesting that protein abundance signatures on NPs are driven by more complex dependencies beyond charge.
Linking NPs’ Physicochemical Properties to Protein Abundance.
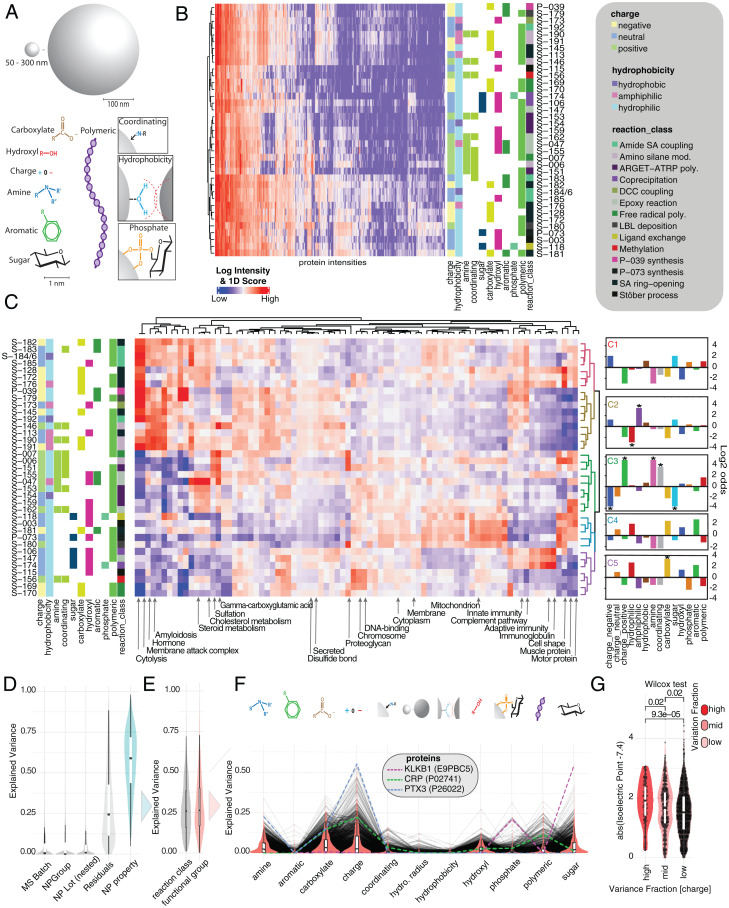
Prompted by the observation that zeta-potential does not fully predict protein corona composition, we further explored the protein corona composition as a function of individual NP properties. We expanded the number of chemically distinct NPs (37 NPs) and interrogated a common pooled plasma, as well as a cohort of human subjects (10 NPs; SI Appendix, Fig. S3), using an automated sample preparation workflow (20) in conjunction with LC-MS/MS. Each NP was annotated to describe its functionalization, including charge; hydrophobicity; and the presence of amine, carboxylate, sugar, phosphate, hydroxyl, polymeric structures, or aromatic groups (Fig. 3A). NPs with exposed amines or pyridyl groups were marked as coordinating. To account for the possibility that reactions necessary to functionalize a particle may not result in complete surface coverage, exposing the functionality of a precursor particle, we also categorized particles according to their reaction class, which is the type of chemical reaction used in the last step of the particle’s synthesis. On the level of individual protein intensities, we observed distinct intensity patterns for groups of NPs (Fig. 3B).
Fig. 3.
Effect of NP surface functionalization on protein corona composition. (A) NPs are classified based on a variety of physicochemical properties and functional groups including charge, polymer, sugar, aromatic systems, phosphates, amines, hydrophobicity, hydroxyl groups, coordinating property, and initial reaction class. (B) Unsupervised hierarchical clustering of median-normalized log10 protein intensities (1% FDR on protein and peptide level). Assay replicates of NP classes are median averaged. Missing values were filtered and imputed according to Materials and Methods. (C) The 1D annotation enrichment scores (heat map color ramp) for NPs. The 1D score was calculated for UniProt keywords as described in Materials and Methods. Enriched annotations are indicated in red; depleted annotations are indicated in blue. NPs are clustered based on the 1D score distributions. The log2–odds ratios of the NPs characteristic for each cluster are depicted as fingerprint diagrams on the right, with starred results indicating significance (P < 0.05) in Fisher’s exact test. (D) Variance decomposition analysis modeling normalized protein intensities as a function of NP’s physicochemical makeup (A). Explained variance by each variable was estimated using a linear mixed effects model and variancePartition package in R. The explained variance in protein intensities across NPs and the unexplained variance (residuals) are depicted as a density distribution. Variances explained for each protein across NP’s reaction class and functional groups are summed (turquoise distribution). (E) NP specific variance broken down into reaction class and functional groups. (F) Functional groups broken down into contribution of individual physicochemical properties. (G) Explained variance for functional group “charge” split into high (explained variance >30%), middle (explained variance <25 and >10%), and low (explained variance <10%). Wilcox test was used to determine P values. y axis depicts the absolute of predicted isoelectric point of each protein – 7.4 (pH of the assay). The larger that value, the more likely the protein has a net charge in the assay and can be affected by NP charge. Inner boxplots report 25 (lower hinge), 50, and 75% quantiles (upper hinge). Whiskers indicate observations equal to or outside hinge ± 1.5 × IQR. Outliers (beyond 1.5 × IQR) are not plotted. Violin plots capture all data points.
We next used a one-dimensional (1D) annotation enrichment analysis to evaluate how these physicochemical properties differentially associate categories of NPs with functional annotations. Hierarchical clustering of NPs based on their 1D enrichment scores yielded five distinct groups of NPs (Fig. 3C). Fisher’s exact test was applied to each of these clusters to highlight the dominant distinguishing NP properties, as a fingerprint of the cluster’s character. Cluster 1 (Fig. 3C, red) is composed of sponge or silica-coated superparamagnetic iron oxide nanoparticles treated with succinic anhydride, which is ring-opened to expose a carboxylate group. Several members of this group (S-182 through S-186) have had other groups (butyl, pyridyl, and hydroxyethyl) subsequently tethered to them via amide coupling, with mixed results in coverage. Cluster 2 (Fig. 3C, yellow) includes the core sponge particle (S-113), as well as several other particles derived from this core, some of which may have hydrophobic or amphiphilic character. Two of the particles (P-039 and S-179) in this cluster have polystyrene surfaces functionalized with acidic/anionic groups (carboxylate and sulfonate). Cluster 3 (Fig. 3C, green) consists almost entirely of amines, with the exceptions being a hydroxyethyl group and an isopropylamide attached via activators regenerated by electron transfer - atom transfer radical polymerization (ARGET-ATRP). Clusters 4 (Fig. 3C, blue) and 5 (Fig. 3C, purple) are composed entirely of hydrophilic groups, including most of the hydroxyl-functionalized particles and all sugar-functionalized ones. These two clusters have similar NP characteristics despite exhibiting some distinct protein enrichment patterns. Clusters 4 and 5 also include several carboxylate-functionalized particles (S-169, S-170, and S-181), the methylated version of amine S-006 (S-156), and a cationic tetraalkylammonium salt particle (S-180).
These clusters of NPs were found to differentially enrich or deplete groups of UniProt keywords. Proteins associated with cytolysis, innate immunity, the membrane attack complex, the complement pathway, amyloidosis, and hormones are heavily enriched by clusters 1 and 2 and depleted by clusters 3, 4, and 5. These keywords are generally enriched for anionic carboxylate-containing particles and depleted for cationic amines. These proteins may also exhibit an affinity for hydrophobic or amphiphilic functionalization, but these observations are confounded by a correlation with reaction class. Proteins associated with cholesterols and steroid metabolism exhibit a similar hydrophobicity pattern but the opposite charge effect: they are enriched by cluster 3 and depleted by several of the anionic particles in cluster 2. Aromatic groups further enhance their enrichment, suggesting the possibility of a pi-stacking effect. The proteins with gamma-carboxyglutamic acid domains follow a charge-driven process, being enriched by all the cationic particles and depleted by most of the anionic ones. The sulfation and proteoglycan proteins follow a similar pattern. DNA-binding and chromosomal proteins show an increased association with hydrophilic clusters 4 and 5, but this association is weaker with the sugar-functionalized NPs. In contrast, the sugar-functionalized particles heavily enrich immunoglobulins and adaptive immunity proteins. Proteins associated with mitochondria, membranes, cell shape, and muscular and motor proteins were enriched by hydrophilic cluster 4 but not cluster 5. Several keywords that exhibited lesser association with protein surface moieties (secreted, disulfide bond, and cytoplasm) showed little enrichment or depletion across the different NPs.
NP research and development can be guided by this type of analysis. In particular, to enrich for specific protein classes or proteins of specific characteristics, NPs should be explored that have fingerprints similar to those of the clusters above, which enrich for respective protein annotations. For example, class architecture topology/fold homologous superfamily (CATH) architectures (25), representing the secondary structures of proteins, are enriched as a function of the charge of the NP surface functionalization (SI Appendix, Fig. S4). In particular, beta-rich secondary structures (e.g., 4 Propeller, Aligned Prism, and Beta Barrel) are associated with NPs containing anionic functional groups, like carboxylate. This pattern aligns with previous observations of lysine and arginine being preferentially exposed in beta-rich structures (26), putatively attracting those to anionic NP functionalizations.
Given the intricate physicochemical makeup of NPs, our next goal was to explore to what degree individual functionalization (e.g., charge, amine groups, or aromatic groups) of NPs affects the protein corona composition and thus abundance of each protein for a respective NP. Across the 37 NPs, some functional characteristics are correlated; for example, the reaction class is not entirely independent from downstream functionalization (SI Appendix, Fig. S5). While confounded variables may obscure the estimated effect sizes, we evaluated the degree to which these pilot data can provide insights into corona formation on a protein-by-protein level.
We conducted variance decomposition to quantify the fraction of observed variance in protein abundance (approximated by intensities) that can be explained by individual covariates (i.e., NP properties) for 32 out of the 37 NPs for which data on the hydrodynamic radius were available (Fig. 3D). On average, more than 50% of the variance in each protein intensity across particles can be explained by this selection of NP properties. A small fraction of the variance is associated with the LC-MS/MS run batch (the defining set of samples run within the same maintenance cycle) and NP lot, and ∼25% of the variance (residuals) remains unexplained (Fig. 3D). NP properties split into reaction class (the final step in functionalization), accounting for ∼20% and the final functionalization, accounting for more than 25% of the variance (Fig. 3E). Among functional groups, charge and carboxylate showed the largest individual contribution to protein intensity differences (Fig. 3F). We depict three proteins that are differentially interrogated by a combination of physicochemical NP characteristics: the inflammation marker and acute phase response protein CRP, the serin-type endopeptidase KLKB1, and the innate immune response protein PTX3. Many proteins are associated with multiple functional groups, which we illustrated for a subset of NP characteristics in a directional network analysis in SI Appendix, Fig. S6. Protein abundances that are driven by charge should on average have isoelectric points (pI) farther away from the pH during protein corona formation. The protein coronas in these studies were generated at pH 7.4. We observed significantly more extreme pIs for those proteins that have a particularly high degree of variance explained by charge (Fig. 3G). In addition, carboxyl functionalization and amine functionalization correlated as expected with higher and lower pI, respectively (SI Appendix, Fig. S6B).
In summary, this analysis indicates that the physicochemical properties of NPs can be predicted, guiding the design of complementary NPs for deep proteome-wide interrogations or tuning NPs to target specific protein classes.
Discussion
One of the great challenges in improving our understanding of the molecular landscape of health and disease is the lack of large-scale proteomics data that could be useful in developing better tests and therapeutics and that could also be used to give functional context to the vast amount of accumulating genomics data. The wide variety of deep proteomics workflows designed to address this challenge usually sacrifice sample throughput for within-sample detection depth. Here we quantify the performance of a multiple NP proteomics workflow that combines scalability and deep proteome coverage.
The depth of a proteome analysis is not only a function of sample preparation but is also influenced by plasma source, the downstream LC-MS/MS method, and the data processing pipeline. Even supposedly similar plasma extraction methods can be a significant source of variation, yielding different protein identification numbers (27). Thus, absolute numbers across workflows and studies cannot be compared directly without providing context. Extensive multistep fractionation, isobaric labeling, and optimized pooling can yield more than 5,000 proteins (19, 22) in plasma, while most deep sample fractionations that combine multiple lengthy depletion steps to remove high-abundance proteins yield 1,000 to 2,000 proteins (18, 28–30). When we compared our five-NP workflow to a depletion and high-H peptide fractionation workflow, depleted and neat plasma using the same pooled plasma injected back-to-back with various gradients and processed with two different search engines, we found the NP had consistently higher workflow performance. Using two different downstream data processing approaches, the five-NP workflow yielded 1,706 to 3,088 protein groups, and deep fractionation yielded 684 to 1,855 protein groups. To provide an independent external reference, we asked a commercial proteomics service facility to perform a deep fractionation and long-gradient acquisition of the same samples. The different versions of internally and externally performed deep workflows using the same plasma pool yielded between 684 (30 min per fraction on DIA) and 1,761 (4 h for each of the nine fractions on DDA) protein groups; the five NPs yielded between 1,706 (30 min per NP on DIA) and 2,014 (2 h per NP on DDA), outperforming neat, depleted, and fractionated plasma even when using considerably shorter gradients and fewer hands-on requirements.
The conceptual difference between a high-pH fractionation deep proteomics workflow and the NP workflow is that the former fractionates each sample at the peptide level according to hydrophobicity. Multiple fractions are pooled (e.g., 24 fractions are pooled nonsequentially into 9 final fractions) to make optimal use of the LC-MS/MS gradient in which peptides are also separated by hydrophobicity. In contrast, NPs deplete and enrich at the protein level, gaining dynamic range for all peptides belonging to the proteoforms bound to each NP. The effective dynamic range compression at the proteoform level might prevent loss of very low-abundance proteins compared to strategies like high pH that require proteins to be retained until peptide fractionation. Our data suggest that NP corona-driven protein-level interrogation is a significantly more efficient strategy to increase depth in complex proteomes compared to high-pH fractionation workflows. An important feature in our development of optimized NPs and panels of NPs is our ability to draw on the extreme diversity of potential chemical modifications that can be designed and engineered into NPs as well as combined into complementary panels to improve overall coverage or performance in a given application. In addition, larger and smaller NP panels (e.g., 2 orthogonal NP) could be designed and engineered to prioritize throughput or coverage of the proteomics workflow depending on study requirements. Given that NPs and conventional deep methods address the dynamic range issue on different levels, future studies aiming for deep proteome mapping could combine the dynamic range compression benefit of NPs with high-pH fractionation to achieve unprecedentedly deep plasma proteomics.
To further optimize and develop new varieties of NPs for deep proteome interrogation, quantifying similarities and dissimilarities between protein corona compositions is key. The large dynamic range of protein abundance in biospecimens (>10 orders of magnitude) combined with the detection range of mass spectrometers (three to four orders of magnitude) and the unique protein–NP interaction results in variable intensity measurements and presence–absence patterns of proteins across NPs. We showed the robustness of each NP’s measurements across replicates of the same sample, while a larger subset of the proteome was captured in combination across many NPs (Fig. 2).
Previous studies have explored how distinct NP properties affect protein corona formation with the goal of understanding NP uptake, blood circulation time, immune system interaction, and recognition (7, 31), as well as protein corona formation ex vivo (32, 33). Recently, Ban et al. aggregated multiple published datasets and employed a random forest model to identify factors in protein corona formation across NP physicochemical properties and experimental conditions (34). A more targeted approach (35) showed the utility of highly specific NPs for targeted interrogation of very low abundance (<1 ng/mL) biomarkers in serum such as the cardiac biomarker troponin I (cTnI). Here, we aimed to identify some of the physicochemical properties that drive protein corona composition at the resolution of individual proteins in our well-defined, automated workflow. Our analysis indicates that altering the bulk physicochemical properties of NPs can be utilized to target specific protein subsets. In contrast to prior work predicting a general inhibition of protein corona formation by negatively charged or long-chained groups (36), our experimentally observed protein intensities did not follow this trend, illustrating the limitations of ab initio computational models in the complex space of NP functional group variants.
Using variance decomposition analysis, we quantified associations between distinct physicochemical properties of NPs and their protein corona makeup at the protein level. While this provides a proof of concept to connect high-level NP functionalization to protein corona composition, some limitations apply to our modeling approach. The model evaluates correlation, not causation within the dataset presented here. There can be hidden properties that correlate with the evaluated covariates and thus drive the observed coefficients. In addition, we did not directly determine the moieties on the NPs that bind specific proteins. However, the correlation between the physicochemical properties of NPs and their protein corona is evidence against the possibility of these proteins coming to a large degree from NP-unrelated sources such as entrapped proteins, protein attachment to well plates, etc. Some effects driving protein corona formation are likely unresolved when looking at bulk NP properties and bulk protein abundance without specifying their individual interaction interface. This includes secondary interactions, such as protein–protein interactions, which may affect protein abundance as a function of the primary protein–NP interaction. Therefore, the determined dependencies must be further explored in larger datasets expanding both the number of NPs and the details of their physicochemical characterization (i.e., quantitative information on functional group density) to establish underlying causalities.
The present study demonstrates the feasibility of designing panels of NPs that together to interrogate classes of protein ligands. We clustered NPs according to their protein-binding behavior and described the fingerprints of some types of NPs that enrich for the binding of specific protein classes. Additional studies employing a larger set of NPs and more detailed and quantitative surface characterizations will enable predictive models mapping out the relation of protein abundance in the corona to the physicochemical properties of NPs. Such studies will also improve the NP engineering and proteomics workflow quality control process by highlighting critical and noncritical NP property ranges at the resolution of individual proteins and protein classes. Recent work on protein structure and surface property prediction such as molecular surface interaction fingerprinting (37) and AlphaFold (38) also presents an intriguing opportunity to identify and understand physicochemical protein properties that drive specific nano–bio interactions. Ultimately, our findings could enable optimization and design of NP and surfaces for interrogating more complete segments of the proteome, prevention or enhancement of specific molecular absorption, or drug delivery in nanomedicine.
Materials and Methods
Synthesis and Characterization of NP Physicochemical Properties.
The NPs used in this study were synthesized as previously described (20). The data from dynamic light scattering, scanning electron microscopy, transmission electron microscopy, and X-ray photoelectron spectroscopy (XPS) were also gathered as previously described.
Biological Samples.
The common plasma sample used in the workflow comparison was pooled from K2 EDTA plasma samples of 153 deidentified healthy and lung cancer patients with various ages, genders, and disease stages. Details of the individual sample acquisition and processing were previously described (20). In brief, samples were collected after confirmed diagnosis of stage 1, 2, 3, or 4 NSCLC but prior to treatment. The samples were not controlled for fed/fasting state.
Sample Preparation.
Protein corona preparation and proteomic analysis.
NPs were synthesized as described previously (20). NP powder was reconstituted in deionized water to a final concentration of 5 mg/mL for all NPs except for SP-339-008 and SP-353-002 (final concentration 2.5 mg/mL) and SP-373-007 (final concentration 10 mg/mL), followed by 10 min sonication and vortexing for 2 to 3 s. To form the protein corona, 100 μL of NP suspension was mixed with 100 μL of plasma samples in microtiter plates. The plates were sealed and incubated at 37 °C for 1 h with shaking at 300 rpm. After incubation, the plate was placed on top of a magnetic collection device for 5 mins to draw down the NPs. The supernatant containing the noncorona, unbound proteins was aspirated by pipetting. The protein corona was washed three times with 200 μL of wash buffer, which contains 150 mM KCl and 0.05% CHAPS in a Tris EDTA buffer with pH of 7.4. Note that EDTA is already present in the samples collected in K2 EDTA tubes.
To digest the proteins bound onto NPs, a trypsin digestion kit (iST 96X, PreOmics) was used according to protocols provided by the vendor. Briefly, 50 μL of Lyse buffer was added to each well and heated at 95 °C for 10 min with agitation at 1,000 rpm. After the plates were cooled to room temperature, trypsin digestion buffer was added, and the plates were incubated at 37 °C for 3 h with shaking at 500 rpm. After stopping the digestion process by addition of the supplied stop buffer, the NPs were removed from the reaction by magnetic collection, and the remaining reaction supernatant was cleaned up with the supplied filter cartridge (styrenedivinylbenzene reversed-phase sulfonate [SDB-RPS]) kit. The peptide was eluted with 75 μL of elution buffer twice and combined. Peptide concentration was measured by a quantitative colorimetric peptide assay kit from Thermo Fisher Scientific.
Plasma depletion.
Plasma samples were subjected to depletion (immunoaffinity-based removal of abundant proteins) using an Agilent 1260 Infinity II Bioinert high-performance liquid chromatography (HPLC) system consisting of autosampler, pumps, column compartment, UV detector, and fraction collector. Plasma depletion was conducted by first diluting 20 μL of plasma to a final volume of 100 μL using Agilent Buffer A plasma depletion mobile-phase. Each diluted sample was filtered through an Agilent 0.22 μ cellulose acetate spin filter to remove any particulates and transferred to a 96-well plate. The plate was then placed in an autosampler and held at 4 °C until further processing. Eighty microliters of the diluted plasma was then injected onto an Agilent 4.6 × 50 mm Human 14 Multiple Affinity Removal System (MARS 14) depletion column housed in the HPLC column compartment at a constant temperature of 20 °C. Mobile-phase conditions used during protein depletion consisted of 100% Buffer A mobile-phase flowing at a rate of 0.125 mL/min. Proteins eluting from the column were detected using the Agilent UV absorbance detector operated at 280 nm with a bandwidth of 4 nm. The early eluting peak for each injection, representing the depleted plasma proteins, was collected using a refrigerated fraction collector with peak intensity–based triggering (200 mAu threshold with a maximum peak width of 3 min). After peak collection, the fractions were held at 4 °C. The sample volume was then reduced to ∼20 μL using an Amicon Centrifugal Concentrator (Amicon Ultra-0.5 mL, 3k molecular weight cut-off [MWCO]) with a centrifuge operating at 4 °C and 14,000 × g. Each depleted sample was then reduced, alkylated, digested, desalted, and analyzed according to the sample preparation and MS analysis protocols described below. During each sample depletion cycle, the MARS 14 column was regenerated with the Agilent Buffer B mobile-phase for ∼4 1/2 min at a flow rate of 1 mL/min and equilibrated back to the original protein capture condition by flowing Buffer A at 1 mL/min for ∼9 min.
Peptide fractionation.
A total of 100 μL of reconstituted peptides was loaded to a Waters XBridge column (2.1 mm × 250 mm, BEH C18, 3.5 mm, 300 Å) using the Agilent 1260 Infinity II HPLC system. The peptides were separated at a flow rate of 350 mL/min using a gradient of 3 to 30% in 30 min, with a total run time of 47 min, and the fractions were collected every 1.5 min. The fractions were then dried using a Speed Vac. Finally, the dried peptides were reconstituted in a solution of 0.1% formic acid (FA) and 3% acetonitrile (ACN), spiked with 5 pmol/mL PepCalMix from SCIEX and concatenated to 9 fractions according to the following scheme: fractions 1, 10, and 19 were pooled; fractions 2 and 11 were pooled; fractions 3 and 12 were pooled; and so on to create 9 concatenated samples.
Outsourced peptide fractionation.
The common plasma sample was sent to a commercial service laboratory with a proteomics facility to perform the deepest plasma proteome workflow available. Briefly, plasma was depleted in triplicate using Top12 immunodepletion columns (Pierce, Thermo Scientific) according to the manufacturer’s protocol. After concentrating proteins using a 5 kDa MWCO spin filter and trypsin digestion, high-pH fractionation was carried out using a Waters XBridge C18 (2.1 mm ID × 150 mm, 3.5 μm) and Agilent 1100 HPLC, yielding 96 fractions in total, which were then concatenated to 12 final fractions.
Mass Spectrometry.
DIA.
LC-MS/MS.
For DIA analyses using sequential window acquisition of all theoretical fragment ion spectra (SWATH), peptides were reconstituted in a solution of 0.1% FA and 3% ACN spiked with 5 fmol/μL PepCalMix from SCIEX. Five μg of peptides in 10 μL of reconstitution buffer was used for each constant mass MS injection. Each sample was analyzed by an Eksigent nanoLC system coupled with a SCIEX TripleTOF 6600+ mass spectrometer equipped with an OptiFlow source using a trap-and-elute method. First, the peptides were loaded on a ChromXP C18CL (0.3 mm ID × 10 mm) trap column and then separated on a Phenomenex Kinetex analytical column (150 mm × 0.3 mm, C18, 2.6 μm, 100 Å) at a flow rate of 5 μL/min using a gradient of 3 to 32% solvent B (0.1% FA, 100% ACN) mixed into solvent A (0.1% FA, 100% water) over 20 min, resulting in a 33-min total run time. The mass spectrometer was operated in SWATH mode using 100 variable windows across the 400 to 1,250 mass-to-charge ratio range.
Data analysis for library generation.
In DIA MS methods, intentionally convoluted MS2 spectra are resolved by algorithmic comparison to predefined prototypical spectra libraries. The samples used to generate the spectral library of this analysis were previously described (20). Briefly, multiple pooled plasma samples processed by Proteograph as well as the common pooled plasma of this study (processed through both Proteograph and deep fractionation) were used to build a spectral library. All the DDA data were first searched against the human UniProt database using the Pulsar search engine in Spectronaut (Biognosys). Then the library, including 3,242 protein groups and 25,773 peptides, was generated using Spectronaut with 1% false discovery rate (FDR) cutoff at peptide and protein level.
DIA raw data processing.
The SWATH data were processed using the Spectronaut analytical software (version 13.8.190930.43655). The default settings were used for the analysis except for quantification, for which no cross-run normalization was chosen. The Q-value cutoff at precursor and protein level was set to 0.01.
We also processed these data by DIA-NN software (24) (version 1.8) in library-free mode. The neural network classifier was set to double-pass mode. The rest of the parameters were set to default. The FDR cutoff at both precursor and protein level (Lib.Q.Value and Lib.PG.Q.Value) was set to 0.01.
DDA.
LC-MS/MS (in house).
The peptide eluates were lyophilized and reconstituted in 0.1% TFA. A 2-μg aliquot from each sample was analyzed by nano LC-MS/MS with Thermo Scientific UltiMate 3000 RSLCnano system interfaced to an Orbitrap Fusion Lumos Tribrid Mass Spectrometer from Thermo Scientific (the LC-MS parameters for 4-h runs are shown in brackets). Peptides were loaded on an Acclaim PepMap 100 C18 (0.3 mm ID × 5 mm) trapping column and eluted over a Waters Acquity M-Class (75 μm × 200 mm) analytic column at 250 nL/min [300 nL/min] using a gradient of 2 to 35% [2 to 30%] acetonitrile over 113 [200] min, for a total time between injections of 125 [240] min. The mass spectrometer was operated in DDA mode, with MS and MS/MS performed at 60,000 [120,000] full width at half maximum (FWHM) resolution and 15,000 [30,000] FWHM resolution, respectively.
LC-MS/MS (outsource).
The peptide eluates were lyophilized and reconstituted in 0.1% TFA. Based on the service facility’s protocol, 20% of each reconstituted sample was analyzed by nano LC-MS/MS with a Waters NanoAcquity HPLC system interfaced to an Orbitrap Fusion Lumos Tribrid Mass Spectrometer from Thermo Scientific. Peptides were loaded on a trapping column and eluted over a 75-μm analytic column at 350 nL/min; both columns were packed with Luna C18 resin (Phenomenex). A 2-h gradient was employed. The mass spectrometer was operated in data-dependent mode, with MS and MS/MS performed in the Orbitrap at 60,000 FWHM resolution and 15,000 FWHM resolution, respectively.
DDA data processing.
MS raw files were processed as described previously (20), in brief, with MaxQuant (v. 1.6.7) and Andromeda (39, 40), searching MS/MS spectra against the UniProtKB human FASTA database (UP000005640, 74,349 forward entries; version from August 2019) employing standard settings. Enzyme digestion specificity was set to trypsin, allowing cleavage N-terminal to proline and up to two miscleavages. Minimum peptide length was set to seven amino acids and maximum peptide mass to 4,600 Da. Methionine oxidation and protein N-terminal acetylation were configured as a variable modification, and carbamidomethylation of cysteines was set as a fixed modification. “Match between runs” was disabled. Identifications were quantified based on protein intensities (only proteins with q value <1%) requiring at least one razor peptide. Proteins that could not be discriminated based on unique peptides were assembled in protein groups. Furthermore, proteins were filtered for a list of common contaminants included in MaxQuant. Proteins identified only by site modification were strictly excluded from analysis.
Database coverage analysis.
To understand in more detail how each workflow covers the database, we divided the database (22) into multiple bins, with stepping size of 0.5 across log10 intensities starting from zero. Then, the percentage of coverage for each workflow was calculated in each bin and plotted against the negative log10 intensities (Fig. 1E).
NP property modeling (10-NP experiment).
We used a previously published dataset comparing 141 healthy and early NSCLC subjects across 10 NPs. These data were median normalized as described previously (20). To compare only valid protein quantifications across subjects and all 10 NPs, we removed subjects and all of their associated samples if for one or more NPs, fewer than 750 proteins were detected. The remaining 157 protein groups were consistently quantified across 45 subjects and all 10 NPs. To determine to what degree individual physicochemical properties correlate with protein abundances comparing profiles across the 157 protein groups, 10 NPs, and 45 subjects, we trained a linear mixed effects model (LMM; lme4) with
setting RMFL = FALSE, which corresponds to a maximum likelihood estimation that models each protein’s intensity of the form
where is an column vector of the observed protein intensities; is an matrix for the constant and fixed effects of ZetaPotential (charge), PolyDispersityIndex (PDI), and HydrodynamicDiameter; and is a column vector of the fixed-effects regression coefficients. is an design matrix for the random effects of subject. is a vector of random effects for subject, which is assumed to be distributed as .
To determine functional annotations associated with the results from the LMM, annotations were matched to UniProt identifiers and enrichments calculated based on the coefficient distributions using the R AnnoCrawler package and implementation of the 1D annotation enrichment (41). To estimate the coefficient stability when building models on subsets of the subjects, the data were split into random (nonoverlapping) sets of 12 subjects. Coefficients calculated with the above-mentioned strategy (this time accepting singular fits) and similarities of model coefficients were quantified using Pearson correlation.
NP property modeling (37-/32-NP experiment).
We used a dataset generated from a single biological sample (PC3) measured across 81 NP lots. The data associated with 10 NP lots were removed due to synthetic uncertainty. MS runs where fewer than 200 protein groups were detected were discarded as outliers. Across the 71 remaining NP lots, 1,559 protein groups were detected, comprising 37 structurally distinct NPs.
The log10 protein intensity data were median normalized as described previously. We filtered for consistent identifications of protein groups by removing protein groups from the entire dataset if they were not detected with at least one NP at ≥50% identification rate throughout assay replicates. In order to estimate the differences between NPs that may deplete proteins below the instrument detection limits, intensities were imputed for protein groups that were not detected in any replicate of a given NP, based on the distribution of the entire population (mean downshift 2.0, width 0.2). The log10 protein group intensities were plotted and clustered (hclust) using ComplexHeatmap (Fig. 3B), after filtering out protein groups that were imputed for more than 75% of the NPs.
For 1D annotation enrichment (Fig. 3C), an intensity difference metric for each protein group was determined by calculating the difference between the median log10 intensity for each NP (e.g., across NP lots) and the median intensity of that protein group across all other NPs. The protein group intensity metrics were associated by their constituent UniProt protein IDs to UniProt keywords, counting each constituent keyword once per protein group. A Wilcoxon–Mann–Whitney signed-rank test for annotation enrichment was then applied, and the results were filtered for a Benjamini–Hochberg FDR <2% (41, 42). The 1D enrichment results were plotted and clustered (hclust) using ComplexHeatmap, and the top five hierarchical clusters were labeled. We used Fisher’s exact test to help identify the enrichment of NP properties based on their clustering in the UniProt keyword analysis.
To quantify the degree to which individual protein intensities correlate with individual NP properties (Fig. 3 D–F) we employed a linear-mixed-effects model and performed variance decomposition analysis (R, variancePartition package). A model was encoded for each protein as
which corresponds to fit a linear mixed effects model of the form
where is an column vector of the observed intensities; is an matrix for the fixed effects of charge, polymeric, sugar, phosphate, amine, hydrophobicity, hydroxyl, coordinating, aromatic, carboxylate, hydrodynamic radius, and reaction class; and is a column vector of the fixed-effects regression coefficients. Polymeric, sugar, phosphate, amine, hydroxyl, coordinating, aromatic, and carboxylate are two-level factors, each encoded as a binary indicator variable for the presence or absence of the factor. Reaction class is a factor with 14 levels and was encoded using dummy coding with 13 binary indicator variables. is an design matrix for the random effects of mass spec batch (MS Batch). is a vector of random effects for MS Batch (MS maintenance cycle), which is assumed to be distributed as . is an design matrix for the random effects of NP group (NPs with the same physicochemical makeup). is a vector of random effects for NP group, which is assumed to be distributed as . is the design matrix of NP lots (a NP synthesis batch) nested in NP group. is a vector of random effects for NP lots nested in NP group, which is assumed to be distributed as . The residual error is a random effect. fitExtractVarPartModel was run for scaled and centered median-normalized protein intensities using “weights = T” and “showWarnings = F.” The latter was used to allow the model to utilize categorical variables as fixed effects. Note that collinearity of covariates and random effects in the model can cause the warning “boundary (singular) fit:.” For this dataset, the random-effect variance estimates for specific proteins can be close to zero, i.e., this protein’s intensity does not vary as a function of a specific random effect, causing a boundary (singular) fit: warning. Note that the model used in SI Appendix, Fig. S6 includes only the following terms: sugar + amine + polymeric + aromatic + phosphate + hydroxyl + carboxylate + reaction_class + (1|MS.Batch) + (1|NPGroup/npLot). For this analysis, we employed the library(lme4) and library(lmerTest) to estimate P values via Satterthwaite's method (43).
Network Analysis.
For network analysis we extracted the coefficients for the LMM discussed above. A pruned network was constructed filtering for coefficients with P value (Satterthwaite’s method) < 0.05 using R libraries (ggraph), (igraph), and (graphlayouts) with layout = “stress.” FDR was estimated using p.adjust() with the Benjamini–Hochberg method. Isoelectric points (pI) of proteins were calculated in R using computePI {seqinr}.
Statistical analysis.
Statistical analysis and visualization were performed using R (v3.5.2) with appropriate packages (44).
Supplementary Material
Acknowledgments
We thank the Center for Advanced Materials Characterization at the University of Oregon for use of their electron microscopy and XPS instruments, the Stanford Nano Shared Facilities at Stanford University for use of their electron microscopy and XPS instruments, and MS Bioworks for their mass spectrometry and sample preparation services. We also thank Moaraj Hasan, Iman Mohtashemi, Sangtae Kim, Harendra Guturu, and Marwin Ko for contributing to discussion.
Footnotes
Competing interest statement: S.F., B.T., T.R.B., P.A.E., M.F., M. McLean, E.M.E., X.Z., V.J.G., T.W., K.R., J.C., M. Mahoney, H.X., E.S.O., C.S., D. Harris, T.L.P., P.M., M.G., R.L., A.S., R.B., J.C.C., S.B., J.E.B., D. Hornburg, and O.C.F. have financial interest in PrognomIQ and Seer. Only Seer was involved in the study design, data collection and analysis, and manuscript writing/editing.
This article is a PNAS Direct Submission.
Data Availability
The mass spectrometry proteomics data for the NSCLC study (SI Appendix, Fig. S3) were deposited to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the Proteomics Identification Database (PRIDE) partner repository (45) with the dataset PXD017052. Data for Figs. 1–3 are available via the PRIDE partner repository (45) with the dataset PXD022285 and PXD028634. Annotations used for annotation enrichment analysis are available as part of the Perseus framework (46). The UniProt FASTA is available at https://www.uniprot.org/ (retrieved 29 August 2019). All other study data are included in the article and/or supporting information.
References
- 1.Bertrand N., et al. , Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 8, 777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J., Kantoff P. W., Wooster R., Farokhzad O. C., Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., et al. , Nano-bio interactions in cancer: From therapeutics delivery to early detection. Acc. Chem. Res. 54, 291–301 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Monopoli M. P., et al. , Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 133, 2525–2534 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Lundqvist M., et al. , The evolution of the protein corona around nanoparticles: A test study. ACS Nano 5, 7503–7509 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Tian X., Chong Y., Ge C., Understanding the nano-bio interactions and the corresponding biological responses. Front. Chem. 8, 446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Cai R., Chen C., The nano-bio interactions of nanomedicines: Understanding the biochemical driving forces and redox reactions. Acc. Chem. Res. 52, 1507–1518 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tenzer S., et al. , Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: A comprehensive quantitative proteomic analysis. ACS Nano 5, 7155–7167 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Xu M., et al. , How entanglement of different physicochemical properties complicates the prediction of in vitro and in vivo interactions of gold nanoparticles. ACS Nano 12, 10104–10113 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Lacerda S. H. D. P., et al. , Interaction of gold nanoparticles with common human blood proteins. ACS Nano 4, 365–379 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Lundqvist M., et al. , Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U.S.A. 105, 14265–14270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walkey C. D., Chan W. C. W., Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 41, 2780–2799 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Cedervall T., et al. , Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. Engl. 46, 5754–5756 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Castagnola V., et al. , Biological recognition of graphene nanoflakes. Nat. Commun. 9, 1577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson N. L., The clinical plasma proteome: A survey of clinical assays for proteins in plasma and serum. Clin. Chem. 56, 177–185 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Crutchfield C. A., Thomas S. N., Sokoll L. J., Chan D. W., Advances in mass spectrometry-based clinical biomarker discovery. Clin. Proteomics 13, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer P. E., Holdt L. M., Teupser D., Mann M., Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13, 942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer P. E., et al. , Plasma proteome profiling to assess human health and disease. Cell Syst. 2, 185–195 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Keshishian H., et al. , Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 12, 1683–1701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blume J. E., et al. , Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat. Commun. 11, 3662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulak N. A., Geyer P. E., Mann M., Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol. Cell. Proteomics 16, 694–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshishian H., et al. , Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol. Cell. Proteomics 14, 2375–2393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biognosys, Spectronaut User Manual (Biognosys, 2020). [Google Scholar]
- 24.Demichev V., Messner C. B., Vernardis S. I., Lilley K. S., Ralser M., DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson N. L., et al. , CATH: An expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 45 (D1), D289–D295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara K., Toda H., Ikeguchi M., Dependence of α-helical and β-sheet amino acid propensities on the overall protein fold type. BMC Struct. Biol. 12, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geyer P. E., et al. , Plasma Proteome Profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol. Med. 11, e10427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geyer P. E., et al. , Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 12, 901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wewer Albrechtsen N. J., et al. , Plasma proteome profiling reveals dynamics of inflammatory and lipid homeostasis markers after Roux-En-Y gastric bypass surgery. Cell Syst. 7, 601–612.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Pernemalm M., et al. , In-depth human plasma proteome analysis captures tissue proteins and transfer of protein variants across the placenta. eLife 8, e41608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilkington E. H., et al. , Profiling the serum protein corona of fibrillar human islet amyloid polypeptide. ACS Nano 12, 6066–6078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh J. Y., et al. , Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat. Commun. 9, 4548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schöttler S., et al. , Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 11, 372–377 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Ban Z., et al. , Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 117, 10492–10499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiambeng T. N., et al. , Nanoproteomics enables proteoform-resolved analysis of low-abundance proteins in human serum. Nat. Commun. 11, 3903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan S., Gupta A., Nandi C. K., Controlling the fate of protein corona by tuning surface properties of nanoparticles. J. Phys. Chem. Lett. 4, 3747–3752 (2013). [Google Scholar]
- 37.Gainza P., et al. , Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 17, 184–192 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Cox J., et al. , Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Hornburg D., AnnoCrawler. Zenodo. https://zenodo.org/record/3939280#.YiELL-jMJ9g. Accessed 4 March 2022..
- 42.Cox J., Mann M., 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 (suppl. 16), S12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuznetsova A., Brockhoff P. B., Christensen R. H. B., lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017). [Google Scholar]
- 44.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020; ). [Google Scholar]
- 45.Vizcaíno J. A., et al. , The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyanova S., Cox J., Perseus: A bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 1711, 133–148 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data for the NSCLC study (SI Appendix, Fig. S3) were deposited to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the Proteomics Identification Database (PRIDE) partner repository (45) with the dataset PXD017052. Data for Figs. 1–3 are available via the PRIDE partner repository (45) with the dataset PXD022285 and PXD028634. Annotations used for annotation enrichment analysis are available as part of the Perseus framework (46). The UniProt FASTA is available at https://www.uniprot.org/ (retrieved 29 August 2019). All other study data are included in the article and/or supporting information.