Significance
Bisphenol A (BPA), found in many plastic products, has weak estrogenic effects that can be harmful to human health. Thus, structurally related replacements—bisphenol S (BPS) and bisphenol F (BPF)—are coming into wider use with very few data about their biological activities. Here, we compared the effects of BPA, BPS, and BPF on human mammary organoids established from normal breast tissue. BPS disrupted organoid architecture and induced supernumerary branching. At a proteomic level, the bisphenols altered the abundance of common targets and those that were unique to each compound. The latter included proteins linked to tumor-promoting processes. These data highlighted the importance of testing the human health effects of replacements that are structurally related to chemicals of concern.
Keywords: bisphenols, mammary gland, organoids, breast cancer, global proteomics
Abstract
Environmental chemicals such as bisphenol A (BPA) are thought to contribute to carcinogenesis through their endocrine-disrupting properties. Due to accumulating evidence about negative human health effects, BPA is being phased out, but in parallel, exposures to replacement chemicals such as bisphenol S (BPS) and bisphenol F (BPF) are increasing. Little is known about their biologic effects, but because of their high degree of chemical relatedness, they may have overlapping as well as distinct actions as compared with BPA. We investigated this theory using a nonmalignant, human breast tissue-derived organoid system and two end points: morphologic and proteomic alterations. At low-nanomolar doses, replacement chemicals—particularly BPS—disrupted normal mammary organoid architecture and led to an increased branching phenotype. Treatment with the various bisphenols (vs. 17-β-estradiol or a vehicle control) produced distinct proteomic changes. For example, BPS up-regulated Cdc42-interacting protein 4, which supports the formation of invadopodia and a mesenchymal phenotype. In summary, this study used a highly physiologically relevant organoid system to provide evidence that replacement bisphenols have protumorigenic effects on the mammary gland at morphologic and proteomic levels, highlighting the importance of studies to evaluate the potential harmful effects of structurally related environmental chemicals.
Bisphenols, of which the most prevalent is bisphenol A (BPA), are environmental chemicals that are used as plasticizers in a variety of goods, including plastic bottles, children's toys, eyeglass lenses, food containers, and some types of thermal paper (e.g., cash register receipts). They leach from these products, contaminating humans (and animals) either directly or indirectly via other environmental media, such as household dust. Thus, in most adults, BPA is detected in serum, tissues, and urine (1, 2). Children (ages 6 to 11) have the highest concentrations of urinary BPA (3, 4). This chemical has structural similarities to estrogen (17-β-estradiol [E2]) and as a result, weakly mimics its activity (5). Hormones and growth factors play an important role in controlling prenatal mammary gland development and later on, the morphologic and functional alterations that occur during puberty, pregnancy, and eventually, menopause. Due to this plasticity, the mammary gland is particularly susceptible to the actions of endocrine-disrupting chemicals (EDCs), such as BPA (6–8). In vivo and in vitro studies have consistently shown that exposures to BPA at crucial developmental stages impair mammary gland development and increase neoplastic transformation (9–12). Treating rats with BPA results in mammary epithelial hyperplasia and enhances proliferation (13), common features of precancerous lesions. Additionally, BPA induces cell cycle progression and increases proliferation of human breast cancer cells in vitro via the up-regulated expression of estrogen-dependent genes (14).
Based on these and other data, BPA has been removed from many commercial products. Most commonly, this chemical of concern is replaced by bisphenol S (BPS) and bisphenol F (BPF) compounds that share close structural similarities with BPA. However, little is known about their endocrine effects and more broadly, their biological activities. Marketing a product as “BPA free” suggests to the consumer that a product is safer, but research shows that replacement bisphenols have adverse effects similar to, or even greater than, BPA. For example, studies in zebrafish, rodents, and human cell culture models show that BPS and BPF have endocrine-disrupting activities. In zebrafish, despite species-specific differences in estrogen receptor (ER) affinity and specificity, BPF and BPS have estrogenic activities similar to BPA (15–17). In rats, exposure to BPF induces uterine growth, which suggests estrogenic effects (18). BPA, BPF, and BPS promote estrogen-dependent cell cycle progression, proliferation, and migration of human MCF-7 breast cancer cells along with epigenetic changes (19, 20). BPS exposure of pregnant and lactating mice limits milk production, suggesting alterations in mammary gland composition (21). In addition to estrogen signaling, BPF affects other endocrine pathways; in rats, oral administration of this compound alters thyroid hormone levels (22).
Current research on bisphenol actions is mainly focused on endocrine effects. It is less well understood whether these chemicals have additional protumorigenic effects independent of their endocrine-disrupting activity. Moreover, tumor development is a multistep process involving heterogeneous cell types and numerous factors, including the potential roles of a variety of environmental chemicals (23, 24). Recapitulating this complexity in an experimental setup is challenging. In this context, tissue organoids are valuable models for understanding the early steps of carcinogenesis. They can be derived from nonmalignant primary tissues ex vivo. When grown for short periods of time in vitro, they maintain many of the genetic and epigenetic features of their normal cognates. Also, organoids have the added advantage of consisting of multiple cell types that are representative of the complexity of the tissue from which they are derived (25, 26).
In this study, we exploited the strengths of the human mammary gland organoid culture system to understand the impact of the BPA replacements, BPF and BPS. Organoids established from nonmalignant human mammary gland tissues were exposed to one of the bisphenols, E2, or the vehicle control. The results showed that BPA replacements, in particular BPS, disrupted organoid architecture, enhanced branching, and caused compound-specific proteomic alterations—effects that were mostly E2 independent. Together, these observations suggested that the mammary gland effects of BPA substitutes should be equally or more concerning than those of the compound they are replacing.
Results
Bisphenols Alter Branching Morphology of Human Mammary Organoids.
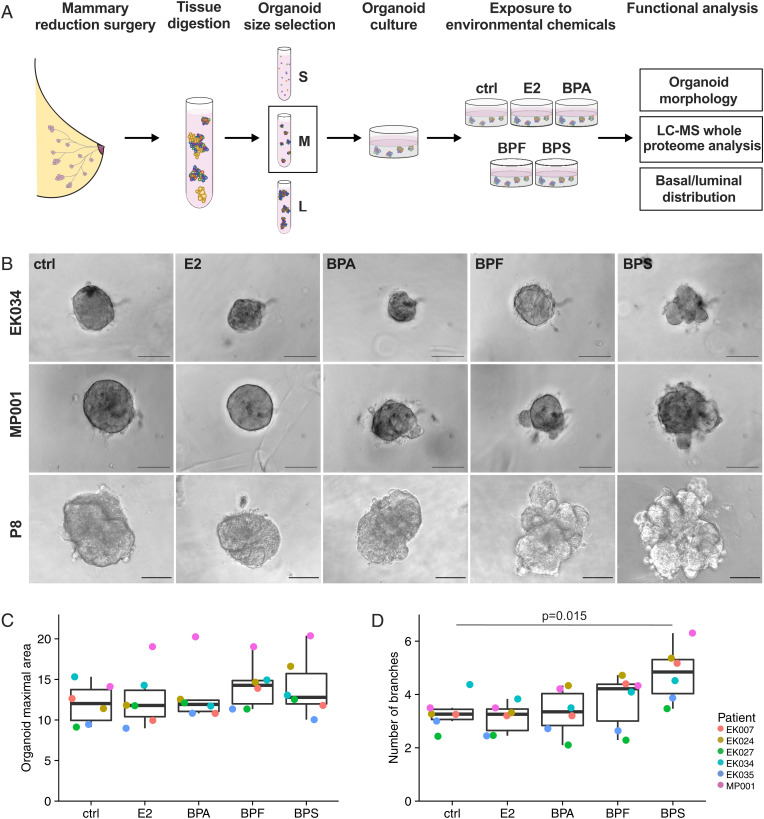
To study the impact of bisphenols on nonmalignant mammary tissue, we established human mammary organoid cultures using tissue obtained from reduction mammoplasty surgeries. We used partially digested tissue fragments to obtain organoids that capture the cellular heterogeneity, tissue structure, and hormone signaling properties of their in vivo counterparts (25). To mimic physiological exposure (see last paragraph of the Discussion), we treated the organoids with a low dose (15 nM) of BPA, BPF, BPS, E2, or vehicle control for 6 d (Fig. 1A). The choice of tissue-derived organoids and physiologic-relevant chemical dosages enabled us to test the effects of bisphenols on human breast tissue in a setting that simulated the in vivo situation (4). First, we analyzed the effect of chemical exposure on the morphology of human mammary organoids in two independent patient cohorts (cohort A with six different individuals and cohort B with four different individuals). Overall, we observed inter- and intrapatient heterogeneity in response to individual chemicals (Fig. 1B and SI Appendix, Fig. S1 A and D). Given the heterogeneity of the patient characteristics (e.g., age, body mass index [BMI]) (SI Appendix, Table S1), differences among donors were not unexpected. Nevertheless, exposure to the replacement chemicals resulted in an altered mammary organoid morphology in both cohorts (Fig. 1B and SI Appendix, Fig. S1 A and D), while overall organoid size, determined by maximal cross-sectional organoid area, was not affected (Fig. 1C and SI Appendix, Fig. S1B). BPS treatment resulted in the strongest increase in the number of organoid branches (cohort A: P = 0.015 [Fig. 1D]; cohort B: P = 0.0005 [SI Appendix, Fig. S1C]). The exposure to BPA and BPF also elevated the degree of branching, with BPF showing a significant increase in the number of branches in cohort B (P = 0.009) (SI Appendix, Fig. S1C). Taken together, as compared with estrogen treatment, bisphenols altered human mammary organoid morphology, with BPS causing the strongest effects in terms of increased branching.
Fig. 1.
Bisphenol exposures alter branching morphology of human mammary organoids. (A) Schematic of the experimental workflow. Nonmalignant primary human mammary tissue from reduction mammoplasty surgeries was digested and size separated, and the medium (M)-sized (vs. small [S] or large [L]) organoids were cultured in Matrigel. They were exposed to the vehicle control (ctrl), E2, or one of the bisphenols (15 nM) for 6 d. The end points analyzed included organoid morphology, the proteome at a global level (liquid chromatography-mass spectrometry [LC-MS]), and the distribution of basal and luminal cells. (B) Representative bright-field images of human breast organoids of different patients after exposure for 6 d to the vehicle control (ctrl), E2, BPA, BPF, or BPS. (Scale bars, 100 μm.) (C) Quantification of the organoid maximal cross-sectional area colored by patient (cohort A). Each dot represents the mean value from the analysis of 5 to 25 organoids per patient per treatment. The median and the interquartile range are shown. The values were not statistically different among the groups. (D) Quantification of the total number of branches per organoid colored by patient (cohort A). Each dot represents the mean number of branches from 5 to 25 organoids per patient per treatment. The median and interquartile range are shown. Results that reached statistical significance using the two-sample Wilcoxon test are noted.
Bisphenol Exposure Resulted in Alterations at the Level of the Proteome.
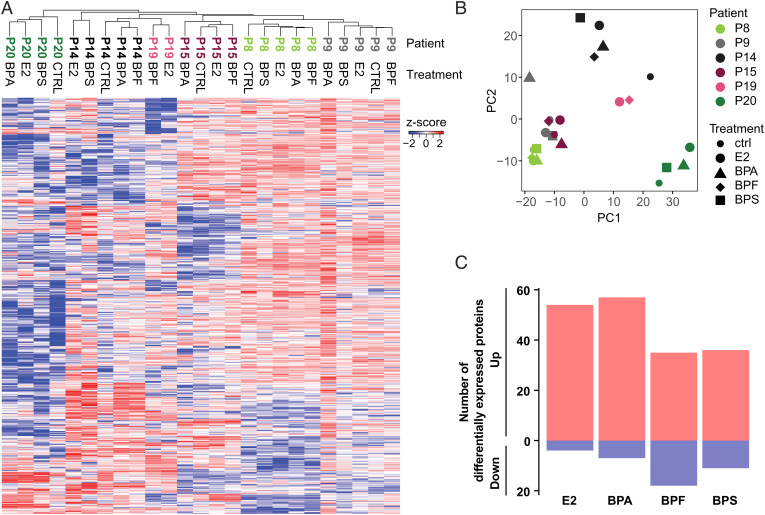
The strong branching phenotype induced by BPS exposure suggested effects beyond mimicking the endocrine functions of estrogen. To measure proteomic alterations caused by these bisphenols, we performed quantitative mass spectrometry (MS) on the treated mammary organoids and compared the relative protein abundances with control organoids. Consistent with the heterogeneity we observed in organoid morphology, unsupervised hierarchical clustering (Fig. 2A and SI Appendix, Fig. S2A) showed sample clustering based on tissue donor rather than chemical treatment. Principal component (PC) analysis of the overall protein abundances (Fig. 2B) confirmed this conclusion. Interestingly, one patient (patient 16) stood out as having a strong up-regulation, across all treatments, of proteins associated with extracellular matrix (ECM) organization and remodeling (SI Appendix, Fig. S2). This donor underwent a previous mammary reduction surgery some years prior to the procedure that enabled sampling tissue for this study (SI Appendix, Table S1). Wound healing involves profound remodeling processes that may explain the observed induction of ECM-related proteins. Considering the unique nature of this sample as a confounding variable, we excluded these data from further analysis.
Fig. 2.
Bisphenol exposures of human mammary organoids produced global proteomic alterations. (A) Unsupervised clustering of the mass spectrometry data from control (CTRL)-, E2-, and bisphenol-treated cultures displayed as a heat map. (B) Principal component (PC) analysis of the DE proteins. The colors denote data from individual patient samples, and the shapes denote the different experimental conditions. (C) Bar graphs showing the total number of significantly DE proteins in the exposure groups relative to organoid cultures that were treated with the vehicle control (P ≤ 0.05, log2 fold change ≥ 2). Blue, down-regulated; red, up-regulated.
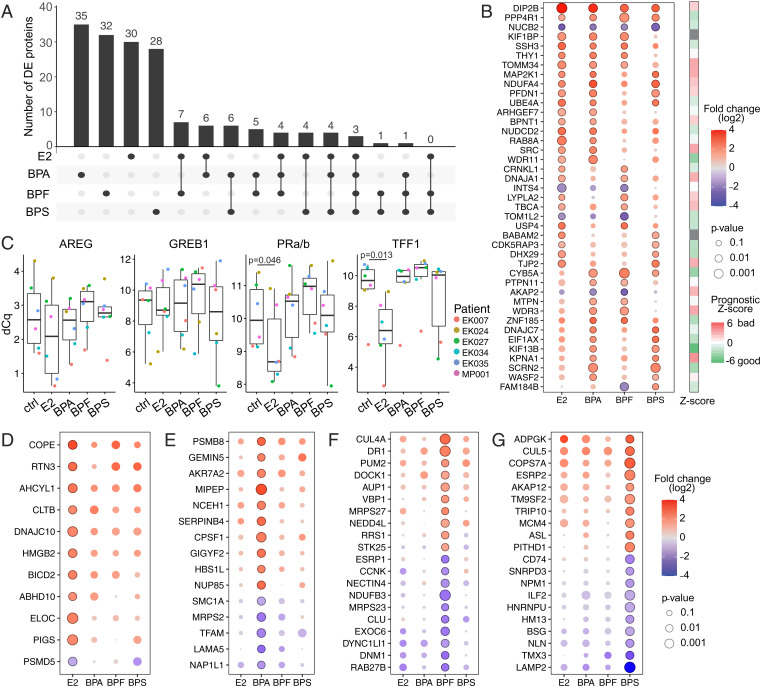
Despite interpatient heterogeneity, 166 proteins were significantly differentially expressed (DE) between at least one of the treatments as compared with the controls across all donor samples (log2 fold change ≥ 2) (Fig. 2C and Dataset S1). Notably, most DE proteins were up-regulated across all treatments. The majority of the significant changes were unique to each treatment (Fig. 3A). Only 41 DE proteins were shared between at least two treatments (Fig. 3B, red–blue scale). The bisphenols, BPA in particular, are described as having endocrine-disrupting effects. However, only about a quarter of the chemically-responsive proteins were also regulated by E2. On the other hand, half of the E2-modulated proteins (28 of 58) were shared with at least one of the three bisphenols (SI Appendix, Fig. S3A). Although not significant, most of the DE proteins showed similar trends in the other treatments (SI Appendix, Fig. S3B). We validated that our organoid culture models maintained responsiveness to E2 by showing transcriptional activation of known E2 target genes using quantitative polymerase chain reaction (qPCR) (27). Expression of progesterone receptors (PR), and transcription termination factor 1 (TTF1) was significantly induced upon E2 but not bisphenol treatments (Fig. 3C).
Fig. 3.
E2 and each bisphenol induced distinct proteome alterations. (A) UpSet plot of the 166 DE proteins identified. Black circles in the matrix indicate the exposures that are part of the intersection, and the bar graph shows the number of DE proteins that are shared among samples at this intersection. (B) The bubble plot shows DE proteins that are shared between different treatments. Color represents the log2 fold change (down-regulation in blue and up-regulation in red), and dot size represents P value. Circles indicate significant P value (P ≤ 0.05). Right shows PRECOG prognostic Z scores derived from breast cancer patients (n = 1,697 patients) (27). Positive scores (red) represent bad prognosis, and negative scores (green) represent good prognosis. (C) Gene expression analysis of E2 target genes (Amphiregulin [AREG], Growth Regulating Estrogen Receptor Binding 1 [GREB1], progesterone receptors [PRa/b], and transcription termination factor 1 [TTF1]) colored by patient (cohort A) using qPCR. Box plots show difference of quantification cycles (dCq); lower levels indicate a higher number of transcripts. Results that reached statistical significance using the two-sample Wilcoxon test are noted. (D–G) The top 10 up- and down-regulated proteins in E2 (D), BPA (E), BPF (F), or BPS (G) that are not shared with the other treatments. Color represents the log2 fold change (down-regulation in blue and up-regulation in red); dot size indicates P value. Circles indicate significant P value (P ≤ 0.05).
Next, we used PRECOG (28) to score the shared DE proteins for their prognostic value over multiple breast cancer studies (n = 1,697 patients) (Fig. 3B, red–green scale). The results showed that shared DE proteins were associated with both good and poor prognoses, without a clear trend in either direction.
DE Proteins across All Treatments.
We were interested in proteins that were DE in response to treatment with E2 and the bisphenols. They included the up-regulation of disco-interacting protein 2 homolog B (DIP2B) and serine/threonine protein phosphatase 4 regulatory subunit 1 (PPP4R1) and the down-regulation of nucleobindin-2 (NUCB2) (Fig. 3B). DIP2B, which was among the most highly DE proteins across all treatments, has been implicated in regulating DNA methylation at a global level (29, 30) and shown to be mutated in early-stage luminal breast and colorectal cancers (31, 32). Changes in DNA methylation have been associated with other environmental exposures, such as cigarette smoke (33) and the flame retardant BDE-47 (34). PPP4R complexed with the catalytic subunit of protein phosphatase 4 (i.e., the PPP4C–PPP4R1 PP4 complex) is thought to play a role in histone deacetylase 3 (HDAC3) activation (35). HDAC3 canonically acts as a transcriptional repressor of transcription factors (36–38) but can also coactivate transcription, for example, of estrogen-related receptor-α in a tissue-dependent fashion (39, 40). HDAC3 also modulates immune responses (41) upon exposure to cigarette smoke (42). Thus, these up-regulated proteins have been suggested to regulate global gene expression through epigenetic mechanisms that could have profound effects on normal cellular functions. NUCB2, another DE protein, was common to all the treatments. Its expression is E2 dependent, up-regulated in ER+ breast cancer, and associated with poor outcomes (43). However, we found that NUCB2 was down-regulated, suggesting other regulatory mechanisms.
Proteins that were DE in all the bisphenol samples but not responsive to E2 treatment included Cytochrome b5 (CYB5A), which was up-regulated. It is involved in the detoxification of aromatic and heterocyclic amine mammary carcinogens found in cigarette smoke (44). Polymorphism of CYB5A was reported to increase breast cancer risk and occurs more frequently in African American than Caucasian women (45). The nuclear transport factor importin-ɑ5 (KPNA1) is significantly up-regulated in mammary organoids treated with either BPA or BPS. KPNA1 is a member of the karyopherin superfamily. Other than their classical role in nucleocytoplasmic transport (46), karyopherins/importins have direct functions in gene regulation (47) and enhance growth factor signaling (48, 49). Altered nuclear transport is found in many cancer types, including breast (50–52), and is associated with aggressive tumor phenotypes (53). Studies determining the substrate specificity of the different importin isoforms are incomplete (54, 55), and the preferred substrates of KPNA1 are yet to be identified. It was recently shown that nuclear import pathways are involved in epigenetic gene silencing (47) and that KPNA1 can directly interact with HDAC1 (56). Thus, up-regulation of KPNA1 could have significant consequences in terms of cellular functions.
Proteome Alterations That Were Distinct to Particular Bisphenol Exposures.
Although similar trends were observed for most proteins across the various exposure groups, the majority of the significantly DE proteins were distinct to a specific treatment (Fig. 3 A and D–G and SI Appendix, Fig. S3B). We observed a strong branching phenotype in the BPS-treated organoids. Cdc42-interacting protein 4 (CIP4; gene: TRIP10), which promotes invasion (57), was significantly up-regulated only by this treatment (Fig. 3G). CIP4 is a potential downstream ER target, responds to growth factor stimuli, and is up-regulated in aggressive ERBB2 (Her2)-positive breast cancer (57, 58). As its name implies, CIP4 interacts with Cdc42, which is up-regulated in BPA-treated mouse mammary organoid cultures (59). CIP4 is involved in E-cadherin trafficking and endocytosis (60). It disrupts cell junctions, promoting migration and invasion of nonmalignant MCF10A mammary epithelial cells (57) and MDA-MB-231 breast cancer cells (61). CIP4 interacts with the protooncogene tyrosine-protein kinase SRC (57), prompting the formation of invadopodia, cellular actin-rich structures necessary for cell migration (61). SRC was up-regulated in the presence of E2 and BPA but not in BPS-treated organoids (Fig. 3B). SRC is a component of the ER pathway (62) and plays an important role in the activation of steroid receptor cross-talk and signaling (63), leading to subsequent protumorigenic cellular downstream processes, such as proliferation and invasion. SRC is overexpressed in various cancers and has been suggested as a therapeutic target (64–66).
Taken together, our data showed that exposure to bisphenols produced distinct changes in the proteome of human nonmalignant mammary organoids and increased the abundance of proteins that regulate gene expression. BPS exposure induced CIP4, a protein involved in cancer cell invasion. Thus, it is possible that bisphenol exposures set in motion processes that increase breast cancer risk.
Bisphenol Treatments Disrupted Mammary Organoid Organization.
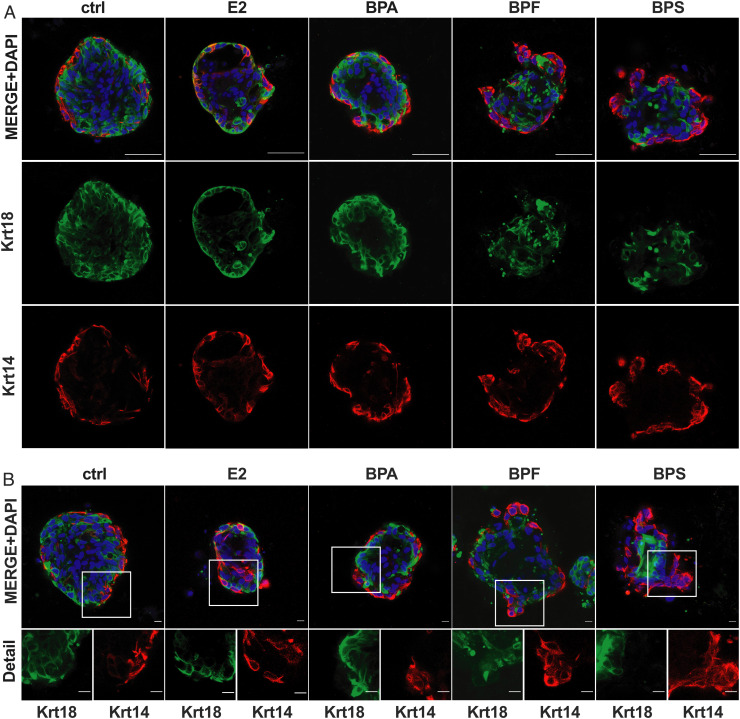
The ducts of normal breast tissue consist of two major epithelial cell lineages; cuboid cells line the lumina (luminal cells) that are surrounded by a myoepithelial layer (basal cells). Luminal cells are thought to be the origin of most breast cancers (67), whereas basal cells are resistant to oncogenic transformation and prevent malignant invasion of transformed luminal cells (68). Recently, it has become evident that an alteration of this organization may contribute to breast cancer risk (69, 70). To further evaluate how bisphenols could contribute to breast cancer risk, we analyzed how treatment with these chemicals affected the organoid’s luminal and basal cell composition and structural organization by immunolocalizing luminal (keratin-18 [Krt18]) and basal (keratin-14 [Krt14]) markers (Fig. 4). All organoids were composed of Krt18+ and Krt14+ cells. Control organoids showed the expected organization, with Krt18+ cells in the center and a layer of Krt14+ cells arranged on the surface. E2 and BPA treatment did not impact the structural organization but slightly increased branching of both Krt18+ and Krt14+ cells. However, organoids treated with BPF and in particular, BPS show a disorganized architecture. Here, we noticed that branches were mostly composed of Krt14+ cells (Fig. 4B). This observation is particularly surprising since basal cells are ER negative (71), suggesting either an E2-independent effect of the bisphenols or an indirect effect of induced paracrine signaling in hormone-responsive luminal cells. Additionally, it was shown in murine mammary organoids using time-lapse microscopy that branching morphogenesis and duct elongation are driven by luminal cells in regions that lack a myoepithelial layer, which later reforms (72). Whether basal cells are the predominant cell type that contributes to branching in the context of BPF and BPS exposures or if different mechanisms of branching that are observed in other organs, such as buckling (73), are involved, could be investigated in future studies. However, the observed BPS-induced branching phenotype may be distinct from physiologic morphogenesis.
Fig. 4.
Bisphenol disrupted human mammary organoid architecture. (A and B) IF staining of human breast organoids. (A) Staining for luminal (green: keratin 18 [Krt18]) and basal (red: keratin 14 [Krt14]) cell markers. (Scale bars, 50 μm.) (B) IF staining as described in A. Image details show the staining of organoid branches for Krt14 and Krt18. (Scale bars, 10 μm.)
Discussion
Our study showed that each bisphenol had distinct effects on the morphology of human mammary gland organoids. Most notably, BPS induced a greater degree of branching than the other chemicals in this class or estrogen. These alterations in fundamental aspects of mammary gland structure were accompanied by unique proteomic signatures. The majority were also distinct from those induced by estrogen. Nevertheless, the commonalities among the DE proteins in the hormone and chemically treated datasets agree with the widely described endocrine-disrupting activity of bisphenols. Together, these findings suggest that, beyond their endocrine effects, the broader biological actions of these and other environmental chemicals should be considered.
In this regard, advanced human tissue culture models play an important role. Toward this end, we used nonmalignant breast tissue to establish human mammary organoids for testing the effects of environmental chemical exposures. They have the advantage of being composed of heterogeneous cell types, including luminal and myoepithelial cells, within a three-dimensional matrix. Thus, they better mimic the architecture of normal mammary glands than breast cancer cell lines grown in a two-dimensional format.
However, our results show that it is important to appreciate the strong interdonor variability in the protein signatures of the organoids and medical history of the patients. For example, a sample from a woman with a previous breast reduction surgery had a unique wound-healing proteome. Also, the organoids in our study were established from reduction mammoplasty tissues donated by patients with high BMI (SI Appendix, Table S1). Obesity enhances ER expression and changes breast composition, increasing the proportion of luminal progenitors to myoepithelial cells (74). Although our model used nonmalignant cells, it may not entirely phenocopy normal, nonobese breast tissue.
Despite these facts, we detected significant bisphenol-associated morphologic alterations in two different patient cohorts including a total of 10 patients. Specifically, BPS exposure induced morphologic changes that are associated with tumorigenic processes, which were less clear at the proteomic level. However, we identified candidate regulators that could be involved. For example, this bisphenol up-regulated expression of CIP4, which plays pleiotropic roles in promoting adhesive and invasive processes that promote tumor progression (57, 61). The fact that we detected protein-level changes in the abundance of this protein strengthens the theory that it is a BPS target with potential functional importance.
The consensus from analyzing human blood samples is that in vivo exposures to unconjugated BPA are in the range of 0.5 to 10 ng/mL, with most studies suggesting an average of ∼1 to 3 ng/mL (75). The maximal BPA concentration of 48.5 ng/mL was measured in urine in children. Some studies investigating EDCs use up to micromolar concentrations and highly E2-responsive cell lines (76, 77). Many factors need to be considered when extrapolating levels of environmental chemicals measured in vivo to concentrations used for in vitro exposures, including the length of time (e.g., chronic vs. acute) and metabolism (e.g., conjugated vs. unconjugated). Thus, we reasoned that the working concentration of bisphenols we employed in the mammary model (15 nM is equivalent to ∼30 to 40 ng/mL depending on the bisphenol) was relevant to human exposures. Significantly, acute treatment for only 6 d induced protumorigenic morphological changes along with protein-level alterations. Thus, chronic bisphenol exposures in vivo could have effects such as those we observed in the organoid model, which might be exacerbated by the numerous other environmental chemicals that circulate in blood. Finally, the results of this study provide evidence that BPA replacement chemicals are not inert, producing morphological changes that could be indicative of increased breast cancer risk. Therefore, BPF and BPS are likely not safer than BPA, and consumer products that contain them should be treated with caution.
Materials and Methods
Chemicals.
E2, BPA, BPF, and BPS were purchased from Sigma-Aldrich and dissolved in dimethyl sulfoxide (DMSO) from ATCC.
Patient Samples.
Benign human mammary gland tissues were obtained from elective mammoplasty reduction surgeries at the University of California, San Francisco (UCSF; cohort A) or Vanderbilt University School of Medicine, which is part of the Cooperative Human Tissue Network (cohort B), in accordance with the institutional review boards’ approval. Tissues were received as deidentified samples, and all subjects provided written informed consent. Medical reports were obtained without personally identifiable information. The samples were kept on ice during handling and processed immediately.
Human Organoid Culture.
Primary human nonmalignant breast organoids were prepared as previously described (78). Briefly, fresh breast tissue was washed with Dulbecco‘s phosphate-buffered saline (DPBS, Gibco) containing 0.1% gentamycin (Gibco) and 1% penicillin/streptomycin (Gibco). Epithelial elements were separated from fat tissue and cut into small pieces. The details of organoid tissue preparation, which differed for each cohort, were driven by how samples were handled at the individual sites. For cohort A, tissue was digested overnight in a spinner flask containing Dulbecco‘s Modified Eagle Medium Nutrient Mixture F-12 (DMEM/F12), 200 U/mL collagenase type III (Worthinton CLS-3), 100 U/mL hyaluronidase (Sigma-Aldrich H3506), 10% charcoal:dextran-stripped fetal bovine serum (FBS, Gemini Bio-Products 100 to 119), and 1× antibiotic–antimycotic (Gibco 15240062). The digests were centrifuged at 16,000 rpm for 10 min; then, the pellets were resuspended in DMEM/F12 and size selected by passing through 100-μm and then, 40-μm cell strainers (ThermoFisher). Tissue from each fraction was collected in cryopreservation media (Roswell Park Memorial Institute [RPMI] 1640, 30% FBS, 10% DMSO). Cohort B tissue was digested in DMEM/F12 (Gibco) containing 10% FBS (Omega), 1% penicillin/streptomycin, 10 μg/mL insulin (Sigma), 2.8 mg/mL collagenase 1 (Gibco), and 0.4 mg/mL hyaluronidase (Sigma) for 12 h at 37 °C. The digests were centrifuged at 600 rpm for 5 min; then, the cell pellets were resuspended in organoid medium [M87A medium without E2 and FBS (79)] and size selected (>40 and <100 μm) by using cell strainers (ThermoFisher). Organoids of cohort A were thawed on the day of the experiment, whereas cohort B was processed immediately without freezing. Organoids were plated in growth factor–reduced, phenol red–free Matrigel (Corning) at a density of 1 organoid per 1 μL. After polymerization, 200 μL organoid medium was added containing the chemicals at a final concentration of 15 nM or 0.005% DMSO (ATCC) as the vehicle control.
Organoid Morphology.
On day 6 of culture, representative organoids from each culture condition were imaged at the cross-section with the largest area with bright-field microscopy using a spinning disk confocal microscope (Zeiss Cell Observer Z1 with a Yokagawa spinning disk, Zeiss LD 20×/NA 0.4 air objective [cohort A]) or a Keyence BZ-X710 All-in-One Fluorescence microscope (cohort B). A branch was defined as a visible protrusion of the organoid body that was in focus that included budding and wrapping structures (80), and the total number of branches was counted at the maximal cross-sectional area. Organoid size was determined by measuring the maximal cross-sectional organoid area using ImageJ/Fiji. The significance of the branching morphology data was determined by using a two-sample Wilcoxon test.
Immunofluorescence Localization.
Organoids from four individuals (UCSF011, UCSF013, UCSF016, EK007) were used in the immunolocalization experiments. After 6 d of culture as described above, they were incubated for 30 min with 25% sucrose (VWR Life Science) prior to fixation for 45 min in 2% PFA (paraformaldehyde, Electron Microscopy Science) at room temperature (RT). Organoids were washed three times for 20 min with phosphate-buffered saline (PBS) containing 7.5% glycine (USB Chemicals) followed by two 10-min washes in PBS at RT and one overnight wash at 4 °C. Permeabilization was accomplished by transfer to 0.5% Triton X-100 (Sigma) for 15 min at RT. Nonspecific immunoreactivity was blocked by incubation with 10% FBS (Omega) or 10% goat serum (ThermoFisher) in immunofluorescence (IF) solution (0.1% bovine serum albumin [BSA], 0.02% Triton X-100, 0.04% Tween-20 in PBS) for 2 h at RT and overnight at 4 °C. Then, the organoids were incubated at 4 °C for 24 h with primary antibodies diluted in IF solution/10% FBS (Krt18 [Dako M7010] 1:100, Krt14 [Abcam ab119695] 1:200). After washing three times for 1 h at RT in IF solution, the organoids were incubated with the appropriate species-specific secondary antibodies diluted in IF solution/10% FBS light protected overnight at 4 °C. Finally, they were washed three times for 1 h at RT in IF solution. For nuclear staining, organoids were incubated with PBS containing diamidino-2-phenylindole (DAPI, Sigma) for 20 min at RT. Representative images were captured with a Keyence BZ-X710 All-in-One Fluorescence microscope or a Leica SP5 laser scanning or laser scanning confocal microscope (Zeiss LSM800) using a Zeiss LD 40×/NA 1.1 water objective.
Mass Spectrometry (MS) Sample Preparation.
Seven biological replicates from each culture condition were subjected to proteomic analysis. Mammary organoid cell pellets were resuspended in 100 μL lysis buffer, which contained 1% sodium dodecyl sulfate (SDS), 75 mM NaCl, 50 mM trishydroxymethylaminomethane (Tris, pH 8.0), and cOmplete mini protease inhibitor mixture (one tablet per 10 mL buffer; Roche). The cells were lysed by sonication (2 s per pulse × 2 pulses, with 15 min on ice between each pulse). Then, the lysates were centrifuged. The volume of the supernatant was reduced using a SpeedVac (0 °C). The protein concentration of each replicate was determined using the bicinchoninic acid (BCA) assay. After volume reduction, an aliquot of each sample (18.75 μL) was loaded onto NuPAGETM 10% Bis-Tris gels (Invitrogen; 1.0 mm, 10 well) and run for 0.5 cm to remove salts in the lysis buffer. The protein bands were cut out of the gels, reduced by incubation with 20 mM Tris 2-carboxyethyl phosphine, alkylated with 40 mM iodoacetamide, and trypsin digested overnight at 37 °C. The resulting peptides were extracted with water/acetonitrile (ACN)/formic acid (FA) (50:50:0.1; vol/vol), dried under vacuum, and resuspended in water/ACN/FA (98:2:0.1; vol/vol) to a final concentration of 0.33 μg/μL.
LC/MS Analysis.
The first step was building the Sequential Window Acquisition of All Theoretical Fragment Ion Mass Spectra (SWATH) library. Aliquots of each digested sample containing 1 μg protein were pooled and separated by high pH reversed-phase high-performance liquid chromatography into 18 fractions using an Agilent 1260 Infinity system fitted with a Zorbax Extent C18 column (4.6 mm inner diameter [i.d.] × 100 mm length). Peptides were eluted at a flow rate of 1 mL/min with a gradient of 0 to 32% solvent B (H2O/ACN/NH4OH; 20:80:0.1; vol/vol) for 50 min. Each fraction was separated using a nanoLC Ultra 2D Plus system (SCIEX) interfaced to a 5600 Triple time of flight (TOF) mass spectrometer (SCIEX). Initially, the peptides were loaded onto a guard column (300 μm i.d. × 5 mm, 5-μm particle size, 300-Å pore size; Acclaim PepMap300 C18; ThermoFisher) and washed with the aqueous loading solvent (0.1% FA) at a flow rate of 2 μL/min for 10 min. Then, the peptides were separated on a heated (35 °C) C18 Acclaim PepMap100 column (75 μm i.d. × 150 mm, 3-mm particle size, 100-Å pore size; Thermo Fisher Scientific). They were eluted with a gradient of 2 to 30% solvent B (solvent A: 98% H2O, 2% ACN, 0.1% FA; solvent B: 100% ACN, 0.1% FA) for 90 min at a flow rate of 300 nL/min. Data-dependent acquisition (DDA) was performed. MS scans from mass-to-charge ratio (m/z) 400 to 1,250 were acquired in positive ion mode followed by MS/MS scans from m/z 100 to 1,500 of the 30 most abundant ions with an exclusion time of 15 s. Proteins were identified using MaxQuant software and the UniProt human database (version 20190501) with isoform sequences. The SWATH spectral library was built based on MaxQuant search results using Skyline (version 19.1) (81).
Second, MS data were acquired from the individual samples. In each case, the peptides were analyzed with the same LC/MS system using the same LC gradient and data-independent (DDI) SWATH acquisition methods. Each SWATH cycle (3.7 s) contained one MS survey from m/z 400 to 1,250 and 72 MS/MS scan windows from m/z 100 to 1,500. The raw data were searched against the spectral library with Skyline. Peptides were identified and quantified with 0.1-Da m/z tolerance using MS/MS transition areas. Proteins were quantitated by summing all detected peptide intensities.
Bioinformatics and Statistics.
A total of 3,256 protein groups were quantified based on Skyline searching. The protein abundance for each sample replicate was normalized by dividing by the total protein level. This approach is based on the assumption that each sample contained the same protein amount. Seven of 29 samples were subjected to repeat analysis to confirm reproducibility. The average Pearson correlation coefficient of the duplicate protein levels was 0.98. In the final analysis, the results from duplicate datasets were averaged. Low-level proteins were removed if they had less than three significant values in each sample group. The final result was 2,598 proteins. The protein level was then log2 transferred, and the missing values were replaced based on the normal distribution. Multiple-sample ANOVA tests were performed with and without patient 16, which results in 1,323 or 657 significant proteins, respectively. Hierarchical clustering and principal component analyses were based on these datasets. Ultimately, patient 16 was removed due to a confounding up-regulation of ECM-related proteins as a result of a previous mammoplasty reduction surgery (SI Appendix, Fig. S2 and Table S1). For per patient comparisons among treatments, a two-sample Student’s t test was performed between each chemical exposure and control culture (DMSO). Determinations of individual up- or down-regulated proteins were based on the results of two-sample Student’s t tests. Data visualization was accomplished using R/Rstudio.
qPCR quantification.
Stimulated cultures were established in triplicate for each patient. After culturing the organoids as described above, replicates from each treatment were combined and extracted from Matrigel by mechanical disruption and dissolution in Cell Recovery Solution (Corning 354253) on ice for 20 min. Then, the tissues were washed three times with ice-cold PBS and pelleted at 400 × g for 5 min. TRIreagent (Zymo Research R2050-1) was added to each sample, and homogenization was achieved by pushing the samples through 30-gauge needles 5 to 10 times. RNA was isolated using a Direct-zol RNA Microprep kit (Zymo Research R2062) and was quantified on a Nanodrop (Thermo Scientific). cDNA conversion was performed with the SuperScript IV First-Strand Synthesis System (Invitrogen 18091050) with poly-dT primer, and the same amount of RNA was used for all treatments of each patient sample. qPCR reactions were performed in triplicates using the PowerUp SYBR Green Master Mix (Applied Biosystems A25742). Primer efficiency for each qPCR target was validated with a five log–range serial dilution of pooled samples from several treatment conditions. The Cq value for each sample was normalized to KRT18 transcript levels to correct for variations in the luminal fraction among conditions and patients (82). Data analysis and statistical evaluation using the two-sample Wilcoxon test were done with R/Rstudio. For primer sequences see SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank our dear mentor and friend, Dr. Zena Werb, who made fundamental contributions to our understanding of mammary gland development and breast cancer biology over the course of her distinguished scientific career. We thank Dr. James Garbe for generously providing M87A medium, Dr. Yi Lui for providing the organoid staining protocol, Dr. Lyndsay Murrow for helpful discussions, and Dr. Michael Mcmaster for editing. This work was supported by European Molecular Biology Organization Long-Term Postdoctoral Fellowship (EMBO ALTF 159-2017 to J.W.), Deutscher Akademischer Austauschdienst Grant 57380613 (to J.H.H.), and California Breast Cancer Research Program Grant 21UB-8011 (to K.W., S.F., and Z.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
The DDA and DDI MS raw data along with the database search files and proteomics data are available in the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository (accession no. MSV000087617).
References
- 1.Vandenberg L. N., et al. , Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Cien. Saude Colet. 17, 407–434 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Caballero-Casero N., Lunar L., Rubio S., Analytical methods for the determination of mixtures of bisphenols and derivatives in human and environmental exposure sources and biological fluids. A review. Anal. Chim. Acta 908, 22–53 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Calafat A. M., Ye X., Wong L. Y., Reidy J. A., Needham L. L., Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ. Health Perspect. 116, 39–44 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC, “Fourth national report on human exposure to environmental chemicals: Updated tables, Volume 2, NHANES 2011–2016” (Rep., Centers for Disease Control and Prevention, Atlanta, GA, 2019).
- 5.Acconcia F., Pallottini V., Marino M., Molecular mechanisms of action of BPA. Dose Response 13, 1559325815610582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gore A. C., et al. , EDC-2: The endocrine society’s second scientific statement on endocrine-disrupting chemicals. Endocr. Rev. 36, E1–E150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton S. E., Endocrine-disrupting compounds and mammary gland development: Early exposure and later life consequences. Endocrinology 147 (6 suppl.), S18–S24 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Rudel R. A., Fenton S. E., Ackerman J. M., Euling S. Y., Makris S. L., Environmental exposures and mammary gland development: State of the science, public health implications, and research recommendations. Environ. Health Perspect. 119, 1053–1061 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., et al. , Pubertal bisphenol A exposure alters murine mammary stem cell function leading to early neoplasia in regenerated glands. Cancer Prev. Res. (Phila.) 7, 445–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandenberg L. N., et al. , Exposure to environmentally relevant doses of the xenoestrogen bisphenol-A alters development of the fetal mouse mammary gland. Endocrinology 148, 116–127 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atlas E., Dimitrova V., Bisphenol S and bisphenol A disrupt morphogenesis of MCF-12A human mammary epithelial cells. Sci. Rep. 9, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez A. L., et al. , Perinatal exposure to bisphenol A or diethylstilbestrol increases the susceptibility to develop mammary gland lesions after estrogen replacement therapy in middle-aged rats. Horm. Cancer 8, 78–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibrahim M. A. A., Elbakry R. H., Bayomy N. A., Effect of bisphenol A on morphology, apoptosis and proliferation in the resting mammary gland of the adult albino rat. Int. J. Exp. Pathol. 97, 27–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., et al. , The estrogenic proliferative effects of two alkylphenols and a preliminary mechanism exploration in MCF-7 breast cancer cells. Environ. Toxicol. 35, 628–638 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Yang F., et al. , Genome-wide identification of the interactions between key genes and pathways provide new insights into the toxicity of bisphenol F and S during early development in zebrafish. Chemosphere 213, 559–567 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Le Fol V., et al. , In vitro and in vivo estrogenic activity of BPA, BPF and BPS in zebrafish-specific assays. Ecotoxicol. Environ. Saf. 142, 150–156 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Moreman J., et al. , Acute toxicity, teratogenic, and estrogenic effects of bisphenol A and its alternative replacements bisphenol S, bisphenol F, and bisphenol AF in zebrafish embryo-larvae. Environ. Sci. Technol. 51, 12796–12805 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Stroheker T., Chagnon M. C., Pinnert M. F., Berges R., Canivenc-Lavier M. C., Estrogenic effects of food wrap packaging xenoestrogens and flavonoids in female Wistar rats: A comparative study. Reprod. Toxicol. 17, 421–432 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Awada Z., et al. , DNA methylome-wide alterations associated with estrogen receptor-dependent effects of bisphenols in breast cancer. Clin. Epigenetics 11, 138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z., Lyu C., Ren Y., Wang H., Role of TET dioxygenases and DNA hydroxymethylation in bisphenols-stimulated proliferation of breast cancer cells. Environ. Health Perspect. 128, 27008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaPlante C. D., Catanese M. C., Bansal R., Vandenberg L. N., Bisphenol S alters the lactating mammary gland and nursing behaviors in mice exposed during pregnancy and lactation. Endocrinology 158, 3448–3461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashihara N., et al. , Subacute oral toxicity study of bisphenol F based on the draft protocol for the “enhanced OECD test guideline no. 407.” Arch. Toxicol. 81, 825–832 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Hiatt R. A., Brody J. G., Environmental determinants of breast cancer. Annu. Rev. Public Health 39, 113–133 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Srivastava V., Huycke T. R., Phong K. T., Gartner Z. J., Organoid models for mammary gland dynamics and breast cancer. Curr. Opin. Cell Biol. 66, 51–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samocha A., Doh H., Kessenbrock K., Roose J. P., Unraveling heterogeneity in epithelial cell fates of the mammary gland and breast cancer. Cancers (Basel) 11, 1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hah N., Kraus W. L., Hormone-regulated transcriptomes: Lessons learned from estrogen signaling pathways in breast cancer cells. Mol. Cell. Endocrinol. 382, 652–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentles A. J., et al. , The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winnepenninckx B., et al. , CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am. J. Hum. Genet. 80, 221–231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell J. T., et al. , DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 12, R10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Encinas G., et al. , Somatic mutations in early onset luminal breast cancer. Oncotarget 9, 22460–22479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Closa A., et al. , Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis 35, 2039–2046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K. W. K., Pausova Z., Cigarette smoking and DNA methylation. Front. Genet. 4, 132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J. F., et al. , Genomic profiling of BDE-47 effects on human placental cytotrophoblasts. Toxicol. Sci. 167, 211–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., et al. , Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 19, 827–839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Kalkum M., Chait B. T., Roeder R. G., The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Ishizuka T., Lazar M. A., The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23, 5122–5131 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozawa Y., et al. , Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 98, 2116–2123 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Emmett M. J., et al. , Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuang Z., et al. , The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 365, 1428–1434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen H. C. B., Adlanmerini M., Hauck A. K., Lazar M. A., Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature 584, 286–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkler A. R., Nocka K. N., Williams C. M. M., Smoke exposure of human macrophages reduces HDAC3 activity, resulting in enhanced inflammatory cytokine production. Pulm. Pharmacol. Ther. 25, 286–292 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Suzuki S., et al. , Nucleobindin 2 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 103, 136–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurian J. R., Chin N. A., Longlais B. J., Hayes K. L., Trepanier L. A., Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem. Res. Toxicol. 19, 1366–1373 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanke K. L., et al. , Polymorphisms in the carcinogen detoxification genes CYB5A and CYB5R3 and breast cancer risk in African American women. Cancer Causes Control 25, 1513–1521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terry L. J., Shows E. B., Wente S. R., Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Dong Q., et al. , Roles of the CSE1L-mediated nuclear import pathway in epigenetic silencing. Proc. Natl. Acad. Sci. U.S.A. 115, E4013–E4022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyamoto Y., Yamada K., Yoneda Y., Importin α: A key molecule in nuclear transport and non-transport functions. J. Biochem. 160, 69–75 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Yamada K., et al. , Cell surface localization of importin α1/KPNA2 affects cancer cell proliferation by regulating FGF1 signalling. Sci. Rep. 6, 21410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler J., et al. , Prosurvival function of the cellular apoptosis susceptibility/importin-α1 transport cycle is repressed by p53 in liver cancer. Hepatology 60, 884–895 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Kau T. R., Way J. C., Silver P. A., Nuclear transport and cancer: From mechanism to intervention. Nat. Rev. Cancer 4, 106–117 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Dahl E., et al. , Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin α2 as a potential novel prognostic marker in breast cancer. Clin. Cancer Res. 12, 3950–3960 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Winkler J., et al. , Cellular apoptosis susceptibility (CAS) is linked to integrin β1 and required for tumor cell migration and invasion in hepatocellular carcinoma (HCC). Oncotarget 7, 22883–22892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Köhler M., et al. , Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell. Biol. 19, 7782–7791 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura M., Thakar K., Karaca S., Imamoto N., Kehlenbach R. H., Novel Approaches for the Identification of Nuclear Transport Receptor Substrates (Elsevier Inc., ed. 1, 2014). [DOI] [PubMed] [Google Scholar]
- 56.Cho N. H., et al. , OpenCell: Proteome-scale endogenous tagging enables the cartography of human cellular organization. bioRxiv [Preprint] (2021). 10.1101/2021.03.29.437450 (Accessed 29 July 2021). [DOI]
- 57.Rolland Y., et al. , The CDC42-interacting protein 4 controls epithelial cell cohesion and tumor dissemination. Dev. Cell 30, 553–568 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Leu Y. W., et al. , Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 64, 8184–8192 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Williams K. E., et al. , Quantitative proteomic analyses of mammary organoids reveals distinct signatures after exposure to environmental chemicals. Proc. Natl. Acad. Sci. U.S.A. 113, E1343–E1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erasmus J. C., et al. , Corrigendum: Defining functional interactions during biogenesis of epithelial junctions. Nat. Commun. 8, 14195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pichot C. S., et al. , Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res. 70, 8347–8356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Migliaccio A., et al. , Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 17, 2008–2018 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Migliaccio A., et al. , Steroid receptor regulation of epidermal growth factor signaling through Src in breast and prostate cancer cells: Steroid antagonist action. Cancer Res. 65, 10585–10593 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Saad F., Lipton A., SRC kinase inhibition: Targeting bone metastases and tumor growth in prostate and breast cancer. Cancer Treat. Rev. 36, 177–184 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Hiscox S., et al. , Dual targeting of Src and ER prevents acquired antihormone resistance in breast cancer cells. Breast Cancer Res. Treat. 115, 57–67 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Mayer E. L., Krop I. E., Advances in targeting SRC in the treatment of breast cancer and other solid malignancies. Clin. Cancer Res. 16, 3526–3532 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Molyneux G., et al. , BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Kim J., et al. , Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc. Natl. Acad. Sci. U.S.A. 109, 6124–6129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murrow L. M., et al. , Changes in epithelial proportions and transcriptional state underlie major premenopausal breast cancer risks. bioRxiv [Preprint] (2020). 10.1101/430611 (Accessed 29 July 2021). [DOI]
- 69.Schnitt S. J., Benign breast disease and breast cancer risk: Morphology and beyond. Am. J. Surg. Pathol. 27, 836–841 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Santagata S., et al. , Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J. Clin. Invest. 124, 859–870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ewald A. J., Brenot A., Duong M., Chan B. S., Werb Z., Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell 14, 570–581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson C. M., On buckling morphogenesis. J. Biomech. Eng. 138, 021005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chamberlin T., D’Amato J. V., Arendt L. M., Obesity reversibly depletes the basal cell population and enhances mammary epithelial cell estrogen receptor alpha expression and progenitor activity. Breast Cancer Res. 19, 128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vandenberg L. N., Hauser R., Marcus M., Olea N., Welshons W. V., Human exposure to bisphenol A (BPA). Reprod. Toxicol. 24, 139–177 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Sengupta S., Obiorah I., Maximov P. Y., Curpan R., Jordan V. C., Molecular mechanism of action of bisphenol and bisphenol A mediated by oestrogen receptor alpha in growth and apoptosis of breast cancer cells. Br. J. Pharmacol. 169, 167–178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pupo M., et al. , Bisphenol A induces gene expression changes and proliferative effects through GPER in breast cancer cells and cancer-associated fibroblasts. Environ. Health Perspect. 120, 1177–1182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Labarge M. A., Garbe J. C., Stampfer M. R., Processing of human reduction mammoplasty and mastectomy tissues for cell culture. J. Vis. Exp. 71, 50011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garbe J. C., et al. , Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 69, 7557–7568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andrew D. J., Ewald A. J., Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol. 341, 34–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacLean B., et al. , Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dunphy K. A., et al. , Inter-individual variation in response to estrogen in human breast explants. J. Mammary Gland Biol. Neoplasia. 25, 51–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanos T., et al. , Progesterone/RANKL is a major regulatory axis in the human breast. Sci. Transl. Med. 5, 182ra55 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DDA and DDI MS raw data along with the database search files and proteomics data are available in the Mass Spectrometry Interactive Virtual Environment (MassIVE) repository (accession no. MSV000087617).