Significance
Neurodegenerative diseases are poorly understood and difficult to treat. One common hallmark is lysosomal dysfunction leading to the accumulation of aggregates and other undegradable materials, which cause damage to brain resident cells. Lysosomes are acidic organelles responsible for breaking down biomolecules and recycling their constitutive parts. In this work, we find that the antiinflammatory and neuroprotective compound, discovered via a phenotypic screen, imparts its beneficial effects by targeting the lysosome and restoring its function. This is established using a genome-wide CRISPRi target identification screen and then confirmed using a variety of lysosome-targeted studies. The resulting small molecule from this study represents a potential treatment for neurodegenerative diseases as well as a research tool for the study of lysosomes in disease.
Keywords: neurodegenerative disease, lysosomes, inflammation, drug discovery, drug development
Abstract
Neurodegenerative diseases affect a rapidly growing number of the aging population globally. These conditions have proven extremely difficult to treat due to our limited understanding of their mechanisms, but they are characterized by protein aggregation, inflammation, lysosomal dysfunction, and neuronal death. Phenotypic drug screens promise to deliver “target agnostic” therapies without being hypothesis limited as with target-based screens. Here, we describe our work to develop and characterize small molecule C381. The compound is a benzyl urea derivative containing a piperidine ring. It is brain penetrant with a ClogP of 3.3 and an oral bioavailability of 48%. We tested the compound in Progranulin−/− mice (a model of lysosomal storage disease and frontotemporal dementia) and the chronic 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model of Parkinson’s disease (PD) where it showed prominent antiinflammatory and neuroprotective effects. In the PD model, C381 restored cognitive function and rescued dopaminergic neuron loss. To identify the target, we performed a genome-wide CRISPR interference (CRISPRi) drug target identification screen, which implicated the lysosome. After validating the screen results with individual knockdown cell lines, follow-up functional studies revealed that C381 physically targets the lysosome, promotes lysosomal acidification, increases breakdown of lysosomal cargo, and improves lysosome resilience to damage. As a first-in-class compound capable of restoring lysosomal function, C381 has the potential both as a therapeutic and as a research compound to better understand lysosomal contributions to disease progression. Together, our work has produced a promising drug candidate for the treatment of neurodegenerative diseases marked by lysosomal dysfunction.
Neurodegenerative diseases represent a severe and growing problem due to our aging population. Millions of people suffer from neurodegenerative diseases in the United States alone. To address this, it is imperative that we work to discover new therapeutic targets to treat these conditions. Despite researchers’ best efforts, this task has proven extremely challenging and aside from recent advancements in the treatment of multiple sclerosis and amyotrophic lateral sclerosis, there are still no Food and Drug Administration approved disease-modifying treatments for any other major neurodegenerative disease (1). Target-based drug discovery has resulted in numerous recent clinical failures, including beta secretase (BACE) 1 inhibitors (2) and monoclonal antibodies (3) for Alzheimer’s disease (AD). The shortcomings of this strategy have led some to suggest a shift toward phenotypic screening for neurodegenerative disease drug discovery (4, 5). Phenotypic screening has the advantage of being “target agnostic,” allowing for discovery of drugs with novel targets and mechanisms of action. This is especially beneficial in the context of neurodegenerative diseases, which are poorly understood and in dire need of new targets. This is the strategy that we employed in this study.
Although the underlying causes for the various neurodegenerative diseases are quite heterogeneous, several common themes emerge from their respective pathologies. These hallmarks include neuroinflammation, toxic protein aggregate accumulation, and neuronal dysfunction or death (6–8). These phenomena are connected potentially through dysfunction of lysosomes—acidic organelles responsible for biomolecule breakdown, nutrient recycling, and other catabolic processes (9–11). We developed a phenotypic screen for identifying small molecule therapeutics for neurodegenerative diseases by targeting two of these hallmarks: Inflammation and neurodegeneration, and the resulting molecule was unexpectedly discovered to act on the lysosome.
This study began with a phenotypic small molecule screen using a Smad-based reporter system based on previous studies showing Smad activation reduced inflammation and neuronal death in neurodegenerative disease models (Fig. 1A) (12–14). The resultant compound after structural optimization of the top hits was C381, which was demonstrated to be both orally bioavailable and brain penetrant. We then tested C381 in various preclinical mouse models of neurodegenerative disease, where it exhibited potent antiinflammatory and neuroprotective activity. In order to probe the mechanism, we performed a genome-wide CRISPR interference (CRISPRi) drug target identification screen, which surprisingly implicated the lysosome. Follow-up functional studies confirmed that C381 targets the lysosome, where it increases acidification and protein breakdown and improves lysosomal resilience to injury.
Fig. 1.
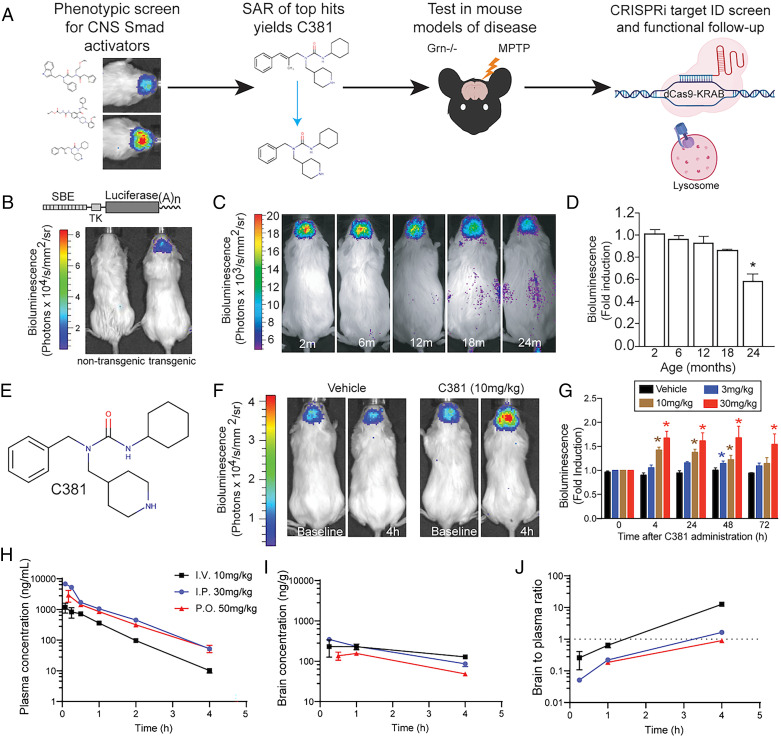
Phenotypic screen for CNS Smad activators yields brain-penetrant compound C381. (A) Graphical summary of work. SAR, structure activity relationship. (B) Reporter construct schematic and example image of transgenic mouse. SBE, Smad binding element; TK, thymidine kinase minimal promoter. (C) Bioluminescence imaging of SBE-luc mice at different ages, showing CNS Smad signaling decreases with age. (D) Quantification of C (one-way ANOVA *P < 0.05). (E) Structure of C381. (F) Bioluminescence imaging of SBE-luc mice treated i.p. with vehicle or 10 mg/kg C381 at baseline and at 4 h postdose. (G) Quantification of bioluminescence changes in C381-treated SBE-Luc mice over 72 h at different doses (two-way ANOVA with repeated measures *P < 0.05). (H) Plasma pharmacokinetics time profile of single-dose administration of C381 to female FVB mice. (I) Brain PK time profile of single-dose administration of C381 to female FVB mice. (J) Brain-to-plasma ratio time profile of single-dose administration of C381 to female FVB mice.
Results
Initial Screening Campaign for Small Molecule Brain Smad Activators.
Utilizing the previously described Smad signaling reporter (SBE-luc) mice (15, 16) (Fig. 1B), we observed that brain Smad signaling decreased with age (Fig. 1 C and D). This finding was consistent with observations in both mice and humans (17). This result, combined with earlier work showing the benefits of increasing Smad signaling in the brain (18–20) and the detriments of inhibition (20–22), served as motivation for a small molecule screen to discover activators of our central nervous system (CNS) Smad reporter. We hypothesized that restoring signaling in this pathway would be therapeutic and antiinflammatory in neurodegenerative disease conditions. A diverse 5,000 compound library was screened using a Smad-binding element (SBE) driven secreted alkaline phosphatase (SEAP) reporter cell line (23) (SI Appendix, Fig. S1A). The cell line was derived from fibroblasts isolated from Tgfb1−/− mice to ensure a TGF-β1–independent mechanism. The three most active compounds from this screen as determined by SEAP release, 5E, 11H, and 3A (SI Appendix, Fig. S1B), were selected for additional testing. Based on activity (SI Appendix, Fig. S1C) and drug-like characteristics, 11H was chosen as the scaffold for further development. A total of 129 derivatives of 11H were synthesized for structure activity relationship (SAR) experiments. The synthesis effort initially focused on modifications of the cyclohexyl ring, but we quickly discovered that a cyclopentyl ring or a methyl group on the cyclohexyl ring substantially reduced or eliminated activity. An isostere modification by changing the carbamide to a thiocarbamide also caused the loss of most of the activity. The various 11H derivatives were tested in vivo, and C381 (Fig. 1E) was chosen as our lead compound based on strong in vivo brain activity, CNS drug-like physicochemical properties, and low toxicity (SI Appendix, Table S1). In 2- to 3-mo-old SBE-luc reporter mice, C381 dose dependently induced Smad reporter activity in the CNS (Fig. 1 F and G). Although this assay did not reveal the direct target of C381, it did show that C381 possessed potentially beneficial CNS bioactivity.
We next characterized C381 by assessing the single-dose pharmacokinetics of the compound in plasma and brain. Female FVB/NJ mice were given C381 by intravenous (i.v., 10 mg/kg), intraperitoneal (i.p., 30 mg/kg), and oral gavage (per os [p.o.], 50 mg/kg) dose routes. The compound was formulated in 10% dimethyl sulfoxide (DMSO):15% Solutol:75% sterile water. Plasma and brain were collected at various time points up to 4 h and analyzed by liquid chromatography–mass spectrometry (LC/MS) for determination of C381 levels (Fig. 1 H and I). Plasma levels after 10 mg/kg i.v. administration peaked (Cmax) at 1,171 ± 415 ng/mL and showed log-linear decay up to 4 h with a short elimination half-life (t1/2) of 0.57 h. The substantial volume (V) of distribution (7.669 L/kg) and fast rate of clearance (Cl, 9.262 L/h/kg) suggested C381 distributed well to tissues and was metabolized rapidly; however, it was not necessarily removed quickly from the body. The extravascular routes of administration (i.p. and p.o.) showed higher Cmax values than i.v. administration due to higher dose levels and robust absorption. C381 was rapidly absorbed, especially for the p.o. group, and oral bioavailability (F) was 48%. The extravascular estimates of V (∼8 to 13 L/kg) and Cl (∼9 L/h/kg) based on F were similar to those after i.v. administration. Brain levels were initially lower than plasma, but did not decline as rapidly up to 4 h, suggesting C381 in the brain is cleared more slowly than from plasma. As a result, the brain-to-plasma concentration ratios increased to >0.5 at time points after ∼0.5 h (i.v.) and ∼1.5 h (p.o. and i.p.) (Fig. 1J). Together, these data suggest C381 is an orally bioavailable and blood–brain barrier (BBB)-permeable activator of CNS Smad reporter activity.
Testing C381 in Preclinical Mouse Models of Neurodegenerative Disease.
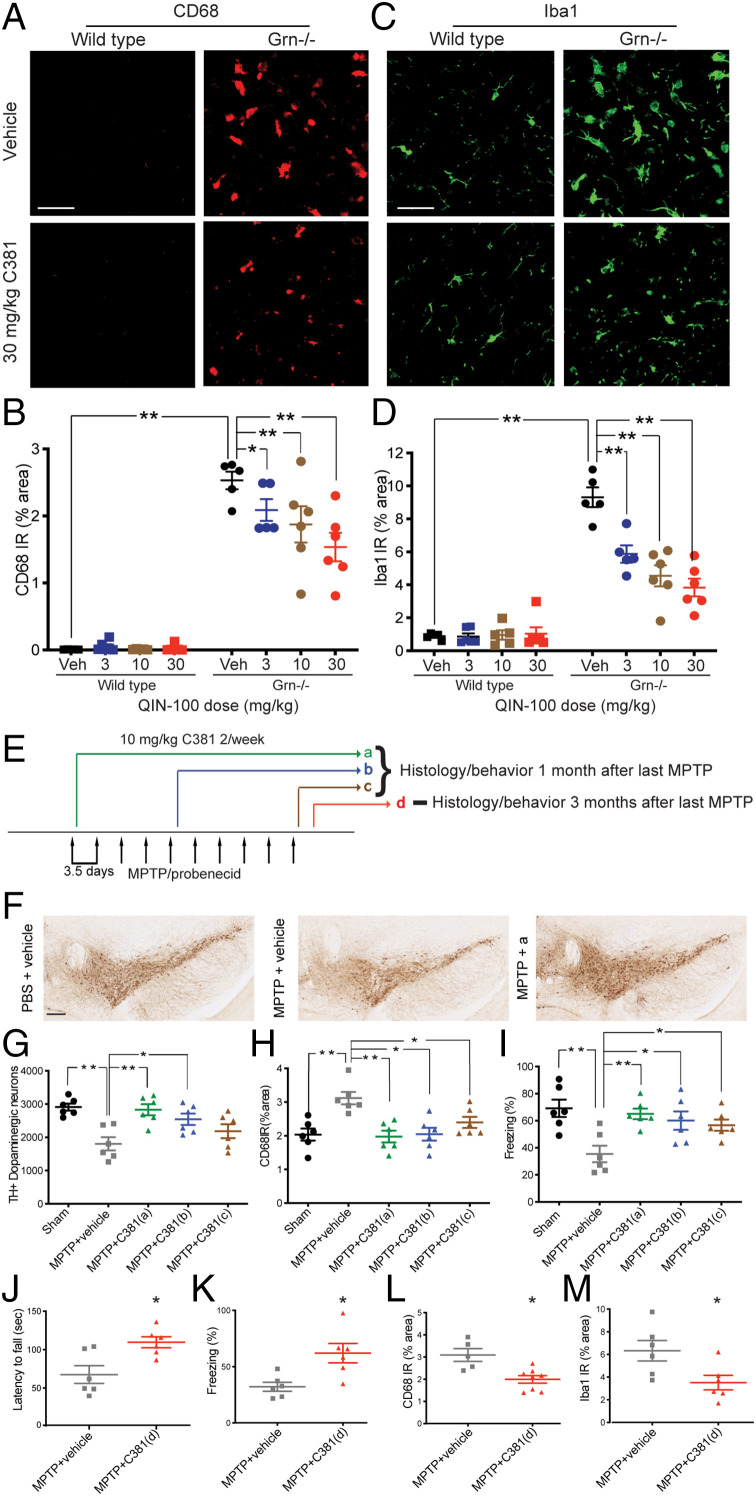
We next tested whether C381 had any beneficial effects in neurodegenerative disease models with a well-described inflammatory component. Frontotemporal dementia (FTD) is the second most common form of dementia in patients under 65 y old (24). The pathology of FTD is heterogeneous, but abnormal inflammation is consistently found in the brains of FTD patients (25). Mutations resulting in haploinsufficiency of progranulin (Grn) cause autosomal dominant familial FTD (26). Grn has been shown to play an important role in the regulation of lysosomal function (27). These FTD patients have less than half of the level of Grn in their plasma compared to healthy individuals or patients with sporadic forms of FTD (28), and their brains are characterized by neurodegeneration, gliosis, and lysosomal dysfunction (29, 30). Complete loss of Grn in homozygous mutants causes a form of neuronal ceroid lipofuscinosis (NCL), a rare lysosomal storage disease. Although these two conditions are distinct, they share many common features, including lysosomal dysfunction (30). Consequently, progranulin-deficient (Grn−/−) mice, while more precisely a model of NCL, are also used as an animal model of FTD and have been utilized as the basis for current FTD treatments in clinical trials (31–33). Histopathologically, aged Grn−/− mice consistently recapitulate many of the pathological features of FTD, including robust gliosis and lysosomal dysfunction (31–33). We saw this model as a good first step in determining whether C381 possessed any antiinflammatory activity. As expected, 12- to 13-mo-old Grn−/− mice showed markedly increased microgliosis vs. age-matched wild-type mice as indicated by CD68 (a lysosomal marker of microglial activation) and Iba1 immunoreactivity (Fig. 2 A–D). Twice weekly i.p. C381 treatments for 1 mo resulted in a dose-dependent reduction in CD68 and Iba1 staining in the Grn−/− mice and no change in the already low levels of CD68 and Iba1 in the wild-type mice (Fig. 2 A–D). This result indicated that C381 exerts antiinflammatory effects on the diseased brain.
Fig. 2.
C381 is antiinflammatory and neuroprotective in preclinical mouse models of neurodegenerative disease. (A) Confocal microscopy images of CD68 immunohistochemistry (IHC) from the thalamus of Grn−/− mice treated twice weekly for 1 mo with C381 or vehicle (30 mg/kg, i.p. twice/week, n = 5 to 6/group). (Scale bar, 12 µm.) (B) Quantification of CD68 immunoreactivity from A (two-way ANOVA *P < 0.05, **P < 0.01). (C) Same as A but for Iba1. (D) Same as B but for Iba1. (E) Treatment and model schematic for MPTP/probenicid PD mice. (F) Representative IHC images of TH staining from SNpc of sham and vehicle or C381-treated MPTP mice. (Scale bar, 200 μm.) (G) Quantification of TH+ neurons remaining in SNpc by treatment group. (H) CD68 immunostaining quantification by treatment group. (I) Freezing behavior in the contextual fear conditioning test by treatment group. (G–I) Statistical test one-way ANOVA *P < 0.05, **P < 0.01. (J) Latency to fall in the rotarod test for treatment group d. (K) Freezing behavior in the contextual fear conditioning test for treatment group d. (L) Quantification of CD68 immunoreactivity in treatment group d. (M) Quantification of Iba1 immunoreactivity in treatment group d. (J–M) t test *P < 0.05.
We next set out to test C381 in the chronic MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) probenicid model of Parkinson’s disease (PD). PD is the second most common neurodegenerative disease, affecting 2 to 3% of the population over age 65 (34). PD is marked by degeneration of dopaminergic neurons, neuroinflammation, and lysosomal dysfunction (35). Having demonstrated that C381 was capable of reducing neuroinflammation and CD68 expression, we thought it may be a good fit for this model. The chronic MPTP model in particular recapitulates many of the features of PD and has been successfully employed in the past for translational PD research (36, 37). We injected 10 mg/kg C381 i.p. twice weekly using three different cotreatment paradigms, starting at the beginning, middle, and end of MPTP/probenecid administration (paradigms a–c in Fig. 2E). One month after the last MPTP and probenecid administration, histopathological analysis revealed that in the mice treated with C381, there were more dopaminergic neurons remaining in the substantia nigra pars compacta (SNpc) compared with phosphate buffered saline (PBS)-injected controls (except paradigm c, Fig. 2 F and G). Remarkably, the protection of dopaminergic neurons by C381 demonstrated that the C381-treated MPTP mice in paradigm a were indistinguishable from the sham control mice. Immunostaining revealed a reduction in CD68 expression in all C381-treated groups, indicating reduced neuroinflammation (Fig. 2H). Mice were also tested for cognitive function with contextual fear conditioning. The mice treated with C381 (all groups) performed significantly better than the vehicle-treated mice, showing improved freezing behavior in the contextual test than the vehicle group (Fig. 2I). To further study the long-term effects of C381, we started C381 injections 3 d after the final MPTP/probenecid administration and continued them until the study endpoint (treatment paradigm d, Fig. 2E). Three months after the last MPTP/probenecid administration, the mice were tested for motor function with the rotarod test and cognitive function with contextual fear conditioning. The mice treated with C381 performed significantly better than the vehicle-treated mice in both tests, showing increased time on the rotarod and improved freezing behavior in the contextual test vs. the vehicle group (Fig. 2 J and K). Additionally, microgliosis assessed by both CD68 and Iba1 immunoreactivity was reduced in the C381-treated mice (Fig. 2 L and M), indicating lower levels of neuroinflammation. Collectively, these preclinical studies showed that C381 possesses neuroprotective and antiinflammatory activity in two separate neurodegenerative disease models in vivo.
C381 Target Identification.
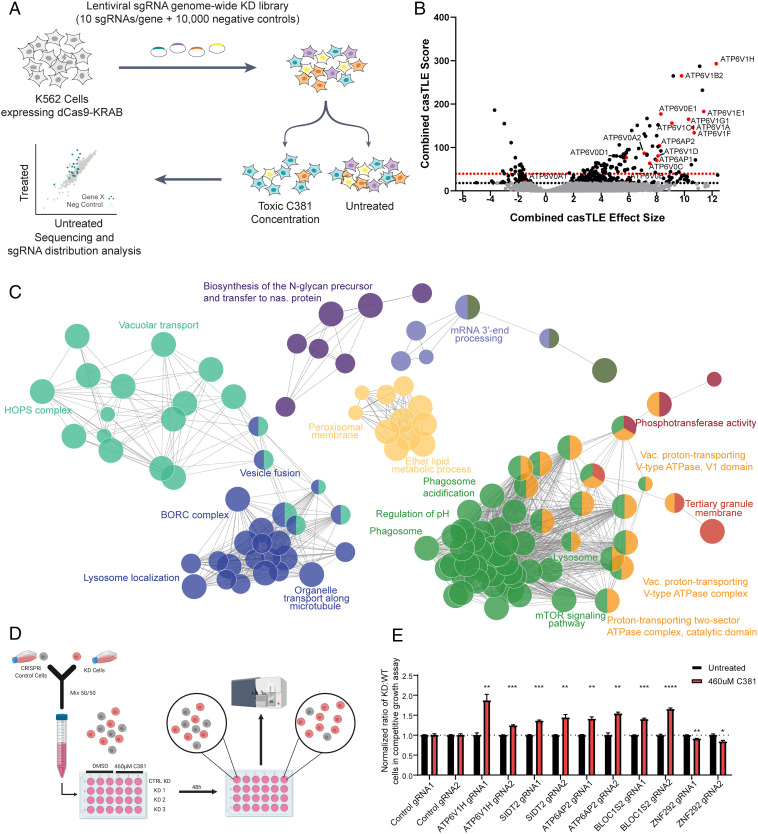
To determine the biological target of C381, we utilized a genome-wide CRISPRi drug target identification “sensitivity” screen (Fig. 3A)—a strategy with a track record of success (38–41). For a sensitivity screen, CRISPRi cells are exposed to toxic levels of a compound of interest. At these high concentrations, the compound will cause cell death, at least in part via overstimulation of its target pathway. In this way, knocking down genes of proteins targeted by the compound will confer either protection or sensitivity, and will be differentially represented in the treated vs. untreated conditions. K562 cells expressing a dCas9-KRAB fusion CRISPRi construct (42) were infected with a lentiviral sgRNA library targeting every gene in the human genome with 10 unique sgRNAs per gene along with ∼10,000 negative control sgRNAs (Fig. 3A), as described previously (43). Cells were maintained at greater than 1,000× coverage of the gRNA library to ensure sufficient representation of each guide (250 million cells for each replicate) and kept at a concentration of 500,000 cells/mL. The screen was carried out in duplicate. We began with a toxic pulse of 460 µM C381 in the treated group, which was found to be the median lethal concentration (LC50) for K562 cells in our preliminary studies. Cells were incubated with the drug for 24 h followed by washing and media replacement to remove C381. The first C381 treatment resulted in a ∼50% reduction in viability, as expected (SI Appendix, Fig. S2). However, when the cells were treated for the second time with 460 µM C381, viability reduction was minimal. This indicated that sgRNAs conferring resistance were already becoming enriched in the treated group. To ensure sufficient selective pressure, we increased the concentrations for the third and fourth pulses to 490 µM and 510 µM, respectively. After four rounds of C381 treatment selection, genomic DNA from the cells was harvested and sequenced to determine the sgRNA distribution in each group (Dataset S1 and Fig. 3B). The protective hits had a much stronger effect size than the sensitizing hits. We hypothesized that the target would likely be a protective hit, as knocking down the target of C381 would most likely confer resistance at high concentrations. Interestingly, pathway and cellular component analysis of the top hits indicated a strong enrichment for lysosomal membrane proteins and pH regulatory machinery, specifically members of the vacuolar-type H+-ATPase (v-ATPase) protein complex (Fig. 3C). Each of the 14 subunits of the v-ATPase complex was a significant hit in the screen (10% false discovery rate [FDR]), along with the two v-ATPase accessory proteins (ATP6AP1 and ATP6AP2) (44). v-ATPase is the proton pump responsible for lysosome acidification, which is necessary for proper function of the resident degradative enzymes as well as maintaining the proton gradient required for transport of various metabolites (45). Dysregulation of lysosomal acidification is exhibited with age and in several neurodegenerative conditions, including FTD and PD (11, 46). Mutations in ATP6AP2, also known as the renin receptor or v-ATPase adapter protein 2, directly cause a rare form of early onset PD (47). Overall, 75% of the enriched gene ontology terms from the protective hits were lysosome/vacuole related (Fig. 3C and SI Appendix, Fig. S3).
Fig. 3.
C381 CRISPRi target identification screen implicates the lysosome and v-ATPase. (A) Schematic of CRISPRi screening process. (B) Volcano plot of screen results. Combined casTLE (Cas9 high-throughput maximum likelihood estimator) score corresponds to statistical confidence. The 10% FDR threshold by the Benjamini–Hochberg test in shown by the dotted black line, and the 10% FDR threshold by the Bonferonni correction is shown by the dotted red line. Combined casTLE effect size shows magnitude and direction of each hit. Positive effect size indicates protective hits, while negative effect size indicates sensitizing hits. Protective hits are genes with sgRNAs enriched in the group treated with C381, and sensitizing hits are genes with sgRNAs depleted in the group treated with C381. Black data points indicate significant hits at the 10% FDR threshold by the Benjamini–Hochberg method. Hits with red data points are v-ATPase subunits. (C) Network representation of enriched gene ontology terms and pathways for protective hits from the CRISPRi screen that were significant at the 10% FDR Bonferonni threshold. Each node (circles) represents one term/pathway. Edges (lines) connect terms with similar gene sets (distance stands in inverse relationship with overlap); further functional clustering (color) was based on kappa statistics. Distance between unconnected nodes is arbitrary. Shown are representative terms per cluster. (D) Schematic for the competitive growth screen validation experiment. A detailed description appears in SI Appendix, SI Materials and Methods. (E) Results of 50/50 competitive growth assay for selection of hits from the screen. Each gene was confirmed with two distinct gRNAs. Each bar represents results from three independent biological experiments. Individual t tests were used to compare treated to untreated for each gRNA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To confirm the results of the screen, we utilized a competitive growth assay with individual knockdown (KD) cell lines. We chose a diverse set of the top gene hits for this assay: Bloc1s2, Atp6v1h, Atp6ap2, Sidt2, and Znf292. To validate the hits, two distinct sgRNAs for each chosen gene along with two negative control sgRNAs (48) were delivered to CRISPRi K562 cells lentivirally along with an mCherry reporter. mCherry+ CRISPRi KD cells were mixed 50/50 with mCherry− CRISPRi cells. Cells were either treated with 460 µM or vehicle, and 48 h later the ratio of mCherry+ to mCherry− cells were assessed via flow cytometry (Fig. 3D). The results of the validation experiment confirmed the screen results (Fig. 3E). Even the weaker sensitizing hit, Znf292, caused a modest increase in C381 toxicity vs. control. The results of the CRISPRi screen, confirmed by the validation experiments, strongly pointed to the lysosome and particularly the lysosomal v-ATPase as the target of C381.
C381 Physically Targets the Lysosome, Promoting Acidification, Activity, and Resilience to Damage.
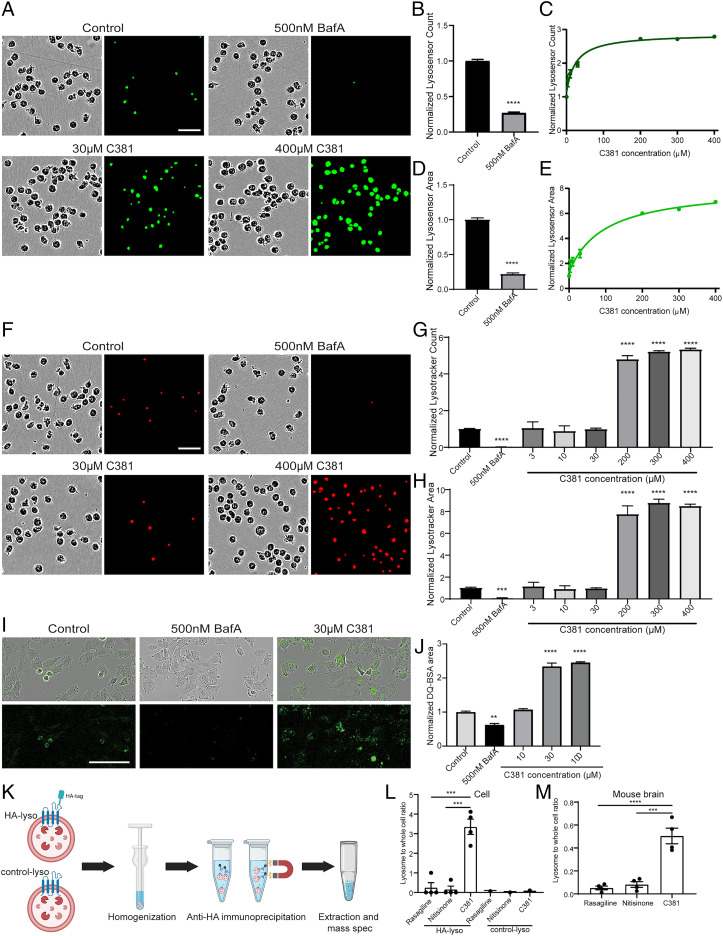
To determine whether C381 functionally targets the lysosome and v-ATPase, we probed the lysosomal effects of C381 using the Lysotracker and Lysosensor chemical probes. Lysotracker preferentially accumulates in acidic organelles and is constitutively fluorescent. Lysosensor is similar in that it also accumulates in acidic organelles but its fluorescence scales with pH - increasing in brightness with lower pH. Using these in tandem allowed us to determine the number and magnitude of lysosomes along with their relative acidity. For a control, we used Bafilomycin A1 (BafA), a known inhibitor of v-ATPase (49, 50). We treated K562 cells with a range of C381 concentrations or 500 nM BafA for 1 h, then stained the cells with Lysotracker Red and Lysosensor Green for 1 h. After washing, we imaged the live cells using the Incucyte S3 system. C381 caused a dose-dependent increase in lysosome acidification as indicated by Lysosensor area and count (Fig. 4 A–E). BafA caused a reduction in Lysosensor signal as expected (Fig. 4 A, B, and D). At the low doses that mimic therapeutic levels, this was a modest induction, but at high doses there was a strong increase in Lysosensor signal (nearly 7× at 400 µM). Lysotracker signal was not significantly changed with therapeutic doses of C381, but at high doses approaching the concentration used in the screen there was a large increase in signal (∼8× by area, Fig. 4 F–H). BafA drastically reduced Lysotracker signal. These results indicated that C381 stimulates lysosomal acidification.
Fig. 4.
C381 promotes lysosome acidification and increases lysosomal proteolysis. (A) Representative Lysosensor Green staining images from K562 cells treated for 1 h with C381 or BafA. (Scale bar, 50 μm.) (B) Lysosome acidification changes shown by count of pH-sensitive Lysosensor Green puncta normalized to confluence in K562 cells treated for 1 h with BafA (t test ****P < 0.0001). (C) Same as in B, but treated with varying concentrations of C381. All concentrations are statistically significant vs. control (one-way ANOVA). (D) Lysosome acidification changes shown by area of pH-sensitive Lysosensor Green staining normalized to confluence in K562 cells treated for 1 h with BafA (t test ****P < 0.0001). (E) Same as in D, but treated with varying concentrations of C381. All concentrations are statistically significant vs. control (one-way ANOVA). (F) Representative Lysotracker Red staining images from K562 cells treated for 1 h with C381 or BafA. (Scale bar, 50 μm.) (G) Overall changes in acidic vesicle number shown by count of Lysotracker Red puncta normalized to confluence in the same cells as in A (t test for BafA; one-way ANOVA for C381 ****P < 0.0001). (H) Overall changes in acidic vesicle area shown by area of Lysotracker Red staining normalized to confluence in the same cells as in A (t test for BafA; one-way ANOVA for C381 ***P < 0.001, ****P < 0.0001). (I) Representative images of changes in DQ-BSA Green breakdown in 8988T cells treated with vehicle, BafA, or C381. (Scale bar, 100 μm.) (J) Quantification of DQ-BSA Green area normalized to cell confluence (one-way ANOVA **P < 0.01, ****P < 0.0001). (K) Schematic outlining Lyso-IP process. Cells expressing an HA tag on lysosomes or control cells were homogenized and Lyso-IP was performed using an anti-HA magnetic bead enrichment protocol. The small molecules from the pulled down lysosomes were then extracted and analyzed by mass spectrometry and compared to the whole cell fraction. (L) Quantification of in vitro lysosomal enrichment of C381 and control drugs nitisinone and rasagiline for both the HA-lyso and control-lyso cells (one-way ANOVA ***P < 0.001). (M) Quantification of in vivo lysosomal enrichment for C381 and control drugs nitisinone and rasagiline in mouse brain. Lysosomes were isolated from one hemibrain with the Thermo Fisher Lysosome Enrichment Kit and compared with the whole cell fraction from the other hemibrain (one-way ANOVA ***P < 0.001, ****P < 0.0001).
We next wanted to determine whether C381 had any functional effects on the lysosome. To probe this, we utilized DQ-bovine serum albumin (BSA). DQ-BSA is conjugated to a quenched dye that becomes fluorescent upon BSA degradation. This means that it only fluoresces once BSA has been degraded by the lysosome. According to our hypothesis that C381 causes vesicular acidification and lysosomal activation, we expected to see an increase in fluorescence signal with treatment, particularly at high concentrations. Cells were treated with C381 or BafA as a control along with DQ-BSA Green. After 2 h of treatment, the cells were washed and imaged on the Incucyte S3. C381 caused a significant increase in normalized DQ-BSA area (Fig. 4 I and J). This indicated that C381 was boosting the levels of DQ-BSA degradation under normal conditions in healthy cells. These results show that C381 is capable of increasing lysosome-mediated protein degradation.
Having demonstrated that C381 modulates lysosomal acidification and activity, we next set out to understand whether this was due to a physical interaction with a lysosomal protein or whether these lysosomal effects were downstream of the true target. To accomplish this, we utilized HEK293 cells expressing a hemagglutinin (HA) tag on TMEM192, a lysosomal membrane protein (HA-lyso cells). The HA tag allows for lysosomal pulldown (Fig. 4K), as described previously (51). Cells were treated for 2 h with C381 and control drugs nitisinone and rasagiline. Nitisinone was chosen based on having a similar molecular weight to C381 (329.48 for C381, 329.23 for nitisinone). Rasagiline was chosen because it has a similar indication to C381 as a BBB-permeable PD drug. After treatment, the lysosomes were pulled down and the lysosomal and whole cell fractions were analyzed for the three drugs via mass spectrometry C381 was significantly enriched in the lysosomal fraction vs. the other two drugs (Fig. 4L). As a control, cells lacking the HA tag (control-lyso cells) were treated and extracted in the same way to ensure that the HA immunoprecipitation itself was not enriching for C381 (Fig. 4L). No enrichment was exhibited in the control-Lyso cells, confirming that C381 was physically targeting the lysosome. We next wanted to determine whether C381 was able to physically target the lysosomes in the brains of mice, where the beneficial in vivo effects were exhibited. Mice were injected (i.p.) with the same three drugs at 50 mg/kg each. Two hours after injection, the mice were killed, the brains were extracted, and the lysosomes were isolated (SI Appendix, SI Materials and Methods). As a control, a portion of the lysate was kept without being enriched for lysosomes in order to generate a “whole cell” fraction. The three drugs were then analyzed in each fraction via mass spectrometry and lysosomal enrichment was assessed. The in vivo experiment results were similar to those from the in vitro experiment, showing strong enrichment of C381 in the lysosomes extracted from the mouse brain (Fig. 4M).
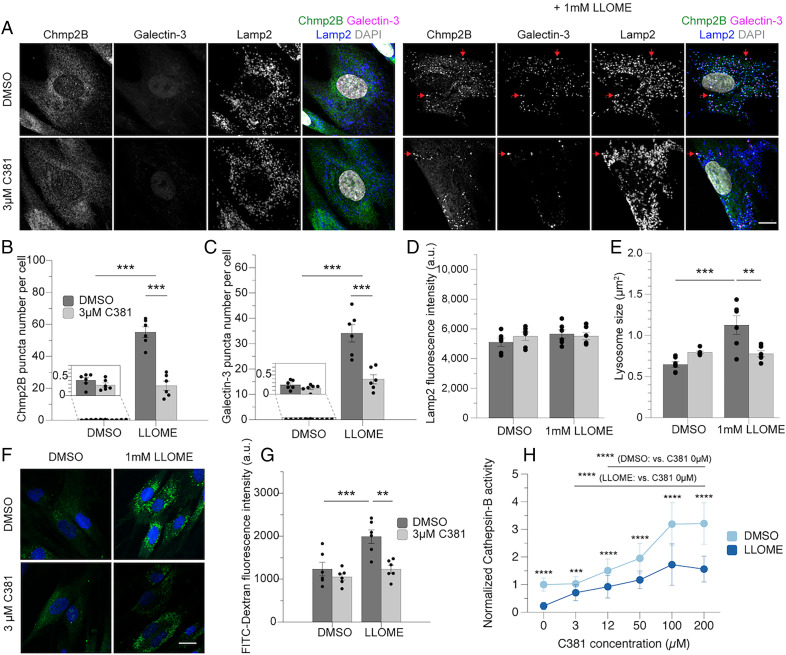
Next, we set out to determine whether C381 was able to provide any protection against lysosomal insults mimicking disease conditions in primary human cells. Human primary dermal fibroblasts were first assessed for sensitivity to C381. C381 was well tolerated in these cells at concentrations up to 100 µM, at which point slight toxicity was exhibited (SI Appendix, Fig. S4 A and B). We next assessed whether C381 could rescue cells from l-leucyl-l-leucine methyl ester (LLOME) toxicity. LLOME is a lysosomotropic agent that severely damages and permeabilizes the lysosomal membrane, causing lysosome alkalization, lysophagy, and cell death (52, 53). Lysosomal membrane permeabilization is a known mechanism of lysosomal pathology in PD (54, 55). Fibroblasts were pretreated with C381 for 24 h and then exposed to 1 or 2 mM LLOME for an additional 24 h. C381 caused a reduction in cell death and an increase in cell viability in the LLOME-treated fibroblasts (SI Appendix, Fig. S4 C and D). These results showed that C381 was protecting cells from LLOME-induced death. We next tested whether C381 was acting to functionally reduce LLOME-mediated lysosomal damage. LLOME causes severe membrane damage to lysosomes, resulting in the activation of ESCRT proteins involved in lysosomal repair and the Galectin proteins involved in lysophagy (52). The ESCRT proteins sense and repair minor membrane damage, but for severe damage the Galectin proteins will bind the inside of the lysosomal membrane and initiate lysophagy (56). As expected, 30 min of 1 mM LLOME resulted in significant recruitment of both Chmp2B (an ESCRT-III protein) and Galectin-3 to Lamp2+ lysosomes (Fig. 5 A–C). However, C381 pretreatment at 3 µM drastically reduced the number of Galectin-3 and Chmp2B puncta by 50% or more (Fig. 5 A–C). These results show that C381 acts to protect lysosomes from membrane damage and to improve cellular resilience to such insults. We also found that C381 did not affect Lamp2 fluorescence intensity, lysosome size, or lysosome number with treatment at baseline (Fig. 5 D and E and SI Appendix, Fig. S4E). In the case of lysosome size, LLOME treatment caused a nearly twofold enlargement of lysosomes, as has been reported previously (53). C381 treatment normalized lysosome size back to baseline levels (Fig. 5E).
Fig. 5.
C381 improves lysosomal resilience to membrane permeabilization and damage. (A) Immunostaining of endogenous Chmp2B, Galectin-3, and Lamp2 in human dermal fibroblasts. Pretreatment of 3 µM C381 for 24 h and then 1 mM LLOME for 30 min. (Scale bar, 20 μm.) Red arrows indicate colocalization of Chmp2B, Galectin-3, and Lamp2. (B–E) Two cell lines in three independent experiments. Each dot represents data from a single cell line in a single experiment. The total number of cells ranges from 51 to 72 per group. (B) Quantification of Chmp2B puncta per cell. (C) Quantification of Galectin-3 puncta per cell. (D) Quantification of average Lamp2 fluorescence intensity per cell in each treatment group. (E) Quantification of lysosomal size in each treatment group. (F) Fluorescence imaging of FITC-dextran–loaded cells treated for 24 h with C381 or DMSO and treated for 30 min with 1 mM LLOME or DMSO. (Scale bar, 20 μm.) (G) Quantification of FITC-dextran fluorescence intensity in each treatment group. Two cell lines were used in three independent experiments. The total number of cells ranges from 109 to 126 per group. (H) Normalized cathepsin-B activity in human dermal fibroblasts treated for 24 h with C381 or DMSO followed by 30-min treatment of 1 mM LLOME or DMSO. The total number of cells ranges from 82 to 112 per group. Statistical analyses were performed with two-way ANOVA and Bonferroni’s post hoc test (**P < 0.01, ***P < 0.001, ****P < 0.0001).
Loss of lysosome acidification is known to be exacerbated in disease conditions, including in the case of lysosomal membrane permeabilization exhibited in PD and in response to lysosomotropic agents (54, 56). We wanted to determine whether C381 could rescue lysosomal alkalization in response to membrane damage. Cells were first loaded with pH-sensitive probe fluorescein isothiocyanate (FITC)-dextran, which accumulates in lysosomes. At the low pH of properly functioning lysosomes, the fluorescence of FITC is quenched. However, upon LLOME treatment, the fluorescence of FITC returns inside the lysosome as acidification is impaired (Fig. 5 F and G). With 3 µM C381 pretreatment, the FITC signal was restored to baseline levels in the presence of LLOME, indicating maintenance of homeostatic lysosomal pH (Fig. 5 F and G). These results show that C381 is capable of maintaining lysosome acidification in the context of lysosomal damage.
The functional consequence of impaired lysosomal acidification is a reduction in the activity of the resident enzymes responsible for breaking down cellular material. One such pH-sensitive enzyme is cathepsin-B. Having demonstrated that C381 helps maintain homeostatic lysosomal pH, we wanted to determine whether this effect corresponded with maintenance of enzyme activity. To test this, we assayed cathepsin-B activity in the presence or absence of LLOME along with a range of C381 concentrations (Fig. 5H). As expected, LLOME treatment reduced cathepsin-B activity by ∼80%. However, with 3 µM C381 treatment in addition to LLOME, cathepsin-B activity was increased more than threefold vs. LLOME alone. This corresponded with the result from the FITC-dextran experiment showing improved pH maintenance with treatment. At higher C381 concentrations, we saw a dose-dependent increase in cathepsin-B activity, eventually surpassing baseline levels. In cells not exposed to LLOME, high concentrations of C381 resulted in a greater than threefold increase in cathepsin-B activity at the highest doses tested. These results indicate that C381 is capable of increasing the activity of a pH-dependent lysosomal enzyme after membrane damage, and even in naive conditions at high concentrations.
C381 Restores Lysosomal Defects in Disease Models In Vivo.
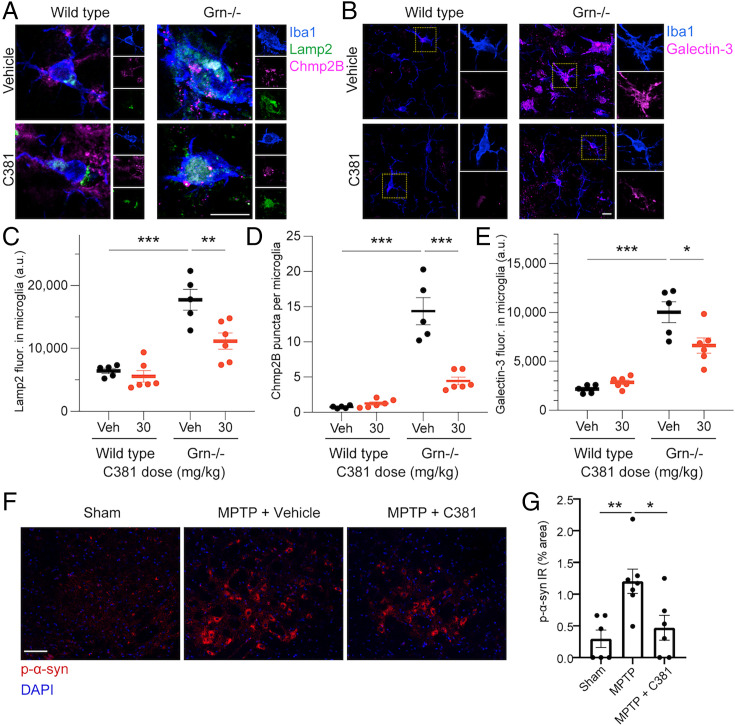
After discovering that C381 targets lysosomes in vitro and in vivo and characterizing its positive effects on lysosomes in cellular models, we set out to revisit our previous in vivo studies to determine whether lysosomes were restored in this context. Lysosomal defects are well established in the Grn−/− model of FTD and NCL, so we first stained brain sections from these mice for the same markers of lysosomes and associated damage we examined in our cellular systems—Lamp2, Chmp2B, and Galectin-3 (Fig. 6 A and B). Consistent with previous reports in both mice and humans (57, 58), we saw an increase in Lamp2 immunoreactivity in thalamic microglia of Grn−/− mice (Fig. 6 A and C). Grn−/− mice treated with C381 showed reduced levels of Lamp2 in microglia, reflecting a rescue of lysosome pathological feature in these mice. Notably, wild-type mice treated with C381 showed no difference in Lamp2 levels in microglia (Fig. 6C). We repeated this analysis with Lamp1 staining, which had parallel results (SI Appendix, Fig. S5 A and B). Likewise, Chmp2B puncta were increased in Grn−/− mice, indicating higher levels of lysosomal damage (Fig. 6 A and D). C381 treatment caused a marked reduction in Chmp2B puncta (Fig. 6D), as in the cellular models. We saw similar results with Galectin-3 staining (Fig. 6 B and E), where there was an up-regulation in the Grn−/− mice that was rescued by C381 treatment. These results demonstrated that C381 treatment reduces lysosomal pathology in the Grn−/− model in vivo.
Fig. 6.
C381 reduces lysosomal pathology in vivo. (A) Fluorescence images of microglia from wild-type and Grn−/− mice for Iba1, Lamp2, and Chmp2B. (Scale bar, 10 μm.) (B) Fluorescence images of microglia from wild-type and Grn−/− mice for Iba1 and Galectin-3. (Scale bar, 10 μm.) (C) Quantification of Lamp2 fluorescence in microglia. (D) Quantification of Chmp2B puncta in microglia. (E) Quantification of Galectin-3 fluorescence in microglia. For C–E, each dot represents one mouse, n = 15 to 28 cells/mouse. (F) Fluorescence images of the SNpc of sham, MPTP, and MPTP + C381 mice for DAPI and p-α-syn. (Scale bar, 50 μm.) (G) Quantification of p-α-syn % area. Statistical analysis performed with one-way ANOVA and Tukey’s multiple comparisons test (*P < 0.05, **P < 0.01, ***P < 0.001).
Like in the Grn−/− model, lysosomal defects have also been reported in the MPTP model of PD (54, 55). Specifically, a causal relationship has been established between lysosomal dysfunction and pathogenic alpha-synuclein (α-syn) accumulation (59). We set out to determine whether C381 treatment in our MPTP model mice reduced accumulation of the pathogenic phosphorylated form of α-syn (p-α-syn). As reported previously (60), we saw increased p-α-syn in the SNpc of the MPTP model mice vs. sham control (Fig. 6 F and G). C381 treatment (paradigm a from Fig. 2E) caused a significant reduction in p-α-syn in the SNpc. These results suggest that C381, through its effects on the lysosome, can reduce the pathogenic accumulation of p-α-syn in PD.
Discussion
Lysosomal dysfunction is increasingly seen as a common thread between neurodegenerative diseases (9, 46, 61, 62). Specifically, lysosomes seem unable to properly acidify in many of these conditions, leading to undegradable biomaterial accumulation as the lysosome-resident proteases and transport channels are unable to function properly (10, 11, 46). The central role of the lysosome in protein degradation, sequestration of intracellular metabolites, and cell signaling make it an extremely attractive therapeutic target for neurodegenerative disease. Many therapies targeted to the lysosome for activation and/or restoration of acidification have been pursued preclinically with positive results (63–66). Unfortunately, many of these have limited translational potential due to their inability to cross the BBB (63, 65) or potential toxicity (64, 65).
Here we report the discovery of a lysosome activator with broad functional benefits. Starting as the brain-penetrant lead compound from a phenotypic screen for brain Smad activators, C381 was revealed to possess potent antiinflammatory and neuroprotective properties in mouse models of FTD and PD. We next probed the mechanism via a genome-wide CRISPRi drug target identification screen that implicated the lysosome. This unexpected discovery was confirmed by follow-up staining and functional testing of lysosomal function with C381 treatment. We then utilized HA-tagged lysosomes to perform lysosome immunopurification (lyso-IP) and confirm that C381 physically targets the lysosome in vitro. We also showed this in vivo using a centrifugation-based lysosomal enrichment kit. C381 was then shown to stimulate lysosomal acidification, increase lysosomal protein breakdown, and improve lysosomal resilience to membrane damage and permeabilization. Due to the widespread lysosomal dysfunction across neurodegenerative diseases, this represents an exciting and promising therapeutic target.
In our in vivo work prior to learning the target of C381, CD68 reduction was a strong observed effect of the drug. At first, we believed this reflected a general antiinflammatory effect, but given that CD68 is a lysosomal marker it is possible this was a direct consequence of C381’s lysosomal activity. Still, the reduction in Iba1 staining in the Grn−/− and MPTP models demonstrates antiinflammatory activity downstream of the drug’s lysosomal effects.
In our in vivo models, microglia had the most prominent lysosomal defects and many of our cell-type–specific readouts came from these cells. Although we demonstrate that C381 has strong effects in microglia, we have no reason to believe that the drug has a cell-type–specific effect. It is possible that the improvement in neuronal survival in the MPTP model is a result of direct action on the neurons rather than a downstream consequence of the drug’s microglial activity. We plan in the future to better characterize the effects of C381 on various cell types, including neurons.
Because of the extremely selective BBB, only a small subset (∼1%) of small molecules are able to penetrate into the brain parenchyma as C381 does (67). Because of this and the generally high attrition rates for CNS drugs, a scoring function has been established for drugs in this space to select for structurally suitable compounds. This multiparameter optimization (MPO) score ranges from 0 to 6 and is based on ClogP, ClogD, total polar surface area, hydrogen bonding donors, molecular weight, and pKa (68). On this 6-point scale, a score of 4 or higher indicates a “desirable” drug. C381 has a CNS MPO score of 4.35.
Our discovery that C381 targets the lysosome and increases lysosomal acidification puts it in a rare class of compounds. These include acidic nanoparticles, EN6, and clioquinol. Acidic nanoparticles have been pursued as therapies for neurodegenerative diseases as they have been shown to restore lysosomal acidification and improve lysosome-mediated degradation of cargo (63, 69). Although they have shown positive results in vitro and with direct injection into the brains of PD mice (63), the translational potential of these nanoparticles is limited, due to their inability to cross the BBB. EN6 is an exciting new small molecule that covalently binds v-ATPase, stimulating lysosome acidification and inducing autophagy via mTORC1 inhibition (65). This compound showed therapeutic potential by promoting lysosome-dependent clearance of TDP-43 aggregates in vitro and by activating autophagy in heart and skeletal muscle in vivo. However, no brain penetrance of the compound was shown and there were other covalent targets of the molecule that have not yet been investigated. The covalent agonist activity of the compound may also represent a translational hurdle from a safety perspective. Clioquinol was originally developed as an antifungal and antiprotozoal drug and is a zinc/copper ionophore. Clioquinol has been tested in multiple models of PD (70–72) and was discovered to have an inverse transcriptional effect to patient-derived LRRK2 mutant–induced neurons (73). While its benefits in PD models were originally ascribed to its chelator activity, it was recently discovered that its primary mechanism is zinc-mediated lysosome activity restoration (64). Clioquinol rescues lysosomal acidification and increases v-ATPase subunit protein levels, both of which contribute to restoring autophagic and lysosomal defects in the LRRK2 mutant model of PD. Unfortunately, clioquinol is only approved for topical use and was found to be neurotoxic when prescribed orally for long periods of time, causing an epidemic of subacute myelooptic neuropathy in Japan (74). Although clioquinol itself will not be translated to the clinic due to its toxicity concerns, similar compounds will likely be pursued to see whether the restorative lysosomal effects can be maintained without the toxicity.
Currently, the link between C381’s Smad activation activity and lysosomal modulation is not clear. Notably, C381 does not seem to activate Smad signaling in various cell lines in vitro, so it may be a CNS-specific response. One potential mechanism of C381 CNS Smad activation is through Beclin-1, one of the few known links between TGF-β/Smad and lysosomal function in the CNS (75, 76). Beclin-1 is important for TGF-β receptor recycling and becomes dysregulated with age and disease (77). Beclin-1 was a significant protective hit from the CRISPRi C381 target identification screen (FDR = 10%, Dataset S1), suggesting that it is involved in the cellular response to C381’s lysosomal effects. Since Beclin-1 is known to increase Smad activation, we speculate that this is a likely mechanism. It is known that with aging and disease, Smad signaling and lysosomal activity both become dysfunctional. In AD, the Smad proteins become mislocalized in the cytoplasm and, despite higher levels of TGF-β1, Smad signaling decreases (78, 79). One explanation for this phenomenon is that these cytoplasmic Smad proteins are marked for degradation but are not being cleared due to lysosomal/autophagic dysfunction. The fact that we show that C381 physically targets the lysosome suggests that the Smad activation effects are downstream of the drug’s main effects.
The precise mechanism of C381’s effects on the lysosome requires further investigation, although it appears to act at least partially through v-ATPases. Interestingly, even drugs resulting from target-based screens are frequently determined to have a mechanism of action independent of their assumed target (40). In order to facilitate the translation of C381 into the clinic, additional investigatory studies on the precise molecular target would be desirable. Our efforts to pull down the target using click chemistry analogs of C381 have been unsuccessful due to loss of activity from the structural perturbation. If it does indeed target the v-ATPase complex, binding studies will be difficult due to its multisubunit composition and difficulties with surface display or recombinant expression. All IND-enabling (Investigational New Drug Application) toxicology studies for C381 have been completed, showing no major concerns. It is our hope to push C381 into clinical trials in the near future, where it can serve as a first-in-class drug for neurodegenerative conditions with lysosomal pathology, such as PD, and help pave the way for other molecules of its kind.
Materials and Methods
SI Appendix contains detailed descriptions of mouse lines and animal experimentation, small molecule screening and pharmacokinetic studies, CRISPRi screen, analysis and validation, mouse brain tissue processing and immunohistochemistry, lysosome isolation and liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis, functional assays for lysosomal acidification and degradation, and cell viability and death assays. Briefly, SBE-luc reporter mice were used for initial screening of C381-dependent Smad activation in the CNS. Two mouse models, Grn−/− and MPTP mice, that have been used to study neurodegenerative diseases associated with lysosomal dysfunction, were tested to evaluate the in vivo effect of C381 on the reduction of neuroinflammation, neurodegeneration, and lysosomal deficit. To determine the biological target of C381, K562 cells expressing a dCas9-KRAB fusion CRISPRi construct were infected with a sgRNA library to enable the whole genome screens. CRISPRi screen results were subsequently validated by competitive growth assays in K562 cells. Analysis of lysosome properties, function, and toxicity were performed in K562 cell, HEK293T cells, and fibroblasts.
Supplementary Material
Acknowledgments
We thank Macy Zardenetta and David Gate for assistance with the CRISPRi screen. We thank Oliver Hahn for help with the pathway analysis of the CRISPRi hits. This work was supported by NIH (NS057496, AG059694, AG064897 to T.W.-C., NS092868 to J.L., and P01AG054407 to J.F.), by the Alzheimer's Drug Discovery Foundation (T.W.-C.) and by a grant from the Michael J. Fox Foundation for Parkinson's Research (J.L.). C.-C.C. was supported by the Life Sciences Research Foundation Postdoctoral Fellowship and the Glenn Foundation for Medical Research Postdoctoral Fellowship in Aging Research.
Footnotes
Reviewers: S.E., The Scripps Research Institute; J.K., Washington University in St. Louis, Washington University Physicians; B.S., Harvard Medical School Children’s Hospital.
Competing interest statement: R.T.V., A.C.Y., J.L., and T.W.-C. are cofounders of Qinotto Inc., a company working to improve drug delivery to the brain that has acquired the rights to C381.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Gribkoff V. K., Kaczmarek L. K., The need for new approaches in CNS drug discovery: Why drugs have failed, and what can be done to improve outcomes. Neuropharmacology 120, 11–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullard A., BACE failures lower AD expectations, again. Nat. Rev. Drug Discov. 17, 385 (2018). [DOI] [PubMed] [Google Scholar]
- 3.van Dyck C. H., Anti-amyloid-β monoclonal antibodies for Alzheimer’s disease: Pitfalls and promise. Biol. Psychiatry 83, 311–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prior M., et al. , Back to the future with phenotypic screening. ACS Chem. Neurosci. 5, 503–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swalley S. E., Expanding therapeutic opportunities for neurodegenerative diseases: A perspective on the important role of phenotypic screening. Bioorganic Med. Chem. 28, 115239 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Wee Yong V., Inflammation in neurological disorders: A help or a hindrance? Neuroscientist 16, 408–420 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Boland B., et al. , Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17, 660–688 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dugger B. N., Dickson D. W., Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9, a028035. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh J. Y., Kim H. N., Hwang J. J., Kim Y. H., Park S. E., Lysosomal dysfunction in proteinopathic neurodegenerative disorders: Possible therapeutic roles of cAMP and zinc. Mol. Brain 12, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik B. R., Maddison D. C., Smith G. A., Peters O. M., Autophagic and endo-lysosomal dysfunction in neurodegenerative disease. Mol. Brain 12, 1–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallings R. L., Humble S. W., Ward M. E., Wade-Martins R., Lysosomal dysfunction at the centre of Parkinson’s disease and frontotemporal dementia/amyotrophic lateral sclerosis. Trends Neurosci. 42, 899–912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tesseur I., Wyss-Coray T., A role for TGF-β signaling in neurodegeneration: Evidence from genetically engineered models. Curr. Alzheimer Res. 3, 505–513 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Liu F. F., et al. , Neuroprotective effects of SMADs in a rat model of cerebral ischemia/reperfusion. Neural Regen. Res. 10, 438–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyss-Coray T., TGF-β pathway as a potential target in neurodegeneration and Alzheimer’s. Curr. Alzheimer Res. 3, 191–195 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Lin A. H., et al. , Global analysis of Smad2/3-dependent TGF-β signaling in living mice reveals prominent tissue-specific responses to injury. J. Immunol. 175, 547–554 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Luo J., Lin A. H., Masliah E., Wyss-Coray T., Bioluminescence imaging of Smad signaling in living mice shows correlation with excitotoxic neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 103, 18326–18331 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Bernhardi R., Cornejo F., Parada G. E., Eugenín J., Role of TGFβ signaling in the pathogenesis of Alzheimer’s disease. Front. Cell. Neurosci. 9, 426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyss-Coray T., et al. , TGF-β1 promotes microglial amyloid-β clearance and reduces plaque burden in transgenic mice. Nat. Med. 7, 612–618 (2001). [DOI] [PubMed] [Google Scholar]
- 19.He Y., et al. , ALK5-dependent TGF-β signaling is a major determinant of late-stage adult neurogenesis. Nat. Neurosci. 17, 943–952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesseur I., et al. , Deficiency in neuronal TGF-β signaling leads to nigrostriatal degeneration and activation of TGF-β signaling protects against MPTP neurotoxicity in mice. J. Neurosci. 37, 4584–4592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brionne T. C., Tesseur I., Masliah E., Wyss-Coray T., Loss of TGF-β 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron 40, 1133–1145 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Tesseur I., et al. , Deficiency in neuronal TGF-β signaling promotes neurodegeneration and Alzheimer’s pathology. J. Clin. Invest. 116, 3060–3069 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesseur I., Zou K., Berber E., Zhang H., Wyss-Coray T., Highly sensitive and specific bioassay for measuring bioactive TGF-β. BMC Cell Biol. 7, 15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratnavalli E., Brayne C., Dawson K., Hodges J. R., The prevalence of frontotemporal dementia. Neurology 58, 1615–1621 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Bright F., et al. , Neuroinflammation in frontotemporal dementia. Nat. Rev. Neurol. 15, 540–555 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Cruts M., et al. , Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Elia L. P., Mason A. R., Alijagic A., Finkbeiner S., Genetic regulation of neuronal progranulin reveals a critical role for the autophagy-lysosome pathway. J. Neurosci. 39, 3332–3344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schofield E. C., et al. , Low serum progranulin predicts the presence of mutations: A prospective study. J. Alzheimers Dis. 22, 981–984 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Mackenzie I. R. A., et al. , The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain 129, 3081–3090 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Ward M. E., et al. , Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci. Transl. Med. 9, eaah5642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cenik B., Sephton C. F., Kutluk Cenik B., Herz J., Yu G., Progranulin: A proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 287, 32298–32306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghoshal N., Dearborn J. T., Wozniak D. F., Cairns N. J., Core features of frontotemporal dementia recapitulated in progranulin knockout mice. Neurobiol. Dis. 45, 395–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arrant A. E., Onyilo V. C., Unger D. E., Roberson E. D., Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J. Neurosci. 38, 2341–2358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poewe W., et al. , Parkinson disease. Nat. Rev. Dis. Primers 3, 1–21 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Senkevich K., Gan-Or Z., Autophagy lysosomal pathway dysfunction in Parkinson’s disease; evidence from human genetics. Parkinsonism Relat. Disord. 73, 60–71 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Langston J. W., The MPTP story. J. Parkinsons Dis. 7, S11–S19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meredith G. E., Rademacher D. J., MPTP mouse models of Parkinson’s disease: An update. J. Parkinsons Dis. 1, 19–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kampmann M., Biohub C. Z., CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 13, 406–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jost M., Weissman J. S., CRISPR approaches to small molecule target identification. ACS Chem. Biol. 13, 366–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jost M., et al. , Combined CRISPRi/a-based chemical genetic screens reveal that rigosertib is a microtubule-destabilizing agent. Mol. Cell 68, 210–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deans R. M., et al. , Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 12, 361–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson M. H., et al. , CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert L. A., et al. , Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toei M., Saum R., Forgac M., Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luzio J. P., Pryor P. R., Bright N. A., Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Colacurcio D. J., Nixon R. A., Disorders of lysosomal acidification—The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korvatska O., et al. , Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum. Mol. Genet. 22, 3259–3268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgens D. W., et al. , Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun. 8, 15178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dröse S., Altendorf K., Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200, 1–8 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Mauvezin C., Neufeld T. P., Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11, 1437–1438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abu-Remaileh M., et al. , Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skowyra M. L., Schlesinger P. H., Naismith T. V., Hanson P. I., Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Repnik U., et al. , L-leucyl-L-leucine methyl ester does not release cysteine cathepsins to the cytosol but inactivates them in transiently permeabilized lysosomes. J. Cell Sci. 130, 3124–3140 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Vila M., Bové J., Dehay B., Rodríguez-Muela N., Boya P., Lysosomal membrane permeabilization in Parkinson disease. Autophagy 7, 98–100 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Dehay B., et al. , Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci. 30, 12535–12544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papadopoulos C., Kravic B., Meyer H., Repair or lysophagy: Dealing with damaged lysosomes. J. Mol. Biol. 432, 231–239 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Götzl J. K., et al. , Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol. Neurodegener. 13, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Götzl J. K., et al. , Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 127, 845–860 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Bourdenx M., Bezard E., Dehay B., Lysosomes and α-synuclein form a dangerous duet leading to neuronal cell death. Front. Neuroanat. 8, 83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu S., et al. , Phosphorylation of tau and α-synuclein induced neurodegeneration in MPTP mouse model of Parkinson’s disease. Neuropsychiatr. Dis. Treat. 16, 651–663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C., Telpoukhovskaia M. A., Bahr B. A., Chen X., Gan L., Endo-lysosomal dysfunction: A converging mechanism in neurodegenerative diseases. Curr. Opin. Neurobiol. 48, 52–58 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Shacka J. J., Roth K. A., Zhang J., The autophagy-lysosomal degradation pathway: Role in neurodegenerative disease and therapy. Front Biosci. 13, 718–736 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Bourdenx M., et al. , Nanoparticles restore lysosomal acidification defects: Implications for Parkinson and other lysosomal-related diseases. Autophagy 12, 472–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallings R., Connor-Robson N., Wade-Martins R., LRRK2 interacts with the vacuolar-type H+-ATPase pump a1 subunit to regulate lysosomal function. Hum. Mol. Genet. 28, 2696–2710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung C. Y.-S., et al. , Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 15, 776–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernandez I., et al. , A farnesyltransferase inhibitor activates lysosomes and reduces tau pathology in mice with tauopathy. Sci. Transl. Med. 11, eaat3005. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pardridge W. M., Drug transport across the blood-brain barrier. J. Cereb. Blood Flow Metab. 32, 1959–1972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wager T. T., Hou X., Verhoest P. R., Villalobos A., Moving beyond rules: The development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 1, 435–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramanathan S., et al. , Theranostic applications of nanoparticles in neurodegenerative disorders. Int. J. Nanomedicine 13, 5561–5576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaur D., et al. , Genetic or pharmacological iron chelation prevents MPTP-induced neurotoxicity in vivo: A novel therapy for Parkinson’s disease. Neuron 37, 899–909 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Finkelstein D. I., et al. , Clioquinol improves cognitive, motor function, and microanatomy of the alpha-synuclein hA53T transgenic mice. ACS Chem. Neurosci. 7, 119–129 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Lei P., et al. , Clioquinol rescues Parkinsonism and dementia phenotypes of the tau knockout mouse. Neurobiol. Dis. 81, 168–175 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Sandor C., et al. , Transcriptomic profiling of purified patient-derived dopamine neurons identifies convergent perturbations and therapeutics for Parkinson’s disease. Hum. Mol. Genet. 26, 552–566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bareggi S. R., Cornelli U., Clioquinol: Review of its mechanisms of action and clinical uses in neurodegenerative disorders. CNS Neurosci. Ther. 18, 41–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Brien C. E., Bonanno L., Zhang H., Wyss-Coray T., Beclin 1 regulates neuronal transforming growth factor-β signaling by mediating recycling of the type I receptor ALK5. Mol. Neurodegener. 10, 69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spencer B., et al. , Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in α-synuclein models of Parkinson’s and Lewy body diseases. J. Neurosci. 29, 13578–13588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pickford F., et al. , The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J. Clin. Invest. 118, 2190–2199 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee H. G., Ueda M., Zhu X., Perry G., Smith M. A., Ectopic expression of phospho-Smad2 in Alzheimer’s disease: Uncoupling of the transforming growth factor-β pathway? J. Neurosci. Res. 84, 1856–1861 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Ueberham U., Ueberham E., Gruschka H., Arendt T., Altered subcellular location of phosphorylated Smads in Alzheimer’s disease. Eur. J. Neurosci. 24, 2327–2334 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.