Significance
Hosts often target the relatively conserved regions in rapidly mutating retroviruses to inhibit their replication. One of these regions is called a primer binding site (PBS), which has to be complementary to the host tRNA to initiate reverse transcription. By analyzing endogenous retroviral elements, we found that host cells use this sequence as a target in efforts to block the expression of viral elements. A specific type of zinc finger protein targets the PBS in a host genome, which not only inhibits the transcription of endogenous viruses but also inhibits the replication of exogenous retroviruses with the same PBS. Thus, our study sheds light on a strategy for searching for host restriction factors targeting retroviruses.
Keywords: KRAB-ZFPs, PBS-Lys, ERVs
Abstract
Eukaryotic genomes harbor sequences derived from the chromosomal integration of ancient viruses, such as endogenous retroviruses (ERVs), which comprise 8% of the human genome. Like exogenous retroviruses, ERVs retain many common functional elements, including the corresponding DNA sequences of transfer RNA (tRNA) primer binding sites (PBSs), which are utilized for reverse transcription initiation by exogenous retroviruses. Here, through a medium-scale analysis of PBS loci positioned within ERVs, coupled with chromatin immunoprecipitation sequencing (ChIP-seq) of Kruppel-associated box zinc finger proteins (KRAB-ZFPs), we identified multiple ZFPs that specifically bind to different PBS loci. Among these, we focused on PBS-Lys, which is utilized by HIV-1, and identified its specific binding proteins to be mouse ZFP961 and human ZNF417/ZNF587. We found that these proteins not only repress ERV transcription but also inhibit retrovirus integration and transcription. Disruption of these ZFPs rendered cells more susceptible to HIV-1 infection. Thus, our research provides a methodology for identifying potential host factors that target retroviruses by ERVs.
A retrovirus uses reverse transcriptase to copy its single-stranded RNA (ssRNA) genome into a double-stranded proviral DNA for replication when infecting host cells. During the initiation phase of retroviral replication, host transfer RNAs (tRNAs), adopted as a primer for retroviral reverse transcriptase, are partially unfolded from their native structure to facilitate the 18 nucleotides at their 3′ termini to be base paired to a specific complementary sequence, termed the primer binding site (PBS), on the viral genomic RNA (1). PBS sequences differ between various retrovirus families (2) reflecting the strong tendency of each retrovirus type to use one specific tRNA for reverse transcriptase priming. For instance, murine leukemia virus (MLV) utilizes tRNAPro (PBS-proline [PBS-Pro]) (3), Mason-Pfizer monkey viruses use tRNA1,2Lys (PBS-Lys1,2) (4), and HIV takes advantage of tRNA3Lys (PBS-Lys3) (5). Extensive biochemical and structural analyses of the reverse transcriptase initiation complex have characterized how precise matching of PBS sequences with their tRNA molecules enables successful priming of reverse transcription (6).
Retroviruses have been infecting mammals for millions of years and have been incorporated into the germline to reshape mammalian genomes as endogenous retroviruses (ERVs) that account for about 8% of the host genomic DNA (7, 8). These ERVs, retaining most viral elements including long terminal repeats (LTRs) and PBS sites, are inactivated by mutations and have been genetically fixed in the genomes of their host species (9). Although incapable of replication, ERVs can still function as transcriptional regulatory elements, for example, in activating the expression of neighboring genes, and collectively exert profound influence on host genome stability (10). Thus, ERVs must be kept in check to prevent their widespread activation and subsequent deleterious impacts on genome stability.
Among mammals, one host cell strategy for coping with ERVs is based on a large family of transcription factors known as Kruppel-associated box zinc finger proteins (KRAB-ZFPs). Specific members of this protein family can recognize PBS sites and initiate heterochromatic silencing by recruiting the KRAB domain binding corepressor KAP1 (also known as TRIM28), the histone methyltransferase SETDB1, and heterochromatin protein 1 (HP1) (11, 12). Perhaps the best characterized ZFP is mouse ZFP809, which can directly recognize PBS-Pro sites in ERVs or integrated proviral DNA, recruiting KAP1 to repress both ERVs (13) and the Moloney murine leukemia virus (MmLV) (14, 15). Therefore, ZFP809 has been considered a restriction factor for MmLV infection. KAP1 has also been reported as a corepressor that mediates the restriction of the PBS-Lys–utilizing retrovirus HIV-1 (16) and forms a repressive complex with PBS-Lys DNA (17). However, the factor directly responsible for recognition of PBS-Lys used by an integrated retroviral DNA is still unknown.
Here, a ChIP-seq analysis led to the identification of a PBS-Lys binding protein, mouse ZFP961, which recruits KAP1 and functions to maintain the heterochromatin state of ERVs. We also identified two human PBS-Lys binding proteins, ZNF417 and ZNF587, by systematically analyzing PBS loci within human ERVs in combination with KRAB-ZFP ChIP-seq data. We further demonstrated that the expression of these ZFPs promotes host cell resistance to PBS-Lys–utilizing HIV-1 pseudovirus. Conversely, knockout of these host proteins increased viral infectivity. Finally, we demonstrate that these ZFPs may influence viral transcription and integration. Thus, our study revealed that mZFP961 and hZNF417/ZNF587 have evolved as species-specific factors for retroviral silencing that help to protect host cells against retroviruses.
Results
Genome-Wide Mapping of ZFP961 Binding Sites Enriched at ERVs Subgroup K.
By analyzing public ChIP-seq data (GSE115287) (18), we inferred that ZFP961 was a potential PBS-Lys binding protein. To investigate the potential repressive activity of ZFP961 toward PBS-Lys, we first used CRISPR-Cas9 to generate a mouse embryonic stem cell (mESC) line harboring a Zfp961 floxed allele that also contained a C-terminal green fluorescent protein (GFP) tag in frame (hereafter referred to as Zfp961-GFPflox/flox) (SI Appendix, Fig. S1A). Since the mESCs that we used for gene editing were derived from Rosa26-CreERT2 mice, treatment of these mESCs with 4-hydroxytamoxifen (4-OHT) enabled the selective depletion of Zfp961 alleles and a subsequent loss of the ZFP961-GFP protein (SI Appendix, Fig. S1 B and C). We were thus able to use this system to determine the consequences from loss of ZFP961 and to experimentally catalog the ZFP961 binding sites across the genome.
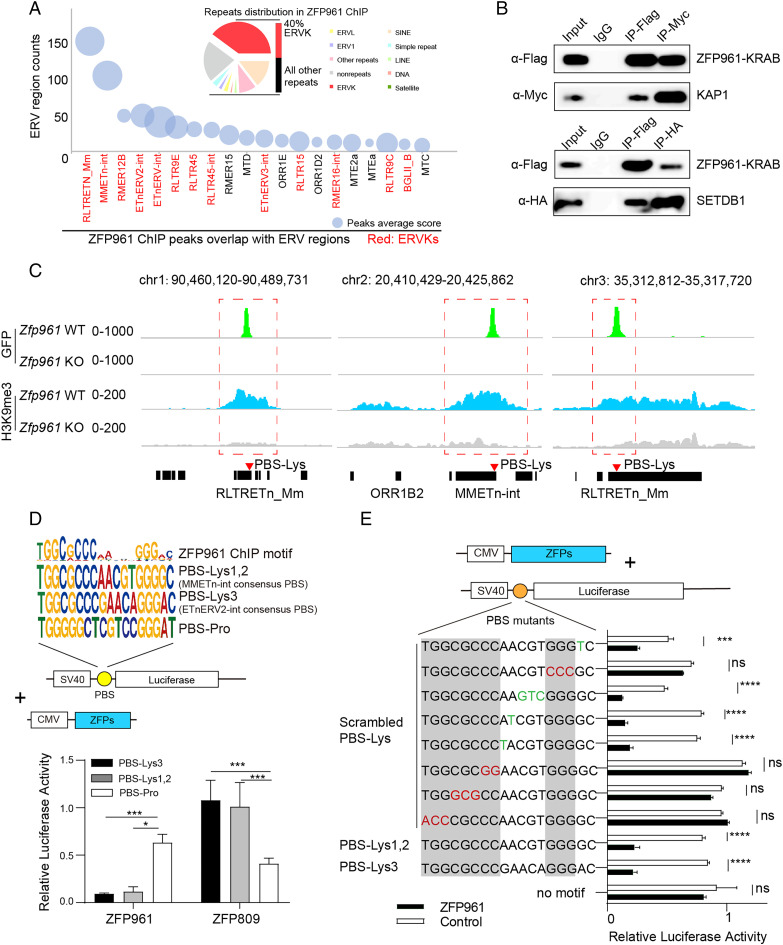
We performed ChIP-seq with anti-GFP antibodies on Zfp961-GFPflox/flox mESCs. Compared to the control Zfp961 knockout (KO) mESCs (treatment of Zfp961-GFPflox/flox ESCs with 4-OHT), we found a total of 413 high-confidence ZFP961 binding peaks [peak score > 500 in model-based analysis of ChIP-seq (MACS) (19)] located with strong enrichment at PBS-Lys–containing ERV subgroup K (ERVK) repeats (according to University of California Santa Cruz RepeatMasker nomenclature) (Fig. 1A). While these comprise only ∼5% of the total repetitive regions in the mouse genome (13), 40% (165 out of 413) of the ZFP961 binding peaks in repeats were of this type (Fig. 1A), clearly highlighting a binding preference of ZFP961 for ERVK regions, including ERVK subfamilies such as RLTRETn_Mm, MMETn-int, and ETnERV2-int (Fig. 1A).
Fig. 1.
ZFP961 preferentially targets PBS-Lys–containing ERVK for repression. (A) The repeat content of ZFP961 ChIP-seq peaks as determined by genomic overlap with RepeatMasker annotation (Upper), and plot of ZFP961 peak numbers and scores at ERV regions (Lower). ERVK regions are marked in red. Bubbles are sized (area) according to average scores of ChIP-seq peaks. (B) Coimmunoprecipitation and Western blotting for Flag-tagged ZFP961-KRAB domain with myc-KAP1 and HA-SETDB1. (C) ChIP-seq signals of ZFP961 and H3K9me3 at three representative ERVK regions. (D) ZFP961 ChIP-seq motifs compared with PBS-Lys1,2, PBS-Lys3, and PBS-Pro (Upper), and relative luciferase activity of HEK293T cells overexpressing ZFP961 or ZFP809 and a SV40 promoter–driven luciferase plasmid containing one of three distinct 18-nt tRNA PBSs, namely, PBS-Lys1,2, PBS-Lys3, or PBS-Pro (control) (Lower). Consensus ZFP961 target motifs were analyzed from high-confidence peaks (peak score >500 in MACS). t test: Error bars indicate SD; *P < 0.05, ***P < 0.001; n = 3. (E) Relative luciferase activity of HEK293T cells overexpressing ZFP961 or control plasmid (pCDNA3.1+) and SV40 promoter–driven reporter containing the PBS-Lys site with the indicated mutations highlighted in red or green. The nucleotides where PBS-Lys3 and PBS-Lys1,2 were identical are marked in gray. t test: Error bars indicate SD; ***P < 0.001, ****P < 0.0001, ns (no significant), P > 0.05; n = 3.
To examine whether ZFP961 and its potential corepressors KAP1 (TRIM28)/SETDB1 physically interact in the vicinity of these targets regions, we overexpressed Flag-tagged ZFP961-KRAB domain (amino acids 1 through 107) with Myc-KAP1 or HA-SETDB1 in HEK293T cells and performed coimmunoprecipitation (co-IP) with α-Flag, α-Myc, or α-HA antibodies, respectively. Western blots confirmed an interaction between ZFP961-KRAB and KAP1/SETDB1 (Fig. 1B). Next, we conducted an integrative analysis to test the cooccupancy of ZFP961, KAP1, and SETDB1, as well as the influence of H3K9me3 and H3K27ac on such loci, at sites in repetitive or nonrepetitive regions. ZFP961 binding was positively correlated with both KAP1 and SETDB1 signals, and there was also an enrichment of H3K9me3 at apparent ZFP961/KAP1/SETDB1 cooccupied sites in repetitive regions (SI Appendix, Fig. S2A). Among the binding sites of ZFP961 in nonrepetitive regions, there was no significant correlation between ZFP961 binding and H3K9me3/H3K27ac signals (SI Appendix, Fig. S2A). In addition, by calculating the density of ChIP-seq reads for H3K9me3 and H3K27ac at different distances from ZFP961 peaks using BEDTools, we found that KO of Zfp961 mainly affected the levels of histone marks closest to ZFP961 binding sites (distance from ZFP961 peaks <1 kb) (SI Appendix, Fig. S2B). H3K9me3 signals were decreased upon depletion of Zfp961 (Fig. 1C and SI Appendix, Fig. S2 A–C), whereas H3K27ac signals were elevated (SI Appendix, Fig. S2 A–C). Interestingly, in addition to these ZFP961 binding sites, we noticed additional KAP1/SETDB1 peaks and broad H3K9me3 marks at some ERVK loci, indicating that multiple KRAB-ZFPs may be responsible for H3K9me3 modifications (SI Appendix, Fig. S2A).
Thus, ZFP961 is a heterochromatin repressor, partnering with KAP1 and SETDB1 to result in pronounced H3K9me3 enrichment and inhibition of H3K27ac at repetitive regions, particularly at those regions characterized by the presence of PBS-Lys sites.
ZFP961 Directly Targets PBS-Lys and Represses Transcription.
We further identified the ZFP961 binding motif in the mouse genome using motif analysis of our ChIP-seq dataset. This analysis revealed a highly significant binding motif comprising 18 base pairs that resembled PBS-Lys (Fig. 1 D, Upper). Interestingly, the target motif showed a lower binding score at the positions where PBS-Lys3 and PBS-Lys1,2 were different, indicating that ZFP961 may have lower DNA binding specificity at this 5-nucleotide (nt) region. ZFP961 could apparently tolerate a variety of diverse DNA sequences present in this region, including GAACA in PBS-Lys3, AACGT in PBS-Lys1,2, and even other sequences (Fig. 1 D, Upper). For example, the mouse ERVK family repeats MMETn-int (PBS-Lys1,2) and ETnERV2-int (PBS-Lys3) are known to use different PBS-Lys subtypes, and all of these repeats were colocalized with ZFP961 in our ChIP-seq dataset (SI Appendix, Fig. S2A).
To verify the binding motifs of ZFP961, we performed luciferase reporter assays using an SV40 promoter–driven luciferase plasmid containing different PBS sequences downstream (Fig. 1 D, Lower). Expression of ZFP961 in HEK293T cells potently repressed reporters with PBS-Lys3 or PBS-Lys1,2, but not with PBS-Pro. Expression of ZFP809 specifically repressed the reporters containing PBS-Pro sequences, consistent with our previous observations of ZFP809 binding and repression of PBS-Pro (13) (Fig. 1 D, Lower). Likewise, ZFP961 repressed reporters with a native ETn LTR/promoter (PBS-Lys) or other types of LTR with PBS-Lys (SI Appendix, Fig. S3A). However, mutation of other types of LTR/PBS (e.g., native intracisternal A-particle (IAP) LTR containing PBS-Phe) or removal of PBS-Lys in promoters abolished the repression activity of ZFP961, compared with control groups (expression of pCDNA3.1+ empty vectors) (SI Appendix, Fig. S3A).
Furthermore, consistent with the aforementioned differential binding specificity across the genome observed from our ChIP-seq profiling, we found that scrambling of the PBS-Lys sequences significantly affected the extent of transcriptional repression of ZFP961 for these various luciferase reporters, with the exception of the middle 5 nt DNA that was discrepant between PBS-Lys3 and PBS-Lys1,2 (Fig. 1E). Also, repression of the reporters was prevented upon deletion of two or three C2H2 zinc fingers (ZFs) of ZFP961 (starting from the third ZF) (SI Appendix, Fig. S3B). Taken together, these results indicate that ZFP961 interacts with PBS-Lys and represses the transcriptional activity of ERVK LTRs by targeting its binding motif.
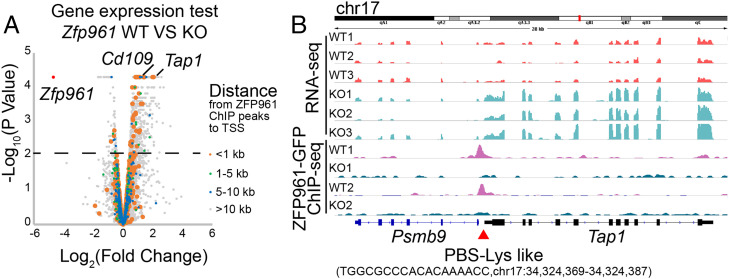
To investigate the functional consequences in the absence of ZFP961 in mESCs, we treated Zfp961-GFPflox/flox cells with 4-OHT to deplete Zfp961 alleles and then performed RNA-seq on three independent Zfp961-GFPflox/flox mESC clones before and after Cre-mediated excision. Strikingly, we found only one gene, Zfp961, was significantly decreased after 4-OHT treatment (Fig. 2A, SI Appendix, Fig. S4A, and Dataset S1). We observed some genes such as Tap1 and Cd109 were up-regulated in KO cells with PBS-Lys–like sites in their promoter regions (distance from the transcription start site [TSS] to ZFP961 peaks <1 kb) that can be directly recognized and targeted by ZFP961 (Fig. 2B and SI Appendix, Fig. S4B). We also measured the expression levels of repetitive elements in Zfp961 wild-type (WT) and KO mESCs and observed remarkable reactivation of one ETnERV2-int locus (chromosome X [chrX]: 89,238,230 to 89,240,479) after Zfp961 KO (SI Appendix, Fig. S4C). This locus (∼2,000 bp in size) contains one PBS-Lys site 382 bp upstream and was shorter than a canonical ETnERV2-int loci (∼7,000 bp). Overlapping with the consensus ETnERV-2-int sequence from the Dfam library, this locus shared some similarity with only 1,083 bp to 2,243 bp of the consensus ETnERV2-int site (part of the gag element) and thus may lack elements essential for other ZFP binding and repression (SI Appendix, Fig. S4D).
Fig. 2.
ZPF961 directly targets PBS-Lys for transcriptional repression. (A) Volcano plot of gene expression in Zfp961 KO mESCs compared with WT mESCs. The colored dots represent genes at different distances from their TSS to ZFP961 peaks. (B) Tracks of RNA-seq and ZFP961-GFP ChIP-seq signals in Zfp961 KO or WT mESCs at the Tap1 locus.
Taken together, ZFP961 specifically bound at PBS-Lys sites and inhibited the transcription of PBS-Lys–containing ERV loci and genes
ZFP961 Represses PBS-Lys–Utilizing Viral Infection.
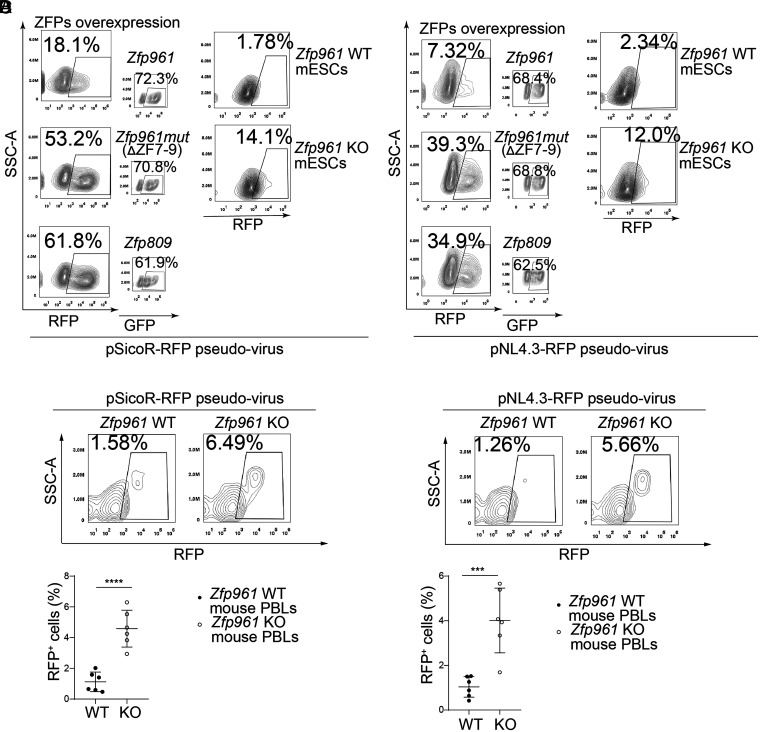
We next sought to confirm the repressive activity of ZFP961 toward a PBS-Lys–utilizing retrovirus. HIV-1, an exogenous lentivirus, is known to use lysine tRNA as a primer to initiate DNA synthesis for reverse transcription (5). We generated RFP reporter HIV-1 replication-defective viral particles (pSicoR-RFP pseudovirus) based on pSicoR-RFP with POL/GAG and vesicular stomatitis virus G protein (VSVG) expression packaging vectors (SI Appendix, Fig. S5A). RFP was thus constitutively expressed under the control of the cytomegalovirus (CMV) promoter, which allowed the detection of integrated virus, regardless of the transcriptional activity of the virus LTR (20). Moreover, we also produced HIV-1 pseudoviral particles by transfecting an LTR-driven RFP-luciferase reporter vector that contained functional POL/GAG and defective ENV (pNL4.3-RFP) alongside a VSVG packaging vector (SI Appendix, Fig. S5A). After transfection with a GFP-fused ZFP961, ZFP961-mutant (ZFP961mut, deletion of amino acids 310 to 393, ΔZFs 7 to 9), or ZFP809, for 6 to 8 h, HEK239T cells were infected with pSicoR-RFP pseudoviruses (without Pol/Gag) or pNL4.3-RFP pseudoviruses (with Pol/Gag) for the next 24 to 48 h, and these cells were then analyzed by flow cytometry (SI Appendix, Fig. S5A). Thus, we were able to measure both the viral infectivity and ZFP transfection efficiency by counting the RFP and GFP cell numbers simultaneously.
After confirming that the transfection efficiencies for each of the groups ranged between 50 and 70% (Fig. 3 A and B), we found that ZFP961 overexpression dramatically inhibited the infectivity of both pSicoR-RFP pseudovirus and pNL4.3-RFP pseudovirus as compared to the ZFP961mut or ZFP809 groups (Fig. 3 A and B). We also tested the endogenous repression effect of ZFP961 on pseudovirus infectivity with Zfp961-GFPflox/flox mESCs (SI Appendix, Fig. S5A). Zfp961-GFPflox/flox mESCs were treated with 4-OHT (Zfp961 KO group) or EtOH (control group) for 48 h and were then subjected to virus infection (SI Appendix, Fig. S5A). Compared to control cells, KO of Zfp961 resulted in a four- to sixfold increase in the viral infectivity (Fig. 3 A and B).
Fig. 3.
ZFP961 potentially inhibits HIV-1 (PBS-Lys) pseudovirus infection. (A and B) FACS (fluorescence-activated cell sorting) analysis to calculate pSicoR-RFP pseudovirus (A) and pNL4.3-RFP pseudovirus (B) infection rates in HEK293T cells overexpressing GFP-tagged ZFP961, ZFP961mut, or ZFP809, as well as in Zfp961 WT and KO mESCs. (C and D) FACS analysis (Upper) and statistical plot (Lower) for the pSicoR-RFP pseudovirus (A) and pNL4.3-RFP pseudovirus (B) infection rate in mouse PBLs before and after Zfp961 KO. t test: Error bars indicate SD; ***P < 0.001, ****P < 0.0001; n = 6.
We also sought to test whether ZFP961 could affect pseudovirus infection in mice. We therefore generated Zfp961 KO mice using CRISPR-Cas9 (SI Appendix, Fig. S5 B and C). We simulated viral infection in mouse lymphocytes using ex vivo pSicoR-RFP pseudovirus and pNL4.3-RFP pseudovirus infection of mouse peripheral blood lymphocytes (PBLs) for 48 h (SI Appendix, Fig. S5A). Flow cytometry data showed that ZFP961 deletion led to an approximately fourfold increase (pSicoR-RFP 1.58 to 6.49%, pNL4.3-RFP 1.26 to 5.66%) in the RFP+ cell ratios, indicating that KO mice were significantly more susceptible to viral infection (Fig. 3 C and D).
Identification of Human KRAB-ZFPs Responsible for PBS Binding.
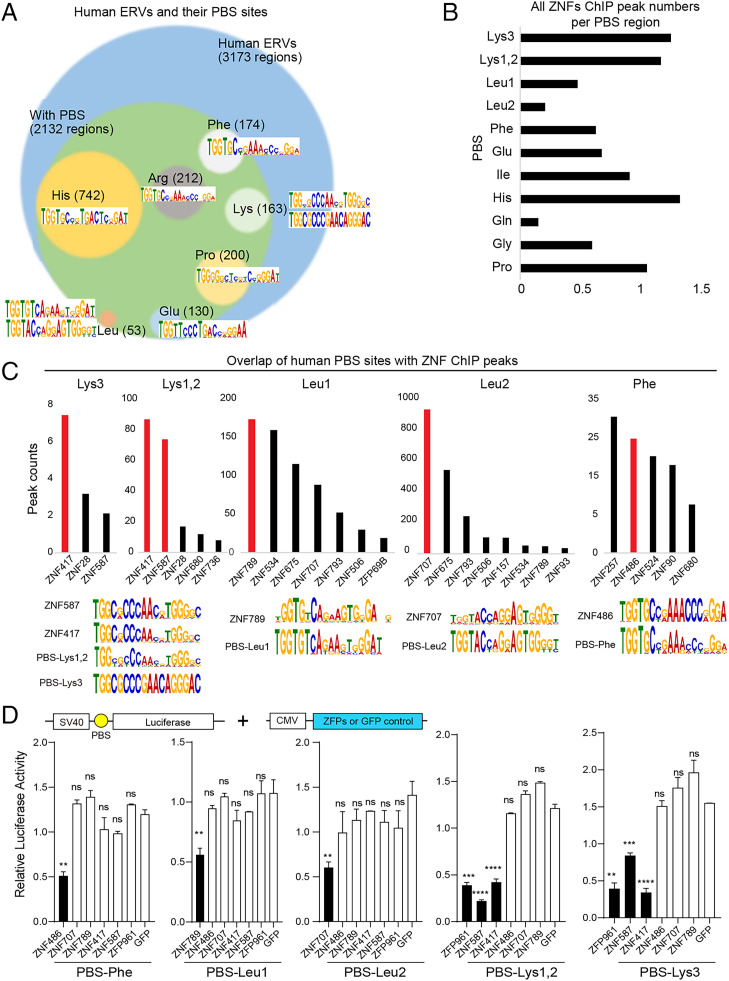
We next expanded our search for PBS-Lys binding ZFPs to the human genome. We extracted various PBS sequences from human ERV published data (21) and calculated PBS motif by multiple enriched motifs for motif elicitation (MEME) (SI Appendix, SI Materials and Methods). Then we can identify PBS motif–containing regions in the human genome by find individual motif occurrences (FIMO) (SI Appendix, SI Materials and Methods). These regions were overlapped with 3,173 human ERV loci. Among them, we found 2,132 PBS sequences in the human genome, including 742 PBS-His, 212 PBS-Arg, 200 PBS-Pro, 174 PBS-Phe, 163 PBS-Lys, 130 PBS-Glu, 53 PBS-Leu, and 458 other types (Gly, Ile, Met, Asn, Gln, Ser, Thr, Val, Trp, and Tyr) (Fig. 4A and Dataset S2). We then integrated and analyzed these PBS-containing regions with published ChIP-seq data (GSE78099 and GSE76496) from 242 KRAB-ZFPs using BEDTools (22, 23). We found that many KRAB-ZFPs were enriched in these PBS-containing regions (Fig. 4B and Dataset S2). Ranking based on peak numbers for individual ZFPs of each PBS type, accompanied by comparisons of each ZFP binding motif with each PBS consensus sequence, identified several candidate ZNF-PBS pairs, including ZNF417/ZNF587 to PBS-Lys, ZNF789/ZNF707 to PBS-Leu, and ZNF486 to PBS-Phe (Fig. 4C). We also confirmed that these KRAB-ZFPs induced silencing of a SV40 promoter–driven luciferase reporter by targeting their corresponding PBS sequences (Fig. 4D).
Fig. 4.
Identification of KRAB-ZFPs targeting human PBS sequences. (A) Human ERVs and their PBS sequences. The circles representing each type of PBS in the human genome are sized (area) according to their relative abundance among all PBS loci. (B) Histogram of the average human ZNF peak numbers per PBS from an analysis of public human ZNF ChIP-seq data (22, 23). (C) Ranking based on the numbers of each ZNF ChIP-seq peaks for each PBS type (Upper), and the consensus motif sequences of ZNFs and PBS loci (Lower). (D) Relative luciferase activity of HEK293T cells overexpressing candidate ZNFs individually, alongside plasmids for SV40 promoter–driven luciferase reporters that contain the corresponding PBS. t test: Error bars indicate SD; **P < 0.01, ***P < 0.001, ****P < 0.0001, ns (no significant), P > 0.05; n = 3.
Human ZNF417 and ZNF587 Target PBS-Lys for Repression.
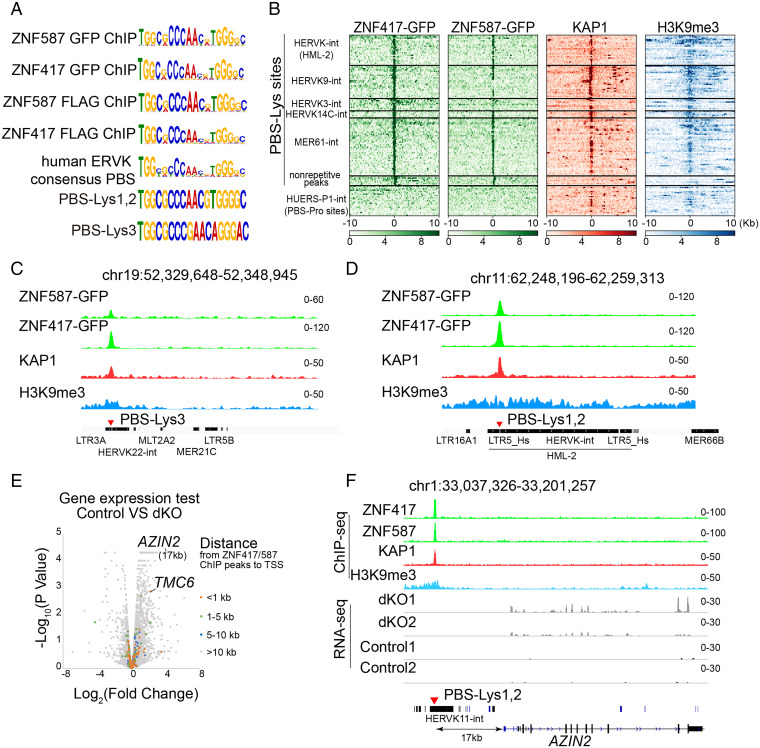
Next, we focused on ZNF417 and ZNF587, the PBS-Lys binding candidate proteins in humans. We found an incredibly high degree of similarity in the amino acid sequences (∼98%) between these two ZFPs. However, neither ZNF417 nor ZNF587 show obvious homology with mouse ZFP961 (SI Appendix, Fig. S6A). We also overexpressed a Flag-tagged KRAB domain (amino acids 15 to 88) of ZNF417/ZNF587 with Myc-KAP1 or HA-SETDB1 in HEK293T cells, and performed co-IP with α-Flag, α-Myc, or α-HA antibodies. Western blotting of co-IP products showed that the KRAB domain of ZNF417/ZNF587 physically interacted with KAP1/SETDB1 (SI Appendix, Fig. S6B), suggesting that ZNF417/ZNF587 recruits the corepressors KAP1 and SETDB1 via the KRAB domain to assemble into a transcriptional repression complex. As expected, ChIP-seq experiments using GFP antibodies in ZNF417/ZNF587-GFP overexpressing HEK293T cells revealed that the binding motifs of both ZNF417 and ZNF587 highly resembled PBS-Lys, consistent with PBS-Lys–utilizing human ERVK loci (Fig. 5A).
Fig. 5.
Human ZNF417 and ZNF587 directly target PBS-Lys–type ERVs. (A) ChIP-seq motifs of ZNF417 and ZNF587 compared with the human ERVK consensus PBS, PBS-Lys1,2, and PBS-Lys3. Consensus sequences of the GFP-tagged ZNF417/ZNF587 target motif were analyzed from our ChIP-seq data. FLAG-tagged ZNF417/ZNF587 ChIP-seq data were adopted from Imbeault et al. (22) and Schmitges et al. (23). (B) Heatmaps of ZNF417/ZNF587, KAP1, and H3K9me3 ChIP-seq data at ERVK-associated PBS-Lys sites and at strong nonrepetitive ZNF417/ZNF587 ChIP-seq peak regions. The 20-kb regions are displayed with the PBS-Lys sequence or with nonrepetitive peaks positioned at the center of the peak regions. PBS-Pro site–containing HUERS-P1-int are shown as control. (C and D) ChIP-seq signals for ZNF417, ZNF587, KAP1, and H3K9me3 at representative genome regions containing PBS-Lys3–type (C) or PBS-Lys1,2–type (D) human ERVKs. (E) Volcano plot of gene expression in ZNF417 ZNF587 dKO cells compared with control K562 cells. The colored dots represent genes at different distances from their TSS to ZFP961 peaks. (F) RNA-seq and ChIP-seq signals in ZNF417 ZNF587 dKO or control K562 cells at the AZIN2 locus.
We also conducted a genome-wide analysis examining ZNF417/ZNF587 and KAP1 cooccupancy at PBS-Lys sites. Heatmaps for repetitive and nonrepetitive regions using our ZNF417 and ZNF587 ChIP-seq data and public KAP1 (SRR3178875) and H3K9me3 (SRR8983692) ChIP-seq data showed that binding of ZNF417 and ZNF587 in human ERVK regions was strongly correlated with KAP1 binding and with the presence of H3K9me3 (Fig. 5B). No such correlations were evident for nonrepetitive regions (Fig. 5B). These genome-scale results indicate that the ZNF417/ZNF587-KAP1 complex executes its repression activity specifically at heterochromatic human ERVK regions. Moreover, we found that ZNF587 preferentially bound PBS-Lys1,2–utilizing human ERVKs (over PBS-Lys3–utilizing human ERVK), whereas ZNF417 bound strongly to both types of human ERVKs (with no significant bias) (Fig. 5 C and D). To confirm the binding preferences of ZNF417/ZNF587 toward PBS-Lys1,2 and PBS-Lys3, we cotransfected SV40 promoter–driven luciferase reporter vectors containing PBS-Lys1,2 or PBS-Lys3 sites with different amounts of ZNF417 and ZNF587 plasmids (ranging from 0 to 500 ng) in HEK239T cells, respectively. The results of these luciferase reporter assays again highlighted that ZNF417 but not ZNF587 preferred to bind to PBS-Lys3–utilizing human ERVKs (SI Appendix, Fig. S6C). Furthermore, we compared the amino acid sequences of each ZF between ZNF417 and ZNF587. Among the 12 ZF repeats, slight differences could be observed in the 4th, 6th, 9th, 10th, and 11th ZFs (SI Appendix, Fig. S6D). To test whether these differences in the ZFs potentially influenced PBS-Lys binding bias between ZNF417 and ZNF587, we replaced those of ZNF587 with the corresponding ZFs of ZNF417, respectively (m4 C329Y, m6 H397Q, m9 H477N/S478C, m10 S495N, and m11 S530A) (SI Appendix, Fig. S6 D and E). Luciferase reporter assays showed that ZNF587 could strongly bind to both PBS-Lys1,2 and PBS-Lys3 when both the sixth and ninth ZFs were replaced, meaning it no longer had binding preferences (SI Appendix, Fig. S6E). The results were consistent with previous studies that the fingerprint amino acid residues at the −1, 2, 3, and 6 positions primarily made base-specific contacts (24). Thus, the binding preference differences between ZNF417 and ZNF587 toward PBS-Lys1,2 and PBS-Lys3 were related to the amino acid differences in their 6th and 9th ZFs.
Using public RNA-seq data of human preimplantation embryos (25), we found that the expression pattern of ZNF417/ZNF587 was mutually exclusive with that of the embryonic genome activation–restricted human ERVKs (SI Appendix, Fig. S7 A and B), suggesting that the repression of human ERVKs by these two ZNFs could exist in the process of preimplantation embryo development in vivo.
We also generated ZNF417 and ZNF587 double KO (dKO) K562 cell lines by disrupting C2H2 domains of the two ZNFs using CRISPR-Cas9 (SI Appendix, Fig. S7 C–E) and investigated the functional consequences by RNA-seq analysis (Fig. 5E and Dataset S3). Some of the genes up-regulated in dKO cells possessed human ERVK elements containing PBS-Lys sites near their promoters that could be directly targeted by ZNF417 and ZNF587, including TMC6 and AZIN2 (Fig. 5 E and F). We also measured the expression levels of repetitive elements in ZNF417 ZNF587 dKO cells. However, we found no remarkable reactivation of human ERVK elements (SI Appendix, Fig. S7F), probably due to the large-scale heterochromatic repression and multiple ZNFs targeting in these regions.
Taken together, these genome-wide results indicate that ZNF417 and ZNF587 function by targeting and binding PBS-Lys sequences and thereby recruiting KAP1/SETDB1 to these sites to establish repressive H3K9me3 modifications.
Human ZNF417 and ZNF587 Are Potential Repressors of PBS-Lys–Utilizing Pseudoviral Infection.
In a typical retrovirus infection cycle, the retrovirus will recruit a specific cognate tRNA to prime its reverse transcription into DNA using its own reverse transcriptase (26). To explore the contribution of specific PBS sites to retroviral infectivity, we mutated the PBS-Lys3 sequence of a GFP-labeled HIV-1 pseudovirus variant into other PBS types (PBS-Pro, PBS-Phe, and the PBS-Lys3 mutant) and then measured the viral titers in HEK293T cells 48 h postinfection by monitoring GFP signals. Both fluorescence imaging and quantification of viral titers revealed dramatically reduced viral infectivity when PBS-Lys3 sequences were mutated to other sequences (SI Appendix, Fig. S8A). These results were consistent with previous studies (27, 28) where mutation of HIV-1 with altered PBS sites corresponding to other tRNA species greatly reduced viral replication efficiency. Previous studies (29) have also shown that not only the viral genome, but also viral proteins, were involved in the selection of correct tRNAs for packaging into an assembling virion.
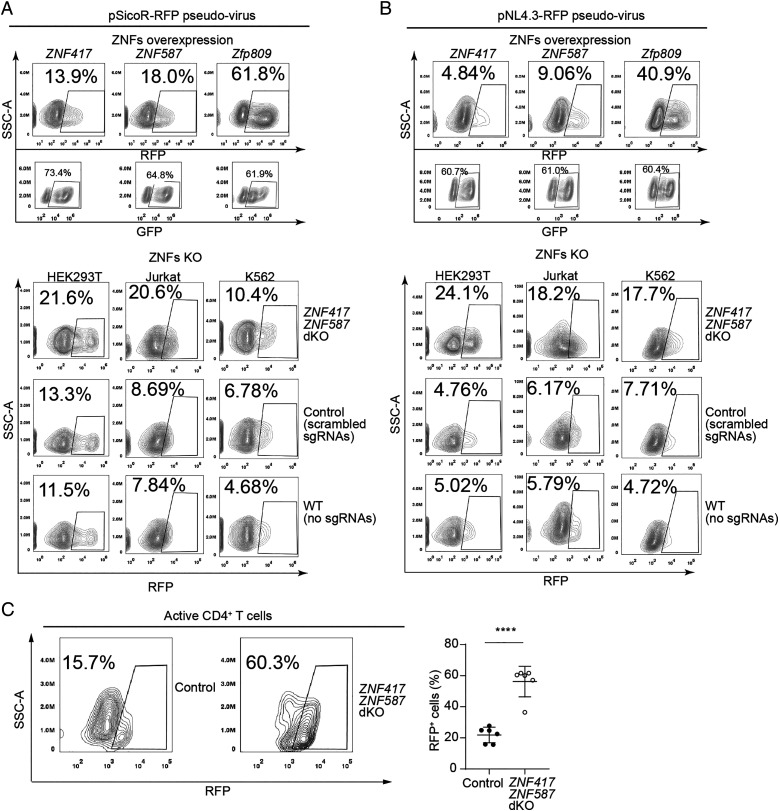
Based on our findings about binding between ZNF417/ZNF587 and PBS-Lys sequences, we next tested whether these two ZFPs may influence PBS-Lys–utilizing virus infection. We transfected GFP-fused ZNF417/ZNF587 into HEK293T cells followed by pSicoR-RFP pseudovirus or pNL4.3-RFP pseudovirus infection. In addition, ZNF417 ZNF587 dKO cells, control cells (scrambled single-guide RNAs [sgRNAs]) (SI Appendix, Fig. S8 B–D), and WT cells (no sgRNAs) were all also infected with pSicoR-RFP pseudovirus or pNL4.3-RFP pseudovirus. On the one hand, flow cytometry analysis counting RFP+ cell ratios revealed that ZNF417/ZNF587 overexpression significantly reduced viral infectivity compared to control cells overexpressing ZFP809 (Fig. 6 A and B, Upper). On the other hand, ZNF417/ZNF587 KO resulted in increased viral infectivity in multiple cell lines, including HEK293T, Jurkat, and K562 cells (Fig. 6 A and B, Lower), compared with control cells or WT cells.
Fig. 6.
ZNF417 and ZNF587 potentially inhibit HIV-1 (PBS-Lys) pseudovirus infection in human cells. (A and B) FACS analysis to calculate pSicoR-RFP pseudovirus (A) and pNL4.3-RFP pseudovirus (B) viral infection rates in HEK293T cells overexpressing GFP-tagged ZNF417, ZNF587, or ZFP809 (Upper), and in ZNF417/ZNF587 dKO, scrambled sgRNA control, and WT cell lines including HEK293T cells, Jurkat cells, and K562 cells (Lower). (C) FACS analysis (Left) and statistical plot (Right) for the infection rate of pNL4.3-RFP pseudovirus in ZNF417 ZNF587 dKO and control human active CD4+ T cells. t test: Error bars indicate SD; ****P < 0.0001; n = 6.
We also validated the infectivity-restricting functions of ZNF417 and ZNF587 against HIV-1 pseudovirus in experiments with primary human immune cells isolated from peripheral blood. We sorted CD4+ T cells from human peripheral blood mononuclear cells for infectivity assays (SI Appendix, Fig. S9 A and B). After incubation with antibodies against CD3 and CD28 and simultaneous stimulation with human interleukin 2 (IL2), over 97.6% of CD4+ T cells were activated (CD4+ CD25+). These activated T cells were nucleofected with plasmids expressing Cas9 and sgRNAs targeting ZNF417 and ZNF587. After confirming ZNF417 ZNF587 dKO, these activated CD4+ CD25+ T cells were subjected to infection challenge with pNL4.3-RFP pseudovirus (SI Appendix, Fig. S9 A–D). The extent of virus infectivity was significantly higher in ZNF417 ZNF587 dKO compared to control cells (Fig. 6C). Thus, ZNF417 and ZNF587 impacted the activity of HIV-1 pseudovirus for primary human immune cells.
ZFP961, ZNF417, and ZNF587 Are Potent Restriction Factors That Repress PBS-Lys–Utilizing Pseudovirus Transcription and Integration.
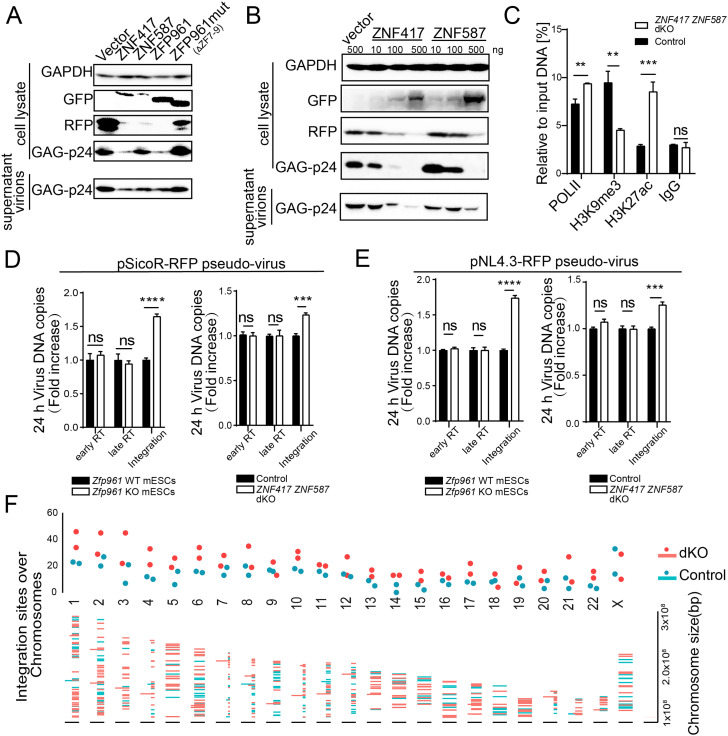
Once retroviruses integrate into a host genome, they can take advantage of host machineries to express viral proteins. Thus, we sought to test whether ZFP961, ZNF417, and ZNF587 affected HIV-1 pseudoviral protein expression. We cotransfected GFP-fused ZFP vectors with the HIV-1 plasmid pNL4.3-RFP containing Pol/Gag and VSVG vector in HEK293T cells, and validated HIV-1 expression by detecting HIV-1 plasmid–specific proteins in cell lysate and supernatant virions with specific antibodies as previously reported (30). Western blotting analysis showed that the expression of HIV-1 GAG protein p24 and the RFP reporter was dramatically repressed by ZFP961 and ZNF417 overexpression, respectively (Fig. 7A), while ZNF587 showed modest repression capacity in viral transcription compared with ZNF417 (Fig. 7 A and B). In addition, when overexpressing these GFP-fused ZFPs in pNL4.3-RFP pseudovirus-infected HEK293T cells, ChIP followed by qPCR revealed that ZFP961 and ZNF417 significantly bound to the LTR of HIV-1 (SI Appendix, Fig. S10A). Moreover, ZNF587 showed a relatively weak capacity for LTR binding compared with ZNF417 and ZFP961 (SI Appendix, Fig. S10A), consistent with our previous luciferase reporter assay results (SI Appendix, Fig. S6C) and ChIP-seq data (Fig. 5C). Luciferase reporter assays further confirmed that expression of these ZFPs in HEK293T cells could potently repress HIV-1 promoter–driven reporters with PBS-Lys but not PBS-Pro, results again highlighting that ZFP961 and ZNF417 but not ZNF587 preferred to target the PBS-Lys3 sites of HIV-1 to repress their transcription (SI Appendix, Fig. S10B). In addition, we performed ChIP in pNL4.3-RFP pseudovirus-infected HEK293T cells using antibodies against RNA polymerase II (POLII), H3K9me3, and H3K27ac. qPCR analysis showed a significant increase of POLII recruitment and H3K27ac levels as well as a dramatic loss of H3K9me3 signals at the HIV-1 LTR in ZNF417 ZNF587 dKO cells, as compared to the levels seen in control cells (Fig. 7C).
Fig. 7.
ZFP961, ZNF417, and ZNF587 limit HIV-1 pseudovirus transcription and integration. (A) Western blot to show the expression of HIV-1 GAG protein p24 and RFP in cell lysate and supernatants (virions) from HEK293T cells cotransfected with pNL4.3-RFP vector, VSVG, and GFP-fused ZNF417, ZNF587, ZFP961, ZFP961mut and control vector. (B) Western blot of HIV-1 GAG protein p24 and RFP expression in cell lysate and supernatants (virions) from HEK293T cells cotransfected with pNL4.3-RFP vector, VSVG, and different amounts of GFP-fused ZNF417 and ZNF587 plasmids, respectively. (C) ChIP assays with antibodies against POLII, H3K9me3, and H3K27ac in ZNF417 ZNF587 dKO or control (generated by scrambled sgRNAs) HEK293T cells, followed by qPCR using primers specific for HIV-1 LTR. Immunoglobulin G (IgG) was used as a control. t test: Error bars indicate SD; **P < 0.01, ***P < 0.001; n = 3. (D and E) Histograms of viral DNA copy numbers in Zfp961 WT and KO mESCs or ZNF417 ZNF587 dKO and control HEK293T cells 12 h postinfection with pSicoR-RFP pseudovirus (D) or pNL4.3-RFP pseudovirus (E). t test: Error bars indicate SD; ***P < 0.001, ****P < 0.0001, ns (no significant) P > 0.05; n = 3. (F) Plot of integration sites over chromosomes. HIV-1 integration events in ZNF417 ZNF587 dKO cells are marked in red, controls in blue.
During the infection process, virus genomic RNA needs to be reverse transcribed into double-stranded DNA first and then integrated into the host genome. Next, we sought to test whether these ZFPs affected virus reverse transcription or integration in experiments using the aforementioned pNL4.3-RFP pseudovirus and pSicoR-RFP pseudovirus. We used previously reported specific primers (31) to test the early and late reverse transcription events from 12 to 96 h postinfection (SI Appendix, Fig. S10C). Viral DNA integration, but not early or late reverse transcription in the first 24 h, was significantly increased upon Zfp961 depletion or ZNF417 ZNF587 dKO (Fig. 7 D and E). However, a significant increase of the first- and double-stranded DNA synthesis of pNL4.3-RFP pseudovirus upon ZFP KO (Zfp961 KO or ZNF417 ZNF587 dKO) was observed after 24 h (SI Appendix, Fig. S10D), when the virus had completed the first round of integration and started self-replication. This may have been partly due to the transcription repression of ZFPs on the viral DNA that had been integrated into the host genome in the first-round replication, thus affecting subsequent viral replication and integration. These results support the hypothesis that ZNF417, ZNF587, and ZFP961 function in viral DNA integration and transcription.
We also performed high-throughput sequencing to further explore the effect of HIV-1 integration inhibition by ZNFs on the host genome. After 48 h of pNL4.3-RFP pseudovirus infection, genomic DNA samples purified from ZNF417 ZNF587 dKO HEK293T cells, as well as control cells, were sheared and subjected to sequencing library preparation based on LTR-linker nested PCR (SI Appendix, SI Materials and Methods). HIV-1 integration site analysis revealed more HIV-1 integration events (dKO: 479 ± 76 vs. control: 271 ± 22) in ZNF417 ZNF587 dKO cells compared to the control group (Fig. 7F and Dataset S4). Additionally, we noticed that viruses moderately tended to integrate into the promoters of genes in ZNF417 ZNF587 dKO cells (SI Appendix, Fig. S10E). The presence of ZNF417 and ZNF587 may be helpful for host cells to maintain genomic stability during viral infection. Taken together, these results suggest that ZNF417, ZNF587, and ZFP961 may function as restriction factors that affect viral infection, not only through sequence-specific transcriptional repression, but also by selectively disrupting the ability of the virus to integrate into a host genome. These ZFPs may play an important role in loading unintegrated HIV-1 DNAs into core and linker histones to form extrachromosomal structures as previously reported (32).
Discussion
Our present study demonstrated that ZFP961 specifically bound to PBS-Lys sites to restrict both endogenous and exogenous retroviral activity by recruiting KAP1 to maintain the heterochromatin state in the mouse genome. Beyond confirming some recently published findings (18, 33), our study deepened our understanding about the species-specific evolution and virus infectivity-restricting functions of PBS-Lys binding proteins by identifying ZNF417/ZNF587 in the human genome.
ERVs are remnants of ancient exogenous retroviruses that have become integrated into host genomes. ERVs occupy a substantial fraction of many mammalian genomes, highlighting the extensive germline invasion by retroviruses (34). Many of these independent invasions occurred after the divergence of particular mammalian orders, so each mammalian order typically has its own distinct ERV composition and history (34). Since mammals have developed KRAB-ZFPs as a defense system to combat endogenous and exogenous retroviruses, each species has its own unique repertoire of KRAB-ZFPs. There is typically a small number of ZFPs shared between closely related species, with many more ZFPs being unique to a given species. Nevertheless, considering the fact that retroviruses must employ a host cell tRNA primer to accomplish their reverse transcription to complete their viral lifecycle, there is relatively strong conservation among retroviruses for each PBS, due to the limited number of tRNA types. Thus, this hypothesis correlates with our observations in PBS-Lys binding proteins, mouse ZFP961 and human ZNF417/ZNF587, which shared low homology (∼27%) in amino acid sequences but exerted the identical biological function by targeting the same DNA sequences. By summarizing the correlation between PBS usage of ERVs and their targeting ZFPs (Dataset S5), we did not find any mouse proteins sharing high homology in amino acid sequences with other human PBS binding proteins such as ZNF486, ZNF789, or ZNF707.
Our ChIP-seq motif calling results showed that both ZNF417 and ZNF587 binding motifs resembled PBS-Lys1,2 rather than PBS-Lys3 (Fig. 5A), suggesting that ZNF417/ZNF587 tended to suppress the PBS-Lys1,2–utilizing ERVs transcriptionally. However, the individual locus in ChIP-seq tracks and in vitro luciferase assays revealed that ZNF417 and ZNF587 have repression capacities on both PBS-Lys sites (Fig. 5 C and D and SI Appendix, Figs. S6C and S10B). We hypothesize that this inconsistency between motif calling and luciferase assay results was due to there being fewer PBS-Lys3 sites compared to PBS-Lys1,2 sites in the human genome. Moreover, compared with ZNF587, ZNF417 exhibited no preference in the binding ability of these two PBS-Lys sites in the repression assays (SI Appendix, Fig. S6C), likely due to the minor differences in amino acid sequences between ZNF417 and ZNF587 (SI Appendix, Fig. S6 D and E).
Previous studies reported that CCCH-ZFP ZAP acts as an HIV-1 restriction factor by directly binding to retroviral RNAs (35, 36). The HIV-1 PBS binding proteins ZNF417/ZNF587 identified in this study are C2H2-ZFPs that can only bind to double-stranded DNA. Moreover, our results showed that these two ZFPs affected the transcription of integrated HIV-1 DNA as well as inhibited the integration of the reverse-transcribed free HIV-1 DNA into the host genome, consistent with the previous finding that KAP1 inhibits HIV-1 integration (16). As KAP1 lacks DNA binding capacity, we speculated that ZNF417/ZNF587 functions together with KAP1 to restrict HIV-1 integration. From this, we suppose that the restriction of MLV activity by ZFP809 might be partially caused by inhibiting the integration of free MLV DNA. ZFP809 is identified as a PBS binding protein that restricts the infectivity of PBS-Pro–utilizing MLVs. However, no human PBS-Lys3 binding protein has been characterized thus far. Our study demonstrates that ZNF417 and ZNF587 represent the human PBS-Lys3 binding proteins that may limit HIV-1 transcription and integration.
Materials and Methods
Detailed information on mouse line maintenance and cell culture and generation of different KO mouse/cell lines is described in SI Appendix, Materials and Methods. Details of the experimental RNA-seq, ChIP-seq, co-IP, Western blotting, luciferase assays, viral production and infection, and quantification of virus integration are provided in SI Appendix, Materials and Methods. All animal experiments were conducted under an animal use protocol approved by the Tongji University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (2018YFA0108900 to P.Y. and 2019YFA0110000 to Y.W.), the National Natural Science Foundation of China (32070652 and 81871164 to P.Y., 32022024 and 31871486 to Y.W. and 31871305 to Z.C.), the Shuguang Program of the Shanghai Education Development Foundation and Shanghai Municipal Education Commission (20SG21 to P.Y.), and the Rising-Star Program of the Shanghai Science and Technology Commission (18QA1404400 to P.Y.). We thank LetPub (https://www.letpub.com/) for linguistic assistance and presubmission expert review.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All datasets that were generated in this study have been deposited in the Gene Expression Omnibus under accession nos. GSE156604, GSE174239, GSE174242, and GSE183046. All other study data are included in the article and/or supporting information. Previously published data were used for this work (National Center for Biotechnology Information Sequence Read Archive; SRR611529, SRR031683, SRR3178875, and SRR8983692).
References
- 1.Marquet R., Isel C., Ehresmann C., Ehresmann B., tRNAs as primer of reverse transcriptases. Biochimie 77, 113–124 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Telesnitsky A., Goff S. P., “Reverse transcriptase and the generation of retroviral DNA” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997). [PubMed] [Google Scholar]
- 3.Harada F., Peters G. G., Dahlberg J. E., The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 254, 10979–10985 (1979). [PubMed] [Google Scholar]
- 4.Waters L. C., Mullin B. C., Bailiff E. G., Popp R. A., Differential association of transfer RNAs with the genomes of murine, feline and primate retroviruses. Biochim. Biophys. Acta 608, 112–126 (1980). [DOI] [PubMed] [Google Scholar]
- 5.Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M., Nucleotide sequence of the AIDS virus, LAV. Cell 40, 9–17 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Larsen K. P., et al. , Architecture of an HIV-1 reverse transcriptase initiation complex. Nature 557, 118–122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoye J. P., Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10, 395–406 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Johnson W. E., Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17, 355–370 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Feschotte C., Gilbert C., Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13, 283–296 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Chuong E. B., Elde N. C., Feschotte C., Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui T., et al. , Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Rowe H. M., et al. , KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Wolf G., et al. , The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 29, 538–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D., Goff S. P., Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf D., Goff S. P., TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Allouch A., et al. , The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9, 484–495 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Wolf D., Hug K., Goff S. P., TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 12521–12526 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf G., et al. , KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. eLife 9, e56337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner N. C., et al. , Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Vargiu L., et al. , Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imbeault M., Helleboid P. Y., Trono D., KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Schmitges F. W., et al. , Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 26, 1742–1752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang P., Wang Y., Macfarlan T. S., The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 33, 871–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L., et al. , Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Jin D., Musier-Forsyth K., Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J. Biol. Chem. 294, 5352–5364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das A. T., Klaver B., Berkhout B., Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys). J. Virol. 69, 3090–3097 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakefield J. K., Wolf A. G., Morrow C. D., Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA(3Lys). J. Virol. 69, 6021–6029 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu W., Ortiz-Conde B. A., Gorelick R. J., Hughes S. H., Rein A., Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71, 6940–6946 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., et al. , Regulation of HIV-1 Gag-Pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell 176, 625–635.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler S. L., Hansen M. S., Bushman F. D., A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7, 631–634 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Geis F. K., Goff S. P., Unintegrated HIV-1 DNAs are loaded with core and linker histones and transcriptionally silenced. Proc. Natl. Acad. Sci. U.S.A. 116, 23735–23742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turelli P., et al. , Primate-restricted KRAB zinc finger proteins and target retrotransposons control gene expression in human neurons. Sci. Adv. 6, eaba3200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gifford R. J., et al. , Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 15, 59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao G., Guo X., Goff S. P., Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297, 1703–1706 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Meagher J. L., et al. , Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc. Natl. Acad. Sci. U.S.A. 116, 24303–24309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets that were generated in this study have been deposited in the Gene Expression Omnibus under accession nos. GSE156604, GSE174239, GSE174242, and GSE183046. All other study data are included in the article and/or supporting information. Previously published data were used for this work (National Center for Biotechnology Information Sequence Read Archive; SRR611529, SRR031683, SRR3178875, and SRR8983692).