Significance
PARP is an important target in the treatment of cancers, particularly in patients with breast, ovarian, or prostate cancer that have compromised homologous recombination repair (i.e., BRCA−/−). This review about inhibitors of PARP (PARPi) is for readers interested in the development of next-generation drugs for the treatment of cancer, providing insights into structure–activity relationships, in vitro vs. in vivo potency, PARP trapping, and synthetic lethality.
Keywords: cancer drugs, inhibitor of Parp, HPF1, synthetic lethality, drug specificity
Abstract
Selective inhibitors of PARP1 and PARP2 (PARP1/2) are used to treat cancer patients with deficiencies in the repair of DNA via homologous recombination. Here we provide a perspective on the reported potencies of the most studied of these inhibitors (olaparib, talazoparib, niraparib, rucaparib, and veliparib) in vitro and in vivo and how these numbers relate to the known structures of these inhibitors bound to the active sites of PARP1 and PARP2. We suggest that the phenomenon of PARP trapping is primarily due to the inhibition of the catalytic activity of PARP1 and that the basis for the higher potency of talazoparib compared to the other inhibitors lies in its more extensive network of interactions with conserved residues in the active site. We also consider the potential role of the recently characterized protein “Histone PARylation Factor 1” (HPF1), which interacts with PARP1/2 to form a shared active site, for the design of the next generation of inhibitors of PARP1/2.
DNA is the genetic material found in every cell and contains the instructions for all processes required for life. DNA damage occurs at frequencies of about 1 million DNA changes per cell per day (1) and is caused by external insults such as ultraviolet (UV) radiation, reactive oxygen species, and toxic chemicals or by internal problems such as replication errors, free radicals, and spontaneous mutations. DNA damage can lead to disease, and in particular, accumulated mutations in DNA often underly tumor formation and cancer progression (2). As a reflection of the importance of maintaining genomic integrity, there exist at least 150 different proteins in the human proteome devoted to DNA repair (3). PARP1 is a key protein involved in the DNA damage response (4–7). PARP1 serves as one of the first responders by detecting single- and double-strand DNA breaks (SSBs and DSBs). Upon binding to damaged DNA, PARP1 becomes activated to utilize NAD+ to add chains of poly(ADP ribose) (PAR) onto itself and other nuclear proteins, especially histones. These PAR chains recruit the appropriate DNA repair machinery, in particular proteins containing PAR-binding motifs, to the sites of DNA damage (8–10). PARP1, the most abundant nuclear protein after histones (11), has many other functions in the nucleus (12), including regulation of replication and transcription via its ability to shape chromatin structure (13), regulation of transcription though direct interactions with transcription factors, and control of cell death via depletion of NAD+ (parthanatos) (14). Surprisingly, the parp1 gene is not essential for cell viability, which led to the discovery of the less abundant yet related PARP2 that assists crucially in the DNA damage response (15–18).
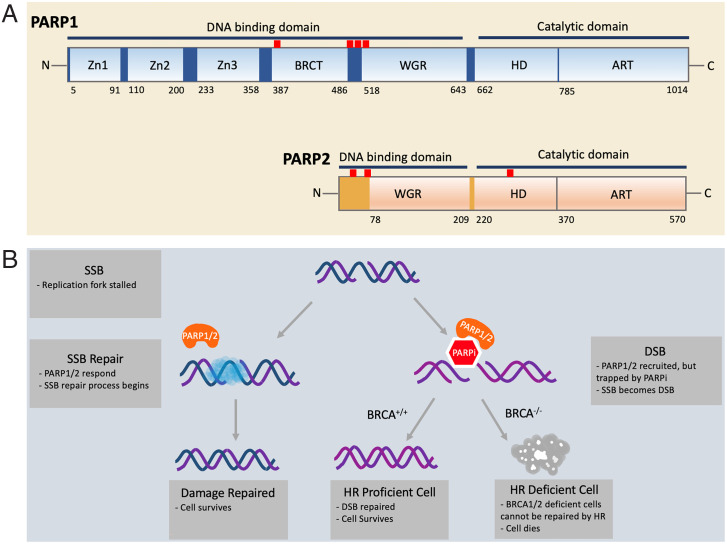
PARP1 and PARP2 both belong to the large class of proteins known as diphtheria-toxin–like ADP ribosyltransferases (ARTs) (19, 20). PARP1 and PARP2 (PARP1/2) both share a highly conserved catalytic domain that harbors the binding sites for NAD+ and the attachment sites for the extending PAR chains (Fig. 1A). The N-terminal domains of PARP1/2, although both responsible for binding DNA, are quite different from each other (Fig. 1A). PARP1 has five known DNA-binding domains, namely three Zn fingers, one BRCT domain, and one WGR domain. The Zn1, (Zn2), Zn3, and WGR domains cooperate in the binding of damaged DNA that leads to the conformational changes required for activation of PARylation (21–23). In contrast, the Zn1, Zn2, Zn3, and BRCT domains cooperate in the binding of intact DNA that does not trigger PARylation (24). PARP2 has only two known DNA-binding domains, namely the unstructured highly positively charged N-terminal region and the WGR domain, which for PARP2 has been shown to mediate the alignment of two DNA ends in close proximity (25–27).
Fig. 1.
(A) Domain structure of PARP1 and PARP2. PARP1/2 contain DNA-binding domains and catalytic domains. In PARP1, the DNA-binding domain includes three zinc fingers (Zn1, Zn2, and Zn3), a breast cancer susceptibility protein-1 C terminus (BRCT) domain, and the tryptophan-glycine-arginine–rich (WGR) domain. The locations of the predominant automodification sites of PARP1 (D387, E488, E491, S499, S507, and S519) are indicated by red uptick bars. In PARP2, the DNA-binding domain includes an unstructured N-terminal region and the WGR domain. The catalytic domains of both PARP1/2 are composed of an alpha-helical subdomain (HD) and the ADP ribosyltransferase subdomain (CAT). The locations of the three known automodification sites of PARP2 (S47, S76, and S281) are indicated by red uptick bars. (B) The mechanism of synthetic BRCA1/2 deficiency and PARPi. During DNA replication, single-strand DNA is vulnerable to breakage. When a single-strand break (SSB) occurs, the replication fork is stalled. PARP1/2 (orange) arrive at the site of the damage and recruit other repair factors (blue cloud). In the presence of PARP1/2 inhibitor (red hexagon), however, PARP1/2 are trapped at the damage site, inhibiting the restart of replication. As a result of unresolved replication stress, a double-strand break (DSB) can arise. Homologous recombination (HR) is one of the most faithful ways to repair a DSB. Cells with functioning BRCA1/2 can carry out HR and restore the genomic integrity, but cancer cells with defective BRCA1/2 cannot. Thus, synthetic lethality selectively kills HR-deficient cancer cells in conjunction with a PARPi.
PARP1/2 gained much clinical interest upon the discovery that targeting these proteins could serve as the basis for treatment of breast cancer through a mechanism known as synthetic lethality (28, 29). Synthetic lethality was first observed by Bridges (30) in 1922 in the “Fly Room” at Columbia University and the term was coined by Dobzhansky (31) in 1946, both studying hidden recessive homologous phenotypes in the fruit fly Drosophila. Whereas various individual mutant genes yielded viable Drosophila on their own, some crossbred homozygotes did not produce viable offspring. The concept of synthetic lethality was more recently expanded from describing interactions between genetic perturbations to also include a combination between a genetic perturbation and treatment with a chemical compound (32). BRCA1 and BRCA2 are known as breast cancer susceptibility genes wherein mutations in one of these resulted in an increase in the risk of developing breast cancer and ovarian cancer in women (33). Although less common, men with germline mutations in BRCA1 or BRCA2 have shown higher risks of developing breast cancer and prostate cancer than noncarriers (34). BRCA1 and BRCA2 are known to be important mediators of homologous recombination (HR), which is one of the major pathways for repairing double-strand breaks in DNA (35).
With this brief background, we can now describe synthetic lethality as it applies to BRCA1/2 and inhibition of PARP1/2 (Fig. 1B). Healthy cells carry at least one copy of the wild-type BRCA gene, which can carry out HR without the assistance of PARP1/2. Introducing an inhibitor of PARP1/2 (PARPi) into these cells does not affect their viability. Tumor cells that are lacking functional BRCA1 or BRCA2 become sensitive to a PARPi because the stalled replication forks, which cannot be restarted without the action of PARP1/2, result in single-strand breaks and eventually in the more deleterious DSB (36). Since repair of these DSBs is mediated by HR, and both BRCA1 and BRCA2 are involved in maintaining genomic integrity through HR (37), BRCA1−/− or BRCA2−/− tumor cells treated with PARPi undergo apoptosis (38–41). This mechanism of synthetic lethality provides the basis for targeted therapy for cancers that are associated with BRCA gene mutations.
There are now four different inhibitors of PARP that are approved for clinical use in breast, ovarian, and prostate cancer with underlying BRCA1/2 deficiencies, namely olaparib (Lynparza; KuDOS Pharmaceutical/AstraZeneca), talazoparib (Talzenna; LEAD Therapeutics/Pfizer), rucaparib (Rubraca; Agouron Pharmaceuticals/Clovis Oncology), and niraparib (Zejula; Merck/Tesaro) (42). These same PARPi, along with veliparib (Abbott/AbbVie) and others, are part of numerous clinical trials for cancers, either as a stand-alone inhibitor or in conjunction with DNA-damaging agents such as chemo- or radiosensitizers (http://ClinicalTrials.gov) or in combination with immunotherapies (43). There have been many excellent and recent reviews of PARPi, from comprehensive coverage of all things PARP (42) to articles that focus more on the structure and activation of PARP (44), PARP trapping (45), structure–activity relationships (46), combination therapies (47), repurposing of PARPi for nononcological diseases (48), and mechanisms of resistance (49). Here we provide our unique view of the field by reviewing the literature from a perspective of measured potencies in vitro and in vivo (Number Crunching) and analysis of interactions between inhibitors and the active sites of PARP1/2 (Structure Gazing). At the close, we provide some insights on new directions in the field of PARPi.
Number Crunching
In Vitro Inhibition of PARPs by PARPi.
Evaluating the potency of small molecule inhibitors is a critical part of any drug discovery effort. The requirements for assays that measure the efficacy of small molecules vary, depending on which step of the discovery process is being undertaken. High-throughput screens of large libraries of compounds that determine relative inhibition values at a fixed concentration of inhibitor need to be robust, cost effective, and amenable to miniaturization and automation. Follow-up screening assays are either similar to the assay used in the high-throughput screen or more often (and more ideally) orthologous and allow for the determination of half-maximal inhibitory concentrations (IC50s). Mechanistic evaluation of inhibitors to determine binding constants (KDs), inhibition constants (KIs), and modes of inhibition (i.e., competitive vs. noncompetitive vs. uncompetitive) requires additional method development and is an essential step in moving inhibitors from the laboratory bench to clinical trials and ultimately patients.
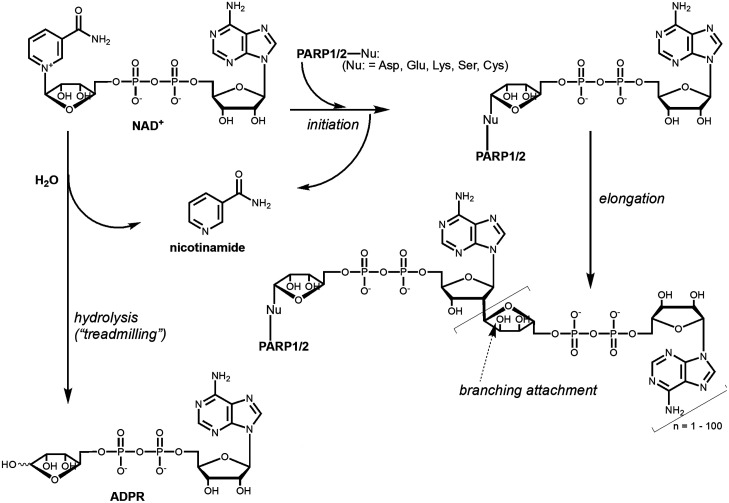
Measuring the activity of PARP1/2 is not a straightforward task because these are very unusual enzymes. Most importantly, PARP1/2 are their own best substrates, which causes two major complications. First, it becomes impossible to independently vary enzyme and substrate concentrations as is typically done to optimize assay conditions for determining enzyme activity and inhibitor potency. Second, every automodification event leads to a changed enzyme. Specifically, in the automodification reaction of PARP1/2, which requires the presence of damaged DNA, ADP-ribose (ADPR) moieties from NAD+ are initially attached onto a variety of amino acid side chains (50) (Fig. 2). These protein-attached ADPRs are then extended to form PAR chains that can make polymers with up to 200 ADPRs (51). In a further complication, branch points in the PAR chains occur approximately every 20 ADPRs (51). Thus, during the course of the reaction with NAD+, every addition of ADPR onto PARP1/2 yields a different form of the enzyme that has lower catalytic activity as the enzyme is modified (52) and reduced binding to DNA once the PAR chains become sufficiently long (53–55). It is likely that automodified PARP1/2 are highly heterogenous, i.e., that every PARP molecule in a reaction mixture carries a different set of modifications. In an additional twist, hydrolysis (“treadmilling,” Fig. 2) can also occur, and this becomes the dominant NAD+-depleting activity at high concentrations of the accessory protein Histone PARylation Factor 1 (HPF1) (below) (56).
Fig. 2.
Chemical mechanism of PARylation by PARP1/2. In an initiation reaction, the ADPR moiety of the NAD+ substrate is attached to an amino acid side chain on PARP1/2. Elongation occurs wherein additional ADPRs are added to the existing ADPR, resulting in chain lengths up to 200. Branching of the PAR chains can also occur at the site indicated by an arrow. Additionally, PARP1 can simply hydrolyze NAD+ to yield free ADPR and nicotinamide in a reaction known as treadmilling.
There are many different approaches to measuring the activity, and thereby the inhibition by small molecules, of PARP1/2. One of the most widely used and most sensitive method assaying PARP1/2 relies on the use of [adenylate32P]-NAD+, which incorporates 32P-ADPR onto protein and thereby allows the facile detection of radiolabeled protein as captured in precipitates, in gels, or on filters (56–62). The availability and suitability as a substrate of biotinylated NAD+ (6-biotin-17-NAD+) allows for a colorimetric assay with detection using streptavidin that is labeled with horseradish peroxidase, although its specificity and/or activity may be altered compared to that of NAD+. Methods amenable to high-throughput screening include scintillation proximity assays (63, 64), chemiluminescent immunoassays (54), capture of NAD+ as a fluorescent adduct (65), and fluorescence polarization (55). Based on the existing literature and our own experiences, the lower limit for reliable detection of the autoPARylation activity of PARP1/2 is best attained using α-32P-NAD+ and is in the range of 20 to 50 nM enzyme. The need for such a relatively high concentration of PARP1/2 is in large part due to the limitation that PARP1/2 is its own best substrate. Having the required concentrations of PARP1/2 above the IC50s and KIs reported for clinically relevant PARPi (10 pM to10 nM; below) runs into the tight-binding limit problem (66). In the tight-binding limit problem, the measured inhibition constant for an inhibitor does not reflect the true binding constant (IC50s and KI), but instead reflects the concentration of active enzyme in solution as all the inhibitor is depleted from solution by tight binding to the protein. Specifically in the case of PARP1, all inhibitors with true binding constants for PARP1 that are significantly below the concentration of PARP1 under the assay conditions will have essentially the same observed affinity, reflecting the concentration of active PARP1. In part to overcome this limitation, we have recently reported a method that allows for the accurate determination of KIs for inhibitors of PARP1 that is sensitive to ∼10 pM (67). Other assays that allow for the determination of subnanomolar binding constants for PARPi include BiaCore (68, 69), although this method suffers from molecular transport issues of the inhibitor across the surface that is used to immobilize the protein (70) and small signal intensities due to the molecular weight ratio of PARPi to PARP1 (∼0.4%).
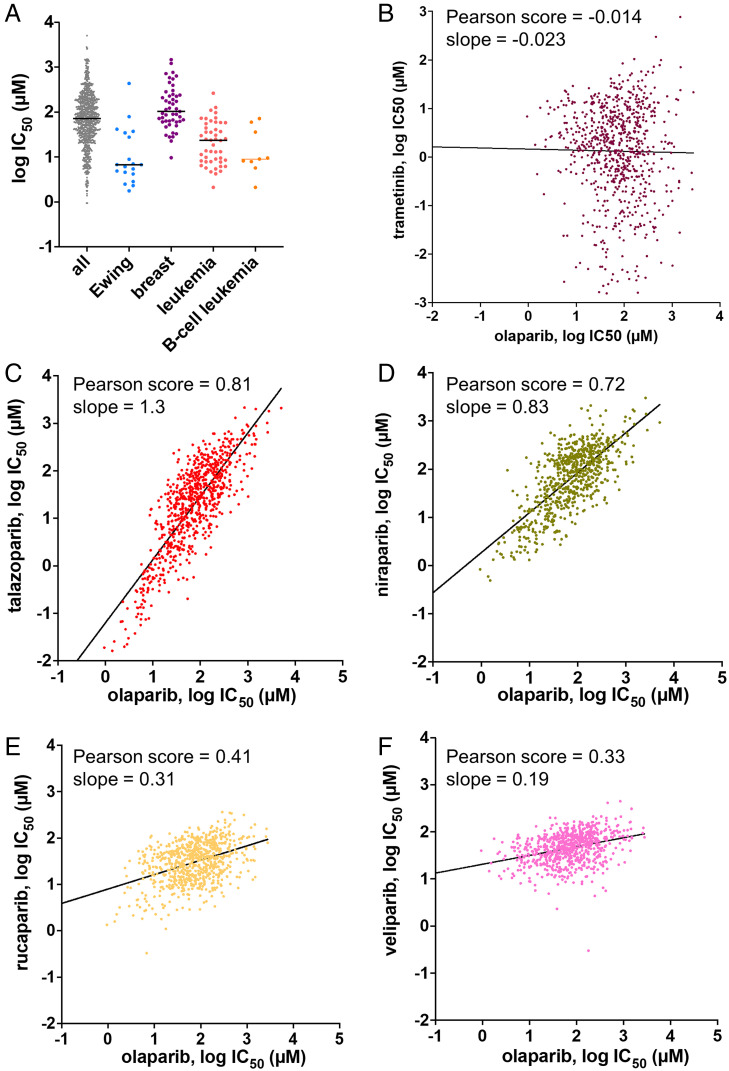
For the “number crunching” of in vitro data, we have combed the literature for reported IC50s and KIs derived from inhibition experiments for various PARPi toward PARP1/2 and compiled these in Fig. 3A (SI Appendix, Table S1). For PARP1, we observe relatively tight clustering of most of the experimental observations, with olaparib, talazoparib, and rucaparib being slightly more potent (median = 0.5 to 1 nM) than niraparib and veliparib (median = 4 to 5 nM) (Fig. 3A). The significant outlier measurements indicated by arrows come from our own experiments using assay conditions developed to overcome the tight-binding limit problem (56) that is likely hampering the determination of true inhibition constants for these more potent inhibitors by most other assay methods. Supporting this interpretation, the higher potency of both talazoparib and rucaparib compared to the other PARPi that we have measured (0.012 and 0.09 nM, respectively) is also partially reflected in BiaCore experiments (0.17 nM or 0.29 and 0.09 nM, respectively) (68, 69). It should be noted that despite the fact that this assay method is based on DNA release, our results do not require the invocation of “PARP trapping,” i.e., an allosteric interaction wherein binding of compounds to the active site leads to tighter binding of DNA to the DNA domains (PARP Trapping). Inhibited PARP1 remains bound to DNA solely because it remains unPARylated. We conclude that it is a common misconception that all PARPi have approximately equal potency. In fact, talazoparib is a significantly more potent PARP1 inhibitor in vitro than all the others, with rucaparib falling between talazoparib and olaparib (67).
Fig. 3.
Summary of inhibition measurements for PARPi with PARP1 and PARP2 and comparison to cell-based data. (A) For PARP1, each reported IC50 and KI value is shown as a point and the line indicates the median value. Our reported measurements for all these inhibitors using a method that avoids the tight-binding limit problem are indicated by black arrows. (B) For PARP2, each reported IC50 and KI value is shown as a point and the line indicates the median value. (C) The ratio of the median IC50 value for BRCT+/+ vs. matched BRCT−/− cells is plotted against the KI as determined in ref. 65, since these values were determined with proper consideration of the tight-binding limit. All raw values for the points in these plots along with literature references can be found in SI Appendix, Tables S1 and S2.
The combined data for inhibition of PARP2 by PARPi yield values for olaparib and rucaparib (median = 0.2 to 0.3 nM) that are noticeably more potent than those for PARP1 (Fig. 3B). In contrast to PARP1, talazoparib is not significantly more potent than olaparib toward PARP2 (median = 0.2 nM). As for PARP1, niraparib and veliparib (median = 2 to 4 nM) are less potent than the other three PARPi. We discuss these differences in apparent affinities of the different PARPi for PARP1 vs. PARP2 further in Structure Gazing.
In Vivo Inhibition of PARPs by PARPi: BRCA−/− vs. BRCA+/+.
We next combed the literature for reports of PARPi in cell-based studies. Comparing different cell-based studies to each other is incredibly tricky. First, as noted in the Introduction, cells deficient in HR are expected to respond with much greater sensitivity to PARPi than cells with functioning HR. Second, different laboratories use different cell lines, some of which may harbor unknown HR deficiencies or even unexpected resistance to PARPi. Third, treatment and analyses differ widely from one study to the next, with incubation of cells with PARPi varying from 5 to 13 d and evaluation methods including cytotoxicity, clonogenic assays, or colony-forming assays. We therefore focus our number crunching of in vivo data on selected studies that compare matched cell lines (BRCA−/− to BRCA+/+ or BRCA+/−, for simplicity BRCA+/+ since loss of HR does not occur in the heterozygous genotype).
The initial descriptions of synthetic lethality with respect to PARPi in 2005 led to the striking observation that BRCA1−/− or BRCA2−/−cell lines displayed a 60- to 1,000-fold greater sensitivity to KU0058684 (a precursor to olaparib) (28) and AG14361 (a precursor to talazoparib) (29) than BRCA+/+ cell lines. This observation has been replicated numerous times with other cell lines and with other PARPi. For example, the potency ratio for PARPi in BRCA−/− to BRCA+/+ is 250- to 300-fold in two different studies (68, 71). For veliparib and niraparib this potency ratio appears to be smaller (10- to 20-fold) (68, 71), consistent with their weaker potency and therefore presumably lower specificity. For rucaparib, the BRCA−/− to BRCA+/+ ratio is an impressive 1,000-fold. For talazoparib the reported ratio is also large but more variable (560- to 10,000-fold). Interestingly, plotting the potency of PARPi as determined by our assay that overcomes the tight-binding limit problem (67) with the ratio (of the median) of these potencies shows a statistically significant correlation (R2 = 0.99; Fig. 3C). This correlation suggests that in vitro inhibition values are more predictive of cell-based efficacies than previously thought (72).
In Vivo Inhibition of PARPs by PARPi: Thousands of Cell Lines.
We also undertook a comparison of different PARPi in the large publicly accessible database Genomics of Drug Sensitivity in Cancer (https://www.cancerrxgene.org; release 8.3) (73). This database lists the analyses of ∼1,000 different cell lines for their response to over 500 different drugs. Although different cell lines have known and unknown underlying deficiencies in HR or other sensitizers to PARPi, several interesting observations can be made by plotting sensitivity to PARPi for different cell types according to tumors from which they were derived. First, and as noted previously (74), cells from Ewing sarcoma harboring the EWS-FLI1 gene translocation are heavily represented in the list of cell lines sorted by sensitivity to inhibition by PARPi (shown for olaparib in Fig. 4A). Unfortunately, this potential lead for treating patients with Ewing sarcoma with PARPi has not panned out in clinical trials due to lack of response to treatment (NCT01583543, NCT01286987, NCT02116777). Second, as expected, most breast cancers are not BRCA−/− and thus do not show any special sensitivity to PARPi (shown for olaparib in Fig. 4A). In fact, many of these cell lines are defective in TP53BP1, which can promote partial restoration of HR and resistance to PARPi (75). Third, leukemias as a whole, and most especially B cell leukemias, are more sensitive to PARPi compared to most other cell lines (shown for olaparib in Fig. 4A) and further investigation of this lead in preclinical and clinical studies seems warranted (76). In addition, further information from this database might be gleaned using advanced computational analyses that take advantage of the complete genomic sequencing of all these cells along with these inhibition data.
Fig. 4.
(A) Distribution of potencies for triggering cell death in 1,000 different cell lines by olaparib. The leftmost column includes all 1,000 cell lines studied in the Genomics of Drug Sensitivity in Cancer. Subsequent columns to the right display cancer-specific cell types wherein the breast cancer cell lines are not predominantly BRCA−/−. The line indicates the median value. The data were obtained from https://www.cancerrxgene.org. (B–F) Comparison of the potency of olaparib to other PARPi in a panel of 1,000 different cell types. The data were obtained from https://www.cancerrxgene.org. The IC50 values for each inhibitor were downloaded in an array and compared to the IC50 values for the other inhibitors for each cell line for which the database contained a value. Each data point represents the log IC50 value of a PARPi (B, trametinib; C, talazoparib; D, niraparib; E, rucaparib; F, veliparib) vs. the log IC50 value of olaparib for a different cell line. To facilitate interpretation and comparisons between the different graphs, the axes are “square” (one log unit is the same length on both x and y axes), and the scales are the same for all five graphs. The black line represents a best-fit linear correlation between the two inhibitors and the Pearson correlation coefficient and slope value are indicated for each graph.
Because this large dataset from thousands of cell lines contains data from different PARPi, it can also reveal interesting aspects related to the targeting and potency of the studied PARPi. We performed a meta-analysis of these data by graphing the measured potency (log IC50) of one inhibitor vs. the potency of another inhibitor, each point representing a different cell line (Fig. 4 B–F). As a negative control, we show a comparison between the PARPi olaparib and trametinib, a 1-nM inhibitor of mitogen-activated protein kinase kinase (MEK). Olaparib and trametinib display no correlation, as expected since they target completely different proteins, pathways, and genetic predispositions (Fig. 4B; Pearson score = −0.014, slope = −0.023). By comparing olaparib and talazoparib (Fig. 4C), we observe a very good correlation of potencies (Pearson score = 0.81; slope = 1.3), suggesting that these two drugs are hitting the same target(s), have the same mechanism of action, and are particularly potent for the same subset of cells. Also, in this comparison one can readily see the 10- to 100-fold greater efficacy of talazoparib compared to olaparib by noting the offset between the x and y axes. This increased potency of talazoparib vs. olaparib is in agreement with our measurement of the KI in vitro (Fig. 3A) (67). A similar correlation of potencies is observed for olaparib vs. niraparib (Pearson score of 0.72, slope = 0.83; Fig. 4D) with about a twofold lower average potency for niraparib vs. olaparib, in agreement with in vitro measurements (Fig. 3A). Surprisingly, rucaparib shows a much weaker correlation with olaparib (Pearson score of 0.41, slope = 0.31; Fig. 4E) despite its apparent equal or greater potency in vitro (Fig. 3A). We speculate that this weaker correlation may be related to the “antitrapper” effect that has been recently described for rucaparib (77) or may be indicative of off-target inhibition against other PARPs or even completely unrelated proteins that also utilize NAD+. The weakest correlation is seen in the comparison of veliparib with olaparib (Pearson score of 0.33, slope = 0.19; Fig. 4E) wherein all cell lines respond similarly and weakly to veliparib (average IC50 of 58 µM), which may help explain the inability of veliparib to make it to the clinic as a stand-alone treatment.
PARP Trapping.
AutoPARylation of PARP1 (and PARP2) leads to the dissociation of these enzymes from DNA due to the charge repulsion between DNA and the PAR chains, which harbor twice the negative charge of DNA or RNA (53–55, 67). PARP trapping in response to treatment of cells with both a DNA-damaging agent (γ-irradiation) and a PARPi (3-aminobenzamide) was first described in 1992 wherein inhibited PARP1 remained bound to DNA due to its inability to perform the PARylation reaction (78). PARP trapping was characterized in much more detail with more potent inhibitors in response to treatment of cells with methyl methanesulfonate. Two different measures of PARP trapping are used, namely increased association of DNA with PARP1 (79) and increased association of PARP1 with chromatin (53). Interestingly, different PARPi have different potencies with respect to their ability to trap PARP1. Talazoparib is 100- to 1,000-fold better at trapping than olaparib and rucaparib, which are both better trappers than veliparib by another factor of at least 5 to 10 (53, 54, 68). PARP trapping has been broadly correlated with in vivo potency and/or toxicity as exemplified in particular for talazoparib. The reason that PARP trapping is more deleterious than persistent SSBs in the absence of PARP1 is that other repair proteins are occluded from the damage sites and the collapse of replication forks that collide with trapped PARP1.
Because it was long assumed that all PARPi have approximately equal potency against PARP1/2 in vitro, much work has gone toward understanding the biochemical basis for why different PARPi have different PARP-trapping potencies. The most likely hypothesis, and the one most pursued by a variety of experimental approaches, is the concept of allosteric coupling between the binding of DNA and PARPi (53, 54). Such an allostery implies that binding of inhibitor leads to tighter binding of DNA, and by thermodynamic necessity, tighter binding of DNA leads to tighter binding of inhibitor. However, despite best efforts using multiple methods, no evidence for such allosteric coupling as a possible cause for the potent trapping of PARPi such as talazoparib has been found (23, 68, 77, 80, 81). Surprisingly, the best evidence for coupling between inhibitor binding in the active site and DNA binding to the Zn fingers exists for rucaparib, niraparib, and veliparib (77), three PARPi that promote (not inhibit) the release of DNA (i.e., they are antitrappers). In contrast, binding of the nonclinical inhibitor EB-47 does lead to tighter DNA binding and suggests that novel PARPi could be designed to better capture this allosteric interaction (77). Based on our measurements of the higher potency of talazoparib compared to the other PARPi in vitro (67), we propose that the origin of PARP trapping is dominated by the inhibition of the activity of PARP1. That is, PARP1 accumulates at sites of DNA damage and in the presence of PARPi remains tightly bound to DNA as there is no autoPARylation to drive dissociation. The reason talazoparib is a much more potent PARP trapper than the other PARPi is that it is a much more potent inhibitor, both in vitro (Fig. 3A) and in cells (Fig. 4B). Consistent with the idea that trapping is intimately linked to the inhibition of PARP activity, it has been recently shown that the major mechanism for the detrapping of PARP1 is its automodification at specific sites of PARylation (82).
Structure Gazing
Interactions of PARPi with the Active Sites of PARP1/2.
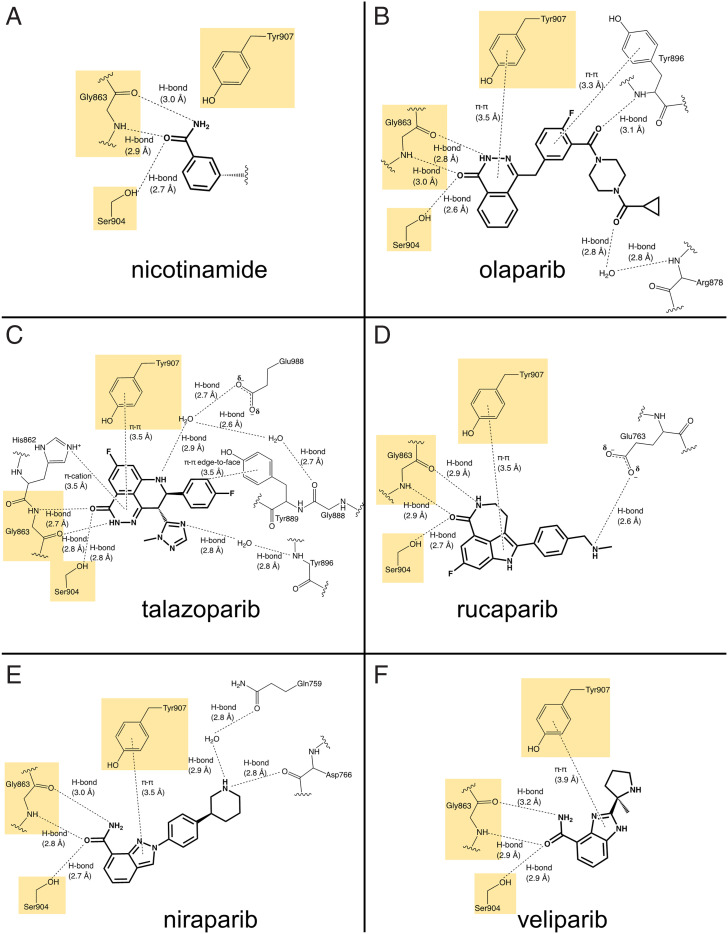
All clinically relevant PARPi are designed to mimic the interactions between the nicotinamide ring of the substrate NAD+ and the active site. This approach grew out of the discovery that nicotinamide analogs such as 3-aminobenzamide were surprisingly potent inhibitors of PARP1 (83). Other early nicotinamide analogs validating this approach were characterized structurally by X-ray crystallography (61). As such, PARPi bind to the catalytic domains of PARP1/2 in competition with NAD+, inhibit PARylation activity, and thus prevent the subsequent release of PARP1/2 from sites of DNA damage. Detailed knowledge of the interactions between PARPi and the active sites of PARP1/2 are available from a multitude of high-resolution crystal structures (SI Appendix, Table S3). For purposes of the descriptions below, we selected the highest-resolution crystal structures of each inhibitor and compared their conserved and unique interactions, both among different inhibitors and between PARP1 and PARP2. For a point of comparison between PARPi and nicotinamide, we rely on the first structure of PARP1 with a full analog of NAD+ (benzamide adenine dinucleotide [BAD]) that was determined in 2018 by the Pascal laboratory (Fig. 5) (84). We focus on the three highly conserved amino acids that play the most important roles in positioning the inhibitors (and the nicotinamide) at the active sites, emphasizing the importance of these interactions for the affinity of PARPi and the nonhydrolyzable BAD. Also, we describe some of the unique interactions of each PARPi that in part explain the different affinities of each inhibitor.
Fig. 5.
Overview of binding pocket for NAD+ in the catalytic domain of PARP1. (Left) The ribbon structure of the catalytic domain of PARP1 is shown in cyan with the nonhydrolyzable NAD+ analog (BAD) shown in green (Protein Data Bank [PDB] ID 6BHV). (Right) The pocket for binding BAD is visualized with the protein surface shown in cyan. Note that the nicotinamide ring sits in the deepest pocket of the active site.
Comparison of all the known structures of PARPi bound to PARP1/2 revealed three conserved interactions that are shared by olaparib, talazoparib, rucaparib, niraparib, and veliparib for PARP1 and PARP2 (Fig. 6 and SI Appendix, Fig. S2). First, there are two hydrogen bonds formed by Gly863 in PARP1 (Gly429 in PARP2) with the bi- or tricyclic ring system of each inhibitor, with the amide nitrogen of Gly863 serving as an H-bond donor and the carbonyl oxygen serving as an H-bond acceptor. Second, Ser904 in PARP1 (Ser470 in PARP2) serves as an H-bond donor to a carbonyl in/on the bi- or tricyclic ring system of each inhibitor. The bidentate interaction of Gly863 and the H bond by Ser904 are the basis for the inhibitor’s mimicry of nicotinamide where these same interactions are made with the exocyclic amide of the nicotinamide (Fig. 6A). Third, each of the inhibitors forms a π–π interaction between its aromatic bi- or tricyclic ring and Tyr907 in PARP1 (Tyr473 in PARP2) with a distance of 3.5 to 3.9 Å (Fig. 6). Although this π–π interaction is not seen with the nicotinamide ring, Tyr907 does form part of the hydrophobic pocket that accommodates the nicotinamide and ribose rings of NAD+ (Fig. 6A). It appears that this π–π interaction contributes significantly to the much higher affinity of PARPi (nM; Fig. 3) compared to nicotinamide (>50 µM) (83) and it may play a significant role in the binding of inhibitors with larger aromatic ring structures (e.g., olaparib) than others with smaller, perhaps less ideally placed aromatic rings (e.g., veliparib) (67).
Fig. 6.
(A–F) Interaction diagrams for PARPi with PARP1. (A) Nicotinamide ring from BAD (PDB ID 6BHV). (B) Olaparib (PDB ID 7KK4). (C) Talazoparib (PDB ID 7KK3). (D) Rucaparib (PDB ID 6VVK). (E) Niraparib (PDB ID 7KK5). (F) Veliparib (PDB ID 7KK6). For simplicity and easier comparison with the PARPi, only the nicotinamide end of BAD is shown here. The full interaction diagram for BAD is included in SI Appendix, Fig. S1. All diagrams were prepared in ChemDraw using the deposited PDB entries indicated.
We next examine some of the differences in how the individual PARPi dock in the catalytic pockets of PARP1/2, starting with olaparib (Fig. 6B). In addition to the aforementioned conserved interactions, olaparib forms two more hydrogen bonds with catalytic domain residues. Tyr896 makes a direct hydrogen bond with its backbone amide as well as a π–π interaction with its aromatic ring, thus clamping olaparib more snugly into the catalytic site of PARP1/2. There is also a hydrogen bond between the terminal carbonyl of olaparib and the backbone amide of Arg878, which is a water-mediated interaction in 7KK4 but seen as a direct interaction in 5DS3. This difference in H bonding is accompanied by a slightly different orientation of the “tail” of olaparib in these two structures, which most likely is not due to the absence of the HD helices in 5DS3, since olaparib bound to PARP2 has both the HD helices and a direct hydrogen bond (SI Appendix, Fig. S2A). In the structure of PARP1 with BAD this Arg878 makes a direct H bond to the adenine ring (SI Appendix, Fig. S1). The high similarity in the active sites of PARP1 and PARP2 and their similar binding modes to olaparib are consistent with their essentially identical in vitro potencies (Fig. 3).
Talazoparib is the most potent of the Food and Drug Administration (FDA)-approved PARPi and its more extensive interactions (compared to olaparib) with the active sites of PARP1 provide clues for its greater affinity (Fig. 6C and SI Appendix, Fig. S2B). First, talazoparib’s tricyclic structure provides excellent conformational rigidity for the conserved interactions with the nicotinamide-binding residues Gly863, Ser904, and Tyr907. Second, talazoparib interacts with the signature His-Tyr-Glu catalytic triad of the ADP ribosyl transferases, namely His862 (His428 in PARP2), Tyr896 (Tyr462 in PARP2), and Glu988 (Glu558 in PARP2). Specifically, His862, depending on its protonation state, forms either a π–π or a π–cation interaction with the tricyclic ring. Second, the amide nitrogen of Tyr896 forms a water-mediated hydrogen bond with a nitrogen in the triazole ring of talazoparib. Third, the side chain of Glu988, which is required for the polymerization activity of PARP1 (85), forms a water-mediated hydrogen bond with nitrogen in the tricyclic ring. In addition, in PARP1, talazoparib makes an edge-to-face π interaction with Tyr889 and a water-mediated hydrogen bond with Gly888 in PARP1. These latter two interactions are not observed in PARP2 and thus these interactions may contribute to talazoparib’s tighter affinity for PARP1 vs. PARP2. Talazoparib, with PARP2, also forms a unique water-mediated hydrogen bond with the helical subdomain residue Glu335. One can speculate that this interaction may hinder the flexibility of the HD subdomain during the initial DNA damage response of PARP1 (21). Overall, the crystal structures of talazoparib bound to PARP1/2 support its tight binding by having a rigid molecular structure, mimicking the bindings of nicotinamide superbly and forming an extensive network of other interactions.
Although the affinity of rucaparib for PARP1 is considered similar (or even tighter) compared to that of olaparib in vitro (Fig. 3A), its weaker potency in vivo is reflected in its structure bound to PARP1 (Fig. 6D; currently, there is no structure available for rucaparib bound to PARP2). In addition to the conserved interactions with Gly863, Ser904, and Tyr907, rucaparib forms only one additional hydrogen bond with the HD-subdomain residue Glu763 at the terminal secondary amine, which was also seen with talazoparib for PARP2 (SI Appendix, Fig. S2B). The seemingly few interactions of rucaparib with PARP1 compared to olaparib (Fig. 6B) and most especially talazoparib (Fig. 6C) may in part be compensated by its rigid molecular structure. The three-ring system in the nicotinamide mimic shares some similarity to talazoparib.
The least potent of the FDA-approved PARPi is niraparib (Fig. 3A). The crystal structure of niraparib is available only for PARP1, but its unique interactions in the active site provide clues for the different characteristics of niraparib. In addition to the usual nicotinamide-mimic interactions of Gly863, Ser904, and Tyr907, niraparib makes two unique interactions with HD-subdomain residues, a water-mediated hydrogen bond with Gln759 and a direct hydrogen bond with Asp766. Because the HD subdomain is conserved in PARP1, PARP2, and PARP3 (21), niraparib is considered one of the most selective inhibitors for PARP1/2 over other PARP family members (86). Thus, although niraparib’s relatively flexible molecular structure and limited interactions may limit its potency, its clamping down of the HD domain, which must be disordered for activation of PARP1 (21), allows niraparib to function as a clinical PARP1/2 inhibitor.
Veliparib has the smallest molecular weight among the PARPi and makes the least number of interactions with active site residues. For PARP1, only the conserved Gly863, Ser904, and Tyr907 interactions are observed (Fig. 6E), although in another structure (2RD6) there is an additional water-mediated hydrogen bond Glu988. Interestingly, veliparib makes two additional interactions with PARP2 (SI Appendix, Fig. S2C), namely water-mediated hydrogen bonds with Glu558 and Glu335, the latter being the same interaction seen with talazoparib (Fig. 5).
Where Do We Go Next?
Because of the success (and limitations) of PARPi in the clinic, the academic and pharmaceutical research communities are actively pursuing alternative applications for existing PARPi as well as for the next generation of PARPi. Below we highlight a few interesting and timely topics.
HPF1.
The past few years have shown some big surprises in the PARP1/2 field resulting from the discovery of HPF1 by Ahel and coworkers (87). Although the mechanism or specificity of PARP1 had been studied for many decades, it turned out that the PARP research community was missing a fundamental component that dictates how this enzyme functions. First, PARylation, which for years was primarily thought to occur on glutamate or aspartate residues (88, 89), is primarily associated with modification of serine residues (7, 90, 91), and this switch in target residue is attributed to HPF1. Second, although it was known that histones can be PARylated by PARP1, reliable in vitro (and in vivo) transPARylation of histones in nucleosomes (on serine residues), instead of autoPARylation of PARP1/2 (on glutamate), was finally observed in the presence of HPF1 (50, 87). Third, the active site of PARP1/2 turned out to be incomplete without the catalytic acid Glu284 contributed by HPF1 to promote PARylation of serines (56, 92, 93). Interestingly, HPF1 may not be the only protein that can redirect the specificity of PARP1 toward histones and serines as PARP1 in Dictyostelium clearly modifies Ser10 and Ser28 in histone H3b but does not have a recognizable HPF1 (94).
PARPi binding to PARP1/2 are not predicted to make a direct interaction with HPF1, despite the close proximity of HPF1 to their active sites. However, we noted that Phe280 from HPF1 forms a π–π stacking interaction with Tyr907 of PARP1 (Tyr473 in PARP2) (92), which forms the key π–π stacking interaction for each of the PARPi (Fig. 6). This suggested to us that some PARPi may bind more potently in the presence of HPF1 than to PARP1/2 alone. In fact, this prediction held true for olaparib for PARP1 wherein the π–π stacking interaction is particularly well oriented, but not for PARP2 (67). The proximity of several residues of HPF1 to the active site of PARP1/2 suggests that next-generation PARPi that capture interactions with HPF1 could have improved specificity and potency. Interestingly, despite the increased in vitro potency of olaparib for PARP1 in the presence of HPF1, deletion of HPF1 sensitizes cells to treatment by olaparib (82). This additional case of synthetic lethality between PARPi and a genetic deletion suggests that further investigation will be required to fully understand the complexities of PARP1/2 and HPF1.
Synthetic Lethality with Proteins Other Than BRCA1/2.
At present, PARPi are approved for use primarily with BRCA1/2-deficient cancers (see the explanation of synthetic lethality above), yet the search is on for finding other predispositions that can be successfully treated with PARPi (HR deficiency-like [“HRDness”]) (Fig. 4A) (95). This approach has had some success in the clinic in the treatment of prostate cancers with DNA repair defects, specifically in those with aberration in the ATM kinase (96). Recently, a number of different approaches have yielded insights into proteins other than BRCA1/2 whose absence or inhibition sensitizes cells to PARPi. CRISPR screens resulted in the discovery of a number of proteins involved in ribonucleotide excision repair, including RNAse H1 and RNAse H2 (97), and nucleotide metabolism including DNPH1 (2′-deoxynucleoside 5′-monophosphate N-glycosidase) and ITPA (inosine triphosphatase) (98). PARPi sensitization is also caused by factors involved in base excision repair such as LIG3, POLB, ALC1, PNKP, XRCC3 (98–100), and the chromatin remodeler SMARCAD1 (101). Interestingly, loss of HPF1 also sensitizes cells to PARPi (82, 87, 98), suggesting that perhaps next-generation PARPi should not capture interactions with HPF1 (as suggested above), but instead compete with the binding of both NAD+ and HPF1. Given the proximity of HPF1 to the active site of PARP1/2, it would seem not that difficult to add suitable chemistry to existing PARPi that would interfere with the docking of HPF1. Combining inhibitors of sensitizer proteins with PARPi opens new possibilities for combination therapies. Mining information from screens such as these with genomic sequencing of patient samples may allow for a rapid expansion for the application of PARPi in patients with the appropriate genetic signature.
Conclusions
With the discovery of the biochemical contribution of HPF1 to the mechanism of the PARP1/2 reaction and the outcome of PARylation, we are entering another era in the drug discovery process for inhibition of PARP1/2. We should be seeking and validating next-generation PARPi that are not mere incremental advances. As the field moves forward, it is important that solid in vitro studies are performed to prevent misguided efforts, an issue not unknown in the PARP field wherein iniparib made it to phase 3 clinical trials before it was discovered not to be a PARPi (102). Additionally, we look forward to further developments in personalized medicine that may allow for a much broader application of these well-tolerated compounds by matching compounds with appropriate cancer patients to maximize therapeutic benefits.
Supplementary Material
Footnotes
Reviewers: I.A., University of Oxford; and A.L., Johns Hopkins University.
The authors declare no competing interest.
Data Availability
Previously published data were used for this work (as specified in the text).
References
- 1.Lodish H., et al. , Molecular Biology of the Cell (Freeman, New York, NY, 2016). [Google Scholar]
- 2.Jeggo P. A., Pearl L. H., Carr A. M., DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 16, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Wood R. D., Mitchell M., Sgouras J., Lindahl T., Human DNA repair genes. Science 291, 1284–1289 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Gibson B. A., Kraus W. L., New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Bai P., Biology of poly(ADP-ribose) polymerases: The factotums of cell maintenance. Mol. Cell 58, 947–958 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Langelier M. F., Eisemann T., Riccio A. A., Pascal J. M., PARP family enzymes: Regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 53, 187–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suskiewicz M. J., Palazzo L., Hughes R., Ahel I., Progress and outlook in studying the substrate specificities of PARPs and related enzymes. FEBS J. 288, 2131–2142 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Teloni F., Altmeyer M., Readers of poly(ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 44, 993–1006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krastev D. B., et al. , Coupling bimolecular PARylation biosensors with genetic screens to identify PARylation targets. Nat. Commun. 9, 2016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasovich M., et al. , Identifying poly(ADP-ribose)-binding proteins with photoaffinity-based proteomics. J. Am. Chem. Soc. 143, 3037–3042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., et al. , PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics 11, 492–500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzese E., et al. , PARP inhibitors in ovarian cancer. Cancer Treat. Rev. 73, 1–9 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Muthurajan U. M., et al. , Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. U.S.A. 111, 12752–12757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David K. K., Andrabi S. A., Dawson T. M., Dawson V. L., Parthanatos, a messenger of death. Front. Biosci. 14, 1116–1128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amé J. C., et al. , PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Schreiber V., et al. , Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277, 23028–23036 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F., Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 329, 18–25 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Chen Q., Kassab M. A., Dantzer F., Yu X., PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 9, 3233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barkauskaite E., Jankevicius G., Ahel I., Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell 58, 935–946 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Hoch N. C., Polo L. M., ADP-ribosylation: From molecular mechanisms to human disease. Genet. Mol. Biol. 43 (suppl. 1), e20190075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawicki-McKenna J. M., et al. , PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell 60, 755–768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eustermann S., et al. , Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell 60, 742–754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudolph J., Mahadevan J., Luger K., Probing the conformational changes associated with DNA binding to PARP1. Biochemistry 59, 2003–2011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudolph J., et al. , The BRCT domain of PARP1 binds intact DNA and mediates intrastrand transfer. Mol. Cell 81, 4994–5006.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obaji E., Haikarainen T., Lehtiö L., Characterization of the DNA dependent activation of human ARTD2/PARP2. Sci. Rep. 6, 34487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilokapic S., Suskiewicz M. J., Ahel I., Halic M., Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin. Nature 585, 609–613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaullier G., et al. , Bridging of nucleosome-proximal DNA double-strand breaks by PARP2 enhances its interaction with HPF1. PLoS One 15, e0240932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farmer H., et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Bryant H. E., et al. , Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Bridges C. B., The origin of variations in sexual and sex-limited characters. Am. Nat. 56, 51–63 (1922). [Google Scholar]
- 31.Dobzhansky T., Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics 31, 269–290 (1946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartwell L. H., Szankasi P., Roberts C. J., Murray A. W., Friend S. H., Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Kuchenbaecker K. B., et al. ; BRCA1 and BRCA2 Cohort Consortium, Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Tai Y. C., Domchek S., Parmigiani G., Chen S., Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 99, 1811–1814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J., Huang J., DNA double-strand break repair pathway choice: The fork in the road. Genome Instab. Dis. 1, 10–19 (2020). [Google Scholar]
- 36.Trego K. S., et al. , Non-catalytic roles for XPG with BRCA1 and BRCA2 in homologous recombination and genome stability. Mol. Cell 61, 535–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell S. N., Kachnic L. A., Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22, 5784–5791 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Scully R., Livingston D. M., In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408, 429–432 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkitaraman A. R., Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Evers B., Helleday T., Jonkers J., Targeting homologous recombination repair defects in cancer. Trends Pharmacol. Sci. 31, 372–380 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Helleday T., The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 5, 387–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtin N. J., Szabo C., Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 19, 711–736 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Vikas P., Borcherding N., Chennamadhavuni A., Garje R., Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front. Oncol. 10, 570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiegel J. O., Van Houten B., Durrant J. D., PARP1: Structural insights and pharmacological targets for inhibition. DNA Repair (Amst.) 103, 103125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pommier Y., O’Connor M. J., de Bono J., Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps17 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y., et al. , The ups and downs of poly(ADP-ribose) polymerase-1 inhibitors in cancer therapy: Current progress and future direction. Eur. J. Med. Chem. 203, 112570 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Yi M., et al. , Advances and perspectives of PARP inhibitors. Exp. Hematol. Oncol. 8, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger N. A., et al. , Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br. J. Pharmacol. 175, 192–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noordermeer S. M., van Attikum H., PARP inhibitor resistance: A tug-of-war in BRCA-mutated cells. Trends Cell Biol. 29, 820–834 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Crawford K., Bonfiglio J. J., Mikoč A., Matic I., Ahel I., Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 53, 64–82 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Alvarez-Gonzalez R., Jacobson M. K., Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry 26, 3218–3224 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Desmarais Y., Ménard L., Lagueux J., Poirier G. G., Enzymological properties of poly(ADP-ribose)polymerase: Characterization of automodification sites and NADase activity. Biochim. Biophys. Acta 1078, 179–186 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Murai J., et al. , Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murai J., et al. , Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurgina T. A., Anarbaev R. O., Sukhanova M. V., Lavrik O. I., A rapid fluorescent method for the real-time measurement of poly(ADP-ribose) polymerase 1 activity. Anal. Biochem. 545, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Rudolph J., Roberts G., Muthurajan U. M., Luger K., HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase. eLife 10, e65773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holtlund J., Jemtland R., Kristensen T., Two proteolytic degradation products of calf-thymus poly(ADP-ribose) polymerase are efficient ADP-ribose acceptors. Implications for polymerase architecture and the automodification of the polymerase. Eur. J. Biochem. 130, 309–314 (1983). [DOI] [PubMed] [Google Scholar]
- 58.Bauer P. I., Hakam A., Kun E., Mechanisms of poly(ADP-ribose) polymerase catalysis; mono-ADP-ribosylation of poly(ADP-ribose) polymerase at nanomolar concentrations of NAD. FEBS Lett. 195, 331–338 (1986). [DOI] [PubMed] [Google Scholar]
- 59.Alvarez-Gonzalez R., 3′-Deoxy-NAD+ as a substrate for poly(ADP-ribose)polymerase and the reaction mechanism of poly(ADP-ribose) elongation. J. Biol. Chem. 263, 17690–17696 (1988). [PubMed] [Google Scholar]
- 60.Ménard L., Thibault L., Poirier G. G., Reconstitution of an in vitro poly(ADP-ribose) turnover system. Biochim. Biophys. Acta 1049, 45–58 (1990). [DOI] [PubMed] [Google Scholar]
- 61.Ruf A., de Murcia G., Schulz G. E., Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry 37, 3893–3900 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Kawaichi M., Ueda K., Hayaishi O., Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J. Biol. Chem. 256, 9483–9489 (1981). [PubMed] [Google Scholar]
- 63.Cheung A., Zhang J., A scintillation proximity assay for poly(ADP-ribose) polymerase. Anal. Biochem. 282, 24–28 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Penning T. D., et al. , Discovery and SAR of 2-(1-propylpiperidin-4-yl)-1H-benzimidazole-4-carboxamide: A potent inhibitor of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer. Bioorg. Med. Chem. 16, 6965–6975 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Putt K. S., Hergenrother P. J., An enzymatic assay for poly(ADP-ribose) polymerase-1 (PARP-1) via the chemical quantitation of NAD(+): Application to the high-throughput screening of small molecules as potential inhibitors. Anal. Biochem. 326, 78–86 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Jarmoskaite I., AlSadhan I., Vaidyanathan P. P., Herschlag D., How to measure and evaluate binding affinities. eLife 9, e57264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudolph J., Roberts G., Luger K., Histone parylation factor 1 contributes to the inhibition of PARP1 by cancer drugs. Nat. Commun. 12, 736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hopkins T. A., et al. , Mechanistic dissection of PARP1 trapping and the impact on in vivo tolerability and efficacy of PARP inhibitors. Mol. Cancer Res. 13, 1465–1477 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Hopkins T. A., et al. , PARP1 trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol. Cancer Res. 17, 409–419 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Goldstein B., Coombs D., He X., Pineda A. R., Wofsy C., The influence of transport on the kinetics of binding to surface receptors: Application to cells and BIAcore. J. Mol. Recognit. 12, 293–299 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Shen Y., et al. , BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 19, 5003–5015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lord C. J., Ashworth A., In the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W., et al. , Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garnett M. J., et al. , Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaspers J. E., et al. , Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pilié P. G., Gay C. M., Byers L. A., O’Connor M. J., Yap T. A., PARP inhibitors: Extending benefit beyond BRCA-mutant cancers. Clin. Cancer Res. 25, 3759–3771 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Zandarashvili L., et al. , Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satoh M. S., Lindahl T., Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 79.Kedar P. S., Stefanick D. F., Horton J. K., Wilson S. H., Increased PARP-1 association with DNA in alkylation damaged, PARP-inhibited mouse fibroblasts. Mol. Cancer Res. 10, 360–368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rudolph J., Mahadevan J., Dyer P., Luger K., Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. eLife 7, e37818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krüger A., Bürkle A., Hauser K., Mangerich A., Real-time monitoring of PARP1-dependent PARylation by ATR-FTIR spectroscopy. Nat. Commun. 11, 2174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prokhorova E., et al. , Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun. 12, 4055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Purnell M. R., Whish W. J. D., Novel inhibitors of poly(ADP-ribose) synthetase. Biochem. J. 185, 775–777 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langelier M.-F., Zandarashvili L., Aguiar P. M., Black B. E., Pascal J. M., NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 9, 844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marsischky G. T., Wilson B. A., Collier R. J., Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 270, 3247–3254 (1995). [DOI] [PubMed] [Google Scholar]
- 86.Thorsell A. G., et al. , Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J. Med. Chem. 60, 1262–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibbs-Seymour I., Fontana P., Rack J. G. M., Ahel I., HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell 62, 432–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wahlberg E., et al. , Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 30, 283–288 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Vyas S., et al. , Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 5, 4426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonfiglio J. J., et al. , Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palazzo L., et al. , Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, 34334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suskiewicz M. J., et al. , HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun F. H., et al. , HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 12, 1028 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brustel J., et al. , Linking DNA repair and cell cycle progression through serine ADP-ribosylation of histones. Nat. Commun. 13, 185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou P., Wang J., Mishail D., Wang C. Y., Recent advancements in PARP inhibitors-based targeted cancer therapy. Precis. Clin. Med. 3, 187–201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mateo J., et al. , DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmermann M., et al. , CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fugger K., et al. , Targeting the nucleotide salvage factor DNPH1 sensitizes BRCA-deficient cells to PARP inhibitors. Science 372, 156–165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hewitt G., et al. , Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol. Cell 81, 767–783.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Juhász S., et al. , The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci. Adv. 6, eabb8626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lo C. S. Y., et al. , SMARCAD1-mediated active replication fork stability maintains genome integrity. Sci. Adv. 7, eabe7804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sinha G., Downfall of iniparib: A PARP inhibitor that doesn’t inhibit PARP after all. J. Natl. Cancer Inst. 106, djt447 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published data were used for this work (as specified in the text).