Significance
The nucleotide-binding oligomerization domain (NOD)-like receptor pyrin domain containing 3 (NLRP3) is a pattern recognition receptor that forms an inflammasome. The cryo-electron microscopy structure of the dodecameric form of full-length NLRP3 bound to the clinically relevant NLRP3-specific inhibitor MCC950 has established the structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation and the mechanism of action of the NLRP3 specific inhibitor. The inactive NLRP3 oligomer represents the NLRP3 resting state, capable of binding to membranes and is likely disrupted for its activation. Visualization of the inhibitor binding mode will enable optimization of the activity of NLRP3 inflammasome inhibitor drugs.
Keywords: inflammasome, NOD-like receptor, NLRP3
Abstract
The nucleotide-binding oligomerization domain (NOD)-like receptor pyrin domain containing 3 (NLRP3) responds to a vast variety of stimuli, and activated NLRP3 forms an inflammasome, which in turn is associated with conditions such as atherosclerosis, Alzheimer’s disease, and diabetes. A multilayered regulatory mechanism ensures proper NLRP3 inflammasome activation, although the structural basis for this process remains unclear. This study aimed to investigate the cryo-electron microscopy structure of the dodecameric form of full-length NLRP3 bound to the clinically relevant NLRP3-specific inhibitor MCC950. The inhibitor binds to the cavity distinct from the nucleotide binding site in the NACHT domain and stabilizes the closed conformation of NLRP3. The barrel-shaped dodecamer composed of the inactive form of NLRP3 is formed mainly through LRR–LRR interactions on the lateral side, and the highly positively charged top and bottom sides composed of NACHT domains provide a scaffold for membrane association. The cryo-electron microscopy structure suggests that oligomerization of NLRP3 is necessary for its membrane association; it is subsequently disrupted for activation, hence serving as a key player in controlling the spatiotemporal NLRP3 inflammasome activation. These findings are expected to contribute to the development of drugs targeting NLRP3 in future.
Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are cytosolic sensors of pathogen- and danger-associated molecular patterns (PAMPs and DAMPs) that induce innate immune responses (1–5). The NLR family pyrin domain containing 3 (NLRP3), which consists of an N-terminal pyrin domain (PYD), a central NAIP, CIITA, HET-E, and TP1 (NACHT) domain, and a C-terminal leucine-rich repeat (LRR) domain, is the most studied member of NLRs that responds to a wide variety of PAMPs and DAMPs and is involved in many physiological and pathological conditions (1–5). Activated NLRP3 recruits ASC and then procaspase-1 to form the NLRP3 inflammasome, which triggers caspase-1 activation that culminates in the maturation of interleukin (IL)-1β and IL-18 for inflammatory responses and cleavage of gasdermin D to induce pyroptosis (1–5). Aberrant activation of the NLRP3 inflammasome is associated with various inflammatory diseases (6, 7), including atherosclerosis, diabetes, obesity (8–10), inflammatory bowel disease (11–13), and Alzheimer’s disease (14–16). Therefore, NLRP3 is an attractive drug target for these diseases, and a number of inhibitors specific to NLRP3 inflammasomes have been developed (17, 18). For example, MCC950 is a recently developed potent inhibitor of NLRP3 (19–22).
NLRP3 inflammasome activation requires two types of stimuli, namely priming (e.g., lipopolysaccharide and tumor necrosis factor) and activation (e.g., nigericin and adenosine triphosphate) stimuli (23). During these processes, many cellular events, including transcriptional up-regulation, posttranslational modification [e.g., phosphorylation (24–28) and ubiquitination (29–33)], cellular localization (34, 35), and protein–protein interactions (36–42), restrict or license NLRP3 activation. In particular, Never in Mitosis A-related kinase 7 (NEK7) has been identified as an essential mediator of NLRP3 activation (37–39). Despite extensive studies, the mechanisms regulating NLRP3 inflammasome activation remain unclear, partly due to the lack of structural information regarding NLRP3. To date, only the structure of the inactive and monomeric form of NLRP3 lacking PYD, in complex with NEK7, has been reported (43).
This study aimed to investigate the structure of full-length NLRP3 in the oligomeric form, which sequesters PYD from self-oligomerization to avoid premature activation. Moreover, this study provides a structural basis for the mechanism of action of the NLRP3-specific inhibitor MCC950.
Results
Cryo-EM Structure of Hexameric Form of PYD-Deleted NLRP3.
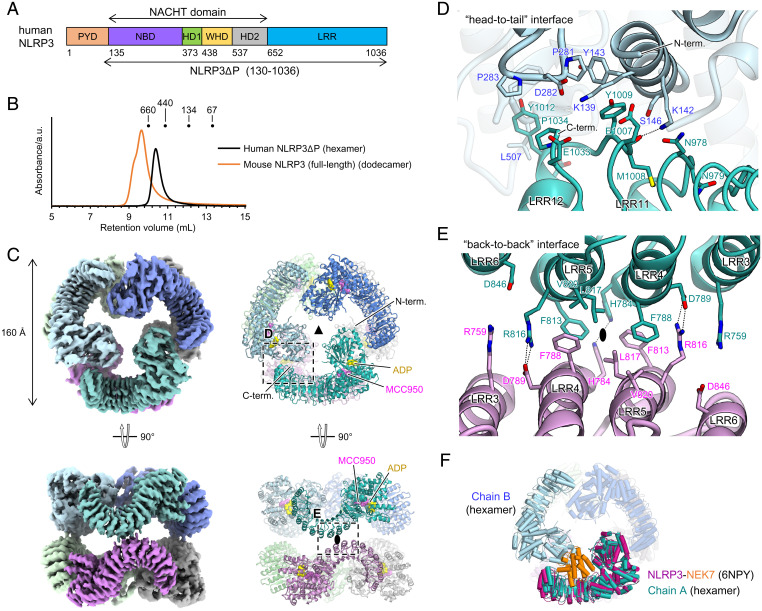
We expressed and purified PYD-deleted recombinant human NLRP3 (residues 130 to 1,036; NLRP3ΔP) (Fig. 1A and SI Appendix, Fig. S1). Addition of the NLRP3 inhibitor MCC950 (19, 20, 22) during protein expression and purification greatly enhanced the stability of NLRP3ΔP. NLRP3ΔP eluted earlier than the monomer, in terms of the expected molecular weight in size-exclusion chromatography (SEC), suggesting that NLRP3ΔP formed oligomers (Fig. 1B). To obtain structural information on the NLRP3ΔP oligomer, we performed cryo-electron microscopy (cryo-EM) of the purified NLRP3ΔP oligomer in the presence of ADP and MCC950. We successfully reconstructed the cryo-EM map of the oligomeric form of NLRP3ΔP to a resolution of 3.2 Å (Fig. 1C and SI Appendix, Figs. S2 and S3 and Table S1).
Fig. 1.
Structure of the human NLRP3ΔP hexamer. (A) Schematic diagram of the domain organization of NLRP3. Residue numbers for human NLRP3 are shown. (B) Size-exclusion chromatography analysis of human NLRP3ΔP and mouse NLRP3 (full length). Elution volumes of the standard proteins thyroglobulin (660 kDa), apoferritin (440 kDa), and bovine serum albumin (134 and 67 kDa) are indicated. (C) Overall structure of the human NLRP3ΔP hexamer. The cryo-EM map (Left) and a ribbon model (Right) are shown. Each protomer is shown in a different color. ADP and MCC950 molecules are shown as yellow and pink spheres, respectively, in the ribbon model. Both threefold and twofold axes are shown. (D and E) Close-up views of the “head-to-tail” (D) and “back-to-back” (E) interfaces. The hydrogen bonds are indicated by dashed lines. The twofold axis is shown in (E). (F) Superposition of the NLRP3-NEK7 complex (PDB 6NPY) onto one protomer in the human NLRP3ΔP hexamer. NLRP3 and NEK7 in the NLRP3-NEK7 complex are shown in dark pink and orange, respectively.
NLRP3ΔP formed a spherical hexamer with a diameter of ∼160 Å (Fig. 1C). The six protomers in the hexamer were arranged with D3 symmetry, where the two trimers, each related by a threefold axis, were also related by a twofold axis perpendicular to the threefold axis (Fig. 1C). Most of the regions of human NLRP3ΔP were resolved in the cryo-EM map, and a structural model was built for residues 135 to 1,034 of human NLRP3ΔP. Several loop regions (residues 130 to 134 [N terminus], 152 to 160 and 180 to 201 [NBD], 452 to 461 and 509 to 515 [WHD], 537 to 553 [HD2], 686 to 725 [LRR], and 1,035 to 1,036 [C terminus]) could not be modeled owing to poor densities (SI Appendix, Fig. S1). Each protomer consisted of the NACHT domain (residues 135 to 651) and the LRR domain (652 to 1,034) (Fig. 1A). The NACHT domain was subdivided into the NBD (residues 135 to 372), HD1 (residues 373 to 437), WHD (residues 438 to 536), and HD2 (residues 554 to 651) (Fig. 1A and SI Appendix, Fig. S1). Two ligand densities within the protomer, corresponding to the bound ADP and MCC950, were observed in the NACHT domain (Fig. 1C and SI Appendix, Fig. S3). Since the protomer in the NLRP3ΔP oligomer was bound to ADP, the structure of NLRP3ΔP represented the inactive and closed form, similar to the previously determined structure of NLRP3ΔP bound to NEK7 (43), with an rmsd value of 2.6 Å (SI Appendix, Fig. S4A). A slight difference in the arrangement of NACHT relative to LRR was observed; distances between the HD2 and the C terminus of LRR were 30 and 35 Å in the NLRP3ΔP protomer of the hexamer and in the NLRP3ΔP bound to NEK7, respectively (SI Appendix, Fig. S4A).
Two interfaces mediated the hexamer formation of NLRP3ΔP, namely the “head-to-tail” interaction between the C terminus of LRR and the NBD and WHD domains through the electrostatic interaction (interface area of 380 Å2) (Fig. 1 C and D and SI Appendix, Fig. S5A) and the “back-to-back” interaction between LRR3 and LRR6 from two protomers around the twofold axis (interface area of 471 Å2) (Fig. 1 C and E). The latter consisted of the central close packing of the hydrophobic residues F788, L817, and F813, which was further strengthened by the peripheral electrostatic interactions mediated by R759, D789, R816, and D846 (Fig. 1 C and E). The hexameric structure was incompatible with NEK7 binding since the NEK7 bound to the concave surface of NLRP3 clashed with the NBD of the adjacent protomer (Fig. 1F).
Inhibitor MCC950 Stabilizes the Closed Conformation of NLRP3.
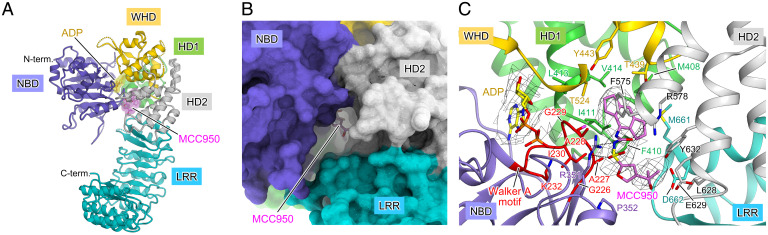
Previously, cryo-EM analysis of human NLRP3ΔP in complex with NEK7 was conducted in the presence of the NLRP3 inhibitor MCC950; however, the binding site and recognition mode of MCC950 were not clarified, due to limited resolution (43). The bound MCC950 was readily identified in the current cryo-EM map of hexameric NLRP3ΔP (SI Appendix, Fig. S3). MCC950 bound to the bottom of the central cavity formed in the NACHT domain (Fig. 2 A and B). The cavity was formed by all of the domains of NLRP3ΔP; NBD, HD2, and LRR formed the entrance and side wall of the cavity, and HD1 and WHD formed the bottom (Fig. 2 A–C). MCC950 and ADP were spatially close, at a distance of ∼8 Å, but were separated by tight packing of NBD, HD1, and WHD at the bottom of the cavity (Fig. 2C). The MCC950 binding cavity was accessible from the NBD-HD2-LRR side, opposite to the ADP-binding cavity (Fig. 2 A and B).
Fig. 2.
MCC950 binds the central cavity in the NACHT domain. (A) Protomer structure of the human NLRP3ΔP hexamer. Each domain is shown in different colors according to the color scheme shown in Fig. 1A. ADP and MCC950 molecules are shown as yellow and pink sticks with semitransparent surfaces, respectively. (B) Surface model of the protomer structure in the human NLRP3ΔP hexamer showing the MCC950 binding cavity formed between NBD, HD2, and LRR. (C) Close-up view of the MCC950 binding site. MCC950 and ADP molecules and the side chains of residues surrounding MCC950 are shown in stick models. Hydrogen bond or ionic interactions are indicated by dashed lines. The Walker A motif is highlighted in red.
The tricyclic hexahydro-s-indacene moiety of MCC950, clearly visible in the cryo-EM map at the bottom of the cavity, formed multiple contacts with the residues A228 (NBD), M408, F410, I411, L413, V414 (HD1), T439, Y443, T524 (WHD), F575, R578, Y632 (HD2), and M661 (LRR) (Fig. 2C). The tricyclic sulfonyl amide moiety was lined up with residues G226, A227, A228, and G229 from the ATP-binding Walker A motif of NBD (Fig. 2C and SI Appendix, Fig. S1). In addition, the amide O atom formed a hydrogen bond with R578 (HD2), and the sulfonyl moiety formed an ionic interaction with R351 (NBD). Although density of the furan moiety was poor, the 2-hydroxypropan and furan moieties formed additional contacts with P352 (NBD), L628, E629, Y632 (HD2), and D662 (LRR).
The closed form of the NACHT domain is generally characterized by ADP-mediated tight packing of the NACHT subdomains. The binding mode of MCC950, where all subdomains of NACHT and LRR are engaged in binding, clearly demonstrated that MCC950 stabilizes the closed form of NACHT in conjunction with ADP (Fig. 2 A and C). Therefore, MCC950 inhibits the opening of NACHT for oligomerization and activation of the NLRP3 inflammasome. Notably, R351, forming ionic interaction with MCC950, is in the sensor 1 motif of NACHT (Fig. 2C and SI Appendix, Fig. S1), which is assumed to be involved in the recognition of the γ-phosphate group of ATP during ATP hydrolysis. Thus, MCC950 binding also prevents ATP binding, which opens the NACHT conformation. This result was consistent with a previous finding of MCC950 closing the NLRP3 conformation and inhibiting NLRP3 activation (19, 20).
Cryo-EM Structure of the Dodecameric Form of Full-Length NLRP3.
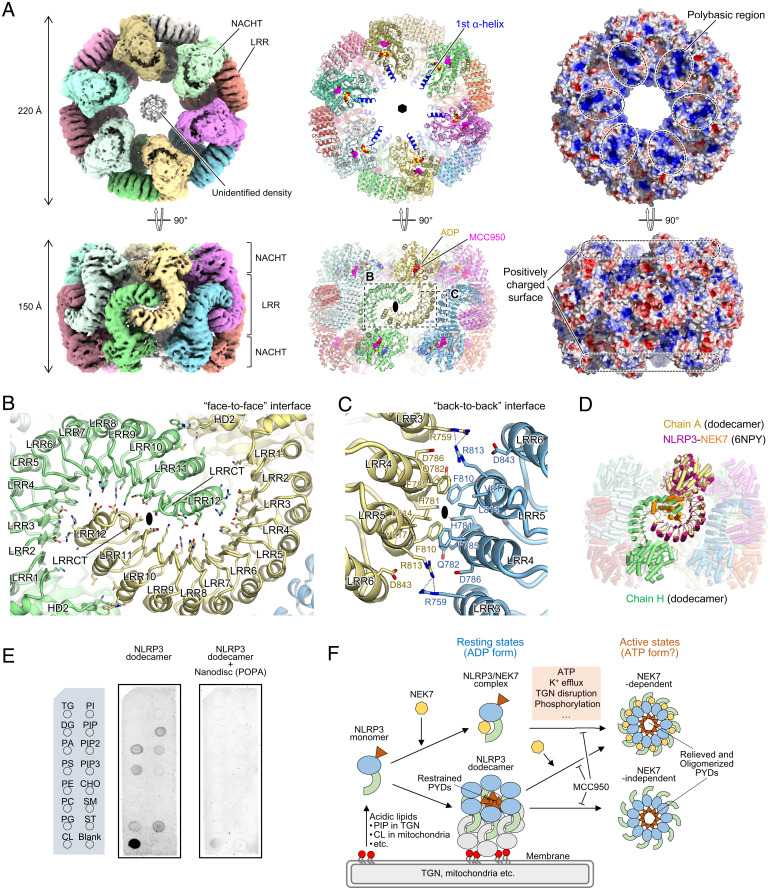
Most of the structural studies on NLRs were based on their signaling domain (e.g., CARD and PYD)-deleted form (43–45); thus, the mechanism underlying the impact of the signaling domain and the following linker region, connected to NACHT, on the structure of NLRs is unknown. Therefore, to study the intact form of NLRP3, we expressed and purified full-length mouse NLRP3 (residues 1 to 1,033) in the presence of ADP and MCC950. NLRP3 eluted earlier than the NLRP3ΔP hexamer in SEC, suggesting that NLRP3 formed a higher-order oligomer (Fig. 1B). We analyzed the structure of the oligomeric form of NLRP3 using cryo-EM. The two-dimensional class-average images clearly showed a sixfold symmetry of the oligomer (SI Appendix, Fig. S2). The resultant three-dimensional reconstruction yielded a 3.6-Å–resolution cryo-EM map with D6 symmetry (Fig. 3A and SI Appendix, Fig. S2 and Table S1). Using the structure of human NLRP3ΔP as a reference, we modeled 12 molecules (dodecamer) of mouse NLRP3 (residues 131 to 1,031), ADP, and MCC950 into the cryo-EM maps.
Fig. 3.
Structure of the full-length mouse NLRP3 dodecamer. (A) Overall structure of the full-length mouse NLRP3 dodecamer. Cryo-EM map (Left), ribbon model (Middle), and electrostatic surface potentials (Right) are shown. Each protomer is shown in a different color. ADP and MCC950 molecules are shown as yellow and pink spheres, respectively, in the ribbon model. Both sixfold and twofold axes are shown in the ribbon model. The first α-helix of the NACHT domain is shown in blue in the ribbon model. (B and C) Close-up views of the “face-to-face” (B) and “back-to-back” (C) interfaces. Hydrogen bonds are indicated by dashed lines. The twofold axes are shown. (D) Superposition of the NLRP3-NEK7 complex (PDB 6NPY) onto one protomer in the mouse NLRP3 dodecamer. NLRP3 and NEK7 in the NLRP3-NEK7 complex are shown in dark pink and orange, respectively. (E) Membrane lipid-binding assay of the NLRP3 dodecamer in the absence or presence of POPA-reconstituted nanodiscs (ND). Layout of the membrane lipid strip sheet (Left) and images of the lipid strip sheet detected by chemiluminescence for the NLRP3 dodecamer (Middle) and NLRP3 dodecamer with POPA-reconstituted ND (Right) are shown. TG, triglyceride; DG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; CL, cardiolipin; PI, phosphatidylinositol; PIP, PtdIns(4)P; PIP2, PtdIns(4,5)P2; PIP3, PtdIns(3,4,5)P3; CHO, cholesterol; SM, sphingomyelin; ST, sulfatide. (F) NLRP3 inflammasome activation model. NEK7-NLRP3 binding and the resting-state oligomerization of NLRP3 (this study) are mutually exclusive events. The NLRP3 dodecamer attaches to the trans-Golgi network (TGN) or mitochondrial membranes via its polybasic region binding to acidic lipids, such as PIP and PA. The NLRP3 inhibitor, MCC950, binds to and stabilizes the closed form of NLRP3, thereby suppressing the disruption of oligomers or structural changes to the open form for NLRP3 activation. Multiple factors are individually or concertedly involved in NLRP3 inflammasome activation, some of which could regulate the activation by directly or indirectly affecting the oligomerized state of NLRP3.
Overall, the dodecamer of NLRP3 formed a barrel-shaped oligomer with a hollow core, in which each of the top and bottom sides was formed by six NACHT domains, and the lateral side was formed by 12 LRR domains (Fig. 3A). The barrel had a diameter of ∼220 Å and a height of 150 Å. The NACHT domains at the top or bottom sides of the barrel made no direct contact with the neighboring NACHT domains; thus, the densities of the NACHT domains were very weak compared with those of the LRR regions, although the cryo-EM density map clearly showed the NLRP3 protomer to be in a closed and inactive conformation (SI Appendix, Fig. S4B). Therefore, the protomer structure in the dodecamer was well aligned with that in the hexamer, yielding an RMSD value of 1.2 Å (SI Appendix, Fig. S4B). Although the current structural model lacked residues 1 to 130, corresponding to PYD and the linker region, unidentified densities were visible at the center of the top and bottom sides of the barrel, possibly originating from some of these regions (Fig. 3A).
The NLRP3 dodecamer is formed mainly via LRR–LRR interactions. Two types of LRR–LRR interface were identified in the dodecamer, namely the “face-to-face” interface (interface area of 1,172 Å2), in which the two LRRs from neighboring protomers interact with their concave surfaces facing each other (Fig. 3 A and B), and the “back-to-back” interface (interface area of 487 Å2), in which the two LRRs interact with their convex surfaces facing each other (Fig. 3 A and C). In the “face-to-face” interface, residues from the concave surface of LRR3 to LRRCT made contact with those of the α-helix of LRR12 and the β-strand of LRRCT (Fig. 3B). Moreover, the loop region of HD2 (residues 615 to 621) made contact with the α-helices of LRR10 and LRR11. This interface showed the electrostatic complementarities in which the negatively charged C-terminal region of LRR domain made contact with the positively charged concave surface of the neighboring LRR (SI Appendix, Fig. S5B). Interestingly, the “back-to-back” interface perfectly overlapped with that observed in the structure of the NLRP3ΔP hexamer (SI Appendix, Fig. S4C). The electrostatic potential distributions on NLRP3 that contribute to the “face-to-face” interface and the residues that contribute to the “back-to-back” interface are highly conserved between human and mouse NLRP3 (SI Appendix, Figs. S1, S4C and S5), suggesting that human NLRP3 also forms oligomers with a similar arrangement. In the dodecameric structure of NLRP3, since the concave surface of LRR, to which NEK7 binds, is occupied by the neighboring LRR, NLRP3 cannot engage with NEK7 (Fig. 3D).
NLRP3 Dodecamer Preferentially Bound to Negatively Charged Membrane.
Interestingly, the top and bottom surfaces of the NLRP3 dodecamer were highly basic (Fig. 3A). Recent evidence indicates that the positively charged N-terminal polybasic region of the NLRP3 (residues 127 to 143 for mouse NLRP3) is required for NLRP3 activation, possibly through membrane association (35). Although our structural model lacked the first half of the polybasic region, the second half formed the first α-helix of the NACHT domain and contributed to the highly basic properties of the top and bottom surfaces of the dodecamer (Fig. 3A and SI Appendix, Fig. S5). We, therefore, hypothesized that the NLRP3 dodecamer is capable of binding to the membrane through the basic regions of NACHT. To examine the membrane-binding capability of the dodecameric form of NLRP3, we conducted a lipid-binding assay using membrane lipid strips (Fig. 3E). As anticipated, the NLRP3 dodecamer bound to acidic lipids, such as phosphatidic acid, phosphatidyl serine, phosphatidyl inositol phosphates, and cardiolipin. These bindings were largely abrogated in the presence of a nanodisc (ND)-containing phosphatidic acid. Therefore, we concluded that the dodecameric form of NLRP3 represented the resting state of NLRP3, which preferentially bound to negatively charged membranes through electrostatic interactions (Fig. 3F).
Discussion
This study revealed the structures of hexameric NLRP3ΔP and dodecameric full-length NLRP3 in the presence of the NLRP3-specific inhibitor MCC950. Although the hexameric structure was generated artificially, possibly due to the absence of N-terminal PYD, the high-quality cryo-EM map enabled us to identify the binding site of MCC950 and determine its inhibitory mechanism for NLRP3. MCC950 was found to bind to the cavity formed between the NACHT subdomains of NLRP3, stabilizing the closed conformation of NLRP3 and suppressing the structural transition from a closed to an open conformation for inflammasome activation. Importantly, MCC950 was found to act in combination with ADP to stabilize the closed conformation. Our findings are consistent with a recently reported study describing a similar binding mode involving MCC950 or its analog to NLRP3 in the crystal structure of the isolated NACHT domain (46) or the cryo-EM structure of the full-length human NLRP3 decamer (47). Visualization of the inhibitor-binding mode will provide opportunity for future optimization and finetuning of the activity of the molecule.
The dodecameric structure of NLRP3 identified in this study should be an inactive form of NLRP3, since each protomer is in the ADP- and inhibitor-bound closed form. However, we postulated that this structure represents a physiologically relevant membrane-bound resting state of NLRP3, which may play an important role in the regulation of NLRP3 inflammasome activation (Fig. 3F). The preference of the NLRP3 oligomer for acidic lipids, specifically for phosphoinositide and cardiolipin (Fig. 3E), is consistent with that in the previous reports describing the NLRP3 association with the negatively charged membrane of dispersed trans-Goldi network (35) or the NLRP3 redistribution to the mitochondria (48). Accumulating evidence has shown that NLRP3 activation is regulated via a multilayered mechanism. While our structural study suggested that the NLRP3 oligomer may represent a resting state capable of membrane binding, incompatibility of the structure for NEK7 binding strongly suggested that the oligomer should be disrupted and reorganized before NLRP3 inflammasome activation (Fig. 3F). Therefore, posttranslational modifications possibly affect the oligomerization of NLRP3, thereby regulating NLRP3 activation.
Phosphorylation of the conserved tyrosine residues at the polybasic region (Y136, Y140, and Y143 for human NLRP3) by Bruton’s tyrosine kinase has been shown to positively regulate NLRP3 activation, possibly through a charge neutralization mechanism (24). These tyrosine residues are located on the positively charged surface of the top or bottom sides of the NLRP3 barrel (Fig. 3A and SI Appendix, Figs. S1, S5 and S6). Thus, phosphorylation of these residues would reduce the affinity for negatively charged membranes and facilitate NLRP3 dissociation from the membrane, which has been proposed to be an essential step for NLRP3 activation (25). Recent reports found that the phosphorylation and subsequent dephosphorylation of S803 were important regulatory mechanisms of NLRP3 activation (49). S803 is located on the concave surface of LRR (LRR5) and is in both the NEK7-binding interface and the “face-to-face” interface in the dodecameric structure of NLRP3 (SI Appendix, Fig. S6). Thus, phosphorylation of S803 could influence both NEK7 binding and dodecamer formation.
The barrel-shaped dodecameric structure of NLRP3 is mainly achieved through LRR–LRR interactions between protomers (Fig. 3 A–C). Although the precise position was not determined in the structure, the PYD and linker region also contributed to the formation of the dodecamer, since NLRP3ΔP formed an artificial hexamer (Figs. 1 and 3). Weak densities were observed at the center of the top or bottom sides of the barrel, which could be due to part of the PYD or the linker region; however, the dimension of the densities (∼40 Å) was insufficient to fully accommodate the six PYDs (Fig. 3A and SI Appendix, Fig. S7). Therefore, the missing PYD and linker region in the current structure were mostly disordered inside and/or outside the barrel (SI Appendix, Fig. S7). Although the disordered PYDs were spatially close to each other, the positional restraint of the NACHT, to which PYD was connected, by forming the dodecamer barrel, would suppress the self-oligomerization of PYDs and hence, subsequent activation of NLRP3.
Recently, two research groups independently reported the structures of the oligomeric form of full-length NLRP3 [decamer for human NLRP3 (47) and 12 to 16 mer for mouse NLRP3 (50)]. Interestingly, there are differences in the oligomeric number between our study and these reports, which may be due to differences in NLRP3 species or sample preparation procedures. The comparison between the human NLRP3ΔP hexamer in this study and the full-length human NLRP3 decamer suggests that the PYDs are essential for the formation of the decamer, and the decamer is a physiologically relevant form of human NLRP3 oligomer. Despite some variation in the oligomerization states, all NLRP3 oligomers share a similar arrangement with conserved “back-to-back” and “face-to-face” interface between neighboring LRRs, suggesting the importance of these interactions for maintaining the inactive oligomer. The results of previous studies complement our findings and support our conclusion that oligomerization of NLRP3 is a physiologically important regulatory mechanism of NLRP3 activation.
Although further work is required to fully understand the complex regulation and activation mechanism of NLRP3, this study has revealed the NLRP3 oligomerization mechanism, which may be crucial to controlling the spatiotemporal activation of NLRP3.
Materials and Methods
Preparation of Recombinant Proteins.
Genes encoding human NLRP3 (residues 130 to 1,036; NLRP3ΔP) and mouse NLRP3 (residues 1 to 1,033) with an N-terminal His×6 FLAG tag followed by precision protease recognition sequence were inserted into the expression vector pFastBac Dual vector (Thermo Fisher Scientific Inc.). For protein expression, ExpiSf9 cells cultured in ExpiSf CD medium (Thermo Fisher Scientific Inc.) were infected with recombinant baculoviruses and incubated in the presence of 5 to 20 μM MCC950 (Angene Chemical) for 60 to 70 h at 27 °C. The cells were collected via centrifugation and disrupted via sonication in a buffer containing 25 mM Hepes-NaOH (pH 7.5), 0.5 M NaCl, 10% glycerol, 1 mM DTT, 25 mM imidazole, 0.1 mM ADP, 50 μM MCC950, and protease inhibitor mixture (Nacalai Tesque). Recombinant proteins were purified from the cleared lysate using Ni-NTA agarose (FUJIFILM Wako). The proteins were eluted using a buffer containing 25 mM Hepes-NaOH (pH 7.5), 0.5 M NaCl, 10% glycerol, 1 mM DTT, 250 mM imidazole, 0.1 mM ADP, and 50 μM MCC950. The eluate was further purified via size-exclusion chromatography (SEC) (ENrich SEC 650 10/300, Bio-Rad) in 25 mM Hepes-NaOH (pH 7.5), 0.2 M NaCl, 1 mM MgCl2, 0.5 mM TCEP, 0.1 mM ADP, and 10 μM MCC950. The peak fractions were collected and concentrated in the SEC buffer containing increased concentrations of ADP (1.0 mM) and MCC950 (50 μM) using an Amicon Ultra centrifugal filter (100-kDa molecular weight cutoff).
Cryo-EM Analysis.
For cryo-EM grid preparation, human NLRP3ΔP hexamer sample (1.9 mg/mL) or mouse NLRP3 dodecamer sample (2.8 mg/mL) was used. Then, 3-μL aliquots of samples were placed onto freshly glow-discharged Quantifoil holey carbon grids (R1.2/1.3, Cu, 300 mesh). After 4.0 s of blotting in 100% humidity at 6 °C, the grid was plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data were collected using a Titan Krios G4i microscope (Thermo Fisher Scientific), running at 300 kV and equipped with a GIF Quantum-LS Energy filter (Gatan) and a Gatan K3 camera in electron-counting mode at the cryo-EM facility of the University of Tokyo, Japan. Imaging was performed at a nominal magnification of 105,000×, which corresponded to a calibrated pixel size of 0.83 Å ⋅ px−1. Each movie was recorded in correlative double sampling (CDS) mode for 5.0 s and subdivided into 64 frames with an accumulated exposure of 68 e– per Å2 on the specimen. Data were acquired by the image-shift method using SerialEM software (51). Cryo-EM data were analyzed using RELION 3.1 (52). Raw movie stacks were motion-corrected using the RELION version of MotionCor2 (53). The contrast transfer function (CTF) parameters were determined using the CTFFIND4 program (54). The data processing workflow is summarized in SI Appendix, Fig. S2. The final resolution was estimated using the gold-standard Fourier shell correlation (FSC) between two independently refined half maps (FSC = 0.143). Local resolution maps were produced using the ResMap program (55). The atomic models of human NLRP3ΔP hexamer and mouse NLRP3 dodecamer were subjected to iterative cycles of manual model adjustment using the COOT program (56) and real-space refinement in the Phenix program (57). EM data processing and refinement statistics are summarized in SI Appendix, Table S1. The cryo-EM maps were deposited in the Electron Microscopy Data Bank. Structural representations were generated using Chimera (58), ChimeraX (59), PyMol (60), or CueMol (http://www.cuemol.org).
Lipid Binding Assay.
For the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA)-reconstituted ND preparation, 4 mg of POPA sodium salt (Avanti, 840857C) dissolved in chloroform was dried up using a gentle stream of nitrogen gas and further dried under high vacuum. Sodium cholate and buffer A (10 mM Hepes-NaOH pH 7.7, 150 mM NaCl, and 1 mM EDTA) were added to the dried lipid film. The lipid was dispersed by vigorous vortexing and then dissolved by continuous sonication at 310 K. The MSP1D1 scaffold protein (homemade) was added to the lipid solution at a final concentration of 133 μM MSP1D1, 30 mM sodium cholate, and 10 mM POPA. The solution was incubated with gentle mixing at ambient temperature for 1 h. The POPA-reconstituted ND was purified by gel filtration, running in buffer A, and concentrated to ∼13 mg/mL using Amicon Ultra centrifugal filters (Merck, 50-kDa MW cutoff). The purified POPA-reconstituted ND was flash-cooled in liquid nitrogen and stored at −70 °C until use.
For the membrane lipid strip assay (Echelon Biosciences Inc., P-6002), the sheets were first blocked using Blocking One (Nacalai Tesque) at ambient temperature for 2 h. The sheets were then gently washed using wash buffer (10 mM Hepes-NaOH pH 7.7, 200 mM NaCl, 1 mM EDTA-K, and 0.01% (wt/vol) Tween-80) and incubated with 2.5 μg/mL NLRP3 dodecamer, 0.5 mg/mL bovine serum albumin (Nacalai Tesque), and 0.05 mM ADP in the absence or presence of 0.05 mg/mL POPA-reconstituted ND at 277 K for 2 h. A partially tag-cleaved form of the NLRP3 dodecamer was used in this assay. After incubation, the sheets were washed five times with wash buffer and incubated with anti–DDDDK-tag mAb-HRP-DirecT (MBL, Cat No. M185-7, Lot No. 009). The signals were detected using Chemi-Lumi One chemiluminescence reagent (Nacalai Tesque).
Supplementary Material
Acknowledgments
We thank Masahide Kikkawa, Haruaki Yanagisawa, Yoichi Sakamaki, and Yoshiaki Kise for the management and support of the Graduate School of Medicine cryo-EM facility at The University of Tokyo. This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology Grant Nos. 19H03164 (U.O.), 20K16274 (H.I.), 20K15730 (Z.Z.), 19H00976 (T.S.); Core Research for Evolutional Science & Technology (CREST), Japan Science & Technology Agency (JST) (T.S.); Sumitomo Dainippon Pharma Co., Ltd. (T.S.); the Takeda Science Foundation (U.O. and T.S.); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (U.O.); the Daiichi Sankyo Foundation of Life Science (U.O.); the Uehara Memorial Foundation (T.S.); and the Naito Foundation (U.O. and T.S.); the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from Japan Agency for Medical Research and Development (AMED) under Grant No. JP21am0101115 (support No. 1570, 1846, 1848).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or supplementary Figures and Table. Cryo-EM maps and related structure coordinates of the human NLRP3ΔP hexamer and mouse NLRP3 (full-length) dodecamer have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession codes EMD-32119 (PDB: 7VTP) and EMD-32120 (PDB: 7VTQ), respectively.
References
- 1.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Guo H., Callaway J. B., Ting J. P., Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamkanfi M., Dixit V. M., Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Tschopp J., Schroder K., NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10, 210–215 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Schroder K., Tschopp J., The inflammasomes. Cell 140, 821–832 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Sharma B. R., Kanneganti T. D., NLRP3 inflammasome in cancer and metabolic diseases. Nat. Immunol. 22, 550–559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangan M. S. J., et al. , Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17, 688 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Masters S. L., et al. , Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 11, 897–904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duewell P., et al. , NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stienstra R., et al. , Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 108, 15324–15329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaki M. H., et al. , The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villani A. C., et al. , Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat. Genet. 41, 71–76 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer C., et al. , Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut 59, 1192–1199 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Heneka M. T., et al. , NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493, 674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halle A., et al. , The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 9, 857–865 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saresella M., et al. , The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol. Neurodegener. 11, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahid A., Li B., Kombe A. J. K., Jin T., Tao J., Pharmacological inhibitors of the NLRP3 inflammasome. Front. Immunol. 10, 2538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Wang H., Kouadir M., Song H., Shi F., Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 10, 128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapia-Abellán A., et al. , MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 15, 560–564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coll R. C., et al. , MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 15, 556–559 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Primiano M. J., et al. , Efficacy and pharmacology of the NLRP3 inflammasome inhibitor CP-456,773 (CRID3) in murine models of dermal and pulmonary inflammation. J. Immunol. 197, 2421–2433 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Coll R. C., et al. , A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley N., Jeltema D., Duan Y., He Y., The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 20, E3328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bittner Z. A., et al. , BTK operates a phospho-tyrosine switch to regulate NLRP3 inflammasome activity. J. Exp. Med. 218, e20201656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., et al. , Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J. Exp. Med. 214, 2671–2693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stutz A., et al. , NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214, 1725–1736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song N., et al. , NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 68, 185–197.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Guo C., et al. , Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 45, 802–816 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Yan Y., et al. , Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160, 62–73 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Han S., et al. , Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 ligase. J. Biol. Chem. 290, 18124–18133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Py B. F., Kim M. S., Vakifahmetoglu-Norberg H., Yuan J., Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Castejon G., et al. , Deubiquitinases regulate the activity of caspase-1 and interleukin-1β secretion via assembly of the inflammasome. J. Biol. Chem. 288, 2721–2733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juliana C., et al. , Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 287, 36617–36622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magupalli V. G., et al. , HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science 369, eaas8995 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J., Chen Z. J., PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan Y., et al. , RACK1 mediates NLRP3 inflammasome activation by promoting NLRP3 active conformation and inflammasome assembly. Cell Rep. 33, 108405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H., et al. , NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid-Burgk J. L., et al. , A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y., Zeng M. Y., Yang D., Motro B., Núñez G., NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abais J. M., et al. , Nod-like receptor protein 3 (NLRP3) inflammasome activation and podocyte injury via thioredoxin-interacting protein (TXNIP) during hyperhomocysteinemia. J. Biol. Chem. 289, 27159–27168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitoma H., et al. , The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39, 123–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu B., et al. , Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670–674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharif H., et al. , Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maekawa S., Ohto U., Shibata T., Miyake K., Shimizu T., Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 7, 11813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Z., et al. , Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Dekker C., et al. , Crystal structure of NLRP3 NACHT domain with an inhibitor defines mechanism of inflammasome inhibition. J. Mol. Biol. 433, 167309 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Hochheiser I. V., et al. , Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature, in press (2022). [DOI] [PubMed]
- 48.Zhou R., Yazdi A. S., Menu P., Tschopp J., A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Niu T., et al. , NLRP3 phosphorylation in its LRR domain critically regulates inflammasome assembly. Nat. Commun. 12, 5862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andreeva L., et al. , NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184, 6299–6312.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrodinger L., The PyMOL Molecular Graphics System v.1.3r1 (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or supplementary Figures and Table. Cryo-EM maps and related structure coordinates of the human NLRP3ΔP hexamer and mouse NLRP3 (full-length) dodecamer have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession codes EMD-32119 (PDB: 7VTP) and EMD-32120 (PDB: 7VTQ), respectively.