Summary
A major challenge in industrial pig production is the prevalence of post-weaning diarrhea (PWD) in piglets, often caused by enterotoxigenic Escherichia coli (ETEC). The increased use of antibiotics and zinc oxide to treat PWD has raised global concerns regarding antimicrobial resistance development and environmental pollution. Still, alternative treatments targeting ETEC and counteracting PWD are largely lacking. Here, we report the design of a pH, temperature, and protease-stable bivalent VHH-based protein BL1.2 that cross-links a F4+ ETEC model strain by selectively binding to its fimbriae. This protein inhibits F4+ ETEC adhesion to porcine epithelial cells ex vivo and decreases F4+ ETEC proliferation when administrated as a feed additive to weaned F4+ ETEC challenged piglets. These findings highlight the potential of a highly specific bivalent VHH-based feed additive in effectively delimiting pathogenic F4+ ETEC bacteria proliferation in piglets and may represent a sustainable solution for managing PWD while circumventing antimicrobial resistance development.
Subject areas: Infection control in health technology, Porcine medicine, Microbiology
Graphical abstract
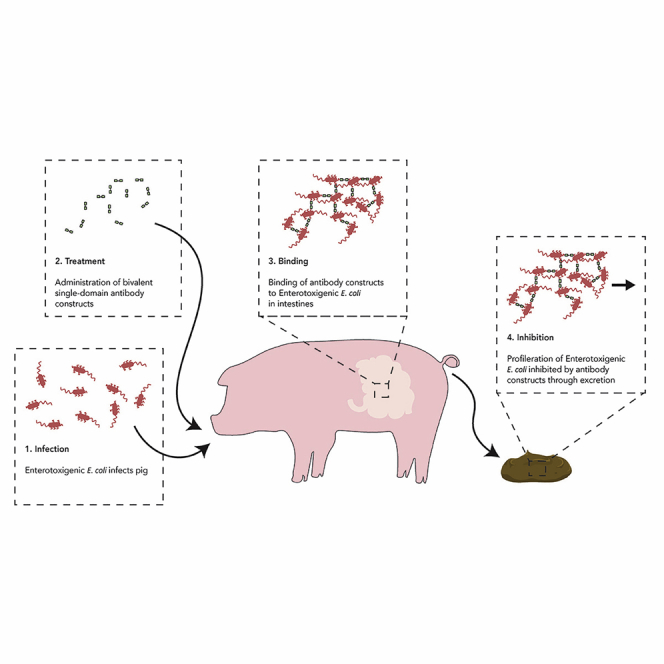
Highlights
-
•
A binding protein was designed as a bivalent VHH construct with a (GGGGS)3 linker
-
•
The protein can cross-link F4+ enterotoxigenic Escherichia coli (ETEC) in vitro
-
•
The protein can prevent adhesion of F4+ ETEC to porcine epithelial cells ex vivo
-
•
The protein can prevent proliferation of F4+ ETEC in piglets
Infection control in health technology; Porcine medicine; Microbiology
Introduction
Enterotoxigenic Escherichia coli (ETEC) is a major cause of illness and mortality in humans and animals. In commercial pig production, ETEC can trigger post-weaning diarrhea (PWD) in newly weaned piglets (Gonzales-Siles and Sjöling, 2016; Gyles and Fairbrother, 2005) because of an immature immune system and an abrupt change in feed and environment. PWD greatly reduces productivity, impacting piglet growth and feed conversion; forcing farmers to administer antibiotics and/or medicinal zinc oxide to the piglets (Gyles and Fairbrother, 2005). PWD poses a great economic burden for pig farmers, while simultaneously harming the environment as the current treatment of PWD accelerates antibiotic resistance development (Johanns et al., 2020; Luppi, 2017; Rhouma et al., 2017) and heavy metal pollution (Johanns et al., 2020). With increased international attention on reducing antibiotic usage and on sustainable farming, legislation is limiting the use of current treatment options. This includes an EU-wide ban on the use of medicinal zinc oxide in pig production from 2022. The development of novel and sustainable solutions for managing ETEC-mediated PWD is therefore warranted (Virdi et al., 2019).
Porcine ETEC strains are characterized by their repertoire of specific fimbrial adhesins and enterotoxins, most of which are carried on plasmids (Gonzales-Siles and Sjöling, 2016). These proteins orchestrate the attachment of the bacterium to the epithelial cells in the gastrointestinal (GI) tract and induce inflammation and fluid loss, respectively (Devriendt et al., 2010; Luo et al., 2015). The most common fimbrial adhesins in porcine ETEC are F4 (also called K88), F5 (K99), F6 (987P), F18, and F41, with the F4 and F18 types being predominant (Luppi et al., 2016; Shepard et al., 2012). The most common enterotoxins in porcine ETEC strains include heat-labile enterotoxin (LT), heat-stable enterotoxins a and b (STa and STb), and Escherichia coli heat-stable enterotoxin (EAST1) (Gonzales-Siles and Sjöling, 2016; Luppi et al., 2016), where the first three are known to be toxic to piglets. In addition, the Shiga-like toxin type 2e, which induces symptoms, such as edema, is specific for the F18 positive (F18+) porcine ETEC strains (Luppi et al., 2016). In combination, these virulence factors mediate pathogenesis (Wang et al., 2020), leading to a reduction in animal welfare and substantial economic losses in industrial pig production (Luppi, 2017).
Previously, it has been reported that IgA-like molecules, based on the fusion of an Fc region with specific camelid single-domain antibody binding domains (VHHs), could prevent an F4+ ETEC strain from adhering to the intestinal lining in piglets, thereby lowering its transit time in the intestine and facilitating its rapid excretion (Virdi et al., 2019). These results are highly promising from a scientific viewpoint, as they demonstrate that ETEC-based virulence can be inactivated in piglets using antibody technology. However, the industrial application of antibody-based molecules in a low-cost market, such as in pig production, sets high demands for the cost of manufacture, which must be low for new products to substitute the current treatment options. Therefore, technological innovation is needed with the scope of developing low-cost solutions for management of ETEC infections in piglets. Here, we demonstrate the utility of a simple concept: A highly specific, pH, temperature, and protease-stable bivalent VHH construct targeting the F4+ ETEC fimbriae to neutralize their binding ability, which, because of its small size and simple structure, is inexpensive to manufacture. We show that intestinal proliferation of F4+ ETEC in weaned piglets can be inhibited via oral administration of such bivalent VHH constructs. Moreover, we elucidate some of the likely underlying mechanisms for inhibition of F4+ ETEC proliferation by the bivalent VHH construct via in vitro and ex vivo investigations. These results point towards a feasible avenue for reducing the risk factors associated with PWD in the industrial setting using low-cost proteins.
Results
Genome assembly of the Danish ETEC isolates AUF4 and AUF18
To ensure that previously identified toxins and fimbriae of F4+ and F18+ ETEC strains were represented in the Danish isolates used for this study, the genomes of two established porcine ETEC isolates, AUF4 and AUF18, were de novo assembled, producing assemblies of 220 (AUF4) and 277 (AUF18) contigs, respectively. The draft assemblies had a frequency of ambiguous bases (N) of 10.82 (AUF4) and 13.18 (AUF18) per 100 kbp, andand the largest produced contigs were 236,509 and 288,553 bp long, respectively. Both assemblies were used in their entirety for analysis, as fragments of plasmids, insertion sequences, and other sequence motifs relating to horizontal gene transfer are essential parts of the ETEC genome carrying resistance genes and virulence factors. Both AUF4 and AUF18 had genome sizes, gene contents, and GC contents comparable to other porcine ETEC genomes (Ren et al., 2014; Shepard et al., 2012; Wyrsch et al., 2015). Both isolates were verified for their F4 and F18 fimbrial genotypes and for the presence of ETEC-related toxins (Table S1) (Carattoli, 2009; Johnson and Nolan, 2009).
Microbially expressed VHH constructs bind F4+ ETEC
Simple VHH constructs were designed for microbial expression based on a previously published proteolytically stable VHH sequence isolated from an immunized llama (Harmsen et al., 2006). The VHH protein was designed as both a monovalent (BL1.1) and bivalent (BL1.2) construct, the latter comprising two VHH domains connected by a Gly-Ser linker (GGGGS)3. The constructs were produced with 6xHis and FLAG-tags or non-tagged for their use in different assays (Figure 1A). To evaluate the stability of the Gly-Ser linker, two bivalent (unrelated) VHH constructs were designed and expressed; one comprising the Gly-Ser linker, and the other comprising the hinge region of the porcine IgG3 as linker (Butler et al., 2009). The enzymatic stability of these bivalent VHH constructs was assessed by subjecting them to pepsin (at pH 3), trypsin (at pH 8), and pancreatin (at pH 8), followed by SDS-PAGE analysis. This revealed that the Gly-Ser linker displayed increased proteolytic stability compared to the hinge region of IgG3 (Figure S1). The Gly-Ser linker variant of BL1.2 was therefore chosen for the remaining part of the study.
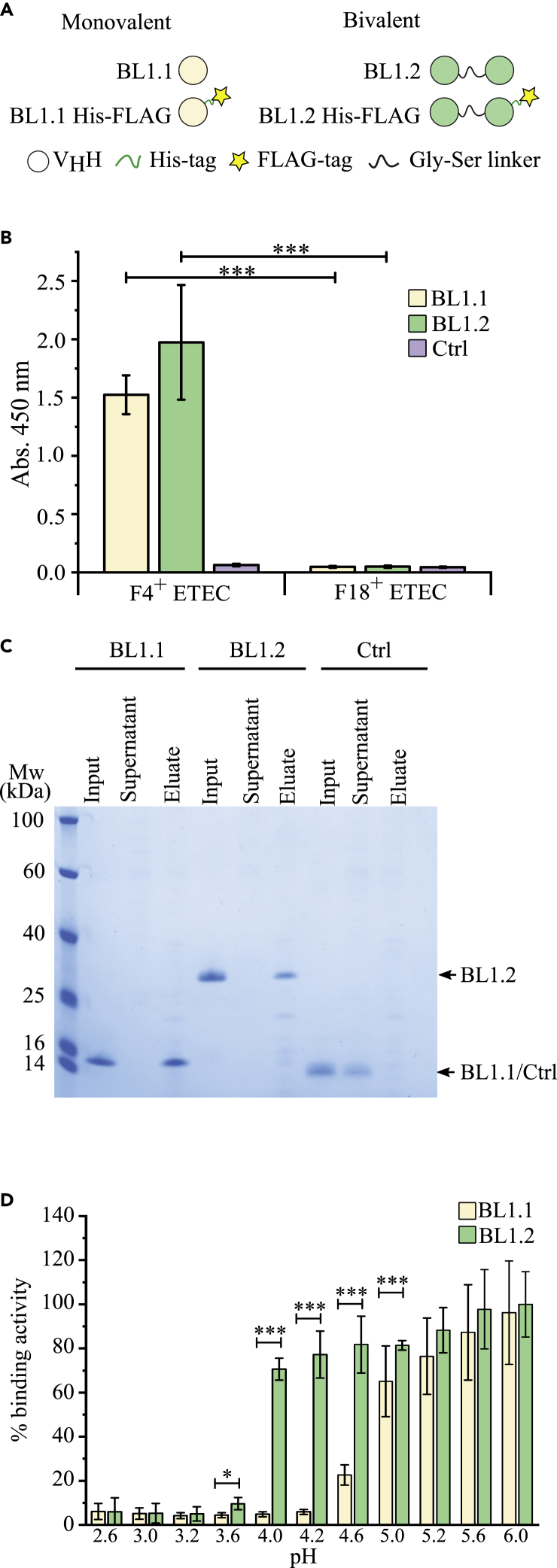
Figure 1.
The bivalent BL1.2 VHH construct outperforms the monovalent BL1.1 construct in binding to F4+ ETEC at pH ranges found in the GI of weaned piglets
(A) Schematic representation of the main VHH constructs used in this study.
(B) Indirect ELISA-based binding analysis of the 6xHis and 3xFLAG tagged monovalent BL1.1 and bivalent BL1.2 constructs to F4+ and F18+ ETEC compared to a control VHH (Ctrl) carrying a 6xHis and 3xFLAG tag.
(C) Representative SDS-PAGE showing “in solution” binding assay of the untagged BL1.1, BL1.2, and a control VHH constructs to F4+ ETEC.
(D) Binding of the 6xHis and 3xFLAG tagged BL1.1 and BL1.2 constructs to F4+ ETEC immobilized on Maxisorp plates at a pH range from 2.6 to 6.0 analyzed by indirect ELISA. Binding analyses (A–D) were performed in independent biological triplicates. Absorbance values (A and D) are presented as mean ± SD (n > 3). Statistics were based on an unpaired two-tailed Student’s t-test (B and D); ∗, p < 0.05; ∗∗∗, p < 0.001.
To confirm that BL1.1 and the Gly-Ser linker-based BL1.2 specifically bind F4+ ETEC, and not the F18+ ETEC serotype, the specificity of the two FLAG-tagged constructs was tested using an indirect ELISA set up with the corresponding bacterial strains immobilized onto the plate. This verified that both BL1.1 and BL1.2 bind F4+, but not F18+ ETEC, and that the control VHH did not bind any of the bacterial strains (Figure 1B). However, as the only F4+ ETEC strain available to us was of the F4ac subtype, it is unknown whether other subtypes, such as F4ab or F4ad, can be bound by BL1.1 and BL1.2. Binding of untagged BL1.1 and BL1.2 to F4+ ETEC was confirmed in solution by incubating the corresponding VHH constructs with F4+ ETEC and visualizing either their depletion from the solution supernatants after pelleting bacteria, or their release from the bacteria after citric acid incubation, respectively, by a Coomassie stained SDS-PAGE. No interaction was seen with the control VHH construct (Figure 1C). The identity of BL1.1 in the bands representing the citric acid eluate was confirmed via mass spectrometry (Table S2).
Protein-protein interactions are, among other biochemical characteristics, typically strongly pH-dependent. We hypothesized that the bivalent BL1.2 construct would exhibit higher binding avidity to F4+ ETEC at lower pH because of the presence of an additional binding domain. To test our hypothesis, we studied the release of BL1.1 and BL1.2 binding to F4+ ETEC at a pH range from 2.6 to 6.0, aiming to resemble conditions present in the GI tract of pre-weaned to post-weaned piglets. Bacteria were immobilized on ELISA plates, incubated with VHH constructs (BL1.1 and BL1.2 at equal amounts), washed and then incubated with buffers at the indicated pH, and after extensive washing, the remaining bound VHH constructs were detected. Strikingly, between pH 5.0 and pH 3.6, BL1.2 binds F4+ ETEC with significantly higher avidity as compared to BL1.1, while above pH 5.0 or below pH 3.6, both constructs show comparable binding to F4+ ETEC (Figure 1D).
Bivalent VHH construct mediates cross-linking of bacteria and prevents F4+ ETEC adhesion to pig enterocytes
Initial binding analyses demonstrated a higher binding avidity of BL1.2 as compared to BL1.1 at lower pH. To unravel if the increased avidity is solely based on BL1.2 binding two epitopes on the same bacterial cell, or if the bivalent VHH construct may also cross-link multiple bacteria by binding epitopes on different bacterial cells, we incubated a suspension of F4+ ETEC with BL1.1 or BL1.2, using phosphate buffered saline (PBS) as control. Based on microscopy, we could demonstrate that the bivalent VHH construct led to the formation of large bacterial aggregates, while PBS and monovalent BL1.1 did not affect the homogeneity of the bacterial suspension (Figure 2A). These findings show that under in vitro conditions, bacteria can be cross-linked via binding of bivalent VHH constructs to epitopes on two separate bacterial cells via each of the VHH domains.
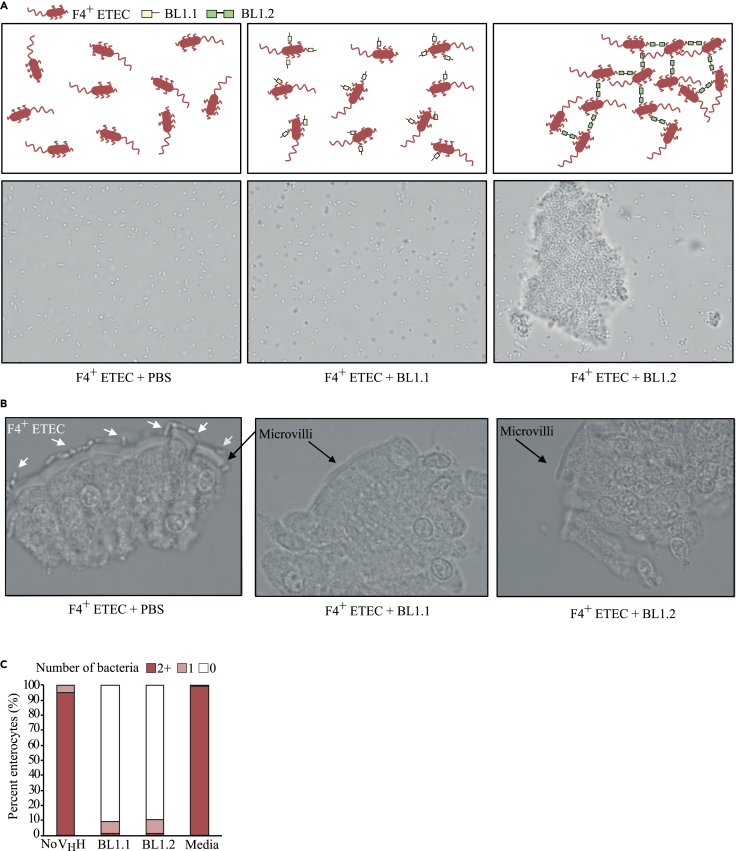
Figure 2.
Bivalent BL1.2 VHH construct induces agglutination of F4+ ETEC in vitro and prevents ETEC adhesion to small intestinal pig enterocytes ex vivo
(A) Schematic representation and microscopy images of F4+ ETEC incubated with PBS, monovalent BL1.1, and bivalent BL1.2 VHH construct.
(B) Microscopy images of freshly isolated small intestinal pig enterocytes incubated with PBS, BL1.1, and BL1.2.
(C) Percentage of pig enterocytes with two or more, one, or without bound F4+ ETEC bacteria, respectively. Enterocytes were added to bacteria exposed to the following treatments: no VHH (PBS), BL1.1, BL1.2, or culture media alone. The number of attached F4+ ETEC bacteria to each pig enterocyte was quantified from counting of 50–150 enterocytes per treatment group.
To test if the constructs are able to inhibit F4+ ETEC adhesion to the villi of small intestinal enterocytes from pigs, we isolated enterocytes, and pre-incubated F4+ ETEC with or without BL1.1 or BL1.2 before addition of the mixture to the cells. The percentage of attachment of F4+ ETEC to the villi of the pig enterocytes was then quantified using microscopy. Indeed, when F4+ ETEC were co-incubated with VHH constructs, bacterial binding to the villi of the enterocytes was greatly reduced (Figure 2C).
Bivalent VHH is stable under GI tract-like temperature, pH, and physiological conditions
A feed product designed to function in the small intestine needs to persist through the digestive tract and remain active in the intestinal environment to inhibit binding of F4+ ETEC to enterocytes. To evaluate pH, temperature, and protease stability, BL1.2 was subjected to stresses akin to those of passing through a pig gut and to conditions in the small intestine, where the product should be able to bind specifically to F4+ ETEC bacteria. BL1.2 was pre-incubated for up to 2 h in fresh pig gastric juice sampled from euthanized pigs, after which its binding to F4+ ETEC was analyzed by an indirect ELISA. The untagged BL1.2 (subjected to gastric juice) was used to coat the wells, after which F4+ ETEC was applied in excess and allowed to bind. Then BL1.1-FLAG was used to quantify the amount of bound bacteria as a measure of ‘functional’ gastric juice treated BL1.2. There was considerable pig-to-pig variability, but compared to neutral pH, treatment of BL1.2 with gastric juice for 60 min or 120 min did not significantly affect the binding to F4+ ETEC (unpaired two-tailed Student’s t-test: p> 0.22 for 60 min and p> 0.24 for 120 min), and the construct retained on average 78% of its ability to bind bacterial cells (Figure S2A). The employed conditions reflected the gastric environment of piglets of 2–3 months of age, and therefore only serve as a proxy for the gastric environment of piglets at weaning (28 days of age), but nonetheless should only represent conditions that are no less harsh than those of post-weaned piglet. Only when the pH was adjusted to 3 (a pH below the stomach pH of 4.4 in adult pigs (Merchant et al., 2011)), binding to bacteria was significantly lower compared to neutral pH, although the construct still retained on average 40% of its ability to bind F4+ ETEC. Subjecting BL1.2 to stomach acid (HCl) at pH 2, 3, or 4 for 1 h at 37°C, before using it to coat an ELISA well, affected the ability of the construct to bind bacteria in a pH-dependent manner. However, at the physiologically relevant pH 4, more than 80% of the construct retained its ability to bind bacteria (Figure S2B). Bile is another component secreted during the passage of the GI tract that could possibly affect the binding of the construct to F4+ ETEC bacteria. However, when we extracted bile from euthanized pigs, and co-incubated the BL1.2 construct and bacteria in increasing doses of pig bile, we found no detrimental effects of pig bile on the binding between construct and bacteria. Rather, the binding between the two was increased by enhancing the bile percentage (Figure S2C). The same phenomenon was observed when the construct was allowed to bind bacteria in the presence of the bile salt sodium deoxycholate (Figure S3).
Determination of the number of VHH molecules binding to a single F4+ ETEC bacterium
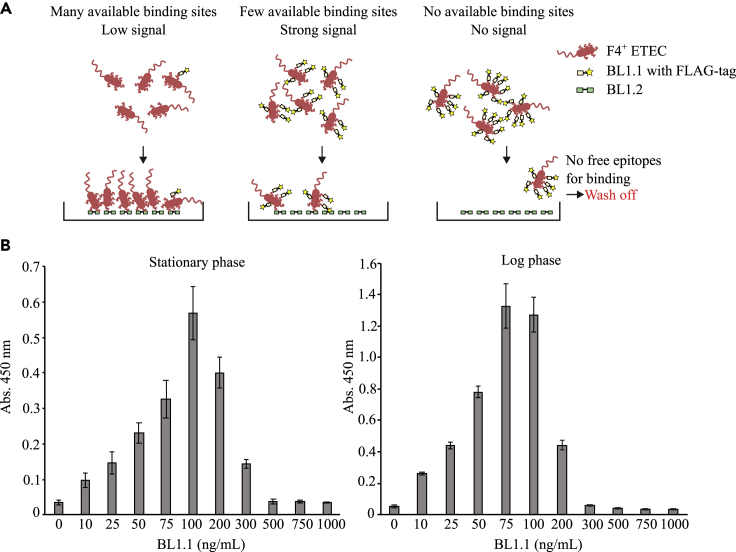
Next, an ELISA-based assay was designed to estimate the number of VHH molecules needed to occupy all relevant fimbrial epitopes present on a single F4+ ETEC bacterium. This estimate is important in order to evaluate the amount of VHH construct needed for in vivo challenge trials (Figure 3), and thus to estimate the cost of using the product in pig production. The estimate was based on binding of the monovalent VHH construct, BL1.1. In this ELISA setup, a fixed number of F4+ ETEC were first co-incubated with increasing amounts of FLAG-tagged BL1.1 before being added to a well coated with a constant amount of untagged BL1.2. With increasing FLAG-tagged BL1.1 concentration, F4+ ETEC bacteria are detected until a cutoff is reached, where all F4+ ETEC fimbriae are bound by FLAG-tagged BL1.1. After this, the signal disappears because there are no fimbriae available for binding to the coated BL1.2 (the FLAG-tagged BL1.1 provides the readout signal). Based on product concentration and CFU of bacterial cultures in these assays, it was estimated that one F4+ ETEC bacterium binds approximately 10,000–15,000 monovalent VHH molecules, as defined using both stationary and log-phase grown cultures. Based on this number, the theoretical total amount needed for inactivating the absolute dose of F4+ ETEC during the course of the in vivo F4+ ETEC challenge trial was calculated to be roughly 7 μg of BL1.2.
Figure 3.
Evaluation of the amount of BL1.1 VHH construct required to occupy all binding epitopes on F4+ETEC
(A) Schematic presentation of the ELISA setup used for determining the number of BL1.1 binding sites on a single F4+ ETEC bacterium.
(B) ELISA-based binding analysis of the monovalent BL1.1 VHH construct to F4+ ETEC cells grown in log or stationary phase. The assay was used to estimate the BL1.1 concentration required to occupy all fimbrial epitopes present on a single F4+ ETEC cell. Binding assays (B) were performed as technical quadruplicates (n = 4), and data are presented as mean + SD.
Bivalent VHH construct mediates rapid clearance of F4+ ETEC challenge in post-weaning piglets
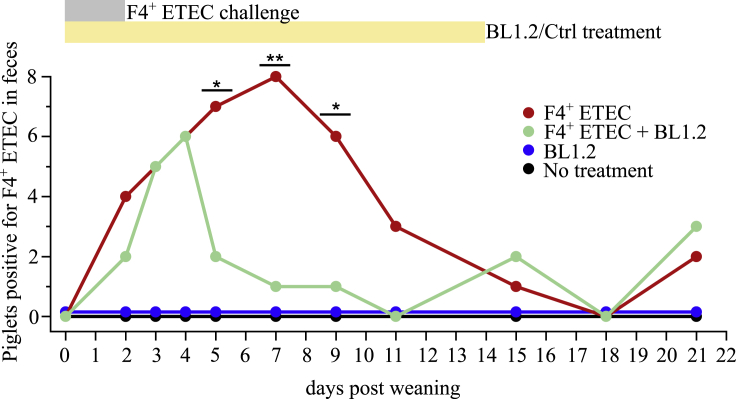
A challenge trial was carried out using pigs, which were genetically susceptible to F4+ ETEC infection because of a genetic variation in the MUC4 gene, as determined by routine genetic screening of sows and offspring (when used for breeding) by Dr. van Haeringen Laboratorium B.V., Wageningen, the Netherlands (Sugiharto et al., 2012). The experiment was designed to test if piglets provided with BL1.2 would hold less intestinal F4+ ETEC bacteria than non-treated controls during the treatment period. F4+ ETEC-susceptible piglets were either challenged with a dose of F4+ ETEC (AUF4) for two consecutive days after weaning (at day 28 of age) or not (here defined as day 1 and 2). The challenged piglets were divided into two groups: One receiving BL1.2 in solution and a control receiving a similar dose of control protein (albumin from chicken egg white). Both groups were dosed twice daily over the course of 14 days, starting on the day of weaning. Pigs were treated with BL1.2 or control protein by oral gavage to ensure even dosage. Fecal samples were collected and monitored for hemolytic bacteria by cultivation, and a subset of colonies was verified by serotyping that confirmed them to be F4+ ETEC. For both challenge groups, the number of piglets with F4+ ETEC increased after experimental exposure to F4+ ETEC, reaching 80% of piglets being positive for F4+ ETEC at day 4 after weaning/after challenge. Of note, all F4+ ETEC treated piglets, except one in the F4+ ETEC + BL1.2 group, tested positive for F4+ ETEC at some point within the first 5 days after challenge, Figure 4, Table S3, while the piglets that were not challenged, but received the BL1.2 construct or control protein, were negative for F4+ ETEC. Importantly, for the F4+ ETEC challenged groups, a statistically significant reduction (at day 5, 7, and 9) in the number of F4+ ETEC positive piglets with BL1.2 administration was observed, which was largest at day 7, where all 8 of the challenged piglets (100%) in the control protein administered group were positive for F4+ ETEC, while only 1 piglet (12.5%) was positive in the BL1.2 administered group (Figure 4). It is worth noting that although homozygotic sows and boars encoding a susceptibility gene (MUC4) to F4+ ETEC infection were used for breeding, for the piglets involved in these experiments, F4+ ETEC infection susceptibility was not assessed. Therefore, it cannot be excluded that some receptor negative piglets could have been present in the different groups.
Figure 4.
The bivalent BL1.2 VHH construct promotes a faster clearance of F4+ ETEC in piglets challenged with F4+ ETEC post-weaning
Piglets in treatment groups “F4+ ETEC” and “F4+ ETEC + BL1.2“ were challenged with 1–1.7 × 109 F4+ ETEC at day one and two post-weaning. The non-challenged groups were provided sodium bicarbonate. BL1.2 or the control protein, chicken egg albumin (Ctrl), which were administered to the “F4+ ETEC” and “no treatment” groups, were provided from day 1–14 via oral gavage. N = 8 for each treatment group. Displayed is the number of piglets tested positive for F4+ ETEC in feces over a period of 21 days post-weaning within each group. Statistical significance between “F4+ ETEC” and “F4+ ETEC + BL1.2” groups were based on a Fisher’s exact test; ∗∗, p < 0.01; ∗, p < 0.05. Source data and specification of positive tested piglets are provided in Table S3.
To assess the effect of F4+ ETEC infection and the benefit of receiving BL1.2, the feed intake, weight gain, diarrhea scores, and basic blood response parameters were evaluated, although the trial was not designed with the purpose of studying these parameters. As expected, no statistically significant differences were observed between the four groups during the duration of the trial (Figure S4 and Table S4).
Discussion
We have developed a simple protein product, BL1.2, which is a bivalent VHH construct that here showed promising results in terms of its ability to reduce in vitro adhesion of F4+ ETEC to freshly isolated pig enterocytes and to enhance the clearance of F4+ ETEC in in vivo challenged piglets. Specifically, BL1.2 was shown in vitro to selectively bind F4+ ETEC, and not F18+ ETEC, to block binding of F4+ ETEC to villi of pig enterocytes, and to be stable under conditions resembling the GI tract of piglets. Moreover, the in vivo challenge results showed a significant reduction in the number of piglets that were infected with F4+ ETEC in the BL1.2 administrated group as compared to the control group after challenge. Based on these in vitro data, we speculate that the in vivo effect of BL1.2 may relate to its ability to inhibit intestinal colonization with F4+ ETEC. However, the in vivo effect of BL1.2 may also be because of its ability to agglutinate bacteria, consistent with other reports where only bivalent antibodies conferred in vivo protection (de Geus et al., 1998; Ma et al., 1990; van Zijderveld et al., 1998). Furthermore, as only the F4ac subtype was employed in this study, it is unknown whether BL1.2 is also effective against other subtypes, such as F4ab or F4ad. The robustness of BL1.2 against potential escape mutants was also not evaluated, but would be relevant for further study.
To provide an early indication of industrial applicability, titration experiments were conducted to evaluate BL1.2 dosage. Based on these experiments, we estimated that a dose of 7 μg of BL1.2 could be sufficient in saturating all binding sites of approximately 2 × 109 CFU of F4+ ETEC. This low dose indicates that the application of a BL1.2-based product could have a low cost-in-use for controlling PWD in the pig industry, even if all binding sites need to be saturated. However, it could also be speculated that a lower dose may suffice, if bacteria are forced to agglutinate by the bivalent construct, as indicated by the ex vivo data. Combined, the data presented here provide proof of concept for the use of a simple and inexpensive bivalent VHH construct as a possible solution for reducing the number of colonizing F4+ ETEC, which could be utilized as an alternative to antibiotics and zinc oxide currently used to manage PWD in piglets.
Previous studies on the use of VHH constructs for prevention of PWD showed a lack of efficacy and attributed this to product degradation in the GI tract (Harmsen et al., 2005; Reilly et al., 1997). However, it could also be speculated that the lack of efficacy could be attributed to the monovalency, and therefore the inability to induce agglutination, for these VHH constructs. Nevertheless, to optimize stability, secretory immunoglobulin A (sIgA)-like constructs, where VHH domains are fused to the porcine IgA Fc region, were constructed, and these constructs showed good efficacy in vivo (Virdi et al., 2013, 2019). Whether the improved efficacy could be attributed to the potentially improved stability or the bivalency of the previously reported sIgA-like constructs is, however, unknown. The significantly lower molecular weight of BL1.2 (∼28 kDa) and very high pH, temperature, and protease stability likely allow for dosing at a similar or lower level than sIgA-like constructs (∼80 kDa), although further testing is needed to define the optimal dose. Studies of other types of antibody-based products, tested for their ability to prevent and treat PWD, include the use of purified pig plasma IgG (Hedegaard et al., 2016, 2017) and spray-dried plasma powder (Niewold et al., 2007). Both these strategies have demonstrated a comparable effect on reducing the risk of PWD to the currently utilized medicinal zinc oxide (Hedegaard et al., 2017). Given the likelihood that specific IgG antibodies exist among the polyclonal mixture of IgGs in these plasma-derived products and are responsible for the observed effect in these cases, it seems likely that even somewhat unstable IgG antibodies may have sufficient stability to exert their effect in the GI tract when administered orally together with scavenger proteins present in feed. In comparison, bivalent VHH constructs are well-defined products with high specificity and affinity and often possess high physicochemical stability (van der Linden et al., 1999). Here, we demonstrate that the bivalent VHH-based protein BL1.2 can endure not only gastric juice treatment, which contains proteases and low pH, but also retain binding functionality under in vivo conditions in the piglet GI tract. As such, this bivalent VHH construct could possibly find utility as a feed additive.
In summary, the simple bivalent VHH construct BL1.2 described here was documented to prevent F4+ ETEC attachment to villi of primary pig enterocytes ex vivo, be suitable for oral administration, and possess in vivo efficacy. This indicates that BL1.2 could find utility as a simple solution for reducing the risk of F4+ ETEC proliferation and development of ETEC-mediated PWD in piglets in industrial pig production.
Limitations of the study
Although the link between F4+ ETEC colonization and negative health impact is well-established for the susceptible of piglets to F4+ ETEC infection, it is relevant to emphasize that during the duration of the trial, no significant differences were detected between challenged piglets receiving BL1.2 and control piglets in regards to weight gain, diarrhea scores, and basic blood response parameters, such as hematocrit percentage and leukocyte numbers. To assess these health parameters better, it is likely that either (1) a larger number of piglets should be enrolled in each group, (2) that a more severe F4+ ETEC challenge should be employed, or (3) that particularly feed intake and weight gain should be monitored for an extended period, as the impact of a milder F4+ ETEC infection might only materialize later in life. Moreover, in regards to feed intake and weight gain, it is important to more thoroughly assess both the feed conversion rate as well as whether the weight gain is attributed to growth or prevention of water loss caused by diarrhea. Finally, productivity parameters could also be interesting to assess in future studies, as not only absolute weight gain and feed conversion, but also time taken to reach full size for a pig, are important parameters for the pig production industry.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Untagged BL1.1 and BL1.2 constructs | Novozymes laboratories, Bagsværd, Denmark | N/A |
| Mouse monoclonal anti-FLAG M2-Peroxidase (HRP) antibody | Sigma | A8592-5X1MG; RRID: AB_439702 |
| Bacterial and virus strains | ||
| E. coli BL21 for cloning and expression | New England Biolabs | C2527H |
| Biological samples | This paper | N/A |
| Porcine gastric juice | This paper | N/A |
| Porcine bile | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| NcoI restriction enzyme | New England Biolabs | R3193S |
| NotI restriction enzyme | New England Biolabs | R3189S |
| T4 DNA ligase | New England Biolabs | M0202S |
| Benzonase Nuclease | Millipore | E1014-25KU |
| cOmplete Protease Inhibitor Cocktail | Roche | 11836145001 |
| r-lysozyme | Sigma Aldrich | L6876-10G |
| SimplyBlue™ SafeStain | Invitrogen | 10432072 |
| LysC (MS grade) | Wako | 121-05063 |
| Trypsin (MS grade) | Promega | V5280 |
| Critical commercial assays | ||
| PureLink™ Genomic DNA Mini Kit | Invitrogen | K182001 |
| Kapa HyperPlus Library Prep Kit | Roche Molecular Systems | KK8514 |
| MiSeq V2 300 Cycles Reagent kit | Illumina | MS-102-2002 |
| Deposited data | ||
| Genome sequence data for the AU-F4 strain | This paper | N/A |
| Genome sequence data for the AU-F18 strain | This paper | N/A |
| Experimental models: Cell lines | ||
| Intestinal villous enterocytes | This paper | N/A |
| Experimental models: Organisms/strains | ||
|
E. coli O149:F4 (9910045-1) Denoted as: F4+ ETEC/AUF4 |
Danish Veterinary Institute (Copenhagen, Denmark) (Frydendahl, 2002) | GenBank: JAKLOV000000000 |
|
E. coli O138:F18 (9910297-2STM) Denoted as: F18+ ETEC/AUF18 |
Danish Veterinary Institute (Copenhagen, Denmark) (Frydendahl, 2002) | GenBank: JAKLOW000000000 |
| F4+ ETEC challenged weaning piglets | University of Aarhus, Foulum, Denmark | N/A |
| Oligonucleotides | ||
| BL1.1 (DNA) | Eurofins Genomics (Ebersberg, German) and Integrated DNA Technologies (Belgium) | N/A |
| BL1.2 (DNA) | Eurofins Genomics (Ebersberg, German) and Integrated DNA Technologies (Belgium) | N/A |
| Ctrl VHH (DNA) | Eurofins Genomics (Ebersberg, German) and Integrated DNA Technologies (Belgium) | N/A |
| Recombinant DNA | ||
| pSANG10-3F | Ahmadi et al. (2020) | N/A |
| Software and algorithms | ||
| SPAdes v. 3.11.1 | http://cab.spbu.ru/software/spades/ | N/A |
| QUAST | http://quast.sourceforge.net/docs/manual.html | N/A |
| SerotypeFinder 1.1 | https://cge.cbs.dtu.dk/services/SerotypeFinder/ | N/A |
| MLST 1.8 | https://cge.cbs.dtu.dk/services/MLST/ | N/A |
| BLASTn | https://blast.ncbi.nlm.nih.gov/Blast.cgi | N/A |
| Q-Exactive instrument | ThermoFisher Scientific | N/A |
| OriginPro version 2021b | OriginLab | N/A |
| Other | ||
| HisTrap FF 1 mL column | GE Life Science | GE17-5319-01 |
| SDS-PAGE (NuPAGE 4-12% gels) | Invitrogen | NP0322BOX |
| C18 trap column | ThermoScientific | 164705 |
| C18 reversed-phase analytical column | ThermoScientific | ES803 |
| Maxisorp plates (Nunc) | ThermoFisher | 44-2404-21 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Andreas Hougaard Laustsen (ahola@bio.dtu.dk).
Materials availability
There are restrictions to the availability of BL1.1 and BL1.2 due to limited production of test article, and product not yet being registered.
Experimental model and subject details
ETEC strains
The porcine ETEC strains, AUF4 and AUF18, were originally isolated from the intestinal content of pigs diagnosed with PWD at the Danish Veterinary Institute (Copenhagen, Denmark) (Frydendahl, 2002), and are routinely used at the Department of Animal Science, Aarhus University. The E. coli O149:F4 (9910045-1) and O138:F18 (9910297-2STM), denoted in this study as F4+ ETEC/AUF4 and F18+ ETEC/AUF18, respectively, have been used as inocula on a routine basis for ETEC challenge trials in pigs during the last decade (Lauridsen et al., 2011; Sugiharto et al., 2014), ensuring their pathogenicity in pigs. These two isolates were used throughout the present study routinely grown O/N in LB medium with shaking (180 rpm) at 37°C; they have been whole-genome sequenced and analyzed for strain and virulence gene repertoire authentication (see separate section on Genome sequencing of the ETEC strains AUF4 and AUF18).
E. coli BL21(DE3)
E. coli BL21(DE3) was routinely used for protein expression. Day 1: O/N culture using cell scrape from glycerol stock into 5 mL LB media + 5 μL kanamycin (50 μg/mL) was grown at 37°C, 250 rpm O/N. Day 2: 1 L of autoinduction media (including 50 μg/mL kanamycin) + 500 μL O/N culture was placed in an incubator for 2 h at 37°C, 200 rpm. Next, the culture was incubated at 25°C for at least 18 h at 200 rpm.
Small intestinal enterocytes from pigs
Freshly isolated intestinal villous enterocytes were prepared as described elsewhere (Moonens et al., 2014). Briefly, from a recently euthanized pig (specific pathogen free Yorkshire-Landrace pigs weighing approximately 30 kg, 2–3 months old), a sample from the mid jejunum was cut out and washed in Krebs-Henseleit buffer (120 nM NaCl, 14 nM KCl, 1 nM KH2PO4, 25 nM NaHCO3, pH 7.4). Krebs-Henseleit buffer with 1% formaldehyde was added and incubated for at least 30 min on ice. Samples were washed in autoclaved PBS (pH 7.4), and, using a glass slide, enterocytes were gently scraped from the mucosa and collected in a sterile tube. Sterile PBS (pH 7.4) was added to the enterocytes, and the material was washed by allowing the cells to pellet by gravity and changing the PBS until the supernatant was clear. Enterocytes were stored at 4°C for up to 6 months.
F4+ ETEC challenged weaning piglets
F4+ ETEC susceptible sows (n = 5) fed with a standard Danish sow diet were used in the study. At weaning (day 28 after birth), their piglets of both sexes (n = 32) were divided randomly into four groups and housed in two seperate rooms with 2 pigs in each pen, giving a total of 16 pens, and hence four replicates of each group. The same feed was provided to all piglets from 3 weeks of age (while suckling) and during the entire experiment. The feed (a standard commercial mixture) was fed twice a day semi-ad libitum and water was provided ad libitum. The groups were: 1) Control (albumin from chicken egg white, Sigma), 2) BL1.2, 3) F4+ ETEC (AUF4) challenge, 4) AUF4 challenge and BL1.2.
Products (BL1.2 or control) were provided orally to each pig twice per day (2 × 200 mg/day of BL1.2), starting during the afternoon on the day of weaning and finishing 14 days after. Products were mixed with non-alcoholic beer (Hvidtøl from Hancock Brewery, Denmark) for ease of administration and provided using gavage. On day 1 and 2 postweaning, piglets were challenged orally with AUF4, 1.05 × 109 (trial day 1) and 1.72 × 109 CFU (trial day 2). Control pigs were gavaged with sodium bicarbonate. Fecal samples from each piglet were collected every day during the first week, every second day during the second week; at all time points directly from the rectum. Samples were analyzed for excretion of total coliforms, hemolytic and non-hemolytic bacteria, and presence of AUF4 was verified by serotyping (5 randomly selected colonies). At the end of the trial, all piglets were euthanized.
The pig in vivo challenge experiment was conducted at Aarhus University, Foulum, Denmark according to a license obtained by the Danish Animal Experiments Inspectorate, Ministry of Food, Agriculture and Fisheries, Danish Veterinary and Food Administration, and animals were followed by proper veterinary surveillance throughout the experiment.
Method details
Genome sequencing of the ETEC strains AUF4 and AUF18
Single colonies of the F4+ ETEC (AUF4) and F18+ ETEC (AUF18) strains were inoculated in LB medium and allowed to grow overnight at 37°C, 200 rpm. Genomic DNA was purified using the PureLink™ Genomic DNA Mini Kit (Invitrogen), and genomic libraries were generated using Kapa HyperPlus Library Prep Kit (Roche Molecular Systems). Genomic libraries were sequenced using the MiSeq V2 300 Cycles Reagent kit (Illumina) as 150 bp paired-end reads. De novo assembly of AUF4 and AUF18 was performed using SPAdes v. 3.11.1 (custom command line options:–careful for reduction of mismatches and short indels) (Bankevich et al., 2012). Assembly statistics were generated using QUAST(Gurevich et al., 2013). The serotype and multi-locus sequence type (MLST) of all isolates were determined in silico using SerotypeFinder 1.1 (%ID: 85%, minimum length: 60%) (Joensen et al., 2015) and MLST 1.8 (typing scheme 1) (Larsen et al., 2012), available from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org). ETEC specific virulence factor genes (Table S5) were identified using BLASTn to verify and outline the virulence gene profile.
Heat inactivation of ETEC strains
The F4+ (AUF4) or F18+ (AUF18) ETEC were grown O/N in LB medium with shaking (180 rpm) at 37°C. The medium was removed, and the bacteria were diluted in PBS to OD600 = 1.0, corresponding to approximately 1 × 109 CFU. The bacteria were heat-inactivated at 56°C for 1 h, and the preparation was stored at 4°C for up to 1 month.
Plasmids and cloning
BL1.1, BL1.2, and Ctrl VHH were ordered as synthetic genes optimized for expression in E. coli from Eurofins Genomics (Ebersberg, German) and Integrated DNA Technologies (Belgium). The genes were subcloned into the pSANG10-3F vector (Martin et al., 2006) under the T7 promoter with an N-terminal pelB leader sequence and C-terminal 6xHis-tag followed by 3xFLAG-tag. The genes were digested using NotI and NcoI restriction enzymes (New England Biolabs) and ligated using T4 DNA ligase (New England Biolabs). BL1.1 and BL1.2 were generated from (Harmsen et al., 2006), sequence K922 (GenBank accession number: AJ810819), and added a Gly-Ser (GGGGS)3 linker to generate the Gly-Ser-linked BL1.2, The control VHH construct consisted of LaM4 binding mCherry from PDB entry 6IR1 (https://doi.org/10.2210/pdb6IR1/pdb). Moreover, to assess the stability of the Gly-Ser linker in comparison with the hinge region of the porcine IgG3, which was previously employed in other studies and suggested to have superior proteolytic stability (Muyldermans, 2021; Saerens et al., 2005; Virdi et al., 2013), an alternative bivalent VHH construct containing the hinge region of the porcine IgG3 (VDIEPPTPICPEICSCPAAEVLG) instead of the Gly-Ser linker was also cloned and expressed as described above.
Protein expression and purification
Expression and purification of recombinant proteins was performed as described in Ahmadi et al. (2020). Briefly, proteins were expressed in E. coli BL21(DE3) using autoinduction media for approximately 18h at 30°C. Cells were harvested by centrifugation (10,000 × g, 30 min, 4°C), resuspended in ice cold 30 mM Tris–HCl pH 8.0, 1 mM EDTA, 20% sucrose (w/v), incubated on ice, and pelleted as before. Pellets were dissolved in ice cold 5 mM MgSO4, incubated on ice, and pelleted. Both buffers were supplemented with 25 U/mL Benzonase Nuclease (Millipore), cOmplete Protease Inhibitor Cocktail (Roche) and 1.5 kU/mL r-lysozyme (Sigma Aldrich). Supernatants were loaded onto a HisTrap FF 1 mL column (GE Life Sciences) connected to an ÄKTA prime plus system (GE Healthcare), and the proteins were eluted using increasing concentrations of imidazole in PBS. Proteins were concentrated and buffer exchanged to PBS using Pierce Protein Concentrator PES, 3K MWCO, centrifugal concentrators, and concentrations were determined at 280 nm, assuming 1 absorbance unit is equivalent to 1 mg/mL of protein.
For the ex vivo characterization experiments, test material of untagged BL1.1 and BL1.2 constructs were produced at Novozymes laboratories, Bagsværd, Denmark. The test product was produced via microbial fermentation with secretory expression, after which the biomass was filtered, and the final test article was delivered as a frozen supernatant containing the protein product for the challenge trial. The test material was confirmed for binding activity before it was used for the trial.
Assessment of linker stability using SDS-PAGE
The two bivalent VHH constructs were incubated at 37°C for 1 h with either pepsin (pH 3, 250 μg/mL), trypsin (125 μg/mL), or pancreatin (100 μg/mL), the two latter at pH 8. After incubation, the samples were analyzed using SDS-PAGE (NuPAGE 4–12% gels, Invitrogen), stained using SimplyBlue™ SafeStain (Invitrogen™), and visually inspected for band intensity and protein construct integrity.
Construct pull-down and citric acid elution assay
1 mL of heat-inactivated F4+ ETEC was pelleted, followed by resuspension in PBS containing BL1.1, BL1.2, or control at 0.07 mg/mL and incubation for 10 min at room temperature (RT). Then, the cell suspensions were pelleted again by centrifugation, washed three times with PBS, after which the bound VHH constructs were eluted by resuspending the cell pellets in 0.1 M citric acid (10 min, RT). Input, citric acid eluate (bound protein), and PBS-VHH-supernatant (non-bound protein) were analyzed by SDS-PAGE (NuPAGE 4–12% gels, Invitrogen) and stained using SimplyBlue™ SafeStain (Invitrogen™). The identity of the protein bands was confirmed with mass spectrometry as described in detail below.
Sample preparation for mass spectrometry
Proteins eluted from F4+ ETEC cell pellets were buffer exchanged (Pierce Protein Concentrator PES, 3K MWCO) into lysis buffer (6 M Guanidinium Hydrochloride, 10 mM TCEP, 40 mM CAA, 100 mM Tris pH 8.5), and protein concentrations were determined using a Bradford assay against a bovine serum albumin standard. Afterwards, samples were processed using a previously established protocol (Kulak et al., 2014). Briefly, samples were diluted 3-fold with 10% acetonitrile, 25 mM Tris pH 8.5 and incubated with LysC (MS grade, Wako) at a LysC:protein ratio of 1:50 at 37°C for 4 h. Next, samples were diluted 1:10 with 10% acetonitrile, 25 mM Tris pH 8.5 and further digested with trypsin (MS grade, Promega) in a dilution of 1:100 at 37°C for 20 h. Enzyme activity was quenched by adding 2% (w/v) trifluoroacetic acid (TFA) to a final concentration of 1% (v/v), and peptides were desalted using in house packed C18 StageTips (Rappsilber et al., 2007). For each sample, 2 discs of C18 material (3M Empore) were packed in a 200 μL tip and activated with 40 μL methanol, followed by 40 μL of 80% (v/v) acetonitrile, 0.1% (w/v) formic acid. Following activation, the tips were equilibrated twice with 40 μL of 1% (w/v) TFA, 3% (v/v) acetonitrile, and the samples were loaded using centrifugation (1800 × g). The tips were subsequently washed twice with 100 μL of 0.1% (w/v) formic acid, before the peptides were eluted into clean 500 μL Eppendorf tubes using 40% (v/v) acetonitrile, 0.1% (w/v) formic acid. The eluted peptides were concentrated in an Eppendorf Speedvac and re-constituted in 1% (w/v) TFA, 2% (v/v) acetonitrile for mass spectrometry analysis.
LC-MS/MS data acquisition
LC-MS/MS analysis was performed on a Q-Exactive Mass Spectrometer (ThermoFisher Scientific) at the DTU Proteomics Core facility. A Thermo EasyLC 1000 HPLC system was used to load the samples (100% Buffer A, 0.1% (w/v) formic acid in water) onto a 2 cm C18 trap column (ThermoFisher 164705), connected in-line to a 50 cm C18 reversed-phase analytical column (Thermo EasySpray ES803), and the peptides were eluted over a 70 minute gradient (6%–60%, 80% (v/v) acetonitrile, 0.1% (w/v) formic acid, 45°C, 250 nL/min). The Q-Exactive instrument (ThermoFisher Scientific) was run using a DD-MS2 top10 method, and full MS spectra were collected at a resolution of 70,000, with an AGC target of 3 × 106 or maximum injection time of 20 ms and a scan range of 300–1750 m/z. For collecting MS2 spectra, parameters were set to a resolution of 17,500, with an AGC target value of 1 × 106 or maximum injection time of 60 ms, a normalized collision energy of 25, and an intensity threshold of 1.7e4. Dynamic exclusion was set to 60 s, and ions with a charge state <2 or unknown were excluded.
ELISA determinations
All ELISA assays were performed on Maxisorp plates (Nunc, ThermoFisher). Binding analyses were performed at least in triplicates and as described in detail below.
Binding of constructs to plate-coated bacteria
Maxisorp plates were coated with heat-inactivated bacteria diluted to an OD600 of 0.1–0.5 in PBS. The plates were washed and blocked with 3% skim milk powder in PBS (3% M-PBS). FLAG-tagged BL1.1, BL1.2, or control VHH were added at 50–100 ng/mL in 3% M-PBS, and bound protein was detected using mouse monoclonal anti-FLAG M2-Peroxidase (HRP) antibody (Sigma, diluted 1:20,000) in 3% M-PBS. Plates were developed by incubating with 3,3′,5,5′-tetramethylbenzidine–peroxide solution, and the reaction was stopped with 2 M H3PO4. Absorbance was measured at 450 nm.
Effect of pH on construct binding to bacteria
Bacteria were coated on Maxisorp plates and incubated with the VHH constructs as described above. After washing off any unbound VHH, acid buffers (200 mM Na2HPO4, 0.1 M citric acid) at pH 2.6 to 6 were added to the wells, and the plates were incubated for approximately 10 min at RT. Acid buffer was removed by pipeting, plates were washed, and the protocol was continued as described above.
Binding of BL1.2 to F4+ ETEC under physiological conditions
Maxisorp plates were coated with BL1.2 without a FLAG-tag (50–500 ng/mL) in PBS and blocked with 3% M-PBS. F4+ ETEC at an OD600 of 0.25 were added to the plate in 3% M-PBS, after which FLAG-tagged BL1.2 (50–100 ng/mL in 3% M-PBS) was added. To ensure that a signal was detected within the dynamic range of the assay, standard curves were run in parallel on the plate. The reaction was developed using the HRP-conjugated-anti-FLAG-tagged antibody, as described in the first ELISA protocol above.
Pre-treatment of BL1.2 with gastric juice
Porcine gastric juice was collected from 4 pigs (specific pathogen free Yorkshire-Landrace pigs weighing approximately 30 kg, 2–3 months old) soon after euthanasia and kept on ice. Gastric juice from piglets in the exact same age group (28 days) as those included in the in vivo trial was unavailable. Pre-treatment was performed within 3 h. The gastric juice samples were cleared by centrifugation, and BL1.2 was added to an approximate concentration of 0.3 mg/mL. pH was adjusted with 1 M HCl using pH strips. Samples were incubated for up to 2 h at 37°C and diluted with PBS, before they were used to coat an ELISA plate. The ELISA was performed using the protocol ‘Binding of BL1.2 to F4+ ETEC under physiological conditions‘ as described above.
Pre-treatment of BL1.2 with HCl
BL1.2 was diluted to an approximate concentration of 0.3 mg/mL. pH was adjusted with 1 M HCl using pH strips to the indicated pH and incubated 1 h at 37°C. After incubation, samples were further diluted with PBS, pH adjusted to pH 7.4, and samples were used to coat a Maxisorp plate. The remaining protocol followed the first ELISA protocol above.
Pre-treatment of F4+ ETEC with bile
Bile was extracted from recently euthanized pigs (6 pigs specific pathogen free Yorkshire-Landrace pigs weighing approximately 30 kg, 2–3 months old) and kept at 4°C until use. Bile was briefly heated to 37°C to dissolve lipid precipitates, then mixed at the different amounts of F4+ ETEC and added to BL1.2 coated Maxisorp plates. Because milk precipitated at the highest concentration of bile, 3% bovine serum albumin in PBS was used instead of skim milk in these experiments.
Assessment of BL1.1-FLAG binding to F4+ ETEC
Maxisorp plates were coated with BL1.2 at approximately 200 ng/mL. Heat-inactivated F4+ ETEC at an OD600 of 0.25 was mixed with increasing concentrations of FLAG-tagged BL1.1 in microcentrifuge tubes. After incubation for at least 15 min, the mix was added to the washed plate, and incubated for 1 h at RT. The plate was washed, and the reaction was developed using the HRP-conjugated-anti-FLAG-tagged antibody, as described in the first ELISA protocol above.
Binding of constructs to F4+ ETEC using microscopy
Approximately 5 × 108 live or heat-inactivated F4+ ETEC were pelleted by centrifugation and resuspended in PBS containing BL1.1 or BL1.2 at a concentration of approximately 50 μg/mL. Samples were analyzed by phase-contrast microscopy at a magnification of 100x.
Adhesion between F4+ ETEC and pig enterocytes
The adhesion assay was performed as previously described (Moonens et al., 2014; Virdi et al., 2013). Briefly, approximately 5 × 108 live or heat-inactivated F4+ ETEC co-incubated with BL1.1, BL1.2 (both at a final concentration of 50 μg/mL), or controls (PBS and culture medium), were added to a slurry of freshly prepared small intestinal enterocytes from pigs (approximately 100 μL 50:50 enterocytes/buffer), and left to adhere with rotation for 1 h at RT. Samples were analyzed by phase-contrast microscopy at a magnification of 100x. For quantification, the number of bound bacteria per enterocyte was counted, quantifying between 50-150 single cells/smaller clusters of enterocytes per treatment group.
Quantification and statistical analysis
Statistical analyses were performed using OrifinPro version 2021b, and statistical differences were determined by unpaired two-tailed Student’s t-test, Fisher-Freeman-Halton test, or Fisher’s exact test as specified in the figure legends and/or in the results section. Statistical parameters, including n and p-values, are reported or indicated in the figures, figure legends, and the results section. The data are expressed as arithmetic means with standard deviations (SD), unless otherwise indicated.
Acknowledgments
We thank Innovation Fund Denmark (InnoBooster program, case number 7041-00260B), and the Danish Ministry of Environment and Food Green Development and Demonstration Programme (GUDP) (case number 34009-19-1585) for financial support. We thank Louise Kruse Jensen (Department of Veterinary and Animal Sciences, Pathobiological Sciences) and her team for kindly donating pig tissue for this study. We thank DTU Proteomics Core for expert assistance in mass spectrometry analysis. We thank Inger Marie Jepsen, Aarhus University, Foulum, for her technical assistance in the in vivo ETEC challenge experiment with pigs and for cultivation and analysis of the E. coli in fecal samples of the pigs. We also thank Stine Lyngby, Technical University of Denmark, for her technical assistance with binding assays.
Author contributions
SWT and AHL designed the VHH constructs. LJ and GMH prepared DNA and genome sequenced ETEC isolates. SWT performed de novo assembly and genome analysis. BKF, SWT, SB, AHL, and CL designed the experiments. BKF, TK, SA, and LL performed the in vitro and ex vivo experiments, and CL performed the in vivo experiment. BKF, MP, TK, and SWT analyzed the data. BKF, MP, TK, SWT, CL, SB, and AHL wrote the manuscript. All authors revised and reviewed the manuscript.
Declaration of interests
SWT and AHL are co-founders, employees, and shareholders in Bactolife ApS, and they are inventors behind the patent application WO2020144164A1. The authors declare no other conflicts of interest.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104003.
Contributor Information
Sandra Wingaard Thrane, Email: swt@bactolife.com.
Susanne Brix, Email: sbp@bio.dtu.dk.
Andreas Hougaard Laustsen, Email: ahola@bio.dtu.dk.
Supplemental information
Data and code availability
-
•
This paper does not report original code.
-
•
Genome sequence data for the AUF4 and AUF18 strains have been deposited at NCBI GenBank and are publicly available at the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ahmadi S., Pucca M.B., Jürgensen J.A., Janke R., Ledsgaard L., Schoof E.M., Sørensen C.V., Çalışkan F., Laustsen A.H. An in vitro methodology for discovering broadly-neutralizing monoclonal antibodies. Sci. Rep. 2020;10:10765. doi: 10.1038/s41598-020-67654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.E., Wertz N., Deschacht N., Kacskovics I. Porcine IgG: structure, genetics, and evolution. Immunogenetics. 2009;61:209–230. doi: 10.1007/s00251-008-0336-9. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Geus B., Harmsen M., van Zijderveld F. Prevention of diarrhoea using pathogen specific monoclonal antibodies in an experimental enterotoxigenic E. coli infection in germfree piglets. Vet. Q. 1998;20(Suppl. 3):S87–S89. [PubMed] [Google Scholar]
- Devriendt B., Stuyven E., Verdonck F., Goddeeris B.M., Cox E. Enterotoxigenic Escherichia coli (K88) induce proinflammatory responses in porcine intestinal epithelial cells. Dev. Comp. Immunol. 2010;34:1175–1182. doi: 10.1016/j.dci.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Frydendahl K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 2002;85:169–182. doi: 10.1016/s0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- Gonzales-Siles L., Sjöling Å. The different ecological niches of enterotoxigenic Escherichia coli. Environ. Microbiol. 2016;18:741–751. doi: 10.1111/1462-2920.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyles C.L., Fairbrother J.M. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005;6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- Harmsen M.M., van Solt C.B., Hoogendoorn A., van Zijderveld F.G., Niewold T.A., van der Meulen J. Escherichia coli F4 fimbriae specific llama single-domain antibody fragments effectively inhibit bacterial adhesion in vitro but poorly protect against diarrhoea. Vet. Microbiol. 2005;111:89–98. doi: 10.1016/j.vetmic.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Harmsen M.M., van Solt C.B., van Zijderveld-van Bemmel A.M., Niewold T.A., van Zijderveld F.G. Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl. Microbiol. Biotechnol. 2006;72:544–551. doi: 10.1007/s00253-005-0300-7. [DOI] [PubMed] [Google Scholar]
- Hedegaard C.J., Strube M.L., Hansen M.B., Lindved B.K., Lihme A., Boye M., Heegaard P.M.H. Natural pig plasma immunoglobulins have anti-bacterial effects: potential for use as feed supplement for treatment of intestinal infections in pigs. PLoS One. 2016;11:e0147373. doi: 10.1371/journal.pone.0147373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard C.J., Lauridsen C., Heegaard P.M.H. Purified natural pig immunoglobulins can substitute dietary zinc in reducing piglet post weaning diarrhoea. Vet. Immunol. Immunopathol. 2017;186:9–14. doi: 10.1016/j.vetimm.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Joensen K.G., Tetzschner A.M.M., Iguchi A., Aarestrup F.M., Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanns V.C., Epping L., Semmler T., Ghazisaeedi F., Lübke-Becker A., Pfeifer Y., Eichhorn I., Merle R., Bethe A., Walther B., et al. High-zinc supplementation of weaned piglets affects frequencies of virulence and bacteriocin associated genes among intestinal Escherichia colipopulations. Front. Vet. Sci. 2020;7:614513. doi: 10.3389/fvets.2020.614513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.J., Nolan L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen C., Vestergaard E.-M., Højsgaard S., Jensen S.K., Sørensen M.T. Inoculation of weaned pigs with E. coli reduces depots of vitamin E. Livest. Sci. 2011;137:161–167. [Google Scholar]
- Luo Y., Van Nguyen U., de la Fe Rodriguez P.Y., Devriendt B., Cox E. F4+ ETEC infection and oral immunization with F4 fimbriae elicits an IL-17-dominated immune response. Vet. Res. 2015;46:121. doi: 10.1186/s13567-015-0264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi A. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porc. Health Manag. 2017;3:16. doi: 10.1186/s40813-017-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi A., Gibellini M., Gin T., Vangroenweghe F., Vandenbroucke V., Bauerfeind R., Bonilauri P., Labarque G., Hidalgo Á. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porc. Health Manag. 2016;2:20. doi: 10.1186/s40813-016-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.K., Hunjan M., Smith R., Kelly C., Lehner T. An investigation into the mechanism of protection by local passive immunization with monoclonal antibodies against Streptococcus mutans. Infect. Immun. 1990;58:3407–3414. doi: 10.1128/iai.58.10.3407-3414.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.D., Rojas G., Mitchell J.N., Vincent K.J., Wu J., McCafferty J., Schofield D.J. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 2006;6:46. doi: 10.1186/1472-6750-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant H.A., McConnell E.L., Liu F., Ramaswamy C., Kulkarni R.P., Basit A.W., Murdan S. Assessment of gastrointestinal pH, fluid and lymphoid tissue in the guinea pig, rabbit and pig, and implications for their use in drug development. Eur. J. Pharm. Sci. 2011;42:3–10. doi: 10.1016/j.ejps.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Moonens K., De Kerpel M., Coddens A., Cox E., Pardon E., Remaut H., De Greve H. Nanobody mediated inhibition of attachment of F18 Fimbriae expressing Escherichia coli. PLoS One. 2014;9:e114691. doi: 10.1371/journal.pone.0114691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. A guide to: generation and design of nanobodies. FEBS J. 2021;288:2084–2102. doi: 10.1111/febs.15515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold T.A., van Dijk A.J., Geenen P.L., Roodink H., Margry R., van der Meulen J. Dietary specific antibodies in spray-dried immune plasma prevent enterotoxigenic Escherichia coli F4 (ETEC) post weaning diarrhoea in piglets. Vet. Microbiol. 2007;124:362–369. doi: 10.1016/j.vetmic.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Reilly R.M., Domingo R., Sandhu J. Oral delivery of antibodies.Future pharmacokinetic trends. Clin. Pharmacokinet. 1997;32:313–323. doi: 10.2165/00003088-199732040-00004. [DOI] [PubMed] [Google Scholar]
- Ren W., Liu G., Yin J., Chen S., Li T., Kong X., Peng Y., Yin Y., Hardwidge P.R. Draft genome sequence of enterotoxigenic Escherichia coli strain W25K. Genome Announc. 2014;2:e00225-15. doi: 10.1128/genomeA.00593-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouma M., Fairbrother J.M., Beaudry F., Letellier A. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017;59:31. doi: 10.1186/s13028-017-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens D., Frederix F., Reekmans G., Conrath K., Jans K., Brys L., Huang L., Bosmans E., Maes G., Borghs G., et al. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal. Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- Shepard S.M., Danzeisen J.L., Isaacson R.E., Seemann T., Achtman M., Johnson T.J. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J. Bacteriol. 2012;194:395–405. doi: 10.1128/JB.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiharto S., Hedemann M.S., Jensen B.B., Lauridsen C. Diarrhea-like condition and intestinal mucosal responses in susceptible homozygous and heterozygous F4R+ pigs challenged with enterotoxigenic Escherichia coli. J. Anim. Sci. 2012;90:281–283. doi: 10.2527/jas.53840. [DOI] [PubMed] [Google Scholar]
- Sugiharto S., Hedemann M.S., Lauridsen C. Plasma metabolomic profiles and immune responses of piglets after weaning and challenge with E. coli. J. Anim. Sci. Biotechnol. 2014;5:17. doi: 10.1186/2049-1891-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden R.H.J., Frenken L.G.J., de Geus B., Harmsen M.M., Ruuls R.C., Stok W., de Ron L., Wilson S., Davis P., Verrips C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzymol. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- van Zijderveld F.G., van Zijderveld-van Bemmel A.M., Bakker D. The F41 adhesin of enterotoxigenic Escherichia coli: inhibition of adhesion by monoclonal antibodies. Vet. Q. 1998;20(Suppl. 3):S73–S78. doi: 10.1080/01652176.1998.9694974. [DOI] [PubMed] [Google Scholar]
- Virdi V., Coddens A., Buck S.D., Millet S., Goddeeris B.M., Cox E., Greve H.D., Depicker A. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc. Natl. Acad. Sci. U S A. 2013;110:11809–11814. doi: 10.1073/pnas.1301975110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi V., Palaci J., Laukens B., Ryckaert S., Cox E., Vanderbeke E., Depicker A., Callewaert N. Yeast-secreted, dried and food-admixed monomeric IgA prevents gastrointestinal infection in a piglet model. Nat. Biotechnol. 2019;37:527–530. doi: 10.1038/s41587-019-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sanz Garcia R., Cox E., Devriendt B. Porcine enterotoxigenic Escherichia coli strains differ in their capacity to secrete enterotoxins through varying YghG levels. Appl. Environ. Microbiol. 2020;86:e00523-20. doi: 10.1128/AEM.00523-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrsch E., Roy Chowdhury P., Abraham S., Santos J., Darling A.E., Charles I.G., Chapman T.A., Djordjevic S.P. Comparative genomic analysis of a multiple antimicrobial resistant enterotoxigenic E. coli O157 lineage from Australian pigs. BMC Genom. 2015;16:165. doi: 10.1186/s12864-015-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
Genome sequence data for the AUF4 and AUF18 strains have been deposited at NCBI GenBank and are publicly available at the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.