Significance
An enduring mystery of development is how its timing is controlled, particularly for development after birth, where timing is highly flexible and depends on environmental conditions, such as food availability and diet. We followed timing of cell- and organism-level events in individual Caenorhabditis elegans larvae developing from hatching to adulthood, uncovering widespread variations in event timing, both between isogenic individuals in the same environment and when changing conditions and genotypes. However, in almost all cases, we found that events occurred at the same time, when time was rescaled by the duration of development measured in each individual. This observation of “temporal scaling” poses strong constraints on models to explain timing of larval development.
Keywords: developmental timing, C. elegans, individual variability, temporal scaling, heterochronic genes
Abstract
It is essential that correct temporal order of cellular events is maintained during animal development. During postembryonic development, the rate of development depends on external conditions, such as food availability, diet, and temperature. How timing of cellular events is impacted when the rate of development is changed at the organism level is not known. We used a unique time-lapse microscopy approach to simultaneously measure timing of oscillatory gene expression, hypodermal stem cell divisions, and cuticle shedding in individual Caenorhabditis elegans larvae, as they developed from hatching to adulthood. This revealed strong variability in timing between isogenic individuals under the same conditions. However, this variability obeyed “temporal scaling,” meaning that events occurred at the same time when measured relative to the total duration of development in each individual. We also observed pervasive changes in timing when temperature, diet, or genotype were varied, but with larval development divided in “epochs” that differed in how event timing was impacted. Yet, these variations in timing were still explained by temporal scaling when time was rescaled by the duration of the respective epochs in each individual. Surprisingly, timing obeyed temporal scaling even in mutants lacking lin-42/Period, presumed a core regulator of timing of larval development, that exhibited strongly delayed, heterogeneous timing. However, shifting conditions middevelopment perturbed temporal scaling and changed event order in a highly condition-specific manner, indicating that a complex machinery is responsible for temporal scaling under constant conditions.
Numerous cellular events that occur during animal development, such as cell division, cell movement, and gene expression, must be tightly coordinated in time to allow formation of a functional organism with a correctly established body plan. However, despite our increasing understanding of the regulation of developmental timing (1–3), how cells in developing organisms measure time and execute events in the correct temporal order remains poorly understood. Moreover, the rate of postembryonic development in animals is affected by external conditions, such as food availability, diet, and temperature. For example, severe dietary restriction extends the duration of larval development in the nematode worm Caenorhabditis elegans as much as tenfold, without apparent defects in development (4). How the timing of individual developmental events is adjusted in response to such changes in the organism-level rate of development is not known.
This question about developmental timing has a parallel in the context of spatial patterning during development. It has been shown that spatial gene expression patterns often scale with organ or embryo size, that is, with the spatial pattern adjusted in each individual organ or embryo so that the spatial features occurred at the same position relative to its overall size (5–8). For example, in Drosophila embryos, gap genes are expressed in bands along the anteroposterior body axis (9, 10). These bands have highly stereotypical positions relative to the embryo’s size, even though this size shows significant variability between individuals (6). Moreover, embryos of closely related species that vary greatly in size exhibit the same number of bands with similar position relative to the size of the embryo (6). Here, we examine whether, analogous to scaling of spatial patterns in development, the timing of development exhibits temporal scaling, meaning that, when the organism-level rate of development is changed, the timing of individual events is adjusted so that they still occur at the same time, when measured relative to the total duration of development. Such a mechanism would ensure the correct synchrony of developmental events even when organism-level timing is changed in an unpredictable manner by shifts in external conditions.
Due to its invariant cell lineage and highly stereotypical development, C. elegans is an ideal model for studying developmental timing. Its postembryonic development consists of four larval stages (L1 to L4) that are separated by a molting event, where a new cuticle is synthesized, and the old cuticle is shed (11). After the final L4 molt, animals enter adulthood. There is a clear periodic aspect to C. elegans development, with molts occurring every ~10 h at 25 °C. Moreover, larval stages are accompanied by oscillatory expression of ∼20% of genes, with peaks occurring once per larval stage (12–14).
Molecular mechanisms that have been proposed for regulation of developmental timing include oscillators, that encode time in periodic changes in protein level, and “hourglass” mechanisms, that record time in the steady accumulation or degradation of proteins (15). Developmental timing has been extensively studied in C. elegans, leading to the discovery of heterochronic genes (2, 3). Heterochronic genes such as lin-14 and lin-28 show expression levels that decrease during larval development, suggestive of an hourglass mechanism (16, 17). Indeed, mutations that perturb the lin-14 and lin-28 temporal expression patterns lead to timing defects, with the identity of events in one larval stage switched with those of a later stage or repeated in subsequent stages (18). At the same time, these mutations otherwise have only limited impact on developmental timing on the organism level, for example, the duration of larval stages. In contrast, the heterochronic gene lin-42 is expressed in an oscillatory manner during development, peaking once every larval stage. In lin-42 mutants, developmental timing is severely perturbed, with strong animal-to-animal variability in larval stage duration (19). The body-wide, oscillatory expression dynamics of lin-42, together with its impact on larval stage duration, makes lin-42 an interesting candidate for a global regulator of developmental timing. Intriguingly, lin-42 is a homolog of Period, an important component of the circadian clock in Drosophila and higher organisms (20). Hence, it has been speculated that lin-42 forms part of an oscillator-based timer that allows cells and organs to read out developmental time (11).
How timing of individual events is impacted by changes in the organism-level rate of development is poorly characterized. Timing of C. elegans larval development is often measured at the population level, by examining the developmental stage of animals sampled from age-synchronized populations. This approach has limited time resolution and does not allow measuring timing of multiple events within the same individual. The latter is a particular problem for mutants such as lin-42, where developmental synchrony between individual animals is lost. However, the alternative approach of following individual animals was, until recently, performed by hand, limiting the number of animals that could be examined. We have recently developed a microscopy approach that allows automated imaging of individual C. elegans larvae during their entire development and at single-cell resolution (21), making it possible to measure the timing of cellular events in many individual larvae. Here, we used this approach to simultaneously measure the timing of three recurring developmental events (oscillatory expression of a molting cycle gene, hypodermal stem cell divisions, and cuticle shedding) in individual C. elegans larvae, both upon changes in environmental conditions (temperature and diet) and in mutants that increased the duration of larval development up to threefold.
Our measurements uncovered strong variability in event timing between individuals, as compared to duration of larval development, even in isogenic animals under identical environmental conditions. Strikingly, this variability obeyed temporal scaling, meaning that events occurred at the same time when rescaled by the total duration of development in each individual. Moreover, changes in average timing between populations that differ in genotype or environmental conditions were not always explained by a simple change in the overall rate of development of the organism. Instead, we found that larval development is divided into distinct epochs, which are sequences of events that, upon variation in conditions or genotype, exhibit changes in timing that are identical, and differ from changes in timing observed for events in other epochs. Yet, this variation in timing between populations also obeys temporal scaling, provided that event times were rescaled by the duration of individual epochs, rather than total duration of development. Surprisingly, we found this was even the case for lin-42 mutants, suggesting that, while lin-42 is crucial for setting the duration of larval stages, it is dispensable for controlling event timing relative to each larval stage. However, condition shifts during larval development perturbed temporal scaling and inverted event order, in a manner that depended on the type of shift. This hints that a complex machinery, potentially requiring coordination of different timing mechanisms, is responsible for temporal scaling under constant conditions.
Overall, our results show that the broad variation observed between individuals, environmental conditions, and genotypes in the timing of cellular events during C. elegans postembryonic development can be captured by the simple concept of temporal scaling, thereby revealing a precise adaptation of cell-level timing to changes in the organism-level rate of development. These observations raise the important question of how temporal scaling is implemented by the molecular mechanisms that control timing of larval development.
Results
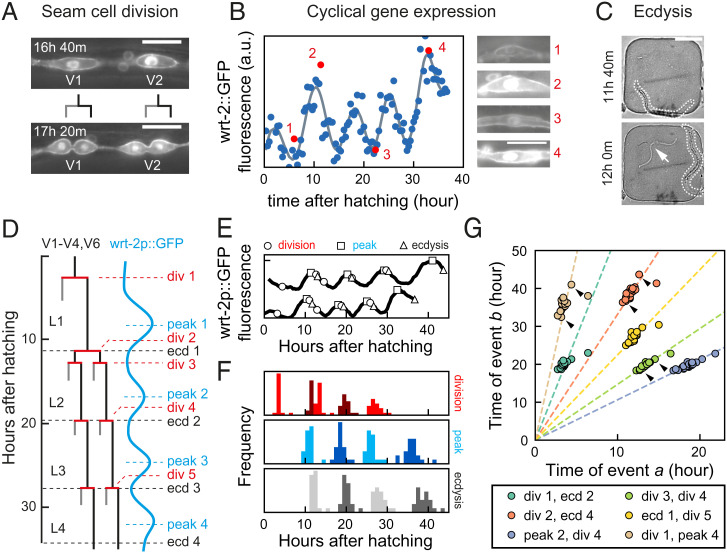
To examine how developmental timing is coordinated at the organism level in developing C. elegans larvae, we measured the timing of multiple developmental events that occurred frequently during larval development: seam cell divisions, oscillatory gene expression, and ecdysis. Seam cells are hypodermal stem cells that divide asymmetrically once every larval stage, yielding one hypodermal cell and one seam cell (Fig. 1 A and D) (22). Seam cells V1 to V4 and V6 undergo an additional symmetric division at the start of L2, doubling their number. Oscillatory gene expression is a pervasive phenomenon in C. elegans, with thousands of gene transcripts oscillating during development with the periodicity of the molting cycle (12–14). We focused on wrt-2, a hedgehog-like protein expressed in seam cells, that peaks in expression once every larval stage (12, 21) (Fig. 1 B and D). Ecdysis is the shedding of the old cuticle at the end of each larval stage (Fig. 1 C and D). By focusing on these three events, we captured qualitatively different developmental processes, while their repetitive nature allowed us to capture many events in a single experiment.
Fig. 1.
Scaling of developmental timing in individual animals. (A–C) Measuring timing of seam cell division, oscillatory gene expression, and ecdysis. (A) Seam cells divide once or twice every larval stage into hypodermal (gray line) or seam cell (black line) daughters. V1- to V6 seam cell division timing was determined using the wrt-2p::GFP::PH; wrt-2p::GFP::H2B fluorescent reporter (wrt-2p::GFP). (Scale bar: 15 µm.) (B) Oscillatory wrt-2 expression was visualized in seam cells of wrt-2p::GFP animals (images 1 to 4, showing posterior V3 seam cell). Time of each wrt-2 peak was determined from the fluorescence averaged over V1 to V6 seam cells (circles) by fitting (gray line, SI Appendix, section S4). Numbers indicate the time points in the side images. (Scale bar: 15 µm.) (C) Ecdysis time was determined by the emergence of the larva (outlined) from the cuticle (arrow). (Scale bar: 100 µm.) (D) Overview of seam cell divisions, wrt-2 peaks, and ecdysis events during larval development. The V5 lineage lacks division 2. (E) Example of variability in timing between two wild-type animals. Markers indicate the timing of divisions (circles), peaks (squares), and ecdyses (triangles). Tracks are running averages with 2-h window size of the data as shown in B, shifted along the vertical axis for clarity. (F) Distribution of events times for standard conditions, wild-type animals fed E. coli OP50 diet at 23 °C (n = 21). (G) Measured times of event pairs , for animals in F. Markers represent single animals. Arrows indicate the examples in E. Times of event pairs show temporal scaling; that is, they cluster along lines of constant , even as individual event times and vary between individuals. Dashed lines are fits .
To accurately measure timing of individual events, we used a time-lapse microscopy approach to follow the full ∼40 h of postembryonic development of individual C. elegans larvae with single-cell resolution (21). Briefly, embryos were placed inside hydrogel chambers filled with sufficient Escherichia coli, as food source, to sustain development into adulthood, while constraining animals to the field of view of the microscope at each stage. By capturing fluorescence and transmitted light images with fast exposure time, we could image developmental dynamics in individual cells inside moving larvae, without immobilization.
To visualize seam cell divisions, we used the strain heIs63[wrt-2p::H2B::GFP,wrt-2p::PH::GFP], with GFP targeted to the nucleus and membrane of seam cells (23). Since seam cell divisions occurred close in time, we defined the time of each round of divisions as the average time at which V1 to V6 cells have divided. Because the heIs63 reporter is under the control of the wrt-2 promoter, it enabled simultaneous measurement of oscillatory wrt-2 expression. Fluorescent images were analyzed automatically using custom-written software to extract the mean GFP fluorescence intensity of seam cells. The time of expression peaks was extracted from the fluorescence profiles, by fitting their dynamics with a combination of Gaussian functions and a linear offset (Fig. 1B and SI Appendix, section S4). Finally, the time of the ecdysis was defined as the time when the shed cuticle was first visible in the transmitted light image (Fig. 1C). For details on data acquisition and analysis, see SI Appendix, sections S1–S3.
Scaling of Developmental Timing in Individual Animals.
We first quantified developmental timing, that is, the time of seam cell division, wrt-2 expression peaks and ecdysis relative to hatching, under standard conditions: wild-type animals fed E. coli OP50 at 23 °C (Fig. 1 E and F). Individual animals showed strong variability in the total duration of development (∼40 h), defined as the time between hatching and L4 ecdysis, with a ∼10-h difference observed between the first and last animal to enter adulthood. We observed similar variability in the timing of all measured events (Fig. 1F). The variability in timing was uncorrelated with the position of each animal in the microchamber array (SI Appendix, Fig. S1 A–G), ruling out that it represented systematic variation due to spatial gradients in temperature and/or chemical environment. Moreover, we found similar timing variability in populations propagated by picking single individuals for 5 to 11 generations (SI Appendix, Fig. S1H), indicating that it did not result from underlying genetic variation between individuals. We therefore concluded that this variability in timing is intrinsic.
For some events, such as the second seam cell division and the first ecdysis, the magnitude of the variability was larger than the average difference in timing. However, we found that the stereotypical event order was maintained in almost all individuals (SI Appendix, Fig. S2A), even for events that took place close together in time, raising the question of how this correct event order was maintained. Interestingly, the variability in timing was strongly correlated: When we plotted event times and measured in the same animal against each other, for different pairs of events , all data points clustered along a line (Fig. 1G and SI Appendix, Fig. S3A). This strong correlation was even present for event pairs widely separated in time, for example, division 1 and peak 4.
A simple argument could explain this observation. As a measure of developmental progression, we first defined the developmental phase , where and are the population-averaged time of event and duration of development under standard conditions (23 °C), respectively, with and 1 corresponding to hatching and adulthood. When we assumed that, for any event , the event time , measured in an individual animal, scales with the total duration of development for that animal, , even as the duration varies significantly between individuals, then the time at which two events and occur within the same individual is related by , independent of total duration of development in that individual. As a result, measurements for individual animals will be clustered along a line of constant , as observed experimentally.
Upon inspection, data for different event pairs scattered around this scaling line (Fig. 1G and SI Appendix, Fig. S3A), although some event pairs exhibited more variation around this line than others. To quantify how closely the experimental data for each event pair adhered to the above scaling relationship, we first fitted the data for each event pair to a line of the form (Fig. 1G) and then calculated, for each data point, the distance to this fitted line (SI Appendix, Fig. S3A). The SD then measured the quality of scaling, with corresponding to all data points exactly on the scaling line. While event pairs that included division 1, peak 4 or ecdysis 4 appeared to exhibit a comparatively lower quality of scaling, we found < 1 h for all event pairs (SI Appendix, Fig. S3C), which was small compared to overall differences in timing observed between individuals.
We could reproduce these observations with a simple, phenomenological model. We assumed that development proceeded with average rate , and each event occurred at a specific average phase , but with animal-to-animal variation in , that remained constant throughout development, and variability in that was uncorrelated between different events (SI Appendix, section S5). If the variability in was sufficiently small, data for event pairs clustered along lines (SI Appendix, Fig. S3 G and H), indicating temporal scaling, with few changes to event order (SI Appendix, Fig. S2C). Increasing variability in reduced the quality of scaling and caused changes in event order, with events taking place close together in time impacted most strongly.
Overall, our results show that the measured changes in timing between individuals can be fully explained by simple rescaling with the total duration of development in each individual. This means that, if an animal executed, for example, its first seam cell division earlier than the rest of the population, it was highly likely to be similarly early in executing all subsequent events for the rest of larval development. This explains how animals maintain correct event order despite the observed variability in timing of individual events.
Scaling of Developmental Timing upon Changes in Temperature.
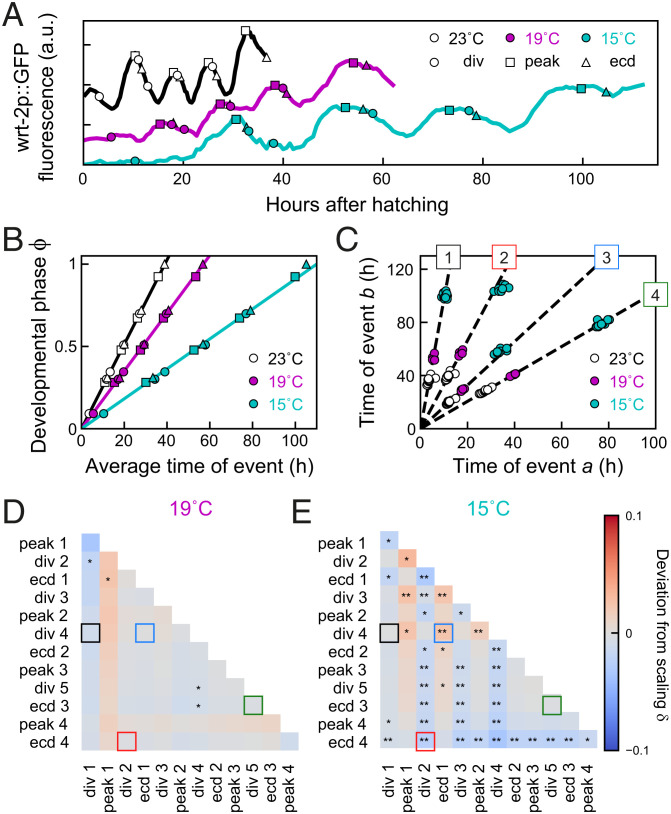
Changes in environmental conditions typically change the duration of development, yet how timing of cell-level events is adjusted to such changes is an open question. To address this, we first measured event timing in individual animals maintained at different temperatures, as the duration of larval development increases with decreasing temperatures (24). As expected, we observed that, as temperature was reduced from the standard temperature of 23 °C to 19 °C and 15 °C, the duration of larval development increased from 39 ± 2 h to 57 ± 1 h and 105 ± 2 h, respectively. Likewise, we found that, as temperature decreased, individual events were delayed more strongly relative to standard conditions (Fig. 2A).
Fig. 2.
Temporal scaling of developmental timing at different temperatures. (A) Examples of timing in individual animals at standard conditions (23 °C, black), 19 °C (magenta), and 15 °C (cyan). (B) Developmental phase as function of time for different temperatures. Developmental progression is modeled as the time evolution of a developmental phase . Each developmental event occurs at a specific phase , where and are the population-averaged time of event and of total duration of development under standard conditions. The phase for standard conditions (black line) increases linearly with rate . Markers indicate events as in A. For 19 °C or 15 °C, the phase increases linearly with the average measured event times, albeit at a lower rate compared to 23 °C. Solid lines are fits to the “Uniform” model (defined in Fig. 3). (C) Measured times for different event pairs: 1, divisions 1 and 4; 2, division 2 and ecdysis 4; 3, ecdysis 1 and division 4; and 4, division 5 and ecdysis 3. Lines are a linear fit to data for standard conditions. Event pair times for different temperatures cluster along the same line; that is, all events occurred at the same time relative to the total duration of development. (D and E) Deviation from scaling for development at (D) 19 °C and (E) 15 °C. In addition, stars indicate the probability that data for standard conditions and 19 °C or 15 °C observe the same scaling relation, as assessed by a Kolmogorov–Smirnov (K-S) test: **P < 0.001, *P < 0.01, and not significant (N.S.) otherwise. At 19 °C, temporal scaling was observed for all event pairs, while some significant deviations were seen at 15 °C. Colored squares correspond to event pairs in C.
To examine the impact of changing temperatures on average event timing, we examined the time evolution of the developmental phase . Our earlier definition ensured that, under standard conditions (23 °C), increases with constant rate (Fig. 2B). If the total duration of development increases or decreases, for example, due to shifting environmental conditions, will change, so that the same developmental phase is reached at a different time compared to standard conditions. When we measured the average time of each seam cell division, wrt-2 peak, and ecdysis for 19 °C and 15 °C, we found that, while the phase indeed increased at a lower rate compared to 23 °C, its increase was still linear in time (Fig. 2B), meaning that all measured differences in average timing were explained by a constant change in the organism-level rate of development, .
When we examined event times in individuals, we found similar variability in timing for animals at 19 °C and 15 °C compared to 23 °C. Moreover, times of event pairs still clustered along lines (Fig. 2C), meaning that variability in timing between individuals was explained by temporal scaling also for these conditions, even though the deviations away from the scaling line were generally stronger than seen for 23 °C (SI Appendix, Fig. S3B): The quality of scaling was decreased for all event pairs, in particular at 15 °C. Interestingly, this increase in variability away from the scaling line was accompanied by more-frequent changes in event order (SI Appendix, Fig. S2B), with most animals at 15 °C reversing order of seam cell divisions and ecdysis in the L1 and L2 larval stages. This result argues against a mechanism that controls event order by causally linking subsequent events, for instance with initiation of seam cell divisions required to trigger the subsequent ecdysis. Finally, we found that, although timing of all events was delayed for 19 °C and 15 °C compared to 23 °C, event pairs clustered along the same line, independent of temperature. This indicates that temporal scaling cannot only explain variability in timing between individuals but also changes in timing between development under different environmental conditions. In the latter case, this means that events occurred at the same relative time, when rescaled with the population-averaged duration of development observed for each temperature, even as development at 15 °C was slowed approximately threefold compared to 23 °C.
We could reproduce these observations in our mathematical model, using the simplest assumption that the observed changes in timing resulted only from a uniformly lowered rate of development (“Uniform” model; Fig. 3A), as seen experimentally (Fig. 2B). Indeed, stochastic simulations showed that the times of event pairs for the “Uniform” model clustered along the same scaling line as for standard conditions, even though the average times for the “Uniform” model were significantly delayed (Fig. 3 B–D). To test this systematically for all event pairs, we defined the deviation of scaling as the signed angle between the lines that fit the data for standard and perturbed conditions (Fig. 3C), with meaning that the line for perturbed conditions has a higher slope than for standard conditions. Here, we used the difference between angles rather than slopes, as the slope diverges when the first event a occurs close to hatching, . For the “Uniform” model, we found that for all event pairs (SI Appendix, section S6).
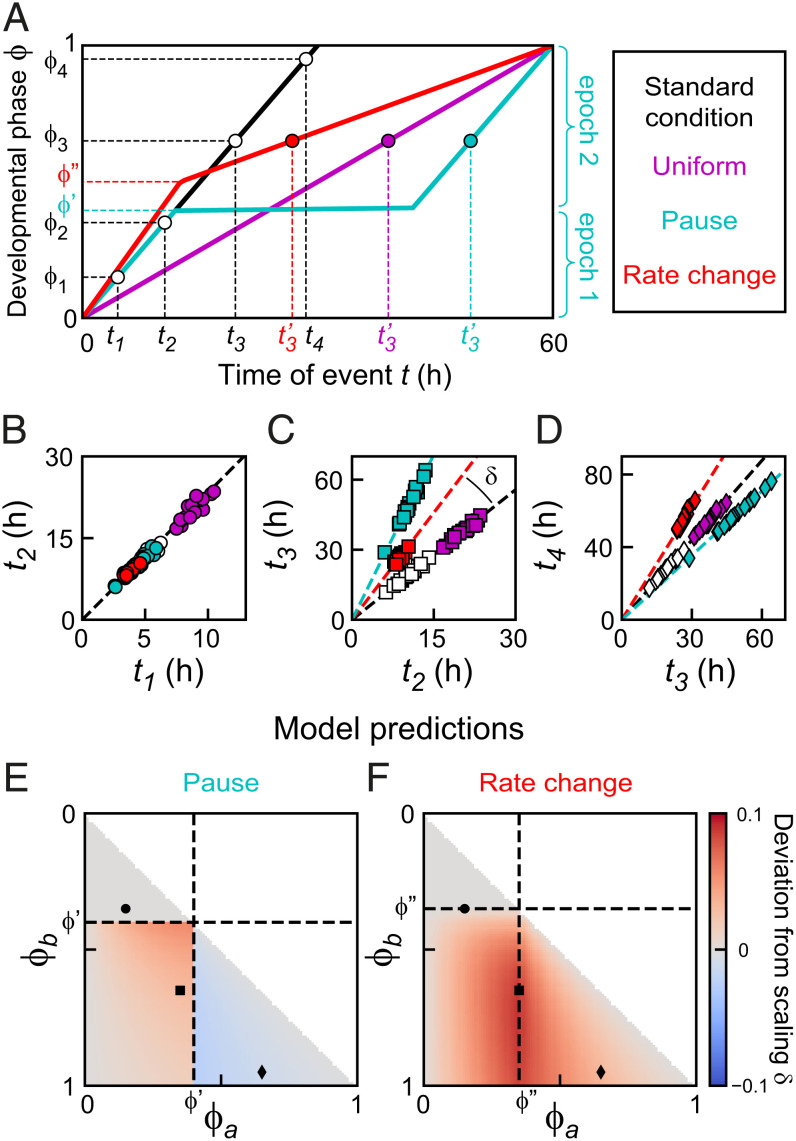
Fig. 3.
Timing models. (A) Developmental phase versus time for three different models that generate the same increase in duration of development: a uniform lower developmental rate (magenta), a single pause at (cyan), or a single change in developmental rate at (red) (SI Appendix, section S5; parameters chosen for clarity). For the “Pause” and “Rate change” models, the discontinuity in developmental rate results in two epochs of constant developmental rate, as illustrated for the “Pause” model. (B–D) Simulated event times for the event pairs indicated in A. Color indicates the timing model. For all models, event pair times cluster along a line; that is, they occur at the same time when rescaled by each individual’s duration of development. For the “Uniform” model, times of event pairs lie along the scaling line for standard conditions (dashed line): that is, they occur at the same time when rescaled by total duration of development. Other models generally deviate from this scaling line. The deviation from scaling is defined as the signed angle between these two lines, as indicated in C. (E and F) Predicted values of for different events pairs and for the (E) “Pause” and (F) “Rate change” models, based on measured parameters (SI Appendix, sections S6 and S8). Markers are the event pairs in B–D. The timing models in A can be distinguished by measuring .
Next, we tested whether this held for our experimental observations. For each event pair and measured in the same individual, we calculated the angle and the deviation from scaling as − , the difference between the average angle for standard (, 23 °C) and perturbed (, 19 °C or 15 °C) conditions, with indicating that the data for standard and perturbed conditions clustered along the same line. In addition, we also used the two-sample Kolmogorov–Smirnov test to estimate the probability that the distributions measured for standard and perturbed conditions were drawn from the same distribution. This analysis showed that most event pairs at 15 °C and 19 °C (Fig. 2 D and E) lie along the same line as data for 23 °C; that is, changes in timing between temperatures are fully captured by temporal scaling with duration of development. At 15 °C, we observed significant deviations from scaling only for event pairs that included divisions 2 to 4 or ecdysis 4. Interestingly, these deviations are consistent with the permutations in event order for 15 °C (SI Appendix, Fig. S2B), indicating that these permutations reflected changes in timing of seam cell divisions, not ecdyses.
Impact of Changes in Diet on Developmental Timing Differs between Epochs.
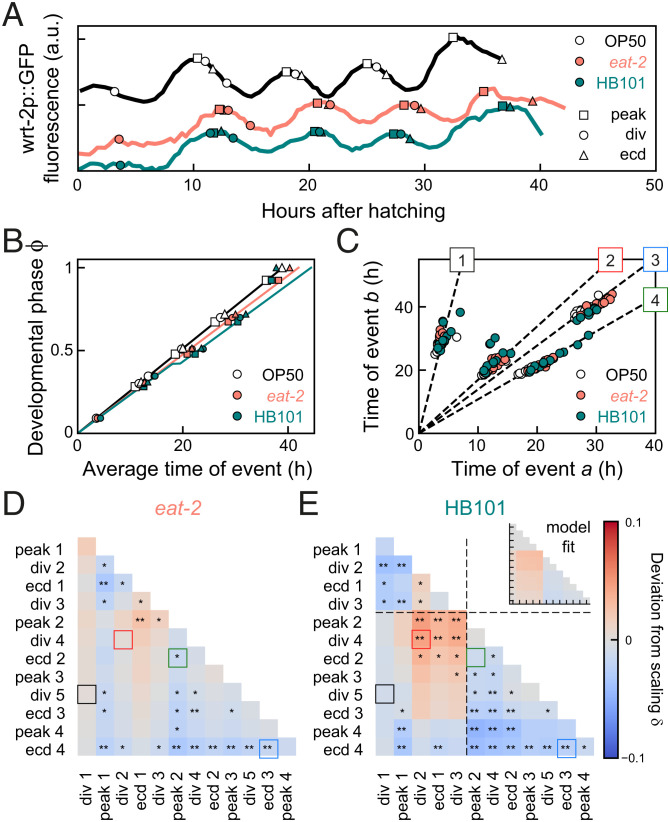
To test whether temporal scaling is also observed under qualitatively different changes in environmental conditions, we studied the impact of changes in food uptake and diet on timing of individual events. Total duration of development can be changed by providing animals with food other than E. coli OP50 (25–27). Here, we used two different approaches. To mimic reduced food uptake, we fed the standard diet, E. coli OP50, to eat-2(ad1113) mutants that exhibit a fivefold decrease in pharyngeal pumping and hence ingest bacteria at a lower rate (28). In addition, we grew wild-type animals on a diet of E. coli HB101, which was reported to have faster larval development (25).
The total duration of development was slightly different in eat-2 mutants (40 ± 2 h) and wild-type animals on HB101 (38 ± 1 h), compared to standard conditions of wild-type animals on OP50 (39 ± 2 h; Fig. 4A). However, we observed more-complex changes in timing when we examined the average timing of seam cell divisions, wrt-2 peaks, and ecdyses (Fig. 4B). For eat-2 mutants, the developmental phase increased linearly with time, with a lower rate compared to standard conditions, consistent with the “Uniform” model. However, for animals fed HB101, the increase of phase in time did not appear to be linear. Instead, development was separated into multiple epochs, that is, sequences of events that differed in how their timing was impacted by changing diet. Events in the first epoch (hatching to division 3) occurred with the same timing as under standard conditions. In the second epoch (peak 2 to ecdysis 3), the phase increased at the same rate, but with a ∼2-h pause compared to standard conditions. In comparison, events in the third epoch (peak 4 and ecdysis 4) were executed much earlier. This shows that the reported earlier entry into adulthood on HB101 (28) did not reflect a general speed-up in larval development, but a more complex combination of pausing and acceleration.
Fig. 4.
Temporal scaling for changes in diet. (A) Developmental timing in individual animals under standard conditions (black) and two conditions that exhibit slower development: wild-type on E. coli HB101 (green) and eat-2 mutants, that have reduced food uptake, on E. coli OP50 (orange). (B) Developmental phase versus time. Solid lines are best fits to the “Uniform” model (eat-2 mutants, orange line) and the “Pause” model (HB101 diet, green line) with a ∼2-h pause between division 3 and peak 2. (C) Measured times for event pairs: 1, divisions 1 and 5; 2, divisions 2 and 4; 3, peak 2 and ecdysis 2; and 4, ecdyses 3 and 4. Lines are the fit for standard conditions. For pairs 2 and 3, animals on HB101 deviate from this scaling line. (D and E) Deviation from scaling for (D) eat-2 mutants and (E) animals fed HB101. Stars indicate the probability that the data observe the scaling relation for standard conditions: **P < 0.001, *P < 0.01, and N.S. otherwise (K-S test). Lines in E indicate the fitted pause time. Inset shows best fit to the “Pause” model. Animals on HB101 exhibit systematic deviations from scaling that match the “Pause” model in sign and magnitude.
The comparatively small differences between standard conditions and animals fed HB1010 raised the question of whether the data for HB101 were indeed not explained by a simple rescaling of time, as for changes in temperature, but required a more complex timing model. We focused on the apparent delay between epochs 1 and 2, and ignored the changes in timing of epoch 3, as the small number of events in that epoch made it difficult to constrain timing models. We constructed a phenomenological model analogous to the “Uniform” model, that incorporated the pause separating epochs 1 and 2 (“Pause” model; Fig. 3A and SI Appendix, section S5). Stochastic simulations of the “Pause” model (Fig. 3 B–D) showed that times of event pairs still clustered along a line, reflecting temporal scaling between individuals. However, this line deviated from the scaling line for standard conditions. Interestingly, the experiments reproduced these simulation results. Data for both eat-2 mutants and fed HB101 still clustered along a line (Fig. 4C), consistent with temporal scaling between individuals, although, compared to standard conditions, the quality of scaling was decreased (SI Appendix, Fig. S3E), and we observed more-frequent permutations of event order for events that occurred close in time (SI Appendix, Fig. S2D). However, for animals fed HB101, but not for eat-2 mutants, some event pairs exhibited small but systematic deviations from the scaling line for standard conditions (e.g., event pairs 2 and 3 in Fig. 4C), as seen in the “Pause” model.
When we calculated the deviation from scaling between the “Standard condition” and “Pause” models, we found that it exhibited a specific pattern (Fig. 3E): While timing of event pairs that both occurred before the developmental pause matched the line for standard conditions, event times clustered along a line with higher slope ( when one event occurred before and the other after the pause, with stronger deviations if both events were close to the pause. When both events occurred after the pause, event times clustered along lines with lower slope (), with stronger deviations when events occurred farther apart in development. When we calculated for all experimentally measured event pairs, we found a clear difference between eat-2 mutants and animals fed HB101 (Fig. 4 D and E). Whereas, for eat-2 mutants, we found for most event pairs, animals fed HB101 showed significant deviations from scaling that resembled, both in magnitude and sign, those predicted by the “Pause” model.
We then compared the performance of the “Uniform” and “Pause” models in reproducing the experimental data as follows (SI Appendix, section S7): We used least-squares fitting to obtain values for the magnitude and timing of the pause that minimized the difference in between experiments and the “Pause" model (Fig. 4E, Inset). The resulting fit accurately reproduced for events in epochs 1 and 2, but underestimated for peak 4 and ecdysis 4, confirming that they form a separate epoch. Next, we fitted the remaining free parameter, the developmental rate , to minimize the difference in the average event time as function of developmental phase (Fig. 4B). Notably, this curve provided an excellent fit to the data in epochs 1 and 2, even though the qualitative shape of this curve depended only on the timing and magnitude of the pause, and was thus fully determined by the fit to . A similar fit of the data in Fig. 4 B, D, and E to the “Uniform” model showed that, while data for eat-2 mutants were best fit by the “Uniform” model, the “Pause” model formed a much better fit for animals fed HB101 (SI Appendix, Fig. S4).
The timing of the pause at the mid-L2 larval stage was notable, as it coincided with the developmental window during which animals, if deprived of food, decide whether to enter dauer, a stress-resistant arrested state (29, 30). We therefore examined whether the pause reflected a transient activation of the dauer regulatory machinery, potentially induced by nutrient stress due to the HB101 diet. DAF-2/insulin signaling controls dauer entry and is triggered by nutrient stress (31). As a measure of insulin activity, we monitored nuclear translocation of DAF-16, as DAF-16 enters the nuclei of many cells both during food deprivation and in dauer (31). We confirmed that our time-lapse imaging approach could detect DAF-16 nuclear translocation in time, as we observed a strong increase in nuclear DAF-16 in the late-L2 stage of daf-2 mutants that enter dauer even with food present (32) (SI Appendix, Fig. S5 A–C). When we examined wild-type animals fed OP50 or HB101, we found that DAF-16 was almost always cytoplasmic, with no difference between the two diets (SI Appendix, Fig. S5 D–F), indicating that the pause in animals fed HB101 was not due to different insulin signaling activity. Insulin signaling activity also controls developmental rate (33), raising the possibility that the variability in timing we observed might result from variation in insulin signaling between individuals. However, our data do not support this, as we observed no correlation between DAF-16 nuclear localization and ecdysis time in individual animals (SI Appendix, Fig. S5G).
Perturbed Developmental Timing and Growth in lin-42 Mutants.
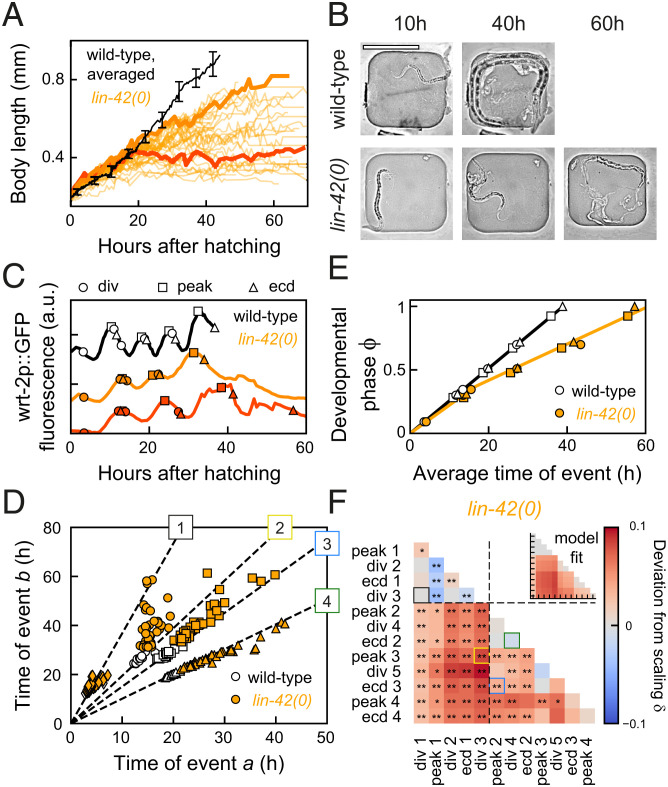
For wild-type animals on HB101 we observed deviations from temporal scaling even as diet had only minor impact on total duration of development. To seek stronger perturbations of developmental timing and, potentially, temporal scaling, we measured event timing in mutants of the heterochronic gene lin-42, for the following reasons. First, lin-42 plays an important role in molting, with mutants showing longer larval stages, strongly reduced synchrony between individuals in progression through larval stages, and frequent developmental arrest, with all these phenotypes increasing in severity as development proceeds (19, 34, 35). Second, lin-42 mutants exhibit heterochronic phenotypes in multiple organs (19, 20, 36, 37), indicating a body-wide role for lin-42. In addition, lin-42 is expressed in an oscillatory manner, peaking once every larval stage (20). This, together with the homology of lin-42 to the circadian clock gene Period, led to the speculation that lin-42 acts as a global developmental timer (11, 19). Finally, lin-42 regulates the expression of many microRNAs, including those involved in timing through the heterochronic pathway, and binds to the promoter of many genes (38–40). Because of this wide-ranging impact on developmental timing and gene expression, lin-42 appeared, to us, to be a prime candidate also for a core component of a potential scaling mechanism. Hence, we examined whether lin-42(0) animals displayed stronger deviations from temporal scaling than observed under changes in diet.
We used the lin-42(ox461) allele that deletes the entire lin-42 locus and shows the strongest perturbation of molting cycle progression (34). Indeed, developmental progression varied strongly between individuals, with animals arresting at different stages of development and frequently skipping seam cell divisions (SI Appendix, Fig. S6A). Most animals skipped the L4 seam cell division and ecdysis, a heterochronic phenotype observed before (19). Moreover, the timing of larval development was highly delayed and variable, as seen, for example, by comparing the time of the L3 ecdysis (42 ± 8 h for lin-42(0), compared to 28 ± 1 h for wild-type animals). In addition, lin-42(0) animals showed reduced growth, as measured by the increase of body length and width over time in individual animals (Fig. 5 A and B and SI Appendix, Fig. S6 B and C). In particular, we observed a fraction of animals that stopped growing completely between the L2 and L4 larval stage, with some reaching body lengths of only 0.3 mm, compared to 0.9 mm for wild-type animals. Surprisingly, all animals that arrested growth appeared to otherwise continue development: They underwent multiple rounds of ecdysis, seam cell divisions, and wrt-2 expression peaks (Fig. 5C). After molting, lin-42(0) animals often remain stuck in their old cuticle, and it was suggested that this interferes with the ability to feed (34). However, we observed growth arrest also in animals that appear to shed their cuticle normally. Moreover, growth-arrested animals also displayed pharyngeal pumping, suggesting that growth arrest was not simply caused by inability to take up food.
Fig. 5.
Growth arrest and temporal scaling in lin-42(0) animals. (A) Body length versus time for standard conditions (black, averaged, error bars are SEM) and lin-42(0) animals (orange lines, individual animals). The lin-42(0) animals exhibited either reduced growth or complete growth arrest (final body length of <0.5 mm) from the mid-L2 stage, ∼20 h after hatching. Thick lines highlight individuals with reduced (light orange) and arrested growth (dark orange). (B) Images of a wild-type (Top) and growth-arrested lin-42(0) larva (Bottom) at different times after hatching. (Scale bar: 200 µm.) (C) Timing in a wild-type larva (black), and in a slow-growing (light orange) and growth-arrested (dark orange) lin-42(0) animal, corresponding to the individuals highlighted in A. The growth-arrested animal executed most events that occur after mid-L2; that is, development was not arrested. (D) Measured times for event pairs: 1, divisions 1 and 3; 2, division 3 and peak 3; 3, peak 2 and ecdysis 3; and 4, division 4 and ecdysis 2. Lines are fits for standard conditions. The lin-42(0) animals show strong variability in timing between individuals and, for event pairs 2 and 3, exhibit clear deviations from the scaling line. (E) Developmental phase versus time in wild-type (black) and lin-42(0) animals (orange). The orange line is the best fit to the “Rate change” model, with wild-type developmental rate until mid-L2, and approximately twofold decreased afterward. (F) Deviation from scaling in lin-42(0) mutants. Stars indicate probability that lin-42(0) data observe the same scaling relation as wild-type: **P < 0.001, *P < 0.01, and N.S. otherwise (K-S test). Inset shows best fit to “Rate change” model. The data match the “Rate change” model, with lines indicating the fitted rate change time.
Temporal Scaling Explains Most Variability in Timing of lin-42 Mutants.
We then asked whether the increased variability in development of lin-42(0) mutants reflected errors in control of developmental timing. When we compared times of event pairs between individuals, variability in timing was indeed dramatically increased compared to wild-type animals (Fig. 5D). Yet, surprisingly, for many event pairs, the data clustered along a line, indicating that this variability in timing scaled between individuals. Indeed, the quality of scaling was comparable to other conditions we examined (SI Appendix, Fig. S3F), with strongly reduced quality only seen for events that occurred in L4, a stage rarely reached by most lin-42(0) animals, and in pairs where one event occurred before peak 2 and the other event after. Moreover, we found that event order was well maintained (SI Appendix, Fig. S2E). Growth-arrested animals did not develop more slowly, and their developmental timing did not exhibit stronger deviations from temporal scaling (SI Appendix, Fig. S6 D–F), suggesting that the timing of development was largely independent of physical growth. Together, these results suggested that the strongly increased variability in timing was not due to intrinsic errors in timing, corresponding to changes in the developmental phase , but rather because of increased variability in the overall rate of development, , between individuals.
To test this hypothesis, we first measured the evolution of the developmental phase in lin-42(0) animals (Fig. 5E). We found that development separated into two epochs that differed in changes to timing: In the first epoch (hatching to division 3), developmental phase increased at the same rate as in wild-type animals, while, in the second epoch (from peak 2 onward), the phase still increased linearly but with a strongly decreased rate compared to wild-type animals. We then used the same procedure as above to fit our experimental data to a model (“Rate change”; Fig. 3A and SI Appendix, section S5) that incorporated the observed change in developmental rate. First, we calculated the deviation from scaling between the data for wild-type and lin-42(0) animals (Fig. 5F). Next, we fitted , as predicted by the “Rate change” model, to the experimental data, thereby obtaining values for two model parameters, the developmental phase at which the rate change occurred and the ratio of the two developmental rates. The resulting fit accurately reproduced the experimental observations (Fig. 5F, Inset), outperforming other models (SI Appendix, Fig. S4A). This showed that the measured deviations from scaling were fully explained by the observed change in the developmental rate. Finally, we obtained the value of the remaining free parameter, the developmental rate of the first epoch, by fitting the phase evolution of the “Rate change” model to that measured experimentally, with, again, good agreement between model and experiments (Fig. 5E). Overall, these results show that most differences in timing between lin-42(0) mutants and wild-type animals are explained by the change in developmental rate in the mid-L2 stage, combined with strongly increased variability in total duration of development between individuals.
However, a subset of event pairs exhibited timing that deviated strongly from temporal scaling, as measured by the scaling quality (SI Appendix, Fig. S3F). The hallmark of scaling is that, if an individual executes one event early compared to the population, then all subsequent events are equally early. Event pairs that showed low scaling quality often violated this rule. For example, some animals that were among the first to execute division 3 exhibited an exceptionally late peak 3, resulting in many points away from the scaling line (Fig. 5D, event pair 2). In general, the durations of the first and second epochs were often poorly correlated in lin-42(0) animals, in particular compared to standard conditions and wild-type animals fed HB101 (SI Appendix, Fig. S7 A–C). Simulations showed that this comparatively weak correlation reproduced the observed deviations from scaling (SI Appendix, Fig. S7 D–G). This suggested that the lack of scaling for these event pairs did not represent timing errors but rather reduced correlation between the developmental rates of the two epochs.
Loss of Epochal Organization upon Shifts in Conditions.
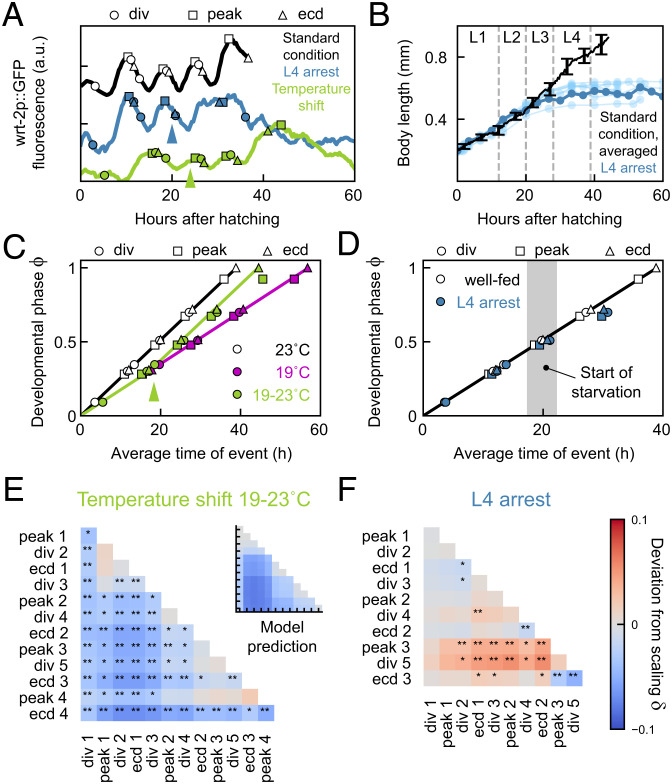
We found that, upon a constant change in external conditions, either all events changed timing in the same manner (temperature, eat-2 mutants) or all events within an epoch did (HB101 diet, lin-42(0) mutants). These coordinated changes in timing ensured that event order was largely maintained, even as the timing of individual events was shifted by many hours. This observation raised the question of whether such coordinated changes in timing between events in an epoch are seen for all condition changes. We addressed this by examining the impact of a qualitatively different kind of condition change, namely, shifting external conditions middevelopment.
We first subjected animals to a shift in temperature from 19 °C to 23 °C in the L2 larval stage. The rate of development increased notably after the shift to higher temperature (Fig. 6 A and C). We tested whether changes in average timing were simply described by development at the rate observed for 19 °C prior to the shift, and the rate for 23 °C after. Indeed, when we used the measured rates for 19 °C and 23 °C and the observed shift time, the “Rate change” model accurately captured the measured average timing for most events, without any free fitting parameters (Fig. 6C). However, a clear exception was the timing of peak 4. Whereas this peak always preceded ecdysis 4 by 3 h at 23 °C, upon temperature shift, this order was reversed, with peak 4 now observed 1 h after ecdysis 4 (SI Appendix, Fig. S2). Other events also showed reversals in order, but these occurred much closer in time than peak 4 and ecdysis 4. When we compared the deviation from scaling between standard conditions and the temperature shift experiment, we found that it strongly resembled the prediction by the “Rate change” model (Fig. 6E). The only exception formed event pairs that included peak 4, consistent with the delayed timing of peak 4 relative to ecdysis 4.
Fig. 6.
Loss of epochal organization upon condition shifts. (A) Developmental timing in a larva at standard conditions (black) and in animals shifted in temperature (green) or food abundance (blue). Arrows indicate time of the shift from 19 to 23 °C (green) or full depletion of food (blue). (B) Body length versus time for standard conditions (black, average, error bars are SEM) and animals shifted to no food (blue). Dark blue line corresponds to the individual in A. Gray lines indicate average ecdysis time for standard conditions. Animals arrest growth directly after depleting food in L3. (C) Developmental phase versus time in animals at 23 °C (black) and 19 °C (magenta), and in animals shifted from 19 to 23 °C (green). Arrow indicates the average time of the temperature shift. Green line is the “Rate change” model, assuming that development progresses at the 19 °C rate prior to the average shift time, and the 23 °C rate after. (D) Developmental phase versus time in well-fed animals (black) and animals shifted to no food (blue). Gray area indicates the time range at which the food supply was depleted. (E and F) Deviation from scaling for animals subjected to shift in (E) temperature or food (F) abundance. Stars indicate probability that the data observes the same scaling relation as standard conditions: **P < 0.001, *P < 0.01, and N.S. otherwise (K-S test). Inset in E shows the prediction of the “Rate change” model shown in C.
Next, we subjected animals to a shift in food abundance. We loaded microchambers with a reduced amount of OP50. Under these conditions, animals developed with normal body growth and timing for early larval development (Fig. 6 B and D) but fully depleted their food supply around the L2 ecdysis, which was accompanied by immediate halting of body length extension. Similar to our observations in lin-42(0) mutants, we found that development proceeded even after growth arrested, as all L3-specific events (peak 3, division 5, and ecdysis 3) were executed. However, we did not observe any subsequent L4-specific events (peak 4 and ecdysis 4), consistent with larvae entering starvation-induced L4 arrest (41). The timing of the L3 events was delayed compared to standard conditions (Fig. 6 A and C), but with clear differences between events. The time of peak 3 was delayed, both because it started later and also due to its increased duration. Division 5 was similarly delayed. However, ecdysis 3 occurred with a timing much closer to that for standard conditions. As a result, the order of events was shifted, so that ecdysis 3 typically preceded peak 3 and division 5 (SI Appendix, Fig. S2). This was further confirmed when we examined the deviation from scaling (Fig. 6F). Whereas the deviation was strong for event pairs including peak 3 and division 5, the deviation was much weaker for ecdysis 3, meaning that the timing of ecdysis 3 was close to that expected for animals on standard conditions.
Our observation that, under a constant change in diet or genotype, events were linked in epochs that changed their timing in a coordinated manner suggested that these events were intrinsically linked in their timing: For example, they might all be controlled by a single timer. However, our observation here, that this epochal organization is not maintained under shifts in conditions, implies that timing of these events is controlled by independent timers and, hence, that their organization in epochs seen under other conditions reflects a precise synchronization in timing.
Discussion
By performing time-lapse imaging on individual C. elegans larvae, we found that timing of seam cell divisions, wrt-2 gene expression oscillations, and ecdysis varied widely, not only when genotype or environmental conditions were changed but even between genetically identical individuals under the same conditions. Surprisingly, we found that all this variation in timing could be explained, to a great extent, by simple, phenomenological timing models that relied on the concept of “temporal scaling.”
In these models, the complexity of developmental progression is reduced to the evolution of a developmental phase in time (Fig. 3). Animal-to-animal variability arises because each animal proceeds through its phase evolution at an intrinsically different rate, giving rise to the strongly correlated variability we measured for timing of event pairs. As a result, timing between individuals exhibits temporal scaling; that is, events occur at the same time when measured relative to the total duration of development in each individual. Varying conditions or genotype results in changes to the evolution of developmental phase. While, in some cases (changes in temperature, impaired food uptake in eat-2 mutants), the rate of phase evolution changed uniformly for all events, in other cases (HB101 diet, lin-42(0) mutants), larval development was divided in multiple epochs that differed in how the evolution of phase is impacted. In the latter case, changes in timing between animals of different genotypes or raised under different conditions could not be explained by simple rescaling with total duration of development. However, event timing did exhibit temporal scaling, when taking into account these epochs; that is, events occur at the same time when measured relative to the duration of each epoch. While these phenomenological models do not provide a molecular mechanism for temporal scaling, they reveal a remarkably simple organization that unifies the pervasive variation in timing seen in our experiments.
It is striking that isogenic animals under identical environmental conditions showed substantial variability in developmental timing. It is unlikely that this reflects systematic variation in environmental conditions, such as temperature, between animals in our experiment setup or genetic variation (SI Appendix, Fig. S1). Similar individual variability was observed recently in the timing of lethargus, as measured by behavioral assays with a similar level of environmental control (42, 43). Overall, these results suggest that this variability in timing is intrinsic, reflecting a variation in developmental rate between individuals. What controls this variation? Recent work showed that animal-to-animal variability in timing of the L4 ecdysis, that is, the transition into adulthood, correlated with age of the mother at fertilization (44), with embryos generated by older mothers developing more rapidly. This was linked to the amount of yolk proteins loaded in each embryo, which increased with the mother’s age, suggesting that the rate of larval development is determined by the status of nutrient stores in the embryo. It is surprising, however, that this variation in rate persists throughout development, particularly as larvae depend, for growth, on feeding rather than internal stores. We observed that the rate of early-larval development was poorly predictive of the late-larval rate for animals at 15 °C and in lin-42(0) mutants (SI Appendix, Fig. S7). For lin-42, this might reflect a role in maintaining the developmental rate at the embryo-set level, but could also reflect that the severity of the lin-42(0) phenotype, as it emerges in the mid-L2 stage, is independent of the early-larval developmental rate.
For wild-type animals fed HB101 and lin-42(0) mutants, we observed discontinuities in the developmental rate that separated larval development into multiple epochs (Figs. 4B and 5E). Interestingly, even though the nature of the discontinuity differed, with a pause for HB101 diet and a rate change for lin-42(0) mutants, in both cases, the transition between the first and second epochs occurred at the mid-L2 larval stage. At this developmental stage, animals may decide to enter the stress-resistant dauer state if the environment is crowded or deprived of food (29, 30), raising the question of whether the change in developmental rate we observed reflected transient activation of dauer decision-making machinery. However, we found that activity of the DAF-2/insulin pathway, a key regulator of entry into dauer (31), did not differ between animals fed OP50 or HB101 (SI Appendix, Fig. S5). This implied that animals did not suffer from stress due to nutrient depletion, making it unlikely that the pause in animals fed HB101 was linked to dauer. We therefore prefer another explanation: C. elegans larvae also shift their metabolism between the L1 and L2 larval stages, from the glyoxylate cycle to the tricarboxylic acid (TCA) cycle (45). The glyoxylate cycle likely allows L1 larvae to use stored lipids as an energy source, potentially rendering their development less dependent on ingestion of food. As a consequence, shifts in diet (from E. coli OP50 to HB101) or inability to ingest or metabolize food in lin-42(0) mutants might only impact developmental timing substantially after shifting to the TCA cycle upon entering the L2 larval stage.
A limit of our study is that it does not provide a mechanistic explanation of temporal scaling. Based on its oscillatory expression and homology to the circadian clock protein Period, lin-42 was proposed to be a core component of a clock controlling C. elegans larval development (11). This hypothesis was supported by the strong loss of developmental synchrony between lin-42(0) individuals. However, we found that this perturbed timing resulted mainly from dramatically increased variability in developmental rate between individuals, while timing of individual events obeyed temporal scaling within each epoch. This result argues against a key role for lin-42 in controlling temporal scaling. One attractive, alternative mechanism to regulate timing in a manner that is synchronized throughout the body and adapts to changes in rate of development under different conditions is to couple developmental timing to physical growth. If each event occurred at a specific body size, it would explain temporal scaling, as changes in conditions that impact the body’s growth rate would naturally lead to concomitant changes in developmental timing. Analogously, cell cycle timing in bacteria and yeast strongly depends on cell size and growth (46–49). Indeed, progression of C. elegans larval development is tightly linked to body size (24), and, under dietary conditions that caused slow growth, larval stages lengthened so that molts occurred at their stereotypical body size (4). Moreover, conditions that do not allow growth, such as starvation, cause developmental arrest at the start of each larval stage (41, 50, 51). It is therefore significant that both lin-42(0) mutants and wild-type animals shifted to no food showed continued development without physical growth (Figs. 5C and 6A). In lin-42(0) mutants, timing is explained by temporal scaling for most animals and event pairs (Fig. 5 E and F), and growth-arrested lin-42(0) individuals showed deviations from scaling similar to lin-42(0) animals that continued growing (SI Appendix, Fig. S6 D–F). Moreover, while wild-type animals shifted to no food showed specific changes in timing that altered the order of events occurring after the shift, the approximate timing of these events was still similar to that found in animals with plentiful food (Fig. 6D). Thus, our observation of continued development with approximately correct timing in growth-arrested animals rules out a mechanism where the execution of a specific event occurs only at a predetermined body size.
Recent studies examined the dependence of developmental timing in C. elegans, fly, and frog embryos on temperature (52–55). Specifically, Kuntz and Eisen (54) found that the timing of fruit fly development scaled uniformly with temperature, similar to our observations for C. elegans larval development. Intriguingly, in these studies, the measured dependence of timing on temperature followed an Arrhenius equation (52–54), which is historically used to describe the temperature dependence of chemical reactions. Temporal scaling was hypothesized to arise because timing of all events follows the same Arrhenius equation, that is, varies by the same factor for a given change in temperature. Because the rates of molecular processes are likely not only controlled by temperature but also by metabolite levels, such thermodynamic mechanisms might also generate temporal scaling upon changes in diet or food uptake, as we observed. However, more-recent measurements disputed the key assumption that all processes follow the same Arrhenius equation (52, 53), raising the question of whether thermodynamic mechanisms by themselves are sufficient to explain temporal scaling, or whether additional active feedback or checkpoint mechanisms are still essential.
Epochs emerged from our data as sets of consecutive events that changed their timing in a coordinated manner upon a constant change in environment or genotype. In particular, events within such epochs generally did not exhibit changes in their relative order, apart from reversals of events that occurred close together in time (SI Appendix, Fig. S2). Hence, our observation of epochs could indicate that these events were controlled by the same timer. This would predict that events in an epoch always show coordinated changes in time, regardless of the type of condition change. However, when we shifted conditions (temperature or food abundance) during larval development, we found that some events were strongly delayed or advanced relative to other events in the same epoch (Fig. 6). This implies that timing of events within an epoch can be controlled independently and that precise tuning and/or active synchronization of timing of each individual event is required to maintain relative order under constant environmental conditions. Whereas the loss of stereotypical order in these experiments did not appear to hamper development, similar loss of synchrony between seam cell divisions and the molting cycle, induced by a nicotinic agonist, caused larval lethality (56), indicating that, in general, maintaining synchrony of developmental events is of vital importance.
One of the most enduring mysteries of development is how its timing is regulated. Whereas we have a deep understanding of how spatial patterns arise during development (8), our understanding of how events like cell division, cell movement, or gene expression are controlled in time is still very limited. Postembryonic development poses a particular challenge, as its rate of progression depends strongly on environmental conditions such as food availability. How timing is adapted on the cellular level in response to such organism-level changes is an open question. On the molecular level, developmental timing is thought to be controlled by either oscillators or “hourglass” mechanisms. In C. elegans, there is evidence for hourglass mechanisms (16, 17) and oscillators (11, 14) controlling timing of larval development in parallel, although it remains unclear how they determine the exact time of cell-level events on the molecular level. A priori, the core characteristic of hourglass and oscillator mechanisms (their decay time and period, respectively) will each be impacted differently by changes in conditions. Temporal scaling is only observed when all decay times and periods are changed in the same way. Hence, our observation of temporal scaling puts strong constraints on possible models that explain developmental timing of C. elegans larval development.
Materials and Methods
Additional information on experimental methods, analysis, and models is provided in SI Appendix, Supplementary Text. Briefly, the following mutants and transgenes were used: heIs63[Pwrt-2::GFP::PH; Pwrt-2::GFP::H2B; Plin-48::mCherry] (23); eat-2(ad1113) (28); lin-42(ox461) (34); zIs356[Pdaf16::daf-16a/b-gfp; rol-6]; stIs10131[elt-7p::H1-wCherry; unc-119(+)]; and daf-2(e1368) (32). For time-lapse experiments, we refer to standard conditions as wrt2p::GFP animals fed E. coli OP50 at 23 °C. For experiments in perturbed conditions, we varied one experimental parameter (genotype, temperature, or diet) while keeping the others unchanged. Division and ecdysis times were recorded by manual annotation, while peak times were determined by fitting to wrt2-p::GFP fluorescence intensity.
Supplementary Material
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (Grant P40 OD010440). We thank Pieter Rein ten Wolde and Tom Shimizu for feedback on the manuscript. This work is part of the research program Vidi with project number 680-47-529, which is financed by the Dutch Research Council (NWO).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All analyzed timing data , analysis scripts and simulation scripts have been deposited in Zenodo (https://doi.org/10.5281/zenodo.6276446) (57).
References
- 1.Ebisuya M., Briscoe J., What does time mean in development? Development 145, dev164368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moss E. G., Heterochronic genes and the nature of developmental time. Curr. Biol. 17, R425–R434 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Rougvie A. E., Control of developmental timing in animals. Nat. Rev. Genet. 2, 690–701 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Uppaluri S., Brangwynne C. P., A size threshold governs Caenorhabditis elegans developmental progression. Proc. Biol. Sci. 282, 20151283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Zvi D., Shilo B. Z., Fainsod A., Barkai N., Scaling of the BMP activation gradient in Xenopus embryos. Nature 453, 1205–1211 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Gregor T., Bialek W., de Ruyter van Steveninck R. R., Tank D. W., Wieschaus E. F., Diffusion and scaling during early embryonic pattern formation. Proc. Natl. Acad. Sci. U.S.A. 102, 18403–18407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houchmandzadeh B., Wieschaus E., Leibler S., Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415, 798–802 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Umulis D. M., Othmer H. G., Mechanisms of scaling in pattern formation. Development 140, 4830–4843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driever W., Nüsslein-Volhard C., A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–93 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Driever W., Nüsslein-Volhard C., The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Monsalve G. C., Frand A. R., Toward a unified model of developmental timing: A “molting” approach. Worm 1, 221–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendriks G. J., Gaidatzis D., Aeschimann F., Großhans H., Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53, 380–392 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kim Dh., Grün D., van Oudenaarden A., Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat. Genet. 45, 1337–1344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meeuse M. W., et al. , Developmental function and state transitions of a gene expression oscillator in Caenorhabditis elegans. Mol. Syst. Biol. 16, e9498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rensing L., Meyer-Grahle U., Ruoff P., Biological timing and the clock metaphor: Oscillatory and hourglass mechanisms. Chronobiol. Int. 18, 329–369 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Moss E. G., Lee R. C., Ambros V., The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Ruvkun G., Giusto J., The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–319 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Ambros V., Horvitz H. R., Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416 (1984). [DOI] [PubMed] [Google Scholar]
- 19.Monsalve G. C., Van Buskirk C., Frand A. R., LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21, 2033–2045 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Jeon M., Gardner H. F., Miller E. A., Deshler J., Rougvie A. E., Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141–1146 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Gritti N., Kienle S., Filina O., van Zon J. S., Long-term time-lapse microscopy of C. elegans post-embryonic development. Nat. Commun. 7, 12500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulston J. E., Horvitz H. R., Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977). [DOI] [PubMed] [Google Scholar]
- 23.Wildwater M., Sander N., de Vreede G., van den Heuvel S., Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 138, 4375–4385 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Byerly L., Cassada R. C., Russell R. L., The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev. Biol. 51, 23–33 (1976). [DOI] [PubMed] [Google Scholar]
- 25.Avery L., Shtonda B. B., Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacNeil L. T., Watson E., Arda H. E., Zhu L. J., Walhout A. J., Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G., Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raizen D. M., Lee R. Y., Avery L., Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141, 1365–1382 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassada R. C., Russell R. L., The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46, 326–342 (1975). [DOI] [PubMed] [Google Scholar]
- 30.Golden J. W., Riddle D. L., The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev. Biol. 102, 368–378 (1984). [DOI] [PubMed] [Google Scholar]
- 31.Murphy C. T., Hu P. J., Insulin/insulin-like growth factor signaling in C. elegans. WormBook 2013 Dec 26, 1–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G., daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Ruaud A. F., Katic I., Bessereau J. L., Insulin/insulin-like growth factor signaling controls non-Dauer developmental speed in the nematode Caenorhabditis elegans. Genetics 187, 337–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edelman T. L., et al. , Analysis of a lin-42/period null allele implicates all three isoforms in regulation of Caenorhabditis elegans molting and developmental timing. G3 (Bethesda) 6, 4077–4086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olmedo M., Geibel M., Artal-Sanz M., Merrow M., A high-throughput method for the analysis of larval developmental phenotypes in Caenorhabditis elegans. Genetics 201, 443–448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrahante J. E., Miller E. A., Rougvie A. E., Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen:green fluorescent protein fusion gene. Genetics 149, 1335–1351 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennessen J. M., Gardner H. F., Volk M. L., Rougvie A. E., Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev. Biol. 289, 30–43 (2006). [DOI] [PubMed] [Google Scholar]
- 38.McCulloch K. A., Rougvie A. E., Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proc. Natl. Acad. Sci. U.S.A. 111, 15450–15455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perales R., King D. M., Aguirre-Chen C., Hammell C. M., LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription. PLoS Genet. 10, e1004486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Wynsberghe P. M., Pasquinelli A. E., Period homolog LIN-42 regulates miRNA transcription to impact developmental timing. Worm 3, e974453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler A. J., Baugh L. R., Sherwood D. R., Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10, e1004426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faerberg D. F., Gurarie V., Ruvinsky I., Inferring temporal organization of postembryonic development from high-content behavioral tracking. Dev. Biol. 475, 54–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mata-Cabana A., et al. , Differential regulation of developmental stages supports a linear model for C. elegans postembryonic development. bioRxiv [Preprint] (2021). 10.1101/2021.05.03.442510. Accessed 4 May 2021. [DOI]
- 44.Perez M. F., Francesconi M., Hidalgo-Carcedo C., Lehner B., Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552, 106–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadsworth W. G., Riddle D. L., Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev. Biol. 132, 167–173 (1989). [DOI] [PubMed] [Google Scholar]
- 46.Cooper S., Helmstetter C. E., Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 31, 519–540 (1968). [DOI] [PubMed] [Google Scholar]
- 47.Johnston G. C., Pringle J. R., Hartwell L. H., Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105, 79–98 (1977). [DOI] [PubMed] [Google Scholar]
- 48.Mitchison J. M., Creanor J., Further measurements of DNA synthesis and enzyme potential during cell cycle of fission yeast Schizosaccharomyces pombe. Exp. Cell Res. 69, 244–247 (1971). [DOI] [PubMed] [Google Scholar]
- 49.Robert L., Size sensors in bacteria, cell cycle control, and size control. Front. Microbiol. 6, 515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baugh L. R., To grow or not to grow: Nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194, 539–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu P. J., Dauer. WormBook: The Online Review of C. elegans Biology. https://www.ncbi.nlm.nih.gov/books/NBK116082/. Accessed 8 August 2007. [DOI] [PMC free article] [PubMed]
- 52.Begasse M. L., Leaver M., Vazquez F., Grill S. W., Hyman A. A., Temperature dependence of cell division timing accounts for a shift in the thermal limits of C. elegans and C. briggsae. Cell Rep. 10, 647–653 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Crapse J., et al. , Evaluating the simple Arrhenius equation for the temperature dependence of complex developmental processes. Mol. Syst. Biol.17, e9895 (2021). [DOI] [PMC free article] [PubMed]
- 54.Kuntz S. G., Eisen M. B., Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genet. 10, e1004293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong J., Amourda C., Saunders T. E., Temporal development of Drosophila embryos is highly robust across a wide temperature range. J. R. Soc. Interface 15, 20180304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruaud A. F., Bessereau J. L., Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development 133, 2211–2222 (2006). [DOI] [PubMed] [Google Scholar]
- 57.O. Filina, B. Demirbas, R. Haagmans, J. S. van Zon, Data set for the article “Temporal scaling in C. elegans larval development.” Zenodo. 10.5281/zenodo.6276446. Deposited 25 February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyzed timing data , analysis scripts and simulation scripts have been deposited in Zenodo (https://doi.org/10.5281/zenodo.6276446) (57).