Significance
High-risk (HR) human papillomaviruses (HPV) from the genus alpha cause anogenital and oropharyngeal cancers, whereas the contribution of HPV from the genus beta to the development of cutaneous squamous cell cancer is still under debate. HR-HPV genomes display potent immortalizing activity in human keratinocytes, the natural target cell for HPV. This paper shows that immortalization of keratinocytes by the beta-HPV49 genome requires the inactivation of the viral E8^E2 repressor protein and the presence of the E6 and E7 oncoproteins but also of the E1 and E2 replication proteins. This reveals important differences in the carcinogenic properties of HR-HPV and beta-HPV but also warrants further investigations on the distribution and mutation frequencies of beta-HPV in human cancers.
Keywords: beta-human papillomavirus, cutaneous squamous cell cancer, E8^E2, immortalization
Abstract
Beta-human papillomaviruses (HPV) have been implicated in the development of cutaneous squamous cell cancer (cSCC) in epidermodysplasia verruciformis (EV) patients and organ transplant recipients. In contrast to high-risk (HR) HPV, which cause anogenital and oropharyngeal cancers, immortalizing activity of complete beta-HPV genomes in normal human keratinocytes (NHK), the natural target cells for HPV, has not been reported. We now demonstrate that the beta-HPV49 wild-type genome is transcriptionally active in NHK but lacks immortalizing activity unless the E8 gene, which encodes the E8^E2 repressor, is inactivated. HPV49 E8− immortalized keratinocytes maintain high levels of viral gene expression and very high copy numbers of extrachromosomal viral genomes during long-term cultivation. Not only disruption of the viral E6 and E7 oncogenes but also of the E1 or E2 replication genes renders E8− genomes incapable of immortalization. E8−/E1− and E8−/E2− genomes display greatly reduced E6 and E7 RNA levels in short-term assays. This strongly suggests that high-level expression of E6 and E7 from extrachromosomal templates is necessary for immortalization. The requirement for an inactivation of E8 while maintaining E1 and E2 expression highlights important differences in the carcinogenic properties of HR-HPV and beta-HPV. These findings strengthen the notion that beta-HPV have carcinogenic potential that warrants further investigations into the distribution of beta-HPV in human cancers.
A significant fraction of human cancers is caused by infections with members of the flavi-, herpes-, hepadna-, papilloma-, polyoma-, or retroviruses (1). Among these, human papillomaviruses (HPV) comprise the largest family with currently 228 classified types (https://www.hpvcenter.se/human_reference_clones/), which are grouped into different genera such as alpha-, beta-, gamma-, mu-, and nu-PV. Persistent infections with a high-risk (HR) HPV (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 58, and 59), which belong to the alpha-PV, can lead to the development of cancer of the anus, cervix uteri, larynx, oropharynx, oral cavity, penis, vulva, and vagina, resulting in 530,000 new cases per year worldwide (2). Therefore, prophylactic vaccines have been introduced to prevent infections with the most prevalent HR-HPV (3). Aside from HR-HPV, HPV from the genus beta have been implicated in the development of cutaneous squamous cell cancer (cSCC) in patients with the rare genodermatosis epidermodysplasia verruciformis (EV) and in organ transplant recipients (OTR) (4–7). Both EV patients and OTR develop disseminated skin warts that can progress to cSCC predominantly in sun-exposed areas (4–7). Beta-HPV have also been found at other body sites such as nasal mucosa, anal canal, oral cavity, genital condylomas, and external genital lesions (8–12). Recent studies also detected beta-HPV DNA in a large fraction of conjunctival squamous cell carcinomas, raising the possibility that beta-HPV may contribute in this instance to carcinoma development (13).
HR-HPV genomes are present as either extrachromosomal elements or integrated into the host chromosomes and are expressed in all precancer and cancer cells, but only viral early but not late genes are transcribed in the majority of cancers (14). cSCC of EV patients maintain high levels of extrachromosomal beta-HPV DNA and only rarely integrated virus sequences and express late viral proteins, suggesting that productive viral replication takes place (6, 15–22). In contrast, beta-HPV copy number decreases from precursor lesions to cSCC to less than one viral genome per tumor cell in OTR, indicating that the majority of tumor cells does not express viral gene products (23). A “hit and run” model has been proposed that suggests that beta-HPV induce precursor lesions at an increased frequency in OTR due to immunosuppression and cause tumor progression in conjunction with ultraviolet ( UV) light but are then no longer required for tumor maintenance. In support of this, UV irradiation has been shown to induce cSCC in beta-HPV38 E6/E7 transgenic mice and Mastomys natalensis PV-infected animals with a mutational signature resembling human cSCC (24, 25).
HR-HPV encode the nonstructural E1, E2, E4, E5, E6, E7, and E8 genes as well the capsid L1 and L2 genes. The E1 helicase and the E2 transcription factor are sequence-specific DNA binding proteins that are essential for viral genome replication by forming an origin recognition complex that recruits the host DNA replication machinery (26, 27). E2 also modulates viral gene expression (27). E8 is mainly expressed as an E8^E2 fusion protein which binds to E2 response elements in the viral genome via its E2 DNA binding domain and acts as a repressor of viral genome replication and gene expression (28). E6 and E7 are the main oncoproteins that bind to a large number of host cell proteins and modulate their activities (29). HR-HPV E6 interact with the E3 ubiquitin ligase E6AP and p53 to induce the proteasomal degradation of p53 and counteract its tumor suppressive activities (29). Key activities of HR-HPV E7 proteins are the interactions with members of the retinoblastoma (pRb) protein family, which serve to deregulate cell cycle progression (29). Among alpha-PV, only HR-HPV (and phylogenetically closely related HPV) genomes can immortalize normal human keratinocytes (NHK), which represent the target cells for HPV in vivo (30–33). NHK can also be immortalized by the heterologous expression of only HR-HPV E6 and E7 (31, 32, 34–36). In line with E6 and E7 mediating immortalization of NHK by HR-HPV, viral genomes, in which E1, E2, E4, E5, or E8 have been inactivated, are capable of immortalization (37–43).
Beta-HPV encode the same set of genes as HR-HPV but lack an E5 gene. Up to now, the heterologous expression of E6 and E7 derived from HPV38 (beta2), 49, 75, or 76 (all beta3) has been shown to immortalize NHK (44–46). HPV49, 75, and 76 E6 proteins have been reported to share some activities with HR-HPV E6, such as binding to the E3 ubiquitin ligase E6AP and p53 to induce the proteasomal degradation of p53 (45, 46). HPV38 E6 also interacts with p53 but does not induce its degradation but rather alters its transcriptional activities (44, 45, 47, 48). Despite these similarities, comparative interactome studies also revealed that major differences exist between HR-HPV and beta-HPV E6 proteins (49, 50), but the contribution of beta-HPV–specific interaction partners to immortalization has not been established. The HPV49 and 76 E7 proteins bind to pRb but, different from HR-HPV, promote its hyperphosphorylation and not degradation (45, 46, 51).
In contrast to HR-HPV, keratinocyte-based model systems supporting beta-HPV genome replication that would allow the analysis of oncogenic properties of complete viral genomes, the viral life cycle and risk factors for cSCC development have not been reported. To generate keratinocytes stably maintaining HR-HPV genomes, NHK are transfected with recircularized viral genomes and a selection marker, and stable cell lines are obtained due to the immortalization capabilities of HR-HPV (37). Using this strategy, we observed that the recircularized reference HPV49 genome, despite reported immortalizing properties of E6 and E7 proteins and being transcriptionally active, does not immortalize NHK. Genetic analyses of HPV49 revealed that the inactivation of E8 and thus presumably a loss of the E8^E2 repressor protein greatly enhances viral replication and gene expression. Remarkably, E8− genomes gain the ability to immortalize NHK, suggesting that the levels of the viral oncoproteins expressed from the wild-type (wt) genome are not sufficient for immortalization. In line with this, immortalization of keratinocytes by HPV49 E8− genomes not only requires expression of the E6 and E7 oncoproteins but also of the E1 and E2 replication and transcription factors whose inactivation greatly decreases E6 and E7 transcription. Our data indicate that the immortalization activity of beta-HPV is restricted at the level of viral gene expression and that this cannot be overcome by integration of viral genomes into host chromosomes but only by inactivation of E8 highlighting important differences between HR-HPV and beta-HPV. Our findings strengthen the notion that beta-HPV have oncogenic potential which warrants further investigations on the distribution of beta-HPV, especially HPV49 and related HPV types, in human precancers and cancers.
Results
HPV49 Transcription in NHK Is Modulated by Early Viral Genes.
Previous studies have reported that the retroviral expression of HPV49 E6 and E7 immortalizes NHK (45, 46). We therefore attempted to generate HPV49-positive human keratinocyte cell lines using a protocol established for HR-HPV (37) by transfecting recircularized HPV49 genomes together with a plasmid encoding a selectable marker. Surprisingly, after drug selection, no actively growing keratinocyte colonies could be obtained. A possible explanation was that HPV49 genomes are not transcriptionally active in NHK and therefore do not express E6 and E7 proteins. Since antibodies for HPV49 proteins are not available, we investigated viral gene expression instead. HPV possess multiple promoters to generate polycistronic transcripts which are then alternatively spliced. No transcript maps for HPV49 are available, but several splice donors (SD) and acceptors (SA) are conserved among animal and human PVs: SDs at the beginning of E1, in E8, and in the 5′ part of the upstream regulatory region (URR) and SAs in front of E2 and in the E2/E4 region. This gives rise to six putative SD–SA combinations: the URR SD linked to the SA upstream of E2 (URR^E2), the URR SD to the SA within E2/E4 (URR^E4), the SD at the beginning E1 to the SA in front of E2 (E1^E2), the SD at the beginning of E1 to the SA in E2/E4 (E1^E4), the SD in E8 to the SA in front of E2 (E8^E2N), and the SD in E8 to the SA within E2/E4 (E8^E2) (Fig. 1A). Putative splice junctions in the HPV49 genome were predicted using the PAVE database [(52) https://pave.niaid.nih.gov] or by sequence alignments. The corresponding copy DNAs (cDNAs) were obtained in plasmid vectors by gene synthesis and used to develop sensitive primer pairs for qPCR (SI Appendix, Table S1). NHK were transfected with recircularized HPV49 genomes, and RNA was isolated 6 d later and then analyzed by qPCR. Melting curve analyses and direct sequencing of qPCR products indicated that all exon borders corresponded to the predicted ones (Fig. 1A). The different spliced transcripts were then quantified in NHK from four different donors transfected with the HPV49 genome 3, 6, and 9 d post transfection (p.t.) (Fig. 1B). This indicated that all splice junctions were expressed at all time points albeit at different levels (Fig. 1B). E1^E4 was the most abundant transcript at all time points. E1^E4 amounts increased 9.5-fold from day 3 to day 6 and then slightly decreased on day 9. E1^E2 amounts were similar as E1^E4 on day 3 but then decreased on day 6 and 9. URR^E4 and E8^E2 levels were ∼eightfold lower than E1^E4 transcripts on day 3. URR^E4 levels stayed relatively constant over time, whereas E8^E2 levels dropped from day 3 to day 9 by twofold. URR^E2 levels were 20-fold lower than E1^E4 on day 3 and then increased 3-fold by day 9. E8^E2N levels were 70-fold lower than E1^E4 levels on day 3 and did not further change. In summary, these results demonstrate that the wt HPV49 genome expresses a variety of spliced transcripts in different amounts for at least 9 d in NHK, suggesting that the lack of immortalization is not due to a block in gene expression.
Fig. 1.
(A) Linear representation of the HPV49 genome. The URR, early genes (E1 to E8) and the late genes L1 and L2 are indicated. Nucleotide positions of identified SD (D) and SA (A) sites are given. Spliced transcripts identified by qPCR and sequencing are indicated by gray bars, and the putative extension of these transcripts to the potential early polyadenylation site is indicated by dashed lines. (B) qPCR analyses of spliced HPV49 transcripts expressed 3, 6, and 9 d p.t. of NHK. PGK1 was used as a reference transcript. The data are derived from four independent experiments using cells from different donors.
To obtain evidence that there is no translational block to viral protein expression in NHK, a genetic analysis was performed. We generated HPV49 E1−, E2−, E6−, E7−, and E8− genomes by inactivating the respective start codons by mutation and transfected them into NHK from four different donors and compared their transcription properties with the wt (Fig. 2A). The most dramatic effects were observed with the E8− genome, where the levels of E1^E4 (170- to 677-fold), URR^E4 (82- to 219-fold), E1^E2 (23- to 267-fold), and E8^E2 transcripts (24- to 297-fold) were significantly increased at all time points. Attempts to quantitate E6 and E7 transcripts in total RNA failed, most likely due to high amounts of transfected DNA templates. To overcome this, messenger RNA (mRNA) was enriched from total RNA using an oligo-dT selection step and analyzed by qPCR. This revealed that E6 and E7 transcripts were present in wt-transfected cells (Fig. 2B). E6 was expressed at lower levels than E7 and both transcripts increased from day 3 to day 6. Furthermore, both E6 and E7 levels were significantly increased in E8− transfected cells (Fig. 2B). Consistent with data for other HPV (38, 41, 42, 53, 54), HPV49 E8− genomes displayed a robust signal of replicated viral genomes 6 d p.t., whereas only a very weak signal could be obtained for wt genomes (Fig. 2C). This confirms that HPV49 E8, most likely expressed as an E8^E2 fusion protein, inhibits gene expression and genome replication comparable to other HPV types (38, 42, 53). The E1− genome displayed significantly reduced E1^E4 and E8^E2 levels and the E2− genome E1^E4, E1^E2, and URR^E4 transcripts. The reduced E1^E4 transcript levels from both E1− and E2− genomes are consistent with a conserved interaction of E1 with E2 to initiate PV genome replication and are therefore most likely due to a reduced replication capacity of E1− and E2− genomes. However, this could not be directly tested as the wt genome does not give rise to a robust replication signal (Fig. 2C). The E2− genome but not the E1− genome had reduced URR^E4 and E1^E2 levels, suggesting that this might be due to the conserved transcription activation function of E2. Interestingly, the E7− genome showed a significant increase of URR^E4, E1^E2, and E1^E4 levels on day 3, pointing to the possibility that E7 may act as an inhibitor of viral gene expression at early time points. In contrast, viral gene expression from the E6− genome was similar to the wt. In summary, the changes in viral transcript levels observed with E1−, E2−, E7−, and E8− genomes strongly suggested that functional viral proteins are expressed, indicating that the lack of immortalization by HPV49 is not due to a block of viral protein expression per se.
Fig. 2.
(A) qPCR analyses of spliced HPV49 transcripts expressed 3, 6, and 9 d p.t. of NHK with HPV49 wt, E1−, E2−, E6−, E7−, or E8− genomes. PGK1 was used as a reference transcript. The data are derived from four independent experiments using cells from different donors. The error bars indicate the SEM. Statistical significance was determined using a ratio-paired t test using the wt as a reference (*P < 0.05; **P < 0.01). (B) qPCR analyses of E6 and E7 transcripts in polyA-enriched RNA in NHK transfected with wt or E8− genomes. The error bars indicate the SEM. Statistical significance was determined using a ratio-paired t test using the wt as a reference (*P < 0.05; **P < 0.01) (C) Southern blot analysis of low–molecular-weight DNA isolated 6 d p.t. of NHK transiently transfected with HPV49 wt and E8− genomes. DNA was digested with DpnI to remove nonreplicated input DNA and EheI to linearize the viral genomes. As a marker (M), 100 pg linearized HPV49 genome was used.
HPV49 E8− Genomes Immortalize NHK.
It was possible that HPV49, despite being transcriptionally active, is not able to express viral protein levels sufficient for immortalization. We therefore repeated immortalization assays with the E8− genome due to its greatly increased transcript and genome replication levels in transient assays and the wt genome as a control in NHK from three different donors. Remarkably, HPV49 E8− transfected NHK formed drug-resistant colonies that were pooled and could be expanded into stable cell lines (Table 1). Growth curves of HPV49 E8− cell lines revealed that the cells continuously proliferated with an average doubling rate of 4.3 d without signs of crisis during the observation period of 300 d (Fig. 3). This strongly indicated that the inactivation of E8 renders HPV49 genomes competent to immortalize NHK.
Table 1.
NHK immortalization assays using HPV49 wt and E8− genomes
| HPV49 genome | Donor No. 1 | Donor No. 2 | Donor No. 3 |
|---|---|---|---|
| wt | − | − | − |
| E8− | + | + | + |
Fig. 3.
Growth curves of HPV49 E8−-positive human keratinocyte cell lines. Cell lines were continuously cultivated for 300 d and growth was recorded.
HPV49 E8− Cell Lines Maintain High Levels of Extrachromosomal Viral DNA and Viral Transcripts.
HR-HPV genomes are present in immortalized keratinocytes either as autonomously replicating extrachromosomal elements or as integrates in host chromosomes. Characterization of the physical state of HPV49 E8− genomes in early (P4 to P7) and late (P36) passage cells by Southern blotting using total cellular DNA revealed several DNA species upon digesting with a restriction enzyme without a recognition sequence in the HPV49 genome (Fig. 4A). These forms are consistent with open circle (oc), closed covalent circle (ccc), and concatenated or integrated (c/i) species of the viral DNA similar to HR-HPV–immortalized keratinocytes (37). Digesting the DNA with a restriction enzyme with a single recognition sequence in HPV49 resulted in the appearance of the linearized form and an almost complete loss of all other forms, indicating that the viral genomes are mainly present as extrachromosomal plasmids. This was further confirmed by an exonuclease V/qPCR assay (55), which revealed that the viral DNA is completely exonuclease V resistant and thus extrachromosomal, whereas resistance of the cellular ACTB gene is less than 6% (Fig. 4B). Quantification of viral copy numbers by qPCR demonstrated that on the average 5,820, 2,975, and 4,525 copies/cell were present (Fig. 4C). Remarkably, these copy numbers are much higher than the 300 to 400 copies/cell determined by a similar approach for HPV16 E8− genomes in immortalized keratinocyte cell lines (41). We next determined viral transcript levels in early and late passage cells, which revealed that all spliced transcripts detected in NHK transiently transfected with wt or E8− genomes were also present in the stable cell lines (Fig. 4D). Similar to short-term assays, the most abundant transcript was E1^E4, followed by E1^E2, URR^E4, E8^E2, URR^E2, and E8^E2N. Furthermore, E7 transcripts were present at levels similar to E1^E2 transcripts. In contrast, E6 transcripts were five times less abundant than E7 transcripts. Consistent with the maintenance of high viral copy numbers in late passage cell lines, all viral transcripts were expressed in late passages at similar amounts as in early passage cells (Fig. 4C). In summary, this reveals that HPV49 E8− genomes are present as high—copy-number plasmids and express different viral transcripts in immortalized cell lines during long-term cultivation. This strongly suggests that immortalization of NHK by HPV49 E8− and their continuous cell growth depends on the presence and expression of the virus comparable to HR-HPV.
Fig. 4.
(A) Southern blot analyses of total cellular DNA isolated from stable HPV49 E8− cell lines No. 1 to 3 using 32P-labeled HPV49 genomes. DNA was digested with NotI (N; a noncutter of HPV49) or SalI (S; a single-cutter of HPV49). The positions of concatemeric/integrated (c/i), open circle (oc), linearized (lin), and closed covalent circle (ccc) forms of the viral DNA are indicated. As a marker (M), 100 pg linearized HPV49 genome was used. (B) Total cellular DNA was incubated with or without exonuclease V, and then the presence of ACTB or HPV49 was determined by qPCR. The data are shown as the fraction resistant to exonuclease V digest. The data are derived from three to four independent DNA preparations. The error bars indicate the SEM. (C) HPV49 genome copy numbers in cell lines No. 1 to 3 were determined in total cellular DNA by qPCR using standard curves for HPV49 and ACTB and are given as viral copies per cell assuming two ACTB copies per cell. The error bars indicate the SEM. (D) qPCR analyses of different spliced HPV49 transcripts expressed in cell lines No. 1 to 3 in early and late passage cells. PGK1 was used as a reference transcript. The error bars indicate the SEM.
The Immortalization Capacity of the HPV49 E8− Genome Depends upon the E1, E2, E6, and E7 Genes.
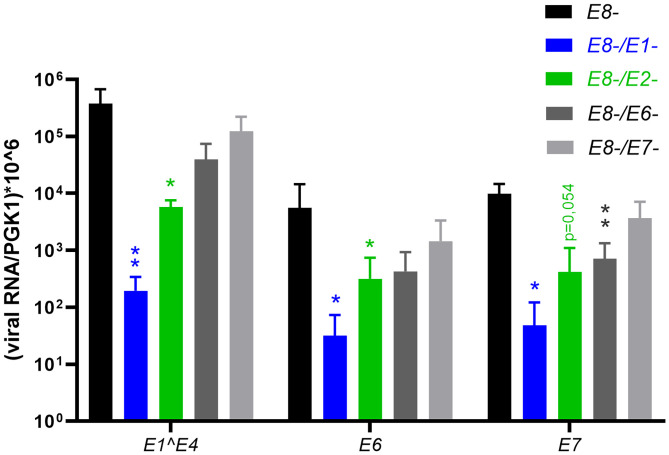
To explore which viral genes are necessary for NHK immortalization E1−, E2−, E6−, and E7− mutations were combined with the E8− mutation in the HPV49 genome. Assessment of these genomes in NHK from three donors and the E8− genome as a positive control surprisingly revealed that not only E6 and E7 but also E1 and E2 are required for immortalization (Table 2). The need for both E6 and E7 indicated that both contribute independently to immortalization and is consistent with the idea that HPV49 E6 and E7 act as oncoproteins. The requirement for both E1 and E2 pointed to the possibility that the effects could be mediated indirectly via reduced genome replication and lower levels of viral gene expression as observed for E1− and E2− genomes (Fig. 2A). Consistent with this interpretation, E6, E7, and E1^E4 transcript levels in transiently transfected NHK expressed from E8−/E1− and E8−/E2− genomes were greatly reduced compared with E8− genomes (Fig. 5). In contrast, E8−/E6− and E8−/E7− genomes showed only minor effects on viral gene expression with the exception of E7 transcripts from E8−/E7− genomes. These data strongly suggest that high copy numbers of extrachromosomal viral genomes are required to express E6 and E7 levels sufficient for immortalization. To test if E6 and E7 modulate growth of HPV49 E8− cell lines, different small interfering RNAs (siRNAs) targeting the HPV49 E6 gene were designed and tested for knock-down of E6 and E7 transcripts. Two siRNAs, si49E6-1 and si49E6-2, significantly reduced E6 and E7 transcript levels (Fig. 6A). Cell numbers were determined 3 d p.t, which revealed reduced numbers by both si49E6-1 and si49E6-2, which were statistically significant for si49E6-1 and showed a trend (P = 0.069) for si49E6-2 (Fig. 6B). Taken together, these data indicate that the growth of HPV49 E8− cell lines is influenced by the E6 and E7 levels.
Table 2.
NHK immortalization assays using HPV49 mt genomes
| HPV49 genome | Donor No. 4 | Donor No. 5 | Donor No. 6 |
|---|---|---|---|
| E8− | + | + | + |
| E8−/E1− | − | − | − |
| E8−/E2− | − | − | − |
| E8−/E6− | − | − | − |
| E8−/E7− | − | − | − |
Fig. 5.
QPCR analyses of HPV49 transcripts expressed 6 d p.t. of NHK with HPV49 E8−, E8−/E1−, E8−/E2−, E8−/E6−, or E8−/E7− genomes. PGK1 was used as a reference transcript. The data are derived from four independent transfections using cells from different donors. The error bars indicate the SEM. Statistical significance was determined using a ratio-paired t test using wt as a reference (*P < 0.05; **P < 0.01).
Fig. 6.
HPV49 E8− cell lines were transfected with different siRNAs (siControl, si49E6-1, and si49E6-2). The amounts of E6 and E7 transcripts in total RNA were measured by qPCR using PGK1 as a reference (A), and cell numbers (B) were determined 3 d p.t. The error bars indicate the SEM. Statistical significance was determined using a one-sample t test using siControl-transfected cells as a reference (*P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
Infections with beta-HPV have been suspected to be involved in the development of cSCC in EV patients and OTR. It is well established that HR-HPV can cause anogenital and oropharyngeal cancers. Consistent with their carcinogenic properties in vivo, HR-HPV, in contrast to low-risk HPV, genomes have been shown to efficiently immortalize NHK. Immortalization requires only the E6 and E7 oncoproteins and is independent from the presence of other early or late genes. A consequence of the inactivation of E1 or E2 are replication-incompetent genomes that integrate into the host genome and hence immortalization of keratinocytes is independent from the physical state of the viral genome and does not require viral replication (30, 37, 39, 56). On the other hand, inactivation of E8^E2 in HR-HPV16 increases viral replication and gene expression but has no influence on cell immortalization, growth, and differentiation properties (38, 41). Taken together, HR-HPV genomes immortalize keratinocytes independently from their replication properties.
The contribution of beta-HPV infections to human cancers is still under debate, and in contrast to HR-HPV, no keratinocyte model using complete viruses has been available allowing the evaluation of oncogenic properties, the viral life cycle, and risk factors for cancer development such as EV susceptibility genes and UV irradiation. Currently, the U2OS cell line is used for beta-HPV genome replication studies, but these cells are already immortalized, not of keratinocyte origin, and do not support the differentiation-dependent viral life cycle (54, 57). Our experiments reveal that HPV49 genomes express spliced URR^E4 and URR^E2 transcripts in both transiently transfected and immortalized keratinocytes. Expression of URR^E4 transcripts has been described in beta-HPV5– and beta-HPV8–positive EV lesions (58, 59) but not in HPV5-positive U2OS cells (54), indicating that cultured keratinocytes mimic the in vivo situation closer than the U2OS cell line. Beta-HPV E6 and E7 functions have been mainly analyzed in human keratinocytes via retroviral expression in the absence of other viral proteins, and this has revealed that beta-HPV E6 and E7 proteins can have immortalizing capabilities and modulate apoptosis, DNA repair, keratinocyte differentiation, and immune responses (60).
HPV49 E6 and E7 proteins have been reported to immortalize efficiently NHK upon retroviral expression (45, 46). Nevertheless, we surprisingly observed that the reference HPV49 genome, in contrast to HR-HPV16, 18, and 31 genomes, which are routinely used in the laboratory to generate immortalized cell lines (41, 42, 56), is incapable of inducing keratinocyte growth and immortalization. Follow-up investigations revealed that the viral genome expresses several spliced transcripts for several days in NHK and that the inactivation of the E1, E2, and E8 genes modulated viral gene expression, consistent with their conserved replication functions in other PV (26–28). These data suggested that the lack of immortalization by HPV49 is neither due to a lack of viral gene expression nor to a block of viral protein expression. Surprisingly, E8− genomes, which replicate to high levels in transient assays and display greatly increased viral transcription, were able to reproducibly immortalize NHK. These cell lines harbor high copy numbers of extrachromosomal viral genomes and express high levels of viral transcripts. The comparison of early with late passages revealed that viral gene expression and the high viral copy numbers are perpetuated in late passages, strongly suggesting that high levels of gene expression are required to maintain the immortalized phenotype. Consistent with this, E8−/E1− or E8−/E2− genomes fail to immortalize NHK and display significantly reduced E6 and E7 transcript levels in short-term assays. While it is possible that E8, which is presumably expressed as an E8^E2 fusion protein, acts as a tumor suppressor independent from its activity as a repressor of viral transcription and replication, we believe that the loss of immortalization activity of E8−/E1− and E8−/E2− genomes favors the explanation that the main reason for the acquisition of immortalization activity by E8− genomes is the increased expression of E6 and E7 transcripts. However, beta-HPV8 E2 has been shown to have oncogenic properties when expressed separately in the skin of transgenic mice and thus it is possible that E2 is not only required for immortalization because of its activity as a replication factor but also as an oncogene (61). The need for a loss of E8 while maintaining E1 and E2 expression points to significant differences between HR-HPV and HPV49. At present, it is unclear why the inefficient expression of E6 and E7 from the HPV49 genome cannot be overcome by integration into the host chromosomes as commonly observed for HR-HPV in tissue culture and in vivo. One possibility is that even highly active cellular promoters do not match the expression levels provided by several thousand copies of viral genomes. On the other hand, it is also possible that the mechanisms driving integration of HR-HPV genomes into host chromosomes while maintaining expression of the E6 and E7 oncogenes is fundamentally different between HR-HPV and beta-HPV. Interestingly, beta-HPV genomes in cSCC from EV patients are present at high copy numbers in an extrachromosomal state, which may indicate that also in these cases high-level viral gene expression is required to maintain the tumor phenotype (6, 15, 18–22). A UV-irradiation–induced increase in HPV8 gene expression in a transgenic mouse model results in enhanced skin tumorigenesis in vivo, providing further evidence that deregulated gene expression can contribute to beta-HPV oncogenicity in vivo (62). CSCC in OTR harbor less than one HPV copy per cell and thus are no longer dependent upon HPV. However, at least a fraction of beta-HPV–positive precancers such as actinic keratoses are positive for late viral protein expression, raising the possibility possible that an interference with E8^E2 could enhance E6 and E7 expression and thus increase the risk of malignant progression (63).
Our data indicate that HPV49 E8^E2 is a crucial regulator of immortalization activity and thus interferences with E8^E2’s repression activity could enhance carcinogenic activity in vivo. In contrast to HR-HPV, complete beta-HPV genome sequences have not yet been systematically analyzed in biopsy material in order to uncover if inactivating E8 mutations occur in vivo and thus could contribute to cancer development. HPV16 E8^E2 transcription is driven by a separate promoter, and studies using the HPV5/U2OS model support the idea that also beta-HPV use a separate promoter for E8^E2 expression, but host cell factors involved in its regulation have not been identified (54, 64). E8^E2 proteins recruit NCoR/SMRT-HDAC3 corepressor complexes to inhibit viral replication via the conserved E8 part (53, 65). It is likely that HPV49 E8^E2 acts by a similar mechanism, and therefore it will be crucial to elucidate how beta-HPV E8^E2 gene expression and protein activity is regulated in order to analyze potential interferences in vivo.
Using this tissue culture model, it is now possible to perform genetic analyses of beta-HPV in order to understand the contributions of interaction partners of E6 and E7, especially beta-HPV specific ones, to both immortalization and viral replication. Furthermore, the hypothesis that the EV susceptibility genes CIB1, TMC6, and TMC8 act as restriction factors for beta-HPV replication in keratinocytes can now be tested (6, 66). It is now also feasible to investigate the effects of UV irradiation, the main risk factor for cSCC development in OTR and EV patients, on beta-HPV replication in a relevant tissue culture model. The immortalizing ability of HPV49 genomes also warrants that the search for HPV49 and related HPV types in keratinocyte-derived precancers and cancers should be intensified.
Materials and Methods
Cell Culture.
NHK were isolated from human foreskin after routine circumcision upon informed consent of patients, which was approved by the ethics committee of the medical faculty of the University Tuebingen (6199/2018BO2) and performed according to the principles of the Declaration of Helsinki. Samples were deidentified prior to use. NHK were isolated and maintained in keratinocyte serum-free medium supplemented with recombinant human epidermal growth factor, bovine pituitary extract, and gentamycin (Thermo Fisher Scientific). For transient transfection experiments, 8 × 104 NHK were seeded into 6-well dishes, 24 h later, cells were transfected with 1 µg recircularized HPV genomes and Fugene HD transfection reagent (Promega) using a ratio of 1:5, and 24 h later, medium was switched from keratinocyte serum-free medium to E-medium (three parts Dulbecco’s modified Eagle’s medium, one part Ham’s F12 supplemented with 5% [vol/vol] fetal bovine serum; 24 µg/l adenine, 0.4 ng/l hydrocortisone, 10 ng/l cholera toxin, 5 μg/l transferrin, 20 pM 3,3′-5-triodo-L-thyronine, 5 ng/l epidermal growth factor, and 5 μg/l insulin), and mitomycin C–treated NIH 3T3 J2 feeder cells were added. To obtain immortalized keratinocyte lines, NHK were transfected with 4 µg recircularized HPV49 genomes and 1 µg pSV2-neo plasmid using Fugene HD. Cells were selected with G418 (150 µg/mL) in the presence of complete E-medium and mitomycin C–treated NIH 3T3 J2 NHP until mock-transfected keratinocytes were dead (8 to 10 d) (37, 41, 42). G418-resistent colonies were expanded as pooled cultures. Cells were maintained continuously for at least 300 d, and splitting dates and dilutions were recorded to generate growth curves. HPV49 E8− cell lines were transfected in 24-well dishes with 30 pmol of control siRNA (siAllstars, Qiagen), si49 E6-1 (GGCUAAUAUUGCUGAGAUAUU), or si49E6-2 (UGUCGUAGGCAUCGAAAUAUU) using RNAiMax (Thermofisher). After 3 d, cells were counted to monitor growth or RNA was isolated to evaluate knock-down of E6 and E7 transcripts by qPCR.
Recombinant Plasmids.
The cloned HPV49 reference genome (NC_001591.1) in pGEM4 (67) was provided by the International HPV Reference Center, Karolinska Institute, Sweden. Plasmids pUC57 HPV49 E1^E4 (nucleotide [nt.] 811 to 926/3,281 to 3,511), pUC57 HPV E1^E2 (nt. 811 to 926/2,635 to 2,790), pUC57 HPV49 URR^E4 (HPV49 nt. 7,505 to 7,560/1 to 62/3,281 to 3,600), pUC57 URR^E2 (nt. 7,505 to 7,560/1 to 62/2,635 to 2,938), pUC57 HPV49 E8^E2 (nt. 1,205 to 1,321/3,281 to 3,600), and pUC57 HPV49 E8^E2N (nt. 1,205 to 1,321/2,635 to 2,938) were made to order by Genscript and used to generate standard curves for qPCR experiments. To generate HPV49 E1−, E2−, E6−, E7−, E8−, E8−/E1−, E8−/E2−, E8−/E6−, and E8−/E7− genomes, the respective ATG start codons were mutated to ACG (E1, E2, E6, E8) or to GTG (E7) by overlap extension PCR and then inserted into wt or E8− genomes by restriction enzyme cloning and then validated by DNA sequencing. All mutations were designed to be silent in overlapping genes (E1− [E7 N100N]; E2− [E1 N591N]; E7− [E6 E138E]; E8− [E1 N127N]).
Southern Blot Analysis.
Total cellular DNA from stable keratinocyte lines was digested with NotI (noncutter for HPV49) or SalI (single cutter for HPV49). Low–molecular-weight DNA was isolated from transiently transfected NHK and digested with DpnI and EheI. Digested DNAs were separated in 0.8% agarose gels. Blotting and hybridization to a 32P-labeled HPV49 probe was carried out as previously described (41). After exposure of the membrane to PhosphoImager screens, signals were visualized using the AIDA software package (Raytest).
qPCR.
HPV49 copy numbers were quantified in total cellular DNA by qPCR using amplicons in the HPV49 L2 gene (HPV49 4973 F: TACCCCACTACGCAACATCA; HPV49 5165 R: AGCTGCTACGTCCCTTTCAA) and the cellular ACTB gene and copy number standards. RNA was isolated from transfected keratinocytes or HPV49 E8− keratinocyte cell lines using the RNeasy mini kit (Qiagen), and cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen). To allow detection of E6 and E7 transcripts in transiently transfected cells, mRNA was enriched from total RNA using the RNeasy Pure mRNA Bead Kit (Qiagen) according to the manufactureŕs instructions. cDNA (50 ng) was analyzed by qPCR in duplicates using a LightCycler 480 and the LightCycler 480 SYBR green I master mix (Roche Applied Science) and 0.3 µM primer pair for HPV49 E1^E4, HPV49 E1^E2, HPV49 URR^E4, HPV49 URR^E2, HPV49 E8^E2, HPV49 E8^E2N, HPV49 E6, HPV49 E7 (SI Appendix, Table S1), or PGK1 (41). Copy numbers were determined by plasmid standards run in parallel.
ExonucleaseV-Resistance Assay.
The assay was adapted from ref. 55 with minor modifications: total cellular DNA (100 ng) was incubated in the presence or absence of 5 U exonuclease V (NEB M0345S) in 1× NEBuffer 4 supplemented with 1 mM ATP for 60 min at 37 °C. Then, the enzyme was inactivated for 10 min at 95 °C. Finally, 10 ng input DNA was measured by qPCR using primers for HPV49 and ACTB.
Supplementary Material
Acknowledgments
This work was supported by a Wilhelm Sander-Stiftung Grant 2018.148.1 to F.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.McLaughlin-Drubin M. E., Munger K., Viruses associated with human cancer. Biochim. Biophys. Acta 1782, 127–150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Martel C., Plummer M., Vignat J., Franceschi S., Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 141, 664–670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiller J. T., Lowy D. R., Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat. Rev. Microbiol. 10, 681–692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandolin L., et al. , Beta human papillomaviruses infection and skin carcinogenesis. Rev. Med. Virol. 30, e2104 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Howley P. M., Pfister H. J., Beta genus papillomaviruses and skin cancer. Virology 479–480, 290–296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orth G., Genetics of epidermodysplasia verruciformis: Insights into host defense against papillomaviruses. Semin. Immunol. 18, 362–374 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Quint K. D., et al. , Human Beta-papillomavirus infection and keratinocyte carcinomas. J. Pathol. 235, 342–354 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Bottalico D., et al. , The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J. Infect. Dis. 204, 787–792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donà M. G., et al. , Incidence, clearance and duration of cutaneous beta and gamma human papillomavirus anal infection. J. Infect. 73, 380–383 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Hampras S. S., et al. , Prevalence and concordance of cutaneous beta human papillomavirus infection at mucosal and cutaneous sites. J. Infect. Dis. 216, 92–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunes E. M., et al. ; HIM Study group, Diversity of beta-papillomavirus at anogenital and oral anatomic sites of men: The HIM Study. Virology 495, 33–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce Campbell C. M., et al. , Cutaneous human papillomavirus types detected on the surface of male external genital lesions: A case series within the HPV Infection in Men Study. J. Clin. Virol. 58, 652–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galati L., et al. , Detection of a large spectrum of viral infections in conjunctival premalignant and malignant lesions. Int. J. Cancer 147, 2862–2870 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Groves I. J., Coleman N., Pathogenesis of human papillomavirus-associated mucosal disease. J. Pathol. 235, 527–538 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Adachi A., Kiyono T., Hayashi Y., Ohashi M., Ishibashi M., Detection of human papillomavirus (HPV) type 47 DNA in malignant lesions from epidermodysplasia verruciformis by protocols for precise typing of related HPV DNAs. J. Clin. Microbiol. 34, 369–375 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgogna C., et al. , Characterization of skin lesions induced by skin-tropic α- and β-papillomaviruses in a patient with epidermodysplasia verruciformis. Br. J. Dermatol. 171, 1550–1554 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Borgogna C., et al. , Characterization of beta papillomavirus E4 expression in tumours from Epidermodysplasia Verruciformis patients and in experimental models. Virology 423, 195–204 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Ostrow R. S., et al. , Human papillomavirus DNA in cutaneous primary and metastasized squamous cell carcinomas from patients with epidermodysplasia verruciformis. Proc. Natl. Acad. Sci. U.S.A. 79, 1634–1638 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfister H., Gassenmaier A., Nürnberger F., Stüttgen G., Human papilloma virus 5-DNA in a carcinoma of an epidermodysplasia verruciformis patient infected with various human papillomavirus types. Cancer Res. 43, 1436–1441 (1983). [PubMed] [Google Scholar]
- 20.Yabe Y., Tanimura Y., Sakai A., Hitsumoto T., Nohara N., Molecular characteristics and physical state of human papillomavirus DNA change with progressing malignancy: Studies in a patient with epidermodysplasia verruciformis. Int. J. Cancer 43, 1022–1028 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Yutsudo M., Shimakage T., Hakura A., Human papillomavirus type 17 DNA in skin carcinoma tissue of a patient with epidermodysplasia verruciformis. Virology 144, 295–298 (1985). [DOI] [PubMed] [Google Scholar]
- 22.Yutsudo M., et al. , Involvement of human papillomavirus type 20 in epidermodysplasia verruciformis skin carcinogenesis. J. Clin. Microbiol. 32, 1076–1078 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenborn S. J., et al. , Human papillomavirus-DNA loads in actinic keratoses exceed those in non-melanoma skin cancers. J. Invest. Dermatol. 125, 93–97 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hasche D., et al. , The interplay of UV and cutaneous papillomavirus infection in skin cancer development. PLoS Pathog. 13, e1006723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viarisio D., et al. , Beta HPV38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog. 14, e1006783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergvall M., Melendy T., Archambault J., The E1 proteins. Virology 445, 35–56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride A. A., The papillomavirus E2 proteins. Virology 445, 57–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreer M., van de Poel S., Stubenrauch F., Control of viral replication and transcription by the papillomavirus E8^E2 protein. Virus Res. 231, 96–102 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Moody C. A., Laimins L. A., Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 10, 550–560 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Dürst M., Dzarlieva-Petrusevska R. T., Boukamp P., Fusenig N. E., Gissmann L., Molecular and cytogenetic analysis of immortalized human primary keratinocytes obtained after transfection with human papillomavirus type 16 DNA. Oncogene 1, 251–256 (1987). [PubMed] [Google Scholar]
- 31.Muench P., et al. , Binding of PDZ proteins to HPV E6 proteins does neither correlate with epidemiological risk classification nor with the immortalization of foreskin keratinocytes. Virology 387, 380–387 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Schütze D. M., et al. , Differential in vitro immortalization capacity of eleven (probable) [corrected] high-risk human papillomavirus types. J. Virol. 88, 1714–1724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodworth C. D., Doniger J., DiPaolo J. A., Immortalization of human foreskin keratinocytes by various human papillomavirus DNAs corresponds to their association with cervical carcinoma. J. Virol. 63, 159–164 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawley-Nelson P., Vousden K. H., Hubbert N. L., Lowy D. R., Schiller J. T., HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 8, 3905–3910 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson J. B., Bedell M. A., McCance D. J., Laiminis L. A., Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J. Virol. 64, 519–526 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Münger K., Phelps W. C., Bubb V., Howley P. M., Schlegel R., The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63, 4417–4421 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frattini M. G., Lim H. B., Laimins L. A., In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. U.S.A. 93, 3062–3067 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lace M. J., Anson J. R., Thomas G. S., Turek L. P., Haugen T. H., The E8–E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J. Virol. 82, 10841–10853 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romanczuk H., Howley P. M., Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. U.S.A. 89, 3159–3163 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott M. L., et al. , Human papillomavirus 16 E5 inhibits interferon signaling and supports episomal viral maintenance. J. Virol. 94, e01582-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Straub E., Dreer M., Fertey J., Iftner T., Stubenrauch F., The viral E8^E2C repressor limits productive replication of human papillomavirus 16. J. Virol. 88, 937–947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stubenrauch F., Hummel M., Iftner T., Laimins L. A., The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74, 1178–1186 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson R., Fehrmann F., Laimins L. A., Role of the E1–E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J. Virol. 79, 6732–6740 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caldeira S., et al. , The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 77, 2195–2206 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornet I., et al. , Comparative analysis of transforming properties of E6 and E7 from different beta human papillomavirus types. J. Virol. 86, 2366–2370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minoni L., et al. , Transforming properties of beta-3 human papillomavirus E6 and E7 proteins. mSphere 5, e00398-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Accardi R., et al. , Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 7, 334–340 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muench P., et al. , Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res. 70, 6913–6924 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Rozenblatt-Rosen O., et al. , Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487, 491–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White E. A., et al. , Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity. J. Virol. 86, 13174–13186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White E. A., et al. , Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. U.S.A. 109, E260–E267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Doorslaer K., et al. , The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 45, D499–D506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dreer M., et al. , Interaction of NCOR/SMRT repressor complexes with papillomavirus E8^E2C proteins inhibits viral replication. PLoS Pathog. 12, e1005556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sankovski E., Männik A., Geimanen J., Ustav E., Ustav M., Mapping of betapapillomavirus human papillomavirus 5 transcription and characterization of viral-genome replication function. J. Virol. 88, 961–973 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myers J. E., et al. , Detecting episomal or integrated human papillomavirus 16 DNA using an exonuclease V-qPCR-based assay. Virology 537, 149–156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karstensen B., et al. , Gene expression profiles reveal an upregulation of E2F and downregulation of interferon targets by HPV18 but no changes between keratinocytes with integrated or episomal viral genomes. Virology 353, 200–209 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Geimanen J., et al. , Development of a cellular assay system to study the genome replication of high- and low-risk mucosal and cutaneous human papillomaviruses. J. Virol. 85, 3315–3329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haller K., Stubenrauch F., Pfister H., Differentiation-dependent transcription of the epidermodysplasia verruciformis-associated human papillomavirus type 5 in benign lesions. Virology 214, 245–255 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Stubenrauch F., Malejczyk J., Fuchs P. G., Pfister H., Late promoter of human papillomavirus type 8 and its regulation. J. Virol. 66, 3485–3493 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tommasino M., The biology of beta human papillomaviruses. Virus Res. 231, 128–138 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Pfefferle R., et al. , The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J. Invest. Dermatol. 128, 2310–2315 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Hufbauer M., et al. , Enhanced human papillomavirus type 8 oncogene expression levels are crucial for skin tumorigenesis in transgenic mice. Virology 403, 128–136 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Borgogna C., et al. , Improved detection reveals active β-papillomavirus infection in skin lesions from kidney transplant recipients. Mod. Pathol. 27, 1101–1115 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Straub E., Fertey J., Dreer M., Iftner T., Stubenrauch F., Characterization of the human papillomavirus 16 E8 promoter. J. Virol. 89, 7304–7313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powell M. L., et al. , NCoR1 mediates papillomavirus E8;E2C transcriptional repression. J. Virol. 84, 4451–4460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Jong S. J., et al. , The human CIB1-EVER1-EVER2 complex governs keratinocyte-intrinsic immunity to β-papillomaviruses. J. Exp. Med. 215, 2289–2310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delius H., Hofmann B., Primer-directed sequencing of human papillomavirus types. Curr. Top. Microbiol. Immunol. 186, 13–31 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.