Significance
A gene regulatory system is an important tool for the engineering of biosynthetic pathways of organisms. Here, we report the development of an inducible-ON/OFF regulatory system using a malO operator as a key element. We identified and modulated sequence, position, numbers, and spacing distance of malO operators, generating a series of activating or repressive promoters with tunable strength. The stringency and robustness are both guaranteed in this system, a maximal induction factor of 790-fold was achieved, and nine proteins from different organisms were expressed with high yields. This system can be utilized as a gene switch, promoter enhancer, or metabolic valve in synthetic biology applications. This operator-based engineering strategy can be employed for developing similar regulatory systems in different microorganisms.
Keywords: inducible expression elements, operator, maltose, synthetic biology, Bacillus subtilis
Abstract
Genetic elements are key components of metabolic engineering and synthetic biological applications, allowing the development of organisms as biosensors and for manufacturing valuable chemicals and protein products. In contrast to the gram-negative model bacterium Escherichia coli, the gram-positive model bacterium Bacillus subtilis lacks such elements with precise and flexible characteristics, which is a great barrier to employing B. subtilis for laboratory studies and industrial applications. Here, we report the development of a malO-based genetic toolbox that is derived from the operator box in the malA promoter, enabling gene regulation via compatible “ON” and “OFF” switches. This engineered toolbox combines promoter-based mutagenesis and host-specific metabolic engineering of transactivation components upon maltose induction to achieve stringent, robust, and homogeneous gene regulation in B. subtilis. We further demonstrate the synthetic biological applications of the toolbox by utilizing these genetic elements as a gene switch, a promoter enhancer, and an ON-OFF dual-control device in biosynthetic pathway optimization. Collectively, this regulatory system provides a comprehensive genetic toolbox for controlling the expression of genes in biosynthetic pathways and regulatory networks to optimize the production of valuable chemicals and proteins in B. subtilis.
Rational reconstruction of metabolic pathways and regulatory systems is one of the important objectives of synthetic biology and metabolic engineering. There have been good examples of successful reprogramming of cellular functions by utilizing different biosensors, genetic circuits, and engineered pathways (1). By employing different genetic elements and genome-editing tools, many new applications, such as cancerous cell detection, memory storage and encryption, biobased production of valuable chemicals, and detection and remediation of toxic chemicals, have been developed to benefit health, the manufacturing industry, and our environment (2, 3). These tools often rely on synthetic gene circuits to perform tunable oscillation of target genes (4), Boolean logic gates to orthogonally regulate input and output signals (5, 6) and genetic elements to adjust the expression levels of many different genes precisely and simultaneously (7–10). In contrast to constitutive gene expression systems, the precise regulation of inducible and repressible gene expression systems would facilitate dynamic gene regulation to achieve optimal production of valuable chemicals while circumventing the burden effect that negatively influences the growth of host cells. An ideal control system should allow rapid and precise regulation of a target gene between the “ON” and “OFF” states or even simultaneous switching of different genes to the ON or OFF state (11–13).
Bacillus subtilis is a well-studied gram-positive model bacterium that has been widely used in industrial biotechnology, particularly for the production of heterologous proteins and chemicals. Unlike the gram-negative model bacterium Escherichia coli, only a few genetic regulatory components are available for use in B. subtilis (14–21), and these components lack high stringency, homogeneity, and a wide dynamic range. Over the years, numerous expression systems and strategies have been developed for B. subtilis to enhance the production of homologous and heterologous proteins (22–25). To date, several gene regulation systems have been developed in B. subtilis based on different types of promoters that can be categorized as inducer-specific promoters (17, 20, 26–28), growth-phase promoters (16, 29), and auto-inducible promoters (19, 30, 31). In many cases, inducible expression systems have also been exploited as gene switches or valves to regulate the metabolic fluxes in host cells to enhance the production of desired products (9, 11, 32–34). Nevertheless, the existing inducible gene regulation systems have their own limitations, and most of them cannot meet the demand for the stringent control of gene expression levels and optimization of protein production simultaneously.
The B. subtilis expression system based on the subtilin-regulated promoter PspaS (SURE) (28) and mannose-induced promoter PmanP (XS) (19) have been widely used in both academic studies and industrial applications. While the two promoter systems are excellent for protein production, it is still unclear whether they are equally useful for more sophisticated metabolic engineering. An optimized maltose-inducible expression system was also reported previously (35), but the system exhibited low levels of target protein production and was strongly repressed in the presence of glucose. A Tet system based on the TetO operator and TetR repressor was also developed in several gram-positive bacteria, including B. subtilis (36, 37), streptomycetes (38), and Clostridium acetobutylicum (39). However, these systems were not further utilized and improved due to the low efficiency and inflexibility of their expression elements. Moreover, their efficacy is also limited due to the lack of reversibility of gene expression. Thus, there has been a great demand for developing a gene expression and regulation system with high efficiency and stringency for sophisticated metabolic engineering in B. subtilis.
Promoters from the pathways for utilizing carbon substrates are always good candidates for the construction of inducible expression systems. These promoters can quickly respond to the inducer, and their strength is tunable with inducer concentration. These carbon substrate inducers are inexpensive and readily available and show positive effects on cell growth. For B. subtilis, maltose is the second preferred carbon source and is transported via the phosphoenolpyruvate-dependent phosphotransferase system (PTS). In our previous study (40), we established a maltose-inducible expression system in B. subtilis by shortening the length of the PmalA promoter and deleting the predicted maltose utilization genes yvdK and malL (41). Nevertheless, the potency of this system is still limited due to the lack of sequence information for the operator (denoted as malO) in the PmalA promoter. For better utilization of B. subtilis in metabolic engineering and synthetic biology studies, stringent expression elements with rapid ON and OFF kinetics, such as the Tet-On/Tet-Off system in eukaryotic cells, are needed.
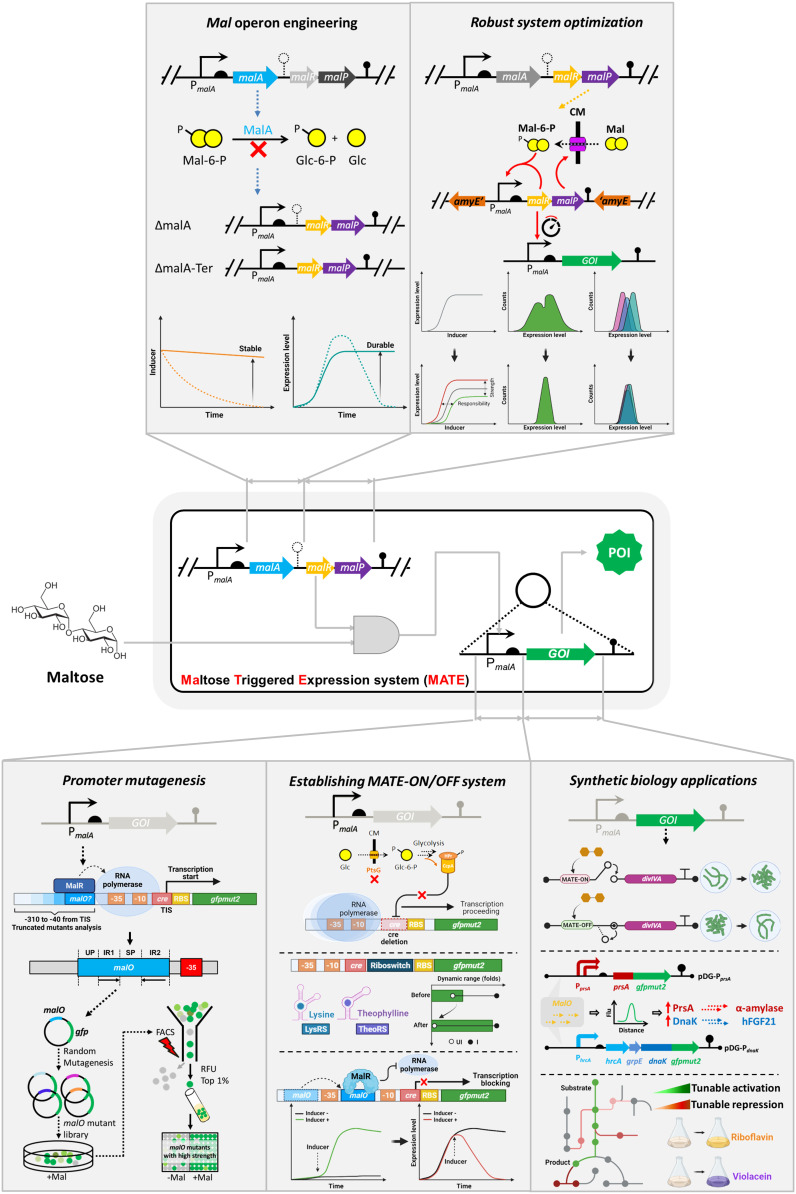
In this paper, we report the development of a malO operator–based genetic toolkit that enables inducible ON and OFF gene regulation with tunable strength in B. subtilis (Fig. 1). To achieve this, the malO operator of the promoter PmalA was identified and analyzed, guiding the subsequent mutagenesis for improved transcriptional activity. The maltose-activated or maltose-repressed genetic element was constructed by replacing or repositioning the wild-type malO operator of the promoter PmalA, generating the maltose-activated MATE-ON system and maltose-repressible MATE-OFF system. Furthermore, the stringency and tolerance of the MATE system were improved, the carbon catabolite repression (CCR) effect was abolished, and the maximal induction fold change was increased to 790-fold without compromising the expression strength. Finally, as a key example of the application potential, the MATE-ON and MATE-OFF systems were employed as a gene switch or metabolic valve to optimize the biosynthesis of riboflavin and violacein in B. subtilis. Taken together, the results indicate that the MATE system developed in this study allows flexible, tunable, homogeneous, and stringent control of gene expression in B. subtilis. This system will serve as a useful tool for a diverse range of metabolic engineering and synthetic biology studies and applications.
Fig. 1.
Schematic representation of strategies in developing the MATE system in B. subtilis. Different expression elements, including the promoter core region, cis-element, translational riboswitch, and chassis cell, were combinatorially optimized to generate the inducible MATE-ON and MATE-OFF systems with robust, stringent, and homogeneous characteristics. The mal operon in B. subtilis, which is responsible for the uptake and hydrolysis of the inducer maltose, was engineered to improve the sensitivity, long-term induction, and homogeneity of our system. The strength of the PmalA promoter was elevated by mutagenesis and high-throughput screening, and the stringency was improved by modifying the cre-element and introducing a translational riboswitch device. Finally, the universality and compatibility of our system were demonstrated by different applications in protein expression and synthetic biology.
Results
Identification of the malO Operator.
Although several transcriptional elements, such as the transcriptional initiation site (TIS), the −35/−10 region and the cre (catabolite repression element)-box of the mal operon, have been identified (42), the malO operator sequence, which is recognized by the transcription factor MalR, has never been identified. Thus, we endeavored to determine the location of the malO operator by gradually truncating the promoter sequence (−310 to −40 from the TIS) upstream of the −35 region of the promoter PmalA (SI Appendix, Fig. S1A). The transcriptional activity of M13 was almost abolished (SI Appendix, Fig. S1B), indicating that the malO operator was likely located between the −112 and −76 positions with respect to the TIS of the promoter. Interestingly, the strength of the fluorescence signal from the M14 and M15 mutants, in which the distance between the potential malO operator and the −35/−10 region of the promoter was shortened, was higher than that from the wild-type promoter. This phenomenon also indicates the potential location of the malO operator because a closer distance might stimulate the activation effect between MalR and RNA, resulting in an increase in reporter gene gfpmut2 expression.
Considering the polymerization of major transcriptional activators and repressors in bacterial cells, the operator sequence is usually found as a trans- or inverted-repeat sequence. To further identify the potential malO sequence between the −112 and −76 positions of PmalA, we searched for the trans- or inverted-repeat sequence in PmalA and then aligned these potential malO sequence regions from B. subtilis, Bacillus licheniformis, Bacillus amyloliquefaciens, Bacillus pumilus, Bacillus clausii, and Staphylococcaceae sp. Finally, a consensus inverted repeat (TTTCCC-N7-GGGAAA) was found in B. subtilis, B. licheniformis, and B. amyloliquefaciens (SI Appendix, Fig. S1C). To further validate whether the conserved sequence found by sequence alignment is the minimal functional sequence of the malO operator, the potential malO sequence was divided into four regions (denoted UP, IR1, Spacer, and IR2), which were analyzed for their roles in promoter activity (SI Appendix, Fig. S2A). The IR1 and IR2 regions of the malO operator were essential for maintaining promoter activity (SI Appendix, Fig. S2B), whereas disrupting the UP region only slightly affected the function of the malO operator. Moreover, the proximal IR2 region (GGGAAA) played a more important role than the distal IR1 region (TTTCCC), which is consistent with the sequence conservation of malO operators in different Bacillus species.
Construction of a Maltose-Activated MATE-ON System.
To develop a PmalA promoter with improved transcriptional activity, a mutagenesis library of the malO operator was constructed and screened with a combination of agar plate and FACS (fluorescence activated cell sorting)-based high-throughput screening method (SI Appendix, Text S1 and Fig. S3, and Fig. 2A). Finally, nine mutants, denoted operator mutants (OM1, OM2, OM6, OM7, and OM11–OM15), were isolated for further experiments (SI Appendix, Fig. S4). To construct a maltose-activated MATE-ON system with higher robustness, GFP expression cassettes harboring different malO mutants were cloned into the high-copy-number plasmid pMA5, testing the expression capacity of the GFP reporter protein. Finally, plasmid pMATE15 with OM15 exhibited the highest level of GFP and was thus selected as the expression vector for further experiments (SI Appendix, Fig. S5). Besides, the maltose utilization pathway was engineered to develop a long-term, continuous, and robust inducible expression system (SI Appendix, Text S2). As expected, the malA-deficient strains showed stronger levels of gene activation than the control strain in the presence of maltose (SI Appendix, Fig. S6A). Notably, the basal expression levels of these malA-deficient strains were also enhanced in the absence of maltose, indicating the occurrence of a leaky expression in these host cells. Similarly, the wild-type strain 1A751 harboring the episomally expressing plasmid also exhibited moderate levels of leaky expression. In comparison to that of the strain harboring the integrative plasmid pDG, the fluorescence induction fold change (maximum/minimum) decreased from 74- to 11-fold in host cells with the high-copy-number expression plasmid. Furthermore, the malA-Ter–deficient strains with malR and malP overexpression exhibited the highest fluorescence intensities and gene expression levels upon induction with maltose. Interestingly, the malA-Ter–deficient strains in which the downstream terminator was removed showed much lower leaky expression than the malA-deficient strains (SI Appendix, Fig. S6A), indicating that a potential transcriptional element might exist in this terminator.
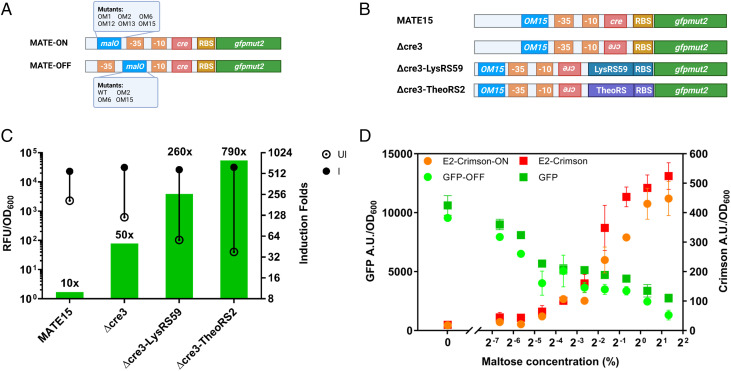
Fig. 2.
Stringency and compatibility evaluation of the MATE system. (A) Schematic illustration of the design of promoters in MATE-ON and MATE-OFF systems. (B) Construction of four different promoter mutants of the MATE-ON system harboring malO-OM15, cre-box mutation, or inserting the ON-type riboswitch LysRS59/TheoRS2. (C) Comparison of the fold induction ratio of the MATE-ON system with different promoter mutants using GFP as a reporter protein. The solid dot and empty dot indicate the fluorescence value that was detected with or without the inducer, respectively. (D) Compatibility of the maltose-activation and repression systems in one cell. Fluorescence reporter genes, E2-crimson and gfpmut2, were controlled by the maltose-activated promoter (MATE-ON) and maltose-repressible promoter (MATE-OFF) simultaneously in one expression vector. Plasmids with only the activated or repressed fluorescence reporter gene were transformed and used as positive controls for each system.
Based on these observations, it was hypothesized that the purported terminator between the malA and malR genes serves as a functional promoter that activates the downstream malR and malP genes in the presence of maltose. To test our hypothesis, this 64-bp terminator-like sequence (denoted Ter) with or without a 300-bp 3′ fragment containing a truncated malA gene was evaluated for promoter activity using pDG. The GFP fluorescence assay (SI Appendix, Fig. S6B) showed comparable promoter activities in these two constructs regardless of maltose induction. This indicates that the functional promoter element is likely located in the 64-bp Ter sequence between the malA and malR genes rather than in the 3′ malA gene. Since the fluorescence signals of this promoter-like element increased upon the induction of maltose, this Ter element is thought to also be activated by the transcription factor MalR. To test this hypothesis, two constructs, namely, pDG-Ter and pDG-malA-Ter, were transformed into a malR-deficient strain of B. subtilis 1A751. Fluorescence assays revealed that these two constructs completely lost their promoter activity in the malR-deficient strain. Furthermore, to identify the functional core region of this Ter promoter, two truncated mutants were constructed, and their promoter activities were determined (SI Appendix, Fig. S6 C and D). The first six nucleotides of the Ter promoter were crucial for maintaining its activity. These six nucleotides are probably part of the −35 region of the Ter promoter based on the consensus sequence of σA promoters. Based on the findings on the stringency and expression capacity in strains with engineered genes of maltose utilization pathway, the 1A751ΔmalA-Ter-MalRP strain was chosen as the host cell for the MATE-ON system.
Optimization of the MATE-ON System in Compatibility, Stringency, Homogeneity, and Reproducibility.
Due to the strong CCR effect by glucose on other sugar metabolism pathways, the activity of our MATE-ON system was also fully disrupted in the presence of glucose. To address that, the cre was fully or partially abolished (SI Appendix, Fig. S7A). Besides, the glucose PTS permease gene ptsG instead of cre-binding protein gene ccpA was deleted due to the characteristics of the B. subtilis cell on glucose and maltose uptake (SI Appendix, Text S3). As a result, the CCR effect was alleviated in our MATE-ON system in the presence of up to 2% (wt/vol) glucose. Interestingly, the disruption of the cre-box in the PmalA promoter also remarkably improved the induction fold change of the MATE-ON system in plasmid pMATE15-Δcre-3 (SI Appendix, Fig. S7 B and C). Based on that, further efforts to improve the stringency of the MATE-ON system were made (SI Appendix, Text S4 and Fig. S8). Finally, the stringency of the system was drastically optimized by employing different ON-type riboswitches, such as lysine-ON riboswitch mutants (LysRS) or synthetic theophylline-activated riboswitch E (TheoRS) (Fig. 2B). As a result, the induction fold change of the optimized MATE-ON system with the theophylline-activated riboswitch reached 790-fold (Fig. 2C) without compromising the robustness of the system (SI Appendix, Fig. S9). Furthermore, the homogeneity, reproducibility, and robustness of our system were also validated by flow cytometry analysis and heterologous protein expression. Results revealed that an ideal homogeneity and reproducibility were observed in our system, and all nine heterologous proteins could be successfully overproduced up to over 60% of the total cellular proteins (SI Appendix, Text S5 and Figs. S10–S12).
Construction of the Maltose-Repressible MATE-OFF System.
The maltose-repressible promoter was developed by repositioning the native malO operator between the −35 and −10 regions of PmalA (Fig. 2A). Thereafter, to achieve an ideal robustness of the repressive system, the −35 and −10 regions of PmalA were mutated to the consensus sequence of the σA promoters to improve the basal activity in the absence of an inducer, generating the prototype of the maltose-repressible MATE-OFF system. Likewise, three maltose-repressive promoter harboring malO operator mutants, OM2, OM6, and OM15, were generated to achieve different repression strengths (SI Appendix, Fig. S13A and Text S2). As shown in the GFP-ssrA fluorescence assay, a remarkable temporal repression of the reporter gene was exhibited in all four constructs of the MATE-OFF system. However, the best repression fold change (about twofold) still needs to be improved (SI Appendix, Fig. S13 B and C). Thus, the repressive regulation of the synthetic PmalA promoter was enhanced by inserting an auxiliary malO operator downstream of the −10 region. In the presence of maltose, this auxiliary operator recruits the transcription factor MalR, resulting in the inhibition of transcriptional initiation via the formation of a DNA loop structure or blocking transcriptional elongation. Two constructs harboring an auxiliary OM6 malO operator downstream of the −10 region at different distances (40 or 50 bp) were generated, which were denoted pMATE-g1-06-1 and pMATE-g1-06-2 (SI Appendix, Fig. S14A). Maximal 6-fold repression of the GFP fluorescence signal was achieved with pMATE-g1-06-1, which is better than those achieved with pMATE-g1-06-2 (2.6-fold) and pMATE-g1 (2-fold). This result suggests that the introduction of an auxiliary operator as a roadblock downstream of the transcription initiation site of the promoter was successful, and a close distance between the auxiliary operator and the TIS guaranteed a better fold repression and a better dynamic range of the GFP expression pattern upon induction (SI Appendix, Fig. S14B). Another attempt to improve the repression fold change of the MATE-OFF system was performed by attenuating the strength of the PmalA promoter. Three mutants, denoted pMATE-g1M1, pMATE-g1M2, and pMATE-g1M3, were generated by site-directed mutagenesis of the −35 region of the maltose-repressible promoter. Among these three mutants, the pMATE-g1M2 construct exhibited the highest repression fold change (26.7-fold). At the cost of the high repression rate, the strength of this MATE-OFF system decreased ∼1.4-fold compared to that of pMATE-g1 (SI Appendix, Fig. S14C). Finally, two versions of the MATE-OFF system, namely, pMATE-g1-06-1 (6-fold repression) with high strength and pMATE-g1M2 (26.7-fold repression) with moderate strength, were constructed for different applications. The second MATE-OFF system is more suitable for stringent repression-mediated gene regulation rather than overexpression of target proteins.
Maltose-Inducible Gene Activation and Repression in a Single Cell.
To corroborate the broad applicability of the MATE systems, we tested whether the MATE-ON and MATE-OFF systems could be compatible and bidirectionally regulated (repressing and activating two different genes simultaneously) in a single cell. To achieve this, plasmid pMATE-AcRg expressing the red fluorescence protein Crimson (driven by the maltose-activated promoter MATE-ON) and green fluorescence protein GFPmut2 (driven by the maltose-repressible promoter MATE-OFF) was constructed. Fluorescence protein genes that were activated or repressed (pMATE-Ac and pMATE-Rg) in two separate plasmids were used as controls. The results indicate that after induction by a gradually increasing concentration of maltose, cells harboring pMATE-AcRg demonstrated increasing green fluorescence and decreasing red fluorescence simultaneously (Fig. 2D). In addition, both the activation and repression systems showed signal strengths comparable to those of the individual expression controls. These results indicated that the MATE-ON and MATE-OFF systems could coexist in a single cell without apparent mutual interference.
Exploitation of the malO Operator as a Promoter Enhancer.
Modification of native promoters to build complex regulatory systems with varying performance is of great interest. To gain insights into the regulatory mechanisms of the cis-elements in promoter activation, we investigated the relationship between operator architecture and promoter activity (SI Appendix, Text S6). We proved that there is an obvious negative correlation between the number of auxiliary operators and the promoter activities (SI Appendix, Fig. S15 A and B), which may be explained by the oligomeric form of the transcriptional activator MalR (SI Appendix, Fig. S16). Moreover, when an auxiliary malO was positioned upstream of the promoter at different distances, an oscillating pattern of promoter activities was observed (SI Appendix, Fig. S15C), and a promoter enhancing phenomenon (1.58- to 2.84-fold) appeared when the auxiliary operator positioned at a specific distance upstreaming of the target promoter (SI Appendix, Fig. S15 C and D). These findings indicate that the malO operator could be exploited as a universal enhancer element for promoters.
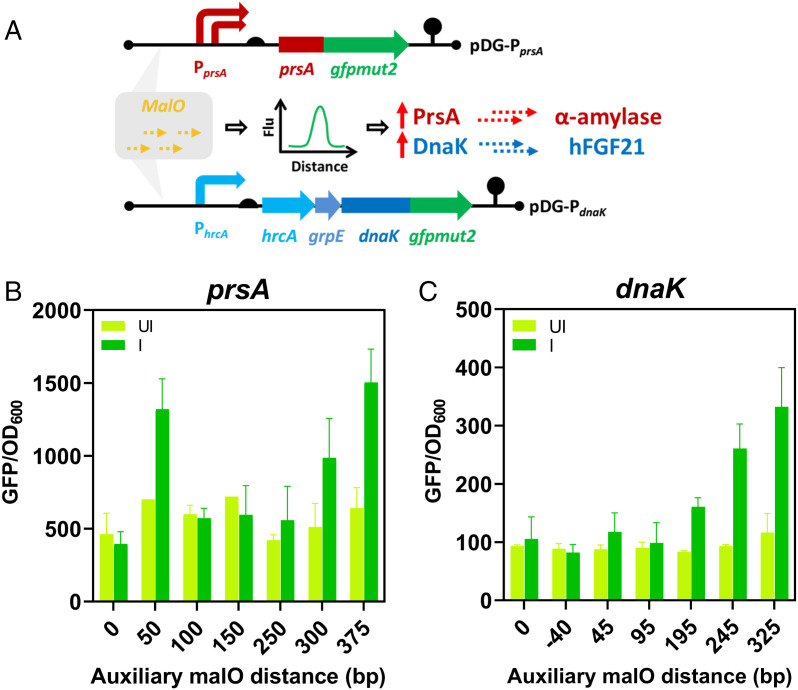
Considering the potential of employing the interaction between malO and MalR as a promoter enhancer, this strategy was further validated in practical applications. Our previous studies have shown that up-regulation of specific chaperones can drastically improve the expression level of target proteins (43, 44). Hence, two proteins whose production was markedly affected by chaperone assistance, alpha-amylase (AmyL) from B. licheniformis and fibroblast growth factor FGF21 from Homo sapiens, were chosen as the target proteins. The two genes, amyL and hFGF21, were constitutively expressed in B. subtilis under the control of the PaprE and PHpaII promoters, respectively. In contrast to employing constitutive or inducible promoters to control the corresponding chaperones PrsA and DnaK, the operator malO was used to enhance the activity of the chaperone’s native promoter. The major benefit of this strategy is that the insertion of malO barely alters the basal activity of the native promoter in the absence of the inducer, avoiding potentially negative side effects on the host cells (45). To achieve this, the activation windows based on the distance between the auxiliary malO and the TATA box of each of the promoters PprsA and PhrcA were determined by measuring the fluorescence of gfpmut2 fused to the C terminus of the protein of interest (Fig. 3A). For the promoter PprsA, the activation window was the malO positioned 50, 300, and 375 nucleotides (nt) upstream of the TATA box, resulting in 1.88-, 1.93-, and 2.35-fold enhancement of promoter activity, respectively, in the presence of maltose (Fig. 3B). For PhrcA, the activation window was the malO positioned 195, 245, and 325 nt upstream of the TATA box, resulting in 1.93-, 2.80-, and 2.86-fold improvement in promoter activity, respectively, upon induction by maltose (Fig. 3C). Moreover, the basal activities of the two synthetic promoters were comparable to those of the respective native forms. After the activation windows were determined, the synthetic promoters with optimized auxiliary malO distances were employed in AmyL- and FGF21-producing strains, controlling the chaperone genes prsA and dnaK in the genome, respectively. Production of amylase and FGF21 was markedly improved due to the up-regulated chaperones (SI Appendix, Fig. S17 A and B). Taken together, operator-mediated activation is a useful strategy to modify the activities of bacterial native promoters with minimal influence on their architecture.
Fig. 3.
Characterization and application of the malO operator in inducible tuning of promoter strength in B. subtilis. (A) Conceptual diagram of the malO operator employed as an inducible enhancer in modifying promoter strength. Alpha-amylase and human FGF21 served as proteins of interest, and their necessary chaperones, PrsA and DnaK, were selected to help fine-tune protein expression levels of the proteins of interest. The effects of different nucleotide distances between the inserted malO operator and the native promoter PprsA (B) and promoter PhrcA (C) were investigated to determine optimal placement of the operator to promote GFP expression with (I) or without (UI) the inducer maltose. Subsequently, the malO operator with the optimal distance was inserted upstream of the promoters PprsA and PhrcA.
Utilizing the MATE System as a Gene Switch.
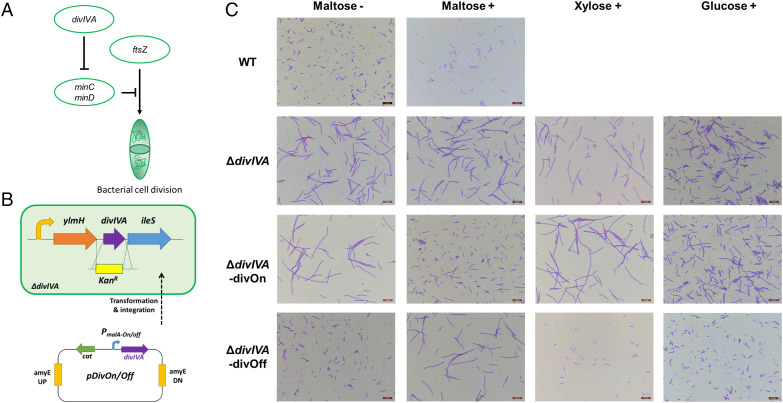
As another potential application of the MATE-ON/OFF system in gene function studies, the expression of physiologically important genes was regulated as an example. The divIVA gene encoding a cell division–initiating protein in B. subtilis was chosen to establish conditional morphology-deficient mutants by using the MATE system. The divIVA gene encodes a coiled-coil protein that has a critical role in the initiation of cell division (Fig. 4A) (46). A distinct filamentary morphology was observed in the divIVA-deficient strain. To explore the effect of inducible regulation of phenotype, a copy of the divIVA gene was placed under the control of the maltose-activated or maltose-repressible promoter and then integrated into the genome of the divIVA-deficient strain at the amyE locus by the pDivOn or pDivOff plasmid (Fig. 4B). The resulting strains, namely, ΔdivIVA-divOn and ΔdivIVA-divOff, were able to express the divIVA gene under different induction conditions, complementing the deficient phenotype and restoring normal morphology. In the absence of the inducer maltose, the extra copy of the divIVA gene was tightly repressed or activated in strains ΔdivIVA-divOn and ΔdivIVA-divOff, leading to the formation of a filamentous or rod-shaped morphology, respectively. Once the inducer maltose was added to ΔdivIVA-divOn or ΔdivIVA-divOff cultures, the expression of divIVA was successfully activated or it repressed divIVA expression, respectively, leading to the switching of the cell appearance from filamentous to rod-shaped or vice versa (Fig. 4C). Besides, the addition of xylose or glucose to the culture medium did not change the morphology of the cells, indicating that the switching of the morphology was not affected by supplementation with a carbon source in the cell culture. Thus, the stringent gene activation or repression characteristics of the MATE system could be utilized as a promising tool for regulating expression of genes including the essential genes.
Fig. 4.
Utilizing the MATE system as a gene switch in controlling cell morphology. (A) The role of DivIVA in regulating cell division in B. subtilis. (B) Cells deficient in the divIVA gene were rescued by integrating a copy of the divIVA gene controlled by the maltose-activated (pDivOn) or -repressible (pDivOff) promoter in different plasmids. (C) MATE-ON and MATE-OFF control of B. subtilis cell morphology. Wild-type (WT), ΔdivIVA, and ΔdivIVA cells harboring a copy of the divIVA gene controlled by the activated or repressible promoter were grown in the presence or absence of the inducer maltose. To exclude the potential effect of maltose as a carbon source on cell growth or morphology, xylose and glucose were supplemented as controls. Cells are shown at 60× magnification. (Scale bar, 10 μm.)
Regulation of Riboflavin Biosynthesis Using the MATE-ON/OFF System.
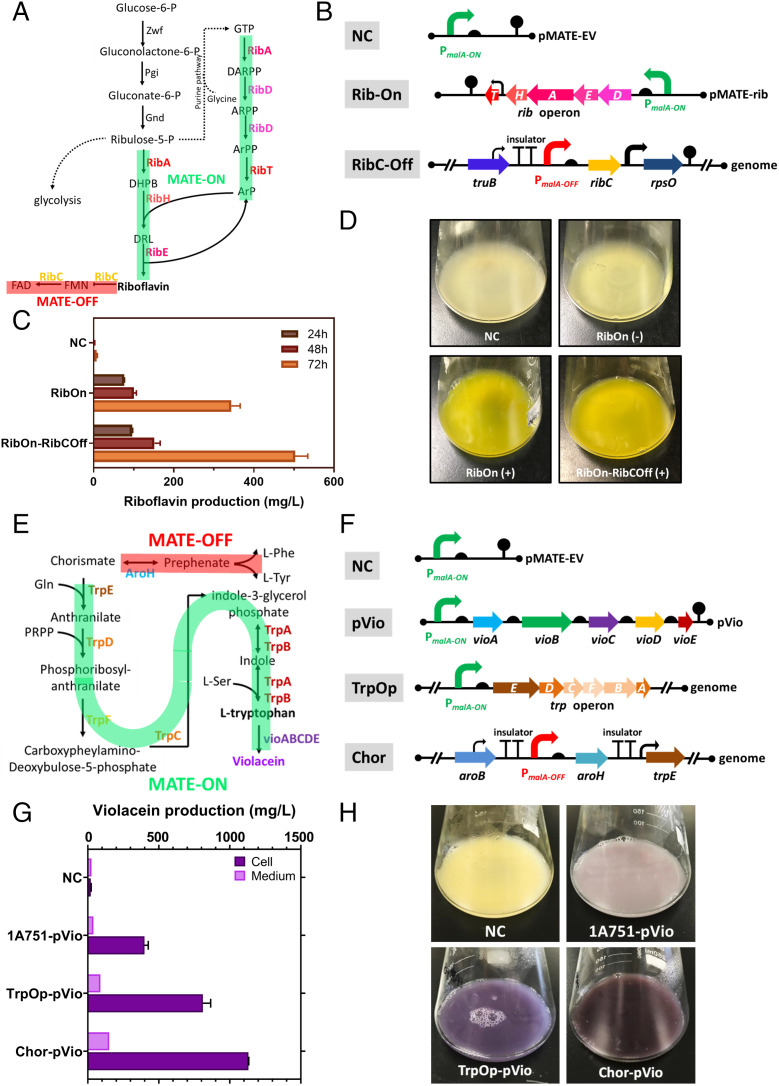
Next, the MATE-ON/OFF system was tested in a metabolic engineering application to see whether it can be used for controlling and maximizing metabolic fluxes toward products of interest in B. subtilis. The biosynthesis of riboflavin (vitamin B2) was chosen as an example. Riboflavin is a crucial micronutrient that is a precursor to the coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) and is required for biochemical reactions in all living cells (47). The biosynthetic pathway of riboflavin was engineered by the MATE-ON/OFF system as follows (Fig. 5A): First, an artificial rib operon comprising five genes, namely, ribD, ribE, ribA, ribH, and ribT, responsible for the biosynthesis of riboflavin from GTP and ribulose-5-P, was constructed in pMATE15 under the control of the CCR-alleviated maltose-activated promoter PmalA-Δcre3, generating the ΔATRP-RibOn strain (Fig. 5B). Second, the native promoter of the ribC gene, which encodes an essential bifunctional flavokinase/FAD synthetase, was replaced by the maltose-repressible promoter PmalA-g1M2 to minimize the conversion of the produced riboflavin to FAD and FMN, generating the ΔATRP-RibOn-RibCOff strain (Fig. 5B). Little to no riboflavin was produced in the ΔATRP-RibOn strain in the absence of maltose, whereas the distinct yellow color representing riboflavin production was observed in the presence of maltose (Fig. 5D). The significant difference in broth color indicated that the biosynthesis of riboflavin was tightly controlled by the MATE-ON system in the ΔATRP-RibOn strain. Finally, the titer of riboflavin in the ΔATRP-RibOn strain was improved from an undetectable level to 343.8 mg/L by simply up-regulating the expression of the rib operon via the MATE-ON system. Furthermore, by combinatorially employing the MATE-OFF system to inducibly down-regulate the expression of the ribC gene in ΔATRP-RibOn-RibCOff, the production of riboflavin was further improved to 503.3 mg/L in 72 h of fermentation (Fig. 5C). This result confirmed that the MATE-ON/OFF system could inducibly regulate the biosynthetic pathway of riboflavin cooperatively, which is a useful characteristic for metabolic engineering applications.
Fig. 5.
Combinatorial optimization of the riboflavin and violacein biosynthesis pathways employing the MATE system in B. subtilis. (A) Schematic of the strategy to optimize the biosynthesis pathway of riboflavin with the MATE-ON and MATE-OFF regulatory system in B. subtilis. (B) Genetic manipulation of the rib operon and ribC gene with elements from the MATE-ON and MATE-OFF system in B. subtilis. (C) Riboflavin production by the engineered and control strains during 72 h of shake flask cultivation. (D) Cultures of riboflavin-producing strains after 72 h. The strain harboring an empty plasmid pMATE15 served as a negative control (NC). The plus and minus characters in parentheses indicate cultivation with and without 1% (wt/vol) maltose as inducer, respectively. (E) Schematic of the strategy to optimize the biosynthesis pathway of L-tryptophan with the MATE-ON and MATE-OFF regulatory system in B. subtilis. (F) Design of the artificial violacein biosynthesis operon (pVio), inducible activated trp operon (TrpOp), and inducible repressive aroH gene (Chor) using elements from the MATE system in B. subtilis. (G) Violacein production by three engineered strains, pVio, TrpOp-pVio, and Chor-pVio, after 48 h of shake flask cultivation. (H) Cultures of violacein-producing strains after 48 h. The strain harboring an empty plasmid pMATE15 served as a negative control.
Fine-Tuning of the Violacein Biosynthetic Pathway Using the MATE System.
After we successfully applied the MATE-ON/OFF system for controlling the native biosynthetic pathway flux, a heterologous biosynthetic pathway that produces violacein was chosen as another example. Since violacein displays a strong inhibitory effect on gram-positive bacteria by penetrating the cytoplasmic membrane of cells (48), successful biosynthesis of violacein in B. subtilis is a good challenge to validate the stringency and robustness of the MATE system; the synthetic pathway of toxic violacein must be controlled precisely and stringently. Another challenge is that the violacein production pathway is encoded by an ∼7.4-kb operon that encodes five heterologous enzymes from Chromobacterium violaceum, including the large (111.3 kDa) rate-limiting enzyme VioB (49).
To construct and optimize the violacein biosynthesis pathway in B. subtilis, an artificial vioABCDE operon was assembled and controlled by the genetic elements from the MATE-ON system (SI Appendix, Text S7). The purple color of the colonies on the agar plate indicated the successful biosynthesis of violacein in B. subtilis in the presence of the inducer maltose (SI Appendix, Fig. S18). Besides, the growth inhibition curve of the violacein-producing strain 1A751-pVio demonstrated that the vioABCDE operon was tightly controlled by the maltose-activated promoter from the MATE-ON system (SI Appendix, Fig. S19). To further improve the production of violacein in B. subtilis, the L-tryptophan pool in the host cell, which is the direct substrate of violacein biosynthesis, needs to be increased together with the vioABCDE artificial operon under the induction of maltose. To achieve that, the L-tryptophan pathway and the shunt pathway for L-phenylalanine and L-tyrosine biosynthesis were regulated by the MATE-ON and MATE-OFF system, respectively (Fig. 5 E and F), generating strains TrpOp-pVio and Chor-pVio (SI Appendix, Text S7). As a result, the concentrations of L-tryptophan in these two engineered strains were improved by 24.2% and 73.7%, respectively, compared with the parent pVio strain (SI Appendix, Fig. S21). The titers of violacein were also improved by 202.6% and 303.1%, respectively, compared with that obtained with the pVio strain (Fig. 5 G and H). Moreover, we also proved that the biosynthetic pathway could be fine-tuned by genetic elements with different strengths from the MATE-ON/OFF system (SI Appendix, Text S7). In our example, five genetic elements from the MATE-ON or MATE-OFF system with different activation or repressive strengths were employed in regulation of the shikimate pathway. As expected, violacein production gradually increased from 139.7 to 341.3 mg/L after 48 h of fermentation when the trp operon was controlled by MATE-ON promoters with increasing activation strength (SI Appendix, Fig. S22A). Of the strains in which the aroH gene was controlled by the MATE-OFF elements with low repression strengths (q12, b6, and g1), lower levels of violacein production were observed compared with those obtained with the elements with strong repression strengths (SI Appendix, Fig. S22B). Taken together, genetic elements from the MATE-ON/OFF system with varying activation and repression strengths can be configured as adjustable valves to precisely modify metabolic fluxes.
Discussion
The availability of well-characterized genetic elements that are able to precisely regulate target genes or pathways is essential for numerous applications in synthetic biology and metabolic engineering. By combinatorial assembly of these elements as a multifunctional toolkit, it is feasible to modify cellular behaviors as desired and exploit the bacteria as workhorses for the production of chemicals and proteins of interest. In contrast to the gram-negative model bacterium E. coli, such elements are lacking in the gram-positive model bacterium B. subtilis, an important bacterium for basic research and industrial applications. Although several genetic elements have recently been developed for B. subtilis, there remain issues associated with these regulatory devices regarding the insufficiency of dual switchable controls, the contradiction between the stringency and robustness of the system, and the practicability of the system in large-scale industrial production. These issues drastically slow down the application of B. subtilis as a universal chassis strain for synthetic biology and metabolic engineering applications.
In this study, we explored the feasibility of exploiting the malO operator, which binds the transcription factor MalR and activates the PmalA promoter, as an inducible ON/OFF genetic toolbox to either activate or repress the expression of target genes in B. subtilis. To achieve this, as the foundation of the MATE-ON/OFF system, the malO operator sequence in B. subtilis was first identified by truncation analysis and homologous sequence alignment in different Bacillus species. By screening a library of malO operator mutants through GFP-based cell sorting, promoter mutants with drastically improved activity were obtained. Previous studies reported that the uptake rate of inducer molecules (50, 51) and the titration of transcription factors (52) are key factors affecting the sensitivity, dynamic range, and homogeneity of inducible expression systems. Thus, the parent strain was engineered to meet the demand for a robust expression system with ideal homogeneity and repeatability. Finally, the maltose-inducible MATE-ON/OFF system was established by replacing or repositioning the native malO operator in the promoter PmalA with selected mutants. Transcription factor MalR was recruiting as activator or competitor for the RNA polymerase, resulting in the transcription initiation or disruption of the target gene in the presence of maltose.
For an inducible regulatory system utilizing sugar or sugar alcohol as inducer, the CCR effect is one of the most barrier that limits the wide application of these systems in industrial fields. Several attempts were made to abolish the CCR effect by eliminating CcpA. However, as a global regulator in carbon catabolism, CcpA occasionally functions as a positive/negative effector of other genes as well (53). Moreover, Moreno et al. reported that the activity of PmalA could not be fully recovered in a ccpA-deficient B. subtilis strain in the presence of maltose and glucose, and the strength of the promoter was lower than that in the wild-type strain in the presence of maltose (54). Thus, another strategy to alleviate the CCR effect by partially or fully removing the cre-box within the PmalA promoter was explored. A previous study reported that the PtsGHI transport system contributes to ∼60% of the glucose transport in B. subtilis (55). In a PtsGHI-deficient strain, glucose was still transported by GlcP and GlcU transporters. Thus, in combination with the effort on the cre-box, the ptsG gene was eliminated to decrease the rate of glucose uptake and slightly improve the efficiency of maltose transport (41). As a result, the CCR effect was fully alleviated even in the presence of 20 g/L glucose.
Unsurprisingly, due to the increasing copy number of cassettes (from a single copy to ∼40 to 50 copies per cell), notable leaky expression was observed in the MATE-ON system. Interestingly, the promoter mutant with the modified cre-box showed an obvious improvement in stringency, and the induction fold change was improved from 2-fold to 45-fold. The GFP assay revealed that the mutation in the cre-box not only improved the system stringency but also slightly affected the activity of the promoter. A similar phenomenon was also observed in a previous study where the mutation in the cre-box could enhance the activity of the promoter by mimicking the UP element and stimulating the interaction between the promoter and RNAP (56). However, in our case, the cre-box was positioned downstream of the −10 region, and the complementary mutation did not change the GC content in this region. One reasonable explanation that needs verification is that an encrypted cis-element was placed in a position that overlapped with the cre-box. Either deletion or complementary mutagenesis would decrease the leaky expression level of the promoter.
Meanwhile, to increase the dynamic range of the MATE-ON system, a transcription–translation dual controlling system was established by intercalating ON-type riboswitches (lysine-ON RS or theophylline-ON RS) downstream of the PmalA promoter. As a result, the induction fold change of the MATE-ON system was improved to 260-fold and 790-fold, respectively. Remarkably, the efforts made in increasing the stringency of our system do not compromise the robustness of the system, which could meet the demands for high stringency and robustness of gene regulation in synthetic biology. For the MATE-OFF system, although the strength of genetic elements was strong enough in the absence of an inducer, the fold change of this repressive system was not ideal. To solve this problem, an auxiliary malO operator was inserted downstream of the −10 region to further inhibit transcription elongation. Finally, via combination with a strategy for attenuating the strength of PmalA, the repression fold change could be improved by up to 26.7-fold and showed ideal dose-dependent repression with the maltose inducer. These results are consistent with the findings from using other tandem operators that reportedly form a complex DNA loop structure by binding the homomultimerized transcription factor at different angles (57), resulting in improved steric hindrance to block transcription initiation and elongation.
To expand the activation effect of the malO operator in other promoters in B. subtilis, characteristics of positioning duplicated operators with different copies or varied distances upstream of the target promoter were explored. By inserting multiple operators ahead of the native PmalA promoter, we expected to observe increasing concentrations of the transcription activator MalR, which is recruited by tandem operators, which would further improve the activity of these synthetic promoters. However, synthetic promoters with more auxiliary operators exhibited lower activity in the presence of maltose, indicating that a potential complex DNA structure might have been formed in the promoter architecture. Repressor tetramers, such as LacI, might bind with auxiliary operators through a DNA loop structure (57–59). Thus, the greater the number of auxiliary operators positioned upstream of the native operator, the greater the possibility of DNA looping to occur in the promoter region, impairing its activity upon induction. Alternatively, this mechanism can be employed as a strategy to rapidly generate a series of promoters with different activities, since the strengths of PmalA and its derived mutants were powerful enough. Furthermore, a distance-dependent activation effect of malO placed upstream of the TATA box in different promoters was determined and exploited in B. subtilis. Similar to the phenomenon found in transcription repressors in E. coli (59), an oscillating pattern of promoter activity was observed in promoter mutants with different distances between auxiliary malO operators. However, no similar mechanism has been reported for transcription activators in bacteria, which motivated us to investigate auxiliary operators with greater distances. We could identify an activation window in a mutant with an auxiliary operator positioned far enough from the core region of the promoter. We then verified its activation potential in other promoters, such as Pr from ribD, PhrcA, and PprsA, and presented a universal strategy of activation in the overexpression of fluorescence protein or chaperones among these promoters in B. subtilis. By utilizing this feature, improvement in the expression levels of multiple genes could be achieved by using just one inducer. Furthermore, consistent basal activity among these promoters in the absence of an inducer is another advantage when optimizing gene regulatory networks in synthetic biology and metabolic engineering studies.
In summary, the MATE-ON/OFF system has several advantages compared to other inducible gene expression systems in B. subtilis. First, the system is very simple and efficient, as it requires only modified promoters with different malO sequences to achieve inducible or repressible regulation of target genes. Second, the CCR effect is fully abolished without compromising promoter strength. Third, both the maltose-inducible and maltose-repressible systems can function simultaneously in a single cell with negligible interference. Fourth, the robust system allows production of target proteins accounting for over 60% of the total cellular proteins and also has a wide dynamic range (790-fold). To the best of our knowledge, this is a demonstration that modification of a single operator, instead of the transcription factor, can positively and negatively regulate target genes within a single cell in response to a single inducer in B. subtilis. We further demonstrated that this system can be a useful toolbox for synthetic biology and metabolic engineering applications. As demonstrated in the examples of producing riboflavin and violacein, the operon and pathways branching from the major biosynthetic pathway could be tightly controlled by the stringent MATE-ON/OFF systems simultaneously upon maltose-mediated induction, resulting in the strong promotion of riboflavin and violacein production in B. subtilis. Therefore, the stringent ON and OFF switch of this system will be a valuable tool for applications that require precise control of gene expression. This system can also be utilized as a gene switch to establish a more complex gene circuit in synthetic biology applications. Moreover, the malO-based ON/OFF approach shown here represents a starting point for systematic exploration of the repository of functional prokaryotic promoters. It is expected that this scalable approach will pave a new way forward to generate more flexible and orthogonal operator-based genetic devices and parts. The availability of such toolboxes will help expedite our attempts to solve complex biological problems and expand the applications of B. subtilis.
Materials and Methods
All of the materials and methods used in this study are detailed in SI Appendix, Materials and Methods, including strains and growth conditions, construction of plasmids, construction of an operator mutant library, high-throughput screening, genome manipulation, homogeneity and reproducibility analysis, standard curve (SI Appendix, Fig. S20), PAGE analysis, and analytical methods.
Supplementary Material
Acknowledgments
S.Y.L. is the chairman of the scientific advisory board of the Tianjin Institute of Industrial Biotechnology. This work was supported by the National Key R&D Program of China (Grant 2020YFA0907800), the National Natural Science Foundation of China (Grant 22178372), and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (grants TSBICIP-KJGG-011 and TSBICIP-CXRC-055).
Footnotes
Reviewers: T.L., Massachusetts Institute of Technology; and T.S., Universitat Greifswald Institut fur Pharmazie.
The authors declare no competing interest.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Niu T., et al. , Engineering a glucosamine-6-phosphate responsive glmS ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of N-acetylglucosamine. ACS Synth. Biol. 7, 2423–2435 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Kotula J. W., et al. , Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U.S.A. 111, 4838–4843 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siuti P., Yazbek J., Lu T. K., Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 31, 448–452 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Stricker J., et al. , A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet J., Yin P., Ortiz M. E., Subsoontorn P., Endy D., Amplifying genetic logic gates. Science 340, 599–603 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Brophy J. A., Voigt C. A., Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morra R., et al. , Dual transcriptional-translational cascade permits cellular level tuneable expression control. Nucleic Acids Res. 44, e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y. J., Kim S. J., Moon T. S., Multilevel regulation of bacterial gene expression with the combined STAR and antisense RNA system. ACS Synth. Biol. 7, 853–865 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Nakashima N., Akita H., Hoshino T., Establishment of a novel gene expression method, BICES (biomass-inducible chromosome-based expression system), and its application to the production of 2,3-butanediol and acetoin. Metab. Eng. 25, 204–214 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Mimee M., Tucker A. C., Voigt C. A., Lu T. K., Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L. B., Zeng A. P., Engineering a lysine-ON Riboswitch for metabolic control of lysine production in Corynebacterium glutamicum. ACS Synth. Biol. 4, 1335–1340 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Gossen M., Bujard H., Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urlinger S., et al. , Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U.S.A. 97, 7963–7968 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan T. T., Schumann W., Development of a glycine-inducible expression system for Bacillus subtilis. J. Biotechnol. 128, 486–499 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Thuy Le A. T., Schumann W., A novel cold-inducible expression system for Bacillus subtilis. Protein Expr. Purif. 53, 264–269 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Lee S. J., Pan J. G., Park S. H., Choi S. K., Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 149, 16–20 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Sun T., Altenbuchner J., Characterization of a mannose utilization system in Bacillus subtilis. J. Bacteriol. 192, 2128–2139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heravi K. M., Wenzel M., Altenbuchner J., Regulation of mtl operon promoter of Bacillus subtilis: Requirements of its use in expression vectors. Microb. Cell Fact. 10, 83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenzel M., Müller A., Siemann-Herzberg M., Altenbuchner J., Self-inducible Bacillus subtilis expression system for reliable and inexpensive protein production by high-cell-density fermentation. Appl. Environ. Microbiol. 77, 6419–6425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toymentseva A. A., Schrecke K., Sharipova M. R., Mascher T., The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb. Cell Fact. 11, 143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dormeyer M., et al. , A novel engineering tool in the Bacillus subtilis toolbox: Inducer-free activation of gene expression by selection-driven promoter decryptification. Microbiology (Reading) 161, 354–361 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Mohamed M. Y. H., Christie G., A system for the expression and release of heterologous proteins from the core of Bacillus subtilis spores. FEMS Microbiol. Lett. 365, FNY270 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zhang K., Su L., Duan X., Liu L., Wu J., High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Fact. 16, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S., Kang Z., Cao W., Du G., Chen J., Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J. Biotechnol. 219, 40–47 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Cui W., et al. , Engineering an inducible gene expression system for Bacillus subtilis from a strong constitutive promoter and a theophylline-activated synthetic riboswitch. Microb. Cell Fact. 15, 199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yansura D. G., Henner D. J., Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 81, 439–443 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P. T., Shaw J. F., Chao Y. P., David Ho T. H., Yu S. M., Construction of chromosomally located T7 expression system for production of heterologous secreted proteins in Bacillus subtilis. J. Agric. Food Chem. 58, 5392–5399 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Bongers R. S., Veening J. W., Van Wieringen M., Kuipers O. P., Kleerebezem M., Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: Strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71, 8818–8824 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panahi R., Vasheghani-Farahani E., Shojaosadati S. A., Bambai B., [Auto-inducible expression system based on the SigB-dependent ohrB promoter in Bacillus subtilis.]. Mol. Biol. (Mosk.) 48, 970–976 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Guan C., et al. , Construction and development of an auto-regulatory gene expression system in Bacillus subtilis. Microb. Cell Fact. 14, 150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng J., et al. , Enhancement of a high efficient autoinducible expression system in Bacillus subtilis by promoter engineering. Protein Expr. Purif. 127, 81–87 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Liu S. D., Wu Y. N., Wang T. M., Zhang C., Xing X. H., Maltose utilization as a novel selection strategy for continuous evolution of microbes with enhanced metabolite production. ACS Synth. Biol. 6, 2326–2338 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Rajkumar A. S., et al. , Engineering of synthetic, stress-responsive yeast promoters. Nucleic Acids Res. 44, e136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., et al. , Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synth. Biol. 7, 1785–1797 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Ming Y. M., Wei Z. W., Lin C. Y., Sheng G. Y., Development of a Bacillus subtilis expression system using the improved Pglv promoter. Microb. Cell Fact. 9, 55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamionka A., Bertram R., Hillen W., Tetracycline-dependent conditional gene knockout in Bacillus subtilis. Appl. Environ. Microbiol. 71, 728–733 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda T., Yamazaki H., Yamashita K., Shinke R., The tetracycline inducible expression of α-amylase in Bacillus subtilis. J. Ferment. Bioeng. 74, 58–60 (1992). [Google Scholar]
- 38.Wang W., et al. , Development of a synthetic oxytetracycline-inducible expression system for streptomycetes using de novo characterized genetic parts. ACS Synth. Biol. 5, 765–773 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Dong H., Tao W., Zhang Y., Li Y., Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: A useful tool for strain engineering. Metab. Eng. 14, 59–67 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Yue J., Fu G., Zhang D., Wen J., A new maltose-inducible high-performance heterologous expression system in Bacillus subtilis. Biotechnol. Lett. 39, 1237–1244 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Schönert S., et al. , Maltose and maltodextrin utilization by Bacillus subtilis. J. Bacteriol. 188, 3911–3922 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto H., Serizawa M., Thompson J., Sekiguchi J., Regulation of the glv operon in Bacillus subtilis: YfiA (GlvR) is a positive regulator of the operon that is repressed through CcpA and cre. J. Bacteriol. 183, 5110–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J., et al. , Enhanced extracellular production of α-amylase in Bacillus subtilis by optimization of regulatory elements and over-expression of PrsA lipoprotein. Biotechnol. Lett. 37, 899–906 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Li D., Fu G., Tu R., Jin Z., Zhang D., High-efficiency expression and secretion of human FGF21 in Bacillus subtilis by intercalation of a mini-cistron cassette and combinatorial optimization of cell regulatory components. Microb. Cell Fact. 18, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martínez-Alonso M., García-Fruitós E., Ferrer-Miralles N., Rinas U., Villaverde A., Side effects of chaperone gene co-expression in recombinant protein production. Microb. Cell Fact. 9, 64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards D. H., Errington J., The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24, 905–915 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Averianova L. A., Balabanova L. A., Son O. M., Podvolotskaya A. B., Tekutyeva L. A., Production of vitamin B2 (Riboflavin) by microorganisms: An overview. Front. Bioeng. Biotechnol. 8, 570828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cauz A. C. G., et al. , Violacein targets the cytoplasmic membrane of bacteria. ACS Infect. Dis. 5, 539–549 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Antônio R. V., Creczynski-Pasa T. B., Genetic analysis of violacein biosynthesis by Chromobacterium violaceum. Genet. Mol. Res. 3, 85–91 (2004). [PubMed] [Google Scholar]
- 50.Hjelm A., et al. , Tailoring Escherichia coli for the l-rhamnose PBAD promoter-based production of membrane and secretory proteins. ACS Synth. Biol. 6, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Kim S. K., Lee D. H., Kim O. C., Kim J. F., Yoon S. H., Tunable control of an Escherichia coli expression system for the overproduction of membrane proteins by titrated expression of a mutant lac repressor. ACS Synth. Biol. 6, 1766–1773 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Wang B., Barahona M., Buck M., Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities. Nucleic Acids Res. 43, 1955–1964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jahreis K., Pimentel-Schmitt E. F., Brückner R., Titgemeyer F., Ins and outs of glucose transport systems in eubacteria. FEMS Microbiol. Rev. 32, 891–907 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Moreno M. S., Schneider B. L., Maile R. R., Weyler W., Saier M. H. Jr., Catabolite repression mediated by the CcpA protein in Bacillus subtilis: Novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39, 1366–1381 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Paulsen I. T., Chauvaux S., Choi P., Saier M. H. Jr., Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: Identification of a novel hexose:H+ symporter. J. Bacteriol. 180, 498–504 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kant S., Kapoor R., Banerjee N., Identification of a catabolite-responsive element necessary for regulation of the cry4A gene of Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 191, 4687–4692 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monteiro L. M. O., Arruda L. M., Silva-Rocha R., Emergent properties in complex synthetic bacterial promoters. ACS Synth. Biol. 7, 602–612 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Becker N. A., Schwab T. L., Clark K. J., Maher L. J. III, Bacterial gene control by DNA looping using engineered dimeric transcription activator like effector (TALE) proteins. Nucleic Acids Res. 46, 2690–2696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker N. A., Peters J. P., Maher L. J. III, Lionberger T. A., Mechanism of promoter repression by Lac repressor-DNA loops. Nucleic Acids Res. 41, 156–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.