Significance
Although plastid division is critical for plant development, how components of the plastid division machinery (PDM) are imported into plastids remains unexplored. A forward genetic screen to identify suppressors of a crumpled leaf (crl) mutant deficient in plastid division led us to find dominant gain-of-function (GF) mutations in TIC236, which significantly increases the import of PDM components and completely rescues crl phenotypes. The defective plastid division phenotypes in crl and tic236-knockdown mutants and CRL-TIC236 association in a functional complex indicate that the CRL-TIC236 module is vital for plastid division. Hence, we report the first GF translocon mutants and unveil CRL as a novel functional partner of TIC236 for PDM import.
Keywords: chloroplast, protein import defect, TIC236, cell death, plastid division
Abstract
TIC236 is an essential component of the translocon for protein import into chloroplasts, as evidenced by the embryonic lethality of the knockout mutant. Here, we unveil a TIC236-allied component, the chloroplast outer membrane protein CRUMPLED LEAF (CRL), absence of which impairs plastid division and induces autoimmune responses in Arabidopsis thaliana. A forward genetic screen exploring CRL function found multiple dominant TIC236 gain-of-function (tic236-gf) mutations that abolished crl-induced phenotypes. Moreover, CRL associates with TIC236, and a tic236-knockdown mutant exhibited multiple lesions similar to the crl mutant, supporting their shared functionality. Consistent with the defective plastid division phenotype of crl, CRL interacts with the transit peptides of proteins essential in plastid division, with tic236-gf mutations reinforcing their import via increased TIC236 stability. Ensuing reverse genetic analyses further revealed genetic interaction between CRL and SP1, a RING-type ubiquitin E3 ligase, as well as with the plastid protease FTSH11, which function in TOC and TIC protein turnover, respectively. Loss of either SP1 or FTSH11 rescued crl mutant phenotypes to varying degrees due to increased translocon levels. Collectively, our data shed light on the links between plastid protein import, plastid division, and plant stress responses.
Chloroplasts evolved from a gram-negative cyanobacterial endosymbiont, with most cyanobacterial genes having been transferred to the host plant genome. Therefore, thousands of nuclear-encoded chloroplast proteins are posttranslationally imported into chloroplasts, orchestrated by outer and inner envelope membrane (OEM and IEM) translocons, respectively termed TOC and TIC. Although an array of translocon proteins has been identified (1, 2), it has been unclear how TOC and TIC are linked across the two envelope membranes separated by an intermembrane space. The recent discovery of the TIC236 protein solved this long-standing question (3). TIC236 is an integral IEM protein associated with TIC components. Its C-terminal domain, located in the intermembrane space, directly interacts with the N-terminal polypeptide transport-associated (POTRA) domains of TOC75-III (hereafter TOC75), the channel protein in the TOC complex. TOC75 and TIC236 are homologs of bacterial TamA (TRANSLOCON ASSEMBLY MODULE A) and TamB, respectively, which together function in bacterial outer membrane protein biogenesis and protein export (3–5).
Plastid division occurs in developing cells to ensure an optimal number of plastids is in place before cell division, requiring the import of a suite of plastid-division machinery (PDM) proteins. The loss of any vital PDM components results in gigantic plastids and a drastically reduced plastid number per cell (6). Surprisingly, several Arabidopsis mutants deficient in plastid division—including crumpled leaf (crl), accumulation and replication of chloroplasts6 (arc6), plastid division2 (pdv2), and filamenting temperature-sensitive z1 (ftsz1)—develop foliar cell death (7), resembling lesion-mimicking mutants (LMM) that exhibit a light-dependent hypersensitive response–like cell death (8, 9). Like LMM, crl and other plastid division mutants constantly up-regulate immune-related genes (7, 10). The gigantic chloroplasts of crl mutants also induce an abnormal cell cycle, with increased endoreduplication activity leading to stunted growth (11). Previous studies have indicated that autoimmune responses, abnormal cell cycle, and growth inhibition are likely mediated by a process called retrograde signaling [i.e., signaling from the gigantic chloroplasts back to the nucleus (7, 10, 11)].
CRL is a nuclear-encoded chloroplast OEM protein. Its short N-terminal region resides in the intermembrane space, followed by a transmembrane domain and a cytosolic chromophore lyase CpcT/CpeT domain characterized from a cyanobacterial CpcT bilin lyase (12). Although the lyase domain retains phycocyanobilin-binding aptitude, there is no apparent correlation between phycocyanobilin-binding ability and crl-induced lesions in Arabidopsis (13), indicating that CRL has gained a divergent function.
Dominant TIC236 Gain-of-Function Mutations Abolish crl-Induced Lesions
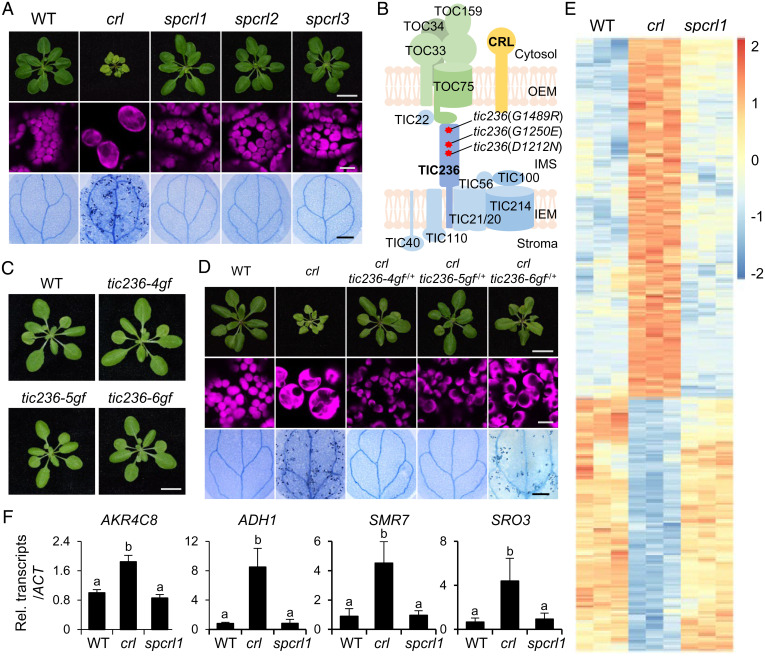
To explore the function of CRL, we performed an ethylmethanesulfonate (EMS) mutagenesis screen to find suppressors of crl (spcrl). The mutagenized Arabidopsis M2 seeds were germinated on soil, and plants showing a wild-type (WT)-like phenotype were selected for further analyses. Among ∼24,000 M2 plants, we found two robust spcrl mutants, namely spcrl1 and spcrl2, whose visible phenotype is nearly indistinguishable from that of WT plants, as well as one (spcrl3) that displayed weaker suppressing efficacy, especially in terms of restoring plastid division (Fig. 1A). Whole-genome sequencing of genomic DNA isolated from each of these spcrl mutants identified putative causal missense mutations in TIC236, specifically TIC236D1212N in spcrl1, TIC236G1250E in spcrl2, and TIC236G1489R in spcrl3 (Fig. 1B). Despite the low protein sequence identity (∼29%) between Arabidopsis TIC236 and Escherichia coli TamB, all three mutated residues are conserved in TamB (SI Appendix, Fig. S1) and across various plant species (SI Appendix, Fig. S2A), highlighting their importance. The phenotypes of tic236(D1212N), tic236(G1250E), and tic236(G1489R) single mutants proved similar to WT plants (Fig. 1C). To distinguish these gain-of-function (tic236-gf) mutants from tic236 knockdown mutants [i.e., tic236-2 (SAIL104-F07, Columbia ecotype) and tic236-3 (RIKEN PST00216, Nossen ecotype) (3)], we renamed tic236(D1212N) as tic236-4gf, tic236(G1250E) as tic236-5gf, and tic236(D1489R) as tic236-6gf, respectively (Fig. 1C). Next, we crossed each tic236-gf mutant with crl to generate all possible genotypes in F2 siblings. PCR-based genotyping of the gf mutants confirmed a dominant effect of the tic236-4gf and tic236-5gf mutations and a less dominant effect of tic236-6gf mutation in rescuing the crl phenotypes (Fig. 1D and SI Appendix, Fig. S2B). Since crl causes constitutive expression of stress-related genes (7, 10, 11), next we conducted global gene expression profiling of spcrl1 relative to the crl mutant. As anticipated based on previous RNA-sequencing (RNA-seq) data (10, 11), stress-related nuclear genes, including cell cycle inhibitor SIAMESE-RELATED 7 (SMR7), showed higher expression in the crl mutant (Fig. 1 E and F and Dataset S1). A total of 393 and 276 genes were found to be up- and down-regulated, respectively, in crl at least twofold (false discovery rate [FDR] < 0.05) compared with WT seedlings. In sharp contrast, their expression was significantly restored in spcrl1 relative to WT seedlings, confirming that the tic236-4gf mutation is epistatic to crl (Fig. 1 E and F and Dataset S1 and S2).
Fig. 1.
Identification of TIC236 gain-of-function mutations in spcrl mutants. (A) The micro and macroscopic phenotypes of the crl suppressors spcrl1, spcrl2, and spcrl3. (Top) Representative images of 21-d-old plants grown on soil. (Scale bar, 1 cm.) (Middle) Confocal images representing chlorophyll autofluorescence (magenta) of mesophyll cells. (Bottom) Cell death in 10-d-old cotyledons, as visualized by TB staining. (Scale bar, 0.5 mm.) (B) Whole-genome sequencing reveals the causative missense mutations in a single genetic locus encoding the TIC236 protein, as indicated by red asterisks. (C) Plant phenotypes of genetically isolated single mutants are shown. (Scale bar, 1 cm.) (D) Different levels of dominance for the D1212N (TIC236-4GF), G1250E (TIC236-5GF), and G1489R (TIC236-6GF) mutations are shown in each tic236-4gf, tic236-5gf, and tic236-6gf heterozygote (−/+) in the crl null mutant background. (Scale bars, 1 cm, 10 μm, and 0.5 mm, respectively.) (E) Heatmap showing the differentially expressed genes in crl versus WT and spcrl1 plants. The genes with at least a twofold change in expression and a FDR of less than 0.05 were selected. The colors of the heatmap represent the z-scores ranging from −2.0 (blue) to 2.0 (red). (F) The relative expression levels of selected crl-induced genes (versus WT) were analyzed in spcrl1 mutant seedlings using qRT-PCR. These genes include AKR4C8 (ALDO-KETO REDUCTASE FAMILY 4 MEMBER C8), ADH1 (ALCOHOL DEHYDROGENASE 1), SMR7 (SIAMESE-RELATED 7), and SRO3 (SIMILAR TO RCD ONE 3). ACTIN2 (ACT2) was used as an internal control. The result is presented as the mean ± SD of three replicates. Lowercase letters indicate statistically significant differences between mean values for each genotype (P < 0.05, ANOVA with post hoc Tukey’s honest significant difference test).
Knockdown of TIC236 Results in crl-Like Phenotypes
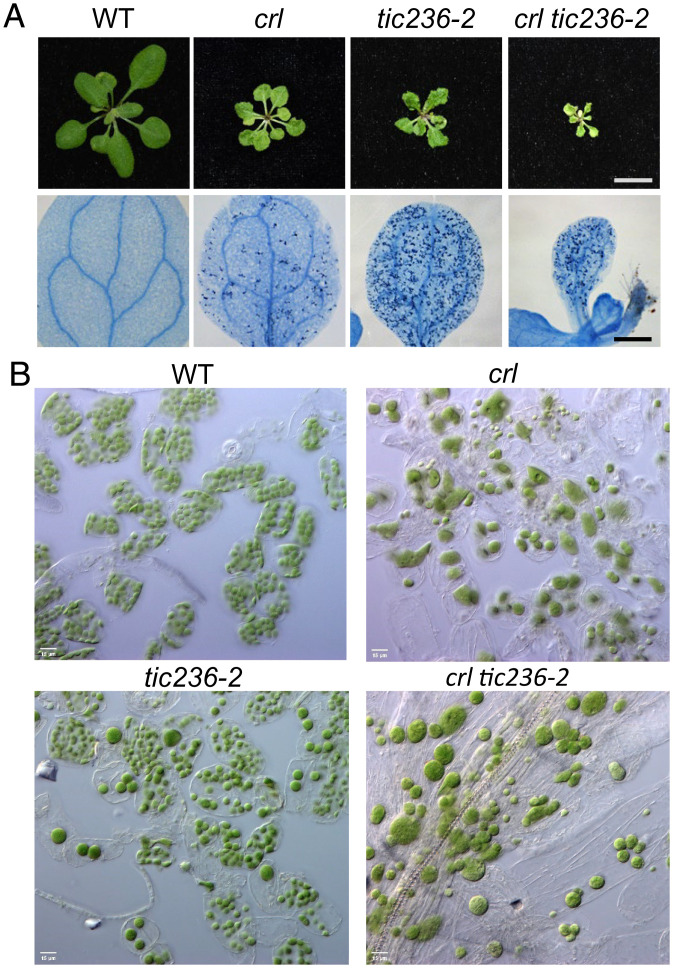
Next, we compared the phenotypes of crl and the knockdown mutant tic236-2. Remarkably, all crl phenotypes, such as growth retardation, localized cell death, and the plastid division defect, were recapitulated in tic236-2 plants (Fig. 2 A and B). It is important to note that whereas CRL mutation causes persistently impaired plastid division, tic236-2 mutant plants exhibit significantly uneven numbers and sizes of plastids per cell (Fig. 2B). Interestingly, tic236-2 also exhibited more foliar cell death than the crl mutant (Fig. 2A), indicating that a combination of reduced general import capability and the plastid division defect seems to potentiate the yet-unknown cause of localized cell death. The crl-induced defect in plastid division was epistatic to tic236-2, as only large chloroplasts were observed in crl tic236-2 double mutant plants (Fig. 2 A and B). This epistatic relationship reveals a fundamental role for CRL in plastid division and a synergistic impact of TIC236 knockdown (e.g., via general import) on crl-induced growth retardation and cell death.
Fig. 2.
The tic236-2 mutant, like the crl mutant, exhibits cell lesion and defective plastid division phenotypes. (A) (Top) Representative images of the rosettes of 21-d-old plants. (Scale bars, 1 cm.) (Bottom) Cell death in 10-d-old cotyledons, as visualized by TB staining. (Scale bar, 0.5 mm.) (B) Petiole mesophyll cells of 17-d-old WT, crl, tic236-2, and crl tic236-2 plants were observed by differential interference contrast optics. (Scale bar, 15 μm.).
CRL Is Associated with TIC236 and Other Translocon Proteins
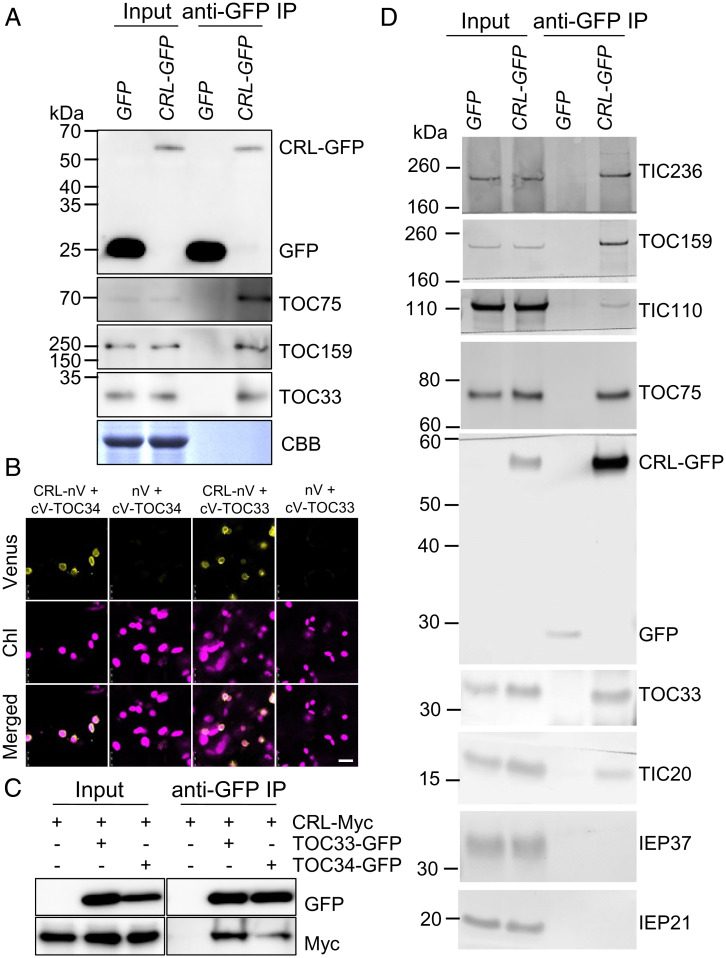
Our discovery of the tic236-gf mutations among the crl suppressors led us to consider CRL as a probable chloroplast translocon–associated protein. Accordingly, we complemented the crl mutant with a GFP-tagged CRL (SI Appendix, Fig. S3A) and coimmunoprecipitated this biologically active CRL-GFP and its accompanying proteins from total cell lysates using anti-GFP antibody–conjugated beads (SI Appendix, Fig. S3B). Cell lysates from WT plants expressing free GFP were used as a control. Eluted proteins were trypsin-digested and subjected to mass spectrometry (MS) analysis. Proteins were counted if at least two unique peptides were detected in any biological sample. The subsequent analysis to find putative CRL-associated proteins revealed a total of 188 proteins, detected with sufficient peptide quantities in at least two CRL-GFP crl samples but detected in only one GFP sample or undetected (Dataset S3). These proteins include TOC75, TOC34, TOC132, and TIC110 (SI Appendix, Fig. S3C). Another six import-related proteins—including TOC33, TOC159, and TIC214—were identified as putative albeit less significant CRL-associated proteins [i.e., detected more in CRL-GFP samples relative to GFP samples (Dataset S4)]. To date, we have not detected endogenous CRL or TIC236 in WT Arabidopsis leaves via our MS-based label-free chloroplast proteome assays, suggesting their lower abundance and/or instability.
We first validated the CRL-TOC interaction by means of coimmunoprecipitation (Co-IP) immunoblotting from total cell lysates using WT plants expressing free GFP as a control (Fig. 3A). Although free GFP was expressed at a high level and efficiently precipitated, it did not associate with the TOC proteins. In contrast, CRL-GFP was coimmunoprecipitated with TOC159, TOC75, and TOC33. Both TOC159 and TOC33 function as the initial receptors in the translocon, and Arabidopsis has two TOC33 family members, TOC33 and TOC34. Although TOC33 and TOC34 can functionally substitute for each other, TOC33 is known to be more important for importing photosynthetic proteins during early chloroplast development, whereas TOC34 is more important for root development (14–16). Next, we used both TOC33 and TOC34 to verify a direct interaction with CRL in vivo using bimolecular fluorescence complementation (BiFC) assays. Whereas no Venus fluorescence signal was detected when the N-terminal half of Venus fluorescence protein (nV) and C-terminal Venus (cV)-tagged TOC33 or TOC34 were coexpressed in Nicotiana benthamiana leaves, coexpression of either cV-TOC33 or cV-TOC34 with CRL-nV generated apparent Venus fluorescence signal in the chloroplast envelope (Fig. 3B). Co-IP and subsequent immunoblotting analyses corroborated their interactions in N. benthamiana leaves (Fig. 3C). To further examine the CRL-translocon association, we also isolated chloroplasts from CRL-GFP–complemented crl plants and GFP-expressing WT control plants. Chloroplasts were lysed by freeze-thawing without any crosslinker treatment, and total membranes were solubilized for immunoprecipitation. As free GFP resides in the nucleus and cytosol, no GFP signal was detected in solubilized chloroplast membranes (Fig. 3D, Input), but a small amount could still be detected after enrichment by immunoprecipitation (Fig. 3D, anti-GFP IP). In addition to the three TOC proteins identified from CRL-GFP chloroplasts, TIC236 was specifically coimmunoprecipitated with CRL-GFP, supporting their genetic interaction. CRL-GFP also associated with TIC20 and TIC110 but not with the nontranslocon proteins IEP37 and IEP21 (17, 18). Together, these data indicate that CRL specifically associates with the chloroplast translocon.
Fig. 3.
CRL is associated with translocon. (A) Co-IP/immunoblot results of CRL-TOC interactions. WT Arabidopsis expressing GFP and CRL-GFP–complemented crl transgenic plants were used. Total proteins were extracted from 5-d-old seedlings and then GFP and CRL-GFP were pulled down. Subsequent immunoblot analysis was conducted using the indicated TOC and TIC antibodies. Coomassie blue (CBB) staining of the SDS-PAGE gels is shown as a loading control (for input samples). (B) BiFC analysis. nV-fused CRL and cV-fused TOC33 (or TOC34) were transiently coexpressed in N. benthamiana leaves. The empty nV vector was used as a negative control. The representative Venus signals, chlorophyll autofluorescence (Chl) signals, and their merged images are shown. (Scale bar, 10 μm.) (C) Co-IP/immunoblot analysis. Different construct combinations, such as CRL-Myc alone, CRL-Myc and TOC33-GFP, or CRL-Myc and TOC34-GFP, were transiently overexpressed in N. benthamiana leaves. After 2 d, GFP-Trap magnetic beads were used to enrich the target and its associated proteins. Subsequent immunoblot analyses were carried out by using anti-GFP and anti-Myc antibodies. (D) Chloroplasts were isolated from WT Arabidopsis expressing GFP and CRL-GFP–complemented crl transgenic plants and hypotonically lysed. Total membranes were solubilized with 1% decylmaltoside and immunoprecipitated with GFP-Trap magnetic particles (Chromotek). Immunoprecipitates were analyzed by SDS-PAGE and immunoblots with antibodies indicated on the right.
GF Mutations Stabilize TIC236 and Enhance Protein Import
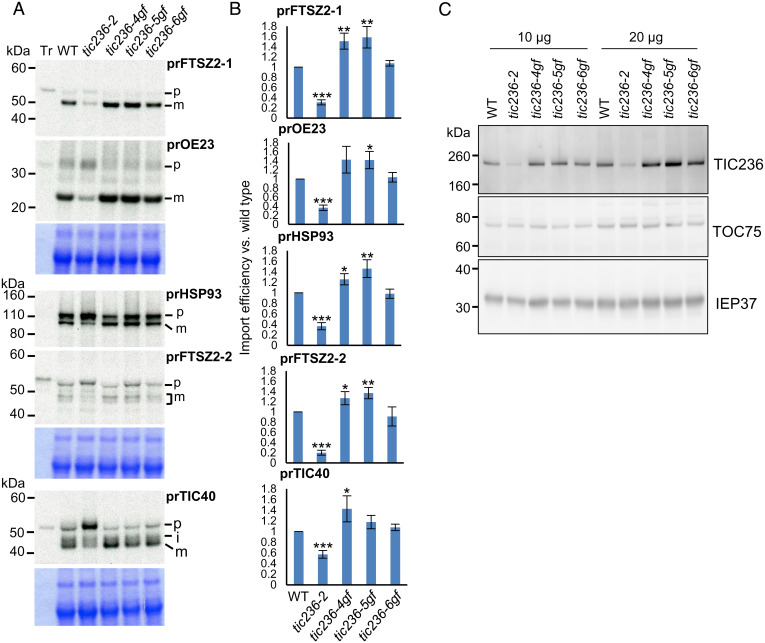
Association of CRL with the translocon suggests that CRL may function in chloroplast protein import and that the tic236-gf mutations may suppress crl-induced lesions by enhancing protein import. We investigated that possibility by means of in vitro import assays. We excluded the crl mutant because of its huge chloroplast size that impedes isolation of intact chloroplasts, essential for in vitro import assays. [35S]methionine-labeled preproteins, including casein lytic proteinase C1 (prCLPC1, also known as prHSP93), translocon at the inner envelope membrane of chloroplasts 40 (prTIC40), oxygen evolving complex subunit 23 kDa (prOE23), and the PDM components prFTSZ2-1 and prFTSZ2-2 were incubated with chloroplasts isolated from each genotype. FTSZ2-1 and FTSZ2-2 are tubulin-like components assembled in the FTSZ-ring, which is one of the contractile rings essential for plastid division (6, 19). FTSZ2-1 and FTSZ2-2 are functionally interchangeable in a dose-dependent manner, and loss of either FTSZ2-1 or FTSZ2-2 negatively affects plastid division (19, 20). The preproteins prOE23, prHSP93, and prTIC40 represent proteins that reside in the thylakoid, stroma, and IEM, respectively. Consistent with a previous report (3), tic236-2 chloroplasts displayed significantly impaired import of all tested preproteins (Fig. 4 A and B). In contrast, we observed enhanced preprotein import for tic236-4gf and tic236-5gf chloroplasts and less so for tic236-6gf chloroplasts (Fig. 4 A and B), in agreement with the respective plant phenotypes (Fig. 1 A and D). It is possible that the increase in TIC236-6GF–caused import was not sufficient to be detected via in vitro import assays, which assesses import within an extremely short time period. However, the slight increase in protein import induced by TIC236-6GF throughout plant development may be sufficient to partially suppress the defects caused by loss of CRL.
Fig. 4.
TIC236-GF mutations increase the import rate of passenger proteins. (A) [35S]Met-labeled preproteins (Tr) were imported into chloroplasts isolated from 14-d-old plants of WT and the four tic236 mutant alleles in 3 mM ATP at room temperature for 10 min. prFtsZ2-1 and prOE23 were coimported, as was prHsp93 and prFtsZ2-2. Reisolated intact chloroplasts were analyzed by SDS-PAGE, and the gels were stained with CBB and dried for fluorography. Equal amounts of proteins were loaded in each lane of the same gel, except for the Tr lane. The region around the chlorophyll a/b binding protein in the CBB-stained gels is shown Below the fluorograph as a loading control. (B) Imported mature proteins were quantified and normalized to that of the WT from the same gel and further corrected by the amount of the chlorophyll a/b binding protein in each lane. Data shown are mean ± SD (n = 3). The significance of P values is shown Above the bars, obtained from two-tailed Student’s t tests. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Chloroplasts isolated from the indicated genotypes were analyzed by immunoblotting using antibodies against proteins indicated on the right.
Assuming that the CRL-TIC236 module is required for importing PDM, we also examined import of the vital PDM components FTSZ2-1 and FTSZ2-2. We observed enhanced import of both those proteins into tic236-4gf and tic236-5gf chloroplasts and again less so in tic236-6gf chloroplasts (Fig. 4 A and B), further evidencing a function for the CRL-TIC236 module in importing PDM for plastid division. The tic236-2 mutant exhibits significantly compromised import aptitude due to reduced levels of TIC236. Therefore, we hypothesized that tic236-gf mutations may enhance preprotein import by stabilizing and hence increasing TIC236 protein level. Indeed, we found that tic236-4gf and tic236-5gf mutations increased the steady-state levels of cognate mutant proteins compared to TIC236 in WT plants (Fig. 4C).
Impaired plastid division has not been reported for the few translocon mutants documented as viable, except for mutants of the chaperonin CPN60 complex that is required for protein import and thylakoid membrane protein insertion (21–23). Since PDM import is a prerequisite for plastid division, we presumed that the CRL-TIC236 module might import some PDM components, whereas the well-established TIC236-harboring TOC/TIC complex acts in universal import, as evidenced by the embryonic lethality of the tic236 null mutant. If this notion is correct, CRL might interact with the transit peptides of PDM components. Our BiFC assays revealed that CRL interacts with the transit peptides of prFTSZ1, prFTSZ2-1, and prFTSZ2-2 but not with that of prRBCS at the OEM (SI Appendix, Fig. S4A). However, coexpression of CRL with the mature form (lacking the transit peptide) of either FTSZ1, FTSZ2-1, or FTSZ2-2 resulted in lack of, or abnormal, Venus signals (foci and rod shapes, SI Appendix, Fig. S4B). This reduction in PDM component import led us to investigate the steady-state levels of FTSZ2 family proteins in the crl mutant relative to WT seedlings by immunoblotting using a rabbit polyclonal anti-FTSZ2 antibody that can recognize both FTSZ2-1 and FTSZ2-2. Note, the predicted molecular masses of mature FTSZ2-1 and FTSZ2-2 proteins are almost indistinguishable (∼45 kDa), and their mature proteins share nearly 85% amino acid sequence identity (24). Seedlings grown on MS medium for 2 and 5 d were analyzed by immunoblotting. Our results indicated a significantly reduced abundance of FTSZ2 family proteins in 2-d-old crl mutant seedlings, whereas levels were comparable in 5-d-old crl and WT seedlings (SI Appendix, Fig. S4C). This result suggests that CRL is critical in facilitating the import of FTSZ2 family proteins in 2-d-old seedlings when plastid division occurs rapidly upon germination. Although FTSZ2 family proteins may still be imported in the absence of CRL and eventually accumulate to sufficient levels, impaired plastid division early in development may render subsequent division more difficult and ultimately give rise to the giant chloroplast phenotype.
Reduced Translocon Turnover Substantially Rescues the crl Phenotypes
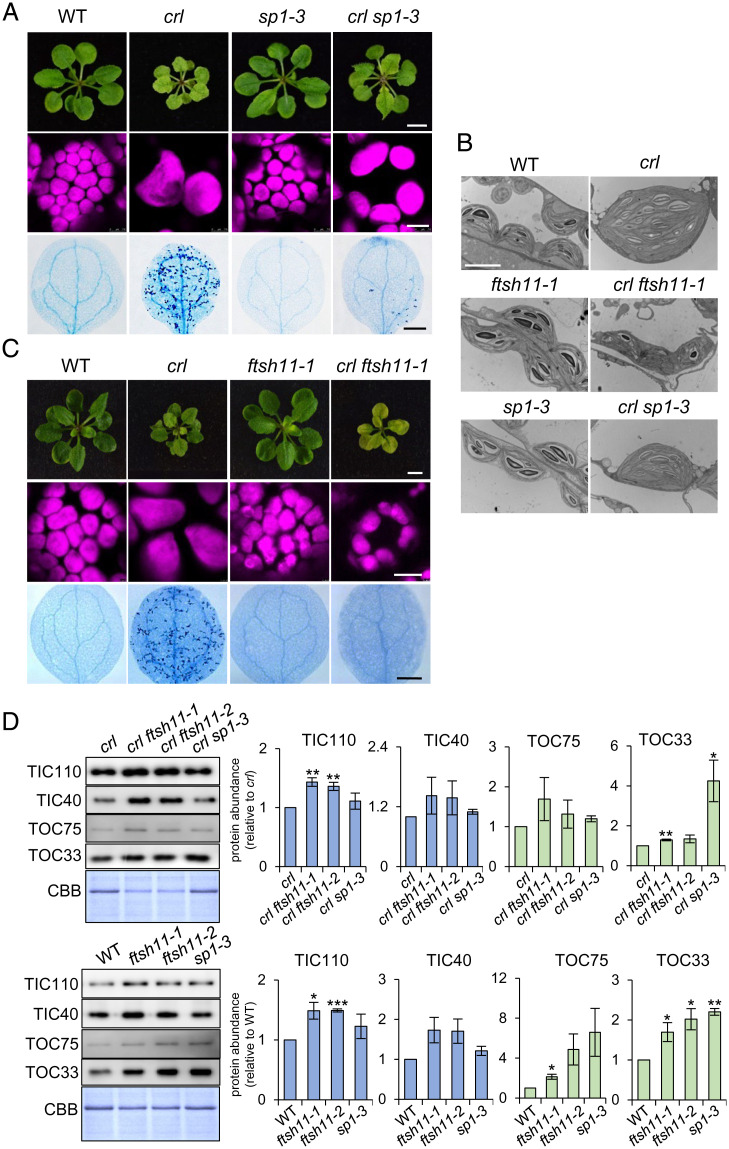
TOC components undergo proteolysis via the ubiquitin–proteasome system (UPS) in a process referred to as chloroplast-associated protein degradation (CHLORAD) (25, 26). The OEM-spanning RING-type ubiquitin E3 ligase [i.e., suppressor of ppi1 locus 1 (SP1)], the TOC75-like protein SP2 that lacks POTRA domains, and the cytosolic AAA+ chaperone CDC48 together drive CHLORAD (27). Loss of SP1 increases the amount of TOC components, whereas SP1 overexpression reduces them. SP1-mediated TOC degradation confers plant tolerance to oxidative stress by decreasing the levels of reactive oxygen species (byproducts of photosynthesis), which arises from decreased import of photosynthesis-associated proteins, demonstrating the physiological importance of SP1 (28). If CRL protein functions in protein import, as evidenced by its association with translocon proteins and the rescued crl phenotypes manifested upon tic236-gf mutations, sp1-driven TOC accumulation may also attenuate the crl-induced phenotypes. Indeed, we found that loss of SP1 partially rescued the crl phenotypes (Fig. 5 A and B). This finding also prompted us to generate crl ftsh11 double-knockout mutants. FTSH11 is a chloroplast envelope-localized AAA+-type protease that may function in TIC turnover (29). Furthermore, it was reported previously that the yeast FTSH11 homolog, YME1, mediates translocon turnover in the mitochondrial intermembrane space (30). FTSH11 also physically interacts with chaperonin 60 (CPN60), the activity of which is required for both plastid division and precursor protein maturation during import (22, 29). The loss of FTSH11 significantly rescued the crl phenotypes, including those of plastid ultrastructure and division, as well as cell death (Fig. 5 B and C, and SI Appendix, Fig. S5 A and B). Moreover, loss of either SP1 or FTSH11 increased the levels of translocon components in both the crl mutant and WT (Fig. 5D). This reverse genetic approach using sp1 and ftsh11 mutants further reinforces the evidence for CRL being a translocon-allied component.
Fig. 5.
Loss of either SP1 or FTSH11 significantly rescues the crl phenotypes. (A and C) Images representing 21-d-old plants (Top), chloroplasts in mesophyll cells (Middle), and cell death in cotyledons (Bottom) are shown. (Scale bars: Top, 5 mm; Middle, 10 µm; Bottom, 0.5 mm.) (B) The chloroplast ultrastructure of 5-d-old seedlings was monitored by transmission electron microscopy. (Scale bar, 5 µm.) (D) From the same plant materials as in A and C, equal amounts of total protein were separated on SDS-PAGE gels and immunoblotted with the antibodies as indicated to detect the relative abundance of TIC and TOC proteins. The quantified protein abundance is shown as mean ± SD of three independent biological repeats after normalization to crl or WT of the same experiments. The significance of P values is obtained from Student’s t tests (two-tailed). *P < 0.05, **P < 0.01, ***P < 0.001. CBB staining of the SDS-PAGE gels is shown as a loading control.
It is also possible that the partially rescued crl phenotypes arising from sp1 and ftsh11 mutations indicate that the crl mutant may have lower translocon levels than WT chloroplasts. However, an immunoblotting analysis revealed no apparent difference in levels of examined translocon protein components, such as TOC33, TOC34, TOC75, TIC40, and TIC110, in crl compared to those in WT and crl suppressors (SI Appendix, Fig. S6).
TIC236 Is Functionally Divergent from Its Ancestral TamB Protein
TIC236 homologs have been identified from all plant species in which they were sought. Mutations in the rice ortholog Substandard Starch Grain4 (SSG4) and the maize ortholog Defective Kernel5 (Dek5) cause plastid abnormality (31). In maize, DEK5 inactivation impairs both plastid division and plastid envelope proteostasis [e.g., reduced protein levels of translocon components and inorganic phosphate transporters (32)], so DEK5 was proposed to have retained its TamB functionality, mostly related to biogenesis of outer membrane proteins. In particular, levels of the OMP85-type β-barrel proteins TOC75 and OEP80 were diminished in dek5 mutants (32). If TIC236 functions in OEM protein biogenesis, we might expect to see reduced levels of translocon components in tic236-2 and increased levels in tic236-gf mutants, respectively. However, the tic236-2 mutant does not exhibit reduced levels of TOC75 (3). Furthermore, all three of our tic236-gf mutant lines displayed comparable levels of TOC75 protein relative to WT plants (Fig. 4C). To further verify that result, we conducted label-free quantitative proteomics analyses using isolated chloroplasts of WT and tic236-gf mutants and found that levels of most of the TOC/TIC components in the tic236-gf mutants remained unchanged relative to WT (SI Appendix, Fig. S7 and Dataset S5).
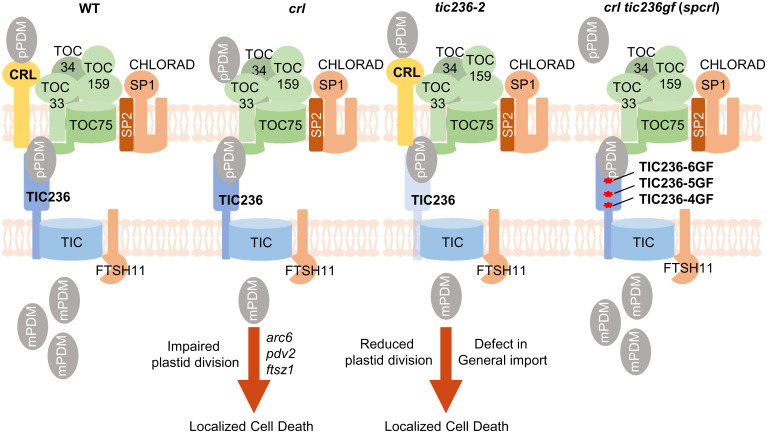
CRL is believed to function in a ROS-triggered chloroplast-to-nucleus retrograde signaling pathway or as a putative PDM component in plastid division (7, 11, 12). However, our study using combined biochemical and forward/reverse genetic approaches has revealed its mutual functionality with the TIC236 protein. Since the tic236 null mutant displays embryonic lethality (3), whereas the crl mutant is viable despite multiple lesions (Fig. 1A), unlike CRL, the function of TIC236 in plastid translocons must be indispensable. The gigantic plastids in both the crl and tic236-2 mutants (Fig. 2B), as well as our evidence of CRL–PDM preprotein interaction (SI Appendix, Fig. S4 A and B), imply that the CRL-TIC236 module imports PDM preproteins at the early phase of cell development. The observed cell death and chloroplast division phenotypes displayed by the crl and tic236-2 mutants should be investigated in yet other translocon mutants to gain further insights into the passenger specificity of the CRL-TIC236 module, as well as into possible spatial–temporal heterogeneity of translocon complexes and its biological relevance. Importantly, our findings also open up a research avenue linking chloroplast dysfunction (especially in certain import pathways) to activation of autoimmune responses (Fig. 6).
Fig. 6.
TIC236-GF mutations rescue the crl mutant by stabilizing TIC236 proteins. In WT, prPDM are imported through the CRL-TIC236-translocon module, enabling plastid division. Either CRL loss or TIC236 knockdown compromises plastid division and induces localized cell death despite remaining unchanged levels of core translocon complex. Similar lesion phenotypes were observed in the canonical plastid division mutants arc6, pdv2, and ftsz1 (7, 10), suggesting that the impaired plastid division results in the activation of autoimmune responses through a yet unknown mechanism. The TIC236-GF mutations overcome CRL deficiency by reinforcing prPDM import with stabilized TIC236-GF proteins. Reduced sp1-driven CHLORAD activity or loss of FTSH11 protease also rescues the crl phenotypes with increased levels of translocon proteins.
In summary, we have reported herein that 1) CRL is a translocon-associated component, absence of which impairs plastid division; 2) knockdown of TIC236 induces multiple lesions and plastid division defects, as also observed in the crl mutant; 3) CRL and TIC236 are likely to interact in the intermembrane space, which is consistent with the rescue of the crl phenotypes by the TIC236-GF mutations; 4) three approaches for increasing translocon component abundance—knockout of either SP1 or FTSH11 and gain-of-function TIC236 mutations—all rescued crl plastid division and cell lesion defects; and 5) TIC236-GF mutations may provide a strategy for engineering the translocon to enhance protein import efficacy.
Materials and Methods
Plant Material and Growth Conditions.
All Arabidopsis seeds used in this study are from the Columbia-0 (Col-0) background. All seed stocks, including ftsh11-1 (SALK_033047), ftsh11-2 (SALK_012285), sp1-3 (SALK_002099), and tic236-2 (SAIL_104-F07), were obtained from the Nottingham Arabidopsis Stock Centre or Arabidopsis Biological Resource Center. The crl null mutant (GABI_714_E08) and the transgenic p35S::CRL-GFP crl lines have been described previously (10, 13). Their genotypes were confirmed by PCR-based analyses using the corresponding primers (SI Appendix, Table S1). Seeds were sterilized in a 5% hypochlorite solution for 3 min, washed five times with sterilized water, and then sown on half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) with 0.5% (wt/vol) sucrose and 0.7% (wt/vol) agar. After stratification at 4 °C for 2 d in the dark, the seeds germinated and grew under either continuous light (CL, 100 µmol ⋅ m−2 ⋅ s−1 at 22 ± 2 °C) or long-day (LD, 22 ± 2 °C, 100 µmol ⋅ m−2 ⋅ s−1 with a 16-h light/8-h dark photoperiod) conditions. N. benthamiana plants were grown under controlled LD conditions. For all transient expression assays, 4-wk-old N. benthamiana plants were used.
EMS Mutagenesis and Whole-Genome Sequencing Analysis.
Under normal growth conditions, we screened the M2 progeny of ∼12,000 M1 crl seeds that had been initially treated with 0.4% (wt/vol) EMS (Sigma-Aldrich) for 8 h as described previously (33). The spcrl1, spcrl2, and spcrl3 mutants were chosen to generate mapping populations. To do that, each spcrl homozygote mutant plant was backcrossed to the parental crl mutant. From the F2 population, at least 50 plants exhibiting WT-like phenotypes were selected for genomic DNA extraction. Genomic DNA isolated from crl mutant plants was used as a control. Genomic DNA (1 μg) isolated using a DNeasy plant mini kit (Qiagen) was used for library construction. Sequencing was performed using a HiSeq2500 (Illumina) sequencer to generate 125–base pair (bp) paired-end reads, as described before (10). The sequencing data were processed in SolexaQA (34) and Cutadapt (version 1.3) software to remove low-quality regions and adapter sequences, respectively. Clean reads were mapped to the TAIR10 genome in BWA-MEM (35) with default parameters. Single nucleotide polymorphisms (SNPs) were called using the “mpileup” function of SAMtools (36). Poor-quality SNPs with a mapping quality < 60 or with a depth < 3 or >200 were filtered out using vcftools (37). Candidate causal mutations were identified using the SHOREmap method (38). The allele frequency and the regions containing a possible causal mutation were analyzed using SHOREmap version 3.0 (39). Mutations within the open reading frame of target genes were considered as potential causative mutations. The mutations were further confirmed by sequencing PCR products using the respective primers listed in SI Appendix, Table S1.
Isolation of tic236 Gain-of-Function Mutants.
We isolated tic236-4gf, tic236-5gf, and tic236-6gf single mutants by backcrossing the corresponding parental lines spcrl1, spcrl2, and spcrl3 to WT plants. Among the F2 segregating population, WT-like plants were chosen for PCR-based genotyping to select lines devoid of transfer DNA insertions in CRL. Next, the same genomic DNA samples were used to amplify DNA fragments covering each of three gain-of-function mutation sites (D1212N, G1250E, and G1489R) using primers listed in SI Appendix, Table S1. The single tic236-gf mutants were selected based on the sequencing results.
Co-IP and MS Analyses.
The complementary DNA (cDNA) (lacking stop codons) of CRL, TOC33, and TOC34 were PCR amplified from WT cDNA and subsequently cloned into pDONR221-Zeo entry vectors by means of a Gateway BP clonase reaction (Invitrogen). To generate C-terminal tag-fused constructs, the purified cDNAs were cloned into destination vectors, including pGWB605 for sGFP or pGWB617 for 4xMyc, by Gateway LR clonase reactions (Invitrogen). Each vector was transformed into Agrobacterium tumefaciens strain GV3101. For transient expression (or coexpression), the suspensions of Agrobacteria carrying different constructs were infiltrated into healthy leaves of 28-d-old N. benthamiana plants, and protein–protein interactions were analyzed after 48 h. Total protein was extracted using a protein extraction (PE) buffer [Tris⋅HCl 50 mM (pH 7.5), NaCl 150 mM, Glycerol 10% (vol/vol), DTT 10 mM, cOmplete protease inhibitor mixture 1 tablet/50 mL (Roche), 1.1% (vol/vol) Nonidet P-40, EDTA 1 mM, Na2MoO4 1 mM, Sodium fluoride (NaF) 1 mM, and Sodium orthvanidate (Na3VO4) 1.5 mM]. After diluting total protein samples to 1 to 2 μg/μL, 20 mg total protein was incubated with 15 μL GFP-TrapMA beads (Chromotek) at 4 °C for 2 h and then washed four times with PE buffer. Finally, the remaining proteins on the washed beads were eluted with 2× sodium dodecyl sulfate (SDS) sample buffer at 95 °C for 10 min. The eluted proteins were subjected to 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gels for immunoblot analyses using anti-GFP (Roche) and anti-Myc (Roche) antibodies.
For Co-IP shown in Fig. 3A, and subsequent immunoblot analysis or Co-IP coupled to MS analysis, rosette leaves from 14-d-old crl mutant plants expressing CRL-GFP under the control of the 35S promoter and WT plants expressing GFP under the control of the 35S promoter were used. The plants were homozygous for the crl mutation and the transgenes. Rosettes were ground to a fine powder in liquid nitrogen and resuspended in IP buffer [Hepes 20 mM (pH 7.5), EDTA 2 mM, EGTA 2 mM, NaCl 100 mM, Glycerol 10% (vol/vol), Triton-X-100 0.2% (vol/vol), Na3VO4 1 mM, NaF 20 mM, and cOmplete protease inhibitor mixture 1 tablet/50 mL (Roche)] at 4 °C for 1 h. After quantification using a Pierce BCA protein assay kit (Thermo Scientific), 10 mg total protein was incubated with 50 μL Dynabeads Protein G (Thermo Scientific) that had been preconjugated with a monoclonal mouse anti-GFP antibody (Roche) at room temperature for 2 h. After washing five times, 10% of the beads were eluted with SDS sample buffer (2×) at 70 °C for 20 min and then loaded on 10% SDS-PAGE gels for subsequent immunoblotting analysis. For Co-IP shown in Fig. 3D, chloroplasts were isolated from the same CRL-GFP–complemented crl and GFP-expressing WT lines grown for 14 d on 1xMS agar media with 2% sucrose (wt/vol). Chloroplasts (0.5 mg chlorophyll/mL in 50 mM Hepes-KOH pH 8.0 and 0.33 M sorbital) without any crosslinker treatment were directly lysed by freeze-thawing three times. Total membranes were collected and solubilized in 50 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 4 mM MgCl2, 1% decylmaltoside (wt/vol), 10% glycerol (vol/vol), and 1× cOmplete protease inhibitor mixture (Roche) as described previously (40). Solubilized membranes were immunoprecipitated with GFP-Trap Magnetic Particles M-270 (Chromotek) and eluted with 1× SDS-PAGE sample buffer. Antibodies against Arabidopsis TIC236 (residues 115 to 266) were produced by cloning the corresponding cDNA into the BglII/XhoI sites of pET22b with an N-terminal His6 tag. Recombinant proteins were purified from E. coli by TALON metal-affinity resins and used to raise antibodies in rabbits. Mouse monoclonal anti-GFP antibodies (Santa Cruz SC-9996) were used for immunoblotting detection of GFP.
In-gel digestion and ensuing MS analysis were performed according to a previous report (41). The obtained mass spectra were compared against the TAIR10 nonredundant database using Mascot Server (version 2.5.1). The parameter settings were prearranged for peptide mass tolerance at 20 ppm, fragment mass tolerance at 0.02 Da, and a maximum of two missed cleavages was allowed. The significance threshold for search results was set at P < 0.05, with an ion score cutoff of 15. Proteins detected at least twice in the CRL-GFP samples but absent or detected only once in the GFP samples were considered potential interacting proteins of CRL (Dataset S3). Among them, translocon and plastid division proteins were selected and shown in Dataset S4.
BiFC Assays.
In addition to the full-length cDNA of CRL, TOC33, and TOC34, cDNA encoding the transit peptide (tp) or mature (m) regions (without the stop codon) of RBCS, FTSZ1, FTSZ2-1, and FTSZ2-2 were cloned into pDONR221-Zeo entry vectors for BiFC analyses. Through Gateway LR reactions (Thermo Scientific), these cDNAs were cloned into the 35S promoter–driven destination vector pDEST to generate tag-fused constructs. The full-length CRL cDNA (lacking the stop codon) was C terminally fused with the N-terminal half of Venus (nV). The full-length TOC33 and TOC34 cDNAs were N-terminally fused with the C-terminal half of Venus (cV). cDNA corresponding to the transit peptides and mature regions of RBCS, FTSZ1, FTSZ2-1, and FTSZ2-2 were C terminally fused with cV. As described previously (42), the N-terminal 79 residues of RBCS were constructed as the transit peptide region. Transit peptide regions of FTSZ1 (1 to 57 aa), FTSZ2-1 (1 to 48 aa), and FTSZ2-2 (1 to 50 aa) were predicted by the ChloroP1.1 server (20, 43, 44). Then, tp-deleted and start codon–appended RBCS, FTSZ1, FTSZ2-1, and FTSZ2-2 constructs were generated. Destination plasmids CRL-nV, cV-TOC33, cV-TOC34, tpRBCS-cV, tpFTSZ1-cV, tpFTSZ2-1-cV, tpFTSZ2-2-cV, mRBCS-cV, mFTSZ1-cV, mFTSZ2-1-cV, and mFTSZ2-2-cV were transformed into Agrobacterium strain GV3101. The Agrobacterium cultures were diluted to the optical density at 600 nm (OD600) of 1.0, then resuspended and washed using infiltration solution [MES 10 mM (pH 5.6), MgCl2 10 mM, and Acetosyringone 150 μM]. Mixtures of the selected strains were infiltrated into 28-d-old N. benthamiana leaves. After transient expression for 48 h, Venus fluorescence was analyzed using confocal laser scanning microscopy (TCS SP8, Leica).
PE and Immunoblot Analysis.
Plant leaf tissues were ground to fine powder in liquid nitrogen and resuspended in IP buffer at 4 °C for 1 h. Protein concentration was determined using a Pierce BCA protein assay kit (Thermo Scientific). Equal concentrations of protein samples were mixed with 4× or 2× SDS loading buffer, denatured at 95 °C for 5 min, and loaded into 10 or 4 to 12% SDS-PAGE gels. GFP-tagged proteins were detected with a mouse anti-GFP antibody (1:5,000; Roche). Rabbit anti-PGL35 antibody was purchased from Agrisera (AS06 116). Immunoblot results were quantified using Image J software (version 1.8.0).
In Vitro Protein Import Assay.
[35S]Met-labeled preproteins were in vitro transcribed/translated using the TNT-coupled wheat germ or reticulocyte lysate system and SP6 or T7 RNA polymerase (Promega). Growth of Arabidopsis seedlings (for 14 d on 1xMS agar media with 2% sucrose), Arabidopsis chloroplast isolation, and import of [35S]-labeled preproteins into isolated chloroplasts were performed as described previously (45). Accession numbers for preproteins are as follows: pea prTIC40 (AY157668), pea prHSP93 (L09547), Arabidopsis prOE23 (At1g06680), Arabidopsis prFTSZ2-1 (At2g36250), and Arabidopsis prFTSZ2-2 (At3g52750).
Chloroplast Size Analysis.
Leaf petioles of 17-d-old seedlings grown on MS medium were excised and fixed in 3.5% glutaldehyde and then prepared for imaging with differential interference contrast microscopes as described previously (46).
Cell Death Determination.
Cell death was assessed via Trypan blue (TB) staining as described previously (10). The TB-stained plants were preserved in 10% (vol/vol) glycerol. Imaging was conducted using a TCS SP8 microscope (Leica Microsystems) and further processed using Leica LAS software (version 4.2.0, Leica Microsystems).
Microscopic Analyses.
Venus and chlorophyll autofluorescence signals were monitored under a confocal microscope at 520 to 600 nm of the emission spectrum with an excitation wavelength of 514 nm (Leica TCS SP8). Representative images were processed using Leica LAS AF Lite software (version 2.6.3, Leica Microsystems). Cotyledons of 5- and 10-d-old seedlings (before and after cell death, respectively) were mostly used for imaging GFP and chlorophyll autofluorescence unless otherwise indicated.
For transmission electron microscopy, cotyledons of 5-d-old seedlings were detached, prefixed, and then rinsed three times using 0.1 M phosphate-buffered saline (PBS) buffer as described previously (10). Then, the samples were postfixed overnight in 1% (vol/vol) osmic acid at 4 °C, washed three times with 0.1 M PBS buffer, dehydrated using a gradient ethanol–acetone series, before being embedded in Spurr’s resin. Ultrathin resin sections (70 nm) were cut using a diamond knife on a Leica UC7 μLtramicrotome, mounted on copper grids, and stained with 2% (wt/vol) uranyl acetate and 0.5% (wt/vol) lead citrate. The stained sections were monitored and photographed using a H7700 transmission electron microscope (Hitachi).
RNA-Seq Library Construction and Data Analysis.
RNA-seq analysis was carried out as described previously (47). Total RNA was extracted from three independent biological replicates of 5-d-old Arabidopsis crl, spcrl1, and WT seedlings using the RNeasy Plant Mini Kit (Qiagen). The isolated RNA was subjected to on-column DNase digestion according to the manufacturer’s instructions. A Nano Photometer spectrophotometer (IMPLEN) was used to verify RNA purity. A Qubit RNA Assay Kit and a Qubit 2.0 Fluorometer (Life Technologies) were employed to determine RNA concentration. An RNA Nano 6000 Assay Kit and Bioanalyser 2100 system (Agilent Technologies) were used to assess RNA integrity for RNA-seq analyses. RNA-seq libraries were built using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolab) based on the manufacturer’s instructions. The RNA-seq libraries were sequenced on an Illumina HiSEq. 2500 platform to generate 100-bp paired-end reads. The raw sequencing data were processed in SolexaQA (version 2.2) to extract paired reads and to remove low-quality reads. The clean reads were mapped to the Arabidopsis genome (TAIR10) using TopHat (48). After mapping, the Python-based software HTseq-count was used to extract raw counts of annotated genes. Differentially expressed genes were identified using the R package edgeR, which uses counts per gene in different samples and performs data normalization using the trimmed mean of M-values method (49). The gene expression data were normalized to transcripts per million according to the total number of mapped clean reads in each library. Genes with at least a twofold change in expression and a FDR of less than 0.05 were deemed to be differentially expressed.
RNA Extraction and qRT-PCR.
Total RNA was prepared using the FastPure Plant Total RNA Isolation Kit (Vazyme). RNA (1 µg) was treated with RQ1 RNase-Free DNase (Promega) and reverse transcribed using HiScript II Q RT SuperMix for qPCR (genomic DNA [+gDNA] wiper) (Vazyme) according to the manufacturer’s instructions. qRT-PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme) and a QuantStudio 6 Flex RT-PCR System (Applied Biosystems). Transcript abundances were calculated using the delta-Ct method (50) and normalized to the transcript levels of ACTIN2 (AT3G18780). The primers used in this study are listed in SI Appendix, Table S1.
Chloroplast Isolation and MS Analysis.
Chloroplasts were isolated from 21-d-old plants as described previously (51, 52), and the extracted total chloroplast proteins were used for subsequent label-free quantitative proteomics analysis (SI Appendix, Fig. S7). Rosette leaves were homogenized in chloroplast isolation buffer (50 mM Hepes-KOH [pH 8.0], 5 mM MgCl2, 5 mM EDTA [pH 8.0], 5 mM EGTA [pH 8.0], 10 mM NaHCO3, and 0.33 M D-sorbitol supplemented with one tablet [per 50 mL] cOmplete protease inhibitor mixture [Roche]) using a Waring blender. After filtering through four-layer Miracloth, the homogenate was centrifuged at 400 × g for 8 min at 4 °C. The pellets were suspended using chloroplast isolation buffer and added onto a two-step Percoll gradient (40:80%). After centrifugation, the enriched chloroplasts between the two Percoll steps were carefully collected and washed twice using HEPES-Sorbitol (HS) buffer (50 mM Hepes-KOH [pH 8.0] and 0.33 M D-sorbitol). The chloroplasts were resuspended in guanidine hydrochloride buffer (6 M guanidine hydrochloride and 100 mM Tris [pH 8.5]). The resuspension was sonicated in an ice bath for 1 min with a pulse of 3 s on and 5 s off, followed by heating at 95 °C for 5 min, and centrifugation at 15,000 rpm for 30 min at 4 °C. The supernatant contained the total chloroplast proteins. Protein concentration was determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific).
For MS analysis, equal amounts of total protein from three independent biological samples were denatured with 10 mM DTT at 56 °C for 30 min. The denatured samples were subjected to alkylation in 50 mM iodoacetamide in the dark for 40 min. The samples were then desalted in 100 mM NH4HCO3 buffer through a Nanosep membrane (Pall Corporation, molecular weight cut-off [MWCO] 10kDa). Desalted proteins were digested using trypsin (40 ng/μL trypsin and 100 mM NH4HCO3, enzyme-to-protein ratio 1:50) at 37 °C for 20 h. The cleaved peptides were then dried in a chilled CentriVap concentrator (Labconco). The peptides were resuspended in 0.1% (vol/vol) formic acid (FA), and subjected to nanoAcquity Ultra Performance LC (Waters) through a 20-mm trap column (C18 5 μm resin, 180 μm inner diameter [I.D.], Waters) with a flow rate of 3 μL/min for 10 min and then eluted to the analytical column (C18 1.7 μm resin, 75 μm I.D., Waters) with a flow rate of 250 nl/min under the following conditions: 140-min gradient from 8 to 25% of solvent B (Acetonitrile, ACN); 15-min gradient from 25 to 40% of solvent B; 5-min gradient from 40 to 90% of solvent B; 5-min washing at 90% of solvent B, and finally equilibration with 97% of solvent A for 15 min (solvent A: 0.1% FA; solvent B: 99.9% ACN/0.1% FA). After analyzing the separated peptides in a Q Exactive Mass Spectrometer (Thermo Electron Finnigan), a full MS survey scan was performed at a resolution of 70,000 at 400 m/z over the m/z range of 300 to 1,800, with an automatic gain controls (AGC) target of 3E6 and a maximum ion injection time (IT) of 30 msec. The top 20 multiply charged parent ions were selected under data-dependent MS/MS mode and fragmented by higher-energy collision dissociation with a normalized collision energy of 27% and an m/z scan range of 200 to 2,000. MS/MS detection was carried out at a resolution of 17,500 with the AGC target value of 5E5 and the maximum IT of 120 msec. Dynamic exclusion was enabled for 30 s.
Label-Free Quantitative Proteomics Analysis.
MaxQuant software (version 1.5.8.3) with an intensity-based absolute quantification (iBAQ) algorithm was used to process and analyze raw MS data as described previously (51, 53, 54). Parent ion and MS2 spectra were compared against the Arabidopsis Information Resource database (https://www.arabidopsis.org/). The precursor ion tolerance was set at 7 ppm, and a fragment mass deviation of 20 ppm was allowed. The detected peptides with a minimum of six amino acids and a maximum of two missed cleavages were assigned. For both peptide and protein identification, the FDR was set to 0.01. The iBAQ intensity value was used as an accurate proxy to calculate protein amounts. Proteins detected with iBAQ intensity values in at least two out of three independent biological samples were considered meaningful.
Statistical Analyses.
Numbers of biological replicates are presented in the figure legends. Statistical analyses were performed by two-tailed Student’s t test or one-way ANOVA with a post hoc Tukey’s honest significant difference test. P's < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank the Core Facility of Genomics and Bioinformatics in Shanghai Center for Plant Stress Biology (PSC) for carrying out whole-genome sequencing analysis and RNA-seq. We thank the Core Facility of Cell Biology in PSC for training students in the use of various microscopes. We also thank the Core Facility of Proteomics in PSC for conducting MS analysis. We thank Masato Nakai (Osaka University) for providing the anti-TOC75 antibody. This research was supported by the Strategic Priority Research Program from the Chinese Academy of Sciences (Grant No. XDB27040102), the 100-Talent Program of the Chinese Academy of Sciences and the National Natural Science Foundation of China (Grant No. 31871397) to C.K., and by grants to H.-m.L. from the Ministry of Science and Technology (Grant No. MOST 110-2326-B-001-016) and Academia Sinica of Taiwan.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Richardson L. G. L., Schnell D. J., Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids. J. Exp. Bot. 71, 1226–1238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwenkert S., Dittmer S., Soll J., Structural components involved in plastid protein import. Essays Biochem. 62, 65–75 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Chen Y. L., et al. , TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Stubenrauch C., et al. , Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat. Microbiol. 1, 16064 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Selkrig J., et al. , Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19, 506–510 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Osteryoung K. W., Pyke K. A., Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65, 443–472 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Šimková K., et al. , The chloroplast division mutant caa33 of Arabidopsis thaliana reveals the crucial impact of chloroplast homeostasis on stress acclimation and retrograde plastid-to-nucleus signaling. Plant J. 69, 701–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruggeman Q., Raynaud C., Benhamed M., Delarue M., To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6, 24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorrain S., Vailleau F., Balagué C., Roby D., Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Li B., et al. , FATTY ACID DESATURASE5 is required to induce autoimmune responses in gigantic chloroplast mutants of Arabidopsis. Plant Cell 32, 3240–3255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudik E., et al. , Chloroplast dysfunction causes multiple defects in cell cycle progression in the Arabidopsis crumpled leaf mutant. Plant Physiol. 166, 152–167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asano T., et al. , A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38, 448–459 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Wang F., et al. , The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function. Plant J. 104, 964–978 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Jarvis P., et al. , An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Constan D., Patel R., Keegstra K., Jarvis P., An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38, 93–106 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Kubis S., et al. , The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15, 1859–1871 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motohashi R., et al. , Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34, 719–731 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Kouranov A., Chen X., Fuks B., Schnell D. J., Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara M., Yoshida S., Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 287, 462–467 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Schmitz A. J., Glynn J. M., Olson B. J. S. C., Stokes K. D., Osteryoung K. W., Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2, 1211–1222 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Kessler F., Blobel G., Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 93, 7684–7689 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki K., et al. , Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 9, 38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klasek L., Inoue K., Theg S. M., Chloroplast chaperonin-mediated targeting of a thylakoid membrane protein. Plant Cell 32, 3884–3901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadirjan-Kalbach D. K., et al. , Allelic variation in the chloroplast division Gene FtsZ2-2 leads to natural variation in chloroplast size. Plant Physiol. 181, 1059–1074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling Q., et al. , Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, 836 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Shanmugabalaji V., Kessler F., CHLORAD: Eradicating translocon components from the outer membrane of the chloroplast. Mol. Plant 12, 467–469 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Ling Q., Huang W., Baldwin A., Jarvis P., Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Ling Q., Jarvis P., Regulation of chloroplast protein import by the ubiquitin E3 Ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adam Z., et al. , The chloroplast envelope protease FTSH11 - Interaction with CPN60 and identification of potential substrates. Front Plant Sci 10, 428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Li L., Jiang H., Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45. J. Cell Biol. 217, 139–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushima R., et al. , Amyloplast-localized SUBSTANDARD STARCH GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 164, 623–636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., et al. , Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. J. Cell Biol. 218, 2638–2658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y., Schumaker K. S., Zhu J. K., EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323, 101–103 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Cox M. P., Peterson D. A., Biggs P. J., SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11, 485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danecek P., et al. ; 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneeberger K., et al. , SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Sun H., Schneeberger K., SHOREmap v3.0: Fast and accurate identification of causal mutations from forward genetic screens. Methods Mol. Biol. 1284, 381–395 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Su P.-H., Li H. M., Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., et al. , Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. U.S.A. 113, E3792–E3800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee D. W., et al. , Molecular mechanism of the specificity of protein import into chloroplasts and mitochondria in plant cells. Mol. Plant 12, 951–966 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Emanuelsson O., Nielsen H., von Heijne G., ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson B. J. S. C., Wang Q., Osteryoung K. W., GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285, 20634–20643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu C. C., Li H. M., Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol. Biol. 774, 339–350 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Pyke K. A., Leech R. M., Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96, 1193–1195 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dogra V., et al. , FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front Plant Sci 8, 1145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Dogra V., Duan J., Lee K. P., Kim C., Impaired PSII proteostasis triggers a UPR-like response in the var2 mutant of Arabidopsis. J. Exp. Bot. 70, 3075–3088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kauss D., Bischof S., Steiner S., Apel K., Meskauskiene R., FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett. 586, 211–216 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Luber C. A., et al. , Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.