Abstract
Malaria is a life-threatening infectious disease primarily caused by the Plasmodium falciparum parasite. The increasing resistance to current antimalarial drugs and their side effects has led to an urgent need for novel malaria drug targets, such as the P. falciparum cGMP-dependent protein kinase (pfPKG). However, PKG plays an essential regulatory role also in the human host. Human cGMP-dependent protein kinase (hPKG) and pfPKG are controlled by structurally homologous cGMP-binding domains (CBDs). Here, we show that despite the structural similarities between the essential CBDs in pfPKG and hPKG, their respective allosteric networks differ significantly. Through comparative analyses of chemical shift covariance analyses, molecular dynamics simulations, and backbone internal dynamics measurements, we found that conserved allosteric elements within the essential CBDs are wired differently in pfPKG and hPKG to implement cGMP-dependent kinase activation. Such pfPKG versus hPKG rewiring of allosteric networks was unexpected because of the structural similarity between the two essential CBDs. Yet, such finding provides crucial information on which elements to target for selective inhibition of pfPKG versus hPKG, which may potentially reduce undesired side effects in malaria treatments.
Keywords: allostery, cGMP, chemical shift covariance analyses, malaria, NMR, PKG, Plasmodium falciparum
Abbreviations: BBR, base-binding region; CBD, cGMP-binding domain; CHESCA, chemical shift covariance analyses; CHESPA, chemical shift projection analysis; cNMP, cyclic nucleotide monophosphate; hPKG, human cGMP-dependent protein kinase; HSQC, heteronuclear single quantum coherence; MD, molecular dynamics; N3A, N-terminal helix bundle; PBC, phosphate-binding cassette; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase; RSD, reduced spectral density; SM, similarity measurement
Allosteric regions, as opposed to highly conserved orthosteric active sites, offer a promising avenue to engineer selectivity between homologous proteins (1, 2, 3, 4, 5, 6). In both Plasmodium falciparum cGMP-dependent protein kinase (pfPKG) and human cGMP-dependent protein kinase (hPKG), the N-terminal regulatory region includes the allosteric regulatory domains, whereas the C-terminal catalytic region spans the kinase active site (7, 8, 9). The regulatory region is responsible for autoinhibition of both PKGs in the absence of cGMP, whereas, in the presence of cGMP, the cGMP-binding domains (CBDs) in the regulatory region are subject to conformational changes that remove the autoinhibition and allow for the activation of PKG. In pfPKG, the regulatory region consists of four CBDs, that is, CBD-A, BCD-B, CBD-C (degenerate), and CBD-D (Fig. 1A) (10). pfPKG CBD-D (i.e., pfD), which is directly connected to the catalytic region through its C-terminal helix, exhibits the highest selectivity and affinity toward cGMP with a Kd of 40 nM and is also the critical domain for autoinhibition and activation of pfPKG (8, 11). In hPKG Iβ, the regulatory region consists of an N-terminal dimerization domain, followed by an autoinhibitory linker region and two CBDs, that is, CBD-A and CBD-B (Fig. 1B). hPKG CBD-B (i.e., hB) is the homologous domain to pfD, with 55% sequence similarity (Fig. 1C) and acts as a critical control region for the autoinhibition and cGMP-dependent activation of hPKG. hB also exhibits higher cGMP versus cAMP selectivity than CBD-A (12, 13), and the conformational dynamics of the isolated pfD and hB domains have been shown to reflect the kinase activities of the respective full-length proteins (14, 15, 16), suggesting that pfD and hB are significant for kinase control.
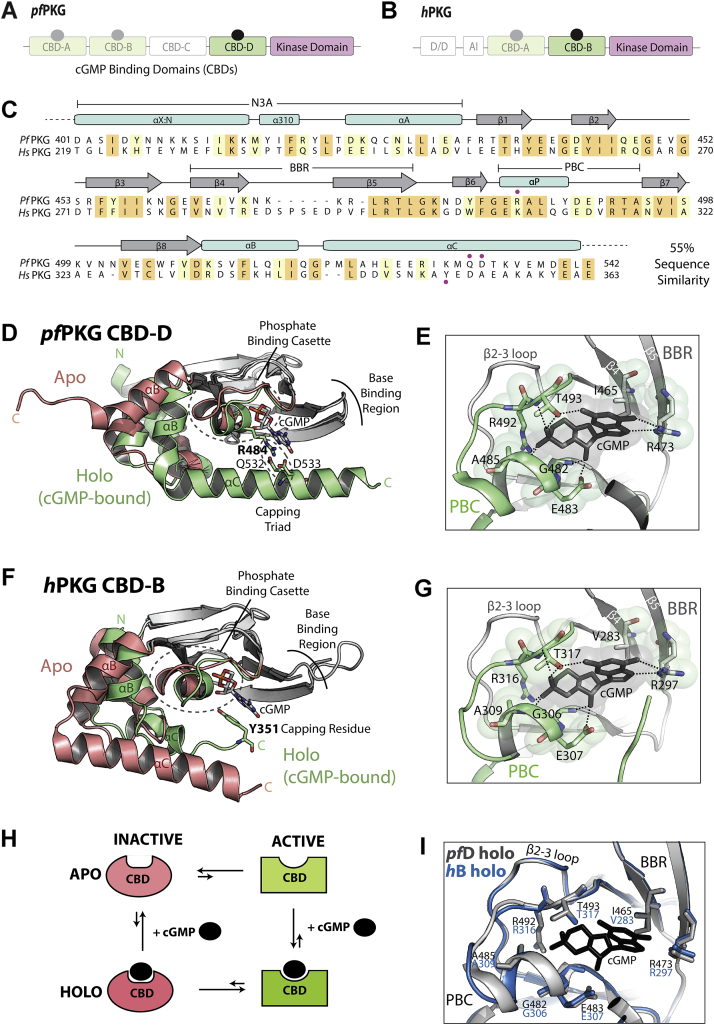
Figure 1.
PfPKG versus hPKG comparison of domain organization, sequence, and structure.A, domain organization of pfPKG, which consists of four cGMP-binding domains (CBDs) followed by a kinase domain. CBD-C is degenerate and does not bind to cGMP. B, domain organization of hPKG, which consists of an N-terminal dimerization/docking domain (D/D), an autoinhibitory sequence (AI), and two tandem CBDs followed by a kinase domain. C, sequence alignment of pfD and hB. Orange highlights indicate identical sequence, and yellow highlights indicate similar residues. Secondary structures are reported on top of the sequences. The red dots on top indicate the capping triad residues for pfPKG, whereas the red dot on the bottom indicates the capping residue for hPKG. D, structures of apo and cGMP-bound holo pfD. The structurally invariant β-subdomain is shown in gray. E, essential cGMP-binding residues in the phosphate-binding cassette (PBC; green) and base-binding region (BBR; gray). F, similar to (D) but for hB. G, similar to (E) but for hB. H, four-state thermodynamic equilibrium of CBD. The autoinhibitory equilibrium of the apo form is coupled to the cGMP-binding equilibrium. I, overlay of pfD (gray) and hB (blue) holo structures of the binding region, exhibiting an RMSD of 0.2 Å in the PBC and 0.1 Å in the BBR. hPKG, human cGMP-dependent protein kinase; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase.
The pfD and hB CBDs are both composed of a contiguous β-subdomain and a noncontiguous α-subdomain (8, 12, 13). The β-subdomain forms a rigid β-barrel that includes the base-binding region (BBR; β4–β5) and the phosphate-binding cassette (PBC) critical for cGMP recognition (Fig. 1, D and F). The PBC and α-subdomain are prone to structural changes upon binding of cGMP. In the apo form, the N-terminal helices (i.e., N3A) are primarily in the “in” conformation, and the PBC and the C-terminal helices (i.e., αB and αC) adopt mainly the “out” conformation (Fig. 1, D and F; red). Upon binding of cGMP, the N3A adopts an “out” conformation, whereas the PBC and C-terminal helices are brought “in” to stabilize the cGMP binding (Fig. 1, D and F; green). Both hB and pfD exist in an equilibrium of inactive and active states in the apo form, which is coupled to the cGMP-binding equilibrium to result in a four-state thermodynamic cycle (Fig. 1H).
Although structurally homologous, a critical difference between pfD and hB is the cGMP capping mechanism (Fig. 1, C, D, and F) (8, 12, 13). In pfD, the cGMP base is capped by R484 in the PBC, which further interacts with Q532 and D533 in αC to bridge the PBC to the C-terminal helix, hence forming a capping triad (Fig. 1D) (8). In hB, the cGMP capping residue, Y351, belongs to αC rather than the PBC (Fig. 1F) (12, 13). Besides the differential capping mechanism, the critical cGMP-anchoring interactions by residues in the PBC and BBR are similar in pfD and hB, exhibiting conserved residues and structure (Fig. 1, E, G, and I).
Here, we map the allosteric networks of both pfD and hB and comparatively analyze them to identify unique elements that differentiate pfPKG from hPKG. NMR methods are best suited to investigate such allosteric networks, as NMR spectroscopy provides atomic-level resolution and sensitivity to dynamics (17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27). To this end, we use comparative chemical shift covariance analyses (CHESCA), which identify subsets of allosterically coupled residues within a domain by monitoring how they respond to a targeted library of allosteric perturbations (28). Since our objective is to define the allosteric networks related to kinase activity, we selected as CHESCA perturbations cGMP analogs that result in a spectrum of kinase activities arising from sampling of different populations of inactive and active states. In addition, to dissect the role of the lid in controlling the allosteric networks, the CHESCA were extended to lidless mutants of pfD and hB (i.e., R484A and Y351A, respectively) (8, 12) and compared with the results of the respective WT CHESCAs.
Based on the aforementioned comparative CHESCA, combined with molecular dynamics (MD) simulations, chemical shift projection analysis (CHESPA), and internal dynamics measurements, our results reveal critical differences between pfD and hB in the wiring of common and essential allosteric elements, including the PBC, β2–β3 loop, N3A, BBR, and C-terminal helices. One of the most significant, yet unexpected, differences is how the BBR is wired to the remaining allosteric network in pfD versus hB, which provides valuable insight to address the selective inhibition of pfPKG versus hPKG and other cyclic nucleotide monophosphate (cNMP)-binding proteins.
Results
Mapping the allosteric networks of the essential CBDs in pfPKG and hPKG through CHESCA
We mapped through CHESCA the allosteric networks of the critical CBDs controlling activation of pfPKG and hPKG, denoted here as pfD and hB, respectively. To enable an unbiased pfD versus hB comparison, both CHESCA relied on the same set of perturbations, that is, the endogenous allosteric effector cGMP (Kd = 51 for pfD (14); 108 nM for hB (16)) and its two phosphorothioate diastereoisomers, Rp-cGMPS (Kd = 66 μM for pfD [Fig. S1B]; 63 μM for hB [Fig. S2A]) and Sp-cGMPS (Kd = 47 μM for hB, Fig. S2B), referred to here as Rp and Sp, respectively. Rp and Sp perturb the hydrogen bonds between the cGMP phosphate and PBC residues that are conserved in pfD and hB (Fig. 1, E and G). Hence, pfPKG and hPKG respond similarly to these cGMP analogs, with Rp acting as a partial agonist/inhibitor and Sp serving as an agonist for both kinases (29, 30, 31) (Fig. 2A). The ability of Rp and Sp to span a wide range of kinase activations at saturation makes these cGMP analogs, together with cGMP and the apo domains, ideally suited as a common perturbation set for the pfPKG and hPKG CHESCAs. All ligands were brought to saturation (Figs. S1 and S2) prior to measuring the chemical shifts utilized in the CHESCA to ensure that pfD and hB differences in CHESCA patterns are not biased by the different cNMP affinities of these domains (8, 14). Using the apo, cGMP, Rp, and Sp set, we then implemented CHESCA for WT pfD and hB as well as their respective lidless mutants R484A pfD and Y351A hB (Fig. 3, A and B).
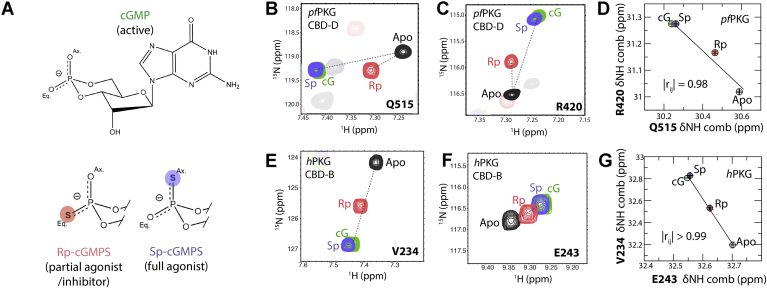
Figure 2.
cGMP analogs used as a perturbation set for the chemical shift covariance analyses (CHESCA).A, covalent structure of cGMP and modifications of cGMP at the ribose-phosphate moiety (Rp-cGMPS and Sp-cGMPS) utilized as a perturbation set to modulate the active–inactive conformational equilibrium of the CBDs. B and C, overlay of 15N–1H HSQC spectra of apo (black), Rp-cGMPS-bound (red), Sp-cGMPS-bound (purple), and cGMP-bound (green) pfD zoomed into Q515 (B) and R420 (C). D, inter-residue combined chemical shift correlations between Q515 and R420 from (B) and (C). E and F, similar to (B) and (C) but for hB residues V234 (E) and E243 (F). G, inter-residue amide 1H–15N combined chemical shift correlations between V234 and E243 from (E) and (F). CBD, cGMP-binding domain; HSQC, heteronuclear single quantum coherence.
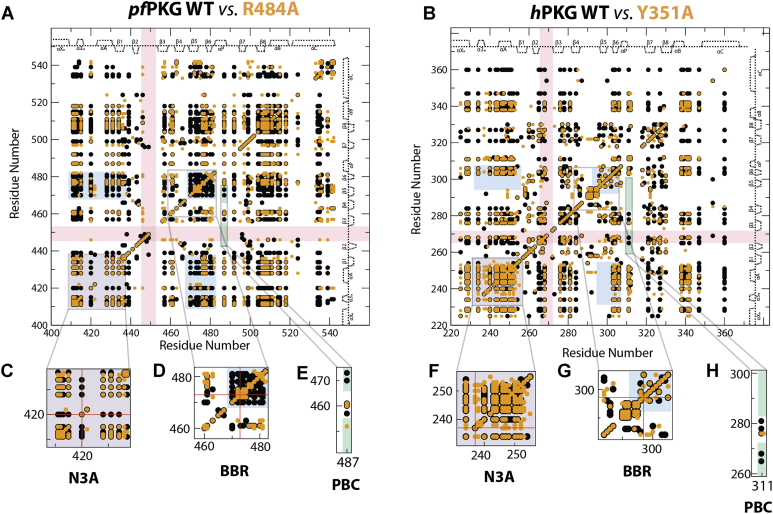
Figure 3.
CHESCA correlation matrices for WT and related capping residue mutants.A, pfPKG and B, hPKG. The cutoff value for the Pearson correlation coefficient was set to 0.95. Color highlights indicate regions that exhibit different patterns of correlations between pfPKG versus hPKG (purple for the N3A; pink for the β2–β3 loop; cyan for the BBR; and green for the PBC). C, expansion of the N3A region from the pfD CHESCA correlation matrix. D, expansion of the BBR region from the pfD CHESCA correlation matrix. E, expansion of PBC residues and their correlations with the BBR and β2–β3 loop of pfD. F, similar to (C) but for hB. G, similar to (D) but for hB. H, similar to (E) but for hB. BBR, base-binding region; CHESCA, chemical shift covariance analyses; hPKG, human cGMP-dependent protein kinase; N3A, N-terminal helix bundle; PBC, phosphate-binding cassette; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase.
CHESCA reveals multiple differences between the allosteric networks of the pfD and hB domains and their lid dependence despite their structural similarities. The comparative pfD versus hB analysis of the allosteric networks mapped through CHESCA for the WT and lidless mutant domains (Fig. 3) reveals several marked yet unexpected pfD versus hB differences in the allostery of these two domains despite their structural similarities (Fig. 1, E, G, and I). Each dot in the CHESCA matrices (Fig. 3) denotes a residue pair exhibiting highly correlated chemical shift variations (Pearson correlation coefficient of >0.95). For example, in pfD, residues Q515 and R420 respond similarly to the perturbation set (Fig. 2, B and C), resulting in a linear inter-residue chemical shift correlation (Fig. 2D), which suggests that these two residues are allosterically coupled. This is because residues that belong to the same allosteric network respond in a similar manner to the CHESCA perturbations, which in the fast exchange regime leads to linearly correlated perturbation-dependent chemical shift changes, as previously shown (28). A similar example is provided by the hB residues V234 and E243 (Fig. 2, E–G), again pointing to another inter-residue coupling. Notably, both correlations (Fig. 2, D and G) rank the cGMP analogs in agreement with the relative degrees of kinase activation, with the Rp partial agonist falling in between the inactive apo and the full agonists (i.e., Sp and cGMP; Fig. 2A), which suggests that the corresponding couplings are relevant for the control of kinase activation.
While both pfD and hB WT matrices (Fig. 3) show correlations throughout the respective domains, selected subregions exhibit notable pfD versus hB differences despite a high degree of sequence homology (Figs. S3 and S4). For example, fewer correlations are observed for the β2–β3 loop in pfD compared with hB (Fig. 3, A and B; pink highlights). On the other hand, the BBR shows more correlations in pfD compared with hB. This applies to both correlations within the BBR and between the BBR and other regions, such as the N3A and BC helices (Fig. 3, A, B, D, and G; blue highlights). Additional distinct pfD versus hB differences are observed for other structurally conserved CBD elements, such as the PBC (Fig. 3, A, B, E, and H) and the N3A (Fig. 3, A, B, C, and F) motifs. Interestingly, the pfD versus hB CHESCA differentials are not limited to the WT correlations but extend also to how such correlations depend on the lid deletion (Fig. 3). In the following sections, we will analyze such subregion-specific CHESCA differences in detail.
pfD versus hB differences in N3A CHESCA patterns reveal differential α-subdomain dynamics in pfPKG versus hPKG
The lid removal through the R484A mutation causes an unexpected loss of intra-N3A correlations in pfD (Fig. 3C). For example, the correlations of R420 with other residues in the N3A are clearly detected in the WT pfD but lost in the R484A mutant (Figs. 3C and 4, A–D). The linear correlations observed for WT pfD (Fig. 4, A and B) are suggestive of a simple active–inactive two-state equilibrium, whereas the nonlinear plots observed for the R484A pfD mutant (Fig. 4, C and D) point to deviations from a two-state equilibrium, suggesting the sampling of additional conformations distinct from the native inactive and active states. The additional conformation is unlikely to arise from the N3A motif itself, which typically adopts a conserved helix–turn–helix arrangement and is subject to rigid-body movements upon cNMP binding (32). However, the chemical shifts of N3A residues report also on changes in the conformation of adjacent structural elements. Specifically, the R420 residue is located at the interface between the N3A and BC helices (Fig. 4E), and therefore, it senses the relative orientation of these two structural units. Hence, a simple but effective explanation of the loss of R420 N3A correlations upon lid deletion (Figs. 3C and 4, A–D) is that in WT pfD the lid ensures that the C-terminal helices are locked in the “in” conformation upon cGMP binding, forcing the N3A in the “out” orientation (Fig. 4F), which is also supported by similarity measurements (SMs) performed on MD simulations of WT pfD (29). In the capping residue mutant (i.e., R484A), on the other hand, the C-terminal helices are no longer stabilized in the “in” conformation and become dynamic, equilibrating between “in” and “out” conformations (Fig. 4G), as confirmed by CHESPA of R484A:cGMP (29). This implies that a third conformation is sampled by the R484A mutant, that is, the N3A “out” and BC “out” state (Fig. 4G), which is absent in the WT, thus explaining why the removal of the lid results in the loss of the correlations detected for R420 in WT pfD (Figs. 3C and 4, A–D).
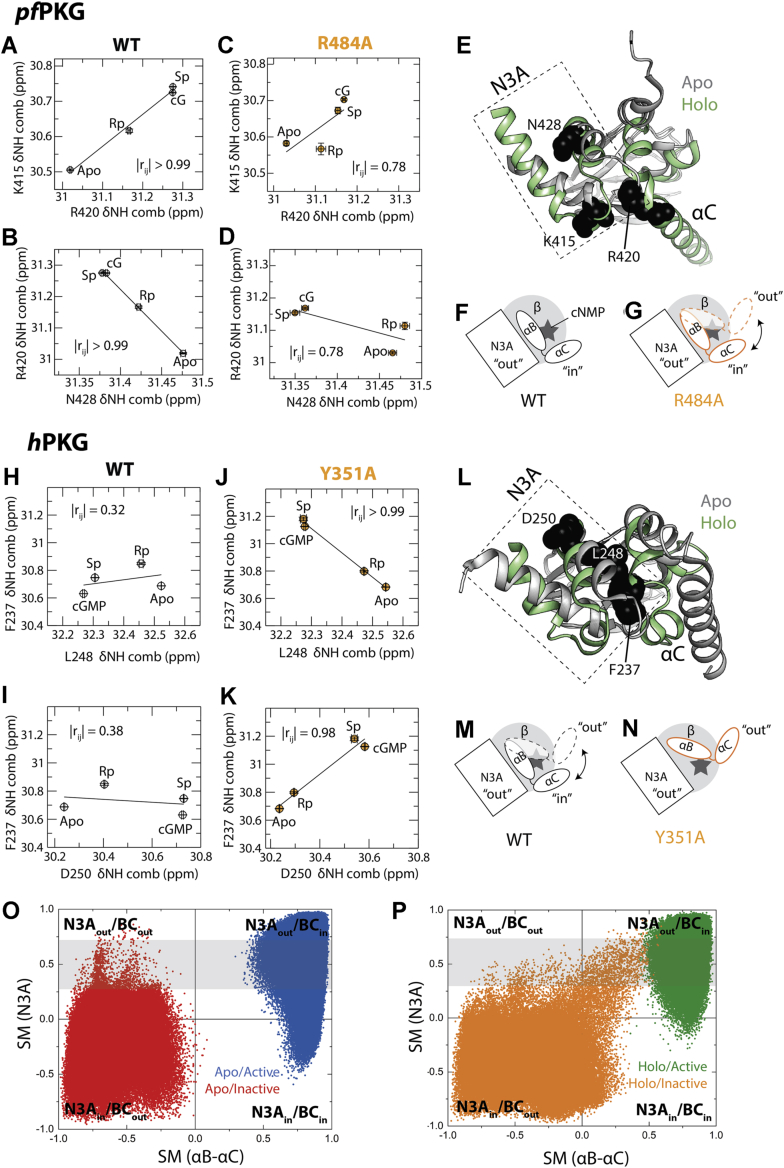
Figure 4.
N3A residues sense different conformational states of the C-terminal helices in pfPKG versus hPKG.A and B, pairwise inter-residue correlation plots for N3A residues in WT pfD. C and D, pairwise inter-residue correlation plots of the same residues from (A) and (B) but for the R484A mutant of pfD. E, residues from (A–D) mapped onto the pfD apo structure (black spheres). These residues span the interface between the N3A and C-terminal helices. F and G, schematics showing the conformations of the C-terminal helices when pfD is bound to a cNMP (gray star), and N3A adopts the “out” position. “β” refers to the β-barrel, represented by a gray circle. For WT (F), the C-terminal helices are locked in the “in” position (29), whereas for the R484A (G), the C-terminal helices sample both “in” and “out” positions because of the loss of the cGMP capping residue (29). H and I, similar to (A) and (B) but for hB. J and K, similar to (C) and (D) but for the hB Y351A mutant. L, similar to (E) but for hB. M and N, similar to (F) and (G) but for hB. For WT (M), the C-terminal helices sample both the “in” and “out” positions, whereas for Y351A (N), the conformational equilibrium of the C-terminal helices shifts to the “out” position because of the loss of the capping residue (Fig. S1). O, N3A versus αB–αC similarity measure (SM) plots for the apo/inactive (red) and apo/active (blue) MD simulations for WT hB. Each quadrant depicts different combinations of conformations for the N3A and αB–αC regions (i.e., “in” or “out”). P, similar to (O) but for the holo/inactive (orange) and holo/active (green) MD simulations. The gray highlights indicate that when the N3A is in the “out” orientation, the B– helices oscillate between the “in” and “out” states. hPKG, human cGMP-dependent protein kinase; MD, molecular dynamics; N3A, N-terminal helix bundle; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase.
A completely different CHESCA pattern is observed for the N3A motif of hB (Fig. 3F). In this domain, a gain of correlation is observed upon going from the WT to the lidless mutant (Y351A). The correlations of F237 with other residues within the N3A are absent in the WT hB but present in the Y351A mutant (Figs. 3F and 4, H–K). Similar to the R420 residue of pfD, the location of F237 of hB allows it to sense the orientation of the BC helices relative to the N3A unit (Fig. 4L), suggesting that in WT hB, the C-terminal helices are dynamic and equilibrate between “in” and “out” conformations, whereas the N3A maintains the “out” arrangement typical of cGMP-bound conformations (Fig. 4M). This hypothesis is supported by previous 15N-relaxation measurements (15), showing enhanced ps–ns and ms–μs dynamics in the lid region of cGMP-bound hB, as well as by MD simulations for the allosteric cycle of WT hB (Figs. 1H, 4, O and P, and S5).
Based on the MD trajectories, we computed SMs, which indicate the conformational or structural similarity, whereby an SM value close to 1 indicates similarity to the active state (i.e., N3Aout or BCin), whereas an SM value close to −1 indicates similarity to the inactive state (i.e., N3Ain or BCout). Each quadrant in the SM plots (Fig. 4, O and P) corresponds to different combinations of the in/out conformational states of the N3A and BC helices. Notably, in the top quadrants, where the N3A adopts the “out” conformation, the BC helices sample both the “in” and the “out” conformations (Fig. 4, O and P; gray highlights), supporting our hypothesis on the conformational dynamics of the BC helices in WT hB. The pfD SM values (29) show that this happens only in hB (Fig. 4M) and not in pfD (Fig. 4F). In the absence of the lid, the C-terminal helices are not recruited to the “in” conformation (Fig. 4N), as is also confirmed by CHESPA of Y351A:cGMP (Fig. S6). Hence, in the lidless hB mutant, the sampling of the third state observed in the WT hB (i.e., N3A “out” and BC “in”) is largely lost, explaining the appearance of CHESCA correlations for F237 upon going from WT to mutant hB. Our MD simulations suggest that these pfD versus hB differences in the lid dynamics when N3A is “out” may also apply to the apo samples (Fig. 4O) (29).
Overall, the pfD versus hB differences in the N3A CHESCA correlations and their lid dependence reveal a reversed dynamic pattern for the adjacent BC helices (Fig. 4, F and G versus Fig. 4, M and N).
The PBC couples to the BBR in pfPKG but to the β2–β3 loop in hPKG
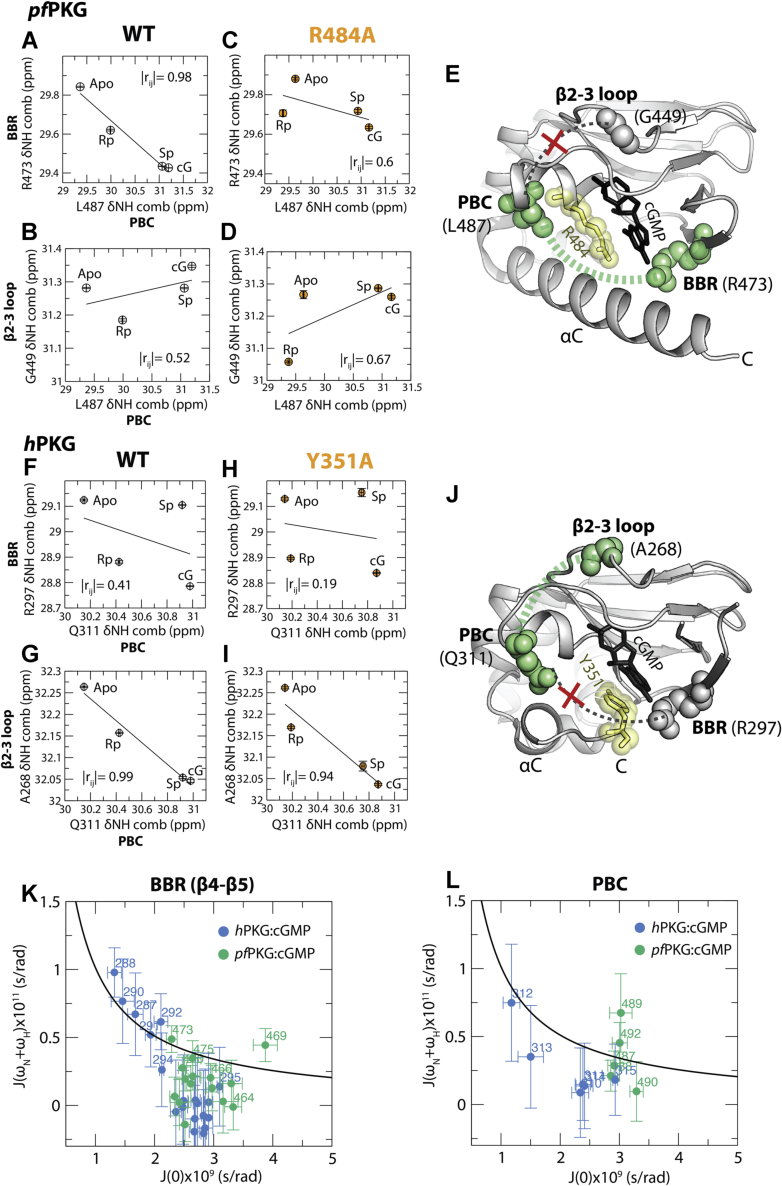
The PBC is a critical region for recognizing and binding cGMP and its analogs (Fig. 1, E and G). Hence, information on how it allosterically couples with other regions of the domain could provide important clues to selectively target pfPKG versus hPKG. Specifically, we evaluated the coupling of the PBC to another critical cGMP-binding region, the BBR, and to the β2–β3 loop, located adjacent to the PBC (Fig. 1, E and G). Although highly similar in structure (Fig. 1I), these regions exhibit clear differences in how they are correlated with other parts of the domain in pfPKG versus hPKG, as mentioned previously. In addition, the PBC in WT pfD exhibits several correlations with residues in the BBR, which are lost in the R484A pfD mutant (Fig. 3E). For example, this is the case for PBC residue L487 (Fig. 3A). Surprisingly, L487 does not exhibit any detectable correlation with the adjacent β2–β3 loop in both WT and the mutant pfD (Figs. 3E and 5, A–D). These observations suggest that in pfD, the PBC is preferentially coupled to the BBR rather than the β2–β3 loop, and that the PBC–BBR allosteric coupling relies on R484 (Fig. 5E).
Figure 5.
The PBC exhibits different correlation patterns with the BBR and β2–β3 loop in pfPKG versus hPKG.A and B, pairwise inter-residue correlation plots of PBC residue (L487) with (A) BBR residue (R473) and (B) β2–β3 loop residue (G449) in WT pfD. C and D, similar to (A) and (B) but for R484A mutant pfD. E, residues from (A–D) mapped onto the pfD holo structure (spheres). The PBC exhibits high correlation with the BBR and low correlation with the β2–β3 loop. Yellow stick/spheres indicate the cGMP capping residue (i.e., R484). F and G, pairwise inter-residue correlation plots of PBC residue (Q311) with (F) BBR residue (R297) and (G) β2–β3 loop residue (G267) in WT hB. H and I, similar to (F) and (G) but for Y351A mutant hB. J, residues from (F–I) mapped onto the hB holo structure (spheres). Unlike pfD, the PBC exhibits high correlation with the β2–β3 loop and low correlation with the BBR. Yellow sticks/spheres indicate the cGMP capping residue (i.e., Y351). K and L, reduced spectral densities for cGMP-bound pfD. K, 2D plot of J(0) versus J(ωH + ωN) for BBR residues of BBR in hB:cGMP (blue) and pfD:cGMP (green). The expected values for a rigid rotor are shown by the black plot. L, similar to (K) but for PBC residues. BBR, base-binding region; hPKG, human cGMP-dependent protein kinase; PBC, phosphate-binding cassette; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase.
Interestingly, for the hB domain, the pattern of PBC–BBR/β2–β3 allosteric coupling is reversed compared with pfD. The PBC residue Q311 in hB does not exhibit correlations with the BBR (i.e., β4–β5) in both WT and mutant hB, whereas it correlates with the adjacent β2–β3 loop in WT but not in the mutant hB (Figs. 3H and 5, F–I), suggesting a scenario opposite to pfD, that is, the PBC is preferentially coupled to the β2–β3 loop rather than the BBR (Fig. 5J). These pfD versus hB differences revealed by CHESCA at the level of allosteric coupling between the PBC and BBR may reflect dissimilarities in the capping mechanisms. For pfD, the capping residue (i.e., R484) is located in the PBC and extends toward the BBR to cap the cGMP guanine base, potentially acting as a bridge in the coupling of the PBC and BBR. On the other hand, the capping residue for hB (i.e., Y351) is located in the αC helix, and thus hB lacks a bridge to couple the PBC to the BBR. In hB, the PBC is then available to couple to the adjacent β2–β3 loop, similar to what was previously observed for other eukaryotic CBDs, such as those of exchange factor directly activated by cAMP, PKA, and hyperpolarization-activated cyclic nucleotide-gated channel (28, 33, 34, 35).
To support the findings on the difference in the coupling of the PBC to the BBR and β2–β3 loop, distances between the PBC and either the BBR or β2–β3 loop were measured from the MD simulations of the pfD and hB in each of the four states of the allosteric cycle (i.e., apo/active, holo/active, holo/inactive, and apo/inactive; Figs. 1H and S7). The PBC:BBR distance measurements show that pfD overall exhibits narrower distance distributions suggestive of a stable PBC–BBR positioning compared with hB (Fig. S7A). Also, pfD exhibits significantly shorter PBC:BBR distances than hB for the holo/inactive and apo/inactive states (Fig. S7A), supporting the proposal of tighter PBC–BBR coupling in pfD versus hB (Fig. 5E). On the other hand, the PBC:β2–β3 distance measurements show that hB overall exhibits both narrower distributions and shorter distances than pfD (Fig. S7B), in agreement with the CHESCA results on the coupling of PBC–β2–β3 in hB (Fig. 5J).
To further probe the PBC–BBR allosteric coupling in pfD versus hB, we complemented our comparative CHESCA with CHESPA (36) in which the NMR chemical shift changes caused by Rp are analyzed relative to the apo and cGMP-bound states (Fig. S8). The Rp phosphorothioate modification directly perturbs the PBC, which interacts with the cyclic phosphate, but not the BBR, which binds the base of cyclic nucleotides instead. Hence, the BBR chemical shift perturbations resulting from the replacement of cGMP with Rp reflect primarily allosteric effects pertaining to how the PBC is coupled to the BBR. As expected based on our CHESCA results, Rp results in greater chemical shift variations in the BBR region for pfD compared with hB (Fig. S8, A and D), consistent with the tighter PBC–BBR allosteric coupling in pfD versus hB. Furthermore, negative fractional activation values (X) are consistently observed for the pfD BBR (Fig. S8B), in agreement with the reduced kinase activation measured for Rp relative to cGMP, thus confirming the functional relevance of the PBC–BBR allosteric coupling unique to pfD.
To further examine the pfPKG versus hPKG difference at the level of the BBR, we complemented our comparative CHESCA and CHESPA with 15N relaxation rates that report on the internal dynamics of pfD and hB (Figs. 5, K and L, S9 and S10) (15). The corresponding reduced spectral densities (RSDs) were computed, revealing higher J(ωH + ωN) and lower J(0) values for selected BBR and PBC regions of hB compared with pfD (Fig. 5, K and L). These observations suggest enhanced BBR/PBC ps–ns dynamics for hB (i.e., quenched ps–ns dynamics for pfD) even in the presence of saturating amounts of cGMP (Fig. 5, K and L). The root mean square fluctuations computed from the MD simulations also show that the BBR and PBC regions are more flexible in hB than pfD, which is a general feature for all four states of the proteins (Fig. S11). The enhanced dynamics observed for both the PBC and BBR in hB relative to pfD are consistent with the reduced coupling between these two cNMP recognition elements in hB versus pfD. Overall, the unique coupling between the PBC and BBR in pfD is significant not only because it offers an opportunity to engineer pfD versus hB selectivity through guanine-based modifications but also because it implies that the pfD BBR is itself part of an extended functional allosteric network, as discussed in the next section.
The BBR is part of the allosteric network controlling kinase activation in pfPKG but not in hPKG
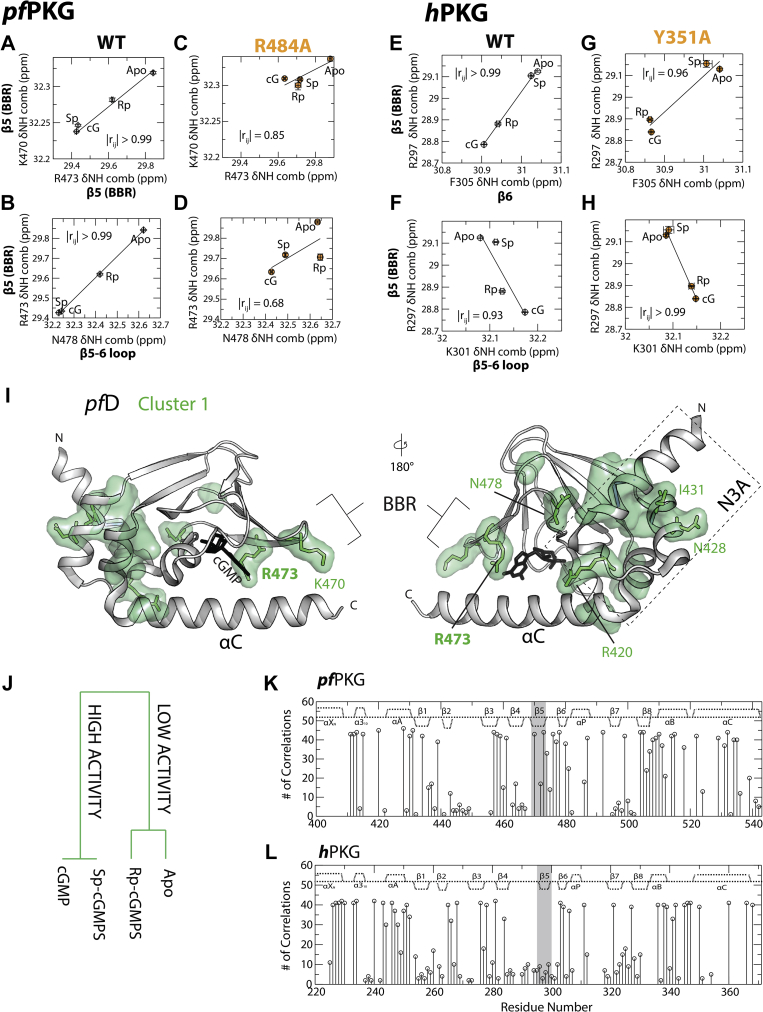
The BBR is a crucial region for binding cGMP and includes highly conserved residues (Fig. 1, E, G, and I). Although this region is not subject to significant pfD versus hB structural changes (Fig. 1I), the CHESCA correlation matrices reveal significant pfD and hB differences at the BBR (Fig. 3, A, B, D, and G). We specifically focused on the conserved Arg residue within the BBR β5 strand, that is, R473 for pfD and R297 for hB (Fig. 1, E and G), as this side chain interacts directly with the guanine base of cGMP (8). First, in pfD, R473 exhibits significant correlations with adjacent residues in β5 and the β5–β6 loop (Figs. 3D and 6, A and B), which are lost in the R484A mutant (Fig. 6, C and D). R473 also exhibits correlations with other parts of the WT pfD domain, such as the N3A, which are again lost in the R484A mutant (Figs. 3A and S12). On the other hand, in hB, the homologous R297 residue exhibits significant correlations with residues in the β5–β6 loop and β6 strand in both WT and the lidless Y351A mutant (Figs. 3G and 6, E–H). Hence, the lid dependence of such BBR correlations differs in pfD and hB. Another pfD versus hB difference pertaining to the BBR is that R297 of hB does not correlate with the N3A in either WT or the lidless mutant (Figs. 3B and S13). Such marked pfD versus hB differences in allosteric couplings suggest distinct roles of the conserved Arg in the BBR of pfD versus hB, despite their structural similarities (Fig. 1, E, G and I).
Figure 6.
The conserved arginine in the BBR is part of an allosteric network for pfPKG but not for hPKG.A and B, pairwise inter-residue correlation plots of the conserved BBR residue and adjacent residues (R473, K470, and N478) in WT pfD. C and D, similar to (A) and (B) but for the R484A mutant pfD. E and F, pairwise inter-residue correlation plots of the conserved BBR residues and nearby residues (R297, K301, and F305) in WT hB. G and H, similar to (E) and (F) but for the Y351A mutant hB. I, map of residues from cluster 1 for pfD shown through a green surface representation. Residues from (A–D) and Fig. S14B are represented through green sticks and are part of cluster 1. J, dendrogram of the agglomerative clustering of the four CHESCA perturbation states (i.e., apo, cGMP-analog-bound, and cGMP-analog-bound states) using residues from cluster 1 (Fig. S14B) of pfD. K and L, residue-specific total numbers of correlations (|rij| > 0.95) in (K) pfD and (L) hB. The β5 strand in the BBR exhibits significant differences in its number of correlations for hPKG versus pfPKG. BBR, base-binding region; CHESCA, chemical shift covariance analyses; hPKG, human cGMP-dependent protein kinase; pfPKG, Plasmodium falciparum cGMP-dependent protein kinase.
To further characterize these differences, agglomerative clustering analysis was performed to more systematically establish whether the conserved BBR Arg in pfD and hB is part of an allosteric network (Figs. 6, I and J, S14 and S15). The dendrogram for the cluster of highly correlated pfD residues that includes R473 is shown in Fig. S14B. Such a dendrogram provides information on the linked residues within the cluster, which are mapped on the structure of cGMP-bound pfD in Figure 6I. Yet, the dendrogram of Fig. S14B does not directly report on the function associated with the specific residue cluster. In order to assign a function to the cluster, a complementary agglomerative clustering analysis was performed relative to the four states (i.e., apo, cGMP-bound, Sp-bound, and Rp-bound states) instead of the residues, using only the chemical shift submatrix containing the residues in the cluster shown in Fig. S14B. The resulting state dendrogram (Fig. 6J) reveals two nodes that clearly separate the states with high kinase activity (i.e., cGMP-bound and Sp-bound states) from those with lower kinase activity (i.e., apo and Rp-bound states), suggesting that the BBR-spanning cluster of Fig. S14B functions as an allosteric activation network for pfD.
A similar agglomerative clustering analysis was performed also for hB (Fig. S15). However, in this case, unlike pfD, the conserved R297 in the BBR of hB is not part of any identified allosteric cluster (Fig. S15). This pfPKG versus hPKG differential is further confirmed by the computation of the residue-specific total numbers of CHESCA correlations in pfD and hB (Fig. 6, K and L). Relative to hB, pfD exhibits a markedly higher number of CHESCA correlations involving β5 within the BBR (Fig. 6, K and L; gray region). This observation further corroborates the notion that the BBR plays a critical role in allosterically coordinating other regions within pfD but not hB. Overall, these results suggest that in pfD but not in hB, the BBR is critical not only to recognize the guanine base of cGMP but also to allosterically control kinase activation.
Discussion
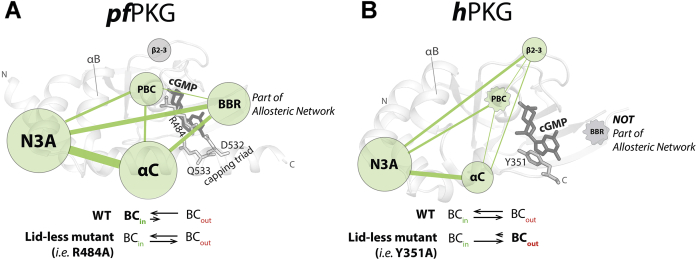
Comparative NMR CHESCA, supported by MD simulations, CHESPA, and backbone dynamic measurements, have revealed unexpected, yet critical, differences in the allosteric networks of the pfD versus hB domains (Fig. 7), despite their structural similarity. At the center of such allosteric rewiring upon going from pfPKG to hPKG is the PBC, which is the primary cyclic phosphate recognition element in both kinases. The PBC switches from being coupled to the β2–β3 loop in hB to being coupled to the BBR in pfD (Fig. 7).
Figure 7.
Comparison of allosteric networks in pfD versus hB.A, in pfD, the BBR is part of the allosteric network. It exhibits correlations with the PBC, where the cGMP capping residue R484 is present. The PBC does not exhibit significant correlations with the β2–β3 loop. The capping triad formed by R484 and Q532–D533 allows for additional interactions between the PBC and αC. In the holo form and when the N3A is in the “out” conformation, the αBC (i.e., BC) helices in WT pfD are stable in the “in” conformation. The lidless R484A mutation causes the BC helices to sample both “in” and “out” conformations. Each region (i.e., N3A, β2–β3, BBR, PBC, and αC) is represented as a node. The size of each node corresponds to the total number of correlations formed with other regions and within itself above the correlation cutoff (i.e., 0.95). The thickness of each line connecting the regions correspond to the total number of correlations formed between the two regions above the correlation cutoff (i.e., 0.95). A line is shown only when the total number of correlations between the two regions exceeds 2. B, in hB, the BBR is not part of the allosteric network. It does not exhibit significant correlations with the PBC, which correlates with the β2–β3 loop instead. The wavy outline of the nodes for the PBC and BBR depict the enhanced ps–ns dynamics in these regions. In the holo form, and when the N3A is in the “out” conformation, the αBC (i.e., BC) helices in WT hB are dynamic and samples both “in” and “out” conformations. In the lidless Y351A mutant, the BC helices are mainly in the “out” conformation relative to the WT. BBR, base-binding region; N3A, N-terminal helix bundle; PBC, phosphate-binding cassette.
Furthermore, in pfD but not hB, the BBR is part of an extended functional allosteric network spanning not only the PBC but also the α-subdomain (i.e., N3A and BC helices). Hence, one of the most unique and notable features of pfPKG emerging from our comparative allosteric analyses is that the BBR is wired differently to the rest of the allosteric network in pfD versus hB. Such pfD versus hB differences extend also to how the allosteric networks of the BBR and N3A motives depend on the deletion of the lid residue side chains.
The drastic pfD versus hB rewiring of allosteric networks involving the BBR likely reflects the different location of the base capping residue side chains in the two domains, that is, Y351 in the αC helix of hB versus R484 in the PBC of pfD (Fig. 1, D and F), as well as marked differences in the underlying dynamics of the pfD and hB domains (Fig. 7). The latter exhibits enhanced ps–ns dynamics relative to the former at the two critical cNMP recognition elements, that is, the PBC and BBR, and the BC helices are subject to differential dynamics in pfD versus hB (Fig. 7). In WT hPKG, the BC helices sample both the “in” and “out” conformations, and this conformational equilibrium shifts toward the “out” orientation upon deletion of the lid (Fig. 7B). On the contrary, in WT pfPKG, the BC helices sample primarily the “in” conformation, and it is only upon deletion of the lid that both “in” and “out” orientations are sampled (Fig. 7A).
Despite the pfD versus hB differences in lid location and dynamics, it is notable that some of the most remarkable rewiring of allosteric networks occurs at structural elements that are highly conserved in hPKG versus pfPKG, such as the BBR (Fig. 1I). For example, the Arg in the β5 strand of the BBR (i.e., R473 in pfD and R297 in hB) forms highly conserved hydrogen bonds to the guanine base of cGMP (Fig. 1, E, G, and I), and yet this very same Arg is also the residue exhibiting some of the most striking hPKG versus pfPKG differences, as discussed previously. Hence, the allosteric rewiring reported here for pfD versus hB was to a large extend unexpected.
The allosteric differences revealed here are likely not limited only to the CBDs of pfPKG and hPKG. The BBR being part of an allosteric network is a feature unique to pfPKG not only relative to hPKG but also to other human cNMP-binding domains, including those of the cAMP-dependent protein kinase (PKA) (34), the exchange factor directly activated by cAMP (28), and hyperpolarization-activated cyclic nucleotide-gated channel (35), in which the BBR is not fully integrated in allosteric clusters. These BBR differences are consistent with the fact that, although the β4 and β5 strands in the BBR define a conserved base recognition element of CBDs, the β4–β5 loop is hypervariable among cNMP-binding domains (37).
The notion emerging here that although the BBR is a critical and conserved base recognition element in CBDs, it is part of a functional allosteric network in pfPKG but not in human cNMP-dependent systems (Fig. 7) bears implications for the selective targeting of pfPKG. For example, it has previously been shown that the base-modified cGMP analog 8-pCPT-cGMP is a partial inhibitor of pfPKG (14) but a full activator of hPKG (38). Our results provide a viable explanation for how 8-pCPT-cGMP selectively inhibits pfPKG. Considering that the BBR is the primary base recognition element (39), base modifications such as 8-pCPT are expected to perturb the BBR (14). Since the BBR is part of a functional allosteric network in pfD, the BBR perturbation caused by 8-pCPT will at least partially disrupt the wiring of residues responsible for activation of pfPKG and hence result in inhibition. On the other hand, the BBR is not part of an allosteric network in hB, hence its direct perturbation is not anticipated to critically affect the activation of hPKG. In addition, the dynamic nature of the BC helices in hPKG could allow for greater tolerance of the active conformation toward the bulky pCPT substituent compared with the more rigid BC helices in pfPKG.
It is important to note the possibility that a domain may not rely solely on a single allosteric network and that multiple concurrent allosteric pathways may coexist, as previously pointed out (40). For example, the N3A motif may be controlled by cNMP binding not only through the lid in the BC helices but also via a PBC–B-helix coupling mediated by a hydrophobic hinge, which has been shown to serve as an essential allosteric pathway in multiple CBDs (3, 6, 35, 41, 42).
Overall, our comparative CHESCA of essential CBDs of pfPKG versus hPKG have revealed unanticipated differences in the wiring of the respective allosteric networks spanning conserved structural elements such as the BC helices, β2–β3 loop, PBC, and BBR. Our results show that allosteric networks may differ even when structures are similar. Hence, mapping allosteric networks is a critical step toward developing selective inhibitors for pfPKG, an attractive antimalarial target with possibly minimal side effects.
Experimental procedures
Expression and purification
P. falciparum PKG CBD-D (401–542) WT and R484A were expressed and purified following protocols published previously (14). Similarly, Homo sapiens PKG Iβ CBD-B (219–369) WT and Y351A were expressed and purified as published previously (15).
NMR spectroscopy
NMR experiments were performed on a Bruker Avance 700-MHz spectrometer equipped with a 5-mm TCI cryoprobe. For the chemical shift–based analyses, two-dimensional 15N–1H heteronuclear single quantum coherence (HSQC) spectra were acquired for apo, cGMP (Millipore–Sigma)-bound, Rp-cGMPS (Biolog)-bound, and Sp-cGMPS (Biolog)-bound pfD or hB. Experiments were recorded using eight scans, a recycle delay of 1.0 s with 128 and 1024 complex points, and spectral widths of 15.9 and 31.8 ppm for the 1H and 15N dimensions, respectively. The acquired spectra were processed using NMRPipe (NIST) (43) and Topspin (Bruker) and analyzed using Sparky (NMRFAM) (44).
For pfPKG, all NMR experiments were acquired at 306 K in the NMR buffer (50 mM Tris, pH 7, 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT) with 5% D2O. For WT, a protein concentration of 100 μM was used for all HSQC experiments. Excess amounts (>2 mM) of the cyclic nucleotides were added to the protein to acquire the HSQC spectra of the cGMP-bound, Rp-bound, and Sp-bound pfD in order to achieve saturated binding, thus minimizing the influence of differing binding affinities on the CHESCA. For R484A, protein and cyclic nucleotide concentrations of 70 μM and 3 mM, respectively, were used. The HSQC assignments were transferred from the WT apo to the R484A apo spectra through spectral comparison. To assign the Rp-bound and Sp-bound pfD, the cyclic nucleotides were titrated into the apo protein, and HSQC spectra were acquired at increasing cyclic nucleotide concentrations. Since the on–off binding exchange was fast in the NMR chemical shift timescale, it was possible to visualize the binding and transfer the assignments from the apo to the Rp-bound or Sp-bound crosspeaks. For Sp binding to WT, binding was slow in the NMR chemical shift timescale, hence, Nz-exchange was used to transfer the assignments from the apo to the Sp-bound peaks.
All NMR spectra for the hPKG Iβ(219–369) CBD-B construct were acquired at 306 K in the NMR buffer (50 mM Tris, pH 7, 100 mM NaCl, and 1 mM DTT) with 5% D2O. Two-dimensional (1H,15N) HSQC NMR spectra were acquired for apo, cGMP-bound, Rp-bound, and Sp-bound samples of WT and Y351A-mutant hB. A protein concentration of 20 μM was used for all HSQC samples. All experiments for cGMP-bound, Rp-bound, and Sp-bound hB were performed with cyclic nucleotide concentrations of >2 mM in order to achieve saturated ligand binding, thus minimizing the influence of differing ligand-binding affinities on the results of the analyses. Assignments for the Y351A-mutant HSQC spectra were obtained via a combination of comparisons with the respective assigned WT HSQC spectra and three-dimensional triple-resonance NMR spectra.
Building of binding isotherms and computation of Kd values were performed following the previously published protocols (3).
Chemical shift analyses—CHESCA and CHESPA
An internal reference (i.e., 15N-labeled N-acetylglycine) was used to align the spectra. CHESCA and related chemical shift correlation analyses were performed as previously described (28, 45), using the apo, cGMP-bound, Rp-bound, and Sp-bound states as a perturbation set. Residues exhibiting a frequency spread across the four states (i.e., apo, Rp, Sp, and cGMP) greater than 10 Hz for the 1H dimension and 5 Hz for the 15N dimension were selected for CHESCA (28). Correlations exhibiting an absolute correlation coefficient of >0.95 were used to illustrate the CHESCA correlation matrices. For the agglomerative clustering, the complete-linkage method and a correlation coefficient cutoff of 0.95 were used. CHESPA was performed as described previously (14, 36), with a combined chemical shift difference cutoff value of 0.05 ppm. Combined chemical shift values were computed as:
Relaxation and RSD analyses
Backbone 15N relaxation experiments (R1, R2, and HN-NOE) were performed with 200 μM pfD bound to cGMP (2 mM) and analyzed using protocols described previously (15, 46). RSDs were computed as previously described (15, 46).
MD simulations
Model preparation
Initial models of hPKG Iβ CBD-B (residues 217–351) were assembled starting from the apo structure (Protein Data Bank code: 4KU8: for the apo/inactive state) and the cGMP-bound structure (Protein Data Bank code: 4KU7: for the holo/active state) (12). The initial apo/active structure was extracted from 4KU8, whereas the starting structure for the holo/active state was reconstructed from 4KU7 by adding missing residues 217 to 222 and 287 to 291 from the apo/inactive structure, with residues 223 to 224 and 285 to 286/292 to 293, respectively, used as superimposition points to align the structures for grafting. Then, energy minimization was conducted for the reconstructed region of the reconstructed holo/active structure using the Amber 16 software (47). The apo/active structure was generated by removing the cGMP from the holo/active model. Finally, the holo/inactive structure was obtained by superimposing the apo/inactive and holo/active structures at the β-barrel region, in order to dock the cGMP into the apo/inactive structure. Energy minimizations were then performed for the side chains of the residues in the BBR and PBC in the reconstructed holo/inactive structure.
MD simulation protocols
For a reliable comparison with the MD simulations of pfD (29), all MD simulations for hB were performed using the Amber 16 software (47) on the Shared Hierarchical Academic Research Computing Network following protocols similar to those described previously (29). The parameters for cGMP were generated using the general Amber force field, and the parameters for the protein are from the Amber FF99SBnmr force field. The TIP3P explicit solvent model was employed and added as a rectangular box with a minimum solute-box edge distance of 12 Å. Each state was equilibrated from energy minimization, followed by 100 ps stepwise temperature simulations from 0 to 306 K with 50 K intervals. In the 0 to 100 K range, we employed the constant volume and temperature (NVT) ensemble, whereas in the 100 to 306 K range, we used the constant pressure and temperature (NPT) ensemble, and the main chain atoms of the protein were restrained with a 5 kcal/mol Å2 force constant. To gradually release the restraint on the main chain atoms, three more 40 ps equilibrations were conducted with 3, 1, and 0 kcal/mol Å2 restraints. Finally, three 1000 ns MD simulations were conducted for each state at 306 K with a weak-coupling algorithm in the NPT ensemble. Structures were saved every 10 ps, and in total, 12 μs of MD trajectories were produced for hPKG. The SMs were computed using the following equation:
and the analyses were implemented as previously described (29, 33). Distances between different regions were computed by using the center of mass of the residues in each region. Root mean square fluctuation values for all four states were measured using their initial models as references (overlaying at their Cα atoms).
Construction of allosteric network graphs
Residues involved in pairwise CHESCA correlations with absolute correlation coefficient >0.95 were assigned to specific structural regions, that is, N3A, β2–β3, BBR, PBC, and αC. Then each pair of regions was analyzed to create a count for the number of connections between and within each region, which were then used in Gephi (Gephi.org) (48) to create a network graph.
Data availability
The NMR chemical shift assignments for cGMP-bound pfD and hB have been deposited in the Biological Magnetic Resonance Data Bank with codes 50203 and 51201, respectively. All remaining data are contained within the article and the supporting information or available upon request from the corresponding author: melacin@mcmaster.ca.
Supporting information
This article contains supporting information (29, 49).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Madoka Akimoto, Dr Rashik Ahmed, Karla Martinez, Hebatallah Mohamed, and Katherine Van for helpful discussions.
Author contributions
J. A. B. and G. M. conceptualization; J. A. B. and G. M. methodology; J. A. B. and G. M. validation; J. A. B., B. V. S., J. H., U. B., and G. M. formal analysis; G. M. investigation; J. A. B., B. V. S., and J. H. data curation; J. A. B. and G. M. writing–original draft; J. A. B., B. V. S., J. H., and G. M. writing–review and editing; J. A. B., J. H., and G. M. visualization; G. M. supervision; G. M. project administration; G. M. funding acquisition.
Funding and additional information
Funding was provided by the Canadian Institutes of Health Research (grant no.: 389522 to G. M.) and the Natural Sciences and Engineering Research Council of Canada grant (grant no.: RGPIN-2019-05990; to G. M.).
Edited by Wolfgang Peti
Supporting information
References
- 1.Cohen P. Protein kinases — the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 2.VanSchouwen B., Boulton S., Melacini G. Mutual protein-ligand conformational selection drives cGMP vs. cAMP selectivity in protein kinase G. J. Mol. Biol. 2021;433:167202. doi: 10.1016/j.jmb.2021.167202. [DOI] [PubMed] [Google Scholar]
- 3.Byun J.A., VanSchouwen B., Parikh N., Akimoto M., McNicholl E.T., Melacini G. State-selective frustration as a key driver of allosteric pluripotency. Chem. Sci. 2021;12:11565–11575. doi: 10.1039/d1sc01753e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byun J.A., VanSchouwen B., Akimoto M., Melacini G. Allosteric inhibition explained through conformational ensembles sampling distinct “mixed” states. Comput. Struct. Biotechnol. J. 2020;18:3803–3818. doi: 10.1016/j.csbj.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byun J.A., Akimoto M., VanSchouwen B., Lazarou T.S., Taylor S.S., Melacini G. Allosteric pluripotency as revealed by protein kinase A. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao H., Mohamed H., Boulton S., Huang J., Wang P., Chen H., Zhou J., Luchowska-Stańska U., Jentsch N.G., Armstrong A.L., Magolan J., Yarwood S., Melacini G. Mechanism of action of an EPAC1-selective competitive partial agonist. J. Med. Chem. 2020;63:4762–4775. doi: 10.1021/acs.jmedchem.9b02151. [DOI] [PubMed] [Google Scholar]
- 7.Baker D.A., Deng W. Cyclic GMP-dependent protein kinases in protozoa. Front. Biosci. 2005;10:1229. doi: 10.2741/1615. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.J., Flueck C., Franz E., Sanabria-Figueroa E., Thompson E., Lorenz R., Bertinetti D., Baker D.A., Herberg F.W., Kim C. Crystal structures of the carboxyl cGMP binding domain of the Plasmodium falciparum cGMP-dependent protein kinase reveal a novel capping triad crucial for merozoite egress. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon T.M., Sheehe J.L., Nukareddy P., Nausch L.W., Wohlfahrt J., Matthews D.E., Blumenthal D.K., Dostmann W.R. An N-terminally truncated form of cyclic GMP–dependent protein kinase Iα (PKG Iα) is monomeric and autoinhibited and provides a model for activation. J. Biol. Chem. 2018;293:7916–7929. doi: 10.1074/jbc.RA117.000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W., Parbhu-Patel A., Meyer D.J., Baker D.A. The role of two novel regulatory sites in the activation of the cGMP-dependent protein kinase from Plasmodium falciparum. Biochem. J. 2003;374:559–565. doi: 10.1042/BJ20030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz E., Knape M.J., Herberg F.W. cGMP binding domain D mediates a unique activation mechanism in Plasmodium falciparum PKG. ACS Infect. Dis. 2018;4:415–423. doi: 10.1021/acsinfecdis.7b00222. [DOI] [PubMed] [Google Scholar]
- 12.Huang G.Y., Kim J.J., Reger A.S., Lorenz R., Moon E.W., Zhao C., Casteel D.E., Bertinetti D., Vanschouwen B., Selvaratnam R., Pflugrath J.W., Sankaran B., Melacini G., Herberg F.W., Kim C. Structural basis for cyclic-nucleotide selectivity and cGMP-selective activation of PKG i. Structure. 2014;22:116–124. doi: 10.1016/j.str.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang G.Y., Gerlits O.O., Blakeley M.P., Sankaran B., Kovalevsky A.Y., Kim C. Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: Insights for cGMP-dependent protein kinase agonist design. Biochemistry. 2014;53:6725–6727. doi: 10.1021/bi501012v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byun J.A., Van K., Huang J., Henning P., Franz E., Akimoto M., Herberg F.W., Kim C., Melacini G. Mechanism of allosteric inhibition in the Plasmodium falciparum cGMP-dependent protein kinase. J. Biol. Chem. 2020;295:8480–8491. doi: 10.1074/jbc.RA120.013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanSchouwen B., Selvaratnam R., Giri R., Lorenz R., Herberg F.W., Kim C., Melacini G. Mechanism of cAMP partial agonism in protein kinase G (PKG) J. Biol. Chem. 2015;290:28631–28641. doi: 10.1074/jbc.M115.685305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell J.C., VanSchouwen B., Lorenz R., Sankaran B., Herberg F.W., Melacini G., Kim C. Crystal structure of cGMP-dependent protein kinase Iβ cyclic nucleotide-binding-B domain: Rp-cGMPS complex reveals an apo-like, inactive conformation. FEBS Lett. 2017;591:221–230. doi: 10.1002/1873-3468.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulton S., Melacini G. Advances in NMR methods to map allosteric sites: From models to translation. Chem. Rev. 2016;116:6267–6304. doi: 10.1021/acs.chemrev.5b00718. [DOI] [PubMed] [Google Scholar]
- 18.Leninger M., Sae Her A., Traaseth N.J. Inducing conformational preference of the membrane protein transporter EmrE through conservative mutations. Elife. 2019;8 doi: 10.7554/eLife.48909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark L.D., Dikiy I., Chapman K., Rödström K.E., Aramini J., LeVine M.V., Khelashvili G., Rasmussen S.G., Gardner K.H., Rosenbaum D.M. Ligand modulation of sidechain dynamics in a wild-type human GPCR. Elife. 2017;6 doi: 10.7554/eLife.28505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan C., Bernard D.N., Bafna K., Gagné D., Agarwal P.K., Doucet N. Ligand-induced variations in structural and dynamical properties within an enzyme superfamily. Front. Mol. Biosci. 2018;5:54. doi: 10.3389/fmolb.2018.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Simone A., Richter B., Salvatella X., Vendruscolo M. Toward an accurate determination of free energy landscapes in solution states of proteins. J. Am. Chem. Soc. 2009;131:3810–3811. doi: 10.1021/ja8087295. [DOI] [PubMed] [Google Scholar]
- 22.Dolgova N.V., Yu C., Cvitkovic J.P., Hodak M., Nienaber K.H., Summers K.L., Cotelesage J.J.H., Bernholc J., Kaminski G.A., Pickering I.J., George G.N., Dmitriev O.Y. Binding of copper and cisplatin to Atox1 is mediated by glutathione through the formation of metal–sulfur clusters. Biochemistry. 2017;56:3129–3141. doi: 10.1021/acs.biochem.7b00293. [DOI] [PubMed] [Google Scholar]
- 23.Verkhivker G.M. Making the invisible visible: Toward structural characterization of allosteric states, interaction networks, and allosteric regulatory mechanisms in protein kinases. Curr. Opin. Struct. Biol. 2021;71:71–78. doi: 10.1016/j.sbi.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Meli M., Pagano K., Ragona L., Colombo G. Investigating the dynamic aspects of drug-protein recognition through a combination of MD and NMR analyses: Implications for the development of protein-protein interaction inhibitors. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piserchio A., Long K., Lee K., Kumar E.A., Abzalimov R., Dalby K.N., Ghose R. Structural dynamics of the complex of calmodulin with a minimal functional construct of eukaryotic elongation factor 2 kinase and the role of Thr348 autophosphorylation. Protein Sci. 2021;30:1221–1234. doi: 10.1002/pro.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Amico R.N., Bosken Y.K., O'Rourke K.F., Murray A.M., Admasu W., Chang C.A., Boehr D.D. Substitution of a surface-exposed residue involved in an allosteric network enhances tryptophan synthase function in cells. Front. Mol. Biosci. 2021;8:679915. doi: 10.3389/fmolb.2021.679915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang H., Banerjee A., Marcus K., Makowski L., Mattos C., Gaponenko V., Nussinov R. The structural basis of the farnesylated and methylated KRas4B interaction with calmodulin. Structure. 2019;27:1647–1659.e4. doi: 10.1016/j.str.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaratnam R., Chowdhury S., VanSchouwen B., Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J., Byun J.A., VanSchouwen B., Henning P., Herberg F.W., Kim C., Melacini G. Dynamical basis of allosteric activation for the Plasmodium falciparum protein kinase G. J. Phys. Chem. B. 2021;125:6532–6542. doi: 10.1021/acs.jpcb.1c03622. [DOI] [PubMed] [Google Scholar]
- 30.Butt E., Van Bemmelen M., Fischer L., Walter U., Jastorff B. Inhibition of cGMP-dependent protein kinase by (Rp)-guanosine 3’,5’-monophosphorothioates. FEBS Lett. 1990;263:47–50. doi: 10.1016/0014-5793(90)80702-k. [DOI] [PubMed] [Google Scholar]
- 31.Song X.-J., Wang Z.-B., Gan Q., Walters E.T. cAMP and cGMP contribute to sensory neuron hyperexcitability and hyperalgesia in rats with dorsal root ganglia compression. J. Neurophysiol. 2006;95:479–492. doi: 10.1152/jn.00503.2005. [DOI] [PubMed] [Google Scholar]
- 32.Akimoto M., Zhang Z., Boulton S., Selvaratnam R., VanSchouwen B., Gloyd M., Accili E.A., Lange O.F., Melacini G. A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP. J. Biol. Chem. 2014;289:22205–22220. doi: 10.1074/jbc.M114.572164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanSchouwen B., Selvaratnam R., Fogolari F., Melacini G. Role of dynamics in the autoinhibition and activation of the exchange protein directly activated by cyclic AMP (EPAC) J. Biol. Chem. 2011;286:42655–42669. doi: 10.1074/jbc.M111.277723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akimoto M., Selvaratnam R., McNicholl E.T., Verma G., Taylor S.S., Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulton S., Van K., VanSchouwen B., Augustine J., Akimoto M., Melacini G. Allosteric mechanisms of nonadditive substituent contributions to protein-ligand binding. Biophys. J. 2020;119:1135–1146. doi: 10.1016/j.bpj.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvaratnam R., VanSchouwen B., Fogolari F., Mazhab-Jafari M.T., Das R., Melacini G. The projection analysis of NMR chemical shifts reveals extended EPAC autoinhibition determinants. Biophys. J. 2012;102:630–639. doi: 10.1016/j.bpj.2011.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson D.A., Akamine P., Radzio-Andzelm E., Madhusudan, Taylor S.S. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 38.Campbell J.C., Henning P., Franz E., Sankaran B., Herberg F.W., Kim C. Structural basis of analog specificity in PKG I and II. ACS Chem. Biol. 2017;12:2388–2398. doi: 10.1021/acschembio.7b00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehmann H., Prakash B., Wolf E., Rueppel A., de Rooij J., Bos J.L., Wittinghofer A. Structure and regulation of the cAMP-binding domains of Epac2. Nat. Struct. Biol. 2003;10:26–32. doi: 10.1038/nsb878. [DOI] [PubMed] [Google Scholar]
- 40.del Sol A., Tsai C.-J., Ma B., Nussinov R. The origin of allosteric functional modulation: Multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selvaratnam R., Mazhab-Jafari M.T., Das R., Melacini G. The auto-inhibitory role of the EPAC hinge helix as mapped by NMR. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehmann H., Arias-Palomo E., Hadders M.A., Schwede F., Llorca O., Bos J.L. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature. 2008;455:124–127. doi: 10.1038/nature07187. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F., Grzesiek S., Vuister G., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao H., Boulton S., Olivieri C., Mohamed H., Akimoto M., Subrahmanian M.V., Veglia G., Markley J.L., Melacini G., Lee W. CHESPA/CHESCA-SPARKY: Automated NMR data analysis plugins for SPARKY to map protein allostery. Bioinformatics. 2021;37:1176–1177. doi: 10.1093/bioinformatics/btaa781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boulton S., Akimoto M., Akbarizadeh S., Melacini G. Free energy landscape remodeling of the cardiac pacemaker channel explains the molecular basis of familial sinus bradycardia. J. Biol. Chem. 2017;292:6414–6428. doi: 10.1074/jbc.M116.773697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Case D.A., Betz R.M., Cerutti D.S., Cheatham T.E., Darden T., Duke R.E. University of California, San Diego; La Jolla, CA: 2016. AMBER16. [Google Scholar]
- 48.Bastian M., Heymann S., Jacomy M. Gephi: An open source software for exploring and manipulating networks. Proc. 3rd Int. AAAI Conf. Weblogs Soc. Media. 2009;3:361–362. [Google Scholar]
- 49.Garcıá de la Torre J., Huertas M., Carrasco B. HYDRONMR: Prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NMR chemical shift assignments for cGMP-bound pfD and hB have been deposited in the Biological Magnetic Resonance Data Bank with codes 50203 and 51201, respectively. All remaining data are contained within the article and the supporting information or available upon request from the corresponding author: melacin@mcmaster.ca.