Abstract
The phosphorylated RNA polymerase II CTD interacting factor 1 (PCIF1) is a methyltransferase that adds a methyl group to the N6-position of 2′O-methyladenosine (Am), generating N6, 2′O-dimethyladenosine (m6Am) when Am is the cap-proximal nucleotide. In addition, PCIF1 has ancillary methylation activities on internal adenosines (both A and Am), although with much lower catalytic efficiency relative to that of its preferred cap substrate. The PCIF1 preference for 2′O-methylated Am over unmodified A nucleosides is due mainly to increased binding affinity for Am. Importantly, it was recently reported that PCIF1 can methylate viral RNA. Although some viral RNA can be translated in the absence of a cap, it is unclear what roles PCIF1 modifications may play in the functionality of viral RNAs. Here we show, using in vitro assays of binding and methyltransfer, that PCIF1 binds an uncapped 5′-Am oligonucleotide with approximately the same affinity as that of a cap analog (KM = 0.4 versus 0.3 μM). In addition, PCIF1 methylates the uncapped 5′-Am with activity decreased by only fivefold to sixfold compared with its preferred capped substrate. We finally discuss the relationship between PCIF1-catalyzed RNA methylation, shown here to have broader substrate specificity than previously appreciated, and that of the RNA demethylase fat mass and obesity-associated protein (FTO), which demonstrates PCIF1-opposing activities on capped RNAs.
Keywords: RNA adenine methylation, N6, 2′O-dimethyladenosine, uncapped RNAs, viral mRNA
Abbreviations: 3mT, 3-methylthymidine; Am, 2′O-methyladenosine; ESI, electrospray ionization; HEK293T, human embryonic kidney 293T cell line; m6A, N6-methyladenine; m6Am, N6, 2′O-dimethyladenosine; m7G, 7-methylguanine; MS, mass spectrometry; MTase, methyltransferase; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; snRNA, small nuclear RNA; VSV, vesicular stomatitis virus
Modified nucleotides in mRNA are particularly enriched within the cap at the 5′ end of eukaryotic mRNAs ((1) and references therein). There are at least four known cap-associated modifications: N7 methylation of the 5′ G yielding 7-methylguanine (m7G), the two ribose 2′O-methyl modifications at the first and second templated nucleotides (Nm), and the N6-methyladenine (m6A) that is found along with 2′O-methylation when the first transcribed nucleotide after the m7G cap is adenine, yielding N6, 2′O-dimethyladenosine (m6Am). In human cells, m7G-capped mRNAs frequently start with 2′O-methylated Am, Gm, Cm or one of the two other adenosine modifications (m6Am or m6A), with Um being seen as well but orders of magnitude less often (2). The dimethylated adenosine m6Am was discovered in 1975 (3), and the enzyme that generates m6Am from Am was biochemically purified soon thereafter (4), but the gene identity (PCIF1) was only revealed recently.
PCIF1 was named as phosphorylated RNA polymerase II CTD interacting factor 1 (5), and it methylates adenosine when A is the first transcribed nucleotide after the m7G cap (6, 7, 8, 9). However, the reported effects of PCIF1-mediated cap-specific adenosine dimethylation vary, even among experiments using the same cell line. Specifically, PCIF1 has been reported to (a) promote the translation of capped mRNAs in human embryonic kidney 293T (HEK293T) cells (6), (b) reduce stability of a subset of m6Am-annotated mRNAs in HEK293T cells while not substantially affecting mRNA translation (7), (c) decrease cap-dependent translation of methylated mRNAs in human malignant melanoma MEL624 cells while not altering mRNA levels or stability (8), or (d) stabilize transcripts in mouse tissues (9). These independent studies jointly indicate that a PCIF1 knockout has limited impact on the mRNA transcriptome. It is possible that PCIF1 plays additional, yet-uncharacterized roles, as it travels from 5′ to 3′ on genes, because of its eponymous direct interaction with the phosphorylated C-terminal domain of RNA polymerase II. While the function of PCIF1 in human cells is still not entirely clear, a 2021 report indicated that PCIF1 methylates viral mRNA and thereby attenuates the antiviral activity of interferon-β (10).
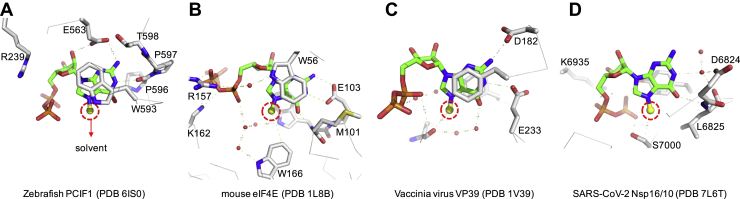
Structurally, human PCIF1 contains an N-terminal pol II CTD-binding WW domain and a large C-terminal methyltransferase (MTase) domain with a canonical Rossmann fold containing a conserved catalytic motif for SAM binding (6). Although a structure is available of human PCIF1 with SAH, lacking the WW domain and the C-terminal end, we do not yet have structural information for human PCIF1 in complex with its RNA substrates. We made use of a structure of the MTase domain of the zebrafish PCIF1 ortholog, which had been crystallized in the presence of SAH and m7G cap (6). Surprisingly, the m7G cap binding by zebrafish PCIF1 (Protein Data Bank: 6IS0 in Fig. 1A) differs from other structurally characterized m7G cap–binding proteins. First, the cap N7 methyl group points to the solvent when bound with zebrafish PCIF1, whereas this methyl group is buried in the protein–substrate interface in three other proteins: the eukaryotic translation initiation factor 4E (11), a viral mRNA cap–specific RNA 2′O-MTase VP39 (12), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp16/10 2′O-MTase (Fig. 1, B–D). This difference may help explain results from a recent study with vesicular stomatitis virus (VSV), in which PCIF1-dependent N6-methylation of VSV mRNA was found to be independent of prior guanine-N7-methylation of the mRNA cap (10). Second, the Watson–Crick pairing edge of the cap guanine does not form the expected three hydrogen bonds when bound to zebrafish PCIF1 (Fig. 1A), whereas eukaryotic translation initiation factor 4E does provide two H-bond acceptors and one H-bond donor to assure sufficient discrimination of this base (Fig. 1B).
Figure 1.
Differences in m7G cap binding. By (A) zebrafish PCIF1, (B) mouse eIF4E, (C) vaccinia virus VP39, and (D) SARS-CoV-2 Nsp16/10. The structures have been aligned so that the m7G is in a similar orientation to highlight similarities and differences. The methyl group at the N7 position points outward to the solvent (zebrafish PCIF1) or inward to the protein (eIF4E, VP39, and Nsp16/10). The guanine ring is stacked against one aromatic residue (zebrafish PCIF1 and SARS-CoV-2 Nsp16/10) or sandwiched between two aromatic residues (eIF4E and VP39). The Watson–Crick pairing edge of the cap guanine involves either two atoms (PCIF1 and Nsp16/10) or three atoms (eIF4E and VP39). eIF4E, eukaryotic translation initiation factor 4E; m7G, 7-methylguanine; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
In this study, we first ask whether human PCIF1 is active in vitro on RNA substrates lacking a cap, since uncapped mRNA molecules do appear and can be recapped by cytoplasmic enzymes (13, 14). At least in 12 yeast species, only a fraction of RNAs were capped with m7G: ∼75% of those having a purine (A or G) at the 5′ end and ∼50% of those starting with a pyrimidine (15). This result implies that a substantial fraction of mRNAs—approximately 25% of even those that start with a purine—are uncapped (at least in yeasts under the growth conditions used). There are also many mammalian mRNA fragments that begin without a cap (16). In addition, during viral infections, some viral genomes generate 5′-triphosphate RNAs without a 5′-cap, either in single-stranded or double-stranded form (17, 18, 19). In summary, cells appear to contain significant amounts of uncapped mRNAs (host or viral) with adenosine in the 5′ position, and PCIF1 could thus play a role in their fates.
Second, we ask whether PCIF1 is active on internal adenosines, particularly internal 2′O-methyladenosine (Am). This question is significant, in part, because the host 2′O-MTase FTSJ3 generates internal 2′O-methylated nucleotides (most of them adenosines) in the HIV RNA genome (20), possibly promoting HIV latency via a pol II-mediated transcription of HIV (21). Interestingly, the most strongly enriched host factor, highly selectively bound to SARS-CoV-2 RNA within infected human hepatocyte Huh7.5 cells, was the nuclearly encoded and mitochondrially localized 2′O-MTase MRM2 (22). Finally, a low but significant level of internal 6mA was observed in a mutant of the RNA virus VSV defective for its own 2′O adenine MTase (VSV-LG4A) (10). PCIF1 has been reported to have ubiquitous nuclear expression (The Human Protein Atlas), but in the Discussion section, we consider its access to the cytoplasm as well.
Results
PCIF1 is active on uncapped RNA molecules
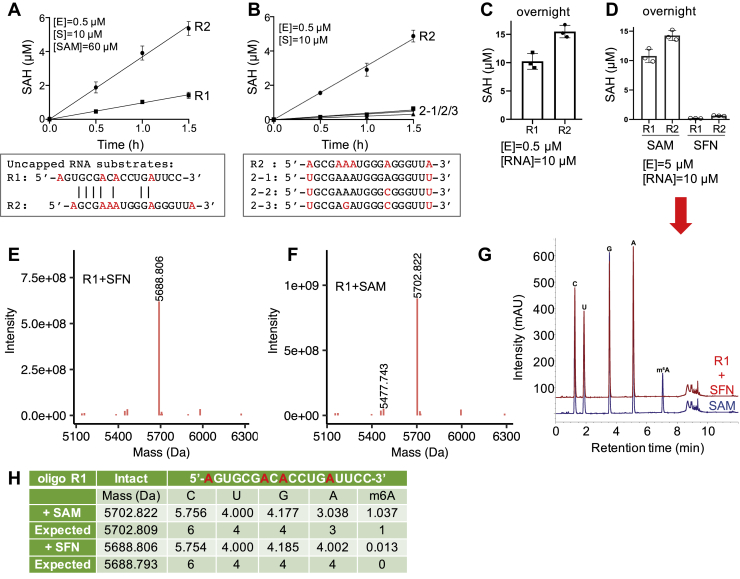
We synthesized two 18-nt RNA molecules with a 5′ adenosine (R1 and R2 in Fig. 2A). Both oligonucleotides contain additional internal Ade residues. Under the conditions established for the capped substrate at pH 8.0 (23), we observed PCIF1 methylation of both uncapped substrates, with higher activity on oligo R2 (Fig. 2A). Replacement of the 5′ A and 3′ A in oligo R2 (with U) resulted in only low residual activity (Fig. 2B).
Figure 2.
PCIF1 activity on the 5′ adenosine of uncapped RNA.A, PCIF1 activity at pH 8.0 on two RNA oligos without caps. B, PCIF1 activity was greatly diminished relative to R2 by A-to-U substitution at the 5′ and 3′ ends. C and D, saturated reactions at two different enzyme concentrations generated one major methylation event in oligo R1, in the presence of SAM or sinefungin (SFN) (N = 3). E and F, deconvoluted ESI spectra from intact mass spectrometry analysis of oligo R1. G, composition of nucleosides (from experiment in D) determined by LC–MS. H, summary of mass spectrometry analysis of oligo R1 (from data in G). The relative abundance of each nucleoside was determined by dividing the UV absorbance by the corresponding extinction coefficient and normalizing to U. ESI, electrospray ionization; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.
Even after saturated overnight reactions, we only observed a single methylation event for oligo R1—specifically, the reaction product SAH had the same molarity as the RNA substrate used in the reaction. In contrast, oligo R2 had between one and two methylation events (Fig. 2C). Increasing the PCIF1 enzyme concentration by 10× did not alter either result (Fig. 2D). As a control, we used sinefungin (a pan methylation inhibitor) in the reaction mixture, instead of methyl donor SAM and, as expected, observed no methylation at all (Fig. 2D).
The four RNA samples shown in Figure 2D were subjected to mass spectrometry (MS) analysis. For the unmodified oligo R1 incubated in the presence of sinefungin, the observed nominal mass and nucleotide compositions matched the calculated molecular mass and expected numbers of nucleotides (Fig. 2, E and G). For the oligo R1 incubated in the presence of SAM, we observed a single major methylation production (∼98%), with the 5′ A having been converted to 6mA (Figs. 2F and S2).
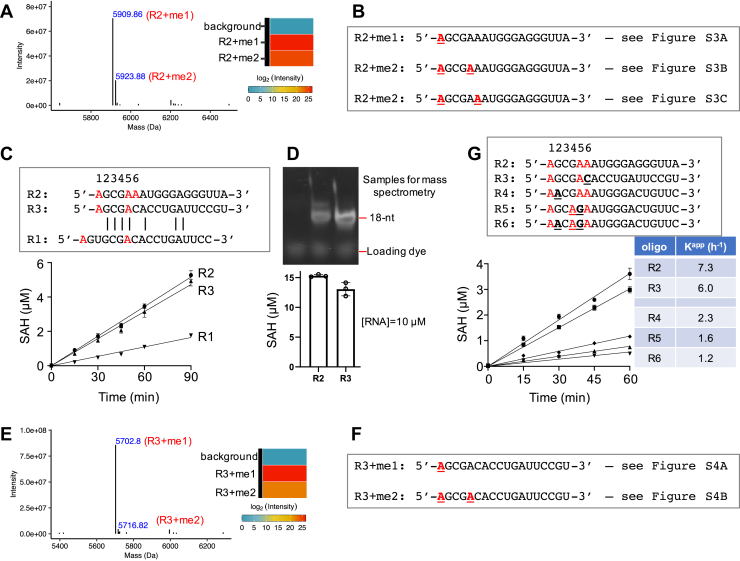
For the oligo R2, MS analysis revealed three methylated species: the fully N6-methylated 5′ A (∼99%), doubly methylated products (estimated to be ∼28%), with the second methylation site at the internal adenosine, three or four residues away from the 5′ end (AGCGAA) (Fig. 3, A and B) but definitely not involving the adenine from the 3′ end (Fig. S3). To confirm the internal methylation sites, we repeated MS analysis in a separate experiment with the R2 oligo and confirmed these observations.
Figure 3.
PCIF1 activity on different RNA oligos.A, deconvoluted ESI spectra from intact mass spectrometry analysis of methylated oligo R2. Aggregated intensity corresponding to each methylation peak is shown to the right. B, summary of methylation sites (A in bold and underlined) with the corresponding tandem mass spectra shown in Fig. S3. C, oligos R2 and R3 have similar activity with the shared 5′-AGCGA sequence, whereas R1 has five residues (as opposed to three) between the first two adenosines. D, saturated reactions on oligos R2 and R3 (bottom) (N = 3). A 15% urea gel showing the methylated oligos used for mass spectrometry analysis (top). E and F, mass spectrometry analysis of methylated oligo R3. G, oligos R4, R5, and R6 have reduced activity, associated with substitutions at nucleotide positions 2, 4, and/or 5. ESI, electrospray ionization; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.
Knowing that the second methylation site is close to the 5′ A in R2, we reduced the distance between the first two adenosines from five residues in R1 to three residues and made a new oligo (R3) that changes the R1 5′-AGUGCGA… to R3 5′-AGCGACA (see sequences in Fig. 3C). This R3 oligo yielded nearly the same PCIF1 activity as did R2 (Fig. 3, C and D). The distance between the two methylated adenosines might reflect the two purine-binding pockets, with one in the active site (adjacent to the SAM-binding site) and the other being the cap-binding site as observed in zebrafish PCIF1 (Fig. 1A). Consistent with R2, we observed methylation primarily at the 5′ adenosine in R3 (Figs. 3E and S4). In addition, a second methylation occurred to an internal adenosine residue, at lower intensity, at position three residues away from the 5′ adenosine (Figs. 3F and S4).
We also realized that, by chance, our design of the 5′ sequence (5′-AGCGA) is somewhat similar to the VSV viral mRNA, in which each gene contains an identical 5′ sequence (5′-AACAG) (24, 25, 26), that is subject to cap-related modifications (10, 27). We thus tested the potential activity of PICF1 on VSV RNA more specifically by synthesizing oligos R4–R6, which contain the VSV AACAG sequence. The results indicate that either one or both substitutions (5′-AGCGA to 5′-AACAG with purine-to-purine changes being underlined) reduced PCIF1 activity (Fig. 3G), suggesting that PCIF1 has some sequence preference for the 5′ end. In a previous study, PCIF1 activity was tested on a number of capped RNA substrates, and the highest activity was on the one containing AGC as the first three residues after the m7G cap (Fig. S4B of Ref. (6)).
PCIF1 activity on 5′ adenosine is enhanced by pre-existing 2′-O methylation
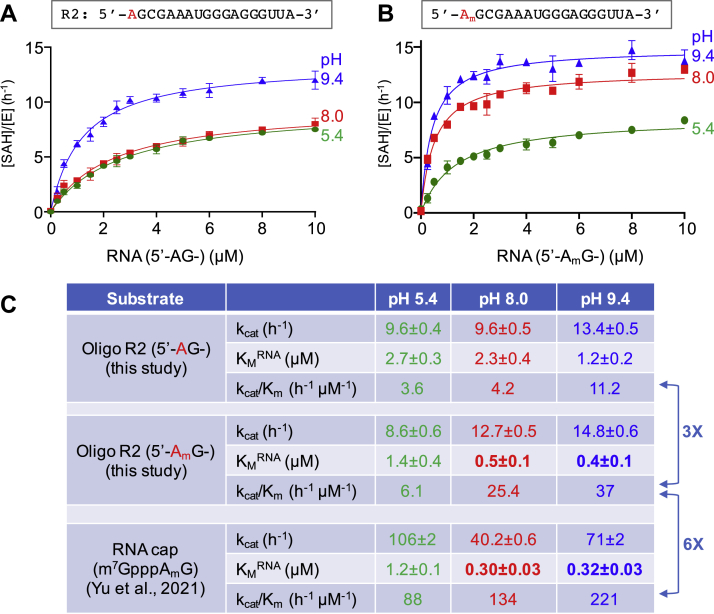
We next measured the PCIF1 kinetic parameters on oligo R2, with and without 2′O methylation, at three different pH values (5.4, 8.0, and 9.4). This pH range was used because we previously characterized PCIF1 activity on a cap analog and observed increasing catalytic efficiency from lower to higher pH that was independent of the buffering agent used (23). By varying concentrations of the RNA substrates, we determined kinetic parameters for reaction rate (kcat), RNA-binding affinity (KM), and catalytic efficiency (kcat/KM) (Fig. 4).
Figure 4.
Kinetic measurements, with varied RNA substrate concentrations, of PCIF1 on 5′-adenosine oligo R2. In the absence (A) and presence (B) of 2′O-methylation. The rate of product SAH formation per enzyme is plotted against RNA concentration. C, summary of kinetic parameters and comparison with a cap analog. Data represent the mean ± SD of two independent determinations, with duplicates assayed for each of three pH values. PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.
We made the following two observations. First, the pH effect shows the same trend as was observed for the cap analog, with the kcat/KM values increasing in the order of pH 5.4 < 8.0 < 9.4, irrespective of 2′O methylation. Second, the reaction rate is fairly similar between the two substrates (kcat ∼10–15 h−1), though the kcat at pH 8.0 is more affected by 2′O methylation than at the other pHs (9.6 h−1 for A methylation versus 12.7 for h−1 for Am methylation).
PCIF1 binds an uncapped 5′-Am oligonucleotide and a cap analog with similar affinities
Given the effects of Am versus A on PCIF1 activity, we next examined Am effects on binding affinity and made the following two observations. First, the binding affinity for the 2′O-methylated substrate is three to five times higher than for the unmodified substrate (KM = 0.5 versus 2.3 μM at pH 8.0 and 0.4 versus 1.2 μM at pH 9.4). This is reminiscent of the observation with the cap analog of tighter PCIF1-binding affinity when Am modification was present (6).
Second, comparing the 5′-Am oligo and the Am modified cap analog, the binding affinities with and without a cap vary over an unexpectedly small range (KM = 0.3–0.5 μM at the two higher pH values). Finally, in contrast to the relatively constant binding affinities, the reaction rates (kcat) are nevertheless higher for the cap analog than the 5′-Am oligo, by three to five times at pH 8.0 and 9.4.
PCIF1 activity on internal adenosine
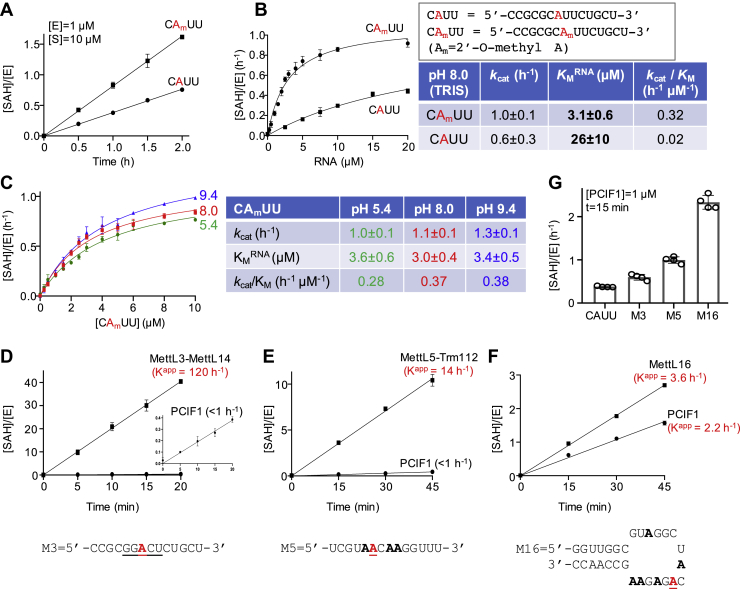
As shown in Figure 3, we observed that a secondary methylation event could occur to an internal adenosine in oligo R2, though at a much reduced rate. We do not know whether PCIF1 has a flanking sequence preference for an internal adenosine, and it might have none; for example, a recent publication on FTSJ3-mediated internal 2′O-adenine methylation revealed no consensus target sequence (20). We designed short 14-mer ssRNA oligos (CAUU and CAmUU in Fig. 5) containing a single internal A and compared the PCIF1 enzymatic activities on the potential substrate (with and without 2′O-methylation) to that of mRNA cap analog and the uncapped version under the same experimental conditions.
Figure 5.
PCIF1 activity on an internal adenosine.A and B, comparison of PCIF1 activity on CAUU with and without 2′O-methylation. C, kinetic measurements of PCIF1 activities by varying concentrations of CAmUU, as a function of pH. D–F, PCIF1 activities on three different RNA substrates, in comparison with respective MTases with the target A in red and underlined. Note that the axes have different scales among the three panels, and the inset in D. G, comparison of relative activity PCIF1 on the four RNA substrates under the same assay conditions (oligos M3, M5, and M16 are shown under D–F) (N = 4). MTase, methyltransferase; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.
First, as expected, we observed increased activity on the 2′O-methylated oligo, compared with the conventional RNA oligo (without any premodifications; Fig. 5A). Second, kinetic measurements suggest that the largest difference lies in the affinity for the premethylated Am substrate, which is greater than eightfold higher than for the unmodified substrate (KM = 3.1 versus 26 μM). In contrast, the reaction rate showed less than 2× variation (kcat = 1.0 versus 0.6 h−1). In other words, PCIF1 had a 16-fold gain in catalytic efficiency (comparing kcat/KM values of 0.32 h−1 μM−1 for CAmUU and 0.02 h−1 μM−1 for CAUU) (Fig. 5B). Third, like the capped and uncapped 5′-A substrates, PCIF1 exhibited the highest catalytic efficiency at pH 9.4, though the difference is much smaller than for the 5′ adenosine (Fig. 5C). We note that the catalytic efficiency of PCIF1 on internal CAmUU is about 100-fold lower than that on uncapped 5′-Am (kcat/KM = 0.38 versus 37 h−1 μM−1) and ∼600 fold lower than that of the cap analog. Thus, the activity of PCIF1 on the internal adenosine is marginal, using this substrate (CAUU) and under the conditions tested, with inefficient enzymatic turnover.
In addition to the CAUU sequence, we tested RNA molecules known to be substrates for other RNA adenosine MTases: a 14-nt oligo containing an mRNA consensus sequence GGACU for MettL3-14 (Fig. 5D), a short linear 14-nt RNA oligo corresponding to the sequence (AACAA) surrounding A1832 of 18S rRNA for MettL5-Trm112 (Fig. 5E), and a 29-nt hairpin oligo corresponding to the hairpin of small nuclear RNA (snRNA) for MettL16 (Fig. 5F). As with CAUU, we observed minimal activity with an apparent Kapp of 1 to 2 h−1. Among these substrates, the hairpin RNA shows the highest activity with a Kapp value of 2.2 h−1, suggesting that more than one adenosine is being methylated in the substrate (ACAGAGAA).
Discussion
PCIF1 has broad substrate specificity
Here, we report that PCIF1 is enzymatically active on an array of RNA substrates, from cap-proximal adenosine to internal adenosine, in 2′O-methylated Am or unmodified A, with sequences derived from mRNA, rRNA, and snRNA. This is a substantially broader substrate range than had been appreciated. Given the apparent significance of RNA 6mA methylation on a variety of human diseases (e.g., (28, 29, 30, 31)), accurate understanding of the activities of RNA MTases is a high priority.
Potential significance of methylation on viral RNA
The potential relevance of PCIF1 methylation of RNA adenosines in viruses, which was first observed in the 1970s (reviewed in Ref. (32) and references therein), is very recently illustrated in VSV. In contrast to the methylation order on cellular mRNA, VSV cap-proximal adenosine methylation proceeds in the opposite order: it requires ribose-2′O-methylation but not guanine-N7 methylation (10). In fact, ribose 2′O methylation of the VSV mRNA cap, catalyzed by the viral MTase (protein L), precedes and facilitates subsequent guanine-N7 methylation (33). The PCIF1-dependent modification of VSV mRNA cap structures may help viral mRNA appear more host like (i.e., for the differentiation between “self” and “non-self” RNA during viral infection), and we show here that PCIF1 binds an uncapped 5′-Am oligonucleotide with approximately the same affinity as that of a cap analog (Fig. 4C).
Like VSV, SARS-CoV, Middle East respiratory syndrome coronavirus, and SARS-CoV-2 have an adenosine as the cap-proximal nucleotide of the nascent mRNA (m7GpppA). It will be interesting to investigate whether SARS-CoV-2 has dimethyladenosine (m6Am) in its m7Gpppm6AmUUAAA cap, and whether PCIF1 is involved in the generation and biological functions of the cap modifications in regulation of SARS-CoV-2 infection (or of other clinically relevant viruses).
PCIF1 is mainly localized to nucleoplasm (The Human Protein Atlas) (9), is associated with active chromatin (at least in HeLa cells) (34), and its protein network comprises mostly factors involved in nascent RNA synthesis by RNA polymerase II (35), where cellular mRNA-capping modifications proceed in an ordered fashion from m7G to Am to m6Am.
The existence of a cytoplasmic pool of PCIF1 has not been reported. In order for PCIF1 to act on RNA from RNA viruses, PCIF1 has to be able to localize to cytoplasm. This might occur via changes in nucleocytoplasmic transport during infection (36), though the nuclear localization signal of PCIF1 has not yet been characterized and does not seem obvious (37). Alternatively, nuclear and cytoplasmic contents mix during mitosis, when nuclear envelope disassembly occurs (38). We also note that the processes of nuclear envelope assembly/disassembly are modulated by some viruses (39).
Fat mass and obesity-associated protein appears to oppose documented activities of PCIF1
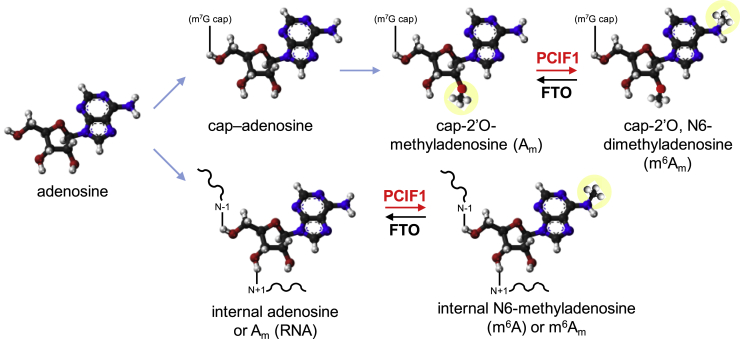
Intriguingly, we note that the methylation activities of PCIF1 in vitro seem to fully oppose to the fat mass and obesity-associated protein (FTO) demethylase activities on N6-methyladenosines (Fig. 6).
Figure 6.
Reactions tested here in which PCIF1 methylates in opposition to the activities of the demethylase FTO (demonstrated by others, seeDiscussionsection). FTO, fat mass and obesity-associated protein; PCIF1, phosphorylated RNA polymerase II CTD interacting factor 1.
FTO has Fe(II)- and α-ketoglutarate-dependent dioxygenase activity (40). FTO was initially characterized as a repair enzyme active on 3-methylthymidine (3mT) in ssDNA (40) or on 3-methyluracil in ssRNA (41). Later, five additional FTO activities were identified: (a) a demethylase that removes the methyl group from internal m6A residues in mRNA (42), (b) a cap-dependent demethylase that preferentially removes the N6 methyl group from m6Am (43), (c) a demethylase active on internal m6A and m6Am in snRNA, and a demethylase of (d) 1-methyladenosine in tRNA (44), and of (e) N6-methyldeoxyadenosine in ssDNA (45).
Besides the important effects on the stability of those mRNAs having m6Am at the 5′ end (at least in HEK293T cells (43)), FTO modulates mRNA splicing, is required for adipogenesis (46), and mediates stem-like properties in colorectal cancer cells (47). Structures have been determined for human FTO, bound to the mononucleotide 3mT (48) or to ssDNA containing an internal N6-methyldeoxyadenosine (45). However, these structures do not explain the preference of FTO for either mRNA caps or for 2′O-methylation. (We have not tested PCIF1 activity on 3mT, 3-methyluracil, or 1-methyladenosine on either RNA or DNA substrates.)
The cytoplasmic pool of FTO impedes cancer stem cell growth (47); specifically, low FTO levels in patient-derived cell lines are associated with high levels of m6Am in mRNA and elevated tumorigenicity and chemoresistance. The hypothesized apparently full complementarity between PCIF1 and FTO activities (Fig. 6) suggests that their expression ratio may be a key regulatory parameter. The present study suggests that another key regulatory parameter is their respective activity levels on their diverse substrates.
Experimental procedures
Recombinant enzymes
The highly purified recombinant enzymes used in this study were recently characterized in our laboratory: human PCIF1 (pXC2055) (23), human MettL3-14 (49, 50), human MettL5-Trm112 (pXC2062-pXC2076), and human MettL16 (pXC2210) (23). The example of purified PCIF1 and its activity is shown in Fig. S1, A–C.
The oligonucleotides were synthesized by Integrated DNA Technologies. The cap analog was purchased from TriLink BioTechnologies (catalog number: N-7113). The sequence of RNA oligo R1 was taken from a study of human RNase H1 complexed with an RNA–DNA hybrid (51). The sequence of RNA oligo R2 was taken from a study of Wilms tumor protein WT1 (52).
Methylation assays
Methylation reactions for LC–MS analyses were carried out in a 60 μl mixture containing 5 μM PCIF1, 10 μM RNA oligo R1 or R2 (Fig. 2A), 60 μM SAM, 60 U recombinant RNasin (an RNase inhibitor; Promega), in a reaction buffer that included 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, and 1 mM DTT. For control samples, 60 μM sinefungin was added instead of SAM. Reactions were performed at room temperature (∼22 °C) with overnight incubation. A 5 μl aliquot of the reaction mixture was serially diluted three times to ensure that the byproduct SAH was in the linear range (Fig. S1, D and E) for the Promega bioluminescence assay (MTase-Glo) (53). The RNA samples after reaction were purified by Monarch RNA Cleanup Kit (New England BioLabs; catalog number: 2040S) using a modified protocol for RNA with length >15 nt.
Kinetic assays were performed in a 20 μl mixture at three pH values (5.4, 8.0, or 9.4), in 20 mM citric acid and 20 mM Bis–Tris propane, 50 mM NaCl, 1 mM DTT, 10 U RNasin, 20 μM SAM, 0.2 μM PCIF1 (for RNA with 5′-adenosine) or 1 μM PCIF1 (for detecting RNA methylation at internal adenosines), with varied RNA concentrations. The reactions proceeded at room temperature for 15 min. The dependence of product SAH formation per enzyme molecule on RNA substrate concentration was analyzed according to Michaelis–Menten kinetics using GraphPad Prism 8 (GraphPad Software, Inc).
Methylation assays of PCIF1 on different RNA oligonucleotides were conducted in the reaction buffer containing 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM DTT, 20 μM or 100 μM SAM, with various PCIF1 and substrate concentrations. Methylation assays of MettL3-MettL14, MettL5-Trm112, and MettL16 were conducted under the optimal conditions for each enzyme as previously characterized in our laboratory (23, 49, 50).
To calibrate SAH concentration and luminescence, the SAH standard solution within the MTase-Glo Methylation Assay Kit (Promega) was subjected to serial twofold dilutions, starting from 4 μM. Luminescence signals from a standard concentration curve of SAH were generated according to the manufacturer’s protocol. An aliquot of 5 μl of the sample was transferred into a low-volume 384-well plate, and the luminescence signal was detected using a Synergy 4 Multi-Mode Microplate Reader (BioTek). A linear regression of the SAH standard was plotted against luminescence (Fig. S1, D and E).
MS
Base composition analysis was initiated by digestion of RNA oligos by treatment at 37 °C for 1 h with the Nucleoside Digestion Mix (NEB; catalog number: M0649S). The resulting nucleoside mixtures, without further purification, were directly analyzed by reversed-phase LC–MS. Nucleoside analysis was performed in duplicate on an Agilent 1290 Infinity II UHPLC equipped with G7117A diode array detector and a 6135XT single quadrupole detector operating in positive electrospray ionization (+ESI) and negative ESI (–ESI) modes. UHPLC was performed using on a Waters XSelect HSS T3 XP column (2.1 × 100 mm, 2.5 μm) with the gradient mobile phase consisting of aqueous ammonium formate (10 mM at pH 4.5) and methanol. MS data acquisition was recorded in total ion chromatogram mode. The relative abundance of each nucleoside was determined by dividing the UV absorbance by the corresponding extinction coefficient
Intact mass analysis and modification mapping were performed by LC–MS/MS on a Vanquish Horizon UHPLC System equipped with a diode array detector and a Thermo Q-Exactive Plus orbitrap mass spectrometer operating under –ESI mode. UHPLC was performed using a Thermo DNAPac RP Column (2.1 × 50 mm, 4 μm) at 70 °C and 0.3 ml/min flow rate with a gradient mobile phase consisting of hexafluoroisopropanol, N,N-diisopropylethylamine aqueous buffer, and methanol. UV detection was performed at 260 nm. Intact mass analysis was performed under full scan mode at a resolution of 70,000 (full width at half maximum) at m/z 200. ESI–MS raw data were deconvoluted using Promass HR (Novatia, Inc). Modification mapping was performed in parallel reaction monitoring mode with a normalized collision energy of 15% at a resolution of 70,000 (full width at half maximum) at m/z 200. The top three ions from each peak were chosen for parallel reaction monitoring analysis. ESI–MS/MS raw data were processed with the Nucleic Acid Search Engine in Open-MS (version 2.6.0) (54).
Data availability
The experimental data that support the findings of this study are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
N. D., E. J. W. and I. R. C. are employees of New England Biolabs, Inc, which is a manufacturer and vendor of molecular biology reagents, including several enzymes and buffers used in this study. This affiliation does not affect the authors’ impartiality, adherence to journal standards and policies, or availability of data. The other authors (D. Y., J. Z., T. W., R. M. B., X. Z., and X. C.) declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Ms Yu Cao for technical assistance as well as Dr Taraneh Hajian and Dr Masoud Vedadi for purified MettL3-14.
Author contributions
T. W., R. M. B., X. Z., and X. C. conceptualization; J. Z. validation; D. Y., N. D., E. J. W., and J. Z. investigation; X. C. writing–original draft; R. M. B. writing–review and editing; I. R. C. and X. Z. supervision; X. Z. project administration; X. C. funding acquisition.
Funding and additional information
This work was supported by US National Institutes of Health (grant no.: R35GM134744; to X. C.); Cancer Prevention and Research Institute of Texas (grant no.: RR160029; to X. C., who is a Cancer Prevention and Research Institute of Texas Scholar in Cancer Research). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karin Musier-Forsyth
Contributor Information
Xing Zhang, Email: XZhang21@mdanderson.org.
Xiaodong Cheng, Email: XCheng5@mdanderson.org.
Supporting information
References
- 1.Cowling V.H. CAPAM: The mRNA cap adenosine N6-methyltransferase. Trends Biochem. Sci. 2019;44:183–185. doi: 10.1016/j.tibs.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamarkin-Ben-Harush A., Vasseur J.J., Debart F., Ulitsky I., Dikstein R. Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. Elife. 2017;6 doi: 10.7554/eLife.21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei C., Gershowitz A., Moss B. N6, O2'-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature. 1975;257:251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- 4.Keith J.M., Ensinger M.J., Moss B. HeLa cell RNA (2'-O-methyladenosine-N6-)-methyltransferase specific for the capped 5'-end of messenger RNA. J. Biol. Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- 5.Fan H., Sakuraba K., Komuro A., Kato S., Harada F., Hirose Y. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 2003;301:378–385. doi: 10.1016/s0006-291x(02)03015-2. [DOI] [PubMed] [Google Scholar]
- 6.Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O., Suzuki T. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363 doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 7.Boulias K., Toczydlowska-Socha D., Hawley B.R., Liberman N., Takashima K., Zaccara S., Guez T., Vasseur J.J., Debart F., Aravind L., Jaffrey S.R., Greer E.L. Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Mol. Cell. 2019;75:631–643.e8. doi: 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sendinc E., Valle-Garcia D., Dhall A., Chen H., Henriques T., Navarrete-Perea J., Sheng W., Gygi S.P., Adelman K., Shi Y. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 2019;75:620–630.e9. doi: 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey R.R., Delfino E., Homolka D., Roithova A., Chen K.M., Li L., Franco G., Vagbo C.B., Taillebourg E., Fauvarque M.O., Pillai R.S. The mammalian cap-specific m(6)Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 2020;32:108038. doi: 10.1016/j.celrep.2020.108038. [DOI] [PubMed] [Google Scholar]
- 10.Tartell M.A., Boulias K., Hoffmann G.B., Bloyet L.M., Greer E.L., Whelan S.P.J. Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-beta. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2025769118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedzwiecka A., Marcotrigiano J., Stepinski J., Jankowska-Anyszka M., Wyslouch-Cieszynska A., Dadlez M., Gingras A.C., Mak P., Darzynkiewicz E., Sonenberg N., Burley S.K., Stolarski R. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5' cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 12.Hodel A.E., Gershon P.D., Shi X., Quiocho F.A. The 1.85 A structure of vaccinia protein VP39: A bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 13.Berger M.R., Alvarado R., Kiss D.L. mRNA 5' ends targeted by cytoplasmic recapping cluster at CAGE tags and select transcripts are alternatively spliced. FEBS Lett. 2019;593:670–679. doi: 10.1002/1873-3468.13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Valle Morales D., Trotman J.B., Bundschuh R., Schoenberg D.R. Inhibition of cytoplasmic cap methylation identifies 5' TOP mRNAs as recapping targets and reveals recapping sites downstream of native 5' ends. Nucleic Acids Res. 2020;48:3806–3815. doi: 10.1093/nar/gkaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z., Lin Z. The origin and evolution of a distinct mechanism of transcription initiation in yeasts. Genome Res. 2021;31:51–63. doi: 10.1101/gr.264325.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malka Y., Steiman-Shimony A., Rosenthal E., Argaman L., Cohen-Daniel L., Arbib E., Margalit H., Kaplan T., Berger M. Post-transcriptional 3 -UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat. Commun. 2017;8:2029. doi: 10.1038/s41467-017-02099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmann G., Patel D.J. Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y. FTSJ3 is an RNA 2'-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019;565:500–504. doi: 10.1038/s41586-018-0841-4. [DOI] [PubMed] [Google Scholar]
- 21.Li Z., Hajian C., Greene W.C. Identification of unrecognized host factors promoting HIV-1 latency. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C., Schmitz C.O., Parker K.R., Woo E., Chang H.Y., Horvath T.L., et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184:2394–2411.e16. doi: 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D., Kaur G., Blumenthal R.M., Zhang X., Cheng X. Enzymatic characterization of three human RNA adenosine methyltransferases reveals diverse substrate affinities and reaction optima. J. Biol. Chem. 2021;296:100270. doi: 10.1016/j.jbc.2021.100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barr J.N., Whelan S.P., Wertz G.W. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stillman E.A., Whitt M.A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J.T., McElvain L.E., Whelan S.P. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J. Virol. 2007;81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moyer S.A., Banerjee A.K. In vivo methylation of vesicular stomatitis virus and its host-cell messenger RNA species. Virology. 1976;70:339–351. doi: 10.1016/0042-6822(76)90276-2. [DOI] [PubMed] [Google Scholar]
- 28.Peng L., Long T., Li F., Xie Q. Emerging role of m(6) a modification in cardiovascular diseases. Cell Biol. Int. 2022 doi: 10.1002/cbin.11773. [DOI] [PubMed] [Google Scholar]
- 29.Worpenberg L., Paolantoni C., Roignant J.Y. Functional interplay within the epitranscriptome: Reality or fiction? Bioessays. 2022;44 doi: 10.1002/bies.202100174. [DOI] [PubMed] [Google Scholar]
- 30.Liu L., Li H., Hu D., Wang Y., Shao W., Zhong J., Yang S., Liu J., Zhang J. Insights into N6-methyladenosine and programmed cell death in cancer. Mol. Cancer. 2022;21:32. doi: 10.1186/s12943-022-01508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An Y., Duan H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer. 2022;21:14. doi: 10.1186/s12943-022-01500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gokhale N.S., Horner S.M. RNA modifications go viral. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahmeh A.A., Li J., Kranzusch P.J., Whelan S.P. Ribose 2'-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugita A., Kuruma S., Yanagisawa N., Ishiguro H., Kano R., Ohkuma Y., Hirose Y. The cap-specific m6A methyltransferase, PCIF1/CAPAM, is dynamically recruited to the gene promoter in a transcription-dependent manner. J. Biochem. 2021;170:203–213. doi: 10.1093/jb/mvab032. [DOI] [PubMed] [Google Scholar]
- 35.Covelo-Molares H., Obrdlik A., Postulkova I., Dohnalkova M., Gregorova P., Ganji R., Potesil D., Gawriyski L., Varjosalo M., Vanacova S. The comprehensive interactomes of human adenosine RNA methyltransferases and demethylases reveal distinct functional and regulatory features. Nucleic Acids Res. 2021;49:10895–10910. doi: 10.1093/nar/gkab900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter T.C. Breaching the barrier-the nuclear envelope in virus infection. J. Mol. Biol. 2016;428:1949–1961. doi: 10.1016/j.jmb.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Lu J., Wu T., Zhang B., Liu S., Song W., Qiao J., Ruan H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021;19:60. doi: 10.1186/s12964-021-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Alvarez A., Cooper J.P. Chromosomes orchestrate their own liberation: Nuclear envelope disassembly. Trends Cell Biol. 2017;27:255–265. doi: 10.1016/j.tcb.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Cohen S., Etingov I., Pante N. Effect of viral infection on the nuclear envelope and nuclear pore complex. Int. Rev. Cell Mol. Biol. 2012;299:117–159. doi: 10.1016/B978-0-12-394310-1.00003-5. [DOI] [PubMed] [Google Scholar]
- 40.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A.P., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia G., Yang C.G., Yang S., Jian X., Yi C., Zhou Z., He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., Gross S.S., Elemento O., Debart F., Kiledjian M., Jaffrey S.R. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A., Jia G., Chen J., He C. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Wei L.H., Wang Y., Xiao Y., Liu J., Zhang W., Yan N., Amu G., Tang X., Zhang L., Jia G. Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2919–2924. doi: 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W., Hao Y.J., Ping X.L., Chen Y.S., Wang W.J., Jin K.X., Wang X., Huang C.M., Fu Y., Ge X.M., et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Relier S., Ripoll J., Guillorit H., Amalric A., Achour C., Boissiere F., Vialaret J., Attina A., Debart F., Choquet A., Macari F., Marchand V., Motorin Y., Samalin E., Vasseur J.J., et al. FTO-mediated cytoplasmic m(6)Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021;12:1716. doi: 10.1038/s41467-021-21758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., Cheng W., Wang J., Feng Y., Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 49.Woodcock C.B., Yu D., Hajian T., Li J., Huang Y., Dai N., Correa I.R., Jr., Wu T., Vedadi M., Zhang X., Cheng X. Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discov. 2019;5:63. doi: 10.1038/s41421-019-0136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu D., Horton J.R., Yang J., Hajian T., Vedadi M., Sagum C.A., Bedford M.T., Blumenthal R.M., Zhang X., Cheng X. Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 2021;49:11629–11642. doi: 10.1093/nar/gkab460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nowotny M., Gaidamakov S.A., Ghirlando R., Cerritelli S.M., Crouch R.J., Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: Insight into HIV reverse transcription. Mol. Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 52.Wang D., Horton J.R., Zheng Y., Blumenthal R.M., Zhang X., Cheng X. Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites. Nucleic Acids Res. 2018;46:3864–3877. doi: 10.1093/nar/gkx1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao K., Zegzouti H., Goueli S.A. Methyltransferase-glo: A universal, bioluminescent and homogenous assay for monitoring all classes of methyltransferases. Epigenomics. 2016;8:321–339. doi: 10.2217/epi.15.113. [DOI] [PubMed] [Google Scholar]
- 54.Wein S., Andrews B., Sachsenberg T., Santos-Rosa H., Kohlbacher O., Kouzarides T., Garcia B.A., Weisser H. A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat. Commun. 2020;11:926. doi: 10.1038/s41467-020-14665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data that support the findings of this study are contained within the article.