Summary
Genetic variants that affect neurological function will often produce changes visible at the level of gross morphology, either of the whole brain or of specific neuronal types. Here we describe how to perfuse and dissect the brain in preparation for Nissl staining. Then we outline steps for culturing mouse primary hippocampal neurons to evaluate dendritic arborization (Sholl analysis).
For complete details on the use and execution of this protocol, please refer to Gennarino et al. (2018).
Subject areas: Cell Biology, Cell culture, Cell isolation, Cell-based Assays, Genetics, Model Organisms, Neuroscience
Graphical abstract

Highlights
-
•
Primary mouse hippocampal neuron isolation and immunohistochemistry
-
•
Transfecting hippocampal neurons with different gene variants
-
•
Sholl analysis of dendritic arborization
-
•
Nissl staining and morphological evaluation of mouse brain
Genetic variants that affect neurological function will often produce changes visible at the level of gross morphology, either of the whole brain or of specific neuronal types. Here we describe how to perfuse and dissect the brain in preparation for Nissl staining. Then we outline steps for culturing mouse primary hippocampal neurons to evaluate dendritic arborization (Sholl analysis).
Before you begin
All research and animal care procedures were approved by the Columbia University Irving Medical Center Institutional Animal Care. Mouse colonies were bred and maintained with standard mouse chow and water ad libitum under a 12 h light / 12 h dark cycle.
You need approval from your Institutional Review Board (IRB) and that of the clinicians with whom you share information about patient variants and clinical data.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| QIAGEN Plasmid Maxi Kit | QIAGEN | Cat# 12162 |
| QIAGEN Plasmid Midi Kit | QIAGEN | Cat# 12143 |
| CalPhos Mammalian Transfection Kit | Takara Bio | Cat# 631312 |
| Chemicals, peptides, and recombinant proteins | ||
| Heat Inactivated-Fetal Bovine Serum (FBS) | Novus Biologicals | Cat# S11150H |
| Hanks’ Balanced Salt Solution (HBSS) | Thermo Fisher Scientific | Cat# 88284 |
| Minimum Essential Media (MEM) | Thermo Fisher Scientific | Cat# 11095080 |
| B-27 Supplement | Thermo Fisher Scientific | Cat# 17504044 |
| L-glutamine 200 mM (100×) | GenDepot | Cat# CA009-010 |
| Neurobasal Medium | Thermo Fisher Scientific | Cat# 21103049 |
| Poly-D-Lysine Solution | Thermo Fisher Scientific | Cat# A003E |
| Laminin I | R&D systems | Cat# 3400-010-02 |
| TrypLE Express Enzyme 1× | Thermo Fisher Scientific | Cat# 12604013 |
| 5-Fluoro-2′-deoxyuridine | MilliporeSigma | Cat# F0503 |
| Uridine | MilliporeSigma | Cat# U3003 |
| Penicillin/Streptomycin | Thermo Fisher Scientific | Cat# 15070063 |
| Triton X-100 | MilliporeSigma | Cat# T8787 |
| VECTASHIELD® Antifade Mounting Medium with DAPI | Vector Lab | Cat# H-1200-10 |
| Sodium pyruvate (100 mM) | Thermo Fisher Scientific | Cat# 11360070 |
| Paraformaldehyde | MilliporeSigma | Cat# 158127-500G |
| 2,2,2 Tribromoethanol | Fisher Scientific | Cat# AAA1870622 |
| 2-Methyl-2-butanol | Fisher Scientific | Cat# AAA18304AP |
| HCl | MilliporeSigma | Cat# 320331-500ML |
| NaOH | MilliporeSigma | Cat# 221465 |
| Normal Goat serum | Abcam | Cat# ab7481 |
| Acetic acid glacial | MilliporeSigma | Cat# 695092-100ML |
| Sucrose | MilliporeSigma | Cat# S0389-500G |
| Tissue-Plus™ O.C.T. Compound Tissue-Plus™ O.C.T. Compound | Fisher Scientific | Cat# 23-730-571 |
| Permount Mounting Medium | Electron Microscopy Sciences | Cat# 17986-01 |
| Nuclease-Free Water (not DEPC-Treated) | Fisher Scientific | Cat# 4387936 |
| Xylene | Fisher Scientific | Cat# X3S-4 |
| Software and algorithms | ||
| Fiji is Just ImageJ (Fiji) version 2.0.0-rc/1.5f | http://imagej.net/ | (Schindelin et al., 2015) |
| Neurolucida 360 | MBF Bioscience, Williston, VT | (Ayata et al., 2018) |
| Imaris | Oxford instruments | Cat# RRID:SCR_007370 |
| Zen2.6 Software | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-core.html | Zeiss |
| Antibodies | ||
| Mouse monoclonal anti-HA | BioLegend | Cat# 901514 |
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970 |
| Donkey anti-mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 594 | Thermo Fisher Scientific | Cat# A32742 |
| Goat anti-Chicken IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat# A32391 |
| Recombinant DNA | ||
| PUM1cDNA expression vector | OmickLink Expression Vector, GeneCopoeia | Cat# pEZ-M02, EX-E2337-M02-10 |
| pEGFP-C1 | Prof. Golenbock's Lab (UMass Chan Medical School, Worcester, Massachusetts 01605, USA) | (Gennarino et al., 2018) |
| Empty expression vector | OmickLink Expression Vector, GeneCopoeia | Cat# EX-NEG-M02 |
| OmickLinkTMExpression Vector pEZ-M02 | GeneCopoeia | Cat# EX-E2337-M02-10 |
| OmickLinkTMExpression Vector | GeneCopoeia | Cat# EX-NEG-M02 |
| Experimental models: Organisms/strains | ||
| B6/129 mixed background Pum1 mouse models. We used male and female mice. Neonatal mice for hippocampal neurons and at 10 weeks of age for Nissl staining | Prof. Haifan Lin’s Lab (Yale School of Medicine, New Haven, Connecticut 06520, USA) | (Gennarino et al., 2018) |
| Other | ||
| SouthernBiotech™ Gelatin Subbed Slides | Fisher Scientific | Cat# OBSLD01BX |
| Corning™ Costar™ Flat Bottom Cell Culture Plates (12-Well) | Corning | Cat# 07-200-82 |
| 70 μm cell strainer | Thermo Fisher Scientific | Cat# 76327 |
| Square coverslip | Thermo Fisher Scientific | Cat# 15-183-88 |
| 3P PVC tubing for perfusion 1.02 mm in diameter | Thermo Fisher Scientific | Cat# NC0279757 |
| Double-Ended Micro-Tapered Stainless-Steel Spatula | Thermo Fisher Scientific | Cat# 2140110 |
| Iris Scissors | VWR | Cat# 470018-890 |
| MeisterHand LISTER Bandage scissors 8" (20.3) | HOSPEQ | Cat# MH50550 |
| FisherbrandTM Standard Dissecting Scissors | Fisher Scientific | Cat# 0895120 |
| Medium Tip dissecting Forceps | VWR | Cat# 470001-846 |
| Premium Superfrost Microscope slides | Fisher Scientific | Cat# 12-544-7 |
| BD Hypodermic Needles, 27G | BD | Cat# 305110 |
| Kimwipes | Fisher Scientific | Cat# 06-666 |
| Embedding mold | Thermo Fisher Scientific | Cat# 50465347 |
| 3P Minipuls Peristaltic Pump | Fisher Scientific | Cat# NC0745002 |
| Leica CM3050S Cryostat | Leica BIOSYSTEMS | Cat# 958846 |
| Fisherbrand™ Slide Drying Bench | Fisher Scientific | Cat# 11-474-470 |
| Zeiss LSM710 | Zeiss | Cat# LSM-170 |
Materials and equipment
| Coating medium | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Laminin 1 mg/mL | 2 μg/mL | 12 μL |
| Poly-D-lysine 10 mg/mL | 0.1 mg/mL | 60 μL |
| Neurobasal Medium | Up to 6 mL | |
| Total | 6 mL | |
Prepare fresh every time.
| Hippocampal plating medium | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Fetal Bovine Serum | 10% | 50 mL |
| L-glutamine | 1% | 5 mL |
| Sodium pyruvate (1 M) | 1% | 5 mL |
| Penicillin/streptomycin | 100 U/mL | 5 mL |
| Neurobasal Medium | Up to 500 mL | |
| Total | 500 mL | |
Store at 4°C for up to 1 month.
| Hippocampal growth medium | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| B-27 supplement | 2% | 1 mL |
| L-glutamine | 1% | 0.5 mL |
| Neurobasal Medium | Up to 50 mL | |
| Total | 50 mL | |
Store at 4°C for up to 1 month.
| Permeabilization/blocking solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Triton X-100 (10%) | 0.3% | 30 μL |
| Normal goat serum (NGS) | 2% | 20 μL |
| PBS 1× | – | 970 μL |
Prepare fresh every time.
| Paraformaldehyde solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Paraformaldehyde (PFA) | 4% | 40 g |
| PBS (Phosphate buffered saline) 20× | 1× | 50 mL |
| HCl 37% and NaOH 5N | To adjust pH 7.4 | |
| H2O milliQ | Up to 1 L | |
| Total | 1,000 mL | |
Note: Add 40 g of PFA in 800 mL of sterile Milli-Q H2O. Stir the mixture at 60°C in a ventilated hood. The PFA powder does not dissolve instantly, so you need to raise the pH of the solution by adding 5N NaOH drop by drop until a clear solution is formed. Cool the solution to room temperature and filter it using 3M filter paper. Add 50 mL of PBS 20× and make sure the pH is 7.4 (you can use 37% HCl to adjust the pH). Then adjust the volume up to 1,000 mL with sterile Milli-Q H2O. Dispense the solution in aliquots of 50 mL. You can store the solution at 4°C for 1–2 weeks, or at −20°C for 4 months. PFA is sensitive to light, protect the solution from direct light to avoid degradation.
CRITICAL: PFA is a confirmed human carcinogen and irritates the eyes, skin, and lungs; prolonged exposure can cause shortness of breath, cough, and lung damage. Prepare under a chemical hood and dispose of it in an appropriate biohazard waste container.
| Avertin solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 2,2,2 Tribromoethanol | 1.25% | 2.5 g |
| 2-methyl-2-butanol 98% | 5 mL | |
| H2O milliQ | Up to 200 mL | |
| Total | 200 mL | |
Note: We used avertin as an injectable anesthetic, as it was allowed in our IACUC protocol. Any IACUC-approved anesthetic can be used.
-
•
On a heated plate (approximately 40°C) dissolve 2.5 g of Tribromoethanol in 5 mL of 2-methyl-2-butanol, stirring vigorously.
-
•
Add distilled water up to a volume of 200 mL.
-
•
Filter the solution through a 0.5 μm.
-
•
Aliquot the solution in brown bottles to protect from light and store at 4°C.
Note: Tribromoethanol degrades to dibromoacetaldheyde and hydrobromic acid; it becomes toxic and can kill the animal before you have a chance to perfuse it. If the pH of the solution is less than 5, assume the solution needs to be prepared again. Once prepared, use the avertin within 14 days, protect the solution from light. Avertin is usually stable for 24 months at room temperature. We suggest to discard the solution if it turns yellow.
CRITICAL: The 2,2,2 Tribromoethanol is hazardous and can cause organ toxicity from inhalation, skin corrosion and irritation, and eye damage. The 2-methyl-2-butanol 98% is highly flammable and harmful in contact with skin, and if inhaled, causing skin damage, respiratory irritation, and dizziness. Please use all the reagents under a chemical hood.
| Cresyl violet solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Cresyl violet powder | 0.1% | 0.1 g |
| Acetic acid (glacial) | 0.25% v/v | 250 μL |
| H2O milliQ | – | 100 mL |
| Total | 100 mL | |
Store at room temperature (around 25°C) and protect the solution from light.
Step-by-step method details
Note: The steps below are only for the protocols we modified from the manufacturer's suggestion. Because we deal with both mouse and human proteins in this protocol, we will distinguish them by referring to the mouse proteins with only the first letter capitalized (e.g., Pum1) and human proteins in all caps (PUM1).
Gross brain morphology with Nissl staining
Animal perfusion
Timing: 10–20 min per animal
Before starting
Pum1 mutant mice were previously generated in a B6/129 mixed background by the University of Connecticut Gene Targeting and Transgenic Facility (Chen et al., 2012).
Note: All work with fixative should be carried out under a chemical fume hood because paraformaldehyde (PFA) is highly toxic.
Prepare the following items:
500 mL bottle with 1× PBS solution.
250 mL bottle with 4% paraformaldehyde (PFA).
500 mL bottle of RNase free distilled water.
Fill the required number of 20 mL vials (one vial per brain) with about 10 mL of 4%PFA.
Prepare the pump:
Prepare the tubing system as in Figure 1A, attaching to one end of the tube a 27G needle.
Figure 1.
Brain perfusion
(A) Minipump shows all the connected tubes.
(B) Glass drop jar to hold the mouse after the anesthesia, before the procedure begins.
(C) The skin is pulled before making a small incision just large enough to insert the scissors and enlarge the gap.
(D) Cut through the diaphragm along the rib cage until you see the heart.
(E) Right atrium and the left ventricle. Insert the needle into the left ventricle and make a small incision on the right atrium.
(F) Cartoon representing how to properly insert the needle in the left ventricle (magnification on the bottom).
(G) Representative image of 5-week old Pum1−/− cerebellum with Nissl staining.
Fill the tubing system connected to the peristaltic pump (Gilson Inc. 3P Miniplus III peristaltic pump, Figure 1A) with 1× PBS until the solution starts coming from the needle drop by drop but not too slowly. The recommended speed is 5 mL/min.
Note: If there is no pump available, it is possible to perfuse the brain using a handheld syringe, but this takes considerable skill to maintain steady pressure at the desired level. We recommend a pump as to produce better, more consistent results.
-
1.
Weigh the mouse in order to calculate the amount of Avertin needed.
-
2.
Anesthetize the mouse by intraperitoneal injection (IP) of 250 mg/kg of avertin (0.5 mL of avertin solution for a 25 g mouse) and place the mouse in a glass jar (or any place with low external sound) for several minutes in order to give the anesthetic time to take effect (Figure 1B).
Note: the age, weight, and general health of the mouse will all influence the amount of time it takes for the anesthetic to work. The IACUC recommends 125–250 mg/kg IP for dosage and estimates 5 min to induce anesthesia, 15–30 min under anesthesia, and 90 min to complete recovery, but the mice might go under anesthesia in 2–3 min or it might take almost 15 min if the animal is old or obese. Therefore, you must check to see whether the mouse has truly lost consciousness by pinching the paws with a forceps — a fully anesthetized animal will not respond. For more details see (Koizumi et al., 2002; Lieggi et al., 2005a, 2005b; Meyer and Fish, 2005; Reid et al., 1999; Tarin and Sturdee, 1972; Zeller et al., 1998).
-
3.
Place the animal on its back on a flat, raised surface (you can use a plastic tray or styrofoam surrounded by a container to collect fluids).
-
4.
Gently tape the animal's paws to a plastic tray (or needle if you use a styrofoam tray) to immobilize them (Figures 1C and 1D).
-
5.
Pull the skin on the abdomen up and make an incision. Insert the scissors and enlarge the gap, making sure to avoid damaging the liver (Figure 1C).
-
6.
Gently make an incision into the diaphragm and along the rib cage to reveal the heart (Figure 1D).
Note: For convenience, incise the sternum, avoiding the lungs. This allows you to bend the upper part of the rib cage to expose the heart (Figure 1D);
-
7.
Carefully insert the pump needle into the left ventricle, making sure not to puncture any other part of the heart.
-
8.
Cut the right atrium, and at the same time, start pumping the PBS 1× solution in at 5 mL/min (Figure 1E and 1F).
Note: Thick blood should be visible. Continue to perfuse with 1× PBS until the liver changes color from dark to light brown and the body structures (paws, for instance) turn white.
-
9.
Perfuse with 15–20 mL of PBS 1×.
-
10.
Stop the pump and quickly transfer the tube in the 4% PFA container.
-
11.
Resume pumping 15–20 mL 4% PFA.
-
12.
As the PFA solution enters the circulatory system, the tail and other body parts will twitch and move.
-
13.
When the paws and tail become inflexible, stop the pump.
-
14.
Remove the needle from heart, flush with water, refill with 1× PBS for the next animal.
-
15.
Decapitate the mouse and cut the skin of the head longitudinally from the neck to between the eyes.
-
16.
With scissors cut the skull starting from the occipital region to the nasal bones.
-
17.
Make a cut between the left and right anterolateral corner of the frontal bone.
-
18.
Using forceps, detach the right and left portion of the skull bone to reveal the brain.
-
19.
Using a spatula scoop the brain away and place it in 10 mL of 4% PFA (Figure 1F).
CRITICAL: Between mice, wash the pump with RNase free distilled water for 3–4 min in order to flush out the residual 4% PFA and then fill the pump with 1× PBS.
Brain preparation
Timing: 3–5 days
-
20.
Incubate brain overnight in 4% PFA-1× PBS solution at 4°C.
-
21.
The next day change to 5% sucrose in 1× PBS and place the vial back at 4°C for 1 h.
-
22.
The next day, change to 10% sucrose in water and place the vial at 4°C for 1 h, the brain should sink to the bottom of the vial.
-
23.
Replace with 20% sucrose in water solution place the vial at 4°C from 2 h to overnight (about 16 h) until the brain will sink at the bottom of the vial.
Note: If the brain does not sink to the bottom of the vial wait until it does sink in the solution.
-
24.
Change to 30% sucrose in water solution and place at 4°C. With 30% sucrose the brain can take longer than one day to sink to the bottom of the vial.
Tissue inclusion and cryostat sectioning
Timing: 1–2 days
-
25.
Remove the brain from 30% sucrose solution, dry the brain as much as you can on a paper towel (we used Kimwipes).
-
26.
Place the brain into a freezing mold, cover with Tissue-Plus™ O.C.T.
-
27.
Place the freezing mold on a flat surface with dry ice.
-
28.
Take the frozen O.C.T. embedded brain out from the mold and section it on Leica CM3050S Cryostat at 30 μm thickness.
-
29.
Mount sections on gelatin-subbed microscope glass slides either directly or collect the slides in a 1× PBS solution in a 12 well plate with no more than 4–5 sections per well.
Note: brain sections in 1× PBS can be stored at 4°C for a short time. If you wish to store them for more than one week, prepare a 1× PBS solution with 0.02% of sodium azide to prevent bacterial growth. This should allow storage at 4°C for about 30 days.
Note: for frozen sectioning, it is also possible to snap-freeze the tissue without the perfusion, but then blood will remain in the tissue. For applications like Nissl staining or immunofluorescence it is important to clear blood from the tissue and fix it with PFA before placing in O.C.T.
-
30.
Dry-mount the slides on the Fisherbrand™ Slide Drying Bench (FisherScientific) at 60°C for several hours or at room temperature overnight.
Nissl staining
Timing: 30–40 min
Prepare 300 mL of ethanol 100%, 95% and 70% in distilled water. Place six glass jars under a chemical fume hood and fill each with one of the following six solutions: 1) 100% 200-proof ethanol, 2) 95% 200-proof ethanol, 3) 70% 200-proof ethanol, 4) distilled water, 5) Cresyl violet solution, 6) Xylene.
-
31.Take the mounted brain slides and immerse them in the following order:
-
a.100% Ethanol for 1–2 min;
-
b.95% Ethanol for 1 min;
-
c.70% Ethanol for 1 min;
-
d.water for 2–3 dips;
-
e.Cresyl violet solution for 2–3 min;
-
f.water for 2–3 dips;
-
g.70% Ethanol for 1 min;
-
h.95% Ethanol for 1 min;
-
i.100% Ethanol for 1 min;
-
j.Xylene for 1 min.
-
a.
Note: immerse the glass slides very carefully to keep the brain slices from sliding out. Always use RNase-free water to treat the slides and to prepare the ethanol solutions.
-
32.
Place 3–4 drops of Permount Mounting Medium (Electron Microscopy Sciences) on the slide.
-
33.
Hold the slide at a slant. Gently, slowly move the coverslip from the bottom edge of the slide to the top in order to allow air to be replaced with Permount. Gently push the cover glass with finger. Wipe the other side of the slide with a Kimwipe and let dry for at least 3 hours to overnight.
-
34.
Images are now ready to be acquired in a bright field microscope (Figure 1G).
Note: If you want to store the slides, please allow 1–2 days for them to dry completely at room temperature before storing them in boxes. It is possible to work with the slides before they are completely dry, but in this case be careful that the cover slip does not fall apart.
Plating and preparing dissociated cultured neurons on coated coverslips for Sholl analysis
Cover slip coating
Timing: 1 h to overnight
-
35.
Under a laminar flow cabinet place one glass cover slip for each well of a 12-well plate.
-
36.
Add 400 μL of coating medium.
-
37.
Incubate at 37°C for 1 h or overnight.
Note: Perform the following steps the day of hippocampal dissociation (see step 40 of tissue harvesting and dissociation section)
-
38.
Wash three times with 4 mL of sterile water.
-
39.
Let dry open under the biosafety cabinet for at least 15 min before seeding.
Tissue harvesting and dissociation
Timing: 1 h and 20 min
Note: Primary hippocampal neurons were dissected from neonatal mice.
CRITICAL: Maintain sterile conditions throughout the entire procedure.
-
40.
Remove the mouse's head and place it in a 10 cm petri dish filled with 5 mL of Hanks’ balanced salt solution (HBSS) 1×.
-
41.
Make a careful incision into the skin and the skull, carefully remove the brain, and transfer it to a 10 cm plate with HBSS 1× (Figure 2A).
-
42.
Separate the two hemispheres (Figures 2B and 2C).
-
43.
Remove the midbrain and meninges.
-
44.
Locate the hippocampus and dissect it using small forceps (Figure 2D).
-
45.
Once four brains are dissected, pool them together in a 15 mL conical tube containing 5 mL of HBSS 1×.
-
46.
Remove the HBSS 1× without aspirating the dissociated hippocampi.
-
47.
Add 1.5 mL pre-warmed TrypLE and pipette up and down with a p1000 in order to mechanically dissociate the tissue.
-
48.
Incubate at 37°C for 15–20 min.
-
49.
During the incubation time aspirate the coating medium from the 12 well plate.
-
50.
Wash 3 times with sterile water.
-
51.
Add plating media in all wells.
-
52.
Remove TrypLE from the 15 mL tube and slowly add 5 mL of HBSS 1×.
-
53.
Centrifuge at 800 g for 5 min.
-
54.
Wash twice with HBSS 1×.
-
55.
Add 750 μL of pre-warmed plating medium to the cells.
-
56.
Dissociate using a p1000 and a p200 pipette, pipetting up and down for 15 times (or until no more cell clumps are visible) (Figure 2E).
-
57.
Put cells through 70 μm cell strainer collecting the cells in a 50 mL conical tube.
-
58.
Transfer the cells in a 1.7 mL Eppendorf tube.
-
59.
Count the cells with a hemocytometer and adjust the volume to 0.5 × 106 cells/mL.
-
60.
Plate ∼3.5 × 105 cells per well in a 12-well plate support (Figure 2F).
-
61.
On the next day (first day in vitro, i.e., DIV1), remove plating media and replace with 1 mL of growth medium.
-
62.
On DIV4, replace half of the medium with fresh medium containing 10 μM final concentration of 5-Fluoro-2′-deoxyuridine/Uridine FdU/U. Repeat this step every 2 days.
Note: We add FdU/U to the growth medium to inhibit the growth of glial cells.
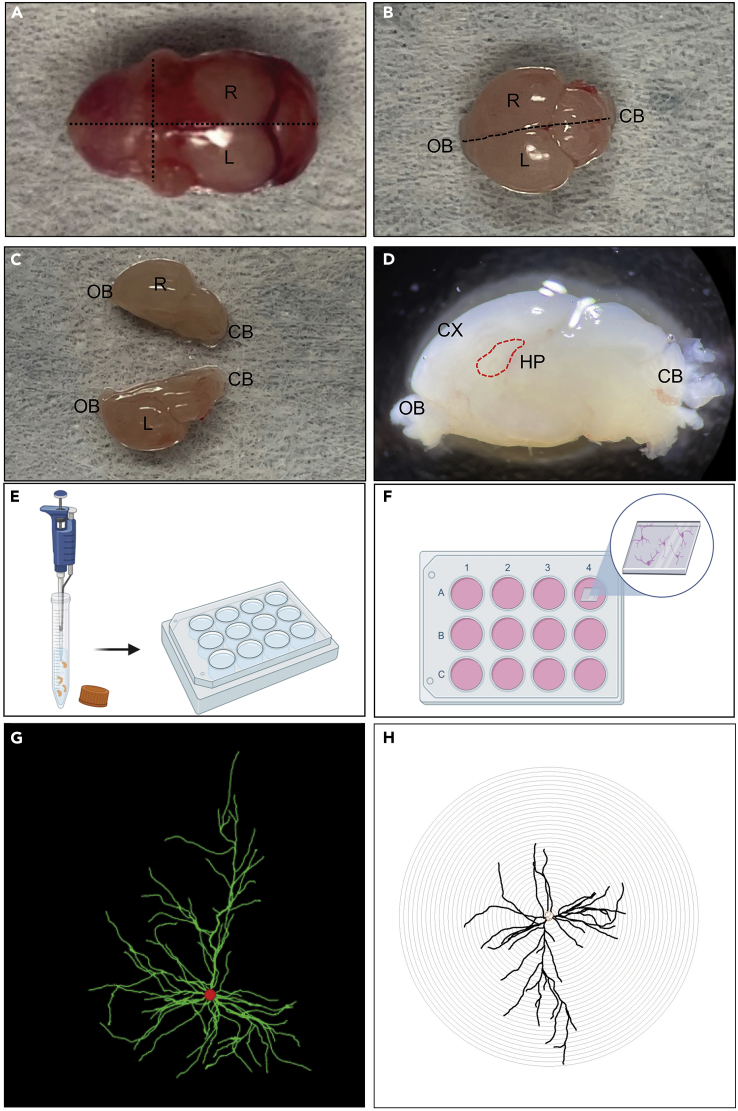
Figure 2.
Dissection of the hippocampus
(A) neonatal (P0) dissected head, the dotted lines indicate where to cut to open the skull.
(B) P0 brain after skull removal. The dotted line indicates where to cut in order to divide the hemispheres.
(C) P0 brain after the symmetrical cut.
(D) Sagittal view of P0 brain. Dotted circle indicates the point to dissect in order to obtain the hippocampus (HP).
(E) Schematic representation of hippocampal cell dissociation after collection.
(F) Schematic representation of primary hippocampal neurons seeded on coated glass coverslip. R: right brain hemisphere, L: left brain hemisphere, CX: cortex, HP: hippocampus, OB: olfactory bulbs, CB: cerebellum.
(G) Representative micrograph of single hippocampal neuron with visible dendrite arborization (in green) and the soma (red circle).
(H) Output of a single neuron by Sholl analysis in order to quantify dendritic arborization.
Transfection of cultured neurons with plasmids
Timing: 60 min
Note: The 3×HA-PUM1 mutants were generated by mutagenesis from wild-type (WT) constructs as described above. Prepare pEGFP-C1 and 3×HA-PUM1 plasmids by DIV9 using an endotoxin-free Plasmid Midi Kit.
-
63.
At DIV9, check the quality of the plated neurons by observing them under a microscope.
Note: At DIV9 the neurons have extended their neurites, healthy neurons have multiple neurites and will be attached to the surface of the coverslip. You may see debris surrounding the cells, which is an indicator of cellular distress and death; if there is a lot of debris, you need to adjust your culture or image acquisition procedures. It is also important to keep the number of glial cells to a minimum, because they will quickly outcompete neurons in growth and make further analysis difficult.
-
64.
For each transfection, prepare Solution A and Solution B (as in the table below) in separate sterile microcentrifuge tubes according to the manufacturer’s instructions (CalPhos Mammalian Transfection Kit, see key resources table).
| Solution A – Transfection mix | |
|---|---|
| Reagent | Amount |
| 2 M Calcium solution | 6.2 μL |
| pEGFP-C1plasmid | 500 ng |
| 3×HA-PUM1plasmid | 500 ng |
| H2O milliQ | Up to 50 μL |
Note: Solution B is a solution of 2× HEPES-buffered saline (2× HBS) provided by the CalPhos Mammalian Transfection Kit.
-
65.
Vortex Solution A while adding Solution B (2× HBS) dropwise (usually takes 30 s–1 min per reaction).
-
66.
Incubate the mixed A+B solution at 24°C–26°C for 30 min.
-
67.
Gently add the A+B solution dropwise to wells of a 12-well plate containing coverslip-plated neurons.
-
68.
Gently move the plate back and forth to evenly distribute the transfection solution.
-
69.
Incubate the plate at 37°C in a 5% CO2 incubator for 8 h to overnight.
-
70.
Replace the medium containing transfection solution with growth medium.
-
71.
24 h later, assess the success of transfection by monitoring pEGFP fluorescence in neurons under a fluorescence microscope.
Note: GFP+ neurons start to become visible under a fluorescence microscope ∼12 h after transfection. The fluorescence will be visible in both soma and neurites.
Immunocytochemistry
Timing: about 20 h
Note: i) The earlier the imaging can be done following mounting, the better. All our imaging was completed within 2 weeks of the initial immunocytochemistry to ensure the best staining; ii) any standardized protocol for imaging can be used. The immunocytochemistry protocol will need to be optimized for the antibodies that you are using and the cells of interest; iii) we imaged with a Zeiss LSM800 confocal microscope and used the Zen2.6 software suite for image acquisition and processing.
-
72.
At DIV14, aspirate growth medium and wash coverslip-plated neurons by adding ice-cold PBS.
Pause point: before immunocytochemistry procedures, inspect all transfected neurons under a fluorescence microscope to ensure they are healthy and attached to the plate surface with minimal cellular debris.
-
73.
Add a solution of 4% PFA in order to fix the cells and incubate for 10 min at room temperature.
-
74.
Aspirate the fixing solution and wash twice neurons by adding ice-cold PBS.
-
75.
Add permeabilization/blocking solution to 12-well plates and incubate at room temperature for 1 h.
-
76.
Aspirate the permeabilization/blocking solution and wash three times for 5 min each, with ice-cold PBS.
-
77.Incubate with primary antibodies as follows:
-
a.prepare 100 μL of primary antibody solution (anti-GFP [diluted 1:1,000] and anti-HA [diluted in 1:100] in blocking solution) per coverslip.
-
b.place a 100 μL drop of primary antibody solution on a parafilm-coated plate for each coverslip.
-
c.retrieve the coverslips using forceps, place onto drops of primary antibody solution, and incubate at 4°C for 16 h.
-
a.
-
78.
Transfer the coverslip-plated neurons to wells of a 12-well plate filled with 1× PBS and wash three times for 10 min each time.
-
79.Incubate with secondary antibodies as follows:
-
a.prepare 100 μL of secondary antibody solution (Goat-anti-mouse-Alexa 488 [diluted 1:500] and Donkey-anti-goat-Alexa 594 [diluted in 1:150] in Blocking solution) for each coverslip. Please note that Alexa 594 is used only if you want to double-stain for HA and PUM1; otherwise, you can just use Alexa 488.
-
b.place 100 μL drop of secondary antibody solution on a parafilm-coated plate for each coverslip.
-
c.retrieve coverslips using forceps, place onto drops of secondary antibody solution, and incubate at 24°C–26°C for 1 h.Note: Cover the plate in aluminum foil after adding the secondary antibodies to avoid exposure to light. Perform all further steps in as little light as possible.
-
d.Transfer the coverslip-plated neurons to wells of a 12-well plate filled with 1× PBS and wash three times for 10 min each time
-
e.prepare microscope slides (Superfrost Plus; Fisher Scientific) and place a drop of VECTASHIELD antifade DAPI mounting medium on the surface of the slide.
-
f.retrieve coverslips from the 12-well plate using forceps and place on slide glasses with neurons facing down.
-
g.seal each coverslip with transparent nail polish.
-
a.
Confocal imaging and analysis
Timing: 2 days
We strongly recommend that you image the samples right after staining because the fluorescence decreases over time. In order to have comparable images, we suggest acquiring all the images over the course of a single confocal session. The stained slides can be stored at 4°C.
-
80.
Acquire Z-stack images with parameter settings using a laser-scanning confocal microscope (Zeiss LSM710) equipped with a 63× objective and Zen2.6 Software (Zeiss).
Note: For these experiments we used the LSM710, but any confocal microscope equipped with a 63× objective can be used.
-
81.
Generate maximum intensity projections for all captured Z-stack files (Figure 2G).
-
82.
Perform neuron tracing and Sholl analysis using Neurolucida 360 (Figure 2H) (MBF Bioscience, Williston, VT, key resources table).
Note: As an internal control, the expression level for each variant was quantified based on the fluorescence signal intensity of the HA tag. Specifically, cells expressing 3×HA-PUM1 were identified and analyzed using the software Imaris (see key resources table). The mean intensity of HA at the cell body was used to quantify PUM1 overexpression for each variant and WT.
Note: To see the final output, you can view all the 212 neurons imaged in (Gennarino et al., 2018) at the following resource website: http://neuromorpho.org/NeuroMorpho_Linkout.jsp?PMID=29474920.
Expected outcomes
The first part of the protocol describes how to perfuse and dissect a mouse brain in preparation for Nissl staining to evaluate brain gross brain morphology. In order to study the morphology of brain and brain regions it is important to master the perfusion steps as well as the cryostat cutting.
The second part of the protocol describes how primary neurons are a great resource to study how different gene variants influence dendritic morphology. We used primary neurons derived from neonatal (P0) mice hippocampi and transduced them with different variants of PUM1 along with a GFP-expressing plasmid. Experimental success requires a large amount of healthy, live-cultured primary neurons, with minimal cell debris and very few glial cells. For Sholl analysis, the signal intensity of GFP in transfected primary neurons needs to be strong enough to visualize the entire dendritic arbor (Gennarino et al., 2018)
Limitations
Ex-vivo culture of neurons can be challenging because primary neurons are very delicate and they have limited lifespan in culture. In addition, the general health of the neurons as well as the amount of debris and glial cells could affect the transfection efficiency. In order to perform Sholl analysis we strongly recommend using a co-transfection with a GFP-expressing plasmid. This will allow quick visualization of transfected neurons and easier analysis readout.
Mouse perfusion, brain dissection and sectioning require expertise that can be developed with practice. We recommend doing a trial run of the entire procedure on wild-type control mice before performing the real experiment.
Troubleshooting
Problem 1
Low viability of seeded neurons.
Potential solutions
It is possible that the procedure to dissociate the primary neurons was too harsh and that this led to cell death. We recommend gently dissociating the cells mechanically, as pipetting too many times can reduce the yield of live cells. We recommend pipetting no more than 15–20 times.
Problem 2
Overgrowth of glial cells.
Potential solutions
If overgrowth of non-neuronal cells is present in the dish, it is very likely that glial cells are contaminating the neuron culture. These cells have the ability to duplicate, it is very important to add 10 μM final of 5-Fluoro-2′-deoxyuridine/Uridine FdU/U to the medium every time it is changed.
Problem 3
High background in Immunofluorescence (IF) experiment.
Potential solutions
High background could be resolved by: i) Titrating the amount of NGS in Blocking solution from 2% up to 10%; ii) Adjusting the amount of secondary antibody used; iii) optimizing the duration of the experiment.
Problem 4
Intensity of the Nissl staining is not satisfactory.
Potential solutions
Repeat the rehydration with ethanol (steps 31a–d) and then restart from step (31e) by increasing the Nissl staining time to 5–7 min. For overstained sections, keep slides in ethanol until they achieve the desired appearance and proceed to the Xylene step (step 31j).
Problem 5
Mouse is not perfused well.
Potential solutions
It can happen that the mouse is not well perfused and this is verifiable from the stiffness of the tail and the overall body after the PFA step. Check for any fluid coming from the nose or the mouth of the animal, which indicates that the operator punctured the lungs. If this happens, consider starting over with a new mouse. If this option is not feasible, consider elongating the post-fix with 4% PFA from overnight to 24 h.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Vincenzo A. Gennarino (vag2138@cumc.columbia.edu).
Materials availability
This study did not generate unique reagents.
Acknowledgments
We thank members of the Gennarino lab for helpful discussions. We also thank V. Brandt for essential input on the manuscript. The Pum1 mouse model was a generous gift of Prof. Haifan Lin of Yale University. This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS; R01NS109858 to V.A.G.) and the Paul A. Marks Scholar Program, Columbia University Vagelos College of Physicians and Surgeons (V.A.G.).
Author contributions
V.A.G. devised the original protocol and wrote the manuscript. N.d.P. wrote the technical sections, edited the manuscript, and re-optimized each step of the protocol. A.C., W.L., and S.B. contributed to the technical sections. A.C. wrote the Nissl staining and perfusion section of the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
No software was generated for this project. All software used in this study is publicly available and links are provided as appropriate in different sections of the methods and key resources table.
References
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T., et al. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049–1060. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zheng W., Lin A., Uyhazi K., Zhao H., Lin H. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr. Biol. 2012;22:420–425. doi: 10.1016/j.cub.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino V.A., Palmer E.E., McDonell L.M., Wang L., Adamski C.J., Koire A., See L., Chen C.A., Schaaf C.P., Rosenfeld J.A., et al. A mild PUM1 mutation is associated with adult-onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell. 2018;172:924–936.e11. doi: 10.1016/j.cell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi T., Maeda H., Hioki K. Sleep-time variation for ethanol and the hypnotic drugs tribromoethanol, urethane, pentobarbital, and propofol within outbred ICR mice. Exp. Anim. 2002;51:119–124. doi: 10.1538/expanim.51.119. [DOI] [PubMed] [Google Scholar]
- Lieggi C.C., Artwohl J.E., Leszczynski J.K., Rodriguez N.A., Fickbohm B.L., Fortman J.D. Efficacy and safety of stored and newly prepared tribromoethanol in ICR mice. Contemp. Top. Lab. Anim. Sci. 2005;44:17–22. [PubMed] [Google Scholar]
- Lieggi C.C., Fortman J.D., Kleps R.A., Sethi V., Anderson J.A., Brown C.E., Artwohl J.E. An evaluation of preparation methods and storage conditions of tribromoethanol. Contemp. Top. Lab. Anim. Sci. 2005;44:11–16. [PubMed] [Google Scholar]
- Meyer R.E., Fish R.E. A review of tribromoethanol anesthesia for production of genetically engineered mice and rats. Lab. Anim. 2005;34:47–52. doi: 10.1038/laban1105-47. [DOI] [PubMed] [Google Scholar]
- Reid W.C., Carmichael K.P., Srinivas S., Bryant J.L. Pathologic changes associated with use of tribromoethanol (avertin) in the Sprague Dawley rat. Lab. Anim. Sci. 1999;49:665–667. [PubMed] [Google Scholar]
- Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin D., Sturdee A. Surgical anaesthesia of mice: evaluation of tribromo-ethanol, ether, halothane and methoxyflurane and development of a reliable technique. Lab. Anim. 1972;6:79–84. doi: 10.1258/002367772781082668. [DOI] [PubMed] [Google Scholar]
- Zeller W., Meier G., Bürki K., Panoussis B. Adverse effects of tribromoethanol as used in the production of transgenic mice. Lab. Anim. 1998;32:407–413. doi: 10.1258/002367798780599811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No software was generated for this project. All software used in this study is publicly available and links are provided as appropriate in different sections of the methods and key resources table.