Abstract
Inborn errors of metabolism have traditionally been viewed as the quintessential single gene disorders; defects in one gene leads to loss of activity of one enzyme causing a metabolic imbalance and clinical disease. However, reality has never been quite that simple, and the classic “one gene-one enzyme” paradigm has been upended in many ways. Multiple gene defects can lead to the same biochemical phenotype, often with different clinical symptoms. Additionally, different mutations in the same gene can cause variable phenotypes, often most dramatic when a disease can be identified by pre-symptomatic screening. Moreover, response to therapy is not homogeneous across diseases and specific mutations. Perhaps the biggest deviation from traditional monogenic inheritance is in the setting of synergistic heterozygosity, a multigenic inheritance pattern in which mutations in multiple genes in a metabolic pathway lead to sufficient disruption of flux through the pathway, mimicking a monogenic disorder caused by homozygous defects in one gene in that pathway. In addition, widespread adoption of whole exome and whole genome sequencing in medical genetics has led to the realization that individual patients with apparently hybrid phenotypes can have mutations in more than one gene, leading to a mixed genetic disorder. Each of these situations point to a need for as much precision as possible in diagnosing metabolic disease, and it is likely to become increasingly critical to drive therapy. This article examines examples in traditional monogenic disorders that illustrates these points and define inborn errors of metabolism as complex genetic traits on the leading edge of precision medicine.
1. Introduction. A trend towards more accurate genetic diagnoses and treatment
Hereditary rare diseases affect approximately 1 in 20 people, but only a minority of patients receive a specific genetic diagnosis [1-4]. Before the advent of next generation sequencing, the diagnostic odyssey typically began with a symptom-based diagnostic approach, moved through screening tests, often by multiple care providers, and resolved with specific gene tests only if sufficient clues were present to allow sequencing of a single gene. Multi-gene panels broadened the scope of clinical sequencing, triggering genetic testing based on a clinical phenotype. More recently, whole exome and whole genome sequencing (WES and WGS, respectively) have become the standard for genetic diagnosis [5]. They have the advantage of bypassing extensive preliminary, non-specific testing, thus leading to more rapid diagnoses, often at a significantly reduced cost and with less emotional turmoil for families. Of note, as many as 5% of patients undergoing WES or WGS provide evidence of being concurrently affected by two genetic disorders rather than a single disorder [6]. In retrospect, these patients may be recognized as having symptoms of both disorders, and in disease specific studies would have inappropriately expanded the phenotype of one or the other of the disorders. Although it has many definitions, precision medicine must start with an accurate diagnosis, and has come to the forefront as a guiding principle for medical treatment largely through recognition of the power of WGS and WES. However, precision medicine has also come to mean the use of genetic information to direct therapy. In most cases to date, this has focused on pharmacogenomics, but also can reflect the use of designer drugs specific to a single mutation in a rare disease (CF example), or targeted to a type of cancer (CAR T) [7,8]. In pharmacogenomics, the simultaneous analysis of genes encoding proteins associated with drug metabolism (pharmacokinetics), drug targets (pharmacodynamics), and drug exposure leading to adverse drug reactions has provided more accurate therapy to treat IEM symptoms [9-11]. In either setting, accurate genetic information and phenotype are critical to therapeutic success.
In many ways, diagnosis and treatment of inborn errors of metabolism (IEMs) presaged the current focus on precision medicine and lead to the description of IEMs as examples of complex genetic traits [12]. Diagnosis was typically through broad based metabolite panels rather than individual tests, and the pathophysiology of the enzyme defect drove therapy. However, directed molecular or enzymatic testing was still necessary as metabolites could be non-specific; for example, methylmalonic acidemia is not one disease by nearly a dozen [13]. Pre-symptomatic screening, especially newborn screening, also identified individuals with a wide variety of disease-causing variants for most disorders, some of which predicted outcome more accurately than the analyte itself, as, for example, with the common C.985A > G mutation in MCAD deficiency. Recently, the field of IEMs has come full circle, with many new disorders identified by WES or WGS rather than metabolite testing.
Identification of patients with single mutations in multiple steps of a metabolic pathway with probable clinical effect was originally recognized in the context of energy deficiency, a phenomenon referred to as synergistic heterozygosity, but has been suggested to play a role in other disorders [14-17]. This article will examine diagnosis and treatment of IEMs in the context of precision medicine and present novel findings in several disorders that broadens the potential scope of the concept of synergistic heterozygosity.
2. Disorders of mitochondrial energy metabolism: new cellular based methods to evaluate synergistic heterozygosity
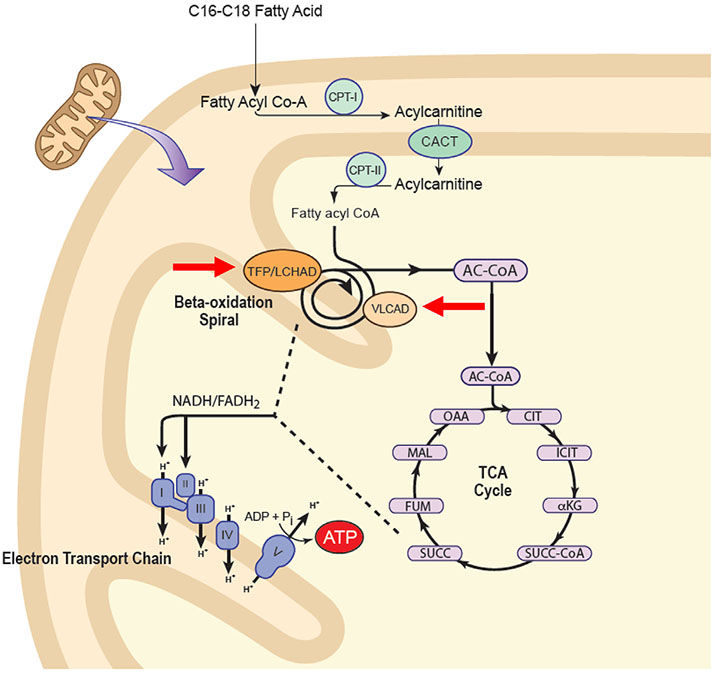
Mitochondrial energy metabolism is a complex process that involves optimum function and efficient interaction of three mitochondrial metabolic cycles: oxidative phosphorylation (OXPHOS), fatty acid oxidation (FAO), and the tricarboxylic acid (TCA) or Krebs cycle (Fig. 1) [18]. OXPHOS is organized into inner mitochondrial membrane supercomplexes that promote substrate channeling and catalytic efficiency, and the enzymes of FAO functionally and physically interact with the supercomplexes to optimize delivery of reducing equivalents from high energy fat stores to OXPHOS [19,20]. It is therefore not surprising that perturbation of energy metabolism is often more complicated than a simple Mendelian trait. A clear example of this phenomenon is seen in the disorders of long chain fatty acid metabolism (LC-FAOD). Here, three main symptoms – hypoglycemia, cardiomyopathy, and rhabdomyolysis – have long been viewed as direct outcomes of decreased cellular ATP levels due to reduction of reducing equivalents from FAO to OXPHOS. However, evidence that a secondary TCA cycle depletion exacerbates this primary energy deficiency led to the development of the novel anaplerotic therapeutic agent triheptanoin that essentially eliminates hypoglycemia in patients with LC-FAODs and greatly improves heart function [18,21,22]. Nevertheless, most patients continue to have episodes of rhabdomyolysis (albeit decreased in number and therapy) in spite of restored of mitochondrial energy homeostasis [18]. Focus is now turning to other possible causes of rhabdomyolysis, most importantly otherwise subclinical inflammation that appears to be induced by the presence of metabolites that accumulate due to the block in FAO. In this regard, we have demonstrated accumulation of increased levels of mitochondrial superoxides in fibroblasts from patients with very long chain acyl-CoA dehydrogenase (VLCAD) deficiency [23]. As in the inflammatory response to pegvaliase in phenylketonuria (PKU), genetic variation in proteins involved at any point the pertinent inflammatory pathway may be relevant in determining the clinical phenotype or response to therapy [24].
Fig. 1.
Possible sites of defects leading to synergistic heterozygosity in disorders of energy metabolism. Multiple enzymes must function together to produce optimum energy output. In the first patient described in the text, heterozygous mutations in ACADVL and mitochondrial HADHA were identified (red arrows). We have previously described patients with symptoms of energy deficiency and heterozygous mutations in fatty acid oxidation and the electron transport chain. Adapted from [61].
Precision medicine in the context of mutation specific therapy is likely to be an important factor in the treatment of LC-FAOD in the coming years. Expression for all of the genes of FAO, as well as many for OXPHOS, is controlled from a single regulatory element for RNA transcription, the PPARδ promotor, while the PPARγ controls an antioxidant pathway that is important in protecting mitochondria against oxidant stress. Thus, treatment of patients with a point mutation in a LC-FAO gene with a PPARδ agonist is a compelling option, leading to both an increase in even a minimally active protein, especially if it has some PPARγ activity leading to induction of the mitochondrial antioxidant pathway [25]. A trial of one such agent is currently in progress (https://www.clinicaltrials.gov/ct2/show/NCT03833128?term=reneo&rank=2). Similarly, increased reactive oxygen species in VLCAD deficient fibroblasts could be reduced with subsequent improved cellular bioenergetics using a potent mitochondrially targeted antioxidant [23].
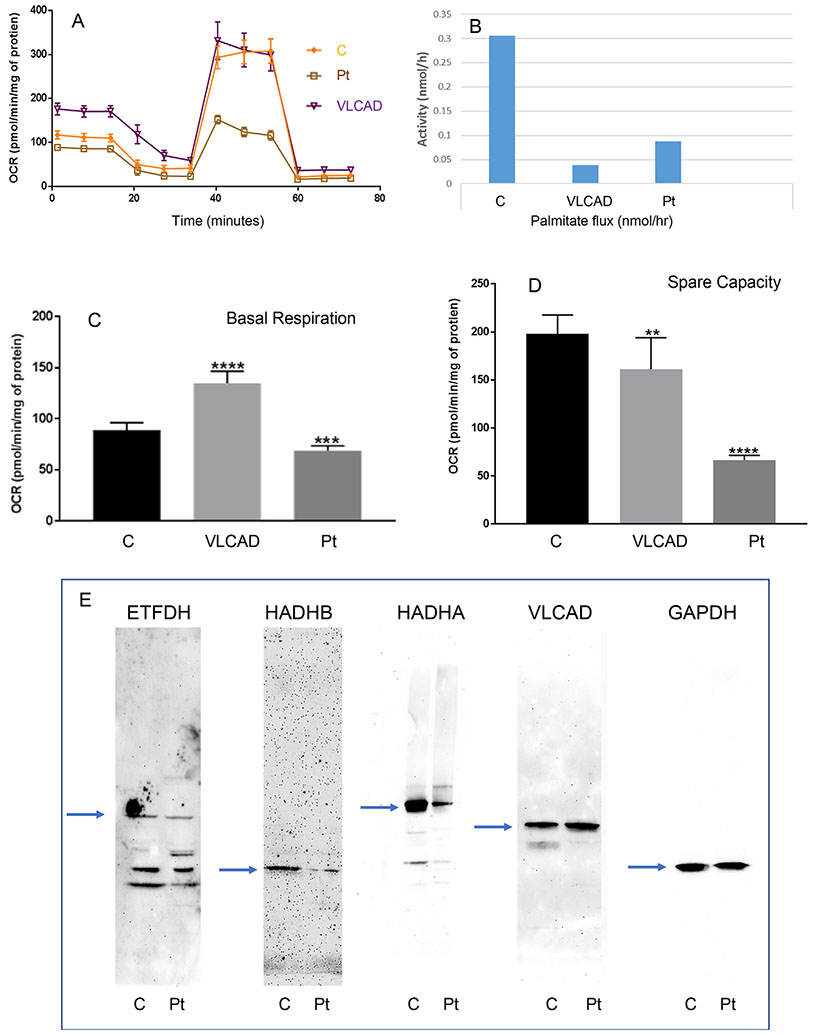
Finally, recognition of a multi-functional energy complex containing both OXPHOS and FAO provides a potential functional explanation for the phenomenon termed synergistic heterozygosity. Originally, patients with clinical symptoms suggestive of energy deficiency but without a specific single diagnosis were shown to have multiple partial defects in both energy pathways [12,14]. Synergistic heterozygosity can theoretically occur due to partial reduction of flux through a metabolic pathway caused by reductions in catalytic efficiency at multiple steps [12]. Alternatively, stability of a multi-enzyme complex may be reduced due to changes in the structure of multiple proteins in the complex, leading to reduction in catalytic efficacy of the whole pathway [19,20]. As an example, we routinely are asked to evaluate cellular energy function in patients with abnormal newborn screens consistent with a LC-FAOD but in whom only a mutation on a single allele in one gene is identified. In this context, cellular bioenergetics are usually normal, and heterozygosity is assumed. However, we have also evaluated cell lines with single mutations in two LC-FAO genes. Fig. 2 shows results from one such cell line from a patient identified with an abnormal newborn screen (elevated C12, C10, C8, C6, C10:1, 5DH + C10OH, C14, C14:1, and C14:2) suggestive of multiple acyl-CoA dehydrogenase deficiency (GA2) but who has remained asymptomatic. Ethylmalonic acid was elevated in urine (36.78 mg/g Cr; normal 0.5–20.2). Molecular studies identified heterozygous mutations in the electron transfer flavoprotein dehydrogenase (ETFDH; c413T > G; pL138R) gene and the gene for the beta subunit of the mitochondrial trifunctional protein (HADHB;c693_696dupCGC; pA233RFS*12). ACADS and ETHE1 variants (other causes of ethylmalonic aciduria) were not identified. The cell line showed appropriately decreased amounts of ETFDH and HADHB protein (but not zero), neither of which alone should affect cellular bioenergetics alone (Fig. 1E). HADHA was also reduced (often seen in the context of HADAB mutations) and VLCAD protein level was normal. Whole cell palmitate oxidation (Fig. 1B) was reduced, not typical of heterozygote cells. Equally striking, the cellular basal respiratory rate and reserve capacity were decreased (Figs. 1A, C, and D), consistent with reduction of OXPHOS activity. As previously described, a VLCAD deficient cell line shows increased basal respiratory rate but decreased reserve capacity, consistent with mitochondrial bioenergetic stress [23].
Fig. 2. Analysis of a synergistic heterozygous cell line.
A. Oxygen consumption rate (OCR) was measured with a Seahorse XFe96 Extracellular Flux Analyzer (Agilent, Santa Clara, CA). Cells were seeded in 96-well Seahorse tissue culture microplates in growth media at a density of 60,000 cells per well. Initial OCR was measured to establish basal respiration (C) followed by injection of oligomycin (an inhibitor of ATP synthase) that reduces OCR, representing ATP turnover. Subsequent injection of 300 nM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, Seahorse XF Cell Mito Stress Test Kit, Santa Clara, CA) dissipates the proton gradient and allows maximum respiration. The rise in OCR upon FCCP addition represents mitochondrial reserve capacity (D). Finally, rotenone and anti-mycin A were added to effectively disable the electron transport chain and inhibiting the total mitochondrial respiration. The remaining OCR represents non-mitochondrial respiration. In all panels, C = control cell line; VLCAD = VLCAD deficient cell line; Pt = cells from the patient with the ETFDH and HADHB mutations. B. Flux through the FAO pathway was quantified by production of 3H2O from [9,10-3H] palmitate. E. Western blots of cellular extracts probed with antibodies to the proteins indicated at the top of each panel. The arrows indicate the expected migration of the native protein (ETFDH, 68 kDa; HADHB, 50 kDa; HADHA, 75 kDa; VLCAD, 48 kDH; GAPDH loading control, 37 kDa).
It has been proposed that cellular FAO flux rather is a better predictor of phenotype than VLCAD activity in cells from patients with VLCAD deficiency [26,27]. However, one of us ((TGJD) was involved in the case of a newborn with VLCAD deficiency who died before newborn screening is traditionally performed in the Dutch screening program. The baby became sick on day one of life with concurrent feeding difficulties and hypothermia and presented to hospital on day two of life in cardiac arrest, where resuscitation was unsuccessful. Postmortem investigations were suspicious for an inborn error of fat metabolism with fatty infiltrate in several organs, and an acylcarnitine profile in cultured fibroblasts was consistent with VLCAD deficiency. Molecular testing demonstrated compound heterozygosity in the ACADVL gene: a paternal c.253dup (p.Asp85Glyfs*19) mutation and a presumed de novo C.469A > G (p.Asn157Asp) variant of unknown significance not found in either parent. Fibroblasts displayed ~45% FAO flux of that measured in control fibroblasts. Thus, the in vivo situation is more complex for neonates. Perinatal physiology transitions include a shift from a passive towards active glucose homeostasis in the liver; from primarily glucose towards free fatty acid driven energy metabolism in the heart; and from passive to active thermogenesis requiring FAO in brown adipose tissue. All of these changes in the newborn must be taken into account and are likely to be driven in some measure by genetic factors. Evaluation of additional examples of such combinations of gene variants in FAO and OXPHOS genes will allow a better understanding of the impact of these effects in patients with pictures of non-classical energy deficiency, and encourage development of therapies targeting stabilization of the energy protein complex rather than focusing on correcting a single enzyme defect. Precision medicine, with application of WES or WGS to identify potential combinations of mutant alleles, is critical to this understanding. This is where the clinic really meets the genetics.
3. Glycogen storage disorders: a new example of synergistic heterozygosity
The glycogen storage diseases (GSDs) are a group of inherited metabolic diseases caused by abnormal degradation or synthesis of glycogen due to diminished or absent enzyme activity [28,29]. The literature describes two different groups of GSDs: hepatic and muscular. Hepatic GSDs are characterized by hypoglycemia, fasting intolerance, and failure to thrive. Hepatomegaly is a cardinal feature of all hepatic GSDs except for glycogen synthase deficiency (GSD 0). GSD type I is a hypoketotic disorder, while GSD 0, III, VI, IX, and XI are ketotic forms of GSD. The readily identified clinical phenotype of hypoglycemia with ketosis, but without identification of biallelic mutations in a gene for one of the enzymes in the glycogen degradation pathway frequently leads to the non-specific diagnosis of “ketotic hypoglycemia” [30]. However, more aggressive evaluation of such patients with WES or WGS has allowed identification of patients with single mutations in multiple steps of the glycogen metabolism pathway, another potential example of synergistic heterozygosity.
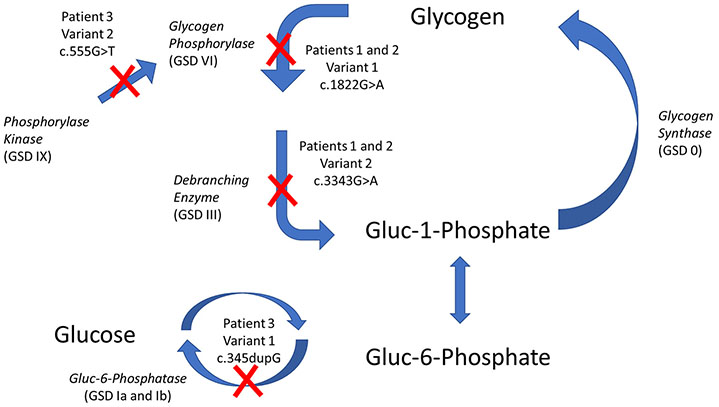
Tables 1 and 2 summarize laboratory, clinical, and molecular findings from three previously unpublished patients. All three patients (patients 1 and 2 were identical twins) presented with a constellation of findings characteristic of a hepatic GSD, including fasting intolerance with ketotic hypoglycemia and mild hepatomegaly; however, a traditional monogenic enzyme deficiency could not be identified. All patients developed overnight ketosis with low prealbumin concentration, characteristic of the ketotic forms of GSD. Additionally, all of the patients responded well to traditional GSD treatment therapies as outlined in the consensus guidelines for the ketotic forms of GSD, and the metabolic abnormalities abated. In each case, the symptoms recurred when treatment was delayed or missed, supporting its benefit. WES analysis of each patient revealed multiple single allelic genetic variants in genes that participate glycogen metabolism (Table 2 and Fig. 3) likely explaining the clinical and biochemical phenotype of GSD. Thus, while functional cellular testing was not available to prove causation, the association of variants in related genes in glycogen metabolism is striking and may represent another example of synergistic heterozygosity. It is worth noting that while each patient presented with symptoms of GSD and was diagnosed based on the phenotypic presentation, traditional focused Sanger sequencing would have missed the diagnosis since only one genetic change was found per gene. The field of genetics is evolving, and precision medicine requires a more comprehensive analysis for complex inheritance patterns including synergistic heterozygosity.
Table 1.
Laboratory and clinical characteristics of the patients with GSD-like symptoms.
| Patient | Clinical symptoms | Pertinent laboratory findings | ||||||
|---|---|---|---|---|---|---|---|---|
| Hypoglycemia | Acidosis | Ketonemia | Hepatomegaly | Hypertriglyceridemia | Additional findings | Liver biopsy | ||
| 1 | Respiratory distress at birth (premature 34 week) | ✓ | ✓ | ✓ | ✓ | ✓ | Ballooning of the hepatocytes ↑Glycogen content ↓ | |
| Hepatomegaly Mild gross and fine motor development delay Failure to thrive | Debranching enzyme activity ↓ Total phosphorylase activity | |||||||
| 2 | Hepatomegaly Failure to thrive | ✓ | ✓ | ✓ | ✓ | ✓ | Neonatal hyperbilirubinemia | |
| 3 | Seizures Short stature Hypotonia Sensory disturbances | ✓ | ✓ | ✓ | ✓ | |||
Table 2.
Genetic Studies in patients with GSD-like symptoms.
| Patient | Variant 1 | Variant 2 | Other variants | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene; (Associated disease) |
Genetic variation | Predicted significance (Polyphen) |
Gene; associated disease |
Genetic variation | Predicted Significance (Polyphen) |
Gene; associated disease |
Genetic variation | Predicted significance (Polyphen) |
|
| 1 and 2 | PYGL; (GSD VI) | p.G608S (c.1822G > A) | Unknown significance | AGL (GSD III) | p.G1115R; (c.3343G > A) | Probably damaging | GBE1 (GSD IV) | p.R190G (c.568A > G) p.I334V (c.1000A > G) p.T507A (C.1519A > G) | BenignBenign Benign |
| 3 | SLC37A4 (GSDI b/c) | p.L116Afs*15 (cc.345dupG) | Deleterious | PHKB (GSD IXb) | p.M185I (c.555G > T) | Unknown significance | None | ||
Fig. 3. Defects in Glycogenolysis Caused by Synergistic Heterozygosity Resulting in a GSD Phenotype.
Defects identified in patients in this study are identified with a red X.
Modifier genes for expression of clinical symptoms in GSDs also need to be considered as clinical studies have documented considerable heterogeneity among GSDIa patients with identical G6PC genotypes [31]. Additionally, experimental studies in liver-specific GSDIa −/− mice have shown that hepatocytes contribute to residual glucose production through α-glucosidase activity, (i.e. glycogen debranching) and/or lysosomal glycogen degradation, providing other points for the impact of gene variants [32]. Indeed, as endogenous glucose production is dominated by the liver, GSDs ultimately offer a less complicated model to study synergistic heterozygosity and modifier gene effects than FAODs due to the involvement of multiple organs and related energy pathways in the latter.
4. Porphyrias: additive disease in an IEM
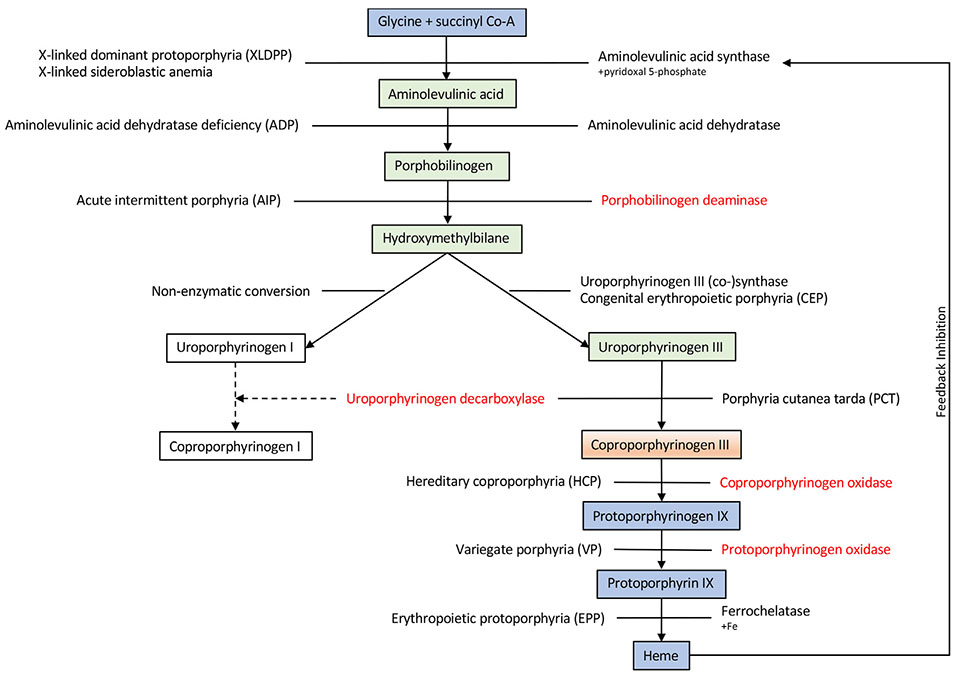
Heme is an important protein synthesized from glycine and succinyl CoA, involved in various systems including respiratory chain cytochromes and cytochrome P450 enzymes (Fig. 4) [33]. The porphyrias are a group of IEMs resulting from enzymatic defects in heme biosynthesis. They are characterized as “erythroid” or “hepatic”, depending on the site of accumulation of abnormal metabolites, and can be primarily classified as “acute” or “cutaneous”, reflecting the major clinical signs and symptoms [34]. The majority of the porphyrias are inherited in an autosomal dominant manner, with most subjects retaining approximately 50% of normal enzyme activity, under most circumstances, resulting in reduced disease penetrance. However, precipitating factors that increase the demand for heme can induce clinical symptoms in individuals with monoallelic mutations. Traditionally, the diagnosis of porphyria is made on the basis of clinical symptoms, characteristic biochemical findings and enzyme assays [35]. However, in some porphyria patients and families, these diagnostic tools reveal simultaneous findings compatible with two different forms of porphyria.
Fig. 4. Heme biosynthetic pathway.
Heme biosynthesis consists of eight enzymatic steps, with the first and the last three steps (Blue) occurring in the mitochondria and the intermediate four steps (Green) occurring in the cytosol. Note that Coproporphyrinogen III is (Orange) produce in the cytosol, however its conversion to Protoporphyrinogen IX occurs in the mitochondrial. If there is an enzymatic defect or inhibitor, metabolites may accumulate, resulting in expression of clinical symptoms. The disorders are listed on the left while the enzymes are listed on the right, except for congenital erythropoietic porphyria (CEP) in which the enzyme and disorder are listed on the right on top of each other; and porphyria cutanea tarda (PCT) in which they are in opposite sites. Aminolevulinic acid synthase (rate limiting enzyme) is induced depending on the bioavailability of free heme pool. *In red are enzymes associated with the corresponding metabolic steps found in dual porphyrias.
At least 15 patients and families have been described in the literature, with biochemical and metabolic testing identifying dominant defects in two enzymes in the heme biosynthetic pathway, leading to combined deficiency of uroporphyrinogen decarboxylase with either porphobilinogen deaminase, coproporphyrinogen oxidase, or protoporphyrinogen oxidase [36-38]. Not surprisingly, patients showed a mixed clinical picture as sufficient flux through the pathway occurred to allow the symptoms related to both defects to manifest. This phenomenon, referred to as dual porphyria, represents yet another example of the potential complex nature of inheritance in IEMs. In addition to the presence of dual porphyrias, a relatively more common scenario is manifestation of an acquired porphyria such as uroporphyrinogen decarboxylase deficiency secondary to liver or renal disease (the most common form of porphyria), preceding the clinical manifestation of a latent or late-onset form of inherited porphyria, most commonly porphobilinogen deaminase deficiency (RBG, unpublished observation).
5. PKU, learning new tricks from an old dog: modifier genes and clinical severity
Complex mechanisms of disease in IEMS exceeds the boundaries of synergistic heterozygosity and extends to interactions of modifying genes. While this topic is covered more broadly in this special issue, one example is useful in considering its role in IEMs. As the most common IEM in many populations, and the one with the longest history of newborn screening, hundreds of mutations have been reported in the phenylalanine hydroxylase (PAH) gene leading to phenylketonuria (PKU) with differing allele frequencies in distinct ethnic groups (see http://www.biopku.org). As a precision medicine tool, there is little doubt that mutation identification in the PAH gene provides some insight into ultimate severity of disease [39-41]. Clearly, null mutations leading to complete lack of protein portend poor phenylalanine (Phe) tolerance and more difficult to control disease. In contrast, those mutations preserving some enzymatic activity are more likely to result in milder disease with a relatively higher Phe tolerance [42]. However genotype/phenotype correlations are imperfect [43]. Rare individuals have been reported with apparently null mutations in the PAH gene who nonetheless are of normal intelligence and show no untoward neurologic effects in spite of a lifetime of blood Phe levels that are consistent with classical PKU [44]. In contrast, siblings with the same mutations may show different disease severity [45]. It is also important to note that mutations in a number of other genes besides PAH either cause hyperphenylalaninemia, including defects in the synthesis or recycling of the PAH biopterin cofactor [46]. Recently, a defect in DNAJC12 has been identified as another hyperphenylalaninemia gene, acting as a co-chaperone protein vital to the proper folding of a group of amino acid hydroxylases including PAH [47-49]. One can imagine that variants in proteins sucha s DNAJC12 could lead to less efficient folding of some mutant PAH proteins, leading to variability in expression of a PAH mutation in different individiuals.
The phenotype-genotype variation seen with PAH mutations is only an extreme example of clinical variation, and raises interesting questions especially regarding the use of the only two drugs approved by the Federal Drug Administration (FDA) for treatment of PKU. Sapropterin is a synthetic biopterin, an essential cofactor for the PAH enzyme, that for most mutations appears to work by acting as a molecular chaperonin that promotes folding of some mutant PAH proteins, thus increasing the residual enzyme activity and reducing the severity of disease [50,51]. One might a priori predict that a response to such a medication would depend solely on the PAH mutation, implying that siblings with the same PAH genotype by definition would respond similarly to treatment with sapropterin. Interestingly, extensive published clinical experience demonstrates otherwise [52,53]. Even in the clinical trials leading to approval of the medication, variability in response independent of genotype was reported except for null mutations leading to complete loss of PAH protein [54,55]. Similarly, patients with PKU, even siblings, show variable responses to treatment with pegvaliase, an enzyme substitution therapy given to patients subcutaneously [56,57].
The factors that mediate genotype/phenotype correlations and response to treatment in patients with PKU remain unknown but are likely to be critical for directing precision treatment of disease. Entry of Phe into the CNS is mediated by a transporter protein (LAT1) that is shared with other large neutral amino acids [58]. Reduced avidity of a variant LAT1 protein to Phe could explain reduced sensitivity to Phe toxicity, and thus explain the identification of patients with classical PKU but with normal cognitive outcome, though this has not been proven [44]. Other proposed mechanisms for variable phenotype include variations in genes regulating transport of phenylalanine across membranes of other cells in the brain, or in those regulating brain vulnerability to high phenylalanine levels.
Other factors that mediate genotype/phenotype correlations and response to treatment in patients with PKU remain unknown but are likely to be similarly variable as the disease. These might include any number of factors ranging from the protein chaperonins as discussed above, to the presence of variant enzymes that allow for alternative metabolism of Phe and thus reduced toxicity in patients with the same PAH mutations, or differential end organ or cell susceptibility. In the case of pegvaliase, such factors such as modulation of immune response to the foreign protein and differences in absorbance or clearance of the enzyme could lead to variation in response to therapy. Identifying the genetic factors that mediate variation in disease phenotype and response to treatment will undoubtedly be a major focus of research in the personalized medicine era, but will require an exhaustive WES, WGS, or epigenomic approach that is likely to require coordination across multiple centers nationally and worldwide. Undirected, broad spectrum metabolomics could also play a role by identifying marker metabolites indicative of alternative metabolic processes that indicate susceptibility or refractivity to pathological outcomes. Ultimately, it must be remembered that with the PAH enzyme inactivated, PKU is the ultimate environmental disorder, with exposure to dietary Phe as the toxin. All current PKU management strategies seek to reduce Phe exposure. In this context, treatment based on reducing the toxicity of Phe rather than lowering it is likely to be a major goal of personalized medicine.
6. Lessons learned and future needs
This admittedly non-exhaustive review of IEMs and novel clinical case presentations demonstrates the need to consider IEMs as examples of complex genetic traits. Biochemical phenotypes are not restricted to defects in a single gene, the need for the coordinated activity of multiple gene products, and the clear involvement of other, non-metabolic genes in modifying clinical symptoms and response to therapy require a new paradigm in studying and treating IEMs. Synergistic heterozygosity is a specific example of a complex pattern of inheritance leading to decreased flux through a metabolic pathway, but modifier genes and digenic inheritance must also be considered. To address this issue, it is first necessary to characterize all patients as completely as possible through broad based metabolomics, WES or WGS, epigenomics, and in vitro or in vivo fluxomics. Currently these modalities are considered research tools, but application to clinical practice will be ubiquitous as we accumulate more undirected data on patients with IEMs. These findings will in turn drive research both on pathophysiology and therapy. With advances in care including next generation gene panels, WES, and WGS, clinicians are faced with interpreting results when genetic changes in multiple genes are identified due to increased use of these tests. Study of IEM patients for genetic heterogeneity and complexity could identify other examples synergistic heterozygosity, and future investigations should examine the various genes involved in biochemical pathways in considering possibilities for this phenomenon. It is also likely that other genetic mechanisms will be identified in such cases, including variations in non-coding regions and ultimately epigenetic alterations [59,60]. Illustrative cases presented here are a reminder that it is critical to look beyond a simple genotype and to treat the clinical phenotype and not just the genotype. Precision medicine, long a hallmark of the diagnosis and treatment of IEMs, will take on ever greater importance, emphasizing the pleiotropic nature of this disorders and their nature as complex genetic traits.
References
- [1].Grosse SD, Dezateux C, Newborn screening for inherited metabolic disease, Lancet 369 (2007) 5–6. [DOI] [PubMed] [Google Scholar]
- [2].Rinaldo P, Lim JS, Tortorelli S, Gavrilov D, Matern D, Newborn screening of metabolic disorders: recent progress and future developments, Nestle Nutr Workshop Ser Pediatr Program 62 (2008) 81–93 (discussion 93-86). [DOI] [PubMed] [Google Scholar]
- [3].Bennett MJ, Rinaldo P, Wilcken B, Pass KA, Watson MS, Wanders RJ, Newborn screening for metabolic disorders: how are we doing, and where are we going? Clin. Chem 58 (2012) 324–331. [DOI] [PubMed] [Google Scholar]
- [4].Wasserstein MP, Caggana M, Bailey SM, Desnick RJ, Edelmann L, Estrella L, Holzman I, Kelly NR, Kornreich R, Kupchik SG, Martin M, Nafday SM, Wasserman R, Yang A, Yu C, Orsini JJ, The New York pilot newborn screening program for lysosomal storage diseases: report of the first 65,000 infants, Genet Med 21 (2019) 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF, Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases, NPJ Genom Med 3 (2018) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, McKnight D, Bai R, Suchy S, Friedman B, Tahiliani J, Pineda-Alvarez D, Richard G, Brandt T, Haverfield E, Chung WK, Bale S, Clinical application of whole-exome sequencing across clinical indications, Genet Med 18 (2016) 696–704. [DOI] [PubMed] [Google Scholar]
- [7].Maiuri L, Raia V, Kroemer G, Strategies for the etiological therapy of cystic fibrosis, Cell Death Differ. 24 (2017) 1825–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jung IY, Lee J, Unleashing the therapeutic potential of CAR-T cell therapy using gene-editing technologies, Mol. Cell 41 (2018) 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mills R, Haga SB, Clinical delivery of pharmacogenetic testing services: a proposed partnership between genetic counselors and pharmacists, Pharmacogenomics 14 (2013) 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Frick A, Benton CS, Scolaro KL, McLaughlin JE, Bradley CL, Suzuki OT, Wang N, Wiltshire T, Transitioning pharmacogenomics into the clinical setting: training future pharmacists, Front. Pharmacol 7 (2016) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Drozda K, Pacanowski MA, Grimstein C, Zineh I, Pharmacogenetic labeling of FDA-approved drugs: a regulatory retrospective, JACC Basic Transl Sci 3 (2018) 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vockley J, Metabolism as a complex genetic trait, a systems biology approach: implications for inborn errors of metabolism and clinical diseases, J. Inherit. Metab. Dis 31 (2008) 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Manoli I, Venditti CP, Disorders of branched chain amino acid metabolism, Transl Sci Rare Dis 1 (2016) 91–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vockley J, Rinaldo P, Bennett MJ, Matern D, Vladutiu GD, Synergistic heterozygosity: disease resulting from multiple partial defects in one or more metabolic pathways, Mol. Genet. Metab 71 (2000) 10–18. [DOI] [PubMed] [Google Scholar]
- [15].Phillips JA 3rd, Poling JS, Phillips CA, Stanton KC, Austin ED, Cogan JD, Wheeler L, Yu C, Newman JH, Dietz HC, Loyd JE, Synergistic heterozygosity for TGFbetal SNPs and BMPR2 mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension, Genet Med 10 (2008) 359–365. [DOI] [PubMed] [Google Scholar]
- [16].Watchko JF, Lin Z, Exploring the genetic architecture of neonatal hyperbilirubinemia, Semin. Fetal Neonatal Med 15 (2010) 169–175. [DOI] [PubMed] [Google Scholar]
- [17].Zangen S, Kidron D, Gelbart T, Roy-Chowdhury N, Wang X, Kaplan M, Fatal kernicterus in a girl deficient in glucose-6-phosphate dehydrogenase: a paradigm of synergistic heterozygosity, J. Pediatr 154 (2009) 616–619. [DOI] [PubMed] [Google Scholar]
- [18].Vockley J, Burton B, Berry GT, Longo N, Phillips J, Sanchez-Valle A, Tanpaiboon P, Grunewald S, Murphy E, Bowden A, Chen W, Chen CY, Cataldo J, Marsden D, Kakkis E, Results from a 78-week, single-arm, open-label phase 2 study to evaluate UX007 in pediatric and adult patients with severe long-chain fatty acid oxidation disorders (LC-FAOD), J. Inherit. Metab. Dis 42 (2019) 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang Y, Mohsen AW, Mihalik SJ, Goetzman ES, Vockley J, Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes, J. Biol. Chem 285 (2010) 29834–29841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Palmfeldt J, Gregersen N, Makhov AM, Conway JF, Wang M, McCalley SP, Basu S, Alharbi H, St C, Croix MJ, Calderon S, Vockley Watkins J, Mitochondrial fatty acid oxidation and the electron transport chain are comprised of a multi-functional mitochondrial protein complex, J. Biol. Chem (2019), 10.1074/jbc.RA119.008680 In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vockley J, Charrow J, Ganesh J, Eswara M, Diaz GA, McCracken E, Conway R, Enns GM, Starr J, Wang R, Abdenur JE, Sanchez-de-Toledo J, Marsden DL, Triheptanoin treatment in patients with pediatric cardiomyopathy associated with long chain-fatty acid oxidation disorders, Mol. Genet. Metab 119 (2016) 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gillingham MB, Heitner SB, Martin J, Rose S, Goldstein A, El-Gharbawy AH, Deward S, Lasarev MR, Pollaro J, DeLany JP, Burchill LJ, Goodpaster B, Shoemaker J, Matern D, Harding CO, Vockley J, Triheptanoin versus trioctanoin for long-chain fatty acid oxidation disorders: a double blinded, randomized controlled trial, J. Inherit. Metab. Dis 40 (2017) 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Seminotti B, Leipnitz G, Karunanidhi A, Kochersperger C, Roginskaya VY, Basu S, Wang Y, Wipf P, Van Houten B, Mohsen AW, Vockley J, Mitochondrial energetics is impaired in very long-chain acyl-CoA dehydrogenase deficiency and can be rescued by treatment with mitochondria-targeted electron scavengers, Hum. Mol. Genet 28 (6) (2019) 928–941, 10.1093/hmg/ddy403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gupta S, Lau K, Harding CO, Shepherd G, Boyer R, Atkinson JP, Knight V, Olbertz J, Larimore K, Gu Z, Li M, Rosen O, Zoog SJ, Weng HH, Schweighardt B, Association of immune response with efficacy and safety outcomes in adults with phenylketonuria administered pegvaliase in phase 3 clinical trials, EBioMedicine 37 (2018) 366–373, 10.1016/j.ebiom.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao XR, Gonzales N, Aronowski J, Pleiotropic role of PPARgamma in intracerebral hemorrhage: an intricate system involving Nrf2, RXR, and NF-kappaB, CNS Neurosci Ther 21 (2015) 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bleeker JC, Kok IL, Ferdinandusse S, de Vries M, Derks TGJ, Mulder MF, Williams M, Gozalbo ER, Bosch AM, van den Hurk DT, de Sain-van der Velden MGM, Waterham HR, Wijburg FA, Visser G, Proposal for an individualized dietary strategy in patients with very long-chain acyl-CoA dehydrogenase deficiency, J. Inherit. Metab. Dis 42 (2019) 159–168. [DOI] [PubMed] [Google Scholar]
- [27].Diekman EF, Ferdinandusse S, van der Pol L, Waterham HR, Ruiter JP, Ijlst L, Wanders RJ, Houten SM, Wijburg FA, Blank AC, Asselbergs FW, Houtkooper RH, Visser G, Fatty acid oxidation flux predicts the clinical severity of VLCAD deficiency, Genet Med 17 (2015) 989–994. [DOI] [PubMed] [Google Scholar]
- [28].Chen MA, Weinstein DA, Glycogen storage diseases: diagnosis, treatment and outcome, Transl.Sci. of Rare Dis 1 (2016) 45–72. [Google Scholar]
- [29].Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS, Glycogen and its metabolism: some new developments and old themes, Biochem. J 441 (2012) 763–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brown LM, Corrado MM, van der Ende RM, Derks TG, Chen MA, Siegel S, Hoyt K, Correia CE, Lumpkin C, Flanagan TB, Carreras CT, Weinstein DA, Evaluation of glycogen storage disease as a cause of ketotic hypoglycemia in children, J. Inherit. Metab. Dis 38 (2015) 489–493. [DOI] [PubMed] [Google Scholar]
- [31].Peeks F, Steunenberg TAH, de Boer F, Rubio-Gozalbo ME, Williams M, Burghard R, Rajas F, Oosterveer MH, Weinstein DA, Derks TGJ, Clinical and biochemical heterogeneity between patients with glycogen storage disease type IA: the added value of CUSUM for metabolic control, J. Inherit. Metab. Dis 40 (2017) 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hijmans BS, Boss A, van Dijk TH, Soty M, Wolters H, Mutel E, Groen AK, Derks TGJ, Mithieux G, Heerschap A, Reijngoud DJ, Rajas F, Oosterveer MH, Hepatocytes contribute to residual glucose production in a mouse model for glycogen storage disease type Ia, Hepatology 66 (2017) 2042–2054. [DOI] [PubMed] [Google Scholar]
- [33].Besur S, Hou W, Schmeltzer P, Bonkovsky HL, Clinically important features of porphyrin and heme metabolism and the porphyrias, Metabolites 4 (2014) 977–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Balwani M, Desnick RJ, The porphyrias: advances in diagnosis and treatment, Blood 120 (2012) 4496–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bonkovsky HL, Maddukuri VC, Yazici C, Anderson KE, Bissell DM, Bloomer JR, Phillips JD, Naik H, Peter I, Baillargeon G, Bossi K, Gandolfo L, Light B, Bishop D, Desnick RJ, Acute porphyrias in the USA: features of 108 subjects from porphyrias consortium, Am. J. Med 127 (2014) 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Akagi R, Inoue R, Muranaka S, Tahara T, Taketani S, Anderson KE, Phillips JD, Sassa S, Dual gene defects involving delta-aminolaevulinate dehydratase and coproporphyrinogen oxidase in a porphyria patient, Br. J. Haematol 132 (2006) 237–243. [DOI] [PubMed] [Google Scholar]
- [37].Harraway JR, Florkowski CM, Sies C, George PM, Dual porphyria with mutations in both the UROD and HMBS genes, Ann. Clin. Biochem 43 (2006) 80–82. [DOI] [PubMed] [Google Scholar]
- [38].Poblete-Gutierrez P, Badeloe S, Wiederholt T, Merk HF, Frank J, Dual porphyrias revisited, Exp. Dermatol 15 (2006) 685–691. [DOI] [PubMed] [Google Scholar]
- [39].Guttler F, Guldberg P, Mutation analysis anticipates dietary requirements in phenylketonuria, Eur. J. Pediatr 159 (Suppl. 2) (2000) S150–S153. [DOI] [PubMed] [Google Scholar]
- [40].Garbade SF, Shen N, Himmelreich N, Haas D, Trefz FK, Hoffmann GF, Burgard P, Blau N, Allelic phenotype values: a model for genotype-based phenotype prediction in phenylketonuria, Genet Med 21 (2019) 580–590. [DOI] [PubMed] [Google Scholar]
- [41].Blau N, Shen N, Carducci C, Molecular genetics and diagnosis of phenylketonuria: state of the art, Expert. Rev. Mol. Diagn 14 (2014) 655–671. [DOI] [PubMed] [Google Scholar]
- [42].Guldberg P, Rey F, Zschocke J, Romano V, Francois B, Michiels L, Ullrich K, Hoffmann GF, Burgard P, Schmidt H, Meli C, Riva E, Dianzani I, Ponzone A, Rey J, Guttler F, A European multi center study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype, Am. J. Hum. Genet 63 (1998) 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Scriver CR, Why mutation analysis does not always predict clinical consequences: explanations in the era of genomics, J. Pediatr 140 (2002) 502–506. [DOI] [PubMed] [Google Scholar]
- [44].van Vliet D, van Wegberg AMJ, Ahring K, Bik-Multanowski M, Blau N, Bulut D, Casas K, Didycz B, Djordjevic M, Federico A, Feillet F, Gizewska M, Gramer, Hertecant JL, Hollak CEM, Jorgensen JV, Karall D, Landau Y, Leuzzi V, Mathisen P, Moseley K, Mungan NO, Nardecchia F, Ounap K, Powell KK, Ramachandran R, Rutsch F, Setoodeh A, Stojiljkovic M, Trefz FK, Usurelu N, Wilson C, van Karnebeek CD, Hanley WB, van Spronsen FJ, Can untreated PKU patients escape from intellectual disability? A systematic review, Orphanet J Rare Dis 13 (2018) 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].DiSilvestre D, Koch R, Groffen J, Different clinical manifestations of hyperphenylalaninemia in three siblings with identical phenylalanine hydroxylase genes, Am. J. Hum. Genet 48 (1991) 1014–1016. [PMC free article] [PubMed] [Google Scholar]
- [46].Opladen T, Hoffmann GF, Blau N, An international survey of patients with tetrahydrobiopterin deficiencies presenting with hyperphenylalaninaemia, J. Inherit. Metab. Dis 35 (2012) 963–973. [DOI] [PubMed] [Google Scholar]
- [47].Anikster Y, Haack TB, Vilboux T, Pode-Shakked B, Thony B, Shen N, Guarani V, Meissner T, Mayatepek E, Trefz FK, Marek-Yagel D, Martinez A, Huttlin L, Paulo JA, Berutti R, Benoist JF, Imbard A, Dorboz I, Heimer G, Landau Y, Ziv-Strasser L, Malicdan MCV, Gemperle-Britschgi C, Cremer K, Engels F, Meili D, Keller I, Bruggmann R, Strom TM, Meitinger T, Mullikin JC, Schwartz G, Ben-Zeev B, Gahl WA, Harper JW, Blau N, Hoffmann GF, Prokisch G, Opladen T, Schiff M, Biallelic mutations in DNAJC12 cause Hyperphenylalaninemia, dystonia, and intellectual disability, Am. J. Hum. Genet 100 (2017) 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Straniero L, Guella I, Cilia R, Parkkinen L, Rimoldi V, Young A, Asselta R, Solda G, Sossi V, Stoessl AJ, Priori A, Nishioka K, Hattori N, Follett J, Rajput A, Blau N, Pezzoli G, Farrer MJ, Goldwurm S, Rajput AH, Duga S, DNAJC12 and dopa-responsive nonprogressive parkinsonism, Ann. Neurol 82 (2017) 640–646. [DOI] [PubMed] [Google Scholar]
- [49].van Spronsen FJ, Himmelreich N, Rufenacht V, Shen N, Vliet DV, Al-Owain M, Ramzan K, Alkhalifi SM, Lunsing RJ, Heiner-Fokkema RM, Rassi A, Gemperle-Britschgi C, Hoffmann GF, Blau N, Thony B, Heterogeneous clinical spectrum of DNAJC12-deficient hyperphenylalaninemia: from attention deficit to severe dystonia and intellectual disability, J. Med. Genet 55 (2018) 249–253. [DOI] [PubMed] [Google Scholar]
- [50].Levy HL, Milanowski A, Chakrapani A, Cleary M, Lee P, Trefz FK, Whitley CB, Feillet E, Feigenbaum AS, Bebchuk JD, Christ-Schmidt H, Dorenbaum A, E Sapropterin Research, Efficacy of sapropterin dihydrochloride (tetrahydrobiopterin, 6R-BH4) for reduction of phenylalanine concentration in patients with phenylketonuria: a phase III randomised placebo-controlled study, Lancet 370 (2007) 504–510. [DOI] [PubMed] [Google Scholar]
- [51].Cunningham A, Bausell H, Brown M, Chapman M, DeFouw K, Ernst S, McClure J, McCune H, O'Steen D, Pender A, Skrabal J, Wessel A, Jurecki E, Shediac R, Prasad S, Gillis J, Cederbaum S, Recommendations for the use of sapropterin in phenylketonuria, Mol. Genet. Metab 106 (2012) 269–276. [DOI] [PubMed] [Google Scholar]
- [52].Trefz FK, Scheible D, Gotz H, Frauendienst-Egger G, Significance of genotype in tetrahydrobiopterin-responsive phenylketonuria, J. Inherit. Metab. Dis 32 (2009) 22–26. [DOI] [PubMed] [Google Scholar]
- [53].Utz JR, Lorentz CP, Markowitz D, Rudser KD, Diethelm-Okita B, Erickson D, Whitley CB, START, a double blind, placebo-controlled pharmacogenetic test of responsiveness to sapropterin dihydrochloride in phenylketonuria patients, Mol. Genet. Metab 105 (2012) 193–197. [DOI] [PubMed] [Google Scholar]
- [54].Gordon P, Thomas JA, Suter R, Jurecki E, Evolving patient selection and clinical benefit criteria for sapropterin dihydrochloride (Kuvan(R)) treatment of PKU patients, Mol. Genet. Metab 105 (2012) 672–676. [DOI] [PubMed] [Google Scholar]
- [55].Stockler-Ipsiroglu S, Yuskiv N, Salvarinova R, Apatean D, Ho G, Cheng B, Giezen A, Lillquist Y, Ueda K, Individualized long-term outcomes in blood phenylalanine concentrations and dietary phenylalanine tolerance in 11 patients with primary phenylalanine hydroxylase (PAH) deficiency treated with Sapropterin-dihydrochloride, Mol. Genet. Metab 114 (2015) 409–414. [DOI] [PubMed] [Google Scholar]
- [56].Harding CO, Amato RS, Stuy M, Longo N, Burton BK, Posner J, Weng HH, Merilainen M, Gu Z, Jiang J, Vockley J, P.-. Investigators, Pegvaliase for the treatment of phenylketonuria: a pivotal, double-blind randomized discontinuation phase 3 clinical trial, Mol. Genet. Metab 124 (2018) 20–26. [DOI] [PubMed] [Google Scholar]
- [57].Thomas J, Levy H, Amato S, Vockley J, Zori R, Dimmock D, Harding CO, Bilder CA, Weng HH, Olbertz J, Merilainen M, Jiang J, Larimore K, Gupta S, Gu Z, Northrup H, P. investigators, Pegvaliase for the treatment of phenylketonuria: results of a long-term phase 3 clinical trial program (PRISM), Mol. Genet. Metab 124 (2018) 27–38. [DOI] [PubMed] [Google Scholar]
- [58].Bik-Multanowski M, Pietrzyk JJ, LAT1 gene variants–potential factors influencing the clinical course of phenylketonuria, J. Inherit. Metab. Dis 29 (2006) 684. [DOI] [PubMed] [Google Scholar]
- [59].Dobrowolski SF, Lyons-Weiler J, Spridik K, Biery A, Breck J, Vockley J, Yatsenko S, Sultana T, Altered DNA methylation in PAH deficient phenylketonuria, Mol. Genet. Metab 115 (2015) 72–77. [DOI] [PubMed] [Google Scholar]
- [60].Dobrowolski SF, Lyons-Weiler J, Spridik K, Vockley J, Skvorak K, Biery A, DNA methylation in the pathophysiology of hyperphenylalaninemia in the PAH(enu2) mouse model of phenylketonuria, Mol. Genet. Metab 119 (2016) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vockley J, Marsden D, McCracken E, DeWard S, Barone A, Hsu K, Kakkis E, Long-term major clinical outcomes in patients with long chain fatty acid oxidation disorders before and after transition to triheptanoin treatment–a retrospective chart review, Mol. Genet. Metab 116 (2015) 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]