The Rab40b/Cul5 complex is an emerging pro-migratory molecular machine. Duncan et al. demonstrate that Rab40b/Cul5 ubiquitylates Rap2 to regulate its localization and activity, ultimately proposing a model by which Rap2 is targeted to the leading-edge plasma membrane to regulate actin dynamics during cell migration.
Abstract
Cell migration is a complex process that involves coordinated changes in membrane transport and actin cytoskeleton dynamics. Ras-like small monomeric GTPases, such as Rap2, play a key role in regulating actin cytoskeleton dynamics and cell adhesions. However, how Rap2 function, localization, and activation are regulated during cell migration is not fully understood. We previously identified the small GTPase Rab40b as a regulator of breast cancer cell migration. Rab40b contains a suppressor of cytokine signaling (SOCS) box, which facilitates binding to Cullin5, a known E3 ubiquitin ligase component responsible for protein ubiquitylation. In this study, we show that the Rab40b/Cullin5 complex ubiquitylates Rap2. Importantly, we demonstrate that ubiquitylation regulates Rap2 activation as well as recycling of Rap2 from the endolysosomal compartment to the lamellipodia of migrating breast cancer cells. Based on these data, we propose that Rab40b/Cullin5 ubiquitylates and regulates Rap2-dependent actin dynamics at the leading edge, a process that is required for breast cancer cell migration and invasion.
Introduction
Cell migration is essential for many normal biological processes including development, wound healing, and the immune response (Franz et al., 2002; Vicente-Manzanares, 2005). On the other hand, it is also critical for the progression of cancer metastasis (Bravo-Cordero et al., 2012). It is well accepted that cell migration requires coordinated changes in membrane trafficking, the actin cytoskeleton, adhesion dynamics, and the targeted secretion of matrix metalloproteinases (MMPs; Ridley et al., 2003; Pollard and Borisy, 2003; Parsons et al., 2010; Murphy and Courtneidge, 2011; Jacob and Prekeris, 2015; Warner et al., 2019). However, the molecular machinery that governs these processes is not fully understood and is a main focus of current studies in the field.
Small monomeric GTPases are tightly regulated molecular switches, cycling between an “active” GTP state and an “inactive” GDP state to facilitate proper protein recruitment to distinct subcellular locations (Reiner and Lundquist, 2018). Members of the Ras GTPase superfamily have been shown to play essential roles in cell migration by coordinating the dynamics of many events described above (Wennerberg et al., 2005; Sadok and Marshall, 2014; Lawson and Ridley, 2018; Gimple and Wang, 2019). Although the Rho family is arguably the most studied in the context of cell migration, it is increasingly clear that other members of the Ras superfamily are equally important for this process. One example is the Rap subfamily (containing Rap1a, 1b, Rap2a, 2b, and 2c), which share >50% sequence identity with Ras proteins (Wennerberg et al., 2005). Rap1 is widely accepted as a key regulator of integrin-mediated cell adhesion and actin reorganization (Kahana and Gottschling, 1999; Bos et al., 2003; Bivona et al., 2004; Bos, 2005; Jeon et al., 2007; Kooistra et al., 2007; Boettner and Van Aelst, 2009). Although Rap1 and Rap2 share ∼60% identity, much less is known about the function of Rap2 GTPases. One of the earliest reported functions of Rap2 was the promotion of B cell migration via integrin-mediated adhesions (McLeod et al., 2002, 2004). This, combined with another initial finding suggesting Rap2 is critical for Xenopus laevis gastrulation, provided early evidence that the Rap2 family may play a crucial role during cell migration across diverse biological systems (Choi and Han 2005). Over the last decade, the field has slowly emerged, where several studies have linked Rap2 to cell polarity, actin cytoskeleton regulation, cancer cell invasion, and most recently, coordination of mechanosensing and Hippo signaling (Taira et al., 2004; Gloerich et al., 2012; Bruurs and Bos, 2014; Di et al., 2015; Meng et al., 2018). However, we still do not fully understand the main function of Rap2 during cell migration, nor the regulation of its subcellular localization or activation.
We previously identified the small monomeric GTPase Rab40b as an important regulator of 3D ECM remodeling, specifically during breast cancer cell migration (Jacob et al., 2013; Jacob et al., 2016). The Rab subfamily of proteins function as master regulators of intracellular membrane traffic (Stenmark, 2009). Rab40 GTPases have a unique extended C-terminal domain that contains the conserved SOCS (suppressor of cytokine signaling) box (Coppola et al., 2019). This ∼40–amino acid motif facilitates binding to Cullin5 (Cul5; Kile et al., 2002; Kamura et al., 2004; Lee et al., 2007; Dart et al., 2015; Yatsu et al., 2015; Linklater et al., 2021; Duncan et al., 2021). Together, this Rab40/Cul5 module, along with RING-box protein Rbx2 and adaptor proteins Elongin B and Elongin C, make up the larger Cullin-RING ligase (CRL5) complex, which canonically regulates protein ubiquitylation (Kile et al., 2002; Petroski and Deshaies, 2005; Linossi and Nicholson, 2012; Okumura et al., 2016; Linklater et al., 2021).
Here, we demonstrate that Rap2 is a substrate of the Rab40b/Cul5 complex and that ubiquitylation of selective Rap2 residues plays a major role in regulating its function. Specifically, we find that Rab40b/Cul5-dependent ubiquitylation regulates targeting of Rap2 to the leading-edge plasma membrane of migrating cells. We demonstrate that inhibition of Rap2 ubiquitylation blocks Rap2 endosome-to-plasma membrane recycling, leading to rapid lysosomal degradation and termination of Rap2 signaling. We also show that Rap2 ubiquitylation is required for its activation. Based on our combined data, we propose a model in which Rab40b is a dual-functioning Rab GTPase, given its coregulation of vesicular MMP trafficking as well as Rap2 spatiotemporal dynamics, and such coregulation plays a key role in driving cell migration.
Results
Rap2 is required for breast cancer cell migration and invasion
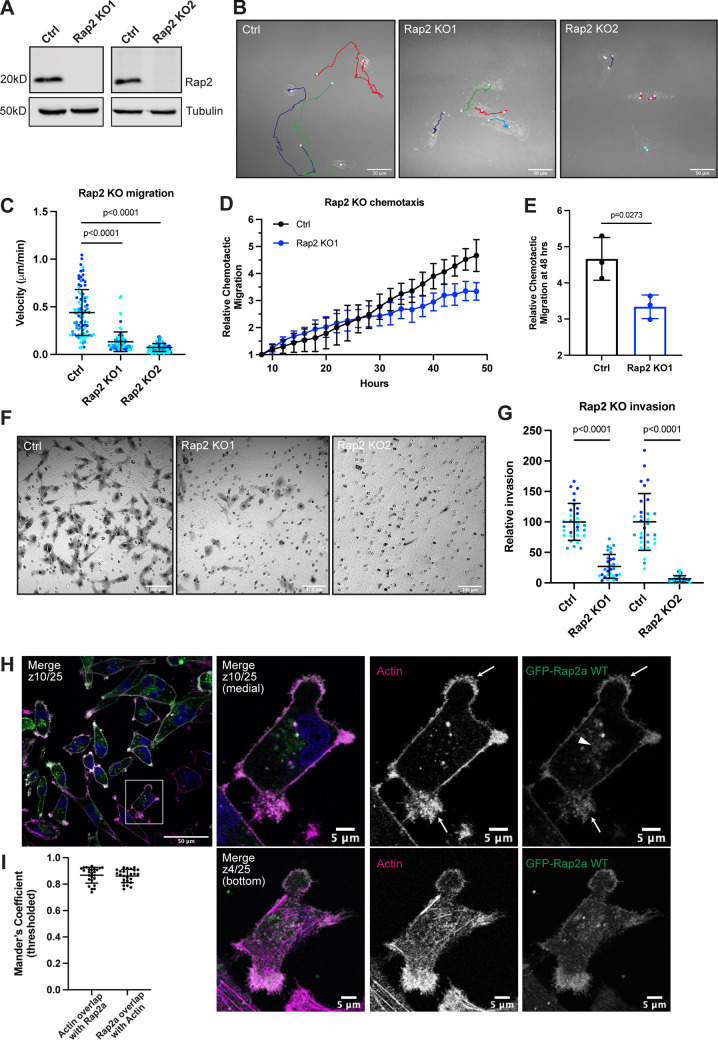
The Rap2 subfamily of small GTPases (Rap2a, b, and c) have been implicated in a variety of biological processes including signal transduction, cell migration, and cell adhesion (Itoh et al., 2007; McLeod et al., 2004, 2002). Although they are suspected to be involved in the pathogenesis of cancer, the exact function of the Rap2 family remains elusive, and existing reports are conflicting. To gain a clearer understanding of Rap2’s role in our breast cancer model system, we made a triple CRISPR/Cas9 knockout (KO) of all three Rap2 isoforms in MDA-MB-231 cells (Rap2 KO). We generated two KO lines (Rap2 KO1 and Rap2 KO2) to diminish the possibility of off-target effects (Fig. 1 A).
Figure 1.
Rap2 is necessary for MDA-MB-231 migration and invasion. (A) Loss of Rap2 confirmed via Western blot. Two different triple CRISPR KOs were made in MDA-MB-231s (see Materials and methods). All Ctrl cells shown in this figure are dox-inducible Cas9 MDA-MB-231s that were used to generate CRISPR lines. 50 µg of lysate was loaded for each sample. (B) Time-lapse 2D migration. Ctrl and Rap2 KO cells were plated on collagen-coated glass dishes and imaged every 10 min for 16 h using a brightfield 40× objective. Representative still images show the last frame of the time-lapse experiment, where the cell body indicates the location of the cell at the last time point. Cells were manually tracked in Fiji using the Manual Tracking plugin. Colors denote individual cell tracks over 16 h. White dots represent the start and finish of each cell track. Scale bars, 50 µm. (C) 2D migration velocity quantification. From the Fiji manual tracking, velocity data (µm/min) was extracted for each individual cell. Three biological replicates were performed for each cell line. For each experiment, 30 cells (color coded) were randomly chosen for velocity tracking (∼3 cells from ∼10 fields of view). Each cell was treated as its own data point (n = 90). One-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Ctrl versus Rap2 KO1, P < 0.0001. Ctrl versus Rap2 KO2, P < 0.0001. (D) Chemotactic cell migration Ctrl versus Rap2 KO1 cells. Cells were imaged every 2 h for 48 h using an IncuCyte S3 instrument. Raw data (the sum area of all migrated cells normalized to the area at time 0) was extracted and averaged for each technical replicate. Data was then adjusted/normalized to the 8-h time point (see Materials and methods). The graph shows relative chemotactic migration over the 48-h time course. Mean ± SD. (E) Chemotactic migration quantification. Three biological replicates were performed, with six technical replicates in each experiment. Statistical analysis (unpaired t test) was performed on the relative chemotactic migration at 48 h. Mean ± SD at 48 h. Ctrl versus Rap2 KO1, P = 0.0273. (F) Boyden chamber invasion assay. Ctrl and Rap2 KO cells were plated in a modified Boyden chamber coated with Matrigel. Cells were allowed 20 h to invade through the Matrigel-coated pores before fixation and crystal violet staining. Inserts were imaged using a brightfield 20× air objective. Representative images are shown. Hollow circles indicate the 8-µm pores. Scale bars, 100 µm. (G) Invasion assay quantification. Three biological replicates were performed, with technical duplicates in each experiment. 5 fields of view were imaged for each Matrigel insert (resulting in 10 fields of view per experiment per condition, color coded). Raw number of cells invaded per field of view were normalized to Ctrl. Rap2 KO1 and Rap2 KO2 cells were analyzed at different times; hence two Ctrl samples. Each field of view was treated as its own data point (n = 30). One-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Ctrl versus Rap2 KO1, P < 0.0001. Ctrl versus Rap2 KO2, P < 0.0001. (H) GFP-Rap2a WT localization. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with phalloidin (magenta) and DAPI (blue). Z-slices are indicated on merged images. In most images, medial z-slices are shown. Here, we also provide a z-slice at the bottom of the cell, to highlight overlap between actin and GFP-Rap2a. Arrows point to example sites of actin and GFP-Rap2a colocalization. The arrowhead indicates the GFP-Rap2a intracellular organelle population. Scale bars, 50 and 5 µm. (I) Colocalization analysis of actin and GFP-Rap2a. Thresholded Mander’s coefficients were calculated using the Fiji Coloc 2 plugin. One biological replicate was performed, with five fields of view and five cells in each field (n = 25). The fraction of actin overlapping with Rap2a (tM1, mean = 0.8678) and the fraction of Rap2a overlapping with actin (tM2, mean = 0.8612) are shown. Mean ± SD. Source data are available for this figure: SourceData F1.
We first asked whether loss of Rap2 leads to any global 2D migration defects, using live-cell imaging. Compared with control cells, Rap2 KO cells exhibit decreased individual cell migration, as indicated by significantly lower velocity (Fig. 1, B and C; and Video 1, Video 2, and Video 3). We next asked whether Rap2 KO cells have defects in chemotactic cell migration. Control and Rap2 KO cells were plated in an IncuCyte ClearView 96-well Chemotaxis Plate with serum-starved medium in the top chamber and full MDA-MB-231 medium (chemoattractant) in the bottom chamber and allowed to migrate toward the chemoattractant for 48 h. As expected, Rap2 KO cells showed decreased chemotactic migration compared with control cells (Fig. 1, D and E). Interestingly, while the Rap2 KO individual cell velocity defect in Fig. 1 C is quite dramatic, we saw only ∼50% inhibition of chemotactic migration at the 48-h time point. This may point at differences in Rap2’s function during individual versus collective cell migration. Nonetheless, these results in sum demonstrate that Rap2 modulates 2D breast cancer cell migration.
Video 1.
MDA-MB-231 Ctrl 2D cell migration. Live 2D migration on collagen-coated glass plate. Ctrl cells are dox-inducible Cas9 MDA-MB-231 cells used to generate the Rap2 CRISPR lines. 10-min intervals, 95 frames. 5 fps. Scale bar, 50 µm.
Video 2.
MDA-MB-231 Rap2 KO1 2D cell migration. Live 2D migration on collagen-coated glass plate. MDA-MB-231 Rap2 KO1 cells. 10-min intervals, 95 frames. 5 fps. Scale bar, 50 µm.
Video 3.
MDA-MB-231 Rap2 KO2 2D cell migration. Live 2D migration on collagen-coated glass plate. MDA-MB-231 Rap2 KO1 cells. 10-min intervals, 95 frames. 5 fps. Scale bar, 50 µm.
We next tested the invasive capability of MDA-MB-231 Rap2 KO cells using a modified Boyden chamber invasion assay. Notably, Rap2 KO cells showed a striking decrease in their ability to invade compared with control cells (Fig. 1, F and G). Taken together, our data demonstrate that Rap2 is necessary for both 2D and 3D migration and invasion in MDA-MB-231s. These data provide strong evidence for the Rap2 subfamily being pro-migratory in breast cancer cells. Additionally, these striking phenotypes convey a clear importance for studying the function and regulation of Rap2 during cell migration.
Rap2 cycles between the lamellipodia plasma membrane and endosomes
To start dissecting how Rap2 may regulate breast cancer cell migration and invasion, we first examined subcellular localization of Rap2 in MDA-MB-231s. In humans, the Rap2 family encompasses three closely related paralogs, Rap2a, Rap2b, and Rap2c, which have >90% sequence identity. All evidence so far suggests that the three Rap2 isoforms have similar functions; thus, for most of our studies, we have focused on the Rap2a isoform and used an MDA-MB-231 cell line stably expressing GFP-Rap2a (Fig. S1 C). As shown in Fig. 1 H, GFP-Rap2a localizes predominantly to the plasma membrane, where it is enriched at lamellipodia ruffles. Importantly, plasma membrane–bound Rap2a clearly colocalizes with actin (Fig. 1 I), consistent with the putative role of Rap2 in regulating actin cytoskeleton dynamics.
Figure S1.
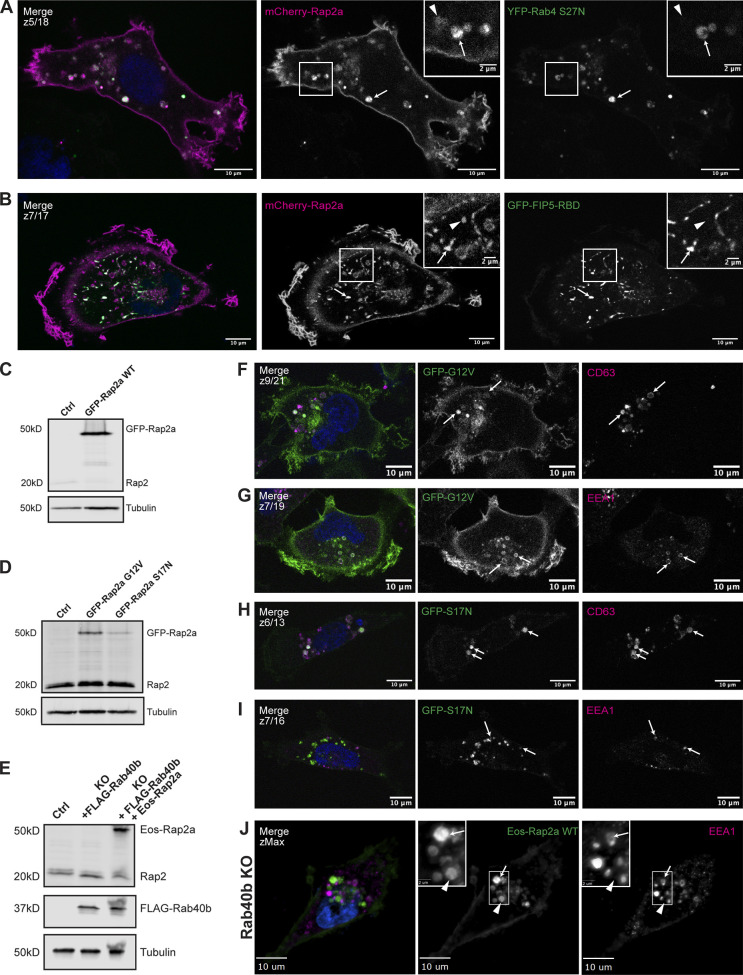
Further characterization of Rap2a subcellular localization in MDA-MB-231 cells and analysis of Eos-Rap2a in Rab40b KO cells. (A) Colocalization of mCherry-Rap2a and YFP-Rab4 S27N. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and YFP-Rab4 S27N, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and YFP-Rab4 S27N overlap. Arrowheads point to mCherry-Rap2a organelles that are not Rab4 S27N positive. Scale bars, 10 and 2 µm. (B) Colocalization of mCherry-Rap2a and GFP-FIP5-RBD. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and GFP-FIP5-RBD, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and GFP-FIP5-RBD overlap. Arrowheads point to mCherry-Rap2a organelles that are not FIP5-RBD. Scale bars, 10 and 2 µm. (C) Western blot showing stable overexpression of GFP-Rap2a WT in MDA-MB-231s (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (D) Western blot showing stable overexpression of GFP-Rap2a-G12V and -S17N in MDA-MB-231s (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (E) Western blot showing generation of rescue line in Fig. 6 E. Rab40b KO cells were first stably transfected with FLAG-Rab40b WT (lentivirus, second column). Then, these cells were transfected with Eos-Rap2a and flow sorted (lentivirus, third column, flow sort instead of selection). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate Rab40b KO CRISPR line. 50 µg of lysate was loaded for each sample. (F) Colocalization of GFP-Rap2a-G12V and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a-G12V were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-G12V and CD63 overlap. Scale bars, 10 µm. (G) Colocalization of GFP-Rap2a-G12V and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a-G12V were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-G12V and EEA1 overlap. Scale bars, 10 µm. (H) Colocalization of GFP-Rap2a-S17N and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a-S17N were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-S17N and CD63 overlap. Scale bars, 10 µm. (I) Colocalization of GFP-Rap2a-S17N and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a-S17N were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of Rap2a-S17N and EEA1 overlap. Scale bars, 10 µm. (J) Eos-Rap2a colocalization with EEA1 in Rab40b KO cells. Rab40b KO MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Widefield microscope. Arrows indicate examples of Eos-Rap2a and EEA1 overlap. Arrowheads point to Eos-Rap2a organelles that are not EEA1 positive. Scale bars, 10 µm, 2 µm. Source data are available for this figure: SourceData FS1.
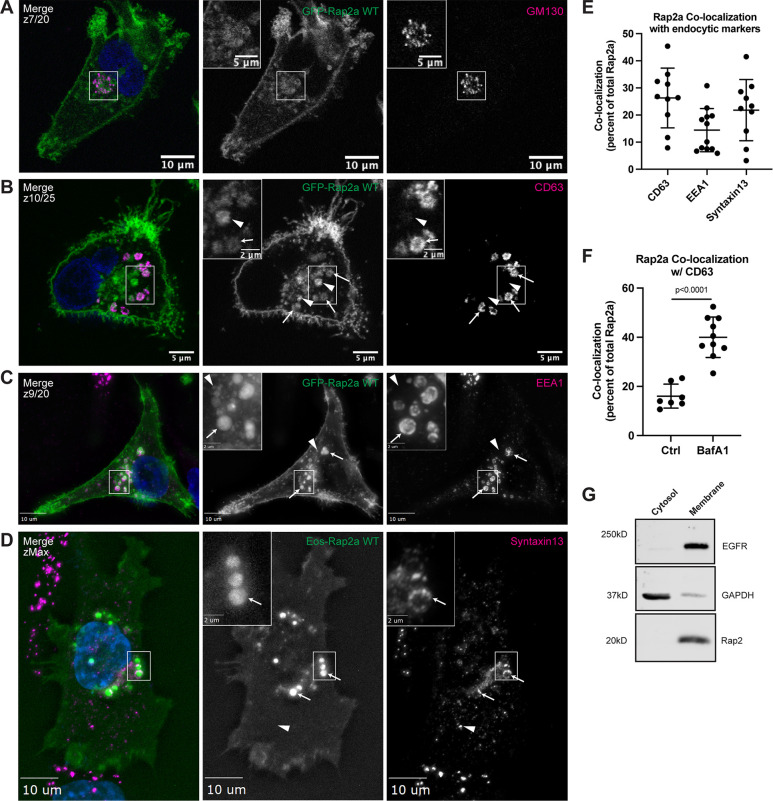
In addition to the plasma membrane population, some of GFP-Rap2a is also observed within intracellular organelles (Fig. 1 H). Since some of the Ras subfamily members, namely HRas and NRas, have been proposed to signal from the Golgi (Hancock, 2003), we wondered about the identity of these Rap2a-containing intracellular organelles. To this end, we costained GFP-Rap2a cells with known endocytic pathway markers including GM130 (Golgi), EEA1 (early endosomes), CD63 (late endosomes/lysosomes), and Syntaxin13 (Rab4 and Rab11 recycling endosomes). While we did observe some Rap2a at the Golgi, consistent with preliminary reports (Pizon et al., 1994), most intracellular GFP-Rap2a was clearly present within the endolysosomal compartment, where it colocalized with both EEA1-positive early endosomes and CD63-positive late endosomes/lysosomes (Fig. 2, A–C and E). Consistent with some of the Rap2 pool being trafficked to lysosomes, we observed an increase in Rap2 colocalization with CD63 when cells were treated with Bafilomycin A1, an inhibitor of lysosomal degradation (Fig. 2 F). Finally, we note that GFP-Rap2a is rarely found in the cytosolic pool. Using cell fractionation analysis, we observed no detectable endogenous Rap2 in the cytosol fraction and robust signal in the membrane fraction, suggesting that Rap2 is predominantly membrane bound in the cell (Fig. 2 G).
Figure 2.
Rap2 localizes to the plasma membrane and endolysosomal compartment in MDA-MB-231s. (A) Colocalization of GFP-Rap2a and GM130. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the Golgi marker GM130 (magenta) and DAPI (blue). Inset shows GFP-Rap2a and GM130 overlap. Scale bars, 10 and 5 µm. (B) Colocalization of GFP-Rap2a and CD63. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of GFP-Rap2a and CD63 overlap. Arrowheads point to GFP-Rap2a organelles that are not CD63 positive. Scale bars, 5 and 2 µm. (C) Colocalization of GFP-Rap2a and EEA1. MDA-MB-231 cells stably expressing GFP-Rap2a were fixed and stained with the early endosome marker EEA1 (magenta) and DAPI (blue). Arrows indicate examples of GFP-Rap2a and EEA1 overlap. Arrowheads point to GFP-Rap2a organelles that are not EEA1 positive. Widefield microscope. Scale bars, 10 and 2 µm. (D) Colocalization of Eos-Rap2a and Syntaxin13. MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with the recycling endosome marker Syntaxin13 (magenta) and DAPI (blue). Arrows indicate examples of Eos-Rap2a and Syntaxin13 overlap. Arrowhead points to Syntaxin13 endosome that is not Rap2a positive. Widefield microscope. Scale bars, 10 and 2 µm. (E) Rap2a colocalization analysis with endolysosomal compartments. 3i SlideBook6 software was used to calculate the percent of total GFP-Rap2a that colocalizes with the markers indicated (see Materials and methods). Two biological replicates were performed, with approximately five cells imaged for each replicate. Mean ± SD. n = 10 for GFP-Rap2a/CD63, n = 12 for GFP-Rap2a/EEA1, n = 10 for Eos-Rap2a/Syntaxin13. (F) Rap2a and CD63 colocalization analysis in Ctrl cells versus cells treated with lysosomal inhibitor Bafilomycin A1. MDA-MB-231 cells stably expressing GFP-Rap2a were treated with either DMSO (Ctrl) or 200 nM Bafilomycin A1 for 16 h. Cells were then fixed and stained for CD63, before colocalization analysis. 3i SlideBook6 software was used to calculate the percent of total GFP-Rap2a that colocalizes with CD63 (see Materials and methods). One biological replicate was performed. Mean ± SD. n = 7 cells for Ctrl, n = 10 cells for BafA1. Unpaired t test. P < 0.0001. (G) Cell fractionation analysis cytosol versus membrane. MDA-MB-231 parental cells were fractionated (see Materials and methods) to determine subcellular distribution of endogenous Rap2. Cytosol and membrane fractions were collected, separated by SDS-PAGE, and subjected to Western blot against known cytosol (GAPDH) and membrane (EGFR) markers. 30 µg of sample was loaded for each fraction. Source data are available for this figure: SourceData F2.
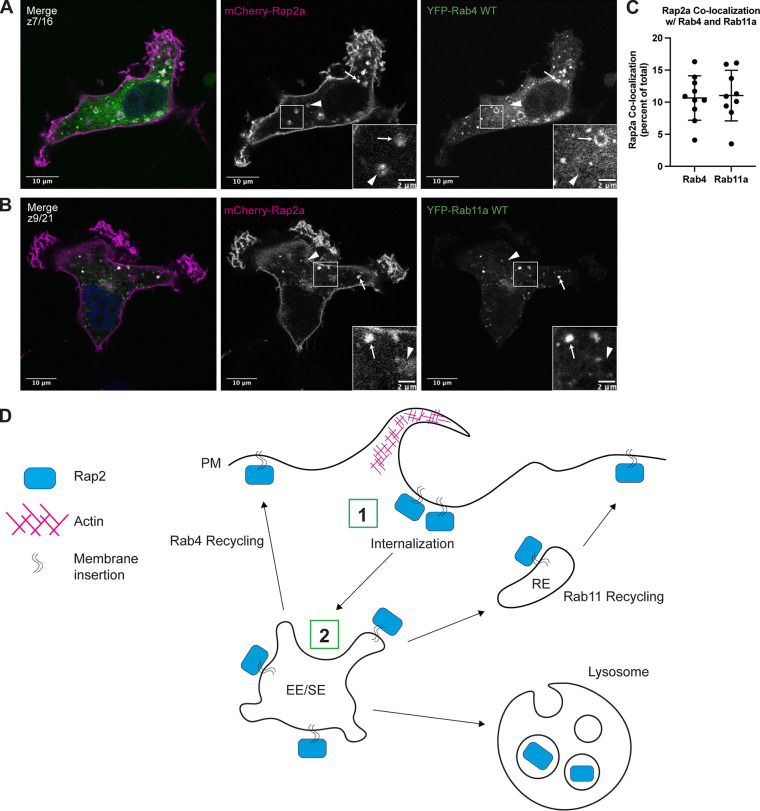
The colocalization with Syntaxin13 suggests that Rap2 may recycle from early endosomes, likely via Rab4 and/or Rab11 recycling endosomes (Fig. 2, D and E; Zerial and McBride, 2001). To investigate this further, we cotransfected MDA-MB-231 cells with mCherry-Rap2a and either YFP-Rab4 or YFP-Rab11a to visualize potential colocalization at recycling endosomes (Fig. 3, A and B). Indeed, we did detect Rap2-Rab4 and Rap2-Rab11 positive organelles, which supports a model in which Rap2 gets recycled to the lamellipodia plasma membrane via the Rab4 and Rab11 pathways (Fig. 3 C).
Figure 3.
Rap2 is dynamically trafficked through the endocytic pathway. (A) Colocalization of mCherry-Rap2a and YFP-Rab4 WT. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and YFP-Rab4 WT, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and YFP-Rab4 WT overlap. Arrowheads point to mCherry-Rap2a organelles that are not Rab4 positive. Scale bars, 10 and, 2 µm. (B) Colocalization of mCherry-Rap2a and YFP-Rab11a WT. MDA-MB-231 cells were transiently cotransfected with mCherry-Rap2a and YFP-Rab11a WT, followed by fixation and staining for DAPI (blue). Arrows indicate examples of mCherry-Rap2a and YFP-Rab11a WT overlap. Arrowheads point to mCherry-Rap2a organelles that are not Rab11a positive. Scale bars, 10 and, 2 µm. (C) Rap2a colocalization analysis with Rab4 WT and Rab11a WT. 3i SlideBook6 software was used to calculate the percent of total mCherry-Rap2a that colocalizes with Rab4 or Rab11a (see Materials and methods). One biological replicate was performed. Mean ± SD. n = 10 for mCherry-Rap2a/YFP-Rab4 WT, n = 9 for mCherry-Rap2a/YFP-Rab11a WT. (D) Model for Rap2 endocytic trafficking. (1) Rap2 is internalized at the lamellipodia plasma membrane via a pinocytosis-like mechanism. (2) Internalization mediates the delivery of Rap2 to EEA1/Rab5-positive early endosomes, where it is sorted into different fates, either recycling to the plasma membrane via the Rab4/Rab11 pathways or targeting to lysosome for degradation. We hypothesize that Rap2 recycling back to the leading edge is needed to sustain cell migration.
To further test whether Rap2 recycles via the Rab4 and Rab11 pathways, we cotransfected MDA-MB-231 cells with mCherry-Rap2a and either YFP-Rab4-S27N (dominant negative) or GFP-FIP5-RBD (C-terminal fragment of Rab11-FIP5 that inhibits Rab11-mediated recycling pathway; Peden et al., 2004; Willenborg et al., 2011). Expression of either mutant delays the exit of recycling cargo from endosomes, thus accumulating cargo proteins in either YFP-Rab4-S27N– or GFP-FIP5-RBD–positive organelles. Consistent with our hypothesis, we observed mCherry-Rap2a present in both YFP-Rab4-S27N (affects Rab4-dependent recycling) and GFP-FIP5-RBD (affects Rab11-dependent recycling) organelles (Fig. S1, A and B). In sum, our data suggest that Rap2 is dynamically trafficked through the endocytic pathway and is likely recycled through early endosomes/recycling pathway back to the leading-edge plasma membrane (Fig. 3 D, trafficking itinerary model for Rap2).
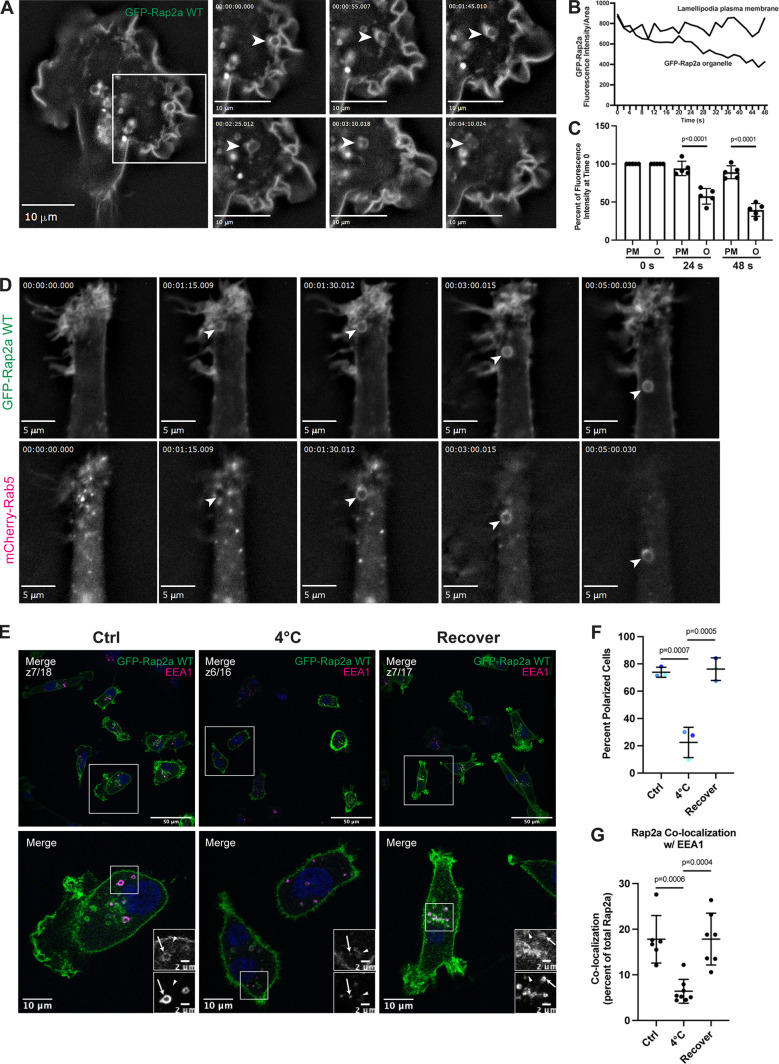
To further define the spatiotemporal properties of Rap2 localization in migrating cells, we next performed time-lapse analysis of GFP-Rap2a-expressing MDA-MB-231 cells. As shown in the Fig. 4 A, GFP-Rap2a was enriched at ruffling lamellipodia, consistent with its proposed role in regulating actin dynamics during cell migration. It is especially clear using live imaging that Rap2 undergoes constant internalization and dynamic trafficking to maintain its enrichment at the leading-edge plasma membrane. We observed that during lamellipodia ruffling, GFP-Rap2a got internalized in large macropinosome-like organelles that originated at lamellipodia and subsequently moved in a retrograde direction toward the retracting end of the cell (Fig. 4 A and Video 4; for additional cell, see Video 5). Importantly, similar pinocytosis-like internalization of leading-edge plasma membrane has been reported before and is known to be required for cell migration (Moreau et al., 2019). Further, GFP-Rap2a–containing organelles can be observed decreasing in fluorescence intensity over time (Fig. 4 B shows quantification for organelle depicted in Fig. 4 A, Video 4, and Fig. 4 C shows quantification for four additional organelles). This hints that Rap2 may be recycled back to the plasma membrane, especially given our previous data suggesting Rap2 may be trafficked through the Rab4/Rab11 recycling pathways. If this is the case, newly internalized GFP-Rap2a organelles should costain with a known early endosome/sorting endosome marker such as Rab5. Indeed, we observed that newly internalized GFP-Rap2a organelles became Rab5 positive soon after internalization (Fig. 4 D and Video 6). In sum, we propose that pinocytosis-like GFP-Rap2a internalization mediates the delivery of Rap2 to EEA1/Rab5-positive early endosomes and that Rap2 is likely recycled back to the leading edge to sustain cell migration (Fig. 3 D).
Figure 4.
Rap2 is enriched at the lamellipodia leading edge, where its internalization and recycling to the plasma membrane are important for cell migration. (A) Live imaging of GFP-Rap2a in MDA-MB-231 cells. Cells stably expressing GFP-Rap2a were plated on a collagen-coated glass dish and imaged every 5 s using a 63× objective. Widefield microscope. Still images are shown. Arrowheads point to an example of GFP-Rap2a internalization over time from the plasma membrane in macropinosome-like vesicles. We also note that GFP-Rap2a organelle decreases in fluorescence intensity over time. See Video 4. Scale bars, 10 µm. (B) Linescan analysis of internalized GFP-Rap2a organelle. A circular line was drawn around the GFP-Rap2a organelle marked in Fig. 4 A, and fluorescence intensity/area was measured at each time point (every 2 s). The same thing was done with a linescan at the plasma membrane. The fluorescence intensity of GFP-Rap2a around the organelle line and at the plasma membrane is plotted as fluorescence intensity/area (µ2) across 48 s. (C) Same analysis as in B, but more organelles quantified. As in B, linescans were drawn both around the GFP-Rap2a organelles (see Materials and methods for criteria) and at the lamellipodia plasma membrane. The intensity of GFP-Rap2a in each pixel along these lines was determined with either ImageJ or 3i Slidebook imaging software. Fluorescence intensity/area (µ2) at 0 s (start of time-lapse), 24 s (half of time-lapse), and 48 s (end of time-lapse) was plotted for all five organelles as a percentage of time 0. Statistical analysis (one-way ANOVA with Tukey’s multiple comparisons test) was used to compare fluorescence intensity changes between the plasma membrane and internalized organelles. Mean ± SD. PM, plasma membrane; O, organelle. PM versus O at 24 s, P < 0.0001; PM versus O at 48 s, P < 0.0001. (D) Live imaging of GFP-Rap2a and mCherry-Rab5 in MDA-MB-231 cells. Cells stably expressing GFP-Rap2a were transiently transfected with mCherry-Rab5, plated on a collagen-coated glass dish, and imaged every 5 s using a 63× objective. Widefield microscope. Still images are shown. Arrowhead points to one example of a GFP-Rap2a internalized organelle over time, which becomes mCherry-Rab5 positive soon after internalization from the plasma membrane. See Video 6. Scale bars, 5 µm. (E) Endocytosis cold block experiment. Three conditions were set up using MDA-MB-231 cells stably expressing GFP-Rap2a: (1) 37°C (Ctrl). (2) 4°C for 60 min. (3) 4°C for 60 min followed by 37°C for 40 min (Recover). Cells were fixed and stained for EEA1 (magenta) and DAPI (blue). Scale bars, 50, 10, and 2 µm. Arrows indicate examples of GFP-Rap2a and EEA1 overlap. Arrowheads point to GFP-Rap2a organelles that are not EEA1 positive. (F) Percentage of polarized cells quantification from cold block experiment in E. Ctrl, 4°C, and Recover cells were scored for polarized versus nonpolarized cells. Briefly, polarized cells had enrichment of Rap2 at the lamellipodia (see Materials and methods). Three biological replicates were performed. Eight fields of view were taken for each n, with approximately six cells in each field. Graph shows percentage of polarized cells for each condition (color coded). Mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test. Ctrl versus 4°C, P = 0.0007; 4°C versus Recover, P = 0.0005. (G) Quantification of colocalization between GFP-Rap2a and EEA1 in Ctrl versus 4°C versus Recover cells. Cells stably expressing GFP-Rap2a were fixed and stained with EEA1. 3i SlideBook6 software was used to calculate the percentage of total GFP-Rap2a that colocalized with EEA1. One biological replicate was performed; n = 6 for Ctrl cells, n = 8 for 4°C cells, n = 7 for Recover cells. One-way ANOVA with Tukey’s multiple comparisons test. Ctrl versus 4°C, P = 0.0006; 4°C versus Recover, P = 0.0004.
Video 4.
GFP-Rap2a live dynamics. Live 2D time-lapse imaging of MDA-MB-231 cells stably expressing GFP-Rap2a. 1-s intervals (plus exposure time), 100 frames. 5 fps. Nearest neighbor deconvolution. Widefield microscope. Scale bar, 10 µm.
Video 5.
GFP-Rap2a live dynamics continued. Same setup as Video 4, providing additional cell. Live 2D time-lapse imaging of MDA-MB-231 cells stably expressing GFP-Rap2a. 5-s intervals (plus exposure time), 51 frames. 5 fps. Nearest neighbor deconvolution. Widefield microscope. Scale bar, 10 µm.
Video 6.
GFP-Rap2a and mCherry-Rab5 live dynamics. Live 2D time-lapse imaging of MDA-MB-231 cells stably expressing GFP-Rap2a, transiently transfected with mCherry-Rab5. 5-s intervals (plus exposure time), 100 frames. 5 fps. Nearest neighbor deconvolution. Widefield microscope. Scale bar, 5 µm. Note that GFP-Rap2a and mCherry-Rab5 do not exactly overlap in the endosomal structure, which is likely due to a delay in image capture because of exposure time.
Finally, to evaluate whether dynamic trafficking of Rap2 is needed for leading-edge formation and ultimate migration, we performed a cold block experiment. Previous work in the membrane trafficking field has shown that incubation of cells at 4°C slows down endocytosis from the plasma membrane. Thus, we incubated GFP-Rap2a–expressing cells at 4°C for 60 min, followed by fixation and staining. Compared with control cells, we observed that 4°C treatment significantly decreased the percentage of polarized cells (cells with defined leading-edge lamellipodia containing enriched GFP-Rap2a), suggesting that rapid endocytosis may be needed to maintain Rap2 lamellipodia enrichment (Fig. 4, E and F). Additionally, we found that GFP-Rap2a colocalized with EEA1 at a lower frequency when the temperature is reduced to 4°C, indicating slower endocytosis and less Rap2 in early endosomes/sorting endosomes (Fig. 4 G). Importantly, both these findings could be rescued when we place 4°C treated cells back at 37°C for 40 min (Recover, Fig. 4, E–G). Altogether, these data support our working model that Rap2 is dynamically trafficked, where quick redistribution and recycling to the leading-edge plasma membrane is important for driving cell migration.
Rap2 subcellular localization and activation state are closely intertwined
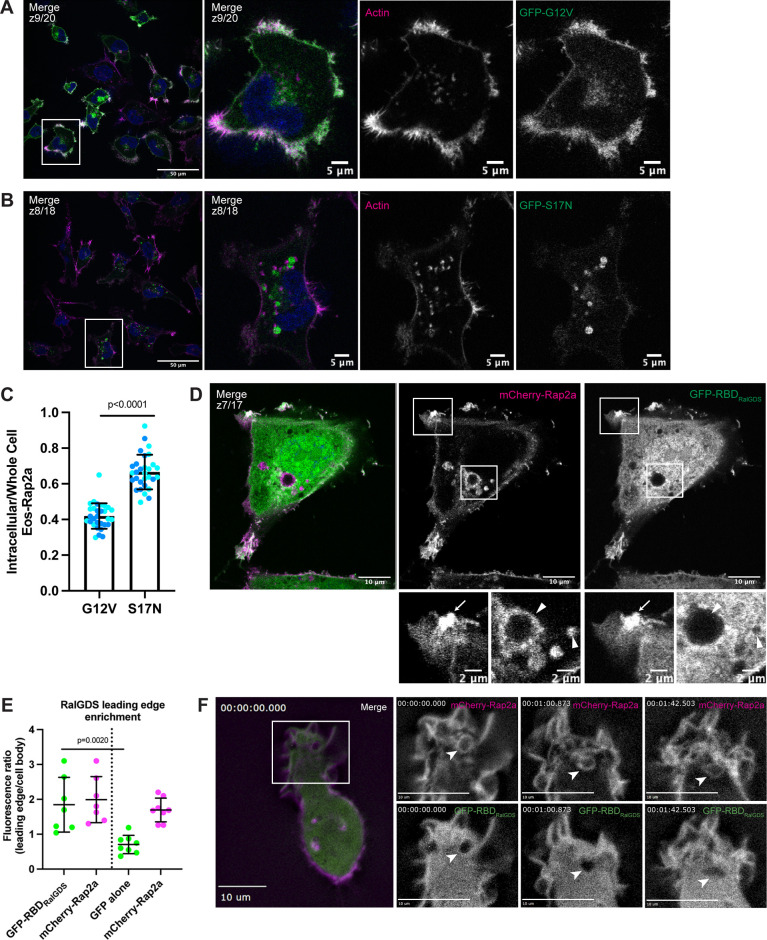
Based on previous Rap1 observations, we speculated that these two populations of Rap2 (plasma membrane versus endolysosomal compartment) might be correlated with nucleotide status and GTPase activity (Ohba et al., 2003; Bivona et al., 2004; Jeon et al., 2007). To test this, we generated stable cell lines expressing either constitutively active (G12V; GTP-bound) or dominant-negative (S17N; GDP-bound) forms of Rap2a (Fig. S1 D) and performed localization analysis (Feig and Cooper, 1988; Gibbs et al., 1989; Feig, 1999). We found that Rap2a-G12V localizes primarily to the lamellipodia plasma membrane, whereas Rap2a-S17N shifted to a more endosome/lysosome localization (Fig. 5, A and B). Consistent with this observation, quantification of the intracellular pool compared to the whole cell (fluorescence signal) revealed a significant difference between Rap2a-G12V and Rap2a-S17N, where the dominant-negative mutant had decreased GFP-Rap2a signal at the plasma membrane (Figs. 5 C and S2 C). Like Rap2a-WT, Rap2a-G12V appeared to traffic between early endosomes, lysosomes, and the lamellipodia plasma membrane (Fig. S1, F and G). However, Rap2a-S17N lost plasma membrane enrichment and was found predominantly in lysosomes and occasionally early endosomes (Fig. S1, H and I). Although there was a difference in expression level of these constructs (Fig. S1 C), we posit that this phenotypic shift of S17N away from the lamellipodia plasma membrane could be due to direct changes in Rap2 activation. There is a possibility that these active/inactive constructs alter cellular dynamics in such a way that their own subcellular localization is indirectly affected. However, because we observed this same phenotype (decreased plasma membrane–bound Rap2) throughout future figures, we would argue that this is a real direct effect on Rap2 subcellular localization.
Figure 5.
Rap2 activation is closely linked with its subcellular localization. (A) GFP-Rap2a-G12V localization. MDA-MB-231 cells stably expressing GFP-Rap2a-G12V were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. (B) GFP-Rap2a-S17N localization. MDA-MB-231 cells stably expressing GFP-Rap2a-S17N were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. (C) Intracellular to whole-cell fraction quantification, Rap2a-G12V versus Rap2a-S17N. To quantify GFP-Rap2a localization changes (i.e., decreased Eos-Rap2a at the plasma membrane in S17N background), intracellular/whole-cell fractions were defined and calculated as the total fluorescence intensity of the intracellular GFP-Rap2a pool divided by the total fluorescence intensity of the whole-cell GFP-Rap2a pool (see Materials and methods). Two biological replicates were performed for each cell line. In each biological replicate, five fields of view were imaged, with 3 cells analyzed from each field (total of 15 cells per n). The two shades of blue represent each set of 15 cells from n1 and n2. Each cell was treated as its own data point, and statistical analysis (unpaired t test) was performed with n = 30 total for each condition. Mean ± SD. G12V versus S17N, P < 0.0001. (D) Colocalization of mCherry-Rap2a and GFP-RBDRalGDS. MDA-MB-231 cells were transiently transfected with mCherry-Rap2a and GFP-RBDRalGDS, then fixed. Arrow points to example of mCherry-Rap2a and GFP-RBDRalGDS overlap at the plasma membrane. Arrowheads point to mCherry-Rap2a intracellular organelles, where GFP-RBDRalGDS is not present. Scale bars, 10 and 2 µm. (E) Quantification of GFP-RBDRalGDS enrichment at the leading edge. In addition to the experiment in 3D, where cells were transfected with mCherry-Rap2a and GFP-RBDRalGDS, another experiment was performed side by side. MDA-MB-231 cells were transfected with mCherry-Rap2a and free GFP (not depicted) to control for free FP localization at the leading edge. Fluorescence intensity ratios (leading edge/cell body, i.e., enrichment at ruffles) were calculated for individual cells (see Materials and methods) in both conditions. One biological replicate was performed, n = 7 cells for GFP-RBDRalGDS condition and n = 8 cells for free GFP condition. Mean ± SD. The two conditions are separated by the dotted line in the center of the graph. Green dots represent GFP-RBDRalGDS or free GFP calculations; magenta represents mCherry-Rap2a. One-way ANOVA with Tukey’s multiple comparisons test. GFP-RBDRalGDS versus GFP alone, P = 0.0020. (F) Live imaging of mCherry-Rap2a and GFP-RBDRalGDS in MDA-MB-231 cells. Cells were transiently transfected with both mCherry-Rap2a and GFP-RBDRalGDS, plated on a collagen-coated glass dish, and imaged every 3 s using a 63× objective. Widefield microscope. Arrowheads point to an example of an mCherry-Rap2a–positive organelle being internalized from the lamellipodia. GFP-RBDRalGDS overlaps with mCherry-Rap2a primarily at the plasma membrane and is not found on the internalized organelle. See Video 7. Scale bars, 10 µm.
Figure S2.
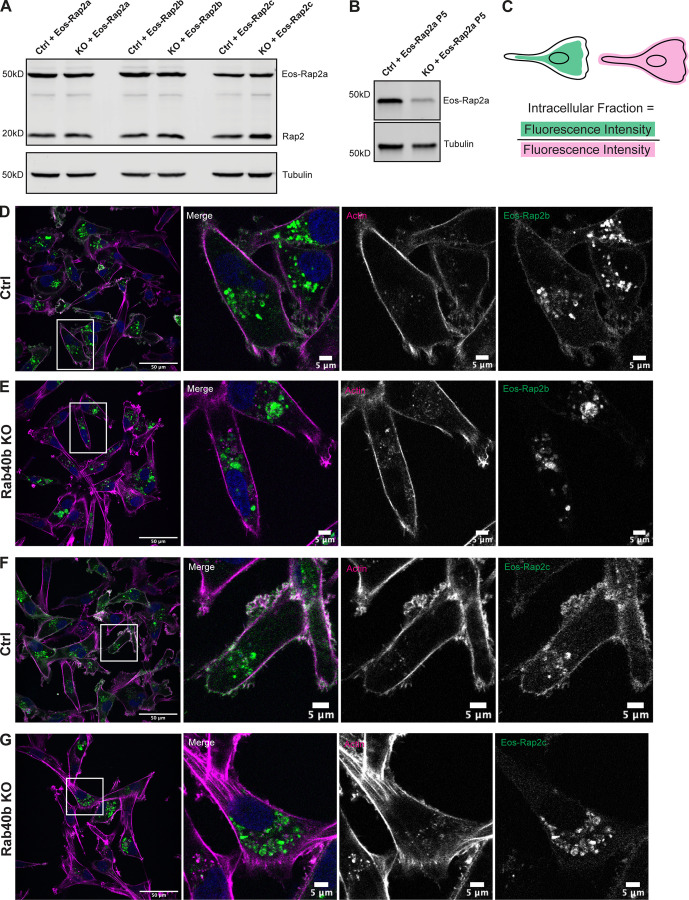
Loss of Rab40b also affects Rap2b and Rap2c subcellular localization. (A) Western blot showing stable overexpression of Eos-Rap2a, Eos-Rap2b, and Eos-Rap2c in MDA-MB-231 Ctrl and Rab40b KO cells (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (B) Western blot showing stable overexpression of Eos-Rap2a in MDA-MB-231 Ctrl and Rab40b KO cells at passage 5 (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. (C) Cartoon representation of intracellular/whole-cell fraction calculation (see Materials and methods). (D) Eos-Rap2b localization in MDA-MB-231 Ctrl cells. MDA-MB-231 cells stably expressing Eos-Rap2b were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. (E) Eos-Rap2b localization in Rab40b KO MDA-MB-231 cells. Rab40b KO cells stably expressing Eos-Rap2b were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. (F) Eos-Rap2c localization in MDA-MB-231 Ctrl cells. MDA-MB-231 cells stably expressing Eos-Rap2c were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. (G) Eos-Rap2c localization in Rab40b KO MDA-MB-231 cells. Rab40b KO cells stably expressing Eos-Rap2c were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 5 µm. Source data are available for this figure: SourceData FS2.
To further investigate how Rap2 localization and activation are interconnected, we visualized Rap2 simultaneously with a known GTP-dependent effector, RalGDS (Spaargaren and Bischoff, 1994; Franke et al., 1997; Nancy et al., 1999; Ohba et al., 2003; Jeon et al., 2007; Liu et al., 2010). Based on our previous data, we hypothesized that a Rap2-effector complex would colocalize at the plasma membrane where the majority of Rap2 is active. MDA-MB-231s were transiently transfected with mCherry-Rap2a and GFP-RBDRalGDS (Ras binding domain of RalGDS) and imaged using fluorescence microscopy. As shown in Fig. 5 D, GFP-RBDRalGDS localized to the leading edge, but was otherwise largely cytosolic. To firmly establish GFP-RBDRalGDS enrichment at ruffling lamellipodia, we used GFP alone as a control and quantified the leading edge to cell body fluorescence ratio (Fig. 5 E). Our statistical analysis gives confidence to the co-occurrence and enrichment of Rap2a and RBDRalGDS at the leading edge. Importantly, mCherry-Rap2a and GFP-RBDRalGDS overlapped at the lamellipodia plasma membrane, but not at intracellular organelles (Fig. 5 D). These observations further support a model in which Rap2 is active at the plasma membrane and inactive when internalized, though still membrane bound. Lastly, to characterize the spatiotemporal properties of Rap2 activation, we performed time-lapse analysis of mCherry-Rap2a concurrently with GFP-RBDRalGDS (Fig. 5 F and Video 7). As shown in Fig. 5 F, mCherry-Rap2a and GFP-RBDRalGDS primarily colocalized at the leading-edge lamellipodia. Over time, when mCherry-Rap2a got internalized from the plasma membrane, we observed that the mCherry-Rap2a organelle was not GFP-RBDRalGDS positive.
Video 7.
mCherry-Rap2a and GFP-RBDRalGDS live dynamics. Live 2D time-lapse imaging of MDA-MB-231 cells transiently expressing mCherry-Rap2a and GFP-RBDRalGDS. 3-s intervals (plus exposure time), 50 frames. 5 fps. Nearest neighbor deconvolution. Widefield microscope. Scale bar, 10 µm.
Overall, these results suggest that endosomal trafficking is critical for Rap2 placement and that Rap2 activation is closely linked to its localization. Importantly, the high co-occurrence of Rap2 with actin supports a model in which Rap2 needs to be localized at the plasma membrane of lamellipodia to control actin dynamics. How Rap2 is continuously recycled between endocytic organelles and the plasma membrane, as well as how Rap2 is specifically targeted and activated at the leading edge of migrating cells, is an important next question.
Targeting of Rap2 to the plasma membrane is dependent on Rab40b/Cul5
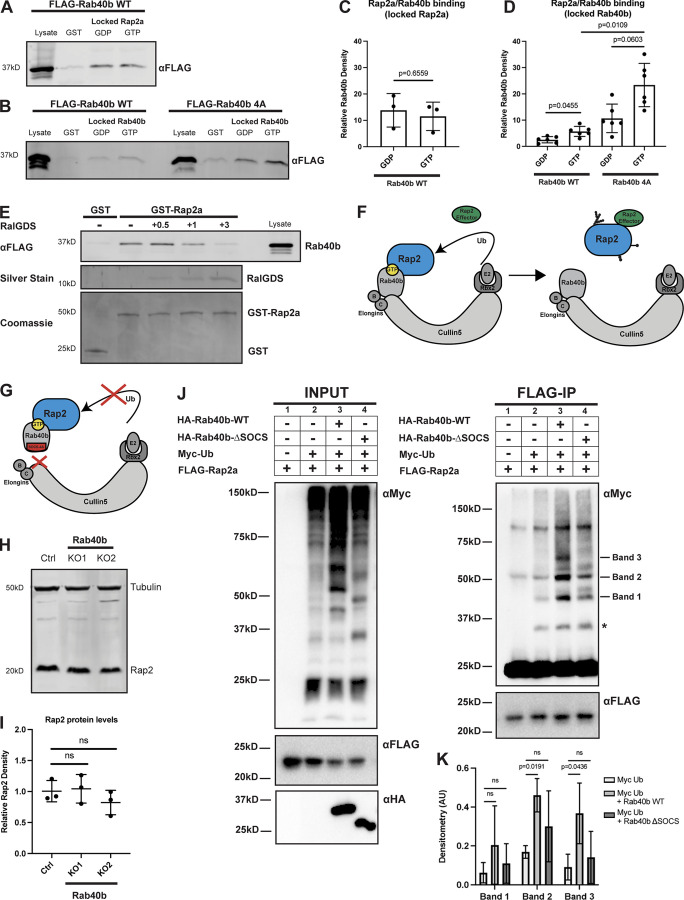
Ubiquitylation of small GTPases has emerged as a possible mechanism for regulating their localization, activation, and signaling (Nethe and Hordijk, 2010; de la Vega et al., 2014; Dohlman and Campbell, 2019). Cumulative data suggest that mono-ubiquitylation regulates localization and signaling of the Ras protein family (Jura et al., 2006). However, how ubiquitylation regulates the Rap family of GTPases, and whether this is different from the Ras proteins, is essentially unknown. We hypothesized that ubiquitylation might serve to modulate spatiotemporal dynamics of Rap2 during cell migration. Importantly, Rap2 has been proposed as a putative substrate of the Rab40c/Cul5 complex in a Xenopus model (Lee et al., 2007). Furthermore, Rab40b KO in MDA-MB-231 cells leads to defects that partially phenocopy Rap2 KO, namely, decreased chemotactic migration and invasion (Linklater et al., 2021). Given these collective studies, we wondered whether the Rab40b/Cul5 complex interacts with and ubiquitylates Rap2 in mammalian cells and what role this ubiquitylation might play in regulating Rap2 localization and activation during mammalian cell migration (Fig. 6 A).
Figure 6.
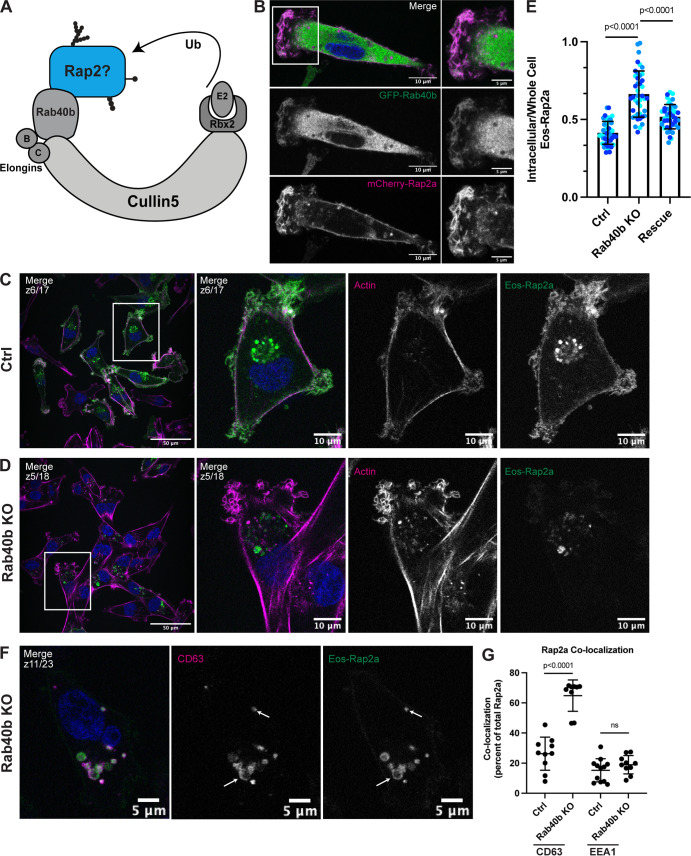
Loss of Rab40b results in a Rap2 sorting defect that leads to decreased Rap2 at the plasma membrane. (A) Rap2 is a putative substrate of the mammalian Rab40b/Cul5 complex. (B) Colocalization of GFP-Rab40b and mCherry-Rap2a. MDA-MB-231 cells stably expressing GFP-Rab40b were transiently transfected with mCherry-Rap2a, then fixed and stained with DAPI (blue). Scale bars, 10 and 5 µm. (C) Eos-Rap2a localization in Ctrl cells. Ctrl cells in this case are MDA-MB-231 parental cells. Eos-Rap2a has the same localization in MDA-MB-231 parental cells versus Cas9 cells (not depicted). MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 10 µm. (D) Eos-Rap2a localization in Rab40b KO cells. Rab40b KO MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 10 µm. (E) Intracellular to whole-cell fraction quantification, Eos-Rap2a in Ctrl versus Rab40b KO versus Rab40b KO Rescue cells. Rab40b KO Rescue cells overexpress FLAG-Rab40b WT (stable population line; see Fig. S1 E). Three biological replicates were performed for each cell line. In each biological replicate, five fields of view were imaged, with 3 cells analyzed from each field (total of 15 cells per n). The three shades of blue represent each set of 15 cells from n1, n2, and n3. Each cell was treated as its own data point, and statistical analysis (one-way ANOVA with Tukey’s multiple comparisons test) was performed with n = 45 total for each condition. Mean ± SD. Eos-Rap2a in Ctrl cells versus Eos-Rap2a in Rab40b KO cells, P < 0.0001; Eos-Rap2a in Rab40b KO cells versus Eos-Rap2a in Rab40b KO Rescue cells, P < 0.0001. ROUT outlier test removed three outliers from Rab40b KO data set. Ctrl cells in this experiment are MDA-MB-231 parental cells. (F) Eos-Rap2a colocalization with CD63 in Rab40b KO cells. Rab40b KO MDA-MB-231 cells stably expressing Eos-Rap2a were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Eos-Rap2a and CD63 overlap. Scale bars, 5 µm. (G) Quantification of colocalization between Rap2a and CD63/EEA1 in Ctrl versus Rab40b KO cells. Ctrl or Rab40b KO cells stably expressing Eos-Rap2a were fixed and stained with either CD63 (late endosomes/lysosomes) or EEA1 (early endosomes). 3i SlideBook6 software was used to calculate the percentage of total GFP-Rap2a that colocalizes with the markers indicated (see Materials and methods). Two biological replicates were performed, with approximately five cells imaged for each replicate. n = 10 Ctrl CD63, n = 9 KO CD63, n = 12 Ctrl EEA1, n = 10 KO EEA1. One-way ANOVA with Tukey’s multiple comparisons test. Ctrl versus Rab40b KO CD63, P < 0.0001; Ctrl versus Rab40b KO EEA1, ns.
To start dissecting whether the Rab40b/Cul5 complex regulates Rap2 during breast cancer cell migration, we first asked whether Rab40b and Rap2 colocalize in MDA-MB-231 cells. We have previously shown that GFP-Rab40b localizes to the leading edge of migrating cells, where it guides actin ruffling at lamellipodia (Jacob et al., 2013; Linklater et al., 2021). Because Rap2 appears to function at the leading edge during cell migration, we hypothesized that Rab40b and Rap2 may colocalize at the plasma membrane. As predicted, we found that GFP-Rab40b and mCherry-Rap2a colocalized at the leading-edge lamellipodia of the cell (Fig. 6 B). The overlap of these two proteins at the plasma membrane support a possible link between Rab40b, Rap2, and cell migration.
Because Rap2 recycling between the endolysosomal compartment and lamellipodia plasma membrane appears to regulate its activity and function, we next wondered whether localization of Rap2 is affected in our Rab40b KO cell line. For this experiment, we used the fluorescent protein Eos, which we found less sensitive to low pH than GFP, thus allowing us to better visualize lysosomal Rap2 (Wiedenmann et al., 2004; Zhang et al., 2012). As shown in Fig. 6 C, Eos-Rap2a in MDA-MB-231 control cells localized to two major populations: plasma membrane–bound and intracellular endolysosomal organelles (Fig. 6 C). Remarkably, Eos-Rap2a levels at the plasma membrane were greatly diminished in Rab40b KO cells, with increased Rap2a accumulation in intracellular organelles (Fig. 6 D). Quantification revealed a higher intracellular to whole-cell Eos-Rap2a fraction in Rab40b KO cells compared with control cells (Figs. 6 E and S2 C). This is not a result of decreased expression, as exogenous levels of Eos-Rap2a were equal in both control and Rab40b KO cell lines (Fig. S2 A). Importantly, we could partially rescue Eos-Rap2a mislocalization with the addition of Rab40b-WT (Figs. 6 E and S1 E).
We again costained these cells with known endocytic pathway markers, namely EEA1 (early endosome) and CD63 (early endosome/late endosome), and measured colocalization with Rap2a. The enriched intracellular Eos-Rap2a in Rab40b KO cells colocalized with both EEA1- and CD63-positive organelles (Figs. 6 F and S1 J). Interestingly, we found that Eos-Rap2a had a higher co-occurrence with CD63 in Rab40b KO cells compared with control cells, whereas EEA1 overlap was unchanged (Fig. 6 G). Given our earlier experiments suggesting that Rap2 is dynamically trafficked and likely recycled to the plasma membrane, this increase in CD63 accumulation suggests that Rab40b/Cul5-dependent ubiquitylation may be required for Rap2 sorting at early endosomes, away from lysosomes, and back to the plasma membrane. This is consistent with the observation that levels of Eos-Rap2a in Rab40b KO cells do decrease over time with passage number, pointing to lysosomal degradation (Fig. S2 B). Importantly, Rab40b depletion also led to similar defects in Eos-Rap2b and Eos-Rap2c subcellular distribution (Fig. S2, A and D–G), suggesting that the three Rap2 isoforms are regulated and function in a similar fashion. We propose, based on these results, that Rab40b regulates Rap2 sorting at early endosomes for recycling to the lamellipodia plasma membrane.
Rap2 is a Rab40b-binding protein
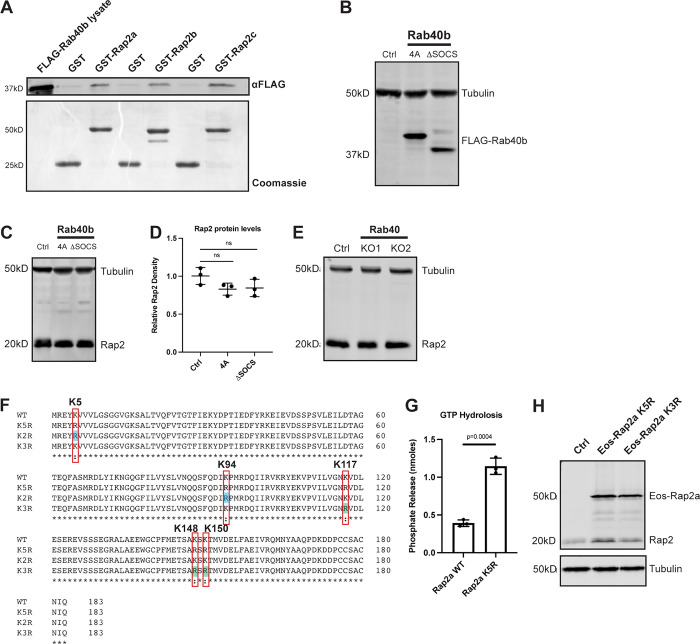
To begin understanding how Rab40b regulates Rap2 localization, we next assessed binding between these two proteins. Interestingly, both Rap2 and Rab40b are small monomeric GTPases, thus raising the question of whether their interaction is dependent on the nucleotide status of Rap2 or Rab40b. To this end, we incubated recombinant GST-Rap2a with MDA-MB-231 cell lysates stably expressing FLAG-Rab40b and analyzed their interaction using GST pull-down assays. Importantly, we found that the binding between these two proteins was independent of Rap2a nucleotide status (Fig. 7, A and C) but was enhanced when Rab40b was loaded with the nonhydrolyzable form of GTP, GMP-PNP (Fig. 7, B and D, left panels). This suggests that Rap2a predominantly binds Rab40b-GTP. We note that the binding was fairly weak, suggesting that the interaction between these proteins is likely quite transient, as would be expected between an enzyme (E3 ligase) and its substrate (Rap2). Additionally, we tested binding between Rab40b and the other Rap2 isoforms and found that Rab40b also interacted with Rap2b and Rap2c, suggesting again that the three Rap2 isoforms may be regulated in a similar manner (Fig. S3 A).
Figure 7.
Rap2 is a Rab40b-binding protein and is ubiquitylated by the Rab40b/Cul5 complex. (A) Rab40b binding to locked Rap2a. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT were incubated with either GST or GST-Rap2a, followed by a GST pull-down assay. Before incubation, GST-Rap2a was loaded with either GDP or GMP-PNP (labeled as GTP for brevity). 25 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (B) Rap2a binding to locked Rab40b. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT or SOCS-4A (left and right, respectively) were incubated with either GST or GST-Rap2a, followed by a GST pull-down. Before incubation, FLAG-Rab40b lysates were loaded with either GDP or GMP-PNP (labeled as GTP for brevity). 25 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (C) Quantification of GST pull-down in A. Three biological replicates were performed. Mean ± SD. GST signal was subtracted from GDP/GTP, and relative Rab40b density was calculated by normalizing to lysate. Unpaired t test. Rab40b WT GDP versus GTP (locked Rap2a), P = 0.6559. GTP = GMP-PNP. (D) Quantification of GST pull-down in B. Six biological replicates were performed. Mean ± SD. GST signal was subtracted from GDP/GTP, and relative Rab40b density was calculated by normalizing to lysate. Brown–Forsythe ANOVA with Dunnett’s T3 multiple comparisons test. Rab40b WT GDP versus GTP (locked Rab40b), P = 0.0455; Rab40b SOCS-4A GDP versus GTP (locked Rab40b), P = 0.0603; Rab40b WT GTP versus Rab40b SOCS-4A GTP (locked Rab40b), P = 0.0109. GTP = GMP-PNP. (E) Competitive binding experiment between FLAG-Rab40b, GST-Rap2a, and untagged RalGDS (RBD). MDA-MB-231 lysates stably expressing FLAG-Rab40b WT (GMP-PNP loaded) were incubated with either GST or GST-Rap2a (GMP-PNP loaded) and increasing concentrations of RalGDS (0.5, 1, and 3 times the amount of GST/GST-Rap2a). 15 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (F) Model summarizing findings in A–D. Rap2 preferentially binds Rab40b-GTP (left). Ubiquitylation of Rap2 by Rab40b/Cul5 and subsequent complex dissociation is necessary for Rap2 to interact with its downstream effector (in a GTP-dependent manner). (G) When the Rab40b/Cul5 complex is disrupted (SOCS-4A mutant), Rab40b binds more strongly to Rap2. We propose that this is due to lack of ubiquitylation/complex dissociation. (H) Rap2 protein levels Ctrl versus Rab40b KO cells. Ctrl and Rab40b KO MDA-MB-231 lysates were probed for αRap2 and αTubulin (loading control). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate CRISPR lines. 50 µg of lysate was loaded for each sample. (I) Quantification of Western blot in H. Three biological replicates were performed. Relative intensity of Rap2 was normalized to the levels of Tubulin and to Ctrl cells. Mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test. (J) Rap2a ubiquitylation in HEK293T cells. HEK293Ts were transfected with pRK5-FLAG-Rap2a ± pRK5-Myc-Ub, pRK7-HA-Rab40b, or pRK7-HA-Rab40b ΔSOCS. After 24 h of transfection, cells were harvested, lysed, and immunoprecipitated with αFLAG. Left column shows lysates (input) probed for Myc, FLAG, and HA. Right column shows FLAG immunoprecipitates probed for Myc and FLAG. Tick marks on the right blot indicate the Rap2-ubiquitin bands that are increased in response to Rab40b addition. Asterisk indicates presumed mono-ubiquitylated form of Rap2 that is not stimulated by Rab40b addition. (K) Quantification of HEK293T ubiquitylation assay in J. Raw densitometry was measured for bands 1, 2, and 3 across four independent experiments. These values (arbitrary units) were then normalized to FLAG-Rap2a input levels and graphed for conditions 2 (Myc-Ub), 3 (Myc-Ub + Rab40b WT), and 4 (Myc-Ub + Rab40b ΔSOCS). Four biological replicates were performed. Mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test. Band 1: Myc-Ub versus Myc-Ub + Rab40b WT, ns; Myc-Ub versus Myc-Ub + Rab40b ΔSOCS, ns; band 2: Myc-Ub versus Myc-Ub + Rab40b WT, P = 0.0191; Myc-Ub versus Myc-Ub + Rab40b ΔSOCS, ns; band 3: Myc-Ub versus Myc-Ub + Rab40b WT, P = 0.0436; Myc-Ub versus Myc-Ub + Rab40b ΔSOCS, ns. Source data are available for this figure: SourceData F7.
Figure S3.
Rap2 protein levels across Rab40 mutant cell lines and generation of Rap2 lysine mutants. (A) Rab40b binding to Rap2a, Rap2b, and Rap2c. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT were incubated with GST, GST-Rap2a, GST-Rap2b, or GST-Rap2c followed by a GST pull-down assay. 25 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (B) Western blot showing stable overexpression of FLAG-Rab40b SOCS-4A and FLAG-Rab40b ΔSOCS in MDA-MB-231s. Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate CRISPR lines. FLAG-Rab40b SOCS-4A and FLAG-Rab40b ΔSOCS were made in a Rab40b KO background. 50 µg of lysate was loaded for each sample. (C) Rap2 protein levels Ctrl versus Rab40b SOCS-4A versus Rab40b ΔSOCS cells. Ctrl, Rab40b SOCS-4A, and Rab40b ΔSOCS cell lysates were probed for αRap2 and αTubulin (loading control). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate CRISPR lines. FLAG-Rab40b SOCS-4A and FLAG-Rab40b ΔSOCS were made in a Rab40b KO background. 50 µg of lysate was loaded for each sample. (D) Quantification of Western blot in C. Three biological replicates were performed. Relative intensity of Rap2 was normalized to the levels of Tubulin and to Ctrl cells. Mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test. (E) Rap2 protein levels, Ctrl versus Rab40 KO cells. Ctrl and Rab40 KO cell lysates (lacking Rab40a, Rab40b, and Rab40c) were probed for αRap2 and αTubulin (loading control). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate CRISPR lines. 50 µg of lysate was loaded for each sample. (F) Protein sequence alignment of Rap2a-WT, K5R, K3R, and K2R. Red boxes denote the five lysines within K5R, blue shades represent K2R, and green shades represent K3R. Alignment made using Clustal Omega. (G) GTP hydrolysis assay Rap2a-WT versus Rap2a-K5R. CytoPhos assay was performed using purified GST-Rap2a-WT and GST-Rap2a-K5R. Colorimetric change at 650 nm was measured and analyzed against a standard curve to determine inorganic phosphate release (nmol). Three biological replicates were performed. Mean ± SD. Unpaired t test. WT versus K5R, P = 0.0004. (H) Western blot showing stable overexpression of Eos-Rap2a-K5R and -K3R in MDA-MB-231s (lentivirus). Ctrl cells are MDA-MB-231 parentals. 50 µg of lysate was loaded for each sample. Source data are available for this figure: SourceData FS3.
We next wondered whether Rap2 can bind its effector proteins while part of a Rab40b/Rap2 complex, or whether these interactions are mutually exclusive. To test this, we performed a competitive binding experiment: FLAG-Rab40b lysates were incubated with GST-Rap2a as before, but with increasing concentrations of the Rap2 effector, RalGDS (Fig. 7 E). Importantly, we discovered that there was indeed competition for Rap2 binding between Rab40b and RalGDS, suggesting that Rap2 cannot bind both proteins simultaneously. Overall, these data suggest that Rap2 dissociation from the Rab40b/Cul5 complex, likely after ubiquitin modification, is required for its interaction with downstream effectors (Fig. 7 F).
To understand the role of Cul5 in regulating Rap2 function, we next asked whether Rab40b binding to Rap2 is dependent on Cul5. To this end, we incubated GST-Rap2a with MDA-MB-231 cells stably expressing a FLAG-Rab40b SOCS-4A mutant, which disrupts binding to Cul5 and formation of the CRL complex (Fig. 7 G; Linklater et al., 2021; Duncan et al., 2021). We found that the SOCS-4A mutation did not block Rap2a and Rab40b binding (Fig. 7 B, right panel), suggesting that Rab40b is able to bind its substrates independently of Cul5 presence. Intriguingly, the Rab40b SOCS-4A mutant binds to Rap2a more strongly than Rab40b WT (Fig. 7 D, right panel). This observation would be consistent with the idea that Rap2 is a Rab40b/Cul5 substrate, since blocking ubiquitylation would likely prevent dissociation between the E3 ligase complex (Rab40b/Cul5) and its substrate protein (Rap2; Fig. 7 G). Furthermore, this fits with the idea that Rap2 must be released from the Rab40b/Cul5 complex to bind its putative effectors (Fig. 7 F).
Rap2 is ubiquitylated by the Rab40b/Cul5 complex
Given that mammalian Rab40b and Rap2 interact, and given our data suggesting that Rap2 is not targeted properly in the absence of Rab40b, we next asked whether the Rab40b/Cul5 complex regulates proteasomal degradation of Rap2 via poly-ubiquitylation. To do this, we measured total protein levels of Rap2 in two different Rab40b KO cell lines as well as two other cell lines expressing either a Rab40b SOCS-4A or Rab40b ΔSOCS mutant (Linklater et al., 2021; Duncan et al., 2021). Importantly, the Rab40b ΔSOCS mutant still contained the C-terminal prenylation site to ensure its ability to associate with membranes (Fig. S3 B). We hypothesized that if the Rab40b/Cul5 complex regulates proteasomal degradation of Rap2, that Rap2 levels should be increased in the absence of Rab40b as well as when the Rab40b/Cul5 complex is disrupted. This is based on the well-accepted doctrine in the field that canonical poly-ubiquitin chains signal for proteasomal degradation via the 26S proteasome (Pickart and Fushman, 2004; Komander and Rape, 2012). Surprisingly, we observed no significant change in Rap2 protein levels across our Rab40b KO cell lines or the Rab40b SOCS mutants (Fig. 7, H and I; and Fig. S3, C and D), suggesting that the Rab40b/Cul5 complex does not mediate canonical ubiquitin-dependent degradation of Rap2. One caveat to the Rab40b KO cell line is the presence of the other Rab40 isoforms, which may compensate for the lack of Rab40b and prevented us from detecting protein level changes. To alleviate this, and since there is also evidence of Rab40c-dependent regulation of Rap2, we measured Rap2 protein levels in two different MDA-MB-231 Rab40 KO cell lines, where cells lack the three Rab40 family members, Rab40a, Rab40b, and Rab40c (Fig. S3 E; Linklater et al., 2021). Importantly, we once again did not detect any significant change in global Rap2 protein levels in the Rab40 KO cell lines, suggesting that the Rab40 GTPases do not mediate proteasomal degradation of Rap2.
At this point, our data postulate that the Rab40b/Cul5 complex binds and regulates Rap2 localization, without affecting global protein levels of Rap2. Additionally, our localization data suggest a potential early endosome sorting defect in Rab40b KO cells, leading to increased targeting of Rap2 to lysosomes. Based on these observations and what we know about the ubiquitin code, we speculated that Rap2 might be subjected to a noncanonical ubiquitin tag that may regulate its sorting and targeting to the lamellipodia plasma membrane (Swatek and Komander, 2016; Akutsu et al., 2016). To begin testing this idea, we used HEK293T cells to test direct ubiquitylation of Rap2 by the Rab40b/Cul5 complex.
Using HEK293T cells, we set up four different transfection conditions: (1) FLAG-Rap2a alone, (2) FLAG-Rap2a + Myc-Ub, (3) FLAG-Rap2a + Myc-Ub + HA-Rab40b WT, and (4) FLAG-Rap2a + Myc-Ub + HA-Rab40b ΔSOCS (Fig. 7 J, left panel). In each case, we immunoprecipitated with anti-FLAG and Western blotted for anti-Myc (Fig. 7 J, right panel). Importantly, we observed stimulation of Rap2 ubiquitylation by Rab40b WT (condition 3). We focused on Rap2-Ub species that were stimulated by Rab40b in all four repeats. First, the Rap2-Ub species labeled as band 1 increased when Rab40b-WT was added but was less enhanced by Rab40b ΔSOCS (condition 4; Fig. 7, J and K). Based on the molecular weight, we hypothesize that band 1 corresponds to a di-Ub Rap2 species. Next, we also saw that bands 2 and 3 were enhanced by the addition of Rab40b-WT but not Rab40b ΔSOCS (Fig. 7, J and K). Again, based on size, we hypothesize that these bands correspond to the addition of three to five ubiquitin moieties on Rap2. One caveat is that band 2 appeared in the FLAG-Rap2a–only control (far left column, condition 1). We believe that band 2 overlaps with a nonspecific band (∼50 kD) that our secondary antibody detects, likely remnants of IgG heavy chain. Overall, when analyzing four independent experiments, these three Rap2-ubiquitin species (band 1, 2, and 3) were consistently stimulated by addition of Rab40b (Fig. 7 K).
Interestingly, we did not see any enhancement of Rap2 mono-ubiquitylation with the addition of Rab40b (∼30 kD band, marked with asterisk). This suggests that Rab40b/Cul5 might work in concert with other E3s, where Rab40b/Cul5 triggers additional ubiquitylation of Rap2 after it is originally primed with ubiquitin by another E3 Ligase. This concept of two E3s acting independently but sequentially to prime and extend ubiquitin marks on protein substrates is established but still incompletely understood (Deol et al., 2019). Finally, we saw little indication of Rab40b-mediated poly-ubiquitylation of Rap2, which would appear as a high molecular weight smear instead of distinct bands. This is consistent with our data showing that loss of Rab40b does not mediate proteasomal targeting of Rap2 (Fig. 7, H and I). Given this, we hypothesize that these nondegradative ubiquitin moieties are likely linkages with a unique downstream signal, possibly to regulate sorting and targeting to the plasma membrane. Although further work will be needed to define the composition of these ubiquitin species (multi-mono, etc.), collectively, these results support Rap2 as a substrate of the Rab40b/Cul5 complex.
Ubiquitylation of Rap2 regulates its subcellular localization
Summarizing our data thus far, we propose that the Rab40b/Cul5 complex regulates Rap2 function via direct ubiquitylation. To start dissecting the molecular consequence of Rap2 ubiquitylation, we sought to identify the specific lysines on Rap2 that are modified by ubiquitin. Previous work on Rap2a in neurons identified four lysines of Rap2a that are putative targets for Nedd4-1–mediated ubiquitylation (K5, K94, K148, and K150; Kawabe et al., 2010). Additionally, the initial study implicating Rap2 as a possible Rab40c/Cul5 substrate proposed that a single lysine mutation at position 117 was sufficient to reduce ubiquitylation of Rap2 (Lee et al., 2007). Thus, we decided to generate a Rap2a-K5R construct (K5, K94, K117, K148, and K150), with the goal of understanding how localization, sorting, and activation are affected by mutation of putative ubiquitylation sites (Fig. S3 F). One potential concern is that mutation of these lysines may alter Rap2 folding and block GTP loading. To refute this possibility, we used an endpoint phosphate assay to measure GTP hydrolysis as a readout for proper Rap2 folding and found that Rap2a-K5R was still capable of binding and hydrolyzing GTP (Fig. S3 G).
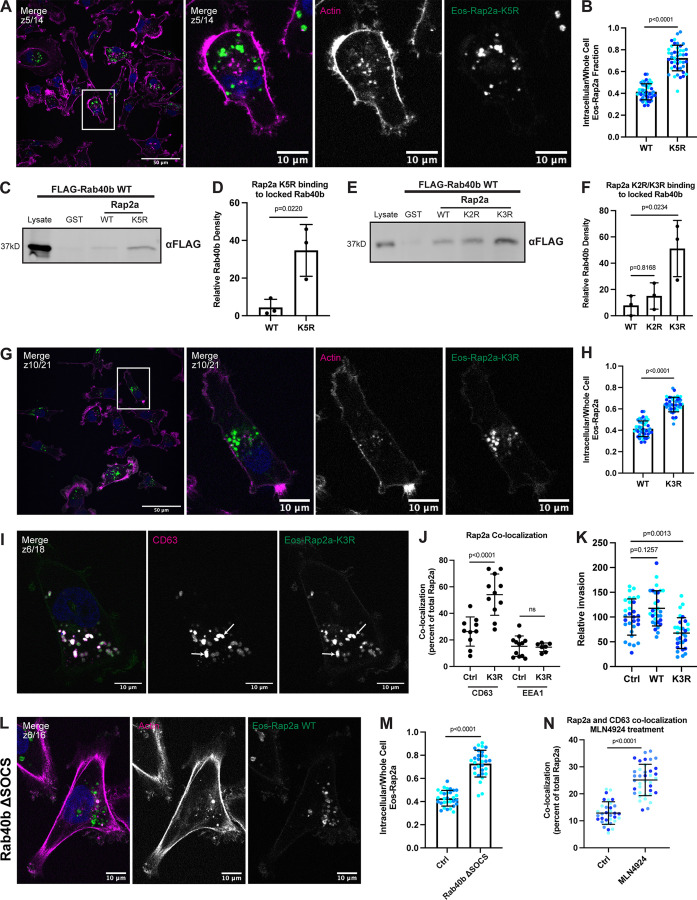
Moving forward, we first asked whether Rap2a-K5R localization is altered compared with Rap2a-WT. To this end, MDA-MB-231 cells stably expressing Eos-Rap2a-K5R (Fig. S3 H) were analyzed by immunofluorescence. We found that Eos-Rap2a-K5R was predominantly localized to the endolysosomal compartment (Fig. 8 A), similar to Eos-Rap2a-WT localization in a Rab40b KO background. Indeed, fluorescence intensity analysis of Rap2a-K5R revealed a higher intracellular to whole-cell fraction compared with Rap2a-WT (Figs. 8 B and S2 C).
Figure 8.
Mutation of five putative ubiquitylation sites within Rap2 affects recycling, subcellular localization, binding to Rab40b, and cell invasion. (A) Eos-Rap2a-K5R localization. MDA-MB-231 cells stably expressing Eos-Rap2a-K5R were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 10 µm. (B) Intracellular to whole-cell fraction quantification, Eos-Rap2a-WT versus Eos-Rap2a-K5R. Three biological replicates were performed for each cell line. In each biological replicate, five fields of view were imaged, with 3 cells analyzed from each field (total of 15 cells per n, color coded). Each cell was treated as its own data point, and statistical analysis (unpaired t test) was performed with n = 45 total for each condition. Mean ± SD. Eos-Rap2a-WT versus Eos-Rap2a-K5R, P < 0.0001. Eos-Rap2a-WT data points are from Fig. 6 E. (C) Rap2a-K5R binding to Rab40b. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT were incubated with GST, GST-Rap2a-WT, or -K5R, followed by GST pull-down. Before incubation, FLAG-Rab40b lysates were loaded with GMP-PNP. 25 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (D) Quantification of GST pull-down in C. Three biological replicates were performed. Mean ± SD. GST signal was subtracted from experimental lanes, and relative Rab40b density was calculated by normalizing to lysate. Unpaired t test. Rap2a-WT versus Rap2a-K5R, P = 0.0220. (E) Rap2a-K2R and Rap2a-K3R binding to Rab40b. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT were incubated with GST, GST-Rap2a-WT, GST-Rap2a-K2R, or GST-Rap2a-K3R followed by a GST pull-down. Before incubation, FLAG-Rab40b lysates were loaded with GMP-PNP. 25 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (F) Quantification of GST pull-down in E. Three biological replicates were performed. Mean ± SD. GST signal was subtracted from experimental lanes, and relative Rab40b density was calculated by normalizing to lysate. One-way ANOVA with Tukey’s multiple comparisons test. Rap2a-WT versus Rap2a-K2R, P = 0.8168; Rap2a-WT versus Rap2a-K3R, P = 0.0234. (G) Eos-Rap2a-K3R localization. MDA-MB-231 cells stably expressing Eos-Rap2a-K3R were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 10 µm. (H) Intracellular to whole-cell fraction quantification, Eos-Rap2a-WT versus Eos-Rap2a-K3R. Three biological replicates were performed for each cell line. In each biological replicate, five fields of view were imaged, with three cells analyzed from each field (total of 15 cells per n, color coded). Each cell was treated as its own data point, and statistical analysis (unpaired t test) was performed with n = 45 total for each condition. Mean ± SD. Eos-Rap2a-WT versus Eos-Rap2a-K3R, P < 0.0001. Eos-Rap2a-WT data points are from Fig. 6 E. (I) Eos-Rap2a-K3R colocalization with CD63. MDA-MB-231 cells stably expressing Eos-Rap2a-K3R were fixed and stained with the lysosomal marker CD63 (magenta) and DAPI (blue). Arrows indicate examples of Eos-Rap2a-K5R and CD63 overlap. Scale bars, 10 µm. (J) Quantification of colocalization between Rap2a and CD63/EEA1 in Ctrl versus K3R cells. MDA-MB-231s stably expressing either Eos-Rap2a WT or Eos-Rap2a K3R were fixed and stained with either CD63 (late endosomes/lysosomes) or EEA1 (early endosomes). 3i SlideBook6 software was used to calculate the percentage of total GFP-Rap2a that colocalizes with the markers indicated (see Materials and methods). Two biological replicates were performed, with approximately five cells imaged for each replicate. n = 10 Ctrl CD63, n = 11 K3R CD63, n = 12 Ctrl EEA1, n = 7 K3R EEA1. One-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Ctrl versus K3R CD63, P < 0.0001; Ctrl versus K3R EEA1, ns. (K) Invasion assay quantification, Ctrl versus Eos-Rap2a WT versus Eos-Rap2a K3R. Three biological replicates were performed, with technical duplicates in each experiment. 5 fields of view were imaged for each Matrigel insert (resulting in 10 fields of view per experiment per condition, color coded). Raw number of cells invaded per field of view were normalized to Ctrl (relative invasion). Ctrl cells are MDA-MB-231 parental cells. Eos-Rap2a WT and K3R are stable overexpressed lines. Each field of view was treated as its own data point (n = 30). One-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Ctrl versus Eos-Rap2a WT, P = 0.1257; Ctrl versus Eos-Rap2a K3R, P = 0.0013. (L) Eos-Rap2a localization in Rab40b ΔSOCS cells. Rab40b ΔSOCS MDA-MB-231 cells expressing Eos-Rap2a were fixed and stained with phalloidin (magenta) and DAPI (blue). Scale bars, 50 and 10 µm. (M) Intracellular to whole-cell fraction quantification, Eos-Rap2a in Ctrl cells versus Rab40b ΔSOCS cells. Two biological replicates were performed for each cell line. In each biological replicate, five fields of view were imaged, with 3 cells analyzed from each field (total of 15 cells per n, color coded). Each cell was treated as its own data point, and statistical analysis (unpaired t test) was performed with n = 30 total for each condition. Mean ± SD. Eos-Rap2a Ctrl versus Eos-Rap2a ΔSOCS cells, P < 0.0001. Eos-Rap2a-WT data points are from Fig. 6 E. (N) Rap2a and CD63 colocalization analysis in Ctrl cells versus cells treated with neddylation/Cul5 inhibitor MLN4924. MDA-MB-231 cells stably expressing GFP-Rap2a were treated with either DMSO (Ctrl) or 300 nM MLN4924 for 24 h. Cells were then fixed and stained for CD63, before colocalization analysis. 3i SlideBook6 software was used to calculate the percentage of total GFP-Rap2a that colocalized with CD63 (see Materials and methods). Three biological replicates were performed, with ∼12 cells imaged for each replicate (color coded for each n). Mean ± SD. n = 33 for Ctrl/DMSO treated cells, n = 38 for MLN4924 treated cells. Unpaired t test. P < 0.0001. Source data are available for this figure: SourceData F8.
We next wondered how mutation of these five lysines might affect Rap2 binding to the Rab40b/Cul5 complex. To test this, we incubated purified GST-Rap2a-K5R with FLAG-Rab40b lysates and performed a GST pull-down assay. Notably, we discovered that Rap2a-K5R bound to Rab40b much more strongly than Rap2a-WT (Fig. 8, C and D). Again, this result phenocopied the increased binding between Rap2a-WT and Rab40b SOCS-4A. If we hypothesize that increased binding is due to lack of dissociation, one prediction is that the Rab40b/Cul5 complex binds Rap2a-K5R but cannot ubiquitylate and dissociate from one another. This would suggest that Rab40b/Cul5 may target at least one or more of the five lysines within the Rap2a-K5R mutant.
The five lysines mentioned above are clustered into two general regions: three near the GTP pocket (K117, K148, and K150) and two removed from the GTP pocket (K5 and K94). Thus, we split the mutations into two groups: K2R (K5, K94) and K3R (K117, K148, K150) to begin dissecting their function (Fig. S3 F). Given the increased binding of Rap2a-K5R to Rab40b, we began with binding assays as a readout of which mutant (K2R or K3R) might house the relevant Rap2 lysines targeted by the Rab40b/Cul5 complex. Notably, we observed a significant increase in the ability of Rap2a-K3R to bind Rab40b compared with Rap2a-WT (Fig. 8, E and F), suggesting that the K3R mutant likely contains the most pertinent Rap2 lysines ubiquitylated by Rab40b/Cul5.
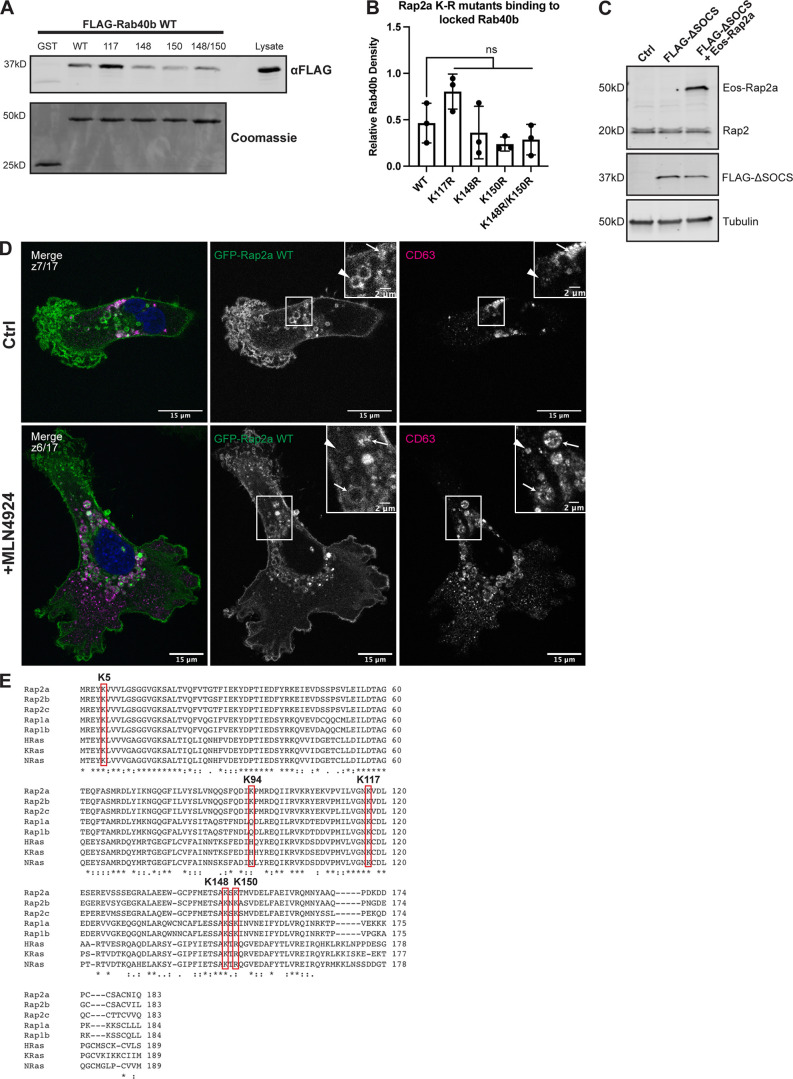
Of course, the goal in any ubiquitylation study is to identify the specific lysine responsible for changes in downstream signaling. Thus, we split the K3R mutant into four individual mutants: K117R, K148R, K150R, and K148R/K150R. We then used our GST-Rap2a and FLAG-Rab40b binding assay to test whether any of these single (or double) mutations could phenocopy the increased GST-Rap2a-K3R binding to Rab40b. Importantly, none of the single mutants recapitulated the increased binding to Rab40b as seen by Rap2a-K3R (Fig. S4, A and B); thus, we hypothesize that Rab40b/Cul5 may ubiquitylate all three lysines (K117, K148, and K150) within Rap2. Indeed, our HEK293T ubiquitylation experiment strongly suggests that Rab40b stimulates the addition of two to five ubiquitin moieties on Rap2, and while it is possible that all of these ubiquitin modifications are conjugated to one single lysine, it is also equally possible for multiple Rap2 lysines to be modified, possibly multi-mono-ubiquitylated.
Figure S4.
Characterization of Rap2 lysine mutants and the role of Cul5 in mediating Rap2 plasma membrane targeting. (A) Rap2a K-R single/double mutants binding to Rab40b. MDA-MB-231 lysates stably expressing FLAG-Rab40b WT were incubated with GST, GST-Rap2a-WT or -K117R, K148R, K150R, or K148R/K150R, followed by a GST pull-down. Before incubation, FLAG-Rab40b lysates were loaded with GMP-PNP. GST alone was used to control for GST binding to Rab40b. 15 µg of lysate input was loaded as a positive control and used to estimate pull-down efficiency. (B) Quantification of GST pull-down in A. Three biological replicates were performed. Mean ± SD. GST signal was subtracted from experimental lanes, and relative Rab40b density was calculated by normalizing to lysate. One-way ANOVA with Tukey’s multiple comparisons test. ns between WT and all individual mutants. (C) Western blot showing generation of cell line in Fig. 8, L and M. Rab40b KO cells were first stably transfected with FLAG-Rab40b ΔSOCS (lentivirus, second column). The cells were then transfected with Eos-Rap2a and flow sorted (lentivirus, third column, flow sort instead of selection). Ctrl cells are dox-inducible Cas9 MDA-MB-231s that were used to generate Rab40b KO CRISPR line. 50 µg of lysate was loaded for each sample. (D) Representative images from MLN4924 experiment in Fig. 8 N. MDA-MB-231 cells stably expressing GFP-Rap2a were treated with either DMSO (Ctrl) or 300 nM MLN4924 for 24 h. Cells were then fixed and stained for CD63 (magenta) and DAPI (blue). Arrows indicate examples of GFP-Rap2a and CD63 overlap. Arrowheads point to GFP-Rap2a organelles that are not CD63 positive. Scale bars, 15 and 2 µm. (E) Protein sequence alignment of Rap2a, Rap2b, Rap2c, Rap1a, Rap1b, HRas, KRas, and NRas. Red boxes denote the five lysines within Rap2a-K5R. Stars indicate fully conserved residues. Alignment made using Clustal Omega. Source data are available for this figure: SourceData FS4.
Moving forward with Rap2a-K3R, we next assessed whether mutation of these three lysines affects localization in a manner similar to the K5R mutant. MDA-MB-231 cells stably expressing Eos-Rap2a-K3R (Fig. S3 H) were analyzed by immunofluorescence. Like the K5R mutant, Eos-Rap2a-K3R was predominately localized within the endolysosomal compartment (Fig. 8, G and H; and Fig. S2 C). Similar to how Rap2a-WT behaves in Rab40b KO cells, Rap2a-K3R had increased presence in lysosomes (CD63) compared with control cells but unchanged colocalization with early endosomes (EEA1; Fig. 8, I and J). This again supports a model in which ubiquitylation of Rap2 is needed for sorting and recycling to the leading-edge plasma membrane. To test this model further, we asked whether recruitment of Rap2 to the plasma membrane is dependent on Rab40b/Cul5 complex formation. To this end, we examined Rap2 localization in a Rab40b ΔSOCS background (Fig. 8 L). Rab40b KO cells stably expressing FLAG-Rab40b ΔSOCS were transfected with Eos-Rap2a WT lentivirus and flow sorted (Fig. S4 C). Consistent with our hypothesis, we discovered that Eos-Rap2a levels at the plasma membrane were noticeably reduced in Rab40b ΔSOCS cells (Fig. 8, L and M). Collectively, these data define a model wherein ubiquitylation by the Rab40b/Cul5 complex is required for sorting and recycling of Rap2 to the leading-edge plasma membrane of migrating breast cancer cells.
Although the mislocalization of Rap2 in Rab40b ΔSOCS strongly suggests that the Rab40b/Cul5 complex is necessary for proper Rap2 targeting to the leading edge, we also tested whether direct inhibition of Cul5 resulted in the same phenotype. To this end, we used MLN4924, a small molecular inhibitor of CRLs (Lan et al., 2016). MDA-MB-231 cells stably expressing GFP-Rap2a were treated with MLN4924, followed by fixation and staining for CD63. Similar to what we observed in Rab40b KO cells, Rab40b ΔSOCS cells, and Rap2a K-R mutant cells, Rap2 accumulated in lysosomes with MLN4924 treatment (Figs. 8 N and S4 D). Additionally, MLN4924 appeared to impact Rap2 enrichment at the leading edge (Fig. S4 D). This suggests that Cul5, and specifically ubiquitylation by Rab40b/Cul5, is needed for dynamic redistribution of Rap2 at the lamellipodia during cell migration.
Finally, to bridge these findings with Rap2’s putative role in regulating cell movement, we wondered whether the K3R mutation would influence cell migration. To this end, we measured the invasiveness of MDA-MB-231 parental cells compared to cells stably expressing either Eos-Rap2a WT or Eos-Rap2a K3R. Remarkably, cells expressing the K3R mutant had decreased invasion overall, suggesting that this mutant may act as a dominant negative (Fig. 8 K).
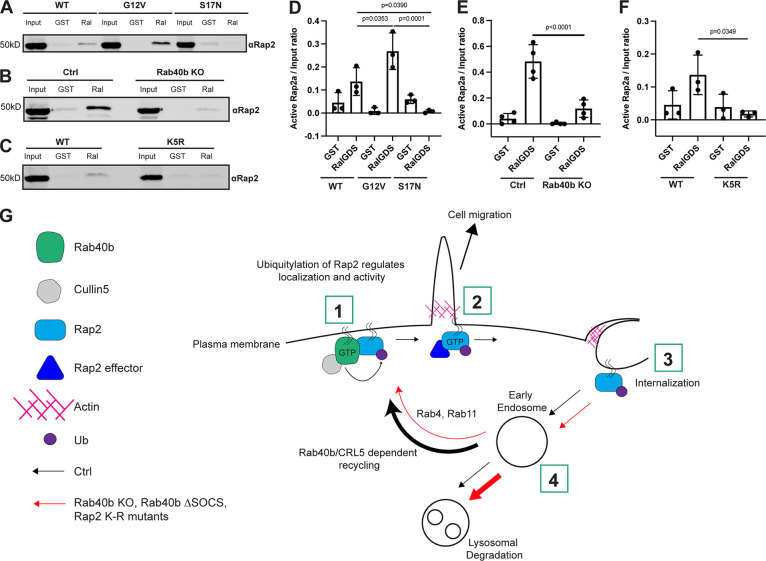
Ubiquitylation of Rap2 is required for its activation
The accumulation of Rap2 in the endolysosomal compartment in Rab40b KO and ΔSOCS cells (Figs. 6 E and 8 M) raises an interesting question about the activation state of Rap2 in these cells, given that the Rap2 dominant-negative mutant localizes almost exclusively to endosomes and lysosomes (Fig. 5 B; and Fig. S1, H and I). To investigate this further, we took advantage of a previously described method to measure Rap2 activation (Spaargaren and Bischoff, 1994; Franke et al., 1997; Nancy et al., 1999; Ohba et al., 2003; Jeon et al., 2007). Specifically, purified GST-RBDRalGDS was incubated with cell lysates expressing various Rap2 mutants, and levels of active Rap2 (bound to GST-RBDRalGDS) were determined by GST pull-down assays followed by Western blotting against Rap2. First, as a proof of principle, MDA-MB-231 cells transfected with GFP- Rap2a-WT, Rap2a-G12V, or Rap2a-S17N were lysed and incubated with GST-RBDRalGDS (Fig. 9 A). We then defined active Rap2a as the fraction of GFP-Rap2a pulled down by GST-RBDRalGDS compared with the input (Fig. 9 D). We found that the results were consistent with the constitutively active and dominant-negative mutations, where Rap2a-G12V was more active and Rap2a-S17N was less active than Rap2a-WT, respectively (Fig. 9 D).
Figure 9.
Activation of Rap2 is regulated by Rab40b and is linked to its ubiquitylation. (A) Activation assay Rap2a-WT, -G12V, and -S17N. MDA-MB-231 parental cells were transiently transfected with GFP-Rap2a-WT, -G12V, or -S17N (pLVX-GFP constructs). Lysates were incubated with either GST or GST-RBDRalGDS, followed by a GST pull-down assay. GST alone was used to control for GST binding to Rap2a. 25 µg of lysate input was loaded as a positive control and used to calculate active levels of Rap2a. (B) Activation assay Rap2a-WT in Ctrl versus Rab40b KO cells. MDA-MB-231 parental cells or Rab40b KO cells were transiently transfected with GFP-Rap2a-WT (pEGFP-construct). Lysates were incubated with either GST or GST-RBDRalGDS, followed by a GST pull-down assay. GST alone was used to control for GST binding to Rap2a. 25 µg of lysate input was loaded as a positive control and used to calculate active levels of Rap2a. (C) Activation assay Rap2a-WT and -K5R. MDA-MB-231 parental cells were transiently transfected with GFP-Rap2a-WT and GFP-Rap2a-K5R (pLVX-GFP constructs). Lysates were incubated with either GST or GST-RBDRalGDS, followed by a GST pull-down assay. GST alone was used to control for GST binding to Rap2a. 25 µg of lysate input was loaded as a positive control and used to calculate active levels of Rap2a. (D) Quantification of activation assay in A. Three biological replicates were performed. Mean ± SD. Active Rap2a/input ratio was defined as active Rap2 signal/lysate signal. One-way ANOVA with Tukey’s multiple comparisons test. GST signal was not subtracted. WT versus G12V, P = 0.0353; WT versus S17N, P = 0.0390; G12V versus S17N, P = 0.0001. (E) Quantification of activation assay in B. Three biological replicates were performed. Mean ± SD. Active Rap2a/Input ratio was defined as active Rap2 signal/lysate signal. One-way ANOVA with Tukey’s multiple comparisons test. GST signal was not subtracted. Ctrl versus Rab40b KO, P < 0.0001. (F) Quantification of activation assay in C. Three biological replicates were performed. Mean ± SD. Active Rap2a/input ratio was defined as active Rap2 signal/lysate signal. One-way ANOVA with Tukey’s multiple comparisons test. GST signal was not subtracted. WT versus K5R, P = 0.0349. (G) Working model for how Rab40b/Cul5 modulates Rap2 subcellular localization and activation via ubiquitylation. (1) Rab40b/Cul5 and Rap2 primarily interact at the plasma membrane, where binding is enhanced when Rab40b is GTP bound. Ubiquitylation by the Rab40b/Cul5 complex causes dissociation. (2) Active Rap2 is then able to interact with its effector proteins at the plasma membrane, where it regulates leading-edge actin dynamics and ultimately drives cell migration. (3) To quickly re-distribute Rap2 to other locations at the lamellipodia plasma membrane, Rap2 is internalized via a pinocytosis-like mechanism. Internalization mediates the delivery of Rap2 to early endosomes. (4) Once at early endosomes, previous ubiquitylation by the Rab40b/Cul5 complex serves as a signal for Rap2 to be recycled back to the leading-edge plasma membrane via the Rab4/Rab11 recycling pathways. Without this ubiquitin signal, Rap2 is sorted to lysosomes for degradation. We hypothesize that Rap2 is continuously recycled to and from the plasma membrane to regulate its localization and activity. Ultimately, we propose that dynamic redistribution of Rap2 is required to maintain its enrichment at the leading edge, where it can regulate actin dynamics and promote cell migration. Source data are available for this figure: SourceData F9.
Next, we asked whether Rap2 activity was affected in cells lacking Rab40b. Indeed, we detected less active Rap2a in Rab40b KO cells compared with control cells, suggesting that the Rab40b/Cul5 complex is required for both Rap2 activation and localization at the plasma membrane (Fig. 9, B and E). Delving further, we next questioned whether Rap2a-K5R activation was altered compared with Rap2a-WT. Again, based on our previous observations and recent studies on Ras regulation, we speculated that mutation of these critical lysines would affect GTP activity. Remarkably, we found that the K5R mutant severely abrogated Rap2 activation, as shown by a lack of Rap2a-K5R binding to GST-RBDRalGDS (Fig. 9, C and F), suggesting that ubiquitylation of these lysines is critical for both Rap2 localization and activation. In conclusion, our data hint at a complex system whereby the Rab40b/Cul5 complex coregulates Rap2 localization and activation. Although it is clear that subcellular distribution and GTP activity are closely intertwined, future investigations are needed to tease apart these pathways and uncover the direct molecular consequences of ubiquitin modification on Rap2.
Discussion
One of the most fundamental questions in cell biology is how cells coordinate highly complex processes such as actin polymerization, membrane trafficking, and adhesion remodeling to drive cell migration. In this study, we set out to characterize the function and regulation of the small GTPase Rap2 during cell migration. Here, we define a novel molecular mechanism by which Rab40b/Cul5-dependent ubiquitylation positively regulates Rap2 subcellular localization, as well as targeting and activation at the leading-edge plasma membrane.
Rap2 subcellular localization and activation are tightly controlled during cell migration
If Rap2 functions by regulating integrin-based adhesion and actin dynamics at the leading edge, one would expect that cells have fine-tuned mechanisms to control the spatiotemporal dynamics of Rap2 in migrating cells. Notably, we found that Rap2 localizes in two primary locations: the lamellipodia plasma membrane and the endolysosomal compartment. Using both fixed and live cell imaging, we discovered that Rap2 is dynamically trafficked through the endocytic pathway. Importantly, we observed that Rap2 is internalized via large macropinosome-like organelles that eventually become Rab5 positive. Once on Rab5 endosomes, Rap2 is then sorted between two pathways, Rab4/Rab11-dependent recycling back to the plasma membrane or lysosomal degradation. Sorting into the Rab4 and Rab11 recycling pathways from early endosomes appears to be a key event that allows rapid and dynamic targeting of internalized Rap2 back to the lamellipodia of migrating cells. Taken together, our collective data suggest a model in which localization of Rap2 is determined by active sorting between lysosomes or recycling to the plasma membrane, thus allowing for dynamic redistribution of Rap2 as cells migrate, to maintain its enrichment at the leading edge (Fig. 9 G).
What is of particular interest is that active (GTP-bound) Rap2 appears to predominantly localize at the plasma membrane, whereas inactive (GDP-bound) Rap2 localizes to endosomes and lysosomes, suggesting that regulation of subcellular distribution may be a key step in modulating activity and function of Rap subfamily members (Rebstein et al., 1993; Ohba et al., 2003; Bivona et al., 2004; Jeon et al., 2007). Consistent with GTP state and localization being interconnected, we show that Rap2 and its known effector RalGDS colocalize at the plasma membrane. There is some evidence that this is common among all Rap2 effectors; however, more work will be needed to uncover additional Rap2 effectors and whether they all localize and function at the plasma membrane (Taira et al., 2004; Machida et al., 2004; Myagmar et al., 2005; Nonaka et al., 2008). Given these cumulative observations, we propose that Rap2 signaling is tightly controlled by regulating its targeting and activation at the leading-edge plasma membrane, and that this is key to driving cell migration. This raises an intriguing question of how cells coregulate recruitment and activation of Rap2 at the plasma membrane and what molecular machinery is involved.
The Rab40b/Cul5 complex modulates Rap2 recycling to the leading-edge plasma membrane via ubiquitylation
Ubiquitylation of Ras GTPases has recently has emerged as an important regulator of their activity and signaling (Jura et al., 2006; Dohlman and Campbell, 2019). Thus, we wondered whether ubiquitylation of Rap2 might play an important role in regulating its spatiotemporal dynamics and activation in migrating cells. Our previous work uncovered the role of Rab40b during breast cancer cell migration and led us to also characterize its interaction with the E3-ubiquitin ligase scaffold protein Cul5 (Jacob et al., 2013; Jacob et al., 2016; Linklater et al., 2021; Duncan et al., 2021). The Rab40 GTPases are unique among other Rabs because of their extended C-terminal SOCS box, which facilitates binding to Cul5, forming a CRL5 ubiquitylation complex. Importantly, our previous work demonstrated that Rab40b depletion partially phenocopies the cell migration and invasion defects in Rap2 KO cells that we report in this study (Linklater et al., 2021). Furthermore, recent evidence suggested that ubiquitylation of the Xenopus ortholog of Rap2 by the XRab40c/Cul5 complex regulates Wnt signaling, although the exact consequences of this ubiquitylation on Rap2 localization, activation, and function remain unclear (Lee et al., 2007). All of these studies combined raise an interesting possibility that Rab40b/Cul5-dependent ubiquitylation may regulate Rap2 localization and activity during mammalian cell migration.
Interestingly, we found that in cells lacking Rab40b, Rap2 is decreased at the plasma membrane and instead accumulates intracellularly, primarily in lysosomes. Consistent with this observation, the levels of ectopically expressed Rap2 decrease with passage number, suggesting that Rap2 undergoes increased lysosomal degradation in Rab40b KO cells (Fig. S2 B). Surprisingly, endogenous levels of Rap2 appear unchanged in Rab40b KO cells. One plausible explanation is that our Rab40b KO cell lines were selected as individual single-cell clones. Therefore, there was ample time for these cells to compensate for loss of Rab40b and perhaps upregulate certain pathways to increase steady-state Rap2 levels. Nonetheless, we can partially rescue the Rap2 lysosomal localization defect with the addition of Rab40b-WT but not with Rab40b-ΔSOCS, a mutant unable to bind Cul5, suggesting that ubiquitylation by the Rab40b/Cul5 complex is necessary for Rap2 recycling to the plasma membrane.
Consistent with Rap2 being a substrate of the Rab40b/Cul5 complex, we discover that Rab40b can induce Rap2 ubiquitylation. We observed little stimulation of Rap2-poly-ubiquitin smear by Rab40b, leading us to believe that the primary Rab40b-driven signal is the addition of smaller ubiquitin species (two to five). Consistent with this, we found no evidence that Rab40b/Cul5-mediated ubiquitylation of Rap2 signals for proteasomal degradation, although additional studies will be needed to fully define the Rab40b-dependent ubiquitin marks on Rap2. Importantly, these findings contrast with a recent study that suggested that Rab40b strictly mediates poly-ubiquitylation of Rap2 in Xenopus embryos (Lee et al., 2007). Whether these differences in Rap2 ubiquitylation are the result of different experimental systems (mammalian versus Xenopus cells) or different Rap2 functions (cell migration versus Wnt signaling) remains to be determined. Finally, since we have no indication that Rab40b/Cul5 stimulates the first mono-ubiquitin mark on Rap2, it is possible that Rab40b/Cul5 works with other E3s, where Rab40b/Cul5 triggers additional ubiquitylation of Rap2 after it is originally primed with ubiquitin by another E3 ligase.
Ubiquitylation at critical Rap2 lysines promotes its plasma membrane targeting
To begin linking the localization defects of Rap2 in Rab40b mutant cells and our evidence suggesting that Rap2 is a substrate of Rab40b/Cul5, we turned to mutation of Rap2 lysines to understand how ubiquitylation regulates Rap2 function. We generated a Rap2a construct with five putative lysines mutated to arginines (K5R, K94R, K117R, K148R, and K150R), since these have been implicated in the regulation of Ras and Rap GTPases (Fig. S4 D). Interestingly, K94 appears to be specific to the Rap2 family and may underlie a unique mechanism to regulate Rap2, whereas K150 is conserved within the Rap1 and Rap2 families (Fig. S4 D). We found that mutation of K5R affects Rap2 subcellular localization in a similar manner to loss of Rab40b. Notably, we observed that Rap2a-K5R is primarily localized within lysosomes. The increased Rap2 in lysosomes suggests that ubiquitylation of Rap2 is required for its recycling to the lamellipodia plasma membrane.
To begin teasing apart the Rap2a-K5R mutant, we separated the mutations into two clusters: K2R (K5 and K94) and K3R (K117, K148, and K150). We found that the K3R mutant phenocopies the localization and Rab40b-binding effect of K5R. Delving further, we split the K3R mutant into single and double mutations. Interestingly, none of the single/double mutants seem to repeat the Rab40b-binding effect we observed with K5R/K3R; thus, we ultimately propose that all three lysines within K3R may be ubiquitylated and regulated by Rab40b/Cul5. However, there are clear caveats with using the binding as a readout for which sites are ubiquitylated, and we cannot fully rule out the possibility that these mutants affect Rap2 and Rab40b binding at a structural level and that a more complex mechanism is at play. Future work will be needed to tease apart exactly how each of these three Rap2 lysines is regulated by Rab40b/Cul5 specifically.
The versatile and complex role of ubiquitylation during protein trafficking
Summarizing so far, we propose that Rab40b/Cul5-mediated ubiquitylation of Rap2 promotes its recycling from early endosomes to the lamellipodia plasma membrane. This is supported by our data demonstrating that mutation of putative Rap2 ubiquitylation sites results in a loss of Rap2 enrichment at the plasma membrane and a concurrent increase in lysosomes. This is quite surprising, as it challenges the current dogma that ubiquitylation of plasma membrane–bound proteins leads to lysosomal degradation. Canonically, ubiquitylation serves as an internalization signal, at which plasma membrane–bound proteins are trafficked and sorted into lysosomes for degradation (Mukhopadhyay and Riezman, 2007; Piper et al., 2014). Our data suggest that ubiquitylation serves as a targeting signal to recycle Rap2 back to the plasma membrane once internalized. While there is at least one recent example of ubiquitin functioning as a recycling signal rather than degradation, this phenomenon remains elusive and poorly studied (Xu et al., 2017). Many future questions remain: Is Rap2 internalized and recycled by two different types of ubiquitin signals? Does Rab40b/Cul5 tag Rap2 right before internalization? What are the readers responsible for recognizing this recycling ubiquitin mark? Is ubiquitin removed by deubiquitylating enzymes (DUBs) before it returns to the plasma membrane to start the cycle again? Although future work will be needed to answer these intriguing questions, our study defines a new pathway that regulates localization and function of Rap2, and possibly Rap1, small GTPases during cell migration.
Differential regulation between Rap and Ras family members
Although the Rap subfamily shares some similarities with the Ras family, our data draw attention to the differences and highlight the need for studying this differential regulation further. First, it is particularly important to compare Rap2 localization with Ras isoform localization: HRas and NRas are localized at both the plasma membrane and the Golgi, whereas KRas predominantly sits at the plasma membrane (Hancock, 2003). The enrichment and active trafficking through the endolysosomal compartment are a unique feature of the Rap family, suggesting distinct regulation and function compared with Ras proteins. Further, our data propose that Rap2 interacts with its effectors predominantly at the plasma membrane, whereas Ras has been shown to signal from the Golgi and the ER (Chiu et al., 2002). Second, Rap2 seems to be regulated by ubiquitin differently than the Ras family. For instance, ubiquitin attached to HRas stabilizes its interaction with endosomes (Jura et al., 2006), whereas we observed that Rap2 ubiquitylation positively controls its membrane targeting and signaling. In both cases, it is apparent that cells need continuous trafficking of Rap and Ras for proper signaling, but the mechanism by which ubiquitylation governs these pathways appears unique.
Despite the differences, previous work in the Ras ubiquitylation field allows us to think more critically about our Rap2 findings and future directions (Baker et al., 2012, 2013; Yin et al., 2020; Thurman et al., 2020). Specifically, mono-ubiquitylation of KRas-K147 impedes GAP-mediated GTP hydrolysis and promotes effector binding (Sasaki et al., 2011). Additionally, mono-ubiquitylation of HRas-K117 enhances the intrinsic rate of nucleotide exchange, promoting activation (Baker et al., 2012, 2013). This site-specific ubiquitylation of Ras isoforms appears to differentially regulate downstream signaling. In the case of Rap2, we would argue that three lysines are putative sites for Rab40b/Cul5-mediated ubiquitylation: K117, K148, and K150. Future work will be aimed at uncovering the functions of each site and how those data compare to previous Ras findings. Of note, both K148 and K150 are located within the HVR, a common region for PTMs, and K150 is specific to the Rap family. Ubiquitylation of this residue could be a novel mechanism to differentially regulate the Rap family proteins.
The complicated link between Rap2 localization and activation
The similarity in localization between our various Rap2 lysine mutants and the dominant-negative mutant is noteworthy and suggests that ubiquitylation may also be critical for Rap2 GTPase function. As mentioned above, there is already precedence for this, as mutation of Ras isoforms at some of these lysines has been shown to affect GTP hydrolysis and GAP binding (Baker et al., 2012, 2013; Dohlman and Campbell, 2019). Notably, we detect less active Rap2 overall in Rab40b KO cells, as well as the K5R mutant. These data argue that ubiquitylation by the Rab40b/Cul5 complex is critical not only for Rap2 recycling to the leading edge, but also for Rap2 activation. However, it remains to be determined whether this occurs via direct regulation of GTP hydrolysis/GAP binding or simply a consequence of Rap2 not being properly targeted to the plasma membrane, where Rap2 GEFs may reside (Gloerich et al., 2012). Being able to tease apart the complex link between plasma membrane targeting and activation will be the focus of future studies. We also want to note that in future studies, it may be beneficial to use an effector more specific to Rap2, especially since RalGDS appears to also bind other Ras and Rap proteins (Spaargaren and Bischoff, 1994; Herrmann et al., 1996). Moreover, unlike binding assays where spatiotemporal information is lost, biosensors may be better suited for measuring Rap activation differences.
The Rab40b/Cul5 complex as a pro-migratory machine
Taken together, our data suggest that the Rab40b/Cul5 complex ubiquitylates Rap2 to regulate its plasma membrane targeting and activity, ultimately promoting breast cancer cell migration (Fig. 9 G). Interestingly, the downstream signaling effect of Rap2 ubiquitylation seems to be dependent on the specific E3 ligase. For instance, Nedd4-1–mediated ubiquitylation of Rap2a inhibits its function and activation, affecting kinase activity and promoting dendrite growth (Kawabe et al., 2010). Similarly, in glioma cells, ubiquitylation of Rap2a by Nedd4-1 inhibits GTP activity, promoting migration and invasion (Wang et al., 2017). Uncovering the differences between these ubiquitylation events is critical to understanding how Rap2 is fine-tuned during cell migration. With Rab40b/Cul5, we establish its role as a pro-migratory molecular machine, where the complex ubiquitylates and regulates a multitude of proteins, many of which directly mediate actin dynamics. Consistent with this model, we have already shown that Rab40b/Cul5 ubiquitylates and regulates the subcellular distribution of another actin regulator, EPLIN (Linklater et al., 2021). Curiously, Rab40b/Cul5 appears to drive poly-ubiquitylation and degradation of EPLIN, whereas here we found it stimulates recycling of Rap2 via a presumed non–poly-ubiquitin mark. How the same Rab40b/Cul5 complex can mediate such different processes, and to what extent these pathways overlap, are exciting future avenues to investigate. Given that ubiquitylation is a fast-acting posttranslational modification pathway, as opposed to transcriptional regulation, this may be an innovative way for cells to quickly change actin and adhesion dynamics at the leading edge. We are now poised to interrogate a number of potential substrates and understand how ubiquitylation may be important for their function, specifically during cell migration.
Materials and methods
Cell culture and generation of lentiviral stable cell lines
MDA-MB-231 cells were cultured in DMEM with 4.5 g/liter glucose, 5.84 g/liter L-glutamine, 1% sodium pyruvate, 1% nonessential amino acids, 1 µg/ml insulin, 1% penicillin/streptomycin, and 10% FBS. HEK293T cells were cultured in DMEM with 4.5 g/liter glucose, 5.84 g/liter L-glutamine, 1% penicillin/streptomycin, and 10% FBS. All GFP-Eos- and FLAG-MDA-MB-231 stable cells lines used in this study were generated using lentivirus transfection (puro). Once lentivirus was collected from HEK293T cells (see Transfections), virus was transferred to MDA-MB-231 target cells and allowed to incubate for 24 h before replacing with fresh MDA-MB-231 medium. Stable population cell lines were then selected using 1 µg/ml of puromycin. Cell lines were routinely tested for mycoplasma. Additionally, all cell lines were authenticated in accordance with ATCC standards.
Generating MDA-MB-231 CRISPR/Cas9 KO cell lines
MDA-MB-231 cells stably expressing Tet-inducible Cas9 (Horizon Discovery Edit-R lentiviral Cas9) were grown in a 12-well dish to ∼75% confluency. Cells were then treated with 1 µg/ml doxycycline (dox) for 24 h to induce Cas9 expression. After 24 h, cells were cotransfected with crRNA:tracrRNA mix and DharmaFECT Duo transfection reagent as described by the Horizon Discovery DharmaFECT Duo protocol available online. Transfected cells were incubated for 24 h, trypsinized, and plated for single clones. Clones were screened through genotyping PCR and Sanger sequencing for Rab40b/Rab40 KOs and Rap2 KOs. For Rap2 KOs, original screening was done via Western blot using the commercially available pan-Rap antibody. gRNAs were purchased from Horizon Discovery (Edit-R Predesigned synthetic crRNA), as well as synthetic tracrRNA (U-002005-xx). All guide RNAs and genotyping primers are listed in Table 2. Rab40 KO clones (used in Fig. S3 E) were generated and genotyped as described previously (Linklater et al., 2021).
Table 2.
Primer sequences
| Primer | Sequence (5′ to 3′) |
| pLVX-Rap2a forward (used to make GFP WT, G12V, S17N, Eos WT) | AAGGCGAATTCCGCGAGTACAAAGTGGTGGTGCTG |
| pLVX-Rap2a reverse (used to make GFP WT, G12V, S17N, Eos WT) | AAGGCTCTAGACTATTGTATGTTACATGCAGAACA |
| pLVX-mEos3.2 forward | AAGGCCTCGAGATGAGTGCGATTAAGCCAGAC |
| pLVX-mEos3.2 reverse | AAGGCGAATTCTCGTCTGGCATTGTCAGGCAA |
| Rap2a-S17N forward (mutagenesis) | GGTAGGCAAAAACGCCCTGACCGTGCAGTTC |
| Rap2a-S17N reverse (mutagenesis) | GAACTGCACGGTCAGGGCGTTTTTGCCTACC |
| pLVX-mCherry-Rap2a forward | GGATCCCGCGAGTACAAAGTGGTGGTGCTG |
| pLVX-mCherry-Rap2a reverse | TCTAGACTATTGTATGTTACATGCAGAACA |
| pLVX-Eos-Rap2b forward | AAGGCGAATTCAGAGAGTACAAAGTGGTGGTACTG |
| pLVX-Eos-Rap2b reverse | TCTAGATCACAGGATCACGCAGGCCGA |
| pLVX-Eos-Rap2c forward | AAGGCGAATTCAGGGAATACAAGGTAGTGGTGTTA |
| pLVX-Eos-Rap2c reverse | TCTAGATTACTGGACAACACAAGTTGTACA |
| pGEX-KG-Rap2a forward | CCGGAATTCTACGCGAGTACAAAGTGGTGGTGCTGGGC |
| pGEX-KG-Rap2a reverse | CCGCTCGAGCTATTGTATGTTACATGCAGAACAGCATGGGTC |
| pLVX-GFP-Rab40b forward | AAGGCGAATTCATGGTGAGCAAGGGCGAGGAGC |
| pLVX-GFP-Rab40b reverse | AAGGCGGATCCTTAAGAAATTTTGCAGCTGTTTCTGGTGC |
| pGPS-FLAG-Rab40b forward (4A and ΔSOCS) | GTTCCAGATTACGCGGTCGACATGAGCGCCCTGGGCAGC |
| pGPS-FLAG-Rab40b reverse (4A and ΔSOCS) | GATGCATGCTCGAGCGGCCGCTTAAGAAATTTTGCAGCTG |
| pLVX-Eos-Rap2aK5R forward | AAGGCGAATTCCGCGAGTACCGCGTGGTGGTGCTG |
| pLVX-Eos-Rap2aK5R reverse | AAGGCTCTAGACTATTGTATGTTACATGCAGAACA |
| pGEX-KG-Rap2aK5R forward | AAGGCGAATTCTACGCGAGTACCGCGTGGTGGTGCTGGGC |
| pGEX-KG-Rap2aK5R reverse | CCGCTCGAGCTATTGTATGTTACATGCAGAACAGCATGGGTC |
| pLVX-Eos-Rap2aK3R forward | AAGGCGAATTCCGCGAGTACAAAGTGGTGGTGCTG |
| pLVX-Eos-Rap2aK3R reverse | AAGGCTCTAGACTATTGTATGTTACATGCAGAACA |
| pGEX-KG-Rap2aK3R forward | CCGGAATTCTACGCGAGTACAAAGTGGTGGTGCTGGGC |
| pGEX-KG-Rap2aK3R reverse | CCGCTCGAGCTATTGTATGTTACATGCAGAACAGCATGGGTC |
| pGEX-KG-Rap2aK2R forward | AAGGCGAATTCTACGCGAGTACCGCGTGGTGGTGCTGGGC |
| pGEX-KG-Rap2aK2R reverse | CCGCTCGAGCTATTGTATGTTACATGCAGAACAGCATGGGTC |
| pGEX-KG-Rap2aK117R,K148R,K150R,K148R/K150R forward (individual mutants) | CCGGAATTCTACGCGAGTACAAAGTGGTGGTGCTGGGC |
| pGEX-KG-Rap2aK117R,K148R,K150R,K148R/K150R reverse (individual mutants) | CCGCTCGAGCTATTGTATGTTACATGCAGAACAGCATGGGTC |
| pGEX-KG-RalGDS-788 forward | CCGGAATTCTAGCGCTGCCGCTCTACAACCAGCAG |
| pGEX-KG-RalGDS-885 reverse | CCGCTCGAGCTAGAACGTCCGCTTCTTGAGGACAAA |
| pEGFP-C2-Rap2a forward | AAGGCCTCGAGCCGCGAGTACAAAGTGGTGGTGCTG |
| pEGFP-C2-Rap2a reverse | AAGGCGAATTCCTATTGTATGTTACATGCAGAACA |
| crRNA-Rab40b Exon3 | TCCAGGGATACTTCAGGCCA |
| crRNA-Rab40b Exon5 | TCTGGCGGCCGAGCAAGGGT |
| Rab40b-Exon3 genotyping forward | CTGACTGGCGGGTGTGTGGTGCACTTGCTG |
| Rab40b-Exon3 genotyping reverse | CCTCCAATCATCCATTAAAGGATGGTAAGTTAAGC |
| Rab40b-Exon5 genotyping forward | GCTAGCATGCCCCCGGAGTCCCCAAGAT |
| Rab40b-Exon5 genotyping reverse | AGGCAGACACTTATCCAGGGGGCCGCTG |
| crRNA-Rap2a-Exon1 | GATGCGCGAGTACAAAGTGG |
| crRNA-Rap2b-Exon1 | CATGAGAGAGTACAAAGTGG |
| crRNA-Rap2c-Exon2 | GGTGAAGGTGAGACTCATGA |
| Rap2a-Exon1 genotyping forward | GCGGTGCTTGTTTATCTCCG |
| Rap2a-Exon1 genotyping reverse | ACGAGGCTGTAGACGAGGAT |
| Rap2b-Exon1 genotyping forward | CCTCCGTTCGGTGGTTTCCG |
| Rap2b-Exon1 genotyping reverse | GACGGCGACGAGTCCACCTC |
| Rap2c-Exon2 genotyping forward | GTTGGGCCCTCTGCTCCTCT |
| Rap2c-Exon2 genotyping reverse | GTCCAGAATTTCCAGCACGG |
CRISPR KO genotyping results
Rap2 KO1: Rap2a allele 1, *GSGGVGKSALTVQFVTGTFIEKYDPTIED FYRKEIEVDSSPSVLEILD (*mutant allele sequence diverges from WT due to frameshift); Rap2a allele 2, *GSGGVGKSALTVQFVTGTFIEKYDPTIEDFYRKEIEVDSSPSVLEILD; Rap2b allele 1, MRE*SALTVQFVTGSFIEKYDPTIEDFYRKEIEVDSSPSVLEILDTAGTEQFASMRDLYIKNGQGFILVYSLVNQQSFQDIKPMRDQIIRVKRYERVPMILVGNKVDLEGEREVSYGEGKALAEEWSCPFMETSAKNKASVDELFAEIVRQMNYAAQPNGDEGCCSACVIL; Rap2b allele 2, MRE*CWARAAWASPRSPCSS; Rap2c allele 1, *VVLGSGGVGKSALTVQFVTGTFIEKYDPTIEDFYRKEIEVDSSPSVLEILDTAGTEQFASMRDLYIKNGQGFILVYSLVNQQSFQDIKPMRDQIVRVKRYEKVPLILVGNKVDLEPEREVM SSEGRALAQEWGCPFMETSAKSKSMVDELFAEIVRQMNYSSLPEKQDQCCTTCVVQ; Rap2 KO2: Rap2a allele 1, *VVLGSGGVGKSALTVQFVTGTFIEKYDPTIEDFYRKEIEVDSSPSVLEILDT; Rap2b allele 1, MREYK*GWCWARAAWASPRSPCSS; Rap2c allele 1, *REYKVVVLGSGGVGKSALTVQFVTGTFIEKYDPTIEDFYRKEIEVDSSPSVLEILD; Rap2c allele 2, *REYKVVVLGSGGVGKSALTVQFVTGTFIEKYDPTIEDFYRKEIEVDSSPSVLEILD; Rab40b KO1: allele 1, MSALGSPVRAYDFLLKFLLVGDSDVGKGEILASLQDGAAESPYGHPAGIDYKTTTI LLDGRRVKLQLWDTS*AREDFVPYSAPTPGAHRV; allele 2, MSALGSPVRAYDFLLKFLLVGDSDVGKGEILASLQDGAAESPYGHPAGIDYKTTTILLDGRRVKLQLWDTSGQGRFCTIFRSYSRGAQGVILVYDIANRWSFDGIDRWIKEIDEHAPGVPKILVGNRLHLAFKRQVPTEQAQAYAERLGVTFFEVSPLCNFNITESFTELARIVLLRHGMDRLWRPSK*C; and Rab40b KO2: allele 1, MSALGSPVRAYDFLLKFLLVGDSDVGKGEILASLQDGAAESPYGHPAGIDYKTTTILLDGRRVKLQLWDTS*AREDFVPYSAPTPGAHRV.
Boyden chamber Matrigel invasion assay
MDA-MB-231 control and Rap2 KO cell lines were grown to ∼70% confluency before setting up the experiment. Cells were washed with PBS, trypsinized, and resuspended in 0.5% serum DMEM (serum-starved MDA-MB-231 medium). While the cell suspension was prepared, Corning Matrigel Invasion Chambers (Corning; #354481) were thawed and equilibrated with 500 μl (top and bottom) of serum-free DMEM for 1 h at 37°C. Next, 750 μl of 10% serum DMEM (full MDA-MB-231 medium) was added to the bottom of the Matrigel Invasion Chambers, and 200,000 cells (in 500 μl) were plated in the top chamber (in 0.5% serum DMEM). Cells were incubated for 20 h at 37°C followed by 4% PFA fixation for 10 min and 0.1% Crystal Violet staining for 10 min. Before fixation, sterile cotton swabs were used to gently scrape and discard noninvaded cells from the inner membrane. After Crystal Violet staining, sterile cotton swabs were again used to soak up excess dye. Matrigel inserts were then rinsed with sterile water and allowed to dry overnight, followed by brightfield imaging with a 20× air objective (Nikon Eclipse Ti2 Inverted A1 Confocal). Three biological replicates were performed, with technical duplicates in each set. For each Matrigel insert, five fields of view were captured, and cells were counted in Fiji (10 data points per condition, per biological replicate). The graph includes all of the data points, but statistical analysis was performed on the average number of cells/field for each biological replicate.
Chemotaxis assay
Chemotaxis cell migration was performed according to the General IncuCyte Chemotaxis Cell Migration Protocol available online. First, the IncuCyte ClearView 96-well plate (4582) was coated (top and bottom) with 1× collagen for 1 h at room temperature. MDA-MB-231 control and Rap2 KO cells were grown to ∼70% confluency before being washed with PBS, trypsinized, and resuspended in 0.5% serum DMEM (serum-starved MDA-MB-231 medium). Serial dilutions were made in 0.5% serum DMEM so that cell suspension concentrations were ∼17 cells/μl. Cells of interest were then seeded at 60 μl/per well (1,000 cells total) in the collagen-coated IncuCyte ClearView plate. This was based on the recommendation by Essen BioScience, having found that 1,000 cells per well for adherent cell types was a reasonable starting point. The IncuCyte plate was then placed on a level surface, where cells were allowed to settle for 30 min at room temperature. Finally, the IncuCyte insert with seeded cells was placed into a prefilled plate with 200 μl of chemoattractant per well. The IncuCyte ClearView plate was then imaged every 2 h for 48 h using an IncuCyte S3 instrument equipped with a 10× objective (CU Cancer Center Cell Technologies Shared Resource). Three biological replicates were performed, with six technical replicates in each set. The IncuCyte Chemotaxis Analysis Software Module (9600-0015) was used to extract “total phase object area normalized to initial top value” (i.e., sum area of migrated cells normalized to time 0 area). The graph shows the raw area (arbitrary units) at each time point (averaged between the six technical replicates). We selected the 8-h time point as time 0, so cells had ample time to adhere to the porous membrane before area measurements were analyzed. This reduced noise in the data and allowed us to express relative chemotactic migration as fold-change over the 8-h time point. Statistical analysis was performed on relative chemotactic migration at 48 h.
Transfections
Polyplus jetPRIME transfection reagent (114-xx) was used for all MDA-MB-231 transient transfections (aside from DharmaFECT Duo reagent, used for generating CRISPR KO cell lines). For a 10-cm dish, 7.5 µg of DNA was mixed with 500 μl of jetPRIME buffer followed by 25 μl of jetPRIME transfection reagent. For a 6-well plate, 2 µg of DNA was mixed with 200 μl of jetPRIME buffer followed by 5 μl of jetPRIME transfection reagent. The transfection mixture was incubated for 10 min at room temperature and added to cells with full-serum medium. Cells were harvested and/or analyzed 24 h after transfection. For a more detailed protocol and guidelines on scaling up or down, see protocol available on the Polyplus transfection website. Lipofectamine 2000 transfection reagent was used for all HEK293T transfections (both for lentivirus generation and for ubiquitin experiments).
Flow cytometry
For cases where Eos-Rap2a was stably expressed in a background already expressing a lentiviral puromycin-resistant construct (Figs. 6 E and 8 M), cell sorting was used to select for Eos-positive cells. For Figs. 6 E and 8 M, Rab40b KO cells were first transfected with FLAG-Rab40b WT or ΔSOCS, respectively, generating a lentiviral stable cell line (populational). Next, these FLAG-Rab40b WT/ΔSOCS cells were transfected with Eos-Rap2a WT via lentivirus and flow sorted. Because of low levels of FLAG-Rab40b WT/ΔSOCS and FLAG-antibody sensitivity issues, we were unable to costain these cells to show both FLAG and Eos; however, our Western blot shows robust Eos-Rap2a signal in both Rab40b WT and Rab40b ΔSOCS cells (Figs. S1 E and 4 C). For flow sorting, cells were washed in PBS, trypsinized, and resuspended in sort buffer (PBS containing 30 mM Hepes, pH 7.4, 1 mM EDTA, and 0.1 g BSA) after lentivirus transfection. Cells were then sorted on a GFP-positive gate using a Beckman Coulter MoFlo XDP100 (CU Cancer Center Flow Cytometry Shared Resource).
Immunofluorescence staining
MDA-MB-231 cells were seeded on collagen-coated glass coverslips and grown in full medium for ≥24 h. Coverslips used for all experiments were no. 1 thickness. Unless otherwise stated, cells were fixed with 4% PFA for 20 min at room temperature and quenched for 5 min at room temperature with quenching buffer (375 mg glycine in 50 ml PBS). Cells were blocked for 30 min at room temperature with block buffer containing PBS, 1 ml FBS, 200 mg saponin, and 0.1 mg BSA. Primary antibodies were diluted in block buffer and incubated for 1 h at room temperature. When costaining for actin, Alexa Fluor 568 phalloidin (a12380) was added with primary antibody solution. See all antibody dilutions in Table 1. Cells were washed with block buffer before adding secondary antibodies (also diluted in block buffer) for 30 min at room temperature. Cells were washed again with block buffer before mounting in Vectashield and sealing with nail polish. During the final wash, Hoechst stain was used at 1:2,000 for 5 min to visualize nuclei.
Table 1.
Antibodies and reagents
| Name | Company, product number, host | Dilution |
|---|---|---|
| FLAG M2 | Sigma F3165, mouse | 1:1,000 |
| Rap2 | BD Biosciences 610215, mouse | 1:1,000 |
| Tubulin | Li-Cor 926-42211, rabbit | 1:2,500 |
| GM130 | BD Biosciences 610822, mouse | 1:100 |
| CD63 | Gift from Andrew Peden (University of Sheffield, Sheffield, UK), mouse | 1:100 |
| EEA1 | Gift from Andrew Peden, rabbit | 1:100 |
| Syntaxin13 | Prekeris Lab (Prekeris et al., 1998), rabbit | 1:100 |
| Ubiquitin P4D1 | Santa Cruz sc-8017, mouse | 1:200 |
| GFP | Invitrogen A-11122, rabbit | 1:100 |
| GAPDH | CST 5174, rabbit | 1:1,000 |
| EGFR | CST 2232, rabbit | 1:1,000 |
| Myc 9E10 | Santa Cruz sc-40, mouse | 1:1,000 |
| HA | Santa Cruz sc-7392, mouse | 1:1,000 |
| FLAG-HRP conjugate | CST 86861, rabbit | 1:1,000 |
| AffiniPure goat anti-mouse IgG, light chain specific | Jackson ImmunoResearch 211-032-171 | 1:1,000 |
| IRDye 680RD anti-mouse secondary | Li-Cor 926-68072 | 1:2,500 |
| IRDye 800CW anti-rabbit secondary | Li-Cor 926-32213 | 1:2,500 |
| Alexa Fluor 568 phalloidin | Thermo Fisher Scientific a12380 | 1:40 |
| Alexa Fluor 488 anti-mouse secondary | Jackson ImmunoResearch 715-545-150 | 1:100 |
| Alexa Fluor 594 anti-mouse secondary | Jackson ImmunoResearch 715-585-150 | 1:100 |
| Alexa Fluor 594 anti-rabbit secondary | Jackson ImmunoResearch 711-585-152 | 1:100 |
| Hoechst 33342 | AnaSpec AS-83218 | 1:2,000 |
| jetPRIME transfection reagent | Polyplus 114-xx | N/A |
| DharmaFECT Duo transfection reagent | Horizon discovery T-2010-xx | N/A |
| Lipofectamine 2000 transfection reagent | Invitrogen 11668027 | N/A |
| tracrRNA | Horizon discovery U-002005-xx | N/A |
| Bafilomycin A1 | Selleckchem S1413 | 200 nM |
| MLN4924 | Selleckchem S7109 | 300 nM |
| Protein G Sepharose 4 Fast Flow | Cytiva 17-0618-01 | 50% |
| Bio-Rad Protein Assay Dye Reagent (Bradford method) | Bio-Rad 5000006 | 1:5 |
| Pierce Silver Stain Kit | Thermo Fisher Scientific 24612 | N/A |
| Intercept TBS Blocking Buffer | Li-Cor 927-60001 | 1:3 |
| CytoPhos Phosphate Assay | Cytoskeleton BK054 | N/A |
Microscopy
Unless otherwise noted, all images were acquired using a Nikon Eclipse Ti2 inverted A1 confocal microscope with a 63× oil objective and a z-step size of 0.5 µm. Time-lapse imaging for Videos 1, 2, and 3 was performed using the same Nikon Eclipse Ti2 inverted A1 confocal, but with a 40× oil objective equipped with a humidified chamber and temperature-controlled stage. Widefield images (noted in figure legends) were acquired with an inverted Zeiss Axiovert 200M microscope using a 63× oil objective, QE charge-coupled device camera (Sensicam), and Slidebook v. 6.0 software (Intelligent Imaging Innovations). Time-lapse imaging for Video 4, Video 5, Video 6, and Video 7 was performed on the same Zeiss Widefield, with a 63× oil objective and a temperature-controlled stage. Images were processed in Fiji or 3i software.
Detailed image analysis for specific experiments
Fig. 1, B and C: Time-lapse imaging was performed using a Nikon Eclipse Ti2 inverted A1 confocal, with a brightfield 40× oil objective equipped with a humidified chamber and temperature-controlled stage. All time-lapses were taken at 10-min intervals with 10–15 z-stacks (z-step size 0.5 µm). Max projections were used for cell tracking (Fig. 1 B shows max). 95 frames were taken, resulting in a total time-lapse of ∼16 h. Videos are 5 frames/s (fps). For tracking, cells were manually tracked using the Manual Tracking plugin in Fiji. A 0.31-µm/pixel constant was used to calculate velocity. Careful effort was made to select the geometric center of the cell when manually tracking. Three biological replicates were performed for each cell line (from three passages). In each biological replicate, ≥10 fields of view (xy positions) were imaged. From each of the 10 xy positions, 2–4 random cells were chosen for manual tracking, totaling 30 cells per biological replicate. For each n, the velocity was averaged for all 30 cells. Statistical analysis (unpaired t test) was performed on the averages of the three biological replicates.
Fig. 1 I: To analyze colocalization between GFP-Rap2a and actin, thresholded Mander’s coefficients (tM) were calculated using the Fiji Coloc 2 plugin. Costes threshold regression was used with point spread function 3.0, Costes randomizations 10. Careful effort was made to pick cells without overexposed signal. One biological replicate was performed, with five fields of view and five cells in each field (n = 25). tM1 was defined as the fraction of actin overlapping with Rap2a (mean = 0.8678), and tM2 was defined as the fraction of Rap2a overlapping with actin (mean = 0.8612).
Fig. 2, E and F; Figs. 3 C, 4 G, and 6 G; and Fig. 8, J and N: To analyze colocalization between GFP/Eos-Rap2a and endolysosomal markers, 3i SlideBook6 software was used to calculate the percentage of total Rap2a that colocalizes with the markers indicated. Briefly, images were fragmented based on fluorescence intensity, and masks were created for both the GFP/Eos-Rap2a channel and the Alexa Fluor 594 channel (endolysosomal marker). Then, the percentage of total Rap2a intensity within the Alexa Fluor 594 channel mask was calculated for individual cells. Fig. 2 E: Two biological replicates were performed, with approximately five cells imaged for each replicate. n = 10 for GFP-Rap2a/CD63, n = 12 for GFP-Rap2a/EEA1, n = 10 for Eos-Rap2a/Syntaxin13. Fig. 2 F: One biological replicate was performed, n = 7 for Ctrl cells, n = 10 for BafA1-treated cells. Fig. 3 C: One biological replicate was performed, n = 10 for mCherry-Rap2a/YFP-Rab4 WT, n = 9 for mCherry-Rap2a/YFP-Rab11a WT. Fig. 4 G: One biological replicate was performed, n = 6 for Ctrl cells, n = 8 for 4°C cells, n = 7 for Recover cells. Fig. 6 G: Two biological replicates were performed, with approximately five cells imaged for each replicate. n = 10 for Ctrl/CD63, n = 9 for Rab40b KO/CD63, n = 12 for Ctrl/EEA1, n = 10 for Rab40b KO/EEA1. Fig. 8 J: Two biological replicates were performed, with approximately five cells imaged for each replicate. n = 10 for Ctrl/CD63, n = 11 for K3R/CD63, n = 12 for Ctrl/EEA1, n = 7 for K3R/EEA1. Fig. 8 N: Three biological replicates were performed, with ∼12 cells imaged for each replicate. n = 33 for Ctrl/DMSO-treated cells, n = 38 for MLN4924-treated cells.
Fig. 4 B: Linescans were drawn both around the designated GFP-Rap2a organelle (arrow in Fig. 4 A) and at the lamellipodia plasma membrane. The intensity of GFP-Rap2a in each pixel along these lines was determined with either ImageJ or 3i Slidebook imaging software. The fluorescence intensity of GFP-Rap2a around the organelle line and at the plasma membrane were plotted as fluorescence intensity/area (µ2) across 48 s.
Fig. 4 C: Same analysis as Fig. 4 B, but more organelles quantified. Graph includes original organelle from Fig. 4, A and B, plus four more organelles from different cells. Criteria for organelle selection were as follows: (1) organelle must be seen being formed at lamellipodia plasma membrane (time 0); and (2) organelle must be visualized in the same plane throughout the entire time-lapse series. So, in sum, only organelles that were seen internalized (time 0) and followed for 48 s (end of time-lapse) in the same plane were traced/quantified. As in Fig. 4 B, linescans were draw both around the GFP-Rap2a organelles (following the criteria above) and at the lamellipodia plasma membrane. The intensity of GFP-Rap2a in each pixel along these lines was determined with either ImageJ or 3i Slidebook imaging software. Instead of plotting the fluorescence intensity/area (µ2) across 48 s for each organelle, the fluorescence intensity of GFP-Rap2a around the organelle line and at the plasma membrane were graphed as the percentage of fluorescence intensity at time 0. Fluorescence intensity/area (µ2) at 0 s (start of time-lapse), 24 s (half of time-lapse), and 48 s (end of time-lapse) were plotted for all five organelles as a percentage of time 0. Statistical analysis (one-way ANOVA) was used to compare fluorescence intensity changes between the plasma membrane and internalized organelles (i.e., organelles do decrease in fluorescence intensity over time).
Fig. 4 F: For the cold-block experiment, polarized versus nonpolarized cells were defined as follows. Polarized cells have an enrichment of Rap2 at the leading-edge plasma membrane; in other words, cells with a polarized Rap2 distribution somewhere along the lamellipodia were scored as polarized. Everything else was scored as nonpolarized. Three biological replicates were performed. Eight fields of view were taken for each n, with approximately six cells in each field. The graph shows the percentage of polarized cells for each condition. Ctrl n1 = 44 polarized/61 total, n2 = 30 polarized/42 total, n3 = 32 polarized/41 total. 4°C n1 = 4 polarized/41 total, n2 = 12 polarized/40 total, n3 = 13 polarized/47 total. Recover n1 = 30 polarized/40 total, n2 = 27 polarized/40 total, n3 = 37 polarized/44 total.
Fig. 5 E: Fluorescence intensity ratios (leading edge/cell body) were calculated for individual cells under two conditions: MDA-MB-231 cells transiently expressing mCherry-Rap2a + GFP-RBDRalGDS or MDA-MB-231 cells transiently expressing mCherry-Rap2a + Free GFP. The two conditions are separated by the dotted line in the center of the graph. Areas/cells of interest were determined and marked using mCherry-Rap2a localization, choosing cells that had a clearly defined Rap2a enrichment/polarization at one of the cell edges. Then, an area was marked at the leading edge and just behind the leading edge (cell body). Background was subtracted, and sum intensity per area was calculated. For each cell, this was done for both the red channel (mCherry-Rap2a) and the green channel (free GFP or GFP-RBDRalGDS). The ratio of fluorescence at the leading edge versus the area behind the edge was then calculated, giving enrichment at the leading edge for each cell. One biological replicate was performed; n = 7 cells for GFP-RBDRalGDS condition and n = 8 cells for free GFP condition.
Figs. 5 C; 6 E; and 8, B, H, and M: To quantify Eos-Rap2a localization changes (i.e., decreased Eos-Rap2a at the plasma membrane), intracellular/whole-cell fractions were calculated. In Fiji, the images were first split into three channels, Eos-Rap2a, actin, and DAPI. The Eos-Rap2a channel was max projected. Using the z-slices of the actin channel to find the cell outlines, whole cells were manually traced with the Fiji polygon tool. Individual whole-cell traces (including area inside) were defined as region of interest 1 (ROI-1). Next, another manual trace was made for the inside of the cell (just inside the plasma membrane marked by actin). Individual intracellular traces (including area inside) were defined as ROI-2. ROI-1 and ROI-2 were individually projected onto the Eos-Rap2a channel. Analyze > Measure was executed on the corresponding Eos-Rap2a cell, then the ROI was moved to an empty region in the same field of view and executed again (background measurement). This sequence was done for both ROI-1 and ROI-2. Integrated density (area × mean fluorescence intensity) was extracted from Analyze > Measure. To generate intracellular/whole-cell fluorescence intensity fractions, background integrated density measurements were first subtracted from ROI-1 and ROI-2. Finally, the intracellular fraction was defined as the fluorescence intensity of ROI-2 (inner cell) divided by the fluorescence intensity of ROI-1 (whole cell; Fig. S2 C). Two to three biological replicates were performed for each experiment (two to three coverslips from at least two different passages, noted in figure legends). In each biological replicate, 5 fields of view were imaged, with 3 cells analyzed from each field (total of 15 cells per n). Criteria for the three cells chosen to analyze were as follows: (1) clearly defined cell outline, (2) no overexposed signal, and (3) enough empty space in the field of view to acquire a corresponding background measurement. Each cell was treated as its own data point, and statistical analysis (unpaired t test or one-way ANOVA with post hoc test) was performed for each condition. The same data points for Eos-Rap2a-WT (in Ctrl cells) were used for Fig. 4 E and Fig. 6, C, I, and K. For each data set, a ROUT outlier test was performed (GraphPad Prism) to identify any outliers. Three outliers were removed from the Eos-Rap2a in Rab40b KO cells data set (Fig. 4 E). No other outliers were found. The same P value was calculated with and without outliers for Fig. 4 E; outliers were removed for purposes of graphing.
Video 6: We note that GFP-Rap2a and mCherry-Rab5 do not exactly overlap in the endosomal structure, which is likely the result of a delay in capturing images due to exposure time.
Antibodies and reagents
See Table 1 for a list of primary and secondary antibodies as well as specific reagents.
DNA constructs
YFP-Rap2a was a gift from Johannes L. Bos (Utrecht University, Utrecht, Netherlands). Rap2a was cloned from the YFP vector into pLVX-puro with an N-terminal GFP tag to generate pLVX-GFP-Rap2a stable cell lines. Rap2a S17N was generated via site-directed mutagenesis using primers in Table 2, and Rap2a G12V was synthesized by Integrated DNA Technologies (IDT) before cloning into pLVX-puro-GFP (N-term). For Eos-Rap2a constructs, mEos3.2-N1 was purchased from Addgene (54525) and then cloned into pLVX-puro. Rap2a was amplified from YFP-Rap2a and cloned into pLVX-mEos3.2, generating an N-terminal Eos tag. Rap2a K5R (K5R, K94R, K117R, K148R, and K150R), K3R (K117R, K148R, and K150R), K2R (K5R and K94R), and single K-R mutations (K117R, K148R, K150R, and K148R/K150R; Fig. S4, A and B) were also synthesized by IDT and subsequently cloned into pLVX-puro-Eos or pGEX-KG. For GST-Rap2a, Rap2a was cloned into pGEX-KG (N-term GST tag). GST-Rap2b and GST-Rap2c pGEX-2T plasmids were purchased from Addgene (55667 and 55668, respectively). For mCherry-Rap2a, mCherry was first cloned into pLVX-puro, then Rap2a was cloned in downstream of mCherry (N-terminal tag). FLAG-Rab40b WT and FLAG-Rab40b SOCS-4A pLVX-puro constructs were generated as previously described (Linklater et al., 2021). To generate the GFP-Rab40b WT construct, Rab40b WT was amplified from the previously used pLVX-puro-FLAG construct and cloned into pLVX-puro-GFP (N-terminal). In Fig. S3 B, Rab40b SOCS-4A and Rab40b ΔSOCS were cloned into the FLAG lentivirus vector pGPS: SOCS-4A was amplified from the previous pLVX-FLAG construct, and Rab40b ΔSOCS was synthesized by IDT and subsequently cloned into FLAG-pGPS. Rab40b ΔSOCS includes residues 1–174 followed by residues 229–278, so Rab40b is missing the SOCS box proper but still contains the C-terminal prenylation site. For RalGDS, GFP-RalGDS was purchased from Addgene (118315). From this construct, amino acid residues 788–884 as well as residue 885 (incorporated via primer) were cloned into pGEX-KG to generate GST-RalGDS (primers listed in Table 2). This 788–885 stretch includes the defined Ras-binding domain (798–885, based on UniProtKB Q12967). For some of the transient transfections (see figure legends), Rap2a was cloned into the mammalian expression vector pEGFP-C2 (N-terminal GFP tag). For ubiquitylation experiments in HEK293T cells, the following mammalian expression vectors were used: pRK5-FLAG-Rap2a, pRK5-Myc-Ub, pRK7-HA-Rab40b, and pRK7-HA-Rab40b ΔSOCS (all cloned from previous constructs). mCherry-Rab5 was purchased from Addgene (49201). YFP-Rab4 WT, YFP-Rab4 S27N, and YFP-Rab11a WT were gifts from Alexander Sorkin (University of Pittsburgh, Pittsburgh, PA). GFP-FIP5-RBD was cloned previously by the Prekeris Lab (Willenborg et al., 2011).
Cell lysis and Western blot analysis
Unless otherwise stated, cells were lysed on ice in buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF. After 30 min, lysates were clarified at 15,000 g in a prechilled microcentrifuge. Supernatants were collected and analyzed via Bradford assay (5000006; Bio-Rad Protein Assay). 50-µg lysate samples were prepared (unless otherwise stated) in 4× SDS loading dye, boiled for 5 min at 95°C, and separated via SDS-PAGE. Gels were transferred onto 0.45-µm polyvinylidene difluoride membrane (IPFL00010), followed by blocking for 30 min in Intercept Blocking Buffer diluted in TBST 1:3 (927-60001). Primary antibodies (made in diluted Intercept Blocking Buffer) were incubated overnight at 4°C. The next day, blots were washed in TBST followed by incubation with IRDye fluorescent secondary antibody (diluted Intercept Blocking Buffer) for 30 min at room temperature. Blots were washed once again with TBST before final imaging on a Li-Cor Odyssey CLx. See Table 1 for primary and secondary antibody product numbers and dilutions.
Cell fractionation
For cell fractionation (Fig. 2 G), MDA-MB-231 parental cells were grown to confluency (∼5 × 10-cm plates). Cells were then trypsinized and washed in PBS before resuspending the cell pellet in 800 μl of 10 mM Hepes, pH 7.4 (200 μl per plate). Pooled cells were lysed with a Dounce homogenizer (20 strokes) and put on ice before adding NaCl to 100 mM final, MgCl2 to 1 mM final, and PMSF at 1 mM. Cells were then spun at 1,000 g for 5 min at 4°C to sediment unbroken cells and nuclei. Supernatant was moved to an ultracentrifuge tube and spun at 100,000 g for 1 h in a fixed-angle TLS-55 ultracentrifuge (Beckman). Supernatant (cytosol fraction) was collected. The remaining pellet was resuspended in 300 μl of ice-cold resuspension buffer (10 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 1% Triton X-100, and 1 mM PMSF). The pellet was broken up with a 1-cm3 syringe needle and then incubated at 4°C for 30 min while rotating. Suspension was then spun at 100,000 g for 1 h in a fixed-angle TLS-55 ultracentrifuge (Beckman). Supernatant (membrane fraction) was collected. The remaining pellet (cytoskeleton fraction) was not collected for this particular experiment. 30-µg protein samples were made for both the cytosol and membrane fraction. Samples were separated via SDS-PAGE and Western blotted for Rap2, GAPDH (cytosol marker), and EGFR (membrane marker).
Cold-block experiment
To slow endocytosis (Fig. 4 E), cells were incubated at 4°C for 60 min. To buffer the medium at low temperature, 20 mM Hepes, pH 7.4, was added. For the Recover condition, cells were incubated at 4°C for 60 min with Hepes buffering, then placed back in the 37°C incubator for 40 min before fixation.
Protein expression and purification
GST-Rap2a was purified from BL21-Gold (DE3) competent cells (230130). Briefly, cultures were grown at 37°C to OD of ∼0.6 and induced with 0.25 mM IPTG final overnight at 16°C. Cells were lysed via French Press in buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 0.1% Tween, 1 mM PMSF, 1 mM MgCl2, and 1 mM 2-mercaptoethanol. Postcentrifugation lysates were incubated with Glutathione beads for 2 h at 4°C and washed with buffer containing 20 mM Hepes, pH 7.4, 300 mM NaCl, and 0.1% Tween. After washing, GST-Rap2a was eluted off the beads using 25 mM Glutathione made in buffer containing 20 mM Hepes, pH 7.4, and 150 mM NaCl. Finally, eluted protein was dialyzed into either PBS or 20 mM Hepes, pH 7.4, and 150 mM NaCl. Protein purity was analyzed via SDS-PAGE, and protein concentration was measured using Bradford assay (Bio-Rad protein assay). GST-Rap2a lysine mutants (K2R, K3R, K5R, and single mutants), GST-RalGDS, GST-Rap2b, and GST-Rap2c were purified using the same protocol.
GST pull-down assays
For GST-Rap2a pull-down assays, MDA-MB-231 cells stably expressing FLAG-Rab40b (WT or 4A) were lysed in buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, and 5 mM iodoacetamide (DUB inhibitor). For GMP-PNP/GDP loading, three steps were followed: (1) add 5 mM EDTA to lysate, incubate for 10 min at room temperature; (2) add 5 mM either GMP-PNP or GDP, incubate for 10 min at room temperature; and (3) add 15 mM MgCl2, incubate for 10 min at room temperature. 0.5 mg of GMP-PNP/GDP loaded lysate was mixed with 10 µg of either GST or GST-Rap2a and incubated for 1 h at room temperature while rotating end-over-end. 75 μl of glutathione beads (50% in PBS) were added and incubated for another 30 min at room temperature while rotating end-over-end. Beads were washed 5× with 1 ml of buffer containing 20 mM Hepes, pH 7.4, 300 mM NaCl, and 0.1% Triton X-100. Protein was eluted with 1× SDS sample loading dye, separated by SDS-PAGE, and analyzed via Western blot. Two thirds of elution was used for Western blot, and one third was used for Coomassie staining to check for presence of GST/GST-Rap2a.
Competition binding assay
MDA-MB-231 cells stably expressing FLAG-Rab40b WT were lysed in buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, and 5 mM iodoacetamide. Both FLAG-Rab40b WT lysates and GST-Rap2a (recombinant) were loaded with GMP-PNP. For GMP-PNP loading, three steps were followed: (1) add EDTA to 5 mM, incubate for 10 min at room temperature; (2) add GMP-PNP to 5 mM, incubate for 10 min at room temperature; and (3) add MgCl2 to 15 mM, incubate for 10 min at room temperature. Once lysates and GST-Rap2a were loaded with GMP-PNP, the following reactions were set up: GST + lysate, GST-Rap2a + lysate, GST-Rap2a + lysate + 5 µg RalGDS, GST-Rap2a + lysate + 10 µg RalGDS, and GST-Rap2a + lysate + 30 µg RalGDS. In all reactions, GST/GST-Rap2a was at 10 µg. RalGDS was untagged. Reactions were incubated for 1 h at room temperature while rotating end-over-end. 75 μl of glutathione beads (50% in PBS) were added and incubated for another 30 min at room temperature while rotating end-over-end. Beads were washed 5× with 1 ml of buffer containing 20 mM Hepes, pH 7.4, 300 mM NaCl, and 0.1% Triton X-100. Protein was eluted with 1× SDS sample loading dye, separated by SDS-PAGE, and analyzed via Western blot. One half of elution was used for Western blot (FLAG), one fourth was used for Coomassie staining to check for presence of GST/GST-Rap2a, and one fourth was used for Silver Stain to analyze RalGDS binding.
GST-RalGDS activation assay
To measure the levels of active Rap2a, we performed GST pull-down assays with GST-RalGDS (see DNA constructs for plasmid details). MDA-MB-231 cells transiently transfected with GFP- or Eos-Rap2a were lysed in buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, and 5 mM iodoacetamide. GMP-PNP/GDP loading was not performed for these assays. 0.5 mg of lysate was mixed with 10 µg of either GST or GST-RalGDS and incubated for 1 h at room temperature. Following this, 75 μl of glutathione beads (50% in PBS) were added, and the lysate/protein/bead solution was incubated for 30 min at room temperature on a nutator. Beads were washed 5× with 1 ml of buffer containing 20 mM Hepes, pH 7.4, 300 mM NaCl, and 0.1% Triton X-100. Protein was eluted with 1× SDS sample buffer, separated by SDS-PAGE, and analyzed via Western blot. Two thirds of elution was used for Western blot, and one third was used for Coomassie staining to check for presence of GST/GST-RalGDS.
GTP hydrolysis assay
CytoPhos Endpoint Phosphate Assay (Cytoskeleton; BK054) was used to measure GTP hydrolysis, specifically the inorganic phosphate (Pi) generated during the enzymatic hydrolysis of GTP. In general, the assay was performed according to the manufacturer’s protocol. GST-Rap2a-WT and -K5R were purified as described above and prepared in a final buffer of 20 mM Hepes, pH 7.4, and 150 mM NaCl (i.e., a nonphosphate buffer). 50 μl of 0.4 mg/ml protein was first loaded with GTP. For GTP loading, three steps were followed: (1) add EDTA to 5 mM, incubate for 10 min at room temperature, (2) add GTP to 1 mM, incubate for 10 min at room temperature, and (3) add MgCl2 to 15 mM, incubate for 10 min at room temperature. Then, 5 μl of GTP-loaded protein was added to 25 μl of Hepes buffer in a 96-well plate (2 µg of protein total). The 96-well plate was incubated at 37°C for 1 h. After 37°C incubation, 70 μl of CytoPhos Reagent was added to each well, resulting in a 100-μl reaction. After 10 min of CytoPhos incubation, colorimetric change at 650 nm was measured using a plate reader. Blank samples were done side by side (50 μl of buffer loaded with GTP). Absorbance from the blank samples was subtracted from Rap2a-positive samples. Three biological replicates were performed, with three technical replicates in each set. A standard curve for Pi release was generated using the phosphate standard supplied in the kit. Phosphate release (nmol) for GST-Rap2a-WT and -K5R was calculated using this standard curve.
HEK293T ubiquitin assays
Briefly, HEK293T cells (∼80% confluency) were transfected with plasmids expressing pRK5-FLAG-Rap2a with or without pRK5-Myc-Ub, pRK7-HA-Rab40b, or pRK7-HA-Rab40b ΔSOCS using Lipofectamine 2000. After 24 h, cells were harvested and lysed in 1 ml lysis buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 10% glycerol) with 1% SDS. Cell lysates were collected and boiled at 95°C for 10 min. Supernatants were then diluted with lysis buffer to reduce SDS concentration to 0.1% and incubated with 5 µg anti-FLAG M2 antibody and 60 μl 50% protein G bead slurry (Cytiva 17-0618-01) overnight. Beads were pelleted by centrifugation and washed three times with lysis buffer plus 0.5 M NaCl. Bound proteins were eluted in 50 μl of 1× SDS sample buffer. Eluates (20 μl) were resolved via SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting. To remove the background IgG heavy chain and light chain after immunoprecipitation, we used an IgG light chain–specific secondary antibody (Jackson; 211-032-171) and FLAG antibody directly conjugated with HRP (CST; 86861S). Blot images were captured using a ChemiDoc MP Imaging system (Bio-Rad).
Statistical analysis
All statistical analyses were performed using GraphPad Prism Software. Unless described otherwise, statistics were performed using an unpaired Student’s t test (two-tailed) or a one-way ANOVA with post hoc test assuming normal Gaussian distribution. In all cases, data were collected from at least from three independent experiments. For microscopy analysis, experiments were performed from at least two different cell passages.
Online supplemental material
Fig. S1 shows further characterization of Rap2a localization including G12V and S17N mutants. Fig. S2 shows the effect of Rab40b KO on Rap2b and Rap2c localization. Fig. S3 examines Rap2 protein levels in various Rab40 KO and mutant cell lines. Additionally, it demonstrates where Rap2 lysine mutants were made within the protein. Fig. S4 provides Rap2 K-R single mutant binding data as well as a sequence alignment highlighting the Rap2 lysine residues of interest in Ras and Rap1 family members. Videos 1, 2, and 3 show 2D cell migration defects in Rap2 KO cells compared with control cells. Videos 4 and 5 are GFP-Rap2a expressing cells, highlighting live dynamics at the leading edge and macropinosome-like internalization. Video 6 shows GFP-Rap2a and mCherry-Rab5 live dynamics, demonstrating that the internalized Rap2a endosome is Rab5 positive. Video 7 presents a cell coexpressing mCherry-Rap2a and GFP-RBDRalGDS, showing that they overlap primarily at the plasma membrane where Rap2a is active.
Supplementary Material
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Acknowledgments
We thank Johannes L. Bos for the gift of YFP-Rap2a.
We thank the University of Colorado Cancer Center Flow Cytometry Shared Resource and Cell Technologies Shared Resource for their assistance with flow sorting and IncuCyte analysis, respectively (P30CA046934). This work was supported by National Institutes of Health (NIH) grant T32-GM008730 to E.D. Duncan, Bolie Scholarship to E.D. Duncan, and NIH grant 1R01-GM122768 to R. Prekeris.
The authors declare no competing financial interests.
Author contributions: E.D. Duncan: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, validation, visualization, writing (original draft, review & editing). K.-J. Han: data curation, methodology (HEK293T ubiquitin experiments). M.A. Trout: data curation, resources (generation of Rap2 K-R single mutants). R. Prekeris: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, supervision, writing (original draft, review & editing).
References
- Akutsu, M., Dikic I., and Bremm A.. 2016. Ubiquitin chain diversity at a glance. J. Cell Sci. 129:875–880. 10.1242/jcs.183954 [DOI] [PubMed] [Google Scholar]
- Baker, R., Lewis S.M., Sasaki A.T., Wilkerson E.M., Locasale J.W., Cantley L.C., Kuhlman B., Dohlman H.G., and Campbell S.L.. 2012. Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 20:46–52. 10.1038/nsmb.2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R., Wilkerson E.M., Sumita K., Isom D.G., Sasaki A.T., Dohlman H.G., and Campbell S.L.. 2013. Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination. J. Biol. Chem. 288:36856–36862. 10.1074/jbc.C113.525691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona, T.G., Wiener H.H., Ahearn I.M., Silletti J., Chiu V.K., and Philips M.R.. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164:461–470. 10.1083/jcb.200311093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner, B., and Van Aelst L.. 2009. Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 21:684–693. 10.1016/j.ceb.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, J.L. 2005. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17:123–128. 10.1016/j.ceb.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Bos, J.L., de Bruyn K., Enserink J., Kuiperij B., Rangarajan S., Rehmann H., Riedl J., de Rooij J., van Mansfeld F., and Zwartkruis F.. 2003. The role of Rap1 in integrin-mediated cell adhesion. Biochem. Soc. Trans. 31:83–86. 10.1042/bst0310083 [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero, J.J., Jose J., Hodgson L., and Condeelis J.. 2012. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 24:277–283. 10.1016/j.ceb.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruurs, L.J.M., and Bos J.L.. 2014. Mechanisms of isoform specific Rap2 signaling during enterocytic brush border formation. PLoS One. 9. e106687. 10.1371/journal.pone.0106687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, V.K., Bivona T., Hach A., Sajous J.B., Silletti J., Wiener H., R.L. Johnson, II, Cox A.D., and Philips M.R.. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343–350. 10.1038/ncb783 [DOI] [PubMed] [Google Scholar]
- Choi, S.-C., and Han J.-K.. 2005. Rap2 is required for Wnt/beta-catenin signaling pathway in Xenopus early development. EMBO J. 24:985–996. 10.1038/sj.emboj.7600571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola, U., Ristoratore F., Albalat R., and D’Aniello S.. 2019. The evolutionary landscape of the Rab family in chordates. Cell Mol. Life Sci. CMLS. 76:4117–4130. 10.1007/s00018-019-03103-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart, A.E., Box G.M., Court W., Gale M.E., Brown J.P., Pinder S.E., Eccles S.A., and Wells C.M.. 2015. PAK4 promotes kinase-independent stabilization of RhoU to modulate cell adhesion. J. Cell Biol. 211:863–879. 10.1083/jcb.201501072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Vega, M., Burrows J.F., and Johnston J.A.. 2014. Ubiquitination: Added complexity in Ras and Rho family GTPase function. Small GTPases. 2:192–201. 10.4161/sgtp.2.4.16707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol, K.K., Lorenz S., and Strieter E.R.. 2019. Enzymatic logic of ubiquitin chain assembly. Front. Physiol. 10:835. 10.3389/fphys.2019.00835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, J., Huang H., Qu D., Tang J., Cao W., Lu Z., Cheng Q., Yang J., Bai J., Zhang Y., et al. 2015. Rap2B promotes proliferation, migration, and invasion of human breast cancer through calcium-related ERK1/2 signaling pathway. Sci. Rep. 5:12363. 10.1038/srep12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman, H.G., and Campbell S.L.. 2019. Regulation of large and small G proteins by ubiquitination. J. Biol. Chem. 294:18613–18623. 10.1074/jbc.REV119.011068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, E.D., Lencer E., Linklater E., and Prekeris R.. 2021. Methods to study the unique SOCS box domain of the Rab40 small GTPase subfamily. Methods Mol. Biol. 2293:163–179. 10.1007/978-1-0716-1346-7_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig, L.A. 1999. Tools of the trade: Use of dominant-inhibitory mutants of ras-family GTPases. Nat. Cell Biol. 1:E25–E27. 10.1038/10018 [DOI] [PubMed] [Google Scholar]
- Feig, L.A., and Cooper G.M.. 1988. Inhibition of NIH 3T3 cell proliferation by a mutant Ras protein with preferential affinity for GDP. Am. Soc. Microbiol. 8:3235–3243. 10.1128/mcb.8.8.3235-3243.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, B., Akkerman J.W., and Bos J.L.. 1997. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J. 16:252–259. 10.1093/emboj/16.2.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz, C.M., Jones G.E., and Ridley A.J.. 2002. Cell migration in development and disease. Dev. Cell. 2:153–158. 10.1016/s1534-5807(02)00120-x [DOI] [PubMed] [Google Scholar]
- Gibbs, J.B., Schaber M.D., Schofield T.L., Scolnick E.M., and Sigal I.S.. 1989. Xenopus oocyte germinal-vesicle breakdown induced by [Val12]Ras is inhibited by a cytosol-localized Ras mutant. Proc. Natl. Acad. Sci. USA. 86:6630–6634. 10.1073/pnas.86.17.6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimple, R.C., and Wang X.. 2019. RAS: Striking at the core of the oncogenic circuitry. Front. Oncol. 9:965. 10.3389/fonc.2019.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.F. 2003. Ras proteins: Different signals from different locations. Nat. Rev. Mol. Cell Biol. 4:373–384. 10.1038/nrm1105 [DOI] [PubMed] [Google Scholar]
- Herrmann, C., Horn G., Spaargaren M., and Wittinghofer A.. 1996. Differential interaction of the Ras family GTP-binding proteins H-Ras, Rap1A, and R-ras with the putative effector molecules raf kinase and ral-guanine nucleotide exchange factor. J. Biol. Chem. 271:6794–6800. 10.1074/jbc.271.12.6794 [DOI] [PubMed] [Google Scholar]
- Lee, R.H.K., Iioka H., Ohashi M., Iemura S.I., Natsume T., and Kinoshita N.. 2007. XRab40 and XCullin5 form a ubiquitin ligase complex essential for the noncanonical Wnt pathway. EMBO J. 26:3592–3606. 10.1038/sj.emboj.7601781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Nelson C.M., Myers C.A., and Bissell M.J.. 2007. Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res. 67:4759–4766. 10.1158/0008-5472.CAN-06-4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, A., and Prekeris R.. 2015. The regulation of MMP targeting to invadopodia during cancer metastasis. Front. Cell Dev. Biol. 3. 10.3389/fcell.2015.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, A., Jing J., Lee J., Schedin P., Gilbert S.M., Peden A.A., Junutula J.R., and Prekeris R.. 2013. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J. Cell Sci. 126:4647–4658. 10.1242/jcs.126573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, A., Linklater E., Bayless B.A., Lyons T., and Prekeris R.. 2016. The role and regulation of rab40b-tks5 complex during invadopodia formation and cancer cell invasion. J. Cell Sci. 129:4341–4353. 10.1242/jcs.193904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, T.J., Lee D.J., Merlot S., Weeks G., and Firtel R.A.. 2007. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 176:1021–1033. 10.1083/jcb.200607072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura, N., Scotto-Lavino E., Sobczyk A., and Bar-Sagi D.. 2006. Differential modification of Ras proteins by ubiquitination. Mol. Cell. 21:679–687.. [DOI] [PubMed] [Google Scholar]
- Kahana, A., and Gottschling D.E.. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell Biol. 19:6608–6620. 10.1128/MCB.19.10.6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R.C., Conaway J.W., Keiichi I., and Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for cul2-rbx1 and cul5-rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055–3065. 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe, H., Neeb A., Dimova K., S.M. Young, Jr., Takeda M., Katsurabayashi S., Mitkovski M., Malakhova O.A., Zhang D.-E., Umikawa M., et al. 2010. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron. 65:358–372. 10.1016/j.neuron.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile, B.T., Schulman B.A., Alexander W.S., Nicola N.A., Martin H.M.E., and Hilton D.J.. 2002. The SOCS box: A tale of destruction and degradation. Trends Biochem. Sci. 27:235–241. 10.1016/s0968-0004(02)02085-6 [DOI] [PubMed] [Google Scholar]
- Komander, D., and Rape M.. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kooistra, M.R.H., Dubé N., and Bos J.L.. 2007. Rap1: A key regulator in cell–cell junction formation. J. Cell Sci. 120:17–22. 10.1242/jcs.03306 [DOI] [PubMed] [Google Scholar]
- Lan, H., Tang Z., Jin H., and Sun Y.. 2016. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 6:24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, C.D., and Ridley A.J.. 2018. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 217:447–457. 10.1083/jcb.201612069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linklater, E.S., Duncan E.D., Han K.J., Kaupinis A., Valius M., Lyons T.R., and Prekeris R.. 2021. Rab40-Cullin5 complex regulates EPLIN and actin cytoskeleton dynamics during cell migration. J. Cell Biol. 220. e202008060. 10.1083/jcb.202008060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linossi, E.M., and Nicholson S.E.. 2012. The SOCS box-adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life. 64:316–323. 10.1002/iub.1011 [DOI] [PubMed] [Google Scholar]
- Liu, C., Takahashi M., Li Y., Dillon T.J., Kaech S., and Stork P.J.S.. 2010. The interaction of Epac1 and Ran promotes Rap1 activation at the nuclear envelope. Mol. Cell Biol. 30:3956–3969. 10.1128/MCB.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, N., Umikawa M., Takei K., Sakima N., Myagmar B.E., Taira K., Uezato H., Ogawa Y., and Kariya K.I.. 2004. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J. Biol. Chem. 279:15711–15714. 10.1074/jbc.C300542200 [DOI] [PubMed] [Google Scholar]
- McLeod, S.J., Li A.H.Y., Lee R.L., Burgess A.E., and Gold M.R.. 2002. The Rap GTPases regulate B cell migration toward the chemokine stromal cell-derived factor-1 (CXCL12): Potential role for Rap2 in promoting B cell migration. J. Immunol. 169:1365–1371. 10.4049/jimmunol.169.3.1365 [DOI] [PubMed] [Google Scholar]
- McLeod, S.J., Shum A.J., Lee R.L., Takei F., and Gold M.R.. 2004. The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization, and Pyk2 tyrosine phosphorylation in B lymphocytes. J. Biol. Chem. 279:12009–12019. 10.1074/jbc.M313098200 [DOI] [PubMed] [Google Scholar]
- Meng, Z., Qiu Y., Lin K.C., Kumar A., Placone J.K., Fang C., Wang K.C., Lu S., Pan M., Hong A.W., et al. 2018. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 560:655–660. 10.1038/s41586-018-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, H.D., Blanch-Mercader C., Attia R., Maurin M., Alraies Z., Sanséau D., Malbec O., Delgado M.G., Bousso P., Joanny J.F., et al. 2019. Macropinocytosis overcomes directional bias in dendritic cells due to hydraulic resistance and facilitates space exploration. Dev. Cell. 49:171–188.e5. 10.1016/j.devcel.2019.03.024 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, D., and Riezman H.. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 315:201–205. 10.1126/science.1127085 [DOI] [PubMed] [Google Scholar]
- Murphy, D.A., and Courtneidge S.A.. 2011. The “ins” and “outs” of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myagmar, B.E., Umikawa M., Asato T., Taira K., Oshiro M., Hino A., Takei K., Uezato H., and Kariya K.-i.. 2005. PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem. Biophysical Res. Commun. 329:1046–1052. 10.1016/j.bbrc.2005.02.069 [DOI] [PubMed] [Google Scholar]
- Nancy, V., Wolthuis R.M., de Tand M.F., Janoueix-Lerosey I., Bos J.L., and de Gunzburg J.. 1999. Identification and characterization of potential effector molecules of the ras-related GTPase Rap2. J. Biol. Chem. 274:8737–8745. 10.1074/jbc.274.13.8737 [DOI] [PubMed] [Google Scholar]
- Nethe, M., and Hordijk P.L.. 2010. The role of ubiquitylation and degradation in RhoGTPase signalling. J. Cell Sci. 123:4011–4018. 10.1242/jcs.078360 [DOI] [PubMed] [Google Scholar]
- Nonaka, H., Takei K., Umikawa M., Oshiro M., Kuninaka K., Bayarjargal M., Asato T., Yamashiro Y., Uechi Y., Endo S., et al. 2008. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem. Biophysical Res. Commun. 377:573–578. 10.1016/j.bbrc.2008.10.038 [DOI] [PubMed] [Google Scholar]
- Ohba, Y., Kurokawa K., and Matsuda M.. 2003. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22:859–869. 10.1093/emboj/cdg087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, F., Joo-Okumura A., Nakatsukasa K., and Kamura T.. 2016. The role of cullin 5-containing ubiquitin ligases. Cell Div. 11:1–16. 10.1186/s13008-016-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, A.A., Schonteich E., Chun J., Junutula J.R., Scheller R.H., and Prekeris R.. 2004. The RCP–rab11 complex regulates endocytic protein sorting. Mol. Biol. Cell. 15:3530–3541. 10.1091/mbc.e03-12-0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski, M.D., and Deshaies R.J.. 2005. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Pickart, C.M., and Fushman D.. 2004. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 8:610–616. 10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Piper, R.C., Dikic I., and Lukacs G.L.. 2014. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harbor Perspect. Biol. 6. a016808. 10.1101/cshperspect.a016808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon, V., Desjardins M., Bucci C., Parton R.G., and Zerial M.. 1994. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci. 107(Pt 6):1661–1670. 10.1242/jcs.107.6.1661 [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/s0092-8674(03)00120-x [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman J., Chen Y.A., and Scheller R. H.. 1998. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143:957–971. 10.1083/jcb.143.4.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebstein, P.J., Weeks G., and Spiegelman G.B.. 1993. Altered morphology of vegetative amoebae induced by increased expression of the Dictyostelium discoideum ras-related gene Rap1. Dev. Genet. 14:347–355. 10.1002/dvg.1020140504 [DOI] [PubMed] [Google Scholar]
- Reiner, D.J., and Lundquist E.A.. 2018. Small GTPases. Wormbook. 2018:1–65. 10.1895/wormbook.1.67.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: Integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sadok, A., and Marshall C.J.. 2014. Rho GTPases: Masters of cell migration. Small GTPases. 5:e29710. 10.4161/sgtp.29710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A.T., Carracedo A., Locasale J.W., Anastasiou D., Takeuchi K., Kahoud E.R., Haviv S., Asara J.M., Pandolfi P.P., and Cantley L.C.. 2011. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 4:ra13. 10.1126/scisignal.2001518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J.T., Horwitz A.R., and Schwartz M.A.. 2010. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaargaren, M., and Bischoff J.R.. 1994. Identification of the guanine nucleotide dissociation stimulator for ral as a putative effector molecule of R-ras, H-Ras, K-Ras, and Rap. Proc. Natl. Acad. Sci. USA. 91:12609–12613. 10.1073/pnas.91.26.12609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark, H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Swatek, K.N., and Komander D.. 2016. Ubiquitin modifications. Cell. Res. 26:399–422. 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira, K., Umikawa M., Takei K., Myagmar B.E., Shinzato M., Machida N., Uezato H., Nonaka S., and Kariya K.-i.. 2004. The Traf2- and nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J. Biol. Chem. 279:49488–49496. 10.1074/jbc.M406370200 [DOI] [PubMed] [Google Scholar]
- Thurman, R., Siraliev-Perez E., and Campbell S.L.. 2020. RAS ubiquitylation modulates effector interactions. Small GTPases. 11:180–185. 10.1080/21541248.2017.1371267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares, M., Webb D.J., and Horwitz A.R.. 2005. Cell migration at a glance. J. Cell Sci. 118:4917–4919. 10.1242/jcs.02662 [DOI] [PubMed] [Google Scholar]
- Gloerich, M., ten Klooster J.P., Vliem M.J., Koorman T., Zwartkruis F.J., Clevers H., and Bos J.L.. 2012. Rap2A links intestinal cell polarity to brush border formation. Nat. Cell Biol. 14:793–801. 10.1038/ncb2537 [DOI] [PubMed] [Google Scholar]
- Wang, L., Zhu B., Wang S., Wu Y., Zhan W., Xie S., Shi H., and Yu R.. 2017. Regulation of glioma migration and invasion via modification of Rap2a activity by the ubiquitin ligase nedd4-1. Oncol. Rep. 37:2565–2574. 10.3892/or.2017.5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, H., Wilson B.J., and Caswell P.T.. 2019. ScienceDirect control of adhesion and protrusion in cell migration by Rho GTPases. Curr. Opin. Cell Biol. 56:64–70. 10.1016/j.ceb.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg, K., Rossman K.L., and Der C.J.. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846. 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- Wiedenmann, J., Ivanchenko S., Oswald F., Schmitt F., Röcker C., Salih A., Spindler K.D., and Nienhaus G.U.. 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA. 101:15905–15910. 10.1073/pnas.0403668101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenborg, C., Jing J., Wu C., Matern H., Schaack J., Burden J., and Prekeris R.. 2011. Interaction between FIP5 and SNX18 regulates epithelial lumen formation. J. Cell Biol. 195:71–86. 10.1083/jcb.201011112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Hankins H.M., MacDonald C., Erlinger S.J., Frazier M.N., Diab N.S., Piper R.C., Jackson L.P., MacGurn J.A., and Graham T.R.. 2017. COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. eLife. 6. e28342. 10.7554/eLife.28342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu, A., Shimada H., Ohbayashi N., and Fukuda M.. 2015. Rab40C is a novel varp-binding protein that promotes proteasomal degradation of varp in melanocytes. Biol. Open. 4:267–275. 10.1242/bio.201411114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, G., Zhang J., Nair V., Truong V., Chaia A., Petela J., Harrison J., Gorfe A.A., and Campbell S.L.. 2020. KRAS ubiquitination at lysine 104 retains exchange factor regulation by dynamically modulating the conformation of the interface. IScience. 23:101448. 10.1016/j.isci.2020.101448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and McBride H.. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117. 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang, M., Chang H., Zhang Y., Yu J., Wu L., Ji W., Chen J., Liu B., Lu J., Liu Y., et al. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods. 9:727–729. 10.1038/nmeth.2021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.