Abstract
The endoplasmic reticulum stress response (ERSR) is activated in various neurodegenerative diseases and/or after CNS traumatic injuries. The ERSR is comprised of three major arms, PERK, IRE-1, and ATF6, with the latter two contributing to the unfolded protein response (UPR). PERK activity overlaps with the integrated stress response (ISR) kinases, PKR, HRI and GCN2 which all signal through, eIF2α, ATF4 and CHOP. All initially attempt to restore ER homeostasis, but if ER stress is unresolved, ATF4/CHOP-mediated cell death is initiated. Here, we investigate the contribution of the inositol-requiring protein-1α (IRE1α)-X-box binding protein-1 (XBP1)-mediated UPR signaling pathway to the pathogenesis of SCI. We demonstrate that deletion of Xbp1 caused an exacerbated ATF4/CHOP signaling in cultured mouse oligodendrocyte (OL) progenitor cells (mOPCs) and enhanced their sensitivity to ER stress. Similar effects were also observed with the Xbp1 pathway inhibitor toyocamycin. Furthermore, OL lineage-specific loss of Xbp1 resulted in enhanced ISR in mice that underwent moderate contusive SCI at the T9 level. Consistently, post-injury recovery of hindlimb locomotion and white matter sparing were reduced in OL Xbp1-deficient mice, that correlated with chronically relative density of OPCs and OLs at the injury epicenter. We conclude that the IRE1-XBP1-mediated UPR signaling pathway contributes to restoration of ER homeostasis in OLs and is necessary for enhanced white matter sparing and functional recovery post-SCI.
Keywords: Oligodendrocytes, ERSR, UPR, ISR, Xbp1, SCI
Introduction
Spinal cord injury (SCI) is a complex pathological condition that results in multiple functional impairments both acutely and chronically (Ahuja et al., 2017; O’Shea et al., 2017). Part of the functional loss after SCI is due to oligodendrocyte (OL) death and white matter damage. Currently, there is no clinically effective treatment available to reduce trauma-induced white matter injury after SCI. Therefore, identification of mechanisms underlying OL death and white matter damage may result in novel molecular targets for therapies to improve functional deficits after SCI.
The proteostasis network includes all the proteins necessary for protein synthesis, folding, and degradation. Integral to proteostasis are the stress response pathways that include the heat shock response (HSR), endoplasmic reticulum stress response (ERSR), integrated stress response (ISR), and the unfolded protein response (UPR) (Labbaddia and Morimoto, 2015). The ERSR, ISR and UPR comprise overlapping signaling modules that respond to a variety of stressors to initially attempt to restore cellular homeostasis and if unsuccessful, initiate cell death (Fig. 1). The ISR is associated with transient phosphorylation of eIF2α (eukaryotic initiation factor 2α) by 4 independent kinases (PERK, GCN2, PKR, HRI) which results in inhibition of general protein synthesis, but increases translation of specific stress-response mRNAs (Schroder et al., 2005; Ron et al., 2007).
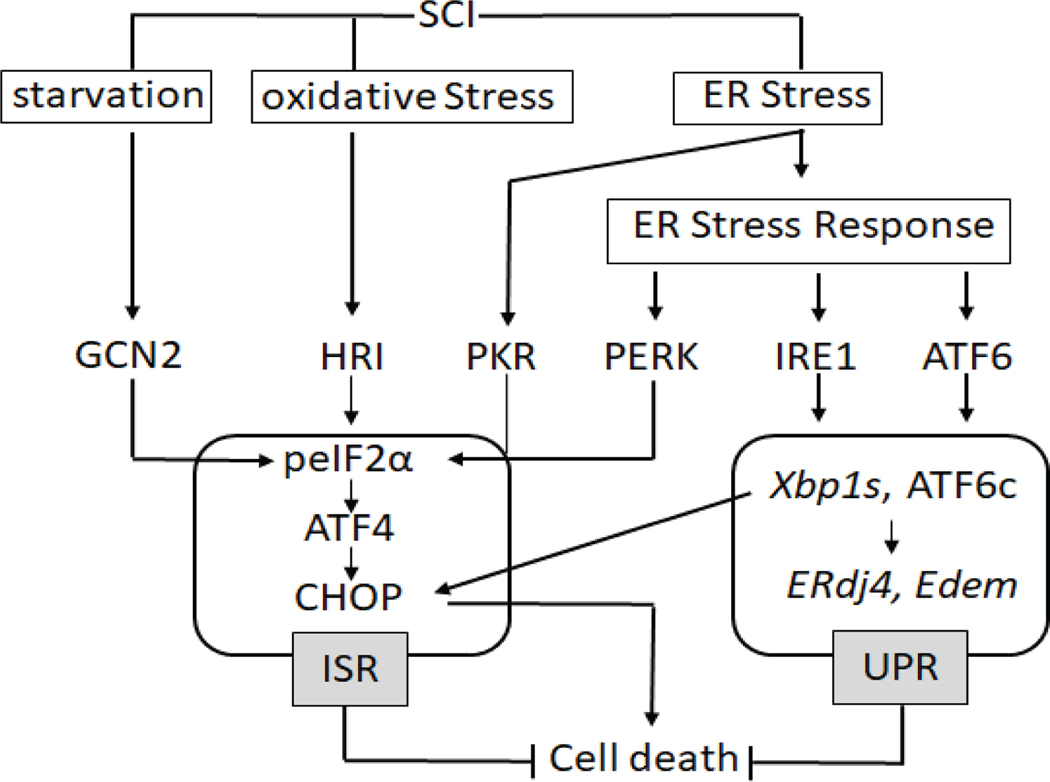
Figure 1: Schematic depicting the ER stress response signaling pathway after SCI.
Traumatic SCI initiates a cascade of cytotoxic events including starvation, oxidative stress and ER stress. A number of protective mechanisms are initiated to maintain cellular homeostasis, that include the integrated stress response (ISR) and the unfolded protein response (UPR). The ERSR initiates PERK-, IRE1-, and ATF6-dependent responses. We define the ISR as encompassing all of the four kinases that phosphorylate eIF2α (PERK, GCN2, HRI and PKR) and the UPR containing its master regulators (IRE1 and ATF6).
The ERSR is an evolutionary conserved cellular mechanism that is activated in response to insults that disrupt ER homeostasis (Schroder et al., 2008; Tabas and Ron, 2011). The primary objective of the ERSR is restoration of ER homeostasis to ensure resumption of cell functions and survival. However, the ERSR may also activate apoptosis if ER homeostasis cannot be timely restored (Ron and Habener,1992). The ERSR has three signaling arms- protein RNA (PKR)-like kinase (PERK), activating transcription factor-6 (ATF6) and oligomerization-driven activation of the ER transmembrane ribonuclease, inositol-requiring protein-1α (IRE1α).
Phosphorylation of eIF2α (peIF2α) results in a temporary block of general protein synthesis to relieve the ER from synthesizing nascent proteins. Concomitantly, translation of ERSR mediators is increased, including activating transcription factor-4 (ATF4). ATF4, together with its downstream target and partner C/EBP (CCAAT enhancer binding protein) homologous protein (CHOP), activates a gene expression program that promotes recovery of protein synthesis but may also initiate apoptosis. Deletion of Chop (Saraswat Ohri et al., 2011) or acute pharmacological enhancement of eIF2α phosphorylation (Saraswat Ohri et al., 2013) attenuated OL apoptosis, enhanced white matter sparing and improved functional recovery after SCI.
The IRE1/XBP1 (X-box binding protein 1) and ATF6 signaling pathways make up the UPR and regulate expression of various pro-homeostatic genes restoring normal function of the ER (Novoa et al., 2001). The UPR is initiated by proteolytic activation of ATF6 and oligomerization-driven activation of IRE1α (Hetz and Papa, 2018). Constitutive deletion of ATF6α modulated the ERSR and decreased OL survival in culture but did not affect locomotor recovery after SCI (Saraswat Ohri et al., 2018). However, such findings do not exclude pathogenic contributions by other UPR mediators such as ATF6-like proteins that may participate in the UPR in a tissue- and cell type-specific manner or the principal UPR effector of IRE1, the transcription factor XBP1 (Hetz and Papa, 2018).
Upon activation, IRE1α removes an unconventional 26-nucleotide intron from Xbp1 mRNA resulting in spliced Xbp1 (Xbp1s). That deletion shifts the open reading frame and enables expression of stable and active XBP1 which stimulates the transcription of numerous UPR effector genes (Yoshida et al., 2001; Hetz and Papa, 2018). Activation of both ATF6 and XBP1 is required for full activation of the transcriptional arm of the UPR (Lee et al., 2002). Xbp1s accumulates in the injured spinal cord tissue in various SCI models (Aufenberg et al., 2005; Penas et al., 2007; Valenzuela et al., 2012). Increased transcript levels of both Xbp1 and Xbp1s are also seen at the injury epicenter after both moderate and severe thoracic contusive SCI (Saraswat Ohri et al., 2011; 2012). Interestingly, worsened or improved hindlimb locomotor recovery was reported after a mouse thoracic hemisection SCI with pan-neural cell deletion of Xbp1 or neuronal/OL overexpression of Xbp1s, respectively (Valenzuela et al., 2012). Here, we address the SCI-induced pathogenic significance of XBP1 specifically in OLs. We inhibit the IRE1-XBP1 pathway in vitro using cultured oligodendrocyte precursor cells (OPCs) and in vivo with OPC/OL-specific deletion of Xbp1 in mice that subsequently underwent moderate contusive SCI.
Materials and Methods
Animals
Animal procedures were performed according to the Public Health Service Policy on Humane Care and Use of Laboratory Animals, Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, 1996), with the approval of the University of Louisville Institutional Animal Care and Use Committee and the Institutional Biosafety Committee. Wild type C57Bl/6 female mice (6–8 weeks) were obtained from Envigo (Indianapolis, IN) and Plp-CreERT2 (proteolipid protein) mice were acquired from Jackson laboratories [Plp-cre - B6.Cg-Tg (Plp1-Cre/ERT) 3Pop/J, catalogue #5975]. Xbp1fl/fl mice were a generous gift from Dr. Kirsten Sigrist at the Harvard School of Public Health. Inducible transgenic mice were generated by crossing Xbp1fl/fl and Plp-CreERT2 mice. Homozygosity was confirmed by genotyping. Tamoxifen was used to restrict the expression of the transgene, Xbp1, primarily from myelinating OLs in the CNS (Wight et al., 1993; Fuss et al., 2001). Plp mRNA is expressed earlier than myelin basic protein (Mbp) mRNA during development, beginning during embryonic life and at early developmental stages of the OL lineage (Wight et al., 1993; Mallon et al., 2002). Tamoxifen dosing: Plp-creERT2+/+:Xbp1fl/fl mice were treated with tamoxifen (1 mg/day for 8 days, i.p. injections) or its vehicle control (sunflower oil) as described (Saraswat Ohri et al., 2018). Ten days after the last tamoxifen/vehicle injection, mice were subjected to SCI at T9 level and analyzed at acute or chronic time-intervals as indicated.
T9 Spinal Cord Injury
Mice were anesthetized by an intraperitoneal (IP) injection of 0.4 mg/g body weight Avertin (2,2,2-tribromoethanol in 0.02 ml of 1.25% 2-methyl-2-butanol in saline, Sigma Aldrich, St Louis, MO). Lacri-Lube ophthalmic ointment (Allergen, Irvine, CA) was used to prevent drying of the eyes. Gentamycin (50 mg/kg; Boehringer Ingelheim, St. Joseph, MO) was administered subcutaneously to reduce infection. A laminectomy was done at the T9 vertebrae and moderate contusion injuries (50 kdyn force/400–600 μm displacement) were performed using the IH impactor (Scheff et al., 2003; Infinite Horizons Inc., Lexington, KY) as described previously (Saraswat Ohri et al., 2011). Sham animals only received T9 laminectomy and were used as experimental controls. Post-surgery, mice received subcutaneous injections of 1 cc of sterile saline and 0.1 cc of gentamycin on the day of surgery and for the next consecutive six days. 0.1 cc buprenorphine was administered subcutaneously on the day of surgery and twice/day for the next 2 days. Mice were placed on a 37°C heating pad until full recovery from anesthesia. Postoperative care included manual expression of bladders twice a day for 7–10 days or until spontaneous voiding returned.
Behavioral Assessment
Open field Basso Mouse Scale (BMS) locomotor analyses were performed prior to injury for each animal to determine the baseline scores and weekly for 6 weeks following SCI exactly as described (Basso et al., 2006; Saraswat Ohri et al., 2011). All raters were trained by Dr. Basso and colleagues at the Ohio State University and were blind to the animal treatment groups.
Spared white matter
Six weeks post-SCI, vehicle- and tamoxifen-treated Plp-creERT2+/+: Xbp1fl/fl mice were transcardially perfused with 4% paraformaldehyde (PFA) and PBS. Dissected spinal cords were submerged in 4% PFA overnight at 4°C, cryoprotected in 30% sucrose for at least 3 days at 4°C, blocked and sectioned serially in 20 μm coronal sections on a cryostat 1 cm rostral and caudal to the injury epicenter. Sections were thaw mounted on microscope slides and stored at −80°C until further use. To detect the extent of spared white matter at the injury epicenter, iron eriochrome cyanine (EC) with an alkali differentiator (Stefanovic et al., 2015) was used to stain myelin as previously described (Scheff et al., 2003; Magnuson et al., 2005). It is an established standard technique that works both for rat and mice spinal cords (Saraswat Ohri et al., 2011, 2013, 2018; Magnuson et al., 2005). Images were obtained using a Nikon Eclipse Ti inverted microscope, and white matter was traced using Nikon Elements software. A code was used to randomize the epicenter sections allowing for unbiased and blinded quantification. The epicenter of each injury was identified visually based on the section with the least amount of spared white matter. Data were normalized to average white matter content in the intact portions of the spinal cord 1 mm caudal or rostral from the injury epicenter.
Immunohistochemistry
Transverse spinal cord sections were immunostained and imaged as previously described (Saraswat Ohri et al., 2011; 2018). Primary antibodies used were APC (mouse, 1:200, Calbiochem) and Olig2 (rabbit, 1:200, Millipore). Species- and isotype-specific secondary antibodies were labeled with Alexa 488 or Alexa 647 (Life Technologies). Hoechst was used to stain the nucleus (1:5,000). Negative controls included appropriate species-specific non-immune Ig subtypes instead of primary antibodies. All images were captured with a Nikon TE 300 inverted microscope equipped with a Spot CCD camera using identical exposure settings. For all tissue assessments, images were photographed with a 10× objective and stitched with Elements™ software during acquisition.
Confocal imaging was performed using an Olympus FV1000 confocal microscope (Olympus America, Central Valley, PA) equipped with 405, 488, 543, and 635 laser lines. Representative confocal stacks (1 μm step size) were captured at 60× magnification and volume rendered using Amira software (Visualization Sciences Group, Burlington, MA).
Image Quantitation
Images were quantified as previously described with slight modifications (Lytle et al., 2007). Cells from coded photomicrographs were counted by an observer blinded to experimental conditions. A reticule of specified area (100×100 μm) was positioned in the ventral-lateral portion of spared white matter, between the tips of the ventral horns (at distal locations) or the border of the spared white matter and lesion (at the epicenter) and the outer perimeter of spinal cord tissue. The area to be counted was identified at 10× and counted at 40× magnification. This area was counted because it was free of lesion at all distances examined. Previous studies indicate that the ventral and ventrolateral areas of the spinal cord are involved in hindlimb function and sparing of these areas contributes greatly to functional recovery (Noga et al., 1991; Loy et al., 2002; Schucht et al., 2002). Labeling was assessed in ventrolateral white matter regions at the epicenter, 1 mm rostral and 1 mm caudal. Three animals were included for each group. For each location, two sections were counted per animal, on both left and right sides. Cells positive for each marker were counted from every alternate box; cells were counted if they were fully contained within the box being counted or touching the top and/or left side of the box. Following cell counts, the ratio of Olig2+/Hoechst and CC1+/Olig2+ was determined.
OPC isolation from mouse cortex
Mouse cortices were dissected from whole brains of wild type and Plp-CreERT2+/+: Xbp1fl/fl mice pups at postnatal day 5–7 (Dincman et al., 2012). Briefly, tissue was dissociated using the Neural Tissue Dissociation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) based on the manufacturer’s instructions. OPC-A media was prepared by adding 2.1 g/L NaHCO3 (Sigma Aldrich) to DMEM-F12 without HEPES (Invitrogen, Carlsbad, CA) and was supplemented with N2 (1%), B27 (2%), Penicillin/Streptomycin (1%, Invitrogen), BSA (0.01%, Sigma), 40 ng/ml FGF2 (Millipore, Billerica, MA), and 20 ng/ml PDGFα (Sigma Aldrich). OPCs were enriched with O4 hybridoma (Developmental Studies Hybridoma Bank, Iowa City, IA) using magnetic cell sorting (MACS) with rat anti-mouse IgM magnetic beads (10% in MACS Buffer). The average mOPC yield obtained was 8–10×106 cells/brain with a viability of 85–95%. 9,000–15,000 cells/cm2 were seeded on a poly-D-lysine (PDL)/laminin-coated 10 cm tissue culture dish, and incubated at 37oC, 5% CO2 for maintenance. Half media changes were performed every alternate day for 4–5 days until mOPCs reached 80–90% confluency. mOPCs were passaged with Accutase (Sigma) and seeded at a density of ∼ 3.9×104 cells/ cm2 into PDL/laminin-coated plates and treated the next day for further analysis. To obtain mature mOLs, passaged WT mOPCs were seeded in differentiation media [DMEM/F12 (No HEPES), 1% N2, 2% B27, 1% P/S, 40 ng/ml T3, 1 ng/ml CNTF, 50 μg/ml insulin] (Dincman et al., 2012; Bankston et al., 2019) and treated on the third day as described below.
MTT assay
Plp-creERT2+/+: Xbp1fl/fl-derived mOPCs were seeded in 96-well plates and allowed to settle for 4–5 hours. CreERT2-mediated deletion of Xbp1 was induced by treating the mOPCs with vehicle or tamoxifen (1μM) for 16 hours. This treatment did not affect OPC number (data not shown). mOPCs were then treated with experimental ER stress inducers, tunicamycin (Tm: 0.025 μg/ml) or thapsigargin (Tg: 2 μM) for 24 hours. Tm blocks N-linked glycosylation (Elbein 1981), while Tg is a non-competitive inhibitor of the ER Ca2+ ATPase (Rogers et al., 1995). Tm and Tg were dissolved in DMSO at concentrations based on manufacturer’s instructions (Calbiochem). The working stock was then diluted in mOPC media. DMSO alone diluted in mOPC media was used as the vehicle control. WT mOPCs and mOLs were treated with various concentrations of toyocamycin (Ty) as indicated for 24 hours. Cell survival was assayed by measuring the conversion of tetrazolium, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma Aldrich] to formazan at a wavelength of 570 nm as previously described (Hetman et al., 1999).
Lactate Dehydrogenase (LDH) release
Samples of conditioned media were collected from Plp-creERT2+/+:Xbp1fl/fl -derived mOPCs, cultured in 96 well plates and treated as detailed above. LDH release was determined using the Pierce™ LDH Cytotoxicity Assay Kit (Life Technologies, Carlsbad, CA) based on the manufacturer’s recommendations.
RNA Extraction, Reverse Transcriptase PCR
Total RNA was extracted from treated mOPCs and SCI epicenter (5 mm) of vehicle- and tamoxifen-treated Plp-creERT2+/+:Xbp1fl/fl mice using Trizol (Invitrogen) based on the manufacturer’s instructions. RNA was quantified by UV spectroscopy and its integrity was confirmed on an ethidium bromide stained formaldehyde agarose gel. cDNA was synthesized with 1 μg of total RNA using the High Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, CA) in a 20 μl reaction volume. As controls, mixtures containing all components except the reverse transcriptase enzyme were prepared and treated similarly. All cDNAs and control reactions were diluted 10× with water before using as a template for quantitative real time.
Quantitative real time polymerase chain reaction (qPCR)
qPCR was performed on an ABI 7900HT Real-time PCR instrument (Applied Biosystems). Briefly, diluted cDNAs were added to Taqman universal PCR master mix (Applied Biosystems) and run in triplicate. Target and reference gene PCR amplification was performed in separate tubes with the following Assay on Demand™ primers (Applied Biosystems): Atf4 (Mm00515324_m1), Atf6 (Mm01295325_m1), Chop (Mm01135937_g1), Edem (Mm00551797_m1), ERdj4 (Mm01622956_s1), Gadd34 (Mm00492555_m1), Gfap (Mm00546086_m1), Grp78 (Mm01333323_g1), neuron-specific enolase (Nse: Mm00468052_m1), Olig2 (Mm01210556_m1), Trib3 (Mm00454879_m1) and Xbp1 (Mm00457359_m1). RNA levels were quantified using the ΔΔCT method. Expression values obtained from triplicate runs of each cDNA sample were normalized to triplicate values for Gapdh (reference gene) from the same cDNA preparation. Transcript levels were compared to their respective levels in sham controls and expressed as fold-changes (Saraswat Ohri et al., 2018).
Xbp1 Splicing
For Xbp1s and ERdj4 amplification, qRT-PCR was performed using RT2 Real-Time SYBR Green Mix (SuperArray Bioscience Corporation) and Xbp1s (forward, ggtctgctgagtccgcagcagg; reverse, aggcttggtgtatacatgg), Erdj4 (forward; gcaatgggagtccttttgaa; reverse, cctggaagtgatgcctttgt) and 18s rRNA (forward, agtccctgccctttgtacaca; reverse, cgatccgagggcctcact) (Herrema et al., 2016) primers. The ΔΔCT method was used for quantification.
Western Blot Analyses
Protein lysates were prepared from treated mOPCs or 5 mm contused spinal cord tissue isolated from sham and injury epicenter of WT and Plp-creERT2+/+: Xbp1fl/fl mice at 24 hour post-injury in protein lysis solution (20 mM Tris, pH-6.8, 137 mM NaCl, 25 mM β-glycerophosphate, 2 mM NaPPi, 2 mM EDTA, 1 mM Na3VO4, 1% Triton X-100, 10% glycerol, protease inhibitor, 0.5 mM DTT, 1 mM PMSF). The BCA kit (Pierce, Rockford, IL) was used to quantify protein levels in the lysates. Equal concentrations of protein were separated on SDS-PAGE gels and transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH) and processed as described previously (Saraswat Ohri et al., 2018). Primary antibodies used were as follows: XBP1s (1:1000; Rbt; Cell Signaling), peIF2α (1:1000; Rbt; Thermofischer), EIF2α (1:1000; Mse; Thermofischer), ATF6c (1:500; Mse, Santa Cruz) and GAPDH (1:5000; Mse; Millipore). Precision plus protein standards (Biorad) were used as loading markers.
Statistical Analyses
A repeated measures analyses of variance (ANOVA) with fixed effects and Bonferroni post hoc t-test was performed to detect differences in BMS scores between vehicle- and tamoxifen-treated Plp-creERT2+/+: Xbp1fl/fl groups over the 6 week testing period. Statistical analysis of qPCR and Western blot data was performed using independent t-test for means with equal or unequal variances or repeated measures ANOVA (one- way or two-way analysis of variance) followed by post-hoc Tukey HSD test. For all other analyses, independent t-tests for means assuming equal variance were performed.
RESULTS
Conditional deletion of Xbp1 in OPC/OL results in enhanced cytotoxicity
We isolated mOPCs from OL-specific Plp-creERT2+/+:Xbp1fl/fl mice to study the cell-autonomous effects of Xbp1 deletion on OPC/OL survival in response to ER stress. To confirm tamoxifen-induced Xbp1 deletion, qPCR analysis from vehicle- and tamoxifen-treated mOPCs was performed. Data showed reduced, but not abolished, induction of Xbp1s and its downstream targets, ERdj4 and Edem (Lee et al., 2003) (Figure 2A), demonstrating partial removal of Xbp1s from mOPCs. Such a partial effect was likely due to limited time of tamoxifen induction that was used to avoid potential side effects of prolonged tamoxifen treatment and/or relatively low expression of Plp-creERT2 in OPCs that is expected to increase as they differentiate to OLs (Mallon et al., 2002). Note that Western blot analysis of XBP1s protein levels paralleled those mRNA reductions (Figure 3A,B).
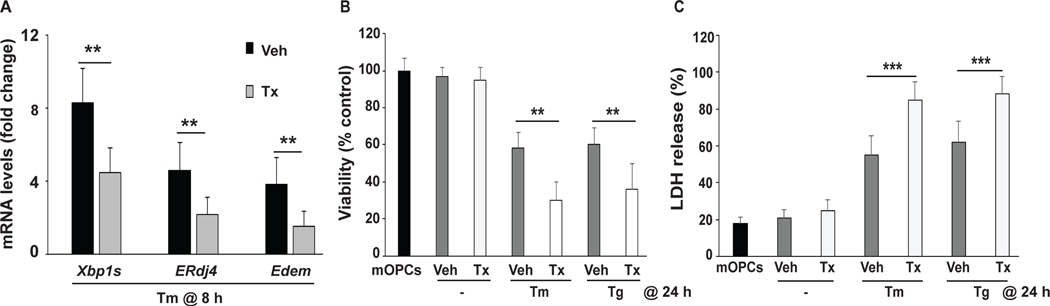
Figure 2: Deletion of Xbp1 potentiates ERSR toxicity in primary mOPCs.
Plp-creERT2+/+:Xbp1fl/fl mice-derived mOPCs were pretreated with vehicle (Veh) or tamoxifen (Tx) to delete Xbp1 and then treated with tunicamycin (Tm) or thapsigargin (Tg) to induce ER stress. A, Total RNA was isolated after 8 hours (h) of Tm and Tg treatment and analyzed for Xbp1s, ERdj4 and Edem transcript levels as indicated. mRNA levels (normalized to Gapdh) were expressed as fold changes compared to levels in the respective control cells. B,C Xbp1 deletion enhanced ER stress-mediated decreases in viable mOPC number (as evaluated by MTT assay, B) and ER stress-induced cell death (as evaluated by LDH release, C) after 24 hours of Tm and Tg treatment. Data are the mean ± SD, (n = 4, **p<0.01, ***p<0.001; Tx vs. Veh).
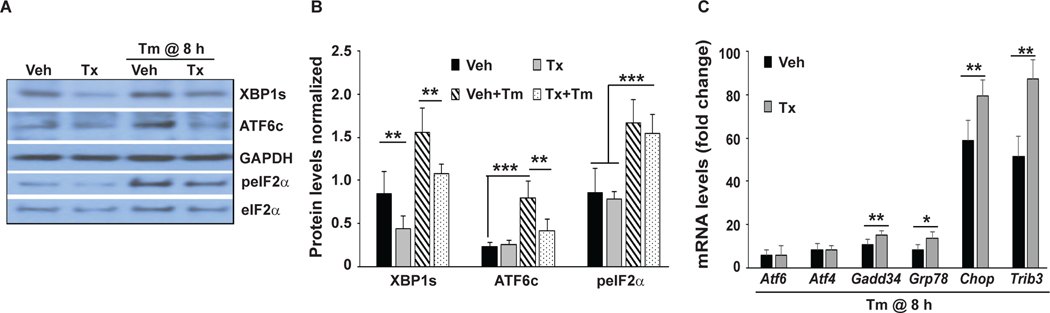
Figure 3: Deletion of Xbp1 in OPCs/OLs enhances the pro-apoptotic signaling of the ISR arm of the ERSR.
Plp-creERT2+/+:Xbp1fl/fl mice-derived mOPCs were pretreated with vehicle (Veh) or tamoxifen (Tx) to delete Xbp1 and then treated with tunicamycin (Tm) to induce ER stress for 8 hours (h). A, Representative Western blots show the activation status of the principal mediators of ERSR (XBP1s, ATF6c, peIF2α). Both under basal conditions and ER stress, moderately reduced levels of XBP1s were observed in Xbp1−/− mOPC cultures. ER stress associated activation of ATF6 cleavage (ATF6c) was reduced while S51 phosphorylation of eIF2α (peIF2α) was unaffected by partial loss of XBP1. B, Quantification of multiple Western blots as in A. C, ER stress-mediated induction of several ISR-response genes is enhanced in Xbp1−/− mOPCs. mRNA levels (normalized to Gapdh) were expressed as fold changes compared with mRNA levels in control cells (vehicle or tamoxifen), respectively. Data represent means ± SD (n = 4, *p<0.05, **p<0.01, ***p<0.001). In C, for all mRNAs quantified, Tm was significantly greater (p<0.01) than control cells.
After 24 hours of treatment, the ER stress inducers Tm or Tg reduced mOPC viability by at least 40%. Cell viability was further decreased by at least 20% in Xbp1-depleted mOPC cultures (Figure 2B). In such cells, increased LDH release further confirmed the compromised survival of Xbp1-deficient OPCs after exposure to ER stress (Figure 2C).
Targeted deletion of Xbp1 from OPC/OL results in enhanced ERSR
To elucidate the underlying mechanism of increased cell death due to reduced Xbp1 levels, we examined the activation of other arms of the ERSR in Xbp1-depleted mOPCs. While Western blot analysis confirmed a significant decrease in XBP1s levels in tamoxifen-treated mOPCs under basal conditions, there was no change in ATF6c or peIF2α levels between the two groups (Figure 3A,B). However, under Tm-induced ER stress, activated ATF6c levels were decreased in tamoxifen-treated mOPCs, suggesting that XBP1s levels may directly regulate ATF6 in a positive feedback loop (Yoshida et al., 2009). In contrast, increased peIF2α levels under Tm-induced ER stress remained unchanged in Xbp1-depleted OPCs (Figure 3A,B). Transcript levels of the ISR genes including Chop/Ddit3, Trib3, and Gadd34/Ppp1r15a as well as the common ISR/UPR-regulated gene Grp78/Hspa5 were all increased by Tm (p<0.001), and they were further elevated in ER- stressed Xbp1-depleted OPCs (tamoxifen-treated mOPCs; Figure 3C). In contrast, while transcript levels of the ERSR-regulated genes Atf6 and Atf4 were increased by Tm treatment, no further change was observed in Xbp1-depleted OPCs. Of note, Chop, Trib3 or Gadd34 have been implicated in cytotoxic responses to ER stress (Saraswat Ohri et al., 2011, 2013). Collectively, these data indicate that in ER-stressed mOPCs, partial deficiency of the UPR-specific transcription factor Xbp1s results in an exacerbated activation of the ISR arm of the ERSR and reduces survival.
Pharmacological inhibition of Xbp1 splicing is sufficient to activate the ISR and compromise viability of mOPCs
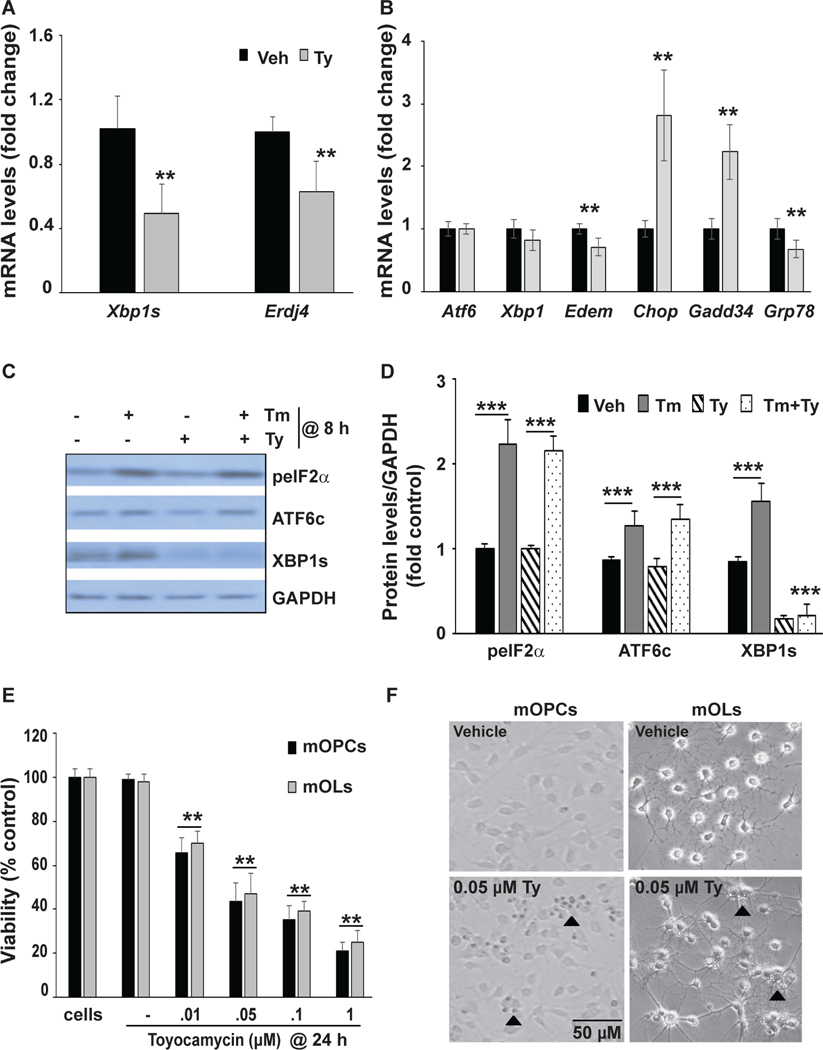
Toyocamycin (Ty) inhibits the RNAse activity of IRE1α that is required for Xbp1 splicing independent of ATF6 cleavage (Ri et al., 2012). Therefore, we used Ty to confirm that the IRE1-XBP1 pathway supports proteostasis and survival of mOPCs. Treatment with Ty for 8 hours reduced baseline transcript levels of Xbp1s and expression of its downstream targets, ERdj4 and Edem (Figure 4A,B). In addition, the pro-apoptotic ISR genes Chop and Gadd34 were upregulated, whereas the pro-homeostatic ISR/UPR gene Grp78 was downregulated (Figure 4B). Ty significantly reduced XBP1s protein levels but had no effects on ATF6c and peIF2α protein levels under basal or stress conditions (Figure 4C,D). Finally, cell survival assays showed a dose-dependent decline in both mOPC and differentiated mOL survival after 24 hours of treatment with Ty (Figure 4E,F). Collectively, these data indicate that IRE1/XBP1 signaling is critical for ER homeostasis and survival of mOPC and mOL under basal culture conditions.
Figure 4: Pharmacological inhibition of Xbp1 splicing induces the ISR and is cytotoxic in mOPCs in the absence of additional ER stress inducers.
A,B Total RNA from WT mOPCs treated with or without toyocamycin (Ty; 0.05 μM) for 8 hours is analyzed by qPCR as indicated. Transcript levels were normalized to Gapdh and expressed as fold changes compared to levels in vehicle control cells. C, Representative Western blots of protein lysates of mOPCs treated with Ty for 8 hours are analyzed for peIF2α, ATF6c and XBP1s in the absence or presence of tunicamycin (Tm). D, Quantitation of multiple, independent Western blots as in C. E, MTT assay shows a dose-dependent decrease in viability of Ty-treated mOPC (black bars) and mature mOL (grey bars) for 24 hours. F, Representative phase contrast micrographs depict declining density of OPC and OL cultures that were exposed to Ty for 24 hours. Numerous shrunk, apoptotic-like cells are present in Ty-treated cultures (arrowheads). Data are means ± SD (n = 4, **p<0.01, ***p<0.001; Ty vs. Veh, Tm+Ty vs. Tm).
Modulation of SCI-associated ERSR by OL-specific deletion of Xbp1
To determine the functional contribution of the XBP1-driven UPR, in OPCs/OLs to the pathogenesis of SCI, Plp-creERT2+/+:Xbp1fl/fl mice were treated with tamoxifen (8 × 1 mg/day, i.p. injections) or its vehicle control (sunflower oil) (Doerflinger et al., 2003; Saraswat Ohri et al., 2018). Western blot confirmed reduced levels of XBP1s in the thoracic spinal cord of tamoxifen-treated mice as determined at day 10 after completion of tamoxifen (Figure 5A,B). Note that the residual XBP1 signal likely comes from other types of spinal cord cells besides OLs. Consistent with this possibility, earlier work identified a ERSR response acutely post-SCI in cells of all 3 neural lineages (neurons, oligodendrocytes, and astrocytes) (Saraswat Ohri et al., 2011; 2013) and in endothelial cells acutely post-SCI (Fassbender et al., 2012).
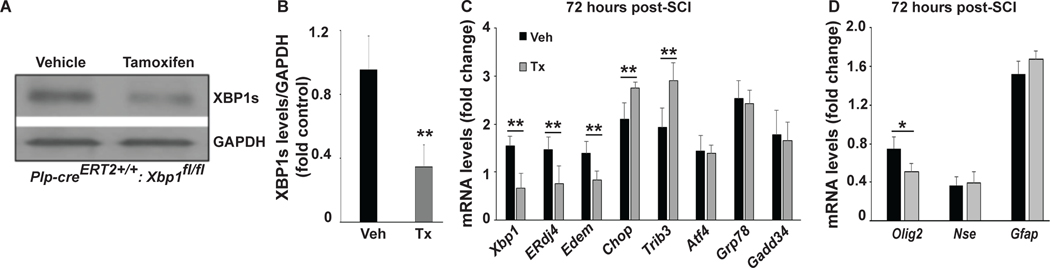
Figure 5: OL-specific deletion of XBP1 attenuates SCI-associated induction of UPR genes while moderately enhancing activation of ISR genes.
Plp-creERT2+/+:Xbp1fl/fl mice were treated with tamoxifen (Tx) or vehicle (Veh) for 8 days. After 10 days, some mice were sacrificed to collect spinal cord tissue to confirm deletion of Xbp1. A, Representative Western blot shows reduced XBP1s protein levels after conditional deletion of Xbp1. B, Quantification of multiple Western blots as in A. Other mice underwent SCI and injury epicenters were collected at 72 hours post-SCI. C, D, Effects of OL-specific deletion of Xbp1 on expression of selected ERSR transcripts (C) and neural cell-specific mRNAs (D). Transcript levels were normalized to Gapdh and expressed as fold changes compared with levels in sham controls (vehicle or tamoxifen), respectively, post-SCI. Data in B-D are the mean ± SD (n =3, *p<0.05, **p<0.01, ***p<0.001; Tx vs. Veh). In C, for all mRNAs quantified, vehicle-treated samples were significantly greater (p<0.01) than sham controls.
Ten days after completion of tamoxifen or vehicle treatment, Plp-creERT2+/+:Xbp1fl/fl mice were subjected to a T9 moderate contusive SCI. At 72 hours post-SCI in vehicle-treated mice, all mRNAs examined were significantly increased relative to respective sham controls (black bars, Figure 5C). However, in tamoxifen-treated mice, Xbp1 mRNA levels and its downstream targets including ERdj4 and Edem were significantly reduced (Figure 5C). In contrast, OL-selective deletion of Xbp1 moderately enhanced the induction of the ISR genes Chop and Trib3 (20 and 30% increase, respectively). Induction of other ERSR genes including Atf4, Grp78 or Gadd34 was unaffected by Xbp1 depletion. In addition, OL lineage-specific Olig2 mRNA levels were significantly lower in tamoxifen-treated mice, while neuronal (Nse) and astrocytic (Gfap) mRNAs were unchanged (Figure 5D). These whole animal data are in line with primary cell culture data (Figure 3C) and suggest that IRE1-XBP1-mediated UPR signaling plays a role in restoration of ER homeostasis and OPCs/OL survival following SCI.
OL-specific deletion of Xbp1 impairs recovery of hindlimb locomotion after thoracic SCI
To understand the consequences of enhanced ERSR due to Xbp1s deletion from OLs after SCI, Plp-creERT2+/+:Xbp1fl/fl mice were analyzed for hindlimb locomotor recovery using the open field BMS scale (Basso et al., 2006). As no baseline BMS difference was observed between these groups at 10 days after completion of tamoxifen treatment, the locomotor circuitry appears to function normally despite an OL lineage-specific deficit of Xbp1 (data not shown). Hence, at least a short term impairment of the IRE1-XBP1 UPR pathway in OLs does not compromise white matter function.
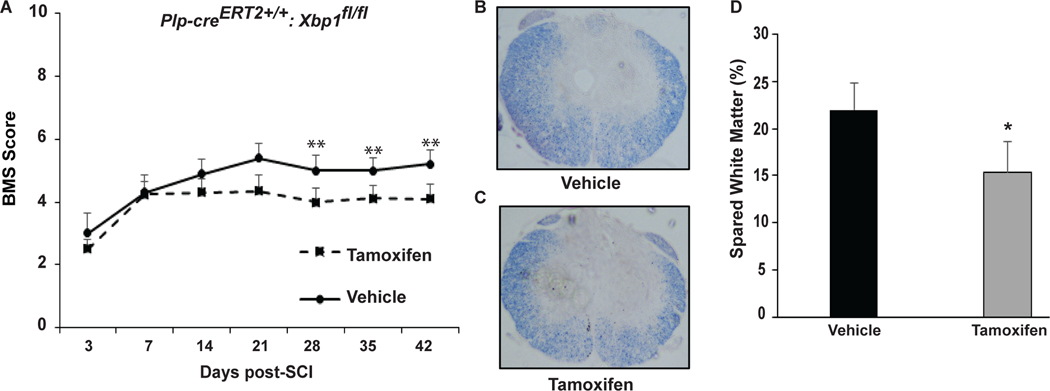
All mice underwent SCI after completion of vehicle or tamoxifen treatment and BMS was evaluated initially on day 3 and 7 post-SCI, and then weekly until week 6. Impact force and displacement of the contusive injuries were not significantly different between groups (data not shown). In the vehicle group, the average BMS score increased from 3.08 at day 3 to 5.33 at day 21 (week 3) and stayed at a similar level through day 42 (Figure 6A). Recovery of hindlimb locomotion was impaired in the tamoxifen group with significantly lower BMS scores at days 28, 35, and 42 (Figure 6A). The average BMS for the vehicle group was 5.33 with most mice recovering plantar stepping (BMS score of 5 or above). However, the average BMS of the tamoxifen group was 4.31 and only 2 out of 7 mice reached a score of 5. Therefore, OL-specific deletion of Xbp1 impaired recovery of hindlimb locomotion after SCI.
Figure 6: OL-specific deletion of Xbp1 impairs recovery of hindlimb locomotion and white matter sparing after SCI.
A, Open field BMS locomotor analyses performed weekly for vehicle- and tamoxifen- treated Plp-creERT2+/+:Xbp1fl/fl mice reveal a significant decrease in locomotor recovery in the latter group. B,C Representative transverse histological sections stained with eriochrome cyanine (EC) from the injury epicenter of Plp-creERT2+/+:Xbp1fl/fl mice treated with (B) vehicle or (C) tamoxifen to identify myelin. D, Quantitative analysis of EC-stained sections of tamoxifen-treated mice show a significant reduction (6.8% ± 3.3%) in spared white matter compared to vehicle-treated mice. Data are the mean ± SD (n = 7/group, *p<0.05).
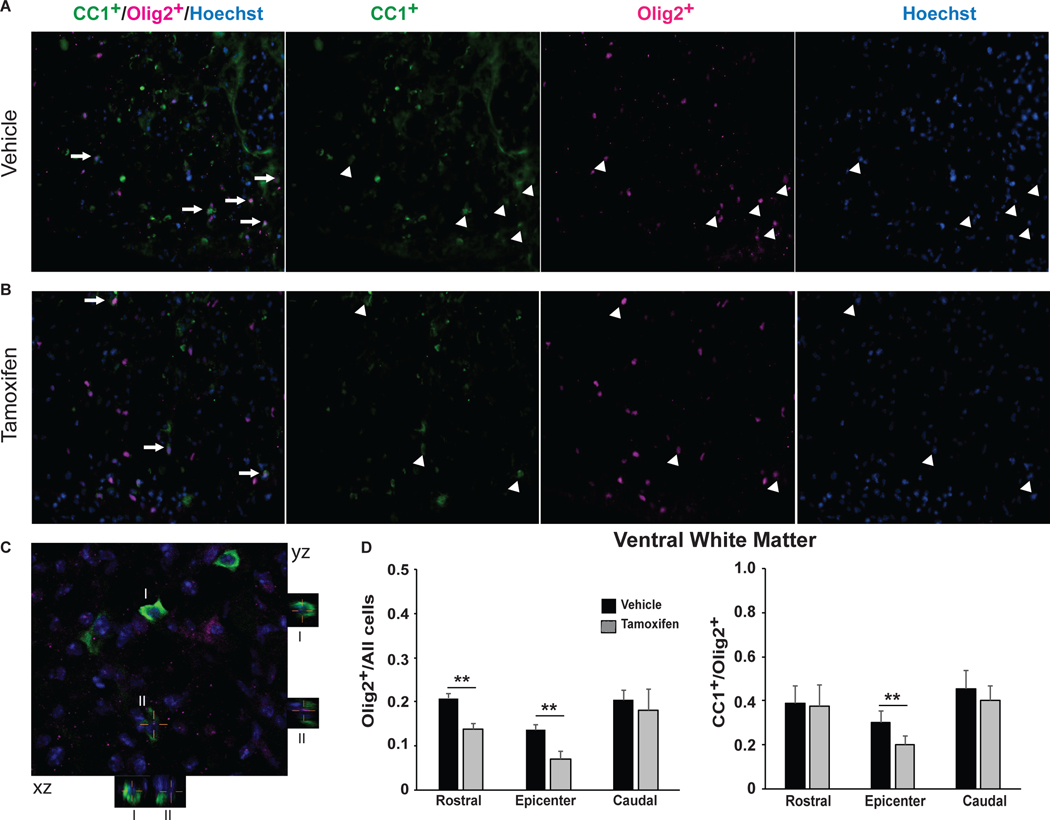
Improved functional recovery correlates with increased spared white matter and OL-specific mRNA transcript levels at 6 weeks post-SCI suggesting reduced OL loss (Saraswat Ohri et al., 2011, 2013; Myers et al, 2018). To determine whether the impaired recovery of hindlimb locomotion in the tamoxifen group is due to an increase in SCI-associated white matter loss, spinal cords were collected from all animals after completion of BMS analysis at week 6 post-SCI and analyzed for epicenter spared white matter. We focused on the white matter as in thoracic contusive SCI, white matter loss, and not neuronal loss, is the principal driver of impaired hindlimb locomotion (Basso et al., 1996; Hadi et al., 2000; Magnuson et al., 2005). Consistent with lower BMS scores, tamoxifen-treated mice had less spared white matter as compared to vehicle-treated mice (15.73% ± 3.35% vs. 22.52% ± 2.30%), respectively as normalized to average white matter content in the intact portions of the spinal cord 1 mm rostral from the injury epicenter (Figure 6B-D). To elucidate the possible underlying cellular mechanism for greater white matter loss in Xbp1-deficient mice, relative content of OL lineage cells was determined at 6 weeks post-SCI. Tamoxifen-treated mice showed a 30–50% reduction in Olig2+ cells at the injury epicenter or 1 mm rostrally respectively, indicating enhanced loss of OL lineage cells (Figure 7A,B,D). Lower ratio of CC1+/Olig2+ cells at the epicenter in tamoxifen-treated Plp-creERT2+/+:Xbp1fl/fl mice suggests greater loss of mature OLs as expected from higher expression of Plp-CreERT2 in those cells (Figure 7A,B,D). Alternatively, reduced OL replacement may account for those effects if Xbp1 is most critical for survival of newly born, differentiating OLs. Collectively, these findings indicate that the IRE1-XBP1 arm of the UPR supports survival and homeostasis of OPC/OLs after SCI and promotes recovery of locomotion by limiting white matter loss.
Figure 7: Xbp1- deficient mice show increased OL lineage cell loss at 6 weeks post-SCI.
Transverse sections (20 μm) were immunostained and imaged under identical conditions. Representative images of vehicle- (A) and tamoxifen- (B) treated Plp-creERT2+/+:Xbp1fl/fl mice show colocalization (A,B) and quantitation of Olig2+/Hoechst and CC1+/Olig2+ (D) cells. Arrows indicate individual colocalized cells. C, Representative confocal image of co-localized CC1+/Olig2+/Hoechst cells that are identified in the xz and yz planes in the merged image. Magnification bar = 15 μm (C), Data (n=3/group) are the mean ± SD. (**p<0.01).
DISCUSSION
There is substantial functional overlap between the 3 arms of the ERSR as well as considerable compensation when one arm is genetically or pharmacologically deleted or inhibited. Adamson et al (2016) demonstrated both unique and overlapping gene expression programs for the 3 signaling arms of the ERSR, with PERK predominantly unique and IRE1 and ATF6 showing some functional redundancy. However, IRE1 was the main driver of UPR responses to ER stress. Shoulders et al (2013) showed both divergent and overlapping gene expression programs initiated by Xbp1s and Atf6 mRNA overexpression. Using a gene deletion, loss of function approach, Lee et al (2003) also showed partially overlapping and redundant activities of XBP1 and ATF6. That in current studies, ATF6 could not compensate for the loss of XBP-1 signaling to prevent OPC/OL survival, either in vitro or in vivo, is consistent with these data.
Results suggest that the IRE1α-XBP1 arm of the UPR contributes to OL survival and locomotor recovery after thoracic SCI. Deletion of Xbp1 in OPCs/OLs resulted in an attenuation of the XBP1-driven UPR and an apparent increase of maladaptive ISR both in primary mOPC cultures under ER stress and injured mouse spinal cords. Such changes were followed by increased cell death under pharmacologically-induced ER stress or greater white matter loss and reduced locomotor recovery after SCI. These observations are in good agreement with previously reported effects of pan-neural cell deletion of Xbp1 in a hemisection model of SCI. In that study, nestin promoter-driven cre recombinase mediated neuronal-, astrocytic- and oligodendrocytic deletion of Xbp1 worsened locomotor recovery following T12-hemisection (Valenzuela et al., 2012). The extent of locomotor deficit appeared earlier and was more robust than that reported here. As we used an OL/OPC-specific cre driver, it is likely that these differences are due to additional pro-homeostatic contributions of the IRE1-XBP1-mediated UPR in neural cell types other than the OL lineage. While neuronal loss is expected to have limited impact on hindlimb locomotion after thoracic SCI (Hadi et al., 2000), astrocyte loss is documented in various SCI models and reactive astrocyte ablation has negative effects on tissue sparing and locomotor recovery (Faulkner et al., 2004). Hence, the UPR in astrocytes may also play a significant role in recovery from SCI.
Although the Plp-Cre driver used in our study has highest expression in mature OLs, it is also expressed at later OPC stages (Wight et al., 1993; Mallon et al., 2002). Therefore, there is a possibility that besides death of mature OLs, Plp-CreERT2-driven deletion of Xbp1 could compromise the remyelination potential after SCI by promoting death or perturbing OPC differentiation. However, as remyelination of the damaged white matter is limited and remyelination inhibition has minimal effect on SCI outcome (Tripathi and McTigue, 2007; Duncan et al., 2018), such a possibility is unlikely. Therefore, OPC/OL Xbp1−/−-mediated enhancement of early post-SCI reduction of the OPC/OL marker Olig2, and the expected cellular specificity of the KO all argue for potentiation of SCI-induced OL death as a main driver of KO effects on white matter pathology and locomotor recovery.
As the loss of function approach used here demonstrates Xbp1 contributions to OL defenses against SCI-associated ER stress, one could also consider a possibility that Xbp1 gain of function may promote OL survival and white matter protection after SCI. Consistent with this possibility, local overexpression of XBP1s in spinal cord neurons and oligodendrocytes enhanced locomotor recovery after T12-hemisection SCI (Valenzuela et al., 2012). Similarly, overexpression of XBP1s protected mouse retinal ganglion cells from axon injury-induced death or autoimmune optic neuritis that modeled multiple sclerosis (Hu et al., 2012; Huang et al., 2017). Whether similar cytoprotective effects could also be achieved in SCI-challenged glia remains to be investigated. If that is the case, the sufficiency of Xbp1 to protect neurons, as well as its demonstrated ability to promote axonal regeneration (Onate et al., 2016), would suggest that XBP1 activation could be a pleiotropic target to improve functional outcomes after other types of SCI and at lumbar and cervical levels where neuronal death is paramount to loss of function.
Consistent with consequences of partial Xbp1 deletion, our data indicate that pharmacological inhibition of the XBP1 activator IRE1 by toyocamycin has negative effects on mOPC and mature mOL survival. Such findings suggest that in cultured OPCs/OLs, Xbp1 is the main downstream effector of IRE1, as seen in transformed cell lines (Adamson et al., 2016; Shoulders et al., 2013, Lee et al., 2003). However, it remains to be seen whether that is also the case in mature OLs that respond to SCI in vivo. Interestingly, besides activating Xbp1 to promote pro-survival UPR, IRE1 also contributes to ER stress-associated cytotoxicity (Hetz and Papa, 2018) upon prolonged IRE1 activation when ER stress homeostasis cannot be restored. The cytotoxic activity of IRE1 may be mediated by IRE1-driven activation of the pro-apoptotic MAP kinase JNK and/or appearance of an IRE1 RNase activity that degrades ER associated mRNAs and switches off the UPR (Hetz and Papa, 2018). An intriguing possibility emerges that increased ER stress toxicity in XBP1-deficient cells is, in fact, mediated by IRE1 that loses its pro-survival effector while still interacting with cytotoxic effectors. Future studies are needed to determine the exact role of IRE1 in SCI pathogenesis.
Collectively, present findings demonstrate that the OL IRE1-XBP1-mediated UPR is a defense mechanism against ER stress and the resulting white matter injury after thoracic contusive SCI. When activity of that pathway is compromised, enhanced cytotoxicity is likely due to hyperactivation of the pro-apoptotic arm of the ISR. Hence, one can wonder whether the XBP1-driven UPR plays a similar role in other pathologies that are associated with ERSR-driven loss of the white matter OLs including MS or Pelizaeus-Merzbacher disease (Lin and Popko, 2009). In addition, therapeutic activation of the XBP1-mediated UPR in OLs appears as an interesting possibility to improve outcome of SCI and other ERSR-associated white matter pathologies.
Main Points.
The IRE1α-XBP1-mediated UPR signaling pathway contributes to restoration of ER homeostasis in oligodendrocytes and thereby limits white matter loss and promotes locomotor recovery post-SCI.
Acknowledgements:
This work was supported by NS045734, GM10350, Norton Healthcare, the Commonwealth of Kentucky Research Challenge for Excellence Trust Fund (SRW, MH) and the Leona M. and Harry B. Helmsley Charitable Trust (SRW, SSO, MH). We thank Christine Nunn for surgical assistance and animal care, Jason Beare and Johnny Morehouse for BMS and TreadScan® analyses, Courtney Shepard for confocal imaging and Darlene Burke for statistical analyses.
Footnotes
The authors declare no conflict of interest.
References
- Adamson B, Norman TM., Jost M, Cho MY., Nunez JK., Chen Y, Villalta JE. Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AA, Gross CA, Dixit A, Parnas O, Regev A, & Weissman JS (2016) A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell, 167, 1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja CS, Wilson JR, Nori S, Kotter Mark RN., Druschel C, Curt A. & Fehlings MG (2017). Traumatic spinal cord injury. Nature reviews, Disease Primers, 3(17018), doi: 10.1038/nrdp.2017.18 [DOI] [PubMed] [Google Scholar]
- Aufenberg C, Wenkel S, Mautes A, Paschen W. (2005) Spinal cord trauma activates processing of XBP1 mRNA indicative of endoplasmic reticulum dysfunction. J Neurotrauma 22, 1018–1024. [DOI] [PubMed] [Google Scholar]
- Bankston AN, Forston MD, Howard RM, Andres KR, Smith AE, Saraswat Ohri S, Bates ML, Bunge MB & Whittemore SR (2019) Autophagy is essential for oligodendrocyte differentiation, survival, and proper myelination. DOI: 10.1002/glia.23646. [DOI] [PubMed] [Google Scholar]
- Basso DM, Beattie MS, Bresnahan JC (1996) Graded histological and locomotor outcomes after spinal cord contusion using the NYU weight-drop device versus transection. Experimental Neurology, 139, 244–256. [DOI] [PubMed] [Google Scholar]
- Basso DM, Fisher LC, Anderson AJ, Jakeman LB, McTigue DM, & Popovich PG (2006). Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. Journal of Neurotrauma, 23, 635–65. [DOI] [PubMed] [Google Scholar]
- Dincman TA, Beare JE, Saraswat-Ohri S, & Whittemore SR (2012). Isolation of cortical mouse oligodendrocyte precursor cells. Journal of Neuroscience Methods, 30, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. (2003) Inducible site-specific recombination in myelinating cells. Genesis, 35: 601–607. [DOI] [PubMed] [Google Scholar]
- Duncan GJ, Manesh SB, Hilton BJ, Assinck P, Liu J, Moulson A, Plemel JR Tetzlaff W. (2018) Locomotor recovery following contusive spinal cord injury does not require oligodendrocyte remyelination. Nature Communications, 9(1),3066, doi: 10.1038/s41467-018-05473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD (1981) The tunicamycins – useful tools for studies on glycoproteins. Trends in Biochemical Sciences, 6, 219–221. [Google Scholar]
- Fassbender JM, Saraswat-Ohri S, Myers SA, Gruenthal MJ, Benton RL, & Whittemore SR (2012) Deletion of endoplasmic reticulum stress-induced CHOP protects microvasculature post-spinal cord injury. Current Neurovascular Research, 9, 274–281. [DOI] [PubMed] [Google Scholar]
- Faulkner JR, Hermann JE, Woo MJ, & Tansey KE (2004) Reactive astrocytes protect tissue and preserve function after spinal cord injury. Journal of Neuroscience 24, 2143–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Afshari FS, Colello RJ, & Macklin WB (2001) Normal CNS myelination in transgenic mice overexpressing MHC class I H-21(d) in oligodendrocytes. Molecular and Cellular Neuroscience, 18, 221–234. [DOI] [PubMed] [Google Scholar]
- Hadi B, Zhang YP, Burke DA, Shields CB, & Magnuson DS (2000) Lasting paraplegia caused by loss of lumbar spinal cord interneurons in rats: no direct correlation with motor neuron loss. Journal of Neurosurgery, 93, 266–275. [DOI] [PubMed] [Google Scholar]
- Herrema H, Zhou Y, Zhang D, Lee J, Hernandez MAS, Shulman GI, & Ozcan U. (2016) XBP1s is an anti-lipogenic protein. The Journal of Biological Chemistry, 291, 17394–17404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, Kanning K, Smith-Cavanaugh JE, & Xia Z. (1999). Neuroprotection by Brain-Derived neurotrophic factor is mediated by extracellular-signal-regulated kinase and phosphatidylinositol-3 kinase. The Journal of Biological Chemistry, 274, 22569–22580. [DOI] [PubMed] [Google Scholar]
- Hetz C,& Papa FR (2018) The unfolded response and cell fate control. Molecular Cell, 69, 169–181. [DOI] [PubMed] [Google Scholar]
- Hu Y, Park KK, Yang L, Wei X, Yang Q, Thielen P, Lee A, Cartoni R, Glimcher LH, Chen DF & He Z. (2012). Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron, 73(3), 445–452. Doi: 10.1016/j.neuron.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Miao L, Liang F, Liu X, Xu L, Teng X., Wang Q, Ridder WH, Shindler KS, Sun Y, & Hu Y. (2017). Neuroprotection by eIF2a-CHOP inhibition and XBP-1 activation in EAE/optic neuritiss. Cell Death and Disease, 8, e2936, doi: 10.1038/cddis.2017.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, & Morimoto RI (2015) The biology of proteostasis in aging and disease. Annual Review of Biochemistry 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, & Kaufman RJ (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes & Development, 16, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, & Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular & Cellular Biology, 23, 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, & Popko B. (2009) Endoplasmic reticulum stress in disorders of myelinating cells. Nature Neuroscience, 12, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy DN, Magnuson DS, Zhang YP, Onifer SM, Mills MD, Cao QL, Darnall JB, Fajardo LC, Burke DA, & Whittemore SR (2002) Functional redundancy of ventral spinal locomotor pathways. Journal of Neuroscience, 22, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JM, & Wrathall R. (2007) Glial cell loss, proliferation and replacement in the contused murine spinal cord. Eur J Neuroscience, 25, 1711–1724. [DOI] [PubMed] [Google Scholar]
- Magnuson DS, Lovett R, Coffee C, Gray R, Han Y, Zhang YP, & Burke DA (2005) Functional consequences of lumbar spinal cord contusion injuries in the adult rat. Journal of Neurotrauma 22, 529–543. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, & Macklin WB (2002). Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. Journal of Neuroscience, 22, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Gobejishvili L, Saraswat-Ohri S, Garrett Wilson C, Andres KR, Riegler AS, Donde H, Joshi-Barve S, Barve S. & Whittemore SR (2019) Following spinal cord injury, PDE4B drives an acute, local inflammatory response and a chronic, systemic response exacerbated by gut dysbiosis and endotoxemia. Neurobiology of Disease, 124, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noga BR, Kriellaars DJ & Jordan LM (1991) The effect of selective brainstem or spinal cord lesions on treadmill locomotion evoked by stimulation of the mesencephalic or pontomedullary locomotor regions. Journal of Neuroscience, 11, 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, & Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol, 153, 1011–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate M, Catenaccio A, Martinez G, Armentano D, Parsons G, Keri B, Hetz C, & Court FA (2016). Activation of the unfolded protein response promotes axonal regeneration after peripheral nerve injury. Scientific Reports, 6, 21709, doi: 10.1038/srep21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea TM, Burda JE, & Sofroniew MV (2017) Cell biology of spinal cord injury and repair. Journal of Clinical Investigation, 127, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penas C, Guzmán MS, Verdú E, Forés J, Navarro X, & Casas C. (2007). Spinal cord injury induces endoplasmic reticulum stress with different cell-type dependent response. Journal of Neurochemistry, 102, 1242–1255. [DOI] [PubMed] [Google Scholar]
- Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, Narita T, Masaki A, Ito A, Ding J, Kusumoto S, Ishida T, Komatsu H, Shiotsu Y, Ueda R, Iwawaki T, Imoto M, & Iida S. (2012) Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer Journal, e79, doi: 10.1038/bcj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TB, Inesi G, Wade R, & Lederer WJ (1995) Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Bioscience Reports 15, 341–349. [DOI] [PubMed] [Google Scholar]
- Ron D, & Habener JF (1992) CHOP, a novel developmentally regulated nuclear protein dimerizes with transcription factors C/EBP and LAP, functions as a dominant-negative inhibitor of gene transcription. Genes & Development, 6, 439–453. [DOI] [PubMed] [Google Scholar]
- Ron D, & Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology, 8, 519–529. [DOI] [PubMed] [Google Scholar]
- Saraswat-Ohri S, Maddie MA, Zhao Y, Qiu MS, Hetman M, & Whittemore SR (2011). Attenuating the endoplasmic reticulum stress response improves functional recovery after spinal cord injury. Glia, 59, 489–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat Ohri S, Maddie MA, Zhang Y, Shields CB, Hetman M, & Whittemore SR (2012) Deletion of the pro-apoptotic endoplasmic reticulum stress response effector CHOP does not result in improved locomotor function after severe contusive spinal cord injury. Journal of Neurotrauma, 29(3), 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat-Ohri S, Hetman M, & Whittemore SR (2013). Restoring endoplasmic reticulum homeostasis improves functional recovery after spinal cord injury. Neurobiology of Disease, 58, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat-Ohri S, Mullins A, Hetman M, & Whittemore SR (2018) Activating Transcription factor-6a modulates the endoplasmic reticulum stress response after spinal cord injury but does not affect locomotor recovery. Journal of Neurotrauma, 35, 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat Ohri S, Bankston AN., Mullins SA, Liu Y, Andres KR Beare JE, Howard RM, Burke DA Riegler AS, Hetman M, & Whittemore SR (2018). Blocking autophagy in oligodendrocytes limits functional recovery after spinal cord injury. Journal of Neuroscience, 38(26), 5900–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff SW, Rabchevsky AG, Fugaccia I, Main JA, & Lumpp JE (2003). Experimental modeling of spinal cord injury characterization of a force-defined injury device. Journal of Neurotrauma, 20, 179–193. [DOI] [PubMed] [Google Scholar]
- Schroder M, & Kaufman RJ (2005). The mammalian unfolded protein response. Annual Review Biochemistry, 74, 739–789. [DOI] [PubMed] [Google Scholar]
- Schroder M. (2008) Endoplasmic reticulum stress responses. Cellular and molecular life sciences, 65, 862–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schucht P, Raineteau O, Schwab ME & Fouad K. (2002) Anatomical correlates of locomotor recovery following dorsal and ventral lesions of the rat spinal cord. Experimental Neurology, 176, 143–153. [DOI] [PubMed] [Google Scholar]
- Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG Wu C, Yates JR Su AI, Kelly JW, Wiseman RL (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep, 3, 1279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic D, Stefanovic M & Lalosevic D (2015) Use of Eriochrome Cyanine R in routine histology and histopathology: is it time to say goodbye to hematoxylin ? Biotech. Histochem 90(6): 461–469. [DOI] [PubMed] [Google Scholar]
- Tabas I, & Ron D. (2011). Integrating mechanisms of apoptosis induced by endoplasmic reticulum stress. Nature Cell Biology, 13, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi R, & McTigue D, (2007) Prominent oligodendrocyte genesis along the border of spinal contusion lesions. Glia, 55(7): 698–711. [DOI] [PubMed] [Google Scholar]
- Valenzuela V, Collyer E, Armentano D, Parsons GB, Court FA, & Hetz C. (2012). Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death and Disease, 16, e272. doi: 10.1038/cddis.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, & Macklin WB (1993). A myelin proteolipid protein-LacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. Journal of Cell Biology, 123, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Odada T, & Mori K, (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell, 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Uemura A, & Mori K (2009). pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Structure and Function, 34, 1–10. [DOI] [PubMed] [Google Scholar]