Abstract
Background/Aim: An early evaluation concerning the effectiveness of supportive oligonucleotide therapy (SOT) in cancer as a monotherapy and in combination with other types of treatment.
Patients and Methods: This study evaluated the clinical condition and performance status (Karnofsky-Index) of 95 patients, post-SOT administration. Furthermore, circulating tumor cells (CTCs) from 47 patients’ pre- and post-SOT administration were measured and analyzed by repeated-measures ANOVA.
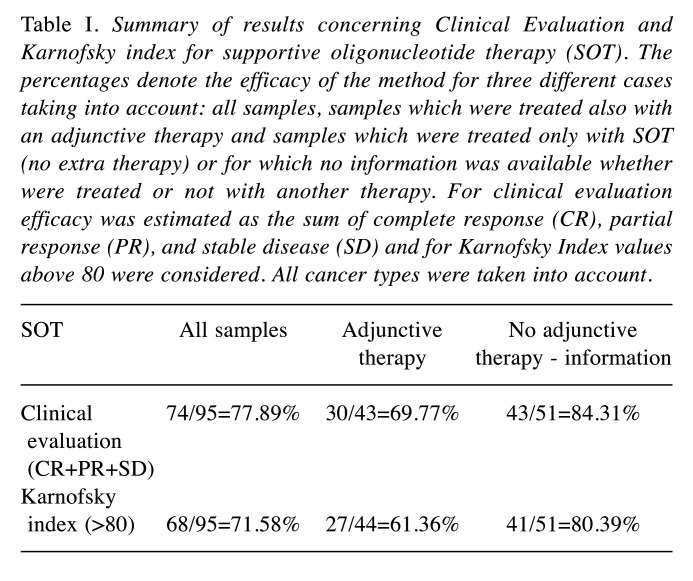
Results: Improvement of the clinical condition was observed in all patients who used SOT (77.89%), SOT in combination with other therapy (69.77%) and SOT as a monotherapy or no information was given concerning another therapy (84.31%). Positive results for Karnofsky-Index were also observed in 71.58%, 61.36%, and 80.39%, respectively. Finally, statistically significant reductions in CTCs were observed for both SOT as a monotherapy and SOT as an adjunctive therapy.
Conclusion: The preliminary results indicate that SOT therapy can be used both as monotherapy as well as in combination with other therapies for cancer.
Keywords: Supportive oligonucleotide therapy, RNA interference, cancer, clinical evaluation, Karnofsky index, CTCs, statistical analysis
It is known that the regulation of the expression of genes involved in fundamental biological processes (apoptosis, proliferation, etc.) can be an alternative treatment option in cancer. Gene expression is controlled through a process called RNA interference (RNAi), where small RNA molecules bind to complementary messenger RNA (mRNA), causing its degradation. Antisense therapy uses oligonucleotides, against a specific mRNA, and contributes to cancer treatment by either silencing genes essential for proliferation or inhibiting the expression of anti-apoptotic proteins. Particularly, RNAi is a normal biological process essential for the regulation of gene expression. Double-stranded RNA (dsRNA) molecules are degraded to shorter dsRNAs, which bind to a specific region of mRNA through the complementarity of bases, causing further degradation of mRNA and therefore no translation (1). The short dsRNAs are derived by cleavage of long dsRNA by Dicer, resulting in the production of microRNAs (miRNAs), short interfering RNAs (siRNAs), and piwi-interacting RNA (piRNA). Both miRNAs and siRNAs then assemble with RNA-induced silencing complex (RISC) and cause gene silencing (2). The applications of RNAi are numerous, including exploration of the function of unknown genes, identification of genes associated with drug resistance, genes involved in signaling transduction pathways, or genes implicated in disease progression or relapse. Furthermore, RNAi has been used in the therapy of different diseases including cancer, viral infections and respiratory diseases (3).
However, the main challenge using RNA molecules is their protection from nucleases. The chemical modifications of siRNA enable an increase in thermal stability, stability against digestion by nucleases, and better bio-distribution (4). In cancer, siRNA molecules contribute usually to the silencing of either oncogenes or anti-apoptotic genes. The present study aimed to identify whether the use of therapy based on modified siRNA molecules, called also supportive oligonucleotide therapy (SOT), could be beneficial for cancer patients, using SOTs designed and produced based on circulating tumor cells’ (CTCs) gene expression profile. The data are based both on follow-up questionnaires received from patients who suffered from cancer and received SOT therapy in combination with other types of therapy or as monotherapy, as well as on CTCs measurements between SOT doses.
Patients and Methods
Samples. The study included 95 cancer patients, both male, and female, representing different types of cancer. The study also included patients both with solid as well as with hematological malignancies. The sample distribution was random including samples from Europe and USA.
All procedures were conducted according to the standards of Safety, Bioethics, and Validation. The study was reviewed and approved by the Bioethical Committee of the Research Genetic Cancer Centre Group. All patients/donors provided written consent for the use of their samples in the present study. The patients retained the right to withdraw their samples until the date when the sample was received at the laboratory and tested.
Blood sample preparation. A total of 20-30 ml of blood were collected from all patients and placed in 50ml sterile falcon tubes (4440100, Orange Scientific, Braine-l’Alleud, Belgium) containing 7 ml of 0.02 M EDTA (E0511.0250, Duchefa Biochemie B.V., Haarlem, The Netherlands). For cancer patients, whole-blood samples were centrifuged for 20 min at 2500×g at room temperature (RT) with 4 ml polysucrose solution (Biocoll separating solution 1077, Biochrom, Berlin, Germany). Mononuclear cells, lymphocytes, platelets, and granulocytes were collected after centrifugation and washed with phosphate-buffered saline (PBS) (P3813, Sigma-Aldrich, Hamburg, Germany). The cells were incubated in lysis buffer (154 mM NH4Cl,31107, Sigma-Aldrich), 10 mM KHCO3 (4854, Merck, Darmstadt, Germany), and 0.1 mM EDTA in deionized water) for 10 min to lyse the erythrocytes. Samples were then centrifuged as above and washed with PBS. Cells were then incubated with pan-cytokeratin beads (recognizing CK4, CK5, CK6, CK8, CK10, CK13, and CK18) (5c-81714, Gentaur, Kampenhout, Belgium) at 4˚C for 30 min. Following incubation, the samples were placed in a magnetic field to collect microbead-bound cells for pan-cytokeratin, which were washed with PBS.
Molecular analysis. Following detection and isolation of CTCs, total RNA was extracted using a RNeasy mini kit (74105; Qiagen, Hilden, Germany). Then, 1 μg extracted RNA was prepared for DNA microarray experiments using an Amino Allyl MessageAmp II kit (AM1753; Ambion, Berlin, Germany). Labeling was performed with an Amersham CyDye post-labeling reactive dye pack (RPN5661; GE Healthcare, Freiburg im Breisgau, Germany). Reference (Cy3-labeled) and patient’s samples (Cy5-labeled) were mixed and co-hybridized using the Human MI ReadyArray platform (HS1100; Microarrays Inc., Huntsville, AL, USA). An HS 400 Pro hybridization station (Tecan, Männedorf, Switzerland) was used for pre-and-post hybridization processes and washing–drying steps. Slides were imaged on an InnoScan 710 (Innopsys, Carbonne, France) scanner with 5-μm resolution. Scanned microarray image data were processed using Array-Pro Analyzer 6.3 software. Data were normalized using background subtraction. Genes that were overexpressed were validated with qPCR reactions. For qPCR, 1 μg of RNA was used for cDNA synthesis and real-time quantitative PCR (RT-qPCR) reactions. For cDNA synthesis, the authors used the PrimeScript RT reagent kit (RR037A; Takara, Saint-Germain-en- Laye, France) and for RT-qPCR the KAPA SYBR Fast master mix universal (×2) (KK4618; Kapa Biosystems, Hamburg, Germany); the final reaction was 20 μl. All the primers for genes of interest and housekeeping genes were designed on Beacon Designer 8. Primer sequences were evaluated using the Basic Local Alignment Search Tool to exclude primers that would amplify undesired genes. Samples were analyzed by PCR as follows: 95˚C for 2 min, followed by 45 cycles of 95˚C for 10 s and 59˚C for 30 s. Upon amplification, a melting curve analysis was performed from 70˚C to 90˚C at 0.5˚C increments for 5 s at each step. PCR data were analyzed by the ΔΔCt method (5). In all reactions, cDNA synthesized from Universal Human Reference RNA (740000-41; Agilent, Santa Clara, CA, USA) was used as a positive control. Additional controls, including no-template and negative controls, were also used in all experiments. The reactions were performed in triplicate.
siRNA molecules against genes associated with essential procedures of tumor progression, such as invasion, apoptosis, migration, etc., were designed following the rules of Reynolds et al. (6). Patients’ CTCs transfected using Xfect reagent (631317; Takara, Beijing, PR China), according to the manufacturer’s instructions and 24-48 h post-transfection cells were validated with cellular and molecular assays.
SOT production – administration. Following the validation of siRNA, modified dsRNA molecules were designed and produced using the Oligomaker48 DNA/RNA Synthesizer. dsRNA molecules were then freeze-dried and tested for the presence of pathogens as well as for the tightness of each tube and send to the clinicians. Upon arrival, SOT was reconstituted in 1 ml of water for injection and dissolved. Then, the solution was placed in a syringe and added 9ml of water for injection. The final solution was applied intravenously.
Questionnaire. The questionnaire used in the present study included information about the patient, date of birth, type of malignancy, and date of last therapy. Then, a clinical evaluation was requested as well as a Karnofsky-Index scale, at least 3 months after SOT administration.
Descriptive statistics. For the descriptive models of the data two response scales were used. The first one was based on the clinical evaluation which categorizes tumor responses into 4 categories: complete response (CR), partial response (PR), stable disease (SD), and progress disease (PD). According to WHO, CR indicates the disappearance of the disease, PR over 50% decrease in target lesions, SD neither partial response nor progressive disease and PD appearance of new lesions, or unequivocal progression of non-target lesions (7). The other scale was based on Karnofsky Performance Scale Index (KI), which classifies patients based on their functional impairment. This scale covers 11 stages from 100 (normal health) to 0 (death). Therefore, the lower the Karnofsky Index, the worse the survival of the patient (8). In this study, we consider as a positive evaluation of SOT therapy the sum of CR, PR, and SD, as well as KI >80. Then, response-based histograms were generated for studying the efficacy of SOT treatment on cancer.
Inferential statistics. For hypothesis testing, one-way repeated-measures ANOVA was used, since it has traditionally been widely applied in assessing differences in non-independent mean values (9). It is the extension of the paired t-test (used to compare the means between two dependent groups), comparing the means between three or more dependent groups (10). This model can be used either (a) to assess differences in an outcome serially measured at different time points in the same subjects or (b) to compare means in which some outcome has been measured in the same subjects under different conditions (e.g., different treatments). The tested null hypothesis is that all related means are equal. Since ANOVA gives an overall p-value, it does not identify which group means are equal or not. This can be achieved by performing post-hoc tests, namely pairwise comparisons of either selected means of interest or all possible combinations of means. This involves testing multiple null hypotheses, and therefore the significance criterion is adjusted accordingly. In this study, we used a Bonferroni adjustment. Finally, since ANOVA is based on the assumption that all sample populations are normally distributed, in order to check the normality of the dataset the Shapiro-Wilk test was utilized (11). The statistical analysis was performed in the R environment (12) using packages such as tidyverse for data manipulation and visualization (13), ggpubr (14) for creating publication-ready plots, and rstatix for statistical analyses (15).
Results
Descriptive statistics.
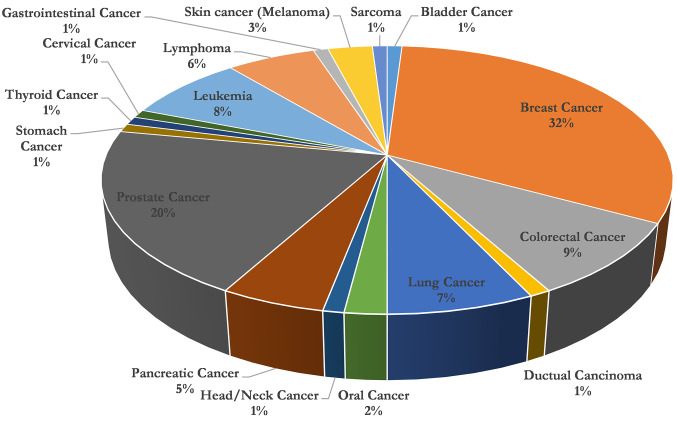
Cancer patients. The data used for the analysis concerned 95 patients aged 41 to 87 years with an average of 59 years, where 46 of them were female and 49 were male respectively. The majority were breast cancer patients (32.00%), followed by prostate cancer patients (20.00%), colorectal cancer (9.00%), and lung cancer (7.00%) The distribution of cancer data is presented in Figure 1. More than half of patients received SOT in combination with other types of therapy (58.00%), while the rest (42.00%) used SOT as monotherapy.
Figure 1. Representation of a percentage cycle chart for cancer cases following supportive oligonucleotide therapy.
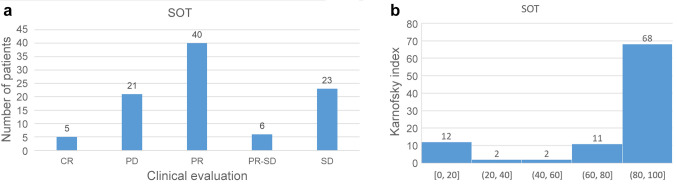
All cancer types. In Figure 2A, a histogram of the clinical evaluation concerning SOT and cancer is presented. As shown in this figure, for the generation of the histogram, all patients were taken into account irrespective of cancer type or whether SOT was combined with another therapy. The study of follow-up questionnaires revealed that for clinical evaluation most answers were PR (40), 23 were SD, 21 PD, 6 PR-SD, and 5 CR. Considering as a positive evaluation of the sum of CR, PR, and SD, the percentage of a positive evaluation of SOT for cancer reaches the value of 77.89% (74/95). Also, Figure 2B shows the histogram of KI. Considering as positive evaluation KI values above 80 (KI >80), corresponding to normal activity and/or normal activity with minor symptoms of a disease, it can be seen that for 68 patients the KI values were above 80 and for 27 patients below 80. Therefore, the percentage of positive evaluation, in this case, was estimated to be 71.58% (68/95).
Figure 2. Histograms of clinical evaluation and Karnofsky Index. (A) Histogram of clinical evaluation of supportive oligonucleotide therapy (SOT) for all samples. (B) Histogram of Karnofsky Index of SOT for all samples.
Similar results were obtained also for sub-datasets considering either patients who have taken another therapy or patients who haven’t or there was no information for this. The results are summarized in Table I. In particular, considering the combination of SOT with another therapy, the results of clinical evaluation showed a percentage of positive evaluation equal to 69.77% (30/43). The respective percentage for KI was 61.36% (27/44). Finally, considering a sample of patients treated with SOT for which no extra therapy was taken or no information was available, the respective percentage for clinical evaluation was 84.31% (43/51), while for KI was 80.39% (41/51). Summarizing, the results indicate that SOT therapy for cancer patients is efficient and can be combined (or not) with other therapies.
Table I. Summary of results concerning Clinical Evaluation and Karnofsky index for supportive oligonucleotide therapy (SOT). The percentages denote the efficacy of the method for three different cases taking into account: all samples, samples which were treated also with an adjunctive therapy and samples which were treated only with SOT (no extra therapy) or for which no information was available whether were treated or not with another therapy. For clinical evaluation efficacy was estimated as the sum of complete response (CR), partial response (PR), and stable disease (SD) and for Karnofsky Index values above 80 were considered. All cancer types were taken into account.
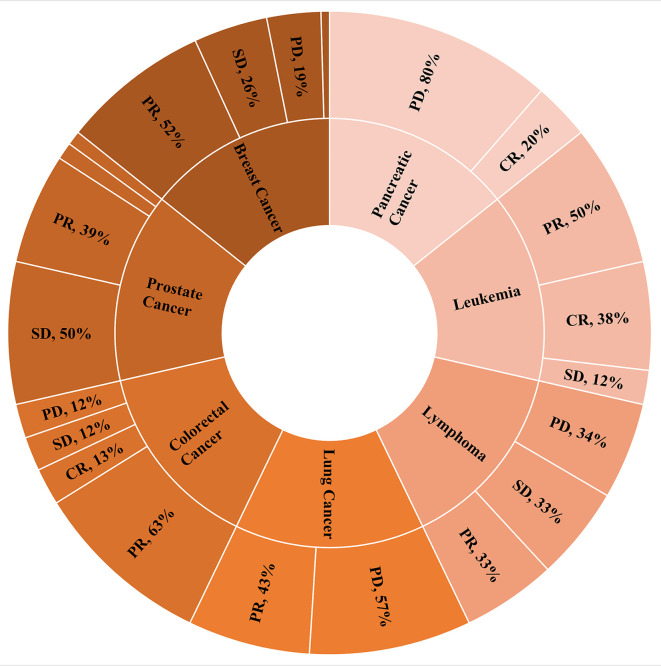
Cancer-specific. Figure 3 presents the distribution of clinical evaluation of patients receiving SOT therapy depending on the type of cancer in a pie chart. SOT response rates indicate that the clinical evaluation of cancer patients receiving SOT therapy varies depending on the type of cancer. From the patient samples, it was observed that the percentage of response to treatment for cancer patients is higher for leukemia (100%), prostate cancer (95%), colorectal (88%), and breast (81%).
Figure 3. Clinical evaluation of patients receiving supportive oligonucleotide therapy (SOT) depending on the type of cancer (showing only percentages above 6%).
Inferential statistics.
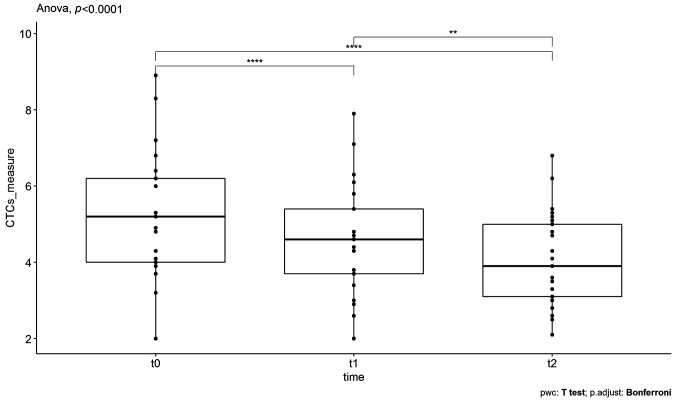
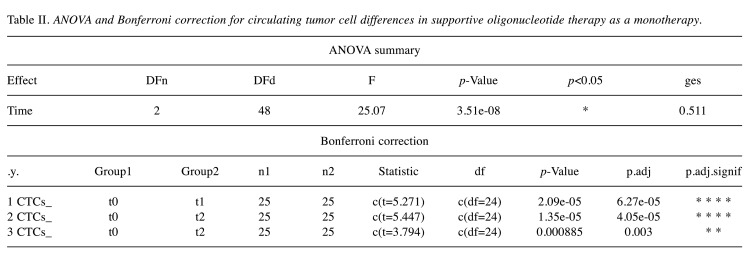
SOT as monotherapy. In the case of SOT as a monotherapy, 25 individuals with a median age of 62 years were measured three times on the same outcome variable (CTCs counts) and the One-Way repeated Measures ANOVA test was applied to investigate statistically significant differences (decreases) of the CTCs between SOT doses in patients. However, ANOVA is based on the assumption that all sample populations are normally distributed. In order to check the normality of the dataset, the Shapiro-Wilk test was utilized. This test gave, as a result, a p-value>0.05 indicating that normality isn’t violated and therefore the application of one-way repeated measures ANOVA is feasible. The statistical results are represented in Figure 4, where the time points of the doses are in the x-axis and CTCs counts in the y-axis. The asterisks denote significance codes, namely ‘****’ stands for p-value ≤0.0001, ‘***’ stands for p-value ≤0.001, ‘**’ stands for p-value ≤0.01, and ‘*’ stands for p-value ≤0.05. At t0, which corresponds to the initial measurement before the first SOT administration, the CTCs were found to have a mean value of 5.23±0.67 for 95% confidence intervals (CI). At t1, which corresponds to CTC measurements after the first administration, the mean value was found equal to 4.63±0.55 for 95%CI. Finally, at t2, which corresponds to the CTC counts after the second administration, the mean value was found to be 4.05±0.47 for 95%CI. As it can be seen, there is a large decrease in the mean value of the CTCs counts after the first and the second administration respectively. Indeed, the application of one-way repeated measures ANOVA revealed that the means of these three groups are not all equal with a p-value<0.05. In particular, the obtained F-statistic value was 25.07 (the comparison is made with F-test) and the amount of variability due to the within-subjects factor (ges) was 0.511. These results are shown in Table II.
Figure 4. The boxplots of circulating tumor cells’ (CTCs) counts, measured for supportive oligonucleotide therapy (SOT) as a monotherapy, at the different time points t0 (before the first SOT administration), t1 (after the first SOT administration), and t2 (after the second SOT administration). The decreases in the mean values of CTCs were found to be statistically significant.
Table II. ANOVA and Bonferroni correction for circulating tumor cell differences in supportive oligonucleotide therapy as a monotherapy.
Finally, to find which pairs of means were significantly different, a multiple comparison test was conducted and the results are also depicted in Table II. Particularly, Table II shows the adjusted p-values (Bonferroni adjustment) corresponding to the multiple pairwise comparisons. As it can be seen, all three pairwise comparisons, namely t0 with t1, t0 with t2, and t1 with t2, revealed significant differences related to decreases in CTCs counts, namely p.adj-value<0.05 for t0-t1 groups’ comparison, p.adj-value<0.05 for t0-t2 groups’ comparison, and p.adj-value<0.05 for t1-t2 groups’ comparison.
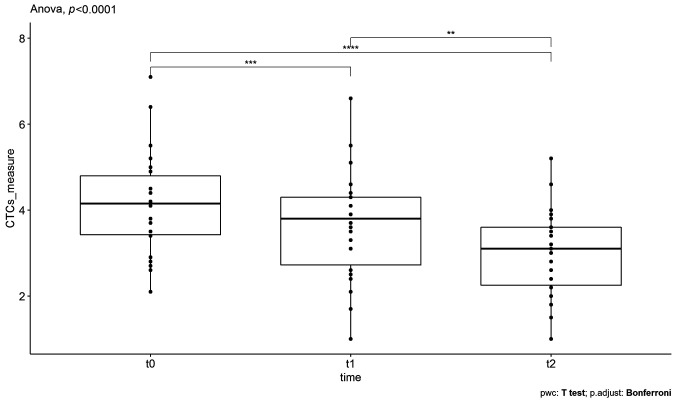
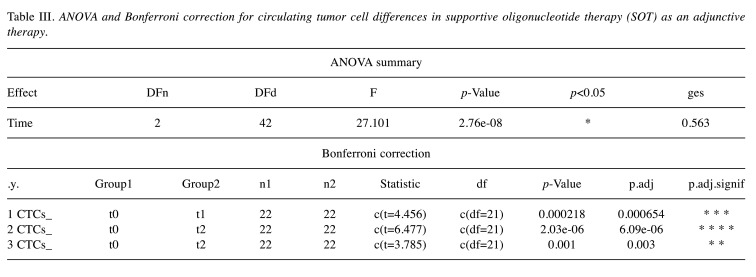
SOT as adjunctive therapy. In the case of SOT as a complementary therapy, the three repeated measures of CTCs from 22 patients with a median age of 67 years were analyzed using the same procedure described previously. In particular, the data were checked for normality using the Shapiro-Wilk test, giving a p-value>0.05. Therefore, the normality assumption was verified. The statistical results are represented in Figure 5 (similar to Figure 4), where the time points of the doses are on the x-axis and CTCs counts on the y-axis. At t0, which corresponds to the initial measurement before the first SOT administration, the CTCs were found to have a mean value of 4.15±0.51 for 95%CI. At t1, which corresponds to CTCs measurements after the first administration, the mean value was found equal to 3.65±0.53 for 95%CI. Finally, at t2, which corresponds to the CTC counts after the second administration, the mean value was found to be 3.01±0.43 for 95%CI. As it can be seen, there is a large decrease in the mean value of the CTC counts after the first and the second administration. Indeed, the application of one-way repeated measures ANOVA revealed that the means of these three groups are not all equal with a p-value <0.05. In particular, the obtained F-statistic value is 27.101 and the amount of variability due to the within-subjects factor (ges) is 0.563. These results are shown in Table III.
Figure 5. The boxplots of circulating tumor cells’ (CTCs) counts, measured for supportive oligonucleotide therapy (SOT) as an adjunctive therapy, at the different time points t0 (before the first SOT administration), t1 (after the first SOT administration) and t2 (after the second SOT administration). The decrease in the mean value of CTCs was found to be statistically significant.
Table III. ANOVA and Bonferroni correction for circulating tumor cell differences in supportive oligonucleotide therapy (SOT) as an adjunctive therapy.
Finally, to find which pairs of means are significantly different, a multiple comparison test was conducted and the results are also depicted in Table III. Particularly, Table III shows the adjusted p-values (Bonferroni adjustment) corresponding to the multiple pairwise comparisons. As it can be seen, all three pairwise comparisons revealed significant differences related to decreases in CTCs counts, namely p.adj-value <0.05 for t0-t1 groups’ comparison, p.adj-value<0.05 for t0-t2 groups’ comparison, and p.adj-value <0.05 for t1-t2 groups’ comparison.
Overall, the statistically significant reduction of the mean CTC’s values in both cases (SOT as a monotherapy as well as a supportive treatment) over time is a strong indication that SOT therapy, appears to play an important role as an effective measure against a further increase of the circulating tumor cells, regardless of any other alternative treatment or intervention (surgery or chemotherapy/radiotherapy).
Discussion
The present study aimed to provide an early evaluation of the use of siRNAs in cancer patients, either as monotherapy or in combination with other types of therapy. These siRNA molecules, SOTs, were designed based on patients’ CTC gene expression profiles and targeted anti-apoptotic proteins. The products were validated with in vitro assays before injection in patients and quality controls performed during and after the production to ensure pathogen-free products.
In particular, the clinical status evaluation of all patients who used SOT therapy for cancer was positive in 77.89%, whereas a positive evaluation was also found based on KI scale since 71.58% of the answers were above 80 on the KI scale. Among patients who used SOT in combination with other therapy, 69.77% of them had a good clinical status evaluation, whereas a positive evaluation was found based on KI scale, since 61.36% of the answers were above 80 according to the KI scale. For patients who used SOT as monotherapy (or no information was given concerning another therapy), the positive clinical status reached 84.31%, whereas 80.39% of the answers were above 80 on the KI scale. As far as the type of cancer is concerned, the clinical status evaluation revealed that the higher response was observed in leukemia (100%), prostate (95.00%), colorectal (88.00%), and breast cancer (81.00%). Finally, statistically significant reductions in CTCs were found over time, both for 25 patients who received SOT as an exclusive treatment (adjusted p-value <0.05), as well as for those (22) who used it as adjunctive therapy or no information was given concerning another therapy (adjusted p-value <0.05). Overall, these preliminary results, based on both descriptive as well as inferential statistics, indicate that SOT can be used either as monotherapy or combined with other therapies to improve patients’ states.
Gene silencing, apart from a normal process of living cells, can be a useful and powerful tool for scientists against different diseases. The use of siRNAs as a therapeutic is very promising since it could target genes that other molecules cannot. These molecules can be used either naked or conjugated with lipids or other molecules (GalNAc). The majority are in clinical trials, at different phases and only a few have been approved by the FDA (16). Onpattro is the first FDA-approved RNAi drug and is a lipid nanoparticle-based short interfering RNA drug designed for polyneuropathies induced by hereditary transthyretin amyloidosis (17,18). Antisense nucleotides have been also used with success in neuromuscular diseases, like Duchenne muscular dystrophy or spinal muscular atrophy, where not many effective therapies have been developed (19). In addition, the siRNA has been used in the hepatitis C virus (HCV), blocking its replication through the destruction of HCV RNA or in other viruses (20). The RNAi therapy can be used also in chronic human virus infections, and the use of nanocarriers enhances the benefit for patients (21). Knockdown of particular proteins, such as selenoprotein K leads to depletion of Borrelia burgdorferi within the tick host Ixodes scapularis, indicating that there is a wide range of therapeutic applications for siRNA molecules (22). Therefore, it can be expanded also to cancer therapy, particularly in cases where overexpression of proteins is involved in tumorigenesis or tumor progression. In most cases, RNAi is used in cancer to silence oncogenes and anti-apoptotic genes. Targeting of Bcl2 anti-apoptotic protein, through degradation of Bcl2 mRNA, is very common in cancer treatment of different types of cancer (23). The modification of siRNA molecules leads to an increase of protection against nucleases, and better pharmacokinetic properties, therefore these molecules can be used in clinical practice (4). The most common modification is the 2’ position of the ribose backbone (2’-O-methyl, 2’-O-methoxyethyl, 2’-deoxy-2’-fluorouridine), or changes of the phosphate linkage of siRNA (24). siRNA molecules can be administered quarterly, to increase the therapeutic effect, without problems for patients, something that is not feasible with small molecular weight molecules or antibodies (25).
Despite the small number of samples considered in this study, we showed that the use of SOTs in cancer can be very beneficial for patients. siRNA molecules are safe and specific on targeting, therefore can be used in cases where other drugs cannot. Even though more samples need to be tested to be used at clinical routine, these preliminary statistical results concerning SOT therapy in cancer are encouraging.
Conflicts of Interest
The Authors report no conflicts of interest in relation to this work.
Authors’ Contributions
IP supervised the study. GB performed analysis of data, drafted the manuscript. AI performed analysis of data, drafted the manuscript. PA carried out the molecular biology assays, drafted the manuscript.
References
- 1.Kim D, Rossi J. RNAi mechanisms and applications. Biotechniques. 2008;44(5):613–616. doi: 10.2144/000112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corey DR. Chemical modification: the key to clinical application of RNA interference. J Clin Invest. 2007;117(12):3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 7.Nishino M, Jagannathan JP, Krajewski KM, O’Regan K, Hatabu H, Shapiro G, Ramaiya NH. Personalized tumor response assessment in the era of molecular medicine: cancer-specific and therapy-specific response criteria to complement pitfalls of RECIST. AJR Am J Roentgenol. 2012;198(4):737–745. doi: 10.2214/AJR.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmermann C. ‘Just give me the best quality of life questionnaire’: the Karnofsky scale and the history of quality of life measurements in cancer trials. Chronic Illn. 2013;9(3):179–190. doi: 10.1177/1742395312466903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schober P, Vetter TR. Repeated measures designs and analysis of longitudinal data: if at first you do not succeed-try, try again. Anesth Analg. 2018;127(2):569–575. doi: 10.1213/ANE.0000000000003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra P, Singh U, Pandey CM, Mishra P, Pandey G. Application of student’s t-test, analysis of variance, and covariance. Ann Card Anaesth. 2019;22(4):407–411. doi: 10.4103/aca.ACA_94_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 12.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/ [Last accessed on November 1, 2021]
- 13.Wickham H, Averick M, Bryan J, Chang W, D’Agostino McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Lin Pedersen T, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. Welcome to the tidyverse. J Open Source Softw. 2019;4(43):1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 14.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.4.0. Available at: https://cran.r-project.org/web/packages/ggpubr/index.html. [Last accessed on June 27, 2020]
- 15.Kassambara A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R package version 0.7.0. Available at: https://CRAN.Rproject.org/package=rstatix. [Last accessed on December 20, 2021]
- 16.Nikam RR, Gore KR. Journey of siRNA: Clinical developments and targeted delivery. Nucleic Acid Ther. 2018;28(4):209–224. doi: 10.1089/nat.2017.0715. [DOI] [PubMed] [Google Scholar]
- 17.Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, Mui BL, Semple SC, Tam YK, Ciufolini M, Witzigmann D, Kulkarni JA, van der Meel R, Cullis PR. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14(12):1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 18.Weng Y, Xiao H, Zhang J, Liang XJ, Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv. 2019;37(5):801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Sardone V, Zhou H, Muntoni F, Ferlini A, Falzarano MS. Antisense oligonucleotide-based therapy for neuromuscular disease. Molecules. 2017;22(4):563. doi: 10.3390/molecules22040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelleher AD, Cortez-Jugo C, Cavalieri F, Qu Y, Glanville AR, Caruso F, Symonds G, Ahlenstiel CL. RNAi therapeutics: an antiviral strategy for human infections. Curr Opin Pharmacol. 2020;54:121–129. doi: 10.1016/j.coph.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Smolic R, Volarevic M, Wu CH, Wu GY. Potential applications of siRNA in hepatitis C virus therapy. Curr Opin Investig Drugs. 2006;7(2):142–146. [PubMed] [Google Scholar]
- 22.Kumar D, Embers M, Mather TN, Karim S. Is selenoprotein K required for Borrelia burgdorferi infection within the tick vector Ixodes scapularis. Parasit Vectors. 2019;12(1):289. doi: 10.1186/s13071-019-3548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YX, Kong CZ, Wang LH, Li JY, Liu XK, Xu B, Xu CL, Sun YH. Ursolic acid overcomes Bcl-2-mediated resistance to apoptosis in prostate cancer cells involving activation of JNK-induced Bcl-2 phosphorylation and degradation. J Cell Biochem. 2010;109(4):764–773. doi: 10.1002/jcb.22455. [DOI] [PubMed] [Google Scholar]
- 24.Lee SJ, Kim MJ, Kwon IC, Roberts TM. Delivery strategies and potential targets for siRNA in major cancer types. Adv Drug Deliv Rev. 2016;104:2–15. doi: 10.1016/j.addr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]