Addicks et al. identify a role for protein acetylation in the regulation of muscle integrity by linking GCN5 acetyltransferase activity to the expression of dystrophin and other genes important for muscle structure. They find that this occurs through GCN5-directed acetylation of the transcriptional repressor YY1.
Abstract
Protein lysine acetylation is a post-translational modification that regulates protein structure and function. It is targeted to proteins by lysine acetyltransferases (KATs) or removed by lysine deacetylases. This work identifies a role for the KAT enzyme general control of amino acid synthesis protein 5 (GCN5; KAT2A) in regulating muscle integrity by inhibiting DNA binding of the transcription factor/repressor Yin Yang 1 (YY1). Here we report that a muscle-specific mouse knockout of GCN5 (Gcn5skm−/−) reduces the expression of key structural muscle proteins, including dystrophin, resulting in myopathy. GCN5 was found to acetylate YY1 at two residues (K392 and K393), disrupting the interaction between the YY1 zinc finger region and DNA. These findings were supported by human data, including an observed negative correlation between YY1 gene expression and muscle fiber diameter. Collectively, GCN5 positively regulates muscle integrity through maintenance of structural protein expression via acetylation-dependent inhibition of YY1. This work implicates the role of protein acetylation in the regulation of muscle health and for consideration in the design of novel therapeutic strategies to support healthy muscle during myopathy or aging.
Introduction
Skeletal muscle is a dynamic tissue that can coordinate structural, metabolic, and contractile adaptations to demands for contractile work. Profound remodeling of muscle can be evoked by physiological signals, including resistance or endurance training, or by pathological signals that arise during disease, myopathy, or aging. Understanding the molecular signaling pathways that regulate muscle are therefore integral to our understanding of motility throughout the lifespan. Essential for muscle integrity, a critical structural link between the actin cytoskeleton and the ECM is maintained by the dystrophin-associated protein complex (DAPC), which stabilizes the sarcolemma during contractions, and transmits force generated by the contractile apparatus to the ECM (Petrof et al., 1993). The DAPC includes intracellular (dystrophin, syncoilin, syntrophin, α-dystrobrevin, and neuronal nitric oxide synthase [nNOS]), transmembrane (β-dystroglycan, sarcoglycans, and sarcospan), and extracellular (α-dystroglycan and muscle-specific laminin) components (Ehmsen et al., 2002). More recently, dysregulation of dystrophin and other DAPC members has been observed in cancer (Acharyya et al., 2005), atrophy (Risson et al., 2009; Swiderski et al., 2021; Chockalingam et al., 2002), and aging (Hughes et al., 2017; Townsend et al., 2011; Hord et al., 2016; Kosek and Bamman, 2008), leading to decreased muscle integrity. Despite their importance, mechanisms underlying regulation of expression of dystrophin and other DAPC members are largely unknown.
Reversible acetylation of proteins is a well-known component of the regulation of gene expression in response to changes in the cellular environment. Lysine acetylation is catalyzed by lysine acetyltransferases (KATs) and can be removed by lysine deacetylases (KDACs). Importantly, dysregulation of KAT and KDAC activity occurs in various diseases, including cancer (Gil et al., 2017), Alzheimer’s disease (Cohen et al., 2011), cardiovascular disease (Li et al., 2020), muscular dystrophy (Consalvi et al., 2011, 2013; Bettica et al., 2016), and aging. Post-translational modifications such as lysine acetylation influence secondary structure, inter- or intramolecular interactions, or catalytic activity. In particular, acetylation of lysine residues on transcription factors is a well characterized post-translational modification that can affect binding to DNA or cofactors for the regulation of gene expression (Narita et al., 2019). In both muscle disease and aging, inhibition of KDAC activity has been shown to result in beneficial effects on muscle (reviewed in McIntyre et al., 2019); however, the mechanisms linking protein acetylation to muscle integrity are poorly understood.
As an evolutionarily conserved KAT, general control of amino acid synthesis protein 5 (GCN5; KAT2A) is capable of acetylating both histone and nonhistone lysines. Acetylation by GCN5 has been shown to stabilize the c-Myc oncoprotein (Patel et al., 2004) or to recruit other KATs for the transcriptional coactivation of tumor suppressor p53 (Barlev et al., 2001), which in each case activates transcription. The acetylation of the metabolic coactivator peroxisome proliferator-activated receptor-γ coactivator (PGC)-1α in liver made the first direct link between GCN5 activity and the inhibition of PGC-1α–driven regulation of gluconeogenesis (Lerin et al., 2006). From these findings arose the hypothesis that GCN5 ablation could enhance PGC-1α–directed mitochondrial biogenesis in muscle. However, recent work could not identify a metabolic role for GCN5 in skeletal muscle (Dent et al., 2017; Svensson et al., 2020).
To interrogate the role of GCN5 acetylation in muscle, we performed an unbiased transcriptomic analysis that identified mouse muscle atrophy (MA) and human muscular dystrophy as the top correlating open-access biosets to that of our muscle-specific mouse knockout (KO) of GCN5 (Gcn5skm−/−) bioset. Additionally, these Gcn5skm−/− mice exhibited a reduction in dystrophin expression that, when assessed in vivo with different experimental models of eccentric muscle contraction, presented with a myopathic phenotype. To explain the regulation of dystrophin and other components of DAPC expression, we discovered a mechanism reliant on GCN5-directed acetylation of the DNA binding zinc finger domain of the transcription factor/repressor Yin Yang 1 (YY1). We conclude that dystrophin and components of the DAPC are positively regulated by GCN5-directed acetylation of YY1.
Results
Transcriptomic analysis reveals the dysregulation of dystrophin and genes associated with the DAPC in Gcn5skm−/− mice
We first set out to generate Gcn5skm−/− mice in order to perform an unbiased transcriptomic analysis that would inform us of potential phenotypes and signaling pathways governed by GCN5 in differentiated muscle. To do this, a conditional mouse KO for GCN5 was generated by flanking exons 5 and 6 of Gcn5 with LoxP sites. Conditional Gcn5-KO mice were then crossed with mice expressing Cre from a transgenic human α-skeletal actin (Hsa) gene promoter (Miniou et al., 1999; Fig. S1), resulting in Gcn5skm−/− mice (Fig. 1 a). Note that Gcn5skm−/− muscle tissue contains residual Gcn5 mRNA from resident nonmuscle cells, vasculature, etc., as is typical of loss-of-function models generated using the HSA-Cre mouse line.
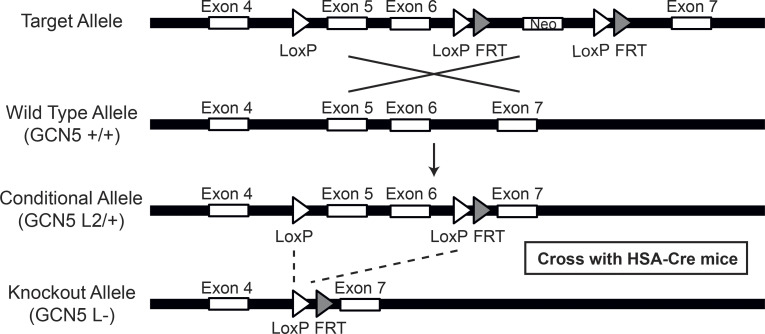
Figure S1.
Schematic of Gcn5 gene targeting and conditional deletion of exons 5 and 6. Maps of the Gcn5 genomic locus, the floxed allele with the neomycin cassette (+neo; target allele) and without the neomycin cassette (−neo; conditional allele). Positions of the exons are indicated. FRT, flippase recognition target.
Figure 1.
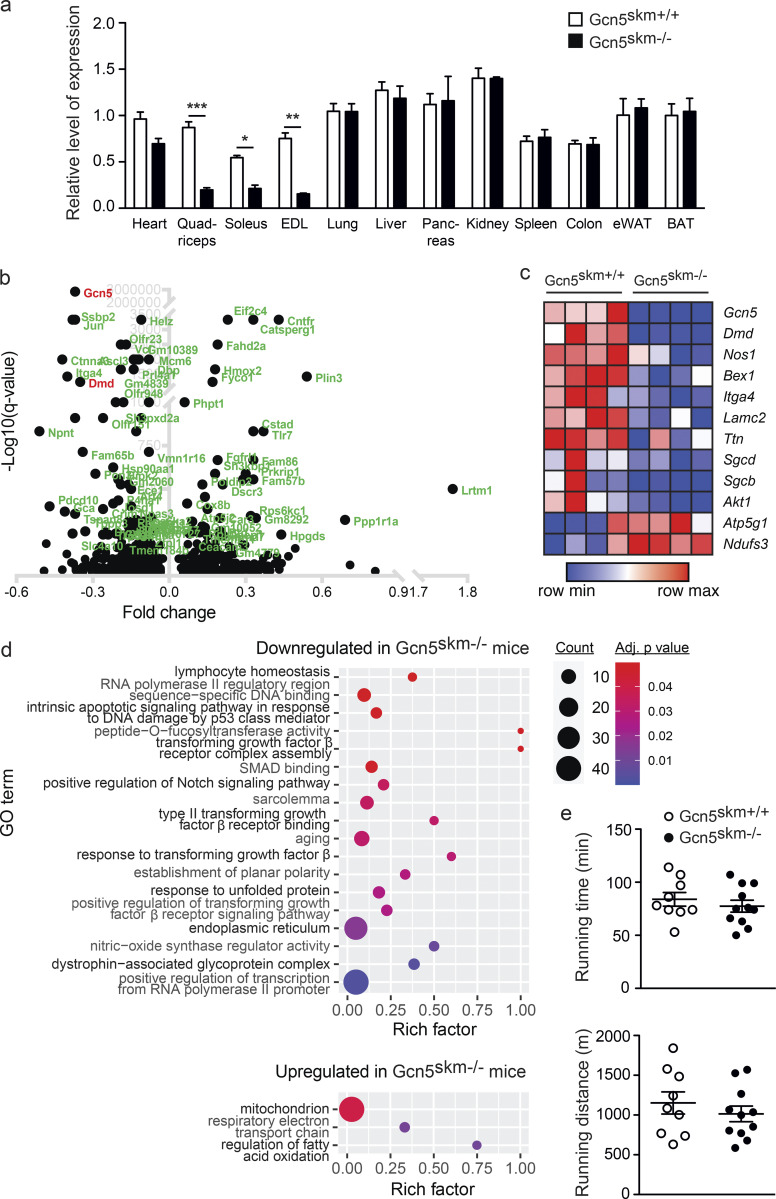
Validation and transcriptomic profiling of Gcn5skm−/− mice. (a) Relative gene expression analysis of Gcn5 in various tissues from 6-mo-old control and Gcn5skm−/− mice as determined by RT-qPCR. Values were normalized to 36b4. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 10–11/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (b) Volcano plot of 1,172 differentially expressed genes from 6-mo-old Gcn5skm−/− gastrocnemius muscle, representing the top 100 most significant genes. (c) Heat map showing dysregulated genes for DAPC and membrane proteins (Dmd, Sgcd, Sgcb, Nos1), ECM proteins (Lamc2, Itga4), energy metabolism proteins (Atp5g1, Ndufs3, Akt1), and Bex1, a marker of muscle regeneration, for 6-mo-old control and Gcn5skm−/− gastrocnemius muscle. n = 4/group. (d) Bubble plots showing the distribution and size of overrepresented GO pathways of the down-regulated gene set and the up-regulated gene set of gastrocnemius muscle from 6-mo-old Gcn5skm−/− mice. n = 4/group. (e) Time and distance run by 4-mo-old Gcn5skm−/− and Gcn5skm+/+ mice on a treadmill at a 5% incline. Data are presented as mean ± SEM. n = 9–11/group. eWAT, epididymal white adipose tissue; BAT, brown adipose tissue.
To further assess effects of loss of GCN5 in muscle, microarray analysis was performed on cDNA libraries generated from gastrocnemius muscle harvested from 6-mo-old Gcn5skm−/− mice and control littermates. cDNAs corresponding to a total of 1,236 microarray features were differentially expressed in the gastrocnemius muscle tissue of Gcn5skm−/− mice, of which 789 were down-regulated and 447 were up-regulated (Data S1). Upon examination, dystrophin (Dmd) was among the most down-regulated genes in Gcn5skm−/− muscle (Fig. 1 b), along with the dysregulation of other DAPC members, including β-sarcoglycan (Sgcb), δ-sarcoglycan (Sgcd), and the structural muscle protein transcript titin (Ttn; Fig. 1 c).
The InnateDB (Breuer et al., 2013) was used for gene ontology (GO) analysis of differentially expressed genes. Several pathways, including DAPC, sarcolemma, NOS regulator activity, and response to TGF-β, were found to be down-regulated in Gcn5skm−/− muscle (Fig. 1 d). Interestingly, in agreement with the hypothesis that Gcn5 negatively regulates mitochondrial biogenesis through acetylation and inactivation of PGC-1α, and despite no observed change in treadmill running (Fig. 1 e), mitochondrial respiratory electron transport chain and regulation of fatty acid oxidation pathways were up-regulated in Gcn5skm−/− muscle (Fig. 1 d).
GCN5 regulates dystrophin protein expression and maintains muscle integrity during eccentric contractions and with age
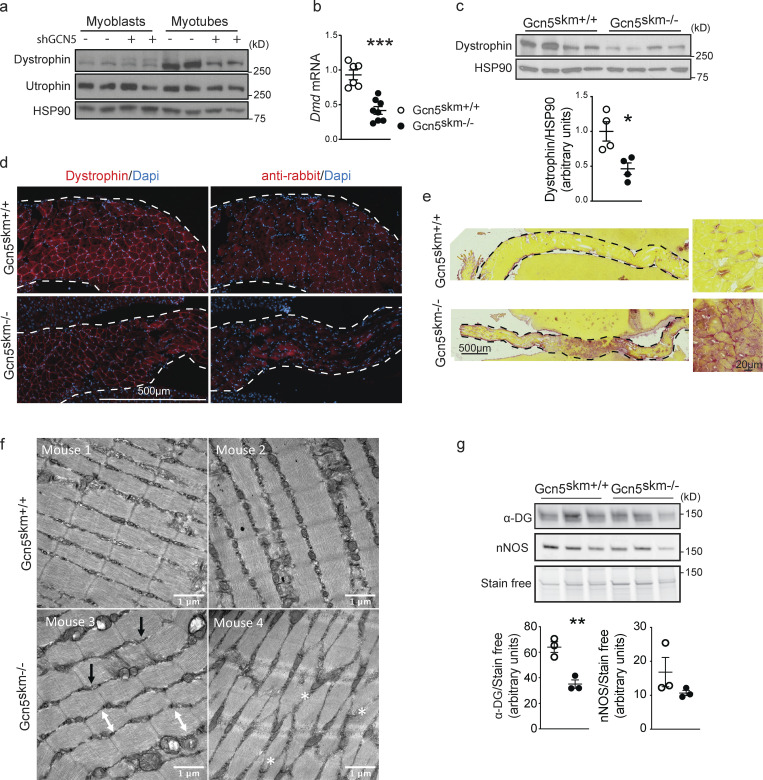
GCN5 regulation of dystrophin protein expression was first confirmed in C2C12 myotubes using adenovirus-driven shRNA constructs to knock down Gcn5 (Fig. 2 a). To confirm the reduction in Dmd gene expression observed in vivo using transcriptomics, we performed both biochemical and histological analyses on 6-mo-old Gcn5skm+/+ and Gcn5skm−/− muscle samples. Quantitative RT-PCR (RT-qPCR) on mRNA isolated from control and Gcn5skm−/− gastrocnemius muscle showed decreased Dmd transcript (Fig. 2 b). Western immunoblotting of tibialis anterior muscle samples and immunostaining of diaphragm muscle samples confirmed that dystrophin protein levels were reduced in Gcn5skm−/− muscle compared with control mice (Fig. 2, c and d). This was complemented by increased fibrosis of the diaphragm and disruption of extensor digitorum longus (EDL) muscle sarcomere organization in Gcn5skm−/− mice (Fig. 2, e and f), all indicating a loss of muscle integrity. In addition, we observed reduced α-dystroglycan protein expression, but no change in nNOS expression, in Gcn5skm−/− quadriceps muscle (Fig. 2 g).
Figure 2.
GCN5 regulates dystrophin protein expression and muscle integrity. (a) Western blot of protein extract from C2C12 myoblasts and myotubes (n = 2/condition) after 4 d of differentiation in the presence or absence of adenovirus-driven shGCN5 using anti-dystrophin and anti-utrophin antibodies, with anti-HSP90 antibodies used as a loading control. (b) Relative expression of Dmd in gastrocnemius muscle from 6-mo-old control and Gcn5skm−/− mice as measured by RT-qPCR relative to housekeeping genes 36b4 and Gapdh. Data are presented as mean ± SEM. n = 8–11/group. ***, P < 0.01 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (c) Western blot of whole protein extract from tibialis anterior muscle of 6-mo-old control and Gcn5skm−/− mice using anti-dystrophin with anti-HSP90 antibodies used as a loading control. Data are presented as mean ± SEM. n = 4/group. *, P < 0.05 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (d) Representative immunofluorescence images of diaphragm muscle sections from 6-mo-old control and Gcn5skm−/− mice using anti-dystrophin (left column) or nonspecific anti-rabbit secondary (right column) antibodies. Images are tile scanned and stitched. Dashed line represents the edge of liver tissue, which was used to mount the diaphragm tissues for sectioning. n = 3/group. Scale bar, 500 μm. (e) Picrosirius red staining of diaphragm muscle sections from 6-mo-old control and Gcn5skm−/− mice. Images were tile scanned and stitched. Dashed line represents the edge of liver tissue that was used on both sides of the diaphragm to help with tissue mounting and sectioning. n = 3/group. Scale bars, 500 μm; inset, 20 μm. (f) 8-wk-old Gcn5skm−/− mice harbor variable abnormal sarcomere structures, including loss of rectangular sarcomere structure (white double-ended arrows), offset z-discs (black arrows), and/or disrupted sarcomeres (white asterisks) as revealed on electron transmission micrographs of EDL muscles. No steps were taken to prevent muscle contraction during sacrifice. (g) Western blot of whole protein extract from left quadriceps muscles of 6-mo-old control and Gcn5skm−/− mice using anti–α-dystroglycan (α-DG) and anti-nNOS antibodies and stain-free protein loading control. Data are presented as mean ± SEM. n = 3/group. **, P < 0.01 versus Gcn5skm+/+ as measured by two-tailed Student’s t test.
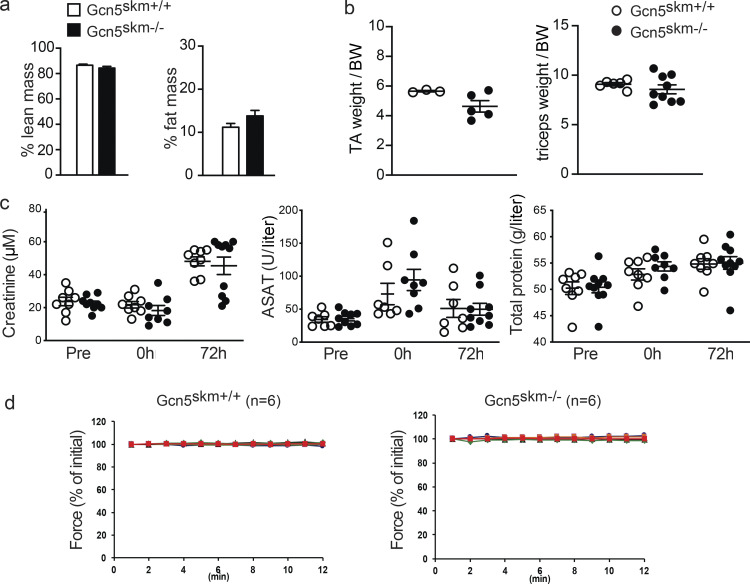
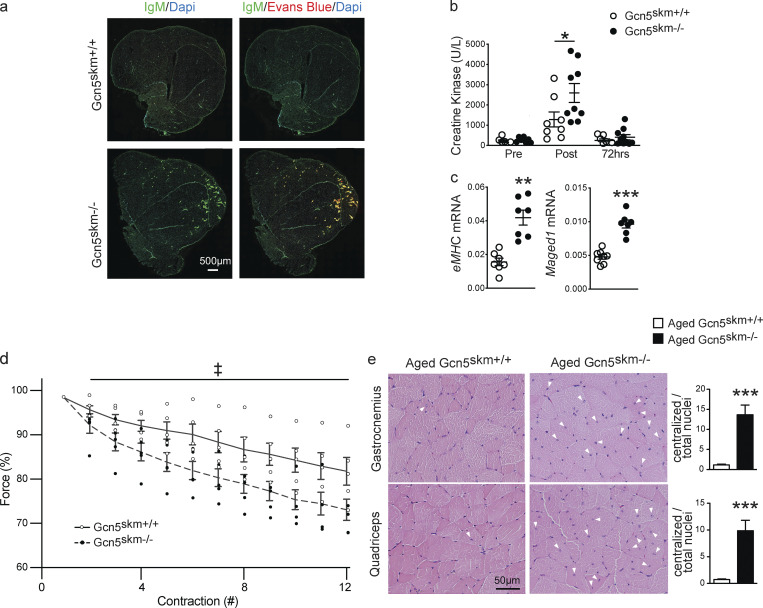
Because Gcn5skm−/− mice have decreased Dmd expression and exhibit a dystrophic gene expression profile, mice were further assessed for an underlying muscle disorder. Gcn5skm−/− mice had no difference in the percentage of lean mass, fat mass, or relative muscle weight compared with controls (Fig. S2, a and b). When 6-mo-old mice were subjected to a moderate protocol of downhill running, a model that can result in muscle damage due to repetitive eccentric muscle contractions (Armstrong et al., 1983), quadriceps muscles of Gcn5skm−/− mice 24 h after running demonstrated higher levels of Evans blue dye (EBD) uptake that overlapped that of anti-IgM immunohistochemical staining (Fig. 3 a), indicating increased permeability of the muscle sarcolemma due to damage (Wooddell et al., 2010; Schmalbruch, 1975). The distance run was the same for all Gcn5skm−/− and control mice; however, immediately following the nonexhaustive downhill running protocol, circulating creatine kinase levels, an early biomarker of muscle damage (Bulfield et al., 1984), were further elevated in Gcn5skm−/− mice compared with their controls (Fig. 3 b), with no significant changes in creatinine, aspartate aminotransferase, or total protein levels (Fig. S2 c). Downhill running in Gcn5skm−/− mice elicited increases in transcript levels for markers of muscle regeneration after 72 h, including embryonic myosin heavy chain (eMhc) and Maged1 (Fig. 3 c), indicating that the damage caused by eccentric contractions triggered a compensatory increase in muscle regeneration in Gcn5skm−/− animals.
Figure S2.
Phenotyping results for Gcn5skm−/− versus Gcn5skm+/+ mice. (a) The percentage lean mass and fat mass for Gcn5skm+/+ and Gcn5skm−/− mice. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 12/group. (b) Tibialis anterior and triceps weights per amount of BW for Gcn5skm+/+ and Gcn5skm−/−. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 3–8/group. (c) Blood creatinine and aspartate aminotransferase (ASAT) levels in units per liter and total blood protein in grams per liter in control and Gcn5skm−/− mice immediately before (Pre), immediately after (0 h), and 72 h after (72 h) downhill run. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 7–9/group. (d) Force produced during a series of contractions after 1 h of equilibrium in apparatus and before initiation of eccentric contraction series for diaphragm strips used in Fig. 4 d. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 5/group.
Figure 3.
GCN5 maintains muscle force and protects from damage following eccentric contractions and with aging. (a) Representative images of immunofluorescent staining of right quadriceps muscles from 6-mo-old control and Gcn5skm−/− mice sacrificed 24 h after downhill running using anti-IgM cross-adsorbed secondary antibody alone (left column) or in combination with EBD fluorescence (right column). Images were tile scanned and stitched. All animals completed the nonexhaustive downhill running protocol. n = 3/group. Scale bar, 500 μm. (b) Plasma creatine kinase levels in 6-mo-old control and Gcn5skm−/− mice measured before (Pre), directly after (Post), and 72 h after (72 h) downhill running. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 7–9/group. *, P < 0.05 by ANOVA with Tukey’s post hoc test versus Gcn5skm+/+. (c) Relative expression of embryonic myosin heavy chain (eMHC) and Maged1 in 6-mo-old control and Gcn5skm−/− gastrocnemius tissues harvested 72 h after downhill running. Data are presented relative to housekeeping genes 36b4 and Gapdh and as mean ± SEM for controls and Gcn5skm−/− mice. n = 7/group. **, P < 0.01; ***, P < 0.001 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (d) Force measurement (percentage of initial contraction) of consecutive eccentric contractions of diaphragm strips harvested from 6-mo-old control and Gcn5skm−/− mice. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 5/group. ‡, main effect of P < 0.0001 by two-way ANOVA for genotype. (e) Representative sections of gastrocnemius and quadriceps muscles from 12-mo-old control and Gcn5skm−/− mice stained with hematoxylin and eosin (brightfield) with examples of centralized nuclei identified with arrowheads. The number of centralized nuclei per total nuclei in control Gcn5skm−/− mice was quantified using three images per section. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 3/group. ***, P < 0.001 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. Scale bars, 50 μm.
To further examine susceptibility to muscle damage and sarcolemma stability in 6-mo-old Gcn5skm−/− mice, the diaphragm, a muscle that exhibits profound progressive pathology in dystrophic animals (Stedman et al., 1991; Addicks et al., 2018), was excised and mounted on a force transducer apparatus while being physiologically maintained ex vivo to measure loss of force during repeated eccentric contractions. Ex vivo eccentric contraction of the diaphragm muscle allows real-time assessment of muscle damage by quantifying muscle force during repeated eccentric contractions (Addicks et al., 2018). To ensure diaphragm strips were correctly prepared and tolerant to repetitive noneccentric contractions, muscles were maintained in vitro for 60 min and then subjected to repeated noneccentric contractions for 12 min with no loss of force (Fig. S2 d). Diaphragm strips from Gcn5skm−/− mice exhibited a more rapid decline in muscle force during a series of eccentric contractions than that in controls (Fig. 3 d), further indicating an intrinsic susceptibility to damage caused by the loss of Gcn5.
When Gcn5skm−/− mice were aged to 12 mo, an increased proportion of centralized nuclei in the gastrocnemius and quadriceps muscles was observed, further indicating a susceptibility to muscle damage (Fig. 3 e).
A transcriptomic meta-analysis of Gcn5skm−/− muscle revealed mouse MA and human muscular dystrophy as the top correlated disease biosets
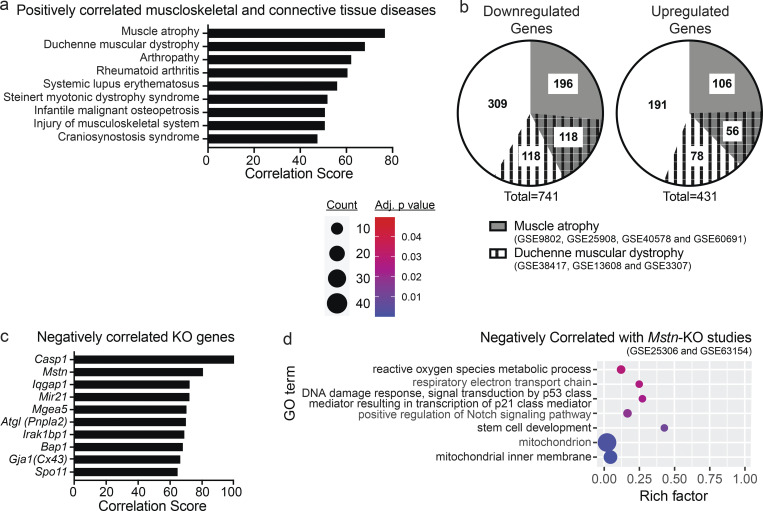
1,236 differentially expressed cDNAs from Gcn5skm−/− gastrocnemius muscles were mapped to 1,171 genes in the Illumina BaseSpace Correlation Engine (BSCE; Kupershmidt et al., 2010) and processed for further meta-analysis with a focus on highly correlated musculoskeletal diseases. The Gcn5skm−/− muscle differentially expressed transcriptomic bioset was positively correlated with nine musculoskeletal diseases, among which MA and Duchenne muscular dystrophy (DMD) had the highest correlation scores (Fig. 4 a). A targeted meta-analysis was performed between the Gcn5skm−/− bioset and 12 individual significantly correlated biosets obtained from 4 mouse MA studies (Mazzatti et al., 2008; Bialek et al., 2011; Jackman et al., 2012; Chua et al., 2015) and 3 significantly correlated biosets obtained from 3 human DMD studies (Bakay et al., 2006; Bachinski et al., 2010; Khairallah et al., 2012). Differentially expressed genes for each study were obtained and correlated with the Gcn5skm−/− bioset (Data S2). Among 741 down-regulated genes in the Gcn5skm−/− bioset, 314 (42.4%) were down-regulated in MA, 236 (31.8%) were down-regulated in DMD, and 118 (15.9%) were down-regulated in both MA and DMD (Fig. 4 b). Among 431 up-regulated genes in the Gcn5skm−/− bioset, 162 (37.6%) were up-regulated in MA, 134 (31.1%) were up-regulated in DMD, and 56 (13.0%) were up-regulated in both MA and DMD.
Figure 4.
Dysregulation of Dmd and DAPC gene expression in Gcn5skm−/− muscle is positively correlated with MA and dystrophy. (a) Bar graph showing all positively correlated musculoskeletal diseases obtained from BSCE by using all differentially expressed genes from the Gcn5skm−/− bioset as an input for the meta-analysis. (b) Pie charts representing commonality among the Gcn5skm−/−, MA, and DMD biosets following a meta-analysis for down-regulated and up-regulated genes. (c) Bar graph showing negatively correlated KO genes obtained from BSCE by using all differentially expressed genes from the Gcn5skm−/− bioset as an input. (d) Bubble plots showing the distribution and size of negatively correlated overrepresented GO pathways between the Gcn5skm−/− differentially expressed gene set and Mstn-KO gene sets (Gene Expression Omnibus accession nos. GSE25306 and GSE63154).
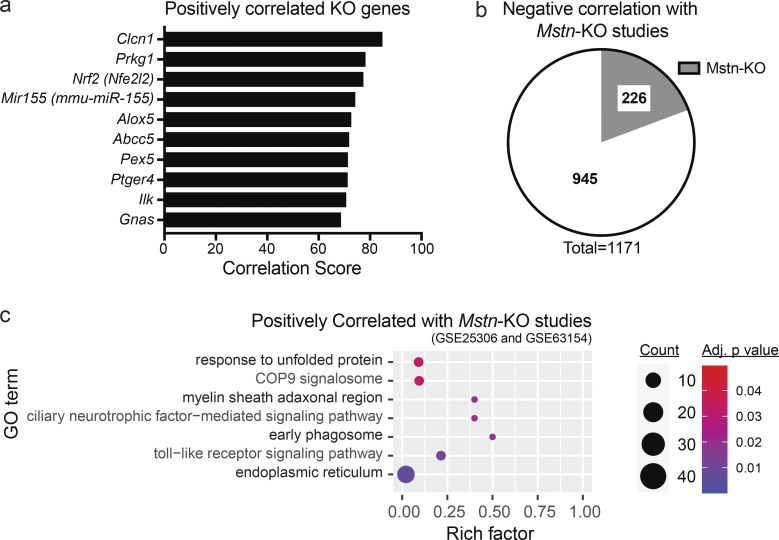
Further supporting an atrophy phenotype, a similar meta-analysis was performed that specifically compared the Gcn5skm−/− muscle bioset with all available mouse KO biosets from any tissue (Figs. 4 c and S3 a). Of those identified, the most negatively correlated bioset from muscle tissue was from a germline Mstn-KO (P < 0.0017), a mouse model that exhibits extensive muscle hypertrophy (Fig. 4 c and Data S3). Of the Gcn5skm−/− muscle bioset of 1,171 up- and down-regulated genes, 226 (19.3%) were found to be negatively correlated with germline Mstn-KO muscle biosets (Rahimov et al., 2011; Yang et al., 2015; Fig. S3 b; Data S3). GO term analysis comparing the Gcn5skm−/− bioset with Mstn-KO biosets was used to examine negatively and positively correlated pathways (Figs. 4 d and S3 c). Notably, the meta-analysis demonstrated a lower Notch signaling pathway gene set expression in the Gcn5skm−/− bioset than in the Mstn-KO biosets, a pathway known to be reduced in muscular dystrophy (Church et al., 2014; Vieira et al., 2015) and important for muscle regeneration (Bi et al., 2016). Several mitochondrial pathways were also negatively correlated, in agreement with increased mitochondrial pathways in Gcn5skm−/− muscle compared with the hypertrophic response in Mstn-KO muscle. In addition, a p53-mediated pathway was negatively correlated and down-regulated in Gcn5skm−/− muscle, which is supported by previous reports for GCN5-directed stabilization of p53. In contrast, the most positively correlated mouse KO biosets (Fig. S3 a) included the muscle-specific sodium channel Clcn1, for which mutations in the gene are associated with myotonia (Pusch, 2002); the remaining positively correlated biosets were not derived from muscle tissue.
Figure S3.
Bioinformatic analysis of the Gcn5skm−/− bioset. (a) Bar graph showing positively correlated KO genes obtained from BSCE by using all differentially expressed genes from the Gcn5skm−/− bioset as an input. (b) Pie chart representing negatively correlated genes between Gcn5skm−/− and Mstn-KO (Gene Expression Omnibus accession nos. GSE25306 and GSE63154) biosets following meta-analysis of the selected studies for down-regulated and up-regulated genes. (c) Bubble plot showing the distribution and size of positively correlated overrepresented GO pathways between the Gcn5skm−/− differentially expressed bioset and Mstn-KO study biosets (Gene Expression Omnibus accession nos. GSE25306 and GSE63154).
YY1 represses Dmd and DAPC expression in mouse myotubes and is correlated to reduced structural gene expression and muscle fiber diameters in humans
To determine the mechanism by which GCN5 might regulate the expression of Dmd and other members of the DAPC, TRANSFAC (transcription factor database; Matys et al., 2006), a manually curated database of eukaryotic transcription factors that includes their genomic binding sites and DNA binding profiles, was queried, and YY1, a Gli-Kruppel zinc finger family protein, was identified as a probable factor to regulate Dmd gene expression (Data S4). Previous work showed that YY1 negatively regulates Dmd expression in muscle myoblasts (Galvagni et al., 1998; Zanotti et al., 2015). Similarly, YY1 was shown to negatively regulate dystrophin Dp71, the smallest protein encoded by the Dmd gene, in hepatic cells (Peñuelas-Urquides et al., 2016). Finally, YY1 appeared to be a potential target of GCN5 acetylation because YY1 was previously shown to be a target of acetyltransferases p300 and p300/CBP-associated factor (PCAF; Yao et al., 2001).
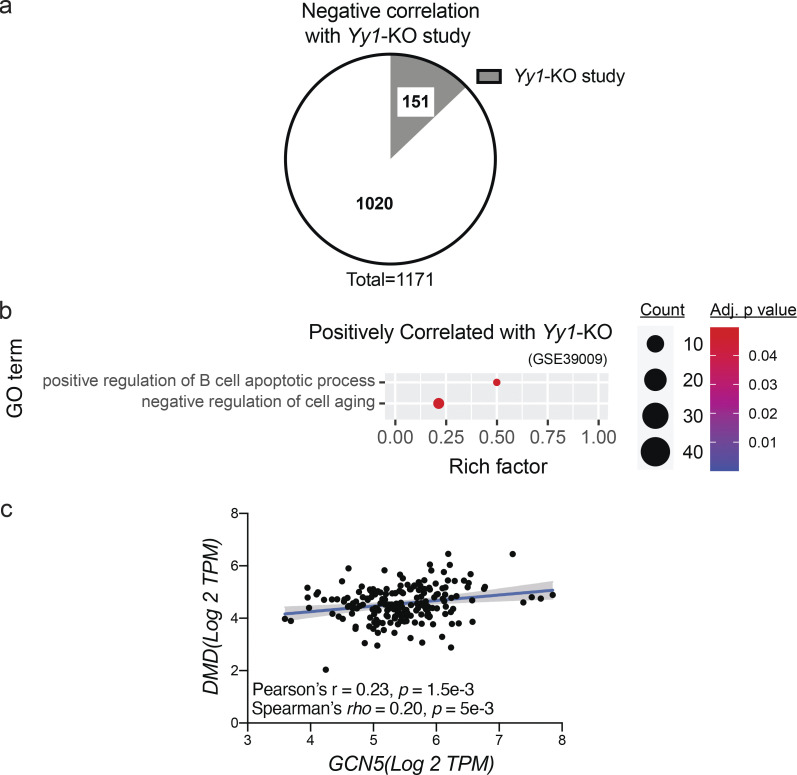
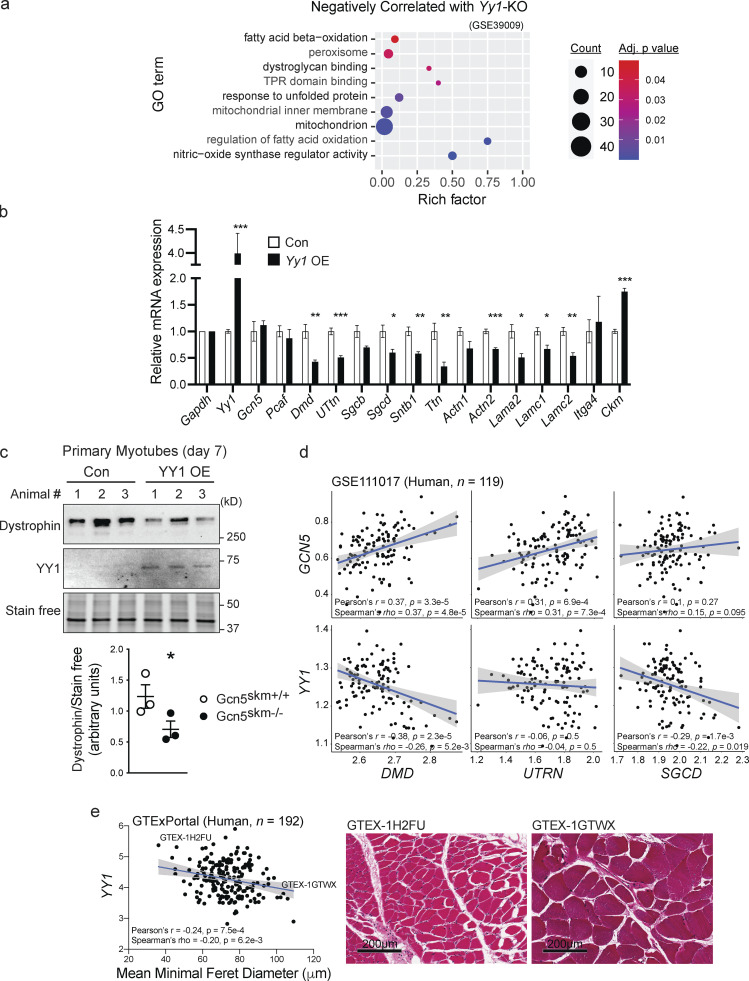
To determine if YY1 was a likely target of GCN5 control, we performed a meta-analysis using BSCE between muscle biosets of Gcn5skm−/− and muscle-specific Yy1-KO mice (Gene Expression Omnibus accession no. GSE39009; Blättler et al., 2012b). This analysis provided a significant (P = 0.0057) negative correlation, with a total of 151 (12.9%) genes being negatively correlated (Fig. S4 a; Data S5). GO term analysis through InnateDB (Breuer et al., 2013) identified dysregulated pathways that were positively (Fig. S4 b) and negatively (Fig. 5 a) correlated between the Gcn5skm−/− and muscle-specific Yy1-KO biosets (Blättler et al., 2012b). These included dystroglycan binding and NOS regulator activity, pathways that matched those reduced in the Gcn5skm−/− versus Gcn5skm+/+ bioset comparison (i.e., dystrophin-associated glycoprotein complex and NOS regulatory activity; Fig. 1 d). These results would imply that GCN5 is negatively correlated to YY1 with respect to directing changes in known markers of muscular dystrophy.
Figure S4.
Correlations between Gcn5skm−/− and Yy1-KO mouse biosets and between GCN5 and DMD gene expression in humans. (a) Pie chart representing negatively correlated genes between Gcn5skm−/− and Yy1-KO (Gene Expression Omnibus accession no. GSE39009) biosets following meta-analysis of the selected studies for down-regulated and up-regulated genes. (b) Bubble plot showing the distribution and size of positively correlated overrepresented GO pathways between the Gcn5skm−/− differentially expressed bioset and the Yy1-KO study bioset (Gene Expression Omnibus accession no. GSE39009). (c) Correlation between GCN5 and DMD gene expression as determined by RNA-seq in human skeletal muscle from the same 192 GTEx samples (GTExPortal database version 8) as in Fig. 4 e.
Figure 5.
YY1 regulates dystrophin and DAPC expression and is negatively correlated to human fiber diameter. (a) Bubble plots showing the distribution and size of negatively correlated overrepresented GO pathways between the Gcn5skm−/− differentially expressed gene set and a Yy1-KO study gene set (Gene Expression Omnibus accession no. GSE39009). TPR, tetratricopeptide repeat. (b) Relative expression of genes in differentiated primary mouse myotubes from four individual empty vector control (Con) or Yy1-overexpressing (Yy1 OE) lentivirus-transduced colonies for Gcn5, Pcaf, DAPC, cell surface, and ECM protein–expressing genes (Dmd, Utrn, Sgcb, Sgcd, Sntb1, Actn1, Actn2, Lama2, Lamc1, Lamc2, Itga4) and muscle-specific genes Ttn and Ckm as determined by RT-qPCR. n = 4 (from four independent colonies selected after lentiviral transduction). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (c) Western blot of dystrophin protein from empty vector control (Con) and YY1-overexpressing (YY1 OE) mouse primary myotubes and stain-free protein loading control. Data are presented as mean ± SEM for controls and Gcn5skm−/− mice. n = 3/group. *, P < 0.05 versus Gcn5skm+/+ as measured by two-tailed Student’s t test. (d) Scatterplots showing correlations of either GCN5 or YY1 transcripts (Z-score of TMM-normalized TPM) with three representative structural muscle transcripts (DMD, UTRN, and SGCD) in human skeletal muscle transcriptome (Gene Expression Omnibus accession no. GSE111017; n = 119). (e) Correlation between mean minimum Feret diameter of human muscle fibers, consisting of 250 measured muscle fibers from 192 GTEx samples (GTExPortal database version 8) and YY1 (Log2 TPM) transcript expression. Representative images of GTEx muscle samples are shown. Scale bars, 200 μm.
To investigate the potential negative relationship between GCN5 and YY1 on the expression of Dmd and other DAPC members, YY1 was overexpressed in primary mouse myotubes in cell culture, and gene expression was assessed through qPCR (Fig. 5 b). Notably, several genes that were down-regulated in Gcn5skm−/− muscle, including Dmd, Sgcb, Sgcd, and Ttn (Fig. 1 c), were also down-regulated by exogenous YY1 in primary myotubes. Overexpression of YY1 did not affect expression of GCN5 or PCAF or muscle-specific creatine kinase (Ckm). Western blotting also confirmed that dystrophin expression is decreased in primary mouse myotubes with exogenous YY1 expression (Fig. 5 c), which is complemented by our observed reduction in myotube dystrophin expression with Gcn5 knockdown (Fig. 2 a).
In the human muscle transcriptome (Gene Expression Omnibus accession no. GSE111017; n = 119), the correlations between transcripts for muscle structural proteins (i.e., DMD, UTRN, and SGCD) and either GCN5 or YY1 were compared. GCN5 exhibits positive correlations with three representative transcripts, DMD, UTRN, and SGCD, although SGCD does not reach statistical significance. Conversely, YY1 shows negative correlations with three genes, but UTRN did not reach significance (Fig. 5 d). Finally, corroborating the positive correlation between Gcn5Skm−/− muscle and MA biosets (Fig. 4, a and b), in human muscle (GTExPortal database version 8; n = 192), YY1 transcript expression was negatively correlated to the mean minimum Feret diameter of muscle fibers (Fig. 5 e). This dataset also showed a positive correlation between GCN5 and DMD transcript expression (Fig. S4 c). These human transcriptomic and phenotype correlations drove us to hypothesize that GCN5 may acetylate and inhibit YY1 to maintain Dmd and DAPC component expression for the maintenance of muscle integrity.
YY1 is an acetylation target of GCN5
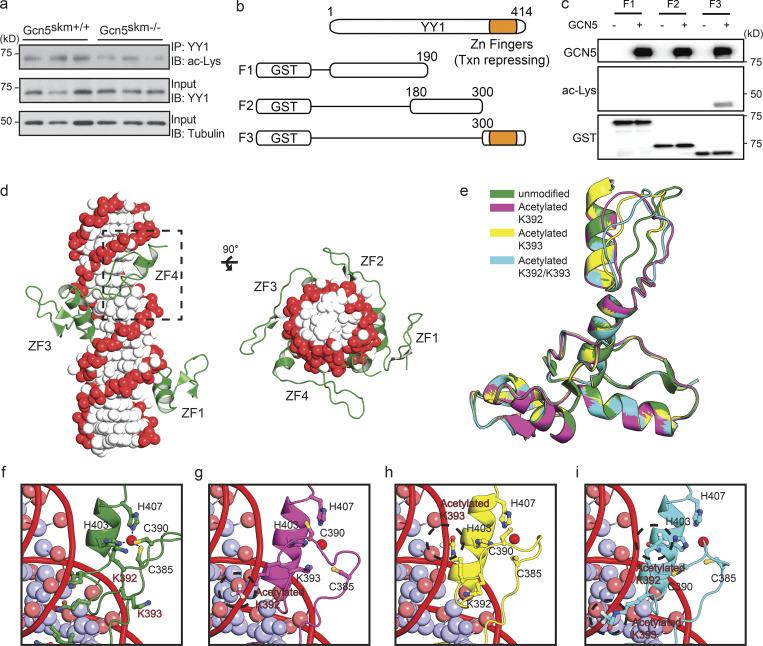
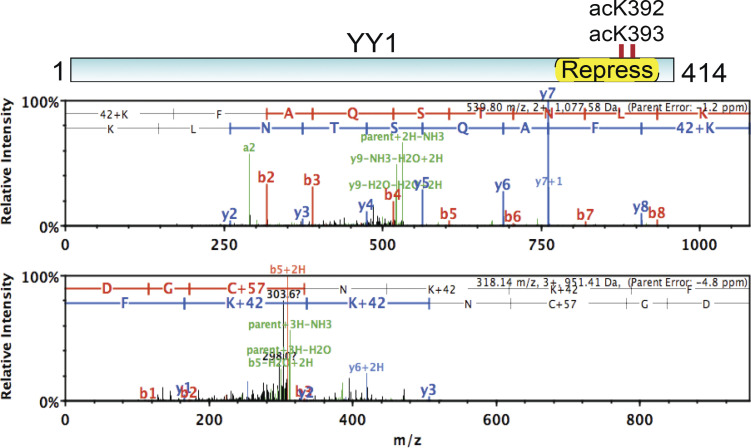
Given that YY1 is known to be acetylated by p300 and PCAF (Yao et al., 2001), we predicted that YY1 may be a target for GCN5 acetylation and regulation. To determine if GCN5 affects YY1 acetylation, YY1 was immunoprecipitated from muscle extracts and probed with anti-acetyl lysine antibody, which revealed decreased acetylation of YY1 protein in Gcn5skm−/− extracts compared with Gcn5skm+/+ controls (Fig. 6 a). This result was further explored in vitro by creating GST-labeled YY1 fragments (F1, aa 1–190; F2, aa 180–300; F3, aa 300–414; Fig. 6 b) to help determine the relative location of GCN5-directed YY1 acetylation. In vitro acetylation of YY1 fragments with GCN5 identified acetylation sites on the C-terminal region of YY1, which includes zinc finger motifs of the YY1 DNA binding domain (F3, aa 300–414; Fig. 6 c). Using nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) of in vitro GCN5 acetylated YY1, we identified two novel acetylation sites (K392 and K393; Fig. S5). A 3D structural analysis of the YY1 DNA binding domain revealed that K392 and K393 reside in an unstructured region of zinc finger 4 outside of the zinc binding site (Houbaviy et al., 1996; Fig. 6, d–i). This model predicts that the positively charged lysine residues of K392/3 interact with the negatively charged phosphate backbone of bound DNA, thereby stabilizing DNA binding in a manner similar to that of lysine residues found in histone proteins. As a result, in this model, neutralization of the positive charge of K392/3 through acetylation could destabilize binding of YY1 to DNA.
Figure 6.
GCN5 acetylates YY1 and alters a predicted model of YY1 binding to DNA. (a) Western blot (IB) of anti-YY1 immunoprecipitated (IP) from controls and Gcn5skm−/− mice probed with anti-acetyl lysine (ac-Lys; top). Control blots for IP YY1 probed with anti-YY1 (middle) and total protein input for IP probed with anti-tubulin (bottom). (b) Schematic showing GST-tagged truncated YY1 used for in vitro acetylation (Fig. 6 c) and MS analysis. (c) In vitro acetylation assay of the in vitro synthesized GST-YY1 fragments incubated with GCN5. YY1 fragments were incubated alone or with GCN5 (top). Acetylation of YY1-fragment (aa 300–414) detected with anti-acetyl lysine (ac-Lys; middle). Input YY1 fragments detected by anti-GST antibody (bottom). (d) Modeled structure of the YY1 zinc finger region bound to DNA. The DNA phosphate backbone and bases are colored red and white, respectively. The human YY1 protein is represented in green. The fourth zinc finger domain is boxed with a dotted line. (e–i) Modeling of the unmodified or acetylated (K392, K393, or K392/393) forms of the fourth zinc finger domain of YY1 and interactions with DNA. The residues involved in coordinating zinc ion, C385, C390, H403, and H407 are shown. The zinc ions are represented by red spheres. The acetylated forms of K392 and K393 are circled with a dotted line.
Figure S5.
Nano-LC-MS/MS analysis showing GCN5-dependent in vitro acetylation of acK392 and acK393 of YY1. m/z, mass-to-charge ratio.
Acetylation of YY1 at K392 and K393 inhibits DNA binding and repression of Dmd
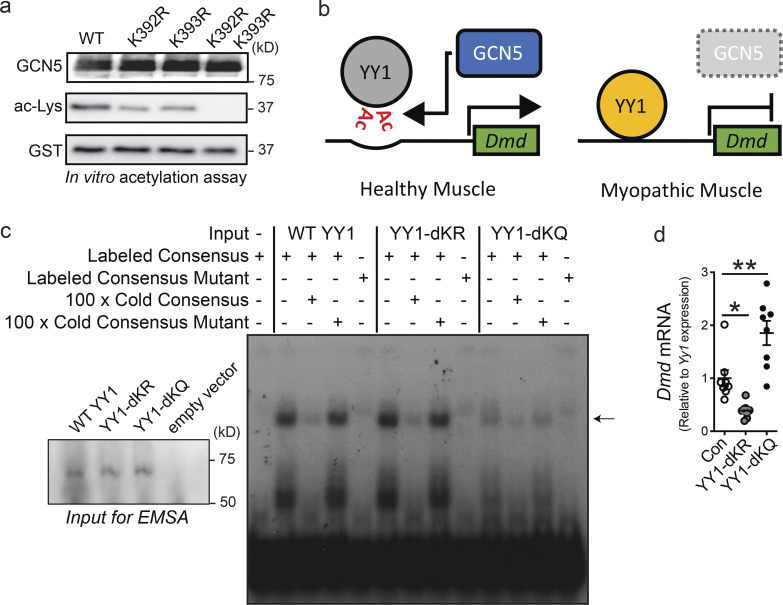
Through the use of in vitro acetylation assays of truncated YY1 fragments, p300 was shown to acetylate YY1 at a site between aa 170 and 200 (Yao et al., 2001). This same work found that PCAF acetylates YY1 at two residues located between aa 261 and 414 (Yao et al., 2001); however, the precise location of acetylation by PCAF was not determined. To expand on our nano-LC-MS/MS analysis and confirm that K392 and K393 are the main acetylation sites of GCN5, both lysines, independently and in tandem, were converted to arginines through site-directed mutagenesis and subjected to in vitro acetylation by GCN5. Mutation of individual lysines resulted in decreased acetylation compared with the WT fragment, while mutation of both lysines resulted in a complete loss of acetylation in vitro (Fig. 7 a).
Figure 7.
Acetylation suppresses YY1 DNA binding and inhibition of Dmd gene expression. (a) Western blot of in vitro acetylation by GCN5 of WT, YY1 (Con), or YY1 with lysines 292 and 293 mutated to arginine individually (K392R or K393R) or together (K392R/K393R) probed with anti-acetyl lysine (ac-Lys). (b) Mechanism for the regulation of YY1 activity by GCN5. Under normal conditions, GCN5 negatively regulates YY1 through acetylation (Ac), and YY1 does not repress Dmd gene expression or other targets. In the absence of GCN5 regulation, YY1 abnormally represses Dmd and other genes, resulting in a dystrophic phenotype. (c) EMSA of in vitro synthesized control YY1 (Con) or YY1 with lysines 392 and 393 mutated to arginine (YY1-dKR) or glutamine (YY1-dKQ). YY1 or YY1 mutants were incubated with radioactively labeled oligonucleotide dimers with consensus YY1 binding site (YY1 Consensus Labeled; lanes 2, 6, and 10), radioactively labeled YY1 consensus oligonucleotide dimers with 100-fold excess unlabeled YY1 consensus oligonucleotide dimers (100 × Cold Consensus; lanes 3, 7, and 11) or with 100-fold excess unlabeled mutated YY1 consensus oligonucleotide dimers (100 × Cold Mutant; lanes 4, 8, and 12), or with labeled mutated YY1 consensus oligonucleotide dimers (YY1 Mutant Labeled; lanes 5, 9, and 13). As a negative control for YY1 consensus binding, transcription-translation reaction without DNA was incubated with radioactively labeled YY1 consensus oligonucleotide dimers (lane 1). Bound oligonucleotide is marked by an arrow. Western blot of in vitro transcribed WT YY1, YY1 392/392 mutants, and empty vector control in inset (lower left) showing uniform expression of YY1 control and mutant proteins used for EMSAs. (d) Dmd mRNA expression in C2C12 myotubes after 4 d of differentiation and transfected for 48 h with FLAG-tagged YY1, YY1-dKR (K392R and K393R), or FLAG-YY1-dKQ (K392Q and K393Q). Data are presented as mean ± SEM. n = 8/group. *, P < 0.05; **, P < 0.01 by ANOVA with Tukey’s post hoc test versus YY1 control.
Because the modeled YY1 DNA structure suggests that neutralization of positive charges on K392 and K393 through acetylation may result in inhibition of DNA binding by YY1, we postulated that GCN5-directed acetylation of YY1 destabilizes the YY1-directed inhibition of Dmd expression (Fig. 7 b). To test whether acetylation of YY1 affects DNA binding, in vitro transcribed full-length WT YY1 and full-length YY1 mutants, which include K392 and K393 tandem mutations that mimic varied acetylation states, were used in electrophoretic mobility shift assays (EMSAs) with labeled YY1 binding consensus oligonucleotide dimers. In agreement with our hypothesis, mutating both K392 and K393 to the uncharged acetylation mimic glutamine (YY1-dKQ) resulted in a loss of DNA binding, while mutating both lysines to the negatively charged arginine (YY1-dKR) retained DNA binding activity (Fig. 7 c). To further establish a role for YY1 in the regulation of Dmd expression, WT YY1 and YY1 mutants were exogenously expressed in C2C12 myoblasts and differentiated for 48 h, after which cells were collected for RT-qPCR analysis. In alignment with our EMSA results, the YY1-dKR mutant, which cannot be acetylated but maintains the negative charge of the WT lysine residues, resulted in decreased Dmd expression (Fig. 7 d). The YY1-dKQ mutant, which does not bind DNA, resulted in increased Dmd expression, likely through a dominant negative effect.
Conclusions
To examine the physiological role of GCN5 in postmitotic skeletal muscle, we crossed Gcn5L2/L2 mice with mice expressing Cre recombinase driven by a transgenic Hsa gene promoter. Initial transcriptomic analysis of the resulting Gcn5skm−/− mice identified reductions in the expression of dystrophin and other DAPC members. Similar to mouse models of muscular dystrophy, such as mdx mice that lack dystrophin expression, GCN5 deficiency in skeletal muscle leads to muscle myopathy that is primarily identified by a susceptibility to eccentric muscle damage.
While GCN5 is recognized as a general regulator of transcription, loss of GCN5 did not appear to have an effect on global transcription, likely due to redundancy with other histone acetyltransferases (Dent et al., 2017). An unbiased analysis of our transcriptomic dataset found multiple down-regulated GO terms related to muscular dystrophy, MA, and aging. This was then further illustrated by a meta-analysis using BSCE which determined that Gcn5Skm−/− muscle best correlated to biosets of rodent MA and human DMD when comparing our bioset with all publicly available data categorized by musculoskeletal and connective tissue diseases. Gcn5Skm−/− muscle exhibited reduced expression of several critical muscle-specific gene transcripts, with the most striking reduction being that of dystrophin. The links between Gcn5Skm−/− and muscular disease were further emphasized by observed negative correlations between the Gcn5Skm−/− bioset and that of KO mice for Mstn, Cx43, and Mir-21, genes that have been linked to playing a negative role in mdx mouse phenotypes (Nouet et al., 2020; Zanotti et al., 2015; Ardite et al., 2012; Dumonceaux et al., 2010; Fig. 4 c). The finding that the Gcn5Skm−/− bioset is negatively correlated with several Mstn-KO biosets complements findings that suggest myostatin pathway inhibition as a potential DMD therapeutic strategy (Campbell et al., 2017). Concomitant with gene expression changes, Gcn5Skm−/− muscle also displayed a myopathic phenotype. Gcn5Skm−/− muscle demonstrated increased fragility with eccentric contraction, as evidenced by release of muscle-specific creatine kinase into the circulation, uptake of EBD into the muscle from the circulation, a histological muscle damage phenotype, and up-regulation of regenerative gene expression, as is also seen in muscle pathology. Using a gold standard assay to assess phenotype in mouse models of muscular dystrophy where muscle integrity is perturbed, we discovered a reduction in diaphragm contraction force produced during repeated eccentric contractions in Gcn5Skm−/− muscle, confirming a hallmark phenotype of mdx mice (Addicks et al., 2018). The data therefore show that GCN5 modulates dystrophin expression and maintains muscle integrity.
Because Dmd was among the most strongly down-regulated genes in Gcn5Skm−/− muscle, we next aimed to determine how GCN5 might modulate Dmd expression and focused on YY1, a known regulator of Dmd (Galvagni et al., 1998). YY1 has been implicated as both a transcriptional repressor, as part of polycomb repressive complex 2, and a transcriptional activator in other contexts (Shi et al., 1997). The Yy1 KO results in embryonic lethality at embryonic day 10.5 due to failure to properly form mesoderm (Donohoe et al., 1999). The muscle-specific KO of Yy1 results in hyperactivation of insulin/insulin-like growth factor signaling, increased insulin/insulin-like growth factor pathway gene expression, and defective mitochondrial morphology and oxidative function (Blättler et al., 2012a, 2012b). Upon further analysis of our Gcn5Skm−/− muscle gene set, the DAPC constituent dystroglycan binding GO term was found to be negatively correlated between genes that are differentially expressed between Yy1-KO muscle and control (Blättler et al., 2012b) and genes that are differentially expressed between Gcn5Skm−/− muscle and control. Supporting this association, overexpression of YY1 in myotubes resulted in decreased expression of Dmd and other DAPC members. Importantly, it was previously found that YY1 is acetylated by both p300 and the GCN5 homologue PCAF and that PCAF-based acetylation resulted in decreased DNA binding of YY1 (Yao et al., 2001). We also found that YY1 acetylation was strongly decreased in Gcn5Skm−/− muscle, suggesting that YY1 is acetylated by GCN5 in vivo and may modulate DNA binding. We then demonstrated that YY1 gene expression was negatively correlated to muscle fiber size in data from the human Genotype-Tissue Expression (GTEx) consortium, indicating that YY1 may have a negative impact on muscle integrity in humans, as we have shown phenotypically in Gcn5Skm−/− mice.
To specifically address whether GCN5 regulates the YY1 via acetylation, we performed an in vitro analysis that demonstrated GCN5-directed acetylation of YY1 at K392 and K393, which are known to locate within the C-terminal zinc finger of four zinc finger DNA binding sites on YY1 (Houbaviy et al., 1996). Indeed, EMSAs performed using control YY1 and YY1 mutants, mimicking acetylated and unacetylated K392/K393, revealed that mutation of the negatively charged lysine residues to uncharged glutamine acetylation mimics resulted in a strong disruption of DNA binding; mutation to negatively charged arginine, which is not acetylated by GCN5 and mimics unacetylated lysine, resulted in stronger DNA binding. This supports the conclusion that acetylation of YY1 by GCN5 at K392/K393 disrupts DNA binding. Indeed, when YY1 mutants were expressed in myotubes, mutation of K392/K393 to glutamine resulted in increased Dmd transcript expression, while mutation to arginine resulted in repression of Dmd expression.
KDAC inhibitors have been found to have beneficial effects for several types of muscle disorders (Minetti et al., 2006; Colussi et al., 2008; Consalvi et al., 2011; Johnson et al., 2013; Pambianco et al., 2016) and have led to phase 2 and phase 3 clinical trials that are currently underway for several forms of muscular dystrophy (Consalvi et al., 2013; Bettica et al., 2016). Our findings might suggest that the YY1 acetylation state in muscle could be maintained by histone deacetylase inhibition to support muscle integrity as a potential additional route of treatment effectiveness. These findings therefore advocate that considerable opportunity remains for a better understanding of the dynamic effects of protein acetylation in the regulation of muscle integrity for the potential treatment of muscle-wasting diseases.
This work identifies a novel role for GCN5 as a regulator of gene expression through acetylation of YY1. As YY1 is a transcriptional regulator (Thomas and Seto, 1999; Meliala et al., 2020), acetylation and repression of YY1 DNA binding may also act to complement the known role of GCN5 in positive regulation of gene expression through acetylation of histones (Grant et al., 1997). Importantly, loss of GCN5 and its repressive acetylation of YY1 at K392/K393 results in YY1-based repression of genes that have critical roles in muscle, most importantly dystrophin, resulting in a phenotype that exhibits some similarities to muscular dystrophy. These findings may be useful for the discovery of new therapeutics for maintaining healthy muscle during muscular dystrophy and other diseases.
Materials and methods
Generation of Gcn5skm−/− mice
For the generation of Gcn5 floxed (Gcn5L2/L2) mice that possess loxP sites flanking exons 5 and 6 of the Gcn5 (KAT2A) gene, genomic DNA covering the Gcn5 locus was amplified from the 129Sv strain by using high-fidelity PCR. The resulting DNA fragments were assembled into the targeting vector designed to insert a loxP site upstream of exon 5 and a second loxP site, followed by an frt-flanked neomycin selection cassette, downstream of exon 6 of the Gcn5 gene. The construct was then electroporated into 129Sv embryonic stem cells. The karyotype was verified, several correctly targeted embryonic stem cell clones were injected into blastocysts from C57BL/6J mice, and resulting chimeric males had the neocassette removed by breeding with ACTFLPe mice on a C57BL/6J background. Progeny were crossed to remove the Flp-expressing transgene, and mice were subsequently bred to C57BL/6J mice for at least 10 generations. Offspring that transmitted the mutated allele, in which the selection marker was excised, and that lost the Flp-expressing transgene (Gcn5L2/WT mice) were selected, mated with HSA-Cre mice, and then further intercrossed to generate HSAcreTg/0/Gcn5L2/L2 mice. A PCR genotyping strategy was subsequently used to identify HSAcreTg/0/Gcn5L2/L2 and HSAcre0/0/Gcn5L2/L2 mice.
General animal phenotyping
Animal experiments were approved by the ethics committee at the University of Ottawa with permit identifier ME-2804 and by the ethics committee of the canton of Vaud, Switzerland, with permit identifier 2285. Phenotyping experiments were performed with validated Eumorphia/EMPReSS standard operating protocols (https://www.eumorphia.org). All mice were maintained in a temperature-controlled (23°C) facility with a 12-h/12-h light/dark cycle with ad libitum access to food and water. A regular chow diet was obtained from Harlan Teklad (diet 2018) and contained 18.6% protein, 44.2% carbohydrate, and 6.2% fat. Body weight (BW) was measured from 8 to 25 wk of age. The mice were fasted 4 h before blood was harvested for subsequent blood measurements and tissues were harvested for protein or RNA isolation and histology (Champy et al., 2004, 2008).
An endurance test was performed using a variable speed belt treadmill enclosed in a plexiglass chamber with a stimulus device consisting of a shock grid attached to the rear of the belt (Panlab). The initial velocity of the belt was 5 m/min with an incline of 5°. The speed was gradually increased by 2 m/min every 12 min. Exhaustion was assumed when mice received more than five shocks (0.1 mA) over a period of 1 min. The test was considered completed after 2 h of running. The distance traveled and time before exhaustion were recorded as maximal running distance and period.
Nonexhaustive downhill running experiments
Mice were placed in a treadmill habituated to a variable speed belt treadmill enclosed in a plexiglass chamber with a stimulus device consisting of a shock grid (Panlab) for 5 min each day for 3 d. On the day of the run, the mice were placed in the treadmill 5 min before the start of the run. The mice were run at a −15° incline (downhill) as follows: 8 m/min for 5 min, 10 m/min for 5 min, and 12 m/min for 30 min. Animals were rested for 5 min, then a second bout was started at 8 m/min for 1 min, 10 m/min for 1 min, and 12 m/min for 28 min. Animals were then rested for another 5 min before their last bout at 8 m/min for 1 min, 10 m/min for 1 min, and 12 m/min for 13 min. All mice completed the run. Mice were injected with EBD 15 min after completion of the run and sacrificed 24 h after the run (as described below).
EBD assessment of muscle damage
Muscle damage was assessed by injecting a 1% solution of EBD into the peritoneal cavity using 1% volume to BW 24 h before sacrifice. EBD was dissolved into PBS and sterilized using filters with a 0.2-µm pore size. Muscle sections from EBD-injected animals were incubated in ice-cold acetone at −20°C for 10 min, washed three times for 10 min with PBS, and mounted with VECTASHIELD Mounting Medium. Microscopic images of fluorescence from EBD-positive muscle fibers were analyzed using ImageJ software.
Eccentric contraction of diaphragm muscle
Eccentric contraction of diaphragm muscle was performed as previously described (Addicks et al., 2018). Briefly, triangular strips were prepared from diaphragm muscles, mounted on a custom-built force measurement apparatus, and maintained under flowing physiological solution (118.5 mM NaCl, 4.7 mM KCl, 2.4 mM CaCl2, 3.1 mM MgCl2, 25 mM NaHCO3, 2 mM NaH2PO4, and 5.5 mM D-glucose, bubbled with 95% O2 and 5% CO2). Muscle mounting was adjusted to generate maximal isometric force when stimulated by a 400-ms train of 0.3-ms square electrical pulses at 10 V and 180 Hz (701C, Electrical Stimulator; Aurora Scientific). Force generation was measured at 5 kHz using a force transducer servomotor system (0.5 N force, 10-mm excursion, 300C, Dual-Mode Muscle Levers; Aurora Scientific). Diaphragm muscle was equilibrated by maintenance in physiological solution for 30 min with stimulated contractions every 100 s, resulting in no decrease in contraction force over the 30-min period. After the 30-min equilibration, muscles were subjected to eccentric contractions by stimulating as above while stretching by 10% of muscle length over a 200-ms period during the last 200 ms of each contraction. Muscle force generation was recorded during a series of 12 eccentric contractions.
Histology
Muscle tissues were harvested from anesthetized mice and immediately frozen in Tissue-TEK O.C.T. compound (Sakura Finetek) using nitrogen-cooled isopentane for 2 min before being stored at −80°C. 8-µm cryosections were fixed with 4% PFA and stained with hematoxylin and eosin, Picrosirius red, or antibodies. For Picrosirius red staining, sections were prepared as previously described (Smith et al., 2016). For immunostaining, sections were fixed in a 4% PFA solution for 10 min, washed with PBS with 0.1% Tween 20 (PBST), and blocked using PBST containing 2% BSA and 5% goat serum for 30 min at RT. This was followed by an overnight application of rabbit anti-dystrophin primary antibody (ab15277; Abcam) at 4°C, followed by washing in PBST and incubation with a secondary anti-rabbit antibody (Life Technologies) or with goat anti-mouse IgM (heavy chain) cross-adsorbed secondary antibody (Alexa Fluor 488A, 21042; Thermo Fisher Scientific). After washing in PBST, tissue sections were mounted using Dako mounting medium.
Creatine kinase measurement
Collected plasma was used for creatine kinase measurements using the Creatine Kinase Flex Reagent Cartridge (Siemens Healthcare Diagnostics), along with creatinine and total protein measurements on the Dimension Xpand Plus Instrument (Siemens Healthcare Diagnostics).
Microscopy
For Fig. 2, d and e, and Fig. 3 a, images were obtained using a Zeiss Celldiscoverer 7 imaging system. Images were obtained using an Axiocam 506 camera and a Plan Apochromat 20×/0.7 NA objective with a 1× tube lens. Emission wavelength/excitation filters used were DAPI, 370 nm–400 nm/465 nm; Alexa Fluor 488, 450 nm–488 nm/517 nm; Alexa Fluor 594, 540 nm–570 nm/594 nm; and Alexa Fluor 647 (EBD), 615 nm–648 nm/668 nm. At least nine focal points were made for each sample, after which individual images were automatically obtained and stitched with the microscope under the control of the Zen acquisition program. For each series of images, settings were reused across samples. All images were taken at RT in the absence of imaging medium (air objectives). Figs. 2 d and 3 a images were unprocessed.
EM
EDL muscles were isolated from male 6-mo-old mice and placed in glass vials containing an RT fixative of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer at pH 7.4 (16537-15; Cedarlane). The vials were placed on a rocker at RT for 30 min, then incubated at 4°C overnight. The muscles were then placed in 1.0% osmium tetroxide for 60 min, rinsed with 0.2 M sodium cacodylate, dehydrated in a series of ethanol solutions, followed by propylene oxide, and then embedded in epoxy resin. Resin blocks were used to create 500-nm sections using an ultramicrotome and stained with toluidine blue. Stained slides were reviewed using light microscopy to identify an area for screening, at which point the resin block was trimmed to a 1-mm hexagonal shape around the tissue of interest. Thin, 60–90-nm sections were created on an ultramicrotome using a diamond knife. The sections were then stained on mesh grids using uranyl nitrate and lead citrate. Images of recognizable sarcomeres for each sample were then acquired on a JEOL JEM-1400Plus 60-kV transmission electron microscope with an indicated magnification of 8,000× and actual magnification of 10,952×.
Genomic profiling
Gene regulatory networks were captured by acquiring mRNA expression patterns using microarrays (Affymetrix Mouse Gene 1.0 ST platform; Zhang et al., 2010) on four samples of gastrocnemius muscles taken from each experimental group. Male 6-mo-old mice were fasted for 5 h before sacrifice. Gastrocnemius skeletal muscle was immediately excised and snap frozen in liquid nitrogen for RNA extraction. TRIzol extraction of total RNA was performed for each sample. Sample concentrations were then measured by NanoDrop and cleaned up with a Qiagen RNeasy kit before preparation for the microarray. Raw microarray data are publicly available in the National Center for Biotechnology Information Gene Expression Omnibus database under accession no. GSE158883.
Correlation engine
To validate our results and find associations by matching and integrating our transcriptomic data with publicly available genomic knowledge, we used Illumina BSCE (https://sapac.illumina.com/products/by-type/informatics-products/basespace-correlation-engine.html). The Gcn5skm−/− versus WT differentially expressed gene set was used as the input bioset and correlated to the 142,219 ontologically tagged biosets comprising 23,529 curated studies on June 29, 2020 (Kupershmidt et al., 2010).
Bioset data stored in the BSCE undergo several preprocessing, quality control, and organization stages. Quality examinations ensure the integrity of the samples and datasets and include evaluations of pre- and postnormalization boxplots, missing value counts, and P value histograms after statistical testing with a false discovery rate analysis to establish whether the number of significantly altered genes is larger than the number anticipated by chance.
The rank-based, nonparametric Running Fisher algorithm implemented within BSCE was used to compare the transcriptomic bioset derived from Gcn5skm−/− versus WT mice with other transcriptomic biosets in the database. The Running Fisher algorithm computes the statistical significance of similarity between ranked fold change values of two bioset gene lists using Fisher’s exact test. This normalized ranking approach enables comparability across data from different studies, platforms, and analysis methods.
Based on the bioset–bioset correlations of disease versus normal experimental designs (31,955 biosets) and associated phenotype tags, the BSCE Disease Atlas was used to identify highly correlated musculoskeletal diseases. The BSCE Knockout Atlas was used to identify highly correlated transcriptomic profiles for available gene KO models. The BSCE Meta-Analysis application was used to aggregate, score, and rank genes across the Gcn5skm−/− versus WT bioset and select highly ranked musculoskeletal disease biosets in BSCE curated studies to assess common and differential RNA expression activities. Overrepresentation GO pathway analysis was performed for the Gcn5skm−/− differentially expressed gene set and the gene sets obtained following meta-analysis (InnateDB; Breuer et al., 2013).
Human minimum Feret diameter and gene expression correlation
The correlation analysis in 119 human skeletal muscles was performed using an RNA-sequencing (RNA-seq) dataset belonging to National Center for Biotechnology Information Gene Expression Omnibus accession no. GSE111017 (Migliavacca et al., 2019). All downloaded datasets were normalized using the trimmed mean of M-values (TMM) normalization method. Finally, all data were converted to Z-scores to avoid batch effects. Pathway enrichment plots and scatterplots were plotted using the ggplot2 R package as the Z-scores of TMM-normalized transcripts per million (TPM).
Images of human cadaver muscle sections and RNA-seq data for corresponding muscle tissue were obtained from publicly available datasets as part of the human GTEx consortium (GTExPortal database version 8). Fiber size quantification was performed in ImageJ for 192 images (∼25% of all images). A total of 250 fibers were measured for minimum Feret diameter per image. The mean minimum Feret diameter for each image was correlated with corresponding YY1 mRNA expression (Log2 TPM). GTEx expression data for YY1 is available at http://www.gtexportal.org/home/gene/YY1. GTEx histology images and pathological notes are available using the GTEx Histology Image Viewer (https://gtexportal.org/home/histologyPage).
EMSA
YY1 and YY1 with lysines 392 and 393 mutated to arginine or glutamine were expressed from plasmid DNA using the TnT Quick Coupled Transcription/Translation System (L1170; Promega) according to the manufacturer’s protocol. Oligonucleotides containing YY1 binding sites (5′-CGCTCCCCGGCCATCTTGGCGGCTGGTGG-3′, 5′-CCACCAGCCGCCAAGATGGCCGGGGAGCG-3′) or mutated sites (5′-CGCTCCGCGATTATCTTGGCGGCTGGTGG-3′, 5′-CCACCAGCCGCCAAGATAATCGCGGAGCG-3′) were suspended in water and annealed by heating to 95°C, then cooling to 21°C at a rate of 0.5°C per minute in a thermocycler. Radiolabeled oligonucleotide dimers were prepared by incubating annealed oligonucleotides with 32P-ATP in the presence of T4 PNK (M0201; New England Biolabs) according to the manufacturer’s protocol and purified using Micro Bio-Spin P-30 Tris Chromatography Columns (7326250; Bio-Rad Laboratories). YY1 was bound to oligonucleotide dimers in a solution of 10 mM Tris, pH 7.5, 60 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 10% glycerol, 0.2 mg/ml BSA, and 50% vol/vol of reticulate lysate reaction in a 15-μl reaction. For each reaction, 50 fmol of labeled probe was added. 5 pmol of unlabeled dimers were added to specified reactions. All reactions also contained 0.1 µg of sonicated salmon sperm DNA (15632011; Thermo Fisher Scientific) to reduce nonspecific binding. Binding reactions were incubated at 25°C for 30 min and loaded directly on a 6% acrylamide nondenaturing Tris-borate gel in the absence of EDTA. Gels were run at 60 V for 1 h, backed with Whatman paper, and dried for 2 h at 80°C in a gel dryer under a vacuum. Dried gels were exposed to film at −80°C overnight with an intensifying screen.
YY1 overexpression in myoblasts
YY1 retrovirus was prepared by transfecting 293T cells with plasmids containing YY1, subcloned from pCMV6-YY1 (Origine #MR206531), under control of the EF1a promoter (parent plasmid was pLV-EF1a-IRES-Neo, a gift from Tobias Meyer, Stanford University School of Medicine, Stanford, CA; Addgene plasmid 85139; http://n2t.net/addgene:85139; RRID:Addgene_85139) and helper plasmids pMDL, pREV, and pVSVG using the calcium chloride method. Myoblasts were infected with an MOI of ∼0.001 by adding viral supernatant at a 1:1 ratio to growth media and incubated overnight in a 35-mm dish. Myoblasts were split to 2 × 10–cm dishes and, after a further 24 h, were selected for neomycin expression by addition of G418. After 4–5 d, isolated individual colonies were transferred to wells in a 48-well dish, expanded, and assessed for YY1 expression. The four highest YY1- and neomycin-expressing colonies (and neomycin-expressing controls), as determined by qPCR, were plated into 35-mm wells and grown to 33% confluence, transferred to differentiation medium (DMEM with 5% horse serum) for 7 d, and collected for qPCR and Western blot analysis.
RNA isolation and qPCR
Cells were collected by trypsinization and centrifugation, then washed once with 10% BSA in PBS and once with PBS. RNA was prepared using the EZ-10 Spin Column Total RNA Miniprep Kit [BS1361(SK8655); BioBasic] according to the manufacturer’s instructions. RNA was quantified using NanoDrop technology (Thermo Fisher Scientific), and, for each sample, 1 µg RNA was converted to cDNA using ProtoScript II reverse transcription (M0368; New England Biolabs) in a 20-μl reaction. 0.5 μl of reverse transcription was used per 10-μl qPCR (Luna Universal qPCR Master Mix).
Total RNA from mouse muscle was extracted using TRIzol and then transcribed to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). Expression of selected genes was analyzed using the LightCycler 480 system (Roche) and SYBR Green chemistry. All qPCR results were presented relative to the mean of 36b4, B2m, and/or Gapdh (comparative cycle threshold method). Primer sequences are specified in Table 1.
Table 1. Primer sets for qRT-PCR analyses.
| Gene | Description | Forward primer | Reverse primer |
|---|---|---|---|
| 36b4 | Ribosomal protein, large, P0 | 5′-AGATTCGGGATATGCTGTTGG-3′ | 5′-AAAGCCTGGAAGAAGGAGGTC-3′ |
| B2m | β2-microglobulin | 5′-TTCTGGTGCTTGTCTCACTG-3′ | 5′-TATGTTCGGCTTCCCATTCT-3′ |
| Gapdh | Glyceraldehyde-3- phosphate dehydrogenase | 5′-TGTGTCCGTCGTGGATCTGA-3′ | 5′-CCTGCTTCACCACCTTCTTGAT-3′ |
| Dmd | Dystrophin exon 1 (dmd) 427 muscle isoform | 5′-TCTCATCGTACCTAAGCCTC-3′ | 5′-CAGTGCCTTGTTGACATTGTTCAG-3′ |
| Utrn | Utrophin | 5′-GCCCTCCCTGCAGATTATTTGG-3′ | 5′-CTGTCCAGTTGACCTTTGATACTCTTC-3′ |
| eMHC | Embryonic myosin heavy chain | 5′-TGAAGAAGGAGCAGGACAC-3′ | 5′-CACTTGGAGTTTATCCACCAGATCC-3′ |
| Maged1 | MAGE family member D1 | 5′-CAAGAGGACCCGCAAGGTT-3′ | 5′-GCCTTTGATCCCCACTGTTG-3′ |
| Yy1 | YY1 transcription factor | 5′-GAAGCAGGTGCAGATCAAGACCC-3′ | 5′-GAGAGGTCAATGCCAGGTATCCC-3′ |
| Gcn5 | KAT2A (general control of amino acid synthesis protein 5-like 2) | 5′-GGAAGGCGCAAGTCCGGG-3′ | 5′-GCTGGAGGTCCATGCGGG-3′ |
| Sgcb | β-sarcoglycan | 5′-ATCCCCATCGATGAGGACCGG-3′ | 5′-CCCATTTGGCCCAATGCGG-3′ |
| Sgcd | Sarcoglycan δ | 5′-CCACAGGAGCACCATGCCC-3′ | 5′-CTGAGTCTCCTTCTAGCTTCAGACCC-3′ |
| Sntb1 | Syntrophin β1 | 5′-ACGGAGCAGACCTGCGGG-3′ | 5′-CATCCAATCTCAGACACCGGG-3′ |
| Ttn | Titin | 5′-GCCAACAGTGGACGATACTCCC-3′ | 5′-CTCTGGATTTCGGCTCCATCCC-3′ |
| Actn1 | Actinin α1 | 5′-CCTGGATCCGGCCTGGG-3′ | 5′-TCTCTGGCTTGGCCAAGCG-3′ |
| Actn2 | Actinin α2 | 5′-CTCGACCCGGCCTGGG-3′ | 5′-TTTCCCCGGTCAGGTTTGGG-3′ |
| Lama2 | Laminin subunit α2 | 5′-CACATGGTGGCAGAGTCCC-3′ | 5′-CCAAAATCCAGTTTCCAGGCC-3′ |
| Lamc1 | Laminin subunit γ1 | 5′-CCAGACTATGCTGGCCGGG-3′ | 5′-CTGGTGTGGAACTTGAGGCG-3′ |
| Lamc2 | Laminin subunit γ2 | 5′-CAGCCTCAGTACCACGCCC-3′ | 5′-CTGCTGTGCCTTCCTTTTCCC-3′ |
| Itga4 | Integrin subunit α4 | 5′-AGGGATAACCAGTGGCTGGG-3′ | 5′-GACGTAGCAAATGCCAGTGGG-3′ |
| Ckm | Creatine kinase, M-type | 5′-GCCATGGCGGCTACAAACCC-3′ | 5′-GCGGAGGCAGAGTGTAACCC-3′ |
| Pcaf | P300/CBP-associated factor | 5′-CGAAGCTGTAGCCATGCCC-3′ | 5′-CTCAAATGGCGGCTTCTTCTCC-3′ |
Western blotting
Cells were collected by trypsinization and centrifugation, then washed once with 10% BSA in PBS and once with PBS. Cells were lysed in cold 0.5% Triton buffer (0.5% Triton X-100, 5 mM Tris, pH 7.6, and 150 mM NaCl), quantified using the Bradford method, and diluted to 0.1 mg/ml with Triton buffer. 5 µg of protein was electrophoresed on a 6% SDS-PAGE gel with 0.05% 2,2,2-trichloroethanol and imaged UV stain free. Proteins were blotted to polyvinylidene difluoride membranes blocked with milk and detected using mouse anti-dystrophin (MAB1645; EMD Millipore), anti-HSP90 (BD Biosciences), anti-YY1 (H-10; Santa Cruz Biotechnology), anti-acetylated lysine (9441; Cell Signaling Technology), anti-GCN5 (sc-20698; Santa Cruz Biotechnology), anti-GST (ab9085; Abcam), anti-nNOS (61-700; Thermo Fisher Scientific), anti–α-dystroglycan (IIH6 C4; Developmental Studies Hybridoma Bank), and anti-mouse HRP secondary antibody (7076; New England Biolabs). Antibody detection reactions were developed by ECL (Advansta) and imaged using the ChemiDoc Touch Imager System (1708370; Bio-Rad Laboratories).
Muscle tissue was snap frozen using liquid nitrogen and pulverized at −80°C. Powdered muscle was then homogenized using the Qiagen TissueLyser for 3 min at 50 Hz in radioimmunoprecipitation assay buffer (20 µl/mg of tissue) with protease inhibitor cocktail (11836153001; Roche). Samples were quantified using the Bradford method. 20 µg of protein was electrophoresed on a 5–8% gradient gel. Proteins were blotted to polyvinylidene difluoride membranes blocked with BSA and were detected using mouse anti-dystrophin (MAB1645; EMD Millipore), anti-HSP90 (BD Biosciences), and anti-acetylated lysine (9441; Cell Signaling Technology) primary antibodies and anti-mouse HRP secondary antibody (7076; New England Biolabs).
Cell culture and treatments of C2C12 myoblasts
C2C12 mouse muscle-derived myoblasts (CRL-1772; American Type Culture Collection) were cultured in DMEM, including 4.5 g/liter glucose, 20% FCS, and 50 µg/ml gentamicin. Differentiation of C2C12 cells into myotubes was induced over 4 d in DMEM, including 4.5 g/liter glucose, 2% horse serum, and 50 µg/ml gentamicin. Cells were tested for mycoplasma using MycoProbe (CUL001B; R&D Systems). Gcn5 knockdown was performed using either adenoviral infection (Addgene) or cell transformation with Gcn5 shRNA (Santa Cruz Biotechnology) using the jetPEI DNA transfection kit (Polyplus) according to the manufacturer’s instructions.
Plasmids and recombinant adenoviruses for in vitro acetylation assays
A plasmid expressing GCN5::FLAG was obtained from Addgene (14106). To generate the plasmid expressing GST::YY1 (pGEX-5X-1-GST-YY1), the coding sequences of three YY1 fragments (F1, 1–190; F2, 180–300; F3, 300–414) were amplified from murine muscle cDNAs and ligated to pGEX-5X-1 plasmids.
In vitro acetylation assays
1 µg of recombinant YY1 protein obtained from the BL21 strain was incubated with 500 ng of recombinant GCN5 in acetylation buffer (150 µM acetyl-coenzyme A, 50 mM Tris-HCl, pH 8, 100 mM NaCl, 10% glycerol, 1 mM PMSF, 1 mM DTT, 1 µg/ml leupeptin, 1 µg/ml pepstatin, and 1 mM sodium butyrate) for 1 h at 30°C. After incubation, samples were resolved with SDS-PAGE sample loading buffer and analyzed by Western blotting or with nano-LC-MS/MS to define the acetylated residues.
MS
Gel lanes were cut into pieces followed by an in-gel digestion with endoproteinase Glu-C or trypsin. These peptide digests were then resuspended and analyzed using nano-LC-MS/MS with an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) coupled to an ultraperformance LC system (Ultimate 3000 RSLC; Thermo Fisher Scientific). Data analysis was performed with Proteome Discoverer (version 1.3), and searches were performed with Mascot and Sequest against a mouse database (UniProt). Data were further processed, inspected, and visualized with Scaffold 4 (Proteome Software).
Structural modeling of acetylated YY1 zinc finger domain
The structural modeling was based on the crystal structure of the DNA-bound form of the human YY1 zinc finger domain (Protein Data Bank accession no. 1UBD; http://www.rcsb.org/pdb/home/home.do). Acetylation at the K392, K393, or K392/K393 residues was introduced using the chemical draw mode of PyMOL (version 1.3; http://www.pymol.org) molecular visualization software. After drawing an acetyl group at the desired position, flexible loop modeling and energy minimization were performed using Modeller software (version 9.13; http://salilab.org/modeller/about_modeller.html). The structures having the lowest energy among the modeled structures were selected for representation. Superimposition of the modeled protein to the unmodified protein was performed for modeling DNA binding modes of acetylated proteins. All structural images were generated using PyMOL.
Statistics
Differences between two groups were assessed using two-tailed t tests. Differences between more than two groups were assessed using one-way ANOVA. To compare the interaction between two factors, two-way ANOVA tests were performed. ANOVA, assessed by Bonferroni’s multiple-comparison test, was used when comparing more than two groups. GraphPad Prism 5 was used for all statistical analyses. All P values <0.05 were considered significant. *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Online supplemental material
Fig. S1 (related to Fig. 1) is a schematic of Gcn5 gene targeting and conditional deletion of exons 5 and 6. Fig. S2 (related to Figs. 2 and 3) demonstrates that Gcn5skm−/− mice have no difference in the percentage of lean mass, fat mass, or relative muscle weight compared with controls and maintain force at equilibrium in diaphragm strips before eccentric contraction. Fig. S3 (related to Fig. 4) includes supplemental metadata analysis figures showing correlations between the Gcn5skm−/− KO bioset and other KO animal biosets with a focus on Mstn-KO biosets. Fig. S4 (related to Fig. 5) includes supplemental metadata analysis figures showing correlations between the Gcn5skm−/− KO bioset and a YY1-KO animal bioset, in addition to showing correlations between GCN5 and DMD transcript expression in humans. Fig. S5 (related to Fig. 6) shows the nano-LC-MS/MS analysis of GCN5-dependent in vitro acetylation of acK392 and acK393 of YY1. Data S1 (related to Fig. 1, b–d) provides microarray features and analysis from control and Gcn5skm−/− mice. Data S2 provides differentially expressed genes and analysis from studies related to Fig. 4 b. Data S3 provides differentially expressed genes and analysis from studies related to Figs. 4 c, S3 b, and S3 c. Data S4 provides TRANSFAC query of factors regulating Dmd. Data S5 provides differentially expressed genes and analysis from studies related to Fig. 5 a and Fig. S4, a and b.
Supplementary Material
Microarray features and analysis from control and Gcn5skm−/− mice. Related to Fig. 1, b–d.
Differentially expressed genes and analysis from studies related to Fig. 4 b.
Differentially expressed genes and analysis from studies related to Figs. 4 c, S3 b, and S3 c.
TRANSFAC query of factors regulating Dmd.
Differentially expressed genes and analysis from studies related to Fig. 5 a and Fig. S4, a and b.
Acknowledgments
The authors are grateful for the technical support of Professor Bernard Jasmin and for resources at the Centre for Neuromuscular Disease, University of Ottawa, especially John Lunde from the facility. The authors are also thankful to Dr. James Flynn from Illumina for advice on in silico analysis, Kelly Sears and Jeannie Mui for sample preparation at the McGill Facility for Electron Microscopy Research, Harry Coenraad from the Children’s Hospital of Eastern Ontario Pathology and Electron Microscopy Department for technical help with sectioning EM samples, and Professor Baptiste Lacoste at the University of Ottawa and the Transmission Electron Microscopy Core Facility for guidance in imaging.
This work was funded by grants from the Canadian Institutes of Health Research (MOP 159455) and the Natural Sciences and Engineering Research Council of Canada Collaborative Research and Training Experience program (Metabolomics Advanced Training and International Exchange program) and discovery grants (RGPIN 2018-06838, DGECR 2018-00012) to K.J. Menzies; the National Natural Science Foundation of China (31871370 and 32000840), the Natural Science Foundation of Guangdong Province (2018A030313655 and 2019A1515011342), Science and Technology Program of Guangzhou (202002030429), to H. Zhang; the Basic Science Research Program of the Korean government (Ministry of Science and ICT grants NRF-2020R1A2C2010964 and 2021R1A5A8029876) to D. Ryu; and the École polytechnique fédérale de Lausanne, the European Research Council (ERC-AdG-787702), the Swiss National Science Foundation (SNSF 31003A_179435), and the Global Research Laboratory grant of the National Research Foundation of Korea (NRF 2017K1A1A2013124) to J. Auwerx. A.E. Green is the recipient of a University of Ottawa Brain and Mind Research Institute uOttawa Eric Poulin Centre for Neuromuscular Disease Scholarship in Translational Research Award.
The authors declare no competing financial interests.
Author contributions: Conception of hypotheses: G.C. Addicks, H. Zhang, D. Ryu, G. Vasam, A.E. Green, J. Auwerx, and K.J. Menzies. Design of the work: G.C. Addicks, H. Zhang, D. Ryu, G. Vasam, A.E. Green, P.L. Marshall, J. Auwerx, and K.J. Menzies. Acquisition of the data: G.C. Addicks, H. Zhang, D. Ryu, G. Vasam, P.L. Marshall, A.E. Green, S. Patel, B.E. Kang, D. Kim, E. Katsyuba, and K.J. Menzies. Drafting of the manuscript: G.C. Addicks, G. Vasam, and K.J. Menzies. Analysis and interpretation of the data: G.C. Addicks, H. Zhang, D. Ryu, G. Vasam, P.L. Marshall, A.E. Green, B.E. Kang, D. Kim, E.G. Williams, J.-M. Renaud, J. Auwerx, and K.J. Menzies. All authors approved the final version of the manuscript.
References
- Acharyya, S., Butchbach M.E.R., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J.E., Fearon K.C.H., Hollingsworth M.A., et al. 2005. Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 8:421–432. 10.1016/j.ccr.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Addicks, G.C., Marshall P., Jasmin B.J., Renaud J.-M., Zhang H., and Menzies K.J.. 2018. Critical assessment of the mdx mouse with ex vivo eccentric contraction of the diaphragm muscle. Curr. Protoc. Mouse Biol. 8:e49. 10.1002/cpmo.49 [DOI] [PubMed] [Google Scholar]
- Ardite, E., Perdiguero E., Vidal B., Gutarra S., Serrano A.L., and Muñoz-Cánoves P.. 2012. PAI-1-regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J. Cell Biol. 196:163–175. 10.1083/jcb.201105013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, R.B., Ogilvie R.W., and Schwane J.A.. 1983. Eccentric exercise-induced injury to rat skeletal muscle. J. Appl. Physiol. 54:80–93. 10.1152/jappl.1983.54.1.80 [DOI] [PubMed] [Google Scholar]
- Bachinski, L.L., Sirito M., Böhme M., Baggerly K.A., Udd B., and Krahe R.. 2010. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve. 42:856–863. 10.1002/mus.21789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakay, M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C., et al. 2006. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 129:996–1013. 10.1093/brain/awl023 [DOI] [PubMed] [Google Scholar]
- Barlev, N.A., Liu L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., and Berger S.L.. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 8:1243–1254. 10.1016/S1097-2765(01)00414-2 [DOI] [PubMed] [Google Scholar]
- Bettica, P., Petrini S., D’Oria V., D’Amico A., Catteruccia M., Pane M., Sivo S., Magri F., Brajkovic S., Messina S., et al. 2016. Histological effects of givinostat in boys with Duchenne muscular dystrophy. Neuromuscul. Disord. 26:643–649. 10.1016/j.nmd.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Bi, P., Yue F., Sato Y., Wirbisky S., Liu W., Shan T., Wen Y., Zhou D., Freeman J., and Kuang S.. 2016. Stage-specific effects of Notch activation during skeletal myogenesis. eLife. 5:e17355. 10.7554/eLife.17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek, P., Morris C., Parkington J., St Andre M., Owens J., Yaworsky P., Seeherman H., and Jelinsky S.A.. 2011. Distinct protein degradation profiles are induced by different disuse models of skeletal muscle atrophy. Physiol. Genomics. 43:1075–1086. 10.1152/physiolgenomics.00247.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blättler, S.M., Cunningham J.T., Verdeguer F., Chim H., Haas W., Liu H., Romanino K., Rüegg M.A., Gygi S.P., Shi Y., et al. 2012a. Yin Yang 1 deficiency in skeletal muscle protects against rapamycin-induced diabetic-like symptoms through activation of insulin/IGF signaling. Cell Metab. 15:505–517. 10.1016/j.cmet.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blättler, S.M., Verdeguer F., Liesa M., Cunningham J.T., Vogel R.O., Chim H., Liu H., Romanino K., Shirihai O.S., Vazquez F., et al. 2012b. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol. Cell. Biol. 32:3333–3346. 10.1128/MCB.00337-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer, K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E.W., Brinkman F.S.L., and Lynn D.J.. 2013. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 41(D1):D1228–D1233. 10.1093/nar/gks1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfield, G., Siller W.G., Wight P.A., and Moore K.J.. 1984. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. USA. 81:1189–1192. 10.1073/pnas.81.4.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, C., McMillan H.J., Mah J.K., Tarnopolsky M., Selby K., McClure T., Wilson D.M., Sherman M.L., Escolar D., and Attie K.M.. 2017. Myostatin inhibitor ACE-031 treatment of ambulatory boys with Duchenne muscular dystrophy: results of a randomized, placebo-controlled clinical trial. Muscle Nerve. 55:458–464. 10.1002/mus.25268 [DOI] [PubMed] [Google Scholar]
- Champy, M.F., Selloum M., Piard L., Zeitler V., Caradec C., Chambon P., and Auwerx J.. 2004. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm. Genome. 15:768–783. 10.1007/s00335-004-2393-1 [DOI] [PubMed] [Google Scholar]
- Champy, M.-F., Selloum M., Zeitler V., Caradec C., Jung B., Rousseau S., Pouilly L., Sorg T., and Auwerx J.. 2008. Genetic background determines metabolic phenotypes in the mouse. Mamm. Genome. 19:318–331. 10.1007/s00335-008-9107-z [DOI] [PubMed] [Google Scholar]
- Chockalingam, P.S., Cholera R., Oak S.A., Zheng Y., Jarrett H.W., and Thomason D.B.. 2002. Dystrophin-glycoprotein complex and Ras and Rho GTPase signaling are altered in muscle atrophy. Am. J. Physiol. Cell Physiol. 283:C500–C511. 10.1152/ajpcell.00529.2001 [DOI] [PubMed] [Google Scholar]
- Chua, J.P., Reddy S.L., Yu Z., Giorgetti E., Montie H.L., Mukherjee S., Higgins J., McEachin R.C., Robins D.M., Merry D.E., et al. 2015. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J. Clin. Invest. 125:831–845. 10.1172/JCI73214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, J.E., Trieu J., Chee A., Naim T., Gehrig S.M., Lamon S., Angelini C., Russell A.P., and Lynch G.S.. 2014. Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy. Exp. Physiol. 99:675–687. 10.1113/expphysiol.2013.077255 [DOI] [PubMed] [Google Scholar]
- Cohen, T.J., Guo J.L., Hurtado D.E., Kwong L.K., Mills I.P., Trojanowski J.Q., and Lee V.M.Y.. 2011. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2:252. 10.1038/ncomms1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi, C., Mozzetta C., Gurtner A., Illi B., Rosati J., Straino S., Ragone G., Pescatori M., Zaccagnini G., Antonini A., et al. 2008. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. USA. 105:19183–19187. 10.1073/pnas.0805514105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consalvi, S., Saccone V., Giordani L., Minetti G., Mozzetta C., and Puri P.L.. 2011. Histone deacetylase inhibitors in the treatment of muscular dystrophies: epigenetic drugs for genetic diseases. Mol. Med. 17:457–465. 10.2119/molmed.2011.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consalvi, S., Mozzetta C., Bettica P., Germani M., Fiorentini F., Del Bene F., Rocchetti M., Leoni F., Monzani V., Mascagni P., et al. 2013. Preclinical studies in the mdx mouse model of Duchenne muscular dystrophy with the histone deacetylase inhibitor givinostat. Mol. Med. 19:79–87. 10.2119/molmed.2013.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, J.R., Martins V.F., Svensson K., LaBarge S.A., Schlenk N.C., Esparza M.C., Buckner E.H., Meyer G.A., Hamilton D.L., Schenk S., et al. 2017. Muscle-specific knockout of general control of amino acid synthesis 5 (GCN5) does not enhance basal or endurance exercise-induced mitochondrial adaptation. Mol. Metab. 6:1574–1584. 10.1016/j.molmet.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe, M.E., Zhang X., McGinnis L., Biggers J., Li E., and Shi Y.. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19:7237–7244. 10.1128/MCB.19.10.7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumonceaux, J., Marie S., Beley C., Trollet C., Vignaud A., Ferry A., Butler-Browne G., and Garcia L.. 2010. Combination of myostatin pathway interference and dystrophin rescue enhances tetanic and specific force in dystrophic mdx mice. Mol. Ther. 18:881–887. 10.1038/mt.2009.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen, J., Poon E., and Davies K.. 2002. The dystrophin-associated protein complex. J. Cell Sci. 115:2801–2803. 10.1242/jcs.115.14.2801 [DOI] [PubMed] [Google Scholar]
- Galvagni, F., Cartocci E., and Oliviero S.. 1998. The dystrophin promoter is negatively regulated by YY1 in undifferentiated muscle cells. J. Biol. Chem. 273:33708–33713. 10.1074/jbc.273.50.33708 [DOI] [PubMed] [Google Scholar]
- Gil, J., Ramírez-Torres A., and Encarnación-Guevara S.. 2017. Lysine acetylation and cancer: a proteomics perspective. J. Proteomics. 150:297–309. 10.1016/j.jprot.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Duggan L., Côté J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640–1650. 10.1101/gad.11.13.1640 [DOI] [PubMed] [Google Scholar]
- Hord, J.M., Botchlett R., and Lawler J.M.. 2016. Age-related alterations in the sarcolemmal environment are attenuated by lifelong caloric restriction and voluntary exercise. Exp. Gerontol. 83:148–157. 10.1016/j.exger.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy, H.B., Usheva A., Shenk T., and Burley S.K.. 1996. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. USA. 93:13577–13582. 10.1073/pnas.93.24.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, D.C., Marcotte G.R., Marshall A.G., West D.W.D., Baehr L.M., Wallace M.A., Saleh P.M., Bodine S.C., and Baar K.. 2017. Age-related differences in dystrophin: impact on force transfer proteins, membrane integrity, and neuromuscular junction stability. J. Gerontol. A Biol. Sci. Med. Sci. 72:640–648. 10.1093/gerona/glw109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman, R.W., Wu C.-L., and Kandarian S.C.. 2012. The ChIP-seq-defined networks of Bcl-3 gene binding support its required role in skeletal muscle atrophy. PLoS One. 7:e51478. 10.1371/journal.pone.0051478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, N.M., Farr G.H. III, and Maves L.. 2013. The HDAC inhibitor TSA ameliorates a zebrafish model of Duchenne muscular dystrophy. PLoS Curr. 5:ecurrents.md.8273cf41db10e2d15dd3ab827cb4b027. 10.1371/currents.md.8273cf41db10e2d15dd3ab827cb4b027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairallah, R.J., Shi G., Sbrana F., Prosser B.L., Borroto C., Mazaitis M.J., Hoffman E.P., Mahurkar A., Sachs F., Sun Y., et al. 2012. Microtubules underlie dysfunction in Duchenne muscular dystrophy. Sci. Signal. 5:ra56. 10.1126/scisignal.2002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek, D.J., and Bamman M.M.. 2008. Modulation of the dystrophin-associated protein complex in response to resistance training in young and older men. J Appl Physiol (1985). 104:1476–1484. 10.1152/japplphysiol.00708.2007 [DOI] [PubMed] [Google Scholar]
- Kupershmidt, I., Su Q.J., Grewal A., Sundaresh S., Halperin I., Flynn J., Shekar M., Wang H., Park J., Cui W., et al. 2010. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 5:e13066. 10.1371/journal.pone.0013066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin, C., Rodgers J.T., Kalume D.E., Kim S.H., Pandey A., and Puigserver P.. 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 3:429–438. 10.1016/j.cmet.2006.04.013 [DOI] [PubMed] [Google Scholar]
- Li, P., Ge J., and Li H.. 2020. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 17:96–115. 10.1038/s41569-019-0235-9 [DOI] [PubMed] [Google Scholar]
- Matys, V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34(90001, Suppl 1):D108–D110. 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzatti, D.J., Smith M.A., Oita R.C., Lim F.-L., White A.J., and Reid M.B.. 2008. Muscle unloading-induced metabolic remodeling is associated with acute alterations in PPARdelta and UCP-3 expression. Physiol. Genomics. 34:149–161. 10.1152/physiolgenomics.00281.2007 [DOI] [PubMed] [Google Scholar]
- McIntyre, R.L., Daniels E.G., Molenaars M., Houtkooper R.H., and Janssens G.E.. 2019. From molecular promise to preclinical results: HDAC inhibitors in the race for healthy aging drugs. EMBO Mol. Med. 11:e9854. 10.15252/emmm.201809854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meliala, I.T.S., Hosea R., Kasim V., and Wu S.. 2020. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics. 10:4183–4200. 10.7150/thno.43481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliavacca, E., Tay S.K.H., Patel H.P., Sonntag T., Civiletto G., McFarlane C., Forrester T., Barton S.J., Leow M.K., Antoun E., et al. 2019. Mitochondrial oxidative capacity and NAD+ biosynthesis are reduced in human sarcopenia across ethnicities. Nat. Commun. 10:5808. 10.1038/s41467-019-13694-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti, G.C., Colussi C., Adami R., Serra C., Mozzetta C., Parente V., Fortuni S., Straino S., Sampaolesi M., Di Padova M., et al. 2006. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 12:1147–1150. 10.1038/nm1479 [DOI] [PubMed] [Google Scholar]
- Miniou, P., Tiziano D., Frugier T., Roblot N., Le Meur M., and Melki J.. 1999. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 27:e27–e30. 10.1093/nar/27.19.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita, T., Weinert B.T., and Choudhary C.. 2019. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20:156–174. 10.1038/s41580-018-0081-3 [DOI] [PubMed] [Google Scholar]
- Nouet, J., Himelman E., Lahey K.C., Zhao Q., and Fraidenraich D.. 2020. Connexin-43 reduction prevents muscle defects in a mouse model of manifesting Duchenne muscular dystrophy female carriers. Sci. Rep. 10:5683. 10.1038/s41598-020-62844-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pambianco, S., Giovarelli M., Perrotta C., Zecchini S., Cervia D., Di Renzo I., Moscheni C., Ripolone M., Violano R., Moggio M., et al. 2016. Reversal of defective mitochondrial biogenesis in limb-girdle muscular dystrophy 2D by independent modulation of histone and PGC-1α acetylation. Cell Rep. 17:3010–3023. 10.1016/j.celrep.2016.11.044 [DOI] [PubMed] [Google Scholar]
- Patel, J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.-J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S., et al. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24:10826–10834. 10.1128/MCB.24.24.10826-10834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñuelas-Urquides, K., Becerril-Esquivel C., Mendoza-de-León L.C., Silva-Ramírez B., Dávila-Velderrain J., Cisneros B., and de León M.B.. 2016. Transcription factors YY1, Sp1 and Sp3 modulate dystrophin Dp71 gene expression in hepatic cells. Biochem. J. 473:1967–1976. 10.1042/BCJ20160163 [DOI] [PubMed] [Google Scholar]
- Petrof, B.J., Shrager J.B., Stedman H.H., Kelly A.M., and Sweeney H.L.. 1993. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA. 90:3710–3714. 10.1073/pnas.90.8.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch, M. 2002. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 19:423–434. 10.1002/humu.10063 [DOI] [PubMed] [Google Scholar]
- Rahimov, F., King O.D., Warsing L.C., Powell R.E., Emerson C.P. Jr., Kunkel L.M., and Wagner K.R.. 2011. Gene expression profiling of skeletal muscles treated with a soluble activin type IIB receptor. Physiol. Genomics. 43:398–407. 10.1152/physiolgenomics.00223.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risson, V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., et al. 2009. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187:859–874. 10.1083/jcb.200903131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalbruch, H. 1975. Segmental fibre breakdown and defects of the plasmalemma in diseased human muscles. Acta Neuropathol. 33:129–141. 10.1007/BF00687539 [DOI] [PubMed] [Google Scholar]
- Shi, Y., Lee J.-S., and Galvin K.M.. 1997. Everything you have ever wanted to know about Yin Yang 1.... Biochim. Biophys. Acta. 1332:F49–F66. 10.1016/S0304-419X(96)00044-3 [DOI] [PubMed] [Google Scholar]
- Smith, L.R., Hammers D.W., Sweeney H.L., and Barton E.R.. 2016. Increased collagen cross-linking is a signature of dystrophin-deficient muscle. Muscle Nerve. 54:71–78. 10.1002/mus.24998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman, H.H., Sweeney H.L., Shrager J.B., Maguire H.C., Panettieri R.A., Petrof B., Narusawa M., Leferovich J.M., Sladky J.T., and Kelly A.M.. 1991. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 352:536–539. 10.1038/352536a0 [DOI] [PubMed] [Google Scholar]
- Svensson, K., Tahvilian S., Martins V.F., Dent J.R., Lemanek A., Barooni N., Greyslak K., McCurdy C.E., and Schenk S.. 2020. Combined overexpression of SIRT1 and knockout of GCN5 in adult skeletal muscle does not affect glucose homeostasis or exercise performance in mice. Am. J. Physiol. Endocrinol. Metab. 318:E145–E151. 10.1152/ajpendo.00370.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski, K., Brock C.J., Trieu J., Chee A., Thakur S.S., Baum D.M., Gregorevic P., Murphy K.T., and Lynch G.S.. 2021. Phosphorylation of ERK and dystrophin S3059 protects against inflammation-associated C2C12 myotube atrophy. Am. J. Physiol. Cell Physiol. 320:C956–C965. 10.1152/ajpcell.00513.2020 [DOI] [PubMed] [Google Scholar]
- Thomas, M.J., and Seto E.. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 236:197–208. 10.1016/S0378-1119(99)00261-9 [DOI] [PubMed] [Google Scholar]
- Townsend, D., Daly M., Chamberlain J.S., and Metzger J.M.. 2011. Age-dependent dystrophin loss and genetic reconstitution establish a molecular link between dystrophin and heart performance during aging. Mol. Ther. 19:1821–1825. 10.1038/mt.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, N.M., Elvers I., Alexander M.S., Moreira Y.B., Eran A., Gomes J.P., Marshall J.L., Karlsson E.K., Verjovski-Almeida S., Lindblad-Toh K., et al. 2015. Jagged 1 rescues the Duchenne muscular dystrophy phenotype. Cell. 163:1204–1213. 10.1016/j.cell.2015.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooddell, C.I., Zhang G., Griffin J.B., Hegge J.O., Huss T., and Wolff J.A.. 2010. Use of Evans blue dye to compare limb muscles in exercised young and old mdx mice. Muscle Nerve. 41:487–499. 10.1002/mus.21527 [DOI] [PubMed] [Google Scholar]
- Yang, X., Koltes J.E., Park C.A., Chen D., and Reecy J.M.. 2015. Gene co-expression network analysis provides novel insights into myostatin regulation at three different mouse developmental timepoints. PLoS One. 10:e0117607. 10.1371/journal.pone.0117607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y.-L., Yang W.-M., and Seto E.. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979–5991. 10.1128/MCB.21.17.5979-5991.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti, S., Gibertini S., Curcio M., Savadori P., Pasanisi B., Morandi L., Cornelio F., Mantegazza R., and Mora M.. 2015. Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim. Biophys. Acta. 1852:1451–1464. 10.1016/j.bbadis.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Gasser D.L., Rappaport E.F., and Falk M.J.. 2010. Cross-platform expression microarray performance in a mouse model of mitochondrial disease therapy. Mol. Genet. Metab. 99:309–318. 10.1016/j.ymgme.2009.10.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray features and analysis from control and Gcn5skm−/− mice. Related to Fig. 1, b–d.
Differentially expressed genes and analysis from studies related to Fig. 4 b.
Differentially expressed genes and analysis from studies related to Figs. 4 c, S3 b, and S3 c.
TRANSFAC query of factors regulating Dmd.
Differentially expressed genes and analysis from studies related to Fig. 5 a and Fig. S4, a and b.