Abstract
Analogues based on the insect cecropin–bee melittin hybrid peptide (CEME) were studied and analyzed for activity and salt resistance. The new variants were designed to have an increase in amphipathic α-helical content (CP29 and CP26) and in overall positive charge (CP26). The α-helicity of these peptides was demonstrated by circular dichroism spectroscopy in the presence of liposomes. CP29 was shown to have activity against gram-negative bacteria that was similar to or better than those of the parent peptides, and CP26 had similar activity. CP29 had cytoplasmic membrane permeabilization activity, as assessed by the unmasking of cytoplasmic β-galactosidase, similar to that of CEME and its more positively charged derivative named CEMA, whereas CP26 was substantially less effective. The activity of the peptides was not greatly attenuated by an uncoupler of membrane potential, carbonyl cyanide-m-chlorophenylhydrazone. The tryptophan residue in position 2 was shown to be necessary for interaction with cell membranes, as demonstrated by a complete lack of activity in the peptide CP208. Peptides CP29, CEME, and CEMA were resistant to antagonism by 0.1 to 0.3 M NaCl; however, CP26 was resistant to antagonism only by up to 160 mM NaCl. The peptides were generally more antagonized by 3 and 5 mM Mg2+ and by the polyanion alginate. It appeared that the positively charged C terminus in CP26 altered its ability to permeabilize the cytoplasmic membrane of Escherichia coli, although CP26 maintained its ability to kill gram-negative bacteria. These peptides are potential candidates for future therapeutic drugs.
Antimicrobial cationic peptides are ubiquitous in nature and are thought to be components of the first line of defense against infectious agents (16). There are four structural classes of cationic peptides: the disulfide-bonded β-sheet peptides, including the defensins; the amphipathic α-helical peptides, such as the cecropins and melittins; extended peptides which often have a single amino acid predominating, e.g., indolicidin; and the loop-structured peptides, like bactenecin (16). Initial interactions of some cationic peptides with gram-negative bacteria are thought to involve binding to surface lipopolysaccharide (LPS) (28, 31). The peptides displace divalent cations that are essential for outer membrane integrity and consequently distort the outer membrane bilayer (26). This allows access to the cytoplasmic membrane, where peptide channel formation has been proposed to occur (21). It is increasingly disputed as to whether peptide channel formation leads to dissolution of the proton motive force and the leakage of essential molecules (9, 19, 37) or whether it is an intermediate step in the uptake of peptide into the cytoplasm, where it inhibits essential functions, e.g., by binding to polyanionic DNA (38).
Cecropins were originally isolated from the immune hemolymph of the North American silk moth Hyalophora cecropia (17). Cecropins have been well studied and characterized with respect to structure and function (12). Based on model membrane studies, the broad spectrum of antimicrobial activity of cecropins has been attributed to its ability to form large pores in bacterial cell membranes (8). A series of hybrid peptides were created, consisting of the amphipathic α-helical N-terminal region of cecropin A and the hydrophobic N-terminal α-helix of the bee venom peptide melittin (35). These hybrids form ion-permeable channels in model lipid membranes (36). To understand the structure-function relationships of these peptides, analogues based on the cecropin (1-8)–melittin (1-18) hybrid (CEME) were studied. The general conclusions from these studies were that the analogues should have a hydrophilic domain and a hydrophobic domain linked by a hinge region (7). A hinge region provides conformational flexibility due to the presence of glycine and proline residues (4). The aromatic residue at position 2 and the α-helical region in the first 11 amino acids have been found to be necessary for antimicrobial action (3). Piers et al. (27) further modified the hybrid peptide CEME (also called MBI-27 [14]) by adding two extra positively charged residues to the C terminus in order to assess the role of ionic charges in interactions with bacteria. The resulting peptide, CEMA (also called MBI-28 [14]), had MICs similar to those of CEME but had an increased ability to permeabilize the outer membranes of gram-negative bacteria and an increased affinity for LPS.
Pseudomonas aeruginosa is a pathogen that is known to colonize the lungs of cystic fibrosis patients. It is believed that the increased salinity of the bronchopulmonary fluids in these patients decreases the efficacy of endogenous cationic peptides of epithelial surfaces, thereby allowing colonization by these bacteria (13, 34). It is because of this that there is an interest in salt-resistant cationic peptides. Lee et al. (20) reported that clavanins, α-helical peptides that derive their cationicity from histidine residues, function in environments with normal and elevated levels of NaCl. This is in contrast to magainin, an α-helical peptide that is susceptible to the presence of 100 mM NaCl (20). In this study we looked at the resistance to the antagonistic effects of salt, the antimicrobial activity, and the mechanism of action of a family of related CEME variants designed here to be slightly different in charge, length, and hydrophobicity and to conform to Edmundson helical-wheel projections (32).
MATERIALS AND METHODS
Materials and bacterial strains.
All peptides were synthesized by N-(9-fluorenyl)methoxycarbonyl chemistry at the Nucleic Acid/Protein Service unit at the University of British Columbia. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phophoglycerol (POPG) were purchased from Northern Lipids Inc. (Vancouver, British Columbia, Canada). o-Nitrophenyl-β-d-galactopyranoside (ONPG) and carbonyl cyanide-m-chlorophenylhydrazone (CCCP) were purchased from Sigma Chemical Co. (St. Louis, Mo.). Bovine serum albumin fraction V lyophilisate was purchased from Boehringer Mannheim (Mannheim, Germany).
Strains used for determining antimicrobial activity included P. aeruginosa PAO1 (15) as well as some antibiotic-resistant strains. P. aeruginosa K385 and 1008OCR01 (nfxB and nalB mutants, respectively) are strains that overproduce specific efflux pumps, resulting in resistance to antibiotics such as norfloxacin (29), cephems, and quinolones (22). P. aeruginosa PAO963 has the nalA phenotype due to a mutation in the locus encoding a DNA gyrase subunit, resulting in nalidixic acid resistance (18, 30). A β-lactamase-derepressed strain (PA-83-48) (6) and an antibiotic-supersusceptible strain (Z61) (5) were also used. Escherichia coli UB1005 and Salmonella typhimurium 14028s and its defensin-supersusceptible phoP phoQ derivative S. typhimurium MS7953s (11) were also used to determine peptide MICs. E. coli ML35, a lactose permease-deficient strain with constitutive cytoplasmic β-galactosidase activity, was used in the cytoplasmic membrane permeabilization assay (21). Mueller-Hinton (MH) broth was used as the general growth medium for all studies reported here.
Preparations of liposomes.
A chloroform solution of lipid (POPC-POPG, 7:3) was mixed with peptides dissolved in methanol. This solution was dried under a stream of N2 in a vacuum to remove the solvent. The resulting lipid-peptide film was rehydrated in 10 mM phosphate buffer (pH 7.0). The suspension was put through five cycles of freeze-thawing to produce multilamellar liposomes, followed by extrusion through 0.1-μm-pore-size double-stacked Poretics membrane filters (AMD Manufacturing Inc., Mississauga, Canada) with an extruder device (Lipex Biomembranes, Vancouver, British Columbia, Canada). A fraction of these liposomes were further extruded with 0.05-μm-pore-size double-stacked Poretics membrane filters.
CD.
Circular dichroism (CD) spectra were measured with a J-720 spectropolarimeter (Jasco, Tokyo, Japan) connected to a Jasco data processor. All samples were in 10 mM sodium phosphate buffer (pH 7.0) and measured in a quartz cell with a 1-mm path length at room temperature. The scanning speed was 10 nm/min (190 to 250 nm), and each spectrum obtained is the average of five scans. The CD spectrum of liposomes alone was subtracted from that of the peptide with liposomes to compensate for light scatter. The α-helical content of the peptides was estimated by the K2D program (2).
MIC.
The MIC of each peptide was determined by using a broth dilution assay modified from the method of Amsterdam (1). Briefly, serial dilutions of each peptide were made in 0.2% bovine serum albumin–0.01% acetic acid solution in 96-well polypropylene (Costar, Corning Incorporated, New York, N.Y.) microtiter plates. Each well was inoculated with 100 μl of the test organism in MH broth to a final concentration of 2 × 104 to 105 CFU/ml. The MIC was taken as the lowest peptide concentration at which growth was inhibited after 18 h of incubation at 37°C. Where indicated, fixed concentrations of NaCl, MgCl2, or sodium alginate (Sigma) were added to each well of the microtiter plate.
Bacterial killing assays.
An overnight culture of E. coli UB1005 was diluted 10−2 in fresh MH broth and allowed to grow to logarithmic (optical density at 600 nm [OD600] of 0.6) or stationary (OD600 of 2.0) phase and then diluted in fresh medium, yielding a working concentration of 108 cells/ml. The peptides or antibiotics were added at four times their MICs, and these suspensions were incubated at 37°C. At regular intervals after peptide addition, samples were removed, diluted, and plated onto MH agar plates to obtain a viable count.
Cytoplasmic membrane permeabilization assay.
The permeabilization of the cytoplasmic membrane of E. coli ML35 by the peptides was determined by their ability to unmask cytoplasmic β-galactosidase to permit hydrolysis of the normally non-membrane-permeative chromogenic substrate ONPG (21). Log-phase bacteria were washed in 10 mM sodium phosphate buffer (pH 7.4). The cells were resuspended in the same buffer with 1.5 mM ONPG. The production of o-nitrophenol over time was monitored spectrophotometrically at 420 nm after the addition of the peptide at four times the MIC (4 μg/ml) or 12.8 μg/ml in the case of CP201 and CP208. This assay was also done in the presence of 100 mM NaCl, 5 mM MgCl2, or 100 μM CCCP.
Animal model.
P. aeruginosa infections of neutropenic mice and protection experiments with antimicrobial peptides were performed exactly as described previously (14).
RESULTS
Peptide design and secondary structure.
In previous studies we had examined the cecropin-melittin hybrid CEME and its variant CEMA. The CEME sequence was modified to increase the α-helical content in the first 14 amino acids, resulting in CP29, using the Edmundson helical-wheel projection (32) to design this and the other peptides described here. This involved substituting hydrophilic amino acids for residues 4, 8, 10, 11, and 14, while residues 6 and 9 were replaced with hydrophobic amino acids. The maintenance of cationic charge was achieved by placing lysine residues at positions 8 and 14. Another related peptide, CP26, had the same first 10 residues as CP29, but the C terminus was predicted to be more hydrophilic because of the addition of an extra positively charged lysine. CP201 was very similar to CP29, with the exception of the flexible hinge region consisting of two glycines in the center of the peptide. CP208 was similar to CP26, with four amino acid replacements, including the replacement of the tryptophan at position 2 with a lysine. In general, these peptides had only slight differences in charge, length, and hydrophobicity (Table 1).
TABLE 1.
Amino acid sequences of antimicrobial cationic peptides used in this study
| Peptide | Amino acid sequence | Length (no. of amino acids) | Charge | % Hydrophobic amino acids |
|---|---|---|---|---|
| CP26 | KWKSFIKKLTSAAKKVVTTAKPLISS | 26 | +7 | 46 |
| CP29 | KWKSFIKKLTTAVKKVLTTGLPALIS | 26 | +6 | 50 |
| CP201 | KWKSFIKNLTKGGSKILTTGLPALIS | 26 | +5 | 42 |
| CP208 | KKKSFIKLLTSAKVSVLTTAKPLISS | 26 | +6 | 42 |
| CEME | KWKLFKKIGIGAVLKVLTTGLPALIS | 26 | +5 | 69 |
| CEMA | KWKLFKKIGIGAVLKVLTTGLPALKLTK | 28 | +6 | 64 |
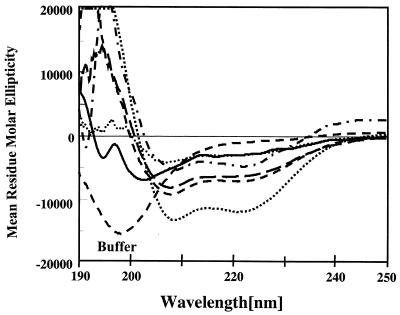
CD spectra were measured in 10 mM sodium phosphate buffer in the presence and absence of POPC-POPG (7:3) liposomes, as well as in the membrane-mimicking environments provided by the addition of sodium dodecyl sulfate (SDS) and trifluoroethanol (TFE; also considered a helix-inducing solvent). The concentrations of peptide and lipid in the buffer were 50 μM and 2 mM, respectively. In buffer, all peptides exhibited spectra characteristic of unordered structure. The spectra of CP26, CEME, CEMA, CP29, and CP201 in the presence of liposomes showed the typical appearance of α-helix-rich structures, with minimal mean residue molar ellipticity values at 207 and 222 nm (Fig. 1). The spectrum of CP208 was essentially that of a random coil. Similar spectra were obtained with both 60- and 90-nm-diameter liposomes, indicating that light scattering by the liposomes did not affect these results. An estimate of percent α-helicity in the various solutions was obtained with the K2D algorithm (2) (Table 2). This program predicted that the peptides contained only α-helix or random-coil secondary structures and no β-sheet structures. In general, CP29 appeared to have the most α-helical structure in the presence of liposomes (approximately 50%); CEMA, CP26, and CP29 were about one-third α-helix; and CP201 and especially CP208 failed to become substantially α-helical. Most researchers have examined α-helicity not in the presence of liposomes but rather in the so-called membrane-mimicking solvents TFE and SDS. Generally speaking, these solvents also revealed that the peptides were α-helical, but the relative α-helicities varied among the different peptides. For example, in SDS, CP208 was as α-helical as CP29. This stresses the importance of the microenvironment in peptide structure formation.
FIG. 1.
CD spectra of cationic peptides in the presence of 90-nm liposomes (POPC-POPG, 7:3) and CP26 in phosphate buffer (random coil). The peptides are represented as follows: CP201, round dots; CP208, solid lines; CEMA, dash-dots; CEME, long dashes; CP26, dashes; and CP29, square dots.
TABLE 2.
α-Helicity in various environments as assessed by CD spectroscopy interpreted according to the K2D algorithm (2)
| Condition | % α-Helix
|
|||||
|---|---|---|---|---|---|---|
| CP26 | CP29 | CP201 | CP208 | CEMA | CEME | |
| Phosphate buffer | 6 | 8 | 8 | 7 | 4 | 7 |
| Liposomesa | 26, 35 | 50, 57 | 20 | 8 | 17, 33 | 29, 30 |
| SDS (40:1) | 42 | 23 | 13 | 26 | 50 | 35 |
| 50% TFE | 30 | 20 | ND | ND | 32 | 25 |
The first number is for 90-nm liposomes, and the second is for 60-nm liposomes. ND, not determined.
Antimicrobial activity.
The MICs of the peptides for selected gram-negative bacteria are shown in Table 3. The MIC was taken as the lowest peptide concentration at which growth was inhibited in a broth dilution assay. CP26 and CP29 were similar in activity to CEMA and CEME. It was of interest to determine if mutations influencing susceptibility to conventional antibiotics had an effect on peptide MICs, especially given the recent observation by Shafer et al. (33) that peptides are influenced by multidrug efflux pathways in Neisseria. Neither nalB nor nfxB, which result in multidrug resistance due to derepression of the MexA MexB OprM and MexC MexD OprJ efflux pathways, respectively, had much effect on peptide MICs. Similarly, little effect was observed for mutants representing the relatively common clinical mutations in DNA gyrase (nalA) or derepression of β-lactamase or the laboratory-derived mutations in Z61 which make the strain supersusceptible to virtually all conventional antibiotics due to outer membrane permeability and efflux defects. Of the other peptides, CP201 had intermediate activity and CP208 had little to no activity against the bacteria tested.
TABLE 3.
Broth dilution MICs of cationic peptides against various gram-negative bacteria
| Species | Strain | Relevant phenotype or genotype | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| CP26 | CP29 | CP201 | CP208 | CEME | CEMA | |||
| P. aeruginosa | PAO1 | Wild type | 2 | 2 | 8 | 64 | 2 | 2 |
| K385 | nfxB | 4 | 4 | 8 | 64 | 2 | 2 | |
| 1008OCR01 | nalB | 2 | 2 | 16 | 128 | 2 | 2 | |
| PAO963 | nalA | 2 | 2 | 8 | 128 | 2 | 2 | |
| PA-83-48 | β-Lactamase derepressed | 4 | 2 | 32 | >128 | 2 | 4 | |
| Z61 | Antibiotic sensitive | 2 | 2 | 32 | 128 | 2 | 4 | |
| E. coli | UB1005 | Wild type | 0.5 | 0.5 | 4 | 32 | 1 | 1 |
| S. typhimurium | 14028s | Parent of MS7953s | 2 | 2 | 32 | >64 | 2 | 4 |
| MS7953s | Defensin sensitive | 0.5 | 0.25 | 2 | 8 | 1 | 1 | |
The best peptides demonstrated modest antimicrobial activity in an animal model. Neutropenic mice (8 to 12 per group) were injected intraperitoneally with 200 to 300 P. aeruginosa M2 cells, leading to 8% survival in control (saline-injected) animals. Intraperitoneal injection, 30 min after the injection of bacteria, of a single dose of 200 μg of CP26 or CP29 led to identical (37%) survival rates after 4 days (P < 0.05 by Fisher’s exact test), results comparable to those achieved by CEME (26.7%) and CEMA (43.3%) (14).
Antimicrobial activity in the presence of salts.
The MICs of the most active peptides for P. aeruginosa were determined in the presence of NaCl, MgCl2, and sodium alginate (Table 4). There was no significant increase in the MICs of CP29, CEME, or CEMA in the presence of 300 mM NaCl, whereas CP26 showed a 16-fold increase in MIC under those conditions. However, CP26 was still resistant (less than a twofold increase in MIC; data not shown) to NaCl antagonism at a NaCl concentration up to 160 mM. The concentration of NaCl in the epithelial cell secretions of a cystic fibrosis patient is about 120 mM (13), and at this concentration all peptides maintained good anti-Pseudomonas activity.
TABLE 4.
Influence of NaCl, MgCl2, and polyanionic alginate on MICs of cationic antimicrobial peptides for P. aeruginosa PAO1
| Additive | Concn | MIC (μg/ml)a
|
|||
|---|---|---|---|---|---|
| CP26 | CP29 | CEME | CEMA | ||
| None | 2 | 2 | 2 | 2 | |
| NaCl | 100 mM | 4 | 2 | 2 | 2 |
| 200 mM | 16 | 2 | 4 | 4 | |
| 300 mM | 32 | 4 | 4 | 4 | |
| MgCl2 | 1 mM | 8 | 4 | 4 | 4 |
| 3 mM | 16 | 16 | 16 | 4 | |
| 5 mM | >32 | 32 | 32 | 32 | |
| Sodium alginate | 0.01% | 2 | 2 | 4 | 4 |
| 0.05% | 8 | 8 | 16 | 32 | |
| 0.1% | 16 | 32 | 64 | 64 | |
These are the average values of three experiments.
The antibacterial activities of CP26, CP29, and CEME were more affected by the presence of divalent cations. At 3 mM MgCl2 (modeling the serum divalent-cation concentrations of 1 mM Mg2+ and 2 mM Ca2+) (1), there was a four- to eightfold increase in MICs. Since the concentration of added Cl− in this case was only 6 mM, and 100 mM Cl− had virtually no effect on MIC in the form of NaCl, it was concluded that the increase in MICs was due to the divalent cation Mg2+. In contrast to these three peptides, CEMA appeared to be relatively resistant to serum divalent-cation concentrations of 3 mM, but not 5 mM, Mg2+. Sodium alginate, a polyanionic polysaccharide related to the mucous exopolysaccharide of P. aeruginosa and intended to be representative of polyanions and polysaccharides present in vivo, antagonized the activities of CEME and CEMA more than those of CP26 and CP29. This effect was probably due to the alginate anion, not the sodium cation. CEMA, which was the most resistant to divalent cations, was the most sensitive to alginate.
Killing assays.
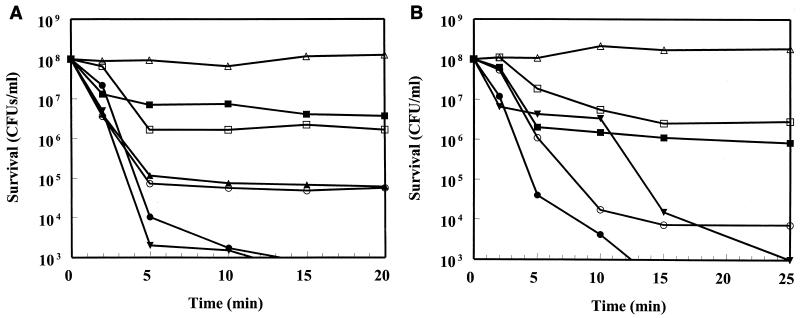
We assessed the ability of the peptides at four times their MICs to kill logarithmic- and stationary-phase E. coli UB1005 in MH medium (Fig. 2A and B, respectively). The peptides killed logarithmic-phase E. coli rapidly by 3 to 5 log orders within 5 min. After 20 min, CP29 and CEME had reduced the number of log-phase bacteria by a total of 6 log orders, whereas CP26 and CEMA showed little reduction after the initial 3-log reduction, although all four peptides had similar MICs for E. coli. In contrast, the conventional antibiotics cationic aminoglycoside gentamicin and β-lactam ceftazidime induced only 2 and 1 log order of killing, respectively. Stationary-phase bacteria in MH broth were killed in a manner similar to log-phase bacteria, with the exception that the kinetics of killing was considerably slower (and for CP29 appeared biphasic) except for CEMA. CP26 (not shown for clarity) was only as efficient at killing as ceftazidime, but the other peptides were more effective than the conventional antibiotics.
FIG. 2.
Survival of logarithmic-phase (A) and stationary-phase (B) E. coli UB1005 in MH broth after addition of fourfold the MIC of peptide, gentamicin, or ceftazidime. This corresponds to 2 μg of CP26, CP29, and ceftazadime per ml, 4 μg of CEME and CEMA per ml, and 0.5 μg of gentamicin per ml. Actual initial concentrations of bacteria ranged from 0.5 × 108 to 2.5 × 108/ml but were corrected to an initial concentration of 1 × 108/ml for clarity. A typical experiment out of three trials is shown. Symbols: ▵, no peptide; ▴, CP26; ▾, CP29; ●, CEME; ○, CEMA; ■, ceftazidime; □, gentamicin. Results for CP26 in panel A were almost superimposable with the ceftazidime results and were thus omitted for clarity. No evidence of bacterial aggregation was observed when viewed under a light microscope or in light-scattering experiments.
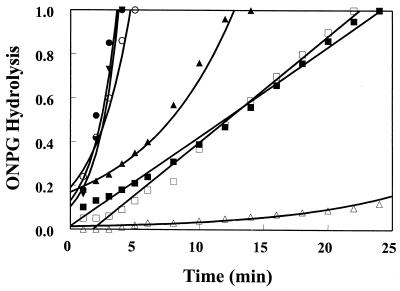
Cytoplasmic membrane permeabilization assay.
The extent of cytoplasmic membrane permeabilization by peptides (at fourfold their MICs) is indicated by the hydrolysis of the chromogenic substrate ONPG by the cytoplasmic enzyme β-galactosidase. The hydrolysis of ONPG was measured spectrophotometrically (Fig. 3). For CEME, CEMA, and CP29, permeabilization was rapid and reached a maximal rate within 1 min. CP26, however, had a considerably lower rate of permeabilization and took 14 min to reach the same level of ONPG hydrolysis as the other three peptides reached in 4 min. CP201 (at threefold its MIC) permeabilized the membrane at a low rate similar to that of another cationic antimicrobial, the lipopeptide polymyxin B, whereas CP208 (at 12.8 μg/ml) demonstrated little to no permeabilizing ability.
FIG. 3.
Permeabilization of cytoplasmic membrane of E. coli ML35 by cationic peptides as determined by their ability to unmask cytoplasmic β-galactosidase. The hydrolysis of ONPG was measured spectrophotometrically at 420 nm. Symbols: ▴, CP26 at 12.8 μg/ml; ●, CEME at 6.4 μg/ml; ○, CEMA at 6.4 μg/ml; ▾, CP29 at 6.4 μg/ml; ■, polymyxin B at 12.8 μg/ml; □, CP201 at 12.8 μg/ml; ▵, CP208 at 12.8 μg/ml.
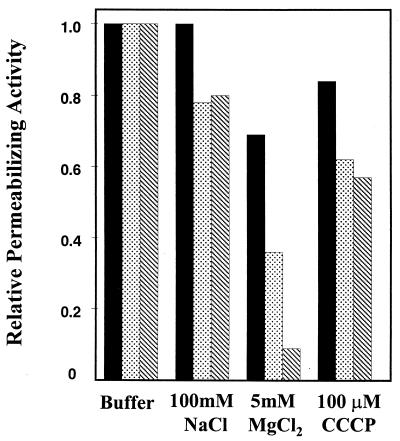
The influence of various factors on cytoplasmic membrane permeabilization was tested for CEME, CP26, and CP29 (Fig. 4). The peptides maintained their abilities to permeabilize the cytoplasmic membrane of E. coli in the presence of NaCl, although CP29 was somewhat more effective. However, in the presence of 5 mM Mg2+, all peptides were affected, and the most salt-sensitive peptide, CP26, lost its ability to permeabilize the cytoplasmic membrane. The peptides were not highly affected by the presence of the uncoupler CCCP at high concentrations, indicating that these peptides can still exert their effects in the absence of a membrane potential, in contrast to, e.g., the indolicidins (10).
FIG. 4.
Effects of NaCl, MgCl2, and CCCP on the cytoplasmic membrane permeabilization activity of cationic peptides CP29 (solid), CEME (stippled), and CP26 (striped).
DISCUSSION
A combination of appropriate chain length, amino acid composition, and positioning of apolar and positively charged residues is required for the antibacterial activity of cationic antimicrobial peptides; however, the exact nature of this combination is still under study. Pathak et al. (24) hypothesized that the amphiphilicity of antimicrobial peptides is the most important factor governing activity, above mean hydrophobicity and α-helix content. In another study it was reported that the antimicrobial activity of the cecropin-melittin hybrid peptides depends on their helical nature (3). Here, we used the already-established cecropin-melittin hybrid peptide, CEME, and CEMA, a variant peptide with two extra amino acids and positive charges in the C terminus, as the templates for further design. Piers et al. (27) showed that CEMA was more efficient at binding the divalent cation binding sites on surface LPS and at destabilizing the outer membrane. However, CEMA did not display notably better bacterial killing than CEME. In this study we showed that the additional four residues at the C terminus of CEMA made it somewhat less α-helical than CEME, which raised the question as to whether the degree of α-helicity is a factor in antibacterial activity. CEME was modified to have an increase in amphipathic α-helical content in its first 14 amino acids, resulting in CP29. This was accomplished by spacing out the lysine and hydrophobic residues to better fit a helical-wheel projection, as well as replacing the glycine residues in the middle of the peptide with threonine, which is more easily accommodated in an α-helix. CP26 was designed by using CP29 as the parent peptide. Its middle hinge region was made more amphipathic and flexible by an added alanine, and the C terminus was modified to become more α-helical and hydrophilic with the addition of an extra positive charge. CP201 was also based on CP29, with one less charge and lower hydrophobicity as a result of the two glycines added to make the central region more flexible, like that of CEME. CP208 was very similar to CP26, with the main difference being the substitution of a lysine for the conserved tryptophan residue at position 2. This peptide was also designed to maintain an α-helical structure.
The CD spectral analysis of these peptides showed that CP29 had the highest helicity of all the peptides. Although CP26 was designed to have a more helical C terminus, the changes around the bend region (i.e., in the vicinity of the proline residue at position 22), including the additional charge, seemed to have a detrimental effect on α-helix formation. CP201 was similar to CP26 in terms of its CD spectra. CP208, however, was essentially a random coil in the presence of liposomes, possibly owing to the lack of the hydrophobic residue tryptophan, which is an amino acid known to be important for interaction of proteins with lipid membranes (23). This was despite the fact that CP208 was designed to have a α-helical structure.
CP26, CEME, CEMA, and CP29 had the same number of amino acids but had charges ranging from +5 to +7, hydrophobic amino acid contents from 46 to 69%, and α-helix contents ranging from 17 to 57% in lipid environments. It was thus of interest to note the similarities and differences between these peptides. All had similar and good activities against the gram-negative bacteria tested and were able to rapidly kill logarithmic-phase bacteria. They also demonstrated similar MICs for antibiotic-resistant mutants of P. aeruginosa, indicating that the mode of action of these α-helical peptides differs from those of conventional antibiotics and that these antibiotics are not effluxed in P. aeruginosa (cf. Neisseria gonorrhoeae [33]). The increase in the α-helicity of CP29 did not make the peptide more active in vitro. The MICs of CP26 were also similar, indicating either that the altered bend region and extra charge did not affect its MIC or that it has a different killing mechanism. CP201, which fell within the ranges of most of the physical properties of the above four peptides, was not a good antibacterial agent, possibly due to the combination of two detrimental factors, namely, lower hydrophobicity (42%) and lower charge (+5). CP208, which also had physical properties similar to those of the four active peptides, had virtually no antibacterial activity, possibly because it lacked a tryptophan residue required for the insertion of the peptide into the lipid membrane. This was supported by the observation that although CP208 was designed to be α-helical, it was unable to form an α-helix upon interaction with liposomes but was able to in the presence of SDS.
The ability to resist salt (NaCl is the most predominant salt in vivo) is important for cationic peptides to function under physiological conditions. In the case of cystic fibrosis, it has been suggested that the susceptibility of epithelial antimicrobial peptides (presumably β-structured defensins) to salt antagonism explains the persistence of chronic P. aeruginosa infections in the lungs of patients with this disease (13). Lee et al. (20) reported the NaCl resistance of the tunicate cationic peptide clavanin in contrast to the α-helical peptides magainin 1 and cecropin P1. It has been observed that extended indolicidins, β-sheet gramicidins, and looped and linear bactenecins are all quite salt (KCl) sensitive (37). Thus, it was of interest to examine the effects of salts on the activities of our α-helical peptides. There were no significant changes in the MICs of CEME, CEMA, and CP29 in the presence of up to 300 mM NaCl, whereas CP26 appeared to be resistant to NaCl only at concentrations up to 160 mM. An NaCl concentration of 120 mM has been reported to be present in the environment of the epithelial cells of a cystic fibrosis patient, which is 30 mM higher than the level of NaCl that antagonizes the activity of epithelial cationic peptides (13). Therefore, even CP26 can be described as NaCl resistant. The differences in activity between CP26 and the other three peptides are presumably related to differences in flexibility and/or hydrophobicity. Interestingly, all peptides were relatively more susceptible to the presence of Mg2+ ions, with a 16-fold increase in their MICs in the presence of 5 mM Mg2+, although CEMA was clearly more resistant to physiologically meaningful (3 mM) divalent cation concentrations. This is unlikely to be due solely to the valency of the positively charged ion. The ionic strength of a MgCl2 solution should be only threefold higher than that of an equivalent NaCl solution, whereas 100-fold-lower Mg2+ concentrations had the same effect as 300 mM Na+. Presumably, these different effects can be explained by differential affinities for a binding site on cells, which we propose here to be on cell surface LPS. Mg2+ has a much higher affinity for binding to LPS than does Na+ (25). The differential effects of divalent and monovalent cations were also demonstrated by the results of the cytoplasmic membrane permeabilization experiments (Fig. 4). However, since interaction with the outer membrane precedes interaction with the cytoplasmic membrane, it is likely that it was at this earlier stage that the peptides were being antagonized. We also examined the antibacterial activities of these peptides in the presence of alginate, a model polyanionic polysaccharide intended to represent those found in vivo. This polyanion reduced the activity substantially at modest concentrations, although CP26 tended to be less affected by this polyanion. In addition to in vitro killing, these peptides demonstrated an ability to work in neutropenic mice.
We have demonstrated here that α-helical peptides with the same general physical properties, but with small differences in hydrophobicity, amphipathicity, charge, and degree of α-helicity, can vary substantially in activity, salt resistance, and permeabilizing ability. CP26 and CP29, which differed by only seven amino acids (four of which were substitutions of one hydrophobic residue for another), had similar MICs and in vivo activities but substantial differences in their resistance to salt antagonism and ability to permeabilize the cytoplasmic membrane. Thus, modest alterations in sequence, and presumably in three-dimensional structure, can result in substantial alterations in a peptide’s properties.
The best α-helical peptides studied here had good activities and were resistant to physiological concentrations of salt. These characteristics may prove to be useful in the design of future therapeutic antibacterial drugs.
ACKNOWLEDGMENTS
This work was financially supported by the Canadian Cystic Fibrosis Foundation through their SPARx program, by the Canadian Bacterial Diseases Network, and by Micrologix Biotech Inc. R.E.W.H. is a recipient of the Medical Research Council of Canada Distinguished Scientist award.
REFERENCES
- 1.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: Williams and Wilkins; 1996. pp. 52–111. [Google Scholar]
- 2.Andrade M A, Chacón P, Merelo J J, Morán F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 3.Andreu D, Merrifield R B, Steiner H, Boman H G. N-terminal analogues of cecropin A: synthesis, antibacterial activity and conformational properties. Biochemistry. 1985;24:1683–1688. doi: 10.1021/bi00328a017. [DOI] [PubMed] [Google Scholar]
- 4.Andreu D, Ubach J, Boman A, Wahlin B, Wade D, Merrifield R B, Boman H G. Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 1992;296:190–199. doi: 10.1016/0014-5793(92)80377-s. [DOI] [PubMed] [Google Scholar]
- 5.Angus B L, Carey A M, Caron D A, Kropinski A M B, Hancock R E W. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer A S, Peters J, Parr T R, Jr, Chan L, Hancock R E W. Role of β-lactamase in in vivo development of ceftazidime resistance in experimental Pseudomonas aeruginosa endocarditis. Antimicrob Agents Chemother. 1987;31:253–258. doi: 10.1128/aac.31.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boman H G, Wade D, Boman A, Wahlin I A, Merrifield R B. Antibacterial and antimalarial properties of peptides that are cecropin-melittin hybrids. FEBS Lett. 1989;259:103–106. doi: 10.1016/0014-5793(89)81505-4. [DOI] [PubMed] [Google Scholar]
- 8.Christensen B, Fink J, Merrifield R B, Mauzerall D. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes. Proc Natl Acad Sci USA. 1988;85:5072–5076. doi: 10.1073/pnas.85.14.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cociancich S, Ghazi A, Hoffman J A, Hetrus C, Letellier C. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J Biol Chem. 1993;268:19239–19245. [PubMed] [Google Scholar]
- 10.Falla T J, Karunaratne D N, Hancock R E W. Mode of action of the antimicrobial peptide indolicidin. J Biol Chem. 1996;271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- 11.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 12.Fink J, Boman A, Boman H G, Merrifield R B. Design, synthesis and antibacterial activity of cecropin-like model peptides. Int J Pep Protein Res. 1989;33:412–421. doi: 10.1111/j.1399-3011.1989.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 13.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human-beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 14.Gough M, Hancock R E W, Kelly N M. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect Immun. 1996;64:4922–4927. doi: 10.1128/iai.64.12.4922-4927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock R E W, Falla T, Brown M. cationic bactericidal peptides. Adv Microb Physiol. 1995;37:136–175. doi: 10.1016/s0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 17.Hultmark D, Steiner H, Rasmusen T, Boman H G. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 18.Inoue Y, Sato K, Fujii T, Hirai K, Inoue M, Iyobe S, Mitsuhashi S. Some properties of subunits of DNA gyrase from Pseudomonas aeruginosa PAO1 and its nalidixic acid-resistant mutant. J Bacteriol. 1987;169:2322–2325. doi: 10.1128/jb.169.5.2322-2325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juretic D, Chan H C, Brown J H, Morell J L, Hendler R W, Westerhoff H. Magainin 2 amide and analogues, antimicrobial activity, membrane depolarization and susceptibility to proteolysis. FEBS Lett. 1989;249:219–223. doi: 10.1016/0014-5793(89)80627-1. [DOI] [PubMed] [Google Scholar]
- 20.Lee I H, Cho Y, Lehrer R I. Effects of pH and salinity on the antimicrobial properties of clavanins. Infect Immun. 1997;65:2898–2903. doi: 10.1128/iai.65.7.2898-2903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehrer R I, Barton A, Daher K A, Harwig S S L, Ganz T, Selsted M E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Investig. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meers P. Location of tryptophans in membrane-bound annexins. Biochemistry. 1990;29:3325–3330. doi: 10.1021/bi00465a025. [DOI] [PubMed] [Google Scholar]
- 24.Pathak N, Salas-Auvert R, Ruche R, Janna M-H, McCarthy D, Harrison R G. Comparison of the effects of hydrophobicity, amphiphilicity and alpha helicity on the activities of antimicrobial peptides. Proteins Struct Funct Genet. 1995;22:182–186. doi: 10.1002/prot.340220210. [DOI] [PubMed] [Google Scholar]
- 25.Peterson A A, Hancock R E W, McGroarty E J. Binding of polycationic antibiotics and polyamines to lipopolysaccharides of Pseudomonas aeruginosa. J Bacteriol. 1985;164:1256–1261. doi: 10.1128/jb.164.3.1256-1261.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson A A, Fesik S W, McGroarty E J. Decreased binding of antibiotics to lipopolysaccharides from polymyxin-resistant strains of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1987;31:230–237. doi: 10.1128/aac.31.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piers K L, Brown M H, Hancock R E W. Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by C-terminal modification. Antimicrob Agents Chemother. 1994;38:2311–2316. doi: 10.1128/aac.38.10.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piers K L, Hancock R E W. The interaction of recombinant cecropin/melittin hybrid peptides with the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1994;12:951–960. doi: 10.1111/j.1365-2958.1994.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 29.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rella M, Haas D. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother. 1982;22:242–249. doi: 10.1128/aac.22.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawyer J G, Martin N L, Hancock R E W. Interaction of macrophage cationic proteins with the outer membrane of Pseudomonas aeruginosa. Infect Immun. 1988;56:693–698. doi: 10.1128/iai.56.3.693-698.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiffer M, Edmundson A B. Use of helical wheels to represent the structures of protein and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafer W M, Qu X-D, Waning A J, Lehrer R I. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith J J, Travis S M, Greenburg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 35.Wade D, Andreu D, Mitchell S A, Silveria A M V, Boman A, Boman H G, Merrifield R B. Antibacterial peptides designed as analogs or hybrids of cecropins and melittin. Int J Pept Protein Res. 1992;40:429–436. doi: 10.1111/j.1399-3011.1992.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 36.Wade D, Boman B, Wahlin B, Drain C M, Andreu D, Boman H G, Merrifield R B. All-d amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci USA. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, M., E. Maier, R. Benz, and R. E. W. Hancock. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry, in press. [DOI] [PubMed]
- 38.Zhang, L., R. Benz, and R. E. W. Hancock. Influence of proline residues on the antibacterial and synergistic activities of α-helical peptides. Biochemistry, in press. [DOI] [PubMed]