Abstract
Background
Myeloid-derived suppressor cells (MDSCs) represent a negative prognostic factor in malignant melanoma. These cells are generated under chronic inflammatory conditions typical of cancer. The transcription factor signal transducer and activator of transcription 3 (STAT3) orchestrates MDSC accumulation and acquisition of immunosuppressive properties. Here we studied STAT3 inhibition by Napabucasin as a way to block MDSC accumulation and activity and its potential to treat malignant melanoma.
Methods
In vitro generated murine MDSC and primary MDSC from melanoma-bearing mice were used to investigate the effects of Napabucasin on MDSC in vitro. The RET transgenic mouse model of malignant melanoma was used to examine Napabucasin therapy efficiency and its underlying mechanisms in vivo. Furthermore, STAT3 activation and its correlation with survival were explored in MDSC from 19 patients with malignant melanoma and human in vitro generated monocytic myeloid-derived suppressor cell (M-MDSC) were used to evaluate the effects of Napabucasin.
Results
Napabucasin was able to abrogate the capacity of murine MDSC to suppress CD8+ T-cell proliferation. The STAT3 inhibitor induced apoptosis in murine MDSC, significantly increased expression of molecules associated with antigen processing and presentation, as well as slightly decreased expression of immunosuppressive factors on these cells. RET transgenic mice treated with Napabucasin showed prolonged survival accompanied by a strong accumulation of tumor-infiltrating antigen-presenting cells and activation of CD8+ and CD4+ T cells. Interestingly, patients with malignant melanoma with high expression of activated STAT3 in circulating M-MDSC showed significantly worse progression-free survival (PFS) than patients with low levels of activated STAT3. In addition, Napabucasin was able to abrogate suppressive capacity of human in vitro generated M-MDSC.
Conclusion
Our findings demonstrate that STAT3 inhibitor Napabucasin completely abrogated the immunosuppressive capacity of murine MDSC and human M-MDSC and improved melanoma-bearing mouse survival. Moreover, patients with malignant melanoma with high expression levels of activated STAT3 in M-MDSC displayed shorter PFS, indicating its role as a promising therapeutic target in patients with malignant melanoma and a predictive marker for their clinical outcome.
Keywords: melanoma, myeloid-derived suppressor cells, tumor microenvironment, immunotherapy
Key messages.
What is already known on this topic
Transcription factor signal transducer and activator of transcription 3 (STAT3) is necessary for survival and proliferation of cancer cells and can promote an immunosuppressive tumor microenvironment by activating myeloid-derived suppressor cells (MDSCs) in malignant melanoma. However, STAT3 inhibition is not yet used as a routine cancer treatment.
What this study adds
We showed that STAT3 inhibition by Napabucasin induced MDSC apoptosis and abrogated their immunosuppressive capacity in mice and humans, prolonged melanoma-bearing mouse survival and led to a shift from pro-tumor to anti-tumor immune cells in the tumor microenvironment. Activated STAT3 in circulating MDSC was also associated with a worse prognosis of patients with melanoma.
How this study might affect research, practice or policy
STAT3 inhibition by Napabucasin should be included in novel combined strategies of tumor immunotherapy to neutralize immunosuppressive capacities of MDSC.
Background
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of immunosuppressive myeloid cells that accumulate under chronic inflammatory conditions typical of cancer.1 In mice, MDSCs are characterized by the expression of CD11b (integrin α-M) and the myeloid differentiation antigen Gr1.2 There are two murine MDSC subpopulations: CD11b+Ly6GhighLy6Clow polymorphonuclear (PMN)-MDSC and CD11b+Ly6G-Ly6Chigh monocytic (M)-MDSC.2 In humans, M-MDSCs are defined as CD33highCD14+HLA-DRlow/− and PMN-MDSC as CD33dimHLA-DRneg CD66b+.2 A third subpopulation, early-stage MDSCs, has been described as HLA-DR-CD33+CD15−CD14− in humans but did not show immunosuppressive capacity.2
MDSC can suppress antitumor immune responses, mainly by inhibiting T-cell and natural killer (NK)-cell activities.1 MDSCs express programmed death ligand 1 (PD-L1), inducing T-cell anergy by the interaction with programmed cell death protein 1 (PD-1) on activated T cells.3 They produce elevated levels of nitric oxide and reactive oxygen species (ROS) by inducible nitric oxide synthase (iNOS) and nicotinamide adenosine dinucleotide phosphate oxidases (NOX).4 Furthermore, the activation of arginase 1 (Arg-1),5 iNOS6 and indolamin-2,3-dioxygenase7 leads to degradation of L-arginine and L-tryptophan, respectively, inducing T-cell starvation. MDSCs produce also prostaglandin E2 (PGE2) by expression of prostaglandin E synthase 2 (PTGES2) and cyclooxygenase 2 (COX2).8 Altogether, these factors create a strongly immunosuppressive tumor microenvironment.1
It has been demonstrated that high MDSC frequencies in patients with malignant melanoma correlated with worse disease outcome.9–11 Moreover, high levels of MDSC have been shown to mediate resistance to immune checkpoint blockade by antibodies against PD-1/PD-L1 and cytotoxic T-lymphocyte-associated protein (CTLA)-4.12 This underlines the importance of MDSC as a therapeutic target in malignant melanoma. Even though several clinical studies blocking MDSC in cancer were initiated, no MDSC-targeting therapy was approved so far.13
MDSCs are generated under a constant presence of inflammatory factors such as interleukin (IL)-6, interferon-γ, IL-1β, granulocyte–macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor (TNF)-α, vascular endothelial growth factor (VEGF) and Toll-like receptor (TLR) ligands.14 It was demonstrated that specifically, signaling via signal transducer and activator of transcription 3 (STAT3) played a pivotal role in MDSC functions.14 Downstream signaling of different cytokine, G protein-coupled, growth factor and tyrosine kinase receptors activates janus tyrosine kinase (JAK), which results in phosphorylation and dimerization of STAT3.15 We have shown that MDSC generation with IL-6 induced upregulation of Arg-1 activity and ROS production in a STAT3-dependent manner, leading to increased immunosuppressive capacity of MDSC.16 The treatment of healthy donor peripheral blood mononuclear cells (PBMCs) with IL-6 for 6 days resulted in an enrichment of CD14+HLA−DR− cells accompanied by an increase of phosphorylated (p)STAT3, ARG1 mRNA expression and ROS production in these cells.17 High pSTAT3 levels in tumor-infiltrating and circulating M-MDSC from patients with head and neck squamous cell carcinoma (HNSCC) were reported to correlate with the upregulation of Arg-1 expression and activity.18 In addition, immunosuppressive CD14+HLA-DRlow/neg cells from patients with melanoma were characterized by upregulated pSTAT3 expression.19 Moreover, CD14+ immunosuppressive myeloid cells from patients with pancreatic cancer expressed STAT3 and ARG1 genes, whereas cells without immunosuppressive capacity from the same patients lacked the STAT3/ARG1 transcriptomic gene signature.20 Also, PD-L1 was described as a target gene of STAT3 and was demonstrated to be upregulated after induction of immunosuppressive myeloid cells by IL-6.21
We have previously shown that the treatment of RET transgenic melanoma-bearing mice with antibodies against IL-6 failed to display antitumor effects.22 Strikingly, it even shortened the survival and accelerated tumor development associated with a decrease in activated CD8+ T cells.22
In the present study, we investigated the potential of the STAT3 inhibitor Napabucasin (also known as BBI608) to restrain MDSC immunosuppression without limiting anti-tumor T-cell response. Napabucasin was initially reported to inhibit STAT3-driven gene transcription and cancer cell stemness.23 We found that Napabucasin inhibited mouse MDSC-mediated immunosuppression in vitro by inducing MDSC apoptosis as well as decreasing the expression of immunosuppressive factors and increasing the expression of molecules involved in the antigen processing and presentation machinery in these cells. In addition, Napabucasin was able to prolong survival of melanoma-bearing mice. In patients with melanoma, elevated frequencies of pSTAT3+ M-MDSC correlated with worse progression-free survival (PFS). Moreover, Napabucasin abrogated T-cell suppression mediated by M-MDSC generated from human monocytes. Therefore, our study suggests STAT3 inhibition by Napabucasin as a valuable therapeutic approach for patients with malignant melanoma.
Methods
Mice
Mice (C57BL/6 background) expressing the human RET oncogene in melanocytes under the mouse metallothionein-I promotor-enhancer24 were provided by Dr I Nakashima (Chubu University, Aichi, Japan). Mice were kept in the animal facility of the University Medical Center (Mannheim, Germany) under specified pathogen-free conditions. Non-transgenic littermates were used as healthy C57BL/6 mice. Both male and female mice at the age of 6–12 weeks were applied for experiments.
Isolation of primary murine cells
Murine bone marrow (BM) cells were isolated using RPMI medium with GlutaMAX (Thermo Fisher) followed by ACK lysis (Thermo Fisher) and filtering through a 40 µm cell strainer (Sarstedt). CD11b+Gr1+ MDSC from the BM of tumor bearing RET transgenic mice were obtained using EasySep mouse MDSC isolation kit (STEMCELL Technologies) according to manufacturer’s instructions. Murine splenic CD8+ T cells were isolated by magnetic-activated cell sorting (MACS, Miltenyi Biotec) according to the manufacturer’s instructions. Single-cell suspensions from murine tumors were obtained by cutting the tumors and filtering through a 100 µm cell strainer (Sarstedt) without enzymatic digestion.
In vitro generation of MDSC from murine BM
The generation of MDSC in vitro was performed as previously described.25 Briefly, 2.5×106 BM cells from healthy C57BL/6 mice were cultured for 4 days in a 10 cm cell culture dish (Corning FALCON) in 10 mL RPMI-1640 medium with GlutaMAX supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, 1 mM sodium pyruvate, 50 µM β-mercaptoethanol, 1 mM Minimal Essential Medium (MEM) non-essential amino acids (all Thermo Fisher), 40 ng/mL GM-CSF and 40 ng/mL IL-6 (both PeproTech, recombinant murine proteins produced in Escherichia coli).
Isolation of human PBMC
Heparinized venous blood samples from untreated patients with melanoma (stages I–IV) were subjected to density gradient centrifugation using Pancoll (1.077 g/mL, PanBiotech). PBMCs were collected, washed with PBS and immediately processed for flow cytometry. Follow-up observations on patients disease progression were done for 2 years.
In vitro generation of human M-MDSC from CD14+ monocytes
PBMCs were isolated as described previously from healthy donor buffy coats. CD14+ monocytes were isolated by MACS (Miltenyi Biotec) according to the manufacturer’s instructions. Monocytes (1×106) were cultured for 4 days in a 12-well plate (Sarstedt) in 1 mL RPMI-1640 medium with GlutaMAX supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin, 10 mM HEPES, 1 mM sodium pyruvate, 50 µM β-mercaptoethanol, 1 mM MEM non-essential amino acids (all Thermo Fisher), 50 ng/mL GM-CSF and 50 ng/mL IL-6 (both Miltenyi Biotec, recombinant human proteins produced in E. coli).
RNA isolation
Total RNA was isolated by RNeasy Mini Kit (Qiagen). DNA digest was performed with on-column RNase-free DNase Set (Qiagen) according to manufacturer’s instructions. RNA concentration was determined using the microplate reader Tecan Infinite M200 and a Nanoquant plate.
Microarray analysis
The Affymetrix GeneChip Mouse Gene V.2.0 ST Array (Thermo Fisher) was used according to the manufacturer’s instructions. A Custom CDF V.22 with ENTREZ-based gene definitions was used to annotate the arrays.26 The raw fluorescence intensity values were normalized by applying quantile normalization and robust multiarray analysis background correction. A batch normalization was used to remove individual mouse variations. Two-way analysis of variance was performed to identify differentially expressed genes using the commercial software package SAS JMP Genomics V.7. A false-positive rate of a=0.05 with false discovery rate correction was set as the level of significance. Gene set enrichment analysis was used to determine whether defined lists (or sets) of genes exhibit a statistically significant bias in their distribution within a ranked gene list.27 Pathways belonging to different cell functions were obtained from public external databases (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg).
Flow cytometry
Cells were treated with 7AAD or fixable viability dye 700 and FcR Blocking Reagent (all BD Biosciences) followed by staining with antibodies (additional file 2, online supplemental tables S1 and S2). For intracellular staining, cells were fixed and permeabilized with eBioscience Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher) according to manufacturer’s instructions. To detect ROS production, we applied CellROX Deep Red Reagent (Thermo Fisher). For apoptosis assay, Annexin V-APC and Annexin V Binding Buffer (both BioLegend) were used according to the manufacturer’s instructions. Acquisition was performed using the 10-color flow cytometer BD FACSLyric and FACSuite software (BD Biosciences). Data were analyzed by FlowJo V.10 software (BD Biosciences). Markers with continuous expression pattern were gated according to the fluorescence minus one (FMO) or isotype control.
jitc-2021-004384supp002.pdf (29.7KB, pdf)
Murine suppression of T-cell proliferation assay
Splenic CD8+ T cells from C57BL/6 mice were stained with 2 nM carboxyfluorescein succinimidyl ester (CFSE) for 5 min at 37°C. T cells were cocultured with in vitro generated MDSC (2:1 ratio) or MDSC isolated from the BM of RET transgenic mice (1:1 ratio) in RPMI-1640 medium with GlutaMAX (supplemented as for MDSC in vitro differentiation) for 72 hours in 96-well round bottom plates (Sarstedt) precoated for 3 hours with anti-CD3 (100 ng/mL, clone 37.51) and anti-CD28 antibodies (50 ng/mL, clone 17A2, both eBioscience). The proliferation of CD8+ T cells was assessed after 72 hours of coculture by measuring CFSE dilution using the BD FACSLyric flow cytometer.
Human suppression of T-cell proliferation assay
CD3+ T cells were isolated from healthy donor PBMC by MACS (Milteny Biotec) and labeled with 10 µM cell proliferation dye eFluor 450 (eBioscience) at room temperature for 20 min. Then T cells were cocultured with in vitro generated MDSC in 100 µl L-lysine and L-arginine free RPMI medium with GlutaMAX (Thermo Fisher) supplemented with 150 µM L-arginine, 0.218 mM L-lysine hydrochloride (both Sigma Aldrich), 10% heat-inactivated FBS and 1% penicillin/streptomycin (both Thermo Fisher) for 4 days in 96-well round bottom plates (Sarstedt) precoated for 2 hours with anti-CD3 (1 µg/mL, clone OKT-3, eBioscience) and anti-CD28 antibodies (2 µg/mL, clone CD28.2, Beckman Coulter). The proliferation of T cells was assessed after 4 days of coculture by measuring proliferation dye eFluor 450 dilution using the BD FACSLyric flow cytometer.
In vitro proliferation assay of RET melanoma cells
Ret melanoma cells established from the RET transgenic melanoma mouse model28 were seeded in flat bottom 96 well plates at a density of 2500 cells/well in RPMI medium with GlutaMAX (Thermo Fisher) supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin (both Thermo Fisher). Cells were treated with 1 µM Napabucasin or 0.01% dimethyl sulfoxide (DMSO) for 0 hour, 24 hours, 48 hours and 72 hours. After the co-culture 10 µl of 12 mM 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) was added to 100 µL medium followed by the incubation for 4 hours. Then formazan was dissolved with DMSO. The absorption was measured at 540 nm (with a reference wavelength of 630 nm) using the microplate reader Tecan Infinite M200.
Mouse in vivo studies
On first signs of tumor growth, RET transgenic mice (with the weight of 16–19 g) were separated into two groups containing equal numbers of males and females. Napabucasin (Cayman Chemical, 20 mg/kg) was injected intraperitoneally for 4 weeks, two times a week, while the control group received adjusted volume of DMSO diluted in PBS. Napabucasin stock solution (20 mg/mL in 100% DMSO) was diluted in PBS to achieve the concentration of 20 mg/kg in 100 µl volume. In the control group, animals received 100 µl of 16%–19% DMSO (V/V, dependent on the mouse weight) in PBS. Mice complying with termination criteria were sacrificed and recorded as deceased. In another set of experiments, mice of the same groups were sacrificed after 3 weeks of therapy, and tumors were isolated for flow cytometric analysis.
Statistical analysis
Statistical analysis of data was performed using GraphPad Prism software on at least three biological replicates. Two groups were compared with paired or unpaired two-tailed Student’s t test assuming Gaussian distribution of the data. Correlation analysis was done by Pearson correlation with two tailed p value. Survival curves were generated using the Kaplan-Meier method, and statistical comparison was done by the log-rank (Mantel-Cox) test. A p value of <0.05 was considered statistically significant.
Results
Napabucasin abrogated MDSC suppressive capacity
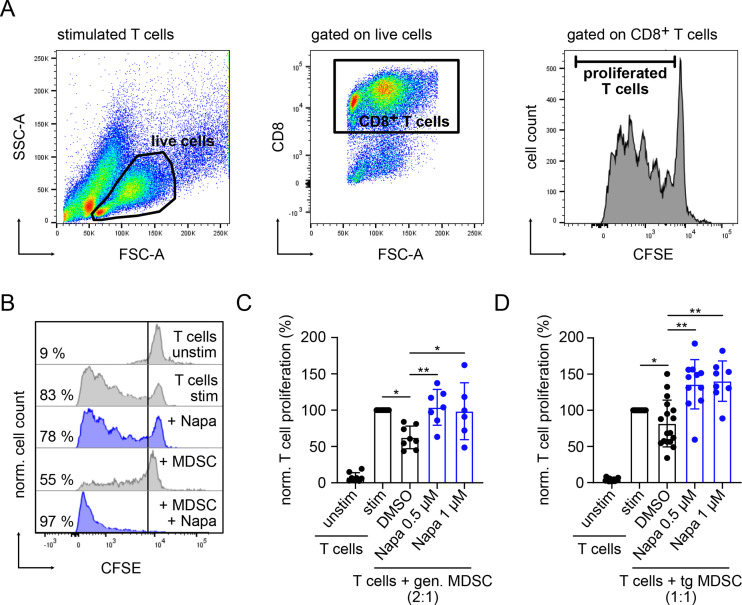
On the co-culture of in vitro generated MDSC with activated CD8+ T cells, MDSC significantly inhibited T-cell proliferation (p<0.05, figure 1A–C). Interestingly, this suppression was completely abrogated by adding the STAT3 inhibitor Napabucasin to the coculture (p<0.01, figure 1B, C). Similarly, MDSC isolated from the BM of RET transgenic melanoma-bearing mice were able to block CD8+ T-cell proliferation, which was also restored by Napabucasin addition (p<0.01, figure 1D). Moreover, Napabucasin could even enhance T-cell proliferation as compared with that of T cells stimulated without MDSC (figure 1C, D). Importantly, Napabucasin did not alter T-cell proliferation at the applied concentrations (0.5 and 1 µM), whereas higher concentrations limited T-cell proliferation (online supplemental figure 1A).
Figure 1.
Napabucasin abrogated immunosuppressive capacity of MDSC in vitro. Suppressive capacity of murine MDSC was determined on the coculture with CFSE-labeled murine-activated splenic CD8+ T cells. After 72 hours of incubation, T-cell proliferation was assessed by CFSE dilution measured by flow cytometry. (A) Gating strategy for proliferated T cells. (B) Representative histograms for the proliferation of unstim and stim T cells incubated alone or in the presence of in vitro gen. MDSC with 1 µM Napa or 0.01% DMSO as control. (C) Cumulative data for T-cell proliferation are presented as the percentage of divided T cells norm. to the respective control of stim T cells alone (mean±SD, n=6–7). (D) MDSCs were isolated from the bone marrow of RET TG melanoma-bearing mice (TG MDSC). Cumulative data for T-cell proliferation in the presence or absence of tg MDSC and with or without Napa are shown as the percentage of divided T cells norm. to the respective control of stim T cells alone (mean±SD, n=8–11). Statistics were performed on not norm. data. *P<0.05, **P<0.01. CFSE, carboxyfluorescein succinimidyl ester; gen., generated; MDSC, myeloid-derived suppressor cell; Napa, Napabucasin; norm., normalized; stim, stimulated; tg, transgenic; unstim, unstimulated.
jitc-2021-004384supp001.pdf (806.9KB, pdf)
Napabucasin induced MDSC apoptosis and changed their immunosuppressive pattern
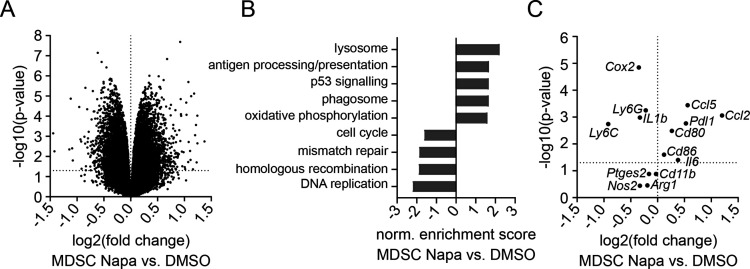
Performing a microarray analysis of generated MDSC treated for 24 hours with Napabucasin or the respective DMSO concentration, we found 1526 genes differentially regulated (p<0.05). Of these genes, 787 were significantly lower expressed in MDSC treated with Napabucasin compared with DMSO-treated control MDSC, and 739 genes showed significantly higher expression (p<0.05, figure 2A). A pathway analysis showed that Napabucasin-treated MDSC displayed significantly downregulated pathways dealing with cell cycle, mismatch repair and DNA replication, related to decreased survival and proliferation (p<0.05, figure 2B). Furthermore, other pathways related to lysosome, phagosome and antigen processing and presentation were significantly upregulated in Napabucasin-treated MDSC (p<0.05, figure 2B). Looking more precisely at the differentially regulated genes, we found that the expression of Il6, Cd80 and Cd86 was significantly upregulated (p<0.05, figure 2C). In contrast, Il1b and Cox2 expression was significantly downregulated (p<0.05), while expression of Ptges2, Nos2 and Arg1 showed a tendency to be decreased (figure 2C).
Figure 2.
Microarray analysis of Napabucasin-treated MDSC. Whole transcriptome of MDSC generated in vitro and treated for 24 hours with 1 µM Napa or 0.01% DMSO was analyzed by microarray (n=3). (A) Volcano plot with differentially expressed genes. Dotted horizontal line indicates significance threshold (p<0.05). (B) Norm. enrichment score of important significantly regulated pathways found by gene set enrichment analysis pathway analysis using Kyoto Encyclopedia of Genes and Genomes pathways. (C) A plot with selected differentially expressed genes important for MDSC phenotype and function is presented. Dotted horizontal line indicates the significance threshold (p<0.05). MDSC, myeloid-derived suppressor cell; Napa, Napabucasin; norm., normalized.
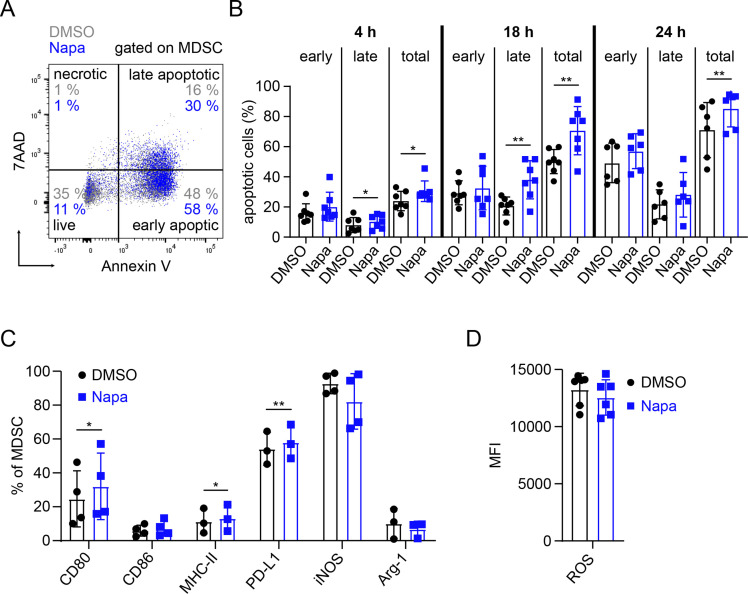
We further investigated the effect of Napabucasin on MDSC cell death, performing 7AAD and annexin V staining and found increased apoptosis of generated MDSC after treatment with Napabucasin compared with the DMSO control (figure 3A). The frequency of total apoptotic cells was significantly higher in MDSC after 4 (p<0.05), 18 and 24 hours (p<0.01) of the treatment with Napabucasin (figure 3B). Furthermore, on 4 (p<0.05) and 18 (p<0.01) hours, the frequency of late apoptotic cells was significantly increased, whereas there were no significant effects on early apoptotic cell frequency at all investigated time points (figure 3B). Importantly, the apoptosis-inducing effect of Napabucasin was found to be specific to MDSC, as it was not observed when treating CD8+ T cells (online supplemental figure S1B).
Figure 3.
Napabucasin promoted in vitro apoptosis and alteration of MDSC phenotype. MDSC generated in vitro were treated with 1 µM Napa or 0.01% DMSO. Apoptosis of MDSC and expression of their markers were measured by flow cytometry. (A) Gating strategy for determination of early (annexin V+7AAD−) and late apoptotic (annexin V+7AAD+), necrotic (annexin V−7AAD+) and live (annexin V−7AAD−) cells is shown. (B) Results are presented as the percentage of early, late and total apoptotic cells among total MDSC on 4, 18 and 24 hours of incubation with Napa (mean±SD, n=6–7). (C) Data are shown as the percentage of MDSC expressing CD80, CD86, MHC-II, PD-L1, iNOS and Arg-1 within total MDSC on 24 hours of culture with Napa (mean±SD, n=3–4). (D) The level of ROS production by MDSC is presented as MFI (mean±SD, n=6). *P<0.05, **P<0.01. Arg-1, arginase 1; iNOS, inducible nitric oxide synthase; MDSC, myeloid-derived suppressor cell; MFI, median fluorescence intensity; MHC-II, major histocompatibility complex class II; Napa, Napabucasin; PD-L1, programmed death ligand 1; ROS, reactive oxygen species.
Analyzing protein expression of markers related to antigen presentation or immunosuppression, we found that Napabucasin induced a significant upregulation of CD80 and MHC class II expression on MDSC (p<0.05, figure 3C). CD86 surface expression remained similar in both groups (figure 3C). In addition, the expression of iNOS and Arg-1 as well as ROS production showed a tendency to be downregulated (figure 3C, D). Surprisingly, PD-L1 surface expression was significantly upregulated after Napabucasin treatment (p<0.01, figure 3C). Gating strategy for studied markers is shown in online supplemental figure S2.
Napabucasin prevented MDSC de novo generation
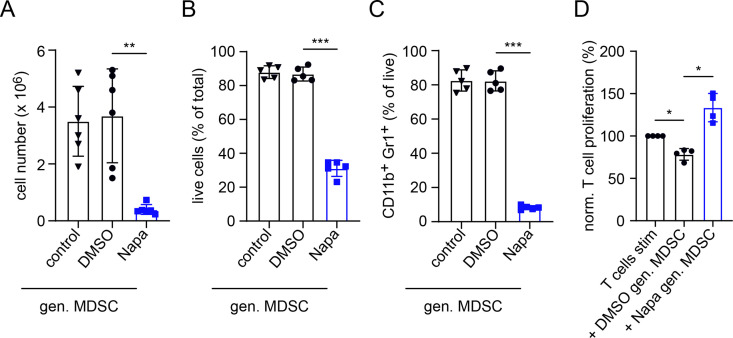
Investigating the generation of MDSC by IL-6 and GM-CSF from murine BM cells in vitro, we observed a significantly decreased cell number after 4 days of culture when Napabucasin was added at the same time as IL-6 and GM-CSF (p<0.01, figure 4A). Furthermore, cell viability and expression of CD11b and Gr1 were significantly decreased after MDSC generation including Napabucasin (p<0.001; figure 4B, C, and online supplemental figure S3). In cultures with DMSO, we found no changes in cell numbers, viability or CD11b+Gr1+ cell frequency after generation (figure 4A–C). Importantly, the remaining viable cells after MDSC generation in the presence of Napabucasin failed to suppress CD8+ T-cell proliferation (p<0.05, figure 4D). Moreover, these Napabucasin-treated MDSCs were even able to increase T-cell proliferation as compared with stimulated T cells cultured without MDSC (figure 4D).
Figure 4.
Inhibition of MDSC generation in vitro by Napaucasin. MDSCs were generated in vitro with IL-6 and GM-CSF (40 ng/mL each, control). Napa (1 µM) or DMSO (0.01%) was added together with cytokines. (A) Data are shown as cell numbers per plate after 4 days of incubation (n=6). Results are presented as the percentage of live (7AAD–) cells within total cells (B) and the percentage of CD11b+Gr1+ cells among total cells (C) measured by flow cytometry (mean±SD, n=5). (D) The function of in vitro generated MDSC was determined on the coculture with CFSE-labeled murine-activated splenic CD8+ T cells. After 72 hours of incubation, T-cell proliferation was evaluated by CFSE dilution measured by flow cytometry. Cumulative data for T-cell proliferation are presented as the percentage of divided T cells norm. to the respective control of stimulated T cells alone (mean±SD, n=4). Statistics were performed on not norm. data. *P<0.05,**P<0.01, ***P<0.001. CFSE, carboxyfluorescein succinimidyl ester; gen., generated; GM-CSF, granulocyte–macrophage colony-stimulating factor; IL, interleukin; MDSC, myeloid-derived suppressor cell; Napa, Napabucasin; norm., normalized.
Napabucasin increased survival of melanoma-bearing mice
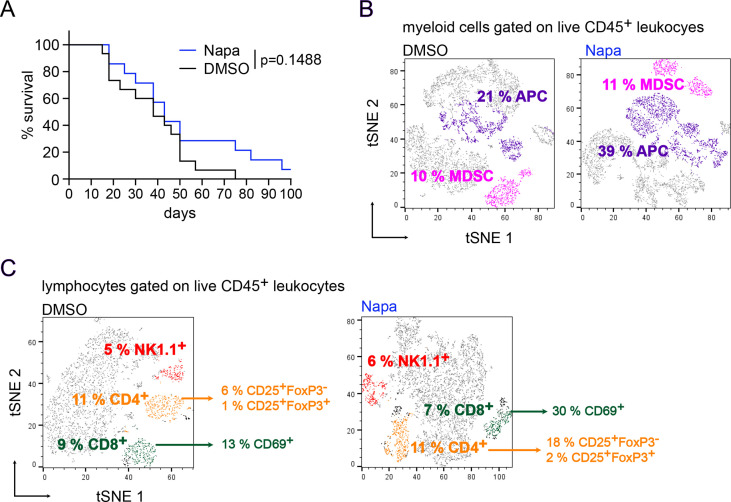
Our in vitro findings that Napabucasin could block MDSC generation and function prompted us to investigate the effects of Napabucasin in melanoma in vivo. To this end, we evaluated the effects of Napabucasin on melanoma progression in RET transgenic mice. Napabucasin-treated mice showed a tendency for prolonged survival, with a median survival of 43 days compared with 38 days in the control group (p=0.1488, figure 5A). Importantly, 1 out of 15 treated mice survived until the end of the experiment (100 days after therapy onset), whereas all non-treated mice were dead by day 75 after the treatment start (figure 5A).
Figure 5.
Effect of Napabucasin in melanoma-bearing mice in vivo. RET transgenic mice with established tumors were injected intraperitoneally with Napa (20 mg/kg) or the corresponding amount of DMSO for 4 weeks, two times per week. (A) Survival of mice is shown as a Kaplan-Meier curve (n=15/group). In another set of experiments, tumor-infiltrating immune cells from Napa-treated (n=2) and DMSO control (n=3) groups were analyzed after 3 weeks of therapy. Results are concatenated, gated on CD45+ live leukocytes and presented as a t-Distributed Stochastic Neighbor Embedding (tSNE) plot. (B) MDSCs were defined as CD11b+Gr1+ cells, whereas APCs were defined as CD11b+Gr1− cells expressing CD11c, CD80, CD86, F4/80 and MHC class II. MDSC and APC frequencies are shown as the percentage among leukocytes. CD11c, CD80, CD86, F4/80 and MHC-II frequencies are shown as the percentage among APCs. (C) Results are presented as the percentage of CD4+ and CD8+ T cells as well as NK1.1+ NK cells within leukocytes. In addition, the percentages of activated CD4+CD25+FoxP3− cells and CD25+FoxP3+ Treg among total CD4+ T cells, as well as activated CD8+CD69+ cells within total CD8+ T cells, are shown. APC, antigen-presenting cell; MDSC, myeloid-derived suppressor cell; MHC-II, major histocompatibility complex class II; Napa, Napabucasin; NK, natural killer; Treg, regulatory T cell.
In a second set of experiments, mice were sacrificed after 3 weeks of therapy to study immune cells in the tumor microenvironment. At this time point, only two out of four mice from the treated group displayed visible tumors, whereas in the control group, macroscopic tumors were found in three out of four mice. However, analyzed tumors showed the comparable tumor weight in both groups. We analyzed the tumor-infiltrating immune cells by flow cytometry. Due to the small amount of analyzed tumors in both groups, statistical analysis was not possible. Therefore, all samples in each group were concatenated and presented as a tSNE plot. In tumors from Napabucasin-treated mice, we found a nearly two-fold elevation of the frequency of CD11b+Gr1− cells, expressing immune stimulatory molecules like MHC class II, CD80 and CD86 as well as F4/80 and CD11c and defined as antigen-presenting cells (APCs) as compared with these cells infiltrating tumors from control group animals (figure 5B). Interestingly, the frequency of CD11b+Gr1+ MDSC, lacking the expression of the aforementioned immune stimulatory molecules, was similar in tumors from both experimental groups (figure 5B). Studying tumor-infiltrating lymphocytes, we detected that frequencies of total CD4+ T cells, CD4+CD25+FoxP3+ regulatory T cells and CD8+ T cells among CD45+ leukocytes were not affected by the treatment (figure 5C). However, frequencies of activated CD69+CD8+ T cells and activated conventional CD4+CD25+FoxP3− T cells were higher than those in non-treated mice (figure 5C). The infiltration of tumors with NK cells was similar in both groups (figure 5C).
Next, we studied also a direct effect of Napabucasin on Ret melanoma cell growth in vitro based on the MTT assay as an indicator of cell viability and proliferation. We found that the treatment of Ret cells with 1 µM Napabucasin for 24, 48 or 72 hours significantly decreased their proliferation as compared with cells treated with 0.01% DMSO (p<0.05, p<0.01, p<0.001, respectively; online supplemental figure S4).
STAT3 inhibition could be beneficial for patients with melanoma
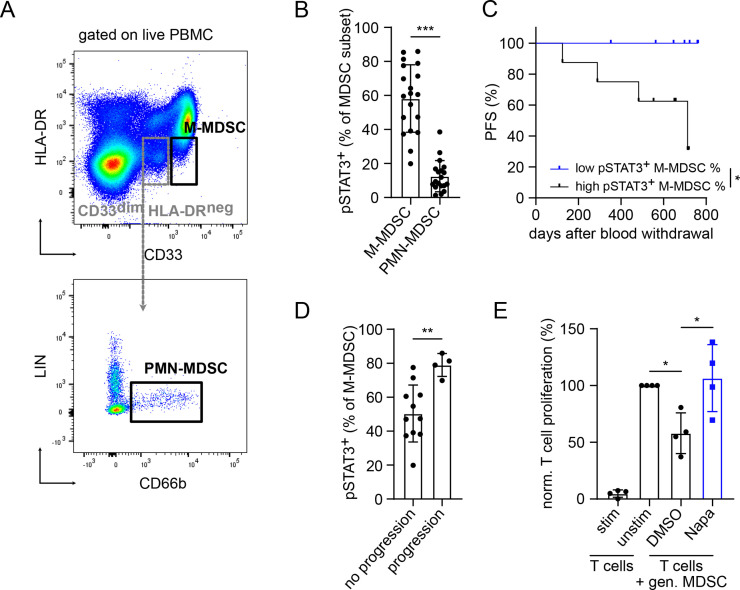
Next, we analyzed the expression of activated STAT3 (defined as pSTAT3) in circulating MDSC from patients with malignant melanoma before the treatment (stages I–IV, individual patient information in online supplemental table S3; overview on patient characteristics and groups in table 1) by flow cytometry (figure 6). Interestingly, we found a significantly higher frequency of pSTAT3+ cells among M-MDSC (CD33highHLA-DRlow/neg) as compared with PMN-MDSC (CD33dimHLA-DRneg CD66b+LINneg) (p<0.001, figure 6B).
Table 1.
Clinical characteristics of patients with malignant melanoma (n=19)
| Characteristics | n |
| Median age, years (range) | 63 (29–84) |
| Sex | |
| Male | 12 |
| Female | 7 |
| Disease stage | |
| I | 6 |
| II | 3 |
| III | 8 |
| IV | 2 |
| Progress after blood withdrawal | |
| No | 11 |
| Yes | 4 |
| Unknown | 4 |
| pSTAT3 (in M-MDSC) classification | |
| High | 8 |
| Low | 7 |
| Stage distribution in ‘pSTAT3 high’ group | |
| I | 1 |
| II | 2 |
| III | 3 |
| IV | 2 |
| Stage distribution in ‘pSTAT3 low’ group | |
| I | 2 |
| II | 1 |
| III | 4 |
| IV | 0 |
| Progressive patients in pSTAT3 high group | |
| Progress | 4 |
| No progress | 4 |
| Progressive patients in pSTAT3 low group | |
| Progress | 0 |
| No progress | 7 |
M-MDSC, monocytic myeloid-derived suppressor cell.
Figure 6.
STAT3 activation in MDSC from patients with melanoma and its inhibition by Napaucasin. PBMC from untreated patients with melanoma (stages I–IV) were analyzed by flow cytometry. (A) Representative dot plots for M-MDSC defined as CD33highHLA-DRlow/neg and PMN-MDSC defined as CD33dim HLA-DRnegCD66b+LIN− are presented. (B) Cumulative data are shown as the percentage of pSTAT3+ cells among respective MDSC subsets (mean±SD, n=19). pSTAT3+ MDSC were gated according to isotype control. (C) PFS of patients with high (>58.2%, n=8) and low (<58.2%, n=7) pSTAT3+ M-MDSC frequencies is shown as a Kaplan–Meier curve. Follow-up period was 2 years. (D) Cumulative data are presented as the percentage of pSTAT3+ M-MDSC within total M-MDSC in patients with (n=11) or without (n=4) progression. (E) Human M-MDSCs were generated in vitro for 4 days with IL-6 and GM-CSF followed by coculture with activated CD3+ T cells labeled with cell proliferation dye eFluor 450 in the presence of 0.5 µM Napa or 0.005% DMSO. On 96 hours of incubation, T-cell proliferation was assessed by proliferation dye dilution measured by flow cytometry. Cumulative data for T-cell proliferation are presented as the percentage of divided T cells norm. to the respective control of stimulated T cells alone (mean±SD, n=4). Statistics were performed on not norm. data. *P<0.05, **P<0.01, ***P<0.001. gen., generated; GM-CSF, granulocyte–macrophage colony-stimulating factor; IL, interleukin; MDSC, myeloid-derived suppressor cell; M-MDSC, monocytic myeloid-derived suppressor cell; Napa, Napabucasin; norm., normalized; PFS, progression-free survival; PMN, polymorphonuclear; STAT3, signal transducer and activator of transcription 3.
To study a possible association between the frequency of circulating pSTAT3+ M-MDSC and the PFS of patients with melanoma, we distributed them into two groups with low and high frequencies of these cells, using the mean value (58.2 %) as a cut-off.11 Eight patients were classified in the ‘pSTAT3 high’ group and seven patients in the ‘pSTAT3 low’ group (table 1). Both groups showed a comparable distribution of patients with respective disease stages. Importantly, we observed no significant differences in the frequency of pSTAT3+ M-MDSC in patients with advanced melanoma (stages III and IV) as compared with patients with stage I+II (online supplemental figure S5). We found that patients with low frequency of pSTAT3+ cells among total M-MDSC displayed significantly longer PFS than patients with high frequency of pSTAT3+ M-MDSC (p<0.05, figure 6C). Moreover, all patients accumulating high amounts of circulating pSTAT3+ M-MDSC were characterized by disease progression (figure 6C). Accordingly, patients with progression showed significantly higher frequencies of pSTAT3+ M-MDSC than those in patients without disease progression (p<0.01, figure 6D). Interestingly, four progressive patients were at the stage III or IV (online supplemental table S3). Finally, M-MDSC generated in vitro from healthy donor CD14+ monocytes on culture with IL-6 and GM-CSF for 4 days could significantly suppress the proliferation of stimulated CD3+ T cells (p<0.05, figure 6E). Importantly, this M-MDSC-mediated suppression of T-cell proliferation was completely abrogated on addition of Napabucasin to the coculture (p<0.05, figure 6E).
Discussion
Signaling via STAT3 has been described to shift the tumor microenvironment into the direction of immunosuppression, promoting the accumulation of MDSC and thereby tumor progression.29 Here we investigated the potential of the STAT3 inhibitor Napabucasin to block MDSC-mediated immunosuppression in malignant melanoma. We observed a complete abrogation of MDSC immunosuppressive capacity on Napabucasin addition to the coculture with stimulated T cells. Napabucasin significantly enhanced MDSC apoptosis and the expression of CD80 and MHC class II molecules. In addition, we found a slight downregulation of iNOS and Arg-1 and a significant decrease in Cox2 gene expression.
In line with our data, it has been recently shown that Napabucasin induced MDSC apoptosis in a caspase 3-dependent manner accompanied by an upregulation of proapoptotic Bcl-2-associated X protein (Bax) and a downregulation of antiapoptotic B-cell lymphoma 2 (Bcl-2).30 Moreover, our microarray analysis of Napabucasin-treated MDSC revealed an increase in oxidative phosphorylation, which is important for mitochondrial ROS production and can induce cell death via the intrinsic apoptosis pathway,31 providing a second mechanism for the induction of MDSC apoptosis by Napabucasin.
STAT3 has been implicated in preventing terminal differentiation and maturation of myeloid cells and shifting these cells to an immunosuppressive phenotype in cancer.32 It has been reported that STAT3 activation resulted in decreased MHC class II expression on APC, especially on dendritic cells (DCs).33 34 Furthermore, STAT3 was demonstrated to be responsible for the downregulation of CD80 and CD86 expression on tolerogenic DC.35 These observations are in accordance with our findings, showing that STAT3 inhibition by Napabucasin led to an upregulation of the expression of CD80 and MHC class II. However, we did not observe any changes in CD86 expression after Napabucasin treatment. Additionally, our microarray analysis highlighted a significant stimulation of the antigen processing and presentation pathways by Napabucasin. Moreover, we found a significant activation of the phagosome and lysosome pathways in MDSC on the treatment. These pathways are known to be implicated in antigen uptake and processing by APC,36 indicating a shift of MDSC phenotype from immunosuppression toward antigen processing and presentation.
STAT3 is playing an important role in the activation of the immunosuppressive capacity of MDSC, especially induced by cytokines such as IL-6.14 Arg-1 and iNOS were shown to be expressed in MDSC in a STAT3-dependent manner.18 20 22 37 However, we observed only a slight decrease in Arg-1 gene and protein expression after the STAT3 inhibition by Napabucasin. In addition, a decrease in Nos2 gene and iNOS protein expression was not statistically significant. It has been shown that Nox2 is responsible for ROS production by MDSC and expressed as a result of STAT3 activation in these cells.4 In line with this observation, we have previously demonstrated that in vitro generated MDSC produce increased levels of ROS by an IL-6/STAT3-dependent mechanism.22 Surprisingly, we failed to observe a downregulation of Nox2 gene expression in Napabucasin-treated MDSC by microarray as well as a significant decrease in ROS production by these cells. The expression of Ptges2 and Cox2 was described to induce production of immunosuppressive PGE2 by MDSC.8 In our experiments, we found a significant downregulation of Cox2 and a slight downregulation of Ptges2 gene expression in Napabucasin-treated MDSC. Although PD-L1 expression on MDSC has been shown to be STAT3-dependent,21 30 we found a significant upregulation of PD-L1 gene and protein expression on the treatment. Since other transcription factors like Jun, NF- κB or Myc also regulate PD-L1 expression,38 it is possible that they are still active after the STAT3 blockade by Napabucasin.
In summary, we did not observe a major downregulation of one specific immunosuppressive marker on MDSC after 24 hours of Napabucasin treatment. Therefore, the abrogation of immunosuppressive capacity could be attributed to the sum of small changes in immunosuppressive pattern together with the significant induction of MDSC apoptosis. Intriguingly, we observed even an increased stimulation of CD8+ T-cell proliferation compared with the activated T cells cultured without MDSC in some cases after Napabucasin treatment. As T-cell proliferation was stimulated by anti-CD3 and anti-CD28 antibodies in the coculture, this increase cannot be fully explained by the upregulation of antigen processing and presentation pathways in MDSC induced by Napabucasin. However, it is plausible that Napabucasin treated MDSC could produce elevated levels of cytokines, resulting in this supplemental stimulation of T-cell proliferation.
In addition to the inhibitory effects of Napabucasin on functionally active MDSC, we demonstrated that its addition during the in vitro generation of MDSC by IL-6 and GM-CSF strongly reduced MDSC generation, and the remaining cells were not able to suppress the proliferation of CD8+ T cells. These results are in accordance with the critical role STAT3 is playing in the accumulation and de novo generation of MDSC.14
It should be mentioned that STAT3 is also important for antitumor immune cells, promoting T-cell activation and memory formation as well as NK-cell cytolytic activity.39–41 We have recently reported that IL-6 blockade in RET transgenic melanoma-bearing mice accelerated tumor growth and significantly shortened survival, accompanied by a decrease in activated CD8+ T-cell infiltration into the tumor.22 However, our current study revealed that STAT3 inhibition by Napabucasin did not affect T-cell survival and proliferation at the concentrations used for the MDSC inhibition. Therefore, we addressed the question if Napabucasin could induce antitumor effects in vivo.
We detected that Napabucasin prolonged survival of RET transgenic melanoma-bearing mice, with 1 out of 15 mice surviving until the end of the experiment, while all untreated mice were already dead. Our data are in agreement with the current literature, reporting that CpG-coupled STAT3 antisense oligonucleotides (ASO) specifically targeting myeloid cells could impair acute myeloid leukemia in immune deficient and immunocompetent mouse models.42 Furthermore, CpG-coupled STAT3 ASO was shown to eradicate the majority of tumors in a pancreatic ductal adenocarcinoma (PDAC) mouse model by decreasing MDSC and increasing CD8+ T-cell frequency.43 Another group described that the administration of STAT3 ASO significantly improved response to radiotherapy and reduced tumor burden in a PDAC mouse model.44 Similar results were obtained with such treatment in a humanized HNSCC mouse model.45 Moreover, the STAT3 inhibitors Stattic and Napabucasin were shown to significantly reduce the growth of mouse colon cancer metastases in the liver by inducing MDSC apoptosis.30 Although we did not find a reduction of melanoma-infiltrating MDSC, the population of APC (CD11b+Gr1− cells, among them CD11c+, CD80+, CD86+, F4/80+ and MHC class II+ cells) was strongly increased after Napabucasin therapy. Furthermore, an increase in activated CD8+CD69+ T cells and activated conventional CD4+CD25+FoxP3− T cells was observed. In contrast to transplanted tumor models, RET transgenic mice develop tumors spontaneously with a different dynamics of tumor development, which more closely resembles the human pathology. This might explain the lack of MDSC reduction on Napabucasin treatment. Due to low cell numbers, we were not able to test the suppressive activity of tumor-infiltrating MDSC after Napabucasin treatment. Thus, it is possible that isolated CD11b+Gr1+ cells, although lacking APC markers, are no longer immunosuppressive after Napabucasin therapy.
Importantly, we also showed that Napabucasin decreased the proliferation of Ret melanoma cells in vitro. This is in line with the data on important role of STAT3 for tumor cell survival, proliferation, angiogenesis, invasiveness and metastasis.46 Since tumors from mice of treated and control groups were of similar size, we suggest that our findings on the effect of Napabucasin on tumor-infiltrating immune cells are independent from its direct effect on the tumor cells. However, for the potential therapeutic targeting of STAT3, it should be better if STAT3 inhibition enhances antitumor immunity in combination with a direct reduction tumor cell growth, leading to improved survival.
Analyzing STAT3 activation status in circulating MDSC from patients with melanoma, we found a higher frequency of pSTAT3 expressing M-MDSC as compared with PMN-MDSC. It is plausible that PMN-MDSC could be more sensitive to the fixation and permeabilization needed for the intracellular staining for pSTAT3 than M-MDSC. This might explain not only lower frequency of pSTAT3+PMN MDSC among total PMN-MDSC but also reduced frequency of PMN-MDSC within PBMC. We demonstrated that a high frequency of M-MDSC expressing pSTAT3 was associated with significantly worse PFS. In line with our findings, immunosuppressive CD14+HLA−DRlow/− MDSCs from patients with malignant melanoma were found to be pSTAT3high.19 Furthermore, in patients with HNSCC, pSTAT3 levels were correlated with an upregulation of Arg-1 expression and MDSC immunosuppressive activity.18 M-MDSC isolated from patients with PDAC showed a high STAT3/ARG-1 gene expression profile as opposed to non-suppressive monocytes.20 Importantly, we found that Napabucasin abrogated the immunosuppressive capacity of human in vitro generated M-MDSC. STAT3 ASO has been shown to exert clinical activity in patients with lymphoma and lung cancer.47 Moreover, Napabucasin in combination with paclitaxel increased survival in three out of nine patients with advanced melanoma in a phase Ib clinical trial.48 The safety and efficacy of Napabucasin as a cancer cell stemness inhibitor was evaluated in patients with cancer in several clinical trials up to phase III.23 It induced only mild adverse events and could suppress metastasis as well as prevent relapse in patients with varying cancer types.23 However, more trials are needed to include Napabucasin into immunotherapeutic strategies for patients with cancer.
Conclusion
Although MDSCs have been shown to correlate with worse prognosis and therapy outcome in different cancer entities (including malignant melanoma), no therapeutic approach for MDSC targeting has been approved for clinical application. We found an impressive ability of the STAT3 inhibitor Napabucasin to abrogate immunosuppressive activity of murine MDSC in vitro without a negative impact on T cells. Moreover, this inhibitor shifted MDSC toward cells with antigen processing and presentation ability as well as increased survival of melanoma-bearing mice. An elevated frequency of human M-MDSC expressing pSTAT3 (as an indicator of STAT3 activation) significantly correlated with decreased PFS of patients with melanoma. Importantly, similar to murine MDSC, in vitro generated human M-MDSC lost their immunosuppressive function on the treatment with Napabucasin. We suggest that STAT3 inhibition by Napabucasin could be a promising strategy to inhibit MDSC functions and to improve current immunotherapeutic approaches for patients with melanoma.
Acknowledgments
We thank Dr Izumi Nakashima (Chubu University, Aichi, Japan) for providing the RET transgenic mice. We thank the staff of the animal facilities of DKFZ Heidelberg and Medical Faculty Mannheim, University of Heidelberg, for the assistance with the animal experiments. We thank the microarray core facility of DKFZ Heidelberg for carrying out the microarray analysis.
Footnotes
RB and AK contributed equally.
Contributors: VU, JU and RB designed the study. RB, AK, FÖ, CT, SL, AL and AS preformed the experiments and the data analysis. VM was responsible for patient enrollment. RB, AK and VU wrote the manuscript. All authors read and approved the manuscript. VU is responsible for the overall content as guarantor.
Funding: This work was supported by the German Federal Ministry of Education and Research–SERPENTINE project in the ERA PerMed network (01KU2017 to VU), the German Research Foundation (project number 259332240/RTG 2099 (to JU and VU) and the German Academic Exchange Service (DAAD to FÖK).
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available on reasonable request. The datasets generated, used and analyzed during the current study are available from the corresponding author. The microarray data generated during the study are available on the GEO repository under the number GSE189147.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human subjects and was approved by the ethics committee of University Medical Center Mannheim, Germany (2010-318N-MA). Subjects gave informed consent to participate in the study before taking part. Murine in vivo studies were approved by the German local authorities (G-40/19, G-73/18) and conducted respecting ethical and legal regulations.
References
- 1.Groth C, Hu X, Weber R, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer 2019;120:16–25. 10.1038/s41416-018-0333-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronte V, Brandau S, Chen S-H, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noman MZ, Desantis G, Janji B, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med 2014;211:781–90. 10.1084/jem.20131916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corzo CA, Cotter MJ, Cheng P, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol 2009;182:5693–701. 10.4049/jimmunol.0900092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 2007;109:1568–73. 10.1182/blood-2006-06-031856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raber PL, Thevenot P, Sierra R, et al. Subpopulations of myeloid-derived suppressor cells impair T cell responses through independent nitric oxide-related pathways. Int J Cancer 2014;134:2853–64. 10.1002/ijc.28622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Du W, Yan F, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol 2013;190:3783–97. 10.4049/jimmunol.1201449 [DOI] [PubMed] [Google Scholar]
- 8.Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol 2012;188:21–8. 10.4049/jimmunol.1101029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan KR, Amaria RN, Ramirez O, et al. Myeloid-derived suppressor cells are associated with disease progression and decreased overall survival in advanced-stage melanoma patients. Cancer Immunol Immunother 2013;62:1711–22. 10.1007/s00262-013-1475-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weide B, Martens A, Zelba H, et al. Myeloid-derived suppressor cells predict survival of patients with advanced melanoma: comparison with regulatory T cells and NY-ESO-1- or melan-A-specific T cells. Clin Cancer Res 2014;20:1601–9. 10.1158/1078-0432.CCR-13-2508 [DOI] [PubMed] [Google Scholar]
- 11.Jiang H, Gebhardt C, Umansky L, et al. Elevated chronic inflammatory factors and myeloid-derived suppressor cells indicate poor prognosis in advanced melanoma patients. Int J Cancer 2015;136:2352–60. 10.1002/ijc.29297 [DOI] [PubMed] [Google Scholar]
- 12.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, et al. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res 2016;22:2908–18. 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber R, Fleming V, Hu X, et al. Myeloid-Derived suppressor cells hinder the anti-cancer activity of immune checkpoint inhibitors. Front Immunol 2018;9:1310. 10.3389/fimmu.2018.01310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol 2011;32:19–25. 10.1016/j.it.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546–58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber R, Groth C, Lasser S, et al. Il-6 as a major regulator of MDSC activity and possible target for cancer immunotherapy. Cell Immunol 2021;359:104254. 10.1016/j.cellimm.2020.104254 [DOI] [PubMed] [Google Scholar]
- 17.Chen M-F, Kuan F-C, Yen T-C, et al. IL-6-stimulated CD11b+ CD14+ HLA-DR- myeloid-derived suppressor cells, are associated with progression and poor prognosis in squamous cell carcinoma of the esophagus. Oncotarget 2014;5:8716–28. 10.18632/oncotarget.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez-Dunddel D, Pan F, Zeng Q, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest 2013;123:1580–9. 10.1172/JCI60083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poschke I, Mougiakakos D, Hansson J, et al. Immature immunosuppressive CD14+HLA-DR-/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-Sign. Cancer Res 2010;70:4335–45. 10.1158/0008-5472.CAN-09-3767 [DOI] [PubMed] [Google Scholar]
- 20.Trovato R, Fiore A, Sartori S, et al. Immunosuppression by monocytic myeloid-derived suppressor cells in patients with pancreatic ductal carcinoma is orchestrated by STAT3. J Immunother Cancer 2019;7:255. 10.1186/s40425-019-0734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfle SJ, Strebovsky J, Bartz H, et al. PD-L1 expression on tolerogenic APCS is controlled by STAT-3. Eur J Immunol 2011;41:413–24. 10.1002/eji.201040979 [DOI] [PubMed] [Google Scholar]
- 22.Weber R, Riester Z, Hüser L, et al. Il-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma. J Immunother Cancer 2020;8:e000949. 10.1136/jitc-2020-000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard JM, Grothey A. Napabucasin: an update on the first-in-class cancer stemness inhibitor. Drugs 2017;77:1091–103. 10.1007/s40265-017-0759-4 [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Takahashi M, Akhand AA, et al. Transgenic mouse model for skin malignant melanoma. Oncogene 1998;17:1885–8. 10.1038/sj.onc.1202077 [DOI] [PubMed] [Google Scholar]
- 25.Marigo I, Bosio E, Solito S, et al. Tumor-Induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity 2010;32:790–802. 10.1016/j.immuni.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 26.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao F, Falk C, Osen W, et al. Activation of p38 mitogen-activated protein kinase drives dendritic cells to become tolerogenic in RET transgenic mice spontaneously developing melanoma. Clin Cancer Res 2009;15:4382–90. 10.1158/1078-0432.CCR-09-0399 [DOI] [PubMed] [Google Scholar]
- 29.Rébé C, Végran F, Berger H, et al. Stat3 activation: a key factor in tumor immunoescape. JAKSTAT 2013;2:e23010. 10.4161/jkst.23010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guha P, Gardell J, Darpolor J, et al. Stat3 inhibition induces Bax-dependent apoptosis in liver tumor myeloid-derived suppressor cells. Oncogene 2019;38:533–48. 10.1038/s41388-018-0449-z [DOI] [PubMed] [Google Scholar]
- 31.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 2014;94:909–50. 10.1152/physrev.00026.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y-L, Banerjee S, White SV, et al. Stat3 in tumor-associated myeloid cells: multitasking to disrupt immunity. Int J Mol Sci 2018;19. 10.3390/ijms19061803. [Epub ahead of print: 19 Jun 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan LLY, Cheung BKW, Li JCB, et al. A role for STAT3 and cathepsin S in IL-10 down-regulation of IFN-gamma-induced MHC class II molecule on primary human blood macrophages. J Leukoc Biol 2010;88:303–11. 10.1189/jlb.1009659 [DOI] [PubMed] [Google Scholar]
- 34.Farren MR, Carlson LM, Netherby CS, et al. Tumor-Induced STAT3 signaling in myeloid cells impairs dendritic cell generation by decreasing PKCβII abundance. Sci Signal 2014;7:ra16. 10.1126/scisignal.2004656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang S, Ristich V, Arase H, et al. Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6--STAT3 signaling pathway. Proc Natl Acad Sci U S A 2008;105:8357–62. 10.1073/pnas.0803341105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harding CV. Class II antigen processing: analysis of compartments and functions. Crit Rev Immunol 1996;16:13–29. 10.1615/CritRevImmunol.v16.i1.20 [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Deng Z, Peng Y, et al. Ascites-derived IL-6 and IL-10 synergistically expand CD14+HLA-DR-/low myeloid-derived suppressor cells in ovarian cancer patients. Oncotarget 2017;8:76843–56. 10.18632/oncotarget.20164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. 10.1016/j.immuni.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh H-M, Yu C-R, Golestaneh N, et al. Stat3 protein promotes T-cell survival and inhibits interleukin-2 production through up-regulation of class O forkhead transcription factors. J Biol Chem 2011;286:30888–97. 10.1074/jbc.M111.253500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegel AM, Heimall J, Freeman AF, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 2011;35:806–18. 10.1016/j.immuni.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cacalano NA. Regulation of natural killer cell function by STAT3. Front Immunol 2016;7:128. 10.3389/fimmu.2016.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Hossain DMS, Duttagupta P, et al. Serum-resistant CpG-STAT3 decoy for targeting survival and immune checkpoint signaling in acute myeloid leukemia. Blood 2016;127:1687–700. 10.1182/blood-2015-08-665604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreira D, Adamus T, Zhao X, et al. Stat3 inhibition combined with CpG immunostimulation activates antitumor immunity to eradicate genetically distinct castration-resistant prostate cancers. Clin Cancer Res 2018;24:5948–62. 10.1158/1078-0432.CCR-18-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oweida AJ, Mueller AC, Piper M, et al. Response to radiotherapy in pancreatic ductal adenocarcinoma is enhanced by inhibition of myeloid-derived suppressor cells using STAT3 anti-sense oligonucleotide. Cancer Immunol Immunother 2021;70:989–1000. 10.1007/s00262-020-02701-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira D, Sampath S, Won H, et al. Myeloid cell-targeted STAT3 inhibition sensitizes head and neck cancers to radiotherapy and T cell-mediated immunity. J Clin Invest 2021;131:e137001. 10.1172/JCI137001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumari N, Dwarakanath BS, Das A, et al. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol 2016;37:11553–72. 10.1007/s13277-016-5098-7 [DOI] [PubMed] [Google Scholar]
- 47.Hong D, Kurzrock R, Kim Y, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 2015;7:314ra185. 10.1126/scitranslmed.aac5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edenfield WJ, Becerra C, Braiteh FS, et al. A phase Ib study of napabucasin plus weekly paclitaxel in patients with advanced melanoma. J Clin Oncol 2017;35:9553. 10.1200/JCO.2017.35.15_suppl.9553 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-004384supp002.pdf (29.7KB, pdf)
jitc-2021-004384supp001.pdf (806.9KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available on reasonable request. The datasets generated, used and analyzed during the current study are available from the corresponding author. The microarray data generated during the study are available on the GEO repository under the number GSE189147.