Abstract
Mitochondria, the primary ATP-producing organelles, are highly abundant in cardiomyocytes. Mitochondrial function readily deteriorates in the presence of stress and, thus, maintenance of mitochondrial quality is essential for sustaining pump function in the heart. Cardiomyocytes under stress attempt to maintain mitochondrial quality primarily through dynamic changes in their morphology, namely fission and fusion, degradation, and biogenesis. Mitophagy, a mitochondria-specific form of autophagy, is a major mechanism of degradation. The level of mitophagy is altered in stress conditions, which, in turn, significantly affects mitochondrial function, cardiomyocyte survival, and death and cardiac function. Thus, mitophagy has been emerging as a promising target for treatment of cardiac conditions. To develop specific interventions, modulating the activity of mitophagy in the heart, understanding how mitochondria are degraded in a given condition is important. Increasing lines of evidence suggest that there are multiple mechanisms by which mitochondria are degraded through mitophagy in the heart. For example, in addition to the well-established mechanism commonly utilized by general autophagy, involving Atg7 and LC3, recent evidence suggests that an alternative mechanism, independent of Atg7 and LC3, also mediates mitophagy in the heart. Here, we describe molecular mechanisms through which mitochondria are degraded in the heart and discuss their functional significance. We also discuss molecular interventions to modulate the activity of mitophagy and their potential applications for cardiac conditions.
Keywords: Mitophagy, Alternative mitophagy, Parkin, Rab9, Drp1
Graphical Abstract
1. Introduction
The heart continuously demands a vast amount of energy for contractions and relaxations. While cardiac mitochondria supply the majority of ATP in the heart at baseline, the damaged mitochondria therein become a source of reactive oxidative species (ROS) and cell-death signalling under pathological conditions.1 The failing heart manifests mitochondrial dysfunction regardless of its aetiology, and mitochondrial dysregulation broadly contributes to the pathophysiology of heart failure. Accordingly, mitochondrial quality control is highly critical in the heart and has been investigated extensively in the last decade.
The mitochondria quality control mechanism is intimately intertwined with mitochondrial dynamics, e.g. fusion and fission, degradation primarily mediated through mitochondria-specific autophagy (mitophagy), and biogenesis.2 The fusion of two mitochondria is mainly mediated by GTPases, mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), and optic atrophy protein 1 (Opa1). Mfn1 and Mfn2 regulate fusion of the outer mitochondrial membranes (OMMs), whereas Opa1 regulates fusion of the inner mitochondrial membranes (IMMs) and cristae remodelling.3 Fission is mainly controlled by dynamin-related protein 1 (Drp1) and its receptors, mitochondrial fission 1 protein (Fis1), mitochondrial division protein 1 (Mdv1), and mitochondrial fission factor (Mff).3 Fission generates uneven daughter mitochondria and those with reduced membrane potential have a reduced probability of fusing again, so that damaged mitochondria are eventually eliminated through mitophagy and replaced through expansion of pre-existing mitochondria (biogenesis).4 When this mechanism is disturbed, the depolarized portion of mitochondria accumulates, thereby leading to global impairment of mitochondrial function.5 Mitophagy is thus considered an essential mechanism for maintaining the quality of mitochondria.
Autophagy is a major mechanism of lysosome-dependent cellular degradation. Although macroautophagy, the most well studied form of autophagy, characterized by the presence of double-membrane vesicles termed autophagosomes, degrades cytosolic materials, and organelles in a non-selective manner, it can also degrade specific targets in a selective manner. The latter is termed selective autophagy and includes mitophagy. Although non-selective autophagy and mitophagy share mechanisms mediating autophagosome formation and degradation, it appears that additional mechanisms are involved in the induction of mitophagy and recognition of mitochondria to be degraded. We have shown recently that autophagy and mitophagy are activated with distinct time courses in the stressed heart, suggesting that mitophagy may be regulated by mechanisms that are distinct from those that regulate non-selective autophagy in the heart. Increasing lines of evidence suggest that mitophagy is involved in modulation of heart disease conditions. Thus, it is important to understand how mitophagy is regulated in response to stress in the heart. In this review, we summarize the molecular mechanisms and the functional significance of mitophagy in the heart at baseline and in response to stress and discuss their clinical implications.
2. Molecular mechanisms of mitophagy
2.1 Ubiquitin-dependent mitophagy in mammalian cells
In mammalian cells, mitophagy is mediated through multiple mechanisms.6,7 Depending upon how damaged mitochondria are labelled for sequestration by autophagosomes, mitophagy in mammalian cells can be broadly classified as either ubiquitin-dependent mitophagy or receptor-dependent mitophagy.7 In the ubiquitin-dependent category, thus far, Pink1- and Parkin-mediated mitophagy has been characterized most intensively.7 Pink1 is a serine/threonine kinase, whereas Parkin is an E3 ubiquitin kinase. Pink1 is anchored at the IMM and degraded by matrix processing peptidase and presenilin-associated rhomboid-like in intact mitochondria.1 In response to depolarization of mitochondrial membrane potential or the unfolded protein response in mitochondria, Pink1 is accumulated in the OMM with its kinase domain facing the cytosol. Stabilization of Pink1 is also regulated by adenine-nucleotide translocator (ANT)-TIM44 mediated inhibition of the presequence translocase TIM23. The effect of ANT upon TIM23 and mitophagy is mediated independently of its nucleotide translocase catalytic activity. How ANT is involved in mitophagy activation under stress remains to be elucidated.8 Evidence suggests that Parkin and Pink1 operate together in a common pathway for mitochondrial quality control.9 Pink1 recruits Parkin to damaged mitochondria through phosphorylation of either ubiquitin at serine 65 or Mfn2 at threonine 111 and serine 442.10 Pink1 also directly phosphorylates and activates Parkin.11,12 Since Parkin can be recruited to peroxisomes or lysosomes when Pink1 is artificially overexpressed and targets these organelles,9 phosphorylation of ubiquitin or the direct interaction between Parkin and Pink1 may be important. In contrast, transgenic expression of an Mfn2 mutant that is not phosphorylated by Pink1 (Tg-Mfn2 AA) in the heart results in lethality with dilated cardiomyopathy,13 suggesting the importance of the Pink1-Mfn2 pathway in the heart at baseline. Parkin ubiquitinates various mitochondrial outer membrane proteins on depolarized mitochondria, including Tom20, VDAC, hexokinase I, Mitofusin, Miro, and MitoNEET/CISD1, whereas LC3-anchor proteins are involved in the recognition of ubiquitinated mitochondrial proteins, including NDP52, Optineurin, NBR1, and p62, by autophagosomes in cultured cells.10 Further investigation is required to establish the significance of most of these mechanisms in mediating mitophagy in the heart in vivo. Compared to the molecular mechanism as to how damaged mitochondria are recognized, how autophagosomes are formed on site remains poorly understood. NDP52 and TBK1 in concert recruit and activate the Ulk1/FIP200 complex independently of AMPK and even in the presence of nutrient rich conditions, which in turn drives targeted autophagosome biogenesis.14
Currently, forced-expression of Parkin and/or CCCP treatment, an intervention that depolarizes mitochondrial membrane potential, is commonly used to investigate Pink1/Parkin-mediated mitophagy in mammalian cells. However, these experimental conditions may not necessarily faithfully reproduce conditions where mitophagy is induced in mammals in vivo.9 Furthermore, Pink 1- and Parkin-dependent mechanisms have never been identified through unbiased screening of mitophagy mediators in mammalian cells.15,16 Thus, more investigation is needed to establish Pink1- and Parkin-dependent mitophagy as the predominant form of mitophagy under various conditions in vivo.
2.2 Receptor-dependent mitophagy in mammalian cells
Several pathways have been reported to regulate mitophagy in a Parkin- or ubiquitin-independent manner. In these mechanisms, damaged mitochondria are directly recognized by autophagosomes via expression of proteins with the LC3-interacting region (LIR) on the OMM; thus, these mechanisms are called receptor-dependent mitophagy. One such example is Fun 14 domain-containing protein 1 (Fundc1), an OMM protein with an LIR, which mediates mitophagy in HeLa cells and MEFs.17 In the resting state, CK2 and Src phosphorylate Fundc1 at serine 13 and tyrosine 18, respectively, and suppress the interaction between LC3 and Fundc1, thereby inhibiting mitophagy. However, under stress conditions, phosphorylation of Fundc1 by Ulk1 at serine 17 and dephosphorylation by PGAM5 at serine 13 promote interaction between LC3 and Fundc1 and induce mitophagy. In other examples of receptor mediated mitophagy, Bnip3- and Nix-mediated mechanisms play a significant role in mediating reticulocyte differentiation.18,19
Besides the aforementioned ubiquitin-dependent mitophagy and receptor-dependent mitophagy, recent evidence suggests that Ulk1-/Rab9-dependent alternative autophagy also plays a major role in mediating starvation- and hypoxia-induced mitophagy in HeLa cells.20 Details concerning alternative autophagy are discussed in Sections 4 and 5.
3. Mitophagy in the heart
3.1 Mitophagy in cardiac homoeostasis and development
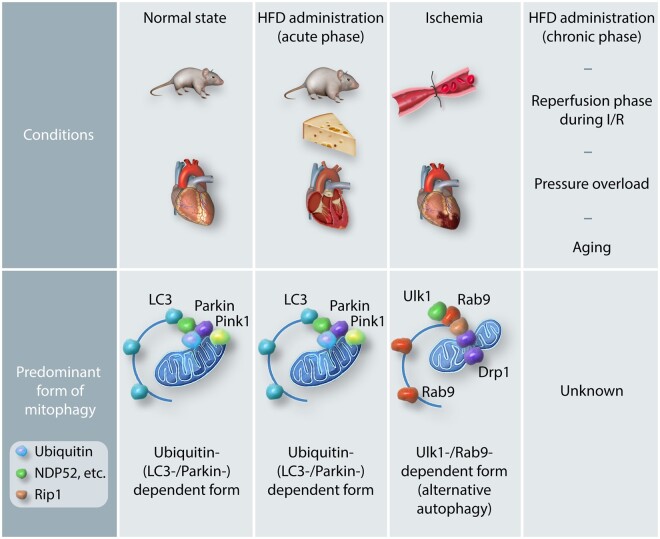
Mitochondria occupy a large percentage of the heart volume and continuously produce ATP and ROS, by-products of respiration, even at baseline. Mitophagy, autophagy targeting mitochondria, is essential for maintaining mitochondrial function and both the energetic and redox homoeostasis in cardiomyocytes. We summarized the cardiac phenotype of animal models of cardiac conditions with genetic manipulations related to the mitophagy pathways (Table 1).
Table 1.
Phenotypes of cardiac disease models with genetic interventions related to the mitophagy pathways
| Target gene | Gene manipulation | Conditions | Mitophagy | Mitochondrial phenotype | Cardiac phenotype | Refs. |
|---|---|---|---|---|---|---|
| Ubiquitin-dependent mitophagy | ||||||
| Parkin | Germ-line KO | Baseline | – | Normal | Normal | 21 |
| Germ-line KO | MI | – | Dysfunction | Increased injury | 21 | |
| Germ-line KO | Baseline | Preserved | Mild dysfunction | Normal | 79 | |
| Germ-line KO | Sepsis | Preserved | Dysfunction | Normal | 79 | |
| Germ-line KO | DOX | Reduced | Dysfunction | Increased injury | 80 | |
| Germ-line KO | IPC | – | – | Increased injury | 28 | |
| Cardiac-specific inducible KO | Baseline (perinatal) | Reduced | Dysfunction | Lethal cardiomyopathy | 13 | |
| Cardiac-specific inducible KO | Baseline (adult) | – | Normal | Normal | 22 | |
| Cardiac-specific TG | Baseline | Increased | Normal | Normal | 22 | |
| Pink1 | Germ-line KO | Baseline | Reduced | Dysfunction | Dysfunction | 81 |
| Germ-line KO | TAC | Reduced | Dysfunction | Increased injury | 81 | |
| Germ-line KO | Exercise | Reduced | – | – | 82 | |
| Cardiac-specific TG | Baseline | – | – | Normal | 83 | |
| Cardiac-specific TG | I/R | – | – | Reduced injury | 83 | |
| Receptor-dependent mitophagy | ||||||
| Bnip3 | Germ-line KO | Baseline | – | – | Normal | 84 |
| Germ-line KO | I/R | – | – | Reduced injury | 84 | |
| Cardiac-specific inducible TG | Baseline | – | – | Dysfunction | 84 | |
| Cardiac-specific inducible TG | MI | – | – | Increased injury | 84 | |
| Nix | Germ-line KO | Baseline | – | – | Dysfunction | 85 |
| Cardiac-specific KO | Baseline | – | – | Normal | 86 | |
| Cardiac-specific KO | TAC | – | – | Preserved function | 86 | |
| Cardiac-specific TG | Baseline | – | – | Dysfunction | 87 | |
| Fundc1 | Cardiac-specific KO | Baseline | – | Dysfunction | Dysfunction | 88 |
| Cardiac-specific KO | MI | – | Dysfunction | Increased injury | 88 | |
| Fusion/fission-related genes | ||||||
| Mfn1/2 | Cardiac-specific KO | Baseline | – | – | Lethal cardiomyopathy | 89 |
| Cardiac-specific KO | Baseline | – | – | Lethal cardiomyopathy | 90 | |
| Cardiac-specific inducible KO | Baseline | – | Dysfunction | Dysfunction | 90 | |
| Cardiac-specific inducible KO | Baseline | – | Dysfunction | Dysfunction | 91 | |
| Cardiac-specific KO | Baseline | – | – | Lethal cardiomyopathy | 92 | |
| Cardiac-specific inducible KO | Baseline | – | Dysfunction | Dysfunction | 92 | |
| Cardiac-specific inducible KO | I/R | – | – | Reduced injury | 93 | |
| Drp1 | Muscle-specific KO | Baseline | – | Dysfunction | Lethal cardiomyopathy | 94 |
| Cardiac-specific KO (homo) | Baseline | – | Dysfunction | Lethal cardiomyopathy | 24 | |
| Cardiac-specific KO (hetero) | Baseline | – | Dysfunction | Dysfunction | 24 | |
| Cardiac-specific inducible KO | Baseline | Increased | Dysfunction | Dysfunction | 91 | |
| Cardiac-specific inducible KO | Baseline | Reduced | Dysfunction | Dysfunction | 25 | |
| Cardiac-specific KO (hetero) | Baseline | Reduced | Dysfunction | Dysfunction | 25 | |
| Cardiac-specific KO (hetero) | I/R | Reduced | Dysfunction | Increased injury | 25 | |
| Cardiac-specific KO (hetero) | TAC | Reduced | Dysfunction | Increased injury | 30 | |
| Opa1 | Germ-line KO (hetero) | Baseline | – | Dysfunction | Normal | 95 |
| Germ-line KO (hetero) | TAC | – | – | Increased injury | 95 | |
DOX, doxorubicin; I/R, ischaemia/reperfusion; IPC, ischaemic preconditioning; KO, knock-out; MI, myocardial infarction; TAC, transverse aortic constriction; TG, transgenic.
In Drosophila melanogaster, knockout of Parkin induces heart failure with mitochondrial abnormality, suggesting that Parkin is essential for maintaining mitophagy and mitochondrial homoeostasis in the heart at baseline.5 Heart-specific deletion of Parkin in mice during the perinatal period with myh6-driven MER-Cre-MER induces lethal cardiomyopathy without postnatal maturation of mitochondria,13 suggesting that Parkin plays a vital role in the maintenance of the mammalian postnatal heart. In contrast, in mice with knockout of Parkin in the germ-line, normal function of the heart and mitochondria is maintained until 12 months of age.21 In this case, germ-line knockout may activate compensatory mechanisms and mask the effect of Parkin-knockout. Similarly, however, tamoxifen-mediated ablation of Parkin at the adult stage in Parkinflox/flox myh6-driven MER-Cre-MER mice22 also did not induce dysfunction in the mouse heart and mitochondria. This suggests that Parkin-dependent mitophagy may not play an essential role in maintaining mitochondrial function in the adult mouse heart. By inference, the abnormal cardiac phenotype observed in Tg-Mfn2 AA13 may be mediated through additional mechanisms besides the suppression of Parkin-mediated mitophagy.
The role of Drp1 in the heart has also been intensively investigated23 homozygous deletion of cardiac Drp1 results in premature death due to heart failure in mice.24,25 Analyses using heterozygous deletion have shown that down-regulation of endogenous Drp1 induces dilated cardiomyopathy with an accumulation of ubiquitinated and dysfunctional mitochondria,24,25 suggesting a pivotal role of Drp1 in mediating mitophagy and mitochondrial homoeostasis in the heart. There have been conflicting reports, however, regarding whether mitophagy can be induced in the heart in the complete absence of Drp1. Although one report showed that mitophagy is mostly eliminated when Drp1 is down-regulated in the heart,25 other reports indicated that mitophagy can be activated without Drp1.22,24 The remaining mitophagy appears to be activated through Parkin-dependent mechanisms and plays an either adaptive or maladaptive role in Drp1 KO hearts. One report suggested that Parkin-mediated mitophagy is critical in maintaining cardiac function in the absence of Drp1 in the heart,24 whereas the other report suggested that simultaneous deletion of Parkin and Drp1 mitigated mitophagy and improved cardiac function, compared to ablation of Drp1 alone.22 In this context, Parkin-mediated mitophagy may exacerbate cardiac dysfunction when mitochondrial division is reduced. Taken together, the induction and functional significance of Parkin-mediated mitophagy relative to Drp1-dependent mitophagy remain to be clarified in the heart. In addition, it will be essential to determine whether the mitophagy observed in Parkin KO mice is Drp1-dependent. If so, it is likely that Parkin and Drp1 mediate distinct forms of mitophagy in the heart. In other cell types, Drp1 has classically been observed to be essential in mediating the separation and mitophagic degradation of depolarized mitochondria.4 However, since mitochondria are relatively fragmented in adult cardiomyocytes under baseline conditions and Drp1 also possesses fission-independent functions, whether or not Drp1 is directly involved in mitophagy in adult cardiomyocytes remains controversial.
A novel function of cardiac-resident macrophages was reported recently.26 It was observed that cardiomyocytes eject LC3-positive membranous particles containing dysfunctional mitochondria and that cardiac macrophages take them up and digest them, thus contributing to mitochondrial homoeostasis in the heart. Each cardiomyocyte is surrounded by five cardiac macrophages, and each macrophage interacts with up to five cardiomyocytes. LC3-positve particles are generated through the autophagic process and preferentially contain dysfunctional mitochondria in cardiomyocytes. The phagocytic receptor, Mertk, plays a significant role in the uptake of these particles by cardiac macrophages. This mechanism plays an important role in maintaining cardiac function both at baseline and during stress, such as ischaemia or isoproterenol-overload. It is unknown, however, whether this mechanism plays a more important role in mediating mitochondrial quality control than mitophagic degradation by lysosomes. The availability of multiple options for the heart to remove damaged mitochondria may allow cardiomyocytes to survive even when one mechanism fails. Subsarcolemmal mitochondria are located close to the cellular surface and, thus, they may be subjected to ejection from cardiomyocytes more frequently than perinuclear or intermyofibrillar mitochondria, although this hypothesis remains to be tested.
3.2 Mitophagy in the heart during ischaemia and reperfusion
Growing evidence suggests that mitophagy is protective for the heart under ischaemic conditions and during post-myocardial infarction cardiac remodelling.3,27 Since cardiac mitochondria are quite vulnerable to oxidative stress during reperfusion, a protective mechanism is essential as the first line of defense. Parkin is up-regulated in the ischaemic border zone from 8 to 48 h after coronary artery ligation.21 Parkin-mediated mitophagy induces ischaemic preconditioning, thereby protecting the heart against recurrent ischaemia.28 Parkin may not prevent acute ischaemic injury without preconditioning, however, since it is not activated at very early time points, such as 4 h, following acute ischaemia.21 The heart chronically undergoes hypertrophy and dilation and its function decreases after myocardial infarction in a process termed cardiac remodelling. Again, mitochondrial dysfunction is commonly observed and precipitates the progression of heart failure after MI.10 Parkin KO mice exhibit more severe cardiac remodelling and higher mortality, but less mitophagy after MI than wild-type mice.21 Thus, Parkin-mediated mitophagy protects the heart during cardiac remodelling after MI. Contributions of Parkin to the overall level of mitophagy and the timing of Parkin-mediated mitophagy during cardiac remodelling remain to be established. Since the eventual outcome is the development of mitochondrial dysfunction and heart failure in many post-MI patients, activation of Parkin-mediated mitophagy alone appears insufficient to prevent the progression of cardiac remodelling.
Although rapid revascularization is essential for minimizing the size of MI, it induces reperfusion injury. In response to 20 min of ischaemia followed by 24 h of myocardial reperfusion, the myocardial injury was comparable between wild-type and Parkin KO mice.28 These results suggest that Parkin may not be involved in mitochondrial quality control mechanisms during myocardial reperfusion. We have shown previously that cardiac-specific heterozygous Drp1 KO mice exhibit more extensive infarction with impairment of mitophagy than wild-type mice in response to 30 min of ischaemia followed by 24 h of reperfusion. Thus, endogenous Drp1 protects the heart against ischaemia/reperfusion (I/R) and this effect may be mediated through activation of mitophagy.25 Further investigation is required to clarify the underlying molecular mechanism of mitophagy and the specific role of mitophagy during myocardial reperfusion.
3.3 Mitophagy in the heart during pressure-overload
Hypertension is a significant comorbidity in patients with heart failure. Among the potential underlying mechanisms of hypertension, ageing, and metabolic stress are commonly associated with an increase in vascular resistance.29 Increasing evidence suggests a significant role of autophagy and mitophagy in cardiomyocytes in mediating the adaptive or maladaptive response of the heart against pressure afterload. A detailed time-course study showed that canonical autophagy is activated by pressure overload within a few hours of transverse aortic constriction when the heart becomes energetically starved due to increased cardiac afterload, and then quickly inactivated within 24 h.30 Interestingly, mitophagy is activated more slowly, 3–5 days after the onset of pressure overload when canonical autophagy is inactivated, suggesting that mitophagy and canonical autophagy are mediated through distinct mechanisms. Dissociation between canonical autophagy and mitophagy was also observed in mouse embryonic fibroblasts in response to hypoxia.20 Currently, the underlying molecular mechanisms by which mitophagy is induced in a manner independent of canonical autophagy during pressure overload remain elusive. The time course of mitophagy corresponds to that of mitochondrial division, but not Parkin-translocation to mitochondria. Down-regulation of Drp1 abolishes mitophagy and boosts cardiac dysfunction. These results imply that Drp1 plays a vital role in mitophagy and cardiac homoeostasis during pressure overload.
Activation of mitophagy in response to pressure overload is also transient, and it is no longer activated after 5 days of pressure overload. The mouse heart manifests mitochondrial dysfunction and heart failure, thereafter. Administration of TAT-Beclin 1, an autophagy-activating peptide that induces dissociation of Beclin 1 from GAPR-1, significantly induces mitophagy and ameliorates heart failure.30 Thus, the inactivation of mitophagy during the chronic phase of pressure overload contributes to the development of heart failure. Reactivation of mitophagy by TAT-Beclin1 does not take place in Drp1 heterozygous knockout mice, again suggesting the importance of Drp1 in mediating mitophagy during pressure overload.
3.4 Mitophagy in the heart during metabolic stress
More than half of diabetes patients develop cardiac abnormality, including hypertrophy and diastolic dysfunction, termed diabetic cardiomyopathy.31 Diabetic cardiomyopathy is accompanied by mitochondrial dysfunction and myocardial accumulation of toxic lipid, including ceramide and diacylglycerol, termed lipotoxicity.31 In the mouse model of type II diabetes, consumption of a high-fat diet (HFD), consisting of 60% saturated fatty acid, up-regulates autophagy in the heart, peaking at 6 weeks. Cardiac-specific knockout of Atg7 (Atg7cKO) or Parkin KO reduces mitophagy by half compared to in wild type, and induces lipid accumulation and cardiac dysfunction after 2 months of HFD consumption, suggesting that mitophagy mediated through autophagy or Parkin-dependent mechanisms protects the heart during the acute phase of HFD consumption. It should be noted that Parkin also plays a significant role in mediating lipid metabolism during HFD consumption. Parkin induces fatty acid uptake by hepatocytes and adipocytes through the ubiquitin-mediated stabilization of the lipid transporter CD36.32 This demonstrates the diverse functions of Parkin besides mitophagy in mediating cellular homoeostasis. Thus, caution should be exercised when interpreting experimental results obtained using Parkin KO mice.
Obesity and metabolic syndrome are often long-term conditions. Our recent study showed that activation of autophagy in response to HFD consumption is transient and starts to decline rapidly after it reaches a peak around 6 weeks. On the other hand, mitophagy is progressively activated even after canonical autophagy is inactivated.33 Thus, canonical autophagy and mitophagy appear to be activated with distinct time courses. Another study showed, however, that the protein level of cardiac Parkin is dramatically decreased after 12 weeks of HFD consumption.34 Thus, whether mitophagy continues to be activated in a Parkin-independent manner during the chronic phase of HFD consumption and, if so, what underlying molecular mechanisms are involved and its functional significance remain to be elucidated.
4. Alternative autophagy
As described above, under some conditions, mitophagy is activated in the heart despite inactivation of non-selective autophagy.30,33 Although the precise molecular mechanism remains elusive, our group recently demonstrated the existence of this unique form of mitophagy, which is mediated by mechanisms similar to the alternative autophagy originally reported by Nishida et al.,35 in the heart and named it alternative mitophagy.35 In the next two sections, we will describe the features of alternative autophagy and mitophagy.
4.1 Origin of alternative autophagosomes
Molecular mechanisms of autophagy, including how autophagosomes are generated, have been extensively investigated.10 Primary mechanisms include autophagosomes formation through conjugation of ubiquitin-like proteins, including LC3 (Atg8), by autophagy-related (ATG) proteins. These mechanisms are also utilized in mitophagy when autophagosomes encapsulating mitochondria are formed. Besides the conventional mechanism of autophagy, another form of autophagy has been reported, called alternative autophagy or Golgi membrane-associated degradation (GOMED).35,36 In alternative autophagy, an autophagosome-like structure with double membranes is induced even in the absence of some ATG proteins and it undergoes lysosomal degradation with its cargo. Although it is hard to distinguish the autophagosome-like structure in alternative autophagy morphologically from autophagosomes in canonical autophagy, it has been proposed that the former originates from an intracellular membrane source distinct from the latter. To date, multiple intracellular membranes have been identified as the origins of the autophagosome membrane in canonical autophagy, including the endoplasmic reticulum (ER), mitochondria, ER-mitochondria contact site, and plasma membrane.37 It has been proposed that the trans-Golgi network is the origin of alternative autophagosomes, based on the following observations: (i) almost all autophagic vacuoles were localized near the Golgi apparatus in cells lacking ATG-conjugation, (ii) the Golgi ministack formation precedes autophagosome formation in these cells, (iii) some isolation membranes extended from the Golgi membrane, (iv) trans-Golgi proteins were detected on alternative autophagosomes and autolysosomes, and (v) the depletion of Golgi proteins inhibited alternative autophagy but not canonical autophagy.37 In addition, Brefeldin A, a specific inhibitor of protein transport from the ER to the Golgi apparatus, significantly suppresses the formation of alternative autophagosomes, but not canonical ones.35 These observations suggest that the trans-Golgi network is a significant membrane source of alternative autophagosomes.
4.2 Molecular mechanisms underlying alternative autophagy
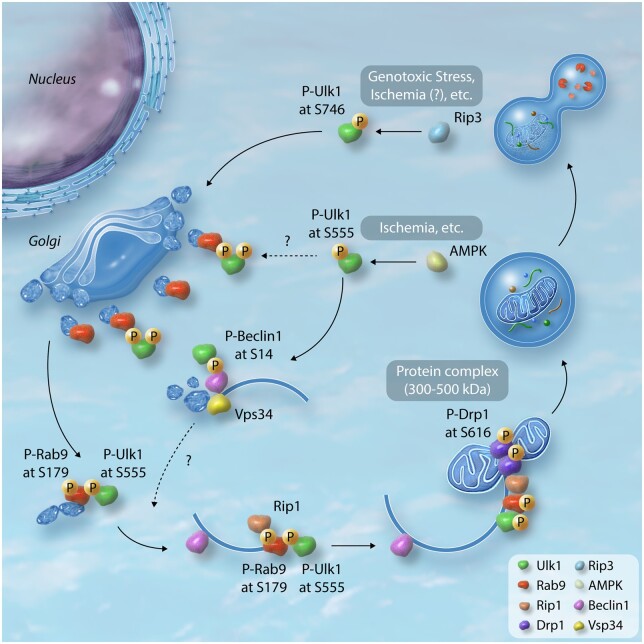
Canonical and alternative forms of autophagy share up-stream molecular machinery, such as the Ulk1-Fip200 complex and phosphatidylinositol 3 kinase (PtdIns3K) complex, as evidenced by the fact that the autophagosome-like structure in cells lacking Atg5, namely alternative autophagy, is abolished by simultaneous silencing of Ulk1, Fip200, Beclin 1, and Vps34 (but not Atg7, Atg12, Atg16, or Atg9).35 Accordingly, alternative autophagy or GOMED is often described as Ulk1-dependent autophagy. Following nutrient starvation or mTOR inhibition, the activated Ulk1 phosphorylates Beclin 1 at serine 14, thereby activating Atg14L-Vps34 complexes.38 The Vps34-Beclin 1 complex serves as a binding partner for several proteins capable of either promoting (Atg14L, UVLAG, Bif1, and AMBRA-1) or inhibiting (Rubicon, Bcl-2, and Bcl-xl) autophagy. Vps34 produces phosphatidylinositol 3-phosphate (PtdIns(3)P), which promotes both canonical autophagy and retrograde trafficking from endosomes to the Golgi apparatus.39 It is thus conceivable that these core complexes located upstream of canonical autophagy regulate Golgi membrane-associated degradation in alternative autophagy as well. Recently, the mechanism by which Ulk1 translocates to the Golgi has been reported using mouse embryonic fibroblasts (MEFs) (Figure 1).40 During genotoxic stress-induced alternative autophagy, Rip3 interacts with Ulk1 in the cytosol and phosphorylates it at serine 746, corresponding to serine 747 of human Ulk1. This phosphorylation plays an essential role in mediating the dissociation of Ulk1 from the Fip200-Atg13 complex and its translocation to the Golgi (Figure 1).40 Interestingly, the serine 746 residue is conserved in Ulk1, but not Ulk2, in higher vertebrates. This process is independent of canonical autophagy or Rip3-mediated necrosis.
Figure 1.
Proposed model of mitophagy mediated by Ulk1-dependent alternative autophagy. Ulk1 forms protein complex with Fip200 and Atg13 at the basal state. Upon stimuli, phosphorylation of Ulk1 at serine 746 by Rip3 allows for its dissociation from the complex and translocation to Golgi.40 This mechanism was found in the MEFs under the genotoxic stress, however, its significance in the heart has yet to be clarified. The trans-Golgi network is proposed to be the origin of alternative autophagosomes. Upon energetic stress, such as ischaemia, multiple sites in Ulk1 are phosphorylated by AMPK. Of these, phosphorylation of serine 555 is important for its translocation to mitochondria in MEFs and skeletal muscle in mice.59,60 Ulk1 phosphorylates Beclin 1 at serine 14, thereby inducing the Beclin 1-Vps34 complex. Vps34 is then activated at phagophore or endosomal membranes, which is pivotal for membrane remodelling, endosomal transport and autophagy.39 Serine 555 phosphorylated Ulk1 acts as a scaffold to assemble a complex comprising Rab9, associated with trans-Golgi membranes. Ulk1 directly phosphorylates Rab9 at serine 179, that facilitates interaction between Rab9 and Rip1 and the consequent phosphorylation of Drp1 at serine 616 in cardiomyocytes.57 Mitochondria labelled with phosphorylated Drp1 are subsequently sequestrated by phagophores assembled via Rab9 in cardiomyocytes.
In mammalian canonical autophagy, ATG-conjugation is vital for autophagosome closure, fusion with lysosomes, and degradation of the inner autophagosomes membrane.41 In alternative autophagy, the membranes that originate from the trans-Golgi network subsequently expand and close through fusion with other membranes from trans-Golgi or endosomal vesicles.35 The GTPase activity of Rab9, an essential molecule for membrane and protein trafficking from the late endosomes to the trans-Golgi network, is required for this process.35 Deletion of Rab9 reduces the number of autophagic vacuoles but not isolation membranes. Rab9 is thus pivotal for the maturation of alternative autophagosomes as a substitute for both ATG-conjugation and LC3. In response to genotoxic stress, Dram1 also participates in the closure of alternative autophagosomes in a p53-dependent manner.42 Whether and how these molecules undergo post-translational modification during the maturation of alternative autophagosomes is currently unknown.
4.3 Biological significance of alternative autophagy
The relevance of alternative autophagy has been reported in various organs, including the heart. Before the discovery of alternative autophagy, autophagic degradation of mitochondria during erythroid maturation was documented in Atg5-depleted cells.43 Further studies have demonstrated the essential role of Ulk1-dependent alternative autophagy in this process.44,45 Interestingly, the dominance of the Ulk1-dependent alternative mechanism in mitochondrial clearance has been reported in foetal definitive erythrocytosis, but not in adult erythrocytosis,45 suggesting that mitophagy is executed through different forms of autophagy in a stimulus- or environment-dependent manner.
Alternative autophagy mediated mitophagy was also found to play a significant role in the development of induced pluripotent stem cells (iPSCs).46 The reprogramming process requires a metabolic switch from oxidative phosphorylation to glycolysis and mitochondrial clearance is involved in this event. An earlier study reported the role of canonical autophagy in this process,47 however, further analysis revealed that this highly efficient reprogramming in MEFs requires Ulk1-/Rab9-dependent alternative autophagy, not Atg5-dependent autophagy, for mitochondrial clearance followed by metabolic reprogramming and iPSCs development.46 These studies agree that the deletion of Ulk1 or Rab9 impairs mitophagy more prominently than that of Atg7 or Atg5 under specific conditions. Ulk1-/Rab9-dependent alternative autophagy is thus neither a leaky phenotype of canonical autophagy nor merely residual autophagic activity detected in ATG-conjugation deficient cells. Rather, alternative autophagy is an important mechanism involved in mitochondrial clearance and cellular homoeostasis.
Substrates other than mitochondria, such as (pro)insulin granules and bacteria, have also been observed in alternative autophagosomes.36,48–50 In pancreatic β cells where unique mechanisms of cellular degradation are utilized,51 (pro)insulin granules can be engulfed by alternative autophagosomes labelled with Syntaxin 6, a marker of the Golgi membrane, when ATG-conjugation is deleted.36 Since canonical autophagy is impaired in diabetic β cells, alternative autophagy may play a critical role in degrading (pro)insulin granules there. Under conditions of infectious disease, the role of autophagy in mediating the recognition and the elimination of intracellular pathogens is well-recognized. Interestingly, some bacteria have been shown to be sequestrated by autophagosomes in cells lacking Atg5.49,50 In the intestinal epithelium, which interfaces with a variety of bacteria, the mitochondrial protein TRIM31 plays an essential role in forming autophagosomes independently of Atg5/Atg7.48 However, the involvement of Rab9 or Syntaxin 6 in these mechanisms has not yet been elucidated.
5. Regulation of mitophagy by alternative autophagy in the heart
Various mechanisms have been identified as mitophagy pathways using cultured cells or animal models with gene manipulation. It is essential to determine which pathway predominantly regulates mitophagy in the heart under disease conditions, e.g. pressure overload, metabolic stress, and I/R, since cardiomyocytes and the mitochondria therein are subject to a variety of stresses. Mice expressing a mitophagy probe allow for the spatial and temporal quantitation of mitophagy in the heart, providing clues about the molecular mechanism mediating mitophagy under specific stress conditions.52
For example, a study with transgenic mice with mCherry-GFP-mtFIS1(101–152) fusion protein (mito-QC mice) showed that the deletion of Pink1 had no effect on mitophagy in the heart at baseline,53 suggesting that mitophagy in the heart is mediated through Pink1-independent mechanisms. However, since mito-QC is expressed on the cytoplasmic side of OMM with a mitochondrial targeting sequence of Fis1 (101–152), it can be degraded more quickly through Parkin- and ubiquitin proteasome-dependent mechanisms compared to mitophagy.54,55 Thus, caution should be exercised when interpreting the result obtained with mito-QC signal.54 Mito-SRAI, a newer mitophagy probe circumvents the potential problem of mito-QC, and, thus, it may allow us to analyse the signalling mechanisms of mitophagy in a more accurate manner.54
Mice expressing another mitophagy probe targeted to mitochondrial matrix, mt-Keima, have also been generated by multiple groups.56,57 Mice generated by crossing heart-specific transgenic mt-Keima mice (Mito-Keima-Tg) with Atg7cKO displayed nearly complete down-regulation of mitophagy in the heart at baseline, but mitophagy was preserved under conditions of starvation or ischaemia compared to in Mito-Keima-Tg without Atg7cKO.57 While Mito-Keima-Tg crossed with Parkin KO exhibited well-preserved mitophagy under energy stress conditions, those with Ulk1cKO showed a significant decrease in mitophagy under conditions of starvation or ischaemia. These results suggest that mitophagy in the heart is executed through canonical autophagy at baseline, but is mediated mainly through Parkin-independent and/or Ulk1-dependent mechanisms under conditions of starvation or ischaemia. Since the mt-Keima signal during starvation and ischaemia was decreased in the presence of Brefeldin A and colocalized with Rab9 puncta, Ulk1-/Rab9-dependent alternative autophagy is thought to be the predominant form of mitophagy under conditions of starvation or ischaemia in the heart.57 Ulk1 directly phosphorylates Rab9 at serine 179 in response to starvation or ischaemia and recruits a fission complex consisting of Ulk1, Rab9, Rip1, and Drp1 to damaged mitochondria (Figure 1). Drp1 is phosphorylated at serine 616 by Rip1, resulting in its activation and mitochondrial fragmentation. Rab9-mediated autophagosomes subsequently engulf the fragmented mitochondria (Figure 1). In this model, it is likely that mitochondrial division occurs simultaneously with autophagosome closure, which is consistent with the recent idea proposed by the Kanki group.58 Interestingly, knock-in mice expressing a phosphorylation-resistant mutant of Rab9, Rab9(S179A), showed severely impaired mitophagy during ischaemia despite the preservation of canonical autophagy flux. This suggests that phosphorylation of Rab9 by Ulk1 is a specific mechanism involved in mitophagy that protects the heart against ischaemia, even in the absence of canonical autophagy. When Ulk1-/Rab9-dependent mitophagy takes place, a large protein complex 300–500 kDa in size, consisting of Ulk1, Rab9, Rip1, Drp1, and possibly other proteins, is formed (Figure 1). The large protein complex may consist of the machinery that forms Rab9-containing autophagosomes, as well as the fission complex, in close proximity to damaged mitochondria. The identity of proteins in the large protein complex and how Rab9-positive autophagosomes are formed and mitophagy is executed remain to be clarified.
6. Pharmacological tools that manipulate mitophagy
Autophagy and mitophagy have been considered attractive targets for pharmacological intervention and many drugs have been identified as autophagy inducers. For example, rapamycin, metformin, and polyphenols, including resveratrol, have been shown to induce autophagy and act protectively in the heart.61,62 These compounds affect multiple cellular functions, and thus, caution must be exercised in determining whether activation of autophagy or mitophagy plays an essential role in mediating their salutary effects in a given condition. Recently, however, we and others have shown more selective and effective interventions, including TAT-Beclin 1, spermidine, and trehalose, inducing autophagy and mitophagy in the heart.30,63,64 Urolithin A also induces mitophagy in Caenorhabditis elegans and C2C12 myoblasts.65 Treatment with spermidine or trehalose is being tested in patients with hypertension or ACS, respectively. The result of a human clinical trial of Urolithin A has been recently published.66 These drugs appear to target proximal mechanisms regulating autophagy and mitophagy, and activation of autophagy and mitophagy plays a key role in mediating their effects (Figure 2).61
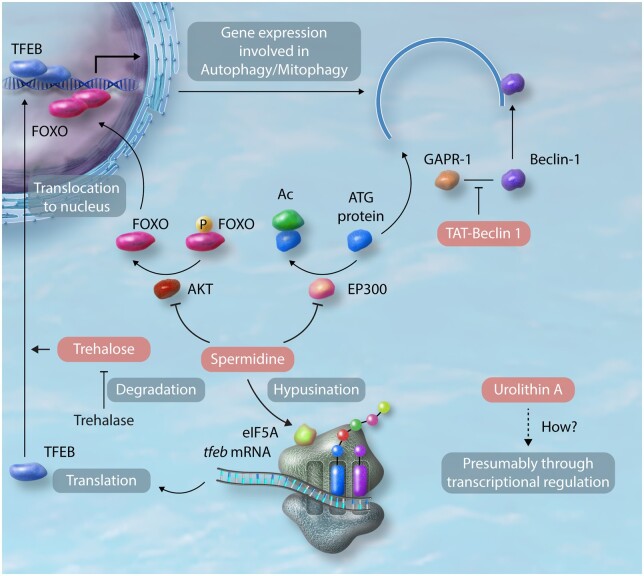
Figure 2.
Molecular mechanisms of the effect of pharmacological interventions on autophagy/mitophagy. TAT-Beclin 1 competitively inhibits the interaction between endogenous Beclin 1 and its negative regulator, GAPR-1, thereby mobilizing Beclin 1 to stimulate autophagy.67 Spermidine stimulates autophagy by inhibiting cytosolic EP300 acetyltransferase.68 Furthermore, spermidine facilitates translocation of FOXO from the cytosol to nucleus and provokes epigenetic reprogramming through inhibition of histone acetyltransferases in the nucleus, thereby activating transcription of autophagy-related genes.69 Spermidine also promotes hypusination of translational factor eIF5A, which uniquely contains the unusual amino acid hypusine, in memory B cells.70 This post-translational modification of eIF5A makes the formation of the first peptide bonds more effective in the translation of TFEB mRNA. Trehalose promotes translocation of TFEB from the cytosol to nucleus, thereby inducing autophagy-related genes in macrophages and cardiomyocytes.64,71 Trehalase expressed in the intestine in mice and humans can degrade Trehalose.64 The detailed mechanism by which Urolithin A induces autophagy/mitophagy has yet to be clarified. Induction of autophagy/mitophagy-related genes and phosphorylation of AMPK were observed after treatment with Urolithin A in C2C12 myoblasts and skeletal muscles in mice and humans.65,66
TAT-Beclin 1 competitively inhibits association of Beclin 1 with its negative regulator, GAPR-1, at the intracellular anchorage site, thereby releasing endogenous Beclin 1 and activating autophagy (Figure 2).67 TAT-Beclin 1 activates autophagy and mitophagy in the heart and attenuates progression of heart failure in the presence of high blood pressure. Spermidine rapidly induces autophagy through inhibition of the acetyltransferase EP300 that primarily deacetylates autophagy-related proteins in cytosol in U2OS cells (Figure 2).68 Spermidine provokes epigenetic reprogramming that activates transcription of autophagy-related genes (Figure 2).69 Spermidine also promotes translation of TFEB mRNA, a master-transcription factor for genes involved in autophagy and lysosomal biogenesis, through hypusination of eIF5A in memory B cells (Figure 2).70 Spermidine improves cardiac function in old hearts and its protective effect is accompanied by stimulation of mitophagy. Trehalose, a natural disaccharide, induces translocation of TFEB from cytosol to nucleus, thereby inducing autophagy and lysosomal biogenesis in macrophages and cultured cardiomyocytes (Figure 2).64,71 Urolithin A induces expression of genes involved in autophagy and mitophagy in skeletal muscles, presumably through transcriptional regulation.63,66 Importantly, the well-established salutary effects of caloric restriction and exercise upon organ functions and ageing are mediated through their ability to stimulate autophagy and mitophagy. Exploring signalling mechanisms and developing small molecules mimicking the effect of caloric restriction or exercise have proven a productive avenue of research and remain promising for further discovery of effective interventions for heart disease.
7. A Clinical perspective on mitophagy in the heart
Recent advancements in cardiovascular medicine have successfully decreased the early mortality in patients with acute coronary syndrome (ACS). However, many survivors of MI develop heart failure due to increased haemodynamic overload and maladaptive remodelling. As a result, the occurrence of chronic heart failure has increased and cardiovascular disease remains the leading cause of death in western countries. Mitochondrial dysfunction is commonly observed in the failing heart and often precedes the transition from the compensated to the decompensated form of heart failure. Mitochondrial dysfunction is also seen in patients suffering heart failure with preserved ejection fraction. Although mitophagy is activated in the stressed heart, the level of mitophagy appears insufficient and mitochondrial dysfunction eventually develops in the heart. As we discussed, mitophagy is activated only transiently after the initiation of pressure overload; it is inactivated thereafter and mitochondrial dysfunction and heart failure follow. Restoring the level of mitophagy could be a rational intervention to delay cardiac decompensation. Currently, the reason mitophagy activation is only transient in the stressed heart remains unclear. We have shown that mammalian sterile 20 like kinase 1 (Mst1), a potent inhibitor of autophagy, is activated 1 week after pressure overload. Mst1 phosphorylates Beclin 1 at threonine 108, thereby inducing Beclin1-Bcl-2 interaction and inhibiting both autophagy and mitophagy.72 It is also possible that proteins involved in autophagy and mitophagy may become depleted after prolonged stress. Thus, it is essential to evaluate whether interventions to reactivate mitophagy would be effective in such a negative environment. Furthermore, whether the aforementioned mitophagy inducers activate mitophagy mediated by the canonical mechanism or the alternative mechanism remains to be clarified.
Modulation of autophagy is used to treat or prevent various diseases. For instance, cancer cells acquire resistance against chemotherapy by activating autophagy. The inhibition of autophagy is thus a therapeutic strategy against this type of cancer, and several drugs are being tested in clinical trials.73 Likewise, autophagy is being investigated as a therapeutic target in the cardiovascular field as well. Earlier studies suggested that the activation of autophagy protects the heart against ischaemia alone but that further activation of autophagy during reperfusion is detrimental.74,75 Although there was a long-standing controversy regarding whether autophagy is a cause of cell death or not, a new form of autophagic cell death, called autosis and morphologically characterized by a concave nucleus and swollen perinuclear space, was identified in the rat brain subjected to hypoxia-ischaemia.76 Interestingly, recent evidence has shown that excessive activation of canonical autophagy during I/R induces autosis and exacerbates cell death in the heart.77 Down-regulation of Rubicon or inhibition of Na+, K+-ATPase, a regulator of autosis, significantly ameliorates cardiac injury during I/R. These results suggest that it is important to maintain the activity of canonical autophagy within an appropriate range during I/R in the heart, where mitochondrial clearance is still required. Accordingly, selective induction of mitophagy, without activating canonical autophagy, may be a novel therapeutic strategy against I/R. Stimulating Ulk1-dependent mitophagy through the phosphorylation of Rab9 may be an attractive approach to applying this strategy. It should be noted that excessive activation of mitophagy induces cell death in doxorubicin-induced cardiotoxicity.78 Thus, it is important to evaluate the levels of autophagy and mitophagy in the heart in a given condition. Currently, it is challenging to evaluate the levels of autophagy and mitophagy in humans in a non-invasive manner. Thus, the development of a convenient, accurate, and non-invasive method for measuring autophagy and mitophagy in the heart in vivo is urgently needed.
Acknowledgements
We thank Daniela Zablocki and Dr Brian Quinn for critical reading of the manuscript. We also thank BioRender for assistance in figure preparations.
Conflict of interest: none declared.
Funding
This work was supported by the Japan Society for the Promotion of Science 19K17601 (T.S.); grants from MSD Life Science Foundation, Cardiovascular Research Fund, Ichiro Kanehara Foundation, and Mochida Memorial Foundation (T.S.); Foundation Leducq Transatlantic Networks 15CBD04 (J.S.); and US Public Health Service Grants HL067724, HL091469, HL138720, HL112330, HL144626, HL150881, and AG23039 (J.S.). J.S. is a recipient of the 2020 Merit Award from the American Heart Association (20 Merit 35120374).
References
- 1. Saito T, Sadoshima J.. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res 2015;116:1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friedman JR, Nunnari J.. Mitochondrial form and function. Nature 2014;505:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forte M, Schirone L, Ameri P, Basso C, Catalucci D, Modica J, Chimenti C, Crotti L, Frati G, Rubattu S, Schiattarella GG, Torella D, Perrino C, Indolfi C, Sciarretta S; Italian Society of Cardiology Working group on Cellular and Molecular Biology of the Heart. The role of mitochondrial dynamics in cardiovascular diseases. Br J Pharmacol 2020;doi: 10.1111/bph.15068. [DOI] [PubMed] [Google Scholar]
- 4. Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS.. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 2008;27:433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW 2nd. Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circ Res 2014;114:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pickles S, Vigie P, Youle RJ.. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 2018;28:R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quiles JM, Gustafsson AB.. Mitochondrial quality control and cellular proteostasis: two sides of the same coin. Front Physiol 2020;11:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoshino A, Wang WJ, Wada S, McDermott-Roe C, Evans CS, Gosis B, Morley MP, Rathi KS, Li J, Li K, Yang S, McManus MJ, Bowman C, Potluri P, Levin M, Damrauer S, Wallace DC, Holzbaur ELF, Arany Z.. The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature 2019;575:375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scarffe LA, Stevens DA, Dawson VL, Dawson TM.. Parkin and PINK1: much more than mitophagy. Trends Neurosci 2014;37:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sciarretta S, Maejima Y, Zablocki D, Sadoshima J.. The role of autophagy in the heart. Annu Rev Physiol 2018;80:1–26. [DOI] [PubMed] [Google Scholar]
- 11. Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM.. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2012;2:120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N.. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2012;2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW.. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 2015;350:aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, Youle RJ.. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell 2019;74:347–362.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanaka K. The PINK1-Parkin axis: an overview. Neurosci Res 2020. Jan 23;S0168-0102(19)30571-1.doi: 10.1016/j.neures.2020.01.006. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 16. Narendra D, Tanaka A, Suen DF, Youle RJ.. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008;183:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, Zhu Y, Zhang X, Li L, Zhang L, Sui S, Zhao B, Feng D.. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep 2014;15:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL.. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008;283:10892–10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I.. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010;11:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirota Y, Yamashita S, Kurihara Y, Jin X, Aihara M, Saigusa T, Kang D, Kanki T.. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy 2015;11:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyan S, Murphy AN, Gustafsson AB.. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem 2013;288:915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, Dorn GW.. Interdependence of Parkin-mediated mitophagy and mitochondrial fission in adult mouse hearts. Circ Res 2015;117:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tong M, Zablocki D, Sadoshima J.. The role of Drp1 in mitophagy and cell death in the heart. J Mol Cell Cardiol 2020;142:138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, Gabrielson K, Kass DA, Iijima M, Sesaki H.. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 2014;33:2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ikeda Y, Shirakabe A, Maejima Y, Zhai P, Sciarretta S, Toli J, Nomura M, Mihara K, Egashira K, Ohishi M, Abdellatif M, Sadoshima J.. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res 2015;116:264–278. [DOI] [PubMed] [Google Scholar]
- 26. Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, Sánchez-Díaz M, Díaz-García E, Santiago DJ, Rubio-Ponce A, Li JL, Balachander A, Quintana JA, Martínez-de-Mena R, Castejón-Vega B, Pun-García A, Través PG, Bonzón-Kulichenko E, García-Marqués F, Cussó L, A-González N, González-Guerra A, Roche-Molina M, Martin-Salamanca S, Crainiciuc G, Guzmán G, Larrazabal J, Herrero-Galán E, Alegre-Cebollada J, Lemke G, Rothlin CV, Jimenez-Borreguero LJ, Reyes G, Castrillo A, Desco M, Muñoz-Cánoves P, Ibáñez B, Torres M, Ng LG, Priori SG, Bueno H, Vázquez J, Cordero MD, Bernal JA, Enríquez JA, Hidalgo A.. A network of macrophages supports mitochondrial homeostasis in the heart. Cell 2020;183:94–109.e123. [DOI] [PubMed] [Google Scholar]
- 27. Bravo-San Pedro JM, Kroemer G, Galluzzi L.. Autophagy and mitophagy in cardiovascular disease. Circ Res 2017;120:1812–1824. [DOI] [PubMed] [Google Scholar]
- 28. Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA.. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One 2011;6:e20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lacolley P, Regnault V, Nicoletti A, Li Z, Michel JB.. The vascular smooth muscle cell in arterial pathology: a cell that can take on multiple roles. Cardiovasc Res 2012;95:194–204. [DOI] [PubMed] [Google Scholar]
- 30. Shirakabe A, Zhai P, Ikeda Y, Saito T, Maejima Y, Hsu CP, Nomura M, Egashira K, Levine B, Sadoshima J.. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation 2016;133:1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakamura M, Sadoshima J.. Cardiomyopathy in obesity, insulin resistance and diabetes. J Physiol 2020;598:2977–2993. [DOI] [PubMed] [Google Scholar]
- 32. Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, Suen DF, Youle RJ, Amar M, Remaley AT, Sack MN.. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest 2011;121:3701–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tong M, Saito T, Zhai P, Oka SI, Mizushima W, Nakamura M, Ikeda S, Shirakabe A, Sadoshima J.. Mitophagy is essential for maintaining cardiac function during high fat diet-induced diabetic cardiomyopathy. Circ Res 2019;124:1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas A, Marek-Iannucci S, Tucker KC, Andres AM, Gottlieb RA.. Decrease of cardiac parkin protein in obese mice. Front Cardiovasc Med 2019;6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S.. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009;461:654–658. [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi H, Arakawa S, Kanaseki T, Miyatsuka T, Fujitani Y, Watada H, Tsujimoto Y, Shimizu S.. Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J 2016;35:1991–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimizu S. Biological roles of alternative autophagy. Mol Cell 2018;41:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL.. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su H, Yang F, Wang Q, Shen Q, Huang J, Peng C, Zhang Y, Wan W, Wong CCL, Sun Q, Wang F, Zhou T, Liu W.. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell 2017;67:907–921.e907. [DOI] [PubMed] [Google Scholar]
- 40. Torii S, Yamaguchi H, Nakanishi A, Arakawa S, Honda S, Moriwaki K, Nakano H, Shimizu S.. Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy. Nat Commun 2020;11:1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N.. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016;354:1036–1041. [DOI] [PubMed] [Google Scholar]
- 42. Nagata M, Arakawa S, Yamaguchi H, Torii S, Endo H, Tsujioka M, Honda S, Nishida Y, Konishi A, Shimizu S.. Dram1 regulates DNA damage-induced alternative autophagy. Cell Stress 2018;2:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA.. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood 2009;114:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB.. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 2008;112:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Honda S, Arakawa S, Nishida Y, Yamaguchi H, Ishii E, Shimizu S.. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun 2014;5:4004. [DOI] [PubMed] [Google Scholar]
- 46. Ma T, Li J, Xu Y, Yu C, Xu T, Wang H, Liu K, Cao N, Nie BM, Zhu SY, Xu S, Li K, Wei WG, Wu Y, Guan KL, Ding S.. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol 2015;17:1379–1387. [DOI] [PubMed] [Google Scholar]
- 47. Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z.. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 2013;13:617–625. [DOI] [PubMed] [Google Scholar]
- 48. Ra EA, Lee TA, Won Kim S, Park A, Choi HJ, Jang I, Kang S, Hee Cheon J, Cho JW, Eun Lee J, Lee S, Park B.. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat Commun 2016;7:11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ.. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog 2009;5:e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T.. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog 2013;9:e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goginashvili A, Zhang Z, Erbs E, Spiegelhalter C, Kessler P, Mihlan M, Pasquier A, Krupina K, Schieber N, Cinque L, Morvan J, Sumara I, Schwab Y, Settembre C, Ricci R.. Insulin granules. Insulin secretory granules control autophagy in pancreatic beta cells. Science 2015;347:878–882. [DOI] [PubMed] [Google Scholar]
- 52. Kaludercic N, Maiuri MC, Kaushik S, Fernandez AF, de Bruijn J, Castoldi F, Chen Y, Ito J, Mukai R, Murakawa T, Nah J, Pietrocola F, Saito T, Sebti S, Semenzato M, Tsansizi L, Sciarretta S, Madrigal-Matute J.. Comprehensive autophagy evaluation in cardiac disease models. Cardiovasc Res 2020;116:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG.. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab 2018;27:439–449.e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Katayama H, Hama H, Nagasawa K, Kurokawa H, Sugiyama M, Ando R, Funata M, Yoshida N, Homma M, Nishimura T, Takahashi M, Ishida Y, Hioki H, Tsujihata Y, Miyawaki A.. Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell 2020;181:1176–1187.e1116. [DOI] [PubMed] [Google Scholar]
- 55. Yoshii SR, Kishi C, Ishihara N, Mizushima N.. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 2011;286:19630–19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T.. Measuring in vivo mitophagy. Mol Cell 2015;60:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saito T, Nah J, Oka SI, Mukai R, Monden Y, Maejima Y, Ikeda Y, Sciarretta S, Liu T, Li H, Baljinnyam E, Fraidenraich D, Fritzky L, Zhai P, Ichinose S, Isobe M, Hsu CP, Kundu M, Sadoshima J.. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest 2019;129:802–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, Saigusa T, Yoshimori T, Sakai Y, Mihara K, Kanki T.. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol 2016;215:649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tian W, Li W, Chen Y, Yan Z, Huang X, Zhuang H, Zhong W, Chen Y, Wu W, Lin C, Chen H, Hou X, Zhang L, Sui S, Zhao B, Hu Z, Li L, Feng D.. Phosphorylation of ULK1 by AMPK regulates translocation of ULK1 to mitochondria and mitophagy. FEBS Lett 2015;589:1847–1854. [DOI] [PubMed] [Google Scholar]
- 60. Laker RC, Drake JC, Wilson RJ, Lira VA, Lewellen BM, Ryall KA, Fisher CC, Zhang M, Saucerman JJ, Goodyear LJ, Kundu M, Yan Z.. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat Commun 2017;8:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren J, Zhang Y.. Targeting autophagy in aging and aging-related cardiovascular diseases. Trends Pharmacol Sci 2018;39:1064–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Frati G, Vecchione C, Sciarretta S.. Novel beneficial cardiovascular effects of natural activators of autophagy. Circ Res 2018;123:947–949. [DOI] [PubMed] [Google Scholar]
- 63. Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Büttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Mühlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F.. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med 2016;22:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sciarretta S, Yee D, Nagarajan N, Bianchi F, Saito T, Valenti V, Tong M, Del Re DP, Vecchione C, Schirone L, Forte M, Rubattu S, Shirakabe A, Boppana VS, Volpe M, Frati G, Zhai P, Sadoshima J.. Trehalose-induced activation of autophagy improves cardiac remodeling after myocardial infarction. J Am Coll Cardiol 2018;71:1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Felix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J.. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med 2016;22:879–888. [DOI] [PubMed] [Google Scholar]
- 66. Andreux PA, Blanco-Bose W, Ryu D, Burdet F, Ibberson M, Aebischer P, Auwerx J, Singh A, Rinsch C.. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat Metab 2019;1:595–603. [DOI] [PubMed] [Google Scholar]
- 67. Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B.. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013;494:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pietrocola F, Lachkar S, Enot DP, Niso-Santano M, Bravo-San Pedro JM, Sica V, Izzo V, Maiuri MC, Madeo F, Mariño G, Kroemer G.. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ 2015;22:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Madeo F, Eisenberg T, Pietrocola F, Kroemer G.. Spermidine in health and disease. Science 2018;359:eaan2788. [DOI] [PubMed] [Google Scholar]
- 70. Zhang H, Alsaleh G, Feltham J, Sun Y, Napolitano G, Riffelmacher T, Charles P, Frau L, Hublitz P, Yu Z, Mohammed S, Ballabio A, Balabanov S, Mellor J, Simon AK.. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol Cell 2019;76:110–125.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, Ali S, Dehestani B, Holloway KB, Micevych PS, Javaheri A, Crowley JR, Ballabio A, Schilling JD, Epelman S, Weihl CC, Diwan A, Fan D, Zayed MA, Razani B.. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun 2017;8:15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, Lim DS, Isobe M, Sadoshima J.. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013;19:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kimmelman AC, White E.. Autophagy and tumor metabolism. Cell Metab 2017;25:1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A.. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation 2012;125:3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J.. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007;100:914–922. [DOI] [PubMed] [Google Scholar]
- 76. Liu Y, Shoji-Kawata S, Sumpter RM Jr, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, Shaw SY, Clarke PG, Puyal J, Levine B.. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA 2013;110:20364–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nah J, Zhai P, Huang CY, Fernandez AF, Mareedu S, Levine B, Sadoshima J.. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest 2020;130:2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wallace KB, Sardao VA, Oliveira PJ.. Mitochondrial determinants of doxorubicin-induced cardiomyopathy. Circ Res 2020;126:926–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Piquereau J, Godin R, Deschenes S, Bessi VL, Mofarrahi M, Hussain SN, Burelle Y.. Protective role of PARK2/Parkin in sepsis-induced cardiac contractile and mitochondrial dysfunction. Autophagy 2013;9:1837–1851. [DOI] [PubMed] [Google Scholar]
- 80. Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S.. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 2013;4:2308. [DOI] [PubMed] [Google Scholar]
- 81. Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW.. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci USA 2011;108:9572–9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, Cai H, Borsche M, Klein C, Youle RJ.. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018;561:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83. Wang K, Zhou LY, Wang JX, Wang Y, Sun T, Zhao B, Yang YJ, An T, Long B, Li N, Liu CY, Gong Y, Gao JN, Dong YH, Zhang J, Li PF.. E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat Commun 2015;6:7619. [DOI] [PubMed] [Google Scholar]
- 84. Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, Dorn GW.. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 2007;117:2825–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dorn GW 2nd. Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Trans Res 2010;3:374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW. 2nd,. Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation 2008;117:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW 2nd. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med 2002;8:725–730. [DOI] [PubMed] [Google Scholar]
- 88. Wu S, Lu Q, Wang Q, Ding Y, Ma Z, Mao X, Huang K, Xie Z, Zou MH.. Binding of FUN14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation 2017;136:2248–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kasahara A, Cipolat S, Chen Y, Dorn GW, Scorrano L.. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and notch signaling. Science 2013;342:734–737. [DOI] [PubMed] [Google Scholar]
- 90. Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, Walsh K.. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res 2012;111:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Song M, Mihara K, Chen Y, Scorrano L, Dorn GW.. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab 2015;21:273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen Y, Liu Y, Dorn GW.. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res 2011;109:1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hall AR, Burke N, Dongworth RK, Kalkhoran SB, Dyson A, Vicencio JM, Dorn GW II, Yellon DM, Hausenloy DJ.. Hearts deficient in both Mfn1 and Mfn2 are protected against acute myocardial infarction. Cell Death Dis 2016;7:e2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, Mizushima N, Mihara K, Ishihara N.. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol Cell Biol 2015;35:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Piquereau J, Caffin F, Novotova M, Prola A, Garnier A, Mateo P, Fortin D, Huynh Le H, Nicolas V, Alavi MV, Brenner C, Ventura-Clapier R, Veksler V, Joubert F.. Down-regulation of OPA1 alters mouse mitochondrial morphology, PTP function, and cardiac adaptation to pressure overload. Cardiovasc Res 2012;94:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]