Abstract
The mechanistic target of rapamycin (mTOR) integrates several intracellular and extracellular signals involved in the regulation of anabolic and catabolic processes. mTOR assembles into two macromolecular complexes, named mTORC1 and mTORC2, which have different regulators, substrates and functions. Studies of gain- and loss-of-function animal models of mTOR signalling revealed that mTORC1/2 elicits both adaptive and maladaptive functions in the cardiovascular system. Both mTORC1 and mTORC2 are indispensable for driving cardiac development and cardiac adaption to stress, such as pressure overload. However, persistent and deregulated mTORC1 activation in the heart is detrimental during stress and contributes to the development and progression of cardiac remodelling and genetic and metabolic cardiomyopathies. In this review, we discuss the latest findings regarding the role of mTOR in the cardiovascular system, both under basal conditions and during stress, such as pressure overload, ischemia, and metabolic stress. Current data suggest that mTOR modulation may represent a potential therapeutic strategy for the treatment of cardiac diseases.
Keywords: mTOR, mTORC1, mTORC2, Rapamycin, Heart disease
Graphical Abstract
1. Introduction
More than two decades have passed since the discovery of the mammalian target of rapamycin (mTOR), recently also renamed ‘mechanistic’ target of rapamycin, by four independent research groups.1–4 Rapamycin, a macrolide possessing antifungal, immunomodulatory, and anti-proliferative properties, inhibits mTOR by interacting with the cytosolic FK506-binding protein of 12 kDa–rapamycin complex (FKBP12).1–3 mTOR has emerged as the central regulator of crucial cellular mechanisms involved in growth and differentiation.5,6 mTOR acts as a nutrient and energy sensor by integrating several external and internal inputs and coordinating the equilibrium between anabolic and catabolic reactions, such as protein synthesis and autophagy, respectively.5–7 mTOR is an evolutionarily conserved serine/threonine kinase of 289 kDa, belonging to the phosphoinositide kinase–related kinase (PIKK) family and homologous of the yeast TOR (DRR) proteins.8,9 mTOR represents the catalytic subunit of two macromolecular complexes, named mTOR complex 1 (mTORC1) and 2 (mTORC2), which also comprise additional accessory, regulatory, and scaffold subunits. mTORC1 is a paramount controller of cellular growth, protein synthesis, nutrient and energy sensing, mitochondrial turnover, and metabolic processes. mTORC2 orchestrates other cellular functions, such as cytoskeletal organization and cell polarity, is less sensitive to rapamycin, and has different or additional regulators and substrates.10–12
Perturbations of mTOR signalling lead to several pathologies, including metabolic syndromes, cancer, and neurodegenerative and cardiovascular diseases.5,6,13 mTOR modulation has been considered as a promising modality for the treatment of a wide variety of diseases and, to date, a number of modulators able to target mTOR have been identified and developed for clinical purposes.
The importance of mTOR signalling in the cardiovascular system has been investigated in multiple studies performed using animal models of mTOR loss of function or gain of function.14,15 Both mTORC1 and mTORC2 are indispensable for pre-natal and post-natal heart development and for cardiac adaption to pressure overload. Genetic deletion of mTOR components produces dramatic cardiac dysfunction in these conditions.14,15 Conversely, persistent or deregulated mTOR activation is maladaptive in several conditions. Partial genetic or pharmacological inhibition of mTOR delays cardiac aging, reduces cardiac damage in response to stress, and mitigates cardiovascular complications related to metabolic and genetic disorders.14,15
Here, we review the latest evidence regarding mTOR pathophysiology in the heart and provide a comprehensive dissection of upstream regulators and downstream substrates of mTORC1 and mTORC2. We also discuss the current therapeutic approaches to modulation of mTOR signalling and their possible translation to human disease.
2. mTORC1 and mTORC2 structure
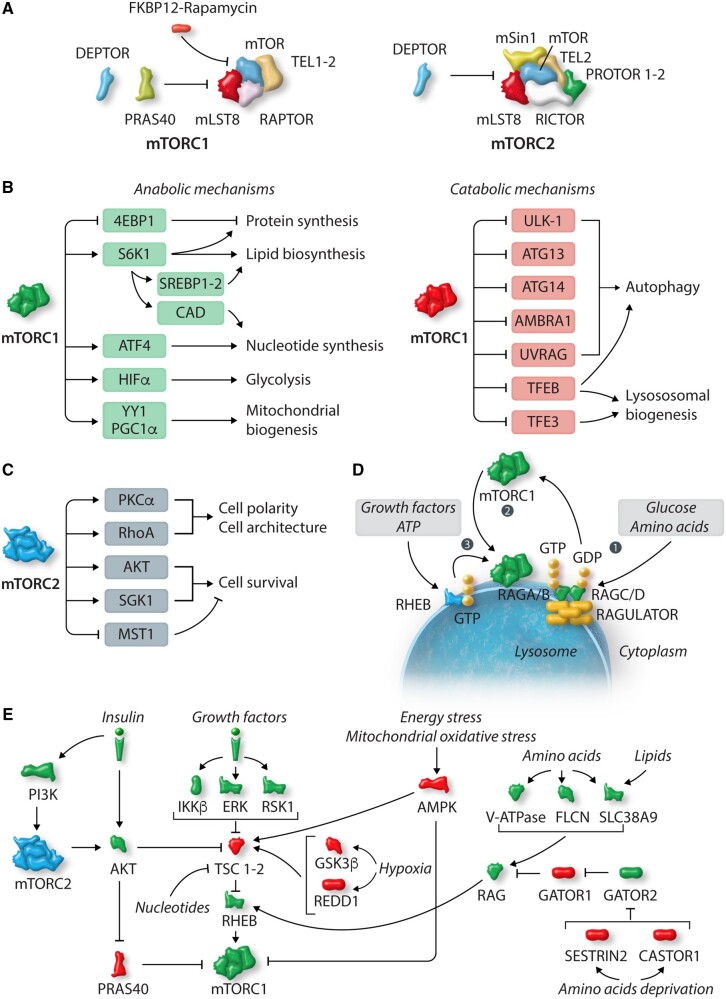
mTOR assembles into two distinct macromolecular complexes by binding to distinct sets of subunits (Figure 1A). Some of these subunits are found in both the mTORC1 and mTORC2 complexes, whereas others are specific to each.
Figure 1.
Overview of mTOR biology. (A) Architecture of mechanistic target of rapamycin (mTOR) complex 1 and 2 (mTORC1-2). (B,C) Molecular mechanisms and substrates modulated by mTORC1 (B) and mTORC2 (C). (D) Schematic representation of mTORC1 activation. mTORC1 activation occurs at the lysosome surface. In response to nutrient signals, Rags is activated (Rag A/B-GTP, Rag C/D-GDP) (1) and mediates mTORC1 translocation to the lysosome surface, in close proximity to RHEB (2). Once at the lysosome, additional inputs (growth factors) are required for RHEB-induced mTORC1 activation (3). (E) Upstream modulators of mTORC1 and mTORC2. Green and red indicate positive and negative regulators of mTOR, respectively. See text for further details. The figure was made using tools provided by Servier Medical Art, among others. Legend: 4E-BP1, eukaryotic translation initiation factor 4E (eIF4E)-binding protein-1; AKT, protein kinase B; AMBRA1, activating molecule in Beclin1-regulated autophagy; AMPK, adenosine monophosphate-activated protein kinase; ATF4, activating transcription factor 4; ATG, autophagy-related gene; CAD, carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase; CASTOR 1-2, cellular arginine sensor for mTORC1 1-2; ERK, extracellular signal-regulated kinase 1/2; GATOR 1-2, GAP activity towards Rags 1-2; DEPTOR, DEP domain-containing mTOR-interacting protein; FKBP12, FK506-binding protein of 12 kDa–rapamycin complex; FLCN, folliculin; GSK3β, glycogen synthase kinase-3β; HIF-1, hypoxia-inducible factor-1α; IKKβ, inhibitor of NF-κB kinase-β; mLST8, mammalian lethal with sec-13 protein 8; mSIN1, mammalian stress-activated protein kinase-interaction protein 1; MST1, mammalian sterile 20-like kinase 1; PI3K, phosphoinositide 3 kinase; PKCα, protein kinase C α; PRAS40, proline-rich AKT substrate 40; PROTOR 1-2, protein observed with RICTOR 1-2; RAG, Ras-related GTPase; RAPTOR, regulatory-associated protein of mTOR; REDD1, regulated in development and DNA damage responses 1; RHEB, Ras homolog enriched in brain; RhoA, Ras homolog gene family, member A; RICTOR, rapamycin-insensitive companion of mTOR; RSK1, p90 ribosomal S6 kinase; S6K1, S6 kinase-1; SGK1, serum and glucocorticoid-induced protein kinase-1; SLC38A9, solute carrier family 38 member 9; SREBP1/2, sterol regulatory element-binding protein 1-2; TEL 1-2, Tel 2 interacting protein 1/2; TFE3, transcription factor E3; TFEB, transcription factor EB; TSC1-2, tuberous sclerosis protein 1/2; ULK1, unc-51-like kinase 1; UVRAG, UV radiation resistance-associated gene; v-ATPase, vacuolar H(+)-adenosine triphosphatase; YY1/PGC-1α, transcription factor yin-yang 1/peroxisome proliferator-activated receptor γ coactivator-1α transcriptional complex.
The core components of mTORC1 are mTOR, mammalian lethal with SEC13 protein 8 (mLST8), and regulatory-associated protein of mTOR (RAPTOR). mLST8 enhances mTOR kinase activity,10,16 while RAPTOR ensures substrate recruitment and drives the subcellular localization of mTORC1, particularly its translocation to lysosomes.17,18 Additional proteins of the mTORC1 complex include the scaffold proteins proline-rich Akt substrate of 40 kDa (PRAS40), DEP domain-containing mTOR-interacting protein (DEPTOR), and Tel 2 interacting protein 1 (TEL2).16,18,19 PRAS40 and DEPTOR are endogenous inhibitors of mTORC1.20–23
The specific subunits of the mTORC2 complex include rapamycin-insensitive companion of mTOR (RICTOR), mammalian stress-activated protein kinase-interaction protein 1 (mSIN1), and protein observed with RICTOR (PROTOR 1/2).24,25 RICTOR is a scaffold protein needed for mTORC2 assembly, function, and substrate recruitment. mSIN1 is a scaffold protein that preserves the mTORC2 complex integrity and kinase activity.26–29 mLST8, TEL2, and DEPTOR are also components of the mTORC2 complex.
3. Mechanisms and substrates regulated by mTOR
Once modulated by different inputs (Figure 1B, C), mTOR transduces these signals via a plethora of substrates involved in the control of fundamental cellular mechanisms. In general, mTOR enhances anabolic processes, such as protein, nucleotide, and lipid synthesis, whereas it represses catabolic processes, such as autophagy.
3.1 mTORC1 functions and substrates
mTORC1 acts as a master regulator of protein synthesis by acting on two major substrates, namely ribosomal protein S6 kinase-1 (S6K1) and eukaryotic translation initiation factor 4E (eIF4E)-binding protein-1 (4E-BP1).
Mitochondrial biogenesis and lipid and nucleotide syntheses represent other anabolic processes regulated by mTORC1, at both the transcriptional and post-transcriptional levels. mTORC1 regulates the transcriptional activity of sterol regulatory element-binding protein 1/2 (SREBP1/2), which promotes lipid and cholesterol synthesis,30 in mouse embryonic fibroblasts (MEFs). SR protein kinase 2 (SRPK2) is another substrate of mTORC1 involved in mediating lipid biosynthesis, through stabilization of enzymes that regulate lipogenesis.31 In addition, mTORC1 enhances mitochondrial biogenesis by promoting the interaction of transcription factor yin-yang 1 (YY1) with peroxisome proliferator–activated receptor γ coactivator-1α (PGC1-α), as demonstrated in mouse skeletal muscle and in C2C12 myotubes.32
mTORC1 maintains the nucleotide pool required for nucleic acid synthesis by triggering de novo purine and pyrimidine synthesis in various human and mouse cells. mTORC1-induced purine synthesis is mediated by activating transcription factor 4 (ATF4)33 in response to growth signals such as insulin. mTORC1 also drives pyrimidine synthesis through S6K-induced activation of carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase (CAD), an enzyme that mediates the first phase of pyrimidine synthesis.34,35
mTORC1 also regulates cellular metabolism. mTORC1 activation promotes a metabolic shift towards glycolysis and enhances the pentose phosphate pathway, through activation of hypoxia inducible factor-1α (HIF-1α) and SREBP1/2, respectively, as observed in a number of human cell lines.30
On the other hand, mTORC1 inhibits catabolic processes, particularly autophagy. Several endogenous components of the autophagy machinery are phosphorylated and negatively regulated by mTOR, including unc-51-like autophagy-activating kinase (ULK-1), autophagy-related (ATG)-13,36–38 ATG14,39activating molecule in Beclin1-regulated autophagy (AMBRA1),40 and UV radiation resistance-associated gene (UVRAG).41 mTORC1 also regulates autophagy at the transcriptional level, inhibiting nuclear localization of transcription factor EB (TFEB) by phosphorylating it at Ser211.42–44 TFEB enhances the transcription of genes involved in lysosomal biogenesis and autophagy. mTORC1 also inhibits lysosomal biogenesis by inactivating transcription factor E3 (TFE3),45 another member of the TFEB family, as reported in adult retinal pigment epithelial cell line-19 (ARPE-19) under nutrient-rich conditions.
3.2 mTORC2 functions and substrates
mTORC2 also mediates crucial cellular functions, although it appears to be involved in the regulation of less broad processes than mTORC1 (Figure 1C). Systemic disruption of either mTORC1 or mTORC2 is embryonically lethal. However, mouse embryos with genetic mTORC2 disruption die in a later stage of development than those with mTORC1 disruption, mainly because of cardiovascular defects. mTORC2 regulates cellular polarity and cytoskeletal organization.25,46 The mechanisms through which mTORC2 regulates cell architecture are not fully understood. However, Protein kinase C-α (PKC-α) and Ras homolog gene family member-A (RhoA) seem to mediate the effect of mTORC2.24,25
mTORC2 is a primary regulator of cell survival through the activation of protein kinase B (AKT), and the subsequent inhibition of forkhead-box (FOXO)-1/3a transcription factors, important regulators of cellular metabolism, growth, and survival.47 mTORC2 promotes activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1), another member of the protein kinase A/protein kinase G/protein kinase C (AGC) family, involved in cell survival, as demonstrated in different human cell lines and in cardiomyocytes.48,49 The regulation of cell survival by mTORC2 is also mediated by crosstalk with the Hippo pathway. The Hippo pathway is involved in the regulation of cell proliferation and apoptosis through inhibition of the pro-growth and survival factor yes-associated protein 1 (YAP1).50 mTORC2 directly inhibits mammalian sterile 20-like kinase (MST1) through phosphorylation of MST1 at Ser438, which in turn inhibits MST1 dimerization, thereby promoting cell survival.51
4. Upstream regulators of mTOR
mTORC1 and mTORC2 sense cellular nutritional and energy status and are activated in response to nutrients and pro-growth signals. mTORC1 activation occurs at the lysosome surface (Figure 1D). Well-coordinated integration of upstream signals deriving from two sets of small G proteins, Ras-related GTPases (Rags) and Ras homolog enriched in brain (RHEB), is required for mTORC1 activation. Rags recruit mTORC1 to lysosomes, bringing it into close proximity with RHEB, and RHEB stimulates mTORC1 kinase activity. In contrast, mTORC1 becomes inactive in response to nutrient starvation, energy stress, hypoxia, or cellular damage (Figure 1E). mTORC2 activity is modulated by growth factors and mTORC1.
4.1 Mechanisms of regulation of mTORC1 activity
In the presence of mitogens and growth factors, mTORC1 is activated, primarily through inhibition of tuberous sclerosis complexes (TSC)1/2. TSC1/2 inhibits mTORC1 by acting as GTPase activating proteins (GAPs) towards RHEB.52,53 Multiple pro-growth signals inhibit TSC1/2.54 Insulin activates mTORC1 through the phosphoinositide 3-kinases (PI3K)/AKT pathway. Insulin stimulates AKT-mediated phosphorylation of PRAS40, which causes dissociation of PRAS40 from mTORC1, thereby activating mTORC1. AKT also phosphorylates TSC2, which in turn dissociates from the surface of lysosomes, thus allowing RHEB-induced activation of mTORC1.55,56 In addition, growth factors inhibit TSC through inhibitor of nuclear factor κB kinase β (IKKβ), the major effector of tumour necrosis factor α (TNFα) signalling. IKKβ interacts with and phosphorylates TSC1 at multiple residues, resulting in its inactivation.57 The RAS/mitogen-activated protein kinase (MAPK) cascade inhibits TSC through extracellular signal-regulated kinase (ERK) or p90 ribosomal S6 kinase (RSK)1.58,59
On the other hand, reduced availability of nutrients and oxygen decreases mTORC1 activity, since anabolic reactions are not advantageous in these conditions. Low amino acid levels are sensed by different sensors, which keep mTORC1 in the inactive state. AMP-activated protein kinase (AMPK) is activated during energy deprivation or mitochondrial oxidative stress and inhibits all pro-growth mechanisms and ATP-consuming processes.60 AMPK inhibits mTORC1 by direct phosphorylation of RAPTOR or by activating TSC2, as demonstrated in MEFs and in HEK293 cells.61,62
Glycogen synthase kinase (GSK)-3β activates TSC2, whereas RHEB is inactivated during energy deprivation.63–65 During hypoxia, regulated in development and DNA damage responses (REDD)-1 is up-regulated and inhibits mTORC1 via TSC1/2,66 whereas hexokinase-II (HK-II) interacts with and deactivates mTORC1 in the presence of low glucose.67 Other cellular stresses, such as amino acid starvation and growth factor removal, as well as hyperosmotic, energetic, and pH stresses, induce lysosomal translocation of TSC1/2, further suggesting that TSC1/2 is critical for mTORC1 inactivation during stresses.68Protein kinase G1 (PKG1) activates TSC2 through phosphorylation at Ser1365 and Ser1366 in cardiac cells undergoing haemodynamic stress, thereby inhibiting mTORC1 and promoting autophagy activation.69
Oxidative stress inactivates mTORC1 via direct redox modifications. Thioredoxin 1 (TRX1) preserves mTORC1 activity by reducing mTOR at Cys1483 in the presence of oxidative stress in cardiomyocytes.70DNA damage-inducible transcript 4-like (DDiT4L), a target of the hypoxia-inducible transcription factor HIF1α, is a negative modulator of mTORC1 during maladaptive cardiac hypertrophy.71 Recently, the Hippo pathway has also emerged as a negative regulator of mTORC1. Stress response protein kinases LATS1 and LATS2 directly phosphorylate RAPTOR at Ser606, as shown in HEK293 cells, thereby reducing RHEB-mediated mTORC1 activation and leading to a reduction of cellular growth and organ size in the presence of growth factors and amino acid stimulation.72
4.2 mTORC1 nutrient sensing
Rags are the main components of the amino acid-sensing machinery that controls mTORC1 recruitment at lysosomal surface. Rags are obligate heterodimers, with RagA and RagB bound with RagC and RagD, respectively. In their active conformation (on-state), RagA/B is bound to GTP whereas RagC/D is bound to GDP. On the other hand, RagA/B is bound to GDP and RagC/D to GTP in their inactive state (off-state). The ‘on-state’ represents the only Rag heterodimer nucleotide configuration that can interact with and recruit mTORC1 at the lysosomal surface, where mTORC1 kinase activity is enhanced by RHEB. Rag heterodimer activity is regulated by several GAPs and guanine nucleotide exchange factors (GEFs) in response to nutrient availability, growth factors, and stress. Rags are anchored to lysosomes through a pentameric complex named Ragulator.73 In the presence of nutrients, Rags are ‘on-state’ and interact with RAPTOR, thereby ensuring lysosomal recruitment of mTORC1 and RHEB-induced activation of mTOR kinase activity.74–79 In low nutrient conditions, Folliculin (FLCN), a GAP for Rag C/D, localizes to lysosomes, where it becomes inactive. Lysosomal Ragulator and FLCN also inhibit the exchange of GDP with GTP in RagA, thereby maintaining the Rag complex in its ‘off-state’.77
Additional signals sense nutrients and amino acids and converge on Rag activation. To date, several amino acid sensors have been identified. GATOR-1 and GATOR-2 complexes act as negative and positive regulators of mTORC1, respectively.80–82 Recently, amino acids derived from lysosomal degradation of proteins were shown to activate mTOR independently of the GATOR–Rag complex, through a mechanism involving homotypic fusion and vacuole protein sorting (HOPS), a tethering complex involved in vesicle fusion.83 SESTRIN-2 inhibits mTORC1 in the presence of low leucine levels.84–86 During amino acid starvation, general control non-derepressible 2 (GCN2) mediates ATF4-induced expression of SESTRIN-2.87 Acetyl-coenzyme A (AcCoA), the final leucine metabolite, was reported to regulate mTORC1 activity by EP300-induced acetylation of RAPTOR. Of interest, both RAPTOR acetylation and AcCoA levels are decreased in tissues of fasted mice.88 Arginine modulates mTORC1 activity through cellular arginine sensor for mTORC1 (CASTOR) and solute carrier family 38 member 9 (SLC38A9), which act as negative and positive regulators of mTORC1, respectively.89–93Vacuolar H(+)-adenosine triphosphatase (v-ATPase) represents another amino acid sensor able to modulate mTORC1 activity. v-ATPase is activated by increased accumulation of amino acids in lysosomes, thereby acting as a positive regulator of mTORC1.94S-adenosylmethionine sensor upstream of mTORC1 (SAMSOR) senses levels of S-adenosylmethionine (SAM), a methyl donor-derived from methionine. Methionine starvation or reduced SAM levels leads to mTORC1 inhibition.95 These results suggest that mTORC1 senses amino acid levels and regulates anabolic processes.
Glucose metabolism also modulates mTOR activity. In vitro inhibition of cardiac glycolytic flux via phosphoglucose isomerase (PGI) inhibition in cardiomyocytes treated with glucose and glutamine correlates with glucose 6-phosphate accumulation and mTOR activation, which, in turn, increases protein synthesis, a hallmark of cardiac hypertrophy.96 In addition, glucose induces mTOR activation by inhibiting branched-chain amino acids (BCAAs) degradation in cardiomyocytes.97 Glucose also activates mTOR through interaction between leucyl-tRNA synthase 1 (LARS1) and RagD, whereas LARS1 releases leucine and fails to activate mTOR under glucose-deficient conditions, as shown in HEK293T cells.98 LARS1 participates in anabolism in the presence of high glucose through the production of leucyl-tRNA and activation of mTOR. Dihydroxyacetone phosphate (DAHP), a membrane-impermeable metabolite involved in lipid synthesis, transmits glucose availability to activate mTORC1.99 These results suggest that glucose participates in anabolism through mTOR activation.
Other metabolites, such as lipids or nucleotides, also regulate mTORC1 activity, although the molecular basis is not completely understood. Fatty acids activate mTORC1 via de novo synthesis of phosphatidic acid.100 Lysosomal low-density lipoprotein (LDL)-derived cholesterol also drives mTORC1 activation through a mechanism involving SLC38A9 and Niemann–Pick C1 protein (NPC1) as positive and negative modulators, respectively.101 Exogenous administration of purine nucleobases activates mTORC1 in MEFs and HeLa cells, in a TSC/RHEB-dependent manner.102 On the other hand, inhibition of purine biosynthesis leads to a reduction in the GTP-bound status of RHEB.103
4.3 Mechanisms of regulation of mTORC2 activity
Growth factors, such as insulin, activate mTORC2 through the PI3K pathway104 (Figure 1E). Phosphatidylinositol 3,4,5-trisphosphate (PIP3), a product of the PI3K pathway, reduces mSIN1 interaction with mTORC2, thereby promoting mTORC2 activation.104,105 TSC1/2 physically interacts with mTORC2, contributing to its activation, independently of RHEB, as demonstrated in multiple human cell lines.106 However, the mechanism through which TSC1/2 activates mTORC2 requires further investigation. Insulin/PI3K/AKT represents the main pathway by which mTORC1 and mTORC2 are interconnected. mTORC1 inhibits insulin/PI3K/AKT through a negative feedback mediated by S6K-induced inactivation of IRS1.107 An adaptor protein, growth factor receptor-bound protein 10 (Grb10), is activated by mTORC1 and inhibits the insulin/IGF-1 receptor.108,109 In addition, S6K1 phosphorylates RICTOR and inhibits mTORC2 in response to mTORC1 activation.110 Conversely, AMPK activates the mTORC2/AKT pathway, thereby promoting cell survival.111
5. mTOR and cardiac diseases
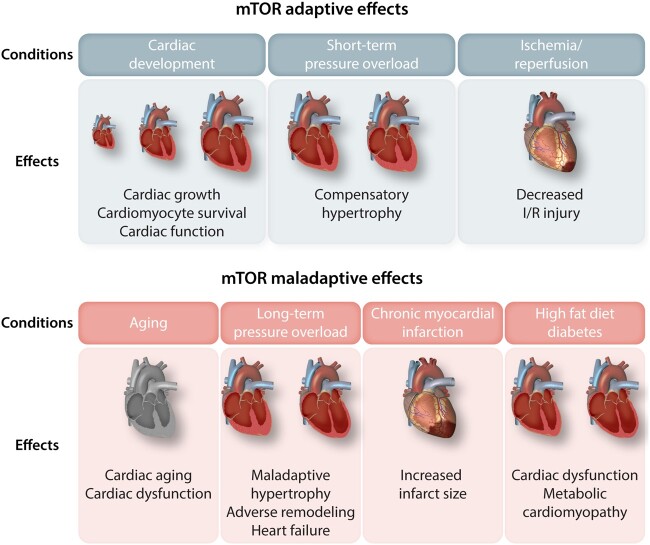
An appropriate balance between anabolism and catabolism, as well as a co-ordinated response to nutrient bioavailability or stress conditions, is required for the maintenance of cardiac function. The role of mTOR signalling has been extensively characterized in pre-clinical models of cardiac stress, such as mice undergoing surgical procedures or metabolic insults. mTOR modulation has both adaptive and maladaptive functions, depending on the type of stress and the level and the duration of its activation.
5.1 Cardiac development
Single systemic deletion of mTOR, RAPTOR, RICTOR, or mLST8 is embryonically lethal, indicating the indispensable role of mTOR signalling in driving embryonic development.47,112,113 Mice with constitutive cardiac deletion of mTOR die in utero or during the perinatal period.114 Embryos of mTOR knockout animals displayed a reduction of cardiomyocyte proliferation and increased apoptosis. Mice with cardiac-specific mTOR deletion during the adult stage showed a reduced lifespan, along with the development of fatal dilated cardiomyopathy. At the molecular level, mitochondrial dysfunction and increased apoptosis and autophagy were observed in the hearts of these mice. Concomitant cardiac deletion of 4E-BP1 improves lifespan, cardiac function, and survival in the mTOR knockout animals.115 Mice with inducible cardiac deletion of RAPTOR during adulthood exhibit decreased survival and cardiac function.116 The role of mTORC1 during heart development was also studied in mice with constitutive cardiac-specific deletion of RHEB. The RHEB knockout animals die during the early post-natal period and show sarcomere derangements and reduced protein synthesis, which are alleviated when 4E-BP1 is concomitantly deleted.117
Constitutive cardiac deletion of RICTOR is not embryonically lethal but leads to impairment of cardiac function at six months of age,51 whereas tamoxifen-induced cardiac-specific RICTOR deletion during adulthood does not affect cardiac growth or function.118 These results indicate that a disruption of mTORC1 and mTORC2 activities is deleterious for cardiac development and function under unstressed conditions, but that mTORC1 appears to be more critical than mTORC2 for the regulation of cardiac homeostasis.
5.2 Cardiac aging
The heart undergoes a series of structural and functional changes during aging119 and modulation of mTOR appears to be a potential strategy to mitigate cardiac complications in the elderly. mTOR expression is enhanced in the senescent heart and rapamycin administration increases lifespan and reverses cardiac dysfunction during aging in mice, along with reduced cardiac expression of genes regulating inflammation, hypertrophy, and contractile function.120–123 However, a recent study showed that mice genetically deficient for the RNA component of telomerase exhibit an over-activation of mTOR in several organs, including the heart. Treatment with rapamycin unexpectedly decreased lifespan in this model, suggesting that mTOR activation is adaptive in the presence of short telomeres.124
A possible approach to reduce mTOR activity during aging is caloric restriction (CR), defined as a low-calorie diet regimen without malnutrition. Restoration of autophagy, in part by mTOR inhibition, is one of the critical mechanisms by which CR delays cardiac aging. CR improves cardiac function and metabolism in the aged heart in mice, by enhancing autophagy and by decreasing markers of senescence along with the reduction of mTORC1 activity.125 A broad class of compounds mimicking CR, termed caloric restriction mimetics (CRMs), has recently emerged as a potentially effective therapeutic tool for delaying age-induced abnormalities.126
GSK-3α is a critical regulator of mTORC1 activity in the aged heart. GSK-3α deletion in mice activates mTORC1 and aggravates cardiac aging, which is accompanied by massive hypertrophy, fibrosis, sarcomere derangement, mitochondrial dysfunction, and impaired autophagy.127 Combined inducible cardiomyocyte-specific deletion of Rho-associated coiled-coil–containing protein kinase (ROCK)1 and ROCK2, two regulators of actin cytoskeleton, reduces cardiac fibrosis in aging, by promoting starvation-induced autophagy and mTOR inhibition. However, the mechanistic link explaining how the decreased activity of ROCK1 and ROCK2 results in mTOR inhibition remains unknown.128
In contrast to the role of mTORC1 in promoting aging, activation of mTORC2 offsets age-induced abnormalities. Systemic heterozygous deletion of RICTOR decreases lifespan in male mice, suggesting that mTORC2 acts as an anti-aging molecule.129 Consistently, RICTOR overexpression in Drosophila slows cardiac aging and up-regulates autophagy. Transforming growth factor β (TGF-β)/INHB/activin dawdle protein, a member of the TGF-β family, inhibits mTORC2 during aging, whereas dawdle knockdown rescues mTORC2 activity and induces cardiac protection.130 Further studies are needed to understand whether this mechanism of regulation of mTORC2 is conserved in mammals.
These results suggest that enhancing mTORC2 activity or decreasing mTORC1 may be a promising strategy to slow cardiac aging.
5.3 Cardiac hypertrophy
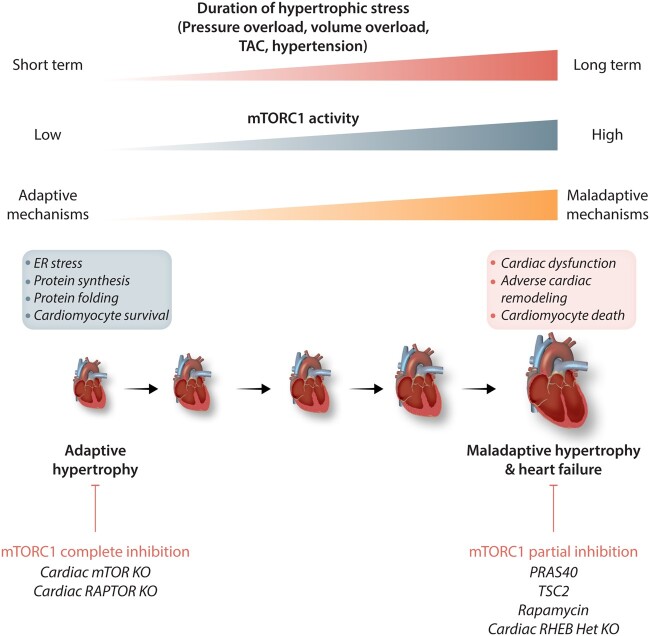
mTOR is a crucial regulator of cardiac hypertrophy and remodelling (Figure 2). mTOR represents a central pathway that is activated by stimuli that trigger cardiac hypertrophy, thereby promoting the development of both adaptive and maladaptive cardiac growth. Cardiac hypertrophy is triggered by mechanical stress, such as pressure or volume overload, and by neuro-hormonal factors, such as angiotensin II and adrenergic stimulation. Angiotensin II (Ang-II) and adrenergic β1/α1 stimulation also activate mTORC1 in the cardiovascular system. In cardiomyocytes in vitro, rapamycin inhibits Ang-II-induced up-regulation of S6K1, but it is unable to reduce atrial natriuretic factor secretion and beta-myosin heavy chain expression in response to Ang-II treatment.131 Similarly, rapamycin inhibits S6K1 activation and protein synthesis induced by isoproterenol or phenylephrine in cultured cardiomyocytes.132,133 These results suggest that G protein-coupled receptors (GPCRs) play a significant role in mTORC1 activation during hypertrophy development.
Figure 2.
mTORC1 modulation during cardiac hypertrophy. mTORC1 mediates both adaptive and maladaptive effects during hypertrophic stress. The figure shows that complete inhibition of mTORC1 is detrimental whereas partial inhibition is protective in response to hypertrophic signals. See text for further details. The figure was made using tools provided by Servier Medical Arts, among others. Legend: ER, endoplasmic reticulum; Het, heterozygous; KO, knockout; mTOR, mechanistic target of rapamycin; mTORC1, mTOR complex 1; PRAS40, proline-rich AKT substrate 40; RAPTOR, regulatory-associated protein of mTOR; RHEB, Ras homolog enriched in brain; TAC, transverse aortic constriction; TSC2, tuberous sclerosis protein 2.
The general consensus regarding the role of mTORC1 in cardiac hypertrophy is that this protein complex mediates both adaptive and maladaptive hypertrophy. Mice with inducible cardiac deletion of mTOR or RAPTOR during adulthood develop marked cardiac dysfunction in response to pressure overload induced by transverse aortic constriction (TAC), without the development of compensatory hypertrophy.115,116 At the molecular level, a reduction of protein synthesis was observed in the knockout mice, due to reduced mTORC1-induced phosphorylation of S6K1 and 4E-BP1.116 Alternatively, X-box binding protein 1 (XBP1), a component of the unfolded protein response (UPR) is down-regulated in pre-clinical models of heart failure and in heart failure patients, whereas its overexpression improves cardiac function in mice. XBP1 promotes adaptive cardiac growth by stimulating mTOR activity at the transcriptional level.134 Since XBP1 senses nutrient availability in metabolic tissues, such as liver and adipose tissue, acting as a positive regulator of anabolism and cell growth, this study may suggest that mTOR activation mediates XBP1 anabolism and cellular growth in response to nutrients. Cardiac-specific overexpression of mTORC1 does not lead to hypertrophy, suggesting that mTORC1 promotes hypertrophy only when hypertrophic insults are present.135 A recent study also highlighted crosstalk between endoplasmic reticulum (ER) stress and mTORC1, which contributes to the development of compensatory hypertrophy. ER stress is activated in mice subjected to severe TAC or undergoing exercise, along with up-regulation of activating transcription factor 6 (ATF6), an enhancer of protein folding. Cardiac ATF6 conditional knockout mice subjected to TAC and analysed after 7 days were unable to develop compensatory cardiac hypertrophy and showed chamber dilatation and cardiac dysfunction. ATF6 knockout leads to RHEB down-regulation, resulting in mTORC1 inhibition. Interestingly, ATF6 induces RHEB expression in the presence of growth factors, but not in response to other activators of ATF6 that do not induce growth, suggesting the stress-specific role of ATF6 in mediating cardiac growth as an adaption to stress.136
Cardiac hypertrophy is also evident in athletes, as an adaptive response of the heart to intense physical activity. mTOR signalling activation is associated with the development of physiological cardiac hypertrophy in response to exercise.137Creb-binding protein/p300–interacting transactivator with ED-rich carboxy-terminal domain (CITED)-4 overexpression was previously found to activate mTORC1 and be sufficient to induce physiological hypertrophy.138 CITED-4 expression was also found to be up-regulated in endurance-exercised mice.139 Modest cardiac dysfunction and dilatation were observed in cardiomyocyte-specific CITED-4 knockout mice subjected to an intensive swim exercise.140 In contrast, cardiac-specific deletion of CITED-4 led to heart failure and cardiac dysfunction in mice in the presence of pressure overload. Reduced mTOR activity, along with increased apoptosis and detrimental autophagy, was observed under these conditions.140
Persistent and excessive mTORC1 activation promotes the transition from adaptive to maladaptive hypertrophy, and partial inhibition of mTORC1 is protective in response to pressure overload. Cardiac-specific heterozygous deletion of the RHEB gene or pharmacological inhibition of mTORC1 improves cardiac remodelling in mouse models of pressure overload and volume overload, such as mitral regurgitation and chronic myocardial infarction.141–145 RAPTOR haploinsufficiency also attenuates heart failure induced by pressure overload or Gαq overexpression. On the other hand, similar beneficial effects are not observed in mice with cardiac-specific overexpression of 4E-BP1, where the lack of compensatory hypertrophy exacerbates cardiomyopathy. The cardioprotective effects of RAPTOR haploinsufficiency may be mediated by 4E-BP1-independent mechanisms, such as the effect of mTORC1 upon mitochondria and metabolism.146 In cancer cells, mTORC1 directly phosphorylates superoxide dismutase 1 at Thr40, thereby inactivating it.147 Thus, the cardioprotective effects of RAPTOR haploinsufficiency may be in part mediated through up-regulation of an antioxidant response. Inhibition of mTORC1 through cardiac-specific PRAS40 overexpression attenuates cardiac hypertrophy and remodelling and ameliorates systolic function during pressure overload.148 TSC2 activation reduces cardiac hypertrophy in response to pressure overload.149 Conversely, FLCN or TSC2 deletion in the heart leads to cardiac hypertrophy and dysfunction by promoting activation of mTORC1.150,151
In mice undergoing pressure overload, PKG activation is sufficient to reduce cardiac hypertrophy. PKG activates TSC2 by phosphorylation at Ser1365 and 1366, resulting in mTORC1 inhibition and activation of protective autophagy.69 This study suggests that PKG signalling acts as a negative regulator of mTORC1 during stress through TSC2 activation. Interestingly, previous work demonstrated that PKG activity is reduced in response to hypertrophic stimuli by oxidation at cysteine 42. It was recently shown that knock-in mice with a PKG redox-dead cysteine 42 to serine mutation show less cardiac hypertrophy and dysfunction in response to pressure overload due to activation of TSC2 and inhibition of mTORC1.152
The p38γ and δ MAPKs contribute to maladaptive hypertrophy during stress by enhancing mTORC1 activity. p38γ and δ MAPKs null mice exhibit reduced mTOR activation and reduced heart growth during the post-natal period whereas they are protected from angiotensin II-induced hypertrophy. p38γ and δ MAPKs phosphorylate the mTOR inhibitory protein DEPTOR and mediate its ubiquitination and degradation.153
Aerobic glycolysis, the so-called Warburg effect, promotes the growth of cancer cells. Increased glucose uptake and intermediates of the glycolytic pathway and its accessory pathways also promote cardiac hypertrophy. In mice undergoing pressure overload, the hexosamine biosynthetic pathway (HBP), the auxiliary pathway of glycolysis, is up-regulated. Overexpression of glutamine: fructose-6-phosphate amidotransferase 1 (GFAT1), a rate-limiting enzyme of the HBP pathway, exacerbates hypertrophy through mTOR activation. GFAT1 activates mTOR through O-GlcNAcylation, a post-translational modification.154 Glucose inhibits BCAA catabolism by down-regulating cAMP response element binding protein (CREB)-induced expression of Krüppel-like factor 15 (KLF15). Accumulation of BCAA, in turn, leads to mTOR activation.97 Other glycolytic intermediates, including glucose-6-phoshate96 and dihydroxyacetone phosphate,99 also activate mTOR. The involvement of these mechanisms in pressure overload-induced cardiac hypertrophy is unknown. In addition, whether mTOR activation induced by glucose metabolites induces adaptive or maladaptive hypertrophy remains to be clarified.mTORC1 is regulated by microRNAs during cardiac remodelling. Cardiac-specific overexpression of microRNA-221 leads to cardiac hypertrophy and dysfunction in mice. MicroRNA-221 suppresses p27, a negative regulator of cyclin-dependent kinase 2 (CDK2), a protein involved in cell cycle regulation. Activation of CDK2 induces mTORC1 activation and reduces autophagy.155 microRNA-99a inhibits autophagy and induces cardiac hypertrophy, by inhibiting GSK3-β and enhancing mTORC1 activity.156 The involvement of endogenous microRNA-221 and micro-99a in cardiac hypertrophy, such as pressure overload-induced hypertrophy, remains to be clarified.
Constitutive cardiomyocyte-restricted deletion of RICTOR induces cardiac dysfunction and dilatation after pressure overload, along with enhanced apoptosis and reduced compensatory hypertrophy. As we discussed earlier, RICTOR knockout activates MST1, a potent inducer of apoptosis. Since MST1 inhibition rescues cardiac dysfunction in RICTOR knockout mice, MST1 acts as the main mediator of mTORC2 modulation during cardiac adaption to stress.51 RICTOR deletion in the heart during adulthood also leads to cardiac dysfunction in mice undergoing TAC.118 The calcium signal transducer 1 (STIM1) in cardiomyocytes in vivo acts as a positive modulator of mTORC2, through direct interaction and phosphorylation of RICTOR, which results in AKT-induced inactivation of GSK3-β. STIM1 silencing in vivo attenuates compensatory hypertrophy through mTORC2 down-regulation and results in increased GSK3-β-induced apoptosis and cardiac dysfunction.157 Overexpression of calcineurin (Cn)Aβ1, a specific Cn isoform, inhibits hypertrophy in response to pressure overload. This is mediated by mTORC2-induced activation of ATF4, which in turn contributes to an enhanced antioxidant defence and improved ATP metabolism by promoting glutathione (GSH) production and decreasing oxidation of mitochondrial proteins.158 This result suggests that mTORC2 activation promotes compensatory cardiac growth and limits maladaptive hypertrophy.
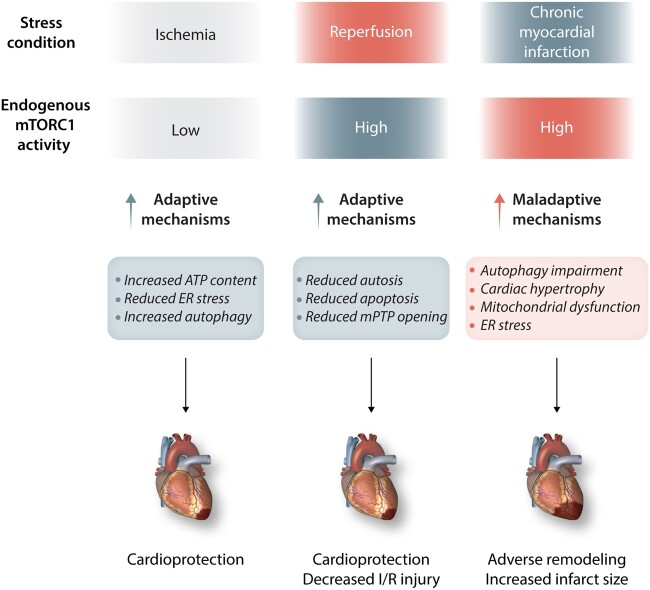
5.4 Acute and chronic myocardial ischemia
Myocardial ischemia or energy stress leads to inactivation of mTORC1 (Figure 3). The latter represents an adaptive response since mTORC1 inhibition allows activation of protective mechanisms, including autophagy, which in turn limits myocardial infarction. RHEB down-regulation and GSK-3β activation contribute to mTOR inhibition in these conditions. RHEB is inhibited in response to ischemia or glucose deprivation, leading to mTORC1 inactivation.64 Forced activation of RHEB induces cell death and ER stress and inhibits autophagy, resulting in increased infarct size. GSK-3β inhibition also leads to mTORC1 activation and autophagy inhibition, contributing to increased myocardial ischemic injury.65 Restoration of autophagy rescues the detrimental effects of either RHEB up-regulation or GSK-3β inhibition.64,65 Caution should be exercised, however, regarding the differential role of mTOR and autophagy during ischemia and reperfusion. mTORC1 inhibition with rapamycin before ischemia reduces ischemia/reperfusion (I/R) injury, whereas it does not confer protection when administered in the reperfusion phase.65,159 Increased production of autophagosomes in the presence of inhibition of autophagosome–lysosome fusion during reperfusion induces a unique form of death, termed autosis, in cardiomyocytes.160 In fact, endogenous mTORC1 is activated during the reperfusion phase and mTORC1 activation during reperfusion limits I/R injury; thus, it appears to be adaptive.65,161 Cardiac overexpression of dominant-negative GSK-3β or systemic heterozygous deletion of GSK-3β decreases I/R injury through mTORC1 activation, which is accompanied by reduced mitochondrial permeability transition pore (mPTP) opening.65 CITED-4 overexpression mitigates I/R injury through mTORC1 activation, along with reduced apoptosis and detrimental autophagy.138 BCAAs administered 30 minutes before ischemia reduce I/R injury through mTOR activation, and these protective effects are abrogated in the presence of rapamycin or in mTOR heterozygous knockout mice,162 where the protective effect of BCAA and mTOR appears to be mediated primarily through their effects during reperfusion. At the molecular level, BCAAs reduced mitochondrial swelling induced by I/R and preserved cell viability in cardiomyocytes undergoing I/R in vitro. In a recent work, the lysosomal-associated transmembrane protein 4B (LAPTM4B) was reported to be down-regulated in the hearts of mice undergoing I/R. Systemic deletion of LAPTM4B (LAPTM4B KO) aggravated I/R injury, whereas its overexpression was beneficial. Mechanistically, LAPTM4B decreased mTORC1 activity and rescued autophagic flux and autophagosomes clearance, through a mTORC1/TFEB-dependent mechanism.163 Rapamycin administration reduced I/R injury in LAPTM4B KO mice. Further studies should be conducted in mice with cardiomyocyte-specific deletion of LAPTM4B.
Figure 3.
mTORC1 modulation during ischemia/reperfusion (I/R) and chronic myocardial infarction. Ischemia reduces mTORC1 activity, conferring cardioprotection. The increased activity of mTOR during reperfusion inhibits maladaptive mechanisms and limits I/R injury. Chronic myocardial infarction enhances mTOR activity, leading to detrimental cardiac effects. See text for further details.
Combined mTORC1 and mTORC2 inhibition abrogates the protective effects of ischemic pre-conditioning, whereas single mTORC1 inhibition does not exert any influence. This suggests that mTORC2 activation is essential for cardioprotection induced by pre-conditioning. The activation of mTORC2 in this condition may be mediated by ribosomal protein S6 (Rps6), a protein needed for protein translation.164mTORC1 activation is detrimental in the heart during chronic ischemia. In rats undergoing myocardial infarction (MI), mTORC1 inhibition using everolimus reduces adverse remodelling and infarct size and stimulates autophagy.143 Another study found that S6K1, an mTORC1 target, is activated in the heart in response to MI in mice, and its pharmacological inhibition attenuates myocardial remodelling and activates AKT.165 AKT activation induced by S6K1 inhibition may be secondary to mTORC2 activation. In mice undergoing MI, loss of RICTOR increases myocardial damage and remodelling, whereas PRAS40 inhibition improves cardiac function and post-infarction remodelling by enhancing the mTORC2-induced activity of AKT.166 This suggests that the induction of a shift from mTORC1 to mTORC2 activation could be a potential strategy to protect the heart from ischemic disease.
5.5 Metabolic cardiomyopathy
As a nutrient sensor, mTOR co-ordinates glucose and lipid metabolism across tissues, including in liver, adipose tissue, and skeletal muscle, with different responses during feeding and fasting cycles. In the presence of nutrients, the insulin level rises, leading to mTOR activation, which in turn promotes nutrient storage by activating lipogenesis, glycogen, and protein synthesis. During fasting, mTOR activity in metabolic tissues decreases, resulting in nutrient mobilization.6 In metabolic disorders, such as obesity and type 2 diabetes, mTOR activity is increased, and accumulating lines of evidence suggest that mTORC1 activation contributes to the development of metabolic cardiomyopathy. In pre-clinical models of metabolic syndrome, the activity of mTORC1 signalling in the heart is increased and its inhibition improves cardiac function, along with the restoration of autophagy.64,167–169 RHEB or AKT activation leads to mTORC1-induced suppression of autophagy in the presence of obesity.170mTORC1 inhibition also attenuates the progression of diabetic cardiac complications. In mouse models of type 2 diabetes, rapamycin or PRAS40 activation improves cardiac function.171,172 In these conditions, reduced oxidative stress is observed, in association with improved glucose metabolism and expression of contractile proteins, such as myosin light chain MLY2, myosin heavy chain 6, and myosin-binding protein C. We previously found that autophagy is inhibited in the heart in response to long-term HFD consumption, due to deregulated activation of the RHEB/mTORC1 signalling pathway. Impairment of autophagy in response to HFD consumption significantly reduced ischemic tolerance and increased infarct size in response to prolonged ischemia. Rapamycin treatment or partial mTOR gene deletion reduced infarct size and apoptosis in mice with HFD-induced metabolic syndrome subjected to acute ischemic injury.64 In a different study, rapamycin treatment was also found to reduce I/R injury in mice with type 2 diabetes through STAT3 and miR-17/20 activation173 and inhibition of prolyl hydroxylase (PHD3), a direct target of miR-17/20. In contrast, type 1 diabetic mice overexpressing a dominant-negative form of mTOR in the heart exhibited severe cardiac dysfunction when compared to mice overexpressing constitutively active mTOR, these results being opposite from those obtained with mTORC1 inhibition in the presence of type 2 diabetes and obesity. It is possible that mTORC2 inhibition contributes to the detrimental effects of dominant-negative mTOR overexpression in this study, although the authors of the study did not observe modulation of mTORC2 in transgenic mice.174
5.6 Genetic and doxorubicin-induced cardiomyopathies
mTOR is activated in genetic cardiomyopathies, such as those caused by mutations in tripartite motif containing 63 (TRIM63), muscle ring finger protein 1 (MuRF1), laminin A/C gene, or genes involved in LEOPARD syndrome.175–178 In pre-clinical models carrying these genetic defects, mTORC1 inhibition reduces cardiomyopathy. A gain-of-function mutation in the RAGC gene is associated with the development of foetal dilated cardiomyopathy, as a result of exacerbated mTOR activation, even in the presence of amino acid deprivation.179 Cardiac deletion of pentatricopeptide repeat domain protein (PTCD1), a protein involved in mitochondrial RNA metabolism, leads to dilated cardiomyopathy, together with transcriptional down-regulation of mitochondrial biogenesis and fatty acid metabolism and up-regulation of mTOR. The down-regulation of mitochondrial RNA assembly induced by loss of PTCD1 leads to increased protein synthesis signalling, suggesting that mTOR is activated as a compensatory response.180 Further studies are needed to clarify whether mTOR inhibition rescues the detrimental effects of PTCD1 inhibition and the mechanistic link between RNA metabolism and transcriptional regulation of mTOR.
The role of mTOR has been explored in cardiotoxicity induced by anthracyclines, such as doxorubicin. mTOR activity is attenuated following doxorubicin treatment, resulting in cardiac atrophy and dysfunction, independently of apoptosis.181Angiotensin-converting enzyme (ACE) inhibition using enalapril reduces cardiac dysfunction in mice treated chronically with doxorubicin, and this is associated with reactivation of mTOR.182 Doxorubicin enhances cardiac expression of phosphoinositide 3-kinase gamma (PI3Kγ), which in turn stimulates the AKT/mTOR/ULK1 pathway and inhibits autophagy.183 Suppression of PI3Kγ reduces doxorubicin-induced cardiac dysfunction by promoting cardiac mitophagy, a selective form of autophagy devoted to mitochondrial digestion. Additional studies are warranted to understand the role of mTORC1 and mTORC2 in doxorubicin-induced cardiotoxicity.
6. Therapeutic prospects: translation to human
The evidence obtained in pre-clinical studies thus far suggests that modulating mTOR signalling may represent a valid therapeutic strategy in a variety of cardiovascular diseases. To date, mTOR inhibitors (Table 1) are widely used in cancer treatment and are being tested in ongoing clinical trials as components of the drug-eluting stent for the treatment of patients with coronary artery diseases (NCT01347554). In these cases, inhibition of mTOR signalling leads to a reduction of cellular proliferation, which is relevant both in cancer cells and in stent restenosis.15,184,185 mTOR inhibition is also efficacious in ameliorating clinical outcomes of patients with idiopathic multicentric Castleman disease (iMCD), a lymphoproliferative disorder characterized by systemic inflammation and organ dysfunction.186
Table 1.
Generations of mTOR inhibitors and their cardiovascular effects
| Name | Generation |
mTORC1
inhibition |
mTORC2
inhibition |
Protective cardiovascular effects in pre-clinical models | References |
|---|---|---|---|---|---|
| Rapamycin | First | Yes | No |
↓Cardiovascular aging ↓Cardiac hypertrophy ↓I/R injury |
121 , 141 , 165 |
| Everolimus |
First (rapalog) |
Yes | no | ↓Adverse cardiac remodelling ↓Infarct size | 143 |
| Sirolimus |
First (rapalog) |
Yes | no | ↓In-stent restenosis | 184 , 185 |
| Torin-1 |
Second (ATP-competitive mTOR kinase inhibitor) |
Yes | yes | ↑Cardiac ischaemic remodelling | 166 |
| RapaLink-1 |
Third (juxtaposition of first- and second-generation inhibitor-binding pockets) |
Yes | yes | Not available | 188 |
The first generation of mTOR inhibitors is rapalogs (rapamycin derivatives), such as sirolimus and everolimus. The second generation of mTOR inhibitors includes ATP-competitive mTOR kinase inhibitors, such as Torin-1, which inhibit both mTORC1 and mTORC2.187 This class also includes mTOR/PI3K dual inhibitors. Unlike rapalogs, second-generation mTOR inhibitors ensure that the feedback loop between AKT and mTORC1 activation is suppressed. Second-generation mTOR inhibitors have exhibited promising results in pre-clinical trials in cancer, although mTOR resistance, due to mutation in mTOR kinase, is often observed. The third class of mTOR inhibitor, called RapaLink-1, was recently developed to overcome mTOR resistance.188 Whether these compounds are also effective in the cardiovascular system remains to be established, since combined inhibition of both complexes may be deleterious during cardiac stress.
Regarding the use of mTOR inhibitors for treating cardiovascular diseases in humans, everolimus was effective in reducing cardiac allograft vasculopathy in the setting of heart transplantation.189,190 The Controlled Level EVERolimus in Acute Coronary Syndromes (CLEVER-ACS) (NCT01529554) clinical trial is evaluating the effects of everolimus on cardiac function and inflammation in patients with ST-elevation myocardial infarction. Another interesting strategy to reduce mTOR activity includes a CR regiment or CRMs. Long-term CR was reported to improve diastolic function in human subjects.191 Ongoing clinical trials are testing these strategies in other heart diseases as well. However, since CR and CRMs also act on other targets besides mTOR, it will be challenging to differentiate the specific role of mTOR modulation from the other effects of CR and CRMs.
7. Conclusion
We have summarized the latest findings regarding the role of mTOR in cardiac pathophysiology (Table 2). mTOR senses the nutrient and energy status and regulates anabolism and catabolism by integrating environmental and intracellular inputs. Recent studies identified the novel determinants of mTOR regulation in heart and vascular cells, both at baseline and during stress. Partial mTOR inhibition attenuates cardiac injury under some conditions, such as during chronic cardiac remodelling, aging, and metabolic disorders. In contrast, complete inhibition of mTOR is detrimental due to the loss of adaptive mechanisms, particularly in response to I/R injury or pressure overload.
Table 2.
mTORC1 activity in cardiac diseases and interventions aimed to reduce disease progression
| Cardiac disease | mTOR activity | Protective/detrimental effects of mTOR inhibition | References |
|---|---|---|---|
|
Cardiac Aging |
Increased | Protective | 120 , 125 |
| Cardiac hypertrophy | Increased |
Protective when partial Detrimental when complete |
|
|
Ischemia/reperfusion Injury |
Decreased during ischemia and increased during reperfusion | Protective before ischemia | 65 , 159 |
|
Myocardial infarction |
Increased | Protective | 143 |
| Metabolic | Increased | Protective | 64 , 171 , 172 |
Some aspects of mTOR signalling remain to be addressed in the near future. First of all, a better comprehension of adaptive and maladaptive mechanisms exerted by mTORC1 and mTORC2 in cardiac diseases is needed to understand how to translate mTOR modulation to the human setting. In particular, the role of mTORC1 as a potential driver of cardiac regeneration after MI should be better elucidated.192 A recent study demonstrated that checkpoint kinase 1 (CHK1) overexpression promotes cardiomyocyte proliferation and improves cardiac function, as well as mTORC1 activation, in mice undergoing permanent MI. Rapamycin blunts cardiomyocyte proliferation induced by CHK1 overexpression in vitro,192 suggesting that mTORC1 activation in the infarcted area, in which mTORC1 activity is usually shut down, could improve myocardial regeneration.
Characterization of upstream modulators and substrates of mTORC1/2 in various stress conditions would be helpful. Since mTORC1 signalling and mTORC2 signalling are interconnected, additional studies on this crosstalk should be conducted, which would clarify when one complex compensates for a reduction in the other, or when mTORC1 and mTORC2 act in a synergistic manner. The pathophysiology of mTORC2 is less well characterized than that of mTORC1. The development of selective mTORC2 inhibitors would help to better comprehend the functions and signalling network of this complex.
Finally, translation of basic mTOR investigation into the clinical setting remains modest. Few studies have investigated mTOR activity in human samples. In aortic samples of patients with abdominal aortic aneurysm (AAA), mTOR is up-regulated compared to in control segments, and this is associated with an impairment of autophagy.193 Further studies should assess the levels of mTOR activity in samples from patients with different cardiovascular diseases. Clarification of this issue is important, since mTOR inhibition does not always lead to protective effects and may also be detrimental. Moreover, mTOR activity increases under some pathologic circumstances whereas it decreases in others, making it difficult to estimate the exact window for therapeutic interventions. Additional studies are also needed to elucidate how polymorphisms or genetic variants of mTOR complexes correlate with cardiovascular diseases. Clinical trials modulating the activities of mTOR should be organized in patients with MI, heart failure, and metabolic diseases.
Acknowledgements
We thank D. Z. for critical reading of the manuscript.
Conflict of interest: none declared.
Funding
This work was supported in part by grants from the Italian Ministry of Research (PRIN 2017N8K7S2_002) and from the Pasteur Institute, Cenci-Bolognetti Foundation to S.S. This work was also supported by Foundation Leducq Transatlantic Networks (Grant 15CBD04) to S.S. and J.S., and US Public Health Service (Grants HL067724, HL091469, HL138720, HL112330, HL144626, HL150881, and AG23039) to J.S. J.S. is a recipient of the 2020 Merit Award from the American Heart Association (20 Merit 35120374).
References
- 1. Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL.. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994;369:756–758. [DOI] [PubMed] [Google Scholar]
- 2. Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH.. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994;78:35–43. [DOI] [PubMed] [Google Scholar]
- 3. Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT.. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem 1995;270:815–822. [DOI] [PubMed] [Google Scholar]
- 4. Chiu MI, Katz H, Berlin V.. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA 1994;91:12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saxton RA, Sabatini DM.. mTOR signaling in growth, metabolism, and disease. Cell 2017;169:361–371. [DOI] [PubMed] [Google Scholar]
- 6. Liu GY, Sabatini DM.. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 2020;21:183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wullschleger S, Loewith R, Hall MN.. TOR signaling in growth and metabolism. Cell 2006;124:471–484. [DOI] [PubMed] [Google Scholar]
- 8. Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN.. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell 1994;5:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP.. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol 1993;13:6012–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP.. mTOR kinase structure, mechanism and regulation. Nature 2013;497:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM.. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006;22:159–168. [DOI] [PubMed] [Google Scholar]
- 12. Laplante M, Sabatini DM.. mTOR signaling in growth control and disease. Cell 2012;149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guertin DA, Sabatini DM.. Defining the role of mTOR in cancer. Cancer Cell 2007;12:9–22. [DOI] [PubMed] [Google Scholar]
- 14. Sciarretta S, Volpe M, Sadoshima J.. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res 2014;114:549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sciarretta S, Forte M, Frati G, Sadoshima J.. New insights into the role of mTOR signaling in the cardiovascular system. Circ Res 2018;122:489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM.. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 2003;11:895–904. [DOI] [PubMed] [Google Scholar]
- 17. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002;110:163–175. [DOI] [PubMed] [Google Scholar]
- 18. Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K.. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002;110:177–189. [DOI] [PubMed] [Google Scholar]
- 19. Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N.. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem 2010;285:20109–20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM.. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 2007;25:903–915. [DOI] [PubMed] [Google Scholar]
- 21. Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH.. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 2007;9:316–323. [DOI] [PubMed] [Google Scholar]
- 22. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM.. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009;137:873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP.. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 2017;552:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004;14:1296–1302. [DOI] [PubMed] [Google Scholar]
- 25. Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN.. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004;6:1122–1128. [DOI] [PubMed] [Google Scholar]
- 26. Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM.. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol 2006;16:1865–1870. [DOI] [PubMed] [Google Scholar]
- 27. Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B.. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006;127:125–137. [DOI] [PubMed] [Google Scholar]
- 28. Yang Q, Inoki K, Ikenoue T, Guan KL.. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 2006;20:2820–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pearce LR, Huang X, Boudeau J, Pawłowski R, Wullschleger S, Deak M, Ibrahim AFM, Gourlay R, Magnuson MA, Alessi DR.. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J 2007;405:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD.. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 2010;39:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, Chavez A, Zhang E, Isik M, Couvillon A, Dephoure NE, Blackwell TK, Yu JJ, Rabinowitz JD, Cantley LC, Blenis J.. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell 2017;171:1545–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P.. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007;450:736–740. [DOI] [PubMed] [Google Scholar]
- 33. Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD.. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 2016;351:728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ben-Sahra I, Howell JJ, Asara JM, Manning BD.. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013;339:1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN.. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 2013;339:1320–1323. [DOI] [PubMed] [Google Scholar]
- 36. Kim J, Kundu M, Viollet B, Guan KL.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N.. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009;20:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ganley IG, Lam Du H, Wang J, Ding X, Chen S, Jiang X.. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009;284:12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan HX, Russell RC, Guan KL.. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 2013;9:1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F.. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013;15:406–416. [DOI] [PubMed] [Google Scholar]
- 41. Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH.. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 2015;57:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM.. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012;5:ra42–ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martina JA, Chen Y, Gucek M, Puertollano R.. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012;8:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J 2012;31:1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R.. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014;7:ra9–ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu L, Das S, Losert W, Parent CA.. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 2010;19:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM.. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006;11:859–871. [DOI] [PubMed] [Google Scholar]
- 48. García-Martínez JM, Alessi DR.. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J 2008;416:375–385. [DOI] [PubMed] [Google Scholar]
- 49. Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A.. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation 2005;111:1652–1659. [DOI] [PubMed] [Google Scholar]
- 50. Ma S, Meng Z, Chen R, Guan KL.. The hippo pathway: biology and pathophysiology. Annu Rev Biochem 2019;88:577–604. [DOI] [PubMed] [Google Scholar]
- 51. Sciarretta S, Zhai P, Maejima Y, Del Re DP, Nagarajan N, Yee D, Liu T, Magnuson MA, Volpe M, Frati G, Li H, Sadoshima J.. mTORC2 regulates cardiac response to stress by inhibiting MST1. Cell Rep 2015;11:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inoki K, Li Y, Xu T, Guan KL.. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003;17:1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J.. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 2003;13:1259–1268. [DOI] [PubMed] [Google Scholar]
- 54. Huang J, Manning BD.. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 2008;412:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD.. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014;156:771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC.. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 2002;10:151–162. [DOI] [PubMed] [Google Scholar]
- 57. Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, He X, Hung JY, Lai CC, Ding Q, Su JL, Yang JY, Sahin AA, Hortobagyi GN, Tsai FJ, Tsai CH, Hung MC.. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 2007;130:440–455. [DOI] [PubMed] [Google Scholar]
- 58. Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP.. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 2005;121:179–193. [DOI] [PubMed] [Google Scholar]
- 59. Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J.. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA 2004;101:13489–13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gonzalez A, Hall MN, Lin SC, Hardie DG.. AMPK and TOR: the Yin and Yang of cellular nutrient sensing and growth control. Cell Metab 2020;31:472–492. [DOI] [PubMed] [Google Scholar]
- 61. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ.. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008;30:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Inoki K, Zhu T, Guan KL.. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003;115:577–590. [DOI] [PubMed] [Google Scholar]
- 63. Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL.. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006;126:955–968. [DOI] [PubMed] [Google Scholar]
- 64. Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G, Sadoshima J.. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation 2012;125:1134–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhai P, Sciarretta S, Galeotti J, Volpe M, Sadoshima J.. Differential roles of GSK-3beta during myocardial ischemia and ischemia/reperfusion. Circ Res 2011;109:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr,. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004;18:2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roberts DJ, Tan-Sah VP, Ding EY, Smith JM, Miyamoto S.. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell 2014;53:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Demetriades C, Plescher M, Teleman AA.. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun 2016;7:10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ranek MJ, Kokkonen-Simon KM, Chen A, Dunkerly-Eyring BL, Vera MP, Oeing CU, Patel CH, Nakamura T, Zhu G, Bedja D, Sasaki M, Holewinski RJ, Van Eyk JE, Powell JD, Lee DI, Kass DA.. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature 2019;566:264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Oka SI, Hirata T, Suzuki W, Naito D, Chen Y, Chin A, Yaginuma H, Saito T, Nagarajan N, Zhai P, Bhat S, Schesing K, Shao D, Hirabayashi Y, Yodoi J, Sciarretta S, Sadoshima J.. Thioredoxin-1 maintains mechanistic target of rapamycin (mTOR) function during oxidative stress in cardiomyocytes. J Biol Chem 2017;292:18988–19000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simonson B, Subramanya V, Chan MC, Zhang A, Franchino H, Ottaviano F, Mishra MK, Knight AC, Hunt D, Ghiran I, Khurana TS, Kontaridis MI, Rosenzweig A, Das S.. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci Signal 2017;10:eaaf5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gan W, Dai X, Dai X, Xie J, Yin S, Zhu J, Wang C, Liu Y, Guo J, Wang M, Liu J, Hu J, Quinton RJ, Ganem NJ, Liu P, Asara JM, Pandolfi PP, Yang Y, He Z, Gao G, Wei W.. LATS suppresses mTORC1 activity to directly coordinate Hippo and mTORC1 pathways in growth control. Nat Cell Biol 2020;22:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mu Z, Wang L, Deng W, Wang J, Wu G.. Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane. Cell Discov 2017;3:17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rogala KB, Gu X, Kedir JF, Abu-Remaileh M, Bianchi LF, Bottino AMS, Dueholm R, Niehaus A, Overwijn D, Fils AP, Zhou SX, Leary D, Laqtom NN, Brignole EJ, Sabatini DM.. Structural basis for the docking of mTORC1 on the lysosomal surface. Science 2019;366:468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM.. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL.. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008;10:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lawrence RE, Fromm SA, Fu Y, Yokom AL, Kim DJ, Thelen AM, Young LN, Lim CY, Samelson AJ, Hurley JH, Zoncu R.. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 2019;366:971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM.. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013;52:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Petit CS, Roczniak-Ferguson A, Ferguson SM.. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013;202:1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM.. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013;340:1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shen K, Valenstein ML, Gu X, Sabatini DM.. Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J Biol Chem 2019;294:2970–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen K, Huang RK, Brignole EJ, Condon KJ, Valenstein ML, Chantranupong L, Bomaliyamu A, Choe A, Hong C, Yu Z, Sabatini DM.. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 2018;556:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hesketh GG, Papazotos F, Pawling J, Rajendran D, Knight JDR, Martinez S, Taipale M, Schramek D, Dennis JW, Gingras AC.. The GATOR-Rag GTPase pathway inhibits mTORC1 activation by lysosome-derived amino acids. Science 2020;370:351–356. [DOI] [PubMed] [Google Scholar]
- 84. Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM.. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016;351:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM.. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016;351:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM.. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 2014;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, Koumenis C, Thompson CB.. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev 2015;29:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Son SM, Park SJ, Lee H, Siddiqi F, Lee JE, Menzies FM, Rubinsztein DC.. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab 2019;29:192–201. e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM.. The CASTOR proteins are arginine sensors for the mTORC1 Pathway. Cell 2016;165:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM.. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 2016;536:229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jung J, Genau HM, Behrends C.. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol 2015;35:2479–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shen K, Sabatini DM.. Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci USA 2018;115:9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G.. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015;519:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM.. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011;334:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, Sabatini DM.. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017;358:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karlstaedt A, Khanna R, Thangam M, Taegtmeyer H.. Glucose 6-phosphate accumulates via phosphoglucose isomerase inhibition in heart muscle. Circ Res 2020;126:60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shao D, Villet O, Zhang Z, Choi SW, Yan J, Ritterhoff J, Gu H, Djukovic D, Christodoulou D, Kolwicz SC Jr, Raftery D, Tian R.. Glucose promotes cell growth by suppressing branched-chain amino acid degradation. Nat Commun 2018;9:2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yoon I, Nam M, Kim HK, Moon HS, Kim S, Jang J, Song JA, Jeong SJ, Kim SB, Cho S, Kim Y, Lee J, Yang WS, Yoo HC, Kim K, Kim MS, Yang A, Cho K, Park HS, Hwang GS, Hwang KY, Han JM, Kim JH, Kim S.. Glucose-dependent control of leucine metabolism by leucyl-tRNA synthetase 1. Science 2020;367:205–210. [DOI] [PubMed] [Google Scholar]
- 99. Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, Lewis CA, Sabatini DM.. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab 2020;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Menon D, Salloum D, Bernfeld E, Gorodetsky E, Akselrod A, Frias MA, Sudderth J, Chen PH, DeBerardinis R, Foster DA.. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J Biol Chem 2017;292:6303–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, Zoncu R.. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 2017;355:1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hoxhaj G, Hughes-Hallett J, Timson RC, Ilagan E, Yuan M, Asara JM, Ben-Sahra I, Manning BD.. The mTORC1 signaling network senses changes in cellular purine nucleotide levels. Cell Rep 2017;21:1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Emmanuel N, Ragunathan S, Shan Q, Wang F, Giannakou A, Huser N, Jin G, Myers J, Abraham RT, Unsal-Kacmaz K.. Unsal-Kacmaz K. purine nucleotide availability regulates mTORC1 activity through the Rheb GTPase. Cell Rep 2017;19:2665–2680. [DOI] [PubMed] [Google Scholar]
- 104. Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, Su B, Wei W.. PtdIns(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov 2015;5:1194–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gan X, Wang J, Su B, Wu D.. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 2011;286:10998–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Huang J, Dibble CC, Matsuzaki M, Manning BD.. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. MCB 2008;28:4104–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shah OJ, Wang Z, Hunter T.. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 2004;14:1650–1656. [DOI] [PubMed] [Google Scholar]
- 108. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM.. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011;332:1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J.. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011;332:1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Julien LA, Carriere A, Moreau J, Roux PP.. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. MCB 2010;30:908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kazyken D, Magnuson B, Bodur C, Acosta-Jaquez HA, Zhang D, Tong X, Barnes TM, Steinl GK, Patterson NE, Altheim CH, Sharma N, Inoki K, Cartee GD, Bridges D, Yin L, Riddle SM, Fingar DC.. AMPK directly activates mTORC2 to promote cell survival during acute energetic stress. Sci Signal 2019;12:eaav3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC.. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. MCB 2004;24:9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S.. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. MCB 2004;24:6710–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhu Y, Pires KM, Whitehead KJ, Olsen CD, Wayment B, Zhang YC, Bugger H, Ilkun O, Litwin SE, Thomas G, Kozma SC, Abel ED.. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS One 2013;8:e54221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang D, Contu R, Latronico MV, Zhang J, Rizzi R, Catalucci D, Miyamoto S, Huang K, Ceci M, Gu Y, Dalton ND, Peterson KL, Guan KL, Brown JH, Chen J, Sonenberg N, Condorelli G.. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest 2010;120:2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shende P, Plaisance I, Morandi C, Pellieux C, Berthonneche C, Zorzato F, Krishnan J, Lerch R, Hall MN, Ruegg MA, Pedrazzini T, Brink M.. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation 2011;123:1073–1082. [DOI] [PubMed] [Google Scholar]
- 117. Tamai T, Yamaguchi O, Hikoso S, Takeda T, Taneike M, Oka T, Oyabu J, Murakawa T, Nakayama H, Uno Y, Horie K, Nishida K, Sonenberg N, Shah AM, Takeda J, Komuro I, Otsu K.. Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem 2013;288:10176–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shende P, Xu L, Morandi C, Pentassuglia L, Heim P, Lebboukh S, Berthonneche C, Pedrazzini T, Kaufmann BA, Hall MN, Ruegg MA, Brink M.. Cardiac mTOR complex 2 preserves ventricular function in pressure-overload hypertrophy. Cardiovasc Res 2016;109:103–114. [DOI] [PubMed] [Google Scholar]
- 119. Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J.. Aging and autophagy in the heart. Circ Res 2016;118:1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA.. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009;460:392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Flynn JM, O'Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, Zykovich A, Mooney SD, Strong R, Rosen CJ, Kapahi P, Nelson MD, Kennedy BK, Melov S.. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell 2013;12:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, DuBois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T.. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep 2013;4:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hua Y, Zhang Y, Ceylan-Isik AF, Wold LE, Nunn JM, Ren J.. Chronic Akt activation accentuates aging-induced cardiac hypertrophy and myocardial contractile dysfunction: role of autophagy. Basic Res Cardiol 2011;106:1173–1191. [DOI] [PubMed] [Google Scholar]
- 124. Ferrara-Romeo I, Martinez P, Saraswati S, Whittemore K, Grana-Castro O, Thelma Poluha L, Serrano R, Hernandez-Encinas E, Blanco-Aparicio C, Maria Flores J, Blasco MA.. The mTOR pathway is necessary for survival of mice with short telomeres. Nat Commun 2020;11:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS.. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 2014;13:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Abdellatif M, Sedej S, Carmona-Gutierrez D, Madeo F, Kroemer G.. Autophagy in cardiovascular aging. Circ Res 2018;123:803–824. [DOI] [PubMed] [Google Scholar]
- 127. Zhou J, Freeman TA, Ahmad F, Shang X, Mangano E, Gao E, Farber J, Wang Y, Ma XL, Woodgett J, Vagnozzi RJ, Lal H, Force T.. GSK-3alpha is a central regulator of age-related pathologies in mice. J Clin Invest 2013;123:1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Shi J, Surma M, Yang Y, Wei L.. Disruption of both ROCK1 and ROCK2 genes in cardiomyocytes promotes autophagy and reduces cardiac fibrosis during aging. FASEB J 2019;33:7348–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lamming DW, Mihaylova MM, Katajisto P, Baar EL, Yilmaz OH, Hutchins A, Gultekin Y, Gaither R, Sabatini DM.. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell 2014;13:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chang K, Kang P, Liu Y, Huang K, Miao T, Sagona AP, Nezis IP, Bodmer R, Ocorr K, Bai H.. TGFB-INHB/activin signaling regulates age-dependent autophagy and cardiac health through inhibition of MTORC2. Autophagy 2020;16:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sadoshima J, Izumo S.. Rapamycin selectively inhibits angiotensin II-induced increase in protein synthesis in cardiac myocytes in vitro: potential role of 70-kD S6 kinase in angiotensin II-induced cardiac hypertrophy. Circ Res 1995;77:1040–1052. [DOI] [PubMed] [Google Scholar]
- 132. Simm A, Schluter K, Diez C, Piper HM, Hoppe J.. Activation of p70(S6) kinase by beta-adrenoceptor agonists on adult cardiomyocytes. J Mol Cell Cardiol 1998;30:2059–2067. [DOI] [PubMed] [Google Scholar]
- 133. Wang L, Proud CG.. Ras/Erk signaling is essential for activation of protein synthesis by Gq protein-coupled receptor agonists in adult cardiomyocytes. Circ Res 2002;91:821–829. [DOI] [PubMed] [Google Scholar]
- 134. Wang X, Deng Y, Zhang G, Li C, Ding G, May HI, Tran DH, Luo X, Jiang DS, Li DL, Wei X, Xu L, Ferdous A, Gillette TG, Scherer PE, Jiang X, Wang ZV.. Spliced X-box binding protein 1 stimulates adaptive growth through activation of mTOR. Circulation 2019;140:566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Shen WH, Chen Z, Shi S, Chen H, Zhu W, Penner A, Bu G, Li W, Boyle DW, Rubart M, Field LJ, Abraham R, Liechty EA, Shou W.. Cardiac restricted overexpression of kinase-dead mammalian target of rapamycin (mTOR) mutant impairs the mTOR-mediated signaling and cardiac function. J Biol Chem 2008;283:13842–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Blackwood EA, Hofmann C, Santo Domingo M, Bilal AS, Sarakki A, Stauffer W, Arrieta A, Thuerauf DJ, Kolkhorst FW, Muller OJ, Jakobi T, Dieterich C, Katus HA, Doroudgar S, Glembotski CC.. ATF6 regulates cardiac hypertrophy by transcriptional induction of the mTORC1 activator, Rheb. Circ Res 2019;124:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kemi OJ, Ceci M, Wisloff U, Grimaldi S, Gallo P, Smith GL, Condorelli G, Ellingsen O.. Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. J Cell Physiol 2008;214:316–321. [DOI] [PubMed] [Google Scholar]
- 138. Bezzerides VJ, Platt C, Lerchenmuller C, Paruchuri K, Oh NL, Xiao C, Cao Y, Mann N, Spiegelman BM, Rosenzweig A.. CITED4 induces physiologic hypertrophy and promotes functional recovery after ischemic injury. JCI Insight 2016;1:e85904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bostrom P, Mann N, Wu J, Quintero PA, Plovie ER, Panakova D, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, Spiegelman BM.. C/EBPbeta controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell 2010;143:1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lerchenmuller C, Rabolli CP, Yeri A, Kitchen R, Salvador AM, Liu LX, Ziegler O, Danielson K, Platt C, Shah R, Damilano F, Kundu P, Riechert E, Katus HA, Saffitz JE, Keshishian H, Carr SA, Bezzerides VJ, Das S, Rosenzweig A.. CITED4 protects against adverse remodeling in response to physiological and pathological stress. Circ Res 2020;127:631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ, Izumo S.. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation 2003;107:1664–1670. [DOI] [PubMed] [Google Scholar]
- 142. McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, Izumo S.. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation 2004;109:3050–3055. [DOI] [PubMed] [Google Scholar]
- 143. Buss SJ, Muenz S, Riffel JH, Malekar P, Hagenmueller M, Weiss CS, Bea F, Bekeredjian R, Schinke-Braun M, Izumo S, Katus HA, Hardt SE.. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. J Am Coll Cardiol 2009;54:2435–2446. [DOI] [PubMed] [Google Scholar]
- 144. Ikeda M, Ide T, Fujino T, Matsuo Y, Arai S, Saku K, Kakino T, Oga Y, Nishizaki A, Sunagawa K.. The Akt-mTOR axis is a pivotal regulator of eccentric hypertrophy during volume overload. Sci Rep 2015;5:15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wu X, Cao Y, Nie J, Liu H, Lu S, Hu X, Zhu J, Zhao X, Chen J, Chen X, Yang Z, Li X.. Genetic and pharmacological inhibition of Rheb1-mTORC1 signaling exerts cardioprotection against adverse cardiac remodeling in mice. Am J Pathol 2013;182:2005–2014. [DOI] [PubMed] [Google Scholar]
- 146. Dai DF, Liu Y, Basisty N, Karunadharma P, Dastidar SG, Chiao YA, Chen T, Beyer RP, Chin MT, Maccoss M, La Spada AR, Rabinovitch PS.. Differential effects of various genetic mouse models of the mechanistic target of rapamycin complex I inhibition on heart failure. Geroscience 2019;41:847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tsang CK, Chen M, Cheng X, Qi Y, Chen Y, Das I, Li X, Vallat B, Fu LW, Qian CN, Wang HY, White E, Burley SK, Zheng XFS.. SOD1 phosphorylation by mTORC1 couples nutrient sensing and redox regulation. Mol Cell 2018;70:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Volkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY, Ornelas L, Joyo E, Thuerauf DJ, Konstandin MH, Gude N, Glembotski CC, Sussman MA.. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci USA 2013;110:12661–12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Morales CR, Li DL, Pedrozo Z, May HI, Jiang N, Kyrychenko V, Cho GW, Kim SY, Wang ZV, Rotter D, Rothermel BA, Schneider JW, Lavandero S, Gillette TG, Hill JA.. Inhibition of class I histone deacetylases blunts cardiac hypertrophy through TSC2-dependent mTOR repression. Sci Signal 2016;9:ra34–ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh H-B, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, Marston Linehan W.. Marston Linehan W. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet 2014;23:5706–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Taneike M, Nishida K, Omiya S, Zarrinpashneh E, Misaka T, Kitazume-Taneike R, Austin R, Takaoka M, Yamaguchi O, Gambello MJ, Shah AM, Otsu K.. mTOR hyperactivation by ablation of tuberous sclerosis complex 2 in the mouse heart induces cardiac dysfunction with the increased number of small mitochondria mediated through the down-regulation of autophagy. PLoS One 2016;11:e0152628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Oeing CU, Nakamura T, Pan S, Mishra S, Dunkerly-Eyring BL, Kokkonen-Simon KM, Lin BL, Chen A, Zhu G, Bedja D, Lee DI, Kass DA, Ranek MJ.. PKG1alpha cysteine-42 redox state controls mTORC1 activation in pathological cardiac hypertrophy. Circ Res 2020;127:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. González-Terán B, López JA, Rodríguez E, Leiva L, Martínez-Martínez S, Bernal JA, Jiménez-Borreguero LJ, Redondo JM, Vazquez J, Sabio G.. p38gamma and delta promote heart hypertrophy by targeting the mTOR-inhibitory protein DEPTOR for degradation. Nat Commun 2016;7:10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tran DH, May HI, Li Q, Luo X, Huang J, Zhang G, Niewold E, Wang X, Gillette TG, Deng Y, Wang ZV.. Chronic activation of hexosamine biosynthesis in the heart triggers pathological cardiac remodeling. Nat Commun 2020;11:1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, Wang Y, Zhao X, Zou Y, Song L, Zhang L, Hui R.. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ 2015;22:986–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li Z, Song Y, Liu L, Hou N, An X, Zhan D, Li Y, Zhou L, Li P, Yu L, Xia J, Zhang Y, Wang J, Yang X.. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ 2017;24:1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Benard L, Oh JG, Cacheux M, Lee A, Nonnenmacher M, Matasic DS, Kohlbrenner E, Kho C, Pavoine C, Hajjar RJ, Hulot JS.. Cardiac Stim1 silencing impairs adaptive hypertrophy and promotes heart failure through inactivation of mTORC2/Akt signaling. Circulation 2016;133:1458–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Padrón-Barthe L, Villalba-Orero M, Gómez-Salinero JM, Acín-Pérez R, Cogliati S, López-Olañeta M, Ortiz-Sánchez P, Bonzón-Kulichenko E, Vázquez J, García-Pavía P, Rosenthal N, Enríquez JA, Lara-Pezzi E.. Activation of serine one-carbon metabolism by calcineurin Abeta1 reduces myocardial hypertrophy and improves ventricular function. J Am Coll Cardiol 2018;71:654–667. [DOI] [PubMed] [Google Scholar]
- 159. Das A, Salloum FN, Durrant D, Ockaili R, Kukreja RC.. Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J Mol Cell Cardiol 2012;53:858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Nah J, Zhai P, Huang CY, Fernandez AF, Mareedu S, Levine B, Sadoshima J.. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest 2020;130:2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J.. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007;100:914–922. [DOI] [PubMed] [Google Scholar]
- 162. Satomi S, Morio A, Miyoshi H, Nakamura R, Tsutsumi R, Sakaue H, Yasuda T, Saeki N, Tsutsumi YM.. Branched-chain amino acids-induced cardiac protection against ischemia/reperfusion injury. Life Sci 2020;245:117368. [DOI] [PubMed] [Google Scholar]
- 163. Gu S, Tan J, Li Q, Liu S, Ma J, Zheng Y, Liu J, Bi W, Sha P, Li X, Wei M, Cao N, Yang HT.. Downregulation of LAPTM4B contributes to the impairment of the autophagic flux via unopposed activation of mTORC1 signaling during myocardial ischemia/reperfusion injury. Circ Res 2020;127:e148–e165. [DOI] [PubMed] [Google Scholar]
- 164. Yano T, Ferlito M, Aponte A, Kuno A, Miura T, Murphy E, Steenbergen C.. Pivotal role of mTORC2 and involvement of ribosomal protein S6 in cardioprotective signaling. Circ Res 2014;114:1268–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Di R, Wu X, Chang Z, Zhao X, Feng Q, Lu S, Luan Q, Hemmings BA, Li X, Yang Z.. S6K inhibition renders cardiac protection against myocardial infarction through PDK1 phosphorylation of Akt. Biochem J 2012;441:199–207. [DOI] [PubMed] [Google Scholar]
- 166. Volkers M, Konstandin MH, Doroudgar S, Toko H, Quijada P, Din S, Joyo A, Ornelas L, Samse K, Thuerauf DJ, Gude N, Glembotski CC, Sussman MA.. Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation 2013;128:2132–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Guo R, Zhang Y, Turdi S, Ren J.. Adiponectin knockout accentuates high fat diet-induced obesity and cardiac dysfunction: role of autophagy. Biochim Biophys Acta 2013;1832:1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Pires KM, Buffolo M, Schaaf C, David Symons J, Cox J, Abel ED, Selzman CH, Boudina S.. Activation of IGF-1 receptors and Akt signaling by systemic hyperinsulinemia contributes to cardiac hypertrophy but does not regulate cardiac autophagy in obese diabetic mice. J Mol Cell Cardiol 2017;113:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Li ZL, Woollard JR, Ebrahimi B, Crane JA, Jordan KL, Lerman A, Wang SM, Lerman LO.. Transition from obesity to metabolic syndrome is associated with altered myocardial autophagy and apoptosis. Arterioscler Thromb Vasc Biol 2012;32:1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Xu X, Hua Y, Nair S, Zhang Y, Ren J.. Akt2 knockout preserves cardiac function in high-fat diet-induced obesity by rescuing cardiac autophagosome maturation. J Mol Cell Biol 2013;5:61–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Volkers M, Doroudgar S, Nguyen N, Konstandin MH, Quijada P, Din S, Ornelas L, Thuerauf DJ, Gude N, Friedrich K, Herzig S, Glembotski CC, Sussman MA.. PRAS40 prevents development of diabetic cardiomyopathy and improves hepatic insulin sensitivity in obesity. EMBO Mol Med 2014;6:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Das A, Durrant D, Koka S, Salloum FN, Xi L, Kukreja RC.. Mammalian target of rapamycin (mTOR) inhibition with rapamycin improves cardiac function in type 2 diabetic mice: potential role of attenuated oxidative stress and altered contractile protein expression. J Biol Chem 2014;289:4145–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Samidurai A, Roh SK, Prakash M, Durrant D, Salloum FN, Kukreja RC, Das A.. STAT3-miR-17/20 signaling axis plays a critical role in attenuating myocardial infarction following rapamycin treatment in diabetic mice. Cardiovasc Res 2020;116:2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Xu X, Kobayashi S, Timm D, Huang Y, Zhao F, Shou W, Liang Q.. Enhanced mTOR complex 1 signaling attenuates diabetic cardiac injury in OVE26 mice. FASEB J 2019;33:12800–12811. [DOI] [PubMed] [Google Scholar]
- 175. Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, Bronson R, Franchini KG, Neel BG, Kontaridis MI.. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest 2011;121:1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, Rabinovitch PS, Kaeberlein M, Kennedy BK.. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med 2012;4:144ra103–144ra103. 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Choi JC, Wu W, Muchir A, Iwata S, Homma S, Worman HJ.. Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J Biol Chem 2012;287:40513–40524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, Marian AJ.. Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res 2012;111:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Long PA, Zimmermann MT, Kim M, Evans JM, Xu X, Olson TM.. De novo RRAGC mutation activates mTORC1 signaling in syndromic fetal dilated cardiomyopathy. Hum Genet 2016;135:909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Perks KL, Rossetti G, Kuznetsova I, Hughes LA, Ermer JA, Ferreira N, Busch JD, Rudler DL, Spahr H, Schondorf T, Shearwood AJ, Viola HM, Siira SJ, Hool LC, Milenkovic D, Larsson NG, Rackham O, Filipovska A.. PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep 2018;23:127–142. [DOI] [PubMed] [Google Scholar]
- 181. Zhu W, Soonpaa MH, Chen H, Shen W, Payne RM, Liechty EA, Caldwell RL, Shou W, Field LJ.. Acute doxorubicin cardiotoxicity is associated with p53-induced inhibition of the mammalian target of rapamycin pathway. Circulation 2009;119:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hullin R, Metrich M, Sarre A, Basquin D, Maillard M, Regamey J, Martin D.. Diverging effects of enalapril or eplerenone in primary prevention against doxorubicin-induced cardiotoxicity. Cardiovasc Res 2018;114:272–281. [DOI] [PubMed] [Google Scholar]
- 183. Li M, Sala V, De Santis MC, Cimino J, Cappello P, Pianca N, Di Bona A, Margaria JP, Martini M, Lazzarini E, Pirozzi F, Rossi L, Franco I, Bornbaum J, Heger J, Rohrbach S, Perino A, Tocchetti CG, Lima BHF, Teixeira MM, Porporato PE, Schulz R, Angelini A, Sandri M, Ameri P, Sciarretta S, Lima-Junior RCP, Mongillo M, Zaglia T, Morello F, Novelli F, Hirsch E, Ghigo A.. Phosphoinositide 3-kinase gamma inhibition protects from anthracycline cardiotoxicity and reduces tumor growth. Circulation 2018;138:696–711. [DOI] [PubMed] [Google Scholar]
- 184. Gallo R, Padurean A, Jayaraman T, Marx S, Roque M, Adelman S, Chesebro J, Fallon J, Fuster V, Marks A, Badimon JJ.. Inhibition of intimal thickening after balloon angioplasty in porcine coronary arteries by targeting regulators of the cell cycle. Circulation 1999;99:2164–2170. [DOI] [PubMed] [Google Scholar]
- 185. Morice MC, Serruys PW, Sousa JE, Fajadet J, Ban Hayashi E, Perin M, Colombo A, Schuler G, Barragan P, Guagliumi G, Molnar F, Falotico R, Lesions R-C-EA.. Randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N Engl J Med 2002;346:1773–1780. [DOI] [PubMed] [Google Scholar]
- 186. Fajgenbaum DC, Langan RA, Japp AS, Partridge HL, Pierson SK, Singh A, Arenas DJ, Ruth JR, Nabel CS, Stone K, Okumura M, Schwarer A, Jose FF, Hamerschlak N, Wertheim GB, Jordan MB, Cohen AD, Krymskaya V, Rubenstein A, Betts MR, Kambayashi T, van Rhee F, Uldrick TS.. Identifying and targeting pathogenic PI3K/AKT/mTOR signaling in IL-6-blockade-refractory idiopathic multicentric Castleman disease. J Clin Invest 2019;129:4451–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS.. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009;284:8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, Barratt DG, Cosulich S, Klinowska T, Rosen N, Shokat KM.. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016;534:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Eisen HJ, Tuzcu EM, Dorent R, Kobashigawa J, Mancini D, Valantine-von Kaeppler HA, Starling RC, Sorensen K, Hummel M, Lind JM, Abeywickrama KH, Bernhardt P, Group RBS.. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. N Engl J Med 2003;349:847–858. [DOI] [PubMed] [Google Scholar]
- 190. Mancini D, Pinney S, Burkhoff D, LaManca J, Itescu S, Burke E, Edwards N, Oz M, Marks AR.. Use of rapamycin slows progression of cardiac transplantation vasculopathy. Circulation 2003;108:48–53. [DOI] [PubMed] [Google Scholar]
- 191. Meyer TE, Kovacs SJ, Ehsani AA, Klein S, Holloszy JO, Fontana L.. Long-term caloric restriction ameliorates the decline in diastolic function in humans. J Am Coll Cardiol 2006;47:398–402. [DOI] [PubMed] [Google Scholar]
- 192. Fan Y, Cheng Y, Li Y, Chen B, Wang Z, Wei T, Zhang H, Guo Y, Wang Q, Wei Y, Chen F, Sha J, Guo X, Wang L.. Phosphoproteomic analysis of neonatal regenerative myocardium revealed important roles of checkpoint kinase 1 via activating mammalian target of rapamycin C1/ribosomal protein S6 kinase b-1 pathway. Circulation 2020;141:1554–1569. [DOI] [PubMed] [Google Scholar]
- 193. Liu S, Huang T, Liu R, Cai H, Pan B, Liao M, Yang P, Wang L, Huang J, Ge Y, Xu B, Wang W.. Spermidine suppresses development of experimental abdominal aortic aneurysms. J Am Heart Assoc 2020;9:e014757. [DOI] [PMC free article] [PubMed] [Google Scholar]