PURPOSE
Deep learning (DL) models have rapidly become a popular and cost-effective tool for image classification within oncology. A major limitation of DL models is their vulnerability to adversarial images, manipulated input images designed to cause misclassifications by DL models. The purpose of the study is to investigate the robustness of DL models trained on diagnostic images using adversarial images and explore the utility of an iterative adversarial training approach to improve the robustness of DL models against adversarial images.
METHODS
We examined the impact of adversarial images on the classification accuracies of DL models trained to classify cancerous lesions across three common oncologic imaging modalities. The computed tomography (CT) model was trained to classify malignant lung nodules. The mammogram model was trained to classify malignant breast lesions. The magnetic resonance imaging (MRI) model was trained to classify brain metastases.
RESULTS
Oncologic images showed instability to small pixel-level changes. A pixel-level perturbation of 0.004 (for pixels normalized to the range between 0 and 1) resulted in most oncologic images to be misclassified (CT 25.6%, mammogram 23.9%, and MRI 6.4% accuracy). Adversarial training improved the stability and robustness of DL models trained on oncologic images compared with naive models ([CT 67.7% v 26.9%], mammogram [63.4% vs 27.7%], and MRI [87.2% vs 24.3%]).
CONCLUSION
DL models naively trained on oncologic images exhibited dramatic instability to small pixel-level changes resulting in substantial decreases in accuracy. Adversarial training techniques improved the stability and robustness of DL models to such pixel-level changes. Before clinical implementation, adversarial training should be considered to proposed DL models to improve overall performance and safety.
INTRODUCTION
Deep learning (DL) algorithms have the promise to improve the quality of diagnostic image interpretation within oncology.1,2 Models generated from DL algorithms have been validated across a variety of diagnostic imaging modalities including magnetic resonance imaging (MRI), computed tomography (CT), and x-ray images with classification accuracy often rivaling trained clinicians.3-9 However, the success of DL models depends, in part, on their generalizability and stability. DL algorithms have been shown to vary output on the basis of small changes in the input data.10,11 Such variability in response to minor changes can signal an instability in the algorithm that could lead to misclassification and problems with generalizability.
CONTEXT
Key Objective
Are deep learning (DL) models trained to classify diagnostic images in oncology more vulnerable to adversarial images than natural images?
Knowledge Generated
We found that DL classification models trained on oncologic images were more susceptible to adversarial images than natural images. Additionally, we found that adversarial image sensitivity can be leveraged to improve DL model robustness.
Relevance
Adversarial images represent a potential barrier to end-to-end implementation of DL models within clinical practice. Nevertheless, adversarial images can also be used to improve the overall robustness of DL models within clinical oncology.
One concerning limitation of DL models is their susceptibility to adversarial attacks. Adversarial images are manipulated images that undergo small pixel-level perturbations specifically designed to deceive DL models.12-15 Pixel-level changes of adversarial images are often imperceptible to humans but can cause important differences in the model output.16-18 The weakness of DL models against adversarial images raises concerns about the generalizability of DL models and the safety of their practical applications in medicine. Adversarial images represent potential security threats in the future, as DL algorithms for diagnostic image analysis become increasingly implemented into clinical environments.19 Additionally, the susceptibility to adversarial attacks provides increasing evidence to the instability of DL models that aim to mimic the classification accuracy of radiologists.
Previous work concerning adversarial images on DL models has largely focused on nonmedical images, and the vulnerability of medical DL models is relatively unknown.18,20 Although techniques to defend against adversarial images have been proposed, the effectiveness of these methods on medical DL models is unclear. Accordingly, we sought to test the effect of adversarial images on DL algorithms trained on three common oncologic imaging modalities. We established the performance of the DL models and then tested model output stability in response to adversarial images with different degrees of pixel-level manipulation. We then tested the utility of techniques to defend the DL models against adversarial images. This research has direct application to the use of DL image interpretation algorithms, as it provides quantitative testing of their vulnerability to small input variations and determines whether there are strategies to reduce this weakness.
METHODS
Ethics Declaration
The research was conducted in accordance with the Declaration of Helsinki guidelines and approved by the Yale University Institutional Review Board (Protocol ID: HIC#2000027592). Informed consent was obtained from all participants in this study.
Data Sets
We examined the behavior of DL algorithm outputs in response to adversarial images across three medical imaging modalities commonly used in oncology—CT, mammography, and MRI. For each imaging modality, a separate DL classification model was trained to identify the presence or absence of malignancy when given a diagnostic image. Each data set was split into a training set and a testing set in a 2:1 ratio.
CT imaging data consisted of 2,600 lung nodules from the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) collection.21 The data set contains 1,018 thoracic CT scans collected from 15 clinical sites across the United States. Lung nodules used for DL model training were identified by experienced thoracic radiologists. The presence of malignancy was based on associated pathologic reports. For patients without pathologic confirmation, malignancy was based on radiologist consensus.
Mammography imaging data consisted of 1,696 lesions from the Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM).22 The CBIS-DDSM contains mammograms from 1,566 patients at four sites across the United States. Mammographic lesions used for DL model training were obtained on the basis of algorithmically derived regions of interest based on clinical metadata. The presence of malignancy was based on verified pathologic reports.
MRI data consisted of brain MRIs from 831 patients from a single-institution brain metastases registry.23 The presence or absence of a malignancy was identified on 4,000 brain lesions seen on MRI. Regions of interest were identified by a multidisciplinary team of radiation oncologists, neurosurgeons, and radiologists. Presence of cancer was identified on the basis of pathologic confirmation or clinical consensus.
To compare the relative vulnerability of DL models trained on oncologic images compared with nonmedical images, two additional DL classification models were trained on established nonmedical data sets. The MNIST data set consists of 70,000 handwritten numerical digits.24 The CIFAR-10 data set includes 60,000 color images of 10 nonmedical objects.25
All images were center-cropped and resized, and pixel values were normalized to the range [0, 1]. For each medical data set, the classes (cancer and noncancer) were balanced, and data were augmented using simple data augmentations: horizontal and vertical flips as well as random rotations with angles ranging between –20° and 20°.
We followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guideline, and a TRIPOD checklist was included (Data Supplement).26
Models
For all DL classification models, we used a pretrained convolutional neural network with the VGG16 architecture.27 Models were fine-tuned in Keras using Stochastic Gradient Descent. Details regarding model architecture and hyperparameter selection for DL model training are provided in the Data Supplement.
Adversarial Image Generation
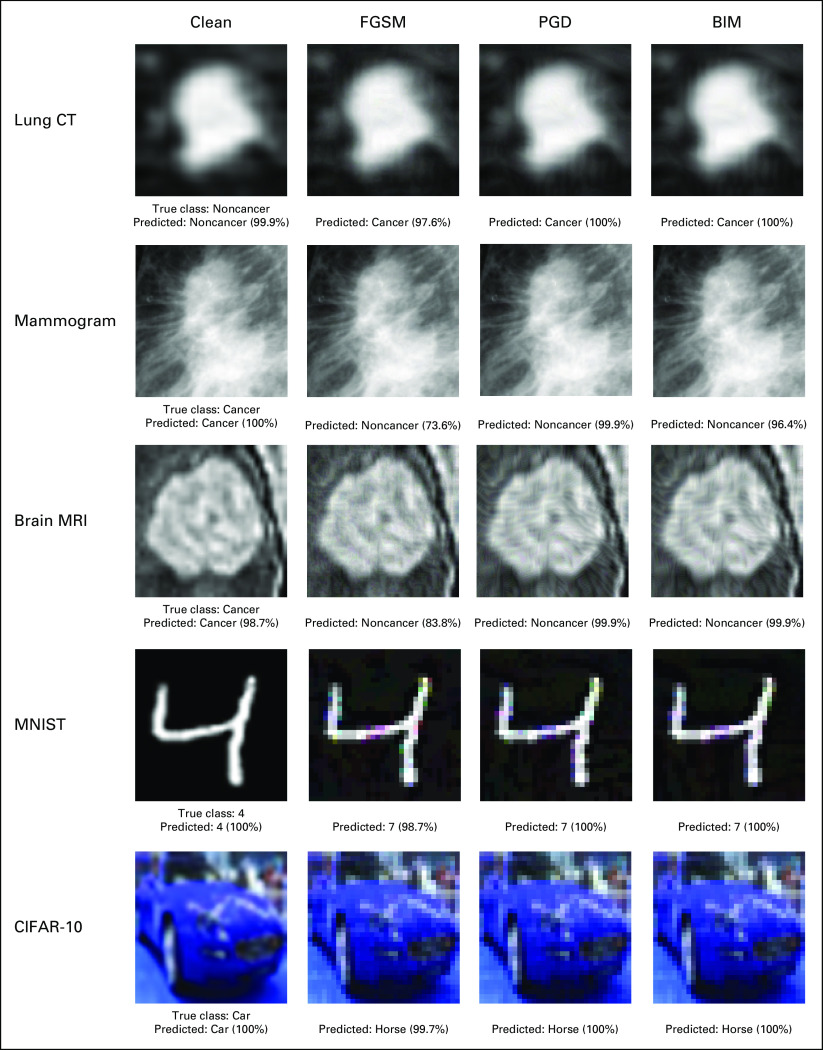
Three commonly used first-order adversarial image generation methods—Fast Gradient Sign Method (FGSM), Basic Iterative Method (BIM), and Projected Gradient Descent (PGD)—were used to create adversarial images on the medical and nonmedical image data sets (Fig 1). Each method aims to maximize the DL model's classification error while minimizing the difference between the adversarial image and original image. All the adversarial image generation methods are bounded under a predefined perturbation size ε, which represents the maximum change to pixel values of an image. Vulnerability to adversarial images was assessed by comparing changes in model performance compared with baseline (without any adversarial images) under various perturbation sizes.
FIG 1.
Examples of clean images and their adversarial counterparts generated using FGSM, PGD, and BIM attack methods. The percentage displayed represents the probability predicted by the model that the image is of a certain class. BIM, Basic Iterative Method; CT, computed tomography; FGSM, Fast Gradient Sign Method; MRI, magnetic resonance imaging; PGD, Projected Gradient Descent.
The single-step FGSM attack perturbs the original example by a fixed amount along the direction (sign) of the gradient of adversarial loss.15 Given input image x, perturbation size, loss function J, and target label y, the adversarial image xadv can be computed as
| (1) |
BIM iteratively perturbs the normal example with smaller step size and clips the pixel values of the updated adversarial example after each step into a permitted range.12
| (2) |
Known as the strongest first-order attack, PGD iteratively perturbs the input with smaller step size and after each iteration, the updated adversarial example is projected onto the ε-ball of x and clipped onto a permitted range.18
| (3) |
Additional information regarding adversarial image generation methods and equation parameters is provided in the Data Supplement.
Susceptibility of DL Models to Adversarial Images
We investigated the DL model performance using FGSM, PGD, and BIM adversarial image generation methods across different levels of pixel perturbation. To evaluate the performance of the DL models on adversarial images, we generated adversarial test sets by applying adversarial attacks on the original clean test sets. We measured relative susceptivity to adversarial images by determining the smallest perturbation ε required for adversarial images to generate a different output. DL models that required larger pixel-level perturbations are likely to be more robust and have higher levels of stability suitable for clinical implementation. Conversely, models that change outputs in response to small pixel-level perturbations are inherently unstable and potentially less generalizable across different clinical settings and patient populations.
Adversarial Training to Improve Model Robustness
One proposed defense mechanism to prevent negative effects of adversarial images is adversarial training, which aims to improve model robustness by integrating adversarial samples into DL model training.18,20 By training on both adversarial and normal images, the DL model learns to classify adversarial samples with higher accuracy compared with models trained on only normal samples. We used a multistep PGD adversarial training to increase the robustness of our DL models against adversarial attacks. In each batch, 50% of training samples were normal images, and the other 50% were adversarial images generated by PGD attack. We used the PGD attack for adversarial training because it is considered the strongest first-order attack, and past research has demonstrated that models adversarially trained with PGD were robust against other first-order attacks.18 The hyperparameters for adversarial training are detailed in the Data Supplement. We investigated the effectiveness of our iterative adversarial training approach on the DL models trained on medical images. We measured the effectiveness of adversarial training by comparing model accuracy on adversarial samples of varying perturbation sizes before and after adversarial training.
Image-Level Adversarial Image Sensitivity and Model Performance
We examined each individual image's adversarial sensitivity, as measured by the level of pixel-level perturbation necessary for DL model prediction to change compared with an unperturbed image. We hypothesized that images requiring smaller pixel-level adversarial perturbations to change DL model predictions were also the images most likely to be misclassified by the model under normal conditions absent of adversarial attack (compared with other clean images). We identified the 20% of images most vulnerable to adversarial perturbation and excluded them from the test set. We then tested the performance of the DL model on the clean version of the reduced test set. If the images most sensitive to adversarial perturbation are also more likely to be misclassified as clean images by the DL model under normal conditions, the model performance on the clean version of the reduced test set would be expected to improve from the model performance on the clean version of the original test.
The proposed networks were implemented in Python 2.7 using TensorFlow v1.15.3 framework.28 Adversarial images were created using the Adversarial Robustness Toolbox v1.4.1.29 The code to reproduce the analyses and results is available online at GitHub.30
RESULTS
Susceptibility of DL Models to Adversarial Images
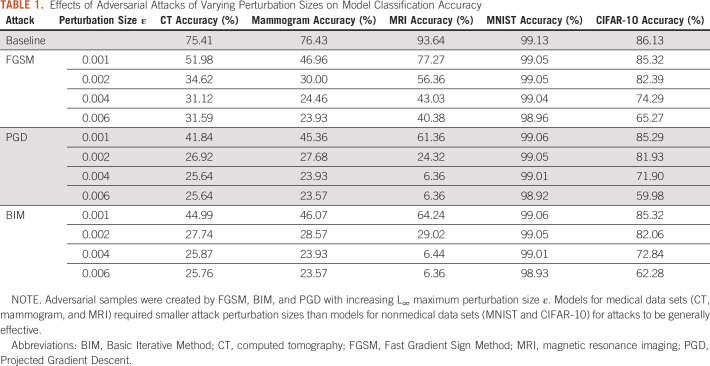
Both medical and nonmedical DL models were highly susceptible to misclassification of adversarial images, resulting in decreases in model accuracy (Fig 2). Medical DL models appeared substantially more vulnerable to adversarial images compared with nonmedical DL algorithms. All three medical DL models required smaller pixel-level perturbations to decrease model accuracy compared with nonmedical DL models (Fig 2). For example, adversarial images generated using the PGD method (perturbation = 0.002) resulted in a DL model accuracy of 26.9% for CT (–48.5% from baseline), 27.7% for mammogram (–48.8% from baseline), and 24.3% for MRI (–61.8% from baseline). By contrast, adversarial images generated using the same methods/parameters did not cause substantial changes in performance for the MNIST (–0.05% from baseline) or CIFAR-10 (–4.2% from baseline) trained models (Table 1). For the medical DL models, adversarial images generated using smaller pixel-level perturbations (ε < 0.004) resulted in misclassification of a majority of images, whereas nonmedical DL models required much larger pixel perturbations (ε > 0.07 for MNIST and ε > 0.01 for CIFAR-10) for similar levels of misclassification (Table 1).
FIG 2.
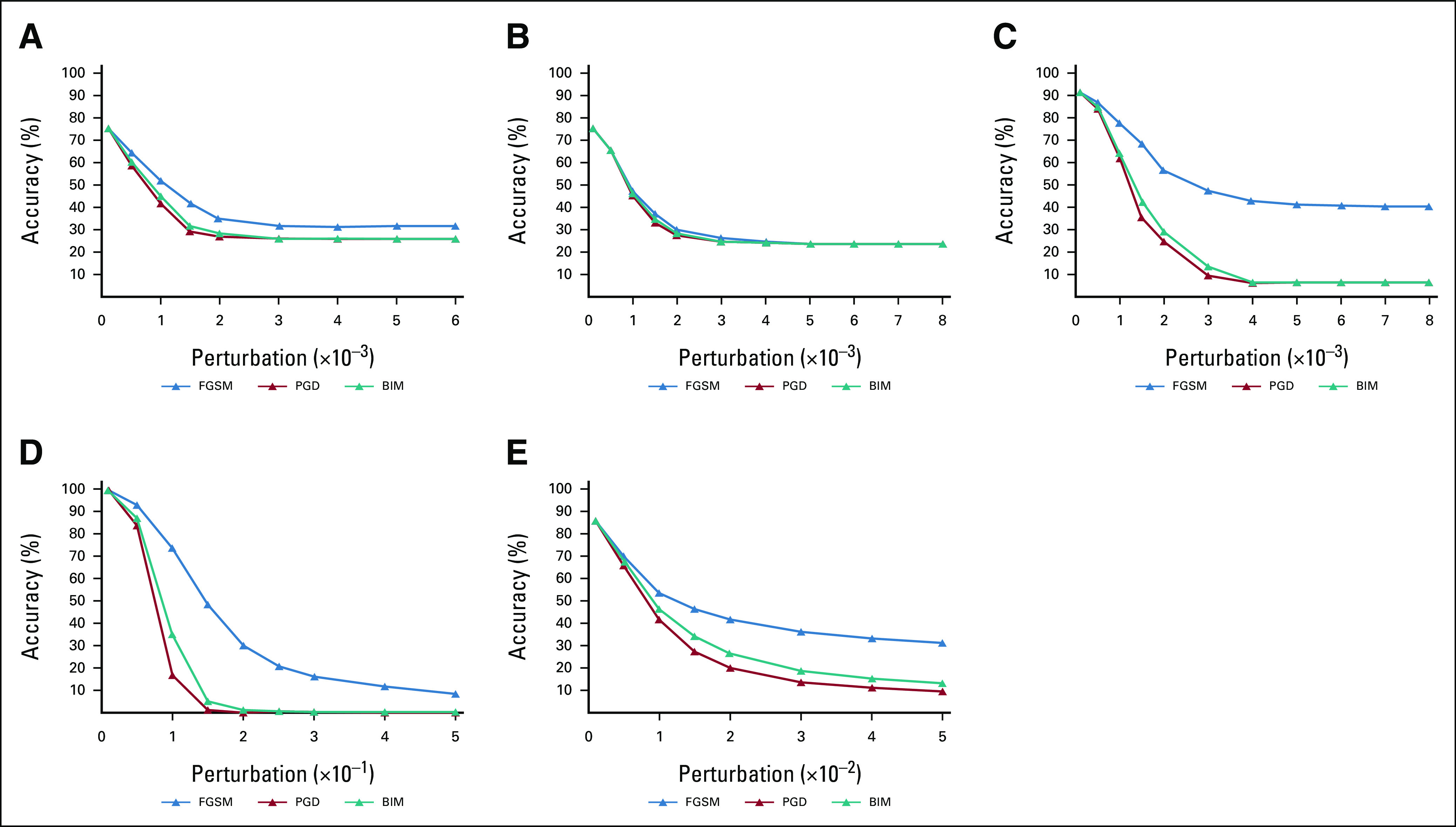
Classification accuracy of VGG16 model on adversarial examples generated by FGSM, BIM, and PGD attacks with increasing L∞ maximum perturbation size ε. Model performance decreased as ε increased for all data sets: (A) lung CT; (B) mammography; (C) brain MRI; (D) MNIST; and (E) CIFAR-10. *Note that the horizontal axis (ε) was scaled to 10–3 for graphs (A) to (C), to 10–1 for (D), and to 10–2 for (E). BIM, Basic Iterative Method; CT, computed tomography; FGSM, Fast Gradient Sign Method; MRI, magnetic resonance imaging; PGD, Projected Gradient Descent.
TABLE 1.
Effects of Adversarial Attacks of Varying Perturbation Sizes on Model Classification Accuracy
Adversarial Training to Improve Model Robustness
Adversarial training led to increased robustness of DL models when classifying adversarial images for both medical and nonmedical images (Fig 3). Compared with baseline trained models, adversarial trained DL models caused absolute accuracy of the model on adversarial images to increase by 42.9% for CT (67.7% v 26.9%), 35.7% for mammogram (63.4% v 27.7%), and 73.2% for MRI (87.2% v 24.3%; Data Supplement). Despite adversarial training, DL models did not reach baseline accuracy, suggesting adversarial training as only a partial solution to improve model robustness. Adversarial training became less effective when attempting to defend against adversarial images that possessed greater pixel perturbations.
FIG 3.
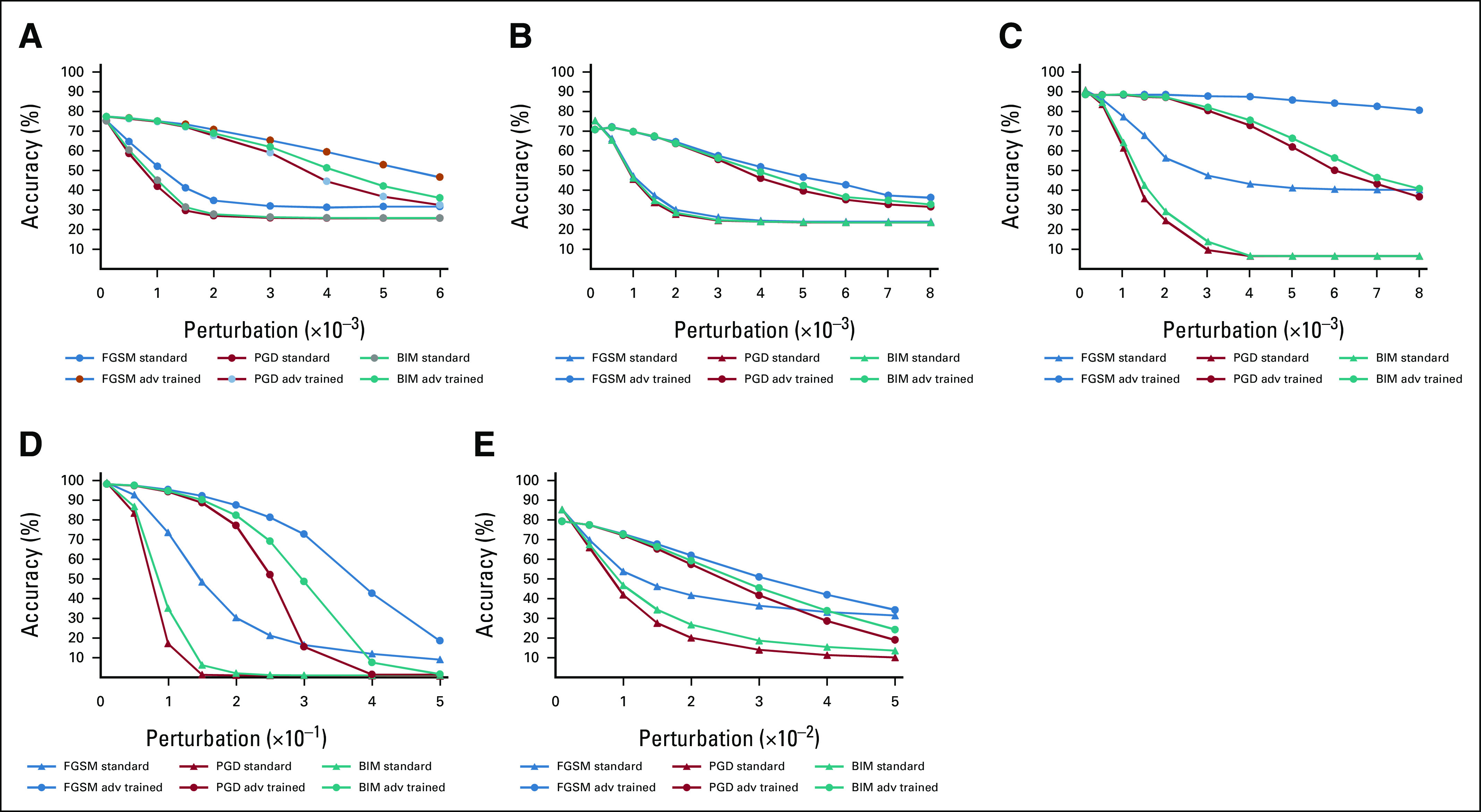
Comparison of model classification accuracy before and after adversarial training on adversarial samples crafted by FGSM, BIM, and PGD with increasing L∞ maximum perturbation size ε. Adversarial training significantly increased model accuracy for data sets: (A) lung CT; (B) mammography; (C) brain MRI; (D) MNIST; and (E) CIFAR-10. *Note that the horizontal axis (ε) was scaled to 10–3 for graphs (A) to (C), to 10–1 for (D), and to 10–2 for (E). BIM, Basic Iterative Method; CT, computed tomography; FGSM, Fast Gradient Sign Method; MRI, magnetic resonance imaging; PGD, Projected Gradient Descent.
Image-Level Adversarial Image Sensitivity and Model Performance
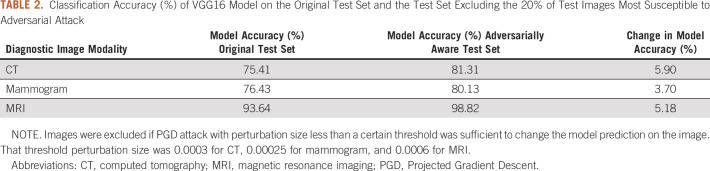
Using image-level adversarial sensitivity, we were able to identify images most at risk for misclassification by the DL models and improve overall model performance across all diagnostic imaging modalities. Excluding the images in which the smallest pixel perturbations changed DL model outputs increased the absolute accuracy of DL models by 5.9% for CT, 3.7% for mammogram, and 5.2% for MRI (Table 2).
TABLE 2.
Classification Accuracy (%) of VGG16 Model on the Original Test Set and the Test Set Excluding the 20% of Test Images Most Susceptible to Adversarial Attack
DISCUSSION
As the role of diagnostic imaging increases throughout clinical oncology, DL represents a cost-effective tool to supplement human decision making and aid in image analysis tasks.31-33 However, instability of DL model outputs can limit the performance and generalizability on large-scale medical data sets and hinder clinical utility. Evaluating a proposed DL model's susceptibility to adversarial images represents a way to identify the most robust DL models versus those at risk for erratic performance. In this study, we found that DL models trained on medical images were particularly unstable to perturbation from adversarial images resulting in significant decreases in expected performance. Moreover, we found diagnostic images within oncology to be more vulnerable to such misclassification compared with DL models trained on nonmedical images. Specifically, compared with nonmedical images, all three diagnostic imaging modalities required substantially smaller perturbation to reduce model performance. Furthermore, we found that adversarial training methods commonly used on nonmedical imaging data sets are effective at improving DL model stability to such pixel-level changes. Finally, we showed that identifying images most susceptible to adversarial image attacks maybe helpful in improving overall robustness of DL models on medical images.
Several recent works have found that state-of-the-art DL architectures perform poorly on medical imaging analysis tasks when classifying adversarial images.14,34-38 Our work extends the findings of previous studies by evaluating performance across three common oncologic imaging modalities used for cancer detection. Additionally, we found that CT, mammography, and MRI images exhibit substantial vulnerability to adversarial images even with small pixel-level perturbations (<0.004). We also show that DL models exhibited different levels of sensitivity to adversarial images across different imaging modalities. Furthermore, although most previous studies used only one fixed perturbation size for adversarial image attack, we varied perturbation size along a broad range to examine the relationship between model performance and attack strength.
In addition, our results corroborate previous work, which showed that DL models trained on medical images are more vulnerable to misclassifying adversarial images compared with similar DL models trained on nonmedical images.14,39 By using MNIST and CIFAR-10 as a control and applying the same attack settings to DL models for all data sets, we determined that DL models for medical images were much more susceptible to misclassifying adversarial images than DL models for nonmedical images. One reason for this behavior could be that medical images are highly standardized, and small adversarial perturbations dramatically distort their distribution in the latent feature space.40,41 Another factor could be the overparameterization of DL models for medical image analysis, as sharp loss landscapes around medical images lead to higher adversarial vulnerability.14
In the past, adversarial training on medical DL models has shown mixed results. In some studies, adversarial training improved DL model robustness for multiple medical imaging modalities such as lung CT and retinal optical coherence tomography.40,42-44 By contrast, Hirano et al45 found that adversarial training generally did not increase model robustness for classifying dermatoscopic images, optical coherence tomography images, and chest x-ray images. The difference in effectiveness of adversarial training can be attributed to differences in adversarial training protocols (eg, single-step v iterative approaches). It is important to note that even in studies where adversarial training showed success in improving model robustness, the results were still suboptimal, as the risk of misclassification increases with perturbation strength even after adversarial training. This is expected as adversarial training, although capable of improving model accuracy on adversarial examples, has limits in effectiveness against strong attacks even on nonmedical image data sets.18
Our work applied an iterative adversarial training approach to DL models for lung CTs, mammograms, and brain MRIs, demonstrating substantial improvement in model robustness for all imaging modalities. The effectiveness of adversarial training was highly dependent on the hyperparameters of adversarial training, especially the perturbation size for attack. Although too-small perturbation sizes limit the increase in model robustness postadversarial training, increasing the perturbation size beyond a certain threshold prevents the model from learning during training, causing poor model performance on both clean and adversarial samples. Our work demonstrated how the performance of the DL model postadversarial training is inversely proportional to the perturbation size of the adversarial samples on which the model is evaluated. Although adversarial training is effective in defending against weaker attacks with smaller perturbation magnitudes, it showed less success with attacks that altered pixels more substantially. Although adversarial training proved successful at improving model performance on adversarial examples, our results were still far from satisfactory. One contributing factor is that medical images have fundamentally differently properties than nonmedical images.14,40 Thus, adversarial defenses well suited for nonmedical images may not be generalizable to medical images.
We also showed that image-level adversarial sensitivity, defined by the level of adversarial perturbation necessary to change image class predicted by model, is a useful metric for identifying normal images most at risk for misclassification. This has potentially useful clinical implications as we can improve the robustness of DL models by excluding such ‘high-risk’ images from DL model classification and instead providing them to a trained radiologist for examination.
There are several limitations to our study. First, we only used two-class medical imaging classification tasks. Thus, our findings might not generalize to multiclass or regression problems using medical images. Given that many medical diagnostic problems involve a small number of classes, our findings are likely still widely applicable to a large portion of medical imaging classification tasks. Our study used only first-order adversarial image generation methods rather than higher-order methods, which have been shown to be more resistant against adversarial training.46 Although most commonly used adversarial image generation methods are first-order, there is still need for additional research on how to defend DL models for medical images against higher-order methods. A final limitation is that we used traditional supervised adversarial training to improve model robustness, whereas other nuanced methods such as semisupervised adversarial training and unsupervised adversarial training exist.40,47,48 Although we demonstrated that supervised adversarial training is an effective method to improve model performance on adversarial examples, an interesting direction for future work would be to compare the utility of supervised adversarial training with that of semisupervised or unsupervised adversarial training on DL models for medical images.
In conclusion, in this work, we used adversarial images to explore the stability of DL models trained on three common diagnostic imaging modalities used in oncology. Our findings suggest that DL models trained on diagnostic images are vulnerable to pixel-level changes, which can substantially change expected performance. Specifically, we found that vulnerability to adversarial images can be a useful method to identify DL models that are particularly unstable in their classifications. Additionally, we found that adversarial image training may improve the stability of DL models trained on diagnostic images. Finally, we found that image-level adversarial sensitivity is a potential way to identify image samples that may benefit from human classification rather than DL model classification. By shedding light on the stability of DL models to small pixel changes, the findings from this paper can help facilitate the development of more robust and secure medical imaging DL models that can be more safely implemented into clinical practice.
Enoch Chang
Research Funding: Radiological Society of North America
James S. Duncan
Patents, Royalties, Other Intellectual Property: Systems, Methods and Apparatuses for Generating Regions of Interest from Voxel Mode Based Thresholds, Publication No: US20190347788A1, application No. 15/978,904. Filed on May 14, 2018, Publication Date: November 14, 2019. Inventors: Van Breugel J, Abajian A, Treli- hard J, Smolka S, Chapiro J, Duncan JS and Lin M. Joint application from Philips, N.V. and Yale University. US Patent 10,832,403 (2020)
Antonio Omuro
Consulting or Advisory Role: Merck, KIYATEC, Ono Pharmaceutical, BTG
Research Funding: Arcus Biosciences (Inst)
Roy Herbst
Leadership: Jun Shi Pharmaceuticals, Immunocore
Consulting or Advisory Role: AstraZeneca, Genentech/Roche, Merck, Pfizer, AbbVie, Biodesix, Bristol Myers Squibb, Lilly, EMD Serono, Heat Biologics, Jun Shi Pharmaceuticals, Loxo, Nektar, NextCure, Novartis, Sanofi, Seattle Genetics, Shire, Spectrum Pharmaceuticals, Symphogen, TESARO, Neon Therapeutics, Infinity Pharmaceuticals, ARMO Biosciences, Genmab, Halozyme, Tocagen, Bolt Biotherapeutics, IMAB Biopharma, Mirati Therapeutics, Takeda, Cybrexa Therapeutics, eFFECTOR Therapeutics, Inc, Candel Therapeutics, Inc, Oncternal Therapeutics, STCube Pharmaceuticals, Inc, WindMIL Therapeutics, Xencor, Inc, Bayer HealthCare Pharmaceuticals Inc, Checkpoint Therapeutics, DynamiCure Biotechnology, LLC, Foundation Medicine, Inc, Gilead/Forty Seven, HiberCell, Inc, Immune-Onc Therapeutics, Inc, Johnson and Johnson, Ocean Biomedical, Inc, Oncocyte Corp, Refactor Health, Inc, Ribbon Therapeutics, Ventana Medical Systems, Inc
Research Funding: AstraZeneca, Merck, Lilly, Genentech/Roche
Harlan M. Krumholz
Employment: Hugo Health (I), FPrime
Stock and Other Ownership Interests: Element Science, Refactor Health, Hugo Health
Consulting or Advisory Role: UnitedHealthcare, Aetna
Research Funding: Johnson and Johnson
Expert Testimony: Siegfried and Jensen Law Firm, Arnold and Porter Law Firm, Martin/Baughman Law Firm
Sanjay Aneja
Sanjay Aneja is an Associate Editor for JCO Clinical Cancer Informatics. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Prophet Consulting (I)
Research Funding: The MedNet, Inc, American Cancer Society, National Science Foundation, Agency for Healthcare Research and Quality, National Cancer Institute, ASCO
Patents, Royalties, Other Intellectual Property: Provisional patent of deep learning optimization algorithm
Travel, Accommodations, Expenses: Prophet Consulting (I), Hope Foundation
Other Relationship: NRG Oncology Digital Health Working Group, SWOG Digital Engagement Committee, ASCO mCODE Technical Review Group
No other potential conflicts of interest were reported.
PREPRINT VERSION
Preprint version available on https://www.medrxiv.org/content/10.1101/2021.01.17.21249704v3
DISCLAIMER
Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology or Conquer Cancer, or Hayden Family Foundation.
SUPPORT
Supported in part by a Career Enhancement Program Grant (PI: S.A.) from the Yale SPORE in Lung Cancer (1P50CA196530) and by a Conquer Cancer Career Development Award (PI: S.A.), supported by Hayden Family Foundation.
AUTHOR CONTRIBUTIONS
Conception and design: Marina Z. Joel, James S. Duncan, Roy Herbst, Sanjay Aneja
Financial support: Sanjay Aneja
Administrative support: Sanjay Aneja
Provision of study material or patients: Sanjay Aneja
Collection and assembly of data: Marina Z. Joel, Sachin Umrao, Sanjay Aneja
Data analysis and interpretation: Marina Z. Joel, Sachin Umrao, Enoch Chang, Rachel Choi, Daniel X. Yang, James S. Duncan, Antonio Omuro, Harlan M. Krumholz, Sanjay Aneja
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Enoch Chang
Research Funding: Radiological Society of North America
James S. Duncan
Patents, Royalties, Other Intellectual Property: Systems, Methods and Apparatuses for Generating Regions of Interest from Voxel Mode Based Thresholds, Publication No: US20190347788A1, application No. 15/978,904. Filed on May 14, 2018, Publication Date: November 14, 2019. Inventors: Van Breugel J, Abajian A, Treli- hard J, Smolka S, Chapiro J, Duncan JS and Lin M. Joint application from Philips, N.V. and Yale University. US Patent 10,832,403 (2020)
Antonio Omuro
Consulting or Advisory Role: Merck, KIYATEC, Ono Pharmaceutical, BTG
Research Funding: Arcus Biosciences (Inst)
Roy Herbst
Leadership: Jun Shi Pharmaceuticals, Immunocore
Consulting or Advisory Role: AstraZeneca, Genentech/Roche, Merck, Pfizer, AbbVie, Biodesix, Bristol Myers Squibb, Lilly, EMD Serono, Heat Biologics, Jun Shi Pharmaceuticals, Loxo, Nektar, NextCure, Novartis, Sanofi, Seattle Genetics, Shire, Spectrum Pharmaceuticals, Symphogen, TESARO, Neon Therapeutics, Infinity Pharmaceuticals, ARMO Biosciences, Genmab, Halozyme, Tocagen, Bolt Biotherapeutics, IMAB Biopharma, Mirati Therapeutics, Takeda, Cybrexa Therapeutics, eFFECTOR Therapeutics, Inc, Candel Therapeutics, Inc, Oncternal Therapeutics, STCube Pharmaceuticals, Inc, WindMIL Therapeutics, Xencor, Inc, Bayer HealthCare Pharmaceuticals Inc, Checkpoint Therapeutics, DynamiCure Biotechnology, LLC, Foundation Medicine, Inc, Gilead/Forty Seven, HiberCell, Inc, Immune-Onc Therapeutics, Inc, Johnson and Johnson, Ocean Biomedical, Inc, Oncocyte Corp, Refactor Health, Inc, Ribbon Therapeutics, Ventana Medical Systems, Inc
Research Funding: AstraZeneca, Merck, Lilly, Genentech/Roche
Harlan M. Krumholz
Employment: Hugo Health (I), FPrime
Stock and Other Ownership Interests: Element Science, Refactor Health, Hugo Health
Consulting or Advisory Role: UnitedHealthcare, Aetna
Research Funding: Johnson and Johnson
Expert Testimony: Siegfried and Jensen Law Firm, Arnold and Porter Law Firm, Martin/Baughman Law Firm
Sanjay Aneja
Sanjay Aneja is an Associate Editor for JCO Clinical Cancer Informatics. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Prophet Consulting (I)
Research Funding: The MedNet, Inc, American Cancer Society, National Science Foundation, Agency for Healthcare Research and Quality, National Cancer Institute, ASCO
Patents, Royalties, Other Intellectual Property: Provisional patent of deep learning optimization algorithm
Travel, Accommodations, Expenses: Prophet Consulting (I), Hope Foundation
Other Relationship: NRG Oncology Digital Health Working Group, SWOG Digital Engagement Committee, ASCO mCODE Technical Review Group
No other potential conflicts of interest were reported.
REFERENCES
- 1. Kann BH, Thompson R, Thomas CR, Jr, et al. Artificial intelligence in oncology: Current applications and future directions. Oncology (Williston Park) 2019;33:46–53. [PubMed] [Google Scholar]
- 2. Aneja S, Chang E, Omuro A. Applications of artificial intelligence in neuro-oncology. Curr Opin Neurol. 2019;32:850–856. doi: 10.1097/WCO.0000000000000761. [DOI] [PubMed] [Google Scholar]
- 3.Siar M, Teshnehlab M.Brain tumor detection using deep neural network and machine learning algorithm. 2019 9th International Conference on Computer And Knowledge Engineering (ICCKE), 2019, pp 363-368.
- 4. Hashemzehi R, Mahdavi SJS, Kheirabadi M, et al. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybernetics Biomed Eng. 2020;40:1225–1232. [Google Scholar]
- 5. Jain G, Mittal D, Thakur D, et al. A deep learning approach to detect Covid-19 coronavirus with x-ray images. Biocybernetics Biomed Eng. 2020;40:1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kann BH, Hicks DF, Payabvash S, et al. Multi-institutional validation of deep learning for pretreatment identification of extranodal extension in head and neck squamous cell carcinoma. J Clin Oncol. 2019;38:1304–1311. doi: 10.1200/JCO.19.02031. [DOI] [PubMed] [Google Scholar]
- 7. Cao H, Liu H, Song E, et al. Dual-branch residual network for lung nodule segmentation. Appl Soft Comput. 2020;86:105934. [Google Scholar]
- 8. Tang Y-X, Tang Y-B, Peng Y, et al. Automated abnormality classification of chest radiographs using deep convolutional neural networks. NPJ Digital Med. 2020;3:70. doi: 10.1038/s41746-020-0273-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digital Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 10. Shaham U, Yamada Y, Negahban S. Understanding adversarial training: Increasing local stability of supervised models through robust optimization. Neurocomputing. 2018;307:195–204. [Google Scholar]
- 11. Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks. arXiv. 2013 arXiv preprint 1312.6199. [Google Scholar]
- 12. Kurakin A, Goodfellow I, Bengio S. Adversarial examples in the physical world. arXiv. 2013 arXiv:1607.02533. [Google Scholar]
- 13. Yuan X, He P, Zhu Q, et al. Adversarial examples: Attacks and defenses for deep learning. arXiv. 2017 doi: 10.1109/TNNLS.2018.2886017. arXiv:1712.07107. [DOI] [PubMed] [Google Scholar]
- 14. Ma X, Niu Y, Gu L, et al. Understanding adversarial attacks on deep learning based medical image analysis systems. Pattern Recognition: 107332. 2020 [Google Scholar]
- 15. Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples. arXiv. 2014 arXiv:1412.6572. [Google Scholar]
- 16. Shu H, Shi R, Zhu H, et al. Adversarial image generation and training for deep neural networks. arXiv. 2020 arXiv:2006.03243. [Google Scholar]
- 17. Tabacof P, Valle E. Exploring the space of adversarial images. arXiv. 2015 arXiv:1510.05328. [Google Scholar]
- 18.Madry A, Makelov A, Schmidt L, et al. Towards deep learning models resistant to adversarial attacks. arXiv. arXiv:1706.06083.
- 19. Finlayson SG, Chung HW, Kohane IS, et al. Adversarial attacks against medical deep learning systems. arXiv. 2018 arXiv:1804.05296. [Google Scholar]
- 20. Ren K, Zheng T, Qin Z, et al. Adversarial attacks and defenses in deep learning. Engineering. 2020;6:346–360. [Google Scholar]
- 21. Armato SG, III, McLennan G, Bidaut L, et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee RS, Gimenez F, Hoogi A, et al. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data. 2017;4:170177. doi: 10.1038/sdata.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang E, Joel M, Chang HY, et al. Comparison of radiomic feature aggregation methods for patients with multiple tumors. medRxiv. 2020 doi: 10.1038/s41598-021-89114-6. 2020.11.04.20226159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lecun Y.THE MNIST DATABASE of handwritten digits. http://yann.lecun.com/exdb/mnist/
- 25. Krizhevsky A, Hinton G. Learning multiple layers of features from tiny images. 2009 [Google Scholar]
- 26. Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 27. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014 arXiv:1409.1556, 2014. [Google Scholar]
- 28. Abadi M, Agarwal A, Barham P, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv. 2016 arXiv:1603.04467. [Google Scholar]
- 29. Nicolae M-I, Sinn M, Tran MN, et al. Adversarial robustness Toolbox v1.0.0. arXiv. 2018 arXiv:1807.01069. [Google Scholar]
- 30.https://github.com/Aneja-Lab-Yale/Aneja-Lab-Public-Adversarial-Imaging
- 31. Kyono T, Gilbert FJ, van der Schaar M. MAMMO: A deep learning solution for facilitating radiologist-machine collaboration in breast cancer diagnosis. arXiv. 2018 arXiv:1811.02661. [Google Scholar]
- 32. Park A, Chute C, Rajpurkar P, et al. Deep learning–assisted diagnosis of cerebral aneurysms using the HeadXNet model. JAMA Netw Open. 2019;2:e195600. doi: 10.1001/jamanetworkopen.2019.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46:e1–e36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paschali M, Conjeti S, Navarro F, et al. Generalizability vs. robustness: Investigating medical imaging networks using adversarial examples. In: Frangi AF, Schnabel JA, Davatzikos C, et al., editors. Medical Image Computing and Computer Assisted Intervention—MICCAI 2018. Cham, Switzerland: Springer International Publishing; 2018. pp. 493–501. [Google Scholar]
- 35. Finlayson SG, Bowers JD, Ito J, et al. Adversarial attacks on medical machine learning. Science. 2019;363:1287–1289. doi: 10.1126/science.aaw4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wetstein SC, González-Gonzalo C, Bortsova G, et al. Adversarial attack vulnerability of medical image analysis systems: Unexplored factors. arXiv. 2020 doi: 10.1016/j.media.2021.102141. arXiv:2006.06356. [DOI] [PubMed] [Google Scholar]
- 37. Asgari Taghanaki S, Das A, Hamarneh G. Vulnerability analysis of chest X-ray image classification against adversarial attacks. arXiv. 2018 arXiv:1807.02905. [Google Scholar]
- 38. Yoo TK, Choi JY. Outcomes of adversarial attacks on deep learning models for ophthalmology imaging domains. JAMA Ophthalmol. 2020;138:1213–1215. doi: 10.1001/jamaophthalmol.2020.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shafahi A, Najibi M, Ghiasi A, et al. Adversarial training for free! arXiv. 2019 arXiv:1904.12843. [Google Scholar]
- 40. Li X, Pan D, Zhu D. Defending against adversarial attacks on medical imaging AI system, classification or detection? arXiv. 2020 arXiv:2006.13555. [Google Scholar]
- 41.Li X, Zhu D.Robust detection of adversarial attacks on medical images. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020, pp 1154-1158.
- 42.Paul R, Schabath M, Gillies R, et al. Mitigating adversarial attacks on medical image understanding systems. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020, pp 1517-1521.
- 43.Vatian A, Gusarova N, Dobrenko N, et al. Impact of adversarial examples on the efficiency of interpretation and use of information from high-tech medical images. 2019 24th Conference of Open Innovations Association (FRUCT), 2019, pp 472-478.
- 44. Hirano H, Koga K, Takemoto K. Vulnerability of deep neural networks for detecting COVID-19 cases from chest X-ray images to universal adversarial attacks. PLoS One. 2020;15:e0243963. doi: 10.1371/journal.pone.0243963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirano H, Minagi A, Takemoto K. Universal adversarial attacks on deep neural networks for medical image classification. BMC Med Imaging. 2021;21:9. doi: 10.1186/s12880-020-00530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li B, Chen C, Wang W, et al. Certified adversarial robustness with additive noise. arXiv. 2018 arXiv:1809.03113. [Google Scholar]
- 47. Chen C, Yuan W, Lu X, et al. Spoof face detection via semi-supervised adversarial training. arXiv. 2020 arXiv:2005.10999. [Google Scholar]
- 48. Uesato J, Alayrac J-B, Huang P-S, et al. Are labels required for improving adversarial robustness? arXiv. 2019 arXiv:1905.13725. [Google Scholar]